Summary
New economically important diseases on crops and forest trees emerge recurrently. An understanding of where new pathogenic lines come from and how they evolve is fundamental for the deployment of accurate surveillance methods. We used kiwifruit bacterial canker as a model to assess the importance of potential reservoirs of new pathogenic lineages. The current kiwifruit canker epidemic is at least the fourth outbreak of the disease on kiwifruit caused by Pseudomonas syringae in the mere 50 years in which this crop has been cultivated worldwide, with each outbreak being caused by different genetic lines of the bacterium. Here, we ask whether strains in natural (non‐agricultural) environments could cause future epidemics of canker on kiwifruit. To answer this question, we evaluated the pathogenicity, endophytic colonization capacity and competitiveness on kiwifruit of P. syringae strains genetically similar to epidemic strains and originally isolated from aquatic and subalpine habitats. All environmental strains possessing an operon involved in the degradation of aromatic compounds via the catechol pathway grew endophytically and caused symptoms in kiwifruit vascular tissue. Environmental and epidemic strains showed a wide host range, revealing their potential as future pathogens of a variety of hosts. Environmental strains co‐existed endophytically with CFBP 7286, an epidemic strain, and shared about 20 virulence genes, but were missing six virulence genes found in all epidemic strains. By identifying the specific gene content in genetic backgrounds similar to known epidemic strains, we developed criteria to assess the epidemic potential and to survey for such strains as a means of forecasting and managing disease emergence.
Keywords: co‐existence, effector repertoire, emerging pathogens, host range
Introduction
There is growing concern worldwide about the emergence of new diseases of crops and forest trees (Elena et al., 2011; Santini et al., 2013). This concern is heightened by changes in climate, land use and commercial networks that could intensify emerging epidemics of plant disease (Bebber et al., 2013; Chakraborty, 2013; Santini et al., 2013). Considerable effort has been invested in elucidating the evolutionary processes that lead to the emergence of new biotypes and pathotypes of pathogens and to their establishment in disease epidemics (Ali et al., 2014; Elena et al., 2011; Gladieux et al., 2008; McCann et al., 2013). An important challenge for the management of upcoming epidemics is to deploy the insight gained from these studies to survey reservoirs of pathogens for the genetic or phenotypic lines with the greatest potential for emergence and to monitor crops that are at the greatest risk for new diseases.
Among the plant pathogens, Pseudomonas syringae has been responsible for a large number of disease emergences. For example, in this century alone, new diseases caused by P. syringae have been reported on over 20 different species of woody plants (Lamichhane et al., 2014). Among the plants that have suffered new diseases caused by this bacterium, kiwifruit has experienced at least four independent emergences of disease caused by different genetic lines of this bacterium over the past 35 years of the mere 50 years in which kiwifruit has been cultivated commercially worldwide. Bacterial canker of kiwifruit was first observed in California in 1980 (Opgenorth et al., 1983). Successively, the disease was reported in Japan (Takikawa et al., 1989), Italy (Scortichini, 1994) and South Korea (Koh and Nou, 2002). In parallel, multiple emergences occurred in China, starting in 1985 and leading to the declaration of the disease in 11 different provinces at present. The recent global kiwifruit canker epidemic, responsible for severe economic losses, appeared in 2008 in Italy (Balestra et al., 2009) and subsequently in the rest of the world, including Spain (Balestra et al., 2011), Portugal (Balestra et al., 2010), Switzerland (European Plant Protection Organization (EPPO), 2011a), Turkey (Bastas and Karakaya, 2012), France (Vanneste et al., 2011), Australia (International Plant Protection Convention (IPPC), 2011), Chile (EPPO, 2011b), Germany (EPPO, 2013) and Slovenia (on declaration). The disease has since been reported in New Zealand (Everett et al., 2011) and is jeopardizing its kiwifruit industry, which represents a main economic revenue for the country (Vanneste, 2012). In addition, leaf spots on kiwifruit have been reported to be caused by strains in yet another new lineage (Vanneste et al., 2013). This progression portends future emergences of new lines of P. syringae pathogenic to kiwifruit.
Recent studies have illustrated the importance of non‐agricultural environments as sources of P. syringae strains pathogenic for plants (Morris et al., 2007). In particular, it has been demonstrated that environmental strains closely related genetically to the tomato speck pathogen (P. syringae pv. tomato) are pathogenic on tomato and have type III secretion system (T3SS) effector repertoires similar to P. syringae pv. tomato (Monteil et al., 2013). This led to the hypothesis that highly aggressive clonal P. syringae pv. tomato populations evolved from a less aggressive environmental strain through a small number of evolutionary events and spread worldwide. In light of this, the emergence of new pathotypes of P. syringae could be relatively simple in terms of the evolutionary processes involved.
It has been proposed that wild Actinidia species could have been the source of the progenitors of the pathogens of cultivated kiwifruit (McCann et al., 2013). However, the ubiquitous nature of P. syringae (Morris et al., 2013) and the link that has been demonstrated between environmental and epidemic populations for other pathovars (Monteil et al., 2013) lead us to wonder about the potential of environmental habitats to harbour pathotypes for future disease outbreaks of kiwifruit canker. To evaluate this potential, we characterized the pathogenicity, fitness, host range and effector repertoire of strains from non‐agricultural habitats that belong to P. syringae phylogroup 1 and compared them with known epidemic strains of P. syringae pv. actinidiae (Psa), also belonging to phylogroup 1, and to other closely related strains known to attack woody crops (Lamichhane et al., 2014). The strains that were characterized here were selected from our collection of environmental strains based on their genetic similarity with Psa strains for various molecular markers. All strains characterized, including the Psa epidemic strains, had broad host ranges. Several environmental strains induced vascular discoloration, slight external symptoms and were equal in their fitness as endophytes to the epidemic strains. The presence of genes in the catechol pathway operon was particularly accurate in predicting the capacity of strains to colonize the kiwifruit vascular system and to cause vascular discoloration. Our approach is novel because, by using a marker for a function suspected to be useful in the colonization of woody tissue, we identified environmental strains that could represent future emerging pathogens for kiwifruit as well as for other woody plant hosts. We propose that the evolution of less aggressive strains towards greater aggressiveness might involve few evolutionary steps, including the acquisition of new pathogenic determinant, such as T3SS effector genes. Genes in the catechol operon could be targeted in surveillance programmes for future epidemic pathogens of kiwifruit and other crops.
Results
Multiple distinct genetic lines of P. syringae strains from freshwater habitats are pathogenic to kiwifruit
Comparison of gene content between publicly available genomes of several P. syringae pathogens of woody plants with those of herbaceous plants revealed an operon with predicted function in the catechol pathway that was present only in pathogens of woody plants. The screening of several P. syringae strains from environmental habitats based on the presence of the catechol operon then allowed us to establish a collection of 10 strains from aquatic habitats and 10 reference strains representing previously described pathovars, in addition to four strains of Psa bv. 1 (representing strains of the first outbreak in Japan and Italy), Psa bv. 3 (current epidemic strains) and Psa bv. 4 (representing the so‐called low virulent strains) (Vanneste et al., 2013). Strains in Psa bv. 2 isolated in Korea were not included in the study. Of the 10 environmental strains, eight had the genes of the catechol operon. Two strains were phylogenetically close to the reference strains, but without the genes in the catechol operon (AF0007, USA0003). All the environmental strains formed different genetic lineages based on multilocus sequence typing (MLST) (Fig. 1). When inoculated on kiwifruit, the strains caused four types of reaction: (i) severe disease with external and internal symptoms (CFBP 7286, CC1676, PA459) (Fig. 2A); (ii) marked but less severe disease with mild external and internal symptoms (AF0015, USA0007, CC1544, USA0001, CSZ0350, CSZ0343, CSZ0761 and SZB0070) (Fig. 2B); (iii) faint canker and/or swelling at the inoculated point (NCPPB 3335 and PseNe107) and faint internal necrosis (NCPPB 2598, PseNe107 and ICMP 18882) (Fig. 2C); and (iv) no external or internal disease symptoms (MAFF 302280PT, DC3000, KN203, MAFF 302273, MAFF 30120, AF0007, USA0003, PaVt10 and 0893_23) (Fig. 2D). All strains causing visible symptoms were successfully re‐isolated from plant tissue. In addition, a few other strains that did not cause any visible symptoms (MAFF 302280PT and 0893‐23) were consistently re‐isolated. Finally, the remaining strains that did not cause any symptoms (DC3000, KN203, MAFF 302273, MAFF 30120 and PaVt10) could not be re‐isolated. In terms of distance from the point of inoculation, Psa bv. 1 and Psa bv. 3 strains were isolated up to 3 cm below the point of inoculation and the environmental strains were isolated up to 1 cm below the point of inoculation. In contrast, strains ICMP 18882 (Psa bv. 4) and MAFF 302280PT (P. syringae pv. morsprunorum) could be isolated only from the point of inoculation, but not beyond. All the bacteria re‐isolated from the inoculated plants had the expected BOX‐PCR profiles.
Figure 1.
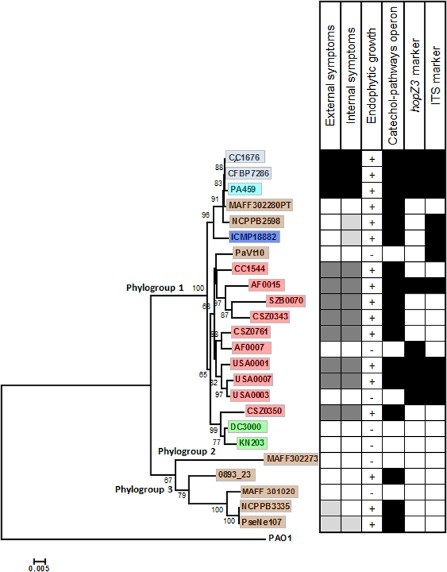
Bayesian tree constructed on the concatenated sequences cts, gyrB, rpoD and gapA (1852 bp) of the strains tested for pathogenicity, endophytic growth and molecular markers. Bootstrap values are indicated at each node. External and internal symptoms are indicated with black boxes (severe disease), dark grey boxes (mild symptoms), light grey boxes (faint symptoms) and white boxes (no symptoms). The presence of the catechol operon involved in aromatic compound degradation, and of hopZ3 and ITS markers, is indicated by black (present) and white (absent) squares. For the catechol operon, all the genes were present for the positive strains. The origin of strains and their biovar affiliation are indicated with colour: light blue for Pseudomonas syringae pv. actinidiae (Psa) bv. 3, turquoise for Psa bv. 1, dark blue for Psa bv. 4, brown for strains isolated from woody hosts, green for strains isolated from herbaceous hosts and red for the environmental strains.
Figure 2.
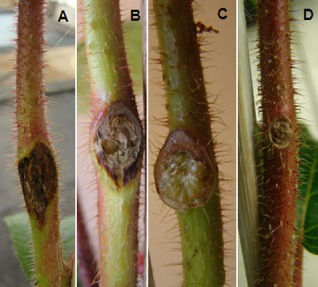
Characteristic external symptoms observed on kiwifruit stems at 30 days post‐inoculation. External necrosis and canker caused by the strain from Actinidia deliciosa CFBP 7286 (A), necrosis caused by the environmental strain AF0015 (B), swelling caused by the strain NCPPB 3335 isolated from Olea europaea (C) and the absence of symptoms on stems inoculated with strain ICMP 18882 isolated from Actinidia deliciosa.
Environmental strains attain population sizes in the vascular tissue of kiwifruit similar to those of epidemic Psa bv. 3 strains and can co‐exist with epidemic strains without disease occurrence
Endophytic growth on kiwifruit, assessed for Psa bv. 3 strain CFPB 7286 and environmental strains USA0007 and CC1544, showed that, when inoculated independently: (i) all strains attained their maximum population size between 3 and 7 days post‐inoculation (dpi) (Fig. 3A,B) and then decreased slightly (Fig. 3A,B); (ii) there was no significant difference (P > 0.1) between the population densities of the environmental strain and CFPB 7286 at all time points; and (iii) strain CFBP 7286 caused severe external and internal necrosis by 21 dpi (Fig. 4A), whereas environmental strains induced mild external and internal symptoms by 21 dpi. Although the inoculum concentration used in this experiment [104 colony‐forming units (CFU)/mL] was much lower than in the other experiments, the symptoms caused by both CFBP 7286 and environmental strains were the same as those observed using an inoculum of 108 CFU/mL. This demonstrates that disease caused by these strains on kiwifruit was not dose dependent in the range of inoculum concentrations tested.
Figure 3.
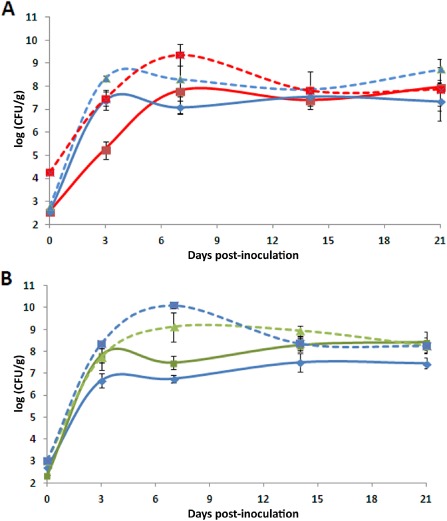
Growth of strains in 3‐week‐old kiwifruit plants. Population densities were determined at the point of inoculation for strains CFBP 7286 (blue) and USA007 (red) of Pseudomonas syringae (A) and for strains CFBP 7286 (blue) and CC1544 (green) (B). Broken lines indicate the growth of each strain independently and full lines represent growth for mixed inoculations. Two replicate experiments were conducted with three independent measurements of population density per time and per replicate experiment. The mean ± standard error at each time point indicated in the figure is based on all six observations pooled over the two replicates, because there were no significant differences in mean density between the two replicate experiments (P < 0.05), except for CFBP 7286 alone at 14 and 21 dpi. In addition, the strains showed the same trend during the two experiments.
Figure 4.

External symptoms recorded on kiwifruit plants 3 weeks after inoculation. The same symptoms were observed on all plants (three per time point). Necrosis observed with strain CFBP 7286 of Pseudomonas syringae (A). No symptoms were recorded on kiwifruit 3 weeks after co‐inoculation with environmental strains (USA0007 and CC1544) and CFBP 7286 (B).
When strains were co‐inoculated, we observed that: (i) for the pair CFBP 7286–CC1544, the population density of strain CFBP 7286 was significantly reduced at 3 and 7 dpi compared with the single inoculation (P ≤ 0.005) and, in contrast, the population density of strain CC1544 was not influenced (P > 0.05) by the presence of CFBP 7286 (Fig. 3B); (ii) for the pair USA0007–CFBP 7286, the population density of CFBP 7286 was reduced significantly (P ≤ 0.05) at 3 and 7 dpi compared with the single inoculation, whereas strain USA0007 was not influenced (P > 0.05) (Fig. 3A); and (iii) for all treatments (co‐ and individual inoculations), bacteria attained and maintained almost the same density (107–108). When strain USA0007 was inoculated in the mixed condition, its initial population density was lower than when it was inoculated individually (Fig. 3A). This difference was maintained up to 7 dpi. However, both environmental strains were able to grow, and their ability to colonize kiwifruit vessels was not influenced by the presence of the epidemic strain CFBP 7286. In addition, we also calculated the level of co‐existence as described by Wilson and Lindow (1994), including the relative yield total (the sum for the two strains of the ratio of population size when they were co‐inoculated to the population size when they were inoculated alone). We observed that the relative yield totals for the pair CFBP 7286–CC1544 were 1.79 and 1.48 at 3 and 7 dpi, respectively, whereas, for the pair USA0007–CFBP 7286, they were 1.58 and 1.68 at 3 and 7 dpi, respectively. Indeed, values of the relative yield total higher than unity indicate co‐existence of the strains. The strains can have a high value of co‐existence even in the presence of decreasing population densities.
The ability of the strains to move into the kiwifruit vessels was reduced for both co‐inoculation experiments, in particular for the co‐inoculation of CFBP 7286 and CC1544, where no bacterial cells were re‐isolated at any time point below or above the point of inoculation (Fig. S1, see Supporting Information). In the co‐inoculation experiments, no symptoms were recorded (Fig. 4B).
During the in vitro test, no effects on growth were observed for the environmental strains when they were co‐inoculated with the CFBP 7286 strain. In contrast, strain CFBP 7286 was significantly inhibited 48 h after inoculation (P ≤ 0.0005) by both environmental strains (Fig. S2, see Supporting Information).
Endophytic growth in kiwifruit is correlated with the presence of a highly conserved operon for the degradation of aromatic compounds that is restricted to phylogroups 1 and 3 of P. syringae
We observed that strains that grew endophytically on kiwifruit were also positive for the presence of the catechol operon. These included strains belonging to Psa bv. 3 (CC1676, CFBP 7286), Psa bv. 1 (PA459), Psa bv. 4 (ICMP 18882) and the environmental strains CC1544, AF0015, SZB0070, CSZ0343, CSZ0761, USA0001, USA0007 and CSZ0350, as well as other strains of P. syringae phylogroup 1 [MAFF 302280PT (pv. morsprunorum), NCPPB 2598 (pv. theae)] and phylogroup 3 [0893_23 (pv. aesculi), NCPPB 3335 (pv. savastanoi), PseNe107 (pv. savastanoi)] (Fig. 1). In contrast, the strains lacking the operon involved in the catechol branch β‐ketoadipate pathway (pv. avellanae strain PaVt10, the environmental strains AF0007 and USA0003, the tomato pathogen DC3000, the pv. maculicola strain KN203 and the pv. mori strain MAFF 302273) were not able to grow endophytically in kiwifruit (Fig. 1). Other molecular markers for Psa (hopZ3) (Balestra et al., 2013) and the internal transcribed spacer (ITS) markers (Rees‐George et al., 2010) were present in only some of the environmental strains and in the epidemic strains causing severe disease symptoms. Among the strains, the presence of hopZ3 and ITS markers was not correlated with disease occurrence and endophytic colonization capacity in kiwifruit (Fig. 1).
Analysis of the draft genomes of the two environmental strains (USA0007 and CC1544), several of the Psa genomes available in GenBank (accession numbers are listed in Table S1, see Supporting Information) and those of P. syringae pv. morsprunorum (strain MAFF 302280PT), P. syringae pv. aesculi (strains 0893_231, NCPPB 3681 and 2250) and P. savastanoi pv. savastanoi (strain NCPPB 3335) revealed that the genes involved in the degradation of phenolic and aromatic compounds, via the catechol branch β‐ketoadipate pathway, are organized in a unique operon. This operon is composed of: (i) a PAS domain S‐box protein that corresponds to the putative promoter; (ii) a protein involved in the meta‐pathway phenol degradation; (iii) a conserved flavoprotein‐oxygenase with a flavin reductase‐like domain; (iv) a flavin adenine dinucleotide (FAD)‐dependent oxido‐reductase; (v) a short‐chain alcohol dehydrogenase involved in the oxidation of different aromatic compounds (such as antibiotics and compounds involved in nitrogen metabolism); and (vi) a dienelactone hydrolase. The latter is responsible for the degradation of chloroaromatic compounds by three different pathways: 4‐chlorocatechol degradation, 3‐chlorocatechol degradation II (ortho) and 3‐chlorocatechol degradation I (ortho) (Sridevi et al., 2012).
Using the primers developed here, a survey conducted on a large collection of strains representing all known phylogroups of P. syringae showed that these genes are present only in P. syringae phylogroups 1 and 3. The absence of the operon from P. syringae strains that are pathogens of herbaceous crops was also confirmed by searching all P. syringae genomes in GenBank with the proteins encoded in this operon. The presence of the operon was also investigated in all the other bacterial genomes available in GenBank. Outside of P. syringae, the complete operon was only found in the genomes of P. pseudoalcaligenes and P. fuscovagine.
The phylogenetic tree constructed on the concatenated sequences forming the operon (Fig. 5) showed the same phylogenetic structure as the MLST tree (Fig. S3, see Supporting Information). To better understand the genetic relationships between the operon and the cts, gapA, gyrB, rpoD housekeeping genes, we made strain‐by‐strain pairwise comparisons of the p distances of the concatenated sequences obtained by the four housekeeping genes with those obtained from the concatenated genes forming the operon. This showed a significant correlation between operon gene sequences and housekeeping genes (Fig. S3). The same tree topology between operon and housekeeping genes, the lack of anomalous nucleotide composition (for example G/C ratio) and the absence of phage integrases, transposons and vestiges of other genetic mobile elements flanking the operon led us to conclude that the operon was not transmitted via horizontal gene transfer, and was probably acquired before the diversification of the P. syringae complex.
Figure 5.
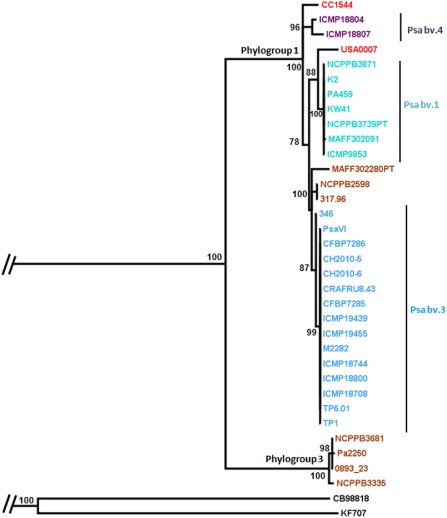
Bayesian tree of the concatenated complete sequences of phenol‐MetA, flavin adenine dinucleotide (FAD)‐dependent oxygenase, short‐chain alcohol dehydrogenase and dienelactone hydrolase. Posterior probabilities are shown at each node. Phylogroups 1 and 3, Pseudomonas syringae pv. actinidiae (Psa) biovars and the names of the strains are indicated on the branches. The tree was rooted with the sequences extracted from the genomes of strain KF707 of Pseudomonas pseudoalcaligenes and strain CEB98818 of Pseudomonas fuscovaginae. The origin of the strains and their biovar affiliation are indicated with colour: light blue for Psa bv. 3, turquoise for Psa bv. 1, dark blue for Psa bv. 4, brown for strains isolated from woody hosts, and red for the environmental strains.
Environmental and epidemic strains of P. syringae pathogenic to kiwifruit have broad host ranges and partially overlapping effector repertoires
Strains CFBP 7286, ICMP 18882, CC1544 and USA0007, characterized for host range, were pathogenic on 16–18 of the 22 host plants tested (Fig. 6). For woody plants, we recorded both external and internal disease symptoms. Some hosts did not bear any external disease symptoms, but were affected internally showing clear vascular necrosis (Fig. 6). All of the inoculated bacterial strains were re‐isolated from vascular tissue at 60 dpi independent of the presence or absence of symptoms, except for strain ICMP 18882, which could not be recovered from Mariana plum (Fig. 6).
Figure 6.

Host range of strains CFBP 7286, ICMP 18882, USA0007 and CC1544. The severity of the disease is expressed with a grey colour scale. For endophytic growth, black boxes indicate that strains grow into the host plant and that they were re‐isolated even in the absence of symptoms. For strains pathogenic on soybean, faba bean, clover, sunflower, geranium and ranunculus, disease was observed only on stems (*). Pseudomonas syringae pv. actinidiae (Psa) bv. 3 is indicated in blue, Psa bv. 4 in dark blue and environmental strains in red.
All woody hosts tested were susceptible to at least one strain (in terms of either internal or external symptoms), whereas some herbaceous plants (spinach and liverwort) did not manifest disease after inoculation with any of the strains. In addition, on hazelnut and oleander, all strains caused only internal symptoms (Fig. 6). The environmental strain CC1544 did not cause external symptoms on woody hosts (except for kiwifruit), but internal symptoms were observed in almost all the plant species tested (Fig. 6).
The presence of type III secretion effector genes in the genomes of two environmental strains (USA0007 and CC1544) and the Psa epidemic strains (accession numbers in Table S1) was determined by blasting the sequences of known effector genes obtained from http://www.pseudomonas‐syringae.org. Psa bv. 3 strains carried 44 full‐length effectors, whereas Psa bv. 4 and Psa bv. 1 strains carried 32 and 40 effectors, respectively (Fig. 7). In contrast, strains USA0007 and CC1544 had 29 and 27 full‐length effector genes, respectively (Fig. 7). Eighteen effector genes were found to be shared among the Psa biovars and environmental strains, whereas four (hopH1, hopZ5, hopAA1‐2, hopAM1‐2) and two (hopF1 and hopE1) effector genes were unique for Psa bv. 3 and Psa bv. 4, respectively (Fig. 7). Three effectors (hopAJ1, hopV1, hopA2) were carried only in the environmental strains. Finally, Psa bv. 4 and the environmental strains lacked six effectors (avrD1, avrRpm1, hopAM1‐1, hopD1, hopQ1‐1, hopBB1) that were present in both Psa bv. 1 and Psa bv. 3.
Figure 7.
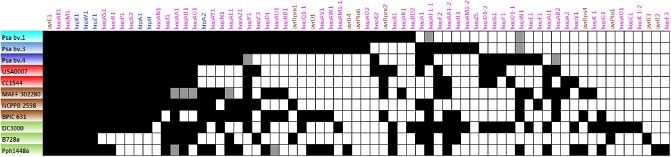
Type III secretion gene repertoires in strains of Pseudomonas syringae. Results for Pseudomonas syringae pv. actinidiae (Psa) bv. 1, Psa bv. 3 and Psa bv. 4 were compressed as the results were identical for all the strains analysed (all the genomes available in GenBank). Strains USA0007 and CC1544 were isolated from river headwaters. Other strains indicated include MAFF 302280 (P. syringae pv. morsprunorum), NCPPB 2598 (P. syringae pv. theae), BPIC 631 (P. syringae pv. avellanae), DC3000 (P. syringae pv. tomato), B728a (P. syringae pv. syringae) and Pph1448a (P. syringae pv. phaseolicola). Black boxes indicate that the genes were found in full length, white boxes indicate the absence of the genes and grey boxes indicate that the genes were truncated. The origin of strains and their biovar affiliation are indicated with colour: light blue for Psa bv. 3, turquoise for Psa bv. 1, dark blue for Psa bv. 4, brown for strains isolated from woody hosts, green for strains isolated from herbaceous hosts and red for the environmental strains.
The hopM1 gene, known to be consistently present in P. syringae phylogroup 1 (Baltrus et al., 2011), was observed in all strains (Fig. 7). Baltrus et al. (2011) previously hypothesized that different allelic forms of hopM1 could influence the host range of the strains from this phylogroup. The same authors showed that phylogenies based on the hopM1 genes split strains from phylogroup 1 into two divergent groups: pv. morspunorum/pv. actinidiae and pv. tomato/pv. lachrymans. In light of this, we investigated the hopM1 phylogenetic relationships among Psa epidemic strains, environmental strains and strains from other pathovars of phylogroup 1. The phylogeny constructed with a Bayesian model showed that environmental strains are divided into at least four different genetic lineages distinct from the epidemic Psa strains (Fig. S4, see Supporting Information). Strain CC1544 isolated from France and strain AF0015 from New Zealand had identical hopM1 alleles, whereas strains isolated from the same water sample in the USA (USA0001 and USA0007) had distinctly different hopM1 alleles. Phylogeny also showed that the hopM1 alleles of Psa bv. 3 and Psa bv. 1 are clonal, whereas that of Psa bv. 4 is genetically related to that of pv. morsprunorum, but distinct from the environmental strains.
Discussion
Our results bolster the concept that non‐agricultural environments are reservoirs of plant pathogens with broad host ranges and complex effector repertories (Goss et al., 2013; Monteil et al., 2013; Morris et al., 2013). Pseudomonas syringae strains from aquatic habitats can colonize and cause symptoms in kiwifruit and other woody and herbaceous plants, and share numerous effectors with epidemic strains of the bacterial canker pathogen, in addition to possessing some unique effectors. The strains pathogenic to kiwifruit characterized here represent clonal lines that are distinct from the lines of the pathogen that have emerged in epidemics over the past several decades. Therefore, the environmental strains characterized here, although showing pathogenic potential on kiwifruit, are unlikely to have contributed to epidemics of bacterial canker seen to date. However, given the numerous processes that can bring environmental strains of P. syringae into encounters with agriculture (Morris et al., 2013), these strains have the potential to contribute to future kiwifruit canker epidemics and, in light of their host ranges, to diseases of herbaceous and other woody species. Contact of environmental strains with kiwifruit plants—either wild or cultivated—might explain the history of emergences of canker in China, where the diversity of genetic lineages implicated in the disease is greater than that in Europe, South America and other countries of Asia‐Pacific (Japan, South Korea and New Zealand). Furthermore, the capacity of the environmental strains to colonize the vascular tissue of woody plants and, most importantly, with low severity of the apparent symptoms, such that this would not lead to phytosanitary interventions, suggests that there are opportunities for co‐existence with epidemic strains and subsequent opportunities for the acquisition of effectors by horizontal gene transfer (as discussed below). Therefore, our work makes an important contribution to the identification of the processes that need to be monitored in order to come closer to predicting new disease emergences.
For the bacterial speck pathogen of tomato, it has been proposed that the epidemic lineage probably emerged from a less aggressive environmental strain through a small number of evolutionary events, such as by the loss of effector genes deleterious for disease development (Monteil et al., 2013). Here, we found that environmental strains had a smaller number of effectors but almost the same host range as that of epidemic Psa strains. However, the environmental strains did not move through the vascular system as efficiently as the epidemic strains. In light of this, we argue that environmental strains could become more highly aggressive by acquiring some effectors via horizontal gene transfer. For horizontal gene transfer to occur, nutrient levels should be sufficiently high in order to support an elevated bacterial density (Dröge et al., 1998). Hence, endophytic growth could be an opportunity to attain the required population density and to ensure the proximity between donor and recipient populations (Manceau et al., 1986). The acquisition of one or more of the six effectors absent from Psa bv. 4 and the environmental strains (avrD1, avrRpm1, hopAM1‐1, hopD1, hopQ1‐1, hopBB1) might enhance aggressiveness. Future studies should focus on the potential horizontal gene transfer between epidemic and environmental strains harbouring the catechol operon to determine the conditions and likelihood of this mechanism contributing to the evolution of environmental strains. To predict the potential for the emergence of new kiwifruit pathogens, it will be necessary to elucidate which combination of effectors would enhance pathogenicity to kiwifruit. For example, avrRpm1 is known to be essential for maximum aggressiveness and for growth of the Brassica pathogen P. syringae pv. maculicola on Arabidopsis (Ritter and Dangl, 1995). Likewise, hopAM1 expressed in P. syringae strains led to an enhanced growth on water‐stressed Arabidopsis plants (Goel et al., 2008). Similar results have been observed on bean, in which hopQ1‐1 of P. syringae pv. phaseolicola suppresses plant defences, but the same effector expressed in P. s. pv. tabaci strains triggers host defences in Nicotiana spp. (Ferrante et al., 2009).
For the current epidemic of bacterial canker of kiwifruit, it has been proposed that wild Actinidia spp. are likely to have been reservoirs of pathogenic strains (McCann et al., 2013). Here, we propose a complementary element to such a scenario, whereby aquatic milieu are reservoirs of a vast diversity of strains, many of which are disseminated across various habitats, via the water cycle in particular, where they can survive and thrive (Morris et al., 2010). This dissemination could bring the strains into contact with plants in a range of biomes, including those in which there are wild relatives of cultivated plants. The small evolutionary steps towards aggressive pathogenicity, such as the acquisition of new genes by horizontal gene transfer or deletion of the genomic determinants deleterious for disease occurrence, could occur across the ensemble of the habitats encountered by the bacteria during the dissemination process. Wild plant hosts could then serve as forces of natural selection for strains having gained in fitness in plant tissues.
Niche overlap is one of the first obstacles to co‐existence via competition for limiting nutritional resources. Therefore, it is not surprising that co‐existence of genetically related lines has rarely been described to occur among epiphytic P. syringae strains, at least not under experimental conditions (Wilson and Lindow, 1994). However, here, we demonstrated the co‐existence of genetically close lines of P. syringae when co‐inoculated in kiwifruit. However, this co‐existence had a cost on bacterial movement and disease development. Lack of symptoms, when the Psa bv. 3 strain was co‐inoculated with the environmental strains, could have been caused by a metabolic cost of competition to the bacteria that reduced the transcription and secretion of proteins involved in disease occurrence in order to maintain growth. The in planta behaviour therefore suggests that, although nutrient availability might not be limiting for the doubling of cells, thereby permitting the strains to avoid marked outcompetition of each other, conditions could nevertheless be limiting for optimal metabolic regulation as a pathogen. The in vitro co‐inoculation also supports this hypothesis, in that the medium is rich, thereby permitting overall co‐existence of environmental and epidemic strains, but leading, perhaps, to the production of secondary metabolites that partially inhibit the growth of the epidemic strain. Whatever the mechanism underlying the co‐existence of environmental and epidemic strains, the high population densities achieved could permit horizontal gene transfer among the different bacterial genotypes, with the consequent acquisition of various genes, including pathogenicity determinants, such as effector genes, that influence bacterial aggressiveness. In this way, weakly pathogenic strains, such as the environmental strains, could rapidly evolve into highly aggressive strains.
Our results also suggest that the metabolic processes associated with the degradation of aromatic compounds are important for fitness in the kiwifruit vascular system. McCann et al. (2013) recently showed that epidemic strains of Psa possess a plasmid with genes involved in aromatic carbon metabolism (genes other than those in the catechol operon), and that these genes are present in other plant pathogens of woody hosts, such as P. syringae pv. aesculi and the xylem pathogen Xylella fastidiosa. Our results showed that strains closely related to kiwifruit‐competent strains from phylogroup 1, but lacking the genes in the operon involved in phenol degradation via the catechol branch β‐ketoadipate pathway, did not colonize kiwifruit plants. These genes are likely to be involved in processes related to the degradation of some lignin intermediates. Lignin contains many phenolic moieties and its degradation can involve different chemical pathways leading to toxic by‐products. Microorganisms commonly use the catechol oxygenase pathway to degrade phenolic compounds (Sridevi et al., 2012) and the genus Pseudomonas is known to possess enzymes in this biochemical pathway (Kang and Park, 1997). By metabolizing lignin intermediates, bacteria avoid the toxic effects during colonization of the plant vascular system, where lignin is an important structural component. The content of phenolic compounds often increases during the attack of plants by different bacterial pathogens (Oi‐Kano et al., 2008). Hence, the ability of P. syringae to degrade phenolic compounds could be a fundamental requisite for endophytic colonization and for pathogenicity. In other bacteria, such as Pseudomonas putida, genes for the degradation of aromatic compounds reside on plasmids or transposons (Williams and Sayers, 1994). By contrast, the catechol operon in P. syringae is highly conserved, and we have shown that its phylogenesis exhibits congruence with housekeeping genes. The maintenance of the operon, in both the P. syringae complex and other Pseudomonas spp. abundantly present in the environment, led us to speculate that the environment at large might be an important driver for the evolution of this operon. In the environment, exposure to aromatic compounds might have been a positive selection criterion for the maintenance of this operon. Exposure to plant degradation products might have further reinforced the selection of this operon (Harwood and Parales, 1996). On encounter with plant vascular tissues, the presence of these genes could then have provided a fitness advantage. Other strains not exposed to the selective forces could have lost the operon. In agreement with this scenario, we observed the presence of this operon only in the P. syringae phylogroup 1 and 3 strains known to attack woody plants (pv. actinidiae, pv. savastanoi, pv. aesculi, pv. morsprunorum and pv. theae). In addition, contact with xenobiotic compounds derived from human activities for more than a century (Díaz, 2004) might have selected bacteria able to use such compounds in their metabolic pathways.
To date, surveillance is one of the most important methods for the control of kiwifruit canker, as well as for other bacterial plant pathogens (Scortichini et al., 2012). Different detection methods have been proposed for Psa identification. The current molecular markers are based on primers that amplify the ITS region (Rees‐George et al., 2010) and the hopZ3 gene (Balestra et al., 2013) identified in strains from the current disease epidemics. In surveys for the presence of the pathogen, the absence of these markers is often a criterion to eliminate a strain from further consideration with regard to its contribution to plant health. As our work illustrates, these detection tools target a narrow range of the ensemble of strains able to grow endophytically in kiwifruit and to cause vascular discoloration. Although such surveillance provides useful information for quarantine measures, it can mask information about synergistic interactions or the emergence of new pathotypes. As a means to estimate the potential for the emergence of new pathotypes, it is also important to survey for the presence of other endophytic P. syringae strains able to colonize kiwifruit vascular tissue. Genes in the catechol operon could provide a tool for this goal. In this way, our work contributes to the creation of a framework of foresight for disease emergence.
Experimental Procedures
Strain isolation and selection
The reference strains used in this study were sourced from public collections or kindly provided by others (Table 1). The strains from different environmental substrates (water, epilithic biofilms) were isolated previously (Morris et al., 2007, 2008), whereas additional strains from kiwifruit and hazelnut were isolated during this study. Isolations were made as described previously (Lamichhane et al., 2013). The environmental strains were selected from an in‐house database of about 1600 strains of P. syringae based on their phylogenetic co‐location in the neighbour‐joining tree constructed on the cts housekeeping gene. Strains that formed clades close to the reference Psa strains CFBP 7286 and KW30 were selected. Finally, all the isolates were tested for the presence of target genes involved in the degradation of aromatic compounds (see below). Phenotypic tests performed on the isolates are listed in Table S2 (see Supporting Information).
Table 1.
Strains used in this study
| Phylogroup | Place of isolation | Pathovar and biovar | Isolated from | Reference | |
|---|---|---|---|---|---|
| Strains | |||||
| CFBP 7286 | 1 | Italy | actinidiae (Psa bv. 3) | Actinidia chinensis | Mazzaglia et al. (2012) |
| CC1676 | 1 | France | actinidiae (Psa bv. 3) | Actinidia deliciosa | This study |
| ICMP 18882 | 1 | New Zealand | actinidiae (Psa bv. 4) | Actinidia chinensis | Everett et al. (2011) |
| KW30 | 1 | Japan | actinidiae (Psa bv. 1) | Actinidia deliciosa | Takikawa et al. (1989) |
| PA459 | 1 | Japan | actinidiae (Psa bv. 1) | Actinidia chinensis | Takikawa et al. (1989) |
| PaVt10 | 1 | Italy | avellanae | Corylus avellana | This study |
| MAFF 302280PT | 1 | USA | morsprunorum | Prunus domestica | Baltrus et al. (2011) |
| NCPPB 2598 | 1 | Japan | theae | Thea sinensis | Sarkar and Guttman (2004) |
| KN203 | 1 | Japan | maculicola | Chinese cabbage | Sarkar and Guttman (2004) |
| DC3000 | 1 | UK | tomato | Tomato | Cuppels (1986) |
| AF0007 | 1 | New Zealand | NA | Water | This study |
| AF0015 | 1 | New Zealand | NA | Water | This study |
| CC1544 | 1 | France | NA | Water | Morris et al. (2008) |
| CSZ0343 | 1 | France | NA | Water | This study |
| CSZ0350 | 1 | France | NA | Water | This study |
| USA0001 | 1 | USA | NA | Water | Morris et al. (2010) |
| USA0003 | 1 | USA | NA | Water | Morris et al. (2010) |
| USA0007 | 1 | USA | NA | Water | Morris et al. (2010) |
| CSZ0761 | 1 | France | NA | Snow pack | This study |
| SZB0070 | 1 | France | NA | Epilithic biofilm | This study |
| MAFF 302273PT | 2 | USA | aceris | Acer sp. | Sawada et al. (1999) |
| 0893_23 | 3 | India | aesculi | Horse chestnut | Green et al. (2010) |
| PseNE107 | 3 | Nepal | savastanoi | Olea europaea | Lamichhane and Varvaro (2013) |
| MAFF 301020 | 3 | Japan | mori | Morus alba | Sarkar and Guttman (2004) |
| NCPPB 3335 | 3 | France | savastanoi | Olea europaea | Rodríguez‐Palenzuela et al. (2010) |
| Strains used as positive controls | |||||
| 41A | 2 | France | syringae | Apricot | This study |
| CFBP 1617 | 2 | USA | aptata | Beta vulgaris | |
| KN10 | 1 | Japan | tomato | Tomato | Sarkar and Guttman (2004) |
| MAFF 301315 | 1 | Japan | lachrymans | Cucumber | Sarkar and Guttman (2004) |
| MAFF 311107 | 4 | Japan | oryzae | Rice | Sarkar and Guttman (2004) |
| CFBP 3637 | 3 | Yemen | phaseolicola | Phaseolus vulgaris | |
| CFBP 3211 | 3 | Yugoslavia | glycinea | Glycine max | |
| CFBP 2067 | 6 | Mexico | helianthi | Helianthus annuus |
NA, not applicable; Psa, Pseudomonas syringae pv. actinidiae.
Identification of genes putatively associated with pathogenesis in woody plants
OrthoMCL (Li et al., 2003) was used to identify protein families present only in the genomes of the woody pathogens Psa 346, CFBP 7285, CFBP 7286, CFBP 7287, K2, KW41, PA459, V and VI, P. syringae pv. morsprunorum MAFF 302280PT and P. savastanoi pv savastanoi, but absent from the genomes of the herbaceous pathogens P. cannabina pv. alisalensis BS91, P. syringae pv. maculicola F1, P. syringae pv. tomato DC3000 and JL1065, P. syringae pv. pisi 1704B, P. syringae 642, P. syringae pv. syringae FF5, P. syringae pv. tabaci ATCC115128, P. fluorescens 0–1 and 5, using the OrthoMCL tool on the website http://pacu.facom.ufms.br/kiwi/orthologsorter/.
Primers, polymerase chain reaction (PCR) conditions, sequencing and phylogenetic analysis
Primers and PCR conditions used to amplify the housekeeping genes gyrB, rpoD and cts (Sarkar and Guttman, 2004) and the gapA gene (Morris et al., 2008) were as described previously. Information about the previously described amplification and sequencing of the marker genes and the catechol operon genes described here is provided in Text S1 (see Supporting Information).
Phylogenetic trees were constructed on the concatenated sequences of cts, gapA, gyrB and rpoD (1852 bp). Several Psa sequences available in GenBank and other sequences of strains belonging to P. syringae phylogroup 1 were added into the analysis. Strains and GenBank accession numbers of the sequences are listed in Table S1, whereas sequences for the strains KN.2 and 346 were extracted from the draft genome available at http://pacu.facom.ufms.br/blast_kiwi/blast.cgi. The two environmental strains USA0007 and CC1544 were sequenced previously (Baltrus et al., 2013). Accession numbers for genomes are AVDY01000000 and AVEI01000000, respectively.
Sequences obtained in this study were deposited in PAMBD (http://genome.ppws.vt.edu/cgi‐bin/MLST/home.pl). We also performed phylogenies on the single and concatenated genes involved in aromatic compound degradation and for hopM1 sequences. For each phylogenetic analysis, PCR amplicons were aligned and cut to obtain sequences of the same length with DAMBE version 5.1.1 (Xia, 2013). Bayesian trees were constructed with the MrBayes program (http://mrbayes.csit.fsu.eduby) (Ronquist and Huelsenbeck, 2003) using 500 000 generations. Maximum likelihood and parsimony phylogeny were created with the Phylip package (http://evolution.genetics.washington.edu/phylip.html).
Pathogenicity test
The strains listed in Table 1 were tested for pathogenicity on kiwifruit plants and for lesion production on various excised fruits. For this test, 3‐month‐old, vegetatively propagated kiwifruit plants (Actinidia deliciosa Liang and Ferguson cv. Hayward) were used. For inoculation on kiwifruit plants, the inoculum consisted of aqueous suspensions of 48‐h bacterial cultures from plates of King's medium B adjusted to 1 × 108 CFU/mL. For all strains inoculated, the fingerprint profiles were determined prior to inoculation by PCR with BOX primers, as described previously (Versalovic et al., 1991), with GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA). Each strain had a unique BOX fingerprint. Inoculation was performed only at a single point. At half the height of the main stem, a leaf petiole was manually detached at the base and 10 μL of bacterial suspension were deposited on the leaf scar, which had been slightly punctured with a sterile hypodermic needle prior to inoculation. The inoculated point was marked with masking tape for later retrieval. Four plants were inoculated for each strain and for the negative control (sterile distilled water) treatment. The inoculated plants were maintained for 35 dpi in the growth chamber at night/day temperatures of 18/23 °C and with a light/dark period of 16/8 h. Plants were watered twice a day, without wetting the leaves, to maintain a constant high humidity. All plants were examined individually for disease symptoms up to 35 dpi and the tests were repeated twice for each strain.
At 35 dpi, disease severity on each plant was scored by considering both the external (necrosis, canker and/or gall) and internal (necrosis) symptoms. Internal disease symptoms on each plant were assessed by excising the subcortical tissues of the woody stem and measuring the length of the necrosis developed along the plant vessels. The following scale was used for disease scoring: no symptoms (apparently healthy external and internal tissues), faint symptoms (external symptoms of <0.20 cm without any internal necrosis), moderate symptoms (both external and internal symptoms of 0.21–1 cm), severe symptoms (both external and internal symptoms of 1.1–3 cm) and highly severe symptoms (both external and internal symptoms of >3 cm).
Re‐isolation of the inoculated strains was performed 5 weeks after inoculation. Re‐isolation of the bacteria from the inoculated woody tissues was made by excising bark with a sterile scalpel; a small fragment of subcortical tissue was removed and placed in a microtube containing 900 μL of phosphate buffer. The suspensions were incubated at room temperature overnight and then three drops of 10 μL were plated onto KB medium supplemented with 50 mg/L of cycloheximide. The putative P. syringae colonies from each isolation point were streaked for purity and their identity was checked by comparing their BOX‐PCR profile with that of the strain used for inoculation.
Endophytic growth in kiwifruit vascular tissue
The capacity for endophytic growth of strain CFPB 7286 (Psa bv. 3) and the two environmental strains USA0007 and CC1544 was evaluated on kiwifruit plants under conditions of single‐ and mixed‐culture inoculation. The inoculum concentration used for this test was 104 CFU/mL for both individual and co‐inoculation treatments. Experiments were performed in the glasshouse under ambient conditions with an approximate temperature of 27 °C during the day (13 h) and 13 °C at night (11 h), and repeated twice. The similarity of behaviour of the strains in the two replicate experiments was verified with a Student's t‐test (described below) and data were pooled for the two replicates for further analysis. More information is provided in Text S1.
Host range determination
The host range of the strains was determined on 13 herbaceous and nine woody plant species. The plant species and the strains used as positive controls are listed in Table S3 (see Supporting Information).
All of the annual plants tested were seed grown and transplanted to fresh medium of decomposed white sphagnum peat. Prunus spp. plants were sourced from commercial nurseries producing plants from seed, re‐potted in the same substrate and maintained in a glasshouse at ambient temperature until inoculation. Annual plants were inoculated 1 month after sowing, whereas 2‐year‐old Prunus spp. and 3‐month‐old hazelnut, oleander and poplar plants were used for inoculation. More information about host range determination is available in Text S1.
Statistical analysis
Data from the endophytic growth assay and competition tests were analysed using a paired Student's t‐test on data expressed as log10 of CFU/g, employing Graphpad software (http://www.graphpad.com/quickcalcs/). Values of P ≤ 0.05 were considered to be statistically significant. The level of co‐existence in the mixed inoculation was calculated by adding the ratio of population size to both co‐inoculated and independent population sizes (Wilson and Lindow, 1994).
Supporting information
Fig. S1 Endophytic population density of CFBP 7286, CC1544 and USA0007 when co‐inoculated and inoculated alone in kiwifruit stem taken above and below the point of inoculation.
Fig. S2 In vitro growth of strains CFBP 7286 (blue), USA0007 (red) and CC1544 (green) of Pseudomonas syringae in 10% strength tryptic soy broth. Full lines indicate the growth of each strain independently, and broken lines represent growth in mixed culture. For strain CFBP 7286, growth in the presence of strain USA0007 is indicated by the dashed line and in the presence of CC1544 by the dotted line. Mean ± standard error at each time point is indicated.
Fig. S3 Phylogenetic comparison between housekeeping genes and the genes forming the catechol operon.
Fig. S4 Bayesian phylogeny of the hopM1 gene. Names of the strains are indicated on the branches and posterior probabilities are indicated at each node. Taxon names are coded with colour: light blue for Pseudomonas syringae pv. actinidiae (Psa) bv. 3, dark blue for Psa bv. 4, brown for strains isolated from woody hosts, green for strains isolated from herbaceous hosts and red for environmental strains. Phylogroups are indicated on the right of the tree based on the housekeeping gene phylogeny (Fig. 1). Phylogroup 1 is formed by three polyphyletic clades, the environmental strains are distributed along different clades and a new clade was observed for the strains AF0015, CC1544 and USA0001.
Table S1 Genome and gene accession numbers of the strains used for genomic analysis.
Table S2 Results of lesion and other phenotypic tests for the strains used in this study.
Table S3 Plant species used in host range tests and bacterial strains used as positive controls.
Text S1 Phylogenetic analysis, pathogenicity and lesion tests and host range determination.
Acknowledgements
The salary of Claudia Bartoli was provided by Tuscia University (Italy). We thank INRA's Plant Health and Environment Department (SPE) for supporting this work. We also thank the glasshouse staff of INRA's Plant Pathology Research unit at Avignon, France, for providing and maintaining all plant materials tested in this study.
References
- Ali, S. , Gladieux, P. , Leconte, M. , Gautier, A. , Justesen, A.F. , Hovmøller, M.S. , Enjalbert, J. and de Vallavieille‐Pope, C. (2014) Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici . Plos Pathog. 10, e1003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra, G.M. , Mazzaglia, A. , Quattrucci, A. , Renzi, M. and Rossetti, A. (2009) Current status of bacterial canker spread on kiwifruit in Italy. Australas. Plant Dis. Notes, 4, 34–36. [Google Scholar]
- Balestra, G.M. , Renzi, M. and Mazzaglia, A. (2010) First report of bacterial canker of Actinidia deliciosa caused by Pseudomonas syringae pv. actinidiae in Portugal. New Dis. Reports, 22, 10. [Google Scholar]
- Balestra, G.M. , Renzi, M. and Mazzaglia, A. (2011) First report of Pseudomonas syringae pv. actinidiae on kiwifruit plants in Spain. New Dis. Reports, 24, 10. [Google Scholar]
- Balestra, G.M. , Taratufolo, M.C. , Vinatzer, B.A. and Mazzaglia, A. (2013) A multiplex PCR assay for detection of Pseudomonas syringae pv . actinidiae and differentiation of populations with different geographic origin. Plant Dis. 97, 472–478. [DOI] [PubMed] [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Romanchuk, A. , Chang, J.H. , Mukhtar, M.S. , Cherkis, K. , Roach, J. , Grant, S.R. , Jones, C.D. and Dangl, J.L. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. Plos Pathog. 7, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus, D.A. , Yourstone, S. , Lind, A. , Guibaud, C. , Sands, D.C. , Jones, C.D. , Morris, C.E. and Dangl, J.L. (2013) Draft genome sequences of phylogenetically diverse suite of Pseudomonas syringae strains from multiple source populations. Genome Announc. 2, e01195‐13. doi: 10.1128/genomeA.01195-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastas, K.K. and Karakaya, A. (2012) First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Turkey. Plant Dis. 96, 452. [DOI] [PubMed] [Google Scholar]
- Bebber, D.P. , Ramotowski, M.A.T. and Gurr, S.J. (2013) Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change, 3, 985–988. [Google Scholar]
- Chakraborty, S. (2013) Migrate or evolve: options for plant pathogens under climate change. Glob. Change. Biol. 19, 1985–2000. [DOI] [PubMed] [Google Scholar]
- Cuppels, D.M. (1986) Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato . Appl. Environ. Microbiol. 51, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz, E. (2004) Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int. Microbiol. 7, 173–180. [PubMed] [Google Scholar]
- Dröge, M. , Pühler, A. and Selbitschka, W. (1998) Horizontal gene transfer as a biosafety issue: a natural phenomenon of public concern. J. Biotechnol. 64, 75–90. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. , Bedhomme, S. , Carrasco, P. , Cuevas, J.M. , Iglesia, F. , Lafforgue, G. , Lalić, J. , Pròsper, A. , Tromas, N. and Zwart, M.P. (2011) The evolutionary genetics of emerging plant RNA viruses. Mol. Plant–Microbe Interact. 24, 287–293. [DOI] [PubMed] [Google Scholar]
- European Plant Protection Organization (2011a) First report of Pseudomonas syringae pv. actinidiae in Switzerland. EPPO Reporting Service n°8 2011/168. Available at www.eppo.org [accessed 14 February 2012].
- European Plant Protection Organization (2011b) First report of Pseudomonas syringae pv. actinidiae in Chile. EPPO Reporting Service n°3 2011/055. Available at www.eppo.org [accessed 14 December 2013].
- European Plant Protection Organization (2013) First report of Pseudomonas syringae pv. actinidiae in Germany. EPPO Reporting Service n°9, 2013/185. Available at www.eppo.org [accessed 17 January 2013].
- Everett, K.R. , Taylor, R.K. , Romberg, M.K. , Rees‐George, J. , Fullerton, R.A. , Vanneste, J.L. and Manning, M.A. (2011) First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in New Zealand. Australas. Plant Dis. Notes, 6, 67–71. [Google Scholar]
- Ferrante, P. , Clarke, C.R. , Cavanaugh, K.A. , Michelmore, R.W. , Buonaurio, R. and Vinatzer, B.A. (2009) Contributions of the effector gene hopQ1‐1 to differences in host range between Pseudomonas syringae pv. phaseolicola and P. syringae pv. tabaci . Mol. Plant Pathol. 10, 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux, P. , Zhang, X.G. , Afoufa‐Bastien, D. , Sanhueza, R.M.V. , Sbaghi, M. and Le Cam, B. (2008) On the origin and spread of the scab disease of apple: out of central Asia. Plos ONE, 3, e1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel, A.K. , Lundeberg, D. , Torres, M.A. , Matthews, R. , Akimoto‐Tomiyama, C. , Farmer, L. , Dangl, J.L. and Grant, S.R. (2008) The Pseudomonas syringae type III effector HopAM1 enhances virulence on water‐stressed plants. Mol. Plant–Microbe Interact. 21, 361–370. [DOI] [PubMed] [Google Scholar]
- Goss, E.M. , Potnis, N. and Jones, J.B. (2013) Grudgingly sharing their secrets: new insight into the evolution of plant pathogenic bacteria. New Phytol. 199, 630–632. [DOI] [PubMed] [Google Scholar]
- Green, S. , Studholme, D.J. , Laue, B.E. , Dorati, F. , Lovell, H. , Arnold, D. , Cottrell, J.E. , Bridgett, S. , Blaxter, M. , Huitema, E. , Thwaites, R. , Sharp, P.M. , Jackson, R.W. and Kamoun, S. (2010) Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum . PloS ONE, 5, e10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, C.S. and Parales, R.E. (1996) The β‐ketoadipate pathway and the biology of self‐identity. Annu. Rev. Microbiol. 50, 553–590. [DOI] [PubMed] [Google Scholar]
- International Plant Protection Convention (2011) Detection of the Asian strain of bacterial canker of kiwifruit in Victoria, Australia. IPPC AUS‐45/1. Available at www.ippc.int [accessed October 2013].
- Kang, M.H. and Park, J.M. (1997) Sequential degradation of phenol and cyanide by a commensal interaction between two microorganisms. J. Chem. Technol. Biotechnol. 69, 226–230. [Google Scholar]
- Koh, Y.J. and Nou, I.S. (2002) DNA markers for identification of Pseudomonas syringae pv. actinidiae . Mol. Cells, 13, 309–314. [PubMed] [Google Scholar]
- Lamichhane, J.R. and Varvaro, L. (2013) Epiphytic Pseudomonas savastanoi pv. savastanoi can infect and cause olive knot disease on Olea europaea subsp. cuspidata. Australas. Plant Pathol. 42, 219–225. [Google Scholar]
- Lamichhane, J.R. , Fabi, A. , Ridolfi, R. and Varvaro, L. (2013) Epidemiological study of hazelnut bacterial blight in central Italy by using laboratory analysis and geostatistics. PloS ONE, 8, e56298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane, J.R. , Varvaro, L. , Parisi, L. , Audergon, J.‐M. and Morris, C.E. (2014) Disease and frost damage of woody plants caused by Pseudomonas syringae: seeing the forest for the trees. Adv. Agron. 126, 235–295. [Google Scholar]
- Li, L. , Stoeckert, C.J., Jr and Roos, D.S. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau, C. , Gardan, L. and Devaux, M. (1986) Dynamics of RP4 plasmid transfer between Xanthomonas campestris pv. corylina and Erwinia herbicola in hazelnut tissues, in planta. Can. J. Microbiol. 32, 835–841. [Google Scholar]
- Mazzaglia, A. , Studholme, D.J. , Taratufolo, M.C. , Cai, R. , Almeida, N.F. , Goodman, T. , Guttman, D.S. , Vinatzer, B.A. and Balestra, G.M. (2012) Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS ONE, 7, e36518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, H.C. , Rikkerink, E.H.A. , Bertels, F. , Fiers, M. , Lu, A. , Rees‐George, J. , Andersen, M.T. , Gleave, A.P. , Haubold, B. , Wohlers, M.W. , Guttman, D.S. , Wang, P.W. , Straub, C. , Vanneste, J. , Rainey, P.B. and Templeton, M.D. (2013) Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. Plos Pathog. 9, e1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil, C. , Cai, R. , Liu, H. , Mechan, L.M. , Leman, S. , Studholme, D. , Morris, C.E. and Vinaterz, B.A. (2013) Non‐agricultural reservoirs contribute to emergence and evolution of Pseudomonas syringae crop pathogens. New Phytol. 199, 800–811. [DOI] [PubMed] [Google Scholar]
- Morris, C.E. , Kinkel, L.L. , Xiao, K. , Prior, P. and Sands, D.C. (2007) Surprising niche for the plant pathogen Pseudomonas syringae . Infec. Gen. Evol. 7, 84–92. [DOI] [PubMed] [Google Scholar]
- Morris, C.E. , Sands, D.C. , Vinatzer, B.A. , Glaux, C. , Guilbaud, C. , Buffière, A. , Yan, S. , Dominguez, H. and Thompson, B.M. (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2, 321–334. [DOI] [PubMed] [Google Scholar]
- Morris, C.E. , Sands, D.C. , Vanneste, J.L. , Montarry, J. , Oakley, B. , Guilbaud, C. and Glaux, C. (2010) Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. mBio, 1, e00107–e00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, C.E. , Monteil, C.L. and Berge, O. (2013) The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu. Rev. Phytopathol. 51, 85–104. [DOI] [PubMed] [Google Scholar]
- Oi‐Kano, Y. , Kawada, T. , Watanabe, T. , Koyama, F. , Watanabe, K. , Senbongi, R. and Iwai, K. (2008) Oleuropein, a phenolic compound in extra virgin olive oil, increases uncoupling protein 1 content in brown adipose tissue and enhances noradrenaline and adrenaline secretions in rats. J. Nutr. Sci. Vitaminol. 54, 363–370. [DOI] [PubMed] [Google Scholar]
- Opgenorth, D. , Lai, M. , Sorrell, M. and Whithe, J.B. (1983) Pseudomonas canker of kiwifruit. Plant Dis. 67, 1283–1284. [Google Scholar]
- Rees‐George, J. , Vanneste, J.L. , Cornish, D.A. , Pushparajah, I.P.S. , Yu, J. , Templeton, M.D. and Everett, K.R. (2010) Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S‐23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 59, 453–464. [Google Scholar]
- Ritter, C. and Dangl, J.L. (1995) The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant–Microbe Interact. 3, 444–453. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Palenzuela, P. , Matas, I.M. , Murillo, J. , López‐Solanilla, E. , Bardaji, L. , Pérez‐Martínez, I. , Rodríguez‐Moskera, M.E. , Penyalver, R. , López, M.M. , Quesada, J.M. , Biehl, B.S. , Perna, N.T. , Glasner, J.D. , Cabot, E.L. , Neeno‐Eckwall, E. and Ramos, C. (2010) Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour‐inducing pathogen of woody hosts. Environ. Microbiol. 12, 1604–1620. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. and Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Santini, A. , Ghelardini, L. , De Pace, C. , Desprez‐Loustau, M.L. , Capretti, P. , Chandelier, A. , Cech, T. , Chira, D. , Diamandis, S. , Gaitniekis, T. , Hantula, J. , Holdenrieder, O. , Jankovsky, L. , Jung, T. , Jurc, D. , Kirisits, T. , Kunca, A. , Lygis, V. , Malecka, M. , Marcais, B. , Schmitz, S. , Schumacher, J. , Solheim, H. , Solla, A. , Szabò, I. , Tsopelas, P. , Vannini, A. , Vettraino, A.M. , Webber, J. , Woodward, S. and Stenlid, J. (2013) Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 197, 238–250. [DOI] [PubMed] [Google Scholar]
- Sarkar, S.F. and Guttman, D.S. (2004) Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70, 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, H. , Suzuki, F. , Matsuda, I. and Saitou, N. (1999) Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49, 627–644. [DOI] [PubMed] [Google Scholar]
- Scortichini, M. (1994) Occurrence of Pseudomonas syringe pv. actinidiae on kiwifruit in Italy. Plant Pathol. 43, 1035–1038. [Google Scholar]
- Scortichini, M. , Marcelletti, S. , Ferrante, P. , Petriccione, M. and Firrao, G. (2012) Pseudomonas syringae pv. actinidiae: a re‐emerging, multi‐faceted, pandemic pathogen. Mol. Plant Pathol. 13, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridevi, V. , Lakshmi, M.V.V. , Manasa, M. and Sravani, M. (2012) Metabolic pathways for the biodegradation of phenol. Int. J. Eng. Sci. Adv. Technol. 2, 695–705. [Google Scholar]
- Takikawa, Y. , Serizawa, S.I. , Takeshi, Y.S. and Goto, M. (1989) Pseudomonas syringae pv. actinidiae pv. nov.: the causal bacterium of canker of kiwifruit in japan. Annu. Phytopathol. Soc. Jpn. 55, 437–444. [Google Scholar]
- Vanneste, J. (2012) Pseudomonas syringae pv. actinidiae (Psa): a threat to the New Zealand and global kiwifruit industry. N. Z. J. Crop Hortic. Sci. 40, 265–267. [Google Scholar]
- Vanneste, J.L. , Poliakoff, F. , Audusseau, C. , Cornish, D.A. , Paillard, S. , Rivoal, C. and Yu, J. (2011) First report of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit in France. Plant Dis. 95, 1311. [DOI] [PubMed] [Google Scholar]
- Vanneste, J.L. , Yu, J. , Cornish, D.A. , Tanner, D.J. , Windner, R. , Chapman, J.R. , Taylor, R.K. , Mackay, J.F. and Dowlut, S. (2013) Identification, virulence and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Dis. 97, 708–719. [DOI] [PubMed] [Google Scholar]
- Versalovic, J. , Koeuth, T. and Lupski, J. (1991) Distribution of repetitive DNA‐sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.A. and Sayers, J.R. (1994) The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas . Biodegradation, 5, 195–217. [DOI] [PubMed] [Google Scholar]
- Wilson, M. and Lindow, S.E. (1994) Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60, 4468–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X. (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 30, 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Endophytic population density of CFBP 7286, CC1544 and USA0007 when co‐inoculated and inoculated alone in kiwifruit stem taken above and below the point of inoculation.
Fig. S2 In vitro growth of strains CFBP 7286 (blue), USA0007 (red) and CC1544 (green) of Pseudomonas syringae in 10% strength tryptic soy broth. Full lines indicate the growth of each strain independently, and broken lines represent growth in mixed culture. For strain CFBP 7286, growth in the presence of strain USA0007 is indicated by the dashed line and in the presence of CC1544 by the dotted line. Mean ± standard error at each time point is indicated.
Fig. S3 Phylogenetic comparison between housekeeping genes and the genes forming the catechol operon.
Fig. S4 Bayesian phylogeny of the hopM1 gene. Names of the strains are indicated on the branches and posterior probabilities are indicated at each node. Taxon names are coded with colour: light blue for Pseudomonas syringae pv. actinidiae (Psa) bv. 3, dark blue for Psa bv. 4, brown for strains isolated from woody hosts, green for strains isolated from herbaceous hosts and red for environmental strains. Phylogroups are indicated on the right of the tree based on the housekeeping gene phylogeny (Fig. 1). Phylogroup 1 is formed by three polyphyletic clades, the environmental strains are distributed along different clades and a new clade was observed for the strains AF0015, CC1544 and USA0001.
Table S1 Genome and gene accession numbers of the strains used for genomic analysis.
Table S2 Results of lesion and other phenotypic tests for the strains used in this study.
Table S3 Plant species used in host range tests and bacterial strains used as positive controls.
Text S1 Phylogenetic analysis, pathogenicity and lesion tests and host range determination.


