Summary
Molecular interaction between the causal agent of blackleg disease, Leptosphaeria maculans (Lm), and its host, Brassica napus, is largely unknown. We applied a deep RNA‐sequencing approach to gain insight into the pathogenicity mechanisms of Lm and the defence response of B. napus. RNA from the infected susceptible B. napus cultivar Topas DH16516, sampled at 2‐day intervals (0–8 days), was sequenced and used for gene expression profiling. Patterns of gene expression regulation in B. napus showed multifaceted defence responses evident by the differential expression of genes encoding the pattern recognition receptor CERK1 (chitin elicitor receptor kinase 1), receptor like proteins and WRKY transcription factors. The up‐regulation of genes related to salicylic acid and jasmonic acid at the initial and late stages of infection, respectively, provided evidence for the biotrophic and necrotrophic life stages of Lm during the infection of B. napus cotyledons. Lm transition from biotrophy to necrotropy was also supported by the expression function of Lm necrosis and ethylene‐inducing (Nep‐1)‐like peptide. Genes encoding polyketide synthases and non‐ribosomal peptide synthetases, with potential roles in pathogenicity, were up‐regulated at 6–8 days after inoculation. Among other plant defence‐related genes differentially regulated in response to Lm infection were genes involved in the reinforcement of the cell wall and the production of glucosinolates. Dual RNA‐sequencing allowed us to define the Lm candidate effectors expressed during the infection of B. napus. Several candidate effectors suppressed Bax‐induced cell death when transiently expressed in Nicotiana benthamaina leaves.
Keywords: blackleg, canola, defence, effector, Leptosphaeria maculans, RNA‐sequencing
Introduction
Blackleg disease is a major cause of yield losses on oilseed rape canola (Brassica napus), the second largest oilseed crop in the world. The disease is caused by the hemibiotrophic fungus Leptosphaeria maculans (Lm), which belongs to the Dothideomycete class of ascomycetes (West et al., 2001). Infection begins with the germination of ascospores (primary source of infection) and asexual spores (pycnidiospores) on the cotyledons and leaves of young B. napus seedlings. After germination, Lm hyphae penetrate through the wounds and stomata, and, as infection progresses, hyphae continue to grow between the cells and into the stem, eventually causing lesions at the base of the stem (stem canker) (Fitt et al., 2006; Rouxel et al., 2011). Disease symptoms on the cotyledons and leaves of Brassica species are visible as the formation of chlorotic tissue surrounding the site of infection, which develops into lesions with pycnidia occurring as black dots on the surface. Blackleg is a challenging disease to study as it exhibits an extremely complex life cycle in the host plant. Lm starts its life cycle as a biotroph during infection of the cotyledons and leaves, as an endophyte whilst growing in the stem and, finally, as a necrotroph leading to the formation of canker (Rouxel and Balesdent, 2005; Rouxel et al., 2011; West et al., 2001). The resistance of B. napus to Lm at the cotyledon stage is race specific. To date, 16 cotyledon resistance (R) genes (Rlm1–Rlm11, RlmS, LepR1–LepR4) have been reported from Brassica species (Raman et al., 2013). Two of these genes, LepR3 and Rlm2, encoding receptor‐like proteins (RLPs), have been cloned (Larkan et al., 2013, 2015). Six Lm avirulence (effector) genes (AvrLm1, AvrLm2, AvrLm4‐7, AvrLm6, AvrLmJ1 and AvrLm11), corresponding to the above‐named R genes, have been cloned (Balesdent et al., 2013; Fudal et al., 2007; Ghanbarnia et al., 2015; Gout et al., 2006; Parlange et al., 2009; Van de Wouw et al., 2014). The genome sequence of the Lm isolate v23.1.3 revealed that the majority of the known and predicted effectors are located within AT‐rich isochores (Rouxel et al., 2011; Van de Wouw et al., 2014). Despite these advances, there has been no comprehensive study describing the regulation of plant defence responses and pathogen virulence genes during the foliar infection of B. napus.
Dual RNA‐sequencing (RNA‐seq) allows accurate gene expression analysis of plant pathogens and their respective hosts simultaneously, and has been applied to investigate biotrophic and necrotrophic fungal–host interactions (Metzker, 2010; Westermann et al., 2012). The recent release of the B. napus cv. Darmor genome sequence (Chalhoub et al., 2014) and the availability of a reference genome for Lm (Rouxel et al., 2011) have provided the opportunity to investigate the molecular interaction in the B. napus–Lm pathosystem. To date, the only RNA‐seq reported for the B. napus–Lm interaction is the study by Lowe et al. (2014). They compared the expression of Lm and B. napus genes at 7 and 14 days after inoculation (dai). Although limited in its scope (only two time points and a total of 164.9 million reads), this work provided a general view of the gene expression profile during the Lm–B. napus interaction (Lowe et al., 2014). These authors, as well as Šašek et al. (2012), reported the up‐regulation of the plant hormones salicylic acid (SA) and ethylene (ET) in B. napus in response to Lm infection. Diverse plant hormones play a central role in the regulation of plant immune responses (Katagiri and Tsuda, 2010; Pieterse et al., 2012). The roles of the plant hormones SA, jasmonic acid (JA) and ET in plant defence have been studied extensively. SA has a major role in resistance against biotrophic pathogens, whereas JA and ET regulate plant defence against necrotrophs (Glazebrook, 2005). Other hormones, such as abscisic acid (ABA), gibberellins (GAs), auxins, cytokinins (CKs), brassinosteroids (BRs) and nitric oxide (NO), have also been reported to function as modulators of the plant immune response (Pieterse et al., 2012).
To obtain a comprehensive gene expression profile for both the pathogen and host during a compatible interaction between B. napus and Lm, RNA prepared from the cotyledon tissues of the susceptible B. napus cv. Topas at 0, 2, 4, 6 and 8 dai was sequenced. In total, 1.38 billion reads for four biological replicates were generated using the Illumina HiSeq‐2500 platform. These data enabled us to define the lifestyle transition of Lm and to reveal the regulation of B. napus defence genes during cotyledon infection. In addition, we present the expression profile of known and candidate Lm effector genes during infection and provide evidence that supports the virulence function of some of the predicted effectors.
Results
Disease progression on B. napus seedlings
One‐week‐old cotyledons of the doubled‐haploid (DH) line B. napus cv. Topas DH16516 were inoculated with pycnidiospores of Lm isolate 00‐100. Under our inoculation protocol, infection initiated by the pycnidiospores continued without any visible symptoms for the first 3–4 dai. At 4 dai, a chlorotic ring surrounding the inoculation site became clearly visible, which later (8 dai) expanded, leading to the formation of a lesion and to tissue collapse around the site of inoculation (Fig. 1A). RNA was prepared from the cotyledon discs surrounding the site of inoculation, sampled at 0, 2, 4, 6 and 8 dai.
Figure 1.
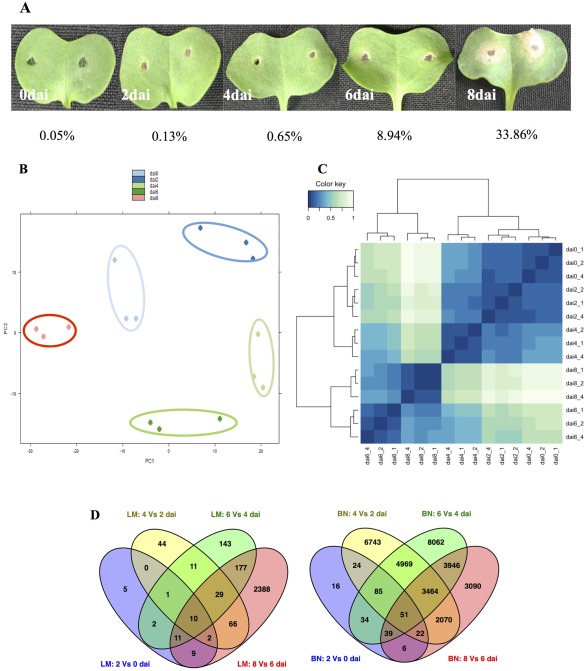
Disease symptoms on cotyledons of the susceptible Brassica napus cv. DH‐Topas 16516 (DHT) infected with Leptosphaeria maculans isolate 00‐100. (A) Cotyledons were photographed at 0, 2, 4, 6 and 8 days after inoculation (dai). The percentages of reads aligned to the genome of L. maculans isolate v23.1.3 for each time point are presented. (B, C) Global evaluation of RNA‐sequencing in L. maculans. Principal component analysis (PCA) (B) and clustering image map (CIM) (C) display a clear distinction among the transcriptome of L. maculans at different time points. (D) Differentially expressed genes (DEGs) that are unique or shared among various time point comparisons in 00‐100 (left, LM) and DHT (left, BN). The numbers of DEGs are noted in each section of the Venn diagrams.
RNA‐sequencing
We applied the dual deep RNA‐seq approach to simultaneously assess genome‐wide expression profiling of B. napus and Lm at five different time points. For each time point, 28.7–115.6 million paired‐end reads were produced and a total of 1.38 billion RNA‐Seq reads were generated for four biological replicates (Table S1, see Supporting Information). In addition, 15.4 million paired‐end RNA‐seq reads were generated from an in vitro‐grown Lm culture. Based on the count distribution, density plots (Fig. S1A, see Supporting Information), clustering of the sample‐to‐sample distances (Fig. S1B) and the MA‐plot (Fig. S1C), the best three of four RNA‐seq replicates were chosen for gene expression analysis. All reads were mapped to the genome sequence of Lm isolate v23.1.3 and the B. napus cv. Darmor reference genome (Chalhoub et al., 2014; Rouxel et al., 2011). The percentages of reads, for each time course, mapped to the pathogen and host genomes are indicated in Fig. 1 and Table S1.
Differentially expressed genes (DEGs) were detected using the DESeq2 package. Genes with a false discovery rate‐Benjamini–Hochberg (FDR‐BH) of less than 0.05 were considered to be differentially expressed. Variability among the samples was determined by preparing a clustering image map (CIM) of the sample‐to‐sample distance matrix and principal component analysis (PCA). The PCA and CIM analysis displayed a clear distinction in the transcriptome of Lm at different time points (Fig. 1B,C).
Lm gene expression profile during infection
The expression profile of 12,469 Lm predicted genes showed that close to 74% of the Lm genes were expressed during the B. napus infection [reads per kilobase of exon per million mapped reads (RPKM) > 2] and 56% were expressed during Lm in vitro growth. A total of 2898 genes showed differential expression (FDR‐BH < 0.05) in at least one time point, with 621 only expressed during host infection (no expression during in vitro growth) (Fig. 1D). Genes with significant changes in expression were determined by pair‐wise comparison of the infection time course (i.e. 2 vs. 0 dai, 4 vs. 2 dai, 6 vs. 4 dai, 8 vs. 6 dai). The total numbers of DEGs for each of these comparisons were 40, 163, 384 and 2692, respectively. A major portion (40%) of the DEGs at the earlier time points during infection (2 vs. 0 dai) included the predicted secreted protein‐encoding genes (SPs), which, in addition to effectors (SP) of unknown function, consisted of the plant cell wall‐degrading enzymes (PCWDEs). The most prevalent PCWDEs at this early stage of infection were the pectin‐degrading enzymes, such as glycoside hydrolase (GH) family 28, GH105 and several pectate lyases (PL1, PL9, PL10, PL11). Pectin is the main component of the cell wall of dicots and, when depolymerized, the plant cell wall becomes more vulnerable and accessible to other PCWDEs (Malinovsky et al., 2014). The Lm genes encoding for chitinases were highly expressed at 2 dai, which is consistent with the role of these enzymes in the growth and development of fungi. The Lm pattern of gene expression at 4 dai was similar to that at 2 dai, with 25% of the total DEGs annotated as potential effectors. In addition to GH and PL classes of Carbohdrate‐Active enZYmes (CAZy), several genes containing carbohydrate‐binding modules (CBMs), including CBM50, which binds chitin (LysM domain‐containing proteins), were differentially expressed at 2 and 4 dai. The expression profiles of Lm genes during the first 4 days of Brassica infection support an initial biotrophic establishment of Lm, which was evident by the relative abundance of effectors (secreted by the pathogen to suppress plant defence) and the reduced number of PCWDEs (largely consisting of pectinases). However, the pattern of DEGs at 6 and 8 dai was more suited to a necrotrophic lifestyle. At these latter time points, predicted secreted proteins constituted a small portion of the total DEGs (15% at 6 dai and 6.9% at 8 dai) and CAZy‐encoding genes at 6 and 8 dai were more abundant and diverse. A notable change in the CAZy profile at 6 and 8 dai was the expansion of GHs and CAZy belonging to the auxiliary activities (AA) group. AAs help GH gain access to the carbohydrates embedded in the plant cell wall (Levasseur et al., 2013).
Other genes that were differentially expressed during infection were genes related to nucleotide and amino acid biosynthesis, starch and sucrose metabolism, nucleotide and amino acid sugar metabolism, and sugar and other transporter families. Differential expression of transporter genes was observed as early as 2 dai and a major facilitator superfamily (MFS) of sugar transporters was highly expressed at this time point. Among the predicted transporter genes with significant changes in expression level, sugar/carbohydrate transporters were prevalent at 4, 6 and 8 dai. There was a significant increase in the numbers and types of transporter genes differentially expressed at 8 dai (amino acid and metal ion transporters) coinciding with the lysis of the host plant tissues and the release of nutrients, as well as toxins and antimicrobial compounds, at this stage.
A search for the genes involved in the biosynthesis of secondary metabolites identified 25 core genes, including polyketide synthases (PKSs), non‐ribosomal peptide synthetases (NRPSs) and dimethylallyltryptophan synthetase/CymD prenyl transferase (DMATS) (Fig. S2, see Supporting Information). Fungal secondary metabolites have diverse functions, including their role as virulence factors in necrotrophic fungi through the production of toxins (Cho, 2015). Using the software SMURF (Khaldi et al., 2010), we predicted 14 PKSs for Lm. The expression of Lm secondary metabolite genes was increased at 6 dai and the majority were highly up‐regulated at 8 dai, which coincides with the necrotrophic stages of Lm during B. napus cotyledon infection. Our results showed significant up‐regulation of PKS6, PKS10, PKS13 and PKS14 at 8 dai, but no transcript was detected for PK2, PK4, PK8, PK11 and PK12 (Fig. S2). Lm PKS10 shows 92% similarity to PKS responsible for the biosynthesis of melanin, a secondary metabolite that contributes to fungal structures and has been reported to play a role in the pathogenicity of human and plant fungi (Elliott et al., 2013; Jacobson, 2000; Scharf et al., 2014). PKS13 and PKS14 have been reported to be required for the production of phomenoic acid, an Lm metabolite that is toxic towards L. biglobosa, another pathogen of B. napus. It has been suggested that the production of phomenoic acid helps Lm to outcompete other fungi in its environmental niche (Elliott et al., 2013). Lm PKS6 has been reported to have an unusual structure by containing both a reductase and a methyl transferase domain not reported previously (Elliott et al., 2013). The expression of the Cladosporium fulvum PKS6 gene has been reported to be reduced during the initial stages of fungal growth, but elevated during the later stages of infection, when conidiophores emerge from the infected plants (Collemare et al., 2014). Another class of enzyme involved in the production of secondary metabolites is the NRPSs. Lm NPRSs have been reported to be involved in the production of the non‐host‐selective phytotoxin, sirodesmin PL. Lm lines with a mutation in an Lm NPRS gene named ‘SirP’ were less virulent during the colonization of B. napus stem, but not cotyledon (Elliotte et al., 2007). We searched the Lm predicted proteins to identify SirP homologues. SirP (jgi.p|Lepmu|2880) was not expressed during any time course in our experiment. Elliotte et al. (2007) detected the expression of this gene in infected cotyledons starting at 10 dai. We also found four genes (jgi.p|Lepmu|1264, jgi.p|Lepmu|1265, jgi.p|Lepmu|1295 and jgi.p|Lepmu|904) that encoded for proteins with significant homology (blastP; E value = 8E‐90, 8E‐51, 3E‐93, 6E‐105) to SirP. All of these four genes encoding for NRPs were differentially expressed during cotyledon infection (Fig. S2); however, their roles in the pathogenicity of Lm have not yet been established.
Defining Lm effectors
All of the Lm effectors identified to date are located within the AT‐rich isochores of the genome (Rouxel et al., 2011; Van de Wouw et al., 2014), with the exception of AvrLm2, which resides in a GC island within the AT‐rich region of the AvrLm1‐2‐6 cluster (Ghanbarnia et al., 2015). Rouxel et al. (2011) identified 122 predicted effectors to be located within the AT‐rich blocks of the Lm genome.
To search for the Lm candidate effectors, we adopted the pipeline (Fig. 2A) that shares common features with the effector predication pipelines described for the filamentous plant pathogens (Guyon et al., 2014; Pedersen et al., 2012; Sperschneider et al., 2015). In total, 668 SPs, 310 of which were small (<300 amino acid) secreted proteins (SSPs), were identified. Included within the SSPs were all the previously cloned AvrLm genes, except for AvrLm1, which was absent from our list of predicted effectors because of the deletion of the 43‐bp N‐terminal fragment (expanding the signal peptide) for the predicted AvrLm1 protein (jgi.p|Lepmu1|10780, Lema_P049660.1; accession number: XM_00385577). Of the 668 SPs, 162 genes had no detectable transcript during infection and 210 genes showed differential expression (Fig. 2B), with 80 being SSPs (Fig. 2C, Table S2, see Supporting Information). These 80 SSPs were considered as the most likely set of Lm effectors. Within the 80 SSPs, expression of the known effector genes AvrLm2, AvrLm6 and AvrLmJ1 was up‐regulated at 4 and 6 dai, and then reduced at 8 dai, confirming the previously reported pattern of expression for these genes (Ghanbarnia et al., 2015). AvrLm1 and AvrLm11 were deleted and AvrLm4‐7 was not expressed in the Lm isolate 00‐100 used in our study. One of the predicted SSPs (Lema_T086540.1) had homology (42% identity) to Six5, an effector from Fusarium oxysporum reported by Ma et al. (2015). Lema_T086540.1 was significantly up‐regulated at 2 and 4 dai but was not expressed during in vitro growth (Fig. S3A, B, see Supporting Information). Sequence alignment revealed the presence of six cysteines that were conserved between Lema_T086540.1 and Six5 (Fig. S3C).
Figure 2.
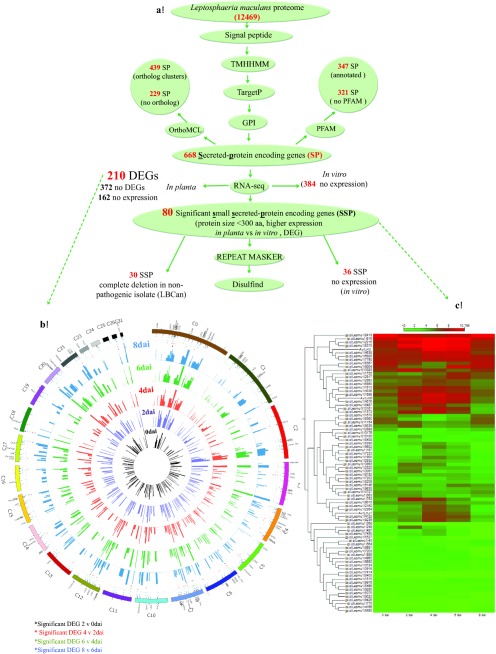
Leptosphaeria maculans (Lm) secretome prediction pipeline. (A) Features used to predict and classify effectors of Lm. aa, amino acid; DEG, differentially expressed gene; GPI, Glycophosphatidylinositol‐anchor; RNA‐seq, RNA‐sequencing; (B) Lm genes encoding predicted secreted proteins (SPs) and their expression levels during different time points. The outermost ring shows 210 of 668 SPs in the Lm genome that were differentially expressed in at least one time point comparison. Circles (from inside to outside) represent the expression of each gene at 0 (black), 2 (purple), 4 (red), 6 (green) and 8 (blue) days after inoculation (dai). Known effectors AvrLm2, AvrLm6 and AvrLmJ1, located on supercontigs 6 and 7 of Lm v23.1.3, are highlighted in red. (C) Heat map of the 80 predicted small secreted protein (SSP)‐encoding genes differentially expressed during infection at different time points. The expression of each gene is based on regularized logarithmic transformation of the average of the best three biological replicates using DESeq2. The Euclidean distance for the distance measure and average linkage for cluster linkage criteria were selected.
Sixteen of the 80 SSPs that showed very low variance among the biological replicates and were highly expressed during infection (Table 1) were selected for transient assay in Nicotiana benthamiana. Four of these SSPs partially or fully suppressed cell death induced by the mouse pro‐apoptotic Bax gene, when co‐expressed in N. benthamiana (Fig. 3A).
Table 1.
A list of 16 highly expressed Leptosphaeria maculans predicted effectors selected for biological assay.
| Pfam | Gene ID | Protein ID | Super contig | Start | End | Protein size | Cys* | DB† | Complete deletion in non‐pathogenic isolate (LBCan)‡ |
|---|---|---|---|---|---|---|---|---|---|
| – | jgi|Lepmu1|616 | Lema_T006160.1 | 0 | 2286679 | 2287371 | 213 | 6 | 3 | Yes |
| – | jgi|Lepmu1|7450 | Lema_T023440.1 | 2 | 76723 | 77286 | 121 | 3 | 0 | Yes |
| – | jgi|Lepmu1|8487 | Lema_T033060.1 | 3 | 287921 | 288676 | 123 | 8 | 4 | Yes |
| – | jgi|Lepmu1|8681 | Lema_T035000.1 | 3 | 1031016 | 1031559 | 124 | 7 | 2 | Yes |
| – | jgi|Lepmu1|9004 | Lema_T038230.1 | 3 | 2336898 | 2337410 | 114 | 6 | 3 | |
| – | jgi|Lepmu1|9010 | Lema_T038290.1 | 4 | 69859 | 70849 | 291 | 12 | >5 | Yes |
| – | jgi|Lepmu1|1613 | Lema_T076380.1 | 10 | 1509440 | 1509941 | 96 | 6 | 3 | Yes |
| – | jgi|Lepmu1|1698 | Lema_T086540.1 | 11 | 92980 | 93502 | 121 | 7 | 3 | Yes |
| – | jgi|Lepmu1|2162 | Lema_T083940.1 | 12 | 646590 | 646977 | 129 | 7 | 3 | Yes |
| – | jgi|Lepmu1|2216 | Lema_T084480.1 | 12 | 827862 | 828642 | 187 | 6 | 3 | Yes |
| – | jgi|Lepmu1|2264 | Lema_T084960.1 | 12 | 946778 | 947374 | 144 | 5 | 2 | |
| – | jgi|Lepmu1|2817 | Lema_T081180.1 | 13 | 1193860 | 1194346 | 104 | 5 | 2 | Yes |
| – | jgi|Lepmu1|2863 | Lema_T081640.1 | 13 | 1352945 | 1353440 | 91 | 9 | 4 | Yes |
| – | jgi|Lepmu1|3097 | Lema_T095640.1 | 14 | 681916 | 682620 | 178 | 10 | >5 | |
| CAP | jgi|Lepmu1|4336 | Lema_T100080.1 | 17 | 427889 | 428585 | 232 | 5 | 0 | |
| – | jgi|Lepmu1|6375 | Lema_T113200.1 | 20 | 228261 | 228840 | 193 | 5 | 1 |
*Cysteine residues.
†DISULFIND (Ceroni et al., 2006) was also used to predict the disulfide bonding (DB) state of cysteines and their disulfide connectivity.
‡Unpublished data.
Figure 3.
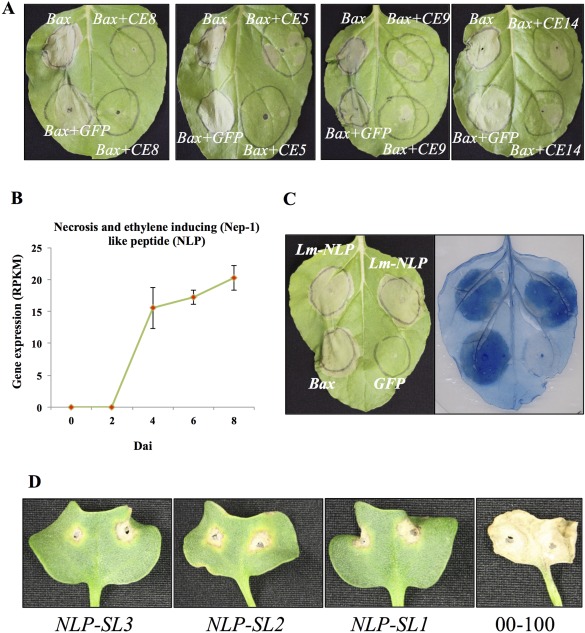
Suppression of BAX‐induced cell death (CD) by Leptosphaeria maculans candidate effectors in Nicotiana benthamiana (Nb) leaves. Agrobacterium tumefaciens (Ag) strains expressing individual L. maculans small secreted proteins (SSPs) and Bax were infiltrated into Nb leaves. Ag carrying pGWB451‐GFP was used as a negative control. BAX‐induced cell death was scored at 4 days after infiltration. (A) Examples of complete suppression of CD by CE8 (jgi|Lepmu1|2216; Lema_T084480.1) and partial suppression of CD by CE5 (jgi|Lepmu1|4336; Lema_T100080.1), CE9 (jgi|Lepmu1|616; Lema_T006160.1) and CE14 (jgi|Lepmu1|9004; Lema_T038230.1) effectors. (B–D) Leptosphaeria maculans necrosis and ethylene‐inducing peptide‐1 (Nep‐1)‐like protein (NLP) (Lm‐NLP) are required for the complete virulence of Lm. (B) Expression of Lm‐NLP at different time points. (C) Transient expression of Lm‐NLP in Nb leaves induces hypersensitive response (HR)‐like cell death. Nb leaves were infiltrated with Ag cultures containing Lm‐NLP, Bax and green fluorescent protein (GFP). Bax and GFP served as positive and negative controls, respectively. Leaves were photographed 5 days after infiltration (left panel) and cell death was visualized by trypan blue staining of the same leaves (right panel). (D) Cotyledons of 7‐day‐old seedlings of Brassica napus cv. DH‐Topas 16516 were inoculated with the L. maculans wild‐type isolate 00‐100 and the three Lm‐NLP silenced (RNAi) lines (NLP‐SL1, NLP‐SL2 and NLP‐SL3), and were photographed at 14 days after inoculation.
The necrosis and ET‐inducing peptide‐1 (Nep‐1)‐like protein (NLP) (Gijzen and Nürnberger, 2006) was also differentially expressed. NLP expression in hemibiotrophic fungi coincides with the initiation of the necrotrophic phase (Qutob et al., 2002; Zaparoli et al., 2011). Expression of Lm NLP (Lm‐NLP) was first detected at 4 dai, increased at 6 dai and remained at the same level at 8 dai (Fig. 3B). This expression pattern provides further evidence for the biotrophic lifestyle of Lm within the first 4 days of cotyledon infection fallowed by a shit to necrotrophism. Lm‐NLP function was confirmed by its ability to cause hypersensitive response (HR)‐like cell death when transiently expressed in N. benthamiana leaves (Fig. 3C). To further determine the role of NLP during Lm infection, we generated transgenic Lm lines expressing an NLP‐RNAi construct. Twenty NLP‐RNAi transformants (Lm‐nlp) were selected and their virulence was examined by inoculation on cotyledons of the susceptible B. napus cv. Topas DH16516. As shown in Fig. 3D, three of the Lm‐nlp lines (NLP‐SL1, NLP‐SL2 and NLP‐SL3) were severely compromised in virulence. Reduction in NLP expression was confirmed by quantitative polymerase chain reaction (qPCR) (Fig. S4A, see Supporting Information). In comparison with wild‐type Lm, Lm‐nlp lines showed a slightly reduced growth rate, but sporulated normally on culture plates (Fig. S4B).
Regulation of plant hormones and defence in B. napus infected by Lm
To define the B. napus hormone defence networks in response to Lm infection, we used hormonometer software (Volodarsky et al., 2009) which compares the input query genes against a database of Arabidopsis genes expressed in response to the application of different hormones. Using the Arabidopsis orthologues of the B. napus DEG as query, we obtained profiles of hormone regulation in B. napus at different time points after infection (Fig. 4A). This comparison revealed a correlation of the B. napus response at the initial phase of infection (2–4 dai) with the gene expression response of Arabidopsis treated with SA, whereas, at the later stages of infection (6 and 8 dai), there was a good correlation with the Arabidopsis gene expression profile in response to the application of JA. Analysis by the software MapMan (Thimm et al., 2004) also showed that upstream genes of the JA pathway were up‐regulated in B. napus at 6–8 dai (Fig. 4B). There was a good correlation throughout all time points with the Arabidopsis genes related to ET. In the case of ABA signalling, the B. napus response at 4 dai correlated with the Arabidopsis response to ABA. Taken together, these data underline the importance of SA, ET and JA pathways, and provide evidence for the involvement of ABA in B. napus in response to Lm infection. Furthermore, the induction of SA in B. napus at 2 and 4 dai substantiates the biotrophic phase of Lm described above.
Figure 4.
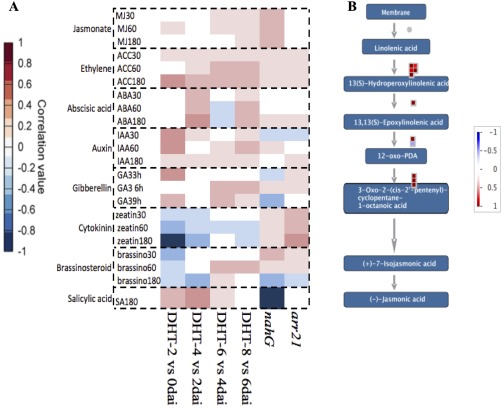
Brassica napus hormonal response to Leptosphaeria maculans infection. (A) A positive correlation between the query (B. napus differentially expressed genes) and the Arabidopsis thaliana genes expressed when treated with various plant hormones is denoted in red, whereas a negative correlation is represented by blue. The transcriptomes of the Arabidopsis mutants arr21 and nahg with increased cytokinin and decreased salicylic acid responses, respectively, are also included. (B) Jasmonic acid (JA) signalling pathway. MapMan output of JA‐related genes that were differentially expressed at 8 vs. 6 days after inoculation (dai) (≥log two‐fold change). Each square represents one gene. Intensity scale: red represents up‐regulated and blue represents down‐regulated genes.
The profile of the B. napus DEGs showed multifaceted defence responses that peaked at 4 dai. The pattern recognition receptors CERK1 (chitin elicitor receptor kinase 1), extracellular RLPs, receptor‐like kinases (RLKs) and wall‐associated kinases (WAKs) were among the differentially expressed cell surface receptors that probably perceive Lm PAMPs (pathogen‐associated molecular patterns) and effectors. It has been shown previously that the Arabidopsis thaliana LRR‐receptor‐like kinase (LRR‐RLK) suppressor of Bir‐1 (AtSOBIR1) interacts with LRR‐RLP resistance genes (Liebrand et al., 2014). We have also reported recently that AtSOBIR1 interacts with the B. napus Rlm2 (Larkan et al., 2015). The expression of five of the six copies of B. napus SOBIR1 homologues peaked at 6 and 8 dai (Fig. S5, see Supporting Information).
Other genes with reported function in plant defence that were differentially expressed included genes encoding for proteases and protease inhibitors, chitinases, peroxidases, transcription factors (WRKY, AP2/EREBP, MYB), genes related to the production of the secondary metabolites and genes involved in plant cell wall reinforcement. The majority of these genes were down‐regulated in the lesion zone surrounding the inoculation site (samples taken at 6 and 8 dai). Among the differentially expressed WRKY transcription factors with a proven role in plant defence were WRKYs 33, 40 and 51.
Lignification of the plant cell wall blocks pathogen entry at the site of infection and is triggered in response to pathogen infection. As depicted in Fig. 5, the expression of the majority of the genes involved at the early steps of lignin biosynthesis [phenylalanine ammonia lyase (PAL), cinnamate 4‐hydroxylase (C4H), 4‐coumarate:CoA ligase (4CL)] increased at 4 dai and remained up‐regulated at 6 and 8 dai.
Figure 5.
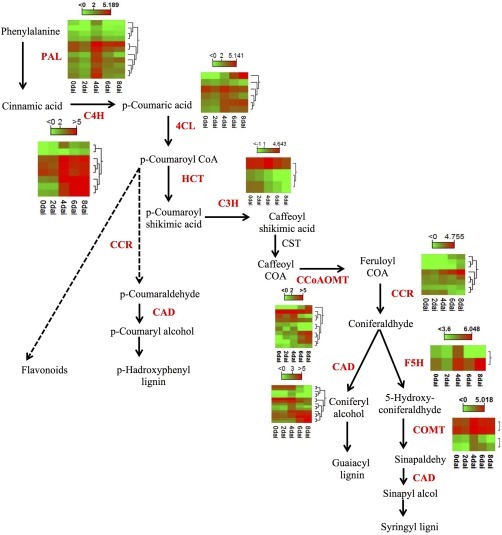
Expression profile of Brassica napus lignin‐related genes at different time points in response to Leptosphaeria maculans infection. Heat map presents the expression of genes ranging from green (low) to red (high). CAD, cinnamyl‐alcohol dehydrogenase; CCoA‐3H, cinnamyl‐alcohol dehydrogenase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; CCR, cinnamoyl‐CoA reductase; C3H, p‐coumarate 3‐hydroxylase; C4H, cinnamate 4‐hydroxylase; 4CL, 4‐coumarate:CoA ligase; COMT, caffeic acid O‐methyltransferase; CST, p‐hydroxycinnamoyltransferase; F5H, ferulate 5‐hydroxylase; HCT, p‐hydroxycinnamoyl‐CoA; PAL, phenylalanine ammonia lyase.
Glucosinolates (GLSs) are sulfur‐ and nitrogen‐containing secondary metabolites found in members of Brassicaceae. The role of GLSs in plant immunity, particularly against necrotrophic plant pathogens, has been well studied (Buxdorf et al., 2013; Laluk and Mengiste, 2010). We found three GLS‐related genes that were significantly up‐regulated at 6 and 8 dai. Two of these genes, the cytochrome P450 (CYP79B2) and SUPERROOT1 (SUR1), are part of the GLS biosynthesis pathway (Fig. S6, see Supporting Information). The expression of another GLS‐related gene, nitrile‐specifier protein 5 (NSP5), was highly up‐regulated at 4 dai and its expression continued to increase at 6 and 8 dai.
Discussion
The genetics of cotyledon resistance to Lm have been studied in detail; however, the genomic research of the B. napus–Lm interaction lags behind. Šašek et al. (2012) examined the expression of several marker genes, associated with various plant hormones, in resistant and susceptible B. napus plants infected with Lm, and reported that the expression of SA‐related genes was induced at the earlier (4–6 dai) stages of B. napus infection, whereas the expression of the genes related to the ET pathway increased at the later stages (8–10 dai). Similarly, Lowe et al. (2014) have reported recently that the marker genes for SA and ABA were up‐regulated at 7 and 14 days after infection of B. napus with Lm, respectively.
Here, we provide a more detailed picture of the global transcriptome that is active in B. napus and Lm during cotyledon infection. The expression profile of the genes related to different plant hormones revealed the importance of SA at the earlier stages of infection (until 4 dai) and the up‐regulation of genes related to the JA pathway at the later stages (6–8 dai) (Figs 5 and 6). Consistent with this, Šašek et al. (2012) reported that the application of benzothiadiazole (BTH), a functional analogue of SA, provided full immunity against Lm infection in B. napus. These authors reported similar results when B. napus was treated with the ET precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) or ABA; however, BTH had a much stronger effect than ACC and ABA in preventing Lm infection.
Figure 6.
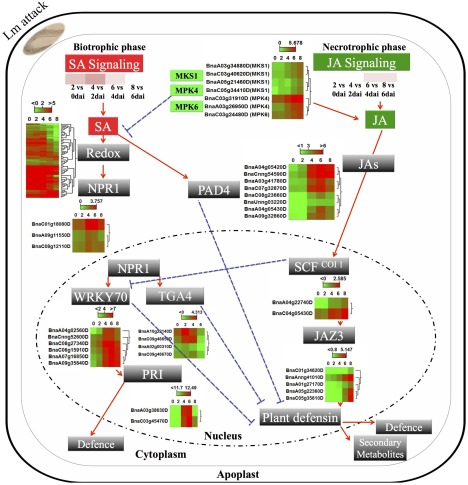
An overview of salicylic acid (SA) and jasmonic acid (JA) signalling in Brassica napus in response to infection with Leptosphaeria maculans. The expression profiles of B. napus genes with homology to known marker genes related to SA and JA pathways are presented as green (low) or red (high). MKS1, MAP kinase substrate 1; MPK4, mitogen‐activated protein kinase 4; MPK6, mitogen‐activated protein kinase 6; SA, salicylic acid; JA, jasmonic acid; NPR1, natriuretic peptide receptor 1; WRKY70, wall‐associated receptor kinase; PR1, pathogenesis‐related protein 1.
Recent large‐scale transcriptomic and genomic studies have revealed the importance of other plant hormones, such as auxin, BRs, GAs and CKs, in plant defence (Robert‐Seilaniantz et al., 2011). The analysis of the expression of plant hormone‐related genes showed that, at 6–8 dai, B. napus genes related to the BR pathway showed a positive correlation with the gene expression profile of Arabidopsis in response to BR (Fig. 5). The role of BRs in B. napus defence against Lm becomes more significant in the light of recent studies revealing the involvement of BRs in PAMP‐triggered immunity (PTI) (Albrecht et al., 2012), and the striking overlap that exists between the PAMP recognition complexes and the plant receptor complexes for the recognition of intercellular filamentous pathogens (Liebrand et al., 2014). Several recent studies have shown that SOBIR1 and BAK1 (BRI1‐associated kinase 1) form a complex with tomato RLPs to recognize C. fulvum, the pathogen of tomato (Postma et al., 2015). Lm shares a high degree of similarity with C. fulvum, in term of its infection process and host plant receptor complex. We have reported recently that a similar mechanism exists for the perception of Lm by B. napus (Larkan et al., 2015, Ma and Borhan, 2015). BAK1 is a co‐receptor for BR and PAMP. In a recent paper, Belkhadir et al. (2012) reported that increased BR signalling antagonizes BAK1‐mediated cell death. It is conceivable that BRs contribute to B. napus defence against Lm in a similar manner as described for PTI (Wang, 2012). Interestingly, BR signalling in response to Lm infection in our experiment was down‐regulated at the initial stage of infection that coincides with the activation of RLP‐RLK receptor complexes, but genes related to the BR pathway were up‐regulated once the lesion had developed (6–8 dai). The up‐regulation of BR‐related genes at the necrotrophic stage of Lm growth could, at least in part, be a result of the production of toxins by Lm at the late stages of infection. Phytotoxins produced by Fusarium strains have been reported to induce BRs (Masuda et al., 2007).
As it is an intercellular pathogen, plant cell surface receptors play a primary role in the initiation of B. napus defence against Lm (Stotz et al., 2014; Larkan et al., 2013, 2015; Ma and Borhan, 2015). Several B. napus receptors were up‐regulated in response to Lm, such as SOBIR1, RLKs containing DUF26 (domain of unknown function26), LRR‐RKs (leucine‐rich repeat receptor kinases) and WAKs. As reported previously, SOBIR1 is a part of the LRR‐RLP recognition complex against Lm (Larkan et al., 2015). DUF26 genes, also known as CRKs (cysteine‐rich receptor kinases), are transcriptionally induced in response to pathogen and SA (Acharya et al., 2007; Kim et al., 2004; Wang et al., 2013, 2014; Wrzaczek et al., 2010). DUF26 transcripts were found to be abundant in the rice apoplast during Magnaporthe oryzae infection (Wang et al., 2014). Overexpression of WAK in Arabidopsis and rice enhances resistance to Botrytis cinerea and M. oryzae, respectively (Wang et al., 2014). We found many transcription factors, including some of the WRKY‐type transcription factors, to be up‐regulated in response to Lm. Several WRKYs have been reported to act as positive and negative regulators of plant defence (Peng et al., 2008; Qiu et al., 2008; Shimono et al., 2007; Wang et al., 2007; Zhang et al., 2008).
Lm is considered to be a hemibiotroph during the foliar infection stage, but no molecular evidence supporting and defining the biotrophic and necrotrophic stages of Lm infection has been provided. We presented several pieces of evidence that defined the hemibiotrophic growth stages of Lm during B. napus cotyledon infection. The biotrophic stage of Lm growth, during the cotyledon infection of B. napus, was marked by the B. napus SA response at 2 and 4 dai, as well as the up‐regulation of the majority of the predicted and known effectors. Other clear evidence was the expression of Lm‐NLP, which was first seen at 4 dai and was at a maximum at 6 and 8 dai. The role of NLP in the induction of necrosis (which is required for necrotrophic growth) was demonstrated by transient assay in tobacco. In addition, we showed that the suppression of NLP expression by RNAi compromised virulence (lesion formation) in Lm. NLP is conserved among prokaryotic and eukaryotic microorganisms and is a well‐known marker for the initiation of necrotrophy in plant‐pathogenic fungi. The production of toxins by fungal plant pathogens is related to the necrotrophic lifestyle. The expression of several of the Lm genes encoding PKSs and NRPSs, with a reported or potential role in the production of toxins in fungi, was upregulated at 6–8 dai (Fig. S2). Some of the Lm toxins, such as sirodesmin, have been reported to play a role in the virulence of Lm during infection of the B. napus stem; however, their role in the development of lesions during cotyledon infection is unclear. The expression of three B. napus genes (identified as CYP79B2, SUR1 and NPS5) related to GLSs was also up‐regulated at the later stages (6–8 dai) of infection. GLSs, mainly found in members of Brassicaceae, are constitutively present in a non‐reactive form, but, on tissue damage and pathogen attack, are hydrolysed, resulting in the production of physiologically active derivatives, such as isothiocyanates (ITCs), thiocyanates and simple nitriles. The role of GLSs in defence against several necrotrophic fungi has been reported (Buxdorf et al., 2013; Stotz et al., 2011). The Arabidopsis CYP79B2/B3 double mutant is more sensitive to the necrotrophic pathogens Alternaria brassicicola and Botrytis cinerea (Buxdorf et al., 2013). SUR1 functions as a C‐S lyase in the GLS pathway, and the Arabidopsis sur1 mutants are impaired in the production of GLSs (Mikkelsen et al., 2004). NPSs, in association with myrosinase, are involved in the hydrolysis of GLSs and the formation of simple nitriles, a toxin with reported function against herbivores (Mumm et al., 2008).
The RNA‐seq conducted in this study allowed us to define a list that includes the most likely Lm effector genes, and the functions of several of these effectors in pathogen virulence and suppression of plant defence were confirmed. Future research on these effector genes should help to identify additional Avr genes and provide insights into the virulence mechanisms of Lm.
Canola breeders have exploited the B. napus resistance against Lm as the most effective and practical approach to control blackleg. Insights into the molecular interaction of B. napus and Lm described here, and additional research, will provide the knowledge and tools required for the genetic improvement of Brassica resistance against blackleg disease.
Experimental Procedures
Plant material and fungal isolate
The B. napus DH line Topas DH16516 and a single‐spore culture of the Lm isolate 00‐100, which possesses three known effectors (AvrLm2, AvrLm6 and AvrLmJ1), were used in this study. Brassica napus cv. Topas has been tested extensively against an international collection of Lm isolates (over 100 isolates) in our laboratory and has been proven to lack race‐specific resistance against blackleg disease (Larkan et al., 2013).
Plant growth and infection conditions
The plant growth and Lm infection conditions have been described previously (Larkan et al., 2013). Plants were grown in a growth chamber at 20 ºC, 16 h light, with a light intensity of c. 450 μmol/m2/s at bench level, and 18 ºC, 8 h dark. For pycnidiospore inoculation, a small wound was made in the centre of each cotyledon of 7‐day‐old seedlings and 10 μL of 2 × 107 spores/mL suspension were applied to each wound (four infection sites per seedling). After inoculation, the seedlings were kept under the same growth conditions and disease progression was monitored.
Total RNA isolation and library preparation for Illumina HiSeq 2500
The cotyledons of 7‐day‐old seedlings were inoculated with isolate 00‐100. Mock inoculation with water served as a negative control. Cotyledon discs of 6 mm in diameter were excised from the infected cotyledons (four biological replicates) at 0, 2, 4, 6 and 8 dai. Twelve discs per sample were ground in liquid nitrogen, and then extracted with TRIzol LS reagent (Invitrogen, Carlsbad, USA) and purified by application to an Ambion mini RNA kit following the manufacturer's instructions. RNA was DNAase treated, quantified by a Qubit fluorometer (Invitrogen, Carlsbad, USA) and checked for quality by an Agilent Bioanalyzer 2100 (Agilent Technologies, Mississauga, Canada). Only samples with RNA integrity numbers above 8.0 were used for sequencing. Sequence reads (100 bp paired‐end) were generated with Illumina TruSeq high‐output version 3 chemistry on a HiSeq 2500 (Illumina, Inc., San Diego, USA) at NRC‐Plant Biotechnology Institute (NRC‐PBI), Saskatoon, SK, Canada.
Read mapping and gene expression analysis
In total, 1.38 billion raw reads were analysed in this study (Table S1). Reads were trimmed, adaptor sequences were removed and reads shorter than 15 bp were discarded (quality score limit, 0.05; trim ambiguous nucleotides <2) using CLC genomics workbench, version 7.5 (QIAGEN, Aarhus, Denmark). Trimmed sequences were mapped to the B. napus and Lm genomes using CLC genomics workbench (Mismatch cost = 2, Insertion cost = 1, Deletion cost = 1, Length fraction = 1, Similarity fraction = 0.97, Maximum number of hits allowed = 1). Brassica napus gene models were downloaded from the Brassica napus Genome Resources – Genoscope and Lm gene models were obtained from the Joint Genome Institute (JGI) (http://genome.jgi.doe.gov /). Total reads per transcript and log2 RPKM were obtained by the RNA‐Seq Analysis function in CLC. The estimate SizeFactors and estimate Dispersions functions in the Bioconductor package DESeq2 v. 1.6.3 (Love et al., 2014) were also used to normalize the raw transcript counts of B. napus and Lm genes, and to identify DEGs using the Benjamini–Hochberg (BH) method for correction of multiple comparisons. We also performed a CIM of the sample‐to‐sample distance matrix and PCA using B. napus and Lm genes to assess the variability among samples.
Functional classification based on Blast2Go and MapMan
Blast2Go‐pro (Conesa and Gotz, 2008) software was used to annotate the DEGs. A Fisher's exact test was used to test the significance of gene ontology (GO) enrichment. KEGG (Kyoto Encyclopedia of Genes and Genomes; Kanehisa and Goto, 2000; Kanehisa et al., 2014) enrichment analysis was performed using the KEGG function of Blast2Go‐pro software to assign predicted pathways for DEGs. DEGs were analysed by MapMan software (Thimm et al., 2004) using log two‐fold changes in expression to visualize metabolic pathways and cellular responses of B. napus to Lm. PCWDEs were defined by conducting a similarity search of the CAZy database (db‐CAN) (Yin et al., 2012).
Hormonometer analysis
The putative A. thaliana orthologues of B. napus DEGs were imported to hormonometer software (Volodarsky et al., 2009) to identify transcriptional similarities between DEGs and Arabidopsis genes related to different plant hormones.
Lm secretome prediction and analysis pipeline
We developed a pipeline to predict and classify effectors of Lm. We first detected the presence of an N‐terminal signal peptide through signalP4.1 (Petersen et al., 2011). Next, we excluded proteins with a predicted transmembrane domain as determined by TMHMM (Krogh et al., 2001). Subsequently, we searched for the presence of subcellular localization signals using TargetP (Emanuelsson et al., 2000). The presence of a glycosylphosphatidylinositol (GPI) anchor was predicted using PredGPI (Pierleoni et al., 2008) to identify proteins anchored to the membrane. We used Pfam (Finn et al., 2014) to verify biological functions enriched in the secretome of Lm. OrthoMCL (Fischer et al., 2011) was used to identify homologous proteins based on sequence similarity by Markov clustering.
Vector constructions
To generate the RNAi constructs, the entire Nep1 open reading frame (ORF) (GenBank accession LEMA_P070010.1) was amplified using the primer pair Nep‐FB/Nep‐RB. The PCR products were introduced into the entry vector pDON/Zeo (Life Technologies, Burlington, Canada) using the gateway protocol described by the manufacturer. For transient expression, pENTR/Zeo::Nep1 was recombined into the binary vector pEarlyGate100 (Earley et al., 2006). Genes for effector candidates were amplified from the B. napus–Lm cDNA library using the corresponding primer combinations (Table S3, see Supporting Information) and recombined via the entry clone pDON/Zeo into the binary vector pGWB414 (Nakagawa et al., 2007). Using the same approach, the mouse Bax gene was amplified with the primer pairs M‐Bax‐FB/M‐Bax‐RB (Table S3), cloned into pDON/Zeo and then recombined into the binary vector pEarlyGate100. All constructs were confirmed by sequencing. All resulting binary plasmids were transformed to the Agrobacterium tumefaciens strain GV3101 (pMP90).
RNAi‐mediated silencing of Lm
The RNAi silencing plasmid pHGYGS::Nlp1 was transformed into A. tumefaciens strain EHA105. Agrobacterium‐mediated transformation of Lm isolate 00‐100 was conducted based on the protocol described by Utermark and Karlovsky (2008). Quantitative PCR (qPCR) was performed as described previously (Ma and Borhan, 2015). qPCR primers are provided in Table S3.
Agrobacterium‐mediated transient assay in N. benthamiana
Agrobacterium‐mediated transient expression of candidate genes in N. benthamiana was performed according to the methods described previously (Ma et al., 2012). Briefly, Agrobacterium was grown to an absorbance of 0.8 at an optical density at 600 nm (OD600 nm) in LB: Luria Broth–mannitol medium supplemented with 20 µm acetosyringone and 10 mm MES (2‐(N‐morpholino) ethanesulfonic acid) (pH 5.6). Cells were pelleted by centrifugation at 2500 g for 20 min and then re‐suspended in infiltration medium (1 × MES; 10 mm MES, pH 5.6, 2% w/v sucrose, 200 µm acetosyringone). Leaves of 4–5‐week‐old N. benthamiana were infiltrated (OD600 = 1) with a needleless syringe and photographed 3–5 days after infiltration. For the suppression of Bax‐mediated cell death, the A. tumefaciens‐containing Bax gene was infiltrated first and, 1 day later, the A. tumefaciens strain carrying the individual effector genes was infiltrated in the same spot. Agrobacterium‐green fluorescent protein (GFP) served as the negative control. For light microscopy observation, leaves were boiled for 5 min in a 1 : 1 mixture of ethanol and 0.33 mg/mL trypan blue in lactophenol and destained overnight in 2.5 g/mL chloral hydrate solution.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 RNA‐sequencing quality assessment. Variation within the four replicates was determined from the following: (A) density plots; red arrows indicate that the density of the counts in the third replicate for the time points 4, 6 and 8 days after inoculation (dai) was distinctly different from that for the same time points in other replicates; (B) clustering of the sample‐to‐sample distances; closely grouped replicates within the clusters are indicated by red circles; (C) the MA plot between two biological replicates per time point; M values (the log fold change), the log of the ratio of level counts for each gene between two biological replicates per time point; A values (the log‐average), the average level counts for each gene across two biological replicates per time point. Genes with similar expression levels in two biological replicates per time point will appear around the horizontal line (y = 0). The third replicates of each time point that were distinctly different from the corresponding points in other replicates are marked by red boxes.
Fig. S2 Expression profiles of Leptosphaeria maculans (Lm) secondary metabolite‐related genes are presented as green (low) or red (high). NS, non‐significant.
Fig. S3 Lema_T086540.1 with homology to the Fusarium effector Six5. (A) Location of the gene in the AT‐rich region of the Leptosphaeria maculans genome. (B) Expression profile of T086540.1 grown in vitro and during Brassica napus infection. (C) Conserved cysteine residues between Six5 and Lema_T086540.1 are marked by red arrows. *Significant at 0.05 probability level. ***Significant at 0.001 probability level.
Fig. S4 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the NLP gene in the wild‐type and NLP silencing line NLP‐SL1. RT‐PCR products obtained from RNA isolated from Brassica napus cv. Topas at 6 days after inoculation with the wild‐type isolate 00‐100 and NLP silencing line NLP‐SL1. NLP gene expression levels are normalized to that of actin. Values are means ± standard error (SE) of triplicate reactions of three independent biological samples. Significant differences are represented by three asterisks (P < 0.001). (B) The NLP‐SL1 silencing line showed a slightly reduced growth rate on V8 medium. Wild‐type L. maculans 00‐100 and one of the NLP silencing lines NLP‐SL1 were grown on a V8 medium plate and photographs were taken at 7 days after inoculation.
Fig. S5 Expression profile of AtSOBIR1 orthologues in Brassica napus.
Fig. S6 Expression profile of Leptosphaeria maculans nitrile‐specifier protein 5 (NSP5), CYP79B2 and SUPERROOT1 (SUR1) at different time points.
Table S1 Summary of RNA‐sequencing (RNA‐seq) reads mapped to the Brassica napus (A) and Leptosphaeria maculans (B) reference genomes.
Table S2 List of small secreted protein (SSP)‐encoding genes as the most likely set of Leptosphaeria maculans predicted effectors that were found to be differentially expressed [false discovery rate‐Benjamini–Hochberg (FDR‐BH) of less than 0.05] based on DESeq2 software.
Table S3 Primer combinations for the amplification of effector candidate‐related genes.
Acknowledgements
We would like to thank Elena Beynon and Colin Kindrachuck for assisting in sample preparation and RNA extraction. Funding for this project was provided by the Agriculture Development Fund, SaskCanola and Western Grain Research Foundation.
References
- Acharya, B.R. , Raina, S. , Maqbool, S.B. , Jagadeeswaran, G. , Mosher, S.L. , Appel, H.M. , Schultz, J.C. , Klessig, D.F. and Raina, R. (2007) Overexpression of CRK13, an Arabidopsis cysteine‐rich receptor‐like kinase, results in enhanced resistance to Pseudomonas syringae . Plant J. 50, 488–499. [DOI] [PubMed] [Google Scholar]
- Albrecht, C. , Boutrot, F. , Segonzac, C. , Schwessinger, B. , Gimenez‐Ibanez, S. , Chinchilla, D. , Rathjen, J.P. , de Vries, S.C. and Zipfel, C. (2012) Brassinosteroids inhibit pathogen‐associated molecular pattern‐triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA, 109, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balesdent, M.H. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A.M. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa . New Phytol. 198, 887–898. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y. , Jaillais, Y. , Epple, P. , Balsemao‐Pires, E. , Dangl, J.L. and Chory, J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe‐associated molecular patterns. Proc. Natl. Acad. Sci. USA, 109, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxdorf, K. , Yaffe, H. , Barda, O. and Levy, M. (2013) The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS One, 8, e70771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni, A. , Passerini, A. , Vullo, A. and Frasconi, P. (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 34, W177–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A. , Tang, H. , Wang, X. , Chiquet, J ., Belcram, H ., Tong, C ., Samans, B ., Corréa, M ., Da Silva, C ., Just, J ., Falentin, C ., Koh, C.S ., Le Clainche, I ., Bernard, M ., Bento, P ., Noel, B ., Labadie, K ., Alberti, A ., Charles, M ., Arnaud, D ., Guo, H ., Daviaud, C ., Alamery, S ., Jabbari, K ., Zhao, M ., Edger, P.P ., Chelaifa, H ., Tack, D ., Lassalle, G ., Mestiri, I ., Schnel, N ., Le Paslier, M.C ., Fan, G ., Renault, V ., Bayer, P.E ., Golicz, A.A ., Manoli, S ., Lee, T.H ., Thi, V.H ., Chalabi, S ., Hu, Q ., Fan, C ., Tollenaere, R ., Lu, Y ., Battail, C ., Shen, J ., Sidebottom, C.H ., Wang, X ., Canaguier, A ., Chauveau, A ., Bérard, A ., Deniot, G ., Guan, M ., Liu, Z ., Sun, F ., Lim, Y.P ., Lyons, E. , Town, C.D ., Bancroft, I ., Wang, X ., Meng, J ., Ma, J ., Pires, J.C ., King, G.J. , Brunel, D ., Delourme, R ., Renard, M. , Aury, J.M ., Adams, K.L ., Batley, J ., Snowdon, R.J ., Tost, J ., Edwards, D ., Zhou, Y ., Hua, W ., Sharpe, A.G. , Paterson, A.H ., Guan, C . and Wincker, P. (2014) Plant genetics. Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Cho, Y. (2015) How the necrotrophic fungus Alternaria brassicicola kills plant cells remains an enigma. Eukaryot. Cell, 14, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collemare, J. , Griffiths, S. , Iida, Y. , Karimi Jashni, M. , Battaglia, E. , Cox, R.J. and de Wit, P.J.G.M. (2014) Secondary metabolism and biotrophic lifestyle in the tomato pathogen Cladosporium fulvum . PLoS One, 9(1), e85877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa, A. and Gotz, S. (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics, 2008, doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elliott, C.E. , Callahan, D.L. , Schwenk, D. , Nett, M. , Hoffmeister, D. and Howlett, B.J. (2013) A gene cluster responsible for biosynthesis of phomenoic acid in the plant pathogenic fungus, Leptosphaeria maculans . Fungal Genet. Biol. 53, 50–58. [DOI] [PubMed] [Google Scholar]
- Elliotte, C.E. , Gardnier, D.M. , Thomas, G. , Cozijnsen, A. , Van de Wouw, A. and Howlett, B.J. (2007) Production of the toxin sirodesmin PL by Leptosphaeria maculans during infection of Brassica napus . Mol. Plant Pathol. 8, 791–802. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O. , Nielsen, H. , Brunak, S. and Heijne, G.V. (2000) Predicting subcellular localization of proteins based on their N‐terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Bateman, A. , Clements, J. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , Heger, A. , Hetherington, K. , Holm, L. , Mistry, J. , Sonnhammer, E.L.L. , Tate, J. and Punta, M. (2014) The Pfam protein families database. Nucleic Acids Res. 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, S. , Brunk, B.P. , Chen, F. , Gao, X. , Harb, O.S. , Iodice, J.B. , Shanmugam, D. , Roos, D.S. and Stoeckert, C.J., Jr . (2011) Using OrthoMCL to assign proteins to OrthoMCL‐DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics, Andreas D. Baxevanis [et al.], Chapter 6, Unit 6 12 11–19. [DOI] [PMC free article] [PubMed]
- Fitt, B.D.L. , Brun, H. , Barbetti, M.J. and Rimmer, S.R. (2006) World‐wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 114, 3–15. [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M.L. , Eckert, M.R. , Cattolico, L. , Bernard‐Samain, S. , Balesdent, M.H. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map‐based cloning of AvrLm6. Mol. Plant–Microbe Interact. 20: 459–470. [DOI] [PubMed] [Google Scholar]
- Ghanbarnia, K. , Fudal, I. , Larkan, N.J. , Links, M.G. , Balesdent, M.H. , Profotova, B. , Fernando, W.G. , Rouxel, T. and Borhan, M.H. (2015) Rapid identification of the Leptosphaeria maculans avirulence gene AvrLm2 using an intraspecific comparative genomics approach. Mol. Plant Pathol. 16, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen, M. and Nurnberger, T. (2006) Nep1‐like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Guyon, K. , Balague, C. , Roby, D. and Raffaele, S. (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genomics, 15, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, E.S. (2000) Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Goto, S. , Sato, Y. , Kawashima, M. , Furumichi, M. and Tanabe, M. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F. and Tsuda, K. (2010) Understanding the plant immune system. Mol. Plant–Microbe Interact. 23, 1531–1536. [DOI] [PubMed] [Google Scholar]
- Khaldi, N. , Seifuddin, F.T. , Turner, G. , Haft, D. , Nierman, W.C. , Wolfe, K.H. and Fedorova, N.D. (2010) SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.T. , Kim, S.G. , Hwang, D.H. , Kang, S.Y. , Kim, H.J. , Lee, B.H. and Kang, K.Y. (2004) Proteomic analysis of pathogen‐responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea . Proteomics, 4, 3569–3578. [DOI] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , von Heijne, G. and Sonnhammer, E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Laluk, K. and Mengiste, T. (2010) Necrotroph attacks on plants: wanton destruction or covert extortion? The Arabidopsis Book, 8, e0136. doi: 10.1199/tab.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan, N.J. , Lydiate, D.J. , Parkin, I.A.P. , Nelson, M.N. , Epp, D.J. , Cowling, W.A. , Rimmer, S.R. and Borhan, M.H. (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor‐like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phyto. 197, 595–605. [DOI] [PubMed] [Google Scholar]
- Larkan, N.J. , Lydiate, D.J. , Yu, F. , Rimmer, S.R. and Borhan, M.H. (2013) Co‐localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol. 14, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan, N.J. , Ma, L. and Borhan, M.H. (2015) The Brassica napus receptor‐like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 13, 983–992. [DOI] [PubMed] [Google Scholar]
- Levasseur, A. , Drula, E. , Lombard, V. , Coutinho, P.M. and Henrissat, B. (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels, 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W. , van den Burg, H.A. and Joosten, M.H. (2014) Two for all: receptor‐associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Love, M.L. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, R.G. , Cassin, A. , Grandaubert, J. , Clark, B.L. , Van de Wouw, A.P. , Rouxel, T. and Howlett, B.J. (2014) Genomes and transcriptomes of partners in plant–fungal interactions between canola (Brassica napus) and two Leptosphaeria species. PLoS One, 9, e103098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. and Borhan, M.H. (2015) The receptor‐like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1‐triggered immunity. Front Plant Sci. 6, 933. DOI=10.3389/fpls.2015.00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Houterman, P.M. , Gawehns, F. , Cao, L. , Sillo, F. , Richter, H. , Clavijo‐Ortiz, M.J. , Schmidt, S.M. , Boeren, S. , Vervoort, J. , Cornelissen, B.J. , Rep, M. and Takken, F.L. (2015) The AVR2‐SIX5 gene pair is required to activate I‐2‐mediated immunity in tomato. New Phytol. 208, 507–518. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Lukasik, E. , Gawehns, F. and Takken, F.L. (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 835, 61–74. [DOI] [PubMed] [Google Scholar]
- Malinovsky, F.G. , Fangel, J.U. and Willats, W.G.T. (2014) The role of the cell wall in plant immunity. Frontiers in Plant Science, doi: 10.3389/fpls.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, D. , Ishida, M. , Yamaguchi, K. , Yamaguchi, I. , Kimura, M. and Nishiuchi, T. (2007) Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana . J. Exp. Bot. 58, 1617–1626. [DOI] [PubMed] [Google Scholar]
- Metzker, M.L. (2010) Sequencing technologies – the next generation. Nat. Rev. Genet. 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D. , Naur, P. and Halkier, B.A. (2004) Arabidopsis mutants in the C‐S lyase of glucosinolate biosynthesis establish a critical role for indole‐3‐acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. [DOI] [PubMed] [Google Scholar]
- Mumm, R. , Burow, M. , Bukovinszkine'kiss, G. , Kazantzidou, E. , Wittstock, U. , Dicke, M. and Gershenzon, J. (2008) Formation of simple nitriles upon glucosinolate hydrolysis affects direct and indirect defense against the specialist herbivore, Pieris rapae . J. Chem. Ecol. 34, 1311–1321. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. and Ishiguro, S. (2007) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Parlange, F. , Daverdin, G. , Fudal, I. , Kuhn, M.L. , Balesdent, M.H. , Blaise, F. , Grezes‐Besset, B. and Rouxel, T. (2009) Leptosphaeria maculans avirulence gene AvrLm4‐7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4‐mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. [DOI] [PubMed] [Google Scholar]
- Pedersen, C. , Themmat, E.V.L. , McGuffin, L. , Abbott, J.C. , Burgis, T.A. , Barton, G. , Bindschedler, L.V. , Lu, X. , Maekawa, T. , Wessling, R. , Cramer, R. , Thordal‐Christensen, H. , Panstruga, R. and Spanu, P.D. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics, 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Bartley, L.E. , Chen, X. , Dardick, C. , Chern, M. , Ruan, R. , Canlas, P.E. and Ronald, P.C. (2008) OsWRKY62 is a negative regulator of basal and Xa21‐mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant, 1, 446–458. [DOI] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , Heijne, G.V. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Pierleoni, A. , Martelli, P.L. and Casadio, R. (2008) PredGPI: a GPI anchor predictor. BMC Bioinformatics, 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Postma, J. , Liebrand, T.W.H. , Bi, G. , Evrard, A. , Bye, R.R. , Mbengue, M. , Joosten, M.H.A.J. and Robatzek, S. (2015) The Cf‐4 receptor‐like protein associates with the BAK1 receptor‐like kinase to initiate receptor endocytosis and plant immunity. bioRxiv doi: 10.1101/019471. [DOI] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Xie, W. , Liu, H. , Li, X. , Xiong, L. and Wang, S. (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant, 1, 538–551. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Raman, H. , Raman, R. and Larkan, N. (2013) Genetic dissection of Blackleg Resistance Loci in rapeseed (Brassica napus L.) In: Plant Breeding from Laboratories to Fields. (Andersen, S. B., ed.). Rijeka, Croatia: InTech, pp. 85–120.
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. and Balesdent, M.H. (2005) The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Mol. Plant Pathol. 6, 225–241. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , van de Wouw, A.P. , Couloux, A. , Dominguez, V ., Anthouard, V ., Bally, P ., Bourras, S ., Cozijnsen, A.J ., Ciuffetti, L.M ., Degrave, A ., Dilmaghani, A ., Duret, L ., Fudal, I ., Goodwin, S.B ., Gout, L ., Glaser, N ., Linglin, J ., Kema, G.H ., Lapalu, N ., Lawrence, C.B ., May, K ., Meyer, M ., Ollivier, B ., Poulain, J ., Schoch, C.L ., Simon, A ., Spatafora, J.W ., Stachowiak, A ., Turgeon, B.G ., Tyler, B.M ., Vincent, D ., Weissenbach, J ., Amselem, J ., Quesneville, H ., Oliver, R.P ., Wincker, P ., Balesdent, M.H . and Howlett, B.J . (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat‐induced point mutations. Nat. Commun. 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šašek, V. , Novakova, M. , Jindrichova, B. , Boka, K. , Valentova, O. and Burketova, L. (2012) Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus . Mol. Plant–Microbe Interact. 25, 1238–1250. [DOI] [PubMed] [Google Scholar]
- Scharf, D.H. , Heinekamp, T. and Brakhage, A. (2014) Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog. 10, e1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Manners, J.M. , Singh, K.B. and Taylor, J.M. (2015) Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11, e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, U.H. , Sawada, Y. , Shimada, Y. , Hirai, M.Y. , Sasaki, E. , Krischke, M. , Brown, P.D. , Saito, K. and Kamiya, Y. (2011) Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate‐derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum . Plant J. 67, 81–93. [DOI] [PubMed] [Google Scholar]
- Stotz, H.U. , Mitrousia, G.K. , de Wit, P.J.G.M. and Fitt, B.D.L . (2014) Effector‐triggered defence against apoplastic fungal pathogens. Trends in Plant Science, 19, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O. , Bläsing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Krüger, P. , Selbig, J. , Müller, L.A. , Rhee, S.Y. and Stitt, M. (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Utermark, J. and Karlovsky, P. (2008) Genetic transformation of filamentous fungi by Agrobacterium tumefaciens . In: Nature Protocol Exchange Nature Publishing Group. URL: http://www.nature.com/protocolexchange/protocols/427.
- Van de Wouw, A.P. , Lowe, R.G. , Elliott, C.E. , Dubois, D.J. and Howlett, B.J. (2014) An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volodarsky, D. , Leviatan, N. , Otcheretianski, A. and Fluhr, R. (2009) HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol. 150, 1796–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Hao, J. , Chen, X. , Hao, Z. , Wang, X. , Lou, Y. , Peng, Y. and Guo, Z. (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Kim, S.G. , Wu, J. , Huh, H.H. , Lee, S.J. , Rakwal, R. , Agrawal, G.K. , Park, Z.Y. , Young Kang, K. and Kim, S.T. (2013) Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics, 13, 1901–1912. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Kwon, S.J. , Wu, J. , Choi, J. , Lee, Y.H. , Agrawal, G.K. , Tamogami, S. , Rakwal, R. , Park, S.R. , Kim, B.G. , Jung, K.H. , Kang, K.Y. , Kim, S.G. and Kim, S.T. (2014) Transcriptome analysis of early responsive genes in rice during Magnaporthe oryzae infection. Plant Pathol. J. 30, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y. (2012) Brassinosteroids modulate plant immunity at multiple levels. Proc. Natl. Acad. Sci. USA, 109, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J.S. , Kharbanda, P.D. , Barbetti, M.J. and Fitt, B.D.L. (2001) Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 50, 10–27. [Google Scholar]
- Westermann, A.J. , Gorski, S.A. and Vogel, J. (2012) Dual RNA‐seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630. [DOI] [PubMed] [Google Scholar]
- Wrzaczek, M. , Brosche, M. , Salojarvi, J. , Kangasjarvi, S. , Idanheimo, N. , Mersmann, S. , Robatzek, S. , Karpiński, S. , Karpińska, B. and Kangasjärvi, J. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor‐like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y. , Mao, X. , Yang, J. , Chen, X. , Mao, F. and Xu, Y. (2012) dbCAN: a web resource for automated carbohydrate‐active enzyme annotation. Nucleic Acids Res. 40, W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaparoli, G. , Barsottini, M.R. , de Oliveira, J.F. , Dyszy, F. , Teixeira, P.J. , Barau, J.G. , Garcia, O. , Costa‐Filho, A.J. , Ambrosio, A.L. , Pereira, G.A. and Dias, S.M. (2011) The crystal structure of necrosis‐ and ethylene‐inducing protein 2 from the causal agent of cacao's Witches' Broom disease reveals key elements for its activity. Biochemistry, 50, 9901–9910. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Peng, Y. and Guo, Z. (2008) Constitutive expression of pathogen‐inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 18, 508–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 RNA‐sequencing quality assessment. Variation within the four replicates was determined from the following: (A) density plots; red arrows indicate that the density of the counts in the third replicate for the time points 4, 6 and 8 days after inoculation (dai) was distinctly different from that for the same time points in other replicates; (B) clustering of the sample‐to‐sample distances; closely grouped replicates within the clusters are indicated by red circles; (C) the MA plot between two biological replicates per time point; M values (the log fold change), the log of the ratio of level counts for each gene between two biological replicates per time point; A values (the log‐average), the average level counts for each gene across two biological replicates per time point. Genes with similar expression levels in two biological replicates per time point will appear around the horizontal line (y = 0). The third replicates of each time point that were distinctly different from the corresponding points in other replicates are marked by red boxes.
Fig. S2 Expression profiles of Leptosphaeria maculans (Lm) secondary metabolite‐related genes are presented as green (low) or red (high). NS, non‐significant.
Fig. S3 Lema_T086540.1 with homology to the Fusarium effector Six5. (A) Location of the gene in the AT‐rich region of the Leptosphaeria maculans genome. (B) Expression profile of T086540.1 grown in vitro and during Brassica napus infection. (C) Conserved cysteine residues between Six5 and Lema_T086540.1 are marked by red arrows. *Significant at 0.05 probability level. ***Significant at 0.001 probability level.
Fig. S4 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the NLP gene in the wild‐type and NLP silencing line NLP‐SL1. RT‐PCR products obtained from RNA isolated from Brassica napus cv. Topas at 6 days after inoculation with the wild‐type isolate 00‐100 and NLP silencing line NLP‐SL1. NLP gene expression levels are normalized to that of actin. Values are means ± standard error (SE) of triplicate reactions of three independent biological samples. Significant differences are represented by three asterisks (P < 0.001). (B) The NLP‐SL1 silencing line showed a slightly reduced growth rate on V8 medium. Wild‐type L. maculans 00‐100 and one of the NLP silencing lines NLP‐SL1 were grown on a V8 medium plate and photographs were taken at 7 days after inoculation.
Fig. S5 Expression profile of AtSOBIR1 orthologues in Brassica napus.
Fig. S6 Expression profile of Leptosphaeria maculans nitrile‐specifier protein 5 (NSP5), CYP79B2 and SUPERROOT1 (SUR1) at different time points.
Table S1 Summary of RNA‐sequencing (RNA‐seq) reads mapped to the Brassica napus (A) and Leptosphaeria maculans (B) reference genomes.
Table S2 List of small secreted protein (SSP)‐encoding genes as the most likely set of Leptosphaeria maculans predicted effectors that were found to be differentially expressed [false discovery rate‐Benjamini–Hochberg (FDR‐BH) of less than 0.05] based on DESeq2 software.
Table S3 Primer combinations for the amplification of effector candidate‐related genes.


