Summary
Cucumber mosaic virus (CMV) has the broadest host range among plant viruses, causing enormous losses in agriculture. In melon, strains of subgroup II are unable to establish a systemic infection in the near‐isogenic line SC12‐1‐99, which carries the recessive resistance gene cmv1 from the accession PI 161375, cultivar ‘Songwhan Charmi’. Strains of subgroup I overcome cmv1 resistance in a manner dependent on the movement protein. We characterized the resistance conferred by cmv1 and established that CMV‐LS (subgroup II) can move from cell to cell up to the veins in the inoculated leaf, but cannot enter the phloem. Immunogold labelling at transmission electron microscopy level showed that CMV‐LS remains restricted to the bundle sheath (BS) cells in the resistant line, and does not invade vascular parenchyma or intermediary cells, whereas, in the susceptible line ‘Piel de Sapo’ (PS), the virus invades all vein cell types. These observations indicate that the resistant allele of cmv1 restricts systemic infection in a virus strain‐ and cell type‐specific manner by acting as an important gatekeeper for virus progression from BS cells to phloem cells. Graft inoculation experiments showed that CMV‐LS cannot move from the infected PS stock into the resistant cmv1 scion, thus suggesting an additional role for cmv1 related to CMV transport within or exit from the phloem. The characterization of this new form of recessive resistance, based on a restriction of virus systemic movement, opens up the possibility to design alternative approaches for breeding strategies in melon.
Keywords: cmv1, Cucumber mosaic virus, host factor, phloem entry, recessive resistance, systemic infection, virus movement
Introduction
More than 30 million tonnes of melon are produced each year worldwide (Faostat, 2013; http://faostat3.fao.org). However, melon production is often seriously affected by diseases produced by plant viruses. Plant viruses are obligate intracellular parasites that exploit the host cellular machinery to establish systemic infections. Following entry into a host cell, viruses move from cell to cell to reach the phloem and spread to distant organs to establish a systemic infection (Hipper et al., 2013). In each cell, viruses hijack the cellular machinery to translate and replicate their genomes. Virus movement depends on one or more virus‐encoded movement proteins (MPs) and often also on other virus‐encoded auxiliary proteins that facilitate virus spread by interaction with host factors (Heinlein, 2015). These host factors may be involved in intracellular transport to plasmodesmata (PDs) or specifically associated with PDs. The cytoskeleton and associated motor proteins play a crucial role in virus movement, in supporting the formation of viral replication complexes (VRCs) and in their movement along the host endomembrane system and across the PD channel (Amari et al., 2014; Harries and Ding, 2011; Harries et al., 2010; Heinlein, 2015; Lazarowitz and Beachy, 1999; Lucas, 2006). Phloem entry requires the ability of the viruses to successfully move through various cell types: from the mesophyll cells through bundle sheath (BS) cells and vascular parenchyma (VP) cells to companion cells (CCs) and, finally, into the phloem sieve elements (SEs) (Hipper et al., 2013). A block in any of these steps prevents systemic movement by restricting the virus to the inoculated leaf. Such a defect in movement may be caused by a lack of an essential host factor, by the inability of virus‐encoded proteins to properly interact with such host‐encoded factors (recessive resistance) or by a specific host resistance response (dominant resistance). As a result of their lack of transcription/translation capacity, SEs are dependent on their neighbouring CCs for maintenance and function, which establishes an obligatory association known as the SE–CC complex (Lough and Lucas, 2006). Virus entry into the SE–CC complex represents a significant barrier to long‐distance virus movement (Ding, 1998; Santa Cruz, 1999; Wang et al., 1998; Wintermantel et al., 1997). This barrier might be determined by the number of PDs that connect the SE–CC complex to the surrounding cells, which relates to the way in which the host plant loads photoassimilates into the phloem. Apoplastic loaders have CCs with only few PD connections to the BS, whereas symplastic loaders, such as cucurbits, have specialized CCs, named intermediary cells (ICs), with numerous PDs in the cell walls shared with BS cells, VP cells or SEs (van Bel et al., 1992; Schmitz et al., 1987; Turgeon et al., 1975). Viruses generally enter the systemic pathway through the SEs of minor veins that anastomose throughout the leaf blade. In Cucumis melo, the vascular bundle of minor veins contains xylem vessels, VP cells, ICs and SEs, surrounded by a layer of specialized mesophyll cells (BS cells). VP cells are characterized by a large vacuole, whereas ICs are rather large cells with small vacuoles and a cytoplasm rich in ribosomes and mitochondria. All of these cell types and the surrounding BS cells are densely connected by PDs. PDs between ICs and SEs, called plasmodesmata pore units (PPUs), are branched on the IC side and thus have a funnel‐like appearance (Schmitz et al., 1987).
Recessive resistance is the most common type of resistance against plant viruses (Kang et al., 2005). Most of the corresponding genes cloned from model and crop species encode translation initiation factors. They affect the accumulation of the virus mainly at the single cell level, although effects on cell‐to‐cell movement have also been reported (for a review, see Truniger and Aranda, 2009). Only few recessive resistances are governed by genes encoding other types of proteins and they all restrict virus multiplication, not systemic virus transport (Amano et al., 2013; Ouibrahim et al., 2014; Yang et al., 2014; Yoshii et al., 2009).
Cucumber mosaic virus (CMV) has the largest host range among plant viruses, being able to infect more than 1200 species of the most economically important families, such as Solanaceae and Cucurbitaceae, and causing severe loses (Edwardson and Christie, 1991). Recessive resistances to CMV are mostly related to the inhibition of cell‐to‐cell or systemic virus movement, or both (Canto and Palukaitis, 2001; Caranta et al., 2002; Dufour et al., 1989; Kang et al., 2005; Kobori et al., 2000; Ohnishi et al., 2011; Sekine et al., 2004; Stamova and Chatelat, 2000; Valkonen and Watanabe, 1999; Yoshii et al., 2004). The genetic inheritance of this trait is usually polygenic, and several quantitative trait loci (QTLs) conferring resistance to CMV have already been described in different systems (Caranta et al., 2002; Chaim et al., 2001; Guiu‐Aragonés et al., 2014; Ohnishi et al., 2011; Palukaitis and Garcia‐Arenal, 2003; Pitrat, 2002). In melon, several QTLs conferring resistance to some strains of CMV have been identified (Guiu‐Aragonés et al., 2014; Pitrat, 2002). In the melon accession PI 161375, cultivar ‘Songwhan Charmi’ (SC), the gene cmv1 confers recessive resistance to CMV strains from subgroup II, but not to strains of subgroup I (Essafi et al., 2009; Guiu‐Aragonés et al., 2015). Resistance to CMV strains of subgroup I requires two additional QTLs (Guiu‐Aragonés et al., 2014). The avirulence determinant of subgroup II viruses has been mapped to the MP gene. The MP is the only viral factor required for resistance, as a chimeric CMV‐LS (subgroup II) derivative carrying the MP of CMV‐FNY (subgroup I) in place of its own MP became virulent in the resistant melon line. Moreover, we have demonstrated previously that the restriction of CMV‐LS systemic infection in SC does not occur at the level of viral replication/translation (Guiu‐Aragonés et al., 2015). Thus, it appears likely that cmv1‐mediated resistance affects virus movement.
Here, we used the near‐isogenic line (NIL) SC12‐1‐99, carrying the cmv1 gene in the background of the susceptible melon accession ‘Piel de Sapo’ (PS) (Essafi et al., 2009), to further characterize the resistance mediated by this gene. We followed the progress of CMV‐LS infection by immunolabelling and grafting analyses, and showed that resistant plants support cell‐to‐cell movement in inoculated leaves, but prevent virus movement from BS cells into VP/IC–SE cells, thus blocking virus entry into the phloem.
Results
CMV‐LS accumulates and moves in the inoculated leaf
We have shown previously that the melon line SC12‐1‐99 exhibits a strain‐specific restriction of systemic accumulation of CMV. In SC12‐1‐99, both CMV‐LS and CMV‐FNY strains were able to accumulate in the inoculated cotyledons, but only CMV‐FNY was able to establish a systemic infection (Guiu‐Aragonés et al., 2015). To investigate whether the resistance conferred by cmv1 against CMV‐LS affects local spread of the virus, we analysed the distribution of CMV‐LS and CMV‐FNY at 3 days post‐inoculation (dpi) in inoculated leaves by tissue printing, followed by hybridization of viral RNA with a virus‐specific probe (Guiu‐Aragonés et al., 2015). As shown in Fig. 1, CMV‐LS and CMV‐FNY showed a similar distribution and localization of viral RNA along and between the veins of the leaves, and no significant difference in the distribution of the respective viral RNAs could be detected between the susceptible PS and resistant SC12‐1‐99 lines. As expected, no signal was detected in the leaves of non‐inoculated control plants. Therefore, both viruses seem to spread through the inoculated leaf and reach the veins in the resistant and susceptible melon lines. This confirms previous observations, indicating that the resistance mediated by cmv1 does not act at the level of virus accumulation (Guiu‐Aragonés et al., 2015) and also that the resistance does not act on mesophyll cell‐to‐cell movement. Interestingly, both strains were also localized along the veins of the resistant parental line SC, which carries at least three genes involved in resistance to CMV (Guiu‐Aragonés et al., 2014). Thus, although providing resistance to both viral strains, these additional genes present in SC do not prevent viral replication or cell‐to‐cell movement in the inoculated leaf.
Figure 1.
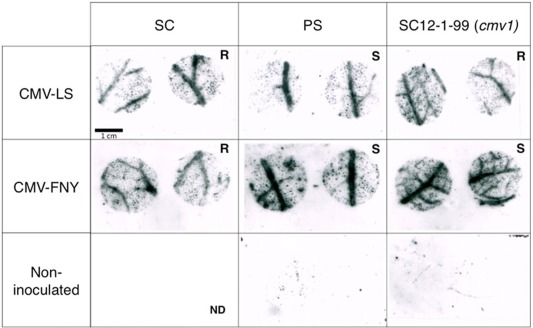
CMV‐LS and CMV‐FNY accumulate in inoculated leaves of the resistant and susceptible melon lines. The first true leaves of the Piel de Sapo (PS), PI 161375 (Songwhan Charmi, SC) and SC12‐1‐99 lines were inoculated with CMV‐LS or CMV‐FNY. Viral RNAs within tissue prints of leaf disc samples (3 days post‐inoculation, dpi) were hybridized with virus‐specific probe. The virus strains accumulate throughout the inoculated leaves irrespective of the presence of the resistance gene(s) in SC and SC12‐1‐99. ND, no data; R, resistant to systemic infection; S, susceptible to systemic infection. CMV, Cucumber mosaic virus.
CMV‐LS cannot be detected in the phloem of the resistant line
The lack of CMV‐LS systemic movement in cmv1 plants might be caused by the inability of the virus to enter the phloem SEs in the inoculated leaves or to exit the phloem in the non‐inoculated leaves. To determine whether or not CMV‐LS can enter the phloem of the resistant SC12‐1‐99 line, plant stems and petioles were cut at 12 dpi and cross‐sections were blotted onto a membrane for virus detection. Figure 2a shows the distribution of CMV in the different sections of the plant. In the susceptible PS line, CMV‐LS developed a systemic infection and, consequently, the virus was detected in all the tissue sections, whereas, in the resistant line SC12‐1‐99, CMV‐LS could not be detected in any of the sections, including the section taken from the petiole of the inoculated leaf. This indicates that the virus is unable to exit from the inoculated leaf and suggests that CMV‐LS might not enter the phloem. In contrast, CMV‐FNY, which infects systemically both melon lines, was detected in all the petiole and stem sections. Together, these observations demonstrate that cmv1 prevents the systemic movement of CMV‐LS by blocking virus entry into the phloem.
Figure 2.
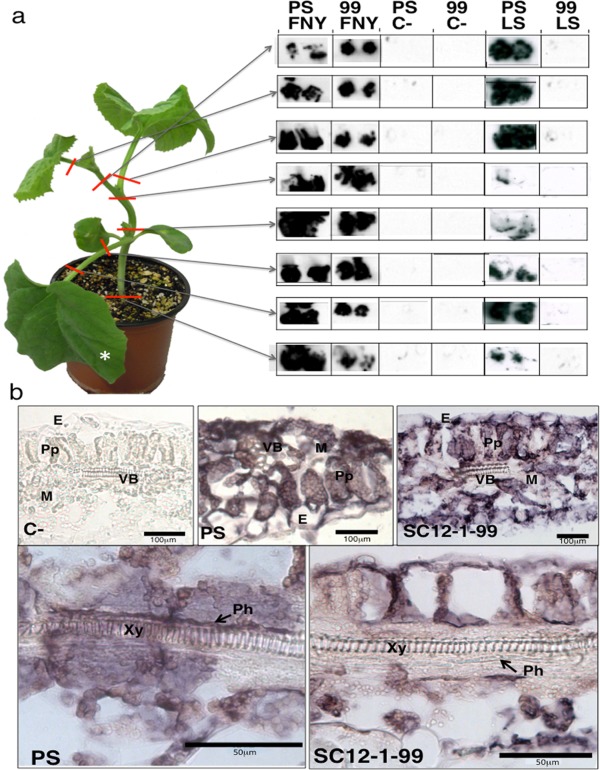
Absence of CMV‐LS from the phloem in the resistant SC12‐1‐99 line. (a) Left: representation of a melon plant and the localization of stem and petiole samples taken at 12 days post‐inoculation (dpi) for tissue printing (right). CMV‐FNY was detected in all samples from the susceptible line (Piel de Sapo, PS) and resistant SC12‐1‐99 line (99), whereas CMV‐LS occurred in samples from the susceptible line (PS), but not in those from the resistant line (99). C‐, samples from non‐inoculated plant. *Inoculated leaf. (b) Light microscopy imaging of the cellular localization of CMV‐LS RNA by in situ hybridization in inoculated leaves of melon plants. The images show longitudinal sections of symptomatic areas in the leaves. The blue/dark colour indicates the presence of viral RNA. Phloem can be observed as a darker blue‐coloured layer above the xylem in PS and as a white layer in SC12‐1‐99. Samples were taken at 6 dpi. C‐, samples from non‐inoculated plant; PS, susceptible line; SC12‐1‐99, resistant line. E, epidermal cells; M, mesophyll cells; Ph, phloem; Pp, palisade parenchyma; VB, vascular bundle; Xy, xylem. CMV, Cucumber mosaic virus.
To test whether CMV‐LS can at least enter the vascular bundle, we investigated the virus distribution in the inoculated leaves by in situ hybridization (ISH) experiments. After inoculation with CMV‐LS, the chlorotic foci produced in the inoculated leaf of both lines were collected at 6 dpi. As illustrated in Fig. 2b, the hybridized sections of both lines showed a specific signal of viral RNA (blue/dark). Viral RNA was detected in epidermal, palisade and mesophyll cells in both lines and was equally distributed in these tissues. Focusing in detail on the vascular system of the minor veins, the virus was detected in the phloem tissues of the susceptible melon line PS as a thin, darker blue line, whereas no viral RNA was detected in the phloem tissues of SC12‐1‐99. This indicates that cmv1 causes an interruption of CMV‐LS movement in a certain cell type surrounding the phloem, and thereby prevents entry of CMV‐LS into the phloem.
cmv1 restricts CMV‐LS movement in BS cells
As light microscopy and ISH on longitudinal and transverse leaf sections did not allow us to identify the specific phloem cell types, we focused our further analysis at the transmission electron microscopy (TEM) level. Minor veins (type V, Fig. 3a) of leaves from the resistant and susceptible melon lines were studied to identify the different cell types of the vascular bundle (Fig. 3b–i). Our observations are in agreement with previous reports on melon vascular anatomy (Schmitz et al., 1987) and also demonstrate that the vascular bundles of both lines share the same anatomy and specific cell morphology. Briefly, in both melon lines, BS cells have large vacuoles and contain a large amount of chloroplasts, mostly with starch, and a large nucleus (Fig. 3b). ICs are always adjacent to SEs. The cytoplasm of the ICs is very dense, containing numerous free ribosomes, and interrupted by many small vacuoles. Mitochondria are quite abundant and distributed all over the cell (Fig. 3f,h). In contrast, we could not observe plastids in any of the studied ICs. VP cells are less dense and are often located between BS cells and ICs, but frequently were not visible in the sections examined. A large number of simple or complex, secondarily modified PDs connect the ICs with the BS cells (Fig. 3h) and VP cells (Fig. 3e,f) and other ICs. These complex PDs seem to occur in clusters at points at which the walls are slightly thickened. Intensive symplastic connection with the BS and VP cells is a characteristic feature of ICs, and we observed no difference in the number and characteristics of PD clusters connecting ICs in both PS and SC12‐1‐99 cell lines. Typical funnel‐shaped PPUs were also present, exhibiting a single pore towards the SEs and complex branching towards the ICs (Fig. 3i). Thus, we did not observe any significant differences in the minor vein morphology between PS and SC12‐1‐99 lines, as can be observed in Fig. 3 by comparing panels (b–f) (SC12‐1‐99) with panels (g, h) (PS). Therefore, it appears unlikely that the ability of CMV‐LS to enter the phloem in PS, but not in SC12‐1‐99, is determined by anatomical differences in phloem architecture.
Figure 3.
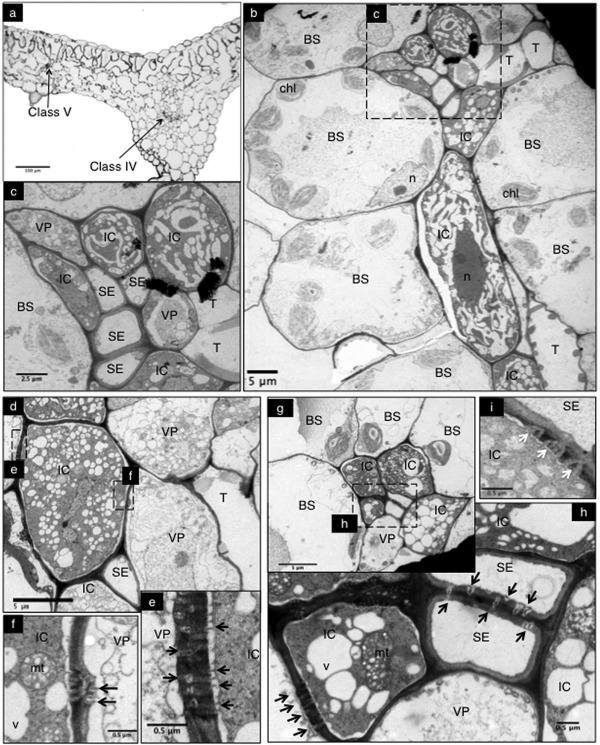
Anatomy of melon minor veins of Piel de Sapo (PS) and SC12‐1‐99 lines. Microscopic study of minor veins in SC12‐1‐99 (a–f, i) and PS (g, h). (a) Light microscopy of a leaf cross‐section including a class IV and a class V minor vein. (b) Corresponding transmission electron microscopy (TEM) image of the class V vein in (a). The minor vein is surrounded by bundle sheath (BS) cells. (c) Magnification of the minor vein of (b), with intermediary cells (ICs) and vascular parenchyma (VP) cells surrounding the sieve elements (SEs). (d) Minor vein in which an IC is symplastically connected with VPs. (e) and (f) are higher magnifications of (d) to observe simple and complex plasmodesmata (PDs) connecting VPs with ICs. (g) Minor vein and enlargement (h) of the PDs connecting SEs and IC–BS. (i) Plasmodesmata pore units (PPUs) connecting ICs and SEs. chl, chloroplasts; mt, mitochondria; n, nucleus; T, tracheid; v, vacuole. Black arrows point to simple or complex PDs and white arrows point to PPUs.
To identify the cellular boundary able to confine CMV‐LS to the inoculated leaves in SC12‐1‐99, we carried out immunogold labelling (IGL) on TEM sections of the inoculated leaves of both PS and SC12‐1‐99 using anti‐coat protein (anti‐CP) antibody. As shown in Fig. 4a, gold particles were observed inside the BS cells of both lines, uniformly distributed in the cytoplasm, whereas virtually no particles were detected in the vacuoles (see PS panels in Fig. 4), indicating a high specificity of labelling. Compared with BS cells, the density of gold particles was reduced in ICs and VP cells. Apparently, a relatively low virus density in ICs and VP cells is sufficient for systemic infection. In contrast, the sections taken from the resistant SC 12‐1‐99 line contained very few gold particles in ICs and VP cells, similar to control samples from non‐inoculated plants. This indicates that cmv1‐mediated resistance correlates with a strong reduction in the viral antigen present in ICs and VP cells (Fig. 4a). To determine whether the different labelling of VP cells and ICs between the resistant SC12‐1‐99 and susceptible PS melon lines was significant, we quantified the gold particles in entire cells of eight ICs and eight VP cells from PS and eight ICs and 10 VP cells from SC12‐1‐99. In order to control for non‐specific labelling, we also counted the gold particles in six ICs and five VP cells in samples from non‐inoculated PS plants. In total, we quantified samples from three grids for each sample type. The results (Fig. 4b) confirm the observations and show the presence of significantly more gold particles in the ICs and VP cells of the susceptible line relative to the corresponding cells of the resistant line [P = 0.0026 (IC); P = 0.0003 (VP)]. The small amount of gold particles found in ICs and VP cells in sections of the resistant line was not significantly different from the number obtained for the same cells in the non‐inoculated control plants [P = 0.14 (IC); P = 0.25 (VP)], indicating that the gold particles found in ICs and VP cells of the resistant line are a result of non‐specific background labelling. Taken together, these data indicate that the barrier to systemic infection by CMV‐LS in the resistant SC12‐1‐99 line is located in the BS cells connecting to either VP cells or ICs.
Figure 4.
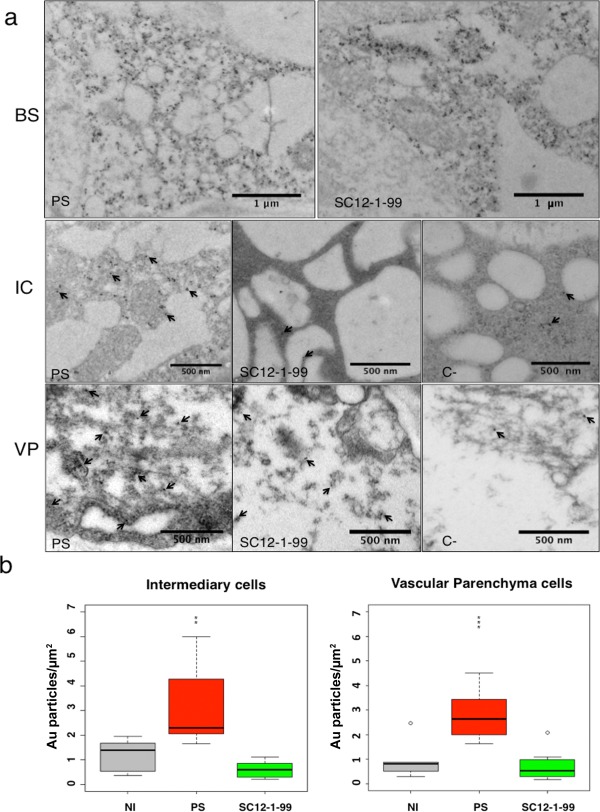
Localization of CMV‐LS coat protein (CP) in the cells of minor veins. (a) Transmission electron microscopic immunocytochemistry of Piel de Sapo (PS) and SC12‐1‐99 lines inoculated with CMV‐LS. Gold particles are detected in bundle sheath (BS) cells, intermediary cells (ICs) or vascular parenchyma (VP) cells. In ICs and VP cells, arrows point to some gold particles (10 nm) in the sections. C‐, negative control from a non‐inoculated PS plant. PS and SC12‐1‐99 are susceptible and resistant lines, respectively. (b) Quantification of gold particles in ICs and VP cells of PS and SC12‐1‐99 lines. Three grids per sample type were used for counting. The numbers of gold particles in ICs and VP cells are represented as Au particles/μm2. The data were analysed using Student's t‐test and were considered to be significant when P ≤ 0.05. Results are represented in a box plot. The bottom and top of the box are the first and third quartiles, respectively. The band inside the box is the median, the second quartile. Outliers or individual points represent the variability outside the upper and lower quartiles. NI, non‐inoculated plants indicating non‐specific gold labelling. Significant differences between PS and SC12‐1‐99 are represented by asterisks: **P ≤ 0.01; ***P ≤ 0.001. CMV, Cucumber mosaic virus.
CMV‐LS does not systemically infect graft‐inoculated SC12‐1‐99 plants
To determine whether CMV‐LS might be able to systemically infect SC12‐1‐99 plants once within the phloem, we grafted healthy SC12‐1‐99 scions onto CMV‐LS‐infected PS rootstocks. Among the 14 grafted plants, 11 did not develop symptoms within 60 days post‐grafting (dpg) and did not contain virus, as shown by the absence of viral RNA in tissue prints (Fig. 5a). Scions of three plants developed systemic, weak symptoms in the leaves proximal to the graft junction, and also contained virus, whereas leaves in the middle and apical parts of the scions remained asymptomatic and free of virus within 60 dpg (Fig. 5b). Similar grafts with susceptible PS scions developed strong systemic virus symptoms in all scions. These observations indicate that SC12‐1‐99 is resistant to CMV‐LS, even when the virus is already in the phloem. Therefore, cmv1 also plays a role in restricting systemic virus movement by suppressing not only phloem entry, but also phloem transport or exit of the virus.
Figure 5.

Graft inoculation of SC12‐1‐99 (scion) on infected CMV‐LS Piel de Sapo (PS) (rootstock). Grafts are represented as the rootstock under the grafted junction (white arrow) and the scion above the junction. (a) Representative healthy, asymptomatic graft. Detection of virus was carried out by tissue printing on distal to proximal leaves from the graft (numbers 1–5), showing that no virus was present in the scion. C+, leaf from CMV‐LS‐infected PS plant. (b) Graft showing very mild symptoms in proximal leaves. Virus was detected by tissue printing on distal to proximal leaves from the graft (numbers 1–5), showing virus in proximal leaves only (leaf 5 and less virus in leaf 4). CMV, Cucumber mosaic virus.
Discussion
Our earlier work has demonstrated that the resistance conferred by cmv1 is effective against strains of subgroup II of CMV, and that the viral determinant for this property maps to the MP (Guiu‐Aragonés et al., 2015). However, it remains unknown whether or not resistance occurs at the level of either cell‐to‐cell or long‐distance movement. Here, we have demonstrated that cmv1 blocks CMV‐LS from entering the phloem at the interface between BS cells and VP cells or ICs in the inoculated tissue, thus preventing the establishment of a systemic infection.
Vein entrance is a crucial step towards systemic infection. The BS cell–phloem interface has been reported as a barrier for systemic virus movement in a number of species resistant to viral infections. This is the case for soybean lines resistant to systemic infection by Cowpea chlorotic mottle bromovirus (CCMV) (Goodrick et al., 1991), transgenic tobacco plants resistant to CMV (Wintermantel et al., 1997), cucumber plants resistant to Tomato aspermy virus (TAV) (Thompson & García‐Arenal, 1998) and Cucumis figarei plants resistant to CMV, the latter showing the restriction to CMV in BS cells at 24 °C, but not at 36 °C (Kobori et al., 2000). BS cells do not differ morphologically from mesophyll cells. Hence, the accumulation of virus in BS cells would suggest a different viral cell‐to‐cell movement mechanism between mesophyll cells, than between BS and phloem cells (Thompson and García‐Arenal, 1998). Accordingly, the TMV MP increases the size exclusion limit of PDs between mesophyll and BS cells, but not in the interface between BS cells and CCs or VP cells in transgenic tobacco plants (Ding et al., 1992). Our study provides evidence of such an alternative mechanism working at the BS cell–phloem interface. It is controlled by cmv1, which acts as a cell‐specific molecular gate allowing the entrance of CMV into the phloem in a manner dependent on the presence of FNY MP. This confirms the importance of the BS cell–phloem boundary in controlling systemic viral infections. To our knowledge, cmv1 is the first recessive resistance gene whose function in supporting viral infection has been localized to a precise cell type, BS cells, where it is able to restrict the development of a systemic infection. Interestingly, the melon resistant accession SC also supports cell‐to‐cell movement and accumulation in the veins of both LS and FNY strains. As SC carries at least three QTLs conferring resistance to several strains of CMV (Guiu‐Aragonés et al., 2014), this indicates that none of the described QTLs impair either the replication or the cell‐to‐cell movement of CMV. Therefore, these QTLs should also be involved in restricting long‐distance movement of the virus in the resistant parent accession, either cooperating with cmv1 in facilitating phloem entry, or having a role in the movement of the virus once it is in the phloem. Thus, again, this points to the importance of the BS cell–phloem interface in determining CMV systemic infection.
The resistance caused by cmv1 is probably manifested by the lack of a phloem‐specific interaction with a factor required for phloem entry and transport. As depicted in the model shown in Fig. 6, the susceptible melon line PS may contain a dominant CMV1 allele encoding a protein variant able to interact with the MP complex of both CMV‐LS (or strains of subgroup II) and CMV‐FNY (or strains of subgroup I) (Fig. 6a), whereas the resistant allele cmv1 may encode a protein variant that can productively interact with the MP complex of CMV‐FNY, but not with that of CMV‐LS (Fig. 6b). Although the cmv1 allele may have evolved to interfere with the interaction with the MP complex, and thereby to cause resistance against CMV‐LS, the virus, in turn, may have evolved variants with mutations that would allow the MP complex to interact again with the altered cmv1 protein and thereby overcome this resistance. This could be the case for CMV‐FNY or a mutant CMV‐LS mimicking the MP of FNY (Fig. 6c), as the alteration of CMV‐LS MP by the introduction of a combination of FNY MP residues in four positions [the group 64–68 (SNNLL to HGRIA) and the point mutations R81C, G171T and A195I] is required to generate an LS virus derivative able to overcome cmv1‐mediated resistance (Guiu‐Aragonés et al., 2015). The characterization of the possible mechanism of resistance conferred by cmv1 will require the identification and cloning of the gene to determine which mutations are present in the resistant allele and whether the encoded protein is able to interact with FNY MP and mutant LS. Following the model depicted in Fig. 6, these putative interactions between MPs and the protein CMV1/cmv1 would be required only in BS cells, and the complex may be involved in either intracellular transport of the virus to the PDs or opening of the PDs. However, our experiments cannot exclude the possibility that the protein CMV1 would somehow be needed in the recipient phloem cells (VP cells and ICs) (Fig. 6) and play some role in either virus entry into these cells or replication coupled with movement to allow the virus to follow the infection. In this case, the boundary for movement would still occur at the interface between BS cells and phloem cells. Viral MPs have been observed at the VRCs and may cooperate in viral RNA transport (for a review, see Heinlein, 2015). In this context, CMV1 would be a host factor that may facilitate the interaction between the MP and VRC. However, this scenario seems more unlikely, as, in this case, the restriction to virus movement should be operating in two cell types (VP cells and ICs) instead of one (BS cells).
Figure 6.
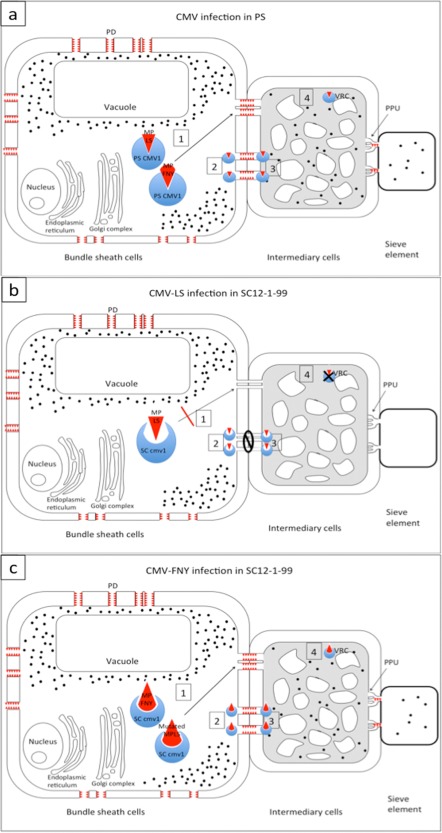
Hypothetical model of CMV‐LS and CMV‐FNY infecting Piel de Sapo (PS) and SC12‐1‐99. The movement protein (MP) (red triangles) opens the size exclusion limit of plasmodesmata (PDs) surrounding the bundle sheath (BS) cells. In all cases, virus (black dots) accumulates in the BS cells. For simplicity, only traffic between BS cells and intermediary cells (ICs) is represented. The model would be the same for traffic between BS cells and vascular parenchyma (VP) cells. (a) Cucumber mosaic virus (CMV) infecting PS. In the BS cells, the protein PS CMV1 can interact directly or indirectly with MPs of both CMV‐LS and CMV‐FNY, leading either to transport to PDs (1) or to the opening of PDs (2), allowing systemic spread of the virus. In (3), the interaction between MP and CMV1 in the ICs can allow viral RNA to enter the ICs. Alternatively, the interaction MP–CMV1 could take place in the viral replication complexes (VRCs) of the ICs, allowing viral replication (4). (b) SC12‐1‐99 infected with CMV‐LS. LS MP would not be able to interact with SC cmv1 protein, impeding all of the above four possibilities. (c) CMV‐FNY or mutated CMV‐LS infection of SC12‐1‐99. FNY MP and mutated LS MP carrying the relevant residues (Guiu‐Aragonés et al., 2015) are able to interact with the SC cmv1 protein and allow any of the four possibilities given above. PPU, plasmodesmata pore units.
Our grafting experiments suggest an additional role for cmv1 as, even if there is a continuous supply of virus from the infected stock, cmv1 is still able to block the systemic infection, suggesting that the virus is unable to move in, or even exit, from the phloem. Thus, cmv1 also has the potential to provide resistance against systemic movement, even in cases in which the virus succeeds in entering the long‐distance pathway. CMV is transmitted by aphids in nature, being able to deposit the virus directly into the phloem of the plant. Therefore, cmv1 could also provide protection to cultures in the field against aphid‐transmitted infections, giving an added value to the deployment of this gene in elite cultivars.
The results reported here demonstrate that cmv1 is the key gene of a new mechanism of recessive resistance based on the restriction of systemic virus transport. Given the severe economic losses produced by CMV, this gene provides opportunities for the design of approaches for breeding strategies in melon that focus on the impairment of CMV systemic movement. The identification of the genes underlying the QTLs that provide resistance to CMV subgroup I strains will further broaden the possibilities for breeding strategies and the range of CMV strains that can be controlled by resistance.
Experimental Procedures
Plants, viruses and inoculations
The genotypes of C. melo used were the Korean accession PI 161375 cultivar SC and the Spanish type PS (line T111), as resistant and susceptible controls, respectively. The NIL SC12‐1‐99 was derived from SC12‐1 (Essafi et al., 2009) and carries an introgression of SC on the linkage group XII which contains the cmv1 gene. Seeds were pre‐germinated and grown as described previously (Guiu‐Aragonés et al., 2015). Virus strains used in this study were CMV‐LS, belonging to subgroup II, and CMV‐FNY, belonging to subgroup I, both provided by Professor P. Palukaitis as infectious clones (Rizzo and Palukaitis, 1990; Zhang et al., 1994). Viral inocula were freshly prepared from infected zucchini squash Chapin F1 (Semillas Fito S.A., Barcelona, Spain) and rub inoculated onto the cotyledons of 7–10‐day‐old melon plants or the first true leaf.
Virus detection
The distribution of CMV in entire leaves of infected plants was studied by molecular hybridization analysis after tissue blotting. Leaves were frozen at −80 °C and blotted onto a nylon membrane as described previously (Díaz‐Pendón et al., 2005). The same plants were also analysed by tissue‐blot hybridization of freshly cross‐sectioned leaf petioles according to Guiu‐Aragonés et al. (2015) using the riboprobe p73, a probe containing partial sequences of CMV‐LS and CMV‐FNY CP genes.
Microscopy
In situ hybridization (ISH)
ISH was performed as described previously (Gosalvez‐Bernal et al., 2008; Javelle et al., 2011). Samples from inoculated leaves were collected at 6 dpi, fixed by vacuum infiltration in a formaldehyde–acetic acid–ethanol (FAA) solution (4% formaldehyde, 5% acetic acid and 50% ethanol in water) and stored overnight at 4 °C. The fixed tissues were then dehydrated in a series of alcohol baths, embedded in paraplast, sectioned in 8‐µm slices and mounted on poly‐l‐lysine‐coated slides. Sections were rehydrated and incubated for 15 min at 37 °C with 1 µg/mL proteinase K. Hybridizations with the riboprobe p73 (Guiu‐Aragonés et al., 2015) were performed overnight at 42 °C with 200 ng/mL of the riboprobe diluted in hybridization buffer (10% dextran sulfate, 50% formamide, 4 × saline sodium citrate buffer (SSC), 1% Denhardt, 100 µg/mL tRNA). After a 0.2 × SSC wash at 42 °C, slides were submitted for 30 min to RNAse A treatment (10 µg/mL at 37 °C). Then, the sections were washed at 42 °C in 0.2 × SSC and incubated for 30 min in blocking solution [100 mm Tris‐HCl, pH 7.5, 150 mm NaCl, 1% bovine serum albumin (BSA), 0.1% Triton]. Sections were incubated with anti‐digoxigenin sheep antibody coupled to alkaline phosphatase (Roche Molecular Biochemicals, Barcelona, Spain), diluted 1 : 1000 in blocking solution. After washing to remove the excess antibody, sections were rinsed in staining buffer (100 mm Tris‐HCl, pH 9.5, 150 mm NaCl, 2.5 mm MgCl2) and incubated for 30 min in staining buffer supplemented with nitroblue tetrazolium and 5‐bromo‐4‐chloro‐3‐indolyl phosphate (NBT/BCIP) (Roche Molecular Biochemicals). The hybridization signal is seen as a blue/dark area in the sections. Negative controls on sections from non‐inoculated plants were carried out for each experiment. Sections were examined using a DMRB LEITZ microscope (Leica Microsystems, Wetzlar, Germany) and photographed with a Leica DFC420C digital colour camera.
Electron microscopy
For TEM, leaf tissue samples (8 × 3 mm2) from PS and SC12‐1‐99 were taken from an inoculated leaf at 7 dpi and from a non‐inoculated leaf as a negative control. Samples were vacuum infiltrated for 2 min in a freshly prepared mixture of 1% (v/v) glutaraldehyde and 3% (v/v) paraformaldehyde (for LR‐White embedding) or 3% (v/v) glutaraldehyde (for Embed 812 embedding) in 0.1 m (pH 7.2) sodium phosphate buffer and incubated for 12 h at 4 °C. Tissue was post‐fixed for 2 h in 0.1% (v/v) osmium tetroxide and stained for 24 h in 2% (w/v) uranyl acetate in 150 mm sodium phosphate buffer (pH 7.2). Samples were then dehydrated through an ethanol series and infiltrated in London Resin White (Sigma‐Aldrich, St Louis, MO, USA) or Embed 812 resin (Electron Microscopy Sciences, Hatfield, PA, USA). Polymerization was performed at 60 ºC for 72 h. For morphological analysis by light microscopy, semi‐thin sections were mounted on slides and stained with toluidine blue. For the ultrastructural study, ultrathin sections of samples embedded in Embed 812 were collected on formvar‐coated copper electron microscopy grids and stained with uranyl acetate and lead citrate. Samples embedded with LR‐White were used for immunocytochemistry. Ultrathin sections were collected on formvar‐coated nickel grids. Grids were incubated with anti‐CP (Loewe® Biochemica GmbH, Sauerlach, Germany) diluted 1 : 5, and then incubated with a secondary antibody anti‐rabbit immunoglobulin G (IgG) conjugated with 10‐nm colloidal gold (Electron Microscopy Sciences) for 1 h using a dilution of 1 : 100. Finally, after washes with phosphate‐buffered saline (PBS), samples were stained for 15 min with uranyl acetate. Controls were made on infected samples incubated without the primary antibody and on sections of non‐inoculated plants. The ultrastructural study was performed with a Jeol 10‐11 with a Gatan camera (Jeol, Tokyo, Japan). Immunogold‐labelled samples were studied in either a Philips Tecnai 12 (FEI, Hillsboro, OR, USA) or Jeol 10‐11 (Jeol) transmission electron microscope.
Quantification and statistical analysis of gold particles
Gold labelling was quantified in entire VP cells and ICs of susceptible (PS) and resistant (SC12‐1‐99) plants. Quantification in the same cell types of non‐inoculated plants was also carried out to determine non‐specific labelling. For each type of sample, sections from three grids were used for quantification.
Morphometric measurements to determine the surface of the cells (μm2) were performed using the free software Image J. The data were analysed using Student's t‐test and considered to be significant when P ≤ 0.05. Results are represented in box plots.
Graft inoculations
PS plants were rub inoculated with sap from CMV‐LS to produce the stocks needed for graft inoculations. Ten days after inoculation, infected plants were used as virus sources by cutting them either below or above the cotyledons, depending on the thickness of the stem.
The scions were 3‐week‐old SC12‐1‐99 (cmv1) and PS plants, used as susceptible control. Scions were collected, given a V shape at the cut end and grafted onto the stock. The grafted region was wrapped with Parafilm M (American National Can, Chicago, IL, USA) and a high humidity (60%–80%) was maintained during the following 2 days to avoid dehydration of the scion. The surviving plants were grown under the conditions described above. The detection of virus was performed by tissue print (see above) as described previously (Guiu‐Aragonés et al., 2015).
Acknowledgements
We thank Fuensanta García for technical support, Peter Palukaitis for CMV‐FNY and CMV‐LS clones, and Laura Pascual for help with graphics and statistics and for critical advice. We also thank Mathieu Erhardt for technical help with ISH and Maria García from the Microscopy Service (SME, SAI) of the University of Murcia, Spain and Dr. Vicente Medina, from Universitat de Lleida, Spain for help with TEM. This work was supported by grants AGL2009‐12698‐C02‐01 and AGL2012‐40130‐C02‐01 from the Spanish Ministry of Science and Innovation. CG‐A was supported by grant BES‐2010‐030274 from the Spanish Ministry of Science and Innovation. The authors do not have any conflicting interests, and all read and agreed the content of the manuscript.
References
- Amano, M. , Mochizuki, A. , Kawagoe, Y. , Iwahori, K. , Niwa, K. , Svoboda, J. , Maeda, T. and Imura, Y. (2013) High‐resolution mapping of zym, a recessive gene for Zucchini yellow mosaic virus resistance in cucumber. Theor. Appl. Genet. 126, 2983–2993. [DOI] [PubMed] [Google Scholar]
- Amari, K. , Di Donato, M. , Dolja, V.V. and Heinlein, M. (2014) Myosins VIII and XI play distinct roles in reproduction and transport of Tobacco mosaic virus . PLoS Pathog. 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel, A.J. , Gamalei, Y.V. , Ammerlaan, A. and Bik, L.P. (1992) Dissimilar phloem loading in leaves with symplasmic or apoplasmic minor‐vein configurations. Planta, 186, 518–525. [DOI] [PubMed] [Google Scholar]
- Canto, T. and Palukaitis, P. (2001) A Cucumber mosaic virus (CMV) RNA 1 transgene mediates suppression of the homologous viral RNA 1 constitutively and prevents CMV entry into the phloem. J. Virol. 75, 9114–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caranta, C. , Pflieger, S. , Lefebvre, V. , Daubeze, A.M. , Thabuis, A. and Palloix, A. (2002) QTLs involved in the restriction of Cucumber mosaic virus (CMV) long‐distance movement in pepper. Theor. Appl. Genet. 104, 586–591. [DOI] [PubMed] [Google Scholar]
- Chaim, A. , Grube, R. , Lapidot, M. , Jahn, M. and Paran, I. (2001) Identification of quantitative trait loci associated with resistance to Cucumber mosaic virus in Capsicum annuum . Theor. Appl. Genet. 102, 1213–1220. [Google Scholar]
- Díaz‐Pendón, J.A. , Fernández‐Muñoz, R. , Gómez‐Guillamón, M.L. and Moriones, E. (2005) Inheritance of resistance to Watermelon mosaic virus in Cucumis melo that impairs virus accumulation, symptom expression, and aphid transmission. Phytopathology, 95, 840–846. [DOI] [PubMed] [Google Scholar]
- Ding, B. , Haudenshield, J.S. , Hull, R.J. , Wolf, S. , Beachy, R.N. and Lucas, W.J. (1992) Secondary plasmodesmata are specific sites of localization of the Tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell, 4, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X.S. (1998) Tobamovirus and potyvirus accumulation in minor veins of inoculated leaves from representatives of the Solanaceae and Fabaceae. Plant Physiol. 116, 125–136. [Google Scholar]
- Dufour, O. , Palloix, A. , Selassie, K.G. , Pochard, E. and Marchoux, G. (1989) The distribution of Cucumber mosaic virus in resistant and susceptible plants of pepper. Can. J. Bot. 67, 655–660. [Google Scholar]
- Edwardson, J.R. and Christie, R.G. (1991) Cucumoviruses In: CRC Handbook of Viruses Infecting Legumes (Edwardson J.R. and Christie R.G., eds), pp. 293–319. Boca Raton, FL: CRC Press. [Google Scholar]
- Essafi, A. , Diaz‐Pendon, J.A. , Moriones, E. , Monforte, A.J. , Garcia‐Mas, J. and Martin‐Hernandez, A.M. (2009) Dissection of the oligogenic resistance to Cucumber mosaic virus in the melon accession PI 161375. Theor. Appl. Genet. 118, 275–284. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations. Statistics division. 2013. http://faostat3.fao.org/browse/Q/QC/E.
- Goodrick, B. , Kuhn, C. and Hussey, R. (1991) Restricted systemic movement of Cowpea chlorotic mottle virus in soybean with nonnecrotic resistance. Phytopathology, 81, 1246–1431. [Google Scholar]
- Gosalvez‐Bernal, B. , Genoves, A. , Navarro, J. , Pallas, V. and Sanchez‐Pina, M. (2008) Distribution and pathway for phloem‐dependent movement of Melon necrotic spot virus in melon plants. Mol. Plant Pathol. 9, 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu‐Aragonés, C. , Monforte, A.J. , Saladié, M. , Corrêa, R.X. , Garcia‐Mas, J. and Martín‐Hernández, A.M. (2014) The complex resistance to Cucumber mosaic cucumovirus (CMV) in the melon accession PI 161375 is governed by one gene and at least two quantitative trait loci. Mol. Breeding, 34, 351–362. [Google Scholar]
- Guiu‐Aragonés, C. , Díaz‐Pendón, J.A. and Martín‐Hernández, A.M. (2015) Four sequence positions of the movement protein of Cucumber mosaic virus determine the virulence against cmv1‐mediated resistance in melon. Mol. Plant Pathol. 16, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries, P. and Ding, B. (2011) Cellular factors in plant virus movement: at the leading edge of macromolecular trafficking in plants. Virology, 411, 237–243. [DOI] [PubMed] [Google Scholar]
- Harries, P.A. , Schoelz, J.E. and Nelson, R.S. (2010) Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol. Plant–Microbe Interact. 23, 1381–1393. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2015) Plant virus replication and movement. Virology, 479–480, 657–671. [DOI] [PubMed] [Google Scholar]
- Hipper, C. , Brault, V. , Ziegler‐Graff, V. and Revers, F. (2013) Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 4, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle, M. , Marco, C.F. and Timmermans, M. (2011) In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. 57, 3328–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yeam, I. and Jahn, M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Kobori, T. , Satoshi, T. and Osaki, T. (2000) Movement of Cucumber mosaic virus is restricted at the interface between mesophyll and phloem pathway in Cucumis figarei . J. Gen. Plant Pathol. 66, 159–166. [Google Scholar]
- Lazarowitz, S.G. and Beachy, R.N. (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell, 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough, T.J. and Lucas, W.J. (2006) Integrative plant biology: role of phloem long‐distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Ohnishi, S. , Echizenya, I. , Yoshimoto, E. , Boumin, K. , Inukai, T. and Masuta, C. (2011) Multigenic system controlling viral systemic infection determined by the interactions between Cucumber mosaic virus genes and quantitative trait loci of soybean cultivars. Phytopathology, 101, 575–582. [DOI] [PubMed] [Google Scholar]
- Ouibrahim, L. , Mazier, M. , Estevan, J. , Pagny, G. , Decroocq, V. , Desbiez, C. , Moretti, A. , Gallois, J.L. and Caranta, C. (2014) Cloning of the Arabidopsis rwm1 gene for resistance to Watermelon mosaic virus points to a new function for natural virus resistance genes. Plant J. 79, 705–716. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. and Garcia‐Arenal, F. (2003) Cucumoviruses. Adv. Virus Res. 62, 241–323. [DOI] [PubMed] [Google Scholar]
- Pitrat, M. (2002) Gene list for melon. Cucurbit Genet. Coop. Rep. 25, 76–93. [Google Scholar]
- Rizzo, T. and Palukaitis, P. (1990) Construction of full‐length cDNA clones of Cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol . Gen. Genet. 122, 249–256. [DOI] [PubMed] [Google Scholar]
- Santa Cruz, S. (1999) Perspective: phloem transport of viruses and macromolecules – what goes in must come out. Trends Microbiol. 7, 237–241. [DOI] [PubMed] [Google Scholar]
- Schmitz, K. , Cuypers, B. and Moll, M. (1987) Pathway of assimilate transfer between mesophyll cells and minor veins in leaves of Cucumis melo L. Planta, 171, 19–29. [DOI] [PubMed] [Google Scholar]
- Sekine, K.T. , Nandi, A. , Ishihara, T. , Hase, S. , Ikegami, M. , Shah, J. and Takahashi, H. (2004) Enhanced resistance to Cucumber mosaic virus in the Arabidopsis thaliana ssi2 mutant is mediated via an SA‐independent mechanism. Mol. Plant–Microbe Interact. 17, 623–632. [DOI] [PubMed] [Google Scholar]
- Stamova, B.S. and Chatelat, R.T. (2000) Inheritance and genetic mapping of Cucumber mosaic virus resistance introgressed from Lycopersicon chilense into tomato. Theor. Appl. Genet. 101, 527–537. [Google Scholar]
- Thompson, J.R. and García‐Arenal, F. (1998) The bundle sheath–phloem interface of Cucumis sativus is a boundary to systemic infection by Tomato aspermy virus . Mol. Plant–Microbe Interact. 11, 109–114. [Google Scholar]
- Truniger, V. and Aranda, M. (2009) Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–159. [DOI] [PubMed] [Google Scholar]
- Turgeon, R. , Webb, J. and Evert, R. (1975) Ultrastructure of minor veins in Cucurbita pepo leaves. Protoplasma, 83, 217–232. [Google Scholar]
- Valkonen, J.P. and Watanabe, K.N. (1999) Autonomous cell death, temperature sensitivity and the genetic control associated with resistance to Cucumber mosaic virus (CMV) in diploid potatoes (Solanum spp). Theor. Appl. Genet. 99, 996–1005. [Google Scholar]
- Wang, H.‐L. , Wang, Y. , Giesman‐Cookmeyer, D. , Lommel, S.A. and Lucas, W.J. (1998) Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long‐distance transport system. Virology, 245, 75–89. [DOI] [PubMed] [Google Scholar]
- Wintermantel, W. , Banerjee, N. , Oliver, J. , Paolillo, D.J. and Zaitlin, M. (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase‐mediated resistance Virology, 231, 248–257. [DOI] [PubMed] [Google Scholar]
- Yang, P. , Lüpken, T. , Habekuss, A. , Hensel, G. , Steuernagel, B. , Kilian, B. , Ariyadasa, R. , Himmelbach, A. , Kumlehn, J. , Scholz, U. , Ordon, F. and Stein, N. (2014) Protein disulfide isomerase like 5‐1 is a susceptibility factor to plant viruses. Proc. Natl. Acad. Sci. USA, 111, 2104–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, M. , Nishikiori, M. , Tomita, K. , Yoshioka, N. , Kozuka, R. , Naito, S. and Ishikawa, M. (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78, 6102–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, M. , Shimizu, T. , Yamazaki, M. , Higashi, T. , Miyao, A. , Hirochika, H. and Omura, T . (2009) Disruption of a novel gene for a NAC‐domain protein in rice confers resistance to Rice dwarf virus . Plant J. 57, 615–625. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Handa, K. and Palukaitis, P. (1994) Mapping local and systemic symptom determinants of Cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 75, 3185–3191. [DOI] [PubMed] [Google Scholar]


