Summary
Verticillium dahliae is a soil‐borne, hemibiotrophic phytopathogenic fungus that causes wilting in crop plants. Here, we constructed a random insertional mutant library using Agrobacterium tumefaciens‐mediated transformation to study the pathogenicity and regulatory mechanisms of V. dahliae. The fungal‐specific transcription factor‐encoding gene Vdpf was shown to be associated with vegetative growth and virulence, with the highest transcript expression occurring during conidia formation in the V991 strain. The deletion mutants (ΔVdpf) and insertion mutants (IMΔVdpf) produced fewer conidia than did the wild‐type (WT) fungi, which contributed to the reduced virulence. Unlike the WT, the complemented strains and IMΔVdpf, ΔVdpf formed swollen, thick‐walled and hyaline mycelium rather than melanized microsclerotia. The ΔVdpf mutants were melanin deficient, with undetectable expression of melanin biosynthesis‐related genes (Brn1, Brn2 and Scd1). The melanin deficiency was related to cyclic adenosine monophosphate (cAMP) and the G‐protein‐coupled signalling pathways in this study. Similar to the WT and complemented strains, the ΔVdpf and IMΔVdpf mutants could also successfully penetrate into cotton and tobacco roots, but displayed reduced virulence because of lower biomass in the plant roots and significantly reduced expression of pathogenicity‐related genes in V. dahliae. In conclusion, these results provide insights into the role of Vdpf in melanized microsclerotia formation, conidia production and pathogenicity.
Keywords: conidiation, fungal‐specific transcription factor, melanized microsclerotia, pathogenicity, Verticillium dahliae
Introduction
Verticillium dahliae is the causal agent of Verticillium wilt, which is the most destructive disease of cotton worldwide in temperate regions. It invades over 200 dicotyledonous plant species, including annual herbaceous plants, perennials and woody species (Bell, 2001; Bhat et al., 2003; Fradin and Thomma, 2006; Klosterman et al., 2009). Therefore, V. dahliae, as an asexual and soil‐borne phytopathogen, is one of the most important species for the study of plant–pathogen interactions.
Verticillium dahliae produces melanized and multicellular microsclerotia that spread via wind, water or plant material. Because of their critical role as long‐term remnants in soil, survival structures represent a significant developmental event in the life cycle of V. dahliae (Wilhelm, 1955). After stimulation by root exudates (Fradin and Thomma, 2006; Schnatho and Mathre, 1966) released in the rhizosphere of host and non‐host plants (Huisman, 1982), microsclerotia germinate and produce one to several hyphae that extend towards the host roots, subsequently entering the parasitic stage by infecting susceptible plants, either at the root tip or at the sites of lateral root formation (Fradin and Thomma, 2006; Schnatho and Mathre, 1966). One‐celled, ovoid/elongate and short‐lived colourless conidia are produced in the vascular tissues and initiate new infection cycles (Schnatho and Mathre, 1966).
Many genes in V. dahliae regulate fungal vegetative growth and pathogenicity (Luo et al., 2014). VDH1 (Klimes and Dobinson, 2003, 2006; Klimes et al., 2008), VdGARP1 (Gao et al., 2010) and VdNLP (Santhanam et al., 2013; Zhou et al., 2012) are required for vegetative growth, whereas Vta2 (Tran et al., 2014), VdSSP1 (Liu et al., 2013), EG‐1 (Maruthachalam et al., 2011; Novo et al., 2006; Valášková and Baldrian, 2006) and VdSNF1 (Tzima et al., 2011) are necessary for successful penetration of host plants. They function in the initial phase of Verticillium infection by encoding putative adhesion‐like cell wall proteins, degrading the cell wall, colonizing xylem vessels and regulating cell wall‐degrading enzyme (CWDE) synthesis (Luo et al., 2014). CPC1 (Timpner et al., 2013) and VdThi4 (Hoppenau et al., 2014) function in adapting to the host's intracellular environment. VMK1 (Rauyaree et al., 2005), VdPKAC1 (Tzima et al., 2010), VdMFS (Kapoor et al., 2009; Maruthachalam et al., 2011; Prasad and Kapoor, 2004) and VdSge1 (Santhanam and Thomma, 2013) play roles in the penetration and necrotrophic phases. All of these cellular growth and differentiation genes that are important for virulence or pathogenicity in V. dahliae are tightly regulated by transcription factors (TFs) through interaction with other TFs or molecular chaperones. Based on the conserved DNA‐binding domains, TFs are generally classified into six types: basic leucine zipper (bZIP) proteins, MYB‐like proteins, MADS‐box proteins, helix–loop–helix proteins, zinc finger proteins and homeobox proteins (Meshi and Iwabuchi, 1995; Pabo and Sauer, 1992). These TFs primarily affect three functional aspects in parasitic fungi. First, they regulate conidiation or appressorium development. For example, C2H2 zinc fingers, which are homeobox TFs (MoHOX1–MoHOX8) in the rice blast fungus Magnaporthe oryzae, are necessary for conidiation and appressorium development, and act via cyclic adenosine monophosphate (cAMP) and Ca2+ signalling and/or mitogen‐activated protein kinase (MAPK) signalling pathways (Kim et al., 2009). Second, TFs regulate melanin biosynthesis in fungi, such as in the maize pathogen Cochliobolus heterostrophus, wherein TF Cmr1 regulates melanin biosynthesis through MAPKs, Chk1 and Mps1 (Eliahu et al., 2007). Third, TFs control fungal pathogenicity. For example, in Alternaria brassicicola, the MAPK encoding gene Amk1 is required for initial colonization and regulates melanin biosynthesis (Cho et al., 2007, 2012), whereas the fungal‐specific TF AbPf2 activates early pathogenesis (Cho et al., 2013). Thus, TFs play a variety of regulatory roles in fungi.
Amongst TFs, Zn(II)2Cys6, C4 and C2H2 are three major classes of zinc finger proteins that have been identified in eukaryotes. The cysteine or histidine residues chelate one or more zinc atoms to mediate sequence‐specific DNA binding. These proteins play diverse roles in sugar metabolism, gluconeogenesis, respiration, amino acid metabolism, vitamin synthesis, mitosis, meiosis, chromatin remodelling, nitrogen utilization, peroxisome proliferation, the stress response and pleiotropic drug resistance (MacPherson et al., 2006). In the V. dahliae genome, 937 zinc finger TF‐encoding genes have been annotated; however, only Vta2 , a C2H2 zinc finger transcription activator, has been shown to regulate the expression of genes encoding putative adhesion‐like cell wall proteins (Tran et al., 2014). Amongst the zinc finger TFs, 16 are Zn(II)2Cys6 fungal‐specific TF domain‐containing proteins. However, the functions of fungal‐specific TFs are unclear, and reports about these TFs are few. In this study, based on Agrobacterium tumefaciens‐mediated transformation (ATMT), which is a powerful methodology for targeted and random mutagenesis (de Groot et al., 1998; Mullins et al., 2001), we identified a gene encoding the Zn(II)2Cys6 fungal‐specific TF (VDAG_08521, Vdpf), and showed that Vdpf was essential for melanized microsclerotia formation and conidia production in V. dahliae. The Vdpf deletion and insertion mutants (ΔVdpf and IMΔVdpf) showed reduced virulence in cotton. Further, we characterized Vdpf by examining the expression patterns of Vdpf, the mechanisms of how Vdpf regulates melanized microsclerotia formation, the effects of melanin deficiency on sensitivity to abiotic stressors, and the importance of the Zn(II)2Cys6 binuclear cluster domain of Vdpf in binding the promoter regions of genes involved in melanin synthesis. Our data provide important insights into fungal‐specific TFs.
Results
Screening and identification of T‐DNA random insertional mutants
Through ATMT, 600 primary hygromycin‐resistant mutants were harvested. Most of the mutant phenotypes, including the growth rate, conidiation, microsclerotial development and pigment alteration, were distinct from the wild‐type (WT) phenotype (Fig. S1, see Supporting Information). Of the mutants, the insertion sites of 41 mutants were successfully identified, and they all caused a phenotype (Table S1, see Supporting Information). Based on the colony phenotypes and primary pathogenicity assays, Vd.M.525, which showed reduced virulence, was used for the remaining studies through homologous recombination. Using high‐efficiency thermal asymmetric interlaced polymerase chain reaction (hiTAIL‐PCR), Vd.M.525 was identified to disrupt the VDAG_08521.1 locus, which carried the Vdpf (poly‐functional factor) gene. Sequencing of the right‐border flanking sequences demonstrated that the HygB resistance cassette disrupted the fungal‐specific TF domain of Vdpf (Fig. 1); this mutant was named IMΔVdpf.
Figure 1.
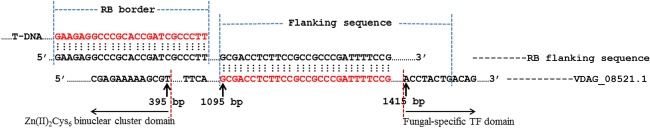
Insertion site of T‐DNA in Vd.M.525 (IMΔVdpf). The T‐DNA cassette was inserted at 1095 bp of the Vdpf (VDAG_08521.1) open reading frame (ORF). The Zn(II)2Cys6 binuclear cluster domain covered the range from 279 to 395 bp, and the fungal‐specific transcription factor (TF) domain covered the range from 1415 to 1715 bp.
Vdpf encodes Zn(II)2Cys6‐type TF
In the V. dahliae genome, 22 genes encode fungal‐specific TF domain‐containing proteins, including 16 genes that encode Zn(II)2Cys6‐type fungal‐specific TFs (Fig. S2, see Supporting Information). Vdpf is one of the genes that encodes the Zn(II)2Cys6‐type TF. A functional domain analysis of the protein indicated that Vdpf (EGY18187.1) contains two conserved domains (Fig. S3A, see Supporting Information). One domain was a typical Zn(II)2Cys6 binuclear cluster domain (Pfam00172: 72–110 residues), in which two zinc atoms are bound to six cysteine residues. This domain showed significant similarity to the Gal4‐like DNA‐binding domain. The other conserved domain, which is known as the fungal‐specific TF domain (Pfam04082: 320–561 residues), functions to assist the Zn(II)2Cys6 cluster in DNA target discrimination. The gene of interest was predicted to encode 833 amino acids, with nuclear localization signal motifs at five locations, i.e. KRRR at 61, RKKK at 78, RRKR at 740, RKRK at 741 and KRKP at 742, as predicted by PSORT (Horton et al., 2007). The gene displayed more than 60% identity with the following fungal‐specific TFs: FGSG_06810 (Fusarium graminearum), FOMG_05053, FOPG_08402, FOXG_02014 (F. oxysporum), ANID_04558 (Aspergillus nidulans), AO090026000614 (A. oryzae), Afu2g02690 (A. fumigatus), ATEG_06908 (A. terreus), ACLA_092100 (A. clavatus), AFL2G_06685 (A. flavus), MGG_07450 (Magnaporthe grisea) and SS1G_00552 (Sclerotinia sclerotiorum) (Fig. S3B). These genes encode proteins that contain a Zn(II)2Cys6 cluster domain and the fungal‐specific TF domain, but the functions and characteristics of these genes have not been studied.
Deletion mutants of the Vdpf gene
To further study Vdpf, deletion mutants (ΔVdpf) were constructed by replacing the coding region of the gene with a hygromycin B (HygB) resistance cassette (Fig. 2A). All of the deletion mutants were confirmed by polymerase chain reaction (PCR), quantitative real‐time reverse transcription‐PCR (qRT‐PCR) and a Southern blotting analysis using gene‐specific probes and sets of PCR primers. Three melanin‐deficient transformants, i.e. ΔVdpf−11, ΔVdpf−12 and ΔVdpf−13, were identified to lack the Vdpf gene. However, the Zn(II)2Cys6 binuclear cluster domain of the Vdpf gene still existed in the five ectopic insertion melanin‐producing mutants and in IMΔVdpf (Fig. 2B), and the fungal‐specific TF domain was absent in both ΔVdpf and IMΔVdpf (Fig. 2C). Outer primers demonstrated the successful deletion of the target gene (Fig. 2D). Southern blot analysis confirmed the insertion of a single copy of the HygB resistance cassette in the ΔVdpf−11, ΔVdpf−13 and IMΔVdpf strains, and the insertion of two copies in the ΔVdpf−12 strain (Fig. 2E). Furthermore, the ΔVdpf−11 mutant was complemented with the WT Vdpf allele with a phleomycin resistance cassette for constitutive expression of the WT Vdpf gene. qRT‐PCR analysis showed that Vdpf was not expressed in ΔVdpf, but exhibited weak expression in IMΔVdpf and was expressed at normal levels in the complemented strain (Fig. 2F). Southern blot analysis using Vdpf probes (Fig. 2E) further confirmed the successful replacement of the Vdpf coding region by the HygB resistance cassette in ΔVdpf. The HygB resistance cassette in ΔVdpf was replaced by the Vdpf allele gene, and normal Vdpf expression showed successful complementation.
Figure 2.
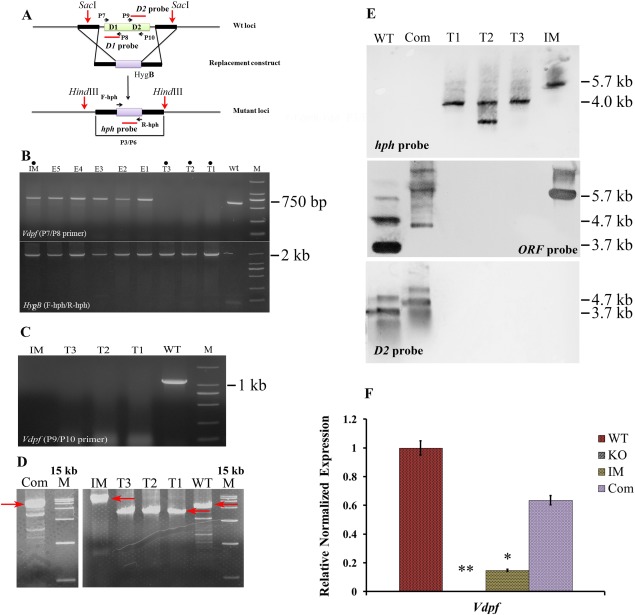
Verification of mutants. (A) The Vdpf (VDAG_08521.1) coding region was replaced with the HygB cassette. (B) Gel electrophoresis of Vdpf and HygB. HygB and the Zn(II)2Cys6 binuclear cluster domain region of Vdpf (VDAG_08521.1) were amplified from all of the mutants with the F‐hph/R‐hph and P7/P8 primers individually. (C) The fungal‐specific transcription factor (TF) domain region amplified with the P9/P10 primer. (D) The mutants were confirmed using nest primers. (E) Southern blotting was performed using the hph, ORF and D2 probes. Four biological replicates (n = 4) were used for this study. (F) Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of Vdpf (VDAG_08521.1) expression in the wild‐type (WT), ΔVdpf, IMΔVdpf and complemented strains. Three biological replicates (n = 3) were used for this study. The dots (•) in (B) indicate the lanes of mutants used in this study. M, 2000‐bp marker in (B) and (C); M, 15 000‐bp marker in (D); T1, ΔVdpf‐11; T2, ΔVdpf‐12; T3, ΔVdpf‐13; IM, insertion mutant; Com, complemented strain; E, EcΔVdpf.
Features of phenotype changes
The knockout mutants (ΔVdpf−11, ΔVdpf−12 and ΔVdpf−13), the random insertion mutant (IMΔVdpf) and the complemented mutant (ΔVdpf:Vdpf‐phleo, Com) were examined for phenotypic changes in growth rate, conidiation, conidia germination, microsclerotia formation and melanin synthesis. The colony morphology of the deletion mutants differed from that of the WT and Com strains (Fig. 3A). For all six strains, the growth rates on potato dextrose agar (PDA) medium were the same (Fig. 3B). However, ΔVdpf−11, ΔVdpf−12 and ΔVdpf−13 produced melanin‐deficient colonies, whereas IMΔVdpf, Com and WT synthesized melanin, and the melanin synthesis of IMΔVdpf was slower and less dense than that of the WT and Com strains. Microscopic observations found that melanized microsclerotia were absent in the ΔVdpf mutants, as observed by the growth of the mutants in both liquid and solid cultures; instead, they formed swollen, thick and hyaline mycelia (Fig. 3C). In addition, the ΔVdpf mutants and IMΔVdpf produced significantly fewer conidia than did the WT strain (P < 0.05; Fig. 3D). The WT allele of the Vdpf gene, under the control of its own promoter, reversed the reduced production of conidia in ΔVdpf mutants. In the WT strain, after 12 h of incubation in potato dextrose broth (PDB), the conidia began to germinate, and the germ tubes formed; at 24 h, hyphal extensions were observed (Fig. S4, see Supporting Information). Measurements of the hyphal extensions at 24 h showed that the hyphal length of the deletion mutants was significantly longer than that of the WT strain (P < 0.05; Fig. 3E,F), whereas their germination rates were not significantly different (P < 0.05; Fig. 3F). Taken together, these results show that Vdpf functions in melanin synthesis, conidiation and microsclerotia formation.
Figure 3.
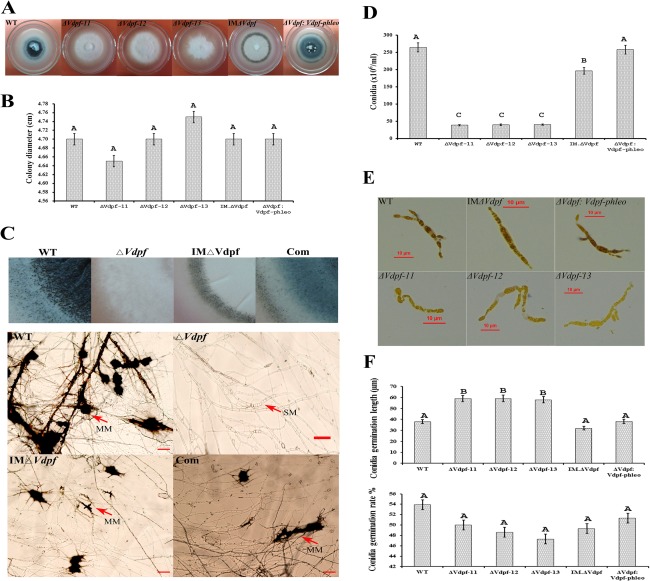
Representative phenotypes of ΔVdpf deletion mutants and the IMΔVdpf insertion mutant. (A) Colony morphology of each strain. (B) Colony diameter comparisons between the mutants and wild‐type (WT) at 20 days post‐inoculation (dpi) on potato dextrose agar (PDA) medium. (C) Morphology of microsclerotia. MM, melanized microsclerotia; SM, specialized mycelium. Scale bars, 20 μm. (D) Conidia produced by each strain. (E, F) Extended hyphae and conidia germination rate/length of WT, ΔVdpf, IMΔVdpf and ΔVdpf:Vdpf‐phleo at 24 h after incubation in potato dextrose broth (PDB). Vdpf, VDAG_08521.1. A one‐way analysis of variance (ANOVA) and Student's t‐test were performed. Different letters indicate a significant difference (P < 0.05). The error bars represent the standard deviation of three replicates.
ΔVdpf mutants are responsive to abiotic stressors as WT V. dahliae
In response to pathogen infection, plants can change the internal environment of the apoplast and the vascular system of cells, including the production of various types of reactive oxygen species (ROS) and phytoalexins, and the alteration of cell pH, on which some cell wall peroxidases are dependent (Bhattacharjee, 2005; Steinberg, 2012). Melanin confers protection against these cellular stress signals in V. dahliae (Kuo and Alexander, 1967). Therefore, to indirectly evaluate the importance of ΔVdpf and IMΔVdpf in managing stressors, we measured the effects of osmolites, acid, alkali and oxygen stress on the vegetative growth of these mutants. When grown on PDA containing 0.7 m KCl, 1.0 m sorbitol or 1.5 mm H2O2, the colony size of ΔVdpf and IMΔVdpf did not vary significantly from those of Com or the WT strains (Fig. S5, see Supporting Information). The colony sizes of the deletion mutants were also comparable with those of the Com strains and the WT fungus under acidic (pH 4.0) and alkaline (pH 12.0) conditions. Interestingly, alkaline conditions stimulated melanin biosynthesis.
Reduced virulence in the ΔVdpf mutants
Cotton (Gossypium hirsutum L.) and tobacco (Nicotiana benthamiana) plants were used to test the pathogenicity of V. dahliae. In 6‐week‐old intact cotton plants, after inoculation for 20 days, infection with the WT V. dahliae caused wilting of leaves. In addition, necrotic leaves fell, whole plants died and the disease index reached 100%. However, in 6‐week‐old intact cotton plants that were infected with the deletion mutants (ΔVdpf and IMΔVdpf), the disease severity was reduced compared with that in the plants which were infected by the WT fungus and the Com strains (Fig. 4A), with a significant decrease in the disease index (P < 0.01) (Table S2, see Supporting Information). Interestingly, ΔVdpf was not as virulent in tobacco as it was in cotton plants, whereas wilting symptoms caused by IMΔVdpf in intact tobacco were comparable with those caused by WT and Com strains (Fig. 4B); however, the occurrence of wilting symptoms caused by IMΔVdpf was slower than those caused by the Com and WT strains. We stained roots to demonstrate whether the ΔVdpf‐dependent reduction in disease severity on cotton was caused by impaired infectivity, including the ability to degrade plant cell walls, infect cortical cells and infect the vascular bundle. The results showed that the ΔVdpf mutants and IMΔVdpf could successfully degrade plant cell walls, infect cortical cells and even infect the vascular bundle, similar to WT V. dahliae (Fig. 4C). To examine whether the reduced virulence was correlated with reduced biomass of the mutant in the plant, the fungal biomass in the roots of the infected plants was determined by PCR and quantitative analysis using Quantity One software. Adj.Vol.INT*mm2 (the quantitative values after the deduction of the background) of WT, Com, control (CK), knockout mutant (KO) and insertion mutant (IM) strains were 61545.74, 60849.64, 0, 29748.77 and 30784.36, respectively, indicating that the biomasses of ΔVdpf and IMΔVdpf were nearly two‐fold less in the roots compared with the biomasses of the WT and Com strains (Fig. 4D). Thus, this suggests that the virulence of V. dahliae may depend on the degree of colonization.
Figure 4.
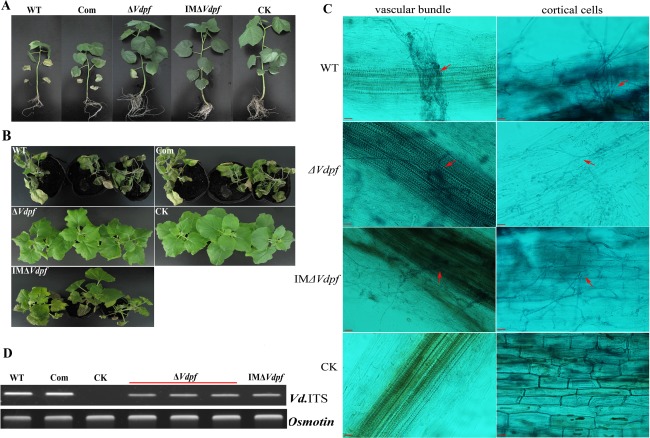
Reduced pathogenicity in ΔVdpf. (A) Necrosis and defoliation in intact cotton plants (Gossypium hirsutum L.) at 20 days post‐inoculation with 4 × 107 conidia of WT, ΔVdpf mutants, IMΔVdpf mutants and complemented strains. (B) Virulence measurements on tobacco plants (Nicotiana benthamiana). (C) Infection of WT, ΔVdpf mutants, IMΔVdpf mutants, complemented strains and CK in root cortical cells and the vascular bundle. (D) Detection of fungal biomass in infected plant roots. WT, wild‐type Verticillium dahlia; ΔVdpf, deletion mutant; IMΔVdpf, insertion mutant; Com, ΔVdpf:Vdpf‐phleo mutant; CK, plant roots inoculated with sterile water. Scale bars, 50 μm. Vdpf, VDAG_08521.1.
Differential expression of vegetative growth‐ and/or pathogenicity‐related genes
To determine whether Vdpf stimulated/inhibited the expression of genes involved in conidial production, microsclerotia formation and pathogenicity in V. dahliae, known vegetative growth‐ and/or pathogenicity‐related genes were assessed by qRT‐PCR. First, we examined the expression of VDH1 (a class II hydrophobin) (Klimes and Dobinson, 2006; Klimes et al., 2008) and Vdgrp1 (glutamic acid‐rich protein 1) (Gao et al., 2010), which have been shown to play important roles in microsclerotia formation. Compared with the WT strain, VDH1 was not expressed in ΔVdpf, and the expression of Vdgrp1 was reduced by 250‐fold (P < 0.05) and 10‐fold in ΔVdpf and IMΔVdpf, respectively (Fig. 5). In contrast, in the IMΔVdpf mutant, the expression of VDH1 was not significantly different from that in the WT strain. Thus, these results explain why the ΔVdpf mutants exhibit the disruption of normal microsclerotia formation.
Figure 5.
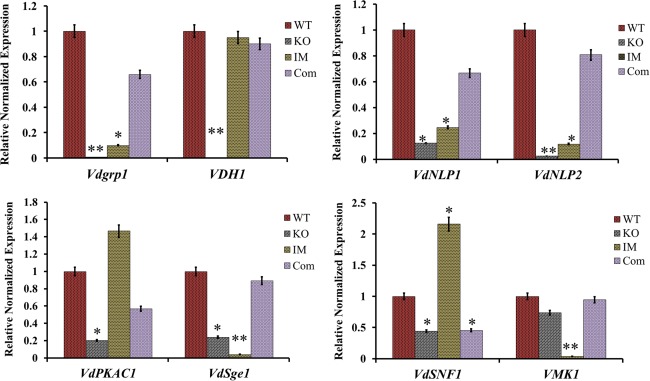
Expression of vegetative growth‐ and pathogenicity‐related genes. The relative transcription abundance of each gene was determined in comparison with the 18srRNA gene transcripts in the same tissue. The y axes illustrate the normalized relative quantity of the transcripts compared with the housekeeping gene 18srRNA. Three biological replicates (n = 3) were used for this study. The error bars indicate the standard deviation. Statistical analysis was performed using t‐test. *P < 0.05. **P < 0.01. WT, wild‐type; KO, knockout mutants; IM, insertion mutants; Com, complemented strains. Vdpf, VDAG_08521.1.
Furthermore, we examined the expression of genes that are directly or indirectly related to pathogenicity in V. dahliae, i.e. VdPKAC1, VdSNF1, VdSge1, VMK1, VdNLP1 and VdNLP2, which have been demonstrated to be the major virulence factors of V. dahliae. Among these genes, VdNLP1 and VdNLP2 have been demonstrated to be the major virulence factors in cotton (Santhanam et al., 2013; Zhou et al., 2012). With the exception of VMK1 expression, which remained unchanged, the expression levels of VdPKAC1, VdSNF1, VdSge1, VdNLP1 and VdNLP2 were decreased by five‐, three‐, four‐, eight‐ and 40‐fold, respectively, in ΔVdpf, and this reduced expression was reversed in the Com strain (Fig. 5). Interestingly, in IMΔVdpf, the expression of the Zn(II)2Cys6 binuclear cluster domain in Vdpf was down‐regulated by seven‐fold compared with the WT strain (Fig. 2), suggesting that the fungal‐specific TF domain strengthens Vdpf transcriptional regulation. Moreover, VdSNF1, which is positively correlated with CWDE synthesis and facilitates penetration in host plants, was up‐regulated by two‐fold; VdPKAC1 [a C subunit of protein kinase A (PKA)], which plays a role in virulence, microsclerotia formation, conidiation and ethylene (ET) production via cAMP‐dependent signalling pathways, was up‐regulated by 1.5‐fold; VMK1 and VdSge1, which promote pathogenicity, were decreased by 27‐ and 25‐fold, respectively; and VdNLP1 and VdNLP2 were down‐regulated by four‐ and eight‐fold, respectively (Fig. 5). These data strongly suggest that Vdpf differentially affects the expression of some vegetative growth‐ and/or pathogenicity‐related genes in V. dahliae and that the fungal‐specific TF domain strengthens this regulatory mechanism. The down‐regulation of these pathogenicity‐related genes in deletion mutants suggests that the reduced pathogenicity caused by deletion mutants is partly correlated with toxicity.
Vdpf gene expression pattern
After culture for 6 h in PDB medium, the conidia of V. dahliae began to form germ tubes. After 12 h of cultivation, the conidia germinated, and hyphae grew after 24 h of cultivation. Verticillium dahliae began to form conidia after 48 h of cultivation. We monitored Vdpf expression at the different stages (Fig. 6A) and found that it was highest at the conidial stage, lowest when the germ tube began to form, and gradually increased during germination and hyphal growth. At the conidium formation stage, Vdpf expression gradually reached nearly the same level as in the conidial phase. This expression pattern could explain why ΔVdpf mutants display reduced numbers of conidia, and cause impaired pathogenicity compared with the WT and Com strains.
Figure 6.
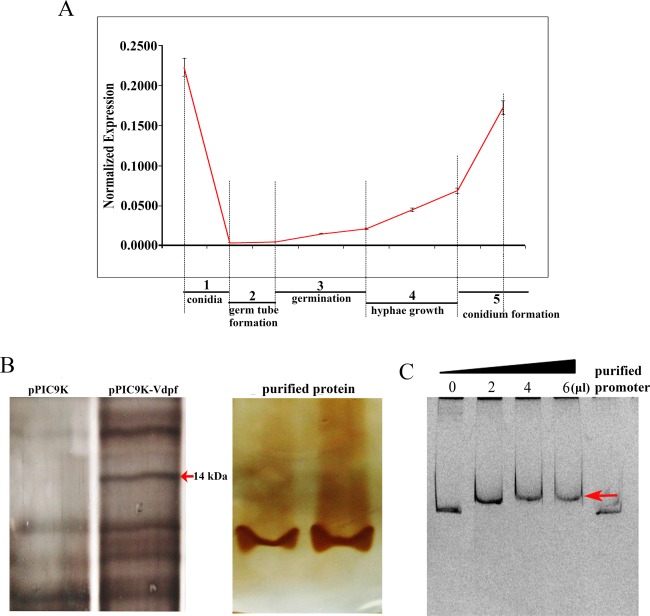
Normalized expression of Vdpf at different stages and in the electrophoretic mobility shift assay (EMSA) of Vdpf. (A) The relative transcript amount at each stage was determined in comparison with the 18srRNA transcripts in the same tissue. The y axes illustrate the relative quantity of the transcripts compared with the housekeeping gene: 18srRNA. Three biological replicates (n = 3) were used for this study. The error bars indicate the standard deviation. The statistical analysis was performed using the t‐test. 0 h, conidia; 6–12 h, formation of germ tube; 12–24 h, conidia germination; 24–48 h, hyphal growth; 48–120 h, conidium formation. (B) The Zn(II)2Cys6 domain expressed in Pichia pastoris (Gs115). The protein expressed in Pichia pastoris was transformed with purified pPIC9k and served as the control. Vdpf (EGY18187.1) was purified using His‐tag. (C) Vdpf binds to its own promoter. A 1000‐bp promoter fragment of Vdpf was used for binding assays with 0, 2, 4 or 6 μL of purified Vdpf. The concentration of purified Vdpf was 513.636 μg/mL. The pure promoter was used as a probe control. The red arrow points to the shift bands. Three biological replicates (n = 3) were used for this study.
Electrophoretic mobility shift assays (EMSAs) confirmed that Vdpf could bind to its own promoter region. The DNA‐binding domain (Zn(II)2Cys6) of Vdpf was overexpressed in Pichia pastoris GS115 and was purified as a His‐tagged protein (Fig. 6B). As shown in Fig. 6C, clear shift bands were observed when the Vdpf promoter was co‐incubated with increasing amounts of the Vdpf protein. Negative control assays showed that no specific shift bands appeared, suggesting that the interaction between Vdpf and its own promoter was involved in Vdpf expression. Through the prediction of TF binding sites using JASPAR, four high‐potential motifs, SUT1 (CGCGGGG), RDS2 (CTCGGGG), CAT8 (CCGGA) and ARG81 (TTGACTCT), to which the GAL4 domain‐containing TFs bind, were discovered.
Effect of Vdpf on the expression of genes involved in melanin synthesis
(+)‐Scytalone, as an indispensable intermediate, has been shown to be produced and converted to melanin by microsclerotia, but not by conidia or hyphae, in V. dahliae; moreover, scytalone dehydratase (SCD1) functions in this process (Bell et al., 1976). Polyketide synthase 1 (PKS1) has been confirmed to be a key enzyme yielding DHN‐melanin in V. dahliae (Wang et al., 2013). Later, PKS18, SCD1, BRN1 and BRN2 were confirmed to be involved in the melanin synthesis pathway in other fungi (Eliahu et al., 2007; Moriwaki et al., 2004; Thompson et al., 2000; Tsai et al., 1999). Through a transcriptome sequencing analysis of melanized and non‐melanized V. dahliae, we showed that the expression of these four genes was up‐regulated in melanized V. dahliae. Two tetrahydroxynaphthalene reductase‐encoding genes, Brn1 and Brn2, which showed significant homology in encoding T3HN reductase and T4HN reductase, respectively, were up‐regulated by nine‐ and 4.8‐fold, respectively (Luo et al., unpublished data). In addition, Scd1 and Pks1, which are the homologues of SCD1 in C. heterostrophus and PKS in Bipolaris oryzae (Eliahu et al., 2007; Moriwaki et al., 2004), were also up‐regulated by 6.6‐ and 1.9‐fold, respectively (Luo et al., unpublished data). In this research, we examined the expression of these four genes (Brn1, Brn2, Pks1 and Scd1) at four time points (0, 18, 48 and 120 h). Brn1, Brn2 and Scd1 were undetectable in the ΔVdpf mutants, whereas Pks1 expression was normal compared with the WT and Com strains (Fig. 7A). Unlike ΔVdpf, the expression of Brn1, Brn2 and Scd1 could be detected in IMΔVdpf; this may be why IMΔVdpf could synthesis melanin, but ΔVdpf could not. These results imply that Vdpf regulates melanin biosynthesis through the direct and indirect regulation of Brn1, Brn2 and Scd1.
Figure 7.
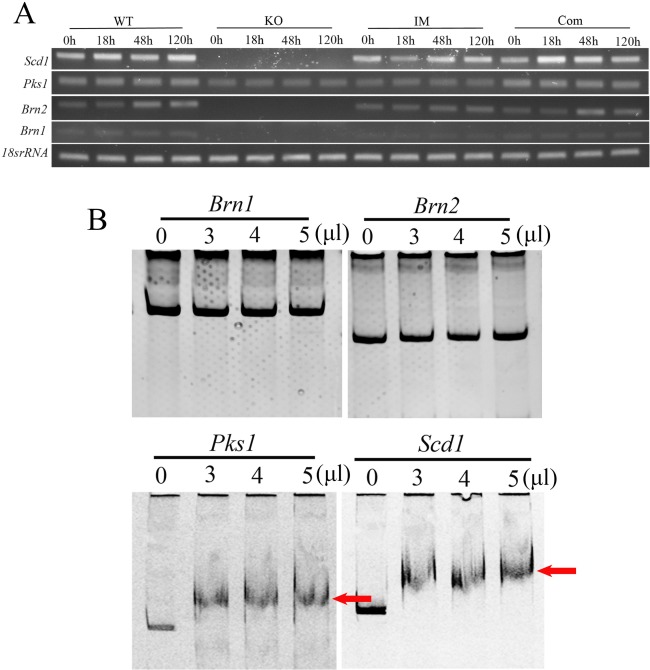
Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) and electrophoretic mobility shift assay (EMSA) of fungal DHN‐melanin biosynthesis‐related genes. (A) RT‐PCR analysed the expression of Pks1, Brn1, Brn2 and Scd1 at 0, 18, 48 and 120 h. WT, wild‐type; KO, knockout mutants; IM, insertion mutants; Com, complemented strains. (B) EMSA for Vdpf (EGY18187.1) and the promoters of melanin synthesis‐related genes. The promoters of Brn1, Brn2, Pks1 and Scd1 were used for the binding assays with 0, 3, 4 or 5 μL of purified Vdpf. The concentration of purified Vdpf was 513.636 μg/mL. The red arrows point to the shift bands. Four biological replicates (n = 4) were used for this study.
Subsequently, we used EMSA to determine whether Vdpf regulates melanin biosynthesis directly by binding to the promoters of Brn1, Brn2, Pks1 and Scd1, or indirectly through signalling pathways, such as the MAPK, cAMP or G‐protein‐coupled signalling pathways. Interestingly, Vdpf could bind to the promoter of Pks1. Apart from this, Vdpf could also bind to the promoter of Scd1, but not to the promoters of Brn1 or Brn2 (Fig. 7B), indicating that unknown signalling pathway(s) may be involved in the regulation of melanin biosynthesis in V. dahliae.
Genes involved in the MAPK, cAMP and G‐protein‐coupled signalling pathways were isolated using a blast search of the Verticillium database (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html). VDAG_04508 (Ac) is the sole adenylate cyclase gene, and VDAG_06474 (Pka) is the only cAMP‐dependent PKA gene in V. dahliae. VDAG_09823 (Mkk1) and VDAG_00874 (Mkh1) encode the MAP kinase kinase MKK1/SSP32 and the MAP kinase kinase kinase, respectively, whereas VDAG_04611 (Gp1), VDAG_02846 (Gp2) and VDAG_09140 (Gp3) encode the same G‐protein‐coupled receptor. Compared with non‐melanized V. dahliae, these genes were differentially expressed in melanized V. dahliae (Luo et al., unpublished data). EMSA showed that Vdpf could bind to the promoters of Ac, Pka, Gp1, Gp2 and Gp3, but not to Mkk1 and Mkh1 (Fig. 8A). RT‐PCR and quantitative analysis revealed that Gp1 was down‐regulated, and the expression of Ac and Mkh1 was undetectable (Fig. 8B). Compared with the WT strain, the expression levels of the other genes were normal in the mutant. These results imply that Vdpf directly regulates cAMP and the G‐protein‐coupled signalling pathways, whereas Mkk1 and Mkh1 are not regulated by this pathway in this assay (Fig. 8C). All of these pathways play roles in the regulation of melanin biosynthesis and may be involved in other processes, such as conidial production and microsclerotial formation.
Figure 8.
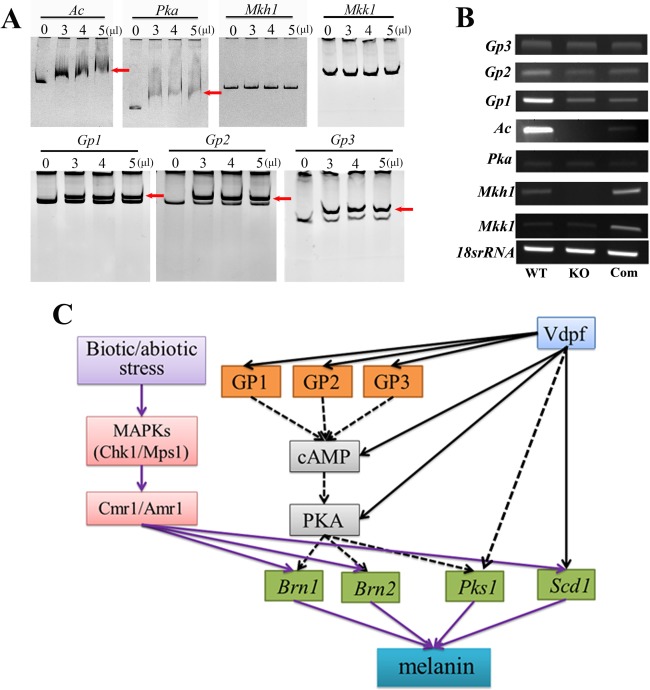
Electrophoretic mobility shift assay (EMSA) and the predicted model for the process of melanin biosynthesis. (A) EMSA for Vdpf (EGY18187.1) and the promoters of Ac, Pka, Mkk1, Mkh1, Gp1, Gp2 and Gp3. Ac represents the adenylate cyclase‐encoding gene, and Pka represents the cyclic adenosine monophosphate (cAMP)‐dependent protein kinase A (PKA)‐encoding gene; these genes are involved in the cAMP signalling pathway. Mkk1 and Mkh1 represent the mitogen‐activated protein (MAP) kinase kinase MKK1/SSP32 gene and the MAP kinase kinase kinase mkh1 gene, respectively, which play important roles in the MAP kinase signalling pathway. Gp1, Gp2 and Gp3 are genes that encode the G‐protein‐coupled receptor; they participate in the G‐protein‐mediated signalling pathway. The binding assays used 0, 3, 4 or 5 μL of purified Vdpf (EGY18187.1). The red arrow points to the shift bands. (B) Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the signalling genes. The expression level was measured by Quantity One software. (C) Deducible signalling pathways in melanin synthesis. Chk1 and Mps1 are two MAP kinase genes involved in the MAP kinase signalling pathway. They have been demonstrated to be involved in melanin synthesis through the transcription factor Cmr1/Amr1. The purple arrows represent known results, and the black arrows represent results in this research study. The broken lines represent unconfirmed results. Three biological replicates (n = 3) were used for this study.
Furthermore, we predicted potential TF binding sites of the promoter region of each gene that could bind with Vdpf using the high‐quality TF binding profile database JASPAR. All of the predictions were performed based on JASPAR CORE fungi. The fungal Zn cluster family of Saccharomyces cerevisiae was selected as the matrix model and the relative profile score threshold was >99%. The probable binding sites and sequence motifs are displayed in Fig. S6 (see Supporting Information). All of the predicted motifs were GAL4 domain‐containing TF binding sites (Badis et al., 2008).
Discussion
In our research, all of the deletion mutant strains of the Vdpf gene consistently displayed a melanin deficiency, a reduction in conidia production, a disability of microsclerotia formation and weakened virulence compared with the WT V. dahliae. Compared with the Com strains, all of these obvious differences also existed, which showed that they were all a result of the loss of Vdpf function. However, the insertion mutant (IMΔVdpf), which destroyed the fungal‐specific TF domain, synthesized melanin and microsclerotia, as did the WT V. dahliae and Com strains. The results illustrate that Vdpf is a polyfunctional factor.
In fungi, Zn(II)2Cys6‐type zinc finger TF proteins are closely linked to various developmental and physiological processes. In Sordaria macrospora, Pro1 plays a role in sexual fruiting body development (Masloff et al., 1999, 2002). In A. flavus, the Zn(II)2Cys6 zinc cluster‐encoding gene family is involved in asexual conidiation, sexual homothallic development and secondary metabolites (Chang and Ehrlich, 2013). In A. nidulans, TamA interacts with LeuB and regulates leucine biosynthesis, NADP‐glutamate dehydrogenase activity and nitrogen metabolism through the regulation of the gdhA gene encoding NADP‐glutamate dehydrogenase (Downes et al., 2013, 2014). In A. oryzae, ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the β‐mannan utilization system (Ogawa et al., 2012). In Bipolaris oryzae, Bmr1, which has two Cys2His2 zinc finger domains and a Zn(II)2Cys6 binuclear cluster domain at the N‐terminal region, is essential for melanin biosynthesis (Kihara et al., 2008). In the rice blast fungus M. oryzae, two Zn(II)2Cys6 TFs, MoCOD1 and MoCOD2, play important roles in conidiation and pathogenicity, and MoCOD1 is also involved in conidial germination and appressorium formation (Chung et al., 2013). Here, our data have shown that one of the Zn(II)2Cys6 TFs of V. dahliae, Vdpf, has important regulatory functions in governing various developmental processes of the fungus. Our studies demonstrate that Vdpf regulates the formation of melanized microsclerotia, which are important for pathogenicity. In addition, Vdpf plays an important role in conidiation and virulence.
For V. dahliae, the disease cycle begins with dormant microsclerotia, which are distributed in the soil or embedded within plant debris and can remain viable for up to 14 years in the absence of a host (Wilhelm, 1955). Melanized microsclerotia play an important role in the disease cycle because they are major sources of inoculum and are the primary long‐term survival structures. Melanin is a dark pigment tissue composed of polymerized phenolic and/or indolic compounds (Nosanchuk and Casadevall, 2003). A wide variety of fungi synthesize DHN‐melanin via the PKS pathway (Bell and Wheeler, 1986). Briefly, the end product, melanin 1,8‐DHN (Tanaka et al., 1991), is synthesized from tetrahydroxynaphthalene (THN) through enzymatic reactions involving BRN1, BRN2, SCD1, and PKS18/PKS1 (Eliahu et al., 2007; Greenblatt and Wheeler, 1986; Moriwaki et al., 2004). All but one of 20 dark brown and black ascomycetous and imperfect fungi appear to make melanin from 1,8‐DHN via this pentaketide pathway. Aspergillus niger and six basidiomycetes do not synthesize DHN and apparently make a different type of melanin. Many studies have examined the pentaketide pathway of melanin synthesis in V. dahliae; these studies have shown that this pathway is substrate specific and occurs during a certain developmental morphology (melanized microsclerotia) associated with the life cycle of the fungus (Bell et al., 1976; Stipanovic and Bell, 1976, 1977; Ten et al., 1977; Tokousbalides and Sisler, 1979; Wheeler et al., 1978). Microsclerotia are the resting structures of V. dahliae and can endure desiccation and cold, thus facilitating the long‐term survival of V. dahliae without its host plant. The dormant microsclerotia germinate after they are stimulated by root exudates (Schnatho and Mathre, 1966). The fungus V. dahliae accumulates dense black melanin granules in the outer wall and surrounding matrix of its microsclerotial cells, but has hyaline hyphae and conidia (Brown and Wyllie, 1970). In V. dahliae, non‐melanized microsclerotia quickly lose their persistence; thus, melanin contributes to the long‐term survival of resting structures in natural soils (Hawke and Lazarovits, 1995; Lockwood, 1960). However, similar to several other fungi (Bolton et al., 2006; Fitzgerald et al., 2004; Thomma, 2003), the role of melanin in Verticillium pathogenicity has not been established (Fradin and Thomma, 2006). Protection by melanin is at least partially a result of the inactivation of the β‐1,3‐glucanase and chitinase enzymes secreted by associated soil microorganisms (Kuo and Alexander, 1967; Potgieter and Alexander, 1966). The TF Cmr1 (C. heterostrophus) (Eliahu et al., 2007) and its homologues Bmr1 (B. oryzae) (Kihara et al., 2008) and Amr1 (Al. brassicicola) (Cho et al., 2012) have been shown to contribute to melanin biosynthesis. In our research, Vdpf, which is not a homologue of Cmr1, was confirmed to play a role in melanin biosynthesis. ΔVdpf mutants were melanin deficient, and this deficiency was a result of the down‐regulation of Brn1, Brn2 and Scd1 (Eliahu et al., 2007). This mutant did not form microsclerotia, but instead exhibited swollen, thick‐walled and hyaline mycelium. However, IMΔVdpf, whose fungal‐specific TF domain was disturbed, was able to synthesize melanized microsclerotia, but at a decreased quantity compared with WT and Com strains. The different phenotypes of ΔVdpf and IMΔVdpf occur because the insertion mutant does not completely silence Vdpf expression. Compared with WT, IMΔVdpf and Com strains, ΔVdpf mutants display reduced pathogenicity in cotton and tobacco, which is contrary to the increased virulence caused by ΔAmr1 mutants of Al. brassicicola (Cho et al., 2012). Because of the successful penetration into the cortical cells and vascular bundle of host plants, melanin deficiency does not affect the infective ability of the mutants.
Pathogenic fungi must evolve a series of strategies to successfully infect their host plants, including overcoming plant defence mechanisms, adapting to the complicated intracellular environments of hosts and conquering adverse environmental changes. ROS are one of the most prominent plant defences (Guo et al., 2011; Molina and Kahmann, 2007). Some factors and genes, such as oxalic acid (Cessna et al., 2000), the YAP1 homologue (Lin et al., 2009; Molina and Kahmann, 2007) and the bZIP TF MoAP1 (Guo et al., 2011), are involved in the mediation of oxidative stress tolerance and suppression of the oxidative burst of the host plant in various phytopathogens. In our studies, oxidative stressors, such as H2O2 and sorbitol, had similar effects on the vegetative growth of ΔVdpf mutants and on the IMΔVdpf, WT and Com strains. No differences were observed in the mutants or the WT strain when exposed to KCl, acidic conditions or alkaline conditions. These results suggest that Vdpf is dispensable for the detoxification of ROS and osmoregulation. In other words, the reduction of virulence in mutants was not regulated through ROS suppression, osmotic pressure or alkalinity or acidity of the inner environment of host plants.
The MAPK and cAMP signalling pathways are two of the primary regulatory pathways in fungi. For example, knockout of the MAPK gene VMK1 demonstrated that MAPK signalling is important for vegetative growth and pathogenicity in V. dahliae (Rauyaree et al., 2005). Moreover, deletion of the PKA subunit C gene, VdPKAC1, results in reduced virulence, conidiation and ET production, but increased development of microsclerotia (Tzima et al., 2010), revealing that the cAMP‐dependent signalling pathway is required for both fungal development and pathogenicity. In our study, we found that cAMP and the G‐protein‐coupled signalling pathways were involved in Vdpf regulation, particularly melanin formation. In C. heterostrophus and Al. brassicicola, melanin biosynthesis is regulated by the MAPK signalling pathway (Cho et al., 2012; Eliahu et al., 2007). Because cAMP and the G‐protein crosslink many signalling pathways, the phenotype of ΔVdpf is highly pleiotropic, which indirectly demonstrates that Vdpf is a polyfunctional factor. However, these findings are not systemic and comprehensive, and a genome‐wide assessment of the roles of signal‐regulated genes and virulence factors may be more valuable for the investigation of the cell growth control mechanisms and pathogenicity of the fungus (Zhang et al., 2008).
In the field, the disease cycle begins with dormant microsclerotia in V. dahliae (Wilhelm, 1955). On germination, microsclerotia produce hyphae that extend towards host roots and infect the susceptible plants either at the root tip or at sites of lateral root formation (Huisman, 1982). After crossing the endodermis, the fungus enters the vascular tissues where it can form conidia (Fradin and Thomma, 2006). These conidia are carried with the sap stream and trapped in pit cavities or at vessel end walls (so‐called trapping sites), where they germinate and penetrate adjacent vessel elements to continue colonization (Fradin and Thomma, 2006). Successful penetration into the cortical cells and vascular bundle of the host plants is the prerequisite for virulence. Root‐staining assays demonstrated that all of the mutants and WT V. dahliae could penetrate into and colonize the hosts, although the fungal biomass of the mutants was lower than that of the WT and Com strains, which illustrated that the reduced virulence of the mutants partly contributed to the reduced conidiation. The reduction in expression of pathogenicity‐related genes in V. dahliae was another contribution to the reduced virulence caused by the deletion mutants in cotton and tobacco plants. VdPKAC1 and VdSNF1 have been demonstrated to be virulence factors in tomatoes and eggplant (Tzima et al., 2010, 2011), VdSge1 is a virulence factor in tomatoes (Santhanam and Thomma, 2013), VMK1 is a virulence factor in cotton and tomatoes (Rauyaree et al., 2005), and VdNLP1 and VdNLP2 are virulence factors in tobacco, Arabidopsis and cotton (Santhanam et al., 2013; Zhou et al., 2012). The loss of Vdpf influenced pathogenicity through reduced conidiation and reduced expression of genes encoding virulence factors, and the fungal‐specific TF domain strengthened the influence. Proteins containing fungal‐specific TF domains are widespread in the phytopathogens Aspergillus, Fusarium, Verticillium and M. oryzae and can be classified into two types: (i) those containing a fungal‐specific TF domain and a DNA‐binding domain; and (ii) those containing only a fungal‐specific TF domain. To date, few studies have investigated the detailed functions of the fungal‐specific TF domain.
In conclusion, Vdpf, which has two conserved domains, the Zn(II)2Cys6 binuclear cluster domain and the fungal‐specific TF domain, is important in the regulation of conidia production, melanized microsclerotia formation and pathogenicity, and the Zn(II)2Cys6 binuclear cluster domain is important in binding the promoter regions of Vdpf and the genes involved in melanin synthesis. These results provide insights into the role of Vdpf in V. dahliae. In future studies, we will endeavour to understand the function of other fungal‐specific TFs in V. dahliae and to investigate the detailed functions of the fungal‐specific TF domain.
Experimental Procedures
Strains and culture conditions
The defoliating type virulent V. dahliae strain, V991 (preserved in our laboratory), was isolated from cotton that originated in Xinjiang, China and was used as a WT strain for transformation in this study; this strain is sensitive to HygB when used at concentrations greater than 30 mg/mL. The strain and its transformants were stored at −80°C in the form of a conidial suspension in 30% glycerol and were reactivated on PDA medium at 25°C. PDB liquid medium was used to prepare the conidia and mycelia. Agrobacterium tumefaciens strain AGL‐1 (preserved in our laboratory) was used to transform the conidia of V. dahliae. Agrobacterium tumefaciens was grown in YEB (0.5% sucrose, 0.1% yeast extract, 1% tryptone, 0.05% MgSO4.7H2O, pH 7.0) agar/liquid medium supplemented with 50 mg/mL of streptomycin. Pichia pastoris GS115 was preserved in our laboratory.
ATMT
ATMT was manipulated as described previously (Maruthachalam et al., 2011) with some modifications. The Ag. tumefaciens strain AGL‐1 containing pPK2 or recombinant vectors was grown at 28°C in PDB medium until the optical density at 660 nm (OD660) reached 0.5. Prior to mixing with an equal volume of a conidial suspension of V991 (107 conidia/mL), the culture was diluted to OD600 = 0.15 in the induction medium (IM) with 200 μm acetosyringone (AS) and cultured for an additional 6 h at 28°C, 200 rpm on an orbital shaker. Then, 250 μL of the mixture were placed onto nitrocellulose filters (pore size, 0.45 μm; diameter, 80 mm; Whatman, Tokyo, Japan) on co‐cultivation medium (CM) for 48 h. Thereafter, the filters were transferred to selective medium containing HygB (50 μg/mL), which acts as a selective agent for V. dahliae transformants, and cefotoxime (500 μg/mL), which eliminates Ag. tumefaciens cells. After 7 days, randomly selected transformants were transferred to fresh PDA medium with 50 μg/mL of HygB.
Identification of positive randomly inserted transformants
The genomic DNA of each transformant was extracted from conidia and mycelia according to the method of Stewart and Via (1993). To ensure that the T‐DNA was inserted in V. dahliae, the transformants were identified by PCR with an hph‐specific primer pair: F‐hph (5′‐ATGCCTGAACTCACCGCGAC‐3′) and R‐hph (5′‐CTATTCCTTTGCCCTCGGAC‐3′). PCR was performed in a 15‐μL mixture containing 5 ng of genomic DNA, 2 × rTaq premix (Takara, Shiga, Japan), 5 pmol of each primer and double‐distilled H2O. The reaction program included 5 min at 95°C; 35 cycles of 1 min at 95°C, 1 min at 60°C and 1.5 min at 72°C; and 10 min at 72°C for the final extension. The hph fragment of the PCR product was 1.02 kb. A hiTAIL‐PCR protocol (Liu and Chen, 2007) was used for the amplification of the sequences flanking the inserted T‐DNA from the positive transformants. The right border (RB) and left border (LB) primers used in this study are shown in Table S3 (see Supporting Information). The PCR products were purified using an E.Z.N.A. Gel Extraction Kit (OMEGA, Shanghai, China) and were sequenced. The T‐DNA insertion sites were detected by blasting with the genome of VdLs.17 (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/Blast.htmL) (Klosterman et al., 2011).
Generation of V. dahliae Vdpf mutants (ΔVdpf)
The VdPf deletion construct was created using fusion PCR (Szewczyk et al., 2006) (Fig. S7A, see Supporting Information). Vdpf deletion mutants were generated by replacing the open reading frame (ORF) with the HygB resistance cassette. First, a 915‐bp Vdpf upstream fragment, a 2018‐bp HygB resistance cassette from the vector pSilent‐1 and an 802‐bp Vdpf downstream fragment were amplified by three sets of primers, i.e. P3/P4, P1/P2 and P5/P6, respectively (Table S4, see Supporting Information). Then, the three fragments were fused to U‐Hph‐D, and U‐Hph‐D was digested by the restriction enzymes HindIII and XhoI, followed by ligation with the binary vector pPZPtk 8.10, which was digested with the same enzymes. The pPZPtk 8.10 vector (donated by Barbara Howlett, Melbourne University, Australia) contains a bidirectional negative selectable marker HSVtk, which was used to select the positive transformants. Finally, the resulting recombinant plasmid, pPZPtk‐U‐Hph‐D, was transformed into the WT as described previously (Maruthachalam et al., 2011). The transformants were screened on PDA supplemented with 50 μm F2du and 50 μg/mL of HygB. The positive transformants and random insertion mutants (IMΔVdpf) were confirmed by PCR screening using P3/P6, F‐hph/R‐hph, P7/P8 (products containing the Zn(II)2Cys6 binuclear cluster domain, D1) and P9/P10 (products containing the fungal‐specific TF domain, D2). qRT‐PCR and Southern hybridization using ORF (includes D1 and D2) and D2 probes were used to identify the deletion mutants, followed by PCR screening. To detect whether HygB was single copy inserted into the mutants, we used F‐hph/R‐hph to create 1021‐bp Southern hybridization probes. Southern hybridization was performed according to the protocol of the DIG High Prime DNA Labelling and Detection Starter Kit I (Roche, Mannheim, Germany). Genomic DNA was digested by HindIII for detection of the hph cassette using Southern blotting, and SacI was used to digest the genomic DNA for detection of Vdpf by Southern blotting. DNA probes were amplified by PCR using PrimeSTAR Max DNA polymerase (Takara), and their accuracies were ensured by sequencing.
Complementation of ΔVdPf mutant strains
The ΔVdpf mutant was complemented by the WT Vdpf allele with its native promoters as described previously (Cho et al., 2012). The complement fragment, which contained the open reading fragment of the Vdpf gene and its native promoter region, was amplified by PCR with primers F‐Vdpf/R‐Vdpf (Fig. S7B). The complementation cassette was linked to the binary vector pPK2 through XbaI and KpnI restriction enzyme digestion. Accordingly, HindIII and EcoRI were used to obtain a 1471‐bp phleomycin resistance cassette as a selectable marker gene from the p480 vector, and the cassette was linked to the same pPK2 as used above. The complement construct was re‐introduced into the ΔVdpf knockout mutant. The complementation mutants (ΔVdpf: Vdpf‐phleo), with a single copy of the Vdpf allele, were confirmed by PCR, and then purified by two rounds of single‐spore isolation in the presence of 100 μg/mL of phleomycin.
Assays for radial growth, microsclerotial formation, conidiation and germination
PDA and PDB media, each containing 50 μg/mL of HygB, were used to study the radial growth, microsclerotial formation and conidiation of the strains. Conidia were harvested from 10‐day‐old cultures, filtered through two layers of lens paper and resuspended to a concentration of 1 × 107 spores/mL in sterile water. Each flask and plate were inoculated with 5 μL of conidial suspension of ΔVdpf, IMΔVdpf, ΔVdpf:Vdpf‐phleo or the WT strain. The PDA plates were incubated at 25°C; subsequently, the colony diameter and colour of the strains were measured and observed every day. For observation of the microsclerotia, sterile cover slips (hydrophobic) were inserted into the PDA plate alongside the inoculation sites. Microsclerotium formation was observed for 20 days. The PDB flasks were incubated at 25°C with continuous agitation at 150 rpm and, on the 10th day, conidial production was calculated. For conidial germination, 102 conidia were inoculated into 1 mL of PDB medium, grown with continuous agitation at 150 rpm and observed for conidial germination at 6, 12, 18, 24, 36 and 48 h. At 24 h, the lengths of the germinated hyphae were measured. The conidia germination rate was calculated using a blood counting chamber at 12 h.
Measurement of the response to stressors
To test the sensitivity of the strains to osmotic stress and oxygen radicals, droplets (5 μL) of conidia suspensions of the WT and mutants were pipetted onto PDA containing 0.7 m KCl, 1.0 m sorbitol or 1.5 mm H2O2. The mutants and WT strains were also cultured at pH 4.0 or pH 12.0 to measure the effects of pH on their growth. Colony diameters were measured at 10 days post‐inoculation (dpi). All of the experiments were repeated three times.
Pathogenicity assays
To prepare the inocula, fungal cultures grown for 7 days in PDB medium were passed through two layers of lens paper to remove the mycelia, and conidial suspensions were adjusted to concentrations of approximately 1 × 107 conidia/mL. Pathogenicity assays of ΔVdpf, IMΔVdpf, the complemented strain and the WT strain were performed in 6‐week‐old intact cotton plants (Gossypium hirsutum L.) using root dip inoculation, as described previously (Fradin et al., 2009), and in 6‐week‐old intact tobacco plants (Nicotiana benthamiana) using soil inoculation. Plants of similar height and with similar‐sized leaves were selected for each treatment. For the root dip inoculation, 4 mL of conidia suspension were added to 200 mL of fresh Hoagland nutrient solution, and the plants were re‐cultivated in this solution for up to 20 dpi, when the disease symptoms appeared. For the non‐inoculation controls, the roots of five plants were immersed in sterile water for 20 min and then re‐cultured in Hoagland nutrient solution. For soil inoculation, the stems close to the soil were first scratched slightly using a sterile needle and then coated with cotton, which absorbed 4 mL of the conidia suspension.
To detect fungal colonization in the infected cotton roots, we used the procedure described by Koske and Gemma (1989), with modifications as follows. Fresh roots were washed with tap water and fixed with FAA (5 mL of formaldehyde, 5 mL of acetic acid and 90 mL of 50% ethanol) for at least 48 h. The roots were then washed several times in running tap water and cut into segments of approximately 1 cm. Subsequently, the roots were incubated in 10% (w/v) KOH for 1 h at 90°C, bleached with fresh alkaline H2O2 solution for 30 min, acidified in 5% glycerol solution for 8 min and stained with 5% Hero blue dark ink (containing 5 mL of blue dark ink and 95 mL of white vinegar) for 3 min at room temperature (approximately 20°C) (Vierheilig et al., 1998). After staining, the roots were rinsed with tap water and preserved in glycerol‐lacto at room temperature (c. 20°C) (Zangaro et al., 2013). The root fragments were examined under a compound‐light microscope (Zeiss Axioskop, Gerbershausen, Germany). The virulence assays on the cotton plants were performed twice.
To more accurately investigate the colonization abilities of the Vdpf mutant compared with the WT strain, the biomass of V. dahliae was estimated by PCR quantification in cotton plants infected with ΔVdpf, IMΔVdpf, ΔVdpf:Vdpf‐phleo and WT, according to Tzima et al. (2011) with slight modifications. DNA was extracted from roots inoculated with ΔVdpf, IMΔVdpf, ΔVdpf:Vdpf‐phleo and WT. In our study, for the detection of fungal strains in cotton plants, the primer pairs Vd‐F (5′‐ATCAGTCTCTCTGTTTATAC‐3′) and Vd‐R (5′‐TAACTACTACGCAAGGAAG‐3′), which were designed based on the internal transcribed spacer ITS1 and ITS2 regions of the 5.8S ribosomal RNA gene (Z29511) of V. dahliae, were used. The cotton osmotin gene, which was amplified using the primer pairs osmotin‐F (5′‐ATCAGTCTCTCTGTTTATAC‐3′) and osmotin‐R (5′‐TAACTACTACGCAAGGAAG‐3′), was used to normalize differences in DNA template amounts. The data were analysed using Quantity One software.
qRT‐PCR, RT‐PCR and gene expression analysis
For RT‐PCR and qRT‐PCR, total RNA was extracted from the VdPf deletion mutant ΔVdPf‐11, IMΔVdpf, ΔVdpf:Vdpf‐phleo and WT V. dahliae using RNAiso Plus reagent (Takara) according to the manufacturer's instructions. Two micrograms of total RNA were used for RT‐PCR with a mixture of oligo (dT) primers and PrimeScript RTase (Takara). Each cDNA was diluted to 100 ng/μL. Thirty cycles of RT‐PCR were run on a Bio‐Rad PTC0200 Peltier Thermal Cycler (Bio‐Rad, Hercules, CA, USA). The 18srRNA gene (Z29511), which contained the stable expression ITS1 and ITS2 regions of the ribosomal RNA genes of V. dahliae, was amplified as an internal control, and its expression values were analysed using a grey‐scale value analysis. For qRT‐PCR, PCR was performed using first‐strand cDNAs as template with iQ™ SYBR Green Supermix (Bio‐Rad). The constitutively expressed 18srRNA gene (Z29511) was used as an endogenous control, and reactions were performed in triplicate. qRT‐PCR was performed on a CFX96™ thermocycler (Bio‐Rad), and the PCR protocol was as follows: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 10 s and annealing temperature for 30 s. The annealing temperature for VdPKAC1, VMK1, VdNLP1, VdNLP2 and Vdgrp1 was 61.7°C, and those for VdSge1 and VdSNF1 were 63.5°C and 65°C, respectively. The housekeeping genes were amplified at an annealing temperature of 64.5°C. The resulting data were analysed with CFX Manager™ software. Normalized expression was calculated as 2–ΔCt using a threshold cycle (Ct), in which ΔCt = (Ctgene – Ct18srRNA). The relative normalized expression between the mutants and WT was determined using the comparative Ct method (2–ΔΔCt), in which ΔΔCt = (Ctgene – Ct18srRNA)mutant – (Ctgene – Ct18srRNA)WT. Each data point was the mean value of three experimental replicate determinations. The gene‐specific primers designed for qRT‐PCR and RT‐PCR are listed in Tables S5 and S6 (see Supporting Information), respectively. The experiment was conducted with three independent biological replicates.
Protein expression and purification
The vector used for the construction of the expression plasmid was pPIC9k, which was digested with EcoRI and NotI. The DNA sequence from the initiation codon to the Zn(II)2Cys6 binuclear cluster domain was cloned using the following primer pairs: F‐Vdpf1 (CCGGAATTCGATCGCATGGGCTTTCAGA) and R‐Vdpf1 (ATTTGCGGCCGCCTACTAATGATGATGATGATGATGTTTGGGAGGGGTACGCTT). The pPIC9k‐Vdpf recombinant plasmid was transformed into P. pastoris GS115, and the subsequent expression procedure was performed according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Pichia pastoris GS115 containing the unmodified plasmid pPIC9k served as a negative control. Protein expression was induced by the addition of 0.5% methanol (100% purity) every 24 h at 30°C for 72 h. The resulting His‐tagged Vdpf was purified on Ni2+ affinity columns using high‐affinity Ni‐charged resin (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. The elution was dialysed overnight and stored at −80°C. The concentration was determined using a BCA protein determination kit, and the purified protein was used for EMSA.
EMSA
DNA fragments (promoter) for the binding activity assays were amplified by PCR from V. dahliae genomic DNA (Table S6). The 2000‐bp upstream fragment of each gene was analysed using online software, such as Promoter 2.0 (Knudsen, 1999), PLACE (Higo et al., 1999; Prestridge, 1991) and Plantcare, to predict the promoter region. Potential TF binding sites were predicted using the online JASPAR database. DNA probes were amplified by PCR using PrimeSTAR Max DNA polymerase (Takara) and their accuracies were ensured by sequencing. EMSA was performed as described previously (Hellman and Fried, 2007). The binding of Vdpf (EGY18187.1) to the DNA probes was performed using 10 μL of a reaction mix containing 1 ng of DNA probes and increasing concentrations of the protein (as indicated in the legend of the corresponding figure). Nuclease‐free water, EMSA buffer [100 mm Tris‐HCl, pH 8.0, 100 mm NaCl, 1 mm dithiothreitol (DTT) and 10% glycerol] and Vdpf were added first. Then, after 10 min of incubation at 30°C, the DNA probes were added, and the samples were incubated at 30°C for another 20 min. The binding system without Vdpf served as a negative control. The reaction mixtures were then subjected to 5% native polyacrylamide gel electrophoresis (PAGE) at 80 V for 2 h in 0.5 × TBE (0.045 mo1/L Tris‐H3BO3, 0.001 mo1/L EDTA). Images of the gels were acquired using Bio‐Rad ChemiDoc MP (Bio‐Rad).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Representative phenotypes of T‐DNA insertional mutants of Verticillium dahliae based on colony growth rate, melanized development, microsclerotial development and pigment alteration on potato dextrose agar (PDA) medium.
Fig. S2 Structure and phylogenetic analysis of fungal‐specific transcription factor (TF) domain‐containing proteins in Verticillium dahliae. Based on protein sequences, the phylogenetic tree was constructed using MEGA5.0 by the neighbour‐joining method and the conservative domains were analysed using ClustalX. One thousand bootstrap replicates were used.
Fig. S3 Structure and phylogenetic analysis of Vdpf (EGY18187) in fungi. (A) Based on the protein sequences of Vdpf (EGY18187), the conservative domains, the Zn(II)2Cys6 binuclear cluster domain and the fungal‐specific transcription factor (TF) domain were analysed using ClustalX. (B) The phylogenetic tree was constructed using MEGA6.0 by the neighbour‐joining method. One thousand bootstrap replicates were used. The sequences were collected from the following organisms: FG, Fusarium graminearum; FO, Fusarium oxysporum; AN, Aspergillus nidulans; AO, Aspergillus oryzae; Afu, Aspergillus fumigatus; AT, Aspergillus terreus; AC, Aspergillus clavatus; AF, Aspergillus flavus; MG, Magnaporthe grisea; SS, Sclerotinia sclerotiorum; UM, Ustilago maydis; SC, Sclerotinia cerevisiae; VD, Verticillim dahliae. The protein IDs are labelled with various colours for better classification.
Fig. S4 The procedure of conidia germination of the wild‐type (WT) at 6, 12, 18, 24, 36 and 48 h.
Fig. S5 Growth comparisons between the mutants and the wild‐type (WT) under pH 4.0/pH 12.0, or in the presence of 0.7 m KCl, 1.0 m sorbitol and 1.5 mm H2O2. Three biological replicates (n = 3) were used for this study. PDA, potato dextrose agar.
Fig. S6 Predication of Vdpf binding sites. (A) Sequence logo of high‐potential binding motifs. (B) Array of potential binding motifs on the promoter regions of genes that could bind with Vdpf (VDAG_08521.1).
Fig. S7 Schematic diagram of the polymerase chain reaction (PCR) strategy used to make each construct. (A) Construct for replacement of the Vdpf (VDAG_08521.1) gene with a hygromycin B transferase (HygB) resistance cassette. (B) Amplification of the wild‐type (WT) allele of the Vdpf (VDAG_08521.1) gene for the construction of complemented strains.
Table S1 Sequence analysis of T‐DNA flanking regions of Verticillium dahliae transformants using the Verticillium group genome database
Table S2 Disease index on individual cotton plants following inoculation with wild‐type Verticillium dahliae ΔVdpf mutants, IMΔVdpf mutants and complemented strains. Plants inoculated with sterile water were used as control.
Table S3 Primers used in high‐efficiency thermal asymmetric interlaced polymerase chain reaction (hiTAIL‐PCR)
Table S4 Primers used for the construction of the replacement cassette
Table S5 Primers used for quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
Table S6 Primers used for quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of melanin biosynthesis‐related genes
Acknowledgements
This work was supported by the National Basic Research Program of China (No. 2014CB138701), the National Natural Science Foundation of China (No. 40103112) and the Natural Science Foundation Project of Chongqing Science Technology Commission (CQ CSTC) (No. 2009BB1123).
Contributor Information
Xingyong Yang, Email: yangxy94@swu.edu.cn.
Jinyan Dong, Email: donjyaa@swu.edu.cn.
References
- Badis, G. , Chan, E.T. , van Bakel, H. , Pena‐Castillo, L. , Tillo, D. , Tsui, K. , Carlson, C.D. , Gossett, A.J. , Hasinoff, M.J. , Warren, C.L. , Gebbia, M. , Talukder, S. , Yang, A. , Mnaimneh, S. , Terterov, D. , Coburn, D. , Li Yeo, A. , Yeo, Z.X. , Clarke, N.D. , Lieb, J.D. , Ansari, A.Z. , Nislow, C. and Hughes, T.R. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Molecular Cell, 32, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A. (2001) Stem and root diseases, Verticillium wilt In: Compendium of Cotton Disease (Kirkpatrick T.L. and Rothrock C.S., eds), pp. 28–31. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Bell, A.A. and Wheeler, M.H. (1986) Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451. [Google Scholar]
- Bell, A.A. , Puhalla, J.E. , Tolmsoff, W.J. and Stipanovic, R.D. (1976) Use of mutants to establish (+)‐scytalone as an intermediate in melanin biosynthesis by Verticillium dahliae . Can. J. Microbiol. 22, 787–799. [DOI] [PubMed] [Google Scholar]
- Bhat, R.G. , Smith, R.F. , Koike, S.T. , Wu, B.M. and Subbarao, K.V. (2003) Characterization of Verticillium dahliae isolates and wilt epidemics of pepper. Plant Dis. 87, 789–797. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal. Curr. Sci. 89, 1113–1121. [Google Scholar]
- Bolton, M.D. , Thomma, B. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Brown, M. and Wyllie, T. (1970) Ultrastructure of microsclerotia of Verticillium albo‐atrum . Phytopathology, 60, 538–542. [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell Online, 12, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P.‐K. and Ehrlich, K.C. (2013) Genome‐wide analysis of the Zn(II)(2)Cys(6) zinc cluster‐encoding gene family in Aspergillus flavus . Appl. Microbiol. Biotechnol. 97, 4289–4300. [DOI] [PubMed] [Google Scholar]
- Cho, Y. , Cramer, R.A. Jr. , Kim, K.‐H. , Davis, J. , Mitchell, T.K. , Figuli, P. , Pryor, B.M. , Lemasters, E. and Lawrence, C.B. (2007) The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of genes encoding hydrolytic enzymes in Alternaria brassicicola . Fungal Genet. Biol. 44, 543–553. [DOI] [PubMed] [Google Scholar]
- Cho, Y. , Srivastava, A. , Ohm, R.A. , Lawrence, C.B. , Wang, K.‐H. , Grigoriev, I.V. and Marahatta, S.P. (2012) Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola . PLoS Pathog. 8, 634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Ohm, R.A. , Grigoriev, I.V. and Srivastava, A. (2013) Fungal‐specific transcription factor AbPf2 activates pathogenicity in Alternaria brassicicola . Plant J. 75, 498–514. [DOI] [PubMed] [Google Scholar]
- Chung, H. , Choi, J. , Park, S.‐Y. , Jeon, J. and Lee, Y.‐H. (2013) Two conidiation‐related Zn(II)(2)Cys(6) transcription factor genes in the rice blast fungus. Fungal Genet. Biol. 61, 133–141. [DOI] [PubMed] [Google Scholar]
- Downes, D.J. , Dayis, M.A. , Kreutzberger, S.D. , Taig, B.L. and Todd, R.B. (2013) Regulation of the NADP‐glutamate dehydrogenase gene gdhA in Aspergillus nidulans by the Zn(II)2Cys6 transcription factor LeuB. Microbiology, 159, 2467–2480. [DOI] [PubMed] [Google Scholar]
- Downes, D.J. , Davis, M.A. , Wong, K.H. , Kreutzberger, S.D. , Hynes, M.J. and Todd, R.B. (2014) Dual DNA binding and coactivator functions of Aspergillus nidulans TamA, a Zn(II)2Cys6 transcription factor. Mol. Microbiol. 92, 1198–1211. [DOI] [PubMed] [Google Scholar]
- Eliahu, N. , Igbaria, A. , Rose, M.S. , Horwitz, B.A. and Lev, S. (2007) Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen‐activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1 . Eukaryot. Cell, 6, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, A. , van Kan, J.A.L. and Plummer, K.M. (2004) Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 41, 963–971. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. and Thomma, B.P. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. , Zhang, Z. , Ayala, J.C.J. , Castroverde, C.D.M. , Nazar, R.N. , Robb, J. , Liu, C.M. and Thomma, B.P. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1 . Plant Physiol. 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Zhou, B.J. , Li, G.Y. , Jia, P.S. , Li, H. , Zhao, Y.L. , Xia, G.X. and Guo, H.S. (2010) A glutamic acid‐rich protein identified in V. dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One, 5, e15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, G. and Wheeler, M. (1986) HPLC analysis of fungal melanin intermediates and related metabolites. J. Liq. Chromatogr. 9, 971–981. [Google Scholar]
- de Groot, M.J.A. , Bundock, P. , Hooykaas, P.J.J. and Beijersbergen, A.G.M. (1998) Agrobacterium tumefaciens‐mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Chen, Y. , Du, Y. , Dong, Y. , Guo, W. , Zhai, S. , Zhang, H. , Dong, S. , Zhang, Z. , Wang, Y. , Wang, P. and Zheng, X. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathog. 7, e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke, M.A. and Lazarovits, G. (1995) The role of melanin in the survival of microsclerotia of Verticillium dahliae . Phytoparasitica, 23, 54. [Google Scholar]
- Hellman, L.M. and Fried, M.G. (2007) Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2, 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppenau, C.E. , Tran, V.‐T. , Kusch, H. , Aßhauer, K.P. , Landesfeind, M. , Meinicke, P. , Popovac, B. , Braus‐Stromeyera, S.A. and Braus, G.H. (2014) Verticillium dahliae VdTHI4, involved in thiazole biosynthesis, stress response and DNA repair functions, is required for vascular disease induction in tomato. Environ. Exp. Bot. 108, 14–22. [Google Scholar]
- Horton, P. , Park, K.‐J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman, O. (1982) Interrelations of root growth dynamics to epidemiology of root‐invading fungi. Annu. Rev. Phytopathol. 20, 303–327. [Google Scholar]
- Kapoor, K. , Rehan, M. , Kaushiki, A. , Pasrija, R. , Lynn, A.M. and Prasad, R. (2009) Rational mutational analysis of a multidrug MFS transporter CaMdr1p of Candida albicans by employing a membrane environment based computational approach. PLoS Comput. Biol. 5, e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, J. , Moriwaki, A. , Tanaka, N. , Tanaka, C. , Ueno, M. and Arase, S. (2008) Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae . FEMS Microbiol Lett. 281, 221–227. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Park, S.‐Y. , Kim, K.S. , Rho, H.‐S. , Chi, M.‐H. , Choi, J. , Park, J. , Kong, S. , Park, J. , Goh, J. and Lee, Y.H. (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae . PLoS Genet. 5, e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimes, A. and Dobinson, K. (2003) Functional characterization of a V. dahliae hydrophobin gene homologue. Can. J. Plant Pathol. 25, 321. [Google Scholar]
- Klimes, A. and Dobinson, K.F. (2006) A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae . Fungal Genet. Biol. 43, 283–294. [DOI] [PubMed] [Google Scholar]
- Klimes, A. , Amyotte, S.G. , Grant, S. , Kang, S. and Dobinson, K.F. (2008) Microsclerotia development in Verticillium dahliae: regulation and differential expression of the hydrophobin gene VDH1 . Fungal Genet. Biol. 45, 1525–1532. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Subbarao, K.V. , Kang, S. , Veronese, P. , Gold, S.E. , Thomma, B.P. , Chen, Z. , Henrissat, B. , Lee, Y.H. , Park, J. , Garcia‐Pedrajas, M.D. , Barbara, D.J. , Anchieta, A. , de Jonge, R. , Santhanam, P. , Maruthachalam, K. , Atallah, Z. , Amyotte, S.G. , Paz, Z. , Inderbitzin, P. , Hayes, R.J. and Heiman, D.I. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, S. (1999) Promoter 2.0: for the recognition of PolII promoter sequences. Bioinformatics, 15, 356–361. [DOI] [PubMed] [Google Scholar]
- Koske, R. and Gemma, J. (1989) A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 92, 486–488. [Google Scholar]
- Kuo, M.‐J. and Alexander, M. (1967) Inhibition of the lysis of fungi by melanins. J. Bacteriol. 94, 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐H. , Yang, S.L. and Chung, K.‐R. (2009) The YAP1 homolog‐mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant–Microbe Interact. 22, 942–952. [DOI] [PubMed] [Google Scholar]
- Liu, S.‐Y. , Chen, J.‐Y. , Wang, J.‐L. , Li, L. , Xiao, H.‐L. , Adam, S.M. and Dai, X.F. (2013) Molecular characterization and functional analysis of a specific secreted protein from highly virulent defoliating Verticillium dahliae . Gene, 529, 307–316. [DOI] [PubMed] [Google Scholar]
- Liu, Y.‐G. and Chen, Y. (2007) High‐efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 43, 649–656. [DOI] [PubMed] [Google Scholar]
- Lockwood, J. (1960) Lysis of mycelium of plant‐pathogenic fungi by natural soil. Phytopathology, 50, 787–789. [Google Scholar]
- Luo, X.M. , Xie, C.J. , Dong, J.Y. , Yang, X.Y. and Sui, A.P. (2014) Interactions between Verticillium dahliae and its host: vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 98, 6921–6932. [DOI] [PubMed] [Google Scholar]
- MacPherson, S. , Larochelle, M. and Turcotte, B. (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthachalam, K. , Klosterman, S.J. , Kang, S. , Hayes, R.J. and Subbarao, K.V. (2011) Identification of pathogenicity‐related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens‐mediated T‐DNA insertional mutagenesis. Mol. Biotechnol. 49, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masloff, S. , Pöggeler, S. and Kück, U. (1999) The pro1 gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics, 152, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masloff, S. , Jacobsen, S. , Pöggeler, S. and Kück, U. (2002) Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 36, 107–116. [DOI] [PubMed] [Google Scholar]
- Meshi, T. and Iwabuchi, M. (1995) Plant transcription factors. Plant Cell Physiol. 36, 1405–1420. [PubMed] [Google Scholar]
- Molina, L. and Kahmann, R. (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell Online, 19, 2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki, A. , Kihara, J. , Kobayashi, T. , Tokunaga, T. , Arase, S. and Honda, Y. (2004) Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae . FEMS Microbiol. Lett. 238, 1–8. [DOI] [PubMed] [Google Scholar]
- Mullins, E.D. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D.M. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Nosanchuk, J.D. and Casadevall, A. (2003) The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5, 203–223. [DOI] [PubMed] [Google Scholar]
- Novo, M. , Pomar, F. , Gayoso, C. and Merino, F. (2006) Cellulase activity in isolates of V. dahliae differing in aggressiveness. Plant Dis. 90, 155–160. [DOI] [PubMed] [Google Scholar]
- Ogawa, M. , Kobayashi, T. and Koyama, Y. (2012) ManR, a novel Zn(II)(2)Cys(6) transcriptional activator, controls the beta‐mannan utilization system in Aspergillus oryzae . Fungal Genet. Biol. 49, 987–995. [DOI] [PubMed] [Google Scholar]
- Pabo, C.O. and Sauer, R.T. (1992) Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- Potgieter, H. and Alexander, M. (1966) Susceptibility and resistance of several fungi to microbial lysis. J. Bacteriol. 91, 1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, R. and Kapoor, K. (2004) Multidrug resistance in yeast Candida . Int. Rev. Cytol. 242, 215–248. [DOI] [PubMed] [Google Scholar]
- Prestridge, D.S. (1991) SCAN, P. S. a computer program that scans DNA sequences for eukaryotic transcription elements. CABIOS, 7, 203–206. [DOI] [PubMed] [Google Scholar]
- Rauyaree, P. , Ospina‐Giraldo, M.D. , Kang, S. , Bhat, R.G. , Subbarao, K.V. , Grant, S.J. and Dobinson, K.F. (2005) Mutations in VMK1, a mitogen‐activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae . Curr. Genet. 48, 109–116. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. and Thomma, B.P. (2013) Verticillium dahliae Sge1 differentially regulates expression of candidate effector genes. Mol. Plant–Microbe Interact. 26, 249–256. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , van Esse, H.P. , Albert, I. , Faino, L. , Nurnberger, T. and Thomma, B. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Schnatho, W.C. and Mathre, D.E. (1966) Host range and differentiation of a severe form of Verticillium albo‐atrum in cotton. Phytopathology, 56, 1155–1161. [Google Scholar]
- Steinberg, C.E. (2012) Stress Ecology: Environmental Stress as Ecological Driving Force and Key Player in Evolution. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- Stewart, C.N. and Via, L.E. (1993) A rapid CTAB DNA isolation technique useful for rapid fingerprinting and other PCR applications. Biotechniques, 14, 748–750. [PubMed] [Google Scholar]
- Stipanovic, R.D. and Bell, A.A. (1976) Pentaketide metabolites of Verticillium dahliae 3. Identification of (–)−3,4‐dihydro‐3,8‐dihydroxy‐1(2H)‐naphthalenone (–)‐vermelone as a precursor to melanin. J. Org. Chem. 41, 2468–2469. [DOI] [PubMed] [Google Scholar]
- Stipanovic, R.D. and Bell, A.A. (1977) Pentaketide metabolites of Verticillium dahliae 2. Accumulation of naphthol derivatives by aberrant‐melanin mutant Brm‐2. Mycologia, 69, 164–172. [PubMed] [Google Scholar]
- Szewczyk, E. , Nayak, T. , Oakley, C.E. , Edgerton, H. , Xiong, Y. , Taheri‐Talesh, N. , Osmani, S.A. and Oakley, B.R. (2006) Fusion PCR and gene targeting in Aspergillus nidulans . Nat. Protoc. 1, 3111–3120. [DOI] [PubMed] [Google Scholar]
- Tanaka, C. , Kubo, Y. and Tsuda, M. (1991) Genetic‐analysis and characterization of Cochliobolus heterostrophus color mutants. Mycol. Res. 95, 49–56. [Google Scholar]
- Ten, L.N. , Tyshtshenko, A.A. , Stepanichenko, N.N. , Gusakova, S.D. , Muhamedjanov, S.Z. , Otroshtshenko, O.S. and Kas'yanenko, A.G. (1977) Metabolites of pathogenic fungus Verticillium dahliae. 6. Pentaketide metabolites and neutral lipids of virulent and avirulent strains. Khim. Prir. Soedin. 5, 632–635. [Google Scholar]
- Thomma, B. (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. [DOI] [PubMed] [Google Scholar]
- Thompson, J.E. , Fahnestock, S. , Farrall, L. , Liao, D.‐I. , Valent, B. and Jordan, D.B. (2000) The second naphthol reductase of fungal melanin biosynthesis in Magnaporthe grisea . J. Biol. Chem. 275, 34 867–34 872. [DOI] [PubMed] [Google Scholar]
- Timpner, C. , Braus‐Stromeyer, S.A. , Tran, V.T. and Braus, G.H. (2013) The Cpc1 regulator of the cross‐pathway control of amino acid biosynthesis is required for pathogenicity of the vascular pathogen Verticillium longisporum . Mol. Plant–Microbe Interact. 26, 1312–1324. [DOI] [PubMed] [Google Scholar]
- Tokousbalides, M.C. and Sisler, H.D. (1979) Site of inhibition by tricyclazole in the melanin biosynthetic‐pathway of Verticillium dahliae . Pestic. Biochem. Physiol. 11, 64–73. [Google Scholar]
- Tran, V.T. , Braus‐Stromeyer, S.A. , Kusch, H. , Reusche, M. , Kaever, A. , Kühn, A. , Valerius, O. , Landesfeind, M. , Aßhauer, K. , Tech, M. , Hoff, K. , Pena‐Centeno, T. , Stanke, M. , Lipka, V. , Braus, G.H. (2014) Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol. 202, 565–581. [DOI] [PubMed] [Google Scholar]
- Tsai, H.‐F. , Wheeler, M.H. , Chang, Y.C. and Kwon‐Chung, K. (1999) A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus . J. Bacteriol. 181, 6469–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima, A. , Paplomatas, E.J. , Rauyaree, P. and Kang, S. (2010) Roles of the catalytic subunit of cAMP‐dependent protein kinase A in virulence and development of the soilborne plant pathogen Verticillium dahliae . Fungal Genet. Biol. 47, 406–415. [DOI] [PubMed] [Google Scholar]
- Tzima, A.K. , Paplomatas, E.J. , Rauyaree, P. , Ospina‐Giraldo, M.D. and Kang, S. (2011) VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell‐wall degradation. Mol. Plant–Microbe Interact. 24, 129–142. [DOI] [PubMed] [Google Scholar]
- Valášková, V. and Baldrian, P. (2006) Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus—production of extracellular enzymes and characterization of the major cellulases. Microbiology, 152, 3613–3622. [DOI] [PubMed] [Google Scholar]
- Vierheilig, H. , Coughlan, A.P. , Wyss, U. and Piché, Y. (1998) Ink and vinegar, a simple staining technique for arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Gao, F. , Huang, J.F. (2013) Cloning and sequence analysis of polyketide synthase gene from Verticillium dahlae of cotton. Xinjiang Agric. Sci. 50, 1724–1729. [Google Scholar]
- Wheeler, M. , Tolmsoff, W. , Bell, A. and Mollenhauer, H. (1978) Ultrastructural and chemical distinction of melanins formed by Verticillium dahliae from (+)‐scytalone, 1,8‐dihydroxynaphthalene, catechol, and L‐3,4‐dihydroxyphenylalanine. Can. J. Microbiol. 24, 289–297. [DOI] [PubMed] [Google Scholar]
- Wilhelm, S. (1955) Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology, 45, 180–181. [Google Scholar]
- Zangaro, W. , Rostirola, L.V. , de Souza, P.B. , de Almeida Alves, R. , Lescano, L.E. , Rondina, A.B. , Nogueira, M.A. and Carrenho, R. (2013) Root colonization and spore abundance of arbuscular mycorrhizal fungi in distinct successional stages from an Atlantic rainforest biome in southern Brazil. Mycorrhiza, 23, 221–233. [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Xu, B. , Wang, Y. , Li, Y. , Qian, Z. , Tang, S. , Huan, S. and Ren, S. (2008) Agrobacterium tumefaciens‐mediated transformation as a tool for insertional mutagenesis in the fungus Penicillium marneffei . Mycol. Res. 112, 943–949. [DOI] [PubMed] [Google Scholar]
- Zhou, B.J. , Jia, P.S. , Gao, F. and Guo, H.S. (2012) Molecular characterization and functional analysis of a necrosis‐ and ethylene‐inducing, protein‐encoding gene family from V. dahliae . Mol. Plant–Microbe Interact. 25, 964–975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Representative phenotypes of T‐DNA insertional mutants of Verticillium dahliae based on colony growth rate, melanized development, microsclerotial development and pigment alteration on potato dextrose agar (PDA) medium.
Fig. S2 Structure and phylogenetic analysis of fungal‐specific transcription factor (TF) domain‐containing proteins in Verticillium dahliae. Based on protein sequences, the phylogenetic tree was constructed using MEGA5.0 by the neighbour‐joining method and the conservative domains were analysed using ClustalX. One thousand bootstrap replicates were used.
Fig. S3 Structure and phylogenetic analysis of Vdpf (EGY18187) in fungi. (A) Based on the protein sequences of Vdpf (EGY18187), the conservative domains, the Zn(II)2Cys6 binuclear cluster domain and the fungal‐specific transcription factor (TF) domain were analysed using ClustalX. (B) The phylogenetic tree was constructed using MEGA6.0 by the neighbour‐joining method. One thousand bootstrap replicates were used. The sequences were collected from the following organisms: FG, Fusarium graminearum; FO, Fusarium oxysporum; AN, Aspergillus nidulans; AO, Aspergillus oryzae; Afu, Aspergillus fumigatus; AT, Aspergillus terreus; AC, Aspergillus clavatus; AF, Aspergillus flavus; MG, Magnaporthe grisea; SS, Sclerotinia sclerotiorum; UM, Ustilago maydis; SC, Sclerotinia cerevisiae; VD, Verticillim dahliae. The protein IDs are labelled with various colours for better classification.
Fig. S4 The procedure of conidia germination of the wild‐type (WT) at 6, 12, 18, 24, 36 and 48 h.
Fig. S5 Growth comparisons between the mutants and the wild‐type (WT) under pH 4.0/pH 12.0, or in the presence of 0.7 m KCl, 1.0 m sorbitol and 1.5 mm H2O2. Three biological replicates (n = 3) were used for this study. PDA, potato dextrose agar.
Fig. S6 Predication of Vdpf binding sites. (A) Sequence logo of high‐potential binding motifs. (B) Array of potential binding motifs on the promoter regions of genes that could bind with Vdpf (VDAG_08521.1).
Fig. S7 Schematic diagram of the polymerase chain reaction (PCR) strategy used to make each construct. (A) Construct for replacement of the Vdpf (VDAG_08521.1) gene with a hygromycin B transferase (HygB) resistance cassette. (B) Amplification of the wild‐type (WT) allele of the Vdpf (VDAG_08521.1) gene for the construction of complemented strains.
Table S1 Sequence analysis of T‐DNA flanking regions of Verticillium dahliae transformants using the Verticillium group genome database
Table S2 Disease index on individual cotton plants following inoculation with wild‐type Verticillium dahliae ΔVdpf mutants, IMΔVdpf mutants and complemented strains. Plants inoculated with sterile water were used as control.
Table S3 Primers used in high‐efficiency thermal asymmetric interlaced polymerase chain reaction (hiTAIL‐PCR)
Table S4 Primers used for the construction of the replacement cassette
Table S5 Primers used for quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
Table S6 Primers used for quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of melanin biosynthesis‐related genes


