Summary
The FTF (Fusarium transcription factor) gene family comprises a single copy gene, FTF2, which is present in all the filamentous ascomycetes analysed, and several copies of a close relative, FTF1, which is exclusive to Fusarium oxysporum. An RNA‐mediated gene silencing system was developed to target mRNA produced by all the FTF genes, and tested in two formae speciales: F. oxysporum f. sp. phaseoli (whose host is common bean) and F. oxysporum f. sp. lycopersici (whose host is tomato). Quantification of the mRNA levels showed knockdown of FTF1 and FTF2 in randomly isolated transformants of both formae speciales. The attenuation of FTF expression resulted in a marked reduction in virulence, a reduced expression of several SIX (Secreted In Xylem) genes, the best studied family of effectors in F. oxysporum, and lower levels of SGE1 (Six Gene Expression 1) mRNA, the presumptive regulator of SIX expression. Moreover, the knockdown mutants showed a pattern of colonization of the host plant similar to that displayed by strains devoid of FTF1 copies (weakly virulent strains). Gene knockout of FTF2 also resulted in a reduction in virulence, but to a lesser extent. These results demonstrate the role of the FTF gene expansion, mostly the FTF1 paralogues, as a regulator of virulence in F. oxysporum and suggest that the control of effector expression is the mechanism involved.
Keywords: effector, FTF, Fusarium wilt, genomic expansion, pathogenicity, transcription factor, virulence
Introduction
Fusarium oxysporum Schlechtend.:Fr. is an anamorphic species complex (Fusarium oxysporum species complex, FOSC) ubiquitous in soils worldwide and able to grow as a saprophyte or by the colonization of plants. The pathogenic strains collectively may infect more than 100 different hosts, many of them important crops (Michielse and Rep, 2009), but the individual isolates are able to infect only one or a few plant species, allowing for classification into host‐specific forms, known as formae speciales (Armstrong and Armstrong, 1981). The comparison between the genomes of the different F. oxysporum formae speciales sequenced to date and those of the two other species of the Fusarium genus, F. graminearum and F. verticillioides, has revealed a well‐conserved core genome in the three species and several lineage‐specific (LS) genomic regions in F. oxysporum (Ma et al., 2010). LS regions are mostly organized into supernumerary chromosomes, which contain genes that are not required for basic metabolic processes and are absent in closely related species. The LS regions of F. oxysporum f. sp. lycopersici include four entire chromosomes (chromosomes 3, 6, 14 and 15), parts of chromosome 1 and 2, and some small scaffolds. It has been shown that the transfer of some LS chromosomes between strains of F. oxysporum may convert a non‐pathogenic strain into a pathogen, thus demonstrating the importance of the LS part of the genome in pathogenicity. These regions show a surprising enrichment of transposable elements and genes predicted to encode secreted proteins, some specifically expressed during plant infection, such as the SIX (Secreted In Xylem) genes (Houterman et al., 2008; Rep et al., 2004).
Among the gene families expanded in the LS genome of F. oxysporum, it is worthwhile to highlight FTF1 (Fusarium transcription factor 1). This gene putatively encodes a Zn(II)2Cys6 transcription factor and was first described in highly virulent (HV) strains of F. oxysporum f. sp. phaseoli (Ramos et al., 2007) as part of a SCAR (Sequence Characterized Amplified Region) designed for in planta detection (Alves‐Santos et al., 2002b). mRNA transcribed from the multiple copies of FTF1, or at least some of them, is abundant in plants colonized by F. oxysporum, but barely detectable in cultures of the fungus (Ramos et al., 2007). The expression level of the gene correlates with the number of copies in strains of F. oxysporum f. sp. phaseoli and the degree of virulence displayed by these strains when colonizing Phaseolus vulgaris L. and Phaseolus coccineus L. plants (Alves‐Santos et al., 2002b; de Vega‐Bartol et al., 2011). Eleven homologues of the gene have been detected in the genome of the tomato pathogen F. oxysporum f. sp. lycopersici strain 4287 (de Vega‐Bartol et al., 2011). Three of these copies (FOXG_16414, FOXG_14257 and FOXG_17458) are located in chromosome 14 close to mini‐clusters of SIX effector‐encoding genes (Schmidt et al., 2013). Recently, it has been shown that FTF1 is highly expressed in root crown and hypocotyl of common bean plants inoculated with an HV strain of F. oxysporum f. sp. phaseoli (Niño‐Sánchez et al., 2015). Furthermore, up‐regulation of FTF1 correlates with the highest level of expression of the effector‐encoding genes SIX1 and SIX6, which takes place when fungal growth is restricted to the vascular system (Niño‐Sánchez et al., 2015). Another gene expansion in F. oxysporum, EBR1 (Enhanced Branching 1), has been shown to be involved in virulence, similar to its counterpart in F. graminearum (Jonkers et al., 2013; Zhao et al., 2011). EBR1 is located in chromosome 7 of F. oxysporum f. sp. lycopersici, and thus is part of the core genome, whereas the other EBR paralogues are located in LS chromosomes. Mutants altered in EBR1 showed reduced growth when grown in culture and reduced virulence against tomato plants (Jonkers et al., 2013).
In this work, we addressed the structure of the FTF gene family of F. oxysporum, and focused on the functional role of the FTF1 paralogues by reducing their expression through gene silencing.
Results
FTF2 is a single copy gene with similarity to FTF1
Early studies on FTF1 showed that HV (when inoculated in common bean plants) and supervirulent (SV, when inoculated in runner bean plants) isolates of F. oxysporum f. sp. phaseoli harboured four and five copies of this gene, respectively (Ramos et al., 2007; de Vega‐Bartol et al., 2011). However, a faint hybridization signal could also be detected in digested DNA of these strains, non‐pathogenic and weakly virulent (WV, when inoculated in common bean plants) strains (Fig. 1), suggesting that all of these strains harbour a second gene able to cross‐hybridize with FTF1. We identified two recombinant phages when a genomic library from the WV strain FOP‐SP4 was hybridized with a probe containing part of the FTF1 promoter and the beginning of the open reading frame (ORF). DNA from both phages was subcloned into pBluescript KS+ and the FTF1 hybridizing region contained in both was sequenced. The coding region of the gene detected in the cloned DNA fragment comprises 3219 bp interrupted by a 49‐bp intron located in the same position as that of the intron found in FTF1. The predicted polypeptide is 1072 amino acids in length and contains the Zn(II)2Cys6 binuclear cluster DNA‐binding motif and a ‘fungal‐specific transcription factor domain’ or ‘medium homology region’ (MHR) (Schjerling and Holmberg, 1996).
Figure 1.
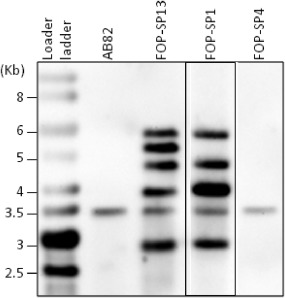
Southern hybridization of HindIII‐digested DNA from different strains of Fusarium oxysporum. The membrane was hybridized with the probe M18B‐M19A which is a segment of the central coding region of the FTF1 (Fusarium transcription factor 1) gene. Size markers are indicated on the left. AB82 is a non‐pathogenic strain, FOP‐SP13 is a supervirulent (SV) strain, FOP‐SP1 is a highly virulent (HV) strain and FOP‐SP4 is a weakly virulent (WV) strain.
Hybridization of several probes derived from the FTF2 coding region to digested genomic DNA from several F. oxysporum strains (non‐pathogenic, WV, HV and SV) showed a single hybridizing fragment in non‐pathogenic and WV strains, and several fragments corresponding to the FTF2 gene and the known copies of the FTF1 gene in the HV and SV strains (data not shown). Sequencing of the DNA insert cloned in one of the recombinant phages revealed an ORF corresponding to a homologue of the BimB gene of Aspergillus nidulans (May et al., 1992) 5 kb upstream of FTF2. Hybridization of the above‐described genomic DNA with a probe derived from the F. oxysporum BimB homologue revealed a single hybridizing fragment of the same size in all the strains analysed (data not shown), indicating that FTF2 is a single copy gene with a genomic location different from that of the FTF1 copies. Further confirmation of the single copy nature of FTF2 was obtained when promoter and ORF fragments of FTF2 were PCR amplified from a set of HV, WV and non‐pathogenic strains. Sequencing of the amplicons revealed that the fragments were 100% identical.
A blast analysis of the 12 available sequenced genomes in FOSC yielded a unique FTF2 homologue in all isolates (Table 1). All the putative proteins identified as FTF2 transcription factors showed similarities higher than 99.7% (in some cases they were identical), indicating that FTF2 is a unique locus in FOSC.
Table 1.
FTF1 (Fusarium transcription factor 1) and FTF2 (Fusarium transcription factor 2) copies in Fusarium oxysporum formae speciales.
| Formae speciales | FTF2 | FTF1a | FTF1b | FTF1c | Truncated |
|---|---|---|---|---|---|
| Pisi | FOVG_10613 | FOVG_18329 | |||
| Fo 47 | FOZG_12687 | FOZG_18011 (1007) | |||
| Fo human | FOYG_11202 | ||||
| radicis‐lycopersici | FOCG_10914 | FOXG_17757 (446) | |||
| lycopersici r. 2 | FOXG_09390 | FOXG_14257 (1070) ‡ | FOXG_17123 (930) | ||
| Chr 9, Sc 11 | Chr 14, Sc 22 | Chr 6, Sc 22 | |||
| FOXG_17458 (1079) | FOXG_16414 (930) | ||||
| Chr 14, Sc 51 | Chr 14, Sc 36 | ||||
| FOXG_14422 (1072)* | FOXG_14000 (930)† | ||||
| Chr. 15, Sc 24 | Chr 6, Sc 21 | ||||
| FOXG_15059 (1072)* | FOXG_17084 (930)† | ||||
| Chr 1, Sc 27 | Chr 6, Sc 41 | ||||
| FOXG_12539 (930)† | |||||
| Chr 3, Sc 18 | |||||
| FOXG_12589 (930)† | |||||
| Chr 3, Sc 18 | |||||
| lycopersici r. 3 | FOWG_05099 | FOWG_17325 (1070) | FOWG_17814 (930) | ||
| FOWG_18009 (1079) | FOWG_17908 (930) | ||||
| FOWG_17740 (930) | |||||
| raphani | FOQG_03479 | FOQG_15325 (1071) | FOQG_19052 (914) | ||
| cubense | FOIG_05174 | FOIG_16560 (1046) | FOIG_16484 (971) | ||
| FOIG_16630 (969) | |||||
| vasinfectum | FOTG_03936 | FOTG_16755 (1072) | FOTG_17956 (969) | FOTG_18226 | |
| FOTG_17879 (1072) | FOTG_18225 | ||||
| FOTG_18080 (1098) | |||||
| arabidopsis | FOXB_12381 | FOXB_19743 | |||
| FOXB_12596 | |||||
| FOXB_18246 | |||||
| melonis | FOMG_08221 | FOMG_18985 (1075) | FOMG_18692 (969) | FOMG_18999 (930) | FOMG_18691 |
| FOMG_19647 | |||||
| conglutinans | FOPG_02091 | FOPG_19962 | |||
| FOPG_18130 | |||||
| FOPG_19108 | |||||
| phaseoli § | JN167165 | phasl14257 | |||
| phasl17458 | |||||
| phasl15059 |
*,†Identical copies.
‡These copies have an exclusive stretch of amino acids between positions 7 and 17.
§Sequenced paralogues, one more FTF1 paralogue has been detected by Southern hybridization; phasl followed by a locus number indicates the homologous locus in the F. oxysporum f. sp. lycopersici genome.
FTF gene expansion
The use of an FTF2 sequence, or the previously described FTF1 sequence, in blast analysis of the available genomes of FOSC, yields, in addition to the FTF2 locus, a number of copies that show a variable degree of similarity and that we collectively denominate as FTF1 (Table 1). The number of FTF1 copies or paralogues found in each strain is variable, ranging from none in the pisi, biocontrol, human, radicis‐lycopersici, arabidopsis and conglutinans isolates, to 10 in the lycopersici race 2 strain (4287). However, the number found in strain 4287 should be approached with caution, as loci FOXG_14422 and FOXG_15059, and loci FOXG_14000, FOXG_17084, FOXG_12539 and FOXG_12589, are identical duplicates. It is worthwhile to highlight that all the copies of FTF1 in F. oxysporum f. sp. lycopersici 4287 map to chromosomes 3, 6, 14 and 15 and scaffold 27 (chromosome 1), which are LS regions in this strain (Ma et al., 2010), whereas FTF2 is located in the core genome (chromosome 9). The analysis of homologies, at both the nucleotide and amino acid levels, between FTF2 and all the FTF1 loci in the different formae speciales shows that FTF2 clusters apart from the FTF1 copies (Figs 2A,C, S1A, see Supporting Information).
Figure 2.
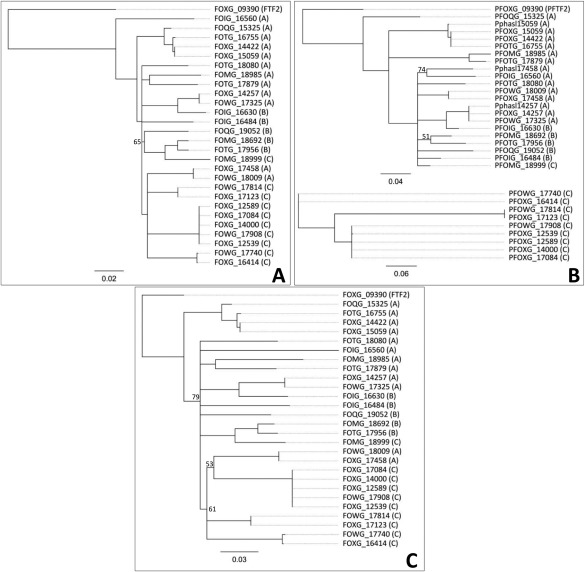
Similarity between the FTF2 (Fusarium transcription factor 2) and FTF1 (Fusarium transcription factor 1) paralogues, and the deduced proteins, found in the Fusarium oxysporum genomes of the different formae speciales. (A) Dendrogram obtained using the alignment of the open reading frames (ORFs). (B) Dendrogram obtained using the alignment of the promoter region (0.5 kb). (C) Dendrogram obtained using the alignment of the deduced proteins. FOXG loci correspond to the f. p. lycopersici race 2 isolate (4287), FOIG loci correspond to the f. sp. cubense isolate, FOQG loci correspond to the f. sp. raphani isolate, FOTG loci correspond to the f. sp. vasinfectum isolate, FOMG loci correspond to the f. sp. melonis isolate and FOWG loci correspond to the f. sp. lycopersici race 3 isolate (see Table 1 for a description of the different loci). The scale indicates the length of branch that represents one substitution per site. Numbers on the branches are bootstrap support values below 80.
The analysis of the proteins putatively encoded by the different FTF1 loci shows a range of variation in length, in contrast with the homogeneity of FTF2, which allows for the classification of three basic types (Table 1). The FTF1a type is 1070–1079 amino acids in length in most cases, similar to the FTF2 deduced protein, and can be found in most formae speciales, including three of the phaseoli paralogues. The FTF1b type is 969–971 amino acids in length and shows a shorter carboxy‐terminus than FTF1a. Paralogues encoding FTF1b are found in the cubense, vasinfectum and melonis isolates. Finally, the FTF1c type of putative protein is 930 amino acids in length, with shorter amino‐ and carboxy‐termini, and is exclusive to f. sp. lycopersici.
Apart from these well‐defined loci, several truncated copies of FTF1 are found in most isolates (see Table 1). The alterations shown by these copies range from small internal deletions or a premature stop codon, as in the raphani and biocontrol isolates, respectively, to large deletions in the amino‐ or carboxy‐terminus, or the central region of the proteins, which most probably preclude a normal function for these putative proteins.
Former expression analysis has suggested that FTF2 and FTF1 respond to different regulatory signals in planta (Niño‐Sánchez et al., 2015). In addition, FTF2 shows a steady low level of transcript accumulation during growth in culture, with no significant differences among strains based on virulence (Fig. S2, see Supporting Information). To gain an insight into the regulation of FTF1 and FTF2, we compared a 500‐bp promoter stretch immediately upstream of the ATG codon (selected as the codon encoding the methionine of the MSG amino acid sequence). The dendrogram in Fig. 2B shows two distinct groups. The promoters of copies FTF1a and FTF1b show a clustering pattern similar to that obtained when comparing the ORFs (Fig. 2A). The promoters of the FTF1c copies cannot be aligned with the promoters of FTF1a and FTF1b, except for FOMG_18999, and therefore they cluster in a different group. It is worthwhile to note that the only FTF1 promoter whose functionality has been demonstrated is Pphasl14257 in FOP‐SP1 (Niño‐Sánchez et al., 2015), which is homologous to the promoter found in the FOXG_14257 copy of strain 4287.
To verify whether the FTF gene family is present in other fungi, blast searches using the FTF1 and FTF2 ORFs, or the deduced polypeptides, as query sequences, were performed against other fungal genomes. A single copy gene homologous to FTF2 was found in the genomes of all the filamentous ascomycetes of the subphyllum Pezizomycotina (Euascomycota) tested, but absent in the genomes of yeasts, basidiomycetes, zygomycetes and chytridiomycetes (Fig. 3). This result indicates that FTF2 is a single copy gene specific to filamentous fungi, whereas the FTF1 paralogues and the genomic expansion are exclusive to FOSC.
Figure 3.
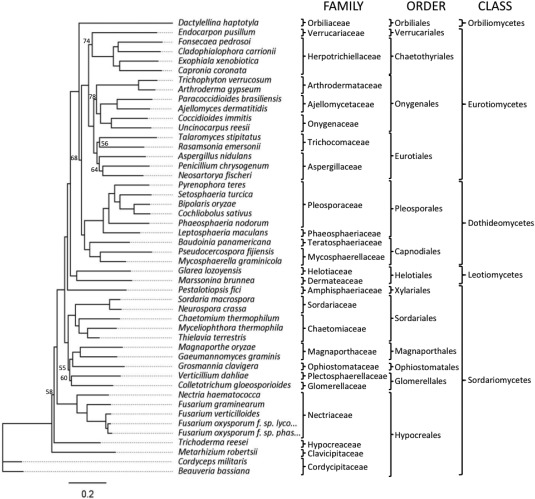
Genetic similarity between the FTF2 (Fusarium transcription factor 2) homologues in different fungal species. The dendrogram was obtained using the open reading frames (ORFs) of the unique FTF2 sequences found in the corresponding fungal genomes, except for the F. oxysporum f. sp. lycopersici and phaseoli ORFs, where FOXG_09390 and GenBank JN167165 were used, respectively. The scale indicates the length of branch that represents one substitution per site. On the right, the names of the species, family, order and class are shown. Numbers on the branches are bootstrap support values below 80.
Analysis of mutants of the FTF genes
The relationship between the number of copies of FTF1 and virulence has been well determined in F. oxysporum f. sp. phaseoli (Alves‐Santos et al., 2002a, b; de Vega‐Bartol et al., 2011) and F. oxysporum f. sp. dianthi (Gómez‐Lama Cabanás, personal communication, Instituto de Agricultura Sostenible (IAS), Cœrdoba, SPAIN). However, the large number of copies found makes the construction of knockout mutants an insurmountable task. The high similarity between FTF2 and FTF1 and among the copies of FTF1 makes this expansion an attractive target for gene silencing based on RNA interference (RNAi) methodology (Fig. S1B). However, the attenuation of gene expression obtained in this way affects FTF2 and the copies of FTF1. To discriminate the contribution of each gene to the virulence phenotype, we first generated null mutants of FTF2 in the FOP‐SP1 strain. As the genome annotation shows that there is an ORF (FOXG_09391) located contiguous to the 3′ end of the coding region of FTF2, a deletion construct was generated using the 5′ end flanking region of FTF2 (which is specific to FTF2) and the 3′ terminus of the FTF2 coding region (which is highly similar between FTF2 and FTF1 copies). This strategy was designed to optimize the probabilities of deleting most of the FTF2 coding region without deleting a FTF1 paralogue or part of the FOXG_09391 coding region, which would result in a double mutation. Several transformants were obtained and subjected to polymerase chain reaction (PCR) and Southern analysis, which verified such a deletion of FTF2, but no alteration of the FTF1 copies (Fig. S3A, see Supporting Information). In addition, reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis was performed to verify wild‐type expression of FTF1 and the lack of FTF2 deleted transcript (data not shown). Two transformants were selected for further analyses (SP1ΔFTF2.1 and SP1ΔFTF2.2). First, it was verified that deletion of FTF2 did not affect growth and sporulation (Table S1 and Fig. S4, see Supporting Information). Then, they were tested for pathogenicity and virulence in inoculation assays of common bean plants. The results in Fig. 4A–D indicate that both transformants show a slight reduction in virulence as measured by the International Center for Tropical Agriculture (CIAT) scale, plant weight and disease index. The virulence reduction reverted to the wild‐type level when the mutation was complemented with the FTF2 wild‐type allele (Figs S3B, 4A–D).
Figure 4.
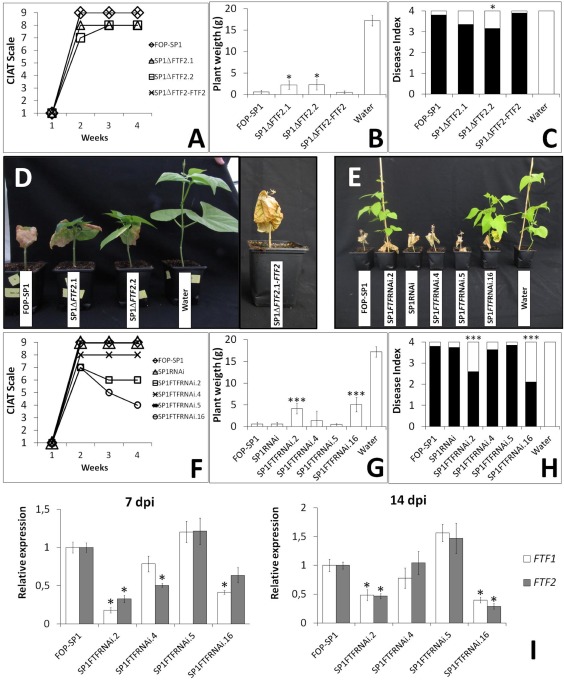
Fusarium wilt induced in common bean plants by Fusarium oxysporum f. sp. phaseoli transformants with reduced expression of FTF (Fusarium transcription factor). Common bean plants 4 weeks after inoculation with FTF2– mutants and complemented mutant (A–D) and inoculation with attenuated expression of FTF (E–H). Severity of Fusarium wilt symptoms as measured by: (A, F) the International Center for Tropical Agriculture (CIAT) scale (the disease index is assigned according to the percentage of chlorotic or necrotic leaves); (B, G) the weight of aerial plant parts 4 weeks after inoculation; (C, H) disease index (assigned according to the number of necrotic vascular vessels and whether or not the plant is dead). (I) Relative expression of the FTF genes in root crown [7 days post‐inoculation (dpi)] and hypocotyl (14 dpi) of colonized plants, as measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). An arbitrary value of 1.0 was used for the transcript level of FTF genes in plants infected with FOP‐SP1. Error bars in each panel indicate the standard deviations in three independent biological experiments. The differences between each transformant and the wild‐type strain in plant weight (B, G), disease index (C, H) and expression of the FTF genes (I) were tested using one‐way analysis of variance (ANOVA) followed by Dunnett's test, and indicated by *P < 0.05 and ***P < 0.001. FOP‐SP1 is a highly virulent (HV) wild‐type strain; SP1FTFRNAi.2, SP1FTFRNAi.4 and SP1FTFRNAi.5 are hygromycin‐resistant transformants; and SP1FTFRNAi.16 is a phleomycin‐resistant transformant harbouring an FTF1 fragment for the induction of gene silencing; SP1RNAi is a hygromycin‐resistant strain which lacks the silencing FTF1 fragment.
For FTF gene silencing, we used a construct based on the strategy developed in Penicilllium chrysogenum and Acremonium chrysogenum (Ullán et al., 2008) to obtain FTFRNAi transformants of the HV phaseoli strain FOP‐SP1 and the lycopersici strain 4287. All the constructs contained a 462‐bp FTF1 fragment to induce gene silencing of FTF1 and FTF2 (Fig. S1B), cloned in binary plasmids harbouring either a gene coding for hygromycin resistance (pFTFRNAi‐Hyg) or phleomycin resistance (pFTFRNAi‐Phleo). The high similarity between the FTF1 and FTF2 ORFs precluded the use of a fragment long enough to induce the specific knockdown of each gene. After transformation of FOP‐SP1, the hygR transformants SP1FTFRNAi.2, SP1FTFRNAi.4 and SP1FTFRNAi.5, the phleoR transformant SP1FTFRNAi.16 and the hygR transformant SP1RNAi, which contains the integrated plasmid backbone without the 462‐bp FTF1 fragment, were selected. Transformants of strain 4287 were obtained in a similar way. Five representative transformants obtained with the silencing construct harbouring the hph gene for hygromycin resistance were selected for further analyses, together with a control transformant obtained with the same construct lacking the silencing‐inducer FTF1 fragment. All the selected transformants were analysed by PCR and Southern hybridizations to verify the integration of the silencing construct or the control construct without the FTF1 fragment (Fig. S3C–G). They were also examined for growth and sporulation in both solid and liquid media. No significant differences could be observed between FTFRNAi transformants and their respective wild‐type strains (Table S1 and Fig. S4).
Silenced FOP‐SP1 transformants were tested for pathogenicty and virulence in common bean plants (P. vulgaris L.) (Fig. 4E–H). All transformants were pathogenic, but differed in virulence. Transformants SP1FTFRNAi.5 and SP1RNAi showed disease indices similar to those induced by the HV strain FOP‐SP1, measured using the CIAT scale, the plant weight of inoculated plants or the disease scale described in Experimental Procedures (Fig. 4F–H, respectively). In addition, the disease progression rate (DPR) was similar. Transformant SP1FTFRNAi.4 showed a slightly reduced disease rating, and transformants SP1FTFRNAi.2 and SP1FTFRNAi.16 showed a significantly reduced disease rating. DPR was also lower for these three transformants (6.25 for SP1FTFRNAi.4, 5 for SP1FTFRNAi.2 and 4.25 for SP1FTFRNAi.16) when compared with FOP‐SP1 (DPR = 7). The virulence reduction was precisely correlated with the attenuation of FTF1 and FTF2 expression, as measured by RT‐qPCR at 7 and 14 days post‐inoculation (dpi) (Fig. 4I). The expression data in Fig. 4I show significant differences in the expression of both genes with respect to normal expression in the wild‐type strain FOP‐SP1. Differences ranged from no reduction in SP1FTFRNAi.5 and the control transformant SP1RNAi to more than 50% for FTF1 in SP1FTFRNAi.16.
In a similar way, FTF‐silenced transformants of the wild‐type 4287 were tested in inoculation assays of tomato plants (Fig. 5). The disease indices estimated for plants inoculated with the silenced transformants were, in all cases, lower than those obtained for plants inoculated with the wild‐type strain 4287 (Fig. 5C), and the plant survival rate was significantly higher (Fig. 5D). When FTF1 and FTF2 expression in the inoculated plants was analysed, the correlation between virulence reduction and attenuated FTF expression was not as clear as in common bean inoculations (Fig. 5E). Most transformants showed a reduction in the expression of both genes, but, in all, the expression of each gene was higher than 65% relative to the expression measured in wild‐type 4287.
Figure 5.
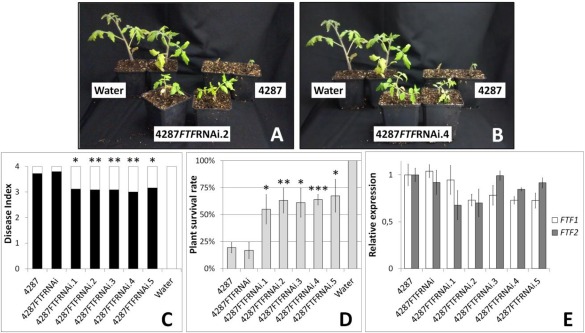
Fusarium wilt induced in tomato plants by Fusarium oxysporum f. sp. lycopersici attenuated transformants. (A, B) Tomato plants 3 weeks after inoculation. Severity of Fusarium wilt symptoms 3 weeks after inoculation, as measured by the disease index (C) and plant survival rate (D). (E) Relative expression of the FTF (Fusarium transcription factor) genes in the root crown of colonized plants at 14 days post‐inoculation (dpi), as measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). An arbitrary value of 1.0 was denoted for the transcript level of FTF genes in plants infected with the wild‐type strain 4287. Error bars indicate the standard deviations obtained in three independent biological experiments. The differences between each transformant and the wild‐type strain in the disease index (C) and plant survival rate (D) were tested using one‐way analysis of variance (ANOVA) followed by Dunnett's test, and are indicated by *P < 0.05, **P < 0.01 and ***P < 0.001. Transformants 4287FTFRNAi.1‐5 and 4287FTFRNAi were obtained with the hph plasmid described in Experimental Procedures.
Common bean and tomato plants inoculated with transformants accumulating low levels of FTF transcripts showed a clear reduction in Fusarium wilt symptoms. Transformants generated with the same construct lacking the FTF1 fragment responsible for the formation of double‐stranded RNAi (dsRNAi) were as virulent as the original wild‐type strains. These results demonstrate that dsRNAi transcribed from both promoters in the transforming constructs induces gene silencing in F. oxysporum. However, the efficiency of gene silencing is greater in the phaseoli strain FOP‐SP1 than in the lycopersici strain 4287. The comparison of the virulence exhibited by the FTF2– mutants and the best FTF attenuated mutants indicates a greater contribution to virulence of functional FTF1 paralogues with respect to FTF2.
FTF1 does not enhance virulence in a WV strain
The virulence of transformants SP1FTFRNAi.2 and SP1FTFRNAi.16, as displayed in common bean inoculation assays, reaches final CIAT scale indices similar to those recorded for WV F. oxysporum f. sp. phaseoli isolates (Alves‐Santos et al., 2002b), although the progression of the disease is different. To test whether FTF1 alone could be responsible for the virulence differences between HV and WV strains, the WV strain FOP‐SP4 was transformed with a functional copy of FTF1 driven by promoter Pphasl14257 (Niño‐Sánchez et al., 2015). Two transformants were selected for inoculation assays. One showed a single integration event, whereas the other showed three integration events, as demonstrated by PCR and Southern analysis (Fig. S5, see Supporting Information). Therefore, the transformants have genetic backgrounds equivalent to FOP‐SP4 plus one or three FTF1 functional native copies. The latter has a more similar copy number to the HV strain FOP‐SP1. The results of inoculation assays of common bean plants showed no significant differences between any of the transformants and the FOP‐SP4 wild‐type (Fig. S6A, see Supporting Information). Transformants of FOP‐SP4 were also obtained with a construct, pPgpdA::FTF1, which harboured the same copy of FTF1 formerly used, driven by the constitutive promoter PgpdA. Six transformants were selected and used to inoculate common bean plants, but none showed significant changes in virulence (Fig. S6B).
To assess whether the above results were the consequence of a lack of transcription of the transformed FTF1 copy in the genetic FOP‐SP4 background, semi‐quantitative RT‐PCR analyses were performed. An FTF1 cDNA band could be amplified from RNAs obtained from plants inoculated with transformants harbouring the FTF1 copy driven either by the native or constitutive promoter at 14 days post‐infection (Fig. S6C). Therefore, the WV strain is able to produce FTF1 transcripts, although with no effect on virulence.
FTF1 is involved in the regulation of virulence factors
The former results indicate that other virulence factors, apart from FTF1, are present in HV and absent in WV strains. It has been shown that the effector genes SIX1 and SIX6 share the same expression pattern as FTF1 during infection of common bean (Niño‐Sánchez et al., 2015). Our analyses of the FOP‐SP4 genome indicate that this WV strain is devoid of both SIX1 and SIX6 genes. Therefore, the virulence of HV strains could be explained if transcription factor FTF1 acts as a positive regulator of effector genes, such as SIX1 and SIX6.
To test this hypothesis, we first analysed the expression of SIX1 and SIX6 in PgpdA::FTF1 transformants of HV strain FOP‐SP1, which constitutively express FTF1. We included in the analysis the gene coding for the transcripton factor SGE1 (Six Gene Expression 1), as it has been proposed to regulate the expression of some SIX genes in F. oxysporum f. sp. lycopersici (Michielse et al., 2009b). RT‐qPCR analyses performed with RNA obtained from three independent transformants grown for 72 h in liquid culture showed a drastic induction of FTF1, which correlated with the induction of the three genes analysed (Fig. 6). These results support those previously obtained, which correlated FTF1, SIX1 and SIX6 expression during host colonization (Niño‐Sánchez et al., 2015).
Figure 6.
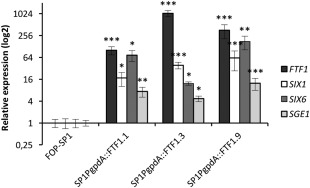
Gene expression analysis in Fusarium oxysporum f. sp. phaseoli PgpdA::FTF1 transformants. Gene expression of the effector‐encoding genes SIX1 (Secreted In Xylem 1) and SIX6 (Secreted In Xylem 6) and the transcription factors SGE1 (Six Gene Expression 1) and FTF1 (Fusarium transcription factor 1) was measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) in three different FTF1 constitutive transformants (SP1PgpdA::FTF1.1, SP1PgpdA::FTF1.3 and SP1PgpdA::FTF1.9). The value 1.0 was denoted for the transcript level of the four genes in the wild‐type strain FOP‐SP1. Error bars indicate the standard deviations in three independent biological experiments. The expression differences between each transformant and the wild‐type strain were tested using one‐way analysis of variance (ANOVA) followed by Dunnett's test, and are indicated by *P < 0.05, **P < 0.01 and ***P < 0.001.
Second, we took advantage of the RNAi transformants obtained from the phaseoli and lycopersici wild‐type strains to compare the expression during host colonization. Figure 7 shows the expression of the three genes analysed in common bean plants (Fig. 7A) and tomato plants (Fig. 7B) inoculated with the same transformants, harbouring the silencing construct, as described previously. The greatest reduction in SIX1 and SIX6 expression (P < 0.05) corresponds to plants inoculated with SP1FTFRNAi.2 (7 dpi) and SP1FTFRNAi.16 (14 dpi), the transformants that showed the minimum expression of FTF1. SP1FTFRNAi.4 also showed a clear reduction in SIX6 expression at 7 dpi (P < 0.05) and intermediate levels of expression of the other genes, in agreement with the intermediate expression shown by the FTF genes (see Fig. 4I). Transformant SP1FTFRNAi.5 showed no reduction in either FTF1 or the three genes analysed. Some of the best silenced transformants in FTF1 derived from the lycopersici wild‐type strain 4287, 4287FTFRNAi.2, 4287FTFRNAi.4 and 4287FTFRNAi.5, also showed the greatest reduction in the expression of the effector genes. The expression of the gene encoding the transcription factor SGE1 was significantly reduced in the best silenced transformants of the phaseoli strain, in a similar manner to the effector genes analysed. This result, together with the induction observed in the transformants constitutively expressing FTF1, demonstrates that SGE1, SIX1 and SIX6 are under positive regulation by FTF1, either directly or indirectly. The results obtained with the FTF‐silenced lycopersici strain indicate that SIX1 and SIX6 are under FTF1 positive regulation, but are less conclusive concerning SGE1.
Figure 7.
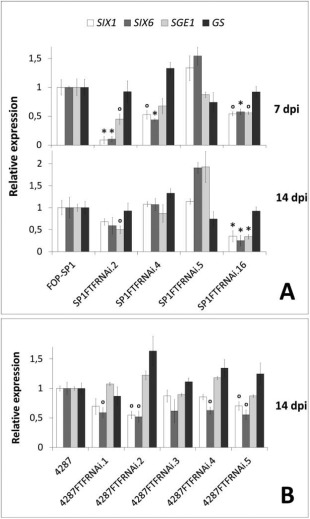
Gene expression analysis in Fusarium oxysporum attenuated transformants. The expression of the effector‐encoding genes SIX1 (Secreted In Xylem 1) and SIX6 (Secreted In Xylem 6), the transcription factor SGE1 (Six Gene Expression 1) and the glutamine synthetase gene (GS), used as control, was measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) in inoculated common bean plants at 7 and 14 days post‐inoculation (dpi) (A) and inoculated tomato plants at 14 dpi (B). FOP‐SP1 and 4287 are F. oxysporum f. sp. phaseoli and lycopersici wild‐type strains, respectively; SP1FTFRNAi.2, SP1FTFRNAi.4, SP1FTFRNAi.5 and 4287FTFRNAi.1–5 are hygromycin‐resistant silenced transformants; SP1FTFRNAi.16 is a phleomycin‐resistant silenced transformant. The value 1.0 was denoted for the transcript level of all genes in the wild‐type strains FOP‐SP1 and 4287. Error bars indicate the standard deviations obtained in three independent biological experiments. The differences between each transformant and the wild‐type strain were tested using one‐way analysis of variance (ANOVA) followed by Dunnett's test, and are indicated by *P < 0.05 and °P < 0.1.
Host plant colonization by silenced strains
In a former study, we showed that WV and HV strains of F. oxysporum f. sp. phaseoli not only differ in the severity of symptoms caused in the host plant (virulence), but also in the colonization pattern: HV strains accumulate more mycelium in infected tissues and are almost exclusively restricted to vascular vessels (Niño‐Sánchez et al., 2015).
To analyse the morphological pattern of host colonization by the silenced transformants, one transformant, SP1FTFRNAi.16, was transformed with a construct harbouring the GFP (green fluorescent protein) gene under the control of the constitutive promoter PgpdA. After checking for the correct integration and expression of GFP (data not shown), one of the SP1FTFRNAi.16‐GFP transformants was selected for inoculation tests. This silenced transformant displayed a colonization pattern resembling that exhibited by the WV strain FOP‐SP4, characterized by extensive growth around the parenchymal cells of the cortex and limited growth inside the vascular vessels (Fig. 8A,B), in contrast with the pattern observed in plants inoculated with FOP‐SP1 (Fig. 8C). The proportion of colonized vessels measured by image analysis was 31.59% ± 4.89%, a value closer to that obtained for FOP‐SP4 (11.39% ± 7.03%) than to that obtained for FOP‐SP1 (81.06% ± 8.8%) (Niño‐Sánchez et al., 2015). The quantification of fungal biomass accumulation in infected plants, measured by RT‐qPCR, showed that silenced transformants accumulate less mycelium than the wild‐type HV strain FOP‐SP1 (Fig. 8B). The largest difference was observed at 7 dpi, in agreement with the observations made in the above‐mentioned study. Again, the phenotype showing the most significant difference from the wild‐type strain was displayed by SP1FTFRNAi.2, one of the most attenuated transformants.
Figure 8.
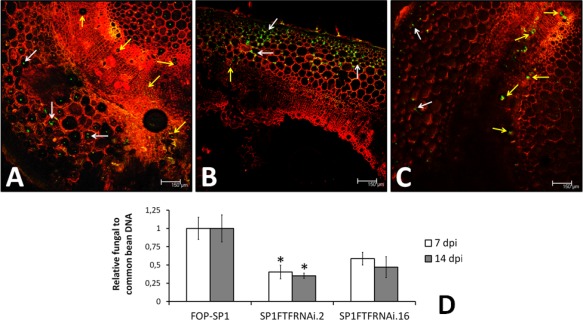
Plant colonization by green fluorescent protein (GFP)‐expressing transformants of Fusarium oxysporum f. sp. phaseoli. Cross‐sections of hypocotyls of common bean plants inoculated with GFP‐expressing transformants derived from the SP1FTFRNAI.16 attenuated strain (A), the weakly virulent (WV) strain FOP‐SP4 (B) (Niño‐Sánchez et al., 2015) and the highly virulent (HV) strain FOP‐SP1 (C) were examined by confocal laser microscopy. Hyphae growing in the parenchyma region are indicated by white arrows, whereas hyphae growing inside the xylem vessels are indicated by yellow arrows. Fungal biomass was quantified by quantitative polymerase chain reaction (qPCR) in plants inoculated with FOP‐SP1 and two attenuated transformants (SP1FTFRNAi.2 and SP1FTFRNAi.16). Fungal DNA relative to common bean DNA was measured by assaying the fungal EF1α gene and the plant actin gene by qPCR using DNA extracted from the root crown at 7 days post‐inoculation (dpi) and hypocotyls at 14 dpi. All measurements were referred to the arbitrary value of unity obtained during FOP‐SP1 colonization. The differences between each transformant and the wild‐type strain were tested using one‐way analysis of variance (ANOVA) followed by Dunnett's test, and are indicated by *P < 0.05.
Both results indicate that FTF knockdown transformants obtained in an HV genetic background show a host colonization pattern similar to that displayed by the WV strain FOP‐SP4, which is devoid of FTF1 copies.
Discussion
The expansion of some gene families encoding transcription factors in F. oxysporum is a striking question that addresses the genetic bases of virulence and host specificity in this species complex. In this study, we analysed the FTF gene expansion, composed of the core genome FTF2 gene and several paralogues of the LS FTF1 gene. FTF2 is well conserved in all filamentous fungi. A single copy of this gene is present in all genomes available to date in FOSC, and also in sets of isolates experimentally found to be non‐pathogenic, WV and HV to common bean plants. The F. graminearum mutant seems to be altered only in ascospore production and discharge (Son et al., 2011), but this phenotype would not be relevant in F. oxysporum as this fungus does not seem to reproduce sexually. We found that FTF2 mutants of F. oxysporum do not display apparent growth or sporulation abnormalities in solid and liquid culture, respectively, but show a slight reduction in virulence. In contrast, the FTF1 paralogues are exclusive to F. oxysporum and can be found in a variable number of copies, which is related both to the forma specialis analysed and the virulence displayed, as shown in this work and in previous reports (Alves‐Santos et al., 2002a; Schmidt et al., 2013; de Vega‐Bartol et al., 2011). Additional lines of evidence point to the role of these paralogues in virulence, such as the expression during plant infection (Niño‐Sánchez et al., 2015; Ramos et al., 2007) and the correlation between the copy number and virulence displayed (de Vega‐Bartol et al., 2011). However, direct confirmation of the role of FTF1 as a virulence factor is lacking because of the experimental problems involving the construction of deletion mutants in all the paralogues. We have successfully used RNA‐mediated gene silencing to attenuate the expression of the FTF genes, and found that mutants most effectively silenced (less than 50% of FTF1 expression) show an important reduction in virulence. Constructs expressing dsRNA complementary to target sequences were obtained by placing a fragment of the FTF1 gene between two promoters in opposite orientations, following the design successfully deployed in other fungi (Ullán et al., 2008). The sense and antisense strands form a dsRNA that triggers gene silencing (Lee et al., 2002; Wang et al., 2003). The presence of a unique NcoI cloning site between the two strong promoters (Ullán et al., 2008) allows for easy insertion of DNA fragments from the genes of interest. This feature implemented in a binary vector harbouring an antibiotic resistance cassette for selection (we have successfully used both the hph and ble genes) results in a system for easy gene silencing in any strain of F. oxysporum. This system would be a useful tool to facilitate the functional analysis of gene expansions, such as that described here, where the deletion of all the copies is not feasible. The correlation between the attenuation of gene expression and reduction in virulence was observed in both the phaseoli and lycopersici transformants, although, for unknown reasons, gene silencing is more effective in transformants obtained from FOP‐SP1 than in those obtained from strain 4287. Silencing efficiency may show variations between strains, as reduced attenuation of gene expression has also been reported in F. oxysporum f. sp. conglutinans 5176, although, in this case, the silencing method relied on the expression of hairpin RNA transgenes (Schumann et al., 2013). In addition, it is worth noting that the number of copies of FTF1 is higher in 4287 than in FOP‐SP1. The reduction in virulence shown by the attenuated mutants is proportionally greater than that shown by the FTF2– mutants, supporting a more important role in virulence for the FTF1 paralogues than for the FTF2 core genome gene.
The best‐described TF gene expansions involved in the pathogenicity of F. oxysporum are EBR1, with the four paralogues EBR2, EBR3, EBR4 and EBR5, and FOW2 (FOXG_06378), with two paralogues, FOXG_12458 and FOXG_21393. The deletion of EBR1 causes a minor reduction in virulence towards tomato plants and reduced radial growth in complete medium (Jonkers et al., 2013), which is also observed in the deletion mutants obtained in F. graminearum (Zhao et al., 2011). The mutant phenotype observed in F. oxysporum f. sp. lycopersici when EBR1 is deleted is substantially restored by a copy of EBR2 placed under the control of the EBR1 promoter. The gene predominantly expressed, both during culture and tomato colonization, is EBR1. When EBR1 is deleted, the majority of transcripts detected during growth in complete medium correspond to EBR3 and, to a lesser extent, to EBR2; the opposite occurs during tomato colonization (Jonkers et al., 2013). Taken together, these results indicate that EBR1 and the paralogues EBR2 and EBR3 have redundant roles during both saprophytic and in planta growth, whereas EBR4 and EBR5 are barely expressed in both conditions. EBR1 is located in the core genome, whereas the copies of EBR2, EBR3 and EBR4 are located in LS chromosomes. Similarly, copy FOXG_06378 of FOW2 is located in the core genome (chromosome 2), whereas the two paralogues are located in LS chromosomes (FOXG_12458 in chromosome 3 and FOXG_21393 in chromosome 6). Deletion of FOW2 in F. oxysporum f. sp. melonis causes a complete loss of pathogenicity (Imazaki et al., 2007), whereas FOW2 insertional mutants in F. oxysporum f. sp. lycopersici show a decrease of 50% relative to the wild‐type level of virulence (Michielse et al., 2009a), which suggests that insertions may have occurred in the paralogues.
None of the above‐described gene expansions have been shown to be involved in the main characteristic of F. oxysporum, its ability to grow and spread through the vascular system of the plant host. On the contrary, the phenotypic features of transformants attenuated in FTF expression strongly suggest a role of the FTF1 paralogues in vascular colonization. In addition to the reduction in virulence demonstrated in the pathogenicity assays, FTF‐silenced transformants show: (i) lower accumulation than HV strains of fungal biomass in infected host tissues; (ii) a change in the colonization pattern, from almost exclusively vascular to a mixture of parenchymal and vascular; and (iii) the majority of fungal biomass (68%) accumulates around the parenchymal cells instead of inside the xylem vessels. These three features are also main differences that distinguish WV strains from HV strains of F. oxysporum f. sp. phaseoli. As shown in previous studies, HV strains are true and almost exclusive vascular colonizers that highly express the FTF1 transcription factor during plant infection, whereas WV strains are better colonizers of parenchyma and lack the FTF1 paralogues (Niño‐Sánchez et al., 2015; Ramos et al., 2007). Furthermore, the presence of extra FTF1 paralogues correlates with enhanced virulence when SV strains of F. oxysporum f. sp. phaseoli infect P. coccineus L. (de Vega‐Bartol et al., 2011). Therefore, we propose that FTF1 paralogues encode a transcription factor required for enhanced virulence and heavy colonization of the host vascular system.
The FTF genes encode transcription factors of the Zn(II)2Cys6 binuclear cluster DNA‐binding type (Todd and Andrianopoulos, 1997). If the expression of FTF1 paralogues, or FTF1 together with FTF2, is needed for effective colonization and spread through the xylem vessels, their most attractive targets would be genes whose products are involved in the vascular lifestyle of F. oxysporum. The best‐known groups of molecules that work as specialized pathogenicity factors in F. oxysporum are the small, cysteine‐rich proteins encoded by the SIX genes (Michielse and Rep, 2009). SIX genes play an important role in the determination of host specificity and the establishment of gene‐for‐gene interactions between F. oxysporum f. sp. lycopersici and its tomato host (Houterman et al., 2008; Rep et al., 2004). We have found direct evidence that strongly suggests that SIX1 and SIX6 genes, both present in the genome of HV phaseoli strains, but absent in the genome of WV strains, are activated by the FTF1 transcription factor. First, mutants that constitutively express FTF1 show enhanced SIX1 and SIX6 expression. Second, FTF‐silenced transformants show reduced expression of both genes. These results support the correlation previously found between FTF1 up‐regulation during host colonization and increased expression of SIX1 and SIX6 (Niño‐Sánchez et al., 2015). It has been reported that SIX expression is dependent on the expression of another transcription factor encoded by the gene SGE1, as the expression of several SIX genes is lost in a deletion mutant of SGE1 (Michielse et al., 2009b). Our results show that SGE1 is also up‐regulated in the phaseoli strain constitutively expressing FTF1, whereas SGE1 expression is down‐regulated in FTF‐silenced strains of F. oxysporum f. sp. phaseoli. Therefore, SGE1 is also likely to be under the control of FTF1, at least in F. oxysporum f. sp. phaseoli, thus making the FTF1 transcription factor a regulator of effector expression.
The variability shown by FTF gene expansion, with up to seven similar genes in strain 4287, is higher than in the case of EBR and FOW expansions. This range of variability, together with the finding that some paralogues are probably not functional because of structural reasons (this work), suggests that the FTF gene family has evolved by gene duplication of an ancestral unique gene, most probably FTF2, and the subsequent accumulation of mutations in the duplicated copies. The FTF1 paralogues are located in LS regions enriched in transposons (Ma et al., 2010; Ramos et al., 2007; Schmidt et al., 2013), whose movement could have originated the duplications. The data presented here demonstrate that some of these mutations have determined a functional specialization of the FTF1 paralogues, which is closely related to the ability to colonize the vascular system.
Experimental Procedures
Fungal strains and culture conditions
The F. oxysporum f. sp. phaseoli strains FOP‐SP1 (HV) and FOP‐SP4 (WV) (Alves‐Santos et al., 1999, 2002a; Armstrong and Armstrong, 1975; Booth, 1971), and the F. oxysporum f. sp. lycopersici race 2 strain 4287 (Di Pietro et al., 1998), were used in this study. All strains were grown as described previously (Alves‐Santos et al., 1999; de Vega‐Bartol et al., 2011). Fungal cultures were established from frozen mycelia stored on 25% glycerol v/v at −80 °C, and incubated at 25 °C with continuous light for 1 week (solid medium) or 5 days at 120–180 rpm (liquid cultures).
Pathogenicity tests
Inoculation of P. vulgaris L. cv. Blanca Riñón with conidia from F. oxysporum strains and transformants was carried out as described previously (Alves‐Santos et al., 1999). After inoculation, the plants were transferred to pots and further incubated in a glasshouse for regular pathogenicity assays, or to 50‐mL Falcon® tubes filled with PGM (Plant Growth Medium) solution, covered with foil and incubated as described previously (Niño‐Sánchez et al., 2015). Inoculation of tomato cultivar Money Maker was performed as described previously (Di Pietro et al., 1998). Plant infection tests were repeated three times in a randomized design and a total of 60 plants were analysed. Disease severity was measured by assessment of the disease symptoms recorded according to the CIAT scale (Alves‐Santos et al., 1999; Pastor‐Corrales and Abawi, 1987), weighing the mass of the aerial part of inoculated plants, quantification of the DPR (García‐Sánchez et al., 2010) and evaluation of the affected vascular vessels according to the tomato disease index (Rep et al., 2004).
Isolation of FTF2
A genomic library was constructed with DNA from strain FOP‐SP4 as described previously (Ramos et al., 2007). To identify the FTF2 gene, 4000 recombinant phages were screened using the probe B285‐A5, which contains part of the promoter and the 5′ coding region of FTF1. DNA purified from the positive phage clones was digested with different restriction enzymes and subjected to Southern blot analysis with several probes derived from the coding region of FTF1. DNA fragments corresponding to hybridization bands were subcloned into pBluescripts KS+ vector and sequenced using the same primers as designed for sequencing of the FTF1 gene (Ramos et al., 2007). The nucleotide sequence of gene FTF2 is available in GenBank under Accession No. JN167165.
Construction of silencing and disruption vectors and generation of mutants
Plasmid pFTF2‐KO was constructed to inactivate the gene FTF2 in FOP‐SP1. To prevent the partial deletion of the FOXG_09391 ORF, contiguous to the 3′ end of FTF2 (locus FOXG_09390), primers Inac‐FTF2‐A3 and Inac‐FTF2‐A4 were designed to PCR amplify a 776‐bp DNA fragment from the 3′ end of the FTF2 coding region. An 844‐bp DNA fragment was amplified from the 5′ flanking region of FTF2 using primers Inac‐FTF2‐O1 and Inac‐FTF2‐O2. Both fragments were PCR amplified using DNA from strain FOP‐SP4 as template and cloned into pRF‐HU2 digested with PacI and Nt.BbvCI USER™ enzymes (Frandsen et al., 2008).
Plasmids pFTFPhleo‐RNAi and pFTFHyg‐RNAi were designed on the basis of the dsRNA expression cassette from pJL43‐RNAi (Ullán et al., 2008) and the binary vector pRF‐HU2 (Frandsen et al., 2008). For plasmid pFTFPhleo‐RNAi construction, first the hygromycin resistance cassette in pRF‐HU2 was excised by digestion with restriction enzymes PacI and Nt.BbvCI. Then, a PCR fragment containing the PpbcC promoter of the Penicillium chrysogenum pcbC gene and the PgpdA promoter of the Aspergillus nidulans glyceraldehyde 3‐phosphate dehydrogenase gene, together with the ble gene under the control of the glutamate dehydrogenase promoter (Pgdh) of Aspergillus awamori, was amplified using pJL43‐RNAi as template and primers RNAi‐U1 and RNAi‐U4. This 3406‐bp amplicon was cloned into pRF‐HU2 digested with PacI and Nt.BbvCI USER™ enzymes (Frandsen et al., 2008), yielding plasmid pPhleo‐RNAi. Primers SilFTF2F‐NcoI and SilFTF2R‐NcoI were used to amplify a 462‐bp fragment, containing a conserved FTF sequence, using DNA from FOP‐SP4 as template. Finally, the 474‐bp amplicon and plasmid pPhleo‐RNAi were digested with restriction enzyme NcoI and ligated using T4 DNA ligase to obtain pFTFPhleo‐RNAi. To obtain pFTFHyg‐RNAi, primers RNAi‐User1 and RNAi‐User2 were used to PCR amplify a 1.7‐kb DNA fragment employing pFTFPhleo‐RNAi as template. This amplicon contains the double‐stranded expression cassette with the FTF sequence cloned in the internal NcoI site. The 1.7‐kb DNA fragment was cloned into pRF‐HU digested with PacI and Nt.BbvCI USER™ enzymes.
Complementation of FTF2– mutants was performed by transformation with plasmid pFTF2, which contains the wild‐type allele FTF2 from strain FOP‐SP4 and the ble gene for phleomycin resistance. First, plasmid pRF‐HU was digested with the enzymes ApaI and ScaI to excise a fragment containing the hph gene. Then, a fragment containing the phleomycin resistance cassette was PCR amplified using primers PhleoF‐ApaI and PhleoR‐ScaI and DNA from plasmid pJL43‐RNAi. This amplicon was ligated to the formerly digested plasmid pRF‐HU, thus replacing the hph gene by the ble gene. Finally, a 4717‐bp amplicon containing the wild‐type allele FTF2 was obtained using primers PromFTF2USER‐F and TerFTF2USER‐R and DNA from strain FOP‐SP4 as template, and ligated to the formerly obtained plasmid digested with PacI and Nt.BbvCI USER™ enzymes.
Plasmid pFTF1 was constructed to express the gene FTF1 in the strain FOP‐SP4 under the control of its native promoter. A DNA fragment was amplified using primers B310User and M40Auser, and DNA from FOP‐SP1 as template. The 4890‐bp amplicon contains the FTF1 paralogue homologous to FOXG_14257 in F. oxysporum f. sp. lycopersici, and contains the ORF together with 670 bp of the promoter region and 1 kb of the 3′ non‐translated region. The amplicon was ligated to pRF‐HU digested with PacI and Nt.BbvCI USER™ enzymes. In a similar way, plasmid pPgpdA::FTF1 was constructed to express the gene FTF1 under the constitutive control of the PgpdA promoter and the TrpC terminator harboured by pRF‐HUE. Primers FTF1UserF and FTF1UserR were used in PCRs with DNA from FOP‐SP1 as template. The amplicon contains the complete ORF of the phaseoli paralogue homologous to FOXG_14257 in F. oxysporum f. sp. lycopersici, and was ligated to pRF‐HUE digested with PacI and Nt.BbvCI USER™ enzymes (Frandsen et al., 2008).
All the constructs were used to genetically transform F. oxysporum employing the Agrobacterium tumefaciens‐mediated transformation procedure as described previously (Mullins et al., 2001; Ramos et al., 2007).
Construction of GFP‐expressing strains
Transformants of F. oxysporum f. sp. phaseoli attenuated in the expression of FTF1 and FTF2 were genetically transformed to express GFP and verified for normal pathogenicity, as described previously (Niño‐Sánchez et al., 2015).
Confocal laser microscopy
The plants inoculated with FOP‐SP1, FOP‐SP4 and the attenuated transformants expressing GFP were maintained in hydroponic cultures and examined each day for a period of 21 days after infection, as described previously (Niño‐Sánchez et al., 2015). The quantification of xylem vessel colonization was performed with the help of ImageJ and carried out as described previously (Niño‐Sánchez et al., 2015).
Nucleic acid manipulations
Genomic DNA was extracted from F. oxysporum mycelium according to the procedures described previously (Alves‐Santos et al., 1999, 2002b; Ramos et al., 2007). RNA was extracted from common bean and tomato plants at different times after inoculation with F. oxysporum strains and transformants. Roots and hypocotyls were cut and immediately frozen at 80 °C and ground in a pestle and mortar under liquid nitrogen. Southern blots were carried out as described previously (Ramos et al., 2007). DNA probes were labelled with digoxigenin‐dUTP (Roche Diagnostics, Basel, Switzerland) by the PCR method using Biotools DNA Polymerase (Biotools SA, Madrid, Spain). Prehybridization, hybridization, washing and detection were performed using a chemiluminescent detection procedure employing CDP‐Star (Roche Diagnostics) according to the manufacturer's recommendations.
Real‐time quantitative analysis of gene expression and fungal biomass quantification
Samples of plants inoculated with F. oxysporum were collected and frozen at −80 °C. Total RNA was extracted and DNAase was treated using the SV Total RNA Isolation System Z3105 (Promega, Madison, WI, USA) according to the manufacturer's recommendations. RNA quantification and cDNA synthesis were performed as described previously (Niño‐Sánchez et al., 2015). The efficiency of each pair of primers was verified as described previously (Niño‐Sánchez et al., 2015). Amplifications were performed in a StepOnePlus™ Real‐Time PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's recommendations. The F. oxysporum EF1α gene and the common bean actin gene were used as endogenous reference genes. The relative expression levels of each gene were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001). Three sets of samples from different infection assays were used (biological replicates), two independent cDNA preparations per biological replicate were obtained and three replicates of each cDNA were analysed to calculate the mean and standard deviation. All the primers used in RT‐qPCR experiments and in the construction of vectors are listed in Table S2 (see Supporting Information).
Fungal biomass in inoculated plant samples was estimated as described previously (Niño‐Sánchez et al., 2015).
Similarity analysis
Similarity searches were performed with the blast program against public DNA/protein databases [National Center for Biotechnology Information (NCBI) and Broad Institute]. The alignment of FTF2 homologous sequences and the comparison of FTF1 and FTF2 sequences were made with the help of the Geneious program (Biomatters, Auckland, New Zealand) and its default parameters (IUB cost matrix, neighbour joining, Tamura–Nei distance model) for MUSCLE/ClustalW and GBLOCKS. Phylogenetic reconstructions were performed using PhyML and evaluated using a bootstrap test with 100 replications. The motifs, regions and domains of FTF2 were characterized at the Broad website (www.broadinstitute.org) and annotated with the Geneious program.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Nucleotide sequence homology between all members of the FTF (Fusarium transcription factor) gene family. (A) Percentages of homology between the open reading frames (ORFs). (B) Percentages of homology between the 462‐bp fragment used for silencing. The letters in parentheses indicate the type of protein deduced from the ORF of each locus.
Fig. S2 Gene expression analysis of FTF2 (Fusarium transcription factor 2) during growth in culture. The expression of the gene FTF2 was measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) in mycelia grown in liquid culture (potato dextrose broth, PDB). FOP‐SP1 and FOP‐SP4 are highly virulent (HV) and weakly virulent (WV) Fusarium oxysporum f. sp. phaseoli wild‐types, respectively. The expression measurements for each strain were referred to the arbitrary value of unity obtained after 6 h of growth. Error bars indicate the standard deviations obtained in three independent biological experiments.
Fig. S3 Southern hybridization of HindIII‐digested DNA from knockout and silenced transformants of Fusarium oxysporum. Digested DNA from knockout mutants obtained from strain FOP‐SP1 and the mutant complemented with the FTF2 (Fusarium transcription factor 2) wild‐type allele, hybridized with probe M18B‐M19A (a fragment of the FTF1 gene) (A, B). Digested DNA from attenuated transformants obtained from strain FOP‐SP1 and hybridized with probe SilFTF2F.NcoI‐SilFTF2R.NcoI which includes the silencing‐inducer FTF1 fragment (C), probe HphF‐HphR which includes the hyg resistance gene (D), and probes HphF‐HphR and PhleoF‐PhleoR, the latter including the phleo resistance gene (E). Digested DNA from attenuated transformants obtained from strain 4287 and hybridized with probe SilFTF2F.NcoI‐SilFTF2R.NcoI (F) and probe HphF‐HphR (G). SP1ΔFTF2.1–6 are knockout mutants obtained by transformation of strain FOP‐SP1; SP1ΔFTF2‐FTF2 is mutant SP1ΔFTF2.1 complemented with a wild‐type FTF2 allele; FOP‐SP1 and FOP‐SP4 are highly virulent (HV) and weakly virulent (WV) strains of F. oxysporum f. sp. phaseoli, respectively; AB82 is a non‐pathogenic F. oxysporum strain; SP1FTFRNAi.2, SP1FTFRNAi.4 and SP1FTFRNAi.5 are attenuated mutants obtained by transformation of strain FOP‐SP1 with the silencing construct containing the hph gene; SP1FTFRNAi.16 is an attenuated mutant obtained by transformation of strain FOP‐SP1 with the silencing construct containing the phleo gene; SP1FTFRNAi was obtained by transformation of FOP‐SP1 with the backbone of the silencing construct lacking the silencing‐inducer fragment; 4287FTFRNAi1–5 are attenuated mutants obtained by transformation of strain 4287 with the silencing construct containing the hph gene; 4287FTFRNAi was obtained by transformation of 4287 with the backbone of the silencing construct lacking the silencing‐inducer fragment; 4287 is an F. oxysporum f. sp. lycopersici race 2 strain.
Fig. S4 Growth in solid culture (potato dextrose agar, PDA) and sporulation in liquid culture (potato dextrose broth, PDB) of FTF2 (Fusarium transcription factor 2) mutants (SP1ΔFTF2.1 and SP1ΔFTF2.2), an FTF2 mutant complemented with the FTF2 wild‐type allele (SP1ΔFTF2‐FTF2), FTF attenuated mutants obtained by transformation of strain FOP‐SP1 (SP1FTFRNAi.2, SP1FTFRNAi.4, SP1FTFRNAi.5, SP1FTFRNAi.16 and SP1FTFRNAi) or strain 4287 (4287FTFRNAi1‐5 and 4287FTFRNAi), and the wild‐type strains FOP‐SP1 and 4287. Measurements were performed after 7 days of growth.
Fig. S5 Southern hybridization of HindIII‐digested DNA from transformants of Fusarium oxysporum. The membrane was hybridized with probe M18B‐M19A which includes the FTF1 (Fusarium transcription factor 1) gene (A) and probe HphF‐HphR which includes the hph gene (B). FOP‐SP4 is a weakly virulent (WV) wild‐type strain; SP4FTF1.1 and SP4FTF1.2 are strains derived from FOP‐SP4 by transformation with a plasmid containing the native FTF1 gene, and contain one copy and three copies, respectively, of FTF1. Size markers (kb) are indicated on the left.
Fig. S6 Fusarium wilt induced in common bean plants by Fusarium oxysporum f. sp. phaseoli strains expressing the FTF1 (Fusarium transcription factor 1) gene. Disease symptoms were measured according to the Center for Tropical Agriculture (CIAT) scale in plants inoculated with transformant strains (SP4FTF1.1 and SP4FTF1.2) harbouring copies of the native FTF1 gene (A) or with transformants harbouring the FTF1 open reading frame (ORF) constitutively expressed by the gpdA promoter (SP4PgpdA::FTF1.1–6) (B). All the transformants were derived from the weakly virulent (WV) FOP‐SP4 strain. (C) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) was carried out using RNA extracted from plant hypocotyls at 14 days post‐inoculation (dpi). FOP‐SP1 is a highly virulent (HV) strain.
Table S1 Growth and sporulation of wild‐types, knockout and attenuated transformants of Fusarium oxysporum.
Table S2 Oligonucleotides used in this study.
Acknowledgements
This research was supported by grant AGL2012‐39876‐C02‐01 from Ministerio de Economía y Competitividad ((MINECO), Spain). J. Niño‐Sánchez and V. Casado‐del Castillo were recipients of fellowships AP2009‐3559 and AP2010‐2742, respectively, from MInisterio de Educación, Cultura y Deporte ((MECD), Spain). V. Tello held a postdoctoral position financed by grant AGL2012‐39876‐C02‐01. We are indebted to Ricardo V. Ullán for the pJL43‐RNAi plasmid and excellent advice on gene silencing, and M. R. Thon and E. P. Benito for critical reading of the manuscript and many valuable suggestions.
References
- Alves‐Santos, F.M. , Benito, E.P. , Eslava, A.P. and Diaz‐Minguez, J.M. (1999) Genetic diversity of Fusarium oxysporum strains from common bean fields in Spain. Appl. Environ. Microbiol. 65, 3335–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves‐Santos, F. , Cordeiro‐Rodrigues, L. , Sayagués, J.M. , Martín‐Dominguez, R. , García‐Benavides, P. , Crespo, M.C. , Díaz‐Mínguez, J.M. and Eslava, A.P. (2002a) Pathogenicity and race characterization of Fusarium oxysporum f. sp. phaseoli isolates from Spain and Greece. Plant Pathol. 51, 605–611. [Google Scholar]
- Alves‐Santos, F.M. , Ramos, B. , García‐Sánchez, M.A. , Eslava, A.P. and Díaz‐Mínguez, J.M. (2002b) A DNA‐based procedure for in planta detection of Fusarium oxysporum f. sp. phaseoli . Phytopathology, 92, 237–244. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.M. and Armstrong, J.K. (1975) Reflections on the Wilt Fusaria. Annu. Rev. Phytopathol. 13, 95–103. [Google Scholar]
- Armstrong, G.M. and Armstrong, J.K. (1981) Formae speciales and races of Fusarium oxysporum and Alternaria alternata In: Fusarium: Disease, Biology and Taxonomy (Cook R., ed), pp. 391–399. Penn State University Press. University Park, Pennsylvania. [Google Scholar]
- Booth, C. (1971) The Genus Fusarium. Kew, Surrey: Commonwealth Mycological Institute. [Google Scholar]
- Di Pietro, A. , García‐Maceira, F. , Huertas‐González, M. , Ruíz‐Roldan, M. , Caracuel, Z. , Barbieri, A. and Roncero, M. (1998) Endopolygalacturonase PG1 in different formae speciales of Fusarium oxysporum . Appl. Environ. Microbiol. 64, 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen, R.J. , Andersson, J.A. , Kristensen, M.B. and Giese, H. (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Sánchez, M. A. , Martín‐Rodrigues, N. , Ramos, B. , de Vega‐Bartol, J.J. , Perlin, M.H. and Díaz‐Mínguez, J.M. (2010) fost12, the Fusarium oxysporum homolog of the transcription factor Ste12, is upregulated during plant infection and required for virulence. Fungal Genet. Biol. 47, 216–225. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathog. 4, e1000061–e1000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki, I. , Kurahashi, M. , Iida, Y. and Tsuge, T. (2007) Fow2, a Zn(II)2Cys6‐type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum . Mol. Microbiol. 63, 737–753. [DOI] [PubMed] [Google Scholar]
- Jonkers, W. , Xayamongkhon, H. , Haas, M. , Olivain, C. , van der Does, H.C. , Broz, K. , Rep, M. , Alabouvette, C. , Steinberg, C. and Kistler, H.C. (2013) EBR1 genomic expansion and its role in virulence of Fusarium species. Environ. Microbiol. 16, 1982–2003. [DOI] [PubMed] [Google Scholar]
- Lee, N.S. , Dohjima, T. , Bauer, G. , Li, H. , Li, M.J. , Ehsani, A. , Salvaterra, P. and Rossi, J. (2002). Expression of small interfering RNAs targeted against HIV‐1 rev transcripts in human cells. Nat. Biotechnol. 20, 500–505. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, L.‐J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, G.S. , McGoldrick, C.A. , Holt, C.L. and Denison, S.H. (1992) The bimB3 mutation of Aspergillus nidulans uncouples DNA replication from the completion of mitosis. J. Biol. Chem. 267, 15 737–15 743. [PubMed] [Google Scholar]
- Michielse, C.B. and Rep, M. (2009) Pathogen profile update: Fusarium oxysporum . Mol. Plant Pathol. 10, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Cornelissen, B.J. and Rep, M. (2009. a) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. Genome Biol. 10, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Manders, E.M.M. , Boas, S. , Olivain, C. , Alabouvette, C. and Rep, M. (2009. b) The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog. 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, E. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Niño‐Sánchez, J. , Tello, V. , Casado‐del Castillo, V. , Thon, M.R. , Benito, E.P. and Díaz‐Mínguez, J.M. (2015) Gene expression patterns and dynamics of the colonization of common bean (Phaseolus vulgaris L.) by highly virulent and weakly virulent strains of Fusarium oxysporum . Front. Microbiol. 6, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor‐Corrales, M.A. and Abawi, G.S. (1987) Reactions of selected bean germ plasms to infection by Fusarium oxysporum f. sp. phaseoli . Plant Dis. 71, 990–993. [Google Scholar]
- Ramos, B. , Alves‐Santos, F.M. , García‐Sánchez, M.A. , Martín‐Rodrigues, N. , Eslava, A.P. and Díaz‐Mínguez, J.M. (2007) The gene coding for a new transcription factor (ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet. Biol. 44, 864–876. [DOI] [PubMed] [Google Scholar]
- Rep, M. , van der Does, H.C. , Meijer, M. , van Wijk, R. , Houterman, P.M. , Dekker, H.L. , de Koster, C.G. and Cornelissen, B.J.C. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Schjerling, P. and Holmberg, S. (1996) Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S.M. , Houterman, P.M. , Schreiver, I. , Ma, L. , Amyotte, S. , Chellappan, B. , Boeren, S. , Takken, F.L.W. and Rep, M. (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum . BMC Genomics, 14, 119–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, U. , Smith, N.A. , Kazan, K. , Ayliffe, M. and Wang, M.‐B. (2013) Analysis of hairpin RNA transgene‐induced gene silencing in Fusarium oxysporum . Silence, 4, 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. , Seo, Y.‐S. , Min, K. , Park, A.R. , Lee, J. , Jin, J.M. , Lin, Y. , Cao, P. , Hong, S.Y. , Kim, E.K. , Lee, S.H. , Cho, A. , Lee, S. , Kim, M.G. , Kim, Y. , Kim, J.E. , Kim, J.C. , Choi, G.J. , Yun, S.H. , Lim, J.Y. , Kim, M. , Lee, Y.H. , Choi, Y.D. and Lee, Y.W. (2011) A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog. 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, R.B. and Andrianopoulos, A. (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21, 388–405. [DOI] [PubMed] [Google Scholar]
- Ullán, R.V. , Godio, R.P. , Teijeira, F. , Vaca, I. , García‐Estrada, C. , Feltrer, R. , Kosalkova, K. and Martín, J.F. (2008) RNA‐silencing in Penicillium chrysogenum and Acremonium chrysogenum: validation studies using β‐lactam genes expression. J. Microbiol. Methods, 75, 209–218. [DOI] [PubMed] [Google Scholar]
- de Vega‐Bartol, J.J. , Martín‐Dominguez, R. , Ramos, B. , García‐Sánchez, M.‐A. and Díaz‐Mínguez, J.M. (2011) New virulence groups in Fusarium oxysporum f. sp. phaseoli: the expression of the gene coding for the transcription factor ftf1 correlates with virulence. Phytopathology, 101, 470–479. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Teckle, E. , Oubrahim, H. , Mieyal, J.J. , Stadtman, E.R. and Chock, P.B. (2003). Stable and controllable RNA interference: investigating the physiological function of glutathionylated actin. Proc. Natl. Acad. Sci. U.S.A. 100: 5103–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Waalwijk, C. , de Wit, P.J.G.M. , van der Lee, T. and Tang, D. (2011) EBR1, a novel Zn(2)Cys(6) transcription factor, affects virulence and apical dominance of the hyphal tip in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 1407–1418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Nucleotide sequence homology between all members of the FTF (Fusarium transcription factor) gene family. (A) Percentages of homology between the open reading frames (ORFs). (B) Percentages of homology between the 462‐bp fragment used for silencing. The letters in parentheses indicate the type of protein deduced from the ORF of each locus.
Fig. S2 Gene expression analysis of FTF2 (Fusarium transcription factor 2) during growth in culture. The expression of the gene FTF2 was measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) in mycelia grown in liquid culture (potato dextrose broth, PDB). FOP‐SP1 and FOP‐SP4 are highly virulent (HV) and weakly virulent (WV) Fusarium oxysporum f. sp. phaseoli wild‐types, respectively. The expression measurements for each strain were referred to the arbitrary value of unity obtained after 6 h of growth. Error bars indicate the standard deviations obtained in three independent biological experiments.
Fig. S3 Southern hybridization of HindIII‐digested DNA from knockout and silenced transformants of Fusarium oxysporum. Digested DNA from knockout mutants obtained from strain FOP‐SP1 and the mutant complemented with the FTF2 (Fusarium transcription factor 2) wild‐type allele, hybridized with probe M18B‐M19A (a fragment of the FTF1 gene) (A, B). Digested DNA from attenuated transformants obtained from strain FOP‐SP1 and hybridized with probe SilFTF2F.NcoI‐SilFTF2R.NcoI which includes the silencing‐inducer FTF1 fragment (C), probe HphF‐HphR which includes the hyg resistance gene (D), and probes HphF‐HphR and PhleoF‐PhleoR, the latter including the phleo resistance gene (E). Digested DNA from attenuated transformants obtained from strain 4287 and hybridized with probe SilFTF2F.NcoI‐SilFTF2R.NcoI (F) and probe HphF‐HphR (G). SP1ΔFTF2.1–6 are knockout mutants obtained by transformation of strain FOP‐SP1; SP1ΔFTF2‐FTF2 is mutant SP1ΔFTF2.1 complemented with a wild‐type FTF2 allele; FOP‐SP1 and FOP‐SP4 are highly virulent (HV) and weakly virulent (WV) strains of F. oxysporum f. sp. phaseoli, respectively; AB82 is a non‐pathogenic F. oxysporum strain; SP1FTFRNAi.2, SP1FTFRNAi.4 and SP1FTFRNAi.5 are attenuated mutants obtained by transformation of strain FOP‐SP1 with the silencing construct containing the hph gene; SP1FTFRNAi.16 is an attenuated mutant obtained by transformation of strain FOP‐SP1 with the silencing construct containing the phleo gene; SP1FTFRNAi was obtained by transformation of FOP‐SP1 with the backbone of the silencing construct lacking the silencing‐inducer fragment; 4287FTFRNAi1–5 are attenuated mutants obtained by transformation of strain 4287 with the silencing construct containing the hph gene; 4287FTFRNAi was obtained by transformation of 4287 with the backbone of the silencing construct lacking the silencing‐inducer fragment; 4287 is an F. oxysporum f. sp. lycopersici race 2 strain.
Fig. S4 Growth in solid culture (potato dextrose agar, PDA) and sporulation in liquid culture (potato dextrose broth, PDB) of FTF2 (Fusarium transcription factor 2) mutants (SP1ΔFTF2.1 and SP1ΔFTF2.2), an FTF2 mutant complemented with the FTF2 wild‐type allele (SP1ΔFTF2‐FTF2), FTF attenuated mutants obtained by transformation of strain FOP‐SP1 (SP1FTFRNAi.2, SP1FTFRNAi.4, SP1FTFRNAi.5, SP1FTFRNAi.16 and SP1FTFRNAi) or strain 4287 (4287FTFRNAi1‐5 and 4287FTFRNAi), and the wild‐type strains FOP‐SP1 and 4287. Measurements were performed after 7 days of growth.
Fig. S5 Southern hybridization of HindIII‐digested DNA from transformants of Fusarium oxysporum. The membrane was hybridized with probe M18B‐M19A which includes the FTF1 (Fusarium transcription factor 1) gene (A) and probe HphF‐HphR which includes the hph gene (B). FOP‐SP4 is a weakly virulent (WV) wild‐type strain; SP4FTF1.1 and SP4FTF1.2 are strains derived from FOP‐SP4 by transformation with a plasmid containing the native FTF1 gene, and contain one copy and three copies, respectively, of FTF1. Size markers (kb) are indicated on the left.
Fig. S6 Fusarium wilt induced in common bean plants by Fusarium oxysporum f. sp. phaseoli strains expressing the FTF1 (Fusarium transcription factor 1) gene. Disease symptoms were measured according to the Center for Tropical Agriculture (CIAT) scale in plants inoculated with transformant strains (SP4FTF1.1 and SP4FTF1.2) harbouring copies of the native FTF1 gene (A) or with transformants harbouring the FTF1 open reading frame (ORF) constitutively expressed by the gpdA promoter (SP4PgpdA::FTF1.1–6) (B). All the transformants were derived from the weakly virulent (WV) FOP‐SP4 strain. (C) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) was carried out using RNA extracted from plant hypocotyls at 14 days post‐inoculation (dpi). FOP‐SP1 is a highly virulent (HV) strain.
Table S1 Growth and sporulation of wild‐types, knockout and attenuated transformants of Fusarium oxysporum.
Table S2 Oligonucleotides used in this study.


