Summary
SDA1 encodes a highly conserved protein that is widely distributed in eukaryotic organisms. SDA1 is essential for cell cycle progression and organization of the actin cytoskeleton in yeasts and humans. In this study, we identified a P hytophthora capsici orthologue of yeast SDA1, named PcSDA1. In P . capsici, PcSDA1 is strongly expressed in three asexual developmental states (mycelium, sporangia and germinating cysts), as well as late in infection. Silencing or overexpression of PcSDA1 in P . capsici transformants affected the growth of hyphae and sporangiophores, sporangial development, cyst germination and zoospore release. Phalloidin staining confirmed that PcSDA1 is required for organization of the actin cytoskeleton. Moreover, 4′,6‐diamidino‐2‐phenylindole (DAPI) staining and PcSDA1‐green fluorescent protein (GFP) fusions revealed that PcSDA1 is involved in the regulation of nuclear distribution in hyphae and sporangia. Both silenced and overexpression transformants showed severely diminished virulence. Thus, our results suggest that PcSDA1 plays a similar role in the regulation of the actin cytoskeleton and nuclear division in this filamentous organism as in non‐filamentous yeasts and human cells.
Keywords: actin cytoskeleton, asexual development, nuclear localization, PcSDA1, Phytophthora capsici, virulence
Introduction
The highly conserved protein encoded by SDA1 was first described in Saccharomyces cerevisiae, where it was found to be essential for cell cycle progression and cell viability (Buscemi et al., 2000). SDA1 plays an essential role in controlling the actin cytoskeleton, which is a common essential structure in all eukaryotes. Organization of the actin cytoskeleton is required for polarized cell growth, cell motility, secretion and endocytosis, and is involved in the preparation of the cells for cytokinesis (Bi et al., 1998). In addition, organization of the actin cytoskeleton has been implicated directly in a surveillance mechanism, called the morphogenesis checkpoint, in eukaryotes (Sia et al., 1996). Accordingly, SDA1 has been reported to be involved in mitosis and nuclear division (Adams et al., 1990), and in cell proliferation (Pardee, 1989; Planas‐Silva and Weinberg, 1997; Zimmerman and Kellogg, 2001). In yeast, SDA1 associates with protein complexes involved in RNA metabolism (Gavin et al., 2002), and might function as a trans‐acting factor in rRNA processing (Ihmels et al., 2002), ribosome biogenesis (Babbio et al., 2004; Fromont‐Racine et al., 2003; Nissan et al., 2002; Takahashi et al., 2003) and protein synthesis (Zimmerman and Kellog, 2001). SDA1 is localized not only in the nucleolus, but also at low levels in the nucleoplasm (Andersen et al., 2002; Buscemi et al., 2000), which is consistent with its involvement in early to intermediate steps of ribosome biogenesis (Babbio et al., 2004). Despite its extensive characterization in S. cerevisiae and human cells, no orthologues have been characterized in filamentous fungi or oomycetes, where the filamentous polynucleate nature of the organisms creates unique challenges for the regulation of the cytoskeleton and nuclear division. Filamentous oomycete pathogens in the genus Phytophthora, such as the pepper pathogen Phytophthora capsici, cause destructive diseases on thousands of plant species (Erwin et al., 1983). Phytophthora capsici often produces long‐lived dormant sexual spores, but has an explosive asexual disease cycle (Erwin and Ribeiro, 1996). In the vegetative stage, P. capsici grows as coenocytic filamentous mycelium. The sporangia are formed on branched sporangiophores that emerge from hyphae. The mature sporangia are easily dislodged and, when immersed in water, can quickly release biflagellate motile zoospores that swim chemotactically towards plants and attack hosts (Lamour et al., 2012). A deeper understanding of the molecular mechanisms of development of sporangia and sporangiophores of P. capsici should advance our understanding of how P. capsici is able to attack its hosts, and thus should facilitate the development of new strategies for disease control. Recent studies have begun to elucidate the molecular mechanisms underlying asexual development in Phytophthora, especially in P. infestans, including the roles of some of the genes involved in interactions with host plants (Ah Fong and Judelson, 2003; Blanco and Judelson, 2005; Gaulin et al., 2002; Kim and Judelson, 2003; Latijnhouwers and Govers, 2003; Latijnhouwers et al., 2004; Maltese et al., 1995; Tani et al., 2004).
In order to characterize the role of SDA1 in a filamentous, multinucleate, non‐fungal eukaryote, we identified an orthologue of the yeast SDA1 gene in P. capsici, which we named PcSDA1. We characterized the role of PcSDA1 at different stages of morphological development and host infection by P. capsici through the use of gene silencing and overexpression. Our results provide insights into the functions and molecular regulation of SDA1 in filamentous organisms, such as P. capsici.
Results
Isolation of PcSDA1 and sequence analysis
The genome sequence of P. capsici was searched using full‐length sequences of the yeast and human SDA1 proteins using the program tblastn with an expected (E) value cut‐off of <10−15. A single gene was detected in both searches which encoded a protein having 40% amino acid identity with human SDA1 and 28% identity with yeast SDA1, in both cases spanning more than 90% of the length of each sequence. As the yeast and human proteins share 35% identity, we concluded that the target, named PcSDA1, was the P. capsici orthologue of SDA1. In support of this conclusion, PcSDA1 was also annotated in the Department of Energy Joint Genome Institute (DOE JGI) database with the pfam05285 domain ‘SDA1’. We cloned PcSDA1 (GenBank number KM275943) from the DNA of P. capsici SD33 using primers designed from the genome sequence (Table S1, see Supporting Information). PcSDA1 encodes a putative 700‐amino‐acid protein (Fig. 1A) with a calculated molecular mass of 80.4 kDa. Three putative bipartite nuclear localization signals were identified in PcSDA1 (Dingwall and Laskey, 1991) starting at residues 254, 582 and 683, respectively (Fig. 1A, C). A highly acidic region (52.9% acidic residues) is also present in the C‐terminal part of the protein, from residue 510 to residue 559 (Fig. 1A). Each of the four other Phytophthora SDA1 sequences, PiSDA1 (P. infestans), PrSDA1 (P. ramorum), PsSDA1 (P. sojae) and PpSDA1 (P. parasitica), also contained three putative nuclear localization signals (Table S2, see Supporting Information, Fig. 1C). Phylogenetic analysis indicated that 24 SDA1 sequences (Table S2) grouped into three clusters (Fig. 1B). PcSDA1 showed a substantial similarity with putative SDA1 proteins from various organisms from the filamentous ascomycete fungus Chaetomium globosum to the oomycete P. parasitica (Fig. 1B, C). The amino acid sequences of SDA homologues group into at least three different clusters. Amino acid sequences of 13 SDA homologues from different fungal species formed one cluster with at least 58% identity (light blue in Fig. 1B). The second cluster comprises the five SDA sequences from animal species with at least 48% identity (AnSDA, Fig. 1B, pale yellow). A third cluster includes the five sequences from Phytophthora species (PhSDA) which are at least 73.8% identical (Fig. 1B, pale green). The PhSDA and animal SDA sequences were, at most, 54.8% similar. In addition, nine SDA1 sequences from oomycete, fungal and animal species displayed many identical residues and conservative substitutions (Fig. S1, see Supporting Information). Therefore, the SDA1 genes encode a highly conserved protein which is widely distributed among eukaryotic organisms. Furthermore, as expected for a highly conserved housekeeping gene, the phylogeny of the SDA1 sequences closely tracks the species taxonomy.
Figure 1.
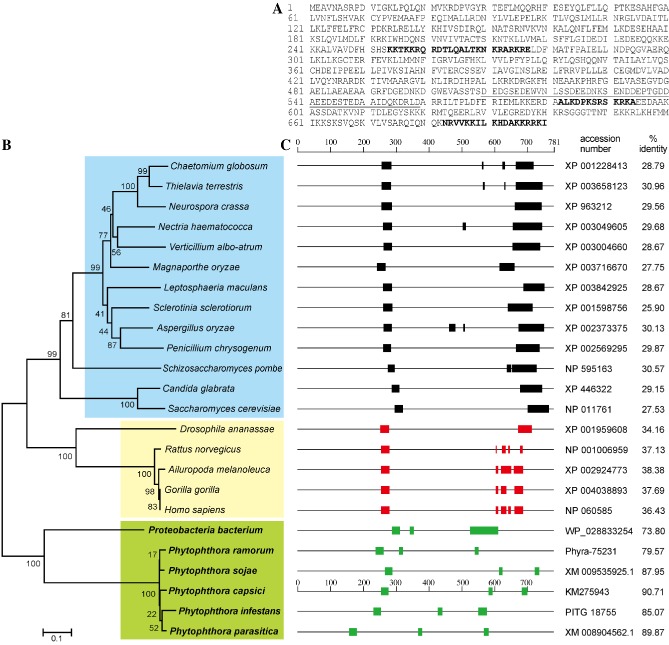
Sequence analysis of PcSDA1. (A) The complete amino acid sequence of PcSDA1 (accession number KM275943). Underlined, acidic region; bold capital letters, putative nuclear localization signals. (B) Phylogenetic analysis of 24 PcSDA orthologues. The phylogram was generated by the neighbour‐joining method as implemented in PAUP* 4.0 Beta. Numbers beside each node indicate bootstrap values as a percentage of 1000 replicates. (C) Schematic diagram comparing the PcSDA1 amino acid sequence and the orthologous sequences in other organisms, obtained using tblastn. The nucleotide sequence accession numbers and the percentage protein identity with PcSDA1 are listed on the right. The P roteobacterium sequence is probably a eukaryotic contaminant (see Experimental procedures). Black, green and red rectangles indicate putative nuclear localization signals.
PcSDA1 gene is differentially expressed during developmental and infection stages
To examine the transcription patterns of PcSDA1 in the five developmental stages (mycelia, sporangia, zoospores, cysts, germination cysts) and during seven time points of infection, we examined the expression of PcSDA1 using real‐time quantitative reverse‐transcribed polymerase chain reaction (qRT‐PCR) (Fig. 2). Transcript levels of PcSDA1 were 2.5‐fold or 3.5‐fold higher in sporangia than in mycelium or germinating cysts. The lowest expression levels were found in zoospores and cysts. During infection, the levels dropped slightly from 1.5 h post‐inoculation (hpi) to 6 hpi, and then climbed three‐fold higher to a peak at 24 hpi, before dropping slightly until 72 hpi. In summary, the transcript patterns of PcSDA1 in five developmental stages suggest that PcSDA1 plays a role during asexual development and, in particular, during mycelial growth, sporangial development and the germination of cysts. In addition, the different transcript patterns of PcSDA1 during infection suggest that PcSDA1 functions throughout infection and, especially, during the later infection stages. These results suggest that PcSDA1 is not only involved in asexual development, but may also play a role during infection.
Figure 2.
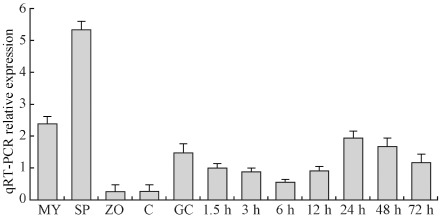
Transcript levels of the PcSDA1 gene. Transcript levels of PcSDA1 were determined by quantitative reverse‐transcribed polymerase chain reaction (qRT‐PCR). RNA was extracted from mycelia (MY), sporangia (SP), zoospores (ZO), cysts (C), germinating cysts (GC) and P hytophthora capsici lesions on the susceptible pepper cultivar 06221 at 1.5, 3, 6, 12, 24, 48 and 72 h post‐inoculation (hpi). Three housekeeping genes, β‐actin, β‐tubulin and U bc of P . capsici, and β‐actin of pepper were used as constitutively expressed endogenous controls and to normalize the expression of PcSDA1. Error bars represent standard errors calculated using three biological replicates for each sample.
Generation of PcSDA1 ‐silenced and overexpression lines
Silencing or overexpression of PcSDA1 in stable transformants created by protoplast transformation (McLeod et al., 2008) was used to test the function of the gene. The transcript levels of PcSDA1 were substantially depressed in five silenced transformants (SiPcSDA1‐1, SiPcSDA1‐2, SiPcSDA1‐3, SiPcSDA1‐4, SiPcSDA1‐5) as measured by RT‐PCR (data not shown) and qRT‐PCR (Figs 3B and S2A, see Supporting Information) assays. Each of these five silenced transformants showed the same phenotypes in hyphal growth, sporangial development and germination, and cyst germination (data not shown). Therefore, we only included two of the silenced transformants (SiPcSDA1‐3 and SiPcSDA1‐4) in subsequent experiments. Five putative overexpression transformants (OPcSDA1‐1, OPcSDA1‐2, OPcSDA1‐3, OPcSDA1‐4, OPcSDA1‐5) were identified by RT‐PCR (data not shown) and qRT‐PCR (Figs 3C and S2B) assays. These five overexpression transformants showed the same phenotypes in these three developmental stages. Thus, we only included two of the overexpression transformants (OPcSDA1‐1 and OPcSDA1‐2) in subsequent experiments.
Figure 3.
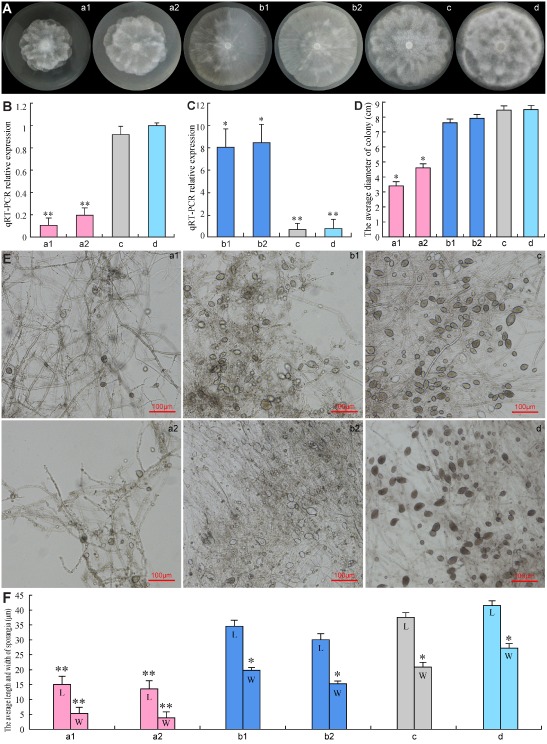
Effect of PcSDA1 silencing or overexpression lines on colony morphology and sporangial size. a1, silenced line SiPcSDA1‐3; a2, silenced line SiPcSDA1‐4; b1, overexpression line OPcSDA1‐1; b2, overexpression line OPcSDA1‐2; c, CK (empty vector transformant); d, WT (wild‐type strain SD33). (A, D) Growth and diameter of the colonies of different strains after 7 days. Colony growth rates were determined from at least 25 plates. Photographs were taken at 7 days. (B, C) Quantitative reverse‐transcribed polymerase chain reaction (qRT‐PCR) analysis of mRNA expression levels of PcSDA1 in mycelia of SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1, OPcSDA1‐2, CK and WT. Error bars represent standard errors calculated using three replicates. (E, F) Variation in sporangia size. L, length; W, width. The average size of sporangia was determined from at least 50 sporangia. Photographs were taken at 9 days after incubation. ‘*’ indicates a significant difference from WT (P < 0.05) using Student's t‐test. ‘**’ indicates a significant difference from WT (P < 0.01) using Student's t‐test. Each experiment was repeated at least three independent times.
The phenotypes of the silenced and overexpression transformants were evaluated in comparison with controls throughout the P. capsici life cycle. The full life cycle of P. capsici is typical of oomycete species, such as Pythium and Phytophthora, as described previously (van West et al., 2003). Comparison of several phenotypes among silenced transformants, overexpression transformants, CK (empty vector) and WT (wild‐type strain SD33) strains indicated that silencing or overexpression of PcSDA1 affected hyphal growth, sporangial development and germination, and cyst germination, as summarized in Table 1 and detailed in the following sections.
Table 1.
Comparison of asexual growth of PcSDA1‐silenced and overexpression lines with two controls
| Phenotype | PcSDA1‐silenced lines† | PcSDA1 overexpression lines† | Controls | |||
|---|---|---|---|---|---|---|
| SiPcSDA1‐3 | SiPcSDA1‐4 | OPcSDA1‐1 | OPcSDA1‐2 | CK | SD33 | |
| Colony diameter (cm)‡ | 3.3 ± 1.0** | 3.7 ± 1.1** | 6.6 ± 2.52* | 7.1 ± 2.1* | 9.1 ± 1.5 | 10.0 ± 1.5 |
| Sporangiophore density (number/cm2)§ | 15.0 ± 3.0** | 12.0 ± 3.0** | 21.6 ± 2.5* | 23.1 ± 2.1* | 29.6 ± 2.2 | 33.5 ± 2.0 |
| Sporangia density (number/cm2)¶ | 16.0 ± 22.5** | 18.5 ± 20.5** | 22.0 ± 20.5* | 26.0 ± 19.6* | 37.5 ± 11.0 | 40.4 ± 6.00 |
| Zoospore release (number) †† | 76.0 ± 3.0** | 70.0 ± 3.05** | 250.3 ± 2.53* | 241.24 ± 2.5* | 375.4 ± 12.5 | 408.6 ± 16.5 |
| Encystment (%)‡‡ | 92.0 ± 0.01** | 87.0 ± 0.01** | 50.3 ± 0.02* | 48.31 ± 0.02* | 30.2 ± 0.04 | 24.5 ± 0.03 |
| Cyst germination (%)§§ | 29.3 ± 0.02** | 25.3 ± 0.01** | 42.3 ± 0.01* | 47.24 ± 0.01* | 85.5 ± 0.01 | 94.3 ± 0.02 |
| Germ tube length of cysts (μm)¶¶ | 19.4 ± 10.0** | 17.5 ± 10.5** | 25.5 ± 10.01* | 21.06 ± 9.02* | 35.1 ± 4.45 | 38.6 ± 3.5 |
†Silenced or overexpression lines derived from SD33. An independent samples t‐test was used to compare each line with SD33; ‘*’ and ‘**’ indicate P < 0.05 and P < 0.001, respectively. Each test was duplicated three times. The values in the table are the mean ± standard error.
‡Based on 7 days of growth in 10% V8 juice agar medium at 25 °C.
§Based on an area of 2 cm2 in colonies grown on 10% V8 juice agar medium for 7 days. Each assay was repeated at least three times.
¶Based on counts of 10 plates made from 8‐day‐old cultures grown in 100‐mm plates at 25 °C.
††Number of 5 × 5‐mm2 colonies releasing zoospores after washing for 24 h at 25 °C. The numbers of zoospores in 10 μL of sterile distilled water were counted.
‡‡Percentage of encysted zoospores in 50 μL of zoospore suspension after 1 h of incubation at 25 °C with 80% humidity.
§§Percentage of cysts forming germ tubes after 2 h of incubation with vortexing to induce encystment at 25 °C, based on counting a minimum of 100 cysts from each strain.
¶¶The length of germ tubes formed from cysts after 8 h in 10% V8 liquid medium at 25 °C, based on averaging a minimum of 100 germinated cysts from each strain.
Influence of PcSDA1 on mycelial growth
To evaluate the requirement for PcSDA1 during mycelial growth, we measured the growth of colonies of two silenced lines (SiPcSDA1‐3 and SiPcSDA1‐4) and two overexpression lines (OPcSDA1‐1 and OPcSDA1‐2) compared with two controls (WT and CK). The colony diameters of SiPcSDA1‐3 and SiPcSDA1‐4 [Fig. 3A, D (a1, a2); Table 1] were significantly smaller than those of either of the controls (WT and CK) after 7 days [Fig. 3A, D (c, d); Table 1] (P < 0.01), whereas the diameters of OPcSDA1‐1 and OPcSDA1‐2 [Fig. 3A, D (b1, b2); Table 1) were nearly the same as those of the two controls (WT and CK). In addition, the colonies of SiPcSDA1‐3 and SiPcSDA1‐4 appeared to be more dense than those of OPcSDA1‐1 and OPcSDA1‐2 and either of the two controls [Fig. 3A (a1, a2, b1, b2, c, d)]. The most conspicuous feature of the SiPcSDA1‐3 and SiPcSDA1‐4 colonies was the fewer number of aerial hyphae than the controls (Fig. S3A, see Supporting Information); rather, the colonies formed a dense hyphal mass around the original site of inoculation [Fig. 3A (a1, a2)]. Although the colonies of OPcSDA1‐1 and OPcSDA1‐2 were less dense [Fig. 3A (b1, b2)] with numerous aerial hyphae (Fig. S3B), they spread quickly from the original site of inoculation when compared with either of the two controls, forming a sparse mycelial mass across the surface of solid V8 medium. In Fig. 3A (b1), however, the mycelium is less dense on the left, but more dense on the right. This might point to the presence of a revertant in the culture as a result of silencing of the transgene.
Influence of PcSDA1 on the number and size of sporangia, and zoospore release
PcSDA1 expression levels influenced the size of sporangia. As shown in Fig. 3E, F (a1, a2, b1, b2, c, d), the sporangial sizes of SiPcSDA1‐3 (length × width, 20 × 17 μm2) and SiPcSDA1‐4 (length × width, 22 × 15 μm2) were considerably smaller than those of CK (length × width, 40 × 25 μm2) and WT (length × width, 44 × 29 μm2) (P < 0.01). However, the sporangial sizes of OPcSDA1‐1 (length × width, 35 × 24 μm2) and OPcSDA1‐2 (length × width, 33 × 22 μm2) were larger than those of SiPcSDA1‐3 and SiPcSDA1‐4 (P < 0.05), but slightly smaller than those of sporangia formed by the controls (P < 0.05). Sporangia of the different strains were induced to differentiate and release zoospores by incubation in Petri's solution at 4 °C for 2 h. Zoospore release was affected by PcSDA1 silencing and overexpression. After 2 h of incubation in chilled Petri's solution, an average of 57%, 58%, 50% and 46% of the sporangia of SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2, respectively, failed to release zoospores, compared with only 6.7% and 8.6% in the controls (CK and WT, respectively). Incubation for longer than 2 h improved the efficiency of zoospore release in all strains, but the release time varied. After 3 h, most sporangia in CK and WT had released their zoospores, but completion of release was delayed to 5 h in SiPcSDA1‐3 and SiPcSDA1‐4 and 4 h in OPcSDA1‐1 and OPcSDA1‐2. In addition, the sizes of the released zoospores were affected by PcSDA1 transcript levels. In the case of SiPcSDA1‐3 and SiPcSDA1‐4, 22% and 26%, respectively, of all zoospores were unusually small. In the case of OPcSDA1‐1 and OPcSDA1‐2, 17% and 20%, respectively, were unusually large, which may have resulted from incomplete cleavage of the sporangial cytoplasm (see below).
Silencing and overexpression of PcSDA1 also affected the total number of sporangia produced by P. capsici at different time points. The numbers of sporangia formed in all strains were counted at 6, 12, 24, 36 and 48 h after induction, as described by Erwin and Ribiero (1996). The number of sporangia gradually increased from 6 to 24 h after induction, reaching a maximum at 24 h, following which the numbers gradually decreased until 48 h (Fig. 4). Compared with the number of sporangia in the control strains, the silenced strains SiPcSDA1‐3 and SiPcSDA1‐4 produced the least sporangia at 12, 24 and 36 h (Fig. 4) (P < 0.01; Wilcoxon rank sum test). At each time point, OPcSDA1‐1 and OPcSDA1‐2 produced fewer sporangia than did the controls, but the numbers of sporangia formed by these two overexpression transformants were larger than those of the two silenced transformants (Fig. 4) (P < 0.01; Wilcoxon rank sum test). These four transformants had few sporangia even 48 h after induction.
Figure 4.
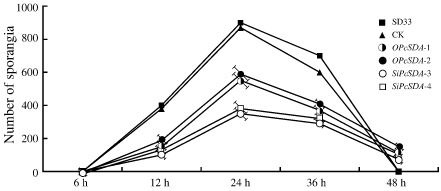
Sporangial production by PcSDA1‐silenced and overexpression lines. The numbers of sporangia were counted at 6, 12, 24, 36 and 48 h after induction of each P hytophthora capsici line into the sporangial forming stage (n > 50). Silenced lines, SiPcSDA1‐3 and SiPcSDA1‐4. Overexpression lines, OPcSDA1‐1 and OPcSDA1‐2. CK, empty vector transformant. WT, wild‐type strain SD33. Each experiment was repeated at least four times on independent occasions. Error bars indicate standard errors from three independent replicates.
Influence of PcSDA1 on sporangiophore and sporangial morphology
We observed the morphology of mycelia and sporangia using differential interference contrast (DIC) microscopy. In culture, sporangiophores arise from hyphae; the sporangiophores then branch and a single sporangium develops at the tip of each branch (Lamour et al., 2012). The sporangia of P. capsici are typically ovoidal to fusiform and have a single terminal papilla. We found that silencing of PcSDA1 had an effect on the frequency of branching of the sporangiophores, and hence the total number of sporangia, as well as the shape and size of the sporangia. The silenced strains produced the fewest branches on sporangiophores among the six strains (P < 0.01), as shown in Fig. 5 (a1‐2, a2‐2, b1‐2, b2‐2, c‐2, d‐2). In contrast, OPcSDA1‐1 and OPcSDA1‐2 produced redundant branches on sporangiophores compared with SiPcSDA1‐3 and SiPcSDA1‐4 and with either of the two controls. Moreover, the sporangia were substantially different in numbers [Fig. 3E (a1, a2, b1, b2, c, d); Table 1] and in shape [Fig. 5 (a1, a2, b1, b2, c, d)] between those produced by the different lines and either of the two controls. The two silenced lines SiPcSDA1‐3 and SiPcSDA1‐4 produced fewer sporangia compared with either of the two overexpression lines and the two controls [Fig. 3 (a1, a2, b1, b2, c, d); Table 1]. Moreover, fewer sporangia were formed by OPcSDA1‐1 and OPcSDA1‐2 than either of the two controls, but they were larger than those of SiPcSDA1‐3 and SiPcSDA1‐4 [Fig. 3E (a1, a2, b1, b2, c, d); Table 1]. In all cases, all sporangia from SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2 were malformed and lacked the distinct apical papillae that are characteristic of P. capsici and readily evident on the sporangia of the two controls [Fig. 5 (a1, a2, b1, b2, c, d)]. The hyphae and sporangia of the two controls were normal under the same conditions, as shown in Figs 3E and 5 (c, d).
Figure 5.
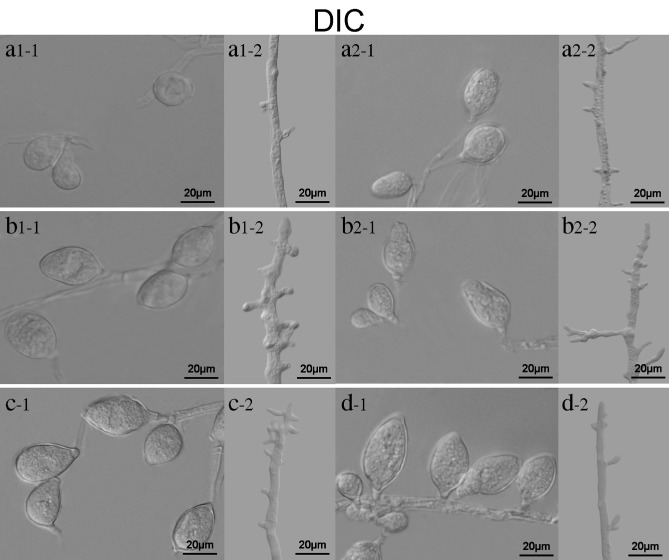
Morphology of sporangia and hyphae in PcSDA1‐silenced and overexpression lines. Sporangia and mycelia were collected from different P hytophthora capsici strains growing on V8 medium plates after 7 days. a1‐1, a1‐2, a2‐1, a2‐2, b1‐1, b1‐2, b2‐1, b2‐2, c‐1, c‐2, d‐1 and d‐2 were observed using differential interference contrast (DIC). a1 and a2, silenced lines SiPcSDA1‐3 and SiPcSDA1‐4; b1 and b2, overexpression lines OPcSDA1‐1 and OPcSDA1‐2; c, CK (empty vector transformant); d, wild‐type strain SD33. Each experiment was repeated at least four times on independent occasions and typical structures are shown.
To further characterize the development of hyphae, sporangiophores and sporangia in all the different lines, we utilized scanning electron microscopy (SEM) and transmission electron microscopy (TEM). In the silenced [Fig. 6A (a1, a2)] and overexpression [Fig. 6A (b1, b2)] lines, the branches on the sporangiophores were substantially different from those of the two controls [Fig. 6A (c, d)]. Sporangiophores of SiPcSDA1‐3 and SiPcSDA1‐4 [Fig. 6A (a1, a2)] produced only a few branches, whereas sporangiophores of OPcSDA1‐1 and OPcSDA1‐2 produced more and shorter branches [Fig. 6A (b1, b2)] compared with either of the two controls [Fig. 6A (c, d)]. In addition, sporangiophores of OPcSDA1‐1 and OPcSDA1‐2 [Fig. 6A (b1, b2)] were obviously thinner compared with those of either of the two silenced lines or the two controls. In order to understand the variation in size and shape of sporangia in silenced or overexpression lines, we examined the ultrastructure of the sporangia within SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2 using TEM. Prior to the induction of sporangial cleavage, there were substantial differences in the cellular structures of sporangia within the two silenced lines [Fig. 6B (a1, a2)] and two overexpression lines [Fig. 6B (b1, b2)] compared with either of the two controls [Fig. 6B (c, d)]. In the lines SiPcSDA1‐3 and SiPcSDA1‐4, the cytoplasm of the sporangia was more disorganized and the organelles were more poorly differentiated (in some cases still fused) [Fig. 6B (a1, a2)]. In the lines OPcSDA1‐1 and OPcSDA1‐2 [Fig. 6B (b1, b2)], however, the sporangia contained numerous lipid bodies with heterogeneous cytoplasm and irregular organization of the organelles compared with the two controls [Fig. 6B (c, d)]. In the controls, there were no significant differences in sporangial cleavage, as shown in Fig. 6B (c, d). Notably, the thickness of the cell wall of the sporangia in the two silenced strains [Fig. 6B (a1, a2)] appeared to be extremely thin at points when compared with either of the four other strains [Fig. 6B (b1, b2, c, d)]. We therefore conclude that PcSDA1 plays an important role during mycelial and sporangial development.
Figure 6.
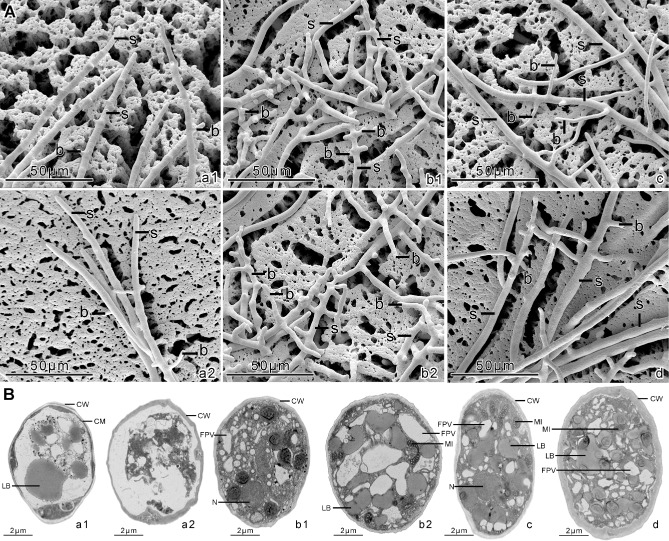
Ultrastructure of sporangia and hyphae in PcSDA1‐silenced and overexpression lines. (A) Scanning electron microscopy of the sporangiophores; b, branches on sporangiophores; S, sporangiophores. (B) Transmission electron microscopy of the sporangia; CW, cellular wall; FPV, vacuoles; LB, lipid bodies; MI, mitochondrion; N, cellular nuclei. a1, silenced line SiPcSDA1‐3; a2, silenced line SiPcSDA1‐4; b1, overexpression line OPcSDA1‐1; b2, overexpression line OPcSDA1‐2; c, CK (empty vector transformant); d, wild‐type strain SD33. Each experiment was repeated at least four times on independent occasions, and typical structures are shown.
PcSDA1 regulation of the actin cytoskeleton
In order to determine whether PcSDA1 plays a role in the regulation of the actin cytoskeleton during mycelial and sporangial development, we examined the actin cytoskeleton in mycelia and sporangia of the PcSDA1‐silenced, overexpression and control lines using phalloidin–rhodamine staining and confocal microscopy. The silenced lines SiPcSDA1‐3 and SiPcSDA1‐4 showed irregularities in the distribution of actin plaques and actin filaments in sporangia and, especially, in mycelia (Fig. 7A, B), compared with the control lines (Fig. 7E, F). In contrast, in the overexpression lines OPcSDA1‐1 and OPcSDA1‐2 (Fig. 7C, D), the actin plaques and actin filaments displayed irregularities in distribution and were brighter and more abundant in both the mycelia and sporangia compared with CK and WT (Fig. 7E, F). In particular, in the mycelia of the overexpression lines, the actin plaques became elongated, clavate and intensely stained [Fig. 7C (c1), D (d1)]. Therefore, PcSDA1 appears to regulate the abundance, distribution and morphology of actin plaques and actin filaments, especially in mycelia. This result is in accordance with the role of SDA1 in Saccharomyces cerevisiae (Buscemi et al., 2000).
Figure 7.
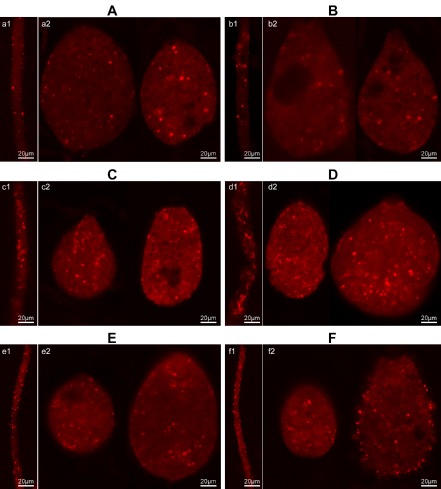
Phalloidin–rhodamine staining of the actin cytoskeleton within sporangia and mycelia of the PcSDA1‐silenced and overexpression lines compared with two controls. (A) Silenced line SiPcSDA1‐3: a1, mycelia; a2, sporangia. (B) Silenced line SiPcSDA1‐4: b1, mycelia; b2, sporangia. (C) Overexpression line OPcSDA1‐1: c1, mycelia; c2, sporangia. (D) Overexpression line OPcSDA1‐2: d1, mycelia; d2, sporangia. (E) CK (empty vector transformant): e1, mycelia; e2, sporangia. (F) Wild‐type strain SD33: f1, mycelia; f2, sporangia. Each experiment was repeated at least four times on independent occasions, and typical structures are shown.
PcSDA1 regulation of nuclear distribution
As actin plays an important role in the regulation of nuclear distribution in filamentous fungi, we investigated whether PcSDA1 regulated nuclear distribution in P. capsici hyphae and sporangia by staining the nuclei with 4′,6‐diamidino‐2‐phenylindole (DAPI). DAPI staining revealed that, in the two control lines, the nuclei were maintained in a regularly spaced distribution [Fig. 8A (c, d)]. In CK or WT, the numbers of nuclei ranged from 9 to 16 in each of 25 sporangia, and from 15 to 20 in each of 14 hyphal segments. In comparison, in the silenced lines SiPcSDA1‐3 and SiPcSDA1‐4, the nuclei were somewhat sparser, irregularly spaced and distorted in shape [Fig. 8A (a1, a2)]. In the two silenced lines, the numbers of nuclei ranged from 8 to 14 in each of 25 sporangia and from 14 to 17 in each of 14 hyphal segments (Table S3, see Supporting Information); these were significantly different than CK and WT (P < 0.001 and P < 0.01, respectively; Wilcoxon rank sum test). In the overexpression lines OPcSDA1‐1 and OPcSDA1‐2, the nuclei were irregularly spaced and much more numerous than in the control or silenced strains, and were also distorted in shape [Fig. 8A (b1, b2)]. In the two overexpression lines, the numbers of nuclei ranged from 30 to 40 in each of 25 sporangia, and from 17 to 20 in each of 14 hyphal segments (Table S3); these were significantly different from CK and WT (P < 0.001 and P < 0.01, respectively; Wilcoxon rank sum test). These results suggest that PcSDA1 influences nuclear distribution, and may also play a role in the regulation of nuclear division or even mitosis in P. capsici.
Figure 8.
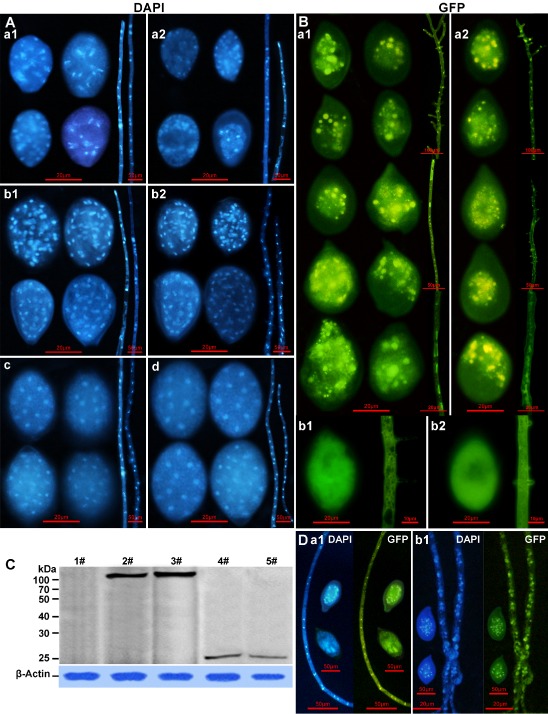
Distribution of nuclei and PcSDA1 within sporangia and mycelia of the PcSDA1‐silenced and overexpression lines compared with two controls. (A) 4′,6‐Diamidino‐2‐phenylindole (DAPI) staining of the nuclear distribution in sporangia and hyphae: a1, silenced line SiPcSDA1‐3; a2, silenced line SiPcSDA1‐4; b1, overexpression line OPcSDA1‐1; b2, overexpression line OPcSDA1‐2; c, CK (empty vector transformant); d, wild‐type strain SD33. (B) Green fluorescent protein (GFP) fluorescence in sporangia and hyphae of the PcSDA1‐GFP overexpression lines: a1, PcSDA1‐GFP‐pHam34‐1; a2, PcSDA1‐GFP‐pHam34‐2; b1, GFP control line GFP‐pHam34‐1; b2, GFP control line GFP‐pHam34‐2. Bright spots inside each structure are nuclei (see D). (C) Western blot analysis reveals that PcSDA1‐GFP protein (108 kDa) is intact and stable in two PcSDA1‐GFP overexpression lines, PcSDA1‐GFP‐pHam34‐1 and PcSDA1‐GFP‐pHam34‐2, using anti‐GFP antibody. Two GFP control lines, GFP‐pHam34‐1 and GFP‐pHam34‐2, were used as controls; expected protein size, 27 kDa. The PcSDA1 protein levels are normalized to the β‐actin loading control using anti‐actin antibody (lower panel); 1#, SD33; 2#, PcSDA1‐GFP‐pHam34‐1; 3#, PcSDA1‐GFP‐pham34‐2; 4#, GFP‐pHam34‐1; 5#, GFP‐pHam34‐2. (D) DAPI staining of sporangia and hyphae from two overexpression PcSDA1‐GFP lines: a1, PcSDA1‐GFP‐pHam34‐1; b1, PcSDA1‐GFP‐pHam34‐2. Each experiment was repeated at least four times on independent occasions and representative structures are shown.
Nuclear localization of PcSDA1
In order to localize PcSDA1 protein in P. capsici, we labelled it by fusion with green fluorescent protein (GFP). We obtained five PcSDA1‐GFP overexpression lines (PcSDA1‐GFP‐pHam34‐1, PcSDA1‐GFP‐pHam34‐2, PcSDA1‐GFP‐pHam34‐3, PcSDA1‐GFP‐pHam34‐4 and PcSDA1‐GFP‐pHam34‐5) and three control GFP expression lines (GFP‐pHam34‐1, GFP‐pHam34‐2 and GFP‐pHam34‐3). There were no visible differences in sporangia and mycelia or other phenotypes among the five PcSDA1‐GFP lines, nor did their phenotypes differ from the untagged overexpression lines. There were no visible differences in sporangia and mycelia or other phenotypes among the three GFP fusion controls, nor did their phenotypes differ from CK or WT. Thus, we selected two PcSDA1‐GFP overexpression lines, PcSDA1‐GFP‐pHam34‐1 and PcSDA1‐GFP‐pHam34‐2, as well as two GFP controls, GFP‐pHam34‐1 and GFP‐pHam34‐2, for further characterization. We used Western blot analysis to confirm that PcSDA1‐GFP protein and GPF control were correctly produced in these four lines (Fig. 8C). PcSDA1‐GFP‐pHam34‐1 and PcSDA1‐GFP‐pHam34‐2 each produced one protein band with a size of 108 kDa, as expected for the PcSDA1‐GFP fusion protein. Moreover, the two GFP controls, GFP‐pHam34‐1 and GFP‐pHam34‐2, produced only a single protein band of 27 kDa, as expected for the GFP protein alone. Sporangia and mycelia from these four lines were examined by fluorescence microscopy. In PcSDA1‐GFP‐pHam34‐1 and PcSDA1‐GFP‐pHam34‐2, GFP fluorescence was concentrated in brightly fluorescent nuclei‐like bodies in the sporangia and hyphae with some diffuse fluorescence in the cytoplasm [Fig. 8B (a1, a2)]. Staining of the sporangia and hyphae with DAPI confirmed that the brightly staining bodies were nuclei (Fig. 8D). Some sporangia exhibited numerous brightly staining nuclei, indicating that most zoospores had not yet been released. In other sporangia, however, only a few GFP‐stained nuclei were observed adjacent to the apical bodies, suggesting that most zoospores had been released. This is consistent with previous observations in Pythium and P. infestans (Ah Fong and Judelson, 2011; Grove et al., 1970). In the controls, the observable GFP fluorescence was evenly distributed in the cytoplasm within GFP‐pHam34‐1 and GFP‐pHam34‐2 lines [Fig. 8B (b1, b2)].
From these results, we conclude that PcSDA1, similar to SDA1 in yeasts and animals, appeared to be mainly localized to nuclei and faintly in the cytoplasm. There was no evidence, however, that PcSDA1 was further localized to nucleoli.
PcSDA1 is required for pathogenicity
To examine the role of PcSDA1 during P. capsici infection of pepper leaves, inoculation assays were performed by inoculating zoospore suspensions from SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2 or the two control strains onto leaves of the susceptible pepper (Capsicum annuum) cultivar (inbred line 06221). Compared with the two controls, the virulence of lines SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2 revealed striking differences. At 1 day post‐inoculation (dpi) with the zoospores of the two controls, the leaves displayed water‐soaked lesions, a typical disease symptom caused by P. capsici. At 3 dpi, the lesions had obviously spread on the leaves and showed large water‐soaked areas (c. 3.0–3.4 cm2) [Fig. 9A, B (c, d]. In contrast, SiPcSDA1‐3 and SiPcSDA1‐4 produced only slight disease symptoms at 2 dpi and had formed significantly smaller lesions at 3 dpi (c. 0.8–1.0 cm2) compared with either of the controls (P < 0.01) [Fig. 9A, B (a1, a2, c, d)]. Similarly, OPcSDA1‐1 and OPcSDA1‐2 caused visible water‐soaked lesions at 2 dpi, which then became only slightly larger (c. 1.6–1.8 cm2) around the site of inoculation at 3 dpi [Fig. 9A, B (b1, b2, c, d)]. We hypothesized that these outcomes might result from fewer germinating cysts and shorter germ tubes in these four mutant strains (Table 1). To explore the reason for the diminished virulence in SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2, a microscopic examination of infection by cysts germinating on a susceptible pepper leaf was performed. We examined cysts germinating at 6, 12 and 24 h after inoculation with zoospores. The cysts of SiPcSDA1‐3 grew much shorter germ tubes on the surface of the leaf than did OPcSDA1‐2 or the two control strains at these three time points [Fig. 9D–F (a1, b1, c, d)]. Thus, it appeared that the germ tubes of SiPcSDA1‐3 had failed to penetrate the epidermal cells of the leaf after 6 or 12 h [Fig. 9D, E (a1)]. In contrast, the germ tubes of OPcSDA1‐2 were much longer and had begun to penetrate the tissue by 12 or 24 h [Fig. 9E, F (b1)]. At 24 h, the germ tubes of both mutants were long and had begun to penetrate the tissue [Fig. 9F (a1, b1)]. However, in both respects, the mutants still remained behind the two control strains at 12 or 24 h, as shown in Fig. 9E, F (c, d). The phenotype of SiPcSDA1‐4 resembled that of SiPcSDA1‐3 and the phenotype of OPcSDA1‐2 resembled that of OPcSDA1‐1 (data not shown). We also monitored the infection potential of mycelia from the six strains on onion (Allium cepa L.) epidermal tissues. As shown in Fig. 9C, the hyphae inoculated onto onion skin remained viable under moist conditions, but differences in hyphal growth occurred among the tested strains. Hyphae of the two controls grew abundantly and easily penetrated into the onion epidermal cells, and were not easily washed off at 3 hpi [Fig. 9C (c, d)]. In contrast, the hyphae of the four mutants SiPcSDA1‐3, SiPcSDA1‐4, SiPcSDA1‐3 and SiPcSDA1‐4 did not penetrate into the onion epidermal cells, even at 5 hpi [Fig. 9C (a1, a2, b1, b2)]. However, the four mutants differed, in that SiPcSDA1‐3 and SiPcSDA1‐4 developed very few hyphae [Fig. 9C (a1, a2)], whereas OPcSDA1‐1 and OPcSDA1‐2 grew very dense hyphae within onion epidermal cells [Fig. 9C (b1, b2)]. These observations suggest that the loss of virulence of the four mutants is not only related to the numbers of germinating cysts, but also to a delayed ability to penetrate epidermal cells.
Figure 9.
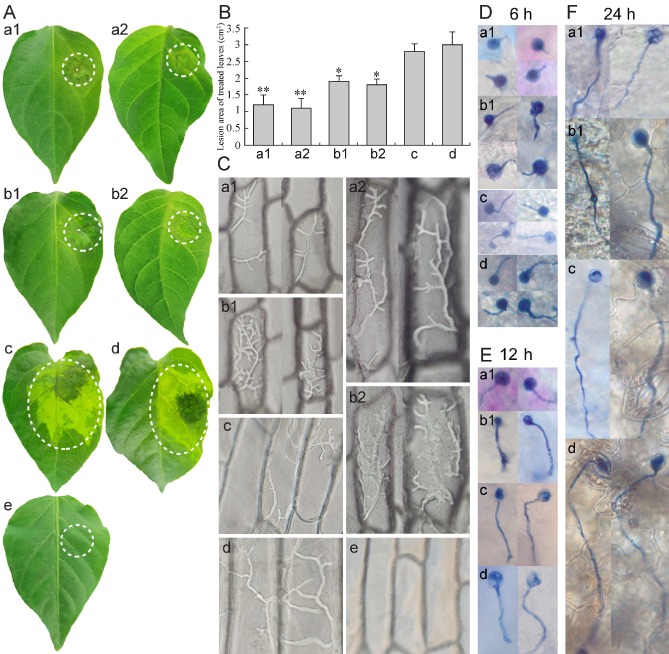
PcSDA1‐silenced and overexpression lines show reduced virulence. (A) Pepper leaflets were inoculated with 2.0 μL of a zoospore suspension (1 × 105 zoospores/mL). Photographs were taken at 3 days post‐inoculation (dpi). (B) Areas of lesions were evaluated at 3 dpi. Bars represent the mean ± standard error from 14 leaves. ‘*’ indicates a significant difference from wild‐type (WT) (P < 0.05) using Student's t‐test. ‘**’ indicates a significant difference from WT (P < 0.01) using Student's t‐test. (C) Microscopic observation of the growth or infection of mycelia on onion epidermal cells after 2 days. These tests were repeated three times with at least 14 onion tissues in each experiment and typical structures are shown. a1, silenced line SiPcSDA1‐3; a2, silenced line SiPcSDA1‐4; b1, overexpression line OPcSDA1‐1; b2, overexpression line OPcSDA1‐2; c, CK (empty vector transformant); d, wild‐type strain SD33; e, distilled water. (D–F) Microscopic observation of cyst germination and germ tubes penetrating the surface of pepper leaves at 6, 12 and 24 h, respectively. Each experiment was repeated three times with at least 14 leaves, and typical structures are shown. a1, silenced line SiPcSDA1‐3; b1, overexpression line OPcSDA1‐1; c, CK (empty vector transformant); d, wild‐type strain SD33.
Discussion
Filamentous organisms, which are common among fungi and oomycetes, face unique cytological challenges, especially those organisms in which multiple nuclei share a common cytoplasm (Seiler and Justa‐Schuch, 2010). The trafficking of vesicles and the transport of nutrients, metabolites, proteins and other macromolecules must be managed over relatively long distances. Local programmes of differentiation, such as polarized hyphal growth, hyphal branching and the formation of a variety of spores, must be established. Transitions between hyphal and unicellular forms must be achieved in organisms that undergo yeast–hyphal transitions or that form unicellular spores. Management of the cytoskeleton and the cell division cycle are key mechanisms required to address these challenges. Thus, the SDA1 protein, which has been identified in S. cerevisiae (Buscemi et al., 2000) and human cells (Babbio et al., 2004) as an important regulator of the actin cytoskeleton and also of the cell division cycle, is potentially a key player in the regulation of the growth and morphology of filamentous organisms, such as fungi (Berepiki et al., 2011; Suei and Garrill, 2008) and oomycetes (Ketelaar et al., 2012; Meijer et al., 2014). However, to date, there have been no studies of this protein outside S. cerevisiae and human cells.
In this study, we have identified the SDA1 gene in the oomycete P. capsici (PcSDA1) and have identified roles in the regulation of growth, sporogenesis, zoospore germination and host infection, using P. capsici transformants underexpressing or overexpressing PcSDA1. The P. capsici orthologue of yeast SDA1 was identified by sequence similarity searches of P. capsici genome sequences. The PcSDA1 gene encodes a protein having 40% amino acid identity with human SDA1 and 28% identity with yeast SDA1, in both cases spanning more than 90% of the length of each sequence. This is similar to the degree of identity (35%) between the yeast and human proteins (Buscemi et al., 2000). Furthermore, the region of PcSDA1 from residue 401 to the C‐terminus was annotated with the SDA1 pfam05285 domain. Thus, PcSDA1 is probably correctly identified as the orthologue of the yeast and human SDA1 genes. PcSDA1 contains three putative nuclear localization signals and an acidic region in the C‐terminus, which is consistent with SDA1 in a wide range of eukaryotic organisms (Andersen et al., 2002; Babbio et al., 2004; Bi et al., 1998; Buscemi et al., 2000). PcSDA1 was expressed most strongly in hyphae, developing sporangia and germinating cysts, and in later infection stages of P. capsici, consistent with an important role in each of these life stages. The two life stages in which PcSDA1 expression is low (zoospores and cysts) are uninucleate, whereas the other stages are multinucleate. Using silenced and overexpression transformants of PcSDA1, we investigated the role of this gene in the regulation of development during each of the four life stages. The silenced lines produced only 10% of the normal levels of PcSDA1 transcripts, whereas the overexpression lines produced 10‐fold higher levels than the wild‐type.
A well‐regulated cytoskeleton and a well‐regulated cell division cycle are essential for polarized hyphal growth. In filamentous fungi (Berepiki et al., 2011), and in the oomycete P. infestans (Ketelaar et al., 2012), disruption of the actin cytoskeleton, for example with latrunculin A, leads to greatly reduced colonial growth rates, loss of polarity of growth and a greatly increased frequency of hyphal branching. In filamentous fungi, the loss of polarity results from disruption of the vesicle trafficking required for the delivery of cell wall constituents to the growing tip (Berepiki et al., 2011). In filamentous fungi, there is also a close connection between the actin cytoskeleton and the regulation of nuclear division, as the actin cytoskeleton is important for correct positioning of nuclei prior to and subsequent to nuclear division (Seiler and Justa‐Schuch, 2010). Similarly, in P. infestans, treatment with latrunculin B causes irregular clustering and spacing of the nuclei (Ketelaar et al., 2012; Meijer et al., 2014). However, there is currently no information in the literature on whether SDA1 plays a role in coordinating nuclear division with the regulation of the actin cytoskeleton in either filamentous fungi or oomycetes.
Here, we observed that silencing of PcSDA1 led to restricted, but dense, colonies during growth in vitro [Fig. 3A (a1, a2)], whereas overexpression led to colony expansion that was nearly the same as the controls, but with significantly lower density of mycelia [Fig. 3A (b1, b2)]. Close inspection of the hyphae revealed that the spacing of hyphal branches was relatively normal in the silenced line [Figs 5 and 6A (a1, a2)], but was highly branched in the overexpression line [Figs 5 and 6A (b1, b2)]. The hyphal tips did not display the swelling characteristic of P. infestans hyphae treated with latrunculin B (Ketelaar et al., 2012). The actin cytoskeleton was significantly irregular in both the silenced and overexpression lines compared with either of the two controls (Fig. 7A–D). Furthermore, the spacing of nuclei was highly irregular in both the silenced and overexpression lines [Fig. 8 (a1, a2, b1, b2)], whereas, in the controls, it was highly regular [Fig. 8A (c, d)]. Thus, the growth and hyphal morphology phenotypes of the PcSDA1‐silenced lines are consistent with the misregulation of the actin cytoskeleton and nuclear division. However, it is not a simple phenocopy of latrunculin B treatment (Ketelaar et al., 2012). It is interesting that not only silencing, but also overexpression, of PcSDA1 causes disruption of the normal growth and hyphal morphology phenotypes. A likely explanation is that the PcSDA1 protein participates in a number of regulatory protein complexes, and thus an excess of the protein disrupts the normal assembly of these complexes. Alternatively, excess PcSDA1 may disrupt the interaction of PcSDA1‐containing regulatory complexes with their normal targets.
The differentiation of multinucleate sporangia by oomycetes, and the subsequent partitioning of these sporangia into a collection of uninucleate motile zoospores, is a complex process requiring precise control of cell polarity, nuclear division, nuclear migration and cytokinesis. As shown in Fig. 8A, in a sporangium just prior to cytokinesis, there is a highly regular spacing of the nuclei. In the chytrid fungus Allomyces macrogynus, disruption of the actin cytoskeleton with cytochalasin D blocked nuclear migration and interfered with membrane formation required for cytokinesis (Lowry et al., 2004). The silenced lines SiPcSDA1‐3 and SiPcSDA1‐4 produced relatively few sporangia [Fig. 3E, F (a1, a2)] that were significantly smaller than those produced by the overexpression lines or either of the two controls [Fig. 3E, F (b1, b2, c, d)]. In contrast, sporangia produced by the overexpression lines OPcSDA1‐1 and OPcSDA1‐2 [Fig. 3E, F (b1, b2)] were nearly equivalent to those of the controls [Fig. 3E, F (c, d)] in size but, like the silenced lines, were far fewer in number (Fig. 4). However, the sporangia produced by the silenced and overexpression lines tended to be misshapen [Fig. 5 (a1‐1, a2‐1, b1‐1, b2‐1)] and their internal structure appeared to be disorganized, especially in the silenced transformants [Fig. 6B (a1, a2, b1, b2)], when compared with the controls [Fig. 6B (c, d)], and the sporangial walls were noticeably thin at points [Fig. 6B (a1, a2)]. DAPI staining revealed that the nuclei in the sporangia and hyphae of the silenced and overexpression lines were highly disorganized [Fig. 8A (a1, a2, b1, b2)] when compared with the controls [Fig. 8A (c, d)]. In the overexpression lines, the sporangia contained many more nuclei, and the nuclei appeared to be continuing to actively divide, suggesting a loss of control of nuclear division in the maturing sporangia [Fig. 8A (b1, b2)]. The disorganized distribution of nuclei in the sporangia also resembled that observed in P. infestans transformants overexpressing the phosphatidylinositol phosphate kinase G4K (Hua et al., 2013). Tagging of PcSDA1 with GFP in two overexpression lines also revealed a highly disorganized distribution of the protein in sporangia and hyphae [Fig. 8B (b1, b2)]. The GFP fluorescence patterns in sporangia and hyphae also appeared identical to the patterns obtained using DAPI [Fig. 8D (a1, b1)], confirming that PcSDA1 is mainly localized to the nuclei and faintly in the cytoplasm. Altogether, these results are consistent with PcSDA1 playing a key role in the regulation of both actin filament organization and mitosis during sporangial maturation in P. capsici.
Infection of host tissue by oomycetes is also a complex morphological and physiological process, requiring several precise changes in cell polarity. As in most Phytophthora species, the mature sporangia of P. capsici readily release motile zoospores under moist conditions. When zoospores reach the surface of host tissue, they shed their flagella, encyst and adhere to the surface. When the cyst germinates to produce a hypha, the germ tube may penetrate the plant directly, or may grow for some distance before differentiating an appressorium to enter the tissue (Fig. 9D–F). In both filamentous fungi and oomycetes, modulation of actin distribution at the tip of invasive hyphae has been shown to be a mechanism to increase the level of intrusive force, without increasing turgor (Suei and Garrill, 2008; Walker et al., 2008). Following entry into host tissues, colonization of these tissues requires the penetration of host cells and the differentiation of haustoria, together with the coordinated delivery of virulence proteins (Lamour et al., 2012). We observed that SiPcSDA1‐3, SiPcSDA1‐4, OPcSDA1‐1 and OPcSDA1‐2 were severely reduced in virulence. SiPcSDA1‐3 and SiPcSDA1‐4 produced extraordinarily small lesions on pepper leaves and an extremely sparse distribution of hyphae within onion epidermal cells [Fig. 9A–C (a1, a2, b1, b2)]. The silenced line SiPcSDA1‐3 also produced very short germ tubes on the surface of the leaf [Fig. 9D–F (a1)]. In contrast, although overexpression lines caused very small lesions on pepper [Fig. 9A (b1, b2)], they produced very dense hyphae within onion epidermal cells [Fig. 9C (b1, b2)] and long germ tubes on the surface of the leaf [Fig. 9D–F (b1)]. These observations are once again consistent with PcSDA1 playing a key role in the regulation of the morphology of the pathogen during each step of infection.
In conclusion, our observations suggest that PcSDA1 plays a similar role in the regulation of the actin cytoskeleton and nuclear division as in yeast and human cells. The role of SDA1 has not been characterized previously in a filamentous fungus or ascomycete. Given the extensive control of cell polarity and morphology required in a filamentous organism, especially one that has multiple cell types and that can penetrate and colonize a host, it is not surprising that the misregulation of SDA1 levels has profound effects on P. capsici. Every cell stage, including hyphae, sporangia, zoospores and infection hyphae, showed strong phenotypes whether SDA1 was silenced or overexpressed. The characterization of the role of SDA1 in P. capsici will add to our knowledge of oomycete morphogenesis, lay the foundations for more detailed studies and identify a potential target of disease control.
Experimental Procedures
Preparation of P . capsici cultures and pepper seedlings
The highly virulent P. capsici strain SD33 was isolated from blighted pepper plants and maintained in our laboratory (Sun et al., 2009). All strains in this study, including transformants, were routinely grown on 10% V8 juice agar medium at 25 °C in the dark (Erwin and Ribeiro, 1996). For the growth assays, all lines were grown on 10% V8 juice agar medium. Radial growth was measured at 5 days. Sporangia and zoospores were obtained as described by Erwin and Ribeiro (1996). Zoospores were induced from 3–4‐day‐old sporangia by washing with sterile distilled water for 24 h at 25 °C, and were then harvested by centrifugation at 4000 g for 4 min. We manually counted the number of zoospores in 10 μL of zoospore suspension. The mycelium (MY), sporangia (SP), zoospores (ZO), cysts (C) and germinated cysts (GC) were obtained as described previously (Hua et al., 2008) with minor modifications. To prepare germinated cysts, zoospores were chilled at −20 °C until some encystment was apparent. Once a few cysts were present, the cultures were warmed to 4 °C and agitated until approximately 90% of zoospores had encysted. The cysts were mixed with an equal volume of 0.2 mg/mL pectin prepared in 40 mm CaCl2 (Kramer et al., 1997) and shaken for 4 h at 23 °C. The cysts and 4‐h‐old germlings were collected by centrifugation and frozen in liquid nitrogen and stored at −80 °C. Alternatively, motile zoospores were vortexed to induce encystment and the germinated cysts were prepared by mixing an equal volume of cyst suspension with 5% V8 broth and incubating at 25 °C with shaking (100 rpm) for 2 h. The samples were immediately frozen in liquid N2 and pulverized for RNA extraction. All determinations for each isolate were performed in at least three replicates. A susceptible pepper cultivar (Capsicum annuum inbred line 06221) was used to evaluate the virulence of different strains. The seedlings of pepper line 06221 were cultivated as described previously (Feng et al., 2014). Briefly, single seedlings at the three‐leaf stage were transplanted into small plastic trays and grown for 14 days before leaves were used in inoculation assays.
Isolation of PcSDA1 and sequence analysis
The SDA1 gene was identified in the P. capsici genome sequence (http://img.jgi.doe.gov/cgi‐bin/w/main.cgi.) by searching a six‐frame translation of the sequence (tblastn) using the full‐length amino acid sequences of the SDA1 protein sequences from S. cerevisiae and humans. The sequences of PsSDA1 (P. sojae) and PrSDA1 (P. ramorum) were downloaded from the DOE JGI website (http://www.jgi.doe.gov/) (Table S2). PiSDA1 (P. infestans) and PpSDA1 (P. parasitica) sequences were downloaded from the Broad Institute website (http://www.broadinstitute.org/) (Table S2). Incorrectly identified introns in the P. infestans and P. sojae National Center for Biotechnology Information (NCBI) accessions were corrected by reference to expressed sequence tags (ESTs) (P. infestans CV934023.1, CV917136.1; P. sojae CF862481.1) and by alignment of the amino acid sequences encoded by the unspliced genome sequence. Sequences missing from the 3′ end of the gene in the P. infestans genome assembly were corrected by reference to EST CV943752.1. Other SDA sequences were downloaded from NCBI‐blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi). One hit was annotated as being from a Proteobacterium bacterium (WP_028833254); however, that sequence contig was probably a eukaryotic contaminant, because the four other genes on the same contig all had eukaryotic genes as their most similar sequence. The candidate amino acid sequences were analysed for conserved functional domains using the online software smart (smart.embl‐heidelberg.de/) and InterProScan (www.ebi.ac.uk/Tools/InterProScan/).
To amplify the PcSDA1 gene from P. capsici SD33 DNA, primers were designed using the Primer Express software version 3.0 (Premier Biosoft International, Palo Alto, CA, USA) based on the PcSDA1 DNA sequence in JGI (Table S1). Genomic DNA of SD33 was extracted from mycelium by the method of Tyler et al. (1995). Reaction parameters for PCR were as follows: 94 °C for 4 min; followed by 40 cycles at 95 °C for 55 s, 55 °C for 40 s and 72 °C for 1 min; and a final extension at 72 °C for 10 min. The 25‐μL PCR included 2.5 μL of DNA template, 0.8 μm gene‐specific primer for each PcSDA1 gene, 12.5 μL of 2 × PCR master mix and 8.5 μL of distilled H2O. The PCR products were cloned into T3‐vector in Escherichia coli DH5α and then sequenced using Foster City, CA, USA of an Applied Biosystems 3730 DNA Analyzer. To verify the sequence of PcSDA1, the sequence of the cloned PCR product was analysed using the GCG software package (Wisconsin Package version 10.0; Genetics Computer Group, Madison, WI, USA). Nucleotide and amino acid sequence homology searches were compared with SDA1 sequences from other organisms using NCBI‐blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi). All sequences obtained are incorporated in Table S2. Multiple alignments of SDA amino acid sequences were aligned using ClustalX 2.0 (Thompson et al., 1997). Phylogenetic trees were generated by neighbour joining, as implemented in PAUP*4.0 Beta with the default parameters. Nodal support of the trees was estimated by bootstrapping, with 1000 pseudoreplicate datasets.
RNA extraction and SYBR green real‐time RT‐PCR assay
To determine the transcript profiles of the PcSDA1 gene in different developmental stages of P. capsici [mycelium (MY), sporangia (SP), zoospore (ZO), cyst (C) and germinating cyst (GC)], the samples were concentrated by centrifugation if needed, and then frozen directly in liquid N2 and stored at −80 °C. To measure the transcript levels of PcSDA1 during the early phases of P. capsici infection, the pepper leaf was inoculated with zoospores of the SD33 strain. Each leaflet was spot incubated with 2.0 μL of zoospore suspension (1 × 105 zoospores/mL) of the relevant strains and then kept in the dark at 25 °C (Feng et al., 2014). Lesion samples were collected at 1.5, 3, 6, 12, 24, 48 and 72 hpi, and immediately placed in liquid nitrogen. Total RNA extraction from frozen lesion tissue or frozen P. capsici tissue, and quantification, were performed as reported previously (Feng et al., 2014). Primers were designed to anneal specifically to PcSDA1 using Primer express software version 3.0 (Premier Biosoft International) based on the PcSDA1 DNA sequence in JGI (Table S1) for SYBR green real‐time PCR. Three housekeeping genes, β‐actin (ID: 189084454), β‐tubulin (ID: 50660663) and Ubc (ID: 23394351), of P. capsici and β‐actin (ID: 45861744) of pepper (Yan and Liou, 2006) were used as constitutively expressed endogenous controls in order to normalize the expression of PcSDA1. The transcript levels of PcSDA1 in P. capsici tissues or lesions were determined using the ICycler IQ RT‐PCR detection system and SYBR primer Script RT‐PCR kit (Bio‐Rad, Richmond, CA, USA). The PCRs and parameters were adjusted slightly from those of Feng et al. (2014). PCRs were as follows: 94 °C for 3 min; followed by 40 cycles at 95 °C for 60 s, 55 °C for 35 s and 72 °C for 1 min; and a final extension at 72 °C for 10 min. The values of the threshold cycles (CT) were ascertained automatically by the instrument, and the fold changes of PcSDA1 were calculated using the equation 2−ΔΔCT according to previous descriptions (PfaffI, 2001). All tests were carried out with at least three replicates.
Construction of recombinant plasmids for stable transformations of P . capsici
We used the plasmid vectors pHam34 and pHspNpt (Feng et al., 2014) to construct transformation plasmids. DNA fragments for the generation of constructs were amplified from DNA of P. capsici SD33 and digested with the restriction enzyme SmaI for cloning into the vector pHam34. Primers are described in Table S1. The sequences of the insertions were verified by DNA sequencing. PcSDA1 was subcloned in the anti‐sense for silencing and in the positive sense for overexpression (Ah Fong et al., 2008). To create the PcSDA1‐GFP fusion, the full‐length GFP coding sequence (723 bp) was amplified from PGFPN vector by PCR with the forward primer 5′‐CACACCCGCCGCGTTAATG‐3′ and reverse primer 5′‐GAAATAATCGAGGACACAGT‐3′ (Bottin et al., 1999; Le Berre et al., 2008). For linking the GFP sequence to PcSDA1, the 3′ end of PcSDA1 was cloned by PCR using the forward primer 5′‐TCCCCCGGGATGGAGGCTGTAAACGCATCT‐3′ and reverse primer 5′‐AAGATTTTAATTAAGGCTAGCACCATGGGCAAGGGCGAGGAACTG‐3′, and the 5′ end of PcSDA1 was cloned by PCR with the forward primer 5′‐CAGTTCCTCGCCCTTGCCCATGGTGCTAGCCTTAATTAAAATCTT‐3′ and reverse primer 5′‐TCCCCCGGGGAAATAATCGAGGACACAGT‐3′ (Bottin et al., 1999; van West et al., 1999). We cloned the strong constitutive HAM34 promoter (550 bp) in front of the PcSDA1 open reading frame (ORF) fused to the GFP (PcSDA1‐GFP‐pHam34 construct) and in front of the GFP alone (GFP‐pHam34 construct) for overexpression of PcSDA1 and GFP genes, respectively, using the primers given in Table S1 (Le Berre et al., 2008). The orientations and structures of the inserts were confirmed by sequencing. For transformation of P. capsici, the preparation of P. capsici protoplasts and the transformation procedure were adjusted slightly from McLeod et al. (2008). Stable transformants, including silenced lines (SiPcSDA1), overexpression lines (OPcSDA1), CK (pHspNpt empty vector) lines, PcSDA1‐GFP‐pHam34 and GFP‐pHam34 lines, were prepared by a co‐transformation strategy (Bottin et al., 1999; McLeod et al., 2008). For transformation, 75 μg of target plasmids and 25 μg of helper plasmid pHspNpt DNA were mixed with 1 mL of P. capsici protoplasts. For preparation of CK (empty vector) transformants, 25 μg of pHspNpt DNA was mixed with 1 mL of protoplasts. Each DNA–protoplast mixture was kept on ice for 5–10 min, and then 1.74 mL of 40% polyethylene glycol 4000 in 0.5 m CaCl2 and 0.8 m mannitol were added slowly. Subsequently, the suspension was gently mixed and placed on ice for 20 min, followed by the addition of 10 mL pea broth liquid medium containing 0.8 m mannitol. This mixture was then poured into a Petri dish that contained 10 mL of pea broth with 50 μg/mL ampicillin and 0.8 m mannitol. After incubation for 14 h at 25 °C, the mixture containing regenerated protoplasts was centrifuged at 10 000 g for 5 min. The supernatant was discarded, and the regenerated protoplast pellets were mixed with 10 mL of pea broth agar (2%) containing 0.8 m mannitol and 30 μg/mL G418 (Sigma: Sigma‐Aldrich Corp. st. Louis. Mo, USA), and plated onto solid pea broth agar containing 30 μg/mL G418. Transformants appeared in the solid medium within 4–10 days at 25 °C under dark conditions and were propagated in pea broth medium containing 30 μg/mL G418 (Sigma). To validate the transformants, transcript levels of PcSDA1 during the growth of mycelium were measured by RT‐PCR and qRT‐PCR, and analysed as described above. Results were obtained from three repeated trials.
Protein extraction and Western blots
The pET28a‐PcSDA1 and pET28a‐GFP constructs were obtained as described by Maier et al. (2015). The cloning junctions were confirmed by DNA sequencing. For expression of PcSDA1 and GFP fusion proteins in E. coli BL21, the BL21 cells carrying the different recombinant plasmids were grown at 37 °C in Luria–Bertani (LB) medium containing 100 mg/mL kanamycin until they reached an optical density at 600 nm (OD600) of 0.5. Expression of PcSDA1 and GFP fusion proteins was induced at an OD600 of 0.8–1.0 by adding 1 mm isopropyl‐β‐thiogalactopyranoside (IPTG), followed by incubation at 37 °C for 12 h, and were then monitored by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and Coomassie blue staining. CK (DNA of pET28a was transformed into E. coli BL21) and E. coli BL21 were used as controls.
Crude proteins were extracted and concentrated from PcSDA1‐GFP‐pHam34, GFP‐pHam34 and SD33 strains as described previously (Wang et al., 2013) and quantified by the method of Bradford (1976). Proteins from the GFP‐pHam34 strain were used as control. For detection of PcSDA1 levels in PcSDA1‐GFP‐pHam34 and GFP‐pHam34, PcSDA1 was used to prepare antibodies in New Zealand white rabbits according to standard protocols (Kothari et al., 2006). Proteins from SD33 strain were used as controls. Approximately 50 μg of total proteins of PcSDA1‐GFP, GFP fusion and PcSDA1 were individually loaded on each lane of a 12% SDS–PAGE gel, and protein gel blot analyses were performed as reported previously (Wang et al., 2013; Yu et al., 2012). Briefly, proteins were transferred from the gel to an Immobi‐lon‐PSQ polyvinylidene difluoride membrane after electrophoresis. The membranes were washed in PBST (PBS with 0.1% Tween 20; PBS: 135 mm NaCl, 3 mm KCl, 1.5 mm KH2PO4 and 8 mm Na2HPO4, pH 7.1) for 2 min and then blocked in PBSTM (PBS with 0.1% Tween 20 and 5% non‐fat dry milk) for 1 h. Mouse monoclonal antibody against GFP or rabbit polyclonal anti‐PcSDA1 was individually added into PBSTM and incubated for 90 min, followed by washing with PBST three times. The membranes were then incubated in PBSTM with a goat anti‐mouse IRDye 800CW (Li‐Cor: 4647 Superior Street Lincoln, Nebraska USA) for 40 min. The membranes were washed three times with PBST and then visualized using a Li‐Cor Odyssey scanner with excitation at 700 and 800 nm. Antibody to β‐actin as normalization endogenous control (mouse, ab6267, 1 : 45 000; rabbit, ab8227, 1 : 5000; Abcam, Cambridge, MA, USA) was used as reported by Zhang et al. (2012). Each experiment was repeated at least three times.
Analysis of colony growth, sporangiophore and sporangial morphology of silenced and overexpressed lines
For growth assays, all strains were subcultured twice on G418‐free 10% V8 juice agar medium. To observe the morphology of sporangia, sporangiophores and hyphae, all strains were individually inoculated into 20 mL of sterile 10% V8 juice in Petri dishes as described by Jia et al. (2009). The colony radius was measured at 1, 3, 5 and 7 days after incubation at 25 °C. Non‐sporulating hyphae were prepared by repeatedly washing 3‐day‐old hyphae with water and incubating them for 1 h at 4 °C until most hyphae developed sporangia. Hyphae and sporangia for microscopy were obtained from liquid cultures. The sizes of sporangia and numbers of zoospores were determined as described by Masago et al. (1977). Samples of sporangia and mycelia at 7 days were collected and stained by trypan blue as described by Dong et al. (2008). Microscopy was performed using an Olympus BX‐53 microscope (Tokyo, Japan). Transmitted light images were collected with DIC optics. Images were overlaid onto DIC images using Adobe Photoshop (SAN jose, California, USA). For green fluorescence images, excitation/emission settings were 460 nm/480–540 nm for GFP. Simultaneous imaging of GFP was achieved with the multitrack channel, using a 500/530 band pass filter. Images were acquired using LSM software (version 4.2 SP1: Carl‐Zeiss‐Promenade 10, 07745 Jena, Germany). Photographs were subsequently processed using the Autolevel and Autocontrast features of Adobe Photoshop 9.0. Each experiment was repeated at least three times.
Fluorescence staining of hyphae and sporangia of different P . capsici lines
Samples of sporangia and hyphae of the different P. capsici lines and CK and WT (SD33) strains were produced as described above. Actin staining was performed by fixing the samples at room temperature in 3.7% formaldehyde for 1 h, washing twice in PBS and then incubating on a rotator for 30 min in PBS containing 0.5% Tween. Afterward, these samples were washed twice in PBS and incubated on a rotator in PBS containing rhodamine–phalloidin at a concentration of 0.3 mm in the dark for 1.5 h. After staining, the tissue samples were washed three times in PBS and then resuspended in PBS containing one‐tenth volume mounting solution (1 mg/mL phenylenediamine, 90% glycerol, 0.1 m Tris‐HCl, pH 8.8); all samples were viewed on a Zeiss LSM 510 confocal microscope (Carl‐Zeiss‐Promenade 10, 07745 Jena, Germany) with an excitation wavelength of 545 nm and a 560–615‐nm band pass emission filter.
In order to visualize nuclei within the sporangia and hyphae of the different P. capsici lines, the mature asexual tissues were stained with the blue fluorescent nucleic acid stain DAPI (Invitrogen: Changning district, Shanghai, China). Sporangia and hyphae were equilibrated briefly with PBS. Approximately 300 μL of DAPI staining solution (300 nm in PBS) was added to the cover slip preparation. After incubation for 2–4 min, the samples were rinsed several times in PBS and then viewed using an Olympus BX53 microscope at 330–385 nm. Images were collected with either DAPI or green fluorescence, and were acquired using LSM software (version 4.2 SP1). Photographs were subsequently processed using the Autolevel and Autocontrast features of Adobe Photoshop 9.0. This assay was repeated at least three times.
SEM
We used SEM to study the morphology of hyphae in each of the P. capsici lines. Hyphae were fixed in 2.5% glutaraldehyde in buffer (0.1 m sodium cacodylate, pH 7.2) for 3 h, washed with buffer and post‐fixed with 2% osmium tetroxide for 2 h. Glutaraldehyde‐fixed samples were post‐fixed in 1% aqueous OsO4, dehydrated using an ascending series of ethanol concentrations (Blanco and Judelson, 2005) and treated in a critical point drier (Balzers Union, Liechtenstein; CPD 020) with liquid CO2. All samples were then mounted on aluminium stubs with two‐sided tape, coated with gold–palladium. Images were obtained with a Phillips XL30 FEG microscope (Phillips, Natick, MA, USA).
TEM
We used TEM to determine whether there were ultrastructural differences between sporangia of silenced, overexpression and control strains. Sporangia were obtained from 10% V8 juice agar medium after 9 days of incubation. They were fixed and dehydrated as described in the previous section. Thin sections (∼70 nm) of LR White‐embedded material were obtained using glass knives on a Reichert microtome (Model OM‐U3; Reichert‐Jung (Leica), Wetzlar, Germany). The sections were collected on formvar‐coated copper grids, stained for 10 min in 2% ethanolic uranyl acetate, and counter‐stained for 5 min with 1.0% lead citrate. Sections were examined with a Philips CM10 transmission electron microscope (Philips Electron Optics, Eindhoven, Germany) at 80 kV and images were captured with a digital camera.
Virulence tests and microscopic observation
For pepper leaf inoculations, the silenced, overexpression and control lines were induced to produce zoospores as described above. Detached pepper leaves at the fifth‐ to sixth‐leaf stage were placed in Petri dishes containing 1.5% (w/v) water agar. Each leaflet was spot incubated with 2.0 μL of a zoospore suspension (1 × 105 zoospores/mL) of the relevant strains and then kept in the dark at 25 °C (Feng et al., 2014). Distilled water was used as a negative control. SD33 and CK were used as positive controls. The sizes of the lesions were measured and photographed at 3 dpi. The mean areas of the lesions were also calculated at 5 dpi. Lesions from 14 treatment leaves per test were measured and differences were evaluated statistically by Student's t‐test. Bars represent the mean ± standard error of 14 leaves (P < 0.05).
In order to microscopically observe the ability of different strains to infect plant cells, we inoculated onion (Allium cepa L.) tissues with mycelia, as described previously (Kebdani et al., 2010). Distilled water was used as a negative control. The inoculated onion tissues were incubated in the dark at 25 °C under 80% humidity for 24 h. The onion epidermis around the inoculation sites was peeled off and used for microscopic observation. Excised lesions of pepper leaves and onion epidermal peels were individually placed into alcohol to decolorize, soaked in 0.5% Coomassie brilliant blue for 2 min and then washed in SDW (sterile distilled water) for 10 min. Pepper leaf lesions or inoculated onion epidermal peels were stained by trypan blue as described by Dong et al. (2008). Germinating cysts growing in the lesions and mycelia developed in the onion epidermal peels were monitored using an Olympus BX53 microscope. Images were obtained with a digital camera. In the case of multiple germ tubes or mycelia, as frequently observed in different strains at 6, 12 or 24 h after inoculation, the longest of the germ tubes was measured. All the tests were repeated three times with at least 14 pepper leaves or 14 onion tissues in each experiment.
Statistical analysis
Data were analysed with Systat 12 (Systat Software Inc., San Jose, CA, USA). Student's t‐test was used for pairwise comparisons of two means. Duncan's multiple range test of least significant difference (LSD) was used for more than two means. P values of 0.05 or 0.01 were used, as indicated.
Supporting information
Fig. S1 Multiple sequence alignment of SDA1 orthologs from Phytophthora, humans, and yeasts. Sequences aligned were based on P. capsici (Phyca11|14866|fgenesh1_pg.PHYCAscaffold_10‐12; KM275943), P. ramorum (Phyra‐75231), P. sojae (XM_009535925.1), P. infestans (PITC_18755), P. parasitica (XM_008904562.1), Saccharomyces cerevisiae (NP_011761.3), Schizosaccharomyces pombe (NP_595163.1), and Homo sapiens (NP_060585.2), Incorrectly identified introns in the P. infestans and P. sojae NCBI accessions were corrected by reference to ESTs (P. infestans CV934023.1, CV917136.1; P. sojae CF862481.1) and by alignment of the amino acid sequences encoded by the unspliced genome sequence. Sequences missing from the 3' end of the gene in the P. infestans genome assembly were corrected by reference to EST CV943752.1. Sequences were aligned using MUSCLE 3.8 using the default parameters. The uppermost and lowermost rows of the alignment indicate amino acids conserved among Phytophthora or all species, respectively. Asterisk indicates identical residues; colon, conservative substitutions; period, partially conservative substitutions. Cyan highlighting indicates residues identical with the Phytophthora consensus sequence.
Fig. S2 qRT‐PCR analysis of transcript levels of PcSDA1 in three other silenced lines and three other over‐expression lines. A: Three other silenced lines a3‐SiPcSDA1‐1, a4‐SiPcSDA1‐2, a5‐SiPcSDA1‐5. B: Three other over‐expression lines b3‐OPcSDA1‐3, b4‐OPcSDA1‐4, b5‐OPcSDA1‐5, c‐CK (empty vector transformant), d‐Wild type strain SD33. Error bars represent standard errors calculated using three replicates. Three housekeeping genes β‐Actin, β‐Tubulin and Ubc of P. capsici and β‐Actin of pepper were used as constitutively expressed endogenous controls and were used to normalize the expression of PcSDA1. Error bars represent standard errors calculated using three biological replicates for each sample.
Fig. S3 Aerial mycelium in colonies of the silenced line, over‐expression line and two controls. The colonies appear to be producing less aerial hyphae in silenced line SiPcSDA1‐3 (A) and over‐expression line OPcSDA1‐1 (B). The colonies appear to be producing abundant aerial hyphae in the two controls, wild type strain SD33 (C) and CK (empty vector transformant) (D). The colonies were cultured for on 10% V8 juice agar for 5 days after placing a 0.5 cm2 plug of inoculum in the center of a 60 mm plate. In all cases, the hyphae had grown to the edge of the plates. .
Table S1 Primers used for all experiments.
Table S2 The SDA1 genes of Phytophthora capsici and other species.
Table S3 Nuclear content of sporangia and hyphae from Phytophthora capsici transformants and wild‐type.
Acknowledgements
We thank Kurt Lamour for kindly uploading the Phytophthora capsici genome sequence. This work was supported by the Special Fund for Agro‐scientific Research in the Public Interest of China (201003004).
References
- Adams, A.E.M. , Johnson, D.I. , Longnecker, R.M. , Sloat, B.F. and Pringle, J.R. (1990) CDC42 and CDC43, two additional genes involved in budding and establishment of cell polarity in the yeast Saccharomyces cerevisiae . J. Cell Biol. 111, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah Fong, A. and Judelson, H.S. (2003) Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus‐like oomycete Phytophthora infestans . Mol. Microbiol. 50, 487–494. [DOI] [PubMed] [Google Scholar]
- Ah Fong, A.M. and Judelson, H.S. (2011) New role for Cdc14 phosphatase: localization to basal bodies in the oomycete Phytophthora and its evolutionary coinheritance with eukaryotic flagella. PLoS ONE, 6, e16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah Fong, A.M.V. , Bormann‐Chung, C.A. and Judelson, H.S. (2008) Optimization of transgene‐mediated silencing in Phytophthora infestans and its association with small‐interfering RNAs. Fungal Genet. Biol. 45, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Andersen, J.S. , Lyon, C.E. , Fox, A.H. , Leung, A.K.L. , Lam, Y.W. , Steen, H. , Mann, M. and Lamond, A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Babbio, F. , Farinacci, M. , Saracino, F. , Carbone, M.L.A. and Privitera, E. (2004) Expression and localization studies of hSDA, the human ortholog of the yeast SDA1 gene. Cell Cycle, 3, 486–490. [PubMed] [Google Scholar]
- Berepiki, A. , Lichius, A. and Read, N.D. (2011) Actin organization and dynamics in filamentous fungi. Nat. Rev. Microbiol. 9, 876–887. [DOI] [PubMed] [Google Scholar]
- Bi, E. , Maddox, P. , Lew, D.J. , Salmon, E.D. , McMillan, J.N. , Yeh, E. and Pringle, J.R. (1998) Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, F.A. and Judelson, H.S. (2005) A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Mol. Microbiol. 56, 638–648. [DOI] [PubMed] [Google Scholar]
- Bottin, A. , Larche, L. , Villalba, F. , Gaulin, E. , Esquerré‐Tugayé, M.‐T. and Rickauer, M. (1999) Green fluorescent protein (GFP) as gene expression reporter and vital marker for studying development and microbe–plant interaction in the tobacco pathogen Phytophthora parasitica var. nicotianae . FEMS Microbiol. Lett. 176, 51–56. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein‐binding dye. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Buscemi, G. , Saracino, F. , Masnada, D. and Carbone, M.L. (2000) The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci. 113, 1199–1221. [DOI] [PubMed] [Google Scholar]
- Dingwall, C. and Laskey, R.A. (1991) Nuclear targeting sequences a consensus? Trends Biochem. Sci. 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Zhang, Z. , Zheng, X. and Wang, Y. (2008) Mammalian proapoptotic bax gene enhances tobacco resistance to pathogens. Plant Cell Rep. 27, 1559–1569. [DOI] [PubMed] [Google Scholar]
- Erwin, D.C. and Ribiero, O.K. (1996) Phytophthora Diseases Worldwide, St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Erwin, D.C. , Bartnicki‐Garcia, S. and Tsao, P.H. (1983) Phytophthora: Its Biology, Taxonomy, Ecology and Pathology, St Paul, MN: American Phytopathological Society. [Google Scholar]
- Feng, B.Z. , Zhu, X.P. , Fu, L. , Lv, R.F. , Storey, D. , Tooley, P. and Zhang, X.G. (2014) Characterization of necrosis‐inducing NLP proteins in Phytophthora capsici . BMC Plant Biol. 14, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont‐Racine, M. , Senger, B. , Saveanu, C. and Fasiolo, F. (2003) Ribosome assembly in eukaryotes. Gene, 313, 17–42. [DOI] [PubMed] [Google Scholar]
- Gaulin, E. , Jauneau, A. , Villalba, F. , Rickauer, M. , Esquerre‐Tugaye, M.T. and Bottin, A. (2002) The CBEL glycoprotein of Phytophthora parasitica var. nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115, 4565–4575. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C. , Bösche, M. , Krause, R. , Grandi, P. , Marzioch, M. , Bauer, A. , Schultz, J. , Rick, J.M. , Michon, A.M. , Cruciat, C.M. , Remor, M. , Höfert, C. , Schelder, M. , Brajenovic, M. , Ruffner, H. , Merino, A. , Klein, K. , Hudak, M. , Dickson, D. , Rudi, T. , Gnau, V. , Bauch, A. , Bastuck, S. , Huhse, B. , Leutwein, C. , Heurtier, M.A. , Copley, R.R. , Edelmann, A. , Querfurth, E. , Rybin, V. , Drewes, G. , Raida, M. , Bouwmeester, T. , Bork, P. , Seraphin, B. , Kuster, B. , Neubauer, G. and Superti‐Furga, G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Grove, S.N. , Bracker, C.E. and Morré, D.J. (1970) An ultrastructural basis for hyphal tip growth in Pythium ultimum . Am. J. Bot. 57, 245–266. [Google Scholar]
- Hua, C. , Wang, Y. , Zheng, X. , Dou, D. , Zhang, Z. , Govers, F. and Wang, Y. (2008) A Phytophthora sojae G protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell, 7, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, C. , Meijer, H.J.G. , de Keijzer, J. , Zhao, W. , Wang, Y.C. and Govers, F. (2013) GK4, a G‐protein‐coupled receptor with a phosphatidylinositol phosphate kinase domain in Phytophthora infestans, is involved in sporangia development and virulence. Mol. Microbiol. 88 (2), 352–370. [DOI] [PubMed] [Google Scholar]
- Ihmels, J. , Friedlander, G. , Bergmann, S. , Sari, O. , Ziv, Y. and Barkai, N. (2002) Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31, 370–377. [DOI] [PubMed] [Google Scholar]
- Jia, Y.J. , Feng, B.Z. and Zhang, X.G. (2009) Polygalacturonase, pectate lyase and pectin methylesterase activity in pathogenic strains of Phytophthora capsici incubated under different conditions. J. Phytopathol. 157, 585–591. [Google Scholar]
- Kebdani, N. , Pieuchot, L. , Deleury, E. , Panabieres, F. , Le Berre, J.Y. and Gourgues, M. (2010) Cellular and molecular characterization of Phytophthora parasitica appressorium‐mediated penetration. New Phytol. 185, 248–257. [DOI] [PubMed] [Google Scholar]
- Ketelaar, T. , Meijer, H.J.G. , Spiekerman, M. , Weide, R. and Govers, F. (2012) Effects of latrunculin B on the actin cytoskeleton and hyphal growth in Phytophthora infestans . Fungal Genet. Biol. 49, 1014–1022. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. and Judelson, H.S. (2003) Sporangia‐specific gene expression in the oomycetous phytopathogen Phytophthora infestans . Eukaryot. Cell, 2, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari, H. , Kumar, P. and Singh, N. (2006) Prokaryotic expression, purification, and polyclonal antibody production against a novel drug resistance gene of Leishmania donovani clinical isolate. Protein Expr. Purif. 45, 15–21. [DOI] [PubMed] [Google Scholar]
- Kramer, R. , Freytag, S. and Schmelzer, E. (1997) In vitro formation of infection structures of Phytophthora infestans is associated with synthesis of stage specific polypeptides. Eur. J. Plant Pathol. 103, 43–53. [Google Scholar]
- Lamour, K.H. , Stam, R. , Jupe, J. and Huitema, E. (2012) The oomycete broad host range pathogen Phytophthora capsici . Mol. Plant Pathol. 13, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. and Govers, F. (2003) A Phytophthora infestans G‐protein subunit is involved in sporangium formation. Eukaryot. Cell, 2, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. , Ligterink, W. , Vleeshouwers, V.G. , van West, P. and Govers, F. (2004) A G‐alpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans . Mol. Microbiol. 51, 925–936. [DOI] [PubMed] [Google Scholar]
- Le Berre, J.Y. , Engler, G. and Panabieres, F. (2008) Exploration of the late stages of the tomato–Phytophthora parasitica interactions through histological analysis and generation of expressed sequence tags. New Phytol. 177, 480–492. [DOI] [PubMed] [Google Scholar]
- Lowry, D.S. , Fisher, K.E. and Roberson, R.W. (2004) Functional necessity of the cytoskeleton during cleavage membrane development and zoosporogenesis in Allomyces macrogynus . Mycologia, 96, 211–218. [PubMed] [Google Scholar]
- Maier, J.A.H. , Ragozin, S. and Jeltsch, A. (2015) Identification, cloning and heterologous expression of active NiFe‐hydrogenase 2 from Citrobacter sp. SG in Escherichia coli . J. Biotechnol. 199, 1–8. [DOI] [PubMed] [Google Scholar]
- Maltese, C.E. , Conigliaro, G. and Shaw, D.S. (1995) The development of sporangia of Phytophthora infestans . Mycol. Res. 99, 1175–1181. [Google Scholar]
- Masago, H. , Yoshikawa, M. , Fukada, M. and Nikanishi, N. (1977) Selective inhibition of Pythium sp. on a medium for direct isolation of Phytophthora sp. from soil and plants. Phytopathology, 67, 425–428. [Google Scholar]
- McLeod, A. , Fry, B.A. , Zuluaga, A.P. , Myers, K.L. and Fry, W.E. (2008) Toward improvements of oomycete transformation protocols. J. Eukaryot. Microbiol. 55, 103–109. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Hua, C. , Kots, K. , Ketelaar, T. and Govers, F. (2014) Actin dynamics in Phytophthora infestans; rapidly reorganizing cables and immobile, long‐lived plaques. Cell. Microbiol. 16, 948–961. [DOI] [PubMed] [Google Scholar]
- Nissan, T.A. , Babler, J. , Petfalski, E. , Tollervey, D. and Hurt, E. (2002) 60S preribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, A.B. (1989) G1 events and regulation of cell proliferation. Science, 246, 603–608. [DOI] [PubMed] [Google Scholar]
- PfaffI, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas‐Silva, M.D. and Weinberg, R.A. (1997) The restriction point and control of cell proliferation. Curr. Opin. Cell Biol. 9, 768–772. [DOI] [PubMed] [Google Scholar]
- Seiler, S. and Justa‐Schuch, D. (2010) Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol. Microbiol. 78, 1058–1076. [DOI] [PubMed] [Google Scholar]
- Sia, R.A.L. , Herald, H. and Lew, D.J. (1996) Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell, 7, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suei, S. and Garrill, A. (2008) An F‐actin‐depleted zone is present at the hyphal tip of invasive hyphae of Neurospora crassa . Protoplasma, 232, 165–172. [DOI] [PubMed] [Google Scholar]
- Sun, W.X. , Jia, Y.J. , Feng, B.Z. , O'Neill, N.R. , Zhu, X.P. , Xie, B.Y. and Zhang, X.G. (2009) Functional analysis of Pcipg2 from the straminopilous plant pathogen Phytophthora capsici . Genesis, 47, 535–544. [DOI] [PubMed] [Google Scholar]
- Takahashi, N. , Yanagida, M. , Fujiyama, S. , Hayana, T. and Isobe, T. (2003) Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22, 287–317. [DOI] [PubMed] [Google Scholar]
- Tani, S. , Yatzkan, E. and Judelson, H.S. (2004) Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans . Mol. Plant–Microbe Interact. 17, 330–337. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL‐X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. , Forster, H. and Coffey, M.D. (1995) Inheritance of avirulence factors and restriction fragment length polymorphism markers in outcrosses of the oomycete Phytophthora sojae . Mol. Plant–Microbe Interact. 8, 515–523. [Google Scholar]
- Walker, C.A. , Köppe, M. , Grenville‐Briggs, L.J. , Avrova, A.O. , Horner, N.R. , McKinnon, A.D. , Whisson, S.C. , Birch, P.R.J. and West, P.V. (2008) A putative DEAD‐box RNA‐helicase is required for normal zoospore development in the late blight pathogen Phytophthora infestans . Fungal Genet. Biol. 45, 954–962. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zhang, K. , Zhu, J. , Song, W.W. , Zhao, L. and Zhang, X.G. (2013) Structure reveals regulatory mechanisms of a MaoC‐like hydratase from Phytophthora capsici involved in biosynthesis of polyhydroxyalkanoates (PHAs). PLoS ONE, 8 (11), e80024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van West, P. , Reid, B. , Campbell, T.A. , Sandrock, R.W. , Fry, W.E. , Kamoun, S. and Gow, N.A. (1999) Green fluorescent protein (GFP) as a reporter gene for the plant pathogenic oomycete Phytophthora palmivora . FEMS Microb. Lett. 178, 71–80. [DOI] [PubMed] [Google Scholar]
- van West, P. , Appiah, A.A. and Gow, N.A.R. (2003) Advances in research on oomycete root pathogens. Physiol. Mol. Plant Pathol. 62, 99–113. [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Yu, X.L. , Tang, J.L. , Wang, Q.Q. , Ye, W.W. , Tao, K. , Duan, S.Y. , Lu, C.C. , Yang, X.Y. , Dong, S.M. , Zheng, X.B. and Wang, Y.C. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196, 247–260. [DOI] [PubMed] [Google Scholar]
- Zhang, R. , Yang, D. , Zhou, C.H. , Cheng, K. , Liu, Z.H. , Chen, L. , Fang, L. and Xie, P. (2012) β‐Actin as a loading control for plasma‐based Western blot analysis of major depressive disorder patients. Anal. Biochem. 427, 116–120. [DOI] [PubMed] [Google Scholar]
- Zimmerman, A.Z. and Kellogg, D.R. (2001) The Sda1 protein is required for passage through Start. Mol. Biol. Cell, 12, 201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Multiple sequence alignment of SDA1 orthologs from Phytophthora, humans, and yeasts. Sequences aligned were based on P. capsici (Phyca11|14866|fgenesh1_pg.PHYCAscaffold_10‐12; KM275943), P. ramorum (Phyra‐75231), P. sojae (XM_009535925.1), P. infestans (PITC_18755), P. parasitica (XM_008904562.1), Saccharomyces cerevisiae (NP_011761.3), Schizosaccharomyces pombe (NP_595163.1), and Homo sapiens (NP_060585.2), Incorrectly identified introns in the P. infestans and P. sojae NCBI accessions were corrected by reference to ESTs (P. infestans CV934023.1, CV917136.1; P. sojae CF862481.1) and by alignment of the amino acid sequences encoded by the unspliced genome sequence. Sequences missing from the 3' end of the gene in the P. infestans genome assembly were corrected by reference to EST CV943752.1. Sequences were aligned using MUSCLE 3.8 using the default parameters. The uppermost and lowermost rows of the alignment indicate amino acids conserved among Phytophthora or all species, respectively. Asterisk indicates identical residues; colon, conservative substitutions; period, partially conservative substitutions. Cyan highlighting indicates residues identical with the Phytophthora consensus sequence.
Fig. S2 qRT‐PCR analysis of transcript levels of PcSDA1 in three other silenced lines and three other over‐expression lines. A: Three other silenced lines a3‐SiPcSDA1‐1, a4‐SiPcSDA1‐2, a5‐SiPcSDA1‐5. B: Three other over‐expression lines b3‐OPcSDA1‐3, b4‐OPcSDA1‐4, b5‐OPcSDA1‐5, c‐CK (empty vector transformant), d‐Wild type strain SD33. Error bars represent standard errors calculated using three replicates. Three housekeeping genes β‐Actin, β‐Tubulin and Ubc of P. capsici and β‐Actin of pepper were used as constitutively expressed endogenous controls and were used to normalize the expression of PcSDA1. Error bars represent standard errors calculated using three biological replicates for each sample.
Fig. S3 Aerial mycelium in colonies of the silenced line, over‐expression line and two controls. The colonies appear to be producing less aerial hyphae in silenced line SiPcSDA1‐3 (A) and over‐expression line OPcSDA1‐1 (B). The colonies appear to be producing abundant aerial hyphae in the two controls, wild type strain SD33 (C) and CK (empty vector transformant) (D). The colonies were cultured for on 10% V8 juice agar for 5 days after placing a 0.5 cm2 plug of inoculum in the center of a 60 mm plate. In all cases, the hyphae had grown to the edge of the plates. .
Table S1 Primers used for all experiments.
Table S2 The SDA1 genes of Phytophthora capsici and other species.
Table S3 Nuclear content of sporangia and hyphae from Phytophthora capsici transformants and wild‐type.


