Summary
Mutable bacterial cells are defective in their DNA repair system and often have a phenotype different from that of their wild‐type counterparts. In human bacterial pathogens, the mutable and hypermutable phenotypes are often associated with general antibiotic resistance. Here, we quantified the occurrence of mutable cells in Pseudomonas viridiflava, a phytopathogenic bacterium in the P. syringae complex with a broad host range and capacity to live as a saprophyte. Two phenotypic variants (transparent and mucoid) were produced by this bacterium. The transparent variant had a mutator phenotype, showed general antibiotic resistance and could not induce disease on the plant species tested (bean). In contrast, the mucoid variant did not display mutability or resistance to antibiotics and was capable of inducing disease on bean. Both the transparent and mucoid variants were less fit when grown in vitro, whereas, in planta, both of the variants and wild‐types attained similar population densities. Given the importance of the methyl‐directed mismatch repair system (MMR) in the occurrence of mutable and hypermutable cells in human bacterial pathogens, we investigated whether mutations in mut genes were associated with mutator transparent cells in P. viridiflava. Our results showed no mutations in MMR genes in any of the P. viridiflava cells tested. Here, we report that a high mutation rate and antibiotic resistance are inversely correlated with pathogenicity in P. viridiflava, but are not associated with mutations in MMR. In addition, P. viridiflava variants differ from variants produced by other phytopathogenic bacteria in the absence of reversion to the wild‐type phenotype.
Keywords: fitness cost, methyl‐directed mismatch repair system, mutation rate, plasticity
Introduction
Phase variation is a reversible process that leads to highly frequent phenotypic switching. Bacteria use phase variation to generate population diversity that increases bacterial fitness under particular environmental conditions (Saunders et al., 2003). This phenotypic plasticity has been linked to different genomic rearrangements and has been extensively described in plant‐associated bacteria. For example, Pseudomonas fluorescens displays phase variation during colonization of the alfalfa rhizosphere with the formation of translucent variants able to colonize the distal part of the roots (Martınez‐Granero et al., 2005, 2006). This phase variation has been reported to be mediated by two site‐specific recombinases, coded by the sss and xerD genes. Also, mutations in the Gac system are related to P. fluorescens phase variation when strains are cultured in vitro (Duffy and Défago, 2000; Natsch et al., 1994). Phase variation is also important in Pseudomonas brassicacearum during root colonization, and seems to be regulated by gacS and gacA genes encoding the expression of sRNA genes (Lalaouna et al., 2012; Pagès et al., 2007).
In addition to plant‐associated bacteria, other forms of genomic plasticity have been described in human pathogenic bacteria. Genomic plasticity can lead to the capacity to colonize new hosts or to become resistant to antibiotic treatments, thereby having a serious impact on human healthcare. Among the various mechanisms underlying genomic plasticity, the occurrence of mutator or hypermutable cells that spontaneously acquire antibiotic resistance has been repeatedly described for human pathogens. Examples include Escherichia coli, Haemophilus influenza, Pseudomonas aeruginosa and Staphylococcus aureus (Marvig et al., 2013; Miller, 2002; Wang et al., 2013; Watson et al., 2004). For all of these pathogens, a defective methyl‐directed mismatch repair system (MMR) regulates the occurrence of mutator/hypermutable antibiotic‐resistant cells. Long‐term experiments on P. aeruginosa chronic lung infections demonstrated the formation of a conspicuous number of mutS, mutL and mutY hypermutators associated with antibiotic resistance (Marvig et al., 2013). Hypermutator bacterial cells can acquire resistance to antibiotics when mutations occur in genes involved in resistance mechanisms. For example, rifampin resistance is induced by a point mutation in the rpoB gene encoding for the β RNA polymerase subunit, causing a change in rifampin binding affinity (Campbell et al., 2001). In addition to antibiotic resistance, hypermutable cells produced by a defective MMR are also linked to resistance to several stress conditions, such as oxidative stresses (Torres‐Barceló et al., 2013). In contrast, antibiotic resistance can bring about fitness costs associated with compensatory mutations that maintain resistance (MacLean et al., 2010). In this light, it is interesting to note that there are no reports of investigations to evaluate the pathogenicity of hypermutable antibiotic‐resistant bacterial cells of human pathogens. In the absence of any particular stress, only a small fraction of bacterial cells are mutators, because mutations in genes linked to important metabolic pathways can be extremely deleterious for bacteria. To avoid the fixation of deleterious mutations into a given bacterial population and the transfer of their associated phenotype to the next generation, mutators restore the original phenotype via additional mutations.
Plant‐pathogenic bacteria manifest types of plasticity which suggest that they are mutators. Pseudomonas viridiflava, a phytopathogenic bacterium in the P. syringae complex with a broad host range, produces variants that differ in proteolytic activity and exopolysaccharide (EPS) production (Bartoli et al., 2014). Spontaneous phenotypic variation has also been observed for the soil‐borne pathogen Ralstonia solanacearum, in which defective EPS variants are produced in response to the down‐regulation of the phcA gene (Brumbley and Denny, 1990). In particular, insertion and deletions in the phcA gene occur both in vitro and in vivo in R. solanacearum strains, leading to phenotypic variants defective in EPS production and with an attenuated aggressiveness on tomato plants (Poussier et al., 2003). This phase variation phenomenon in R. solanacearum is, however, reversible when variants are grown in planta, whereas no reversions were observed in in vitro conditions. The reversion observed in planta leads to the restoration of the more aggressive EPS‐producing wild‐type with the non‐mutated phcA restored by secondary mutations on the mutated phcA (Poussier et al., 2003). In addition, variants of Xanthomonas oryzae pv. oryzae, occurring during the stationary phase in in vitro culture, deficient in EPS production, are less aggressive, although they maintain a certain level of pathogenicity (Rajeshwari and Sonti, 2000). Recently, phenotypic variations have also been described in Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbits (Shrestha et al., 2013). The authors demonstrated that phenotypic variants of A. citrulli are different from the parental wild‐types with regard to biofilm formation, swimming mobility and aggressiveness on melon plants.
For the plant pathogens mentioned above, the impetus for the study of phenotypic variation was the observation of changes in colony type in culture—mucoid and nonmucoid types in particular—and phenotypes commonly associated with phase variation, such as motility and EPS production. All the studies demonstrated that variants were often associated with reduced aggressiveness on the plant species tested. However, the resistance of variants to antibiotics and their mutation rate relative to their wild‐type counterparts have not been examined to date in plant‐pathogenic bacteria. In contrast, the impetus for many of the studies of the occurrence of hypermutable pathogens of humans has been the emergence of clinical bacterial strains under conditions in which antibiotics are in continuous use. This has led to the assumption that the use of antibiotics is the main stimulating factor for the formation of resistant cells and for the evolution of secondary mutations able to compensate for the fitness cost of antibiotic resistance (Lenski, 1998). Nevertheless, some authors have proposed that the environment could have an important role in selecting mutator phenotypes, especially in human pathogens that are ubiquitous in the environment, such as P. aeruginosa or Vibrio cholera (Faruque et al., 2004; Rau et al., 2010).
Although several types of phenotypic variant have been reported for various plant‐pathogenic bacteria, mutability or hypermutability per se has not yet been described. Moreover, none of the studies described above have shown a correlation between antibiotic resistance and mutator phenotypes, or a correlation of high mutation rate with pathogenicity. For such reasons, it is difficult to understand whether there is a parallel between the mutator antibiotic‐resistant cells produced in human pathogens during host infection and the phenotypic variants produced by phytopathogenic bacteria. Recently, it has been demonstrated that isolates of Botrytis cinerea acquire resistance to pyrrolnitrin when grown in vitro with lethal doses of this antibiotic (Ajouz et al., 2010). The resulting resistant isolates presented a high fitness cost manifested as attenuated aggressiveness on tomato plants and apple fruits (Ajouz et al., 2010). These results clearly demonstrate that the benefit of resistance to pyrrolnitrin is associated with fitness costs in terms of pathogenicity. However, fungi have different molecular mechanisms than bacteria for antibiotic resistance, and the comparison with hypermutable antibiotic bacterial cells might not be appropriate.
In a recent study, we have demonstrated that the plant pathogen P. viridiflava produces phenotypic variants in terms of proteolytic activity and EPS production (Bartoli et al., 2014). Here, we examined whether transparent and mucoid variants were associated with phenotypes also found in human bacterial pathogens. We demonstrated that naturally occurring transparent variants display a mutator phenotype both in vitro and in planta, and also have general antibiotic resistance. We also found that this mutator phenotype is inversely correlated with pathogenicity. Interestingly, the mutator P. viridiflava phenotype was not related to mutations in genes that code for components of the MMR. Our study suggests that the formation of antibiotic‐resistant cells seems to be a common trait of mutator cells, but the molecular mechanisms that underline it are not the same in all bacteria and probably not strictly related to mutations in MMR genes. Furthermore, the inability of the mutator antibiotic‐resistant cells to induce disease raises the question of whether, in human pathogens, these types of mutant are actually responsible for disease or only for persistence of the pathogen.
Results
Pseudomonas viridiflava variants do not revert and the temporal dynamics of their wild‐types is strain dependent
The difference in proteolytic activity of the two colony variants (mucoid and transparent) of P. viridiflava was used to characterize the dynamics and reversion of the variants produced by strains BS0005, PVB‐H and CC1582. This experiment was performed mainly to understand the behaviour of the variants in vitro and whether they were able to revert. The mucoid variant was proteolytic, producing halos on tryptone soya agar (TSA) medium supplemented with skimmed milk (see Experimental procedures), whereas the transparent variant lacked this activity. During growth, all colonies of the wild‐type inocula were mucoid, but they were able to produce transparent colonies during in vitro growth. In contrast, neither mucoid nor transparent variants reverted during the in vitro incubation period. The mucoid colonies of the mucoid variants were different from those of the wild‐type as they did not produce transparent cells. Nevertheless, during the 48 h of in vitro growth, the rate of occurrence of detectable levels of transparent variants was different from one strain to another. In particular, in strains BS0005 and PVB‐H, the transparent variants were observed when the population size was 106 colony‐forming units (CFU)/mL (Table S1, see Supporting Information), whereas, for strain CC1582, the transparent variants were observed when the bacterium attained a population size of 109 CFU/mL (Table S1). Likewise, the ratio of transparent to mucoid variants also varied among strains, and globally we observed the highest rate of transparent variants for strain BS0005 (Table S1). Taken together, these observations lead us to conclude that: (i) the wild‐type state is able to spontaneously produce two phenotypic variants, a mucoid and a transparent variant, and the phenotype of the wild‐type before long‐term culture is mucoid; (ii) both of the variants are unable to revert and, as such, they can be considered as stable mutants, and the original phenotype cannot be restored under normal culture conditions; and (iii) the mucoid variants are different from the wild‐type because of their inability to produce transparent variants.
Transparent variants are antibiotic‐resistant mutators
In several human bacterial pathogens, variants are associated with a high mutation rate (>10−8 mutants per generation) and the ability to resist several antibiotics. We wondered whether, in P. viridiflava, the mucoid or transparent variant was associated with the phenotypes previously observed for human bacterial pathogens, namely with antibiotic resistance. The investigation of mutation frequencies between wild‐type, mucoid and transparent variants revealed three interesting results. First, mucoid variants did not generate any rifampicin‐resistant mutants. Second, the transparent variants showed higher mutation levels than the wild‐types. The transparent variant of strain BS0005 was characterized by a mutation frequency of 1.10 × 10−7 when measured in terms of the frequency of rifampicin‐resistant lines, twice as frequent and significantly greater than that of its wild‐type counterpart (4.92 × 10−8) (P < 0.05). In strain PVB‐H, although the mutation frequency was 1.06 × 10−7 for the transparent variant, being over six times greater than that of the wild‐type (2.50 × 10−8), these frequencies were not statistically significant (P > 0.05). Concerning strain CC1582, there was a two‐fold difference in the mutation frequencies for the transparent and wild‐types (1.20 × 10−7 and 5.80 × 10−8, respectively), and this difference was not statistically significant (P > 0.05). Third, when the rifampicin‐resistant mutants obtained from the wild‐types were transferred to tubes containing gelatin, mutants were unable to liquefy the gelatin, confirming that the antibiotic‐resistant mutants were characterized by a transparent phenotype. Indeed, when transferred to TSA medium supplemented with skimmed milk (TSAM), the rifampicin‐resistant mutants had the same phenotype as that of the transparent variants. These results corroborated the observation that, for mucoid variants, rifampicin‐resistant mutants were not detected, possibly because the occurrence of these mutants for the mucoid variants was below the detection threshold. In agreement with a previous study (Hall and Henderson‐Begg, 2006), we suggest that transparent cells are in fact mutators, but are weak mutators rather than hypermutators.
To investigate whether the mutator transparent phenotype of P. viridiflava was also associated with other antibiotic resistance, we tested the susceptibility to several antibiotics of both mucoid and transparent variants for strains BS0005, PVB‐H and CC1582. In all cases, we found that all cells of transparent variants showed resistance to amoxicillin, ampicillin, kanamycin and rifamycin. In contrast, the mucoid variants did not grow in the presence of these antibiotics. Finally, all the strains were inherently resistant to carbenicillin and cephalotin, and susceptible to gentamicin, streptomycin and tetramycin, and no differences were observed between the mucoid and transparent variants.
Mucoid and transparent variants are less fit than their wild‐type counterparts in vitro but not in planta
To determine whether differences in fitness exist between the mucoid and transparent variants and the wild‐types, we assessed growth kinetics both in vitro and in bean and cantaloupe plants. Statistically significant differences were observed in vitro between the variants and their wild‐type counterparts. Overall, both the mucoid and transparent variants of each strain tested were significantly less fit when grown in vitro compared with their wild‐type counterparts. In particular, the fitness difference (measured as a growth difference among strains and variants) between wild‐types and their variants was highly significant for all strains at 14 h post‐inoculation (hpi) (P < 0.005) and at 17, 20, 24 and 42 hpi (P < 0.0005) (Fig. 1). In contrast, no significant differences in terms of growth were observed between mucoid and transparent variants (Fig. 1A–C).
Figure 1.
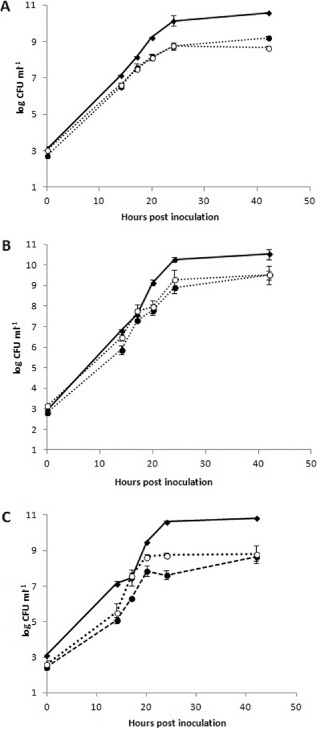
Fitness of the strains, inferred as differences in the in vitro growth of wild‐types and of mucoid and transparent variants, for strains BS0005 (A), PVB‐H (B) and CC1582 (C). Wild‐types are indicated with full lines and phase variants with dotted lines (●, mucoid; ○, transparent). Standard errors are indicated for each mean. CFU, colony‐forming units; hpi, hours post‐inoculation.
In contrast with the results observed in vitro, no statistically significant differences (P > 0.05) were observed on bean plants in terms of population size between wild‐types and their variants for all of the strains tested. The mucoid and transparent variants were able to attain the same population density as that of their wild‐type counterparts at 3, 10 and 14 days post‐inoculation (dpi) (Fig. 2A–C), demonstrating that there was no fitness cost (in terms of growth of the strains and variants tested) of the mucoid and transparent variants. The only exception was for strain BS0005, in which the growth of the transparent variant in bean plants was higher than that of the mucoid variant and wild‐type counterpart at 7 and 14 dpi (P < 0.005) (Fig. 2A). When the ratio between mucoid and transparent colonies was monitored in the wild‐types of the three strains tested, we observed that the BS0005 and PVB‐H wild‐types produced transparent variants at 3 dpi when their population density was 106 CFU/g (Table S1). In contrast, the CC1582 wild‐type strain did not produce any transparent variant during host infection (Table S1). It should be noted that the total population size in bean plants did not exceed about 108 CFU/g, suggesting that such a population size for the CC1582 strain was not sufficient to observe variants for this strain. Concerning strain BS0005, the maximum number of transparent variants was observed at 7 dpi, where 15.0% of the population consisted of transparent variants. Likewise, for strain PVB‐H, the peak of transparent variants was detected at 3 dpi, and the transparent variants represented 35.5% of the total bacterial population. An analysis of growth kinetics was also performed on cantaloupe seedlings and plants, as we have demonstrated previously a correlation between phase variation and aggressiveness on these seedlings (Bartoli et al., 2014). None of the variants and wild‐types of the strains tested grew on cantaloupe seedlings and plants; their population size was constant during the experiment (data not shown). In both cantaloupe seedlings and plants, the wild‐types did not produce transparent variants. In addition, the wild‐type strains induced disease only on seedlings, but not on 1‐month‐old plants (data not shown). These results suggest that the aggressiveness reaction on cantaloupe seedlings is only a hypersensitive reaction and not associated with bacterial multiplication in host cells.
Figure 2.

Fitness of the strains, inferred as differences in the in planta growth of wild‐types and of mucoid and transparent variants in 1‐month‐old bean plants, for strains BS0005 (A), PVB‐H (B) and CC1582 (C). Wild‐types are indicated with full lines and phase variants with dotted lines (●, mucoid; ○, transparent). Standard errors are indicated for each mean. CFU, colony‐forming units; dpi, days post‐inoculation.
Mucoid variants and wild‐types are pathogenic to bean plants, whereas transparent mutators do not induce disease
Although we observed an increase in bacterial population size in bean plants when we inoculated plants with wild‐type and variant counterpart suspensions at 105 CFU/mL, we did not observe any disease symptoms. By contrast, disease symptoms were observed with more concentrated suspensions (108 CFU/mL). To investigate in more detail the effect of the same inoculum density on pathogenicity, the wild‐types and their variants were separately inoculated at 108 CFU/mL. In this case, all the wild‐types and mucoid variants induced disease on bean plants, whereas the transparent variants were unable to cause disease (Fig. 3A–C). In particular, both the wild‐type and mucoid variants caused severe (stem necrosis > 1 cm) and moderate (stem necrosis < 1 cm) disease symptoms for strains BS0005 and PVB‐H, respectively. However, for strain CC1582, disease severity was high (necrosis > 1 cm) for the wild‐type and moderate for the mucoid variant.
Figure 3.

Disease symptoms on bean stems caused by the wild‐type (A) and the mucoid variant (B) of strain BS0005, 7 days after inoculation with bacterial suspensions at 108 colony‐forming units (CFU)/mL. The transparent BS0005 variant did not cause any symptoms (C). The behaviour of wild‐type and of mucoid and transparent variants was the same for all the strains tested.
For all strains, the population size of the transparent variants was lower than that of their wild‐type and mucoid counterparts (P < 0.0005) (Fig. 4). Furthermore, strain CC1582 was unable to produce transparent variants during the colonization of bean plants, whereas, for strains BS0005 and PVB‐H, 9% and 33%, respectively, of the colonies were transparent variants.
Figure 4.
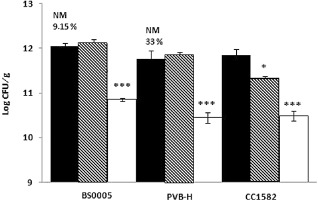
Population density of the wild‐type (filled bars) and of mucoid (hashed bars) and transparent (open bars) variants during the disease assay on bean plants for strains BS0005, PVB‐H and CC1582. Population density was determined 7 days after inoculation. Standard errors are indicated for each mean. Statistically significant differences of the means between wild‐type and mucoid or transparent variants (NM) for the same strain are expressed as: *P < 0.05; ***P < 0.0005. Percentage values on the top of the bars indicate the total percentage of transparent variants of each strain. CFU, colony‐forming units.
Mutations in genes for DNA repair are not involved in the production of mutable antibiotic‐resistant transparent cells
To investigate whether the formation of transparent mutable cells in P. viridiflava might be caused by a defective MMR or genes related to this system, we sequenced the mutS, mutL, mutT, recA and recX genes of mucoid, transparent, wild‐types and rifampicin mutants. We found no differences in gene sequences between the wild‐types and variants or mutants. In E. coli, it has been shown that genes in the MMR are under horizontal gene transfer (HGT) and that they have been lost and acquired at different times during the evolution of this bacterial species, influencing the genome plasticity of the bacterium (Denamur et al., 2000). We investigated the putative HGT in the MMR genes of P. viridiflava. We built the phylogeny of the MMR genes of the strains BS0005, PVB‐H, CC1582, TA0043 and UASW0038 and those of other P. syringae phylogroups (Fig. S1, see Supporting Information). Bayesian phylogeny demonstrated that the P. viridiflava strains, in phylogroup 7 of the P. syringae complex, did not acquire the MMR genes via HGT from other phylogroups (Fig. S1). Indeed, HGT did not occur in any of the strains of the P. syringae phylogroups used in our phylogeny. Taken together, these results suggest that the formation of the variants in P. viridiflava is not regulated by mutation or HGT in MMR genes.
Discussion
Phase variation is the genetically programmed switching from one phenotype to another for a given trait. The occurrence of phase variants is related to processes that mediate gene expression in response to particular environmental conditions or can also occur spontaneously (Henderson et al., 1999; Meyers and Bull, 2002). Unlike bona fide mutants, phase variants are able to restore their original phenotype (‘reversion’) when conditions are appropriate, although there are examples in which reversion has not been found (Henderson et al., 1999). In addition, phase variants occur at frequencies higher than 10−5 switches per cell per generation, unlike spontaneous mutations which have been reported to occur with an average frequency of 10−7 mutations per cell per generation (Allegrucci and Sauer, 2008). Here, we have shown that the phase variants spontaneously produced by P. viridiflava are unable to revert after one to several days of growth in synthetic medium or after being inoculated into a host plant. The occurrence of the transparent and mucoid variants was different among the strains tested, with the highest value observed for strains BS0005 and PVB‐H, in which transparent variants occurred when the total population size was 106 CFU/mL. These results led us to suggest that the mucoid and transparent variants produced by P. viridiflava are actually programmed mutants rather than bona fide phase variants. In contrast with our results, the phytopathogenic bacteria R. solanacearum, A. citrulli and X. oryzae have been reported to restore their EPS‐producing aggressive phenotype after growth in host plants (Poussier et al., 2003; Rajeshwari and Sonti, 2000; Shrestha et al., 2013), suggesting that P. viridiflava displays a different phenomenon from that of other plant‐pathogenic bacteria.
Programmed mutation in the absence of particular stimuli or environmental stress allows the generation of variation by increasing the probability that individuals will express one or more phenotypes that could be adapted to future environmental conditions (Beaumont et al., 2009; Meyers and Bull, 2002). Given the stable behaviour of the mucoid and transparent variants and their frequency, we can assume that, in P. viridiflava, there is a constitutive mechanism that stimulates the production of mutants, even without any apparent specific stimuli.
Another possible scenario could be that the production of these mutants is regulated by stressful conditions, such as nutritional limitations. For example, E. coli mutants are produced under limited amino acid availability during the stationary phase (Poole, 2012). In biofilms of Streptococcus pneumoniae, the development of non‐phase variation colony variants was related to high hydrogen peroxide (H2O2) conditions (Allegrucci and Sauer, 2008). However, our results from in vitro experiments support the idea that P. viridiflava produces mutants without any stress induction, because no mutators were detected in the stationary phase, but they occurred during the exponential growth phase in both in vitro and in vivo conditions. Another hypothesis with regard to the basal mechanisms that regulate the formation of mutator bacterial cells suggests a possible role for cell–cell communication (quorum sensing). For example, high mutation rate and phase variation in E. coli are related to population density and are regulated by the quorum‐sensing regulating gene luxS (Krasovec et al., 2014). Here, as the mutator transparent cells could not be detected until the bacterial population density was 106 CFU/mL, a similar phenomenon might also be involved in the formation of P. viridiflava mutator cells. However, we attempted to induce variation with stationary phase supernatants from in vitro culture (data not shown), but the variants did not occur before the culture suspensions reached the densities described here.
In several human pathogens, spontaneous mutants arising during in vitro culture or during host infection have a mutator phenotype associated with resistance to antibiotics (Marvig et al., 2013; Wang et al., 2013; Watson et al., 2004). Here, we investigated whether the mucoid and transparent variants were also characterized by a mutator phenotype resulting in resistance to antibiotics. As observed previously for human bacterial pathogens, we found that only the transparent variants had a frequency of mutation leading to rifampicin resistance higher than that of the wild‐type, and this also resulted in a greater frequency of resistance to a range of antibiotics. In human clinical strains, antibiotic‐resistant cells are associated with chronic diseases, and it has been assumed that these mutants are positively selected for by the use of antibiotics (Lenski, 1998). However, for a plant pathogen, we can question the advantage of resistance to antibiotics. Antibiotic resistance is known to provide fitness advantages in soil, because of the abundance of antibiotic‐producing organisms in soil (Chait et al., 2011). Although P. viridiflava is a ubiquitous bacterium isolated from different environments, including those related to the water cycle (Bartoli et al., 2014), it has not been isolated from soil. However, its ability to degrade pectin and the presence of genes for putrescine found in its genome (Bartoli et al., 2014) suggest that it might be able to live in soil, or at least in environments teaming with other microorganisms. A recent study has shown how P. syringae is transported through the soil to the water table during snow melt and rain run‐off (Monteil et al., 2013). This trajectory could transport P. viridiflava to localizations in soil strata that have not yet been explored, where it could survive. Water habitats are also rich in antibiotics because of the use of antimicrobial compounds in agriculture, run‐off from medical environments and the presence of antibiotic‐producing organisms (Vaz‐Moreira et al., 2014). Antibiotic resistance can occur in surprising habitats, such as in the sources of mineral and spring waters (Vaz‐Moreira et al., 2014). This suggests that the population dynamics of antibiotic‐resistant strains are not only related to the presence of antibiotics in contaminated waters, but also to other mechanisms, such as the presence of microbial competitors (antagonists) that could serve as a positive selective force for resistance. In this context, the selection of mutations that lead to antibiotic resistance could occur outside of situations in which antibiotics are deployed by humans. Given the wide presence of P. viridiflava in fresh waters (Morris et al., 2010), these habitats could be reservoirs in which transparent antibiotic‐resistant cells can proliferate. In support of this hypothesis, we also isolated fixed transparent P. viridiflava cells from different environments (data not shown). In addition, intrinsic antibiotic resistance (comprising the ensemble of non‐acquired genes that influence the susceptibility to antibiotics) has also been demonstrated in different bacterial communities (Baquero et al., 2013; Fajardo et al., 2008). It is tempting to speculate that the transparent phase and the associated antibiotic resistance serve P. viridiflava during the degradation of plant tissues via pectolytic activity when competition with other bacteria could be intense. This adaptation would distinguish P. viridiflava from the rest of the P. syringae complex, where we have not observed this same type of phenotypic variation and where soft rot capacity is absent.
Our results demonstrate that both mucoid and transparent variants are less fit in vitro, but not in planta, compared with their wild‐type counterparts, suggesting that they are more adapted to plant tissue conditions. Hypermutable P. aeruginosa cells isolated from patients with cystic fibrosis showed a fitness cost both in vitro and in the host (Montanari et al., 2007). In contrast, hypermutable carbapenem‐resistant P. aeruginosa mutants had a fitness advantage when inoculated in mice, raising questions about the adaptive advantage of antibiotic‐resistant bacteria (Skurnik et al., 2013). This latter observation is in agreement with our results, in that P. viridiflava variants showed no loss in fitness in bean plants. Reactive oxygen species (ROS) are produced in plants after pathogen infection as a defence response (Kotchoni and Gachomo, 2006). It is possible that P. viridiflava variants survive better than their wild‐type counterparts when ROS are produced by the host. However, preliminary results of resistance to 0.5%–2% H2O2 did not show any apparent differences between the wild‐types and variants (data not shown). Longer term experiments in host plants are necessary to better elucidate the population dynamics of P. viridiflava variants and their effective role in ensuring infection of the host. Transparent cells were defective in causing disease in bean plants compared with the wild‐type and mucoid counterparts. As these transparent variants are unable to produce EPSs (Bartoli et al., 2014), the lack of disease could be related to the absence of EPSs. Indeed, EPSs are known to be an important virulence factor in several phytopathogenic bacteria, such as Erwinia amylovora, R. solanacearum and X. campestris (Denny et al., 1988; Milling et al., 2011; Oh and Beer, 2005). Indeed, in both R. solanacearum and X. oryzae, EPS‐negative variants were less aggressive and, contrary to the results reported here for P. viridiflava, also showed a fitness cost in planta. Understanding the mechanisms underlying the occurrence of these transparent non‐pathogenic variants could be an important point in the development of new disease control strategies based on the enhancement of the occurrence of non‐pathogenic variants. It is also worth noting that, when high doses of bacteria (108 CFU/mL) were inoculated, the final bacterial population density was 100‐fold higher than when lower doses (105 CFU/mL) were inoculated. As, in both cases, strains attained their plateau, an explanation for such a phenomenon is that the plant elicits a programmed defence response after a given time post‐inoculation; the response would be time dependent rather than dependent on the bacterial density.
Mutability associated with antibiotic resistance, as described in bacterial pathogens of humans, is linked to a defective MMR caused by mutations in genes for proteins that are involved in the MMR (Marvig et al., 2013; Oliver et al., 2000; Wang et al., 2013; Watson et al., 2004). Here, we showed that neither the mut genes nor the rec genes were mutated in the transparent mutator cells. From this, we can speculate that the modulation of gene expression could be involved in a defective MMR in P. viridiflava strains. For example, in Bacillus subtilis, the over‐expression of the mutS gene decreases the mutation rate of this bacterium (Pedraza‐Reyes and Yasbin, 2004). However, other mechanisms could underline the occurrence of this phenomenon. In human cells, for example, small RNAs regulate the expression of mutL affecting the occurrence of tumour cells (Mao et al., 2012). As the MMR is highly conserved in all organisms from prokaryotes to eukaryotes (Schofield and Hsieh, 2003), the small RNAs may regulate the expression of MMR genes in bacteria, and thus be related to the mutator phenotype. These mechanisms could be revealed by the sequencing of the total mRNA of mucoid and transparent P. viridiflava cells.
Here, we have demonstrated that there is a clear trade‐off between mutation rate, antibiotic resistance and pathogenicity in P. viridiflava strains. We have shown how phytopathogenic bacteria, in a similar way to human bacterial pathogens, experience high mutation rates linked to the acquisition of mutated loci that alter phenotypic traits, such as resistance to antibiotics. The latter trait suggests that plant‐pathogenic bacteria could be used as model organisms in experimental evolution studies investigating the evolution of mutable loci associated with antibiotic resistance. In addition, the comparison of whole genomes of P. viridiflava strains and their variant counterparts with those of other P. syringae complex strains could help to elucidate whether this process is frequent in P. viridiflava and not in the rest of P. syringae, and why such a combination of phenotypes is associated with transparent P. viridiflava cells. This could elucidate the evolutionary history of P. viridiflava relative to the other P. syringae phylogroups, and whether the regulation of phenotypes associated with transparent variants is a stochastic event or a programmed process related to the ecology of this bacterial species.
Experimental Procedures
Bacterial strains and isolation of variants
Three P. viridiflava strains were used in this study: strain BS0005 was isolated from leaves of kiwifruit plants (Actinidia deliciosa) showing symptoms of bacterial necrosis in Italy in 2008 (Bartoli et al., 2014); strain PVB‐H was isolated from leaf spots of basil (Ocimum basilicum) in Hungary in 2012 (Végh et al., 2012); and strain CC1582 was isolated from an epilithic biofilm in a creek in France in 2006 (Morris et al., 2010). Strains were stored at −20 °C in phosphate buffer supplemented with 40% glycerol. Prior to further analysis, strains were streaked on 10% strength TSA (TSA/10) (Difco, Detroit, MI, USA) and incubated at 26 °C for 3 days.
The presence of phenotypic variation has been described previously in all three strains (Bartoli et al., 2014). The phase variants were characterized by two different morphologies, a mucoid and a transparent colony type, based on their ability to produce EPSs. The mucoid variant degrades gelatin indicating proteolytic activity, whereas the transparent variant does not (Bartoli et al., 2014). Given the difference in proteolytic activity, TSA/10 medium supplemented with skimmed milk (10%) (TSAM) was used in all experiments for the rapid discrimination of the variants. Mucoid variants produced a transparent halo around the colony on TSA/10 skimmed milk medium, whereas the transparent variant did not produce any halo and had a flat and transparent morphology.
To obtain phase variants, the wild‐type strains were grown in tubes containing 10% tryptic soya broth (TSB/10) for 24 h at 26 °C. Bacterial cultures were then serially diluted and plated on TSAM, and the plates were incubated at room temperature. After 48 h, the plates were checked and the phase variants were selected on the basis of their morphology on TSAM. Three colonies of each variant were picked with sterilized toothpicks, transferred into a new TSB/10 tube and incubated for 48 h. The bacterial cultures were serially diluted, plated and checked for purity for each variant to confirm a fixed phenotype. Ten of these stable variants for each strain were stored at −20 °C in phosphate buffer supplemented with 40% glycerol. To verify the stability of the variants, they were grown in TSB/10 for four successive periods of 1 week. At the end of each week of incubation, the cultures were checked for mucoid and transparent variants by dilution plating on TSAM, and then 1 mL of the culture was transferred to 9 mL of fresh TSB/10. These three stable variants of each strain were used for the further experiments described below.
Antibiotic resistance assay and determination of mutation frequencies
The antibiotic resistance of the strains was tested on amoxicillin (50 mg/L), ampicillin (50 mg/L), carbenicillin (100 mg/L), cephalotin (50 mg/L), gentamicin (50 mg/L), kanamycin (50 and 100 mg/L), rifamycin (50 mg/L), streptomycin (50 and 100 mg/L) and tetramycin (25 and 50 mg/L). An aliquot (10 μL at 105 CFU/mL) of bacterial suspension was plated on King's B (KB) medium (King et al., 1954) supplemented with an antibiotic. In addition, bacterial suspensions were also plated on KB without antibiotics. Five plates were used for each of the variants and their wild‐type counterparts. The plates were incubated at 26 °C and scored for bacterial growth at 5 days after inoculation.
The mutation frequencies of the wild‐types as well as the mucoid and transparent variants were assayed on 100 mg/L rifampicin plates as described previously (Besier et al., 2008). These frequencies were determined from cultures of stable variants after 24‐h periods of growth in 15 replicate tubes of 10 mL of TSB/10. Cultures from each tube were plated on five KB plates supplemented with rifampicin. The same bacterial cultures were also dilution plated on KB without rifampicin to quantify the total bacterial population size. Colonies were counted after 3 days of incubation at 26 °C. The mutation frequency was determined by dividing the density of cells resistant to rifampicin by the total population. The experiment was repeated three times and strains with mutation frequencies > 10−8 were considered to be mutators according to Prunier et al. (2003). Finally, all the rifampicin mutants were transferred into gelatin tubes to determine their proteolytic activity. The gelatin degradation test was performed by adding 15% of gelatin (Difco) into nutrient agar. The medium was then autoclaved and transferred into sterilized 1.5‐mL Eppendorf tubes. Bacterial colonies were inoculated into the gelatin tubes with sterilized toothpicks and incubated for 2 days at 26 °C. Following incubation, tubes were stored at 4 °C for 15 min. Lack of solidification of the medium after cooling indicated that the gelatin had been liquefied.
In vitro and in planta fitness assays and disease severity test
To investigate the putative differences in fitness, growth curve assays for each strain were performed on wild‐types, mucoid and transparent variants both in vitro and in planta. For the in vitro test, 102 CFU/mL bacterial suspensions were inoculated into four tubes of TSB/10 and incubated on a rotary shaker at 150 rpm for 48 h at 26 °C. At 14, 17, 20, 24 and 42 hpi, 100 μL of bacterial cultures were taken and serially diluted and plated on TSAM. Plates were incubated at room temperature and colonies were counted after 48 h of incubation. The experiment was repeated three times and the data were averaged to determine the total bacterial population at each time point. For wild‐type strains, the possible occurrence of transparent variants as well as potential reversion of mucoid and transparent treatments were monitored.
Fitness in planta was evaluated on cantaloupe (Cucumis melo var. cantalupensis Naud. cv. Védrantais) at the cotyledon stage and on 1‐month‐old plants, and on 1‐month‐old bean plants (Phaseolus vulgaris cv. Pinto). Six cantaloupe seedlings and four bean and four cantaloupe plants for each time point and for each treatment (wild‐type, mucoid and transparent variants) were used. Cantaloupe seedlings were inoculated with bacterial suspensions at 105 CFU/mL, as described previously (Morris et al., 2008), and the stems of cantaloupe and bean plants were inoculated with bacterial suspensions at 105 CFU/mL, as described by Bartoli et al. (2015). Plants and seedlings were incubated in a growth chamber at night/day temperatures of 18 °C/23 °C and with a light/dark period of 16 h/8 h. Re‐isolations were made as described by Bartoli et al. (2015) for up to 7 days for seedlings and up to 14 days for plants by serially diluting and plating on TSAM. Plates were incubated at room temperature for 48 h and colonies were counted. Bacterial populations were expressed as CFU per gram of sample for wild‐types, and the proportions of mucoid and transparent variants were calculated for each time point.
Disease severity was assessed on 1‐month‐old bean plants to evaluate whether the wild‐type, mucoid and transparent variants showed different levels of aggressiveness. Twenty microlitres of bacterial suspensions at 108 CFU/mL for each treatment were inoculated into the stems at three different points on each of four plants per mutant or wild‐type for each strain. Plants were incubated as described above. Disease severity was scored at 4 and 7 dpi based on the intensity of necrosis that developed on the inoculated stems. The following scale was used: 0, no symptoms; <1 cm length, moderate symptoms; >1 cm length, severe symptoms. Re‐isolations were made from all the plants, and the percentages of mucoid and transparent variants were calculated from plants inoculated with the wild‐types. No symptoms were detected on plants inoculated with sterilized distilled water.
mutS, mutL, mutT, recA and recX gene sequencing and analysis
The entire mutL (1965 bp), mutT (948 bp), recA (1065 bp) and recX (423 bp) genes and a part of mutS (750 bp) were sequenced to investigate whether the hypermutable transparent variants were associated with mutations involved in a defective MMR. Primers and polymerase chain reaction (PCR) conditions are described in Supporting Information Methods S1. Bayesian phylogeny was made using Mr Bayes software (Ronquist and Huelsenbeck, 2003).
Statistical analysis
Student's t‐tests were performed to compare wild‐type, mucoid and transparent variants using GraphPad online software (http://www.graphpad.com/). For comparison of mutation frequencies between wild‐types and transparent variants, we used the Mann–Whitney U‐test. P < 0.05 was considered to be statistically significant.
Supporting information
Fig. S1 Bayesian phylogeny constructed on the concatenated cts, gyrB, gapA and rpoD housekeeping genes and mutS, mut, mutT, recA and recX genes. Names of phylogroups are indicated according to Berge et al. (2014).
Table S1 Occurrence of transparent variants during growth in vitro and in bean plants.
Methods S1 Protocol for mut and rec gene amplification and sequencing.
Acknowledgements
We thank the technicians of the Experimental Installation team at INRA in Avignon, France for providing us with the plant material used in the pathogenicity tests. We thank the anonymous reviewers for constructive criticisms that helped to improve the quality of this article. The PhD fellowship for Claudia Bartoli was provided by Tuscia University.
References
- Ajouz, S. , Nicot, P.C. and Bardin, M. (2010) Adaptation to pyrrolnitrin in Botrytis cinerea and cost of resistance. Plant Pathol. 59, 556–566. [Google Scholar]
- Allegrucci, M. and Sauer, K. (2008) Formation of Streptococcus pneumoniae non‐phase‐variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J. Bacteriol. 190, 6330–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero, F. , Tedim, A.P. and Coque, T.M. (2013) Antibiotic resistance shaping multi‐level population biology of bacteria. Front. Microbiol. 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli, C. , Berge, O. , Monteil, C. , Guilbaud, C. , Balestra, G.M. , Varvaro, L. , Jones, C. , Dangl, J.L. , Baltrus, D.A. , Sands, D.C. and Morris, C.E. (2014) The Pseudomonas viridiflava phylogroups in the P. syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity‐related traits. Environ. Microbiol. 16, 2301–2315. [DOI] [PubMed] [Google Scholar]
- Bartoli, C. , Lamichhane, J.R. , Berge, O. , Guilbaud, C. , Varvaro, L. , Balestra, G.M. , Vinatzer, B.A. and Morris, C.E. (2015) A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: the example of kiwifruit canker. Mol. Plant Pathol. 16, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, H.J.E. , Gallie, J. , Kost, C. , Ferguson, G.C. and Rainey, P.B. (2009) Experimental evolution of bet hedging. Nature, 462, 90–93. [DOI] [PubMed] [Google Scholar]
- Berge, O. , Monteil, C.L. , Bartoli, C. , Chandeysson, C. , Guilbaud, C. , Sands, D.C. and Morris, C.E. (2014) A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS ONE. doi: 10.1371/journal.pone.0105547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besier, S. , Zander, J. , Kahl, B.C. , Kraiczy, P. , Brade, V. and Wichelhaus, T.A. (2008) The thymidine‐dependent small‐colony‐variant phenotype is associated with hypermutability and antibiotic resistance in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 52, 2183–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbley, S.M. and Denny, T.P. (1990) Cloning of wild‐type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J. Bacteriol. 172, 5677–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, E.A. , Korzheva, N. , Mustaev, A. , Murakami, K. , Nair, S. , Goldfarb, A. and Darst, S.A. (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell, 104, 901–912. [DOI] [PubMed] [Google Scholar]
- Chait, R. , Vetsigian, K. and Kishony, R. (2011) What counters antibiotic resistance in nature? Nat. Chem. Biol. 8, 2–5. [DOI] [PubMed] [Google Scholar]
- Denamur, E. , Lecointre, G. , Darlu, P. , Tenaillon, O. , Acquaviva, C. , Sayada, C. , Sunjevaric, I. , Rothstein, R. , Elion, J. , Taddei, F. , Radman, M. and Matic, I. (2000) Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell, 103, 711–721. [DOI] [PubMed] [Google Scholar]
- Denny, T.P. , Makini, F.W. and Brumbley, S.M. (1988) Characterization of Pseudomonas solanacearum Tn5 mutants deficient in extracellular polysaccharide. Mol. Plant–Microbe Interact. 1, 215–223. [Google Scholar]
- Duffy, B.K. and Défago, G. (2000) Controlling instability in gacS‐gacA regulatory genes during inoculant production of Pseudomonas fluorescens bio‐control strains. Appl. Environ. Microbiol. 66, 3142–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo, A. , Martínez‐Martín, N. , Mercadillo, M. , Galán, J.C. , Ghysels, B. , Matthijs, S. , Cornelis, P. , Wiehlmann, L. , Tümmler, B. , Baquero, F. and Martinez, J.L. (2008) The neglected intrinsic resistome of bacterial pathogens. PLoS ONE, 3, e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque, S.M. , Chowdhury, N. , Kamruzzaman, M. , Dziejman, M. , Rahman, M.H. , Sack, D.A. , Nair, G.B. and Mekalanos, J.J. (2004) Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera‐endemic area. Proc. Natl. Acad. Sci. USA, 101, 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L.M.C. and Henderson‐Begg, S.K. (2006) Hypermutable bacteria isolated from humans—a critical analysis. Microbiology, 152, 2505–2514. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R. , Owen, P. and Nataro, J.P. (1999) Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33, 919–932. [DOI] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kotchoni, S.O. and Gachomo, E.W. (2006) The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 31, 389–404. [DOI] [PubMed] [Google Scholar]
- Krasovec, R. , Belavkin, R.V. , Aston, J.A.D. , Channon, A. , Aston, E. , Rash, B.M. , Kadirvel, M. , Forbes, M. and Knight, C.G. (2014) Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell–cell. Nature. doi: 10.1038/ncomms4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna, D. , Fochesato, S. , Sanchez, L. , Schmitt‐Kopplin, P. , Haas, D. , Heulin, T. and Achouak, W. (2012) Phenotypic switching in Pseudomonas brassicacearum involves GacS‐ and GacA‐dependent Rsm small RNAs. Appl. Environ. Microbiol. 78, 1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R.E. (1998) Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1, 265–270. [PubMed] [Google Scholar]
- MacLean, R.C. , Hall, A.R. , Perron, G.G. and Buckling, A. (2010) The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Genet. 11, 405–415. [DOI] [PubMed] [Google Scholar]
- Mao, G. , Lee, S. , Ortega, J. , Gu, L. and Li, G.‐M. (2012) Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 22, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martınez‐Granero, F. , Capdevila, S. , Sanchez‐Contreras, M. , Martın, M. and Rivilla, R. (2005) Two site‐specific recombinases are implicated in phenotypic variation and competitive rhizosphere colonization in Pseudomonas fluorescens . Microbiology, 151, 975–983. [DOI] [PubMed] [Google Scholar]
- Martınez‐Granero, F. , Rivilla, R. and Martin, M. (2006) Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 72, 3429–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig, R.L. , Johansen, H.K. , Molin, S. and Jelsbak, L. (2013) Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 9(9), e1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, L.A. and Bull, J.J. (2002) Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 17, 551–557. [Google Scholar]
- Miller, K. (2002) Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J. Antimicrob. Chemother. 49, 925–934. [DOI] [PubMed] [Google Scholar]
- Milling, A. , Babujee, L. and Allen, C. (2011) Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defence responses in wilt‐resistant tomato plants. PLoS ONE. doi: 10.1371/journal.pone.0015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari, S. , Oliver, A. , Salerno, P. , Mena, A. , Bertoni, G. , Tümmler, B. , Cariani, L. , Conese, M. , Döring, G. and Bragonzi, A. (2007) Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology, 153, 1445–1454. [DOI] [PubMed] [Google Scholar]
- Monteil, C.L. , Lafolie, F. , Laurent, J. , Clement, J.‐C. , Simler, R. , Travi, Y. and Morris, C.E. (2013) Soil water flow is a source of the plant pathogen Pseudomonas syringae in subalpine headwaters. Environ. Microbiol. doi: 10.1111/1462-2920. [DOI] [PubMed] [Google Scholar]
- Morris, C.E. , Sands, D.C. , Vinatzer, B.A. , Glaux, C. , Guilbaud, C. , Buffière, A. , Yan, S. , Dominguez, H. and Thompson, B.M. (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2, 321–334. [DOI] [PubMed] [Google Scholar]
- Morris, C.E. , Sands, D.C. , Vanneste, J.L. , Montarry, J. , Oakley, B. , Guilbaud, C. and Glaux, C. (2010) Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe and New Zealand. mBio, 1, e00107–e00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsch, A. , Keel, C. , Pfirter, H.A. , Haas, D. and Défago, G. (1994) Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60, 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.S. and Beer, S.V. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. [DOI] [PubMed] [Google Scholar]
- Oliver, A. , Cantòn, R. , Campo, P. , Baquero, F. and Blàzquez, J. (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288, 1251–1253. [DOI] [PubMed] [Google Scholar]
- Pagès, D. , Sanchez, L. , Conrod, S. , Gidrol, X. , Fekete, A. , Schmitt‐Kopplin, P. , Heulin, T. and Achouak, W. (2007) Exploration of intraclonal adaptation mechanisms of Pseudomonas brassicacearum facing cadmium toxicity. Environ. Microbiol. 9, 2820–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza‐Reyes, M. and Yasbin, R.E. (2004) Contribution of the mismatch DNA repair system to the generation of stationary‐phase‐induced mutants of Bacillus subtilis . J. Bacteriol. 186, 6485–6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, K. (2012) Stress responses as determinants of antimicrobial resistance in Gram‐negative bacteria. Cell, 20, 227–234. [DOI] [PubMed] [Google Scholar]
- Poussier, S. , Thoquet, P. , Trigalet‐Demery, D. , Barthet, S. , Meyer, D. , Arlat, M. and Trigalet, A. (2003) Host plant‐dependent phenotypic reversion of Ralstonia solanacearum from non‐pathogenic to pathogenic forms via alterations in the phcA gene. Mol. Microbiol. doi: 10.1046/j.1365-2958.2003.03605. [DOI] [PubMed] [Google Scholar]
- Prunier, A.‐L. , Malbruny, B. , Laurans, M. , Brouard, J. , Duhamel, J.‐F. and Leclercq, R. (2003) High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187, 1709–1716. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, S. and Sonti, R.V. (2000) Stationary‐phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 4797–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, M.H. , Hansen, S.K. , Johansen, H.K. , Thomsen, L.E. , Workman, C.T. , Nielsen, K.F. , Jelsbak, L. , Høiby, N. , Yang, L. and Molin, S. (2010) Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ. Microbiol. 12, 1643–1658. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. and Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saunders, N.J. , Moxon, E.R. and Gravenor, M.B. (2003) Mutation rates: estimating phase variation rates when fitness differences are present and their impact on population structure. Microbiology, 149, 485–495. [DOI] [PubMed] [Google Scholar]
- Schofield, M.J. and Hsieh, P. (2003) DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57, 579–608. [DOI] [PubMed] [Google Scholar]
- Shrestha, R.K. , Rosenberg, T. , Makarovsky, D. , Eckshtain‐Levi, N. , Zelinger, E. , Kopelowitz, J. , Sikorski, J. and Burdman, S. (2013) Phenotypic variation in the plant pathogenic bacterium Acidovorax citrulli . PLoS ONE, 8, e73189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik, D. , Roux, D. , Cattoir, V. , Danilchanka, O. , Lu, X. , Yoder‐Himes, D.R. , Han, K. , Guillard, T. , Jiang, D. , Gaultier, C. , Guerin, F. , Aschard, H. , Leclercq, R. , Mekalanos, J. , Lory, S. and Pier, G.B. (2013) Enhanced in vivo fitness of carbapenem‐resistant oprD mutants of Pseudomonas aeruginosa revealed through high‐throughput sequencing. Proc. Natl. Acad. Sci. USA, 110, 20 747–20 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Barceló, C. , Cabot, G. , Oliver, A. , Buckling, A. and MacLean, R.C. (2013) A trade‐off between oxidative stress resistance and DNA repair plays a role in the evolution of elevated mutation rates in bacteria. Proc. R. Soc. B 280, 20130007. doi: 10.1098/rspb.2013.0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz‐Moreira, I. , Nunes, O.C. and Manaia, C.M. (2014) Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol. Rev. doi: 10.1111/1574-6976.12062. [DOI] [PubMed] [Google Scholar]
- Végh, A. , Hevesi, M. , Némethy, Z. and Palkovics, L. (2012) First report of bacterial leaf spot of basil caused by Pseudomonas viridiflava in Hungary. Plant Dis. 96, 141. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Wu, C. , Shen, J. , Wu, Y. and Wang, Y. (2013) Hypermutable Staphylococcus aureus strains present at high frequency in subclinical bovine mastitis isolates are associated with the development of antibiotic resistance. Vet. Microbiol. 165, 410–415. [DOI] [PubMed] [Google Scholar]
- Watson, M.E. , Burns, J.L. and Smith, A.L. (2004) Hypermutable Haemophilus influenzae with mutations in mutS are found in cystic fibrosis sputum. Microbiology, 150, 2947–2958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Bayesian phylogeny constructed on the concatenated cts, gyrB, gapA and rpoD housekeeping genes and mutS, mut, mutT, recA and recX genes. Names of phylogroups are indicated according to Berge et al. (2014).
Table S1 Occurrence of transparent variants during growth in vitro and in bean plants.
Methods S1 Protocol for mut and rec gene amplification and sequencing.


