Summary
Taxonomic status
Bacteria; Phylum Proteobacteria; Class Gammaproteobacteria; Order Xanthomonadales; Family Xanthomonadaceae; Genus Xanthomonas; Species Xanthomonas euvesicatoria, Xanthomonas vesicatoria, Xanthomonas perforans and Xanthomonas gardneri.
Microbiological properties
Gram‐negative, rod‐shaped bacterium, aerobic, motile, single polar flagellum.
Host range
Causes bacterial spot disease on plants belonging to the Solanaceae family, primarily tomato (Solanum lycopersicum), pepper (Capsicum annuum) and chilli peppers (Capsicum frutescens).
Disease symptoms
Necrotic lesions on all above‐ground plant parts.
Distribution
Worldwide distribution of X. euvesicatoria and X. vesicatoria on tomato and pepper; X. perforans and X. gardneri increasingly being isolated from the USA, Canada, South America, Africa and Europe.
A wide diversity within the bacterial spot disease complex, with an ability to cause disease at different temperatures, makes this pathogen group a worldwide threat to tomato and pepper production. Recent advances in genome analyses have revealed the evolution of the pathogen with a plethora of novel virulence factors. Current management strategies rely on the use of various chemical control strategies and sanitary measures to minimize pathogen spread through contaminated seed. Chemical control strategies have been a challenge because of resistance by the pathogen. Breeding programmes have been successful in developing commercial lines with hypersensitive and quantitative resistance. However, durability of resistance has been elusive. Recently, a transgenic approach has resulted in the development of tomato genotypes with significant levels of resistance and improved yield that hold promise. In this article, we discuss the current taxonomic status, distribution of the four species, knowledge of virulence factors, detection methods and strategies for disease control with possible directions for future research.
Keywords: bacterial spot, pepper, tomato, Xanthomonas
How the Taxonomy of Bacterial Spot Xanthomonads Has Changed over the Years
Bacterial spot was first discovered on tomato in South Africa in 1914, and named Bacterium vesicatorium by Doidge (1920). The disease was later identified in Indiana by Gardner and Kendrick (1921) and they were prepared to propose the name B. exitiosum, but deferred to Doidge's designation. The two strains varied noticeably in amylolytic activity, but were treated as one pathogen. Gardner and Kendrick (1923) later discovered that B. vesicatorium also caused a leaf spot of pepper (Capsicum annuum), but Higgins (1922) was the first to fully describe the disease. After its initial discovery, B. vesicatorium was thought to be the only causal agent of bacterial spot when it was reclassified several times to Pseudomonas vesicatoria, Phytomonas vesicatoria and Xanthomonas campestris pv. vesicatoria (Xcv) (Dye, 1978; Jones et al., 1998b; Stevens, 1925).
Bacterial spot strains were considered as one group until the mid‐1990s, when independent studies demonstrated that Xcv consisted of two genetically and phenotypically distinct groups (A and B) (Stall et al., 1994; Vauterin et al., 1995). These two groups are also diverse in terms of their fatty acid profiles, serology, carbon utilization assays and DNA restriction enzyme digestion profiles. Strains belonging to the same group had DNA homology values of >70% and <46% with strains in the other group (Stall et al., 1994). Therefore, Vauterin et al. (1995) proposed that the two groups should be reclassified as X. axonopodis pv. vesicatoria (A) and X. vesicatoria (B). Group A strains, unlike group B strains, are primarily negative for starch hydrolysis and pectolytic activity (Bouzar et al., 1996). More recently, two additional genetically distinct xanthomonads have been isolated from tomato. Šutic (1957) isolated an organism in 1953 in former Yugoslavia that he named Pseudomonas gardneri. Later, it was suggested to be synonymous with X. vesicatoria (Dye, 1966); however, results of DNA–DNA hybridization indicated that these organisms were genetically distinct (DeLey et al., 1978). A fourth genetically distinct pathogen was isolated from tomato in Florida in the 1990s and was determined to be a new race of Xcv (Jones et al., 1995, 2000). These two strains were later characterized as groups D (‘X. gardneri’) and C (X. axonopodis pv. vesicatoria), respectively, based on DNA–DNA hybridization, pathogenicity tests, 16S rRNA and restriction fragment analysis of the entire genome (Jones et al., 2000). Based on the results of DNA–DNA hybridization, Jones et al. (2004) determined that strains within groups C and D were distinct Xanthomonas species, and that bacterial spot is incited by four distinct species: X. euvesicatoria (A), X. vesicatoria (B), X. perforans (C) and X. gardneri (D).
However, the taxonomy for the bacterial spot pathogens may not be fully resolved. Young et al. (2008) conducted multilocus sequence analysis (MLSA) using four housekeeping genes (dnaK, fyuA, gyrB and rpoD), which revealed that X. gardneri appears to be synonymous with X. cynarae, which causes a bacterial spot disease on capitulum bracts of artichokes (Cynarascolymus) (Trébaol et al., 2001). Whole‐genome comparison indicated that X. gardneri and X. cynarae should be considered as a single species (M. Jacques and J. B. Jones, unpublished data). In addition, the MLSA results suggested a close phylogenetic relationship between X. perforans and X. euvesicatoria. Almeida et al. (2010) suggested the use of six housekeeping genes (fusA, gapA, gltA, gyrB, lacF and lepA) to distinguish between Xanthomonas species. Using these genes for the comparison of strains, multiple recombination events between X. euvesicatoria and X. perforans strains were observed (Timilsina et al., 2015). Therefore, the taxonomic position of these three bacterial spot species will be further evaluated and could change in the future.
Host Range and Race Structure
The host range of bacterial spot xanthomonads includes a wide range of plants belonging to the Solanaceae family, but mainly tomato (Solanum lycopersicum), cherry tomato (Solanum lycopersicum var. cerasiforme), currant tomato (L. pimpinellifolium), pepper (Capsicum annuum), chilli peppers (Capsicum frutescens), C. baccatum, C. anomalum, C. chinensis and C. pubescens (Baker et al., 2014; Sahin and Miller, 1998). Additional solanaceous hosts have been suggested (Dye et al., 1964; Elliott, 1951); however, at least one study has demonstrated that some of these other genera are not hosts for the bacterium (Laub and Stall, 1967).
As plant resistance was identified in pepper and tomato genotypes, pathogenic races were identified based on differential reactions. Currently, four tomato races and 11 pepper races have been identified (Stall et al., 2009). Among the four species, various pepper and tomato races of X. euvesicatoria, X. vesicatoria and X. perforans (tomato only) have been widely reported (Horvath et al., 2012; Jones et al., 1995; Kebede et al., 2014). Xanthomonas gardneri has also been associated with pepper and tomato bacterial spot from different regions of the world. In 2010, an X. perforans strain was isolated from a pepper field in Florida (J. B. Jones, unpublished data), suggesting recent host range expansion. Strains within X. euvesicatoria, X. vesicatoria and X. gardneri that are pathogenic only on tomato, only on pepper, and on both tomato and pepper have been identified. Avirulence (Avr) genes that restrict the strains to pepper or tomato include avrBs4 (resulting in a hypersensitive response on both tomato and C. pubescens) and avrBsT (resulting in a hypersensitive response on pepper) (Ballvora et al., 2001; Minsavage et al., 1990). With both Avr genes being plasmid borne, the loss of a plasmid allows a strain to become pathogenic on that host (Canteros et al., 1991; Minsavage et al., 1990). Apart from specific races, host–pathogen combination is important for pathogen aggressiveness, e.g. X. euvesicatoria was found to be more aggressive on pepper than on tomato (Ignjatov et al., 2010).
Distribution of Xanthomonads
Xanthomonas spp. are widely distributed in different geographical regions probably because of contaminated seed (Kebede et al., 2014). Two species, X. euvesicatoria and X. vesicatoria, historically have had a worldwide distribution and, more recently, the other two species have been increasingly isolated. Different species of bacterial spot‐causing xanthomonads have been isolated from tomato‐growing regions/countries (Bouzar et al., 1999; Jones et al., 2005; Hamza et al., 2010; 2012; Jibrin et al., 2014; Kebede et al., 2014; Kim et al., 2010; Quezado‐Duval et al., 2004; Stall, 1995; Stoyanova et al., 2014; Timilsina et al., 2015; Vauterin et al., 1995).
Changes in pathogen populations have been documented. In Florida, bacterial spot of tomato has changed considerably over the years, even though there has been no dramatic change in the tomato cultivars used in commercial production fields during the last decade. Prior to 1991, tomato race 1 (T1) strains of X. euvesicatoria were the only known causal agent of bacterial spot of tomato (Jones et al., 1995). A field survey in 1991 recovered a low number of tomato race 3 (T3) strains of X. perforans in Florida, with the T1 strains being the prevalent bacterial spot pathogen (Jones et al., 1995). The prevalence of X. perforans T3 strains increased from 1991 to 1994 (Jones et al., 1998a) as a probable result of inhibitory bacteriocins produced by X. perforans strains that targeted T1 strains of X. euvesicatoria (Hert et al., 2005). In the late 1990s, tomato race 4 (T4) strains of X. perforans were identified based on the differential reaction on Xv3 and RXopJ4 (Astua‐Monge et al., 2000a, b; Sharlach et al., 2013; Stall et al., 2009). A survey conducted in 2006 in different tomato production fields throughout Florida revealed a dramatic shift in pathogen populations, with 100% of the strains being X. perforans, 77% of which were T4 and the remainder were T3 (Horvath et al., 2012). A similar situation was reported for tomato production areas in Ohio and Michigan, with an increase in X. gardneri strains where X. euvesicatoria T1 was previously the prevalent bacterial species on tomato (Ma et al., 2011). In Canada, X. euvesicatoria and X. gardneri are the prevalent species and, recently, X. perforans strains have been isolated (Cuppels et al., 2006; Cândido et al., 2008). A recent study by Timilsina et al. (2015) has indicated an increase in the geographical distribution of X. gardneri.
Molecular characterization has revealed species diversity of bacterial spot xanthomonads in Europe, Asian countries, such as Taiwan and India, and African countries, such as Ethiopia, Nigeria, Tanzania and various islands in the southwest Indian Ocean region (Hamza et al., 2012; Kebede et al., 2014; Mbega et al., 2012; Stoyanova et al., 2014). Although there have been reports of bacterial spot from different parts of the world, in many cases, placement into the actual species has not been determined. A map showing the distribution of xanthomonads associated with bacterial spot is presented in Fig. 1.
Figure 1.
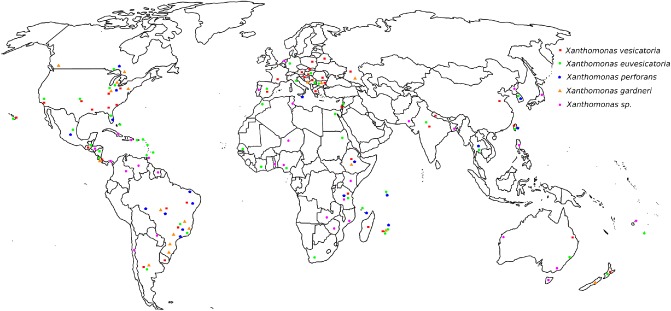
Current worldwide distribution of Xanthomonas causing bacterial spot of tomato and pepper.
Epidemiology
Seed can serve as an important inoculum source, especially in seedling production facilities, where high temperatures, high relative humidity, high plant density and overhead irrigation create an ideal environment for disease development and spread. It has been speculated that even a low incidence of seed transmission can introduce exotic strains (Pohronezny et al., 1992). Xanthomonads can also survive epiphytically in the tomato phyllosphere. Epiphytic populations in latent infection under favourable conditions could be an important factor in the dissemination of a pathogen (McGuire et al., 1991). The spread of the pathogen on transplants as a result of latent infections or epiphytic populations has been considered as a major source of outbreaks. Epiphytic survival on weeds has been noted, although at low population levels (Jones et al., 1986). The bacterium can also persist in crop residue for extended periods of time. In more temperate climates, the bacteria generally survive in crop residue for less than 2 years, but, in tropical and subtropical regions, the bacterium survives only a few months (Jones et al., 1986). Bacteria can also survive on volunteer pepper/tomato plants, and in soil in association with crop residue. However, it cannot survive for more than several days in soil after crop residue decomposes (Jones et al., 1986).
Disease is favoured by various temperatures and by high precipitation, depending on the particular bacterial species (Araújo et al., 2010). Xanthomonas gardneri has been found primarily in cooler temperature regions (Jones et al., 1998b). In addition, one study has shown that X. gardneri caused more disease at 20 °C than did other bacterial spot xanthomonads (Araújo et al., 2010).
The bacteria are disseminated within a field by wind‐driven rain droplets, wounding caused during field operations (grafting, clipping, tying, harvesting, spraying of pesticides) and aerosols (Lindemann and Upper, 1985; McInnes et al., 1988). On immigration onto the leaf surface, the bacterium colonizes the phyllosphere, which has been visualized using confocal laser scanning microscopy (Potnis et al., 2014; Zhang et al., 2009). The bacterium can readily be observed in depressions between epidermal cells, around stomata (Fig. 2), trichomes, lenticels and hydathodes, through all of which the bacterium penetrates the host. Zhang et al. (2009) observed low‐level type III secretion system (T3SS) gene (hrp, hypersensitive response and pathogenicity) expression on the leaf surface as early as 24 h post‐inoculation using an HrpG‐GFP (green fluorescent protein) reporter [HrpG is an important regulator of T3SSs as well as other systems, including the type II secretion system (T2SS)], where cells had not yet entered the leaf. This suggests that the HrpG regulon is not up‐regulated on the leaf surface on immigration prior to invasion. Once the bacterium reaches the stomata, a high level of Hrp expression is observed (Wilson et al., 2006; Zhang et al., 2009). The bacterial spot pathogen enters the leaf apoplast through the stomata, followed by growth in the substomatal chamber and multiplication in intercellular spaces (Fig. 3). The later stages of disease involve widespread cell death, tissue necrosis and egress of Xanthomonas to the leaf surface, increasing phyllosphere population levels (Fig. 3).
Figure 2.
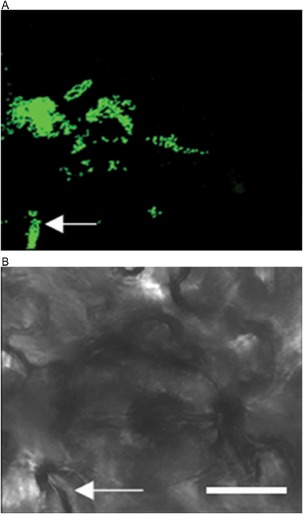
Colonization of bacterial spot Xanthomonas perforans in the tomato phyllosphere. On immigration to the leaf surface, X. perforans colonizes around the stomata (indicated by arrow) and in the grooves of the epidermal cells as seen in this confocal laser scanning micrograph. Five‐week‐old tomato plants were dip inoculated with 106 colony‐forming units (cfu)/mL of green fluorescent protein (GFP)‐labelled X. perforans. The images were captured on day 3 post‐inoculation. (A) GFP channel. (B) Bright field. White bar, 25 μm.
Figure 3.
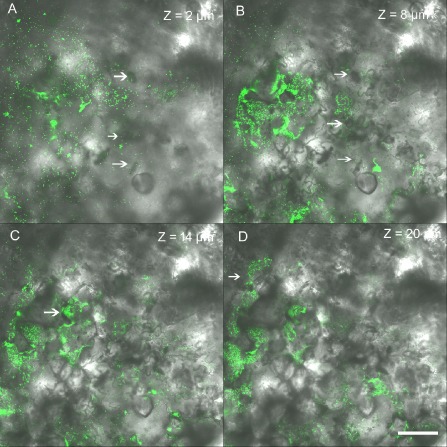
Colonization of bacterial spot Xanthomonas perforans in the tomato phyllosphere. Bacterium enters though stomata (A), followed by growth in the substomatal chamber (B) (indicated by arrow). Bacterium multiplies in the intercellular spaces (C and D). Representative photomicrographs showing green fluorescent protein (GFP)‐labelled virulent X. perforans aggregates that are part of the lesion along a Z stack of 20 μm overlaid with Nomarski differential interference contrast images (40× magnification). Z values represent the distance in micrometres from the abaxial (lower) leaf surface. White bar, 25 μm.
Disease Symptoms
Bacterial spot of tomato and pepper is characterized by necrotic lesions on the leaves, stems, petals and flowers, and fruit (Jones et al., 1991; Fig. 4A). During the initial stages of symptom development, circular water‐soaked lesions appear, which later dry and turn dark brown to black with a wet to greasy appearance (Vallad et al., 2004). Sometimes, halos are present around the spots. Primary lesions coalesce, resulting in extensive necrosis and a blighted appearance. The lesions may have a water‐soaked appearance during favourable conditions with rainy weather and when dew is present. In the case of X. perforans, lesions on the leaves often develop a shot‐hole appearance when conditions favour rapid bacterial growth (Stall et al., 2009, Fig. 4A). Lesions caused by X. gardneri have a characteristic water‐soaked appearance. On fruit, the lesions have a raised, scabby appearance (Fig. 4B). Xanthomonas euvesicatoria and X. vesicatoria have been associated with fruit lesions. For years, X. perforans has not been associated with fruit lesions although, recently, strains have been isolated from fruit (G. E. Vallad, University of Florida, unpublished data). Recently, X. gardneri has also been associated with deep, large fruit lesions that are problematic for the processing of tomatoes. Fruit lesions caused by X. gardneri begin as large, water‐soaked spots that become crusty and may take on a star‐shaped appearance as the epidermal tissue surrounding the lesion cracks (Ma et al., 2011; Miller, 2012). In pepper production fields where bacterial spot is prevalent, defoliation is commonly observed; furthermore, shedding of blossom and young fruit often occurs. Fruit quality is affected because of the presence of lesions and sunscald as a result of defoliation (Ritchie, 2000). A reduction in fruit quality and additional fruit loss caused by secondary post‐harvest pathogens can result in significant yield reductions.
Figure 4.

Bacterial spot disease symptoms on the leaves (A) and fruits (B) of pepper and tomato. On the leaves, dark brown to black lesions with a necrotic blighted appearance are seen. Classic shot‐hole lesions are observed on tomato leaves infected with Xanthomonas perforans (shown as enlarged image). Scabby and raised lesions are seen on the fruit.
Molecular Aspects of Host–Pathogen Interaction
Bacterial spot xanthomonads possess a collection of virulence factors that are involved in different stages of disease development. On contact with host surfaces, external signals activate two‐component signalling cascades in xanthomonads. A few of these, such as RpfC/RpfG (quorum sensing), RavS/RavR (He and Zhang, 2008; He et al., 2009), CorS/CorR (Qian et al., 2008a, b) and PhoPQ (Lee et al., 2008) have been studied for their role in virulence in xanthomonads (Qian et al., 2008a). The HrpG/HrpX response regulator system, which controls the activation of T2SS and T3SS, has been very well studied in X. euvesicatoria (Koebnik et al., 2006). These systems orchestrate virulence gene expression.
The Hrp cluster encoding the T3SS of Xanthomonas was first characterized in X. euvesicatoria (Bonas et al., 1991). The protein effectors that are secreted and translocated by T3SS into the host cell are determinants of pathogenicity and host range for the pathogen. Bacterial spot xanthomonads contain at least 45 effectors based on computational analysis, a few of which have been experimentally confirmed to be secreted by T3SS (Potnis et al., 2011) and studied for their role in disease development. Core effectors that play essential roles at different stages of disease development include AvrBs2, XopD, XopF1, XopK, XopL, XopN, XopQ, XopR, XopX, XopZ1 and XopAD. Some of these core effectors, XopD, XopL and XopN, are virulence factors that interfere with pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) (Kim et al., 2009, 2013; Singer et al., 2013). PTI is activated early in response to infection and is a major contributor in restricting the growth of a pathogen. XopN suppresses PTI responses by interacting directly with tomato 14‐3‐3 isoform TFT1 and TARK1 (tomato atypical receptor‐like kinase), and influences symptom development and pathogen growth. XopN mutants of X. euvesicatoria display attenuated growth in planta as well as impaired symptom development (Kim et al., 2009). The effector AvrBs2 contributes to the virulence and fitness of the pathogen (Gassmann et al., 2000; Kearney and Staskawicz, 1990). Two other core effectors, XopX and XopZ, have been identified as virulence factors in other xanthomonads (Kim et al., 2009; Metz et al., 2005).
Several plant pathogen effectors are known to exhibit mimicry of host proteins to interfere with eukaryotic cellular processes. Examples include XopD, XopL and AvrBs3. In X. euvesicatoria, XopD mimics enzymes of the small ubiquitin‐related modifier (SUMO) pathway. This effector is a SUMO protease that suppresses host transcription, promotes pathogen growth and suppresses host defence responses at the late stages of tissue colonization. XopD suppresses ethylene levels by desumoylating the ethylene‐responsive transcription factor, SIERF4 (Kim et al., 2013). Another effector, XopL, exhibits E3 ubiquitin ligase activity and contains a novel structural fold that interacts with plant ubiquitination system components (Singer et al., 2013) and suppresses PTI. It interferes with ubiquitination, a key eukaryotic cell function, with an XL box and leucine‐rich repeat (LRR) domain, and subverts plant immunity. AvrBs3, which belongs to the transcription activator‐like (TAL) effector family, mimics a host transcription activator by binding and activating upa20, a master cell size regulator (Kay et al., 2007). A homologue of avrBs3, avrHah1, which is found in X. gardneri, is responsible for enhanced water‐soaking phenotype and virulence (Schornack et al., 2008). These TAL effectors have been well studied for their contribution to virulence by activating host genes, resulting in pathogen spread and disease (Marois et al., 2002).
Comparative genomic analyses of the four Xanthomonas spp. associated with bacterial spot have revealed unique variable effectors. These four Xanthomonas spp. all contain XopJ family effectors (i.e. avrBsT, xopJ, avrXv4 and avrRxv) that might be redundant in function. Xanthomonas euvesicatoria contains two effectors belonging to the XopJ family, xopJ1 and xopJ3 (avrRxv). Xanthomonas vesicatoria and X. perforans possess avrBsT and avrXv4, respectively; recently collected X. perforans contains both effectors, whereas X. gardneri has none (Potnis et al., 2011; Thieme et al., 2005). The effector AvrXv4 is involved in PTI suppression, whereas the effector avrBsT encodes acetyl transferase that targets microtubule‐associated protein, interferes with lipid signalling and activates the hypersensitive response. AvrBsT has also been shown to interfere with effector‐triggered immunity activated by AvrBs1 through an SNF1‐related kinase (Szczesny et al., 2010). Effector XopJ from X. euvesicatoria is involved in the inhibition of protein secretion by targeting vesicle trafficking, thereby interfering with cell wall‐based defences (Bartetzko et al., 2009). Xanthomonas vesicatoria and X. gardneri may have acquired unique effectors via horizontal gene transfer, possibly from other Xanthomonas species or other genera. Next‐generation sequencing analyses have revealed that the two tomato pathogens, X. gardneri and Pseudomonas syringae pv. tomato, which both prefer lower optimal temperature for infection (Araújo et al., 2010), share a number of type III effectors, including xopAO, xopAS and avrBs1, which are absent from other bacterial spot xanthomonads. The acquisition of these effectors in X. gardneri might be one of the factors causing the unusual aggressiveness of these strains on tomato (Potnis et al., 2011). Interestingly X. gardneri is more closely related to X. campestris pv. campestris, a weed and cruciferous pathogen, based on whole‐genome phylogeny, as well as phylogeny based on important pathogenicity clusters (Potnis et al., 2011), suggesting the possible evolution of this tomato pathogen from wild populations found on weeds or the plant‐associated environment.
Cell wall‐degrading enzymes, such as cellulases, polygalacturonases, xylanases and proteases, are secreted by the T2SS. The role of the T2SS in contributing to virulence has been studied in X. euvesicatoria. Xylanases and proteases, under the control of HrpG and HrpX, are secreted via the T2SS Xps system (Szczesny et al., 2010). Another T2SS cluster, known as the Xcs system, does not contribute to virulence, but can partially complement xps mutants. Substrate specificities for T2SS differ among xanthomonads (Szczesny et al., 2010), possibly based on the preference of host tissues.
The lipopolysaccharide (LPS) biosynthesis cluster, defined by a 14–26‐kb region flanked by etfA and metB, is variable in terms of identity and size across the sequenced xanthomonads (Patil et al., 2007). However, in a comparative study for this cluster among sequenced xanthomonads, variation in LPS cluster was not associated with host or tissue specificity (Lu et al., 2008). Interestingly, among bacterial spot xanthomonads, pepper pathogens possess an identical LPS cluster, whereas X. perforans has a hybrid LPS cluster, sharing similarity with X. citri. The possible role of LPS O‐antigen in pepper pathogenicity has been studied, although it contributes to pepper pathogenicity by increasing in planta growth when expressed with two other pepper pathogen‐specific genes, XCV1839 and XopG effector (Potnis et al., 2011). A secreted lipolytic enzyme encoded by lipA of X. euvesicatoria has been shown to be involved in virulence during interaction of the pathogen with tomato (Tamir‐Ariel et al., 2012).
Recently, a transcriptomics approach has been used in X. euvesicatoria 85‐10 to identify the transcriptional start sites and the small RNAs that could be involved in virulence. This study suggested a role for the sx12 small RNA in the virulence of X. euvesicatoria in pepper, proposing that it interfered with the complex interactions between the pathogen and the host (Schmidtke et al., 2012). More recently, a regulatory small RNA, sx13, has been shown to be involved in the regulation of HrpG/HrpX. The small RNA sx13 has also been shown to regulate motility and signal transduction pathways, thereby contributing to the virulence of X. euvesicatoria (Schmidtke et al., 2012).
Detection and Identification
The bacteria are readily isolated from infected tissue by streaking onto nutrient agar, sucrose peptone or yeast–dextrose–calcium carbonate (YDC) agar, and are characterized by yellow, mucoid and convex colonies (Doidge, 1921). The yellow colour is caused by the presence of xanthomonadins, a unique class of brominated, aryl‐polyene, water‐insoluble pigments present in the outer membrane (Stephens and Starr, 1963). The bacteria are straight, Gram‐negative rods with single polar flagella. Various biochemical and physiological tests have been used to characterize these bacteria. The bacteria are catalase positive and oxidase negative, and can be identified to species by various molecular techniques. During the reclassification of Xanthomonas species, Jones et al. (2004) used several techniques other than DNA sequencing to characterize the species. These techniques included the determination of amylolytic and pectolytic activity, protein patterns with sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS‐PAGE) (Bouzar et al., 1994), serological reactions with a panel of monoclonal antibodies using an enzyme‐linked immunosorbent assay (Bouzar et al., 1994) and pulsed‐field gel electrophoresis patterns (Jones et al., 2000). Strains of X. euvesicatoria and X. gardneri are weakly amylolytic and pectolytic, whereas X. vesicatoria and X. perforans strains show strong amylolytic and pectolytic activity (Jones et al., 2004). All four species exhibit distinct reaction patterns with the selected monoclonal antibodies, together with distinct protein profiles (Jones et al., 2004). A 25–27‐kDa protein was present in three Xanthomonas species, except for X. euvesicatoria strains. In addition, Xanthomonas strains can also be differentiated by inoculation onto race‐specific resistant tomato and pepper genotypes. Moreover, various research studies on the diagnosis and identification of xanthomonads causing bacterial spot of pepper and tomato have been carried out. Kuflu and Cuppels (1997) developed a diagnostic DNA probe, but only for the diagnosis of X. euvesicatoria and X. vesicatoria. Later, Cuppels et al. (2006) developed PCR primer sets based on the probes that can distinguish between the four species of bacterial spot xanthomonads.
Researchers have evaluated many different methods to diagnose and identify the bacterial spot pathogens. Rademaker et al. (2000) compared the results of amplified fragment length polymorphism (AFLP) analysis with those of DNA–DNA hybridization involving 80 Xanthomonas strains belonging to 20 DNA–DNA homology groups. The AFLP analysis provided phylogenetic results that were comparable with the DNA–DNA hybridization studies. Therefore, AFLP analysis was useful in differentiating species of Xanthomonas, such as the bacterial spot pathogens. Koenraadt et al. (2009) designed polymerase chain reaction (PCR) primers specific to the bacterial spot pathogens based on unique banding patterns resulting from AFLP analysis. DNA was extracted from strains representing the four bacterial spot species and digested with restriction enzymes. Only DNA fragments that were present in every strain belonging to one bacterial species and not in any other species were selected for sequencing. This allowed for four species‐specific PCR primers to be designed. These primers were able to accurately amplify all of the tested bacterial spot strains. However, further validation of the primers for X. perforans and X. gardneri was necessary because the number of strains tested for these two species was limited. These primers are particularly useful in seed assays (Koenraadt et al., 2009). Araújo et al. (2012) further validated these primers using DNA extracted from 30 X. euvesicatoria, 30 X. vesicatoria, 50 X. gardneri and 50 X. perforans strains collected in Brazil. After validation, they designed a multiplex PCR assay involving Koenraadt's four species‐specific primer sets. This multiplex PCR assay effectively differentiates and identifies the tested Brazilian strains from the four bacterial spot species.
Since the mid‐1990s, researchers have explored the use of diagnostic methods based on regions of hrpB, specifically hrpB2, for the bacterial spot pathogens. hrpB is part of the hrp gene cluster (Bogdanove et al., 1996; Noël et al., 2002). In addition, hrpB2 is essential for type III protein secretion and pathogenicity (Rossier et al., 2000). Leite et al. (1994) were the first to explore a diagnostic method based on the hrp gene cluster. PCR primers based on different regions of the hrp gene cluster were created, and restriction enzyme analysis (REA) was used to detect 28 different X. campestris pathovars. Unique banding patterns were observed for different xanthomonads. In addition, this method was also effectively utilized to detect the bacterial spot pathogen Xcv from tomato and pepper seed washings (Leite et al., 1995). Obradovic et al. (2004b) expanded on this work by developing PCR primers designed to amplify a 420‐bp fragment of hrpB2 from the four Xanthomonas spp. associated with bacterial spot. Following digestion of the amplified PCR products with three restriction enzymes (CfoI, TaqI and HaeIII), each species produced unique gel patterns that were used for their identification. Strayer et al. (2014) also utilized hrpB2 by developing a multiplex quantitative PCR to simultaneously detect all four bacterial spot pathogens. Four species‐specific probes and two primer sets were created based on the hrpB2 sequence of each species. This method effectively and accurately identified pure cultures of 72 strains representing all four pathogens. It is an improvement over the other gel‐based detection techniques because it saves time and materials when processing large sample numbers.
As Xanthomonas spp. can be disseminated via contaminated seed and plant material, the detection of bacterial spot in tomato or pepper seeds and seedlings is critical for the reduction of potential inoculum sources. Bacterial spot pathogens can be isolated and detected in soil, seed and plant material via a semi‐selective culture medium called Tween. The pathogen's lipolytic enzymes utilize the fatty acid ester, Tween‐80, as a substrate, which causes the formation of a white crystalline halo or precipitate around yellow bacterial spot colonies (McGuire and Jones, 1989; McGuire et al., 1986). In addition, the International Seed Federation (International Seed Federation, 2011; 2013) describes similar protocols for the detection of Xanthomonas spp. in tomato and pepper seed. These protocols involve the use of a modified Tween Medium B (mTMB) in combination with another semi‐selective medium (CKTM) to isolate xanthomonads from seed (Sijam et al., 1991, 1992). Identification of the bacterial spot pathogens is completed on the basis of colony morphology on YDC and pathogenicity assays (ISF, 2011; 2013).
Integrated Pest Management of Bacterial Spot
Xanthomonas strains are either endemic or introduced into field production areas via infested seed or contaminated transplant material (Jones et al., 1986; Sijam et al., 1991). Thus, the primary management strategy should include the use of pathogen‐free, certified seed or disease‐free transplant material (Ritchie, 2000). Preventative measures, such as treating seed with hot water, can significantly reduce disease inoculum in seeds. In addition, removal of potential inoculum sources, such as volunteer plants and infected host plants, should be carried out on a timely basis. Field isolation from infected host plants in close proximity, accompanied by sanitation, physical removal and disposal of diseased crop material, and crop rotation with non‐hosts, should be followed for disease management (Goode and Sasser, 1980; Ritchie, 2000).
Chemical control has been a primary focus in pest management strategies for bacterial spot of tomato and pepper. Copper‐based bactericides have been used extensively for disease control. The grower standard for many years has been copper‐based bactericides in combination with mancozeb or maneb (ethylene‐bis‐dithiocarbamates) (Conover and Gerhold, 1981; Marco and Stall, 1983). Copper‐based bactericides became the standard in part because of the emergence of streptomycin resistance in xanthomonads in the 1960s (Thayer and Stall, 1961). Unfortunately, copper‐tolerant xanthomonads have been present since the 1960s and were described by Marco and Stall (1983). In the 1980s, it was observed that copper–mancozeb combinations resulted in improved bacterial spot control because of the increased release of soluble copper when compared with copper alone (Conover and Gerhold, 1981; Marco and Stall, 1983). Therefore, it became the grower standard. However, disease control with copper–mancozeb is not effective when copper‐tolerant strains are present together with optimal weather conditions for disease development (Jones and Jones, 1985; Obradovic et al., 2004a).
As indicated, disease management strategies using chemicals composed of metal ions such as copper have been explored. More recently, researchers have evaluated the use of different types of nanomaterials to control bacterial spot of tomato caused by X. perforans. Paret et al. (2013) evaluated a nanoscale version of the light‐activated molecule TiO2. TiO2 was evaluated alone or doped with either silver (TiO2/Ag) or zinc (TiO2/Zn). TiO2/Ag and TiO2/Zn significantly reduced the bacterial populations in vitro. In glasshouse and field trials, TiO2/Zn‐treated plants showed significantly reduced disease severity when compared with those treated with copper–mancozeb. Unfortunately, phytotoxicity was observed, which limits the commercialization of this product. In addition, Ocsoy et al. (2013) developed a silver‐based nanocomposite, termed Ag‐dsDNA‐GO. In this composite, silver ions accumulate in the major groove of dsDNA attached to graphene oxide (GO). Ag‐dsDNA‐GO showed high antibacterial activity at low concentrations (16 ppm) in vitro. In this study, Ag‐dsDNA‐GO at 100 ppm provided bacterial spot control at levels comparable with those of copper–mancozeb‐treated plants. This was also confirmed in multiple glasshouse studies conducted by A. Strayer et al. (University of Florida, unpublished data). However, these results need to be further validated in field trials before this composite can be considered as a feasible alternative.
The plant activator Acibenzolar‐S‐methyl (ASM) (Actigard 50WG, Syngenta Crop Protection, Greensboro, NC, USA), which activates systemic acquired resistance (SAR), has been extensively studied. Although ASM treatment for the first 6 weeks reduced disease symptoms, application throughout the season at weekly intervals was not efficient (Roberts et al., 2008). Effective disease control was reported when ASM was applied in combination with other biological and chemical control measures (Graves and Alexander, 2002; Obradovic et al., 2005), and has been suggested as a possible alternative to excessive reliance on copper bactericides for rotation (Louws et al., 2001). In addition, ASM is not recommended for a full season application because of its effects on the plant and associated decreased yield (Graves and Alexander, 2002; Romero et al., 2001). It has been suggested that there is a fitness cost in expressing SAR by plants in the absence of a pathogen (Louws et al., 2001). Recent field studies examining ASM application rates and frequency have led to improved usage patterns to maximize disease control, whilst minimizing the impact on plant yields (Huang et al., 2012).
Antibiotics and molecular additives have also been evaluated for bacterial spot disease control. Streptomycin and kasugamycin have been labelled for use against bacterial spot in the glasshouse. Streptomycin‐resistant xanthomonads were reported very early after extensive use in the field (Thayer and Stall, 1962). A comparative study showed that kasugamycin was as effective as standard copper–mancozeb treatment for the control of bacterial spot (Vallad et al., 2010). It also significantly increased the total marketable yield of tomato when used together with other bactericides, such as Kocide (Ivors et al., 2006). Worthington et al. (2012) examined the use of a small molecule additive, 2‐aminoimidazole (2AI), in combination with copper for the management of bacterial spot. It is an analogue of the marine sponge natural product oroidin and lacks bactericidal activity alone. In vitro, 2AI, in combination with copper, was shown to suppress copper resistance in X. euvesicatoria and decreased biofilm formation. In field experiments, this combination reduced bacterial spot disease and increased bell pepper fruit yields.
Efforts to identify biological control strategies for the control of bacterial spot have been extensive. Bacteriophages have been widely studied for the control of bacterial spot disease in tomato and pepper and other crops (Fig. 5; Balogh et al., 2003). Foliar applications of bacteriophage have provided effective control of disease when compared with a copper–mancozeb standard (Flaherty et al., 2000; Momol et al., 2008; Obradovic et al., 2004a). However, the efficacy of bacteriophage depends on the environmental factors that determine phage survival in plants (Iriarte et al., 2007). Other biological agents have shown limited efficacy in controlling the disease when applied alone, but research has begun to focus on improved integration into disease management programmes (Fravel, 2005). During an evaluation of 50 different biological agents, Cellulomonas turbata BT1 showed the highest reduction in disease severity and Pseudomonas syringae Cit7 gave the most consistent control (Byrne et al., 2005). In addition to these two bacterial species, various plant growth‐promoting rhizobacteria (PGPRs) and Bacillus pumilis have shown effectiveness in suppressing bacterial spot in some field trials (Ji et al., 2006).
Figure 5.
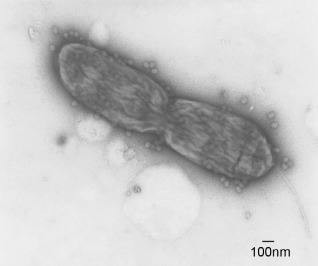
Foliar applications of bacteriophage have provided effective control of bacterial spot disease on tomato and pepper. The figure shows bacteriophage of X. vesicatoria attached to dividing cell of Xanthomonas. Photograph courtesy of Dr Botond Balogh, Nichino America, Inc.
Host resistance has been a major goal of pepper and tomato breeding programmes. Conventional resistance (R) gene‐mediated bacterial spot resistance relies on known Avr gene–R gene interactions. However, genetic resistance may be ineffective because of shifts in the bacterial populations that emerge even before resistant cultivars are deployed (Gassmann et al., 2000). These race shifts may impact the durability of plant resistance. Quantitative or multigenic resistance to bacterial spot has also been identified, and appears to be more durable than plant resistance associated with hypersensitive reactions.
Six dominant R genes have been identified in pepper: Bs1; Bs2; Bs3 (Stall et al., 2009); Bs4C (Strauß et al., 2012); Bs7 (Potnis et al., 2012); and BsT (Minsavage et al., 1990). They interact with avrBs1, avrBs2, avrBs3, avrBs4C, avrBs7 and avrBsT, respectively, to provide a hypersensitive type of resistance. Although other R genes have been identified using classical Avr–R gene studies, Bs4C, present in pepper (C. pubescens PI235047), was identified on the basis of transcriptome profiling with next‐generation sequencing (RNA‐seq), and was expressed only in the presence of avrBs4 (Strauß et al., 2012). The Bs4C gene was determined using bioinformatics to encode a structurally unique R protein. Among quantitative resistances, various levels of resistance to bacterial spot have been found in pepper accessions with two to three recessive R genes (Jones et al., 2002; Riva et al., 2004). Efforts to transfer these quantitative resistances from various PI accessions into commercial varieties led to backcross populations with various levels of resistance; additional intercrossing of plants specifically from PI163192 and PI271322 backcross lines resulted in a level of resistance higher than either parent under field conditions (Jones et al., 2002). This additive resistance was found to be governed by the recessive genes bs5 and bs6. Several seed companies are currently incorporating these recessive genes into the commercial cultivars (Stall et al., 2009).
Four sources of resistance have been identified in tomato: Hawaii 7998 derived Rx1, Rx2 and Rx3 (Yang et al., 2005); Xv3 (Astua‐Monge et al., 2000a); RXopJ4 (Sharlach et al., 2013); and Bs4 (Bonas et al., 1993). These four sources interact with AvrRxv (Whalen et al., 1993), AvrRxv, AvrXv3, XopJ4 and AvrBs4, respectively, to elicit a hypersensitive response. These tomato resistance genes were first identified in Hawaii 7998 (H7998), Hawaii 7981 (H7981) and S. pimpinellifolium accession PI128216, respectively (Wang et al., 2011; Whalen et al., 1993; Yang and Francis, 2007). Another approach in tomato has been to identify genes that confer resistance to multiple bacterial spot races. Two loci, QTL‐11 (from H7998, on chromosome 11) and QTL‐3 (from PI114490), are associated with resistance to multiple races (Hutton et al., 2010; Yang et al., 2005). Quantitative resistance to bacterial spot in tomato lines has been found to various degrees and against various races. PI114490 has shown resistance against races T1, T2 and T3, which represent three bacterial spot species; therefore, there is considerable interest to introgress this quantitative resistance into commercial lines (Scott et al., 2006).
Transgenic resistance offers another promising means for the control of bacterial spot. Recent technologies, such as TALEN‐based genome editing, hold the potential to engineer disease resistance given the well‐studied interactions of AvrBs3‐like proteins with pepper Bs3 alleles (Romer et al., 2009). Transgenic tomato plants expressing the Arabidopsis pattern recognition receptor (PRR) EF‐Tu receptor (EFR), a class of immune receptor, conferred resistance to X. perforans strains (Lacombe et al., 2010). The search for a core effector essential to pathogen virulence/fitness and its corresponding plant target led to the deployment of the Bs2 gene from pepper, a very close relative of tomato that interacts in a gene‐for‐gene manner, with the AvrBs2 effector, and resistance against bacterial spot was observed in tomato expressing Bs2 originally cloned from the pepper gene (Tai et al., 1999). Recently, a tomato transgenic Bs2 overexpression line expressed in VF 36, and designated VF 36‐BS2, and other transgenic lines carrying Bs2, have been tested for durable resistance in field trials against bacterial spot pathogens. These lines conferred a high level of resistance against field populations of X. perforans in Florida, under high disease pressure, in the absence of copper pesticides. In addition, these transgenic lines had double the fruit yields of those lines lacking the Bs2 transgene (Horvath et al., 2012). However, some rare mutant bacterial strains with mutations in avrBs2 were isolated that could overcome the resistance by losing the ability to be recognized by the Bs2 transgene. In the future, transgenic plants with multiple targets for such core effectors might reduce the rate of mutations (Dangl et al., 2013).
Future Prospects
Diversity among the bacterial spot xanthomonads has been observed in various studies. Given this diversity, very little is known about the pathogenicity factors of the strains from a worldwide collection. Cost‐effective, next‐generation sequencing methods would be useful in providing a genetic snapshot of these diverse strains from different parts of the world. Recent MLSA studies have indicated differences within a single species. A genome of a single strain cannot provide the correct representation of an entire species and, in some cases, not even of a regional population; therefore, sequencing many bacterial spot strains would also help in better defining the core virulence factors of bacterial spot xanthomonads. Finding targets for these core factors would offer candidates for the development of transgenic varieties. Current efforts for control of the disease have pointed towards several strategies that could be employed in conjunction. Rotation and a combination of various chemical bactericides have been suggested in response to the current expansion of chemically tolerant bacterial spot populations. The identification of potential new bactericides or chemical enhancers to manage bacterial spot disease is equally important. Nanoparticles, such as TiO2 and Ag‐dsDNA‐GO, appear to have the potential to effectively manage bacterial spot disease. The nano‐form of other bactericidal heavy metals, such as copper, could be an area to explore in the future.
Acknowledgement
The authors would like to thank Dr Dave Ritchie for providing photographs of diseased pepper plants and helpful suggestions.
References
- Almeida, N.F. , Yan, S. , Cai, R. , Clarke, C.R. , Morris, C.E. , Schaad, N.W. , Schuenzel, E.L. , Lacy, G.H. , Sun, X. and Jones, J.B. (2010) PAMDB, a multilocus sequence typing and analysis database and website for plant‐associated microbes. Phytopathology, 100, 208–215. [DOI] [PubMed] [Google Scholar]
- Araújo, E.R. , Pereira, R.C. , Moita, A.W. , Ferreira, M.A.S.V. , Café‐Fiho, A.C. and Quezado‐Duval, A.M. (2010) Effect of temperature on pathogenicity components of tomato bacterial spot and competition between Xanthomonas perforans and X. gardneri. III International symposium on tomato diseases. Acta Hort. 914, 39–42. [Google Scholar]
- Araújo, E.R. , Costa, J.R. , Ferreira, M.A.S.V. and Quezado‐Duval, A.M. (2012) Simultaneous detection and identification of the Xanthomonas species complex associated with tomato bacterial spot using species‐specific primers and multiplex PCR. J. Appl. Microbiol. 113, 1479–1490. [DOI] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Minsavage, G.V. , Stall, R.E. , Davis, M.J. , Bonas, U. and Jones, J.B. (2000a) Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant‐inducible avirulence gene. Mol. Plant–Microbe Interact. 13, 911–921. [DOI] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Minsavage, G.V. , Stall, R.E. , Vallejos, C.E. , Davis, M.J. and Jones, J.B. (2000b) Xv4–vrxv4: a new gene‐for‐gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii . Mol. Plant–Microbe Interact. 13, 1346–1355. [DOI] [PubMed] [Google Scholar]
- Baker, R. , Bragard, C. , Candresse, T. and van der Wolf, J. (2014) Scientific Opinion on the pest categorisation of Xanthomonas campestris pv. vesicatoria (Doidge) Dye. EFSA J. 12(6), 3720. [Google Scholar]
- Ballvora, A. , Pierre, M. , van den Ackerveken, G. , Schronack, S. , Rossier, O. , Ganal, M. , Lahaye, T. and Bonas, U. (2001) Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol. Plant–Microbe Interact. 14, 629–638. [DOI] [PubMed] [Google Scholar]
- Balogh, B. , Jones, J. , Momol, M. , Olson, S. , Obradovic, A. , King, P. and Jackson, L.E. et al (2003) Improved efficacy of newly formulated bacteriophages for management of bacterial spot on tomato. Plant Dis. 87, 949–954. [DOI] [PubMed] [Google Scholar]
- Bartetzko, V. , Sonnewald, S. , Vogel, F. , Hartner, K. , Stadler, R. , Hammes, U. and Bornke, F. (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall‐associated defense responses. Mol. Plant–Microbe Interact. 22, 655–664. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Beer, S.V. , Bonas, U. , Boucher, C.A. , Collmer, A. , Coplin, D.L. , Cornelis, G.R. , Huang, H.C. , Hutchenson, S.W. , Panopoulos, N.J. and Van Gijsegem, F. (1996) Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20, 681–683. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Schulte, R. , Fenselau, S. , Minsavage, G.V. , Staskawicz, B.J. and Stall, R.E. (1991) Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant–Microbe Interact. 4, 81–88. [Google Scholar]
- Bonas, U. , Conrads‐Stauch, J. and Balbo, I. (1993) Resistance in tomato to Xanthomonas campestris pv vesicatoria is determined by alleles of the pepper‐specific avirulence gene avrBs3. Mol. Gen. Genet. 238, 261–269. [DOI] [PubMed] [Google Scholar]
- Bouzar, H. , Jones, J.B. , Minsavage, G.V. , Stall, R.E. and Scott, J.W. (1994) Proteins unique to phenotypically distinct groups of Xanthomonas campestris pv. vesicatoria revealed by silver staining. Phytopathology, 84, 39–44 [Google Scholar]
- Bouzar, H. , Jones, J.B. , Somodi, G.C. , Stall, R.E. , Daouzli, N. , Lambe, R.C. , R. Felix Gastelum and R. Trinidad Correa (1996) Diversity of Xanthomonas campestris pv. vesicatoria in tomato and pepper fields of Mexico. Can. J. Plant Pathol. 18, 75–77. [Google Scholar]
- Bouzar, H. , Jones, J.B. , Stall, R.E. , Louws, F.J. , Schneider, M. , Rademaker, J.L.W. , de Bruijn, F.J. and Jackson, L.E. (1999) Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: evidence for common lineages within and between countries. Phytopathology, 89, 328–335. [DOI] [PubMed] [Google Scholar]
- Byrne, J. , Dianese, A. , Ji, P. , Campbell, H. , Cuppels, D. , Louws, F. et al (2005) Biological control of bacterial spot of tomato under field conditions at several locations in North America. Biol. Control, 32, 408–418. [Google Scholar]
- Cândido, E.S. , Pereira, J.L. , Quezado‐Duval, A. , Noronha, E. , Krüger, R.H. and Quirino, B.F. (2008) Xanthomonas gardneri exoenzymatic activity towards plant tissue. World J. Microbiol. Biotechnol. 24, 163–170 [Google Scholar]
- Canteros, B. , Minsavage, G. , Bonas, U. , Pring, D. and Stall, R. (1991) A gene from Xanthomonas campestris pv. vesicatoria that determines avirulence in tomato is related to avrBs3. Mol. Plant–Microbe Interact. 4, 628–632. [DOI] [PubMed] [Google Scholar]
- Conover, R.A. and Gerhold, N.R. (1981) Mixtures of copper and maneb or mancozeb for control of bacterial spot of tomato and their compatibility for control of fungus diseases [Phytophthora infestans, Stemphylium solani, Xanthomonas campestris pv. vesicatoria, Florida]. Proc. Fla. State Hort. Soc. 94, 154–156. [Google Scholar]
- Cuppels, D. , Louws, F.J. and Ainsworth, T. (2006) Development and evaluation of PCRbased diagnostic assays for the bacterial speck and bacterial spot pathogens of tomato. Plant Dis. 90, 451–458. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLey, J. (1978) Modern molecular methods in bacterial taxonomy: evaluation, application, prospects. Proc. Int. Conf. Plant Pathol. Bacteriol . 4th Angers, pp. 347–357.
- Doidge, E.M. (1920) A tomato canker. J. Dep. Agric. Union S. Afr. 1, 718–721. [Google Scholar]
- Doidge, E.M. (1921) A tomato canker. Ann. Appl. Biol. 7, 407–430. [Google Scholar]
- Dye, D.W. (1966) Cultural and biochemical reaction of additional Xanthomonas species. N. Z. J. Sci. 9, 913–919. [Google Scholar]
- Dye, D.W. (1978) Genus IX. Xanthomonas. Dowson 1939. N. Z. J. Agric. Res. 21, 153–177. [Google Scholar]
- Dye, D.W. , Starr, M.P. and Stolp, H. (1964) Taxonomic clarification of Xanthomonas vesicatoria based upon host specificity, bacteriophage sensitivity, and cultural characteristics. Phytopathol. Z. 51, 394–407. [Google Scholar]
- Elliott, C. (1951) Manual of Bacterial Plant Pathogens. Waltham, MA: Chronica Botanica, 186. [Google Scholar]
- Flaherty, J.E. , Jones, J.B. , Harbaugh, B.K. , Somodi, G.C. and Jackson, L.E. (2000) Control of bacterial spot on tomato in the greenhouse and field with H‐mutant bacteriophages. HortScience, 35, 882–884. [Google Scholar]
- Fravel, D. (2005) Commercialization and implementation of biocontrol 1. Annu. Rev. Phytopathol. 43, 337–359. [DOI] [PubMed] [Google Scholar]
- Gardner, M.W. and Kendrick, J.B. (1921) Bacterial spot of tomato. J. Agric. Res. 21, 123–156. [Google Scholar]
- Gardner, M.W. and Kendrick, J.B. (1923) Bacterial spot of tomato and pepper. Phytopathology, 13, 307–315. [Google Scholar]
- Gassmann, W. , Dahlbeck, D. , Cjesnokova, O. , Minsavage, G.V. , Jones, J.B. and Staskawicz, B.J. (2000) Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 182, 7053–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, M.J. and Sasser, M. (1980) Prevention—the key to controlling bacterial spot and bacterial speck of tomato. Plant Dis. 64, 831–834. [Google Scholar]
- Graves, A. and Alexander, S. (2002) Managing bacterial speck and spot of tomato with acibenzolar‐S‐methyl in Virginia. Plant Health Prog. doi: 10.1094/PHP-2002-0220-01-RS. [DOI] [Google Scholar]
- Hamza, A. , Robene‐Soustrade, I. , Jouen, E. , Gagnevin, L. , Lefeuvre, P. , Chiroleu, F. et al (2010) Genetic and pathological diversity among Xanthomonas strains responsible for bacterial spot on tomato and pepper in the southwest Indian Ocean region. Plant Dis. 94, 993–999. [DOI] [PubMed] [Google Scholar]
- Hamza, A. , Robene‐Soustrade, I. , Jouen, E. , Lefeuvre, P. , Chiroleu, F. , Fisher‐Le Saux, M. et al (2012) MultiLocus Sequence Analysis‐ and Amplified Fragment Length Polymorphism‐based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst. Appl. Microbiol. 35, 183–190. [DOI] [PubMed] [Google Scholar]
- He, Y.W. and Zhang, L.‐H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiol. Rev. 32, 842–857. [DOI] [PubMed] [Google Scholar]
- He, Y.‐W. , Boon, C. , Zhou, L. and Zhang, L.‐H. (2009) Co‐regulation of Xanthomonas campestris virulence by quorum sensing and a novel two‐component regulatory system RavS/RavR. Mol. Microbiol. 71, 1464–1476. [DOI] [PubMed] [Google Scholar]
- Hert, A. , Roberts, P. , Momol, M. , Minsavage, G. , Tudor‐Nelson, S. and Jones, J. (2005) Relative importance of bacteriocin‐like genes in antagonism of Xanthomonas perforans tomato race 3 to Xanthomonas euvesicatoria tomato race 1 strains. Appl. Environ. Microbiol. 71, 3581–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, B.B. (1922) The bacterial spot of pepper. Phytopathology, 12, 501–517. [Google Scholar]
- Horvath, D.M. , Stall, R.E. , Jones, J.B. , Pauly, M.H. , Vallad, G.E. , Dahlbeck, D. et al (2012) Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE, 7, e42036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.‐H. , Vallad, G.E. , Zhang, S. , Wen, A. , Balogh, B. , Figueiredo, J.F.L. , Behlau, F. , Jones, J.B. , Momol, M.T. and Olson, S.M. (2012) Effect of application frequency and reduced rates of acibenzolar‐S‐methyl on the field efficacy of induced resistance against bacterial spot on tomato. Plant Dis. 96, 221–227. [DOI] [PubMed] [Google Scholar]
- Hutton, S.F. , Scott, J.W. , Yang, W. , Sim, S.‐C. , Francis, D.M. and Jones, J.B. (2010) Identification of QTL associated with resistance to bacterial spot race T4 in tomato. Theor. Appl. Genet. 121, 1275–1287. [DOI] [PubMed] [Google Scholar]
- Ignjatov, M. , Gasić, K. , Ivanović, M. , Šević, M. , Obradović, A. and Milošević, M. (2010) Characterisation of Xanthomonas euvesicatoria strains pathogens of pepper in Serbia. Pesticidi i Fitomedicina, 25, 139–149. [Google Scholar]
- International Seed Federation (ISF) (2011) Methods for the detection of Xanthomonas spp. in tomato seed. Available at http://www.worldseed.org/isf/detection_methods.html [accessed 13 January 2015].
- International Seed Federation (ISF) (2013) Methods for the detection of Xanthomonas spp. in pepper seed. Available at http://www.worldseed.org/isf/detection_methods.html [accessed 13 January 2015].
- Iriarte, F. , Balogh, B. , Momol, M. , Smith, L. , Wilson, M. and Jones, J. (2007) Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl. Environ. Microbiol. 73, 1704–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivors, K. , Milks, D. and Holmberg, C. (2006) Evaluation of products for control of bacterial spot on tomato. In: 22nd Annual Tomato Disease Workshop, North Carolina State University, Fletcher, NC: p. 29. [Google Scholar]
- Ji, P. , Campbell, H. , Kloepper, J. , Jones, J. , Suslow, T. and Wilson, M. (2006) Integrated biological control of bacterial speck and spot of tomato under field conditions using foliar biological control agents and plant growth‐promoting rhizobacteria. Biol. Control, 36, 358–367. [Google Scholar]
- Jibrin, M.O. , Timilsina, S. , Potnis, N. , Minsavage, G. , Shenge, K.C. , Akpa, A. et al (2014) First report of Xanthomonas euvesicatoria causing bacterial spot disease in pepper in northwestern Nigeria. Plant Dis. 98, 1426. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. and Jones, J.P. (1985) The effect of bactericides, tank mixing time and spray schedule on bacterial leaf spot of tomato. Proc. Fla. State Hort. Soc. 98, 244–247. [Google Scholar]
- Jones, J. , Stall, R. , Scott, J. , Somodi, G. , Bouzar, H. and Hodge, N. (1995) A third tomato race of Xanthomonas campestris pv. vesicatoria . Plant Dis. 79, 395–398. [Google Scholar]
- Jones, J. , Bouzar, H. , Somodi, G. , Stall, R. , Pernezny, K. , El‐Morsy, G. et al (1998a) Evidence for the preemptive nature of tomato race 3 of Xanthomonas campestris pv. vesicatoria in Florida. Phytopathology, 88, 33–38. [DOI] [PubMed] [Google Scholar]
- Jones, J. , Stall, R. and Bouzar, H. (1998b) Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36, 41–58. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Pohronezny, K.L. , Stall, R.E. and Jones, J.P. (1986) Survival of Xanthomonas campestris pv. vesicatoria on tomato crop residue, weeds, seeds, and volunteer tomato plants. Phytopathology, 76, 430–434. [Google Scholar]
- Jones J.B., Jones J.P., Stall R.E. and Zitter T.A. (eds) (1991) Compendium of Tomato Diseases. St. Paul, MN: APS Press. [Google Scholar]
- Jones, J.B. , Bouzar, H. , Stall, R. , Almira, E. , Roberts, P. , Bowen, B. et al (2000) Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 50, 1211–1219. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Minsavage, G.V. , Roberts, P.D. , Johnson, R.R. , Kousik, C.S. , Subramanya, S. and Stall, R.E. (2002) A non‐hypersensitive resistance in pepper to the bacterial spot pathogen is associated with two recessive genes. Phytopathology, 92, 273–277. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Lacy, G.H. , Bouzar, H. , Stall, R.E. and Schaad, N.W. (2004) Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27, 755–762. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Lacy, G.H. , Bouzar, H. , Minsavage, G.V. , Stall, R.E. and Schaad, N.W. (2005) Bacterial spot‐worldwide distribution, importance and review. Acta Hort. 695, 27–34. [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Kearney, B. and Staskawicz, B.J. (1990) Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2 . Nature, 346, 385–386. [DOI] [PubMed] [Google Scholar]
- Kebede, M. , Timilsina, S. , Ayalew, A. , Admassu, B. , Potnis, N. , Minsavage, G.V. et al (2014) Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur. J. Plant Pathol. 140, 677–688. [Google Scholar]
- Kim, J.G. , Li, X. , Roden, J.A. , Taylor, K.W. , Aakre, C.D. , Su, B. , Lalonde, S. , Kirik, A. , Chen, Y. , Baranage, G. , McLane, H. , Martin, G.B. and Mudgett, M.B. (2009) Xanthomonas T3S effector XopN suppresses PAMP‐triggered immunity and interacts with a tomato atypical receptor‐like kinase and TFT1. Plant Cell, 21, 1305–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.G. , Stork, W. and Mudgett, M.B. (2013) Xanthomonas type III effector XopD desumolyates tomato transcription factor SIERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe, 13, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Olson, T.N. , Peffer, N.D. , Nikolaeva, E.V. , Park, S. and Kang, S. (2010) First report of bacterial spot of tomato caused by Xanthomonas gardneri in Pennsylvania. Plant Dis. 94, 638. [DOI] [PubMed] [Google Scholar]
- Koebnik, R. , Kruger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenraadt, H. , Van Betteray, B. , Germain, R. , Hiddink, G. , Jones, J.B. and Oosterhof, J. (2009) Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Hort. 808, 99–102. [Google Scholar]
- Kuflu, K.M. and Cuppels, D.A. (1997) Development of diagnostic DNA probe for xanthomonads causing bacterial spot of peppers and tomatoes. Appl. Environ. Microbiol. 63, 4462–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe, S. , Rougon‐Cardoso, A. , Sherwood, E. , Peeters, N. , Dahlbeck, D. , van Esse, H.P. , Smoker, M. , Rallapalli, G. , Thomma, B. , Staskawicz, B.J. , Jones, J.D.G. and Zipfel, C. (2010) Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. [DOI] [PubMed] [Google Scholar]
- Later, C. et al (2006) developed PCR primer sets based on the probes that can distinguish between the four species of bacterial spot xanthomonads.
- Laub, C.A. and Stall, R.E. (1967) An evaluation of Solanum nigrum and Physalis minima as suspects of Xanthomonas vesicatoria . Plant Dis. Rep. 51, 659–661. [Google Scholar]
- Lee, S. , Jeong, K. , Han, S. , Lee, S. , Phee, B. , Hahn, T. and Ronald, P. (2008) The Xanthomonas oryzae pv. oryzae PhoPQ two‐component system is required for avrXa21 activity, hrpG expression and virulence. J. Bacteriol. 190, 2183–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, R.P. , Minsavage, G.V. , Bonas, U. and Stall, R.E. (1994) Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria . Appl. Environ. Microbiol. 60, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, R.P. , Jones, J.B. , Somodi, G.C. , Minsavage, G.V. and Stall, R.E. (1995) Detection of Xanthomonas campestris pv. vesicatoria associated with pepper and tomato seed by DNA amplification. Plant Dis. 79, 917–922. [Google Scholar]
- Lindemann, J. and Upper, C.D. (1985) Aerial dispersal of epiphytic bacteria over bean plants. Appl. Environ. Microbiol. 50, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louws, F. , Wilson, M. , Campbell, H. , Cuppels, D. , Jones, J. , Shoemaker, P. et al (2001) Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Dis. 85, 481–488. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Patil, P.B. , Van Sluys, M. , White, F.F. , Ryan, R. , Dow, M. , Rabinowicz, P. , Salzberg, S.L. , Leach, J.E. , Sonti, R. , Brendel, V. and Bogdanove, A.J. (2008) Acquisition and evolution of plant pathogenesis‐associated gene clusters and candidate determinants of tissue‐specificity in Xanthomonas . PLOS ONE, DOI: 10.1371/journal.pone.0003828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Lewis Ivey, M. and Miller, S. (2011) First report of Xanthomonas gardneri causing bacterial spot of tomato in Ohio and Michigan. Plant Dis. 95, 1584. [DOI] [PubMed] [Google Scholar]
- Marco, G.M. and Stall, R.E. (1983) Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria that differ in sensitivity to copper. Plant Dis. 67, 779–781. [Google Scholar]
- Marois, E. , Ackerveken, G. and Bonas, U. (2002) The Xanthomonas type III effector protein avrBs3 modulates plant gene expression and induces cell hypertrophy in susceptible host. Mol. Plant–Microbe Interact. 15, 637–646. [DOI] [PubMed] [Google Scholar]
- Mbega, E.R. , Mabagala, R. , Adriko, J. , Lund, O.S. , Wulff, E.G. and Mortensen, C.N. (2012) Five species of xanthomonads associated with bacterial leaf spot symptoms in tomato from Tanzania. Plant Dis. 96, 760. [DOI] [PubMed] [Google Scholar]
- McGuire, R.G. and Jones, J.B. (1989) Detection of Xanthomonas campestris pv. vesicatoria in tomato In: Detection of bacteria in seed and other planting material (Saettler A.W., Schaad N.W. and Roth D.A., eds), Am Phytopathol. Soc. pp. 59–62. [Google Scholar]
- McGuire, R.G. , Jones, J.B. and Sasser, M. (1986) Tween media for semiselective isolation of Xanthomonas campestris pv. vesicatoria from soil and plant material. Plant Dis. 70, 887–891. [Google Scholar]
- McGuire, R.G. , Jones, J.B. , Stanley, C.D. and Cszinsky, A.A. (1991) Epiphytic populations of Xanthomonas campestris pv. vesicatoria and bacterial spot of tomato as influenced by nitrogen and potassium fertilization. Phytopathology, 81, 656–660. [Google Scholar]
- McInnes, T.B. , Gitaitis, R.D. , McCarter, S.M. , Jaworski, C.A. and Phatak, S.C. (1988) Airborne dispersal of bacteria in tomato and pepper transplant fields. Plant Dis. 72, 575–579. [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C.Q. , Al Sady, B. , Clark, E.T. and Staskawicz, B.J. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Miller, S.A. (2012) Bacterial spot‐ A new old problem in the Midwest processing tomatoes. The tomato magazine. 4–5. [Google Scholar]
- Minsavage, G.V. , Dahlbeck, D. , Whalen, M.C. , Kearny, B. , Bonas, U. , Staskawicz, B.J. and Stall, R.E. (1990) Gene‐for‐gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria–pepper interactions. Mol. Plant–Microbe Interact. 3, 41–47. [Google Scholar]
- Momol, T. , Jones, J. , Olson, S. , Obradovic, A. , Balogh, B. and King, P. (2008) Integrated management of bacterial spot on tomato in Florida. Retrieved from http://ufdc.ufl.edu/IR00003017/00001 on 2nd May, 2014.
- Noël, L. , Thieme, F. , Nennstiel, D. and Bonas, U. (2002) Two novel type III‐secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic, A. , Jones, J.B. , Momol, M.T. , Balogh, B. and Olson, S.M. (2004a) Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Dis. 88, 736–740. [DOI] [PubMed] [Google Scholar]
- Obradovic, A. , Mavridis, A. , Rudolph, K. , Janse, J.D. , Arsenijevic, M. , Jones, J.B. , Minsavage, G.V. and Wang, J.F. (2004b) Characterization and PCR‐based typing of Xanthomonas campestris pv. vesicatoria from peppers and tomatoes in Serbia. Eur. J. Plant Pathol. 110, 285–292. [Google Scholar]
- Obradovic, A. , Jones, J. , Momol, M. , Olson, S. , Jackson, L. , Balogh, B. et al (2005) Integration of biological control agents and systemic acquired resistance inducers against bacterial spot on tomato. Plant Dis. 89, 712–716. [DOI] [PubMed] [Google Scholar]
- Ocsoy, I. , Paret, M.L. , Ocsoy, M.A. , Kunwar, S. , Chen, T. , You, M. and Tan, W. (2013) Nanotechnology in plant disease management: DNA‐directed silver nanoparticles on graphene oxide as an antibacterial against Xanthomonas perforans . ACS Nano 7, 8972–8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret, M.L. , Vallad, G.E. , Averett, D.R. , Jones, J.B. and Olson, S.M. (2013) Photocatalysis: effect of light‐activated nanoscale formulations of TiO2 on Xanthomonas perforans and control of bacterial spot of tomato. Phytopathology, 103, 228–236. [DOI] [PubMed] [Google Scholar]
- Patil, P.B. , Bogdanove, A.J. and Sonti, R.V. (2007) The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 7, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohronezny, K. , Stall, R.E. , Canteros, B.I. , Kegley, M. , Datnoff, L.E. and Subramanya, R. (1992) Sudden shift in the prevalent race of Xanthomonas campestris pv. vesicatoria in pepper fields in South Florida. Plant Dis. 76, 118–120. [Google Scholar]
- Potnis, N. , Krasileva, K. , Chow, V. , Almeida, N.F. , Patil, P.B. , Ryan, R.P. , Sharlach, M. , Behlau, F. , Dow, J.M. , Momol, M.T. , White, F.F. , Preston, J.F. , Vinatzer, B.A. , Koebnik, R. , Setubal, J.C. , Norman, D.J. , Staskawicz, B.J. and Jones, J.B. (2011) Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics, 12, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis, N. , Minsavage, G. , Kennon Smith, J. , Hurlbert, J.C. , Norman, D. , Rodrigues, R. , Stall, R.E. and Jones, J.B. (2012) Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene‐for‐gene interaction in Pepper. Mol. Plant–Microbe Interact. 25, 307–320. [DOI] [PubMed] [Google Scholar]
- Potnis, N. , Soto‐Arias, J.P. , Cowles, K. , van Bruggan, A. , Jones, J.B. and Barak, J.D. (2014) Xanthomonas perforans colonization influences Salmonella enterica in the tomato phyllosphere. Appl. Environ. Microbiol. 80, 3173–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W. , Han, Z. , Tao, J. and He, C. (2008a) Genome‐scale mutagenesis and phenotypic characterization of two‐component signal transduction systems in Xanthomonas campestris pv. campestris ATCC33913. Mol. Plant–Microbe Interact. 21, 1128–1138. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Han, Z. and He, C. (2008b) Two component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol. Plant–Microbe Interact. 21, 151–161. [DOI] [PubMed] [Google Scholar]
- Quezado‐Duval, A.M. , Leite Jr, R.P. , Truffi, D. and Camargo, L.E. (2004) Outbreaks of bacterial spot caused by Xanthomonas gardneri on processing tomato in central‐west Brazil. Plant Dis. 88, 157–161. [DOI] [PubMed] [Google Scholar]
- Rademaker, J.L. , Hoste, B. , Louws, F.J. , Kersters, K. , Swings, J. , Vauterin, L. , Vauterin, P. and de Bruijn, F.J. (2000) Comparison of AFLP and rep‐PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50, 665–677. [DOI] [PubMed] [Google Scholar]
- Ritchie, D.F. (2000) Bacterial spot of pepper and tomato. Plant Health Instructor. doi: 10.1094/PHI-I-2000-1027-01. [DOI] [Google Scholar]
- Riva, E.M. , Rodrigues, R. , Pereira, M.G. , Sudre, C.P. and Karasawa, R. (2004) Inheritance of bacterial spot disease in Capsicum annuum L. Crop Breed. Appl. Biotechnol. 4, 490–494. [Google Scholar]
- Roberts, P. , Momol, M. , Ritchie, L. , Olson, S. , Jones, J. and Balogh, B. (2008) Evaluation of spray programs containing famoxadone plus cymoxanil, acibenzolar‐S‐methyl, and Bacillus subtilis compared to copper sprays for management of bacterial spot on tomato. Crop Prot. 27, 1519–1526. [Google Scholar]
- Romer, P. , Strauss, T. , Hahn, S. , Scholze, H. , Morbitzer, R. , Grau, J. , Bonas, U. and Lahaye, T. (2009) Recognition of AvrBs3‐like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150, 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, A. , Kousik, C. and Ritchie, D. (2001) Resistance to bacterial spot in bell pepper induced by acibenzolar‐S‐methyl. Plant Dis. 85, 189–194. [DOI] [PubMed] [Google Scholar]
- Rossier, O. , Van den Ackerveken, G. and Bonas, U. (2000) HrpB2 and HrpF from Xanthomonas are type III secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38, 828–838. [DOI] [PubMed] [Google Scholar]
- Sahin, F. and Miller, S.A. (1998) Resistance in Capsicum pubescens to Xanthomonas campestris pv. vesicatoria pepper race 6. Plant Dis. 82, 794–799. [DOI] [PubMed] [Google Scholar]
- Schmidtke, C. , Findeiss, S. , Sharma, C.M. , Kuhfuss, J. , Hoffmann, S. , Vogel, J. , Stadler, P. and Bonas, U. (2012) Genome‐wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 40, 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , Minsavage, G.V. , Stall, R.E. , Jones, J.B. and Lahaye, T. (2008) Characterization of AvrHah1, a novel AvrBs3‐like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 178, 546–556. [DOI] [PubMed] [Google Scholar]
- Scott, J.W. , Hutton, S.F. , Jones, J.B. , Francis, D.M. and Miller, S.A. (2006) Resistance to bacterial spot race T4 and breeding for durable and broad resistance to other races. Rep. Tomato Genet. Coop. 56, 33–36. [Google Scholar]
- Sharlach, M. , Dahlbeck, D. , Liu, L. , Chiu, J. , Jiménez‐Gómez, J.M. , Kimura, S. , Koenig, D. , Maloof, J.N. , Sinha, N. , Minsavage, G.V. , Jones, J.B. , Stall, R.E. and Staskawicz, B.J. (2013) Fine genetic mapping of RXopJ4, a bacterial spot disease resistance locus from Solanum pennellii LA716. Theor. Appl. Genet. 126, 601–609. [DOI] [PubMed] [Google Scholar]
- Sijam, K. , Chang, C.J. and Gitaitis, R.D. (1991) An agar medium for the isolation and identification of Xanthomonas campestris pv. vesicatoria from seed. Phytopathology, 81, 831–834. [Google Scholar]
- Sijam, K. , Chang, C.J. and Gitaitis, R.D. (1992) A medium for differentiating tomato and pepper strains of Xanthomonas campestris pv. vesicatoria . Can. J. Plant Pathol. 14, 182–184. [Google Scholar]
- Singer, A.U. , Schulze, S. , Skarina, T. , Xu, X. , Cui, H. , Eschen‐Lippold, L. , Egler, M. , Srikumar, T. , Raught, B. , Lee, J. , Scheel, D. , Savchenko, A. and Bonas, U. (2013) A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 9 DOI: 10.1371/journal.ppat.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stall, R.E. (1995) Xanthomonas campestris pv. vesicatoria. In: Pathogenesis and Host Specificity in Plant Diseases: Histological, Biochemical, Genetic and Molecular Bases. Vol. 1 Prokaryotes, (Singh U.S., Singh R.P. and Kohmoto K., eds), pp. 167–184. Tarrytown, NY: Pergamon/Elsevier. 321. [Google Scholar]
- Stall, R.E. , Beaulieu, C. , Egel, D. , Hodge, N.C. , Leite, R.P. , Minsavage, G.V. , Bouzar, H. , Jones, J.B. , Alvarez, A.M. and Benedict, A.A. (1994) Two genetically diverse groups of strains are included in Xanthomonas campestris pv. vesicatoria . Int. J. Syst. Bacteriol. 44, 47–53. [Google Scholar]
- Stall, R.E. , Jones, J.B. and Minsavage, G.V. (2009) Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 47, 265–284. [DOI] [PubMed] [Google Scholar]
- Stephens, W.L. and Starr, M.P. (1963) Localization of carotenoid pigment in the cytoplasmic membrane of Xanthomonas juglandis . J. Bacteriol. 86, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, F.L. (1925) Plant disease Fungi In: Plant disease fungi. (Steven F.L. ed), pp. 1871–1934, New York, The Macmillan company; Print. [Google Scholar]
- Stoyanova, M. , Vancheva, T. , Moncheva, P. and Bogatzevska, N. (2014) Differentiation of Xanthomonas spp. causing bacterial spot in Bulgaria based on biolog system. Int. J. Microbiol. doi: 10.1155/2014/495476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauß, T. , van Poecke, R.M.P. , Strauß, A. , Römer, P. , Minsavage, G.V. , Singh, S. , Wolf, C. , Strauß, A. , Kim, S. , Lee, H.‐A. et al (2012) RNA‐seq pinpoints a Xanthomonas TAL‐effector activated resistance gene in a large‐crop genome. Proc. Natl. Acad. Sci. USA, 109, 19 480–19 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, A. , Paret, M.L. , Jones, J.B. and Jeyaprakash, A. (2014) Multiplex qPCR assay for detecting the four causal agents of bacterial spot of tomato. 2014 APS‐CPS Joint Meeting. Minneapolis, MN, American Phytopathological Society. [Google Scholar]
- Šutic, D. (1957) Tomato bacteriosis. Posebna Izd. Inst. Zasht. Bilja, Beograd [Spec. Edit. Inst. Plant Prot., Beograd]. 6, 1–65. English Summary. Rev. Appl. Mycol 36, 734–735.
- Szczesny, R. , Büttner, D. , Escolar, L. , Schulze, S. , Seiferth, A. and Bonas, U. (2010) Suppression of the AvrBs1‐specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on an SNF1‐related kinase. New Phytol. 187, 1058–1074. [DOI] [PubMed] [Google Scholar]
- Tai, T.H. , Dahlbeck, D. , Clark, E.T. , Gajiwala, P. , Pasion, R. , Whalen, M.C. , Stall, R.E. and Staskawicz, B.J. (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA, 96, 14 153–14 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir‐Ariel, D. , Rosenberg, T. , Navon, N. and Burdan, S. (2012) A secreted lipolytic enzyme from Xanthomonas campestris pv. vesicatoria is expressed in planta and contributes to its virulence. Mol. Plant Pathol. 13, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer, P.L. and Stall, R.E. (1961) A survey of Xanthomonas vesicatoria resistance to streptomycin. Proc. Fla. State Hort. Soc. 75, 163–165. [Google Scholar]
- Thayer, P.L. and Stall, R.E. (1962) A survey of Xanthomonas vesicatoria resistance to streptomycin. Florida Agricultural Experiment Stations Journal Series No. 1523. Florida State Horticultural Society. 163–165. [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Büttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klösgen, U. , Patschkowski, T. , Rückert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorhölter, F.J. , Weber, E. , Pühler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timilsina, S. , Jibrin, M.O. , Potnis, N. , Minsavage, G.V. , Kebede, M. , Schwartz, A. , Bart, R. , Staskawicz, B. , Boyer, C. , Vallad, G.E. , Pruvost, O. , Jones, J.B. and Goss, E.M. (2015) Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri . Appl. Environ. Microbiol. 81(4), 1520–1529. doi: 10.1128/AEM.03000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trébaol, G. , Manceau, C. , Tirilly, Y. and Boury, S. (2001) Assessment of the genetic diversity among strains of Xanthomonas cynarae by randomly amplified polymorphic DNA analysis and development of specific characterized amplified regions for the rapid identification of X. cynarae . Appl. Environ. Microbiol. 67, 3379–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallad, G. , Pernezny, K. and Momol, T. (2004) A Series on Disease in the Florida Vegetable Garden: Tomato. Retrieved on 26th Feb, 2015 from http://edis.ifas.ufl.edu/pp121. University of Florida, IFAS Extension. [Google Scholar]
- Vallad, G.E. , Pernezny, K.L. , Balogh, B. , Wen, A. , Figueiredo, J.F.L. , Jones, J.B. et al (2010) Comparison of kasugamycin to traditional bactericides for the management of bacterial spot on tomato. HortScience, 45, 1834–1840. [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. and Swings, J. (1995) Reclassification of xanthomonas . Int. J. Syst. Bacteriol. 45, 472–489. [Google Scholar]
- Wang, H. , Hutton, S.F. , Robbins, M.D. , Sim, S.C. , Scott, J.W. , Yang, W. , Jones, J.B. and Francis, D.M. (2011) Molecular mapping of hypersensitive resistance from tomato ‘Hawaii 7981’ to Xanthomonas perforans race T3. Phytopathology, 101, 1217–1223. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C. , Wang, J.F. , Carland, F.M. , Heiskell, M.E. , Dahlbeck, D. , Minsavage, G.V. , Jones, J.B. , Scott, J.W. , Stall, R.E. and Staskawicz, B.J. (1993) Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol. Plant–Microbe Interact. 6, 616–627. [DOI] [PubMed] [Google Scholar]
- Wilson, M. , Moss, W. , Zhang, Y. and Jones, J. (2006) Chapter 12: molecular interactions at the leaf surface: Xanthomonas and its host In: Microbial Ecology of Aerial Plant Surfaces (Bailey M.J., Lilley A.K., Timms‐Wilson T.M. and Spencer‐Phillips P.T.N., eds), pp. 181–190. Wallingford, Oxfordshire: CABI. [Google Scholar]
- Worthington, R.J. , Rogers, S.A. , Huigens, R.W. III , Melander, C. and Ritchie, D.F. (2012) Foliar‐applied small molecule that suppresses biofilm formation and enhances control of copper‐resistant Xanthomonas euvesicatoria on pepper. Plant Dis. 96, 1638–1644. [DOI] [PubMed] [Google Scholar]
- Yang, W. and Francis, D.M. (2007) Genetics and breeding for resistance to bacterial diseases in tomato: prospects for marker‐assisted selection In: Genetic Improvement of Solanaceous Crops. Volume 2: Tomato (Razdan M.K. and Mattoo A.K., eds), pp. 379–419. Enfield, NH: Science Publishers. [Google Scholar]
- Yang, W. , Sacks, E.J. , Lewis Ivey, M.L. , Miller, S.A. and Francis, D.M. (2005) Resistance in Lycopersicon esculentum intraspecific crosses to race T1 strains of Xanthomonas campestris pv. vesicatoria causing bacterial spot of tomato. Phytopathology, 95, 519–527. [DOI] [PubMed] [Google Scholar]
- Young, J.M. , Park, D.C. , Shearman, H.M. and Fargier, E. (2008) A multilocus sequence analysis of the genus Xanthomonas . Syst. Appl. Microbiol. 31, 366–377. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Callaway, E. , Jones, J.B. and Wilson, M. (2009) Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur. J. Plant Pathol. 124, 379–390. [Google Scholar]


