Summary
Plant disease resistance (R) proteins that confer resistance to viruses recognize viral gene products with diverse functions, including viral suppressors of RNA silencing (VSRs). The P0 protein from poleroviruses is a VSR that targets the ARGONAUTE1 (AGO1) protein for degradation, thereby disrupting RNA silencing and antiviral defences. Here, we report resistance against poleroviruses in Nicotiana glutinosa directed against Turnip yellows virus (TuYV) and Potato leafroll virus (PLRV). The P0 proteins from TuYV (P0T u), PLRV (P0PL) and Cucurbit aphid‐borne yellows virus (P0CA) were found to elicit a hypersensitive response (HR) in N. glutinosa accession TW59, whereas other accessions recognized P0PL only. Genetic analysis showed that recognition of P0T u by a resistance gene designated RPO1 (Resistance to POleroviruses 1) is inherited as a dominant allele. Expression of P0 from a Potato virus X (PVX) expression vector transferred recognition to the recombinant virus on plants expressing RPO1, supporting P0 as the unique Polerovirus factor eliciting resistance. The induction of HR required a functional P0 protein, as P0T u mutants with substitutions in the F‐box motif that abolished VSR activity were unable to elicit HR. We surmised that the broad P0 recognition seen in TW59 and the requirement for the F‐box protein motif could indicate detection of P0‐induced AGO1 degradation and disruption of RNA silencing; however, other viral silencing suppressors, including the PVX P25 that also causes AGO1 degradation, failed to elicit HR in N. glutinosa. Investigation of P0 elicitation of RPO1 could provide insight into P0 activities within the cell that trigger resistance.
Keywords: hypersensitive response, poleroviruses, silencing suppressor
Introduction
The Polerovirus genus includes aphid‐transmitted viruses that infect a wide range of commercially important vegetable and small grain cereal crops, causing significant annual losses in crop yields worldwide (Stevens et al., 2005; Taliansky et al., 2003; Zhang et al., 2009). Viruses of this genus share in the organization of their 5.6–5.8‐kb single‐stranded RNA genomes, which distinguish these species from members of Enamovirus and Luteovirus, the two other genera of the family Luteoviridae (Stevens et al., 2005). Although the 3′ block of viral coding sequences is similar within the family, the Luteovirus genome is distinctly related to the Tombusviridae in the genome 5′ coding sequence block, whereas the Polerovirus and Enamovirus genomes share a relationship to the Sobemovirus genus in this region, which includes the first open reading frame (ORF) that encodes a viral suppressor of RNA silencing (VSR), designated P0 (Stevens et al., 2005; Taliansky et al., 2003).
The P0 proteins from both Turnip yellows virus (TuYV) and Cucurbit aphid‐borne yellows virus (CABYV) have an F‐box‐like domain that interacts with the Arabidopsis thaliana SKP1 homologues ASK1 and ASK2, suggesting that P0 functions in SCF E3 ubiquitin ligase complexes (Pazhouhandeh et al., 2006). However, P0 does not facilitate proteasome‐mediated degradation as expected and directs the autophagic degradation of Arabidopsis ARGONAUTE (AGO) proteins (Baumberger et al., 2007; Bortolamiol et al., 2007; Derrien et al., 2012). ARGONAUTE1 (AGO1) is an endonuclease that binds small interfering RNAs (siRNAs) generated by Dicer‐like cleavage of viral double‐stranded RNAs (Baumberger et al., 2007; Bortolamiol et al., 2007). P0 specifically prevents the de novo association of AGO1 with siRNAs (Csorba et al., 2010). The role of P0 proteins in E3 ubiquitin ligase complexes is novel and interesting because of the potential role of K63‐linked ubiquitylation of AGO1 for targeting to the autophagic machinery, rather than the K48‐linked polyubiquitylation typically implicated in proteasome targeting (Derrien et al., 2012).
VSRs represent a contact point between viruses and innate host defence machinery analogous to the microbial effectors that target basal defences to promote infection (Pumplin and Voinnet, 2013). Plants have evolved a large repertoire of intracellular resistance (R) proteins with nucleotide‐binding and leucine‐rich repeat (NB‐LRR) domains that mediate effector‐triggered immunity (ETI); NB‐LRR proteins may recognize the activity of microbial effector proteins on targets that function in plant innate immunity or act as decoy proteins that mimic these targets to induce ETI (Block and Alfano, 2011). Viral proteins, including VSRs, have similarly been found to be elicitors of plant immunity through NB‐LRR proteins (Zvereva and Pooggin, 2012). VSRs that target plant AGO proteins may also elicit ETI in some genetic backgrounds. The 2b protein from Cucumber mosaic virus (CMV2b) inhibits AGO1 slicer activity in vivo in susceptible Nicotiana benthamiana, whereas 2b from the related species Tomato aspermy virus (TAV2b) elicits a robust host response in Nicotiana tabacum cultivar Samsun when expressed from Tobacco mosaic virus (TMV) (Li et al., 1999; Zhang et al., 2006). In A. thaliana, the Turnip crinkle virus (TCV) protein P38, which serves as the virus coat protein (CP), also inhibits AGO1 activity by binding via a GW residue repeat that mimics GW motifs from host RNA‐induced silencing complex (RISC) proteins that normally interact with AGO1 (Azevedo et al., 2010) and function as elicitors [formerly Avirulence (Avr) determinant] for the A. thaliana RCY1 locus encoding the NB‐LRR protein HRT in resistant ecotype Di‐17 (Zhao et al., 2000). The 25‐kDa movement protein (MP) from Potato virus X (PVX) targets AGO1 for degradation and elicits a hypersensitive response (HR) in potato cultivars expressing the Nb gene (Chiu et al., 2010; Malcuit et al., 1999). Although most known gene‐for‐gene interactions with VSRs induce resistance accompanied by HR, the P19 protein of tombusviruses, which inhibits RNA silencing by sequestering siRNAs, was recently shown to elicit extreme resistance to Tomato bushy stunt virus (TBSV) in some Nicotiana species (Sansregret et al., 2013).
We undertook the present study to identify disease resistance sources against poleroviruses in Nicotiana species, as no resistance genes have been described for this genus to date. We screened a number of Nicotiana accessions for Polerovirus resistance using the infectious binary vector clone of TuYV‐FL1, because of its broad host range, which includes the model plants Arabidopsis thaliana and several Nicotiana species (Brault et al., 2003). We identified N. glutinosa accessions that showed resistance against poleroviruses TuYV‐FL1 and Potato leafroll virus (PLRV), with phenotypes ranging from HR to extreme resistance in different accessions. Resistance appeared to be inherited as a single, dominant gene, designated RPO1 (Resistance to POleroviruses 1), with the P0 protein functioning as the Polerovirus elicitor of immune responses and cell death.
Results
Nicotiana glutinosa carries a resistance gene recognizing poleroviruses
A small‐scale screen of Nicotiana species was conducted to find accessions with potential resistance to poleroviruses based on the appearance of HR following infiltration of Agrobacterium tumefaciens carrying the infectious TuYV‐FL1 binary vector clone (pBIN‐TuYV) (Brault et al., 2003). The TuYV‐FL1 isolate was originally designated as a Beet western yellows virus (BWYV) isolate, but was reclassified in 2002 as a species distinct from bona fide BWYV; previous reports using the same clones as in our study retained the former designation (Baumberger et al., 2007; Bortolamiol et al., 2007; Csorba et al., 2010; Derrien et al., 2012; Pazhouhandeh et al., 2006). Within the species N. glutinosa, accessions were identified with different visible responses on leaves in the infiltration patch. Nicotiana glutinosa accession TW59 displayed a confluent cell death response within the infiltration patch beginning at 2 days post‐infiltration (dpi) that resembled an HR (Fig. 1A). The remaining six of seven accessions tested were phenotypically similar, and TuYV infiltration sites typically were indistinguishable from sites infiltrated with Agrobacterium carrying the empty pBIN61 vector, although, in some experiments, necrosis in part of the infiltration patches was observed with a delayed onset compared with the rapid uniform HR‐like necrosis observed for TW59 (Fig. S1, see Supporting Information). The N. glutinosa TW59 line is morphologically distinct from the remaining six accessions that share morphological characteristics and HR phenotypes for TuYV. Accession TW61 was selected as representative of these six N. glutinosa genetic backgrounds and used for further analysis (Fig. 1A ).
Figure 1.
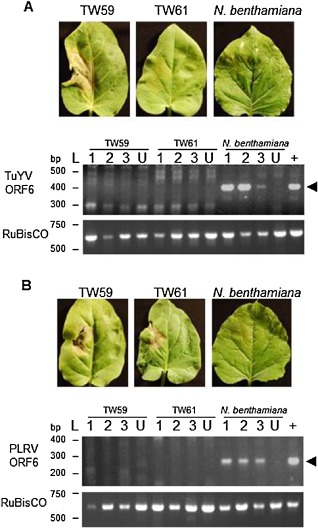
Nicotiana glutinosa is resistant to Turnip yellows virus (TuYV) and Potato leafroll virus (PLRV). Nicotiana glutinosa leaves of 4‐week‐old plants were agroinfected with Agrobacterium carrying the infectious clone of TuYV‐FL1. (A) Local infiltrated leaves of accession TW59 demonstrated a hypersensitive response (HR) against TuYV (left). Local leaves of TW61 (centre) and N. benthamiana (right, the susceptible control) appeared to be unresponsive. Representative leaves from three experiments performed in triplicate are shown. RNA from non‐inoculated upper leaves of N. benthamiana and N. glutinosa accessions TW59 and TW61 agroinfected with TuYV and control, uninfiltrated plants (U) was extracted for reverse transcription‐polymerase chain reaction (RT‐PCR). ORF6 of TuYV was chosen for PCR amplification, as it is included in all viral RNAs (genomic and subgenomic). The positive controls (+) were RT‐PCRs performed on RNA extracted from the local leaves of N. benthamiana infected with TuYV. The ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RuBisCO) large subunit was amplified to confirm RNA integrity. The expected size of the TuYV RT‐PCR product of 402 bp (arrowhead) was observed only in N. benthamiana, although some non‐specific products were amplified in all N. glutinosa samples, including uninfected leaves, with the large number of amplification cycles used. (B) To determine whether N. benthamiana and N. glutinosa accessions TW59 and TW61 displayed resistance against PLRV, leaves from the three plants were agroinfected with PLRV. Leaves were monitored for signs of cell death and photographed at 6 days post‐infiltration (dpi). To detect systemic spread of the virus, RNA was extracted from upper leaves of N. benthamiana and N. glutinosa accessions TW59 and TW61 agroinfected with pBIN‐PLRV. RT‐PCR targeting ORF6 of PLRV and the RuBisCO large subunit was performed on RNA from non‐inoculated upper leaves of plants infected by agroinfiltration, from control uninfected plants (U) and from infiltrated leaves of N. benthamiana (+). The expected size of the PLRV RT‐PCR product of 292 bp (arrowhead) was observed only in N. benthamiana, although some non‐specific products were amplified in all N. glutinosa samples, including uninfected leaves, with the large number of amplification cycles used.
To determine whether the cell death seen in the infiltration patch was indicative of resistance to TuYV, reverse transcription‐polymerase chain reaction (RT‐PCR) of the ORF6 region found in all viral RNAs (genomic and subgenomic) was performed to detect signs of systemic spread of the virus 10 days after agroinfiltration of the TuYV infectious clone. Although viral transcripts were detected in N. benthamiana, a susceptible species (Pfeffer et al., 2002), we were unable to amplify ORF6 from cDNA from N. glutinosa accessions TW59 and TW61 (Fig. 1A), indicating that both accessions were able to restrict viral spread to systemic tissues, although they differed in whether or not cell death was elicited by agroinfiltration of the infectious TuYV clone. TuYV RNA could be amplified from leaves at 2 dpi (Fig. S2, see Supporting Information) before the onset of HR, and transcripts of the ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RuBisCO) large subunit could be amplified from cDNA from non‐inoculated upper leaves (Fig. 1A), indicating that RNA extracted from N. glutinosa contained no inhibitors of the RT‐PCRs. More sensitive quantitative RT‐PCR also failed to amplify virus genome from non‐infected leaves of N. glutinosa (data not shown), substantiating the results seen with conventional RT‐PCR.
To determine whether the resistance activity against TuYV observed in N. glutinosa could protect against other poleroviruses, N. glutinosa accessions TW59 and TW61 and N. benthamiana were agroinfected with PLRV, a relative of TuYV in the same genus. HR‐like necrosis was detected on both TW59 and TW61 local leaves, but not on N. benthamiana (Fig. 1B). To verify that the observed HR‐like necrosis phenotypes in the N. glutinosa plants were accompanied by a resistance response against PLRV, RNA was extracted from non‐inoculated upper leaves at 10 dpi for RT‐PCR to amplify the PLRV ORF6. Similar to the results with TuYV, only N. benthamiana showed signs of PLRV ORF6 in the upper non‐inoculated leaves, whereas TW59 and TW61 had no viral RNAs detected by conventional RT‐PCR (Fig. 1B) or quantitative RT‐PCR (data not shown). PLRV RNA could be amplified from agroinfiltrated leaf tissue at 2 dpi (Fig. S2). The presence of cell death in agroinfected leaves and the lack of detectable PLRV RNAs in TW59 and TW61 non‐inoculated upper leaves suggest that both plants displayed the same resistance and cell death responses against PLRV.
To characterize the cell death observed in response to TuYV and PLRV, infiltrated leaves were examined for hydrogen peroxide accumulation by 3,3′‐diaminobenzidine (DAB) staining and induction of pathogenesis‐related (PR) gene expression. Hydrogen peroxide detection preceded cell death in TW59 leaf patches infiltrated with TuYV and PLRV, and in TW61 leaf patches infiltrated with PLRV (Fig. S3, see Supporting Information). PR‐1 transcript levels and PR‐2 protein levels showed some elevation at 2 dpi in TuYV‐challenged TW59 and PLRV‐challenged TW61 (Figs S4 and S5, see Supporting Information), but the differences in levels were only modest as Agrobacterium tumefaciens itself induces PR gene expression (Pruss et al., 2008). The phenotypes resembled other virus elicitors of resistance, suggesting that the cell death is HR‐type necrosis.
The P0 protein is an elicitor of immune responses in N. glutinosa
Several viral elicitors of corresponding R proteins have cellular functions as VSRs (Azevedo et al., 2010; Giner et al., 2010; Li et al., 1999). On the basis of these findings, we tested the P0 protein of TuYV (P0Tu) for such activity in N. glutinosa. Infiltration of the seven N. glutinosa accessions to transiently express P0Tu using the Cauliflower mosaic virus (CaMV) 35S promoter resulted in the appearance of HR on accession TW59 only (Table 1) . For comparison, plants were infiltrated with a construct expressing the TMV P50 protein that elicits the responses mediated by the N. glutinosa Toll interleukin 1 receptor (TIR)‐NB‐LRR protein N, and also with the combination of the coiled‐coil (CC)‐NB‐LRR protein Rx from potato and its elicitor, the PVX CP. Accession TW59 alone presented an HR elicited by P0Tu by 3 dpi, whereas P50 induced an HR within 2 days of agroinfiltration in the six accessions that did not respond to P0Tu (Table 1 and Fig. 2A). Two accessions (TW61 and TW66) were selected for further genetic analysis as representatives of the six lines that were unresponsive to P0Tu. We crossed pollen from TW59 plants onto TW61 and TW66 to examine the inheritance of HR in the progeny. Both P50 and P0Tu elicited HR on all F1 progeny examined, showing dominant inheritance of the N gene and a resistance (R) gene for TuYV, hereafter called RPO1. When F2 progeny were examined, P0Tu recognition was inherited in approximately a 3:1 ratio, as was P50 recognition (Table 1), demonstrating the expected Mendelian inheritance pattern of dominant R genes. The two R genes appear to be unlinked, as HRs segregated independently (Fig. 3).
Table 1.
Elicitation of hypersensitive response (HR) by P0T u in Nicotiana glutinosa is conferred by a single dominant locus
| Accession | Generation | HR P0Tu | HR P50TMV |
|---|---|---|---|
| TW58 | P | 0/12 | 12/12 |
| TW59 | P | 12/12 | 0/12 |
| TW61 | P | 0/12 | 12/12 |
| TW63 | P | 0/12 | 12/12 |
| TW64 | P | 0/12 | 12/12 |
| TW65 | P | 0/12 | 12/12 |
| TW66 | P | 0/12 | 12/12 |
| TW59 × TW61 | F1 | 30/30 | 30/30 |
| F2 | 35/47a | 35/47 | |
| TW59 × TW66 | F1 | 26/26 | 26/26 |
| F2 | 27/40b | 33/40 |
Chi‐squared analysis confirms that proportions fit the expected 3:1 ratio for a single gene trait with a χ2 value of 0.05 and P = 0.823.
Chi‐squared analysis confirms that proportions fit the expected 3:1 ratio for a single gene trait with a χ2 value of 1.2 and P = 0.273.
Figure 2.
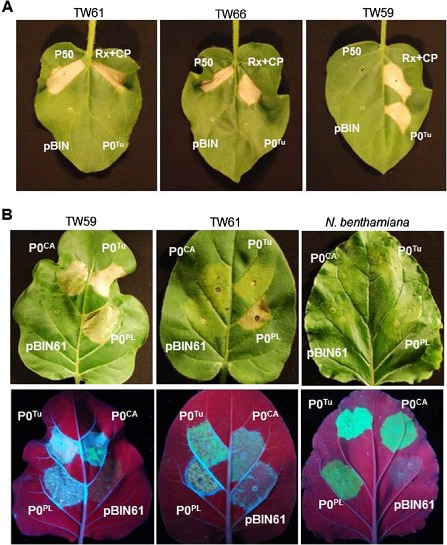
P0 proteins elicit hypersensitive response (HR)‐like cell death in Nicotiana glutinosa. (A) Transient over‐expression of the P0T u protein elicited cell death in Turnip yellows virus (TuYV)‐recognizing N. glutinosa line TW59. Leaves were infiltrated with Agrobacterium carrying binary vectors for transient expression of P0T u, the Tobacco mosaic virus (TMV) helicase (P50) recognized by the N gene and Rx plus its elicitor, the Potato virus X coat protein (PVX CP), or empty vector pBIN61. Leaves were photographed 5 days after infiltration. (B) To determine whether the P0 proteins from other viruses could also elicit HR in N. glutinosa, leaves from N. benthamiana and N. glutinosa accessions TW59 and TW61 were co‐infiltrated with the P0 proteins and green fluorescent protein (GFP). Agroinfiltrated leaves were photographed under white light to detect cell death and under UV light to show GFP accumulation enabled by P0 viral suppressor of RNA silencing (VSR) activity at 6 days post‐infiltration (dpi). The pBIN61 empty vector was co‐infiltrated with pBIN‐GFP as a negative control for HR induction and silencing suppression.
Figure 3.
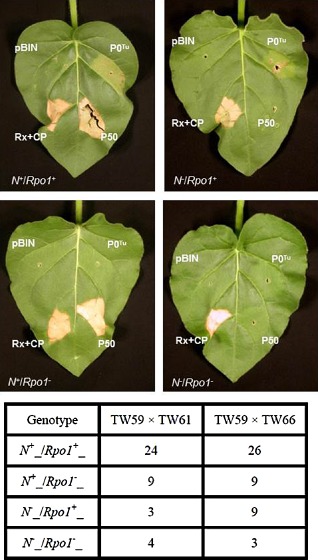
N and RPO1 (Resistance to POleroviruses 1) segregate independently in Nicotiana glutinosa. F 2 progeny of crosses between N. glutinosa accessions were agroinfiltrated for transient expression of the Tobacco mosaic virus (TMV) P50 and P0T u. Rx plus Potato virus X coat protein (PVX CP) and empty pBIN61 vector were infiltrated on the same leaves as positive and negative controls for hypersensitive response (HR) induction, respectively. Representative leaves of the four possible elicitor recognition phenotypes from TW59 × TW66 F 2 offspring are shown. Segregation ratios based on the number of plants presenting the four possible phenotypes (shown below representative leaves for trials with 40 and 47 individuals, respectively) were close to the 9:3:3:1 distribution expected for independently segregating genes that govern simple dominant traits, with a high level of statistical confidence (P > 0.99 and P > 0.90 for TW59 crossed with TW66 and TW61, respectively).
In order to determine whether the RPO1 activity could provide broad recognition of poleroviruses, the P0 proteins from three poleroviruses (TuYV, PLRV and CABYV) were infiltrated into the leaves of N. glutinosa TW59 and TW61 and N. benthamiana to test for HR elicitation. P0CA and P0Tu share 27% amino acid sequence identity (64% similarity), whereas P0PL shows greater divergence from the two yellows poleroviruses, exhibiting only 19% amino acid identity (47% similarity) to P0Tu. Agrobacterium tumefaciens carrying the green fluorescent protein (GFP) gene was co‐infiltrated with the P0 constructs to indirectly monitor whether P0 suppression of silencing was functional in each experiment, as determined by the accumulation of GFP protein in the infiltrated sites at 6 dpi. The P0CA and P0PL proteins elicited HR‐like necrosis on TW59 leaves typically beginning 3 days after agroinfiltration; only P0PL elicited cell death in TW61 (Fig. 2B). The cell death patterns for P0Tu and P0PL mirrored those observed on leaves infiltrated with infectious virus clones of TuYV and PLRV, further supporting P0 as an elicitor of HR‐type necrosis and suggesting it to be the elicitor of resistance to infectious Polerovirus through RPO1. This was further supported by the detection of hydrogen peroxide preceding cell death in TW59 patches infiltrated with all three P0 proteins and TW61 patches infiltrated with P0PL (Fig. S3), as well as elevated PR‐2 levels in plants infiltrated with P0 proteins compared with the controls (Fig. S5). In TW61, PR‐2 was similarly induced (Fig. S5) by P0PL, which produces necrosis, and P0Tu, which does not; this is consistent with our observation that TW61 shows extreme resistance to TuYV (Fig. 1A) and our suggestion that P0 is the elicitor of Rpo1.
As deletion of P0 from poleroviruses renders the viruses non‐infectious (Sadowy et al., 2001), we tested whether P0 is the Polerovirus elicitor by cloning the P0 genes into PVX to determine whether RPO1 recognition could be transferred to a heterologous virus. PVX has been used previously to identify the TBSV proteins P19 and P22 as HR elicitors in N. tabacum and N. clevelandii, respectively (Scholthof et al., 1995). Both N. glutinosa and N. benthamiana are susceptible to wild‐type PVX infection, which causes mild mosaic symptoms in both plants. PVX virus was rub inoculated onto leaves of N. glutinosa accessions TW59 and TW61, and N. benthamiana, and monitored for HR development and PVX CP accumulation. At 9 days post‐infection, cell death was observed on the local leaves of TW59 inoculated with PVX‐P0Tu, PVX‐P0PL and PVX‐P0CA as HR foci (Fig. 4A). As expected, inoculation of TW61 with PVX‐P0PL induced lesions on local leaves. Necrotic lesions were also observed on the leaves of TW61 inoculated with PVX‐P0Tu and PVX‐P0CA, and on N. benthamiana local leaves inoculated with PVX‐P0Tu, PVX‐P0PL and PVX‐P0CA; however, the appearance of chlorotic foci and their progression to tissue collapse and desiccation on N. benthamiana lagged behind that on N. glutinosa.
Figure 4.
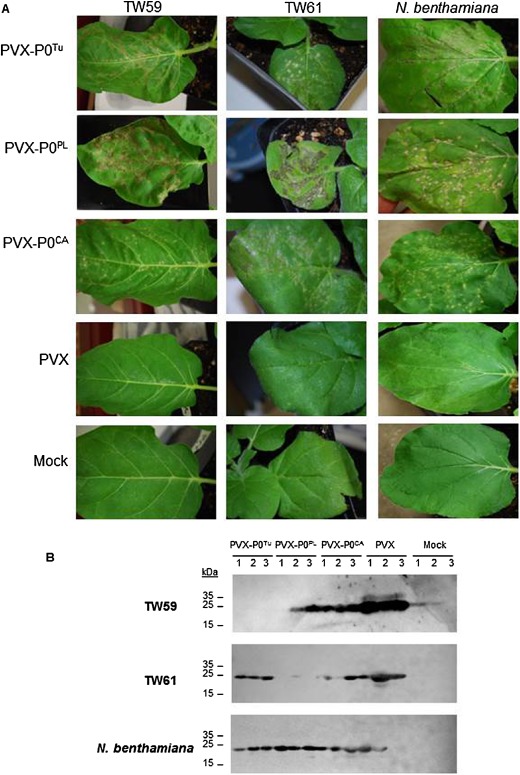
Heterologous expression of P0 by Potato virus X (PVX) confers avirulence on Nicotiana glutinosa. (A) Plants were rub inoculated with wild‐type (WT) PVX or PVX constructs heterologously expressing P0 proteins. At 9 days post‐infiltration (dpi), local leaves were monitored for cell death. Cell death was observed in local leaves of all plants infected with PVX constructs heterologously expressing P0 protein. Mock‐inoculated plants were rub inoculated with the same phosphate buffer as used to prepare virus stocks. (B) The presence of PVX in inoculated leaves was determined through immunoblotting to detect the PVX coat protein (CP). TW59 plants infected with PVX‐P0T u and TW61 plants infected with PV‐P0PL had greatly reduced or no CP accumulation in the inoculated leaves, suggesting that hypersensitive response (HR)‐like cell death observed on these leaves accompanied effective local resistance responses.
As HR does not always correspond to complete virus resistance, proteins were extracted from local leaves to detect the PVX CP. No CP was detected in TW59 local leaves infected with PVX‐P0Tu or in most TW61 local leaves infected with PVX‐P0PL (Fig. 4B and Table 2). Although TW59 recognized P0CA and P0PL expressed from pBIN61 (Fig. 2B) and cell death was observed in TW59 local leaves inoculated with PVX‐P0PL or PVX‐P0CA (Fig. 4A ), PVX CP was still detected in these leaves (Fig. 4B and Table 2), suggesting that PVX can spread sufficiently fast to escape the HR induced by these P0 proteins, which can prevent the Polerovirus from spreading.
Table 2.
P otato virus X coat protein (PVX CP) levels in plants inoculated with PVX‐P0T u, PVX‐P0PL and PVX‐P0CA
| TW59* | TW61* | Nicotiana benthamiana * | |
|---|---|---|---|
| Local | |||
| PVX‐P0Tu | 0/9 | 6/9 | 9/9 |
| PVX‐P0PL | 7/9 | 0/9 | 9/9 |
| PVX‐P0CA | 9/9 | 9/9 | 9/9 |
| PVX | 9/9 | 9/9 | 9/9 |
| Mock | 0/9 | 0/9 | 9/9 |
| Systemic | |||
| PVX‐P0Tu | 1/9 | 9/9 | 9/9 |
| PVX‐P0PL | 7/9 | 9/9 | 9/9 |
| PVX‐P0CA | 9/9 | 9/9 | 9/9 |
| PVX | 9/9 | 9/9 | 9/9 |
| Mock | 0/9 | 0/9 | 0/9 |
*Three trials were performed in total with each plant infected in triplicate for a total of nine plants.
Plants were also examined for the systemic spread of virus to assay resistance to PVX expressing P0. Non‐inoculated upper leaves of several inoculated TW61 and N. benthamiana plants revealed necrotic lesions, indicating that PVX‐P0Tu, PVX‐P0PL and PVX‐P0CA were able to spread systemically to upper leaves regardless of the cell death in local leaves, with trailing necrosis into the upper parts of the plant (Fig. 5A). Nicotiana benthamiana plants displayed a severe systemic necrosis phenotype that resulted in the death of the plant around 2 weeks post‐infection. In contrast, TW59 upper non‐inoculated leaves displayed the lowest levels of necrotic lesions when inoculated with PVX expressing the P0 proteins. Most notably, PVX‐P0Tu caused no lesions on non‐inoculated upper leaves of most TW59 plants, suggesting that P0Tu expression was sufficient to elicit resistance to PVX expressing P0Tu in TW59. Immunoblotting further confirmed that PVX carrying P0 proteins spread to upper non‐inoculated leaves in all nine TW61 plants; however, PVX CP was undetected in eight of nine TW59 plants inoculated with the PVX‐P0Tu construct, indicating that P0Tu could elicit resistance responses against PVX in this accession (Fig. 5B, Table 2). The extent of resistance to PVX‐P0Tu in TW59 and to PVX‐P0PL in TW61 was investigated further using real‐time PCR. Using this more sensitive assay, PVX RNA could be detected locally and systemically in all plants, even in the asymptomatic leaves of TW59 plants infected with PVX‐P0Tu, albeit at significantly lower relative levels than seen in the susceptible N. benthamiana plants (Fig. S6, see Supporting Information). Although N. glutinosa accessions showed resistance to both TuYV and PLRV, their specificity for the PVX‐P0 constructs differed, but was consistent with the observations that P0Tu elicits the most rapid and robust HR‐like response on TW59, and P0PL is the only one of the three P0s that elicits HR‐like cell death on TW61 that is similar in strength and timing of onset.
Figure 5.
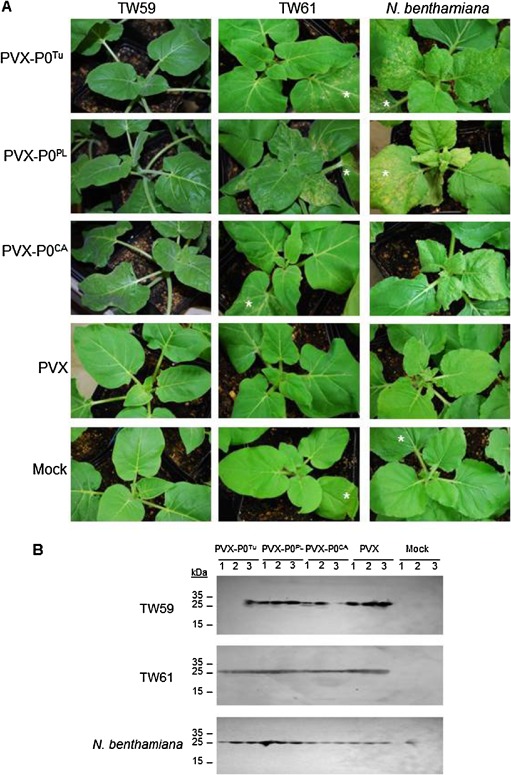
Non‐inoculated upper leaves of TW59 plants inoculated with Potato virus X (PVX) clones expressing P0T u demonstrate resistance. (A) Upper leaves of Nicotiana glutinosa accessions TW59 and TW61 and N. benthamiana plants were photographed 9 days after rub inoculation with the PVX constructs. Wild‐type PVX without heterologous protein expression (PVX) and mock inoculated plants in which leaves were rub inoculated with phosphate buffer were included as controls. Asterisks indicate the inoculated leaves that are in view. (B) PVX was detected systemically in plants at 9 days post‐infection by immunoblotting with anti‐PVX coat protein (CP) antibodies. TW61 failed to show resistance for all constructs, whereas some resistance was seen in TW59 to PVX expressing P0s (see Table 2).
Recognition by RPO1 requires P0T u F‐box protein function
The recognition of pathogens has been shown in different model systems to occur either by direct binding of cognate R proteins by pathogen elicitors or by indirect action on another host protein that may be guarded by its R protein (Jones and Dangl, 2006). To test whether RPO1 detects P0 activity either by generally monitoring silencing suppression or by guarding AGO1 and detecting P0‐mediated degradation, we transiently expressed VSRs from other viruses to test for HR elicitation in N. glutinosa. The two VSRs, CMV2b and TCV P38, were included as VSRs that interfere with AGO1 function through binding (Azevedo et al., 2010; Zhang et al., 2006), whereas the PVX P25 protein was included as a VSR that causes loss of AGO1 accumulation (Chiu et al., 2010). Other than the specific P0 interactions on N. glutinosa TW59 or TW61, none of the VSRs tested elicited cell death, indicating that perturbations in RNA silencing or AGO1 function are not sufficient to elicit RPO1 (Fig. 6). In addition, co‐infiltration of N. glutinosa accession TW59 with P0Tu and A. thaliana AGO1, AGO2 or AGO4 had no effect on the development of HR (data not shown).
Figure 6.
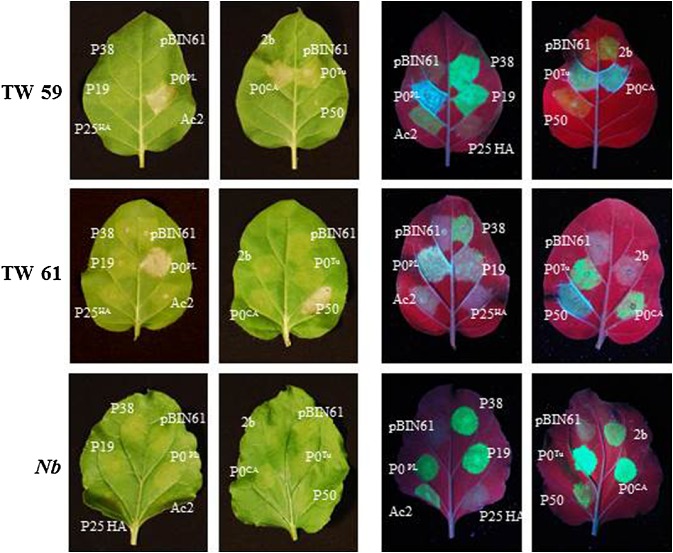
ARGONAUTE1 (AGO1)‐targeting viral suppressors of RNA silencing (VSRs) from other viruses failed to induce hypersensitive response (HR)‐like cell death in Nicotiana glutinosa. Plants were co‐infiltrated with green fluorescent protein (GFP) and VSR genes cloned into pBIN61 to test whether HR induction in N. glutinosa could be elicited generally by VSRs targeting AGO1. P0 proteins and Tobacco mosaic virus (TMV) P50 were included as positive controls for HR‐like cell death on TW59 (all P0 proteins) and TW61 (P0PL and P50). Potato virus X (PVX) P25 has been shown previously to target AGO1 for degradation (Chiu et al., 2010), whereas Cucumber mosaic virus (CMV) 2b and Turnip crinkle virus (TCV) P38 inhibit AGO1 activity through binding (Azevedo et al., 2010; Zhang et al., 2006). Leaves were photographed at 6 days post‐infiltration (dpi) under white (left) and UV (right) light to visualize HR‐like cell death and VSR activity through GFP accumulation.
To further test whether P0 activity could trigger HR, we tested a mutant in the P0Tu F‐box motif to determine whether its activity was required for the elicitation of RPO1‐mediated HR. The mutation of two amino acids of the F‐box motif (L63A, P64A) has been shown previously to abolish P0Tu VSR activity and the interaction between P0 and the ASK1 component of the SCF ligase complex (Pazhouhandeh et al., 2006). As the addition of P38 as a secondary silencing suppressor was seen to stabilize the P0TuLP1 mutant expression levels at 5 dpi (Pazhouhandeh et al., 2006), a time point at which transient expression in the absence of a VSR is seen to be silenced, P0TuLP1 was also tested for HR induction on co‐expression of P38. The F‐box motif mutant was unable to elicit HR, even in the presence of the secondary silencing suppressor (Fig. 7A).
Figure 7.
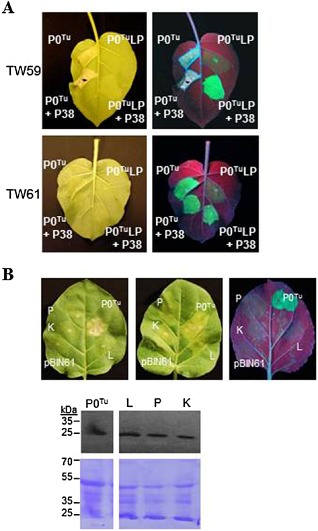
The F‐box function of P0T u is required for RPO1 (Resistance to POleroviruses 1) function. (A) Transient expression of P0T u failed to elicit a cell death response in resistant Nicotiana glutinosa (left) when a mutation was introduced into the F‐box motif (P0LP1). The LP1 mutation abolishes P0 function as a suppressor of gene silencing in N. glutinosa, seen by the co‐expression of P0 and green fluorescent protein (GFP) in leaves of susceptible plants (right). (B) Three new P0T u F‐box mutants, designated L (L60A and L63A), P (P64G) and K (R57K and L60A), were cloned with a carboxy‐terminal haemagglutinin (HA) tag for detection on immunoblots. Agrobacterium carrying wild‐type and mutant P0T u constructs or empty pBIN61 vector was co‐infiltrated with pBIN61‐GFP and examined at up to 6 days post‐infiltration (dpi). Mutants were all impaired in both hypersensitive response (HR) elicitation on TW59 and silencing suppression when visualized under UV light at 6 dpi, but all P0T u proteins accumulated to similar levels when detected on immunoblots with anti‐HA horseradish peroxidase (HRP). The Coomassie brilliant blue‐stained blot is shown for comparison of protein loading.
Three additional mutants in the F‐box motif were generated and cloned into pBIN61 in frame with a carboxy‐terminal haemagglutinin (HA) tag, designated mutants L (L60A and L63A), P (P64G) and K (R57K and L60A). The wild‐type HA‐tagged P0Tu elicited HR similar to the untagged protein, beginning at 2 dpi. All three F‐box mutants showed loss of silencing suppression in N. benthamiana and were unable to elicit HR in N. glutinosa TW59, despite accumulating to levels similar to that of the functional wild‐type protein (Fig. 7B).
Discussion
The continuing identification of viral proteins that function to suppress RNA silencing demonstrates that this mechanism is a significant defence barrier against viral infection (Pumplin and Voinnet, 2013). Overcoming this defence mechanism is crucial for successful infection by RNA viruses, including the poleroviruses: in the context of TuYV, mutation of the P0 protein in its F‐box motif, which abolishes the suppressor of gene silencing activity in a GFP transient expression assay, renders the virus incapable of infecting an otherwise susceptible host, N. benthamiana (Pazhouhandeh et al., 2006). Given the importance of RNA silencing as a first line of defence within the cell, it can be easily envisioned that this critical mechanism for virus‐targeted innate immunity could be under surveillance by a second line of defence, namely ETI (Sansregret et al., 2013). The evolution of resistance genes in plants that detect VSRs is a counterpart of the ETI evolved in plants directed against microbial effectors secreted into cells to dampen innate immunity. In some cases, it appears that effectors bind directly to plant R proteins and could potentially activate them through this binding; however, the activity of other effectors on a target host protein has been shown to occur indirectly via the activation of a cognate R protein that has the targeted protein under surveillance (Collier and Moffett, 2009; Dodds and Rathjen, 2010). In the latter case, monitoring the activity of an effector could represent a more stringent barrier for viruses to overcome, as loss of elicitor activity would reduce virus pathogenicity and fitness on both resistant and susceptible plants.
In the case of P0, we surmised that the low sequence identity among the three proteins from different poleroviruses suggests that a shared activity by these P0 proteins triggers RPO1 in TW59 and allows for resistance to both TuYV and PLRV in both TW59 and TW61, albeit with phenotypes presenting differently in terms of HR accompanying resistance. Loss of P0Tu VSR activity by mutation of the F‐box‐like motif is accompanied by loss of its elicitation of HR, implicating P0 function in an E3 ubiquitin ligase complex as a potential trigger of ETI. Taken together, the elicitor functions demonstrated for VSRs targeting AGO1, the related role of Polerovirus P0 proteins in targeting AGO1 to suppress antiviral defences and the observation that the P0 protein is encoded by the most rapidly evolving ORF of the Polerovirus genus (Krueger et al., 2013) all suggest that P0 is involved in a molecular arms race with plant hosts and is a likely elicitor of ETI in some plant species.
The presentation of HR during infection versus extreme resistance, which is not accompanied by cell death, is well documented for viruses, and is dependent on a number of factors, including plant genetic background (Moffett, 2009). Differences in phenotype when comparing virus infection versus transient expression of the elicitor protein have also been observed, as exemplified by the Rx protein, which mediates extreme resistance to PVX, whereas the PVX CP, its elicitor, induces HR when expressed by agroinfiltration (Bendahmane et al., 1999), or by a similar difference seen between TBSV infection versus expression of the TBSV P19 protein in tobacco (Sansregret et al., 2013). The differences observed in HR‐like necrosis induced by TuYV and PLRV infection versus transient over‐expression of their P0 genes, together with the differences in resistance involving HR versus extreme resistance, are consistent with reports for other viral elicitors of plant immune responses that vary in cell death phenotype. In the case of our PVX constructs expressing P0 proteins, N. benthamiana showed systemic necrosis responses leading to death of the plant typical of a susceptible interaction and distinct from the HR‐associated necrosis that can limit virus spread (Mandadi & Scholthof, 2013). In N. glutinosa, only resistance that was incomplete was observed in the case of PVX‐P0Tu on TW59 when non‐inoculated upper leaves were examined. RPO1 may be an effective immune receptor to activate defences and eliminate poleroviruses, but potexviruses, such as PVX, which are not phloem limited, are possibly able to spread more rapidly systemically than resistance can be activated. However, RT‐PCR detection of low levels of PVX‐P0 virus in the non‐inoculated upper leaves of N. glutinosa plants did not determine whether a functional P0 protein was retained in the virus which was able to spread throughout the resistant plants from the inoculated leaves. The reduced systemic spread of PVX from TW61 leaves inoculated with PVX‐P0PL and from TW59 leaves inoculated with PVX‐P0Tu supports our proposal of P0 as the Polerovirus elicitor of immune responses on N. glutinosa. It would be interesting to establish whether or not these resistance activities in accessions TW59 and TW61 are determined by different alleles at the same locus.
We suggest that a potential candidate for P0 targeting for indirect detection by RPO1 may be AGO1, as it is targeted for degradation in the presence of P0 (Baumberger et al., 2007; Bortolamiol et al., 2007). P0 has been shown to interact with A. thaliana AGO1 both in vitro and in vivo (Bortolamiol et al., 2007). We speculate that this interaction could resemble the association of the A. thaliana protein RIN4 with the NB‐LRR protein RPS2: RPS2 is activated on degradation of RIN4 caused by the elicitor of RPS2, the Pseudomonas syringae AvrRpt2 protein (Mackey et al., 2003). However, none of the AGO1‐perturbing VSRs tested induced HR, suggesting that AGO1 elimination per se is not responsible for RPO1 activation (Baumberger et al., 2007; Bortolamiol et al., 2007; Chiu et al., 2010). Indeed, if loss of AGO1 through degradation induced by P25 or P0 was sufficient to activate RPO1, it would be expected that N. glutinosa would have shown resistance to PVX infection rather than be fully susceptible, as seen in our experiments, as the gene encoding P25 is present in the PVX genome. However, we do not rule out the possibility that RPO1 recognizes an interaction specific to the P0–AGO1 association.
In addition to AGO1, P0Tu has been shown to cause degradation of AGO2, AGO4, AGO5, AGO6 and AGO9 (Baumberger et al., 2007). P25, however, was shown to interact with AGO1, AGO2, AGO3 and AGO4, but only AGO1 accumulation was affected by the presence of P25 (Chiu et al., 2010). AGO2 represents another likely candidate for RPO1 surveillance, as this AGO isoform has been shown to play a major role in antiviral resistance (Alvarado and Scholthof, 2011). It remains to be determined whether P0Tu, P0CA and P0PL have the same or different specificities for N. glutinosa AGO proteins. Our present observations maintain the possibilities that: (i) RPO1 proteins encoded by different alleles survey alternative AGO proteins that are degraded in the presence of P0 proteins; or (ii) RPO1 interacts directly with P0 proteins and divergent amino acid residues determine the different recognition specificities seen on different N. glutinosa accessions.
The lack of effective resistance in many field crops to poleroviruses presents an opportunity for the deployment of a dominant R gene, such as RPO1, as a transgene to protect important commercial varieties. Conventional breeding has achieved a reduction in sugar beet losses to TuYV; however, resistance traits available in the sugar beet germplasm are limited to quantitative trait loci (Stevens et al., 2005). Further improvements in sugar beet resistance to TuYV by conventional plant breeding are unlikely with reliance on multigenic, additive traits that must be introduced without loss of the original commercial traits of the crop cultivar. Dominant resistance has been shown in a Lactuca virosa accession IVT 280, which was followed up with a report of extreme resistance and, possibly, immunity to TuYV in Lactuca species in a field trial; however, the molecular determinants of this resistance have yet to be identified (Maisonneuve et al., 1991). PLRV is an important pathogen of potato that can reduce tuber yields by up to 50% in virus‐infected plants, with known resistance limited to quantitative trait loci (Taliansky et al., 2003). Some resistance has been identified in wild potato relatives; one resistance locus in Solanum etuberosum, Rlretb, has recently been mapped, allowing the introgression of resistance into commercial potato varieties with marker‐assisted breeding (Kelley et al., 2009), although this presents the challenge of maintaining the characteristics of the original commercial variety. So far, resistance by transformation of popular potato varieties has been accomplished by transgenic expression of PLRV ORFs for pathogen‐derived resistance (Kawchuk et al., 1990); however, this approach lacks broad specificity for the poleroviruses that could be achieved if RPO1 could provide protection from species within the genus that diverge at the nucleotide level. Given the ease of transient expression in Nicotiana species, this genus may provide a source of disease resistance genes to multiple pathogens (Vega‐Arreguin et al., 2014).
Although we cannot exclude the possibility that the activities seen against TuYV and PLRV in TW59 and TW61 are caused by different, genetically linked R genes, RPO1 could represent natural resistance to different Polerovirus species through the recognition of the most rapidly evolving protein encoded in the virus genome (Krueger et al., 2013). RPO1 resistance may prove to be broadly durable in the field against poleroviruses if P0 activity or amino acid residues that are absolutely required for virulence are necessary to trigger ETI; sequence changes that do not alter this function are unlikely to allow escape from detection, whereas mutants in P0 that block RPO1 detection by eliminating this virulence activity will face negative selection. The elucidation of how P0s trigger RPO1‐mediated ETI could also provide an insight into the virulence function of this protein, allowing us to further understand Polerovirus pathogenesis.
Experimental Procedures
Plant materials and agroinfiltration
Wild‐type N. glutinosa accessions (obtained from the USDA National Genetic Resources Program) and N. benthamiana seeds were sown in Sunshine Mix #1 soil, covered with vermiculite and grown under constant fluorescent light at 22 °C. After 2 weeks, individual plants were transferred into pots and allowed to grow for another 2 weeks before agroinfiltration. The plants were placed in growth chambers with a 16 h : 8 h light–dark cycle at 22 °C for several days before agroinfiltration to acclimatize to the photoperiod. P0Tu F‐box mutants were generated by overlapping PCR using primers to introduce mutations and restriction sites for cloning into the pBIN61 vector linearized by XbaI and BamHI digestion for fusion with the HA epitope tag at the C‐terminal end, as described previously (Sacco et al., 2007). All pBIN clones were electroporated into Agrobacterium tumefaciens C58C1 containing the helper plasmid pCH32. Overnight A. tumefaciens cultures were diluted to an optical density at 550 nm (OD550) of 0.1 in agroinfiltration buffer [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES) buffer and 10 mm MgCl2 at pH 5.6] plus 150 μm of acetosyringone for individual or co‐infiltrations. After agroinfiltration, the plants remained in the growth chambers for up to 2 days before being returned to constant light for phenotype development. Plant leaves that were co‐infiltrated with P0 constructs and GFP were digitally photographed (Nikon, Tokyo, Japan) at 6 dpi under white light to record symptoms of HR and illuminated with Blak Ray B‐100 long‐wave ultraviolet lamps (UVP) in the dark to observe for the presence or absence of GFP fluorescence. Hydrogen peroxide accumulation was detected by staining with 1 mg/mL DAB dissolved in 10 mm disodium phosphate buffer, pH 7, with 0.05% (v/v) Tween 20 for 4 h in the dark (Thordal‐Christensen et al., 1997). Leaves were destained by boiling in ethanol and stored in 70% (v/v) glycerol until photographed.
RNA isolation and RT‐PCR detection
To detect for the presence of virus in the plants, 0.1 g samples of leaf tissue from plants agroinfected with PLRV or TuYV were collected after 2 days (local) or 10 days (systemic) and extracted using the RNeasy Plant Mini Kit (Qiagen, Venlo, The Netherlands). The RNA samples were reverse transcribed using a SuperScript III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA) employing the manufacturer's protocol. Standard PCR was performed using 0.5 μm of both forward and reverse primers targeting the ORF6 of TuYV (TuYVORF6f, 5′‐AACCTCAACCGATCAGAAC‐3′; TuYVORF6r, 5′‐AACCTCAACCGATCAGAAC‐3′) or PLRV (PLRVORF6f, 5′‐CTGACTTTTCTCCGAAAG‐3′; PLRVORF6r, 5′‐GATGAGACACCATACGTG‐3′). Control PCRs were performed on the RuBisCO large subunit using primer pair RbcLf (5′‐ATGTCACCACAAACAGAGACTAAAGC‐3′) and RbcLr (5′‐GTAAAATCAAGT CCACCRCG‐3′) to verify the integrity of RNA and cDNA synthesis. The tubes were placed in an MJ mini gradient thermal cycler (Bio‐Rad, Hercules, CA, USA) for PCR, with the following programme used: one cycle of 98 °C for 1 min, 40 cycles of 98 °C for 15 s, 45 °C for 30 s, 72 °C for 1 min, and one cycle of 72 °C for 5 min. Real‐time PCR was conducted using the SensiMix SYBR No‐ROX Kit (Bioline, Taunton, MA, USA) with 0.1 μm primers to amplify actin (ActF, 5′‐AGTGGTCGTACAACTGGTATTG‐3′; ActR, 5′‐GCAAGGTCCAAACGAAGAATG‐3′), TuYV ORF6 (Tu‐ORF6F, 5′‐GGGCAAGGTATGAACGGATTA‐3′; TuORF6R, 5′‐GGGAAGGGTCAGGTTGTATC‐3′), PLRV ORF6 (PL‐ORF6F, 5′‐ACGCAACGATAACACCTACC‐3′; PL‐ORF6R, 5′‐CGGGACCAACGAGAAAGAAT‐3′), PR‐1 (PR‐1F, 5′‐GTGGGTCGATGAGAAACAGTAT‐3′; PR‐1R, 5′‐GAACCCTAGCACATCCAACA‐3′) or PVX (PVXfor, 5′‐GAAGTATGCTCCAGTGGTATGG‐3′; PVXrev, 5′‐GGTTGGTGACTCCATTGAAGA‐3′). PCRs were performed using the Bio‐Rad C1000 Thermal Cycler with the following amplification protocol: one cycle of 95 °C for 10 min, 45 cycles of 95 °C for 15 s, 53 °C for 15 s, 72 °C for 15 s, and one cycle of 95 °C for 10 s. Melting curves of PCR amplicons were obtained immediately following amplification with temperatures ranging from 65 to 90 °C, with 0.5 °C increment increases in temperature every 5 s.
PVX constructs and mechanical infection of N. glutinosa
PVX constructs heterologously expressing P0 proteins were cloned by ligating P0 genes PCR amplified with the primer pair PVXMCSfor (5′‐CATCGATTCCCGGGCATTTCATTTGGAGAGGAC‐3′) and PVXMCSrev (5′‐GTCGACCCGGGAGAGAGACTGGTGATTTCAG‐3′), which adds a ClaI restriction site at the 5′ end and a SalI restriction site at the 3′ end of the PCR products for cloning into the MCS of the vector pGR107 between the SalI and ClaI sites (Lu et al., 2003). Agrobacterium tumefaciens strain GV3101 with plasmid pSoup was transformed by electroporation with the vectors, and then infiltrated into N. benthamiana as described above. Reconstituted PVX viruses were extracted from plant sap using 50 mm potassium phosphate buffer, pH 7.2. The PVX extracts were rub inoculated onto the plant leaves dusted with carborundum, with mock‐infected plants rub inoculated with potassium buffer.
Immunoblotting
Total proteins from agroinfiltrated plants or PVX‐infected tissues were extracted in 2.5 mL extraction buffer [GTEN (10% (v/v) glycerol, 25 mm tris(hydroxymethyl)aminomethane (Tris), pH 7.5, 1 mm ethylenediaminetetraacetic acid (EDTA), 150 mm NaCl), 10 mm dithiothreitol (DTT), protease inhibitor cocktail and 2% (w/v) polyvinylpolypyrrolidone (PVPP)] per gram of tissue. The homogenates were centrifuged at 13 000 g for 10 min at 4 °C; 100 μL of the homogenate were mixed with 25 μL of 5 × sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) loading buffer and 6.75 μL of β‐mercaptoethanol, and boiled for 5 min at 100 °C prior to gel loading. Samples were electrophoresed through denaturing 12.5% or 15% polyacrylamide gels and then transferred to poly(vinylidene difluoride) (PVDF) membranes. The blocked membranes were incubated with anti‐PVX CP (Agdia Inc., Elkhart, IN, USA) diluted to 1:2000 in Tris‐buffered saline containing 0.1% (v/v) Tween 20 (TBS‐T) for 1 h, followed by rabbit anti‐mouse immunoglobulin G (IgG) horseradish peroxidase (HRP)‐conjugated antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) diluted to 1:5000 in TBS‐T for 1 h, with appropriate washing. For detection of PR‐2 (Class I β‐1,3‐glucanase), immunoblotting was similarly performed with rabbit polyclonal antibodies (Agrisera, Vännäs, SWEDEN) diluted to 1:2000, followed by mouse anti‐rabbit HRP conjugate (Santa Cruz Biotechnologies) diluted to 1:5000. HA‐tagged P0BW proteins were incubated with anti‐HA HRP‐conjugated antibodies (Sigma, St. Louis, MO, USA) diluted to 1:5000 in TBS‐T for 1 h. Pierce ECL Western Blotting Substrate (Thermo Scientific, Waltham, MA, USA) reagents were applied to the membranes, and chemiluminescence was documented using the Kodak (Rochester, NY, USA) image station 4000MM Pro Imager.
Supporting information
Fig. S1 Turnip yellows virus (TuYV)‐induced hypersensitive response (HR)‐like cell death in Nicotiana glutinosa accession TW59. Nicotiana glutinosa leaves of 4‐week‐old plants were infiltrated with Agrobacterium carrying the infectious clone of TuYV‐FL1 or with empty pBIN61 vector. TW59 underwent HR‐like cell death beginning at 2 days post‐infiltration, whereas other N. glutinosa accessions demonstrated mild yellowing or some chlorosis with small patches of necrosis that appeared by 6 dpi, as shown by leaves representative of these two phenotypes from accessions TW61 and TW66.
Fig. S2 Polerovirus detection in Nicotiana glutinosa‐agroinfected leaves. Nicotiana glutinosa accessions TW59 and TW61 were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 (A, B) or Potato leafroll virus (PLRV) (C, D) or with empty pBIN61 vector. RNA was extracted from leaves at 48 h post‐infiltration for real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) amplification to confirm that the virus could be amplified from RNA extracted from N. glutinosa. No inherent inhibition of the RT‐PCR using RNA from this plant as template was observed. Plants were assayed in duplicate with the standard deviation determined for three technical replicates and PCRs were normalized to actin.
Fig. S3 Detection of hydrogen peroxide accumulation elicited by P0 in Nicotiana glutinosa. Leaves from N. glutinosa accessions TW59 and TW61 were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 or Potato leafroll virus (PLRV) (A, B) or the P0 genes (C, D) together with empty pBIN61 vector. The Tobacco mosaic virus (TMV) P50 gene was included as a positive control for TW61, which carries the N gene. Leaves were removed from plants 40 h (A, B) or 50 h (C, D) post‐infiltration and immersed in 3,3′‐diaminobenzidine (DAB) stain for 4 h in the dark prior to ethanol destaining and storage in 70% glycerol before being photographed.
Fig. S4 Pathogenesis‐related 1 (PR‐1) gene induction in Nicotiana glutinosa induced by poleroviruses. Nicotiana glutinosa leaves were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 (A) or Potato leafroll virus (PLRV) (B) or with empty pBIN61 vector. RNA was extracted from leaves at 48 h post‐infiltration for real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) amplification to detect PR‐1 gene induction. Higher levels of PR‐1 transcripts were observed in TW59 agroinfected with TuYV and in TW61 agroinfected with PLRV, the two combinations that demonstrate the most rapid and robust hypersensitive responses (HRs). Plants were assayed in duplicate with three technical replicates, and transcripts were normalized to actin.
Fig. S5 Pathogenesis‐related 2 (PR‐2) protein accumulation in Nicotiana glutinosa induced by poleroviruses. Nicotiana glutinosa leaves were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 or Potato leafroll virus (PLRV), with P0 genes or with empty pBIN61 vector. The Tobacco mosaic virus (TMV) P50 gene was included as a positive control on TW61, which carries the N gene, and non‐infiltrated leaves were used as negative controls (Cont). Proteins were extracted from leaves at 48 h post‐infiltration, immunoblotting with PR‐2 (Class I β‐1,3‐glucanase) rabbit polyclonal antibodies (Agrisera), followed by mouse anti‐rabbit horseradish peroxidase (HRP) conjugate. The blot was stained with Coomassie brilliant blue following visualization using ECL chemiluminescence. The positions of the closest protein standards from the PageRuler Plus Prestained Protein Ladder (Thermo Scientific) are given on the immunoblots.
Fig. S6 Detection of Potato virus X (PVX) carrying P0 in local and systemic leaves of resistant Nicotiana glutinosa. Virus stocks of PVX or PVX carrying P0Tu or P0PL were rub inoculated onto N. benthamiana (NB) or N. glutinosa accession TW59 or TW61. Local leaves (A, B) and non‐inoculated upper leaves (C, D) were collected at 9 days post‐inoculation for RNA extraction and real‐time polymerase chain reaction (PCR) detection of PVX. Leaves from uninfected N. glutinosa plants were included as negative controls for non‐specific amplification. PVX‐specific PCRs were normalized with actin, and relative levels are shown for three biological replicates for PVX and two biological replicates for the controls, and error bars show standard deviations for three technical replicates.
Acknowledgements
We are grateful to Veronique Ziegler‐Graff for providing clones for the expression of P0Tu and P0CA, and the Turnip yellows virus (TuYV) infectious virus clone, and Stewart Gray for an infectious Potato leafroll virus (PLRV) clone. We thank Nilay Patel and members of his laboratory, Christopher Ott and Chiaokai Wen, for assistance with quantitative polymerase chain reaction. This work was funded by the California State University Program for Education and Research in Biotechnology and the US National Science Foundation (#1122256). The authors have no conflicts of interest to declare.
References
- Alvarado, V.Y. and Scholthof, H.B. (2011) AGO2: a new Argonaute compromising plant virus accumulation. Front Plant Sci. 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, J. , Garcia, D. , Pontier, D. , Ohnesorge, S. , Yu, A. , Garcia, S. , Braun, L. , Bergdoll, M. , Hakimi, M.A. , Lagrange, T. and Voinnet, O. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen‐encoded GW repeat protein. Genes Dev. 24, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. and Alfano, J.R. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolamiol, D. , Pazhouhandeh, M. , Marrocco, K. , Genschik, P. and Ziegler‐Graff, V. (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621. [DOI] [PubMed] [Google Scholar]
- Brault, V. , Bergdoll, M. , Mutterer, J. , Prasad, V. , Pfeffer, S. , Erdinger, M. , Richards, K.E. and Ziegler‐Graff, V. (2003) Effects of point mutations in the major capsid protein of beet western yellows virus on capsid formation, virus accumulation, and aphid transmission. J. Virol. 77, 3247–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, M.H. , Chen, I.H. , Baulcombe, D.C. and Tsai, C.H. (2010) The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, S.M. and Moffett, P. (2009) NB‐LRRs work a ‘bait and switch’ on pathogens. Trends Plant Sci. 14, 521–529. [DOI] [PubMed] [Google Scholar]
- Csorba, T. , Lozsa, R. , Hutvagner, G. and Burgyan, J. (2010) Polerovirus protein P0 prevents the assembly of small RNA‐containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472. [DOI] [PubMed] [Google Scholar]
- Derrien, B. , Baumberger, N. , Schepetilnikov, M. , Viotti, C. , De Cillia, J. , Ziegler‐Graff, V. , Isono, E. , Schumacher, K. and Genschik, P. (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA, 109, 15 942–15 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Giner, A. , Lakatos, L. , Garcia‐Chapa, M. , Lopez‐Moya, J.J. and Burgyan, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kawchuk, L.M. , Martin, R.R. and McPherson, J. (1990) Resistance in transgenic potato expressing the Potato leafroll virus coat protein gene. Mol. Plant–Microbe Interact. 3, 301–307. [Google Scholar]
- Kelley, K.B. , Whitworth, J.L. and Novy, R.G. (2009) Mapping of the potato leafroll virus resistance gene, Rlr (etb), from Solanum etuberosum identifies interchromosomal translocations among its E‐genome chromosomes 4 and 9 relative to the A‐genome of Solanum L. sect. Petota . Mol. Breed. 23, 489–500. [Google Scholar]
- Krueger, E.N. , Beckett, R.J. , Gray, S.M. and Miller, W.A. (2013) The complete nucleotide sequence of the genome of Barley yellow dwarf virus‐RMV reveals it to be a new Polerovirus distantly related to other yellow dwarf viruses. Front. Microbiol. 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.W. , Lucy, A.P. , Guo, H.S. , Li, W.X. , Ji, L.H. , Wong, S.M. and Ding, S.W. (1999) Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D.C. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alonso, J.M. , Ecker, J.R. and Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Maisonneuve, B. , Chovelon, V. and Lot, H. (1991) Inheritance of resistance to Beet western yellows virus in Lactuca‐virosa L. Hortscience, 26, 1543–1545. [Google Scholar]
- Malcuit, I. , Marano, M.R. , Kavanagh, T.A. , De Jong, W. , Forsyth, A. and Baulcombe, D.C. (1999) The 25‐kDa movement protein of PVX elicits Nb‐mediated hypersensitive cell death in potato. Mol. Plant–Microbe Interact. 12, 536–543. [Google Scholar]
- Mandadi, K.K. and Scholthof, K.B. (2013) Plant immune responses against viruses: how does a virus cause disease? Plant Cell, 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, P. (2009) Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 75, 1–33. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Marrocco, K. , Lechner, E. , Berry, B. , Brault, V. , Hemmer, O. , Kretsch, T. , Richards, K.E. , Genschik, P. and Ziegler‐Graff, V. (2006) F‐box‐like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. , Dunoyer, P. , Heim, F. , Richards, K.E. , Jonard, G. and Ziegler‐Graff, V. (2002) P0 of Beet western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76, 6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pruss, G.J. , Nester, E.W. and Vance, V. (2008) Infiltration with Agrobacterium tumefaciens induces host defense and development‐dependent responses in the infiltrated zone. Mol. Plant–Microbe Interact. 21, 1528–1538. [DOI] [PubMed] [Google Scholar]
- Pumplin, N. and Voinnet, O. (2013) RNA silencing suppression by plant pathogens: defence, counter‐defence and counter‐counter‐defence. Nat. Rev. Microbiol. 11, 745–760. [DOI] [PubMed] [Google Scholar]
- Sacco, M.A. , Mansoor, S. and Moffett, P. (2007) A RanGAP protein physically interacts with the NB‐LRR protein Rx, and is required for Rx‐mediated viral resistance. Plant J. 52, 82–93. [DOI] [PubMed] [Google Scholar]
- Sadowy, E. , Maasen, A. , Juszczuk, M. , David, C. , Zagorski‐Ostoja, W. , Gronenborn, B. and Hulanicka, M.D. (2001) The ORFO product of Potato leafroll virus is indispensable for virus accumulation. J. Gen. Virol. 82, 1529–1532. [DOI] [PubMed] [Google Scholar]
- Sansregret, R. , Dufour, V. , Langlois, M. , Daayf, F. , Dunoyer, P. , Voinnet, O. and Bouarab, D. (2013) Extreme resistance as a host counter‐counter defense against viral suppression of RNA silencing. PLoS Pathog. 9, e1003435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Scholthof, H.B. , Scholthof, K.B. and Jackson, A.O. (1995) Identification of tomato bushy stunt virus host‐specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell, 7, 1157–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, M. , Freeman, B. , Liu, H.Y. , Herrbach, E. and Lemaire, O. (2005) Beet poleroviruses: close friends or distant relatives? Mol. Plant Pathol. 6, 1–9. [DOI] [PubMed] [Google Scholar]
- Taliansky, M. , Mayo, M.A. and Barker, H. (2003) Potato leafroll virus: a classic pathogen shows some new tricks. Mol. Plant Pathol. 4, 81–89. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z.G. , Wei, Y.D. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Vega‐Arreguin, J.C. , Jalloh, A. , Bos, J.I. and Moffett, P. (2014) Recognition of an Avr3a homologue plays a major role in mediating non‐host resistance to Phytophthora capsici in Nicotiana species. Mol. Plant–Microbe Interact. 27, 770–780. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Lin, Z. and Xin, Z. (2009) Research progress in BYDV resistance genes derived from wheat and its wild relatives. J. Genet. Genomics, 36, 567–573. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , DelGrosso, L. , Yigit, E. , Dempsey, D.A. , Klessig, D.F. and Wobbe, K.K. (2000) The amino terminus of the coat protein of Turnip crinkle virus is the AVR factor recognized by resistant arabidopsis. Mol. Plant–Microbe Interact. 13, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Zvereva, A.S. and Pooggin, M.M. (2012) Silencing and innate immunity in plant defense against viral and non‐viral pathogens. Viruses, 4, 2578–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Turnip yellows virus (TuYV)‐induced hypersensitive response (HR)‐like cell death in Nicotiana glutinosa accession TW59. Nicotiana glutinosa leaves of 4‐week‐old plants were infiltrated with Agrobacterium carrying the infectious clone of TuYV‐FL1 or with empty pBIN61 vector. TW59 underwent HR‐like cell death beginning at 2 days post‐infiltration, whereas other N. glutinosa accessions demonstrated mild yellowing or some chlorosis with small patches of necrosis that appeared by 6 dpi, as shown by leaves representative of these two phenotypes from accessions TW61 and TW66.
Fig. S2 Polerovirus detection in Nicotiana glutinosa‐agroinfected leaves. Nicotiana glutinosa accessions TW59 and TW61 were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 (A, B) or Potato leafroll virus (PLRV) (C, D) or with empty pBIN61 vector. RNA was extracted from leaves at 48 h post‐infiltration for real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) amplification to confirm that the virus could be amplified from RNA extracted from N. glutinosa. No inherent inhibition of the RT‐PCR using RNA from this plant as template was observed. Plants were assayed in duplicate with the standard deviation determined for three technical replicates and PCRs were normalized to actin.
Fig. S3 Detection of hydrogen peroxide accumulation elicited by P0 in Nicotiana glutinosa. Leaves from N. glutinosa accessions TW59 and TW61 were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 or Potato leafroll virus (PLRV) (A, B) or the P0 genes (C, D) together with empty pBIN61 vector. The Tobacco mosaic virus (TMV) P50 gene was included as a positive control for TW61, which carries the N gene. Leaves were removed from plants 40 h (A, B) or 50 h (C, D) post‐infiltration and immersed in 3,3′‐diaminobenzidine (DAB) stain for 4 h in the dark prior to ethanol destaining and storage in 70% glycerol before being photographed.
Fig. S4 Pathogenesis‐related 1 (PR‐1) gene induction in Nicotiana glutinosa induced by poleroviruses. Nicotiana glutinosa leaves were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 (A) or Potato leafroll virus (PLRV) (B) or with empty pBIN61 vector. RNA was extracted from leaves at 48 h post‐infiltration for real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) amplification to detect PR‐1 gene induction. Higher levels of PR‐1 transcripts were observed in TW59 agroinfected with TuYV and in TW61 agroinfected with PLRV, the two combinations that demonstrate the most rapid and robust hypersensitive responses (HRs). Plants were assayed in duplicate with three technical replicates, and transcripts were normalized to actin.
Fig. S5 Pathogenesis‐related 2 (PR‐2) protein accumulation in Nicotiana glutinosa induced by poleroviruses. Nicotiana glutinosa leaves were infiltrated with Agrobacterium carrying the infectious clone of Turnip yellows virus (TuYV)‐FL1 or Potato leafroll virus (PLRV), with P0 genes or with empty pBIN61 vector. The Tobacco mosaic virus (TMV) P50 gene was included as a positive control on TW61, which carries the N gene, and non‐infiltrated leaves were used as negative controls (Cont). Proteins were extracted from leaves at 48 h post‐infiltration, immunoblotting with PR‐2 (Class I β‐1,3‐glucanase) rabbit polyclonal antibodies (Agrisera), followed by mouse anti‐rabbit horseradish peroxidase (HRP) conjugate. The blot was stained with Coomassie brilliant blue following visualization using ECL chemiluminescence. The positions of the closest protein standards from the PageRuler Plus Prestained Protein Ladder (Thermo Scientific) are given on the immunoblots.
Fig. S6 Detection of Potato virus X (PVX) carrying P0 in local and systemic leaves of resistant Nicotiana glutinosa. Virus stocks of PVX or PVX carrying P0Tu or P0PL were rub inoculated onto N. benthamiana (NB) or N. glutinosa accession TW59 or TW61. Local leaves (A, B) and non‐inoculated upper leaves (C, D) were collected at 9 days post‐inoculation for RNA extraction and real‐time polymerase chain reaction (PCR) detection of PVX. Leaves from uninfected N. glutinosa plants were included as negative controls for non‐specific amplification. PVX‐specific PCRs were normalized with actin, and relative levels are shown for three biological replicates for PVX and two biological replicates for the controls, and error bars show standard deviations for three technical replicates.


