Summary
Rice stripe virus (RSV) is the type species of the genus Tenuivirus and represents a major viral pathogen affecting rice production in East Asia. In this study, RSV p2 was fused to yellow fluorescent protein (p2‐YFP) and expressed in epidermal cells of Nicotiana benthamiana. p2‐YFP fluorescence was found to move to the nucleolus initially, but to leave the nucleolus for the cytoplasm forming numerous distinct bright spots there at later time points. A bimolecular fluorescence complementation (BiFC) assay showed that p2 interacted with fibrillarin and that the interaction occurred in the nucleus. Both the nucleolar localization and cytoplasmic distribution of p2‐YFP fluorescence were affected in fibrillarin‐silenced N. benthamiana. Fibrillarin depletion abolished the systemic movement of RSV, but not that of Tobacco mosaic virus (TMV) and Potato virus X (PVX). A Tobacco rattle virus (TRV)‐based virus‐induced gene silencing (VIGS) method was used to diminish RSV NS2 (encoding p2) or NS3 (encoding p3) during RSV infection. Silencing of NS3 alleviated symptom severity and reduced RSV accumulation, but had no obvious effects on virus movement and the timing of symptom development. However, silencing of NS2 abolished the systemic movement of RSV. The possibility that RSV p2 may recruit or manipulate nucleolar functions to promote virus systemic infection is discussed.
Keywords: fibrillarin, nucleolus, Rice stripe virus, systemic movement
Introduction
The nucleolus is a plurifunctional subnuclear structure that may harbour as many as 4500 proteins (Boisvert et al., 2007; Boulon et al., 2010; Shaw and Brown, 2012). The interaction of animal viruses with the nucleolus has been studied extensively since the beginning of the 1990s. Most, if not all, well‐studied animal viruses have been found to have a nucleolar phase in their infection cycle. A link between the ability to interact with the nucleolus and the outcome of viral infection has been established in many cases (Hiscox, 2007; Salvetti and Greco, 2014). In recent years, diverse plant viruses belonging to different genera or families have also been found to encode one or multiple proteins localizing to the nucleolus (Martínez and Daròs, 2014; Rossi et al., 2014; Taliansky et al., 2010; and references therein). In the nucleolus, Groundnut rosette virus (GRV, an umbravirus) open reading frame 3 (ORF3) interacts with fibrillarin and recruits this nucleolar protein to form infectious ribonucleoproteins (RNPs) in the cytoplasm. This is crucial for the systemic movement of GRV (Kim et al., 2007a, b). Fibrillarin is also essential for the systemic movement of Potato leafroll virus (PLRV, a polerovirus), but the mechanism by which PLRV uses fibrillarin remains unclear (Kim et al., 2007b). The nucleolar localization of NIa/Vpg plays an important role in Potato virus A (PVA, a potyvirus) replication, pathogenicity and systemic movement. This is partially explained by the fact that NIa/Vpg may need to enter the nucleolus to suppress RNA silencing (Rajamäki and Valkonen, 2009). NIa/Vpg also interacts with fibrillarin. However, the depletion of fibrillarin reduced PVA accumulation, but had no significant effects on PVA systemic movement. For most other plant viruses, the rationale for their nucleolus targeting is currently unclear. However, a growing number of correlative studies have indicated that the nucleolus may play a role in many steps in the infection cycle of these viruses (Martínez and Daròs, 2014; Rossi et al., 2014; Taliansky et al., 2010; and references therein).
Rice stripe virus (RSV) is the type species of the genus Tenuivirus, which has not been assigned to any family (King et al., 2012). RSV is transmitted by the small brown planthopper (SBPH) (Laodelphax striatellus Fallén) in a persistent and circulative–propagative manner (Falk and Tsai, 1998). In Far East Asia, RSV represents one of the most important viral pathogens severely affecting rice production (Cho et al., 2013, Hibino, 1996; Wei et al., 2009). Recently, RSV has been reported to occur in Vietnam, a country in South‐East Asia (Ren et al., 2013).
The genome of RSV comprises four single‐stranded RNA (ssRNA) segments, named RNA1–RNA4, respectively, in decreasing order of size. RNA1–RNA4 are encompassed separately by nucleocapsid proteins to form distinct filamentous RNPs (Ramírez and Haenni, 1994). RNA1 is of negative sense and encodes a 337‐kDa protein (pc1) considered to be the RNA‐dependent RNA polymerase (RdRp) associated with RSV virions (Toriyama et al., 1994). RNA2–RNA4 use an ambisense coding strategy, each containing two non‐overlapping ORFs, one in the 5′ half of the viral RNA (the genes they encode are named NS2–NS4 and the proteins they encode are named p2–p4, respectively) and the other in the 5′ half of the viral complementary RNA (the genes they encode are named NSVc2–NSVc4 and the proteins they encode are named pc2–pc4, respectively; Falk and Tsai, 1998; Kakutani et al., 1990, 1991; Takahashi et al., 1993; Zhu et al., 1991). pc4 (32 kDa) is a movement protein which may also play a role in RSV pathogenicity (Xiong et al., 2008; Xu and Zhou, 2012; Zhang et al., 2012). p4 (20.5 kDa) is the major non‐structural protein which often accumulates to very high levels in RSV‐infected plants (Kong et al., 2014; Lin et al., 1998). pc3 (35 kDa) is the nucleocapsid protein (Hayano et al., 1990; Kakutani et al., 1991). p3 (23.9 kDa) is a silencing suppressor (Xiong et al., 2009). pc2 (94 kDa) can be cleaved into two proteins, pc2‐N and pc2‐C, which target the Golgi apparatus and the endoplasmic reticulum (ER), respectively, in cells of Nicotiana benthamiana (Yao et al., 2014; Zhao et al., 2012). p2 (22.8 kDa) has weak silencing suppressor activities (Du et al., 2011).
Owing to the large genome, the unusual coding strategy, the absolute insect transmission and the use of monocots as its host, RSV is resistant to genetic manipulations. As a result, studies on RSV–host interaction are difficult and our knowledge of the mechanisms by which RSV infects its host remains poor. The situation is beginning to change after the discovery that RSV can infect N. benthamiana and Arabidopsis thaliana under laboratory conditions (Sun et al., 2011 and Xiong et al., 2008). Recently, N. benthamiana has been shown to be a feasible model plant to investigate the roles of host factors in RSV infection (Jiang et al., 2014; Kong et al., 2014).
At least three RSV proteins, namely p3, pc3 and p4, may localize to the nucleus (Lian et al., 2014; Xiong et al., 2009). This suggests that RSV may have a nuclear phase in its replication cycle. However, this possibility has never been explored. In this article, we present our data suggesting that RSV p2 may recruit or manipulate nucleolar functions to promote virus systemic movement in N. benthamiana.
Results
RSV p2 moves to and through the nucleolus when expressed individually in epidermal cells of N. benthamiana
To study the subcellular localization of RSV p2, a fusion of p2 with yellow fluorescent protein (YFP; p2‐YFP) was created by cloning the p2 ORF into the plasmid vector pEarleyGate 101 (Earley et al., 2006). The recombinant plasmids were agroinfiltrated into epidermal cells of N. benthamiana leaves. The expression of the fusion protein was confirmed by Western blotting using commercially available antibodies against green fluorescent protein (GFP, Fig. 1G). YFP fluorescence was observed at 24, 48, 60, 72 and 84 h post‐infiltration (hpi) by confocal laser scanning microscopy (CLSM).
Figure 1.
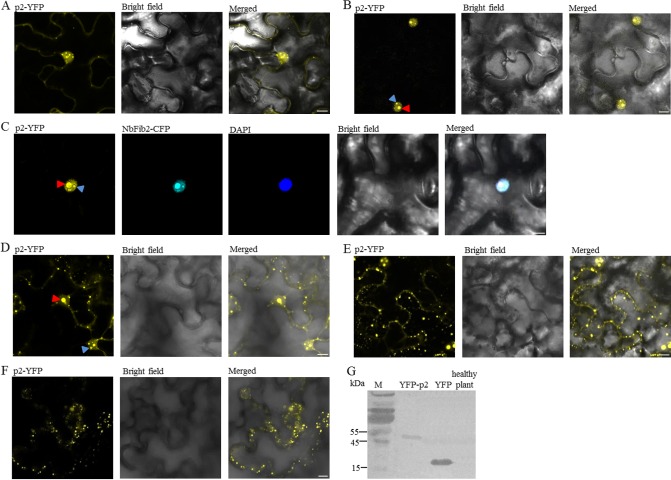
Subcellular localization of Rice stripe virus (RSV) p2 in epidermal cells of Nicotiana benthamiana (A–F) and Western blotting confirming the expression of the fusion protein p2‐yellow fluorescent protein (p2‐YFP, G) at 48 h post‐infiltration (hpi). p2‐YFP was expressed individually (A, B, D, E and F) or co‐expressed with NbFib2 tagged with cyan fluorescent protein (CFP, C) in leaves of N. benthamiana by agroinfiltration. Fluorescence was observed at 24 (A), 48 (B, C), 60 (D), 72 (E) and 84 hpi (F) by confocal laser scanning microscopy. Cells co‐expressing p2 and NbFib2 were stained with 4,6‐diaminophenylindole (DAPI) to show the nucleus (C). Possible nucleoli or Cajal bodies described in the text are designated with red and blue arrows, respectively. Scale bars, 10 μm.
At 24 hpi, YFP fluorescence was found in both the nucleus and the cytoplasm. In the cytoplasm, most of the fluorescence was diffuse. In the nucleus, however, the fluorescence accumulated and formed multiple bright spots (Fig. 1A). At 48 hpi, YFP fluorescence was found to localize predominantly in the nucleus, being concentrated in structures that resembled the nucleolus and some other small subnuclear structures reminiscent of Cajal bodies (Fig. 1B; Cioce and Lamond, 2005). The nucleolar localization of p2‐YFP was confirmed by its co‐localization with fibrillarin (NbFib2 of N. benthamiana, GenBank accession: AM269909), a marker protein of the nucleolus (Fig. 1C; Barneche et al., 2000; Kim et al., 2007a, b). By 60 hpi, in addition to the intense fluorescence signals in the nucleolus, many small bright spots of unknown nature were found in the cytoplasm near the plasma membrane and in perinuclear regions (Fig. 1D). By 72 hpi, the numbers of small spots in the cytoplasm had increased (Fig. 1E). Fluorescence was also apparent in the nucleus. However, in most cells, the single, large fluorescent body observed at 60 hpi had changed into two to four smaller ones (Fig. 1D, F). By 84 hpi, numerous small spots were found in the cytoplasm near the plasma membrane. Faint fluorescence was observed from the nucleus, but, in most cases, the fluorescence did not accumulate in the nucleolus (Fig. 1E). A fusion protein with YFP at the N‐terminus of p2 (YFP‐p2) and fusion proteins tagging p2 with cyan fluorescent protein (CFP) were also used to study the cellular localization of p2. The same results were obtained (data not shown).
p2 interacts with fibrillarin
Many nucleolus‐targeting viral proteins interact with one or multiple nucleolar proteins (Hiscox, 2007; Salvetti and Greco, 2014). For plant viruses, at least five nucleolus‐targeting proteins from Poa semilatent virus (PSLV; Semashko et al., 2012), Potato mop‐top virus (PMTV; Wright et al., 2010), GRV (Kim et al., 2007a, b), Beet black scorch virus (BBSV; Wang et al., 2012) and PVA (Rajamäki and Valkonen, 2009), respectively, have been shown to interact with fibrillarin. A bimolecular fluorescence complementation (BiFC) assay was carried out to investigate the interaction between p2 and NbFib2 (Walter et al., 2004). To do this, the full‐length ORF of p2 was cloned into the vector pEarleyGate 201‐YC and that of NbFib2 into pEarleyGate 201‐YN. Transformed Agrobacterium tumefaciens EHA105 carrying each of these constructs was mixed and infiltrated into the leaves of N. benthamiana. Strong YFP fluorescence derived from the reconstitution of the YFP fluorophore was observed at 48 hpi, indicating the interaction of p2 and NbFib2 in living plant cells (Fig. 2). Similar results were obtained when p2 was fused with the N‐terminal fragment of YFP and NbFib2 with the C‐terminal fragment of YFP. By contrast, fluorescence was not detected in leaf cells co‐infiltrated with pEarleyGate 201‐YC/pEarleyGate 201‐YN, pEarleyGate 201‐YC‐p2/pEarleyGate 201‐YN or pEarleyGate 201‐YC/pEarleyGate 201‐YN‐NbFib2 (data not shown). As shown in Fig. 2, the interaction between p2 and NbFib2 occurred in the nucleus. Similar BiFC experiments showed that p2 also interacted with fibrillarin of rice (GenBank accession: AK103477) and Arabidopsis (GenBank accession: AAG10153; data not shown).
Figure 2.
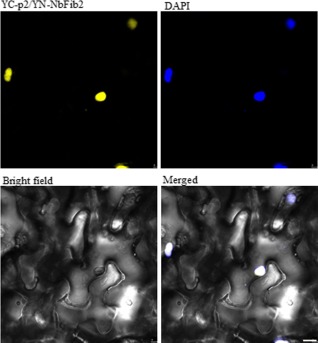
Bimolecular fluorescence complementation (BiFC) assay showing interaction between p2 and NbFib2. In the figures shown here, the full‐length open reading frame (ORF) of p2 was cloned into the vector pEarleyGate 201‐YC (YC‐p2) and that of NbFib2 into pEarleyGate 201‐YN (YN‐NbFib2). The recombinant plasmids were introduced into epidermal cells of Nicotiana benthamiana by agroinfiltration. Fluorescence was observed at 48 h post‐infiltration by confocal laser scanning microscopy. The nucleus was stained with 4,6‐diaminophenylindole (DAPI). Scale bars, 10 μm.
Fibrillarin depletion affects the nucleolar localization and cytoplasmic distribution of p2
The interaction with fibrillarin has been shown to be required for the nucleolar localization of GRV ORF3 (Kim et al., 2007b). To test the effects of fibrillarin on p2 localization, we down‐regulated the expression of NbFib2 in N. benthamiana using a Tobacco rattle virus (TRV)‐based virus‐induced gene silencing (VIGS) method in the manner described previously by Kim et al. (2007b) and Rajamäki and Valkonen (2009).
At 12 days post‐inoculation (dpi), plants inoculated with TRV‐NbFib2 could be separated into three groups according to symptoms: Group I, severely dwarfed plants with arrested growth; Group II, plants with slightly shortened stems, mild leaf chlorosis and negligible leaf deformity; Group III, plants with very mild leaf chlorosis, but without detectable leaf deformity (Fig. 3A). The three groups of plants showed >90%, 45%–60% and <10% reduction in NbFib2 expression, respectively, as demonstrated by real‐time reverse transcription polymerase chain reaction (RT‐PCR, Fig. 3B).
Figure 3.
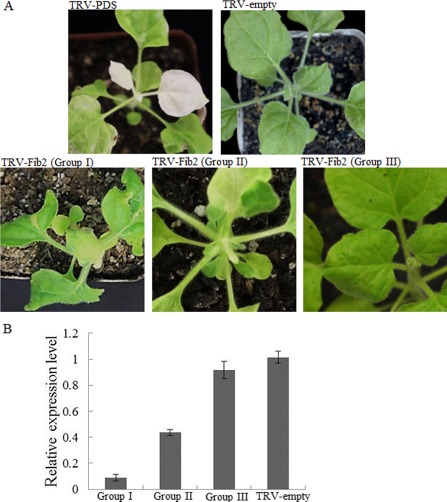
Virus‐induced gene silencing of NbFib2. (A) Symptoms of Nicotiana benthamiana infected by Tobacco rattle virus (TRV), TRV‐PDS and TRV‐Fib2 as indicated. For plants inoculated with TRV‐Fib2, representative plants belonging to each of the three groups reported in the text are shown. (B) Varying silencing efficiencies of NbFib2 in representative plants belonging to each of the three groups shown in (A). For real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) detecting the expression of NbFib2, three plants belonging to each group were pooled as one sample and the experiments were performed in triplicate.
YFP‐p2/p2‐YFP was expressed in representative plants belonging to each of the three groups and in plants inoculated with TRV alone using agroinfiltration. The YFP fluorescence was observed at 2 and 3 days after agroinfiltration (dai).
As shown in Fig. 4A, B, the localization of p2‐YFP in plants inoculated with TRV alone or in Group III plants was similar to that in wild‐type plants, i.e. fluorescence accumulated in the nucleolus initially (2 dai), but left the nucleolus for the cytoplasm and formed numerous small bright bodies there at 3 dai (data not shown). In Group II plants, a nucleolar accumulation of p2‐YFP was not found at 2 dai. Instead, fluorescence was found in both the nucleus and the cytoplasm (Fig. 4C). The distribution of fluorescence did not show any detectable change at 3 dai (data not shown). In Group I plants, p2‐YFP fluorescence showed a diffuse localization in both the nucleus and the cytoplasm at 2 dai (Fig. 4D). In addition, fluorescence did not show any detectable change at 3 dai (data not shown). These results suggest that fibrillarin may play a role in both the nucleolar localization and the appropriate cytoplasmic distribution of p2.
Figure 4.
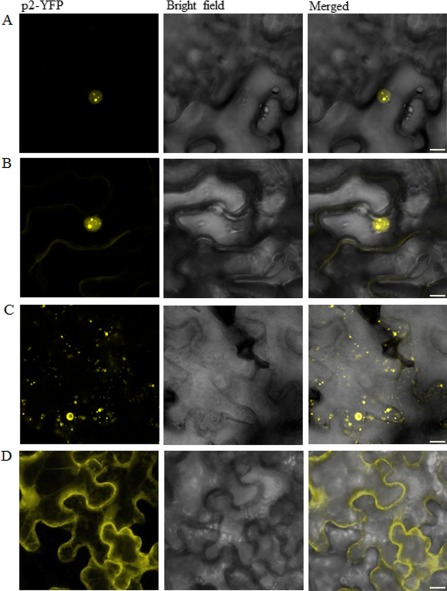
Subcellular localization of Rice stripe virus (RSV) p2 fused with yellow fluorescent protein (YFP) in epidermal cells of Nicotiana benthamiana inoculated with empty Tobacco rattle virus (TRV) (A) and N. benthamiana belonging to Group III (B), Group II (C) and Group I (D) as described in the text. The YFP fluorescence was observed 2 days after agroinfiltration. Scale bars, 10 μm.
Fibrillarin depletion abolishes the systemic movement of RSV in N. benthamiana
Previously, it has been reported that the knock‐down of fibrillarin abolishes the systemic movement of GRV and PLRV. Fibrillarin depletion did not prevent the systemic movement but reduced the accumulation of PVA (Kim et al., 2007b; Rajamäki and Valkonen, 2009). To investigate the role of fibrillarin in RSV infection, crude extracts of RSV‐infected rice were used to mechanically inoculate Group II plants in the manner described previously (at 10 days after TRV‐NbFib2 inoculation; Jiang et al., 2014; Xiong et al., 2008). Nicotiana benthamiana plants previously infected by empty TRV were used as controls. The inoculated plants were monitored daily for symptoms associated with RSV infection. The accumulation of RSV was detected by RT‐PCR and Western blotting at 22 dpi of RSV using antibodies against RSV p4, the major non‐structural protein of RSV, whose accumulation correlates with RSV replication and host symptom development (Kong et al., 2014; Lin et al., 1998).
In control plants, symptoms including leaf chlorosis, yellowing and downward leaf curling appeared at 12–15 dpi of RSV. The symptoms persisted for months before the death of the plants (Fig. 5A). RSV could be readily detected in both inoculated and systemic leaves (Fig. 5C). This indicates that TRV does not affect RSV infection of N. benthamiana. A similar observation has been made recently by Jiang et al. (2014).
Figure 5.
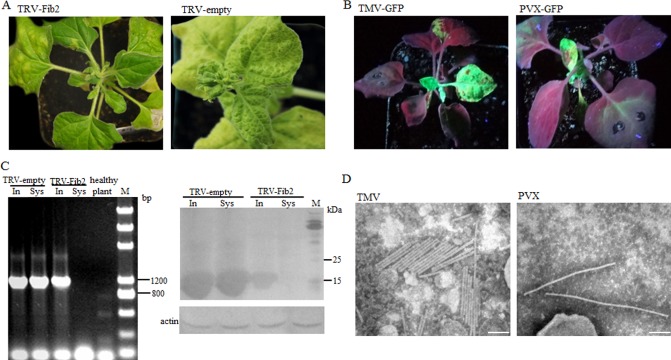
Effects of NbFib2 depletion on Rice stripe virus (RSV) infection. NbFib2‐silenced Nicotiana benthamiana plants belonging to Group II were inoculated with RSV (A, left), Potato virus X‐green fluorescent protein (PVX‐GFP) (B, right) or Tobacco mosaic virus‐GFP (TMV‐GFP) (B, left), as described in the text. Nicotiana benthamiana previously inoculated with empty Tobacco rattle virus (TRV) was used as a control (A, right). Nicotiana benthamiana plants are photographed at 22 days after virus inoculation. The accumulation of RSV 22 days after virus inoculation in inoculated (In) or upper (Sys) leaves was detected by reverse transcription‐polymerase chain reaction (RT‐PCR, C, left) and Western blotting (C, right). The presence of PVX‐GFP and TMV‐GFP was visualized under UV light (B), and the presence of intact PVX and TMV virions in systemically infected leaves was demonstrated by electron microscopy (D; scale bars, 100 nm).
In fibrillarin‐silenced N. benthamiana, symptoms associated with RSV infection were not observed either at this time or later (Fig. 5A). RT‐PCR and Western blotting failed to detect RSV in newly developed leaves (Fig. 5C). However, RSV accumulated in inoculated leaves (Fig. 5C). The accumulation level was comparable with, although obviously lower than, that in control plants (Fig. 5C). Fifteen N. benthamiana plants were used in the experiment; the experiment was carried out in triplicate and the results were reproducible. This suggests that fibrillarin depletion prevents the systemic movement of RSV in N. benthamiana.
Kim et al. (2007b) have demonstrated that the silencing of fibrillarin does not inhibit the normal cellular functions necessary for general virus infection. To confirm this under our experimental conditions, Group II N. benthamiana plants were inoculated with Tobacco mosaic virus‐GFP (TMV‐GFP) and Potato virus X‐GFP (PVX‐GFP). In plants infected by either virus, GFP fluorescence was observed as early as 9 dpi of virus (Fig. 5B). TMV and PVX were readily detectable in the leaves by RT‐PCR (data not shown). Intact virions of TMV and PVX could also be observed by electron microscopy (Fig. 5D). Thus, fibrillarin silencing under our experimental conditions does not disturb the cellular functions necessary for general virus infection.
p2 is involved in systemic movement of RSV in N. benthamiana
The above results strongly suggest that RSV p2 may function in virus systemic movement. To test this, a TRV vector containing a partial sequence of NS2 was created and used to infect N. benthamiana as described above. Nicotiana benthamiana plants infected by empty TRV or TRV carrying a partial sequence of NS3 were used as controls. Throughout our experiments, N. benthamiana infected by TRV‐NS2 or TRV‐NS3 showed no obvious differences from those infected by TRV alone (data not shown). Thus, it seems unlikely that TRV‐NS2 or TRV‐NS3 may target host genes important for normal physiology. The upper leaves of these plants were inoculated with RSV at 12 dpi of TRV/TRV‐NS2/TRV‐NS3. Symptoms associated with RSV infection were monitored daily and the accumulation of RSV was detected at 22 dpi of RSV as described above.
TRV‐NS3‐ and TRV‐treated N. benthamiana plants were similar to each other in terms of the timing of symptom development, i.e. symptoms appeared at 12–15 dpi of RSV in the top leaves and persisted for months (Fig. 6A). The accumulation of RSV was readily detectable in both inoculated and systemic leaves in TRV‐NS3‐treated plants (Fig. 6B), although the symptoms were obviously milder and the accumulation of RSV was lower in these plants when compared with the control (TRV‐treated plant; Fig. 6A, B). The accumulation of the gene NS3 in inoculated leaves of TRV‐NS3‐treated plants was detected by real‐time RT‐PCR. The results showed that the expression of NS3 was reduced by about 60%.
Figure 6.
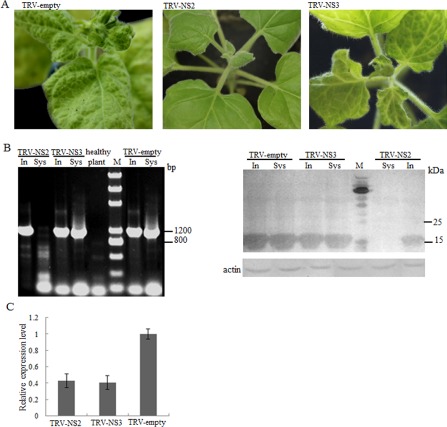
Effects of NS2 silencing on Rice stripe virus (RSV) infection. Nicotiana benthamiana plants were inoculated with either Tobacco rattle virus (TRV), TRV‐NS2 or TRV‐NS3, as indicated, 12 days before RSV inoculation. The symptoms shown in the figure (A) were photographed 22 days after RSV inoculation. The accumulation of RSV in inoculated (In) and upper (Sys) leaves of TRV‐NS2/TRV‐NS3‐treated N. benthamiana was detected by reverse transcription‐polymerase chain reaction (RT‐PCR) (B, left) and Western blotting (B, right), 24 days after RSV inoculation. The accumulation of the gene NS2 in inoculated leaves of TRV‐NS2‐treated plants at 22 days post‐inoculation of RSV was detected by real‐time RT‐PCR (C).
Symptoms were not observed even 40 days after RSV inoculation on newly emerged leaves of plants treated with TRV‐NS2 (Fig. 6A). Only four of the 15 plants developed symptoms at 60 dpi of RSV. As shown in Fig. 6B, RSV was not detected in systemic leaves. However, RSV accumulated in inoculated leaves (Fig. 6B). The accumulation of the gene NS2 in inoculated leaves was detected by real‐time RT‐PCR. The results showed that the expression of NS2 was reduced by about 55% (Fig. 6C). Thus, p2 of RSV may indeed play a role in the systemic movement of RSV.
Discussion
Overall, we found that: (i) RSV p2 trafficks between the nucleolus and the cytoplasm; (ii) p2 interacts with fibrillarin and fibrillarin is important for the normal cellular distribution (and the dynamics of distribution) of p2; (iii) fibrillarin is important for the systemic movement of RSV; and (iv) p2 may play a role in RSV systemic movement. It should be noted that the localization experiments and experiments showing the interaction between p2 and fibrillarin reported here were conducted out of context of viral infection. Further experiments are needed to demonstrate the dynamics of p2 localization and the interaction between p2 and fibrillarin in the process of RSV infection.
Although many plant viruses have been shown to interact with fibrillarin, RSV is the third plant virus shown to require fibrillarin for systemic infection after GRV and PLRV (Kim et al., 2007a, b). The mechanism by which RSV uses fibrillarin is unclear at present. GRV has been shown to redirect fibrillarin to the cytoplasm and to use fibrillarin to form infectious RNPs (Kim et al., 2007a, b). It is possible that RSV may recruit a small proportion of fibrillarin to function in the cytoplasm. However, a more plausible possibility is that RSV may use fibrillarin in a different manner, as we found that the interaction between p2 and fibrillarin occurred exclusively in the nucleus (Fig. 2). In our co‐localization studies of p2 and fibrillarin, fibrillarin was never found outside the nucleus. Instead, p2 was trapped in the nucleolus when NbFib2 was overexpressed (Fig. 1C and data not shown). Moreover, fibrillarin was not found in the cytoplasm when it was expressed as a fusion with YFP in RSV‐infected N. benthamiana (data not shown). In this context, it is interesting to note that PVA requires fibrillarin for enhanced replication, but not for systemic movement (Rajamäki and Valkonen, 2009). This suggests that the same host protein may be used by different viruses for different purposes, or for the same purposes but in a different way.
The fact that an interaction partner of p2 is involved in RSV systemic movement implicates a role of p2 in this process. This cannot be tested directly because of the resistance of RSV to genetic manipulation. However, RSV has a segmented genome and uses independently transcribed, non‐overlapping mRNAs to produce proteins (Falk and Tsai, 1998; Kormelink et al., 2011; Nguyen and Haenni, 2003; Wu et al., 2013). Because of this, we used RNA interference (RNAi) to investigate the role of p2 in RSV infection. Although RNAi has been widely used to study the gene functions of some animal viruses, especially reoviruses and some negative‐strand RNA viruses, similar technologies have been used rarely in studies of plant viruses (Bitko and Barik, 2001; Kobayashi et al., 2006). One may argue that small interfering RNAs (siRNAs) derived from NS2 may target RSV genomic RNAs. Currently, we cannot rule out this possibility. However, genomic RNAs of negative‐strand RNA viruses, such as RSV, are coated with nucleocapsid proteins. Nucleocapsid proteins of negative‐strand RNA viruses may block siRNA‐targeted recognition and degradation, as suggested by Ahlquist (2002), and demonstrated by the fact that synthetic siRNAs against Respiratory syncytial virus induce the degradation of viral mRNA, but not nucleocapsid‐coated genomic RNA, in cultured human cells (Bitko and Barik, 2001). Recent work by Shimizu et al. (2011) supports the feasibility of RNAi in the study of RSV gene functions. The researchers expressed RNAi constructs targeting different RSV genes in rice. Transgenic rice showed very different levels of resistance to RSV depending on the RSV genes targeted. Importantly, the levels of resistance in plants expressing RNAi constructs targeting RSV genes expressed from the same genomic RNAs varied from being immune to no enhanced resistance. This strongly suggests that the RNAi constructs target specific genes of RSV rather than genomic RNAs. We anticipate that RNAi can be used in more sophisticated ways to study gene functions of RSV in the future.
The finding that p2 may function in the systemic movement of RSV is also consistent with observations made by Shimizu et al. (2011). In their studies, among the 80 transgenic rice plants expressing an RNAi construct targeting NS2, the appearance of symptoms was delayed by 2–35 days in 48 (60%) plants. Twenty (25%) plants never exhibited symptoms prior to harvest. Also, this is consistent with our previous finding that p2 has silencing suppressor activities (Du et al., 2011). A number of silencing suppressors of plant viruses play a role in virus systemic movement (Díaz‐Pendón and Ding, 2008; Hipper et al., 2013).
The fact that both fibrillarin and p2 are required for the systemic movement of RSV and the finding that p2 requires fibrillarin to target the nucleolus point to a scenario in which RSV p2 may manipulate or recruit nucleolar functions to promote virus systemic infection. For example, p2 may need to enter the nucleolus to undergo necessary modifications or processing for appropriate cytoplasmic distribution. The processing or modification may be important for the normal function of p2. Alternatively, p2 may need to recruit some unknown factors localized in the nucleolus for its functions in the cytoplasm. It is also possible that p2 may enter the nucleolus to disturb certain host functions which normally restrict viral infection. However, further studies are needed to provide direct evidence showing that p2 needs to target the nucleolus to promote RSV systemic movement.
Overall, the results of this study are consistent with the emerging notion that interaction with the nucleolus is a pan‐virus phenomenon, even for plant viruses (Taliansky et al., 2010). In addition, the data presented here deepen our understanding of RSV–host interaction and provide potential targets for the design of new anti‐viral strategies to control RSV.
Experimental Procedures
Plasmid construction
Total RNA was extracted from N. benthamiana leaves and RSV‐infected rice leaves using an EasyPure Plant RNA Kit manufactured by Bejing Transgen Biotech Co. Ltd. (Beijing, China). Reverse transcription was carried out using a Fast Quant RT Kit with gDNase (Tiangen Biotech Co., Ltd., Beijng, China). NbFib2 and RSV‐NS2 (GenBank accession: AM269909 and EF198702) were amplified by PCR using the primers listed in Table S1 (see Supporting Information). The vectors (pEarleyGate 101‐p2/NbFib2, pEarleyGate 102‐p2/NbFib2, pEarleyGate 201‐YC‐p2/NbFib2 and pEarleyGate 201‐YN‐p2/NbFib2) used in cellular localization and BiFC were generated using Gateway recombination technology (Invitrogen China, Shanghai, China) by inserting RSV‐NS2 and NbFib2 into the entry vector pDONR 221 (Karimi et al., 2002). For VIGS assays, RSV‐NS2, RSV‐NS3 (GenBank accession: EF493242) and NbFib2 were amplified by the primers listed in Table S1. The PCR products were digested with EcoRI and BamHI, and ligated into the TRV vector pTRV2 digested with the same enzymes. All the plasmid constructs used in this study were confirmed by sequencing (Takara, Dalian, China).
Agrobacterium‐mediated transient expression
Agrobacterium tumefaciens strains EHA105 or GV3101 carrying the genes of interest were grown separately to an optical density at 600 nm (OD600) of 0.8 at 28 °C on Luria–Bertani liquid medium supplemented with 50 μg/μL of rifampicin and 50 μg/μL of kanamycin. The cultures were centrifuged at 12 000 g for 1 min and resuspended in induction medium [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.6, 10 mm MgCl2 and 150 μm acetosyringone]. In co‐localization and BiFC assays, A. tumefaciens strains containing different constructs were mixed in equal volume. In VIGS assays, A. tumefaciens carrying pTRV2‐NbPDS, pTRV2‐NS2, pTRV2‐NbFib2 or empty pTRV2 was mixed with an equal volume of TRV1. The mixtures of the bacterial cultures were incubated at room temperature for 3 h, and then infiltrated into fully grown upper leaves. Six‐week‐old N. benthamiana plants were used for the experiments.
Confocal imaging analysis
The fluorophores in CFP and YFP were excited at 458 and 514 nm, and images were taken using BA480–495‐ and BA535–565‐nm emission filters, respectively.
Quantitative RT‐PCR
The total RNAs of the upper leaves of NbFib2‐ or NS2‐silenced plants were extracted and RT‐PCRs were performed as described previously. The primer pairs NS2‐qPCRF/NS2‐qPCRR, Fib2 qPCRF/Fib2‐qPCRR, NS3‐qPCRF/NS3‐qPCRR and 18S‐qPCRF/18S‐qPCRR were used to detect the expression of NS2, NbFib2 and NS3. 18S RNA of N. benthamiana was used as a reference gene (Eppendorf China, Shanghai, China, AG 22331 Real‐Time PCR System, Hamburg No. 5345 018780). The 20‐μL PCR included 1.0 μL of RT product, 10 μL of SYBR qPCR Mix, 0.8 μL of primers (Table S1) and 9.2 μL of diethylpyrocarbonate (DEPC) water; the reactions were incubated in a 96‐well optical plate at 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min (TOYOBO, Shanghai, China, No. QPS‐201). The Ct data were determined using default threshold settings. The threshold cycle (Ct) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
Virus inoculation
For RSV inoculation assays, a Jiangsu isolate of RSV (LS‐JSJJ03, EF198702, EF198682, EF198733, EF198702), maintained in our laboratory through transmission using L. striatellus, was employed in this study. RSV inoculation was performed as described previously using crude extracts of RSV‐infected rice (Zhang et al., 2012).
PVX‐GFP and TMV‐GFP were inoculated by agroinfiltration. GFP fluorescence was observed under a hand‐held, 100‐W, long‐wave UV lamp (UV Products, Upland, CA, USA). Then, fluorescent leaves were collected and ground. The grinding solution was dipped on the copper grid with a Formvar membrane for 1–2 min, washed using double distilled water, stained with 2% uranyl acetate and observed under an H‐7650 electron microscope (Hitachi, Japan).
Detection of RSV accumulation
Inoculated and upper non‐inoculated leaves of N. benthamiana infected by RSV were collected individually. RT‐PCR using a primer pair amplifying the RSV coat protein (CP) was employed to detect RSV.
For Western blotting, inoculated and non‐inoculated leaves were homogenized in 2 mL of extraction buffer containing 50 mm phosphate (pH 8.0), 10 mm tris(hydroxymethyl)aminomethane (Tris) (pH 8.0), 500 mm NaCl, 0.1% Tween 20, 0.1% Nonidet P40 (NP‐40), 0.1% β‐mercaptoethanol, 1 mm phenylmethylsulfonylfluoride (PMSF) and one‐quarter of a Roche Protease inhibitor cocktail MINI tablet. The crude extracts were centrifuged at 12 000 g for 10 min. The supernatant was transferred into a new centrifuge tube and centrifuged at 12 000 g for another 15 min; 10 μL of supernatant were mixed with 2 μL of 5 × sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) loading buffer. Proteins in the extracts were separated by electrophoresis by 12% SDS‐PAGE at 80 V for 1 h and then at 120 V for another 40 min. Proteins in the gels were transferred onto polyvinylidene difluoride (PVDF) membranes by electrophoresis cell at 60 V for 1 h (Beijing WoDe Life Sciences Instrument Company, Beijing, China) and probed with RSV‐P4 polyclonal antibody. The polyclonal antibody was a goat–anti‐rabbit immunoglobulin G (IgG) conjugated with alkaline phosphatase (Sigma, St. Louis, MO, USA) and used at 1:10 000 (v/v) dilution. Proteins on the membrane were visualized by nitroblue tetrazolium–5‐bromo‐4‐chloroindol‐3‐yl phosphate (NBT‐BCIP) solution (Shanghai Promega Biological Products, Co, Ltd Shanghai, China).
Supporting information
Table S1 Primers used in this study.
Acknowledgements
We are grateful to Professor Xinzhong Cai (Zhengjiang University, Hangzhou, China) for providing the TRV vector and plasmid pTRV2‐NbPDS, and to Dr Aiming Wang (AAFC‐Southern Crop Protection and Food Research Centre, Canada) for providing the Gateway vectors for localization and BiFC studies. This work was supported by grants from the National Basic Research Program 973 (2014CB138402, 2014CB138403 and 2010CB126203), the Natural Science Foundation of China (31401715, 31171821 and 31272018), the Key Project of the National Research Program of China (2012BAD19B03) and the Doctoral Fund of the Ministry of Education of China (20113515110001). The authors have no conflicts of interest to declare.
References
- Ahlquist, P. (2002) RNA‐dependent RNA polymerases, viruses, and RNA silencing. Science, 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- Barneche, F. , Steinmetz, F. and Echeverri′a, M. (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana . J. Biol. Chem. 275, 27 212–27 220. [DOI] [PubMed] [Google Scholar]
- Bitko, V. and Barik, S. (2001) Phenotypic silencing of cytoplasmic genes using sequence‐specific double‐stranded short interfering RNA and its application in the reverse genetics of wild type negative‐strand RNA viruses. BMC Microbiol. 1, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert, F.M. , van Koningsbruggen, S. , Navascues, J. and Lamond, A.I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. [DOI] [PubMed] [Google Scholar]
- Boulon, S. , Westman, B.J. , Hutten, S. , Boisvert, F.M. and Lamond, A.I. (2010) The nucleolus under stress. Mol. Cell 40, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, W.K. , Lian, S. , Kim, S.M. , Park, S.H. and Kim, K.H. (2013) Current insights into research on Rice stripe virus . Plant Pathol. J. 29, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce, M. and Lamond, A.I. (2005) Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21, 105–131. [DOI] [PubMed] [Google Scholar]
- Díaz‐Pendón, J.A. and Ding, S.W. (2008) Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326. [DOI] [PubMed] [Google Scholar]
- Du, Z. , Xiao, D. , Wu, J. , Jia, D. , Yuan, Z. , Liu, Y. , Hu, L. , Han, Z. , Wei, T. , Lin, Q. , Wu, Z. and Xie, L. (2011) p2 of rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor[J]. Molecular Plant Pathology, 12, 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Falk, B.W. and Tsai, J.H. (1998) Biology and molecular biology of viruses in the genus Tenuivirus . Annu. Rev. Phytopathol. 36, 39–163. [DOI] [PubMed] [Google Scholar]
- Hayano, Y. , Kakutani, T. , Hayashi, T. and Minobe, Y. (1990) Coding strategy of rice stripe virus: major nonstructural protein is encoded in viral RNA segment 4 and coat protein in RNA complementary to segment 3[J]. Virology, 177, 372–374. [DOI] [PubMed] [Google Scholar]
- Hibino, H. (1996) Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 34, 249–274. [DOI] [PubMed] [Google Scholar]
- Hipper, C. , Brault, V. , Ziegler‐Graff, V. and Revers, F. (2013) Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 4, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox, J.A. (2007) RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Lu, Y. , Li, K. , Lin, L. , Zheng, H. , Yan, F. and Chen, J. (2014) Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol. Plant Pathol. 15, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T. , Hayano, Y. , Hayashi, T. and Minobe, Y. (1990) Ambisense segment 4 of rice stripe virus: possible evolutionary relationship with phleboviruses and uukuviruses (Bunyaviridae). J. Gen. Virol. 71, 1427–1432. [DOI] [PubMed] [Google Scholar]
- Kakutani, T. , Hayano, Y. , Hayashi, T. and Minobe, Y. (1991) Ambisense segment 3 of rice stripe virus: the first case of a virus containing two ambisense segments. J. Gen. Virol. 72, 465–468. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Inzé, D. and Depicker, A. (2002) GATEWAY™ vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Ryabov, E.V. , Kalinina, N.O. , Rakitina, D.V. , Gillespie, T. , MacFarlane, S. , Haupt, S. , Brown, J.W. and Taliansky, M. (2007a) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , MacFarlane, S. , Kalinina, N.O. , Rakitina, D.V. , Ryabov, E.V. , Gillespie, T. , Haupt, S. , Brown, J.W. and Taliansky, M. (2007b) Interaction of a plant virus‐encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA, 104, 11 115–11 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, A.M.Q. , Adams, M.J. , Lefkowitz, E.J. and Carstens, E.B. (2012) Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Vol. 9 Elsevier. [Google Scholar]
- Kobayashi, T. , Chappell, J.D. , Danthi, P. and Dermody, T.S. (2006) Gene‐specific inhibition of reovirus replication by RNA interference. J. Virol. 80, 9053–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Wu, J. , Lu, L. , Xu, Y. and Zhou, X. (2014) Interaction between Rice stripe virus disease‐specific protein and host PsbP enhances virus symptoms. Mol. Plant 7, 691–708. [DOI] [PubMed] [Google Scholar]
- Kormelink, R. , Garcia, M.L. , Goodin, M. , Sasaya, T. and Haenni, A.L. (2011) Negative‐strand RNA viruses: the plant‐infecting counterparts. Virus Res. 162, 184–202. [DOI] [PubMed] [Google Scholar]
- Lian, S. , Cho, W.K. , Jo, Y. , Kim, S.M. and Kim, K.H. (2014) Interaction study of rice stripe virus proteins reveals a region of the nucleocapsid protein (NP) required for NP self‐interaction and nuclear localization. Virus Res. 183, 6–14. [DOI] [PubMed] [Google Scholar]
- Lin, Q.T. , Lin, H.X. , Wu, Z.J. , Lin, Q.Y. and Xie, L.H. (1998) Accumulations of coat protein and disease‐specific protein of Rice stripe virus in its host. J. Fujian Agric. Univ. 27, 257–260. [Google Scholar]
- Martínez, F. and Daròs, J.A. (2014) Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 88, 10 725–10 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M. and Haenni, A.L. (2003) Expression strategies of ambisense viruses. Virus Res. 93, 141–150. [DOI] [PubMed] [Google Scholar]
- Rajamäki, M.L. and Valkonen, J.P. (2009) Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna‐like Potato virus A in Nicotiana species. Plant Cell, 21, 2485–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, B.C. and Haenni, A.L. (1994) Molecular biology of tenuiviruses, a remarkable group of plant viruses. J. Gen. Virol. 75, 467–475. [DOI] [PubMed] [Google Scholar]
- Ren, C. , Cheng, Z. , Miao, Q. , Fan, Y. and Zhou, Y. (2013) First report of rice stripe virus in Vietnam. Plant Dis. 97, 1123. [DOI] [PubMed] [Google Scholar]
- Rossi, M. , Genre, A. and Turina, M. (2014) Genetic dissection of a putative nucleolar localization signal in the coat protein of ourmia melon virus . Arch. Virol. 159, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Salvetti, A. and Greco, A. (2014) Viruses and the nucleolus: the fatal attraction. Biochim. Biophys. Acta, 1842, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semashko, M.A. , González, I. , Shaw, J. , Leonova, O.G. , Popenko, V.I. , Taliansky, M.E. , Canto, T. and Kalinina, N.O. (2012) The extreme N‐terminal domain of a hordeivirus TGB1 movement protein mediates its localization to the nucleolus and interaction with fibrillarin. Biochimie, 94, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Shaw, P. and Brown, J. (2012) Nucleoli: composition, function, and dynamics. Plant Physiol. 158, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T. , Nakazono‐Nagaoka, E. , Uehara‐Ichiki, T. , Sasaya, T. and Omura, T. (2011) Targeting specific genes for RNA interference is crucial to the development of strong resistance to Rice stripe virus . Plant Biotechnol. J. 9, 503–512. [DOI] [PubMed] [Google Scholar]
- Sun, F. , Yuan, X. , Zhou, T. , Fan, Y. and Zhou, Y. (2011) Arabidopsis is susceptible to Rice stripe virus infections. J. Phytopathol. 159, 767–772. [Google Scholar]
- Takahashi, M. , Toriyama, S. , Hamamatsu, C. and Ishihama, A. (1993) Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNAsegment 2. J. Gen. Virol. 74, 769–773. [DOI] [PubMed] [Google Scholar]
- Taliansky, M.E. , Brown, J.W.S. , Rajamäki, M.L. , Valkonen, J.P.T. and Kalinina, N.O. (2010) Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv. Virus Res. 77, 119–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama, S. , Takahashi, M. , Sano, Y. , Shimizu, T. and Ishihama, A. (1994) Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses . J. Gen. Virol. 75, 3569–3580. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. , Blazevic, D. , Grefen, C. , Schumacher, K. , Oecking, C. , Harter, K. and Kudla, J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhang, Y. , Xu, J. , Shi, L. , Fan, H. , Han, C. , Li, D. and Yu, J. (2012) The R‐rich motif of Beet black scorch virus P7a movement protein is important for the nuclear localization, nucleolar targeting and viral infectivity. Virus Res. 167, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T.Y. , Yang, J.G. , Liao, F.L. , Gao, F.L. , Lu, L.M. , Zhang, X.T. , Li, F. , Wu, Z.J. , Lin, Q.Y. , Xie, L.H. and Lin, H.X. (2009) Genetic diversity and population structure of rice stripe virus in China. J. Gen. Virol. 90, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Wright, K.M. , Cowan, G.H. , Lukhovitskaya, N.I. , Tilsner, J. , Roberts, A.G. , Savenkov, E.I. and Torrance, L. (2010) The N‐terminal domain of PMTV TGB1 movement protein is required for nucleolar localization, microtubule association, and long‐distance movement. Mol. Plant–Microbe Interact. 23, 1486–1497. [DOI] [PubMed] [Google Scholar]
- Wu, G. , Lu, Y. , Zheng, H. , Lin, L. , Yan, F. and Chen, J. (2013) Transcription of ORFs on RNA2 and RNA4 of Rice stripe virus terminate at an AUCCGGAU sequence that is conserved in the genus Tenuivirus . Virus Res. 175, 71. [DOI] [PubMed] [Google Scholar]
- Xiong, R. , Wu, J. , Zhou, Y. and Zhou, X. (2008) Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82, 12 304–12 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, R. , Wu, J. , Zhou, Y. and Zhou, X. (2009) Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus . Virology, 387, 29–40. [DOI] [PubMed] [Google Scholar]
- Xu, Y. and Zhou, X. (2012) Role of rice stripe virus NSvc4 in cell‐to‐cell movement and symptom development in Nicotiana benthamiana . Front. Plant Sci. 3, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, M. , Liu, X. , Li, S. , Xu, Y. , Zhou, Y. , Zhou, X. and Tao, X. (2014) Rice stripe tenuivirus NSvc2 glycoproteins targeted to the Golgi body by the N‐terminal transmembrane domain and adjacent cytosolic 24 amino acids via the COP I‐ and COP II‐dependent secretion pathway. J. Virol. 88, 3223–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Pei, X. , Wang, Z. , Jia, S. , Guo, S. , Zhang, Y. and Li, W. (2012) The Rice stripe virus pc4 functions in movement and foliar necrosis expression in Nicotiana benthamiana . Virology, 425, 113–121. [DOI] [PubMed] [Google Scholar]
- Zhao, S.L. , Dai, X.J. , Liang, J.S. and Liang, C.Y. (2012) Surface display of rice stripe virus NSvc2 and analysis of its membrane fusion activity[J]. Virologica Sinica, 27, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Hayakawa, T. , Toriyama, S. and Takahashi, M. (1991) Complete nucleotide sequence of RNA3 of rice stripe virus: an ambisense coding strategy. J. Gen. Virol. 72, 763–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used in this study.


