Summary
Leptosphaeria maculans causes stem canker of oilseed rape (Brassica napus). The APSES transcription factor StuA is a key developmental regulator of fungi, involved in morphogenesis, conidia production and also more recently described as required for secondary metabolite production and for effector gene expression in phytopathogenic fungi. We investigated the involvement of the orthologue of StuA in L. maculans, LmStuA, in morphogenesis, pathogenicity and effector gene expression. LmStuA is induced during mycelial growth and at 14 days after infection, corresponding to the development of pycnidia on oilseed rape leaves, consistent with the function of StuA described so far. We set up the functional characterization of LmStuA using an RNA interference approach. Silenced LmStuA transformants showed typical phenotypic defects of StuA mutants with altered growth in axenic culture and impaired conidia production and perithecia formation. Silencing of LmStuA abolished the pathogenicity of L. maculans on oilseed rape leaves and also resulted in a drastic decrease in expression of at least three effector genes during in planta infection, suggesting either that LmStuA regulates, directly or indirectly, the expression of several effector genes in L. maculans or that the infection stage in which effectors are expressed is not reached when LmStuA expression is silenced.
Keywords: conidiation, effectors, Leptosphaeria maculans, morphogenesis, oilseed rape, regulation
APSES transcription factors (TFs) are specific to fungi and are characterized by highly conserved basic helix–loop–helix (bHLH) DNA‐binding domains (Ramirez‐Zavala and Dominguez, 2008) that bind to a specific StuAp response element with the consensus sequence (A/T)CGCG(T/A)N(A/C) (Dutton et al., 1997). APSES TFs are known to regulate morphological changes during asexual and sexual reproduction (Doedt et al., 2004; Miller et al., 1992; Ohara and Tsuge, 2004; Scherer et al., 2002; Sheppard et al., 2005). In addition to regulating the genes involved in morphogenesis, APSES TFs also control the expression of genes encoding metabolic enzymes (Doedt et al., 2004; Sheppard et al., 2005), secreted enzymes (Korting et al., 2003) and cell wall proteins (Sohn et al., 2003). StuA from Aspergillus nidulans was the first APSES TF to be described (Miller et al., 1992), and is required for conidiation and perithecia formation. StuA has been characterized in several phytopathogenic fungi and has been demonstrated to be required for the formation of aerial hyphae, efficient conidiation and the formation of perithecia (IpCho et al., 2010; Nishimura et al., 2009; Ohara and Tsuge, 2004; Tong et al., 2007). Its involvement in pathogenicity has also been investigated. In Fusarium oxysporum, deletion of FoStuA has a major effect on sporulation processes, but does not result in decreased virulence (Ohara and Tsuge, 2004), whereas, in Fusarium graminearum, deletion of FgStuA greatly reduces pathogenicity on wheat heads and the expression of secondary metabolites (Lysøe et al., 2011), and, in Fusarium culmorum, deletion of FcStuA leads to a complete loss of pathogenicity towards wheat stem base and root tissues (Pasquali et al., 2013). In Glomerella cingulata, StuA is required for the generation of appressorial turgor pressure and full pathogenicity on intact apple fruit (Tong et al., 2007). In Magnaporthe oryzae, StuA is involved in the mobilization of lipid droplets and glycogen from conidia to appressoria, and is therefore required for appressorium‐mediated infection (Nishimura et al., 2009). In Stagonospora nodorum, StuA has been reported to be required for pathogenicity, for the production of proteins involved in central carbon metabolism and of a metabolite mycotoxin (alternariol), and for the expression of SnTox3 encoding a host‐selective proteinaceous toxin (IpCho et al., 2010).
Leptosphaeria maculans is the hemibiotrophic fungus responsible for stem canker of oilseed rape (Brassica napus). Leptosphaeria maculans is present all over Brassica‐growing areas, except China (Dilmaghani et al., 2009; Fitt et al., 2006; West et al., 2001). Its complex life cycle includes phases of saprophytism, endophytism and necrotrophy, demonstrating remarkable plasticity. Following sexual reproduction on stem debris, leaf infections by ascospores cause phoma leaf spots, supporting asexual multiplication. The life cycle of the pathogen is completed by a lengthy symptomless colonization phase during which the pathogen grows from the leaf lesions along the petiole to the stem, where cankers develop at the end of the growing season (Rouxel and Balesdent, 2005). The L. maculans genome has an isochore‐like structure with GC‐equilibrated isochores (average of 51% GC) which are gene rich, whereas AT‐rich isochores (33.9% GC) are mostly devoid of active sequences and are made up of mosaics of intermingled and degenerated repeated elements (Rouxel et al., 2011). One hundred and twenty‐two genes encoding putative effectors have been predicted in AT‐rich isochores (Rouxel et al., 2011), including the five avirulence genes identified so far in L. maculans: AvrLm1, AvrLm6, AvrLm4‐7, AvrLm11 and AvrLmJ1 (Balesdent et al., 2013; Fudal et al., 2007; Gout et al., 2006; Parlange et al., 2009; Van de Wouw et al., 2013). Seventy‐three per cent of these effector genes share the same expression profile: a very low expression in axenic culture and a drastic increase in expression during primary leaf infection, with a peak of expression at 7 days post‐inoculation (dpi). In Soyer et al. (2014), we showed that local loosening of the chromatin structure in the environment of effector genes is a prerequisite for the induction of their expression during infection of oilseed rape by L. maculans. We therefore hypothesized that the relaxation of the chromatin structure allows the action of one or several TFs, not yet identified, to efficiently coordinate effector gene expression during infection. Here, we report the identification and functional characterization of LmStuA, the L. maculans orthologue of StuA. We aimed to obtain an understanding of the role of LmStuA in fungal development, primary leaf infection of oilseed rape and expression of effector genes. We generated L. maculans transformants in which the expression of LmStuA was decreased, and analysed hyphal morphogenesis and growth, conidiation, perithecia formation and pathogenicity. The effect of LmStuA silencing on the expression of effectors was also investigated during plant infection.
Four genes encoding TFs with an APSES DNA‐binding domain were retrieved from the genome of L. maculans. Among them, a StuA orthologue (GenBank accession number CBY0240) was identified by a BiDirectional Best Hit using blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1990) with the amino acid sequence of StuA from S. nodorum as a reference (IpCho et al., 2010). Functional domains have been identified using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan). LmStuA encodes a protein of 645 amino acids that shares 82% identity with StuA of S. nodorum and contains the typical APSES domain (IPR003163) within its sequence. To validate the automated annotation of LmStuA, rapid amplification of cDNA ends‐polymerase chain reaction (RACE‐PCR) analysis was performed. Untranslated regions (UTRs), transcriptional start and stop sites, and intron positions were annotated following PCR amplification and sequencing of the 3′ and 5′ ends of cDNA using a Creator SMART cDNA Library Construction Kit (Clontech, Palo Alto, CA, USA), according to the manufacturer's recommendations, and using LmStuA‐5UTRL1, LmStuA‐5UTRL2, LmStuA‐3UTRU1 and LmStuA‐3UTRU2 as specific primers (Table S1, see Supporting Information). Three introns of 51, 54 and 49 bp were identified in the coding sequence of LmStuA (Fig. S1, see Supporting Information). Amplification of the 3′UTR region of the gene produced a unique band of about 1000 bp that was cloned and sequenced. The 3′UTR showed a polyadenylation site 994 bp downstream of the stop codon. Amplification of the 5′UTR region of the gene produced two bands of about 1000 and 1300 bp that were cloned and sequenced. The sequencing of these two bands showed the presence of two lengthy introns in the 5′UTR sequence and that the 5′ end was located at either 2479 or 1258 bp upstream of the start codon (Fig. S1). This is consistent with StuA transcripts of A. nidulans (Miller et al., 1992) and G. cingulata (Tong et al., 2007), which correspond to two different mRNAs including long 5′ leaders, and suggests a post‐transcriptional control of LmStuA expression. Post‐transcriptional controls have been identified in fungi, in particular for genes involved in nutrition (Hinnebusch, 1997; Sachs, 1998). Yeast GCN4 expression was one of the first examples of translational control in eukaryotes. This gene encodes a TF involved in the activation of the genes required for amino acid availability (Hinnebusch, 1997). Another example is the CreA gene involved in catabolic repression regulation. This gene is regulated at both the transcriptional and post‐transcriptional level. Protein degradation is suspected to occur in the presence of derepressing carbohydrates (Strauss et al., 1999). More recently, environmental pH has been reported to be involved in transcription and post‐transcriptional processing of the protease BcACP1 of Botrytis cinerea (Rolland et al., 2009).
LmStuA expression was investigated using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in the sequenced isolate v23.1.3 (Rouxel et al., 2011). Total RNA was extracted from mycelium grown for one week in Fries liquid medium, and from infected leaf tissues as described previously (Fudal et al., 2007). Three time points important for host invasion during primary infection were investigated: 3 dpi, when conidia have germinated and the fungus has just penetrated the leaves; 7 dpi, when the fungus is still growing asymptomatically and effector genes reach their peak of expression; 14 dpi, when the necrotrophic symptoms appear and pycnidia differentiate. qRT‐PCR was performed as described previously (Fudal et al., 2007). For each condition tested, three different RNA extractions from three different biological samples and two reverse transcriptions for each biological replicate were performed. The primers used for qRT‐PCR are described in Table S1. C t values were analysed as described by Muller et al. (2002) for expression profile analysis. Elongation factor 1α (EF1α) was used as a constitutive reference gene. The expression of actin relative to EF1α was used as a control. The gene expression level of actin relative to EF1α showed no significant variations among the different in planta and in vitro conditions (data not shown). The expression of LmStuA relative to EF1α peaked at 14 dpi, correlating with the development of symptoms and conidia production, and was also high during mycelial growth in axenic culture (Fig. 1). This expression profile is consistent with that of StuA from S. nodorum and F. oxysporum. In these species, StuA is expressed during vegetative growth and conidiation, and, in S. nodorum, StuA expression increases during the late stages of infection correlating with the production of pycnidia (IpCho et al., 2010; Ohara and Tsuge, 2004).
Figure 1.
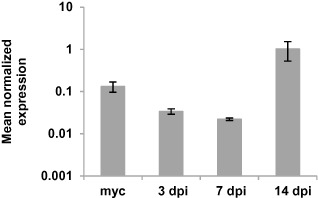
Expression of LmStuA during in vitro growth of Leptosphaeria maculans and oilseed rape infection. The expression of LmStuA was analysed in the strain v23.1.3 by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR); 3, 7 and 14 dpi, RT‐PCR products obtained from RNA isolated from oilseed rape cotyledons (Westar), 3, 7 and 14 days post‐infection; myc, RT‐PCR product obtained from RNA isolated from mycelial culture. RNA extracted from uninfected cotyledons and water was used as a negative control. Gene expression levels are relative to elongation factor 1α (EF1α). Each data point is the average of three biological replicates (extractions from different biological material) and two technical replicates (two RT‐PCRs). Standard error of the mean normalized expression level is indicated by error bars.
LmStuA was silenced by RNA interference (RNAi). Indeed, in L. maculans, knock‐out strategies were found to be particularly inefficient because of the need for large flanking homologous regions and rare homologous recombination events (Gardiner and Howlett, 2004), and RNAi was demonstrated to be an efficient alternative strategy (Fudal et al., 2007). Vector pPZPnat1‐LmStuA for RNAi‐mediated silencing of the LmStuA gene was constructed as described by Fudal et al. (2007). The pJK11 vector contains a G. cingulata gpdA promoter fragment and an A. nidulans trpC terminator fragment, separated by a multiple locus site in which inverted repeats of coding sequence of LmStuA have been cloned using the primers described in Table S1. The expression cassette was excised by digestion with SpeI and XhoI, and inserted into the binary vector pPZPnat1 digested with SpeI and XhoI, creating the vector pPZPnat1‐LmStuA. Agrobacterium tumefaciens‐mediated transformation of v23.1.3 from L. maculans was performed as described previously (Gout et al., 2006). Transformants were then plated onto minimal medium complemented with nourseothricin (50 mg/L) and cefotaxime (250 mg/L). After transformation of L. maculans with the pPZPnat1‐LmStuA vector, 19 transformants resistant to nourseothricin were recovered. Residual expression of LmStuA was assessed by qRT‐PCR during axenic culture. LmStuA expression in the transformants was never lower than 21% compared with the wild‐type (WT) strain. Three transformants, s.LmStuA‐21, s.LmStuA‐49 and s.LmStuA‐64, with reduced expression of StuA of 21%, 49% and 64%, respectively, compared with the WT strain (Table 1), were selected for further analysis.
Table 1.
Effect of LmStuA silencing on growth, conidia production, fertility and pathogenicity of L eptosphaeria maculans
| Isolate | Relative expression level (%)a | Radial growth (mm)b | Conidia/mLc | Sexual fertility (number of perithecia)d | Pathogenicity on cotyledonse |
|---|---|---|---|---|---|
| Wild‐type | |||||
| v23.1.3 | 100 | 51.6 ± 0.7 | 1.97 × 108 | Yes (>5) | 3.54 ± 0.72 |
| Transformants | |||||
| s.LmStuA‐21 | 21 | 43 ± 1.8f | 2.17 × 107 | No (0) | 1 ± 0.00f |
| s.LmStuA‐49 | 49 | 31 ± 0.75f | 4.33 × 107 | No (<5) | 1 ± 0.00f |
| s.LmStuA‐64 | 64 | 51.3 ± 0.6 | 2.83 × 107 | Yes (>5) | 1 ± 0.00f |
LmStuA expression was assessed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) with RNA isolated from mycelial culture. Expression is relative to the elongation factor 1α (EF1α) expression level and to the expression of LmStuA in the wild‐type isolate v23.1.3 (2−ΔΔ C t method).
Radial growth (mm) at 8 days post‐inoculation on V8 agar plates.
Conidiation (number of conidia/mL). Conidia were collected from five plates per transformant.
Fertility was measured by crossing v23.1.3 and the selected silenced LmStuA (s.LmStuA) transformants with the compatible isolate v23.1.2. Crosses were considered as fertile if more than five perithecia ejecting ascospores were produced per plate.
Pathogenicity was assayed 14 days after inoculation on the susceptible cultivar Westar of Brassica napus. Results are expressed as the mean score using the IMASCORE rating scale, comprising six infection classes (ICs), where IC1–IC3 correspond to resistance and IC4–IC6 to susceptibility (Balesdent et al., 2006).
P < 0.05. Disease scores and radial growth of each isolate were analysed using analysis of variance (ANOVA). Transformants were compared with v23.1.3 by a Dunnett multiple comparison test (α = 0.05). All statistical analyses were performed using XLStat 7.5 software.
We investigated axenic growth, conidiation and perithecia formation of the transformants s.LmStuA‐21, s.LmStuA‐49 and s.LmStuA‐64 compared with the WT strain. Growth assays were performed by the deposition of a 5‐mm plug at the centre of a 90‐mm Petri dish containing 25 mL of V8 juice agar. Radial growth was measured at 8 days after incubation in a growth chamber at 25 °C, with three replicates. s.LmStuA‐21 and s.LmStuA‐49, but not s.LmStuA‐64, grew significantly more slowly than the WT strain on V8 medium (Table 1). Compared with the WT strain, s.LmStuA mycelium appeared to be stunted, less aerial and embedded in the solid medium (Fig. 2A). We observed hyphal morphology on minimal MMII medium and noticed that the hyphae were less branched in the two transformants with the highest level of silencing (Fig. 2B). The hydrophobicity of s.LmStuA and WT isolates was assessed by depositing a drop of water at the surface of the mycelium. In the s.LmStuA transformants and the WT strain, the drop of water persisted at the surface of the mycelium and did not penetrate the hyphae, even after 10 min. Highly sporulating cultures were obtained on V8 juice agar medium, as described previously (Ansan‐Melayah et al., 1995). Conidiation was affected in the three transformants compared with the WT strain (Table 1), with an average concentration of 3 × 107 conidia/mL for the transformants compared with more than 1 × 108 conidia/mL for the WT strain. The formation of perithecia was assessed in the three transformants and the WT strain. In vitro crosses were performed as established previously (Gall et al., 1994). Using v23.1.2 as a compatible strain, five crosses were performed per transformant, and crosses were considered to be fertile when more than five perithecia ejecting ascospores were produced per plate. Transformants s.LmStuA‐21 and s.LmStuA‐49 were affected in their fertility, whereas s.LmStuA‐64 was as fertile as the WT strain v23.1.3 (Table 1). The APSES family of TFs controls a wide range of biological processes, and the deletion or mutation of StuA leads to pleiotropic defects in fungi. Accordingly, the silencing of LmStuA triggered multiple defects in L. maculans. Silenced LmStuA mycelia showed morphological defects consistent with the defects usually associated with StuA mutations or deletions, as in F. oxysporum, A. nidulans, F. graminearum and F. culmorum, with less aerial mycelium and stunted colonies (Lysøe et al., 2011; Ohara and Tsuge, 2004; Pasquali et al., 2013). Since the observation of Clutterbuck (1969), showing that StuA mutants of A. nidulans are altered in conidiophore development, conidia production has also systematically been shown to be reduced in all fungi in which this gene has been mutated. StuA has also been reported to be required for the formation of perithecia in F. graminearum and G. cingulata (Lysøe et al., 2011; Tong et al., 2007), and for mating in U. maydis (García‐Pedrajas et al., 2010).
Figure 2.
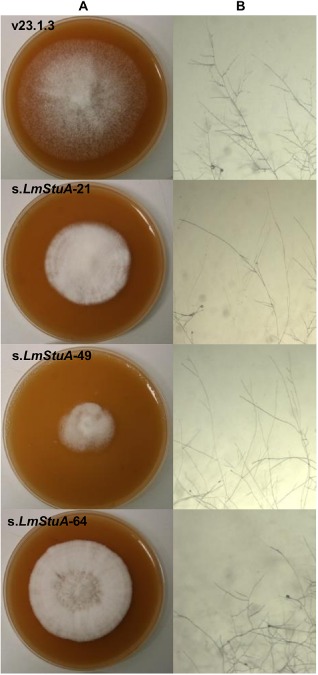
Effect of LmStuA silencing on hyphal morphology of Leptosphaeria maculans. Transformants silenced for LmStuA and the v23.1.3 isolate were grown on V8 medium (A) and minimal medium MMII (B) agar plates. Observations of hyphae on MMII were made at ×4.
We examined the pathogenicity of the transformants by inoculating a susceptible cultivar of B. napus (Westar). Pathogenicity assays were performed on cotyledons of 15‐day‐old plantlets, as described by Balesdent et al. (2006). Symptoms were scored on 10–12 plants at 14 dpi using the IMASCORE rating scale comprising six infection classes (ICs) (Balesdent et al., 2006), with two biological replicates. Although conidia production was low for the transformants, we were able to obtain the concentration used for pathogenicity assays (107 conidia/mL). Typical sporulating lesions were observed for the WT. In contrast, all silenced LmStuA transformants failed to cause disease and induced a typical black resistance response at the point of inoculation. The symptoms did not evolve with time to cause disease on the leaves. This phenotype is consistent with that which has been described in other phytopathogens, such as G. cingulata, U. maydis, M. oryzae, S. nodorum, F. graminearum and F. culmorum (García‐Pedrajas et al., 2010; IpCho et al., 2010; Lysøe et al., 2011; Nishimura et al., 2009; Pasquali et al., 2013; Tong et al., 2007), whereas StuA is considered to be dispensable for pathogenicity in F. oxysporum (Ohara and Tsuge, 2004). Thus, except for StuA of F. oxysporum, the importance of StuA in pathogenicity seems to be conserved among phytopathogenic fungi, whatever their pathogenic behaviour, including dimorphic transition (U. maydis) or the requirement for appressoria to penetrate host leaves (i.e. M. oryzae and G. cingulata). However, the role of LmStuA in L. maculans pathogenicity could be indirect and mainly caused by morphological defects which block fungal development after spore germination.
We investigated the expression of three effector genes by qRT‐PCR in the WT isolate and in the three selected silenced transformants at 7 dpi (corresponding to the peak of expression of effector genes in the WT strain). Silencing of LmStuA completely abolished the expression of AvrLm1, AvrLm6 and AvrLm4‐7, or drastically reduced their expression at 7 dpi compared with the WT strain (Table 2). Effector genes are major pathogenicity determinants of L. maculans. Notably, the loss of AvrLm4 function is associated with a fitness cost: avrLm4 virulent isolates produce fewer lesions of smaller diameter than do AvrLm4 avirulent isolates (Huang et al., 2006). Silenced LmStuA transformants showed pathogenicity defects which may be explained by this decrease in effector gene expression. In F. graminearum and F. culmorum, StuA is required for the expression of secondary metabolite gene clusters (Lysøe et al., 2011; Pasquali et al., 2013). In contrast, the production of secondary metabolites in F. oxysporum was not affected in the FoStuA mutants (Ohara and Tsuge, 2004) and, as these mutants showed no pathogenicity defect, we can hypothesize that there is an association between the effects triggered by StuA on the production of effector/secondary metabolites and pathogenicity. Interestingly, and closer to the L. maculans model, effector gene expression was also reduced in the SnStuA mutant of S. nodorum (IpCho et al., 2010), a closely related Dothideomycete relying on proteinaceous effector production to induce necrotrophic symptoms. Thus, it seems that the role of StuA in regulating effector gene expression is conserved. However, whether or not StuA regulates effector gene expression directly remains unclear. Indeed, both SnStuA and LmStuA mutants showed impaired growth and sporulation (IpCho et al., 2010; this study); thus, it is also likely that in planta development of the fungi is blocked at a step at which the expression of effector genes is not yet initiated. In LmStuA mutants, the amount of effector gene expression at 7 dpi is lower than the expression of effector genes at 3 dpi in the WT isolate (Fudal et al., 2007; Gout et al., 2006; Parlange et al., 2009), supporting the hypothesis that parasitic growth is blocked early in the leaves.
Table 2.
Effect of LmStuA silencing on the expression of selected effector genes AvrLm1, AvrLm4‐7 and AvrLm6 in planta
| Isolate/transformant | Mean normalized expression during plant infectiona | ||
|---|---|---|---|
| AvrLm4‐7 | AvrLm1 | AvrLm6 | |
| v23.1.3 | 3.0 | 1.7 | 4.6 |
| s.LmStuA‐21 | 2.9 × 10−3 | b | b |
| s.LmStuA‐49 | 7.6 × 10−2 | 2.3 × 10−1 | 9.8 × 10−2 |
| s.LmStuA‐64 | 1.2 × 10−3 | b | b |
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of expression was performed in the wild‐type strain v23.1.3 and in three transformants silenced for LmStuA at 7 days post‐infection of oilseed rape leaves. Gene expression levels are relative to elongation factor 1α (EF1α) and calculated as described by Muller et al. (2002). Each value is the average of two biological replicates (two extractions from different biological material) and two technical replicates (two RT‐PCRs).
No expression detected.
We have shown that LmStuA is required for normal growth, perithecia formation, pathogenicity on oilseed rape leaves and expression of effector encoding genes in L. maculans. LmStuA silencing triggers drastic effects on the morphogenesis and pathogenicity of L. maculans, indicating that it may affect a large number of genes and pathways. Further work is required to identify the set of genes regulated directly by LmStuA.
Supporting information
Fig. S1 Nucleotide sequence of the genomic region encoding LmStuA and amino acid sequence of the corresponding predicted protein. 5′ and 3′ untranslated regions of the gene are indicated as red and dark green, respectively. Introns are indicated as green.
Table S1 List of primers used in this study.
Acknowledgements
The authors wish to thank B. Auclair and M. Willigsecker (INRA Bioger, Grignon, France) for plant management; L. Coudard for the management of fungal strains; and A. Gautier and J. Linglin for assistance in sequencing. J. L. Soyer was funded by Young Scientist Funding by INRA.
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ansan‐Melayah, D. , Balesdent, M.H. , Buée, M. and Rouxel, T. (1995) Genetic characterization of AvrLm1, the first avirulence gene of Leptosphaeria maculans . Phytopathology, 85, 1525–1529. [Google Scholar]
- Balesdent, M.H. , Louvard, K. , Pinochet, X. and Rouxel, T. (2006) A large scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur. J. Plant Pathol. 114, 53–65. [Google Scholar]
- Balesdent, M.H. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A.M. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence toward Brassica rapa . New Phytol. 198, 887–898. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A.J. (1969) A mutational analysis of conidial development in Aspergillus nidulans . Genetics, 63, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilmaghani, A. , Balesdent, M.H. , Didier, J.P. , Wu, C. , Davey, J. , Barbetti, M.J. , Li, H. , Moreno‐Rico, O. , Phillips, D. , Despeghel, J.P. , Vincenot, L. , Gout, L. and Rouxel, T. (2009) The Leptosphaeria maculans/Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 58, 1044–1058. [Google Scholar]
- Doedt, T. , Krishnamurthy, S. , Bockmuhl, D.P. , Tebarth, B. , Stempel, C. , Russell, C.L. , Brown, A.J. and Ernst, J.F. (2004) APSES proteins regulate morphogenesis and metabolism in Candida albicans . Mol. Biol. Cell, 15, 3167–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton, J.R. , Johns, S. and Miller, B.L. (1997) StuAp is a sequence‐specific transcription factor that regulates developmental complexity in Aspergillus nidulans . EMBO J. 16, 5710–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitt, B.D.L. , Brun, H. , Barbetti, M.J. and Rimmer, S.R. (2006) World‐wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 114, 3–15. [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M.L. , Eckert, M.R. , Cattolico, L. , Bernard‐Samain, S. , Balesdent, M.H. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map‐based cloning of AvrLm6 . Mol. Plant–Microbe Interact. 20, 459–470. [DOI] [PubMed] [Google Scholar]
- Gall, C. , Balesdent, M.H. , Robin, P. and Rouxel, T. (1994) Tetrad analysis of acid phosphatase, soluble protein patterns, and mating type in Leptosphaeria maculans . Phytopathology, 84, 1299–1305. [Google Scholar]
- García‐Pedrajas, M.D. , Baeza‐Montañez, L. and Gold, S.E. (2010) Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain. Mol. Plant–Microbe Interact. 23, 211–222. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. and Howlett, B.J. (2004) Negative selection using thymidine kinase increases the efficiency of recovery of transformants with targeted genes in the filamentous fungus Leptosphaeria maculans . Curr. Genet. 45, 249–255. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the dothideomycete Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. (1997) Translational regulation of yeast GCN4: a window of factors that control initiator–tRNA binding to ribosome. J. Biol. Chem. 272, 21 661–21 664. [DOI] [PubMed] [Google Scholar]
- Huang, Y.J. , Evans, N. , Fitt, B.D.L. , Li, Z.Q. , Rouxel, T. and Balesdent, M.H. (2006) Fitness cost associated with loss of the AvrLm4 avirulence function in Leptosphaeria maculans (phoma stem canker of oilseed rape). Eur. J. Plant Pathol. 114, 77–89. [Google Scholar]
- IpCho, S.V.S. , Tan, K.C. , Koh, G. , Gummer, J. , Oliver, R.P. , Trengove, R.D. and Solomon, P.S. (2010) The transcription factor StuA regulates central carbon metabolism, mycotoxin production and effector gene expression in the wheat pathogen Stagonospora nodorum . Eukaryot. Cell, 9, 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korting, H.C. , Hube, B. , Oberbauer, S. , Januschke, E. , Hamm, G. , Albrecht, A. , Borelli, C. and Schaller, M. (2003) Reduced expression of the hyphal‐independent Candida albicans proteinase genes SPA1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 52, 623–632. [DOI] [PubMed] [Google Scholar]
- Lysøe, E. , Pasquali, M. , Breakspear, A. and Kistler, H.C. (2011) The transcription factor FgStuAp influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 54–67. [DOI] [PubMed] [Google Scholar]
- Miller, K.Y. , Wu, J. and Miller, B.L. (1992) StuA is required for cell pattern formation in Aspergillus . Genes Dev. 6, 1770–1782. [DOI] [PubMed] [Google Scholar]
- Muller, P.Y. , Janovjak, H. , Miserez, A.R. and Dobbie, Z. (2002) Processing of gene expression data generated by quantitative real‐time RT‐PCR. Biotechniques, 32, 1372–1379. [PubMed] [Google Scholar]
- Nishimura, M. , Fukada, J. , Moriwaki, A. , Fujikawa, T. , Ohashi, M. , Hibi, T. and Hayashi, N. (2009) Mstu1, an APSES transcription factor, is required for appressorium‐mediated infection in Magnaporthe grisea . Biosci. Biotechnol. Biochem. 73, 1–8. [DOI] [PubMed] [Google Scholar]
- Ohara, T. and Tsuge, T. (2004) FoSTUA, encoding a basic helix‐loop‐helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia and chlamydospores, in the fungal plant pathogen Fusarium oxysporum . Eukaryot. Cell, 2, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlange, F. , Daverdin, G. , Fudal, I. , Kuhn, M.L. , Balesdent, M.H. , Blaise, F. , Grezes‐Besset, B. and Rouxel, T. (2009) Leptosphaeria maculans avirulence gene AvrLm4‐7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4‐mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. [DOI] [PubMed] [Google Scholar]
- Pasquali, M. , Spanu, F. , Scherm, B. , Balmas, V. , Hoffmann, L. , Hammond‐Kosack, K.E. , Beyer, M. and Migheli, Q. (2013) FcStuA from Fusarium culmorum controls wheat foot and root rot in a toxin dispensable manner. PLoS ONE, 8, e57429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez‐Zavala, B. and Dominguez, A. (2008) Evolution and phylogenetic relationships of APSES proteins from Hemiascomycetes. FEMS Yeast Res. 8, 511–519. [DOI] [PubMed] [Google Scholar]
- Rolland, S. , Bruel, C. , Rascle, C. , Girard, V. , Billon‐Grand, G. and Poussereau, N. (2009) pH controls both transcription and post‐transcriptional processing of the protease BcACP1 in the phytopathogenic fungus Botrytis cinerea . Microbiology, 155, 2097–2105. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. and Balesdent, M.H. (2005) The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Mol. Plant Pathol. 6, 225–241. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , van de Wouw, A.P. , Couloux, A. , Dominguez, V. , Anthouard, V. , Bally, P. , Bourras, S. , Cozijnsen, A.J. , Ciuffetti, L.M. , Degrave, A. , Dilmaghani, A. , Duret, L. , Fudal, I. , Goodwin, S.B. , Gout, L. , Glaser, N. , Linglin, J. , Kema, G.H. , Lapalu, N. , Lawrence, C.B. , May, K. , Meyer, M. , Ollivier, B. , Poulain, J. , Schoch, C.L. , Simon, A. , Spatafora, J.W. , Stachowiak, A. , Turgeon, B.G. , Tyler, B.M. , Vincent, D. , Weissenbach, J. , Amselem, J. , Quesneville, H. , Oliver, R.P. , Wincker, P. , Balesdent, M.H. and Howlett, B.J. (2011) Effectors diversification within compartments of the Leptosphaeria maculans genome affected by RIP mutations. Nat. Commun. 2, 202. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, M.S. (1998) Post transcriptional control of gene expression in filamentous fungi. Fungal Genet. Biol. 23, 117–125. [DOI] [PubMed] [Google Scholar]
- Scherer, M. , Wei, H. , Liese, R. and Fischer, R. (2002) Aspergillus nidulans catalase‐peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell, 1, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, D.C. , Doedt, T. , Chiang, L.Y. , Kim, H.S. , Chen, D. , Nierman, W.C. and Filler, S.G. (2005) The Aspergillus fumigatus StuA protein governs the up‐regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell, 16, 5866–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, K. , Urban, C. , Brunner, H. and Rupp, S. (2003) EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47, 89–102. [DOI] [PubMed] [Google Scholar]
- Soyer, J.L. , El Ghalid, M. , Glaser, N. , Ollivier, B. , Linglin, J. , Grandaubert, J. , Balesdent, M.H. , Connolly, L.R. , Freitag, M. , Rouxel, T. and Fudal, I. (2014) Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans . PLoS Genet. 10, e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, J. , Horvath, H.K. , Abdallah, B.M. , Kindermann, J. , Mach, R.L. and Kubicek, C.P. (1999) The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post‐transcriptional level. Mol. Microbiol. 32, 169–178. [DOI] [PubMed] [Google Scholar]
- Tong, X. , Zhang, X. , Plummer, K.M. , Stowell, K.M. , Sullivan, P.A. and Farley, P.C. (2007) GcSTUA, an APSES transcription factor, is required for generation of appressorial turgor pressure and full pathogenicity of Glomerella cingulata . Mol. Plant–Microbe Interact. 9, 1102–1111. [DOI] [PubMed] [Google Scholar]
- Van de Wouw, A.P. , Lowe, R.G. , Elliott, C.E. , Dubois, D.J. and Howlett, B.J. (2013) An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J.S. , Kharbanda, P.D. , Barbetti, M.J. and Fitt, B.D.L. (2001) Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 50, 10–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Nucleotide sequence of the genomic region encoding LmStuA and amino acid sequence of the corresponding predicted protein. 5′ and 3′ untranslated regions of the gene are indicated as red and dark green, respectively. Introns are indicated as green.
Table S1 List of primers used in this study.


