Summary
Three hundred and ninety‐four sweet potato accessions from Latin America and East Africa were screened by polymerase chain reaction (PCR) for the presence of begomoviruses, and 46 were found to be positive. All were symptomless in sweet potato and generated leaf curling and/or chlorosis in Ipomoea setosa. The five most divergent isolates, based on complete genome sequences, were used to study interactions with Sweet potato chlorotic stunt virus (SPCSV), known to cause synergistic diseases with other viruses. Co‐infections led to increased titres of begomoviruses and decreased titres of SPCSV in all cases, although the extent of the changes varied notably between begomovirus isolates. Symptoms of leaf curling only developed temporarily in combination with isolate StV1 and coincided with the presence of the highest begomovirus concentrations in the plant. Small interfering RNA (siRNA) sequence analysis revealed that co‐infection of SPCSV with isolate StV1 led to relatively increased siRNA targeting of the central part of the SPCSV genome and a reduction in targeting of the genomic ends, but no changes to the targeting of StV1 relative to single infection of either virus. These changes were not observed in the interaction between SPCSV and the RNA virus Sweet potato feathery mottle virus (genus Potyvirus), implying specific effects of begomoviruses on RNA silencing of SPCSV in dually infected plants. Infection in RNase3‐expressing transgenic plants showed that this protein was sufficient to mediate this synergistic interaction with DNA viruses, similar to RNA viruses, but exposed distinct effects on RNA silencing when RNase3 was expressed from its native virus, or constitutively from a transgene, despite a similar pathogenic outcome.
Keywords: begomovirus, RNase3, sweet potato, Sweet potato chlorotic stunt virus, synergism
Introduction
Sweet potato [Ipomoea batatas (L.) Lam] is a perennial plant and the sixth most important crop on which the world depends for its food security (FAO, 2013). Because sweet potato is vegetatively propagated by taking cuttings from a previous crop (directly or from sprouted tubers), the build‐up of virus infections over generations is a major constraint and contributes to severe losses and cultivar decline (Clark et al., 2012). More than 30 viruses have been reported to infect sweet potato, and half are recently described DNA viruses belonging to the families Geminiviridae and Caulimoviridae (Clark et al., 2012). Geminiviruses (family Geminiviridae) are plant viruses that have a circular, single‐stranded DNA genome encapsidated within twinned isometric particles (Fauquet and Stanley, 2003). They are grouped into four genera based on insect vector, host range and genome organization (Fauquet and Stanley, 2003). Members of the genus Begomovirus are transmitted by whiteflies, have single or bipartite component genomes and infect dicotyledonous plants. Within the genus Begomovirus, the sweet potato‐infecting viruses are monopartite, but are distinct from other monopartite begomoviruses, forming a phylogenetically unique lineage, and are referred to as sweepoviruses as a group (Albuquerque et al., 2012; Esterhuizen et al., 2012; Fauquet and Stanley, 2003; Wasswa et al., 2011). Sweepovirus taxonomy is complex and the currently sequenced sweepoviruses have been suggested to correspond to up to 17 different species based on nucleotide sequence identities (Albuquerque et al., 2012). However, the occurrence of frequent recombinants and the lack of any complementing biological differences between suggested species currently render these classifications of little practical use and, in this paper, we refer to them simply as sweepoviruses.
Despite the lack of obvious symptoms associated with sweepovirus infections, yield reductions of between 10% and 80% have been reported in infected sweet potato plants (Clark and Hoy, 2006; Ling et al., 2010). As they can occur at relatively high incidences in crops, they may be responsible for considerable crop losses on a global scale.
Studies with several other sweet potato‐infecting viruses have shown that most can cause severe synergistic disease complexes when co‐infected with Sweet potato chlorotic stunt virus (SPCSV; genus Crinivirus, family Closteroviridae), leading to increased yield losses. These include RNA viruses of the genera Potyvirus, Ipomovirus, Carlavirus and Cucumovirus, as well as otherwise symptomless DNA viruses of the genera Cavemovirus and Solendovirus (Cuellar et al., 2011b; Karyeija et al., 2000; Mukasa et al., 2006; Untiveros et al., 2007). The best studied of these synergistic diseases is that between SPCSV and Sweet potato feathery mottle virus (SPFMV; genus Potyvirus, family Potyviridae), which has been shown to be mediated by the SPCSV‐encoded RNase3 protein, which can also mediate synergistic disease with two other unrelated RNA viruses (Cuellar et al., 2009). RNase3 is a double‐stranded RNA (dsRNA)‐specific class 1 RNA endoribonuclease III that can digest long and short dsRNA, and functions as an RNA silencing suppressor (RSS) (Cuellar et al., 2009; Weinheimer et al., 2014). RNase3 catalytic activity is required for its RSS activity (Cuellar et al., 2009), implicating RNA cleavage in the process of RSS suppression as well as synergistic disease induction. Although the exact mechanism of RNase3 action has not yet been elucidated, it is clear that it is able to mediate increased susceptibility of sweet potato to a wide range of viruses (Cuellar et al., 2009).
However, synergism between SPCSV and sweepoviruses has not yet been reported. Wasswa et al. (2011) reported that a Ugandan sweepovirus isolate was not obviously synergized by co‐infection with SPCSV; however, our own observations with our reference isolate StV1 seemed to indicate that this is not the case for all strains. Therefore, in the current study, we screened a selection of sweet potato germplasm for the presence and variability of sweepoviruses, and selected the six most diverse isolates for complete sequencing and co‐infection experiments with SPCSV to determine whether synergism with SPCSV is a general phenomenon for sweepoviruses as it is with other viruses. Small interfering RNA (siRNA) profiles in single and mixed infections were also determined with the reference isolate StV1, and compared with those of the well‐studied interaction between SPCSV and SPFMV.
Results
Virus detection and host symptoms
During standard virus indexing of germplasm material at the International Potato Center (CIP), Lima, Peru, generally between 10% and 20% of samples were found to be infected with begomoviruses (data not shown). We took advantage of one of the batches of Latin American sweet potato accessions (329 genotypes; Table S1, see Supporting Information), going through virus indexing to characterize the amplified begomovirus nucleotide sequences identified in 39 accessions. In addition, we screened by polymerase chain reaction (PCR) a collection of 65 (symptomless; Table S1) sweet potato plants collected in East Africa, seven of which were found to be positive for begomoviruses. Symptomatology was recorded among the sweet potato accessions from the Americas in the indicator plant Ipomoea setosa. No large variation in symptoms was observed among plants infected with begomovirus, but they could be broadly characterized into three categories: typical upwards leaf curling, chlorosis, or both were observed in indicator plants with all isolates that were positive to sweepoviruses by PCR (Table 1); in several cases, plants were co‐infected by other viruses; nevertheless, the infected sweet potato plants themselves were all symptomless. None of the samples used in this work were found to be co‐infected with SPCSV.
Table 1.
Sweepovirus isolates used in this study
| Isolate name | Symptoms in Ipomoea setosa b | Co‐infected with | Country of origin | Sequence accession number |
|---|---|---|---|---|
| Arg33 | C,RU | Argentina | KC253260 | |
| Arg34 | C,RU,D | Argentina | KC253261 | |
| Arg35 | R,C,RD,RU,D,Vc | SPFMV | Argentina | KC253262 |
| Arg36 | C,RU,D | Argentina | KC253263 | |
| Arg37 | RU | Argentina | KC253264 | |
| Arg38 | R,C,Cp,RU,D,Vc,IVC | SPFMV | Argentina | KC253265 |
| Col22 | RU,Vc,Np,Ln | Colombia | KC253252 | |
| Col9 | C,RU | Colombia | KC253242 | |
| Cub5a | C | Cuba | KC253236 | |
| Cub42 | C,IVC | Cuba | KC288164 | |
| Dom13 | C,RU,IVC | Dominican Republic | KC253244 | |
| Dom2 | C,RU | Dominican Republic | KC253239 | |
| Ecu8 | C,Cp,RU,Vc | SPFMV, SPVG | Ecuador | KC253241 |
| Gtm16 | C,RU | Guatemala | KC253247 | |
| Gtm17 | C,RU | SPVCV | Guatemala | KC253248 |
| Gtm19 | C,RU,D,IVC | Guatemala | KC253250 | |
| Gtm20 | C,RU | Guatemala | KC253251 | |
| Jam12a | C,RU | Jamaica | KC253235 | |
| Jam23 | C,RU | SPVCV | Jamaica | KC253253 |
| Jam24 | C | Jamaica | KC253254 | |
| Mex31a | C,RU,D | Mexico | KC253237 | |
| Mex32 | C,RU,D | Mexico | KC253259 | |
| Mex39 | C,RU,D,Np | SPCV | Mexico | KC288161 |
| Nic25 | C,RU,D | Nicaragua | KC253255 | |
| Pam18 | C,RU,D,IVC | Panama | KC253249 | |
| Pan14 | C,RU,D | Panama | KC253245 | |
| Pan15 | RD,RU,D,Ld | Panama | KC253246 | |
| Per10a | C,RU | Peru | KC253233 | |
| Per6a | C,RU | Peru | KC253234 | |
| Per7 | C,RU | Peru | KC253240 | |
| Pri21 | C,RU,D,IVC | Puerto Rico | KC288165 | |
| Pry11 | C | Paraguay | KC253243 | |
| Pry26 | Cp,RU,D,Vc | SPFMV, SPVG | Paraguay | KC253256 |
| Pry27 | C,RU | Paraguay | KC253257 | |
| Pry29 | RU | Paraguay | KC253258 | |
| Pry30 | C,RU | Paraguay | KC288166 | |
| Pry40 | C,RU | Paraguay | KC288162 | |
| StV1a | RD,RU,D,Ld | St. Vincent and the Grenadines | KC253238 | |
| StV41 | D, IVC | St. Vincent and the Grenadines | KC288163 | |
| Tza13 | nt | Tanzania | KC288167 | |
| Tza16 | nt | SPMMV | Tanzania | KC288169 |
| Uga15 | nt | Uganda | KC288168 | |
| Uga19 | nt | Uganda | KC288170 | |
| Uga29 | nt | SPFMV | Uganda | KC288171 |
| Uga34 | nt | Uganda | KC288172 | |
| Uga37 | nt | Uganda | KC288173 |
Genomes were fully sequenced and isolates used in synergism studies.
Symptoms on leaves: C, chlorosis; Cp, chlorotic point; D, dwarfing; IVC, interveinal chlorosis; Ld, leaf deformation; Ln, leaf necrosis; Np, necrotic point; nt, not tested; R, rugosity; RD, roll down; RU, roll up; Vc, vein clearing.
Sweepovirus sequence variability and characterization
The sequences of the PCR fragments obtained using the universal sweepovirus primers SPG1 and SPG2 (Li et al., 2004) of 46 isolates identified in this study were determined and compared with those available in GenBank by Pairwise Aligned Sequence Comparison (PASC) and phylogenetic analysis. Phylogenetic analysis using our sequences and others available in GenBank showed that the isolates sequenced in this study spanned the variability found in sweepoviruses, except for the cluster corresponding to Ipomoea yellow vein virus and Sweet potato leaf curl canary virus (data not shown).
The complete genomes of the most divergent sweepoviruses based on the analysis of the partial sequences described above, which were found to be free of any other virus detectable by index grafting to I. setosa, were selected for complete genome sequencing together with our reference isolate. These included isolates StV1 (Saint Vincent), Mex31 (Mexico), Cub5 (Cuba), Jam12 (Jamaica), Per6 and Per10 (Peru). These isolates were then also used in double infection studies with SPCSV‐m2‐47 as described below. Complete genome comparison confirmed that all six viruses were quite different from each other, with less than 89% nucleotide identity over their genome, except for Jam12 and Cub5 (91.7%), and StV1 and Per10 (93.1%). Isolates StV1, Per‐6 and Per‐10 were most similar to the International Committee for the Taxonomy of Viruses (ICTV) approved species Sweet potato leaf curl virus (93%, 93% and 98% identity to the type isolate, respectively), whereas Jam‐12 and Cub‐5 were most similar to Sweet potato leaf curl Georgia virus (91% and 95% similarity to the type isolate, respectively) and Mex‐31 was most similar to Sweet potato leaf curl South Carolina virus (93% identity to the type isolate). Alignment and phylogenetic analysis using 108 complete sweepovirus genome sequences available from GenBank confirmed that the sequenced isolates were well distributed among the known sweepovirus variability (Fig. 1). All sequences were deposited in the GenBank database (see Table 1 for GenBank accession numbers).
Figure 1.
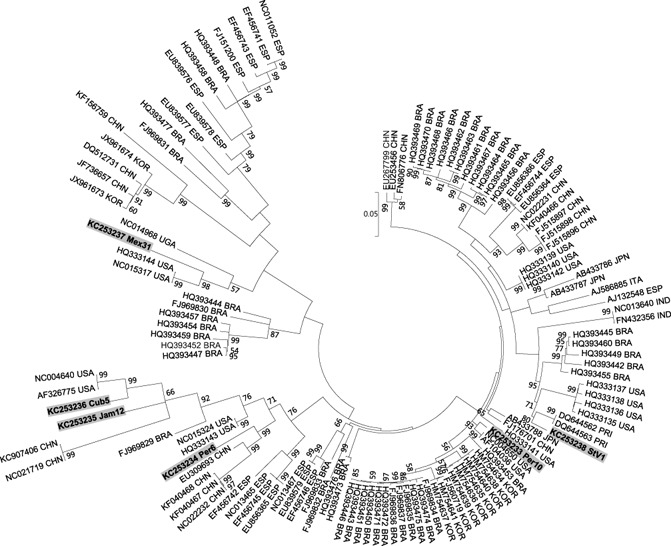
Maximum likelihood phylogenetic tree based on complete genome sequences of 123 isolates. Sequences are indicated with their accession numbers and country of origin; sequences determined in this study are in bold and highlighted. Country codes: BRA, Brazil; CHN, China; Cub, Cuba; ESP, Spain; IND, India; ITA, Italy; Jam, Jamaica; JPN, Japan; KOR, South Korea; Mex, Mexico; Per, Peru; PRI, Puerto Rico; StV, Saint Vincent and the Grenadines; UGA, Uganda; USA, USA.
Synergistic interaction of sweepoviruses with SPCSV in sweet potato
Sweepovirus isolates StV1, Mex31, Cub5, Jam12, Per6 and Per10 were used as inoculum for synergistic interaction studies in sweet potato cultivar ‘Huachano’. Co‐infection with SPCSV led to symptoms of upward leaf curling transiently around 3–4 weeks post‐inoculation only in isolate StV1. This was repeated when plants were cut back and symptoms occurred again in the re‐growth after about 3–4 weeks. None of the other five isolates caused any visible symptoms at any time point. No increase in the severity of SPCSV symptoms was observed (i.e. purpling/yellowing of older leaves or stunting) in combination with any of the sweepovirus isolates. Signal intensity analysis of DNA dot‐blot hybridization (Fig. S1, see Supporting Information) from plants infected with the six sweepovirus isolates showed a significantly higher accumulation of the viruses in plants co‐infected with SPCSV (Fig. 2) in all cases, except isolate Cub5, for which the difference was not significant (P < 0.05). The extent and time point of manifestation of this difference varied considerably between the different isolates, but could be generally divided into three categories. Hybridization signals of isolates StV1 and Mex31 were significantly different from those of the remaining isolates and showed the overall highest virus titres and distinct titre development: sweepovirus titres initially increased much more rapidly in mixed infected plants than in singly infected plants during the first 3 weeks, reaching a peak between 4 and 5 weeks post‐inoculation and another peak at 10 weeks post‐inoculation. However, isolates Per6 and Per10 co‐infected with SPCSV showed an increase in titre during the first 6 weeks (primary infection) relative to singly infected plants and, after cutting back at 6 weeks, the re‐grown sweet potato plants (secondary infection) reached a peak in titre after 1 week, followed by a reduction. Finally, isolates Cub5 and Jam12, although significantly different from each other, showed much lower titres and smaller differences between single and mixed infected plants during primary infection, whereas differences in secondary infection were more pronounced, especially in Jam12. In general, the severity of symptoms caused in the indicator plant I. setosa correlated positively with the titres determined in sweet potato, with StV1 and Mex31 showing the most rapidly developing and severe symptoms in I. setosa, and Cub5 the mildest symptoms. Isolate StV1 showed the strongest signals of all isolates tested and, except for the first week, showed the largest differences in titre between singly and doubly infected plants throughout the experiment. It was also the only isolate to induce symptoms typical of begomovirus infection, upward leaf curling (Fig. 3a), although this only happened transiently during the third and fourth weeks after infection, and again 4 weeks after cutting back the plants. This occurred in all plants co‐infected with SPCSV and coincided with the time of maximum virus accumulation in the plants (Fig. 2).
Figure 2.
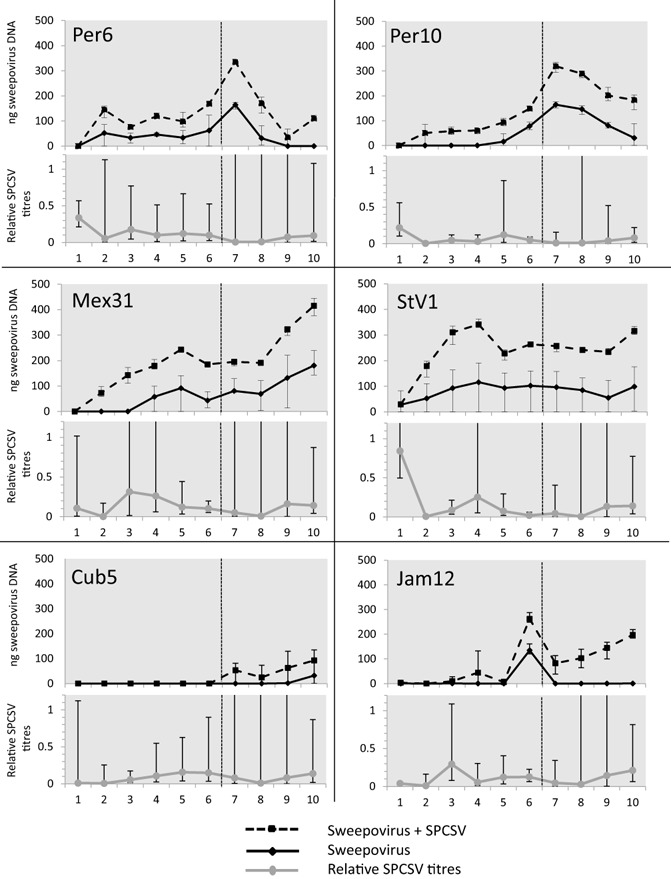
Sweepovirus titres and relative Sweet potato chlorotic stunt virus (SPCSV) titres from single and dually infected plants with different sweepovirus isolates. For each sweepovirus isolate, the titres of sweepovirus, determined by signal intensity analysis of DNA dot‐blots, are shown in the top graph in single (full line) and mixed (dotted line) infection with SPCSV m2‐47. The bottom graph indicates the titres of SPCSV (grey line) in dual infection with sweepovirus isolates relative to single SPCSV infection, as determined by real‐time qualitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Horizontal axis indicates weeks post‐inoculation of sweepovirus and the dotted vertical line through the graphs indicates the time point at which plants were cut back and left to re‐grow. Error bars indicate the range of minimum and maximum values.
Figure 3.
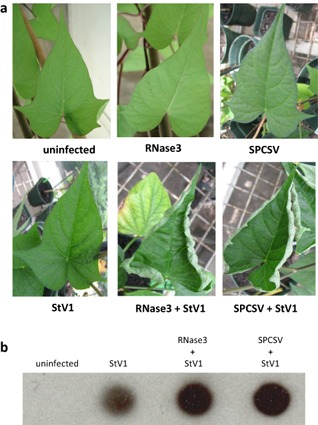
Symptoms and increased sweepovirus titres in Sweet potato chlorotic stunt virus (SPCSV) co‐infected and RNase3‐expressing transgenic sweet potato plants 10 weeks after inoculation. (a) Phenotype of typical leaves from uninfected non‐transgenic, uninfected RNase3‐transgenic and SPCSV‐infected non‐transgenic sweet potato cultivar Huachano (top), and StV1‐infected non‐transgenic, StV1‐infected RNase3‐transgenic, and SPCSV and StV1 dual‐infected non‐transgenic plants of the same cultivar (bottom). (b) DNA dot‐blot of uninfected and StV1‐infected non‐transgenic, StV1‐infected RNase3‐transgenic and SPCSV and StV1 dual‐infected non‐transgenic plants.
Although the dot‐blot indicated increased titres of sweepoviruses in dual infections, qualitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of the SPCSV Hsp70 gene region (RNA2) in the same plants indicated that average SPCSV RNA titres were significantly reduced (P < 0.01) compared with those in SPCSV single infection in all combinations (Fig. 2, Table S2, see Supporting Information). This was confirmed by triple antibody sandwich‐enzyme‐linked immunosorbent assay (TAS‐ELISA) tests (detecting the coat protein) with the isolate StV1, where a decrease in SPCSV antigen was observed (data not shown).
To test whether the observed increases in sweepovirus titres could be mediated by RNase3 alone, as has been shown for RNA viruses, transgenic plants expressing RNase3 (Cuellar et al., 2009) were inoculated with StV1. The infected plants developed typical leaf curling symptoms in the same temporary fashion as seen for non‐transgenic plants co‐inoculated with SPCSV and StV1, and a similar increase in virus titres (Fig. 3). Similarly, infection with two unrelated DNA viruses, Sweet potato collusive virus (SPCV; genus Cavemovirus, family Caulimoviridae) and Sweet potato vein clearing virus (SPVCV; genus Solendovirus, family Caulimoviridae) also reproduced symptoms similar to those caused by co‐infection with SPCSV, and resulted in increased viral titres (Fig. S2, see Supporting Information).
Analysis of virus‐derived siRNA from singly and doubly infected, and RNase3‐transgenic, sweet potato plants
Raw siRNA sequence reads of the seven different samples analysed in this study are available at https://research.cip.cgiar.org/confluence/display/cpx/CIP.sweetpotato.2014. Comparison of siRNA sequences determined from uninfected, StV1‐, SPCSV‐, and SPCSV and StV1‐infected plants showed notable changes in the relative amounts of siRNAs corresponding to each virus, as well as the regions to which they mapped, particularly in the case of SPCSV (Figs 4, 5 and S3, see Supporting Information). The relative number of siRNA reads corresponding to SPCSV increased by more than three‐fold from 7500/million reads to 23 708/million reads, with most of the increase corresponding to RNA1 (Fig. 4). Relative amounts of begomovirus‐specific siRNA did not change, beyond the variation found between individual samples of the same treatment (±0.1%), in doubly infected plants relative to singly infected plants (1.1‐fold; from 82 032/million reads to 90 275/million reads). StV1‐specific siRNAs also mapped to similar positions in the genome, and no obvious differences could be observed (Fig. 5a). This was in stark contrast with the mapping of reads to the SPCSV genome, where a dramatic change could be observed in the relative amounts and positions to which the siRNAs mapped on the genome (Fig. 5b). This was characterized by a several‐fold increase in siRNAs corresponding to SPCSV (Fig. 4), a reduction in 21‐nucleotide siRNA (reduced from 37% to 15.8% of all siRNAs corresponding to SPCSV) and a corresponding increase in 22‐ and 23‐nucleotide siRNAs (increased from 39.6% to 47.1% and 12.8% to 26.5%, respectively, of all siRNAs corresponding to SPCSV), as well as a near‐disappearance of siRNAs matching to the 5′ regions of SPCSV (Fig. 5b), in plants co‐infected with isolate StV1 relative to single SPCSV infection. To determine whether the effect of change in siRNA mapping to SPCSV was specific to the synergism between SPCSV and StV1, or a general response found in synergistic interactions with other viruses, we sequenced siRNAs from plants infected with SPCSV and SPFMV. No reduction in the mapping of siRNAs to the 5′ region was observed in these plants (Fig. 5b), nor was there a change in the total amount of siRNAs corresponding to SPCSV (Fig. 4).
Figure 4.
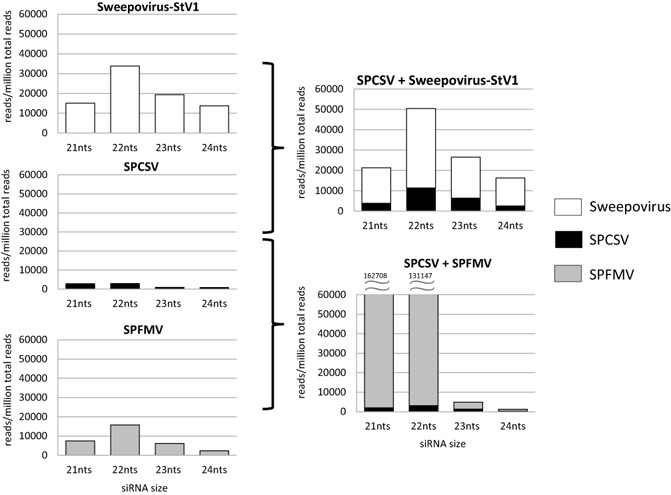
Distribution of small interfering RNA (siRNA) size classes corresponding to sweepovirus (white bars), Sweet potato chlorotic stunt virus (SPCSV) (black bars) and Sweet potato feathery mottle virus (SPFMV) (grey bars) in single and dually infected plants. Vertical axis shows the number of siRNA reads per million total reads, and the horizontal axis indicates the size class of siRNA. Numbers above the 21‐ and 22‐nucleotide (nt) siRNAs in the right bottom graph indicate the number of siRNAs corresponding to SPFMV, which exceed the scale of the graph. Analysis of the total number of all small RNA reads is provided in Fig. S3.
Figure 5.
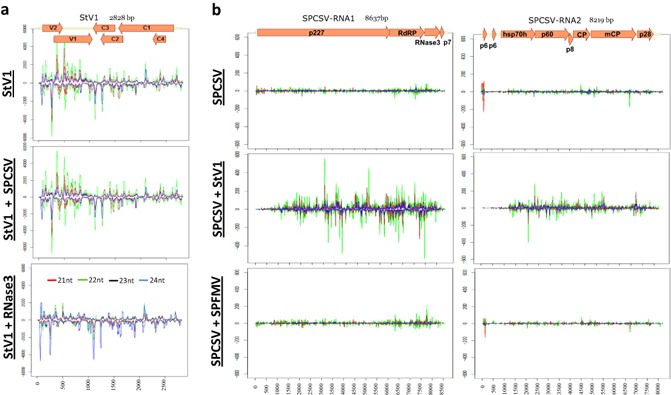
Coverage of viral genomes by different small interfering RNA (siRNA) size classes. (a) Coverage of sweepovirus StV1 genome in single (top graph) or dual (middle graph) infection with Sweet potato chlorotic stunt virus (SPCSV) or in RNase3‐transgenic plants (bottom graph). (b) Coverage of SPCSV genome by different siRNA size classes in single (top graph) or dual infection (middle graph) with StV1 or Sweet potato feathery mottle virus (SPFMV) (bottom graph). Vertical axis shows number of siRNA reads per million total reads, and horizontal axis indicates nucleotide position on the genome. Red, green, black and blue lines indicate 21‐, 22‐, 23‐ and 24‐nucleotide RNAs, respectively. A schematic representation of the virus genome is provided at the top with named box arrows representing the locations of the respective encoded genes.
When comparing the distribution of StV1‐specific siRNAs between StV1 and SPCSV co‐infected plants and StV1‐infected RNase3‐expressing plants, however, a surprising difference was noted, in that a several‐fold reduction in 21‐ and 22‐nucleotide siRNAs could be observed in RNase3‐expressing plants relative to SPCSV co‐infected plants and also StV1 singly infected plants (Fig. 5b).
Discussion
Until the beginning of this century, most surveys of sweet potato viruses did not mention begomoviruses (Valverde et al., 2007). Nevertheless closely related begomoviruses have now been reported from all over the world, including North America (Lotrakul et al., 1998, 2002, 2003), the Mediterranean (Banks et al., 1999; Briddon et al., 2006; Cohen et al., 1997; Lozano et al., 2009), Asia (Bi and Zhang, 2012; Luan et al., 2007; Onuki et al., 2000), South America (Albuquerque et al., 2012; Fuentes and Salazar, 2003; Paprotka et al., 2010; Rodríguez‐Pardina et al., 2012) and Africa (Miano et al., 2006; Wasswa et al., 2011). Our report corroborates the common occurrence of sweepoviruses and reveals clear synergistic interactions of sweet potato begomoviruses with SPCSV, another widely distributed virus and mediator of several synergistic interactions (Cuellar et al., 2011b; Karyeija et al., 2000; Mukasa et al., 2006; Untiveros et al., 2007).
Few reports exist on synergistic interactions between RNA and DNA viruses. We have shown previously that SPCSV can interact with members of the Caulimoviridae family of reverse transcribing viruses (Cuellar et al., 2011b), and here we show that this is also the case for sweepoviruses. However, our data also show that the extent of this synergism varies considerably between different sweepovirus isolates and that, in the majority of cases, it is not associated with clear symptoms. This observation may explain why such interactions have not been noticed previously. Nevertheless, the fact that five of six diverse viruses tested showed clear and significant increase in titres in co‐infection with SPCSV indicates that this may be a common phenomenon among sweepoviruses. As has been shown for other synergistic interactions with SPCSV (Cuellar et al., 2008, 2011b; Mukasa et al., 2006), we also found that an increase in titre of the synergized virus is associated with a corresponding decrease in titre of SPCSV relative to single infection (Fig. 2). It is not yet clear why SPCSV titres are reduced in synergistic interactions, but it may simply be a result of competition for limited resources of the two co‐infecting viruses in infected cells, where the association is favouring one over the other.
Nevertheless, when we analysed siRNA targeting of SPCSV in single relative to dual infection with sweepovirus isolate StV1, we were able to detect a striking difference in the relative amount and distribution of siRNA reads. Total siRNAs corresponding to SPCSV increased several fold (Fig. 4), and 22‐ and 23‐nucleotide siRNAs increased relative to 21‐nucleotide siRNA, whereas siRNAs matching to the 5′ regions of SPCSV nearly disappeared (Fig. 5b), in plants co‐infected with isolate StV1 relative to single SPCSV infection. This effect was apparently specific to the interaction of SPCSV with StV1, as similar changes were not observed in the interaction between SPCSV and SPFMV (Figs 4 and 5b).
The specific changes in the distribution and relative quantity of SPCSV‐specific siRNAs as a result of co‐infection with StV1 suggest a modified and increased targeting of SPCSV by the plant's RNA silencing system. A plausible explanation is that this is the result of interference by StV1 with the host's RNA silencing machinery. Although this may seem counterintuitive, it has become clear in recent years that different branches of the RNA silencing system in plants compete with each other for access to cellular machinery (Jauvion et al., 2012). RSS suppression by sweepoviruses is more likely to affect pathways inhibiting the replication of DNA viruses in the nucleus, and this may not necessarily benefit, or even be detrimental to, replicating RNA viruses in the cytoplasm. Two RNA viruses with similar replication strategies, however, are less likely to compromise each other's silencing suppression. This alone may explain why similar changes are not seen in the interaction between SPCSV and SPFMV when compared with the interaction between SPCSV and StV1. In addition, although the tissue tropism of sweepoviruses has not yet been determined, many begomoviruses are phloem limited, similar to SPCSV, and, if this is also the case for sweepoviruses, the effects of the two viruses on each other's replication may be expected to be more evident than in the case in which tissue tropism is distinct, such as for SPFMV and SPCSV (Karyeija et al., 2000).
Although RSSs have not yet been reported for sweepoviruses, a number of studies have reported up to three RSS proteins encoded by single and multipartite begomoviruses, including the (homologues of) V2, C2 and C4 proteins (Amin et al., 2011; Buchmann et al., 2009; Chellappan et al., 2005; Vanitharani et al., 2004; Zhang et al., 2011; Zrachya et al., 2007). These RSSs have been found to suppress silencing at both the transcriptional and post‐transcriptional level, but not all homologous proteins from different viruses have RSS activity or necessarily function in similar ways (Amin et al., 2011; Raja et al., 2010). Thus, although it can be expected that sweepoviruses encode RSS proteins, it is not possible to deduce their identity or how they will function on the basis of knowledge from other begomoviruses. However, we can use our observations regarding the relative changes in siRNA distributions to speculate as to which components of the RNA silencing machinery might be affected. The phenotype of reduced siRNA targeting of the 5′ region and increased targeting towards the 3′ region of SPCSV is reminiscent to that found in cucumber mosaic virus (CMV; Genus Bromovirus, family Bromoviridae)‐infected RDR1‐defective Arabidopsis (Wang et al., 2010). This may indicate that StV1 interferes with RDR1 function. RDR proteins could be expected to fulfil an important role in antiviral silencing against geminiviruses, as they do not normally produce dsRNA. Nevertheless, one must be careful when extrapolating conclusions from one specific model system to sweet potato because marked differences have been found between different geminivirus host combinations (Akbergenov et al., 2006; Miozzi et al., 2013; Rodríguez‐Negrete et al., 2009).
Previously, we have demonstrated that RSS encoded by SPCSV (RNase3) is responsible for the enhanced accumulation of co‐infecting RNA viruses in synergistic interactions mediated by SPCSV (Cuellar et al., 2009). Although the exact mechanism of RNase3 function has not been fully elucidated, its dsRNase activity is essential for silencing suppression as well as enhanced accumulation of viruses in transgenic plants (Cuellar et al., 2009; Kreuze et al., 2005). RNase3 has little substrate specificity in vitro, processing both long and short dsRNA, including siRNAs and pre‐miRNAs (Cuellar et al., 2009; Kreuze et al., 2005; Weinheimer et al., 2014). Nevertheless, its target in plants must be specific, as transgenic plants are phenotypically completely normal, except for their extreme susceptibility to viruses (Cuellar et al., 2009). We hypothesize that the same mechanism is involved in SPCSV synergisms with RNA and DNA viruses. Indeed, infection of RNase3 transgenic sweet potato plants with StV1 (Fig. 3), as well as SPCV and SPVCV (Fig. S2), provoked the characteristic symptoms observed in plants co‐infected with SPCSV. Surprisingly, however, siRNA distribution patterns of StV1 were perceptibly different in plants constitutively expressing RNase3 relative to those from plants co‐infected with SPCSV, in that 21‐ and 22‐nucleotide siRNAs were strongly reduced in RNase3 plants. This suggests that, despite the biologically similar outcomes of enhanced StV1 viral titres and symptom induction, clear differences occur in how RNA silencing is affected in either situation. We offer two possible explanations for this discrepancy: (i) constitutive over‐expression of RNase3 in all plant cells results in a distinct effect of RNase3 on the silencing pathway relative to phloem‐specific expression; or (ii) RNase3 function is modulated by other SPCSV‐encoded proteins to limit its effect to certain sites in the silencing pathway. It is intriguing that, in spite of its constitutive expression in all plant cells, RNase3 does not cause visible collateral effects on sweet potato. Future analysis of siRNA sequences in RNase3 plants infected with different types of virus may shed more light onto the exact target and mechanism of the RNase3‐provoked susceptibility to viruses.
Although we did not analyse the potential effect on yield of the different virus combinations in the current study, the strong increase in sweepovirus titres found in some interactions suggests that yield impacts could be expected, and this should be a priority for future studies. Indeed, other studies have shown significant impacts of sweepovirus infection on the yield of sweet potato, despite being largely symptomless (Clark and Hoy, 2006; Ling et al., 2010). In addition, increases in sweepovirus titres in plant tissues could lead to an increased rate of transmission of the virus by its vector, contributing to more rapid virus spread.
Experimental Procedures
Virus isolates
The 39 begomovirus isolates described in this study (Tables 1 and S1) were identified in sweet potato accessions from Central and South America (Mexico, Guatemala, Cuba, Jamaica, Nicaragua, Dominican Republic, Saint Vincent and the Grenadines, Colombia, Ecuador, Peru, Argentina, Paraguay, Panama and Puerto Rico), after indexing by grafting onto the indicator plant I. setosa followed by PCR (see below), during routine virus indexing performed at CIP (329 accessions). These accessions were either collected by CIP or acquired from other collections between 1986 and 1994. Accessions collected by CIP were established under an insect‐proof screenhouse before being transferred in vitro, where they were maintained as part of CIPs global sweet potato collection. Samples acquired from other sources were obtained in vitro, or as vine cuttings or roots, and, in the latter cases, were established and introduced in vitro as described for the CIP‐collected materials. An additional seven virus isolates were identified by PCR screening from 65 sweet potato genotypes collected from different regions of East Africa (Uganda, Kenya, Tanzania) and originally maintained under field conditions for breeding purposes, and subsequently transferred in vitro for transfer to CIP's sweet potato collection at CIP‐Lima. Metadata of the accessions in which sweepoviruses were identified are provided in Table S1. For synergism experiments, SPCSV isolate m2‐47, lacking the p22 gene (Cuellar et al., 2008, 2011a) and maintained in I. setosa, was used. The SPCV and SPVCV isolates used have been described in Cuellar et al. (2011b).
DNA amplification, cloning and sequence analyses
The Saint Vincent and Grenadines isolate (StV1) was isolated from sweet potato accession CIP400025. The accession has been tested for 10 viruses by ELISA, and grafting onto I. setosa. To amplify begomovirus‐specific fragments from different sweet potato accessions (Table 1), a simple and quick method of DNA extraction using sodium hydroxide was used to prepare template DNA for PCR (Wang et al., 1993). Shoots were collected from in vitro plantlets and homogenized in 0.5 m NaOH buffer in a ratio of 1:5 (tissue : buffer). The samples were centrifuged at 12 000 g for 10 min to spin down the debris. After a spin down, samples were diluted 100 times with Tris‐HCl (100 mm, pH 8) and 1 μL of leaf extract was used directly for PCR in a 25‐μL reaction employing the 2 × phusion polymerase readymade master mix (Finnzymes, Helsinki, Finland) and sweepovirus‐specific primers SPG1 (5′‐CCCCKGTGCGWRAATCCAT‐3′) and SPG2 (5′‐ATCCVAAYWTYCAGGGAGCTAA‐3′) (Li et al., 2004), designed to amplify a 901‐bp region encompassing partial AC1 and AC2 open reading frames (ORFs).
For cloning of the selected begomovirus genomes, total DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) protocol (see below), followed by separation of low‐molecular‐weight DNA using a plasmid isolation protocol (alkaline lysis) and the Wizard miniprep kit (Promega, Madison, WI, USA). The quality and amount of DNA were checked by agarose gel electrophoresis and spectrophotometry using a Nanodrop analyser (ND‐1000, Thermo Fisher Scientific, Wilmington, Delaware, USA), respectively. In the case of isolates Per10 and Jam12, 5 μg of low‐molecular‐weight DNA was used for the amplification of circular DNA using Phi29 polymerase (New England Biolabs, Ipswich, Massachusetts, USA) reaction with a 5× excess of random hexamer primers, according to the manufacturer's instructions. The amplified DNA was then linearized using SmaI for Per10 and StuI for Jam12, resulting in the expected 2.7‐kb fragment. Isolates Per6, Mex31, Cub5 and StV1 were amplified by inverse PCR using a set of degenerate primers designed on the basis of a previously amplified and sequenced region (Bego‐F, 5′‐CTGRCCTCCTCTAGCAGATCKCC‐3′; Bego‐R, 5′‐GARCCTGCKCCTGGATTGCAGAGR‐3′), resulting in the expected 2.3‐kb fragment.
The PCR and digested Phi29 amplified products were separated by agarose gel electrophoresis and then excised and purified using a gel extraction kit (Promega). The fragments were cloned into pGEM‐T easy vector (Promega). Transformation of Escherichia coli DH5α was performed by heat shock at 42 °C for 90 s. Using blue–white screening, putative transformants were screened and confirmed by restriction analysis using EcoRI enzyme prior to sequencing. The samples were then prepared for sequencing (Macrogen, Seoul, South Korea) using SP6 and T7 primers and a set of specifically designed internal primers.
Sequence alignments and phylogenetic analysis were performed using MEGA5.1 (Tamura et al., 2011). Alignments were performed using Muscle and phylogenetic trees were generated after calculating the best‐fitting model: maximum‐likelihood method with the general time‐reversible model using gamma‐distributed rates (with five discrete gamma categories) with invariant sites.
Synergistic interaction of sweepoviruses with SPCSV in sweet potato
Sweet potato cultivar ‘Huachano’ (accession CIP420065), obtained from the germplasm collection of CIP, was used as a rootstock for the graft inoculation of isolates StV1, Per‐6, Per‐10, Jam‐12, Cub‐5 and Mex‐31 with or without SPCSV. SPCSV isolate m2‐47 (Untiveros et al., 2007) was maintained in sweet potato cv. Huachano through cuttings, and all sweepovirus isolates were maintained in I. setosa plants by serial grafting. Nodes from the middle part of virus‐infected source plants were used as scions to graft inoculate sweet potato. Four‐week‐old cuttings of sweet potato cv. ‘Huachano’ were side grafted in the middle of the plant with SPCSV or healthy sweet potato scions. Two weeks later, two nodes above the initial graft, plants were grafted with healthy or sweepovirus‐infected I. setosa scions, thus generating plants infected with SPCSV alone, SPCSV plus sweepovirus, sweepovirus alone and mock inoculated. Three plants per treatment were inoculated and the formation of a graft union was confirmed. Plants were cut back below the graft unions 6 weeks after the last graft inoculation and left to re‐grow for four more weeks. The development of symptoms was recorded every week after inoculation, and total nucleic acid was extracted (see below for methods) from 10‐mm leaf discs from a combination of three leaves collected from the apex, middle and bottom part of each plant at 1–10 weeks post‐inoculation for dot‐blot detection of begomovirus and real‐time qRT‐PCR detection of SPCSV. Leaf samples were stored at −20 °C. TAS‐ELISA for SPCSV was carried out as described previously (Karyeija et al., 2000), 4 weeks after cutting back infected plants, only on plants with single SPCSV infection and mixed infection of SPCSV and isolate StV1. In a separate experiment, three replicates of the RNase3 transgenic sweet potato ‘Huachano’ event R3 (Cuellar et al., 2009) and three non‐transgenic plants were also infected with StV1 and tested by DNA dot‐blot, together with non‐transgenic plants infected with SPCSV and StV1, 4 weeks after cutting back infected plants.
Dot‐blot hybridization and signal quantification
For detection by dot‐blot hybridization, total DNA was purified using the CTAB method (Doyle and Doyle, 1987). Frozen leaf tissues (250 mg) were processed immediately by grinding in 2 mL of CTAB buffer [2% CTAB, 100 mm Tris‐HCl, pH 8.0, 20 mm ethylenediaminetetraacetic acid (EDTA), 1.4 m NaCl, 1.0% Na sulfite and 2.0% Polyvinylpyrrolidone average molecular weight 40,000 (PVP‐40)] using a polypropylene sack. The homogenate was centrifuged at 10 000 g for 10 min and the supernatant (750 μL) was transferred to a 1.5‐mL microcentrifuge tube and mixed with an equal volume of chloroform–isoamyl alcohol (24:1). The mixture was centrifuged at 12 000 g for 10 min and the aqueous phase (500 μL) was transferred to a new 1.5‐mL microcentrifuge tube before mixing in 550 μL of isopropanol. The mixture was incubated on ice for 10 min and centrifuged at 12 000 g for 10 min at 4 °C. The pellet was washed with 70% ethanol, centrifuged at 12 000 g for 5 min, air dried and dissolved in 100 μL of Nuclease free water (NFW). Five micrograms of total DNA in a total volume of 200 μL were used for hybridization. Standards of 125, 50 and 25 ng of plasmid DNA containing the region corresponding to the probe used were also added in duplicate to each membrane tested to normalize and quantify the results. Digoxigenin (DIG)‐labelled probes encompassing the Rep gene region (AC1) of StV1 were synthesized by PCR using primers SPG1 and SPG2 (Li et al., 2004), Taq polymerase (Promega) and DIG‐labelled deoxynucleotides (Roche, West Sussex, UK). Total DNA from infected plants was transferred to a nylon membrane (Hybond‐N; Amersham Biosciences AB, Amersham, Freiburg, Germany) using a Bio‐Dot SF Cell (Bio‐Rad, Richmond, California, USA), cross‐linked by UV irradiation (50 mJ) in a cross‐linking oven (Stratagene, La Jolla, California, USA), prehybridized for 90 min at 65 °C in 0.02% sodium dodecylsulfate (SDS), 5 × SSC (750 mm NaCl, 75 mm sodium citrate), 50% formamide, 2% (w/v) N‐lauroylsarcosine, and then hybridized in the same solution at 65 °C for 16 h after addition of the DIG‐labelled probe. After hybridization, membranes were washed twice in 2 × SSC and 1% SDS at room temperature for 15 min, incubated for 30 min with anti‐DIG antibodies conjugated with alkaline phosphatase, and washed twice with maleate buffer with 0.3% Tween‐20. The reaction was developed using chemiluminescent substrate disodium 3‐(4‐methoxyspiro(1,2‐dioxetane‐3,2′‐(5′‐chloro)tricyclo[3.3.1]decan)‐4‐yl)phenyl phosphate (CSPD) (Roche, Penzberg, Germany) and Omat‐S film (Kodak, Rochester, New York, USA). Signal intensities of the hybridized spots were measured from developed films using Gel Doc equipment in conjunction with Quantity One software (Bio‐Rad) under white light. The signal intensity was determined using a volume circle tool, ensuring that circles were all the same size and covered each spot exactly, with global background subtraction and avoiding overexposed pixels. The concentration of viral DNA inside each circle was then determined using a regression curve based on the volumes of the plasmid standards within each membrane using the Volume Analysis Report and Volume Regression Curve within Quantity One. The estimated viral concentrations (in nanograms) were then employed for statistical analysis using the SAS statistical package. Membranes were stripped and hybridized using rDNA‐specific probes (amplified using primers: Ribosomal F, 5′‐ACAGCAGAACGACCAGAGAACGC‐3′; Ribosomal R, 5′‐GCACGCTAGGTACGACCACCACT‐3′) to confirm equal loading of DNA between samples. Initially, a repeated measures analysis of variance was performed revealing highly significant (P < 0.0001) probability of interactions between time point, isolate and treatment. Subsequently an analysis of variance was performed for each time point to determine the effect of treatments and isolates at each time point. A full analysis of variance considering time and isolate as factors was also performed to determine the global effect of single versus dual infection for each isolate.
Real‐time qRT‐PCR for detection of SPCSV
The same samples as described for the dot‐blot hybridizations above were used to extract RNA employing the CTAB method described previously, but modified to precipitate total RNA by the addition of an equal volume of 4 m LiCl rather than isopropanol, and overnight incubation at 4 °C followed by centrifugation at 10 000 g for 20 min. The pellet was washed with 70% ethanol as described above and re‐dissolved in 100 μL of NFW.
A TaqMan real‐time RT‐PCR assay was then used to detect SPCSV. One‐step real‐time RT‐PCR assays were performed using the TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in a 25‐μL final reaction volume containing 2 U of Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV RT), 300 nm of each primer, 100 nm of the probe and 2 μL of template RNA. The following thermal cycling conditions were used: 42 °C for 30 min (cDNA synthesis), 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Primers for SPCSV were SPCSV‐Uni‐E‐F (5′‐CGGAGTTTATTCCCACYTGTYT‐3′) and SPCSV‐Uni‐E‐R (5′‐GGGCAGCCYCACCAA‐3′) and the probe was SPCSV‐Uni‐E‐P (5′‐[FAM]‐TCTGTCACGGCTACAGGCGACGTG‐[TAMRA]‐3′), corresponding to the Hsp70h region on RNA2. Cytochrome oxidase (COX) was used as internal reference gene employing the primers COX‐F (5′‐CGTCGCATTCCAGATTATCCA‐3′) and COX‐R (5′‐CAACTACGGATATATAAGAGCCAAAACTG‐3′), and probe COX‐P (5′‐[VIC]‐TGCTTACGCTGGATGGAATGCCCT‐[TAMRA]‐3′).
Results were analysed with MxPro QPCR Software and statistically significant differences between single and mixed infections were determined for each time point/virus combination as well as each virus combination over all time points, using the Relative Expression Software Tool (REST) v2.0.12 (Qiagen GmbH, Hilden, Germany).
siRNA sequencing and analysis
Total RNAs were extracted from three leaves each of healthy, SPCSV‐infected, StV1‐infected, SPCSV + StV1‐infected, SPFMV‐infected and SPCSV + SPFMV‐infected ‘Huachano’ plants, as well as StV1‐infected RNase3 transgenic ‘Huachano’ plants (Cuellar et al., 2009) at several months after inoculation using TRIZOL reagent. siRNAs were purified from 4% agarose gel and sent for library preparation and Illumina sequencing (Provider: Fasteris Life Sciences SA, Plan‐les‐Ouates, Switzerland).
Reads were mapped to the corresponding genomes using MAQ software, and results were visualized using a custom script (Fuentes et al., 2012) and Microsoft Excel (bar charts).
Supporting information
Fig. S1 (a) DNA dot‐blot results of sweepoviruses used to generate the graphs in Fig. 2. (b) Results of the same blots with ribosomal DNA probe, demonstrating equal loading.
Fig. S2 Symptoms of pararetroviruses [Sweet potato vein clearing virus (SPVCV) and Sweet potato collusive virus (SPCV)] in transgenic sweet potato cultivar ‘Huachano’ expressing the RNase3 gene of Sweet potato chlorotic stunt virus (SPCSV). (a, c) Non‐transgenic ‘Huachano’ infected with SPVCV and SPCV, respectively. (b, d) Transgenic ‘Huachano’ plants expressing RNase3 and infected with SPVCV and SPCV, respectively. (e) Dot‐blot detection of SPVCV in transgenic versus non‐transgenic sweet potato ‘Huachano’ plants. N.I., not infected plant; F, detection of SPCV by Nitrocellulose membrane enzyme‐linked imunosorbent assay (NCM‐ELISA); 1, 2, non infected ‘Huachano’ plant; 3, 4, SPCV infected ‘Huachano’ plants; 5, 6, SPCV in RNase3‐transgenic plants; 7, 8, SPCV in co‐infection with SPCSV.
Fig. S3 Distribution of total small RNA size classes corresponding to SvT1 (white bars), Sweet potato chlorotic stunt virus (SPCSV, black bars), Sweet potato feathery mottle virus (SPFMV, dark grey bars) and other sequences (light grey bars) in wild‐type plants singly and dually infected with StV1 and SPCSV or RNase3‐transgenic plants infected with StV1 (a), wild‐type plants singly and dually infected with SPFMV and SPCSV (b), and non‐infected wild‐type plants (c). The vertical axis shows the total number of small RNA reads, and the horizontal axis indicates the size class of siRNA, and the exact numbers are tabulated below the graph, including the sums and grand total.
Table S1 Origin of plants, collection date, genotype name and germplasm accession numbers of plants screened for sweepoviruses in this study.
Table S2 Relative expression of Sweet potato chlorotic stunt virus (SPCSV) in mixed infection with six sweepovirus isolates, relative to single infection, over all time points based on Relative Expression Software Tool (REST) analysis of qualitative reverse transcription‐polymerase chain reaction (qRT‐PCR) results.
Acknowledgements
We are grateful to all members of the Virology Unit at the International Potato Center (CIP), Lima, Peru. Thanks are due to Silver Tumwegamire and Willmer Perez for providing access to sweet potato samples from East Africa, and Neil Boonham and Ian Adams for the design of real‐time PCR assays. Financial support from British Biological Sciences Research Council (BBSRC)/Department for international Development (DFID) (grant BBF0040281) and International Centre for Genetic Engineering and Biotechnology(BBSRC)‐The Academy of Sciences for the Developing World (TWAS)‐United Nations Educational, Scientific and Cultural Organization / International Basic Sciences Programme (UNESCO/IBSP) Joint Programme on Capacity Building in Basic Molecular Biology (Contract No.: CRP/101005) is acknowledged.
References
- Akbergenov, R. , Si‐Ammour, A. , Blevins, T. , Amin, I. , Kutter, C. , Vanderschuren, H. , Zhang, P. , Gruissem, W. , Meins, F. Jr , Hohn, T. and Pooggin, M. (2006) Molecular characterization of geminivirus‐derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque, L.C. , Inoue‐Nagata, A.K. , Pinheiro, B. , Resende, R.O. , Moriones, E. and Navas‐Castillo, J. (2012) Genetic diversity and recombination analysis of sweepoviruses from Brazil. Virol. J. 9, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, I. , Hussain, K. , Akbergenov, R. , Yadav, J.S. , Qazi, J. , Mansoor, S. , Hohn, T. , Fauquet, C.M. and Briddon, R.W. (2011) Suppressors of RNA silencing encoded by the components of the Cotton Leaf Curl Begomovirus‐BetaSatellite Complex. Mol. Plant–Microbe Interact. 24, 973–983. [DOI] [PubMed] [Google Scholar]
- Banks, G.K. , Bedford, I.D. , Beitia, F.J. , Rodriguez‐Cerezo, E. and Markham, P.G. (1999) A novel geminivirus of Ipomoea indica (Convolvulacae) from southern Spain. Plant Dis. 83, 486. [DOI] [PubMed] [Google Scholar]
- Bi, H. and Zhang, P. (2012) Molecular characterization of two sweepoviruses from China and evaluation of the infectivity of cloned SPLCV‐JS in Nicotiana benthamiana . Arch. Virol. 157, 441–454. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. , Bull, S.E. and Bedford, I.D. (2006) Occurrence of Sweet potato leaf curl virus in Sicily. Plant Pathol. 55, 286. [Google Scholar]
- Buchmann, R.C. , Asad, S. , Wolf, J.N. , Mohannath, G. and Bisaro, D.M. (2009) Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome‐wide reductions in cytosine methylation. J. Virol., 83, 5005–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan, P. , Vanitharani, R. and Fauquet, C.M. (2005) MicroRNA‐binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA, 102, 10 381–10 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, C.A. and Hoy, M.W. (2006) Effects of common viruses on yield and quality of Beauregard sweetpotato in Louisiana. Plant Dis. 90, 83–88. [DOI] [PubMed] [Google Scholar]
- Clark, C.A. , Davis, J.A. , Abad, J.A. , Cuellar, W.J. , Fuentes, S. , Kreuze, J.F. , Gibson, R.W. , Mukasa, S.B. , Tugume, A.K. , Tairo, F.D. and Valkonen, J.P.T. (2012) Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis. 96, 168–185. [DOI] [PubMed] [Google Scholar]
- Cohen, J. , Milgram, M. , Antignus, Y. , Pearlsman, M. , Lachman, O. and Loebenstein, G. (1997) Ipomoea crinkle leaf curl caused by a whitefly‐transmitted gemini‐like virus. Ann. Appl. Biol. 131, 273–282. [Google Scholar]
- Cuellar, W.J. , Tairo, F. , Kreuze, J.F. and Valkonen, J.P.T. (2008) Analysis of gene content in Sweet potato chlorotic stunt virus RNA1 reveals the presence of p22 RNA silencing suppressor in only few isolates: implications to viral evolution and synergism. J. Gen. Virol. 89, 573–582. [DOI] [PubMed] [Google Scholar]
- Cuellar, W.J. , Kreuze, J.F. , Rajamäki, M.‐L. , Cruzado, K.R. , Untiveros, M. and Valkonen, J.P.T. (2009) Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. USA, 106, 10 354–10 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar, W.J. , Cruzado, R.K. , Fuentes, S. , Untiveros, M. , Soto, M. and Kreuze, J.F. (2011a) Sequence characterization of a Peruvian isolate of Sweet potato chlorotic stunt virus: further variability and a model for p22 acquisition. Virus Res. 157, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar, W.J. , De Souza, J. , Barrantes, I. , Fuentes, S. and Kreuze, J.F. (2011b) Distinct cavemoviruses interact synergistically with sweet potato chlorotic stunt virus (genus Crinivirus) in cultivated sweet potato. J. Gen. Virol. 92, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–19. [Google Scholar]
- Esterhuizen, L.L. , Heerden, S.W. , Rey, M.E.C. and Heerden, H. (2012) Genetic identification of two sweet‐potato‐infecting begomoviruses in South Africa. Arch. Virol. 157, 2241–2245. [DOI] [PubMed] [Google Scholar]
- FAO (2013) FAOSTAT http://faostat.fao.org.
- Fauquet, C.M. and Stanley, J. (2003) Geminivirus classification and nomenclature: progress and problems. Ann. Appl. Biol. 142, 165–189. [Google Scholar]
- Fuentes, S. and Salazar, L.F. (2003) First report of Sweet potato leaf curl virus in Peru. Plant Dis. 87, 98. [DOI] [PubMed] [Google Scholar]
- Fuentes, S. , Heider, B. , Tasso, R.C. , Romero, E. , Zum Felde, T. and Kreuze, J.F. (2012) Complete genome sequence of a potyvirus infecting yam beans (Pachyrhizus spp.) in Peru. Arch. Virol. 157, 773–776. [DOI] [PubMed] [Google Scholar]
- Jauvion, V. , Rivard, M. , Bouteiller, N. , Elmayan, T. and Vaucheret, H. (2012) RDR2 partially antagonizes the production of RDR6‐dependent siRNA in sense transgene‐mediated PTGS. PLoS ONE, 7, e29785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyeija, R.F. , Kreuze, J.F. , Gibson, R.W. and Valkonen, J.P.T. (2000) Synergistic interactions of a potyvirus and a phloem‐limited crinivirus in Sweet potato plants. Virology, 269, 26–36. [DOI] [PubMed] [Google Scholar]
- Kreuze, J.F. , Savenkov, E.I. , Cuellar, W. , Li, X. and Valkonen, J.P.T. (2005) Viral Class 1 RNase III involved in suppression of RNA silencing. J. Virol. 79, 7227–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R.H. , Salih, S. and Hurtt, S. (2004) Detection of geminiviruses in sweetpotato by polymerase chain reaction. Plant Dis. 88, 1347–1351. [DOI] [PubMed] [Google Scholar]
- Ling, K.‐S. , Jackson, D.M. , Harrison, H. , Simmons, A.M. and Pesic‐VanEsbroeck, Z. (2010) Field evaluation of yield effects on the U.S.A. heirloom sweetpotato cultivars infected by Sweet potato leaf curl virus. Crop Prot. 29, 757–765. [Google Scholar]
- Lotrakul, P. , Valverde, R.A. , Clark, C.A. , Sim, J. and De La Torre, R. (1998) Detection of a geminivirus infecting Sweet potato in the United States. Plant Dis. 82, 1253–1257. [DOI] [PubMed] [Google Scholar]
- Lotrakul, P. , Valverde, R.A. , Clark, C.A. , Hurtt, S. and Hoy, M.W. (2002) Sweetpotato leaf curl virus and related geminiviruses in sweetpotato. Acta Hort. (ISHS), 583, 135–141. [Google Scholar]
- Lotrakul, P. , Valverde, R.A. and Clark, C.A. (2003) Properties of a begomovirus isolated from Sweet potato [Ipomoea batatas (L.) Lam.] infected with Sweet potato leaf curl virus. Rev. Mex. Fitopatol. 21, 128–136. [Google Scholar]
- Lozano, G. , Trenado, H.P. , Valverde, R.A. and Navas‐Castillo, J. (2009) Novel begomovirus species of recombinant nature in sweet potato (Ipomoea batatas) and Ipomoea indica: taxonomic and phylogenetic implications. J. Gen. Virol. 90, 2550–2562. [DOI] [PubMed] [Google Scholar]
- Luan, Y.S. , Zhang, J. , Liu, D.M. and Li, W.L. (2007) Molecular characterization of sweet potato leaf curl virus isolate from China (SPLCV‐CN) and its phylogenetic relationship with other members of the Geminiviridae. Virus Genes, 35, 379–385. [DOI] [PubMed] [Google Scholar]
- Miano, D.W. , LaBonte, D.R. , Clark, C.A. , Valverde, R.A. , Hoy, M.W. , Hurtt, S. and Li, R. (2006) First report of a begomovirus infecting sweetpotato in Kenya. Plant Dis. 90, 832. [DOI] [PubMed] [Google Scholar]
- Miozzi, L. , Pantaleo, V. , Burgyan, J. , Accotto, G.P. and Noris, E. (2013) Analysis of small RNAs derived from tomato yellow leaf curl Sardinia virus reveals a cross reaction between the major viral hotspot and the plant host genome. Virus Res. 178, 287–296. [DOI] [PubMed] [Google Scholar]
- Mukasa, S.B. , Rubaihayo, P.R. and Valkonen, J.P.T. (2006) Interactions between a crinivirus, an ipomovirus and a potyvirus in coinfected sweetpotato plants. Plant Pathol. 55, 458–467. [Google Scholar]
- Onuki, M. , Honda, Y. and Hanada, K. (2000) Geminate particle morphology of Sweet potato leaf curl virus in partially purified preparation and its serological relationship to two begomoviruses by Western blotting. J. Gen. Plant Pathol. 66, 182–184. [Google Scholar]
- Paprotka, T. , Boiteux, L.S. , Fonseca, M.E.N. , Resende, R.O. , Jeske, H. , Faria, J.C. and Ribeiro, S.G. (2010) Genomic diversity of sweet potato geminiviruses in a Brazilian germplasm bank. Virus Res. 149, 224–233. [DOI] [PubMed] [Google Scholar]
- Raja, P. , Wolf, J.N. and Bisaro, D.M. (2010) RNA silencing directed against geminiviruses: post‐transcriptional and epigenetic components. Biochim. Biophys. Acta, 1799, 337–351. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Negrete, E.A. , Carrillo‐Tripp, J. and Rivera‐Bustamante, R.F. (2009) RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83, 1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Pardina, P. , Luque, A. , Nome, C. , López‐Colomba, E. , Fuentes‐Delgado, S. and Feo, L. (2012) First report of Sweet potato leaf curl virus infecting sweet potato in Argentina. Australas. Plant Dis. Notes, 7, 157–160. [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untiveros, M. , Fuentes, S. and Salazar, L.F. (2007) Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla‐, cucumo‐, ipomo‐, and potyviruses infecting sweet potato. Plant Dis. 91, 669–676. [DOI] [PubMed] [Google Scholar]
- Valverde, R.A. , Clark, C.A. and Valkonen, J.P.T. (2007) Viruses and virus disease complexes of sweetpotato. Plant Viruses, 1, 116–126. [Google Scholar]
- Vanitharani, R. , Chellappan, P. , Pita, J.S. and Fauquet, C.M. (2004) Differential roles of AC2 and AC4 of Cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Qi, M.Q. and Cutler, A.J. (1993) A simple method of preparing plant‐samples for PCR. Nucleic Acids Res. 21, 4153–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.B. , Wu, Q. , Ito, T. , Cillo, F. , Li, W.X. , Chen, X. , Yu, J.L. and Ding, S.W. (2010) RNAi‐mediated viral immunity requires amplification of virus‐derived siRNAs in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 107, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasswa, P. , Otto, B. , Maruthi, M.N. , Mukasa, S.B. , Monger, W. and Gibson, R.W. (2011) First identification of a sweet potato begomovirus (sweepovirus) in Uganda: characterization, detection and distribution. Plant Pathol. 60, 1030–1039. [Google Scholar]
- Weinheimer, I. , Boonrod, K. , Moser, M. , Wassenegger, M. , Krczal, G. , Butcher, S.J. and Valkonen, J.P. (2014) Binding and processing of small dsRNA molecules by the class 1 RNase III protein encoded by sweet potato chlorotic stunt virus. J. Gen. Virol. 95, 486–495. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Chen, H. , Huang, X. , Xia, R. , Zhao, Q. , Lai, J. , Teng, K. , Li, Y. , Liang, L. , Du, Q. , Zhou, X. , Guo, H. and Xie, Q. (2011) BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation‐mediated gene silencing in Arabidopsis. Plant Cell, 23, 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrachya, A. , Glick, E. , Levy, Y. , Arazi, T. , Citovsky, V. and Gafni, Y. (2007) Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus‐Israel. Virology, 358, 159–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (a) DNA dot‐blot results of sweepoviruses used to generate the graphs in Fig. 2. (b) Results of the same blots with ribosomal DNA probe, demonstrating equal loading.
Fig. S2 Symptoms of pararetroviruses [Sweet potato vein clearing virus (SPVCV) and Sweet potato collusive virus (SPCV)] in transgenic sweet potato cultivar ‘Huachano’ expressing the RNase3 gene of Sweet potato chlorotic stunt virus (SPCSV). (a, c) Non‐transgenic ‘Huachano’ infected with SPVCV and SPCV, respectively. (b, d) Transgenic ‘Huachano’ plants expressing RNase3 and infected with SPVCV and SPCV, respectively. (e) Dot‐blot detection of SPVCV in transgenic versus non‐transgenic sweet potato ‘Huachano’ plants. N.I., not infected plant; F, detection of SPCV by Nitrocellulose membrane enzyme‐linked imunosorbent assay (NCM‐ELISA); 1, 2, non infected ‘Huachano’ plant; 3, 4, SPCV infected ‘Huachano’ plants; 5, 6, SPCV in RNase3‐transgenic plants; 7, 8, SPCV in co‐infection with SPCSV.
Fig. S3 Distribution of total small RNA size classes corresponding to SvT1 (white bars), Sweet potato chlorotic stunt virus (SPCSV, black bars), Sweet potato feathery mottle virus (SPFMV, dark grey bars) and other sequences (light grey bars) in wild‐type plants singly and dually infected with StV1 and SPCSV or RNase3‐transgenic plants infected with StV1 (a), wild‐type plants singly and dually infected with SPFMV and SPCSV (b), and non‐infected wild‐type plants (c). The vertical axis shows the total number of small RNA reads, and the horizontal axis indicates the size class of siRNA, and the exact numbers are tabulated below the graph, including the sums and grand total.
Table S1 Origin of plants, collection date, genotype name and germplasm accession numbers of plants screened for sweepoviruses in this study.
Table S2 Relative expression of Sweet potato chlorotic stunt virus (SPCSV) in mixed infection with six sweepovirus isolates, relative to single infection, over all time points based on Relative Expression Software Tool (REST) analysis of qualitative reverse transcription‐polymerase chain reaction (qRT‐PCR) results.


