Summary
Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor (SNARE) proteins play critical and conserved roles in membrane fusion and vesicle transport of eukaryotic cells. Previous studies have shown that various homologues of SNARE proteins are also important in the infection of host plants by pathogenic fungi. Here, we report the characterization of a SNARE homologue, FgVam7, from Fusarium graminearum that causes head blight in wheat and barley worldwide. Phylogenetic analysis and domain comparison reveal that FgVam7 is homologous to Vam7 proteins of Saccharomyces cerevisiae (ScVam7), Magnaporthe oryzae (MoVam7) and several additional fungi by containing a PhoX homology (PX) domain and a SNARE domain. We show that FgVam7 plays a regulatory role in cellular differentiation and virulence in F. graminearum. Deletion of FgVAM7 significantly reduces vegetative growth, conidiation and conidial germination, sexual reproduction and virulence. The ΔFgvam7 mutant also exhibits a defect in vacuolar maintenance and delayed endocytosis. Moreover, the ΔFgvam7 mutant is insensitive to salt and osmotic stresses, and hypersensitive to cell wall stressors. Further characterization of FgVam7 domains indicate that the PX and SNARE domains are conserved in controlling Vam7 protein localization and function, respectively. Finally, FgVam7 has been shown to positively regulate the expression of several deoxynivalenol (DON) biosynthesis genes TRI5, TRI6 and TRI101, and DON production. Our studies provide evidence for SNARE proteins as an additional means of regulatory mechanisms that govern growth, differentiation and virulence of pathogenic fungi.
Keywords: DON, endocytosis, Fusarium graminearum, plant infection, SNARE protein
Introduction
Fusarium graminearum (teleomorph: Gibberella zeae) is an important pathogen causing Fusarium head blight (FHB) or head scab disease on wheat and barley (Bai and Shaner, 2004; Goswami and Kistler, 2004; McMullen et al., 1997). Under favourable conditions, F. graminearum spreads rapidly, resulting in severe grain yield losses. In addition to yield loss, infected cereals are often contaminated with deoxynivalenol (DON) and zearalenones, which are mycotoxins extremely toxic to humans and livestock (Desjardins et al., 1996; McMullen et al., 1997). FHB is difficult to control, similar to other plant diseases caused by Fusarium species, and the current means is primarily dependent on azole fungicides that exhibit many negative traits (Beyer et al., 2006; Paul et al., 2008). Therefore, there is an urgency to identify and develop novel and effective control strategies for FHB. Previous studies have shown that growth, differentiation and pathogenicity are regulated by various regulatory pathways in F. graminearum, and a number of pathogenicity‐related genes, including those encoding trichothecene mycotoxins, are also contributory factors to the virulence of the fungus (Fan et al., 2013; Han et al., 2007; Hou et al., 2002; Jenczmionka et al., 2003; Jenczmionka and Schafer, 2005; Lee et al., 2002; Lu et al., 2003; Seong et al., 2005, 2006; Shim et al., 2006; Son et al., 2011; Urban et al., 2003; Wang et al., 2011; Yu et al., 2008; Zheng et al., 2012; Zhou et al., 2010). However, membrane transport events associated with mycotoxin production, as well as growth, differentiation and pathogenicity, remain poorly understood.
Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor (SNARE) proteins are core protein machinery for intracellular membrane fusion in eukaryotic cells (Chen and Scheller, 2001). Despite their different sizes and structures among many organisms, the SNARE proteins share a conserved structure, called the SNARE domain, that consists of 60–70 amino acids arranged in heptad repeats (Pratelli et al., 2004; Sutton et al., 1998). SNAREs mediate membrane fusion during all trafficking steps of the intracellular communication process, including the secretory pathway, and have been studied extensively in mammals and plants, and the yeast Saccharomyces cerevisiae (Burri and Lithgow, 2004; Burri et al., 2003; Sanderfoot et al., 2000). The availability of genomic sequences has also facilitated the identification and characterization of SNARE proteins in filamentous fungi. A total of 21 putative SNARE proteins was found in Aspergillus oryzae, and their cellular distribution pattern based on enhanced green fluorescent protein (eGFP) fusion was also determined (Kuratsu et al., 2007). UmYup1, a SNARE protein homologue, is known to mediate endocytic recycling through early endosomes and is required for hyphal morphogenesis and pathogenesis in the corn smut fungus Ustilago maydis (Fuchs and Steinberg, 2005; Wedlich‐Soldner et al., 2000). Previously, we have identified two SNARE proteins, MoVam7 and MoSec22, from the rice blast fungus Magnaporthe oryzae, and have found that MoVam7, required for vacuolar morphology and endocytosis, plays a critical role in hyphal growth, conidial formation and virulence (Dou et al., 2011). MoSec22 has a role similar to MoVam7 and is also critical for conidiophore development in M. oryzae (Song et al., 2010). These findings suggest that conserved SNARE proteins are important for normal growth and development of infection‐related morphogenesis events in phytopathogens.
Similar to A. oryzae, F. graminearum contains 21 putative SNARE proteins and, of these, GzSyn1 and GzSyn2 have been reported to be involved in hyphal growth, sexual development and virulence (Hong et al., 2010). Here, we characterized FgVam7 as a homologue of fungal Vam7 proteins in F. graminearum. We found that deletion of FgVAM7 results in reduced conidiation and impaired sexual reproduction. The ΔFgvam7 mutant also exhibits a significant reduction in vegetative growth and the production of DON mycotoxins. In addition, FgVam7 plays a role in the response to various stresses and is required for pathogenicity. The involvement of FgVam7 in growth and differentiation suggests that FgVam7‐mediated membrane transport underlies most of the development processes relevant to plant infection by F. graminearum.
Results
Identification of FgVam7 in F. graminearum
Previous studies by others and by us have shown that fungal Vam7 proteins play important roles in the regulation of morphogenesis and development (Balderhaar and Ungermann, 2013; Dou et al., 2011; Wedlich‐Soldner et al., 2000). To identify the Vam7 protein in F. graminearum, ScVam7 and MoVam7 protein sequences were used in a blast search of the F. graminearum genomic database (http://mips.helmholtz‐muenchen.de/genre/proj/FGDB/). One putative Vam7 protein, named FgVam7, was identified. Phylogenetic analysis revealed that FgVam7 is well conserved with a high amino acid sequence identity to Vam proteins of other fungi (Fig. S1A, see Supporting Information). It shares 24% amino acid similarity to S. cerevisiae Vam7, 51% to M. oryzae MoVam7, 53% to Neurospora crassa NcVam7, 41% to U. maydis UmVam7 and 43% to Aspergillus fumigatus AfVam7. Domain prediction reveals that FgVam7 contains an N‐terminal PhoX homology (PX) domain and a C‐terminal SNARE domain, similar to other Vam7 proteins (Fig. S1B).
FgVam7 plays a crucial role in vegetative growth and conidiation
The conservation of amino acid sequence and domain composition suggests that FgVam7 exhibits conserved cellular functions. To explore the role of FgVam7 in vegetative growth and conidiation, the ΔFgvam7 mutant and the complemented transformant ΔFgvam7/FgVAM7 were generated and, together with the wild‐type PH‐1 strain, were cultured on V8 juice agar plates at 25 °C for 3 days. The ΔFgvam7 mutant showed a significantly reduced growth rate and a distinct colony morphology with fewer aerial hyphae. To determine whether its growth defect was medium dependent, we examined the vegetative growth of the ΔFgvam7 mutant on yeast extract–glucose medium (YEG), complete medium (CM) and minimal medium (MM), and found that the growth rate and aerial hyphae were reduced significantly (Fig. 1A,B). We further examined the ability of the ΔFgvam7 mutant to produce conidia, and found that conidial production was also decreased significantly. The number of conidia in the ΔFgvam7 mutant was reduced to approximately 25%, compared with that of the wild‐type (Fig. 1C). These results suggest that FgVam7 plays an important role in hyphal growth and conidiation.
Figure 1.
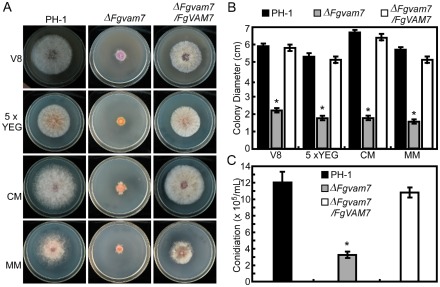
Defects of the ΔFgvam7 mutant in vegetative growth and conidiation. (A) Colonies of wild‐type PH‐1, ΔFgvam7 mutant and the ΔFgvam7/FgVAM7 complemented strains on various media for 3 days. CM, complete medium; MM, minimal medium; YEG, yeast extract–glucose. (B) Statistical analysis of the colony diameter of the indicated strains. (C) Quantification of the conidia produced by the indicated strains. Error bars represent the standard deviation from three independent experiments and asterisks indicate statistically significant differences (P < 0.01).
FgVam7 regulates the polarity of conidial germination
To examine the role of FgVam7 in conidial germination, conidia of PH‐1, the ΔFgvam7 mutant and ΔFgvam7/FgVAM7 were inoculated in liquid yeast extract–peptone–dextrose (YEPD) medium and observed for germination. It was found that over 80% conidial germination of the wild‐type and complemented transformant showed multipolarity with the production of two or more germ tubes in 4 h or 6 h, compared with 16% and 22% of that of the ΔFgvam7 mutant (Fig. 2, Table 1). Over 84% of the conidia of the ΔFgvam7 mutant showed a unipolar germination pattern, producing only one germ tube even in 8 h (Fig. 2, Table 1). These results indicate that FgVam7 plays a role in conidial germination by controlling germ tube polarity in F. graminearum.
Figure 2.
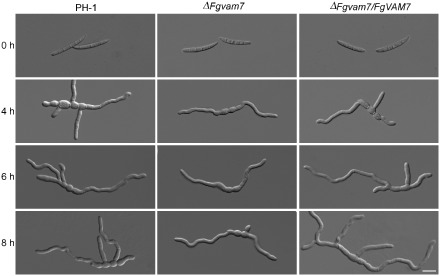
Conidial germination of PH‐1, the ΔFgvam7 mutant and the ΔFgvam7/FgVAM7 complemented strain in liquid yeast extract–peptone–dextrose (YEPD) medium. Examination was performed at 4, 6 and 8 h. Bar, 20 μm.
Table 1.
Conidial germination of the ΔFgvam7 mutant
| Strain | Multipolarity (%) | Unipolarity (%) | ||||
|---|---|---|---|---|---|---|
| 4 h | 6 h | 8 h | 4 h | 6 h | 8 h | |
| PH‐1 | 82.0 ± 5.1A* | 85.0 ± 8.0A | 86.0 ± 5.1A | 18.0 ± 5.1B | 15.0 ± 8.1B | 14.0 ± 4.0B |
| ΔFgvam7 | 15.0 ± 7.0B | 16.0 ± 5.1B | 22.0 ± 6.0B | 85.0 ± 8.1A | 84.0 ± 8.0A | 78.0 ± 5.1A |
| ΔFgvam7/FgVAM7 | 81.0 ± 4.8A | 83.0 ± 6.0A | 85.0 ± 5.0A | 19.0 ± 4.1B | 17.0 ± 8.0B | 15.0 ± 4.1B |
*The same letter indicates no significant difference. Different letters mark statistically significant differences (P < 0.01).
FgVam7 has an important role in female fertility
To test the role of FgVam7 in sexual development, the wild‐type PH‐1, ΔFgvam7 mutant and complemented strains were inoculated on carrot agar plates. After 10 days of inoculation, PH‐1 and the complemented strains formed numerous perithecia, whereas the ΔFgvam7 mutant formed less than one‐half of the perithecia compared with the wild‐type (Fig. 3A). When the perithecia were dissected, 75% were mature and normal (type 1) and 25% had few asci (type 2) in the ΔFgvam7 mutant, compared with 90% of type 1 and 10% of type 2 found in the wild‐type (Fig. 3B). In addition, ascospores produced by the ΔFgvam7 mutant were the same as those produced by the wild‐type (Fig. 3C). However, further vitality tests showed that only 62.5% of ascospores of the ΔFgvam7 mutant germinated and formed colonies, in comparison with 93.5% in the wild‐type. These results suggest that FgVam7 plays an important role in perithecia formation, ascus development and ascospore vitality.
Figure 3.
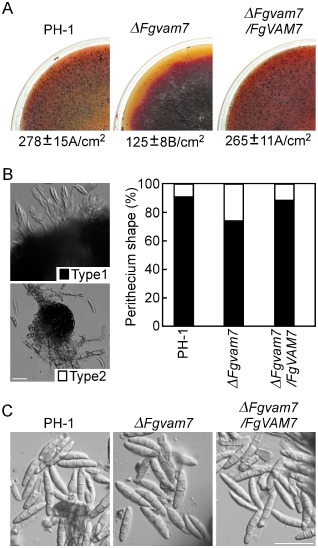
The ΔFgvam7 mutant was defective in sexual reproduction. (A) Self‐crossing cultures of the wild‐type PH‐1, ΔFgvam7 mutant and complemented strains. Black dots on the surface are perithecia. (B) Ascospores oozing from the perithecia and percentages of different perithecia types. Type 1, mature perithecia with numerous asci. Type 2, perithecia with few asci. (C) The morphology of the ascospores from the indicated strains. Bar, 100 μm.
FgVam7 is essential for host colonization and virulence
To determine whether FgVam7 plays a role in plant infection, we first evaluated the colonization of the ΔFgvam7 mutant on tomato fruits. After incubation at 25 °C for 4 days, water‐soaked and collapsed lesions were observed on tomato infected by the wild‐type PH‐1 and complemented mutant strains. In contrast, the ΔFgvam7 mutant caused small necrotic lesions under the same conditions (Fig. 4A). To confirm this result, we point inoculated the flowering wheat heads with conidial suspensions. The wild‐type PH‐1 strain caused typical necrotic symptoms in the inoculated and nearby spikelets at 14 days post‐inoculation (dpi). No disease developed on the wheat heads inoculated with the conidia of the ΔFgvam7 mutant under the same conditions. Spikelets close to the inoculation site remained healthy at 14 dpi (Fig. 4B). Based on these results, we conclude that FgVam7 is required for the colonization of plants and virulence.
Figure 4.
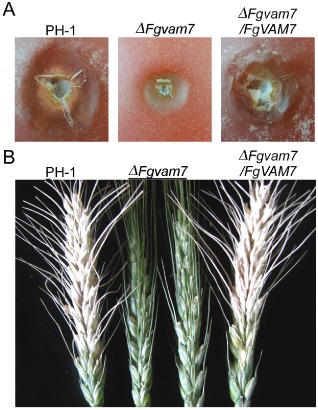
The ΔFgvam7 mutant was defective in disease spread. (A) Infection assay with tomato fruits. Conidial suspensions of PH‐1, ΔFgvam7 and ΔFgvam7/FgVAM7 were injected into tomato fruits as described in Experimental procedures. Photographs were taken at 4 days post‐inoculation (dpi). (B) Infection assays with flowering wheat heads. 10 μL conidial suspensions (1 × 106 conidia/mL) of the indicated strains were injected into the floret in the sixth spikelet of the flowering wheat head. Typical heads were photographed at 14 dpi.
FgVam7 modulates DON production and the expression of trichothecene synthase genes
To explore the role of FgVam7 in mycotoxin DON production, diseased wheat kernels from the inoculated spikelets were isolated and the DON content was measured. The production of DON and 3‐acetyl‐deoxynivalenol (3‐ADON) was reduced significantly in the ΔFgvam7 mutant. Only 2.0 ppm DON and 10.0 ppm 3‐ADON were detected in the wheat kernels infected by the ΔFgvam7 mutant, compared with 3.2 ppm DON and 21.0 ppm 3‐ADON detected in the wild‐type PH‐1 strain (Fig. 5A). To confirm this finding, the expression of the trichothecene synthase genes TRI5 and TRI6 and the trichothecene acetyltransferase gene TRI101, which are involved in DON biosynthesis, was detected by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The expression level of these three genes was reduced significantly in the ΔFgvam7 mutant (Fig. 5B), suggesting that FgVam7 is involved in the modulation of TRI5, TRI6 and TRI101 expression, which affects DON and 3‐ADON synthesis.
Figure 5.
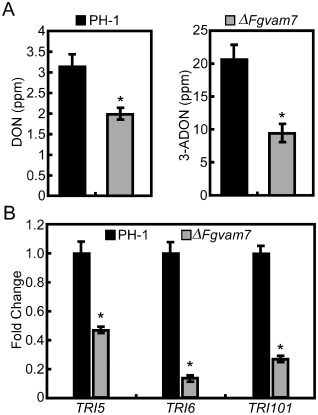
Deoxynivalenol (DON) production and the expression of DON biosynthesis genes. (A) DON and 3‐acetyl‐deoxynivalenol (3‐ADON) were measured as a means of fungal biomass quantification. (B) Expression levels of TRI5, TRI6 and TRI101 were measured by real‐time quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Following normalization against TUB2 expression, the relative expression levels in the ΔFgvam7 mutant were presented as fold changes in comparison with those of the wild‐type PH‐1 (artificially set to unity). The mean value and standard deviations were calculated with the data of three biological replicates. Asterisks indicate statistically significant differences (P < 0.01).
FgVam7 regulates vacuole morphogenesis and endocytosis
Our previous study showed that MoVam7 is involved in vacuole morphogenesis and endocytosis of M. oryzae (Dou et al., 2011). To determine whether FgVam7 has a similar function, we examined the vacuole morphology using transmission electron microscopy. Over 85% of the vacuoles of the ΔFgvam7 mutant were small and fragmented, with numerous irregular spheroids observed in both conidial and hyphal cells. In contrast, one single large vacuole was seen in 80% of the cells of the wild‐type (Fig. 6A). To determine whether the defect in vacuole morphology affects endocytosis, FM4‐64 staining was performed. Wild‐type cells rapidly take up the FM4‐64 dye within 1 min of exposure, whereas no clear staining pattern was observed in the ΔFgvam7 mutant cells at the same time point. However, the normal staining pattern was observed 2 min after exposure, indicating that FgVam7 plays an important role in maintaining normal morphology, but only causes delayed endocytosis (Fig. 6B).
Figure 6.
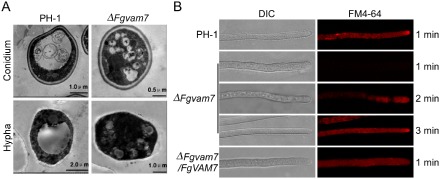
The ΔFgvam7 mutant was defective in vacuole morphogenesis and endocytosis. (A) The vacuole structure of the ΔFgvam7 mutant conidium and young hypha shown by transmission electron microscopy. (B) FM4‐64 staining assay. Strains were grown for 1 day on PDA (46 g potato dextrose agar powder in 1 L double‐distilled H2O)‐overlaid microscope slides before the addition of FM4‐64. Photographs were taken at the indicated periods. DIC, differential interference contrast.
FgVam7 plays a crucial role in response to various stressors
As the ΔFgvam7 mutant showed defects in vacuolar morphology and endocytosis timing, we examined the response of the mutant to several environmental stressors. Strains were cultured on CM plates with NaCl, sorbitol (salt and osmotic stress), H2O2 (oxidative stress) and Calcofluor white (CFW), sodium dodecylsulfate (SDS) and Congo red (CR) (cell wall stressors). The growth rate of the ΔFgvam7 mutant was increased significantly on the plates with 1 m NaCl and 1 m sorbitol (Fig. 7A,B). In contrast, growth was strongly decreased in the presence of 0.05% CFW and 0.01% SDS (Fig. 7C,D). No significant changes were observed on the plates with 5 mm H2O2 and 0.05% CR (Fig. 7C,D). These findings indicate that FgVam7 may have a role in the maintenance of cell wall integrity and osmoregulation.
Figure 7.
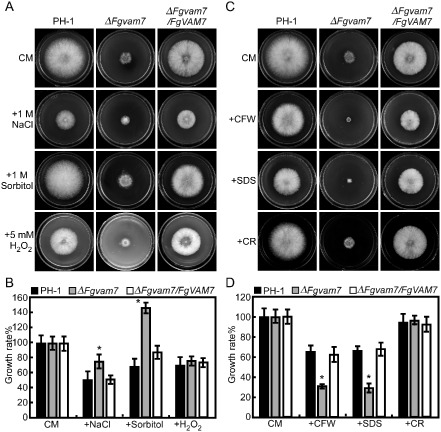
Defects of the ΔFgvam7 mutant in response to various stressors. (A) The wild type PH‐1, ΔFgvam7 mutant and complemented ΔFgvam7/FgVAM7 mutant strains were cultured on medium supplemented with hyperosmotic and oxidative stressors. (C) The indicated strains were cultured on plates with cell wall antagonists. (B, D) The growth rates (artificially set to 100% on CM plates) of the indicated strains under various stress conditions. Error bars represent the standard deviation from three independent experiments and asterisks indicate statistically significant differences (P < 0.01). CFW, Calcofluor white; CM, complete medium; CR, Congo red; SDS, sodium dodecylsulfate.
Expression and subcellular localization of FgVam7‐GFP fusion proteins
In S. cerevisiae, the Vam7‐GFP fusion protein was detected in the cytoplasm and vacuolar membranes (Sato et al., 1998). To detect the expression and localization pattern of the FgVam7‐GFP proteins in F. graminearum, an FgVAM7‐GFP fusion construct was generated and transformed into the ΔFgvam7 mutant. Unlike ScVam7‐GFP, strong GFP signals were observed mainly in the vacuoles of the vegetative hyphae and conidia. Further, CMAC (7‐amino‐4‐chloromethylcoumarin: vacuole tracker) staining analysis also showed the vacuolar localization of the FgVAM7‐GFP protein (Fig. 8).
Figure 8.
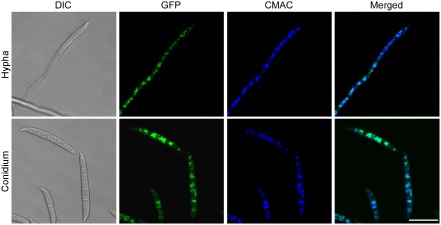
Expression and localization of FgVAM7‐GFP in Fusarium graminearum. Vegetative hyphae and conidia expressing the FgVAM7‐GFP fusion construct were examined. The same field was examined under differential interference contrast (DIC) and epifluorescence microscopy [green fluorescent protein (GFP) or 7‐amino‐4‐chloromethylcoumarin (CMAC) staining]. Bar, 20 μm.
Secretome comparison between the ΔFgvam7 mutant and wild‐type PH‐1
Because SNARE proteins are known to be involved in secretion transport, we identified the differentially expressed secretory proteins in the ΔFgvam7 mutant to explore the role of FgVam7 in protein secretion. Through two‐dimensional gel electrophoresis (2‐DE) examination, 11 protein spots were positively identified to be differentially expressed in the ΔFgvam7 mutant (Fig. 9, Table 2). Further analysis found that FGSG_00184, FGSG_01596 and FGSG_00314 are glycoside hydrolases required for normal spore wall assembly, whereas FGSG_06549 and FGSG_03544 are chitin deacetylases involved in the biosynthesis of cell wall components, FGSG_03687 is a carboxylesterase, FGSG_03816 is a gluconolaconase involved in carbohydrate acid metabolism, and FGSG_01346 is an enolase that catalyses the conversion of 2‐phosphoglycerate to phosphoenolpyruvate. Finally, FGSG_07608 is a 5′/3′‐nucleotidase SurE, and FGSG_03850 is a mitochondrial cytochrome‐c peroxidase involved in the response to oxidative stress (Table 2). As some of these proteins are related to the degradation or biosynthesis of the cell wall, their identification indicates that FgVam7 may play a role in the regulation of the secretory proteins important in the infection and colonization of plants.
Figure 9.
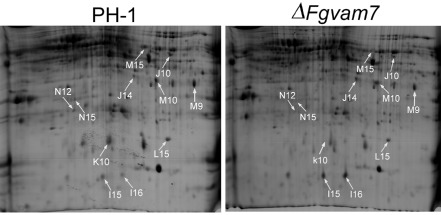
Identification and confirmation of differentially expressed proteins from wild‐type PH‐1 and ΔFgvam7 mutant strains. Results of two‐dimensional gel electrophoresis (2‐DE) and matrix‐assisted laser desorption/ionization time‐of‐flight tandem mass spectrometry (MALDI‐TOF‐TOF MS) assays. The numbers with arrows indicate the differentially expressed and identified protein spots.
Table 2.
Identification of differentially expressed secretory proteins in the ΔFgvam7 mutant
| Spot no.b | Gene locusc | Protein named | Signal P | Protein scoree | SC (%)f | MPg | Mutant/wild‐typeh | Cellular locations |
|---|---|---|---|---|---|---|---|---|
| M9 | FGSG_00184 | Glycoside hydrolase, CRR1 | Y | 194 | 9 | 3 | 0.42a | Extracellular |
| M10 | FGSG_06549 | Chitin deacetylase, CDA1 | Y | 123 | 4 | 4 | 0.36a | Extracellular |
| M15 | FGSG_03816 | Gluconolaconase | Y | 302 | 12 | 3 | 0.004a | Vacuolar |
| N15 | FGSG_03687 | Carboxylesterase | Y | 191 | 6 | 2 | 0.09a | Extracellular |
| K10 | FGSG_01346 | Enolase | Y | 486 | 10 | 3 | 0.44a | Peroxisomal |
| J10 | FGSG_03544 | Chitin deacetylase, CDA1 | Y | 114 | 11 | 2 | 192.5a | Extracellular |
| J14 | FGSG_07608 | 5′/3′‐Nucleotidase SurE | Y | 69 | 3 | 1 | 2.67a | Extracellular |
| L15 | FGSG_01596 | Glycoside hydrolase | Y | 384 | 13 | 5 | 3.54a | Extracellular |
| N12 | FGSG_00314 | Glycosyl hydrolase | Y | 57 | 4 | 1 | 2.88a | Extracellular |
| I15 | FGSG_03850 | Peroxidase/catalase 2 | Y | 327 | 3 | 4 | 1.40a | Mitochondrial |
| I16 | FGSG_03850 | Peroxidase/catalase 2 | Y | 481 | 3 | 5 | 8.31a | Mitochondrial |
Indicates significant difference between mutant and wild‐type.
Numbering corresponds to the two‐dimensional gel electrophoresis (2‐DE) in Fig. 9.
Gene locus from Fusarium database.
Names of the proteins obtained via MASCOT software from the Fusarium database.
MOWSE score probability for the entire protein.
The sequence coverage of the identified proteins.
The total number of identified peptides.
The protein abundance ratio (mutant/wild‐type).
Dissection of FgVam7 SNARE and PX domain function
FgVam7 contains a PX and a SNARE domain similar to MoVam7. To dissect the PX domain function, we transformed the FgVam7SNARE (SNARE domain only) and FgVam7 (full‐length) vectors into the ΔFgvam7 mutant. We also transformed the MoVam7SNARE‐ (SNARE domain only), MoVam7PX+FgVam7SNARE‐ (PX domain and SNARE domain chimera), FgVam7PX+MoVam7SNARE‐ (PX domain and SNARE domain chimera) and MoVam7‐GFP (full‐length) fusion vectors into the ΔFgvam7 mutant strain. The resulting transformants were screened by GFP signal and characterized. Phenotype analysis revealed that FgVam7SNARE, but not MoVam7SNARE, fully restored the conidiation and virulence defects, and partially restored the growth defect (Fig. 10A–D). FgVam7PX+MoVam7SNARE and MoVam7 did not restore any defects of the ΔFgvam7 mutant. Interestingly, the inclusion of the PX domain from MoVam7 (MoVam7PX+FgVam7SNARE) appeared to partially suppress FgVam7SNARE function (Fig. 10A–D). To further examine whether the heterologous PX interferes with Vam7 function, we introduced the MoVam7SNARE (SNARE domain only), FgVam7PX+MoVam7SNARE (PX domain and SNARE domain chimera) fusion vectors into the ΔMovam7 mutant strain. Indeed, although MoVam7SNARE partially suppresses the defect of ΔMovam7 in conidiation and virulence, FgVam7PX+MoVam7SNARE shows the same defect as the ΔMovam7 mutant (Fig. S3, see Supporting Information). These findings indicate that the SNARE domains of FgVam7 and MoVam7 are important for protein function, but share no cross‐species functions, in contrast with the PX domain which interferes heterologously with Vam7 function.
Figure 10.
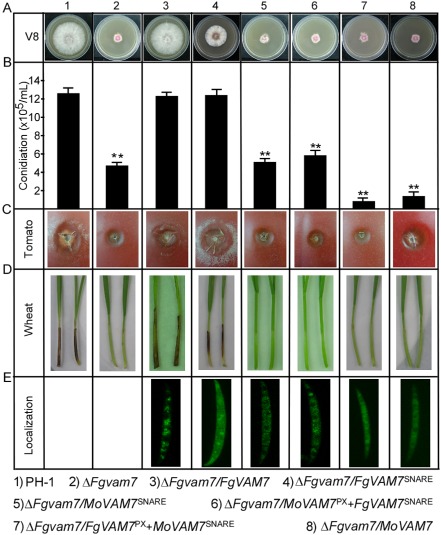
The N‐ethylmaleimide‐sensitive factor attachment protein receptor (SNARE) and PhoX homology (PX) domain functional dissection. (A) Growth and colony morphology of the indicated strains on V8 plates. Photographs were taken after incubation at 25 °C for 3 days. (B) Quantification of the conidia produced by the indicated strains. Error bars represent the standard deviation from three independent experiments and asterisks indicate a significant difference between the indicated strains (P < 0.01). (C) Infection assay with tomato fruits. Conidial suspensions of the indicated strains were injected into tomato fruits as described in Experimental procedures. Photographs were taken at 4 days post‐inoculation (dpi). (D) Infection assay with wheat seedlings. Conidial suspensions were inoculated onto the wheat coleoptiles as described previously (Zhang et al., 2012). Photographs were taken at 10 dpi. (E) Cellular localization of different proteins in conidia.
We then observed the protein localization in each transformant and attempted to examine whether the localization pattern was related to protein function. Compared with FgVam7‐GFP, which mainly localizes in vacuoles, MoVam7SNARE‐, MoVam7‐ and MoVam7PX+FgVam7SNARE‐GFP were mainly detected in the cytoplasm, but not the vacuoles. Although FgVam7SNARE‐GFP was found in the cytoplasm and as cytoplasmic punctate structures, FgVam7PX+MoVam7SNARE‐GFP was detected only in the cytoplasm (Fig. 10E). These results indicate that the FgVam7 PX domain is required for correct localization and full function.
Discussion
SNARE proteins play central roles in membrane fusion and vesicle transport, and are well conserved in eukaryotic organisms (Dou et al., 2011; Sato et al., 1998; Song et al., 2010; Wedlich‐Soldner et al., 2000). Previous studies of fungal Vam7 proteins, including MoVam7, ScVam7 and UmYup1, have shown that they are bi‐domain proteins containing an N‐terminal PX domain and a C‐terminal SNARE domain (Dou et al., 2011). Not surprisingly, FgVam7 also contains the PX and SNARE domains which are highly homologous to the aforementioned proteins, further highlighting the conserved nature of the Vam7 proteins. However, FgVam7 exhibits unique functions as it cannot complement the ΔMovam7 mutant (Fig. S3) and, likewise, MoVam7 cannot rescue the defects of the ΔFgvam7 mutant (Fig. 10).
The PX domain specifically binds to phosphatidylinositol‐3‐phosphate (ptdIns‐3‐P) and plays a role in the assembly of multiprotein complexes at specific membrane surfaces (Yu and Lemmon, 2001). In S. cerevisiae and U. maydis, the PX domain is known to be required for Vam7 function (Sato et al., 1998; Wedlich‐Soldner et al., 2000). In the present study, the PX domain of FgVam7 showed a different regulation mechanism in the localization and functions of the protein in F. graminearum.
SNARE proteins are involved in fungal growth, development and virulence. Yeast ScVam7 is essential for vacuole morphogenesis (Sato et al., 1998). UmYup1 is required for hyphal morphogenesis, endocytic membrane fusion and polar growth (Wedlich‐Soldner et al., 2000). MoVam7 and MoSec22 play crucial roles in hyphal growth, conidiation, vacuole morphology, virulence and endocytosis (Dou et al., 2011; Song et al., 2010). We have shown here that FgVam7 plays roles similar to those of UmYup1, MoVam7 and MoSec22, and is involved in polar growth, conidiogenesis, pathogenesis and endocytosis, indicating that the proteins functioning in membrane trafficking are important for the correct regulation of infection‐related morphogenesis in different fungi. Disruption of UmYup1 in U. maydis altered morphogenesis and polar growth. In addition, deletion of MoVAM7 and MoSEC22 prevented plant infection (Dou et al., 2011; Song et al., 2010). Similarly, the ΔFgvam7 mutant showed a defect in polar growth and no virulence on wheat heads. Although the 60% reduction in growth rate of the ΔFgvam7 mutant may contribute to defects in plant infection, other important pathogenicity factors are probably affected in the mutant, as its virulence was completely abolished. In addition, DON is an important virulence factor in F. graminearum, and DON production was also reduced in the ΔFgvam7 mutant. Further, qRT‐PCR assays suggested that FgVam7 modulates DON production by regulating the expression of three trichodiene synthase genes TRI5, TRI6 and TRI101. Because these three genes are not located in the same TRI cluster, FgVAM7 probably modulates TRI5, TRI6 and TRI101 through different pathways.
In addition, vesicles and vacuoles are known to contain enzymes associated with secondary metabolism in plants and fungi, including enzymes involved in the biosynthesis of alkaloids (Ziegler and Facchini, 2008), flavonoids, the non‐ribosomal peptide cyclosporin (Hoppert et al., 2001), the β‐lactam antibiotic penicillin (Lendenfeld et al., 1993) and the polyketide aflatoxin (Hong and Linz, 2008; 2009; Lee et al., 2004). These reports indicate that the SNARE FgVam7 protein could be involved in vesicle and endosome transport affecting the synthesis and storage of mycotoxins.
In M. oryzae, the ΔMovam7 and ΔMosec22 mutants are more sensitive to cell wall stressors and show a defect in cell wall integrity (Dou et al., 2011; Song et al., 2010). In our results, the ΔFgvam7 mutant was also sensitive to cell wall stressors, suggesting that FgVam7 plays a role in cell wall integrity. However, it showed increased tolerance to salt and osmotic stresses. The cause of this conflict remains unclear and awaits future studies. As the ΔFgvam7 mutant showed a defect in vacuole morphology and delayed endocytosis, it is reasonable to speculate that it will display an altered response to various environmental stresses. In addition, MoVam7 is known to be involved in conidial morphology (Dou et al., 2011), whereas the deletion of FgVam7 in F. graminearum did not affect conidial shape. The discrepancies suggest that Vam7 proteins have distinct roles among different fungi.
In S. cerevisiae, Vam7 interacts with Vam3, which function together at a common step in the vacuolar protein sorting (VPS) pathway. The ΔScvam7 and ΔScvam3 mutants show very similar phenotypes (Darsow et al., 1997; Sato et al., 1998; Srivastava and Jones, 1998). ScVam3‐GFP is localized to vacuolar membranes and is required at a late step during the transport of multiple proteins to the vacuole, whereas ScVam7‐GFP is detected in both the cytoplasm and on vacuolar membranes. In U. maydis, Yup1‐GFP is localized to vesicles that show rapid salutatory motion along the microtubules (Wedlich‐Soldner et al., 2000). Nevertheless, unlike ScVam7‐ and UmYup1‐GFP, FgVam7‐GFP is localized mainly in the vacuoles of F. graminearum. The diverse localization patterns of Vam7 proteins indicate that Vam7 may play distinct roles at different stages during exocytosis or endocytosis, such as membrane fusion or vacuole assembly and sorting. FgVam7 may preferentially target components involved in endosome and vacuole development. The deletion of FgVam7 compromised morphogenesis, resulting in many small and fragmented, perhaps immature, vacuoles and endosomes, which may underlie the other cellular functions of FgVam7. In addition, we identified a series of secretory proteins regulated by FgVam7 that were related to cell wall assembly or other functions. Further characterization of these proteins will help us to better understand the regulatory mechanisms of SNARE proteins during pathogen–plant interactions.
Taken together, our results indicate that the SNARE protein FgVam7 plays a pleiotropic role in F. graminearum, and is involved in hyphal and polar growth, asexual and sexual development, stress response and plant infection. Further insight into the role of SNARE proteins will be provided by the identification and characterization of targets of FgVam7 during the cycles of exocytosis and endocytosis.
Experimental Procedures
Fungal strains and cultures
The F. graminearum strain PH‐1 was used as the wild‐type strain. All strains used in this study were cultured on V8 juice agar plates. For genomic DNA and RNA isolation, fungal strains were grown in 50 mL liquid YEPD medium (3 g yeast extract, 10 g peptone, 20 g dextrose in 1 L double‐distilled H2O) at 25 °C, 175 rpm for 36 h, and the hyphal mass was harvested and lyophilized. For conidiation, mycelial agar blocks were inoculated into 100 mL carboxymethylcellulose (CMC) liquid medium (15 g CMC, 1.0 g NH4NO3, 1.0 g KH2PO4, 0.5 g MgSO4.7H2O, 1.0 g yeast extract in 1 L double‐distilled H2O) and incubated at 25 °C, 150 rpm for 5 days. Growth assay was performed on V8, 5 × YEG, CM and MM media incubated at 25 °C for 3 days. For stress assay, strains were cultured on CM with different concentrations of NaCl, KCl, sorbitol, CFW, SDS and H2O2, and incubated at 25 °C for 3 days. Protoplast preparation and fungal transformation were performed as described by Hohn and Desjardins (1992). TB3 medium (0.3% yeast extract, 0.3% casamino acid, 20% sucrose) supplemented with 250 μg/mL hygromycin B (Roche, San Francisco, CA, USA) and 250 μg/mL bleomycin (Invitrogen, Carlsbad, CA, USA) was used for the selection of transformants. Conidiation and detached leaf infection assays were performed as described previously (Song et al., 2010; Zhang et al., 2011). For sexual reproduction, aerial hyphae of 7‐day‐old carrot agar cultures of the indicated strains were pressed down with 300 μL of sterile 0.1% Tween‐20, as described previously (Zheng et al., 2012).
Targeted gene deletion and complementation
The FgVAM7 gene deletion mutant ΔFgvam7 allele was generated with the split marker approach with the primers 1F/2R, 3F/4R, HYG/F‐HY/R and YG/F‐HYG/R (Fig. S2A, see Supporting Information). After transformation of wild‐type PH‐1 protoplasts, hygromycin‐resistant transformants were screened by PCR with primers 5F/6R and confirmed by Southern blot analysis. When hybridized with the FgVAM7 gene probe amplified with primers 5F/6R, the expected band was detected in PH‐1, but not in the ΔFgvam7 mutant. When hybridized with the HPH probe amplified with primers FL1111/FL1112, the expected band was detected in the ΔFgvam7 mutant, but absent in PH‐1 (Fig. S2B). For complementation, a 1.1‐kb fragment containing the entire FgVAM7 gene driven by the RP27 promoter was amplified with primers FL14257/FL14258 and inserted into pYF11‐tagged GFP (bleomycin resistance). The resulting constructs were sequenced and transformed into the protoplast of the ΔFgvam7 mutant. The primers are listed in Table S1 (see Supporting Information).
Tomato fruits and wheat infection assays
To assay the colonization of the ΔFgvam7 mutant on wounded tomatoes, cluster‐type ripened tomato fruits were surface sterilized with absolute ethanol and washed with sterile distilled water. Conidia of PH‐1 and ΔFgvam7 mutant strains were collected and suspended in sterile distilled water containing 0.01% Tween‐20 at a concentration of 107 conidia per millilitre; 10 μL of conidial suspension were injected into tomatoes at 1–2 cm depth and incubated in a chamber (97% relative humidity, 26 °C, 14 h light and 10 h dark), as described previously (Di Pietro et al., 2001). The infection assay on flowering wheat inflorescences was also performed as described previously (Seong et al., 2005) with minor modifications. Approximately 6‐week‐old (anthesis) wheat cultivar Norm with flowering heads was used for infection assays. A floret in the sixth spikelet of the flowering wheat head was inoculated by injecting 10 μL of the conidial suspension (1 × 106 conidia/mL) in 0.01% (v/v) Tween‐20; 10 μL of 0.01% Tween‐20 served as a control. For each treatment, nine wheat heads were inoculated. Infected spikelets in each head were counted and analysed at 2 weeks after inoculation.
qRT‐PCR analysis
RNA samples were isolated from vegetative hyphae of PH‐1 and the ΔFgvam7 mutant, and used for first‐strand cDNA synthesis. RT2 PCR Real‐Time SYBR Green/ROX PCR master mix (TaKaRa, Dalian, China) was used for RT‐PCR assays. Primer pairs TRI5QF/TRI5QR, TRI6QF/TRI6QR and TRI101QF/TRI101QR were used to amplify the TRI5, TRI6 and TRI101 genes, respectively. The relative quantification of each transcript was calculated by the 2–ΔΔCT method (Livak and Schmittgen, 2001) with the F. graminearum β‐tubulin gene TUB2 as the internal control. The primers are listed in Table S1.
Generation of the FgVAM7‐, MoVAM7‐, domain‐ and domain chimera‐GFP fusion constructs
To generate the FgVAM7‐, MoVAM7‐, SNARE domain‐ and PX domain chimera‐GFP fusion constructs, the fragments of the FgVAM7, MoVAM7 and domain regions driven by the RP27 promoter were amplified and cloned into pYF11 by the yeast gap repair approach (Bruno et al., 2004). The resulting plasmids were confirmed by sequencing analysis to contain the in‐frame fusion constructs. Zeocin‐resistant transformants were isolated after transformation of the ΔFgvam7 mutant with the constructs. The resulting transformants were examined for GFP signals under an epifluorescence microscope. The primers are also listed in Table S1.
DON assay
The wild‐type and ΔFgvam7 mutant strains were incubated on solid grain (10 g) medium in the dark for 3 weeks at 20 °C and then at 10 °C for DON and 3‐ADON production. Trichothecene standards, including DON and 3‐ADON, were purchased from Sigma‐Aldrich (St. Louis, MO, USA). The method of trichothecene assay, including extraction, clean‐up and derivatization (Goswami and Kistler, 2005), was used with modifications. Grain (200 g) was randomly selected and ground for 5 min in an all‐purpose grinder, and 20 g of finely ground sample were treated for 1 h with 80 mL acetonitrile [acetonitrile–water, 84/16 (v/v)] in a shaker. A 4‐mL extract was eluted through a 1‐g clean‐up column containing C‐18 and aluminium oxide (3 : 1, w/w). The elution was dried and then dissolved in a mixture of trimethylsilylimidazole‐trimethylchlorosilane (100 : 1). The trimethylsilyl derivations of DON were identified and quantified using a Thermo Trace GC Ultra capillary column gas chromatograph equipped with an auto injector (AS3000), electron capture detector and MN Permabond SE‐54‐DB‐17 (Macherey‐Nagel, Bethlehem, PA, USA). The injection port and detector temperatures were 220 °C and 300 °C, respectively. The column temperature was 100 °C for 1 min, 15 °C/min to 250 °C for 5 min. The carrier gas was helium at a flow rate of 0.8 mL/min. Toxin extraction and analysis were repeated three times and the mean values were used for data analysis.
Light microscopy assays
To examine endocytosis, strains were grown for 1 day on PDA (46 g potato dextrose agar powder in 1 L double‐distilled H2O)‐overlaid microscope slides before being stained with N‐3‐triethylammoniumpropyl‐4‐p‐diethylamino‐phenyl‐hexatrienyl pyridinium dibromide (FM4‐64) (Molecular Probes, Eugene, OR, USA) following the procedures described previously (Fischer‐Parton et al., 2000). Photographs were taken under a confocal laser scanning microscope. Staining with CMAC (Molecular Probes) was performed as described previously (Shoji et al., 2006). Mycelia were cultured in liquid YEPD medium for 12 h and conidia were harvested from CMC medium cultured for 3 days. Confocal microscopy was performed on a Zeiss Axiovert 200 M microscope equipped with a Zeiss (Oberkochen, Germany) LSM 710 META system using 403/1.2 NA and 633/1.2 NA C‐Apochromat water immersion objectives.
Protein extraction and 2‐DE
Total secreted proteins were isolated from the mycelia of the ΔFgvam7 mutant and wild‐type PH‐1 strains after 4 days of incubation in liquid MM. For 2‐DE, the protein extracts were dried in vacuum and dissolved in 800 μL lysis solution containing 7 m urea, 2 m thiourea, 4% (w/v) 3‐[(3‐cholamidopropyl)dimethylammonio]‐1‐propanesulfonate (CHAPS), 65 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonylfluoride (PMSF) and 0.5% (v/v) biolytes (Bio‐Rad, Hercules, CA, USA). Insoluble materials were removed by centrifugation and the protein concentration of the sample was quantified using the Bradford method. About 1200 μg protein was separated by loading the sample on an 18‐cm pH 4–7 nonlinear gradient IPG strip (GE Healthcare, Pittsburgh, PA, USA). The second electrophoretic dimension was by 12% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). The signal was observed by coomassie brilliant blue (CBB) G‐250. The gel image was digitalized with a gel scanner (Powerlook 2100XL, UMAX, Shanghai, China), and analysed using the PDQuest™ software package (Version 7.2.0; Bio‐Rad). Spots were detected, matched and normalized on the basis of the total density of gels with the parameter of percentage volume according to the software guide. For each protein spot, the mean relative volume (RV) was computed at every sampling, and the spots showing a mean RV that changed by more than 1.5‐fold and P < 0.05 in different stages were considered to be differentially expressed.
In‐gel digestion and matrix‐assisted laser desorption/ionization time‐of‐flight tandem mass spectrometry (MALDI‐TOF‐TOF MS) analysis
Protein spots with differential expression patterns were manually excised from gels and processed according to the standard method or as described by Wang et al. (2014). Following trypsin (Promega, Madison, WI, USA) digestion, the peptides were extracted twice with 0.1% trifluoroacetic acid (TFA) in 50% acetonitrile, pooled and lyophilized, before dissolving in 5 mg/mL α‐cyano‐4‐hydroxycinnamic acid (CHCA) containing 0.1% TFA and 50% acetonitrile. MALDI‐TOF‐TOF MS analyses were conducted using the Bruker Daltonics Ultraflex MALDI‐TOF‐TOF™ analyser (Leipzig, Germany). All spectra of proteins were submitted to a database search using the online MASCOT program (http://www.matrixscience.com) against the F. graminearum genome database. The search parameters were as follows: 0.15‐Da mass tolerance for peptides and 0.25‐Da mass tolerance of TOF‐TOF fragments; one allowed trypsin miscleavage; carbamidomethyl of cysteine (Cys) as a fixed modification; and oxidation of methionine (Met) and pyro‐glutamic acid (pyro‐Glu) formation of N‐terminal glutamine (Gln) and Glu as variable modification. Only significant hits, as defined by MASCOT probability analysis (P < 0.05), were accepted.
Supporting information
Fig. S1 Phylogenetic and structural analysis of FgVam7 and its homologues. (A) Phylogenetic analysis of FgVam7 and its homologues. The phylogenetic tree was drawn using Clustal W 1.83. ScVam7 (Saccharomyces cerevisiae, NP_011303.1), MoVam7 (Magnaporthe oryzae, XP_001907727.1), NcVam7 (Neurospora crassa, XP_957713.1), UmYup1 (Ustilago maydis, XP_761553.1) and AfVam7 (Aspergillus fumigatus, XP_749396.1). (B) Structural analysis of FgVam7 and its homologues. aa, amino acid; PX, PhoX homology domain; SNARE, Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor.
Fig. S2 Targeted deletion of the FgVAM7 gene in Fusarium graminearum. (A) Schematic diagram of the FgVAM7 gene and gene replacement construct. (B) Southern blots of EcoRI‐digested genomic DNA of the wild‐type PH‐1 and ΔFgvam7 mutant strains hybridized with the FgVAM7 and HPH probes, respectively.
Fig. S3 Functions of the SNARE (Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor) and PX (PhoX homology) domains of MoVam7 in conidiation and virulence of Magnaporthe oryzae. (A) Growth and colony morphology of the indicated strains on SDC (straw decoction and corn medium) plates. Photographs were taken after incubation at 28 °C for 7 days. (B) Conidiation assays of the indicated strains. Photographs were taken at 24 h post‐inoculation (hpi) induced under light. Error bars represent the standard deviation from three independent experiments and asterisks indicate significant difference (P < 0.01). (C) Pathogenicity assays of the indicated strains on detached barley leaves. Photographs were taken at 5 days post‐inoculation. (D) Green fluorescent protein (GFP) signal observation in hyphae and conidia of different strains under confocal microscopy.
Table S1 Primers used in this study.
Acknowledgements
This research was supported by the National Basic Research Program of China (Grant No. 2013CB127800), Natural Science Foundation of Jiangsu (Grant No. BK2012362, HZ) and the specially appointed professorship (Jiangsu, China). We thank Ping Wang of Louisiana State University Health Sciences Center, New Orleans, LA, USA for critical review of the manuscript.
References
- Bai, G.H. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. [DOI] [PubMed] [Google Scholar]
- Balderhaar, H.J. and Ungermann, C. (2013) CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J. Cell Sci. 126, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Beyer, M. , Klix, M.B. , Klink, H. and Verreet, J.A. (2006) Quantifying the effects of previous crop, tillage, cultivar and triazole fungicides on the deoxynivalenol content of wheat grain—a review. J. Plant Dis. Protect. 113, 241–246. [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J.R. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryot. Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri, L. and Lithgow, T. (2004) A complete set of SNAREs in yeast. Traffic, 5, 45–52. [DOI] [PubMed] [Google Scholar]
- Burri, L. , Varlamov, O. , Doege, C.A. , Hofmann, K. , Beilharz, T. , Rothman, J.E. , Sollner, T.H. and Lithgow, T. (2003) A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA, 100, 9873–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.A. and Scheller, R.H. (2001) Snare‐mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2, 98–106. [DOI] [PubMed] [Google Scholar]
- Darsow, T. , Rieder, S.E. and Emr, S.D. (1997) A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins, A.E. , Proctor, R.H. , Bai, G.H. , McCormick, S.P. , Shaner, G. , Buechley, G. and Hohn, T.M. (1996) Reduced virulence of trichothecene‐nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant–Microbe Interact. 9, 775–781. [Google Scholar]
- Di Pietro, A. , Garcia‐Maceira, F.I. , Meglecz, E. and Roncero, M.I.G. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Dou, X.Y. , Wang, Q. , Qi, Z.Q. , Song, W.W. , Wang, W. , Guo, M. , Zhang, H.F. , Zhang, Z.G. , Wang, P. and Zheng, X.B. (2011) MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae . PLoS ONE, 6, e16439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Urban, M. , Parker, J.E. , Brewer, H.C. , Kelly, S.L. , Hammond‐Kosack, K.E. , Fraaije, B.A. , Liu, X. and Cools, H.J. (2013) Characterization of the sterol 14alpha‐demethylases of Fusarium graminearum identifies a novel genus‐specific CYP51 function. New Phytol. 198, 821–835. [DOI] [PubMed] [Google Scholar]
- Fischer‐Parton, S. , Parton, R.M. , Hickey, P.C. , Dijksterhuis, J. , Atkinson, H.A. and Read, N.D. (2000) Confocal microscopy of FM4‐64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198, 246–259. [DOI] [PubMed] [Google Scholar]
- Fuchs, U. and Steinberg, G. (2005) Endocytosis in the plant‐pathogenic fungus Ustilago maydis . Protoplasma, 226, 75–80. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2005) Pathogenicity and in planta mycotoxin accumulation among members of the Fusarium graminearum species complex on wheat and rice. Phytopathology, 95, 1397–1404. [DOI] [PubMed] [Google Scholar]
- Han, Y.K. , Kim, M.D. , Lee, S.H. , Yun, S.H. and Lee, Y.W. (2007) A novel F‐box protein involved in sexual development and pathogenesis in Gibberella zeae . Mol. Microbiol. 63, 768–779. [DOI] [PubMed] [Google Scholar]
- Hohn, T.M. and Desjardins, A.E. (1992) Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris . Mol. Plant–Microbe Interact. 5, 249–256. [DOI] [PubMed] [Google Scholar]
- Hong, S.Y. and Linz, J.E. (2008) Functional expression and subcellular localization of the aflatoxin pathway enzyme Ver‐1 fused to enhanced green fluorescent protein. Appl. Environ. Microbiol. 74, 6385–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.Y. and Linz, J.E. (2009) Functional expression and sub‐cellular localization of the early aflatoxin pathway enzyme Nor‐1 in Aspergillus parasiticus . Mycol. Res. 113, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.Y. , So, J. , Lee, J. , Min, K. , Son, H. , Park, C. , Yun, S.H. and Lee, Y.H. (2010) Functional analyses of two syntaxin‐like SNARE genes, GzSYN1 and GzSYN2, in the ascomycete Gibberella zeae . Fungal Genet. Biol. 47, 364–372. [DOI] [PubMed] [Google Scholar]
- Hoppert, M. , Gentzsch, C. and Schorgendorfer, K. (2001) Structure and localization of cyclosporin synthetase, the key enzyme of cyclosporin biosynthesis in Tolypocladium inflatum . Arch. Microbiol. 176, 285–293. [DOI] [PubMed] [Google Scholar]
- Hou, Z.M. , Xue, C.Y. , Peng, Y.L. , Katan, T. , Kistler, H.C. and Xu, J.R. (2002) A mitogen‐activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant–Microbe Interact. 15, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Jenczmionka, N.J. and Schafer, W. (2005) The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Curr. Genet. 47, 29–36. [DOI] [PubMed] [Google Scholar]
- Jenczmionka, N.J. , Maier, F.J. , Losch, A.P. and Schafer, W. (2003) Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head‐blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43, 87–95. [DOI] [PubMed] [Google Scholar]
- Kuratsu, M. , Taura, A. , Shoji, J. , Kikuchi, S. , Arioka, M. and Kitamoto, K. (2007) Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae . Fungal Genet. Biol. 44, 1310–1323. [DOI] [PubMed] [Google Scholar]
- Lee, L.W. , Chiou, C.H. , Klomparens, K.L. , Cary, J.W. and Linz, J.E. (2004) Subcellular localization of aflatoxin biosynthetic enzymes Nor‐1, Ver‐1, and OmtA in time‐dependent fractionated colonies of Aspergillus parasiticus . Arch. Microbiol. 181, 204–214. [DOI] [PubMed] [Google Scholar]
- Lee, T. , Han, Y.K. , Kim, K.H. , Yun, S.H. and Lee, Y.W. (2002) Tri13 and Tri7 determine deoxynivalenol‐ and nivalenol‐producing chemotypes of Gibberella zeae . Appl. Environ. Microbiol. 68, 2148–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendenfeld, T. , Ghali, D. , Wolschek, M. , Kubicekpranz, E.M. and Kubicek, C.P. (1993) Subcellular compartmentation of Penicillin biosynthesis in Penicillium‐chrysogenum—the amino‐acid precursors are derived from the vacuole. J. Biol. Chem. 268, 665–671. [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(–Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, S.W. , Kroken, S. , Lee, B.N. , Robbertse, B. , Churchill, A.C. , Yoder, O.C. and Turgeon, B.G. (2003) A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc. Natl. Acad. Sci. USA, 100, 5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen, M. , Jones, R. and Gallenberg, D. (1997) Scab of wheat and barley: a re‐emerging disease of devastating impact. Plant Dis. 81, 1340–1348. [DOI] [PubMed] [Google Scholar]
- Paul, P.A. , Lipps, P.E. , Hershman, D.E. , McMullen, M.P. , Draper, M.A. and Madden, L.V. (2008) Efficacy of triazole‐based fungicides for Fusarium head blight and deoxynivalenol control in wheat: a multivariate meta‐analysis. Phytopathology, 98, 999–1011. [DOI] [PubMed] [Google Scholar]
- Pratelli, J. , Sutter, J.U. and Blatt, M.R. (2004) A new catch in the SNARE. Trends Plant Sci. 9, 187–195. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A. , Assaad, F.F. and Raikhel, N.V. (2000) The Arabidopsis genome. An abundance of soluble N‐ethylmaleimide‐sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T.K. , Darsow, T. and Emr, S.D. (1998) Vam7p, a SNAP‐25‐like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 18, 5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, K. , Hou, Z.M. , Tracy, M. , Kistler, H.C. and Xu, J.R. (2005) Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum . Phytopathology, 95, 744–750. [DOI] [PubMed] [Google Scholar]
- Seong, K. , Li, L. , Hou, Z.M. , Tracy, M. , Kistler, H.C. and Xu, J.R. (2006) Cryptic promoter activity in the coding region of the HMG‐CoA reductase gene in Fusarium graminearum . Fungal Genet. Biol. 43, 34–41. [DOI] [PubMed] [Google Scholar]
- Shim, W.B. , Sagaram, U.S. , Choi, Y.E. , So, J. , Wilkinson, H.H. and Lee, Y.W. (2006) FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum . Mol. Plant–Microbe Interact. 19, 725–733. [DOI] [PubMed] [Google Scholar]
- Shoji, J.Y. , Arioka, M. and Kitamoto, K. (2006) Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae . Eukaryot. Cell, 5, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. , Seo, Y.S. , Min, K. , Park, A.R. , Lee, J. , Jin, J.M. , Lin, Y. , Cao, P. , Hong, S.Y. , Kim, E.K. , Lee, S.H. , Cho, A. , Lee, S. , Kim, M.G. , Kim, Y. , Kim, J.E. , Kim, J.C. , Choi, G.J. , Yun, S.H. , Lim, J.Y. , Kim, M. , Lee, Y.H. , Choi, Y.D. and Lee, Y.W. (2011) A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog. 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.W. , Dou, X.Y. , Qi, Z.Q. , Wang, Q. , Zhang, X. , Zhang, H.F. , Guo, M. , Dong, S.M. , Zhang, Z.G. , Wang, P. and Zheng, X.B. (2010) R‐SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae . PLoS ONE, 5, e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A. and Jones, E.W. (1998) Pth1/Vam3p is the syntaxin homolog at the vacuolar membrane of Saccharomyces cerevisiae required for the delivery of vacuolar hydrolases. Genetics, 148, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R.B. , Fasshauer, D. , Jahn, R. and Brunger, A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Urban, M. , Mott, E. , Farley, T. and Hammond‐Kosack, K. (2003) The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4, 347–359. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Zhang, S. , Hou, R. , Zhao, Z. , Zheng, Q. , Xu, Q. , Zheng, D. , Wang, G. , Liu, H. , Gao, X. , Ma, J.W. , Kistler, H.C. , Kang, Z. and Xu, J.R. (2011) Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum . PLoS Pathog. 7, e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. , Shen, G. , Gong, J.J. , Shen, D.Y. , Whittington, A. , Qing, J. , Treloar, J.S. , Boisvert, S. , Zhang, Z.G. , Yang, C. and Wang, P. (2014) Noncanonical G beta Gib2 is a scaffolding protein promoting cAMP signaling through functions of Ras1 and Cac1 proteins in Cryptococcus neoformans . J. Biol. Chem. 289, 12 202–12 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich‐Soldner, R. , Bolker, M. , Kahmann, R. and Steinberg, G. (2000) A putative endosomal t‐SNARE links exo‐ and endocytosis in the phytopathogenic fungus Ustilago maydis . EMBO J. 19, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.Y. , Seo, J.A. , Kim, J.E. , Han, K.H. , Shim, W.B. , Yun, S.H. and Li, Y.W. (2008) Functional analyses of heterotrimeric G protein G alpha and G beta subunits in Gibberella zeae . Microbiology, 154, 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.W. and Lemmon, M.A. (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3‐phosphate. J. Biol. Chem. 276, 44 179–44 184. [DOI] [PubMed] [Google Scholar]
- Zhang, H.F. , Tang, W. , Liu, K.Y. , Huang, Q. , Zhang, X. , Yan, X. , Chen, Y. , Wang, J.S. , Qi, Z.Q. , Wang, Z.Y. , Zheng, X.B. , Wang, P. and Zhang, Z. (2011) Eight RGS and RGS‐like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae . PLoS Pathog. 7, e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.W. , Jia, L.J. , Zhang, Y. , Jiang, G. , Li, X. , Zhang, D. and Tang, W.H. (2012) In planta stage‐specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell, 24, 5159–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D. , Zhang, S. , Zhou, X. , Wang, C. , Xiang, P. , Zheng, Q. and Xu, J.R. (2012) The FgHOG1 pathway regulates hyphal growth, stress responses, and plant infection in Fusarium graminearum . PLoS ONE, 7, e49495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.Y. , Heyer, C. , Choi, Y.E. , Mehrabi, R. and Xu, J.R. (2010) The CID1 cyclin C‐like gene is important for plant infection in Fusarium graminearum . Fungal Genet. Biol. 47, 143–151. [DOI] [PubMed] [Google Scholar]
- Ziegler, J. and Facchini, P.J. (2008) Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59, 735–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic and structural analysis of FgVam7 and its homologues. (A) Phylogenetic analysis of FgVam7 and its homologues. The phylogenetic tree was drawn using Clustal W 1.83. ScVam7 (Saccharomyces cerevisiae, NP_011303.1), MoVam7 (Magnaporthe oryzae, XP_001907727.1), NcVam7 (Neurospora crassa, XP_957713.1), UmYup1 (Ustilago maydis, XP_761553.1) and AfVam7 (Aspergillus fumigatus, XP_749396.1). (B) Structural analysis of FgVam7 and its homologues. aa, amino acid; PX, PhoX homology domain; SNARE, Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor.
Fig. S2 Targeted deletion of the FgVAM7 gene in Fusarium graminearum. (A) Schematic diagram of the FgVAM7 gene and gene replacement construct. (B) Southern blots of EcoRI‐digested genomic DNA of the wild‐type PH‐1 and ΔFgvam7 mutant strains hybridized with the FgVAM7 and HPH probes, respectively.
Fig. S3 Functions of the SNARE (Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor) and PX (PhoX homology) domains of MoVam7 in conidiation and virulence of Magnaporthe oryzae. (A) Growth and colony morphology of the indicated strains on SDC (straw decoction and corn medium) plates. Photographs were taken after incubation at 28 °C for 7 days. (B) Conidiation assays of the indicated strains. Photographs were taken at 24 h post‐inoculation (hpi) induced under light. Error bars represent the standard deviation from three independent experiments and asterisks indicate significant difference (P < 0.01). (C) Pathogenicity assays of the indicated strains on detached barley leaves. Photographs were taken at 5 days post‐inoculation. (D) Green fluorescent protein (GFP) signal observation in hyphae and conidia of different strains under confocal microscopy.
Table S1 Primers used in this study.


