Summary
Resistance in rice cultivars to the rice blast fungus Magnaporthe oryzae is complex and is controlled by both major genes and quantitative trait loci (QTLs). We undertook a genome‐wide association study (GWAS) using the rice diversity panel 1 (RDP1) that was genotyped using a high‐density (700 000 single nucleotide polymorphisms) array and inoculated with five diverse M. oryzae isolates. We identified 97 loci associated with blast resistance (LABRs). Among them, 82 were new regions and 15 co‐localized with known blast resistance loci. The top 72 LABRs explained up to 98% of the phenotypic variation. The candidate genes in the LABRs encode nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) resistance proteins, receptor‐like protein kinases, transcription factors and defence‐related proteins. Among them, LABR_64 was strongly associated with resistance to all five isolates. We analysed the function of candidate genes underlying LABR_64 using RNA interference (RNAi) technology and identified two new resistance alleles at the Pi5 locus. We demonstrate an efficient strategy for rapid allele discovery using the power of GWAS, coupled with RNAi technology, for the dissection of complex blast resistance in rice.
Keywords: genome‐wide association study (GWAS), host resistance, QTL, rice blast, SNP
Introduction
Rice blast, caused by the fungal pathogen Magnaporthe oryzae, is a devastating disease that reduces rice yields by 10%–30% (Skamnioti and Gurr, 2009). The development of broad‐spectrum and durable resistance to the disease is one of the major efforts in rice breeding (Zhang, 2007). Molecular mapping for rice blast resistance (R) genes started in the late 1980s, when the first restriction fragment length polymorphism (RFLP) map was constructed (McCouch et al., 1988). To date, over 80 rice blast R genes have been mapped in the rice genome, and about 21 of these have been cloned over the past three decades (Liu et al., 2014). Except for Pid2, which encodes a lectin protein kinase, all blast R genes belong to the nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) immune receptor family. Mapping of quantitative trait loci (QTLs) for partial resistance was first reported in the early 1990s. For example, 10 QTLs were identified in the durable resistant cultivar Moroberekan (Wang et al., 1994). Since then, numerous studies have reported the identification of QTLs in different genetic backgrounds and with the inoculation of diverse M. oryzae isolates. A meta‐analysis detected 165 meta‐QTLs that were derived from the initial dataset of 347 QTLs (Ballini et al., 2008). Two new recent studies have identified more blast QTLs (Ashkani et al., 2013; Jia and Liu, 2011). Among them, only two, i.e. pi21 and Pb1, have been cloned; these encode a proline‐rich protein and an NBS‐LRR protein, respectively (Fukuoka et al., 2009; Hayashi et al., 2010). All of the R genes or QTLs cloned to date were isolated using a classical biparental map‐based cloning strategy, which is laborious and time consuming. Because mapping resolution in conventional mapping populations is limited, fine mapping is required, and it usually takes 4–5 years to clone a single R gene. In addition, most broad‐spectrum resistance against M. oryzae is conditioned by both major genes and QTLs, and little is known about the diversity of alleles at any of the known blast R genes or QTLs. Thus, the complex genetic architecture of resistance to M. oryzae cannot be easily dissected using conventional mapping strategies in which only two alleles are represented in a population and a single fungal isolate is used to evaluate the progeny.
Genome‐wide association studies (GWASs) were first used for the genetic investigation of human diseases (Wei et al., 2009), but have been extensively used to dissect complex traits in plant species in recent years (Brachi et al., 2011; Buckler et al., 2009; Huang et al., 2010; Jia et al., 2013; Morris et al., 2013; Zhao et al., 2011). In rice, GWAS has been used to identify genes and QTLs associated with traits related to agronomic performance, grain quality, abiotic stress tolerance and domestication (Famoso et al., 2011; Huang et al., 2010), but there are relatively few reports of GWAS being used to identify loci associated with disease resistance (Wang et al., 2015; Zhao et al., 2011). Although several different panels of germplasm have been used for GWAS in rice, currently the only publically available set of purified lines for which genotypes are also freely available is the rice diversity panel 1 (RDP1) (Eizenga et al., 2014). RDP1 consists of 420 diverse rice cultivars collected from 82 countries, and this collection provides a well‐balanced representation of the five major subpopulations of Oryza sativa (Zhao et al., 2011). Previously genotyped with only 44 000 single nucleotide polymorphisms (SNPs), this panel was recently genotyped with a high‐density rice array (HDRA) that assays 700 000 high‐quality SNP loci and provides approximately 1 SNP/570 bp throughout the rice genome (S. R. McCouch et al., unpublished data). At this level of SNP coverage, the most important factor limiting GWAS resolution is the size of the diversity panel or, more specifically, the number of recombination breakpoints represented in RDP1. Thus, an expanded set of purified accessions, the rice diversity panel 2 (RDP2), has recently been developed and genotyped with HDRA to further enhance the power and resolution of association mapping in rice (S. R. McCouch et al., unpublished data).
RDP1 and RDP2, in combination with the HDRA genotyping dataset, are an important resource for the rice community and represent an expandable, open‐source GWAS platform for rice. The germplasm can be used by groups throughout the world to phenotype any trait(s) of interest. By integrating the HDRA dataset with information about different phenotypes generated by researchers working in different environments, multi‐trait and multi‐environment models of plant performance, plant development and response to environment can be developed. These models provide the basis for a deepening of our understanding of basic biological processes and for immediate applications in rice improvement.
To efficiently dissect the genetic architecture of blast resistance, we used five geographically diverse M. oryzae isolates to inoculate the RDP1 accessions. Based on the GWAS results, we identified candidate genes underlying the loci associated with blast resistance (LABRs) and undertook more detailed genetic and functional analysis to confirm the identity of a gene underlying a major R locus that was associated with resistance to all five isolates. We validated our findings molecularly in transgenic rice by silencing two candidate genes in two different resistance backgrounds. These experiments led to the identification of two new R alleles that conferred broad‐spectrum resistance to M. oryzae. Our study demonstrates the power of combining GWAS and RNA interference (RNAi) technologies as a rapid and powerful strategy for deciphering the genetics of blast resistance and for pinpointing novel R alleles in the rice genome.
Results
Phenotypic variation for resistance to M. oryzae in RDP1
Five blast isolates collected in China, Colombia, India, the Philippines and South Korea were used to evaluate blast resistance in RDP1. Simple sequence repeat (SSR) marker analysis showed that the isolates were genetically diverse (Fig. 1a). A total of 362, 331, 312, 335 and 333 rice cultivars from RDP1 were inoculated with isolates R01‐1, RB22, 75‐1‐127, 0‐249 and P06‐6, respectively (Table S1, see Supporting Information). The distribution of blast disease resistance scores for each of the five isolates is shown for RDP1 as a whole and for each subpopulation independently in Fig. 1b,c and Fig. S1 (see Supporting Information). For all five isolates, the distributions were skewed in favour of resistance (0–2) rather than susceptibility (7–9) scores. When inoculated with isolate R01‐1, for example, 172 of the RDP1 cultivars had scores of 0–2, but only 71 had scores of 7–9 (Fig. 1b). When mean resistance scores were plotted for each of the subpopulations individually, the tropical japonica population was consistently the most resistant [P < 0.0001; mean ± standard deviation (SD) resistance score, 1.83 ± 2.02] to R01‐1 compared with the other subpopulations, and the temperate japonica population was significantly more susceptible (P < 0.0001; mean ± SD resistance score, 5.74 ± 2.15) (Fig. 1c). The other four subpopulations were close to the mean blast score of all 372 tested cultivars. When resistance scores were compared across isolates, 60 accessions were classified as highly resistant and 16 were highly susceptible to all five M. oryzae isolates (Table S2, see Supporting Information).
Figure 1.
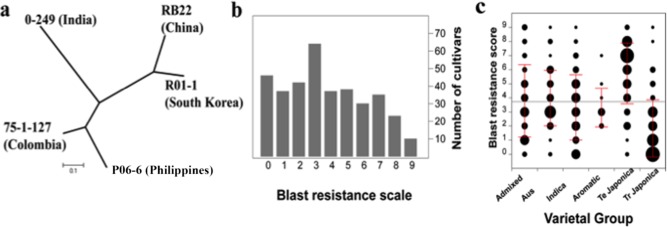
Phylogenetic tree of the five blast isolates and distribution of blast resistance scores to R01‐1 in the rice diversity panel 1 (RDP1) and six subpopulations. (a) Phylogenetic tree of the five isolates constructed using 11 pairs of simple sequence repeat (SSR) markers (see details in Experimental procedures). (b) Distribution of blast resistance scores to isolate RO1‐1 in RDP1. (c) Distribution of blast resistance scores to isolate RO1‐1 in the six subpopulations. The area of the black circle represents the number of varieties and the red lines represent the standard deviation (SD) of the blast resistance scores. Te, temperate; Tr, tropical.
Pair‐wise correlation analysis indicated that the pathogenicities of the five isolates were positively correlated in every case. Correlation was highest between isolates R01‐1 and RB22 (R = 0.64, P < 0.0001). The correlation coefficients of other pair‐wise comparisons ranged from 0.21 to 0.43 (P < 0.0001) (Table S3, see Supporting Information). The relationship among the five isolates based on the pathogenicity result is consistent with that obtained from the SSR analysis (Fig. 1a).
Identification of LABRs in the rice genome using GWAS
Using the 700‐K SNP dataset (S. R. McCouch et al., unpublished data), we identified 97 non‐redundant LABRs associated with resistance to the five M. oryzae isolates (Table S4, see Supporting Information). These LABRs were distributed on all 12 rice chromosomes (Figs 2a and 3). The quantile–quantile (Q–Q) plots for the GWAS results are shown in Fig. 2b, and indicate that the model is well fitted to the data. Among the 97 LABRs, 82 were new, nine were located in the intervals containing previously mapped R loci, and six were located in the regions with previously cloned R genes (Figs 2a and 3). Detailed information about the 15 LABRs that contained previously mapped or cloned R genes is provided in Table S5 (see Supporting Information). Twelve of the LABRs were associated with resistance to two of the fungal isolates, two were associated with resistance to three isolates, seven were associated with resistance to four isolates, and LABR_64 was associated with resistance to all five isolates (Table S4).
Figure 2.
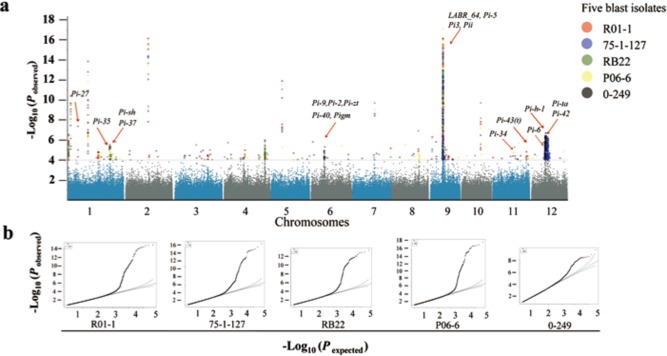
Genome‐wide association scan for loci associated with blast resistance (LABRs) against five isolates using rice diversity panel 1 (RDP1). (a) The Manhattan plots of LABRs on 12 rice chromosomes. The genomic coordinates are displayed along the x‐axis and the logarithm of the odds (LOD) score for each single nucleotide polymorphism (SNP) is displayed on the y‐axis. The LOD score of each dot represents a transformed P value, –log10 P. The red arrows indicate that the identified LABRs are co‐localized with previously mapped or cloned resistance (R)‐gene regions. (b) Quantile–quantile (Q–Q) plots for the genome‐wide association results to the five isolates.
Figure 3.
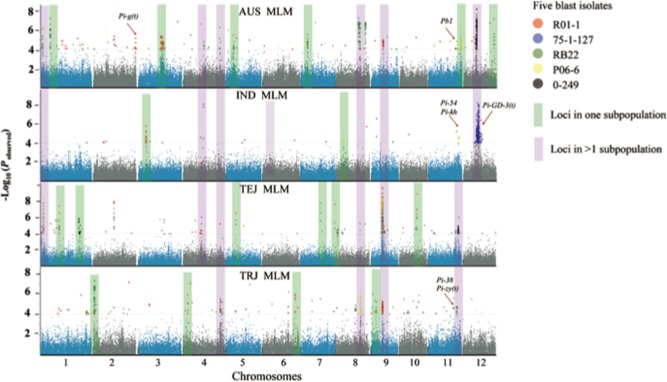
Genome‐wide association scan of blast resistance across subpopulations to five isolates. AUS, aus; IND, indica; TEJ, temperate japonica; TRJ, tropical japonica. The coloured bands indicate the resistance‐associated regions. Purple‐coloured bands show the loci detected in at least two of the subpopulations. Green‐coloured bands show the loci only detected in one of the subpopulations. MLM, mixed linear model.
When the reference Nipponbare genomic sequence (MSU V7.0) underlying the 97 LABRs was searched using gene ontology (GO) terms to identify candidate R genes, a detailed analysis of the annotations in the regions led to the identification of 116 R or R‐related candidate genes (Table S4). These 116 candidate genes could be classified into eight main functional groups of genes: NBS‐LRR genes (34 genes), receptor‐like protein kinase (RLK) genes (17 genes), transcription factor genes (12 genes), ubiquitination‐related genes (seven genes), oxidase/oxidoreductase genes (five genes), DNA‐binding genes (five genes), protein phosphorylation‐related genes (four genes) and heat shock protein genes (four genes) (Fig. 4a). This result indicated that a variety of genes with different biological functions were associated with both qualitative and quantitative resistance to M. oryzae.
Figure 4.
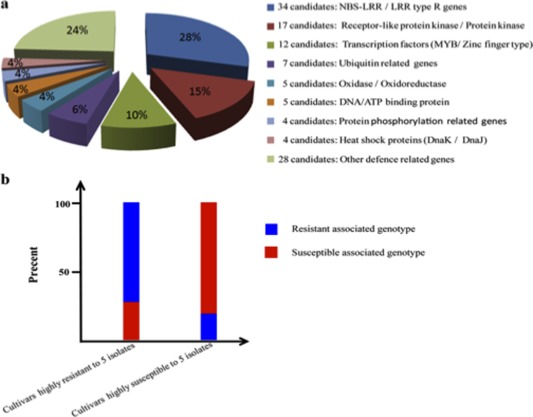
Classification of the 116 candidate resistance (R) or R‐related genes in the 97 loci associated with blast resistance (LABRs) and the single nucleotide polymorphism (SNP) genotype frequency in the highly resistant and highly susceptible cultivars. (a) Classification of the 116 candidate genes. NBS‐LRR, nucleotide‐binding site leucine‐rich repeat. (b) The SNP genotype frequency (in the 26 selected loci: LABR_3, 8, 11, 13, 14, 26, 27, 31, 36, 44, 45, 46, 56, 61, 64, 67, 69, 70, 72, 74, 82, 85, 91, 92, 93, 95) in the highly resistant and highly susceptible cultivars to the five isolates.
Next, we compared the SNP genotypes across each of the 26 selected LABRs with relatively high minor allele frequency (MAF) value (LABR_3, 8, 11, 13, 14, 26, 27, 31, 36, 44, 45, 46, 56, 61, 64, 67, 69, 70, 72, 74, 82, 85, 91, 92, 93, 95) in the 60 rice cultivars that were highly resistant to all five isolates and the 16 rice cultivars that were highly susceptible to the five isolates. Interestingly, the 60 highly resistant cultivars had 72.20% resistance‐associated SNP genotypes in the 26 loci when the most significant SNP in each region was selected to represent the genotype of cultivars. In contrast, the 16 highly susceptible cultivars had only 19.17% resistance‐associated SNP genotypes in the 26 loci (Fig. 4b). These results indicate that, at the LABRs identified in this study, the 60 highly resistant rice varieties are enriched for resistance alleles, whereas the 16 highly susceptible varieties are enriched for susceptibility alleles.
We used a simple additive model in the Statistical Analysis System (SAS) system to determine whether the genotypes of the 97 LABRs were correlated with the blast resistance phenotypes. The 72 LABRs with the most significantly associated SNPs from the total of 97 LABRs (P < 0.05, Table S6, see Supporting Information) were included in the models. The analysis showed that the 72 LABRs explained 36.38%–98.23% of the phenotypic variation in rice resistance to the five M. oryzae isolates (Table S7, see Supporting Information). Among the 72 LABRs, some loci were associated with resistance to more than one isolate; for example, LABR_4 (Chr. 1: 2 459 749–2 547 317) was associated with resistance to four isolates and LABR_64 (Chr. 9: 9 489 046–9 729 496) was associated with resistance to all five isolates (Table S4). These results indicate that some LABRs may contain multiple R or R‐related genes, or may contain genes that provide broad‐spectrum resistance to more than one fungal isolate.
As shown in Fig. 3, GWAS was employed to analyse the association between SNPs and blast resistance to the five isolates in four of the subpopulations of O. sativa (aus, indica, temperate japonica and tropical japonica). Three major types of LABR to R01‐1 were identified when the results from the subpopulations were compared with those from the entire population. Type 1 LABRs were detected only in RDP1 as a whole. Type 2 LABRs were detected in both RDP1 as a whole and subpopulation/s (Fig. 3, highlighted in purple). Type 3 LABRs were detected only in the subpopulations (Fig. 3, highlighted in green). These results indicate that some LABRs are subpopulation specific, which might be determined by the presence/absence or different frequencies of specific alleles in one or more subpopulations (Famoso et al., 2011). Surprisingly, LABR_64 was the only locus that was detected for all five isolates in both RDP1 as a whole and in all of the individual subpopulations, except for the indica subpopulation when analysed independently (Figs 2 and 3).
Sequence analysis of the LABR_64 locus among resistant cultivars
LABR_64 on chromosome 9 was of interest for further analysis because of the broad spectrum of blast isolates to which it conferred resistance and because of the significance of the GWAS result. Using the 44‐K SNP dataset, we identified 16 SNPs with a logarithm of the odds (LOD) score ≥ 4.0 that were located within the 220‐kb target region associated with LABR_64 (Fig. 5a). Using the same phenotypic data in combination with the HDRA (700‐K SNP) dataset, we identified 82 SNPs with a LOD score ≥ 4.0 in the same 220‐kb target region (Fig. 5b). There are 33 annotated genes within this region in the Nipponbare genome (Fig. 5c). Among them are two NBS‐LRR genes, referred to in this study as LOC_Os09g15840 and LOC_Os09g15850 (their alleles in resistant cultivars are LABR_64‐1 and LABR_64‐2), which were identified as the most likely candidate blast R genes because of their sequence similarity to several previously cloned R genes that were known to be co‐located in the same target region: Pi5, Pi3, Pi15 and Pii (Hittalmani et al., 2000; Pan et al., 2003; Wang et al., 1994). None of the significant SNPs identified by the 44‐K SNP dataset were located within the coding regions of either of the NBS‐LRR candidate genes. On the contrary, using the 700‐K SNP dataset, five SNPs [SNP‐9.9663382 (P = 3.71 × 10−15), SNP‐9.9665626 (P = 1.07 × 10−5), SNP‐9.9665702 (P = 8.60 × 10−16), SNP‐9.9667603 (P = 2.51 × 10−7) and SNP‐9.9668044 (P = 7.21 × 10−5)] with a peak value of P = 8.60 × 10−16 were located within LABR_64‐1, and 10 SNPs [SNP‐9.9680445 (P = 2.90 × 10−12), SNP‐9.9682378 (P = 2.09 × 10−14), SNP‐9.9682420 (P = 4.19 × 10−16), SNP‐9.9682585 (P = 7.01 × 10−16), SNP‐9.9682753 (P = 4.66 × 10−16), SNP‐9.9682888 (P = 1.01 × 10−16), SNP‐9.9684972 (P = 2.52 × 10−6), SNP‐9.9687566 (P = 1.82 × 10−5), SNP‐9.9687903 (P = 4.08 × 10−6) and SNP‐9.9688188 (P = 3.47 × 10−5)] with a peak value of P = 1.01 × 10−16 were located within LABR_64‐2 (Fig. 5b,c). These results illustrate the power of the HDRA mapping data to narrow down the target region from 220 kb to less than 20 kb, a significant improvement in mapping resolution. On the basis of these results, we inferred that both LABR_64‐1 and LABR_64‐2 are strong candidate genes associated with blast resistance. This is consistent with the model proposed for the Pi5 locus, whereby resistance is conferred by dual NBS‐LRR genes.
Figure 5.
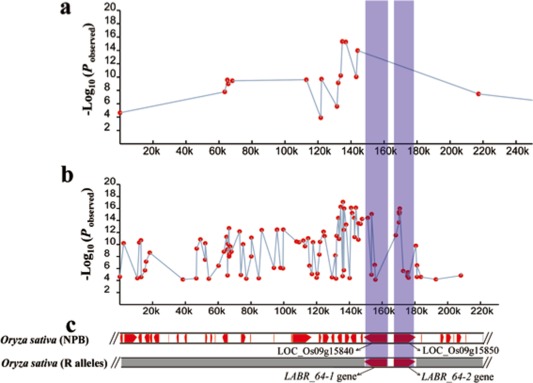
Comparative analysis of the blast resistance‐associated single nucleotide polymorphism (SNP) maps identified by the 44‐K and 700‐K datasets at the LABR_64 locus. (a) The blast resistance‐associated SNP map identified by the 44‐K dataset. Each red dot indicates one associated SNP. (b) The blast resistance‐associated SNP map identified by the 700‐K dataset. (c) The gene annotation of the 220‐kb region in Nipponbare. The purple bands indicate the LABR_64‐1 and LABR_64‐2 regions.
Using the sequences of the cloned R genes Pi5‐1 and Pi5‐2 as references, we designed nine primer pairs (Table S8, see Supporting Information) to amplify the sequence across the two candidate genes in the 24 resistant and susceptible cultivars. Among them, 12 resistant cultivars with the resistance‐associated SNP genotype at the LABR_64 locus and 12 susceptible cultivars with the susceptibility‐associated SNP genotype at the LABR_64 locus were randomly selected from the diversity panel. At the LABR_64‐1 locus, amplification was obtained from both the resistant and susceptible cultivars when the primer pairs LABR_64‐1_p1, p3 and p4 were used (Fig. S2, see Supporting Information). However, the expected band was only obtained in the resistant cultivars when the primer pair #LABR_64‐1_p2 was used. At the LABR_64‐2 locus, the expected fragments were only amplified from the 12 resistant cultivars, but not from the 12 susceptible cultivars, when the four primer pairs LABR_64‐2_p2, 3, 4 and 5 were used (Fig. S3, see Supporting Information). In contrast, amplification was obtained from both the resistant and susceptible cultivars when the primer pair #LABR_64‐2_p1 was used. These results suggest that the sequence in the resistant cultivars at both the LABR_64‐1 and LABR_64‐2 loci is conserved, but that, in the susceptible cultivars, there have been major structural changes in the DNA caused by various kinds of mutation.
We then cloned and sequenced the fragments amplified from the resistant cultivars at both loci. Most of the regions in the susceptible cultivars could not be amplified. When the sequences from the resistant cultivars for the Pi‐5 locus were compared, we identified two gene family members, namely our candidate genes LABR_64‐1 and LABR_64‐2. The DNA sequences among the resistant cultivars were highly similar (99.5–100% identity) at both loci. However, when the sequences were compared using blast, no DNA sequence identity between the two loci was detected. Similarly, at the protein level, the amino acid sequences are highly similar among the resistant cultivars at both loci (Figs S4 and S5, see Supporting Information), but the identity between the two loci is quite low (29.8%) (Fig. S6, see Supporting Information). These results confirmed that the alleles at LABR_64‐1 and LABR_64‐2 are highly conserved among the resistant cultivars.
Both LABR_64‐1 and LABR_64‐2 genes are essential for LABR_64‐mediated blast resistance
To confirm the function of the candidate blast resistance genes underlying LABR_64‐1 and LABR_64‐2, and to expedite the utility of this work for immediate applications in plant breeding, we selected two elite, high‐performing resistant Japonica cultivars, i.e. 312007 and 301279, for RNAi knockdown experiments. We sequenced the LABR_64‐1 (Genbank accession: KM016793 and KM016794) and LABR_64‐2 (Genbank accession: KM016807 and KM016808) genes in both cultivars and compared their sequences with those of the previously characterized blast‐resistant genes Pi5‐1, Pi5‐2, Pi15 and Pii. When compared with the Pi5‐1 gene, originally identified in the tropical japonica cultivar Moroberekan (Wang et al., 1994), the sequence of LABR_64‐1 in cultivar 312007 was found to differ by five non‐synonymous SNPs, leading to five predicted amino acid differences in the protein product. However, the DNA sequence and predicted protein product of LABR_64‐1 in 312007 were exactly the same as those previously reported for Pi15 and Pii (Fig. 6a). The LABR_64‐2 protein in 312007 contained three amino acid substitutions and a large deletion in the C‐terminal region when compared with Pi5‐2 in Moroberekan (Fig. 6b). In cultivar 301279, LABR_64‐1 had a large insertion and LABR_64‐2 had a large deletion, both in C‐terminal regions, in comparison with Pi5‐1 and Pi5‐2, respectively (Fig. 6a,b). Single amino acid differences between LABR_64‐1 or LABR_64‐2 and Pi15 and Pii were also found in 301279. These results demonstrate that both 312007 and 301279 contain previously undescribed alleles at the LABR_64/Pi5/Pi15/Pii locus.
Figure 6.
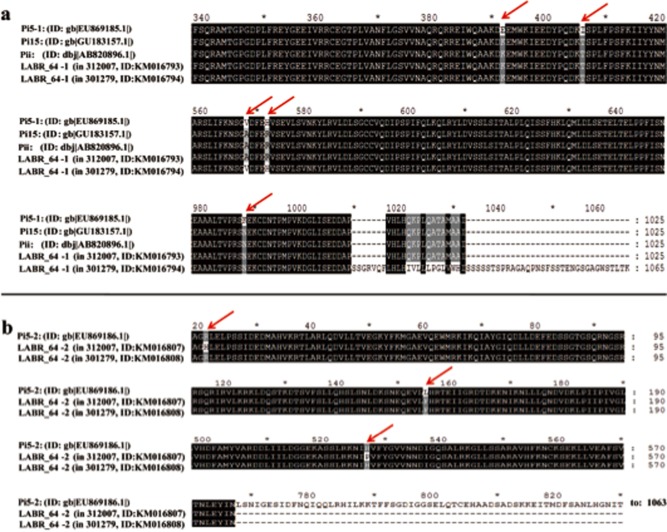
Protein sequence alignment in the allelic region of Pi5‐1, Pi15, Pii and LABR_64. (a) Protein sequence differences among Pi5‐1, PI15, Pii and LABR_64‐1 (in 312007 and 301279). Only the polymorphic regions are shown. The red arrow indicates the single amino acid polymorphic regions. (b) Protein sequence differences among Pi5‐2 and LABR_64‐2 (in 312007 and 301279).
Two RNAi constructs targeting a specific region in each LABR_64 candidate gene (106–316 bp in LABR_64‐1 and 547–897 bp in LABR_64‐2) were made and transformed into the calli of 312007 and 301279 (see RNAi sequence information in Experimental procedures). Over 20 independently transformed lines were obtained for each construct. We confirmed the presence of the transgene in the T1 generation by polymerase chain reaction (PCR), and the gene expression level of the target genes in the T2 generation by reverse transcription (RT)‐PCR (Fig. 7a,b). The T1 plants were punch inoculated and the T2 plants were spray inoculated with R01‐1. At least three transgenic lines for each RNAi construct were identified and used for blast inoculations. The segregation of resistant and susceptible plants (1R : 3S) in the T2 generation was consistent with the presence of a single copy of a dominant transgene conferring susceptibility in the three selected lines (chi‐squared goodness‐of‐fit test, P > 0.75).
Figure 7.
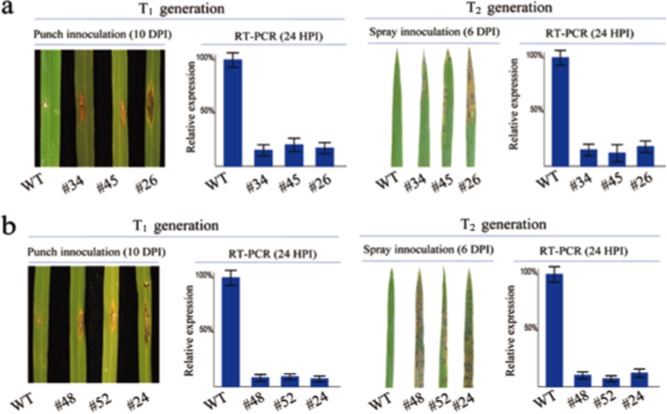
Blast disease evaluations of the RNA interference (RNAi) transgenic lines of the LABR_64‐1 and LABR_64‐2 genes in the T1 and T2 generations and wild‐type (WT) 312007. (a) Punch and spray inoculation and gene expression results of the LABR_64‐1 RNAi lines in the 312007 background. The plants were inoculated with R01‐1. Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis was carried out using the total RNA isolated at 24 h after inoculation (HPI). (b) Punch and spray inoculation and gene expression results of the LABR_64‐2 RNAi lines in the 312007 background. The plants were inoculated with R01‐1. RT‐PCR analysis was carried out using the total RNA isolated at 24 HPI. DPI, days post‐inoculation.
The punch inoculation of the T1 plants in the 312007 background with isolate R01‐1 showed large disease lesions in the three tested transgenic lines (#34, #45 and #26) compared with the wild‐type, which was consistent with the reduced expression level of LABR_64‐1 (Fig. 7a, left panel). The spray inoculation of transgenic T2 lines with isolate R01‐1 showed similar results (Fig. 7a, right panel). When LABR_64‐1 was silenced in the second resistant cultivar, 301279, inoculation with isolate R01‐1 in this genetic background again produced a susceptible response (data not shown). Similarly, when LABR_64‐2 was silenced in either the 312007 or 301279 background, three transgenic lines (#48, #52 and #24) in both the T1 and T2 generations showed high susceptibility to R01‐1 (Fig. 7b). Together, these results clearly demonstrate that the genes underlying LABR_64‐1 and LABR_64‐2 in both japonica cultivars 312007 and 301279 are homologues of Pi5/Pi15/Pii, and that the two cultivars carry novel functional alleles that confer resistance against M. oryzae.
Discussion
Various genomic platforms have been used for GWAS in plants (Brachi et al., 2011; Buckler et al., 2009; Huang et al., 2010; Jia et al., 2013; Morris et al., 2013; Zhao et al., 2011). To facilitate the public use of the new technology for gene mapping and cloning, one of the important requirements for such a platform is the easy access of both the SNP dataset and mapping population. Zhao et al. (2011) demonstrated the value of GWAS using the publically available RDP1 and a 44‐K SNP dataset to locate agronomic traits in rice. Using the same RDP1 germplasm, but taking advantage of the increased resolution of the HDRA (700‐K SNP) genotyping dataset, we identified 97 LABRs involved in resistance to five M. oryzae isolates. Eighty‐two of the LABRs were located in genomic regions in which no previously identified blast R genes or QTLs had been mapped, whereas 15 co‐localized with previously identified blast resistance loci that encoded NBS‐LRR‐type R proteins, proteins involved in the recognition and signalling of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (Zhang and Zhou, 2010) or pathogenesis‐related proteins. One of the most significant peaks in GWAS identified a major resistance locus, LABR_64, which was associated with high levels of resistance for all five blast isolates in RDP1 as a whole and in individual subpopulations. We compared the 97 LABRs with 10 known blast resistance QTLs from the OryGenesDB database and found that nine LABRs were co‐located in the four known QTL regions, suggesting an overlap between the LABRs and previously reported QTLs in the rice genome. Further fine mapping of these LABRs in the future will enable us to pinpoint the exact physical locations of these QTLs.
To select candidate resistance genes from the LABR_64 locus for functional analysis, we compared the Manhattan plots showing results from GWAS using the same phenotypic dataset in combination with the 44‐K SNP chip and the 700‐K SNP chip. On the 44‐K SNP map, both LABR_64‐1 and LABR_64‐2 were located in a region of SNP paucity and no major peak was observed directly above these two candidate genes. Using the 700‐K SNP map, five significant SNPs were identified across the region carrying LABR_64‐1 (the most significant was SNP‐9.9665702, P = 8.60 × 10−16), and 10 significant SNPs were identified across LABR_64‐2 (the most significant was SNP‐9.9682888, P = 1.01 × 10−16). Compared with the 44‐K SNP dataset, which defined a 220‐kb GWAS LABR, use of the HDRA SNP dataset in combination with exactly the same phenotypic data allowed us to narrow the LABR to approximately 20 kb.
Genetic complementation with a wild‐type genomic fragment of the target gene and RNA silencing of the expression of the target gene in transgenic plants are the two main methods for gene functional confirmation. The former strategy requires the cloning of the coding and promoter regions of the target gene for complementation, which is laborious and time consuming. In this study, we used the latter, because it is easy to make an RNAi construct and silencing efficiency is high in rice. Our sequence analysis revealed that the two candidate genes LABR_64‐1 and LABR_64‐2 in the resistant cultivars 312007 and 301279 are new alleles at the previously identified complex Pi5/Pi15/Pii locus. The RNAi silencing of LABR_64‐1 and LABR_64‐2 in the two cultivars confirmed the function of both genes against M. oryzae. It takes about 2–3 months to evaluate the resistance of the entire RDP1 to a single blast isolate in growth chambers and 5–6 months to obtain adult transgenic plants for punch inoculation. Therefore, the strategy developed in this study by combining GWAS and RNAi reduced the time required to map and clone a new R gene from 4–5 years to less than 1 year, and can be easily applied to other complex traits in rice and other crop plants.
The identification of 116 candidate genes underlying the 97 LABRs provides a large number of well‐supported candidate R genes that can be targeted by RNAi technology for the future validation of the results of this study, as demonstrated here. It is possible that, because we are aligning sequences using the Nipponbare genomic sequence as the reference, we will fail to discover novel gene duplications that might confer new forms of resistance or susceptibility in diverse germplasm resources. Thus, our estimate of how many genes underlie these LABRs could be an underestimate. This would be particularly likely if the resistance phenotype were coming from an indica or an aus donor, where structural variation and genomic rearrangements occur throughout the genome. This is a consequence of the large divergence time separating the ancestors of indica and aus from those of temperate japonica (i.e., Nipponbare), and has been well documented in relation to numerous traits of agronomic importance (Gamuyao et al., 2012; Hattori et al., 2009; Yanagihara et al., 1995; Yang et al., 2006; Zhai et al., 2011).
Magnaporthe oryzae is one of the 10 most important fungal pathogens of crop plants (Dean et al., 2012). Its genome undergoes rapid evolution as a result of the movement of transposable elements and the rearrangement of repetitive sequences, causing frequent breakdown of host resistance in rice cultivars (Valent, 1990; Valent and Khang, 2010). However, the process of co‐evolution between M. oryzae and its host plant has enabled rice to develop sophisticated genetic mechanisms to help avoid the loss of resistance. Molecular mapping has shown that broad‐spectrum resistance to M. oryzae is controlled by multiple R genes and many smaller effect QTLs (Liu et al., 2013). For example, the well‐known, durably resistant upland rice cultivar, Moroberekan, contains not only two R genes, Pi5 and Pi7, but 10 additional QTLs that confer partial resistance (Wang et al., 1994). In this study, we identified 97 LABRs that co‐localize with 116 resistance‐related candidate genes. Gene annotation analysis classified these genes into eight functional groups, each having a different function in plant immune responses. The largest group consisted of 34 NBS‐LRR genes that are associated with effector‐triggered immunity (ETI), whereas the rest are genes involved in the recognition and signalling of PAMP‐triggered immunity (PTI) or basal resistance. This study supports the conclusion that rice plants utilize a large number of genes that play diverse roles in the mediation of innate immunities against M. oryzae.
Among the genes involved in PTI recognition, signalling and defence activation, the largest group contains 17 genes that encode putative RLKs or receptor‐like proteins (RLPs). The characterized pattern recognition receptors (PRRs) in rice are RLKs or RLPs (Akamatsu et al., 2013; Chen et al., 2014; Hu et al., 2005; Liebrand et al., 2013). The genes involved in the regulation of Ca2+ content and the generation of reactive oxygen species (ROS), such as calmodulin‐related genes and oxidase/peroxidase/reductase genes, were also identified in this study. Recent research has demonstrated that calmodulin‐related proteins regulate the function of target proteins involved in plant responses to pathogen attack. Rapid increases in the intracellular Ca2+ concentration contribute to the oxidative burst, nitric oxide (NO) production, the hypersensitive response, the accumulation of salicylic acid and the induced expression of pathogenesis‐related genes (Poovaiah et al., 2013). Oxidase genes are directly related to the generation of ROS, which is one of the main responses in plant PTI. For example, the Arabidopsis RLK BIK1, a component of the PRR FLS2‐mediated immune pathway, directly phosphorylates the NADPH oxidase RbohD at specific sites to enhance ROS generation (Li et al., 2014). The involvement of other genes that encode transcription factors, ubiquitination‐related proteins, heat shock proteins and phosphorylation‐related genes in plant immunity has also been well documented in rice (Liu et al., 2013). Although the function of these candidate genes in conferring resistance to M. oryzae needs to be further confirmed, the identification of GWAS peaks that co‐localize with these genes suggests that there is abundant natural variation in the rice gene pool that can be harnessed to enhance the basal resistance to M. oryzae and to improve the spectrum and durability of resistance in new cultivars. Detailed genetic and biochemical analysis of these genes is likely to provide new insights into the complex mechanisms and intricate regulatory networks that collectively provide rice with resistance to blast disease.
Our results demonstrate that the LABR_64 locus on rice chromosome 9 is associated with broad‐spectrum blast resistance in several different subpopulations of rice. The locus confers resistance to all five M. oryzae isolates tested, and occurs as one component of a complex, multi‐locus disease resistance strategy in diverse landraces of rice. LABR_64 co‐localizes with the previously identified Pi5 locus, initially identified from the durably resistant cultivar Moroberekan (Zhang and Zhou, 2010). Pi5 resistance requires two NBS‐LRR genes (Lee et al., 2009), which were also found in the R genes Pia (Okuyama et al., 2011) and Pikm (Ashikawa et al., 2008). A recent study has demonstrated that the R gene pair RGA4 and RGA5, both located within LABR_64, can recognize both M. oryzae effectors Avr‐Pia and Avr1‐CO39 by direct binding (Cesari et al., 2013). These results suggest that the unique, dual R‐gene structure and ability to recognize multiple Avr genes might explain the broad‐spectrum resistance of the locus. In addition, the identification of two new alleles at the Pi5/Pi15/Pii/LABR_64 locus in this study offers new opportunities for rice blast control because different alleles are effective against diverse M. oryzae populations. Further, the markers and information from this study can be immediately utilized by plant breeders to identify and target the novel allele complex at the Pi5/Pi15/Pii/LABR_64 locus for introgression into diverse genetic backgrounds in which broad‐spectrum blast disease resistance is required. The study aptly bridges the utilization of high‐resolution GWAS for blast‐resistant locus discovery, the use of open‐source plant materials, genotypic data and bioinformatics tools to rapidly identify candidate genes associated with well‐resolved GWAS‐LABRs, the application of RNAi transgenic strategies for candidate gene validation, and the open deployment of information so that the breeding community can make immediate use of the findings.
Experimental Procedures
Plant and fungal materials
Seeds of the 420 O. sativa accessions that comprise RDP1 were provided by the Genetic Stocks–Oryza (GSOR) Collection, USDA ARS Dale Bumpers National Rice Research Center, Stuttgart, AR, USA. We grew all the accessions in a glasshouse at the Ohio State University in the summer of 2011 and only harvested sufficient seed from about 390 of the accessions for blast inoculation. The five M. oryzae isolates were collected from the following rice‐growing countries: R01‐1 (South Korea), RB22 (China), 75‐1‐127 (Columbia), 0‐249 (India) and P06‐6 (Philippines), as described by Zhou et al. (2006). These isolates were used in this study because they are genetically and geographically diverse. They are stable in culture and sporulate well. Isolate P06‐6 from the Philippines has been used in many previous mapping studies (Qu et al., 2006; Zhu et al., 2012). The five isolates were genotyped using 11 pairs of SSR markers reported by Kaye et al. (2003) (Fig. S7, see Supporting Information).
Evaluation of blast resistance phenotypes
Rice seedlings and fungal cultures were prepared for spray inoculation as described by Park et al. (2012). About 15 seedlings per cultivar were used for blast inoculations, and the experiment was performed three times. The 0–9 scoring system was used for the evaluation of blast symptoms, as described previously (Zhu et al., 2012): 0, no infection; 1, small closed lesions; 2, small lesions/grey centres; 3, small elliptical lesions/heavy borders; 4, expanding elliptical lesions; 5, lesion area 10%–25%; 6, lesion area 26%–50%; 7, lesion area 51%–75%; 8, lesion area 76%–90%; 9, lesion area >90%. The mean score of three replications was used for GWAS. Blast punch inoculation followed the original method, as described previously (Ono et al., 2001), with slight modifications. A 10‐µL volume of a spore suspension (5 × 105 spores/mL) was applied to slightly punctured sites of leaves on plants that were about 6 weeks old. Lesion size was recorded 9–10 days after inoculation. The infection ratio was calculated as described previously (Kawano et al., 2010).
Genome‐wide association analysis
Initially, GWAS was performed on RDP1 accessions using the publicly available 44 000‐SNP dataset (MAF > 0.05) (Zhao et al., 2011), Tassel3.0 software (http://www.maizegenetics.net/index.php?option=com_content&task=view&id=89&Ite-mid=119) and the mixed linear model (MLM). MLM (Yu et al., 2006) uses a joint kinship matrix (K) and population structure (Q) model. This model can be described in Henderson's matrix notation (Henderson, 1975) as y = Xβ + Zu + e, where y is the vector of observations, β is an unknown vector containing fixed effects, including genetic marker and population structure (Q), u is an unknown vector of random additive genetic effects from different background QTLs for individuals/cultivars, X and Z are the known design matrices, and e is the unobserved vector of the random residual. The ‘Q + K’ approach improves statistical power compared with ‘Q’ only (Henderson, 1975). To control type 1 error, we considered only regions that had more than two SNPs with P < 1.0 × 10−4 within a 200‐kb genomic window for subsequent analysis. The integrated Manhattan plots map was drawn using PERL (Christiansen et al., 2012) and its scalable vector graphics (SVG) module. Subsequently, GWAS was performed using the 700‐K SNP data (HDRA6.4). We used EMMAX (Kang et al., 2010) to fit a standard linear mixed model, y = μ + Xb + Zu + e, where y is a vector of phenotypes, μ is the mean, b is the additive allele effect, X is a matrix of the genetic variants, Z is a matrix connecting phenotypes to the plants, u is a vector of additive effects [∼N(0, σ 2 G), where σ 2 is the additive genetic variance and G is the identity by descent (IBS) kinship matrix] and e is a vector of random residuals. Three Principal Component (PC) covariates were added to the model when the entire RDP1 was analysed together and no covariates were used when each subpopulation was processed independently. Manhattan and Q–Q plots were produced using R package, https://cran.r-project.org/web/packages/qqman/.
Bioinformatics analysis of LABRs in the rice genome
We used the rice genome sequence version of MSU V7.0 as the reference. All subsequent analysis was performed manually using the sequence alignment software blast and blat as well as PERL scripts. First, we determined the genomic positions of the 97 LABRs identified by GWAS and obtained the sequences from the reference genome; we then aligned all the predicted genes to these sequences to determine which genes are mapped to the LABRs. Second, we expanded our search for candidates using the GO and orthology searches, targeting all R and R‐related genes in rice based on protein and/or sequence similarity with known R, signalling and defence‐related genes.
RNAi constructs and rice transformation
The specific nucleotide sequences of the LABR_64‐1 and LABR_64‐2 candidate genes for the RNAi experiments are shown in Fig. S8 (see Supporting Information). We performed blastn searches in the National Center for Biotechnology Information (NCBI) databases to make sure that the two RNAi fragments had <80% DNA sequence identity with any rice genes and the 21‐bp fragments from the two sequences were not 100% to any region of the other rice genes. The primer sequences used for the preparation of the two RNAi constructs are listed in Table S8 (primers 10 and 11). The pANDA vector (Miki and Shimamoto, 2004) was used to construct the RNAi plasmids. The RNAi plasmids were transferred into rice calli using Agrobacterium‐mediated transformation (Qu et al., 2006). The T1 transgenic plants were confirmed by hygromycin‐resistant screening, PCR and RT‐PCR using the primers listed in Table S8. Primers 12 and 13 were used for PCR screening of the transgenic plants, and primers 14 and 15 were used for real‐time PCR analysis of target gene expression in the transgenic plants.
Gene Accession Numbers Deposited in Genbank
LABR_64‐1 allelic gene in resistant cultivar of 312007, Genbank accession number: KM016793.
LABR_64‐1 allelic gene in resistant cultivar of 301279, Genbank accession number: KM016794.
LABR_64‐1 allelic gene in resistant cultivar of 301068, Genbank accession number: KM016795.
LABR_64‐1 allelic gene in resistant cultivar of 301081, Genbank accession number: KM016796.
LABR_64‐1 allelic gene in resistant cultivar of 301138, Genbank accession number: KM016797.
LABR_64‐1 allelic gene in resistant cultivar of 301156, Genbank accession number: KM016798.
LABR_64‐1 allelic gene in resistant cultivar of 301379, Genbank accession number: KM016799.
LABR_64‐1 allelic gene in resistant cultivar of 312013, Genbank accession number: KM016800.
LABR_64‐1 allelic gene in resistant cultivar of 301189, Genbank accession number: KM016801.
LABR_64‐1 allelic gene in resistant cultivar of 301411, Genbank accession number: KM016802.
LABR_64‐1 allelic gene in resistant cultivar of 301177, Genbank accession number: KM016903.
LABR_64‐1 allelic gene in resistant cultivar of 301060, Genbank accession number: KM016804.
LABR_64‐1 allelic gene in resistant cultivar of 301278, Genbank accession number: KM016805.
LABR_64‐1 allelic gene in resistant cultivar of 301246, Genbank accession number: KM016806.
LABR_64‐2 allelic gene in resistant cultivar of 312007, Genbank accession number: KM016807.
LABR_64‐2 allelic gene in resistant cultivar of 301279, Genbank accession number: KM016808.
LABR_64‐2 allelic gene in resistant cultivar of 301068, Genbank accession number: KM016809.
LABR_64‐2 allelic gene in resistant cultivar of 301081, Genbank accession number: KM016810.
LABR_64‐2 allelic gene in resistant cultivar of 301138, Genbank accession number: KM016811.
LABR_64‐2 allelic gene in resistant cultivar of 301156, Genbank accession number: KM016812.
LABR_64‐2 allelic gene in resistant cultivar of 301379, Genbank accession number: KM016813.
LABR_64‐2 allelic gene in resistant cultivar of 312013, Genbank accession number: KM016814.
LABR_64‐2 allelic gene in resistant cultivar of 301189, Genbank accession number: KM016815.
LABR_64‐2 allelic gene in resistant cultivar of 301411, Genbank accession number: KM016816.
LABR_64‐2 allelic gene in resistant cultivar of 301177, Genbank accession number: KM016817.
LABR_64‐2 allelic gene in resistant cultivar of 301060, Genbank accession number: KM016818.
LABR_64‐2 allelic gene in resistant cultivar of 301278, Genbank accession number: KM016819.
LABR_64‐2 allelic gene in resistant cultivar of 301246, Genbank accession number: KM016820.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The distribution of blast resistance in the diversity panel and subpopulations to isolates 75‐1‐127, RB22, 0‐249 and P06‐6.
Fig. S2 Polymerase chain reaction (PCR) amplification of the LABR_64‐1 sequence in the resistant and susceptible cultivars. The PCR products were separated on a 1.0% agarose gel.
Fig. S3 Polymerase chain reaction (PCR) amplification of the LABR_64‐2 sequence in the resistant and susceptible cultivars. The PCR products were separated on a 1.0% agarose gel.
Fig. S4 Protein sequence alignment in the allelic region of Pi5‐1 and LABR_64‐1 in the 12 resistant cultivars with the resistance (R) single nucleotide polymorphism (SNP) genotype. Only polymorphic regions between the two proteins are shown. The red arrows indicate the single amino acid polymorphic regions.
Fig. S5 Protein sequence alignment in the allelic region of Pi5‐2 and LABR_64‐1 in the 12 resistant cultivars with the resistance (R) single nucleotide polymorphism (SNP) genotype. Only polymorphic regions between the two proteins are shown. The red arrows indicate the single amino acid polymorphic regions.
Fig. S6 Protein sequence alignment between LABR_64‐1 and LABR_64‐2 in 301068. The two proteins share 29.8% sequence identity and 43.9% sequence similarity.
Fig. S7 Polyacrylamide gel electrophoresis (PAGE) of the five isolates using 14 pairs of primers reported by Kaye et al. (2003). The simple sequence repeat (SSR) markers highlighted in yellow were used for the construction of the phylogenetic tree; 1–5 represent the five isolates RB22, R01‐1, P06‐6, 0‐249 and 75‐1‐127, respectively. #SSR‐1, Pyrms 7&8; #SSR‐2, Pyrms 15&16; #SSR‐3, Pyrms 33&34; #SSR‐4, Pyrms 43&44; #SSR‐7, Pyrms 63&64; #SSR‐8, Pyrms 81&82; #SSR‐9, Pyrms 83&84; #SSR‐10, Pyrms 87&88; #SSR‐11, Pyrms 93&94.
Fig. S8 LABR_64‐1 and LABR_64‐2 RNA interference (RNAi) construct maps. (a) LABR_64‐1 RNAi construct; the RNAi fragment is highlighted in yellow. (b) LABR_64‐2 RNAi construct; the RNAi fragment is highlighted in blue. The gene‐specific fragment (without any sequence similarity to other genes in the rice genome) was cloned into the pANDA vector by the Gateway cloning method. The β‐glucuronidase (Gus) gene was used as the linker for two RNAi fragments.
Table S1 Blast disease scores of rice cultivars with five isolates.
Table S2 List of the cultivars highly resistant or susceptible to all five isolates.
Table S3 Pearson correlation coefficients of the pathogenicity of the five isolates.
Table S4 The regions associated with blast resistance to the five Magnaporthe oryzae isolates.
Table S5 The 15 loci associated with blast resistance (LABRs) that were co‐localized with known blast resistance loci.
Table S6 The top 72 loci associated with blast resistance (LABRs).
Table S7 The percentage of the phenotypic variation explained by the 72 loci associated with blast resistance (LABRs).
Table S8 Primers used in this study.
Acknowledgements
This research was supported by grants from the 973 Project (2012CB114005) of the Ministry of Science and Technology China, the National Natural Science Foundation of China (31201476), International cooperation project of National Natural Science Foundation of China (31461143019) and the USAID‐IRRI Linkage project. We thank the Genetic Stocks Oryza (GSOR) collection laboratory, the USDA Dale Bumpers National Rice Research Center, for providing the seeds of the Rice Diversity Panel 1.
References
- Akamatsu, A. , Wong, H.L. , Fujiwara, M. , Okuda, J. , Nishide, K. , Uno, K. , Imai, K. , Umemura, K. , Kawasaki, T. , Kawano, Y. and Shimamoto, K. (2013) An OsCEBiP/OsCERK1‐OsRacGEF1‐OsRac1 module is an essential early component of chitin‐induced rice immunity. Cell Host Microbe, 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Ashikawa, I. , Hayashi, N. , Yamane, H. , Kanamori, H. , Wu, J. , Matsumoto, T. , Ono, K. and Yano, M. (2008) Two adjacent nucleotide‐binding site‐leucine‐rich repeat class genes are required to confer Pikm‐specific rice blast resistance. Genetics, 180, 2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkani, S. , Rafii, M.Y. , Rahim, H.A. and Latif, M.A. (2013) Genetic dissection of rice blast resistance by QTL mapping approach using an F3 population. Mol. Biol. Rep. 40, 2503–2515. [DOI] [PubMed] [Google Scholar]
- Ballini, E. , Morel, J.‐B. , Droc, G. , Price, A. , Courtois, B. , Notteghem, J.‐L. and Tharreau, D. (2008) A genome‐wide meta‐analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant–Microbe Interact. 21, 859–868. [DOI] [PubMed] [Google Scholar]
- Brachi, B. , Morris, G.P. and Borevitz, J.O. (2011) Genome‐wide association studies in plants: the missing heritability is in the field. Genome Biol. 12, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E.S. , Holland, J.B. , Bradbury, P.J. , Acharya, C.B. , Brown, P.J. , Browne, C. , Ersoz, E. , Flint‐Garcia, S. , Garcia, A. , Glaubitz, J.C. , Goodman, M.M. , Harjes, C. , Guill, K. , Kroon, D.E. , Larsson, S. , Lepak, N.K. , Li, H. , Mitchell, S.E. , Pressoir, G. , Peiffer, J.A. , Rosas, M.O. , Rocheford, T.R. , Romay, M.C. , Romero, S. , Salvo, S. , Villeda, H.S., H. Silva, H.S. , Sun, Q. , Tian, F. , Upadyayula, N. , Ware, D. , Yates, H. , Yu, J. , Zhang, Z. , Kresovich, S. , and McMullen, M.D. (2009) The genetic architecture of maize flowering time. Science, 325, 714–718. [DOI] [PubMed] [Google Scholar]
- Cesari, S. , Thilliez, G. , Ribot, C. , Chalvon, V. , Michel, C. , Jauneau, A. , Rivas, S. , Alaux, L. , Kanzaki, H. , Okuyama, Y. Morel, J.‐B. , Fournier, E. , Tharreau, D. , Terauchif, R. , and Thomas Kroj, T. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Zuo, S. , Schwessinger, B. , Chern, M. , Canlas, P.E. , Ruan, D. , Zhou, X. , Wang, J. , Daudi, A. , Petzold, C.J. , Heazlewoodc, J.L. , and Ronalda, P.C. (2014) An XA21‐associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant, 7, 874–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen, T. , Foy, B.D. and Wall, L. (2012) Programming Perl‐Unmatched Power for Text Processing and Scripting: Covers Version 5.14. O'Reilly: Sebastopol, CA, USA. [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizenga, G.C. , Ali, M. , Bryant, R.J. , Yeater, K.M. , McClung, A.M. and McCouch, S.R. (2014) Registration of the rice diversity panel 1 for genomewide association studies. J. Plant Regist. 8, 109–116. [Google Scholar]
- Famoso, A.N. , Zhao, K. , Clark, R.T. , Tung, C.‐W. , Wright, M.H. , Bustamante, C. , Kochian, L.V. and McCouch, S.R. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome‐wide association analysis and QTL mapping. PLoS Genet. 7, e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, S. , Saka, N. , Koga, H. , Ono, K. , Shimizu, T. , Ebana, K. , Hayashi, N. , Takahashi, A. , Hirochika, H. , Okuno, K. and Yano, M. (2009) Loss of function of a proline‐containing protein confers durable disease resistance in rice. Science, 325, 998–1001. [DOI] [PubMed] [Google Scholar]
- Gamuyao, R. , Chin, J.H. , Pariasca‐Tanaka, J. , Pesaresi, P. , Catausan, S. , Dalid, C. , Slamet‐Loedin, I. , Tecson‐Mendoza, E.M. , Wissuwa, M. and Heuer, S. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature, 488, 535–539. [DOI] [PubMed] [Google Scholar]
- Hattori, Y. , Nagai, K. , Furukawa, S. , Song, X.‐J. , Kawano, R. , Sakakibara, H. , Wu, J. , Matsumoto, T. , Yoshimura, A. , Kitano, H. , Matsuoka, M. , Mori, H. and Ashikari, M. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature, 460, 1026–1030. [DOI] [PubMed] [Google Scholar]
- Hayashi, N. , Inoue, H. , Kato, T. , Funao, T. , Shirota, M. , Shimizu, T. , Kanamori, H. , Yamane, H. , Hayano‐Saito, Y. , Matsumoto, T. , Yano, M. and Takatsuji, H. (2010) Durable panicle blast‐resistance gene Pb1 encodes an atypical CC‐NBS‐LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510. [DOI] [PubMed] [Google Scholar]
- Henderson, C.R. (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics, 31, 423–447. [PubMed] [Google Scholar]
- Hittalmani, S. , Parco, A. , Mew, T.V. , Zeigler, R.S. and Huang, N. (2000) Fine mapping and DNA marker‐assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100, 1121–1128. [Google Scholar]
- Hu, H. , Xiong, L. and Yang, Y. (2005) Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta, 222, 107–117. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Wei, X. , Sang, T. , Zhao, Q. , Feng, Q. , Zhao, Y. , Li, C. , Zhu, C. , Lu, T. , Zhang, Z. , Li, M. , Fan, D. , Guo, Y. , Wang, A. , Wang, L. , Deng, L. , Li, W. , Lu, Y. , Weng, Q. , Liu, K. , Huang, T. , Zhou, T. , Jing, Y. , Li, W. , Lin, Z. , Buckler, E.S. , Qian, Q. , Zhang, Q.‐F. , Li, J. and Han, B. (2010) Genome‐wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. [DOI] [PubMed] [Google Scholar]
- Jia, G. , Huang, X. , Zhi, H. , Zhao, Y. , Zhao, Q. , Li, W. , Chai, Y. , Yang, L. , Liu, K. , Lu, H. , Zhu, C. , Lu, Y. , Zhou, C. , Fan, D. , Weng, Q. , Guo, Y. , Huang, T. , Zhang, L. , Lu, T. , Feng, Q. , Hao, H. , Liu, H. , Lu, P. , Zhang, N. , Li, Y. , Guo, E. , Wang, S. , Wang, S. , Liu, J. , Zhang, W. , Chen, G. , Zhang, B. , Li, W. , Wang, W. , Li, H. , Zhao, B. , Li, J. , Diao, X. and Han, B. (2013) A haplotype map of genomic variations and genome‐wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961. [DOI] [PubMed] [Google Scholar]
- Jia, Y. and Liu, G. (2011) Mapping quantitative trait loci for resistance to rice blast. Phytopathology, 101, 176–181. [DOI] [PubMed] [Google Scholar]
- Kang, H.M. , Sul, J.H. , Service, S.K. , Zaitlen, N.A. , Kong, S.‐y. , Freimer, N.B. , Sabatti, C. and Eskin, E. (2010) Variance component model to account for sample structure in genome‐wide association studies. Nat. Genet. 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, Y. , Akamatsu, A. , Hayashi, K. , Housen, Y. , Okuda, J. , Yao, A. , Nakashima, A. , Takahashi, H. , Yoshida, H. , Wong, H.L. , Kawasaki1, T. and Shimamoto, K. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe, 7, 362–375. [DOI] [PubMed] [Google Scholar]
- Kaye, C. , Milazzo, J. , Rozenfeld, S. , Lebrun, M.H. and Tharreau, D. (2003) The development of simple sequence repeat markers for Magnaporthe grisea and their integration into an established genetic linkage map. Fungal Genet. Biol. 40, 207–214. [DOI] [PubMed] [Google Scholar]
- Lee, S.‐K. , Song, M.‐Y. , Seo, Y.‐S. , Kim, H.‐K. , Ko, S. , Cao, P.‐J. , Suh, J.‐P. , Yi, G. , Roh, J.‐H. and Lee, S. , An, G. , Hahn, T.‐R. , Wang, G.‐L. , Ronald, P. and Jeon, J.‐S. (2009) Rice Pi5‐mediated resistance to Magnaporthe oryzae requires the presence of two coiled‐coil‐nucleotide‐binding‐leucine‐rich repeat genes. Genetics, 181, 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Li, M. , Yu, L. , Zhou, Z. , Liang, X. , Liu, Z. , Cai, G. , Gao, L. , Zhang, X. and Wang, Y. Chen, S. and Zhou, J.‐M. (2014) The FLS2‐associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe, 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Liebrand, T.W. , van den Berg, G.C. , Zhang, Z. , Smit, P. , Cordewener, J.H. , America, A.H. , Sklenar, J. , Jones, A.M. , Tameling, W.I. , Robatzek, S. , Thomma, B.P. , and Joosten, M.H. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA, 110, 13 228–13 228 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Liu, J. , Ning, Y. , Ding, B. , Wang, X. , Wang, Z. and Wang, G.‐L. (2013) Recent progress in understanding PAMP‐ and effector‐triggered immunity against the rice blast fungus Magnaporthe oryzae . Mol. Plant, 6, 605–620. [DOI] [PubMed] [Google Scholar]
- Liu, W.D. , Liu, J.L. , Ning, Y.S. , Ding, B. , Wang, X.L. , Wang, Z.L. , et al. (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R. , Kochert, G. , Yu, Z. , Wang, Z. , Khush, G. , Coffman, W. and Tanksley, S. (1988) Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76, 815–829. [DOI] [PubMed] [Google Scholar]
- Miki, D. and Shimamoto, K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495. [DOI] [PubMed] [Google Scholar]
- Morris, G.P. , Ramu, P. , Deshpande, S.P. , Hash, C.T. , Shah, T. , Upadhyaya, H.D. , Riera‐Lizarazu, O. , Brown, P.J. , Acharya, C.B. , Mitchell, S.E. , Harrimane, J. , Glaubitze, J.C. , Bucklere, E.S. and Kresovicha, S. (2013) Population genomic and genome‐wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA, 110, 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama, Y. , Kanzaki, H. , Abe, A. , Yoshida, K. , Tamiru, M. , Saitoh, H. , Fujibe, T. , Matsumura, H. , Shenton, M. , Galam, D.C. , Undan, J. , Ito, A. , Sone, T. and Terauchi, R. (2011) A multifaceted genomics approach allows the isolation of the rice Piat‐blast resistance gene consisting of two adjacent NBS‐LRR protein genes. Plant J. 66, 467–479. [DOI] [PubMed] [Google Scholar]
- Ono, E. , Wong, H.L. , Kawasaki, T. , Hasegawa, M. , Kodama, O. and Shimamoto, K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA, 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q.H. , Hu, Z.D. , Takatoshi, T. and Wang, L. (2003) Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. Acta Bot. Sin. 45, 871–877. [Google Scholar]
- Park, C.‐H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. , Valente, B. and Wang, G.‐L. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah, B.W. , Du, L.Q. , Wang, H.Z. and Yang, T.B. (2013) Recent advances in calcium/calmodulin‐mediated signaling with an emphasis on plant–microbe interactions. Plant Physiol. 163, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S. , Liu, G. , Zhou, B. , Bellizzi, M. , Zeng, L. , Dai, L. , Han, B. and Wang, G.‐L. (2006) The broad‐spectrum blast resistance gene Pi9 encodes a nucleotide‐binding site‐leucine‐rich repeat protein and is a member of a multigene family in rice. Genetics, 172, 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti, P. and Gurr, S.J. (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150. [DOI] [PubMed] [Google Scholar]
- Valent, B. (1990) Rice blast as a model system for plant pathology. Phytopathology, 80, 33–36. [Google Scholar]
- Valent, B. and Khang, C.H. (2010) Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 13, 434–441. [DOI] [PubMed] [Google Scholar]
- Wang, G.‐L. , Mackill, D.J. , Bonman, J.M. , McCouch, S.R. , Champoux, M.C. and Nelson, R.J. (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics, 136, 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Jia, MH. , Ghai, P. , Lee, FN. and Jia, Y. (2015) Genome wide association of rice blast disease resistance and yield related components of rice. Mol. Plant–Microbe Interact. (ja), 28, 1383–1392. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Wang, K. , Qu, H.‐Q. , Zhang, H. , Bradfield, J. , Kim, C. , Frackleton, E. , Hou, C. , Glessner, J.T. , Chiavacci, R. Stanley, C. , Monos, D. , Grant, S.F.A. , Polychronakos, C. and Hakonarson, H. (2009) From disease association to risk assessment: an optimistic view from genome‐wide association studies on type 1 diabetes. PLoS Genet. 5, e1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara, S. , McCouch, S.R. , Ishikawa, K. , Ogi, Y. , Maruyama, K. and Ikehashi, H. (1995) Molecular analysis of the inheritance of the S‐5 locus, conferring wide compatibility in Indica/Japonica hybrids of rice (O. sativa L.). Theor. Appl. Genet. 90, 182–188. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Feng, Z. , Zhang, X. , Jiang, K. , Jin, X. , Hang, Y. , Chen, J.‐Q. and Tian, D. (2006) Genome‐wide investigation on the genetic variations of rice disease resistance genes. Plant Mol. Biol. 62, 181–193. [DOI] [PubMed] [Google Scholar]
- Yu, J. , Pressoir, G. , Briggs, W.H. , Bi, I.V. , Yamasaki, M. , Doebley, J.F. , McMullen, M.D. , Gaut, B.S. , Nielsen, D.M. , Holland, J.B. , Kresovich, S. and Buckler, E.S. (2006) A unified mixed‐model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. [DOI] [PubMed] [Google Scholar]
- Zhai, C. , Lin, F. , Dong, Z. , He, X. , Yuan, B. , Zeng, X. , Wang, L. and Pan, Q. (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189, 321–334. [DOI] [PubMed] [Google Scholar]
- Zhang, J. and Zhou, J.M. (2007) Strategies for developing green super rice. Proc. Natl. Acad. Sci. USA, 104, 16 402–16 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q.F. (2010) Plant immunity triggered by microbial molecular signatures. Mol. Plant , 3, 783–793. [DOI] [PubMed] [Google Scholar]
- Zhao, K. , Tung, C.‐W. , Eizenga, G.C. , Wright, M.H. , Ali, M.L. , Price, A.H. , Norton, G.J. , Islam, M.R. , Reynolds, A. , Mezey, J. , McClung, A.M. , Bustamante, C.D. and McCouch, S.R. (2011) Genome‐wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa . Nat. Commun. 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. , Qu, S. , Liu, G. , Dolan, M. , Sakai, H. , Lu, G. , Bellizzi, M. and Wang, G.‐L. (2006) The eight amino‐acid differences within three leucine‐rich repeats between Pi2 and Piz‐t resistance proteins determine the resistance specificity to Magnaporthe grisea . Mol. Plant–Microbe Interact. 19, 1216–1228. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Chen, S. , Yang, J. , Zhou, S. , Zeng, L. , Han, J. , Su, J. , Wang, L. and Pan, Q. (2012) The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theor. Appl. Genet. 124, 1295–1304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The distribution of blast resistance in the diversity panel and subpopulations to isolates 75‐1‐127, RB22, 0‐249 and P06‐6.
Fig. S2 Polymerase chain reaction (PCR) amplification of the LABR_64‐1 sequence in the resistant and susceptible cultivars. The PCR products were separated on a 1.0% agarose gel.
Fig. S3 Polymerase chain reaction (PCR) amplification of the LABR_64‐2 sequence in the resistant and susceptible cultivars. The PCR products were separated on a 1.0% agarose gel.
Fig. S4 Protein sequence alignment in the allelic region of Pi5‐1 and LABR_64‐1 in the 12 resistant cultivars with the resistance (R) single nucleotide polymorphism (SNP) genotype. Only polymorphic regions between the two proteins are shown. The red arrows indicate the single amino acid polymorphic regions.
Fig. S5 Protein sequence alignment in the allelic region of Pi5‐2 and LABR_64‐1 in the 12 resistant cultivars with the resistance (R) single nucleotide polymorphism (SNP) genotype. Only polymorphic regions between the two proteins are shown. The red arrows indicate the single amino acid polymorphic regions.
Fig. S6 Protein sequence alignment between LABR_64‐1 and LABR_64‐2 in 301068. The two proteins share 29.8% sequence identity and 43.9% sequence similarity.
Fig. S7 Polyacrylamide gel electrophoresis (PAGE) of the five isolates using 14 pairs of primers reported by Kaye et al. (2003). The simple sequence repeat (SSR) markers highlighted in yellow were used for the construction of the phylogenetic tree; 1–5 represent the five isolates RB22, R01‐1, P06‐6, 0‐249 and 75‐1‐127, respectively. #SSR‐1, Pyrms 7&8; #SSR‐2, Pyrms 15&16; #SSR‐3, Pyrms 33&34; #SSR‐4, Pyrms 43&44; #SSR‐7, Pyrms 63&64; #SSR‐8, Pyrms 81&82; #SSR‐9, Pyrms 83&84; #SSR‐10, Pyrms 87&88; #SSR‐11, Pyrms 93&94.
Fig. S8 LABR_64‐1 and LABR_64‐2 RNA interference (RNAi) construct maps. (a) LABR_64‐1 RNAi construct; the RNAi fragment is highlighted in yellow. (b) LABR_64‐2 RNAi construct; the RNAi fragment is highlighted in blue. The gene‐specific fragment (without any sequence similarity to other genes in the rice genome) was cloned into the pANDA vector by the Gateway cloning method. The β‐glucuronidase (Gus) gene was used as the linker for two RNAi fragments.
Table S1 Blast disease scores of rice cultivars with five isolates.
Table S2 List of the cultivars highly resistant or susceptible to all five isolates.
Table S3 Pearson correlation coefficients of the pathogenicity of the five isolates.
Table S4 The regions associated with blast resistance to the five Magnaporthe oryzae isolates.
Table S5 The 15 loci associated with blast resistance (LABRs) that were co‐localized with known blast resistance loci.
Table S6 The top 72 loci associated with blast resistance (LABRs).
Table S7 The percentage of the phenotypic variation explained by the 72 loci associated with blast resistance (LABRs).
Table S8 Primers used in this study.


