Summary
CfAvr4, a chitin‐binding effector protein produced by the Dothideomycete tomato pathogen Cladosporium fulvum, protects the cell wall of this fungus against hydrolysis by secreted host chitinases during infection. However, in the presence of the Cf‐4 immune receptor of tomato, CfAvr4 triggers a hypersensitive response (HR), which renders the pathogen avirulent. Recently, several orthologues of CfAvr4 have been identified from phylogenetically closely related species of Dothideomycete fungi. Of these, DsAvr4 from Dothistroma septosporum also triggers a Cf‐4‐dependent HR, but CaAvr4 and CbAvr4 from Cercospora apii and Cercospora beticola, respectively, do not. All, however, bind chitin. To identify the region(s) and specific amino acid residue(s) of CfAvr4 and DsAvr4 required to trigger a Cf‐4‐dependent HR, chimeric and mutant proteins, in which specific protein regions or single amino acid residues, respectively, were exchanged between CfAvr4 and CaAvr4 or DsAvr4 and CbAvr4, were tested for their ability to trigger an HR in Nicotiana benthamiana plants transgenic for the Cf‐4 immune receptor gene. Based on this approach, a single region common to CfAvr4 and DsAvr4 was determined to carry a conserved proline residue necessary for the elicitation of this HR. In support of this result, a Cf‐4‐dependent HR was triggered by mutant CaAvr4 and CbAvr4 proteins carrying an arginine‐to‐proline substitution at this position. This study provides the first step in deciphering how Avr4 orthologues from different Dothideomycete fungi trigger a Cf‐4‐dependent HR.
Keywords: Avr4 effectors, Cf‐4 immune receptor, hypersensitive response
Introduction
Leaf mould disease of tomato (Solanum lycopersicum) is caused by the Dothideomycete fungal pathogen Cladosporium fulvum (syn. Passalora fulva and Fulvia fulva) (Thomma et al., 2005). Following penetration of tomato leaves through open stomata, the fungus colonizes the extracellular (apoplastic) space between mesophyll cells, where it secretes a plethora of small cysteine (Cys)‐rich effector proteins (Stergiopoulos and de Wit, 2009; de Wit et al., 2009a,b, 2012). In a compatible interaction, these effectors function as virulence factors involved in the promotion of successful infection, typically by modulating the host defence responses (Bolton et al., 2008; van Esse et al., 2007, 2008; de Jonge et al., 2010; Laugé et al., 1997; Mesarich et al., 2014; Ökmen et al., 2013). However, in an incompatible interaction, a C. fulvum effector or its modulated host virulence target is recognized by a corresponding Cf immune receptor of tomato (direct or indirect recognition, respectively) (de Wit et al., 2009b; Wulff et al., 2009a). This recognition triggers a hypersensitive response (HR), a localized form of cell death that arrests pathogen growth at the infection site (Heath, 2000). All Cf immune receptors identified to date belong to the receptor‐like protein (RLP) class of cell surface receptors, which possess extracytoplasmic leucine‐rich repeats (eLRRs), a membrane‐spanning domain and a short cytoplasmic tail (Kruijt et al., 2005). Current evidence suggests that these eLRRs are responsible for the direct or indirect recognition of C. fulvum effector proteins targeted to the host apoplast (van der Hoorn et al., 2001; Seear and Dixon, 2003; Wulff et al., 2001, 2009b).
A particularly well‐studied C. fulvum effector protein is Avr4 (hereafter referred to as CfAvr4), which triggers an HR on recognition by the Cf‐4 immune receptor of tomato (Joosten et al., 1994; Thomas et al., 1997). Following secretion into the tomato apoplast, the 135‐amino‐acid CfAvr4 pre‐pro‐protein is processed at its N‐ and C‐termini by fungal and/or host proteases to a mature protein of 86 amino acids with eight Cys residues (Joosten et al., 1994, 1997). These Cys residues form four intramolecular disulfide bonds (Cys40–70, Cys50–56, Cys64–109 and Cys86–101; Fig. 1) which, with the exception of Cys50–56, are required for the conformational stability of CfAvr4 in the protease‐rich environment of the tomato apoplast (van den Burg et al., 2001, 2003). Based on this disulfide bonding pattern, as well as preliminary nuclear magnetic resonance (NMR) spectra, it was determined that CfAvr4 carries a peritrophin‐A (IPR002557)/chitin‐binding type 2 (ChtBD2; SM00494) domain of carbohydrate‐binding module family 14 (CBM14) (van den Burg et al., 2003, 2004; Chang and Stergiopoulos, 2015). As the name suggests, this domain binds chitin (β‐1,4‐linked N‐acetylglucosamine), a major structural component of fungal cell walls that is absent from plants, and a target for hydrolysis by plant chitinases as part of their basal defence response (Grover, 2012). Indeed, in vitro polysaccharide affinity precipitation assays confirmed that CfAvr4 specifically binds chitin (van den Burg et al., 2003, 2006), whereas in situ fluorescence studies demonstrated that, during the colonization of susceptible tomato, CfAvr4 binds to exposed chitin present in the cell wall of C. fulvum infectious hyphae (van den Burg et al., 2006). Remarkably, CfAvr4 has been shown to protect the cell wall of several fungal species, including Trichoderma viride, Fusarium solani f. sp. phaseoli, Fusarium oxysporum f. sp. lycopersici and Plectosphaerella cucumerina, against hydrolysis by basic tomato chitinases in vitro (van den Burg et al., 2006; van Esse et al., 2007). The same experiment could not be performed with C. fulvum, however, as this fungus is insensitive to basic tomato chitinases in vitro (van den Burg et al., 2006). This is probably caused by a matrix of glucans and proteins, which renders the cell wall chitin of C. fulvum inaccessible to both chitinases and CfAvr4 during in vitro growth (de Wit and Kodde, 1981; de Wit and Roseboom, 1980). Nevertheless, because the cell wall chitin of C. fulvum was found to be exposed to basic plant chitinases in planta, it was proposed that CfAvr4 protects the cell wall of this fungus against the deleterious effects of basic tomato chitinases during host colonization (van den Burg et al., 2006). In support of this proposed counter‐defensive virulence function, the CfAvr4 gene was found to be expressed only during infection of tomato (Joosten et al., 1997; Mesarich et al., 2014), when the fungus is exposed to host chitinases. Furthermore, Arabidopsis thaliana plants heterologously expressing CfAvr4 were shown to be more susceptible to chitinous pathogens, including Botrytis cinerea and P. cucumerina, than control plants not expressing this gene, whereas the susceptibility of tomato to non‐chitinous plant pathogens, including Phytophthora brassicae and Pseudomonas syringae pv. tomato, remained unchanged (van Esse et al., 2007). In accordance with this important role in protection, silencing of CfAvr4 gene expression resulted in reduced virulence of C. fulvum during infection of susceptible tomato (van Esse et al., 2007).
Figure 1.
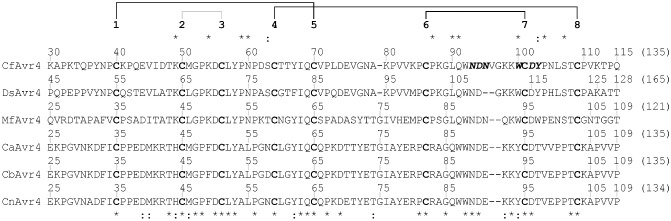
Avr4 is a small, cysteine‐rich, chitin‐binding effector protein produced by Cladosporium fulvum and other Dothideomycete fungi. The mature CfAvr4 protein sequence from C. fulvum was aligned to its corresponding region in DsAvr4 (Dothistroma septosporum), MfAvr4 (Mycosphaerella fijiensis), CaAvr4 (Cercospora apii), CbAvr4 (Cercospora beticola) and CnAvr4 (Cercospora nicotianae) using Clustal Omega (Sievers et al., 2011). Conserved (*) and physicochemically similar (:) amino acid residues shared between CfAvr4, DsAvr4 and MfAvr4, which are not present or are less similar in CaAvr4, CbAvr4 and CnAvr4, are shown above the alignment. Conserved and physicochemically similar amino acid residues shared between all Avr4 orthologues are shown below the alignment. Cysteine residues are highlighted in bold, and are sequentially numbered above the protein sequences. The disulfide bonding pattern of CfAvr4 (van den Burg et al., 2003) is shown above the cysteine residue numbering scheme, with those disulfide bonds conserved as part of the carbohydrate‐binding module family 14 (CBM14) motif indicated in black. Amino acid residues of CfAvr4 putatively involved in chitin binding (van den Burg et al., 2004) are highlighted in bold italics.
Recently, several orthologues of CfAvr4 have been identified from phylogenetically closely related species of Dothideomycete fungi. Examples include DsAvr4 from Dothistroma septosporum (the red band needle blight pathogen of pine), MfAvr4 from Mycosphaerella fijiensis (the black leaf streak pathogen of banana), and CaAvr4 from Cercospora apii, CbAvr4 from Cercospora beticola and CnAvr4 from Cercospora nicotianae (the leaf spot pathogens of celery, sugar beet and tobacco, respectively) (Fig. 1) (Stergiopoulos et al., 2010; de Wit et al., 2012). Avr4 was thus suggested to be a ‘core’ effector with a conserved biological function to promote the virulence of multiple plant‐pathogenic fungi on distantly related host species (Stergiopoulos et al., 2010). In support of this classification, MfAvr4 was shown to specifically bind chitin and to provide protection against basic plant chitinases in vitro, indicating that this protein is a functional orthologue of CfAvr4 (Stergiopoulos et al., 2010). Notably, it was also shown that DsAvr4 and MfAvr4, but not CbAvr4 and CnAvr4, trigger an HR in Nicotiana benthamiana plants transgenic for the Cf‐4 immune receptor gene (Cf‐4–N. benthamiana) using an Agrobacterium tumefaciens transient transformation assay (ATTA) (Stergiopoulos et al., 2010; de Wit et al., 2012), a result that could be replicated in tomato plants carrying the Cf‐4 gene (Stergiopoulos et al., 2010). Collectively, these results demonstrate that some, but not all, orthologues of CfAvr4 are capable of triggering a Cf‐4‐dependent HR.
In this study, we set out to characterize in more detail the interaction between CfAvr4 and Cf‐4, as well as DsAvr4 and Cf‐4, by identifying the region(s) and specific amino acid residue(s) of CfAvr4 and DsAvr4 required to trigger a Cf‐4‐dependent HR. For this purpose, chimeric and mutant proteins, in which specific protein regions or single amino acid residues, respectively, were exchanged between (i) CfAvr4 and CaAvr4 or (ii) DsAvr4 and CbAvr4, were tested for their ability to trigger an HR in Cf‐4–N. benthamiana using an ATTA. Based on this approach, a single region common to CfAvr4 and DsAvr4 was determined to carry a conserved proline (Pro) residue necessary for HR elicitation. Potential roles for the identified Pro residue in triggering a Cf‐4‐dependent HR are discussed.
Results
CaAvr4 does not trigger a Cf‐4‐dependent HR
As shown by Stergiopoulos et al. (2010), as well as by de Wit et al. (2012), orthologues of CfAvr4 can be tested for their ability to trigger an HR in Cf‐4–N. benthamiana plants using an ATTA. Based on this approach, we set out to determine whether CaAvr4 from Ce. apii can, like CfAvr4, DsAvr4 and MfAvr4, trigger a Cf‐4‐dependent HR. As expected, the positive control, CfAvr4, but not the negative control, CbAvr4, triggered an HR (Fig. 2). An HR, however, was not triggered by CaAvr4 (Fig. 2).
Figure 2.

The CaAvr4 protein from Cercospora apii does not trigger a Cf‐4‐dependent hypersensitive response. The CaAvr4 protein from Ce. apii, together with the CfAvr4 and CbAvr4 proteins from Cladosporium fulvum and Cercospora beticola, respectively, were tested for their ability to trigger a hypersensitive response in Nicotiana benthamiana plants transgenic for the Cf‐4 immune receptor gene of tomato using an Agrobacterium tumefaciens transient transformation assay (ATTA). Leaves were photographed at 7 days post‐agroinfiltration. The ATTA was repeated three times with reproducible results. +, positive control; −, negative control.
The Cys6–Cys7 region of CfAvr4 and DsAvr4 carries one or more amino acid residues required to trigger a Cf‐4‐dependent HR
We reasoned that CfAvr4, DsAvr4 and MfAvr4 should share one or more conserved or physicochemically similar amino acid residues required to trigger a Cf‐4‐dependent HR. Based on this reasoning, CaAvr4, CbAvr4 and CnAvr4, which do not trigger an HR in the presence of Cf‐4, should not share this conservation or similarity. A total of 10 conserved and two physicochemically similar amino acid residues shared between CfAvr4, DsAvr4 and MfAvr4, but absent from CaAvr4, CbAvr4 and CnAvr4, were identified by protein sequence alignment (Fig. 1). Seven of these residues were found to reside within the region spanning Cys residues 6 and 8 (Cys6–Cys8), which also carries the six amino acid residues putatively involved in chitin binding (van den Burg et al., 2004).
To identify the region(s) of CfAvr4 and DsAvr4 carrying one or more amino acid residues required to trigger a Cf‐4‐dependent HR, chimeric CfAvr4 and DsAvr4 proteins, in which all stretches of amino acid sequence bordered by Cys residues (Cys1–Cys2, Cys2–Cys3, Cys3–Cys4, Cys4–Cys5, Cys5–Cys6, Cys6–Cys7 and Cys7–Cys8) were exchanged with their corresponding regions from CaAvr4 and CbAvr4, respectively, were tested for their ability to trigger an HR in Cf‐4–N. benthamiana plants using an ATTA. In addition, chimeric CfAvr4 and DsAvr4 proteins, in which the Cys6–Cys8 region was exchanged with its corresponding region from CaAvr4 and CbAvr4, respectively, were also tested. As expected, an HR was triggered by the positive controls, CfAvr4 and DsAvr4, but not by the negative controls, CaAvr4 and CbAvr4 (Fig. 3). Chimeric CfAvr4 and DsAvr4 proteins carrying the Cys1–Cys2, Cys2–Cys3, Cys3–Cys4, Cys4–Cys5, Cys5–Cys6 or Cys7–Cys8 region of CaAvr4 and CbAvr4, respectively, also triggered an HR (Fig. 3). With the exception of the HR triggered by CfAvr4‐CaCys5–Cys6 (moderate HR), the strength of this HR was visually similar to that triggered by the corresponding positive control (Fig. 3). In contrast, an HR was not triggered by chimeric CfAvr4 and DsAvr4 proteins carrying the Cys6–Cys7 or Cys6–Cys8 region from CaAvr4 and CbAvr4, respectively (Fig. 3). Together, these results suggest that one or more of the four conserved amino acid residues shared between the Cys6–Cys7 region of CfAvr4, DsAvr4 and MfAvr4, but absent from CaAvr4, CbAvr4 and CnAvr4, are required to trigger a Cf‐4‐dependent HR.
Figure 3.
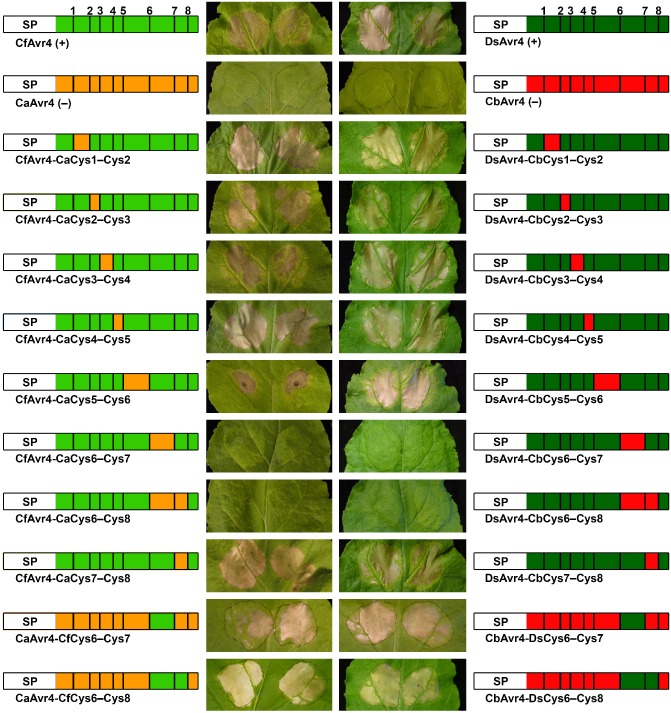
The Cys6–Cys7 region of CfAvr4 and DsAvr4 is required to trigger a Cf‐4‐dependent hypersensitive response. Chimeric proteins were generated by exchanging specific regions of protein sequence between CfAvr4 and CaAvr4, as well as DsAvr4 and CbAvr4. Each was tested for its ability to trigger a hypersensitive response in Nicotiana benthamiana plants transgenic for the Cf‐4 immune receptor gene of tomato using an Agrobacterium tumefaciens transient transformation assay (ATTA). Leaves were photographed at 7 days post‐agroinfiltration. ATTAs were repeated three times with reproducible results. A schematic representation of each protein is shown beside each photograph. SP, Pathogenesis‐Related 1A (PR1A) signal peptide of Nicotiana tabacum. Cysteine residues are represented by bold vertical bars, and are sequentially numbered. +, positive control; −, negative control.
To further confirm these results, chimeric CaAvr4 and CbAvr4 proteins, in which this stretch of amino acid sequence was exchanged with its corresponding region from CfAvr4 and DsAvr4, respectively, were tested for a gain of HR‐triggering ability in Cf‐4–N. benthamiana using an ATTA. At the same time, chimeric CaAvr4 and CbAvr4 proteins, in which the Cys6–Cys8 region was exchanged with its corresponding region from CfAvr4 and DsAvr4, respectively, were also tested. Indeed, all four chimeric proteins were able to trigger an HR of similar visual intensity to CfAvr4 and DsAvr4 (Fig. 3). These results confirm that one or more of the four conserved amino acid residues shared between the Cys6–Cys7 region of CfAvr4, DsAvr4 and MfAvr4, but absent from CaAvr4, CbAvr4 and CnAvr4, are required to trigger a Cf‐4‐dependent HR.
A conserved Pro residue in CfAvr4 and DsAvr4 is required to trigger a Cf‐4‐dependent HR
We set out to determine which of the four conserved amino acid residues shared between the Cys6–Cys7 region of CfAvr4, DsAvr4 and MfAvr4, but absent from CaAvr4, CbAvr4 and CnAvr4, are required to trigger a Cf‐4‐dependent HR. For this purpose, we employed an approach of site‐directed mutagenesis to generate mutant CfAvr4 proteins carrying one of the four corresponding variable amino acid residues from CaAvr4 (CfAvr4‐Pro87Arg, CfAvr4‐Leu90Gln, CfAvr4‐Gln91Trp and CfAvr4‐Trp100Tyr), and tested their ability to trigger an HR in Cf‐4–N. benthamiana plants using an ATTA. As expected, an HR was triggered by the positive control, CfAvr4, but not by the negative control, CaAvr4 (Fig. 4). Of the four mutants tested, CfAvr4‐Leu90Gln, CfAvr4‐Gln91Trp and CfAvr4‐Trp100Tyr triggered an HR of comparable strength to that elicited by the positive control (Fig. 4). In contrast, an HR was not triggered by CfAvr4‐Pro87Arg (Fig. 4). Next, to determine whether the corresponding Pro residue in DsAvr4 (Pro102; Fig. 1) is also required to trigger a Cf‐4‐dependent HR, a DsAvr4‐Pro102Arg mutant was tested. As expected, an HR was triggered by the positive control DsAvr4, but not by the negative control CbAvr4 (Fig. 4). The DsAvr4‐Pro102Arg mutant, however, was unable to trigger an HR (Fig. 4). Together, these results suggest that Pro87 of CfAvr4 and Pro102 of DsAvr4 are required to trigger a Cf‐4‐dependent HR.
Figure 4.
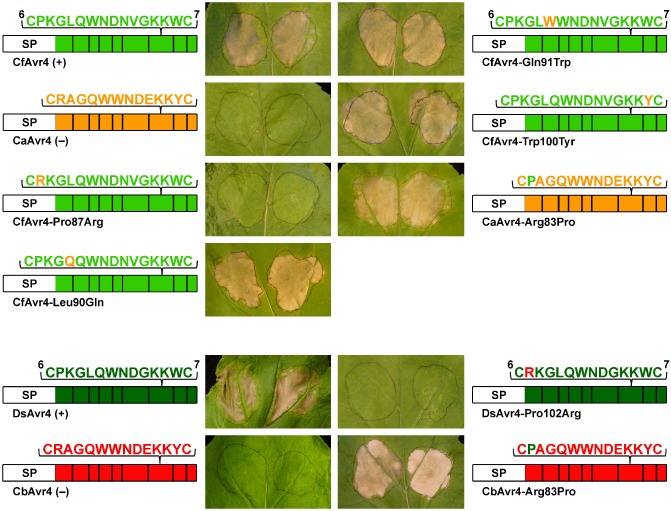
A conserved proline residue in CfAvr4 and DsAvr4 is required to trigger a Cf‐4‐dependent hypersensitive response. Mutant proteins were generated by exchanging single amino acid residues between the Cys6–Cys7 region of CfAvr4 and CaAvr4, as well as DsAvr4 and CbAvr4, using site‐directed mutagenesis. Each mutant was tested for its ability to trigger a hypersensitive response in Nicotiana benthamiana plants transgenic for the Cf‐4 immune receptor gene of tomato using an Agrobacterium tumefaciens transient transformation assay (ATTA). Leaves were photographed at 7 days post‐agroinfiltration. ATTAs were repeated three times with reproducible results. A schematic representation of each protein is shown beside each photograph. SP, Pathogenesis‐Related 1A (PR1A) signal peptide of Nicotiana tabacum. Cysteine residues are represented by bold vertical bars, with cysteine residues 6 and 7 numbered. +, positive control; −, negative control.
To further confirm these results, the reciprocal mutants of CaAvr4 and CbAvr4, carrying Pro87 of CfAvr4 and Pro102 of DsAvr4, respectively (CaAvr4‐Arg83Pro and CbAvr4‐Arg83Pro), were tested for a gain of HR‐triggering ability in Cf‐4–N. benthamiana plants using an ATTA. Indeed, both were able to trigger an HR (Fig. 4). These results therefore confirm that Pro87 of CfAvr4 and Pro102 of DsAvr4 are required to trigger a Cf‐4‐dependent HR.
To determine whether the conserved Pro residue plays a role in protein stability, we set out to establish whether wild‐type (WT), chimeric and mutant Avr4 proteins, with or without an N‐terminal His6‐FLAG tag, can be differentially detected in concentrated apoplastic fluid samples harvested from leaves of WT N. benthamiana plants (i.e. in the absence of Cf‐4) following an ATTA. This was carried out by Western blotting with one of several Avr4‐specific antibodies or, in the case of His6‐FLAG‐tagged proteins, an anti‐His6 or anti‐FLAG antibody. Unfortunately, however, no WT, chimeric or mutant Avr4 proteins could be detected (results not shown). Although frustrating, this result was not surprising, given that the monitoring of Avr4 protein levels in planta has, in the past, proven to be technically challenging (e.g. van Esse et al., 2006). Furthermore, as shown previously by van Esse et al. (2006), affinity tags, such as His6‐FLAG, are frequently removed from the N‐ or C‐terminus of C. fulvum effectors in the apoplast of N. benthamiana and tomato, preventing their detection. In an attempt to circumvent this issue, we heterologously produced His6‐FLAG‐tagged WT, chimeric and mutant Avr4 proteins using Pichia pastoris, and incubated these proteins with apoplastic fluid harvested from WT N. benthamiana or tomato. An effort was then made to detect these proteins by Western blotting using the antibody set mentioned above. Unfortunately, however, this approach was also unsuccessful (results not shown). Consequently, a conclusion regarding the stability of WT, chimeric and mutant Avr4 proteins in the leaf apoplast of N. benthamiana, and thus a role for the identified Pro residue in promoting this stability, could not be made.
WT and mutant Avr4 proteins bind chitin
It has been shown previously using an in vitro affinity precipitation assay that CfAvr4 and MfAvr4 specifically bind chitin (van den Burg et al., 2006; Stergiopoulos et al., 2010). To determine whether DsAvr4, CaAvr4 and CbAvr4, as well as the mutant proteins CfAvr4‐Pro87Arg, DsAvr4‐Pro102Arg, CaAvr4‐Arg83Pro and CbAvr4‐Arg83Pro, also bind chitin, all were heterologously produced by P. pastoris in liquid culture, and their affinity for chitin (crab‐shell chitin), as well as another insoluble polysaccharide, chitosan (deacetylated chitin), was examined using an in vitro affinity precipitation assay. Like CfAvr4 (positive control) and MfAvr4 (Stergiopoulos et al., 2010), all WT and mutant Avr4 proteins readily precipitated in the presence of chitin, but not of chitosan (Fig. 5), confirming that they bind chitin.
Figure 5.
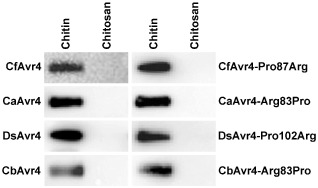
Wild‐type (WT) and mutant Avr4 proteins bind chitin. An in vitro affinity precipitation assay was carried out to determine whether His6‐FLAG‐tagged WT and mutant CfAvr4, CaAvr4, DsAvr4 and CbAvr4 proteins heterologously produced by Pichia pastoris can bind to the insoluble polysaccharides chitin and chitosan. Following incubation with each insoluble polysaccharide and subsequent centrifugation, His6‐FLAG‐tagged WT or mutant Avr4 protein bound to the pelleted insoluble polysaccharide fraction was resolved by Tricine–sodium dodecylsulfate–polyacrylamide gel electrophoresis (TSDS–PAGE), and visualized by Western blotting using anti‐FLAG (α‐FLAG) antibody. Affinity precipitation assays were repeated twice with reproducible results.
Discussion
During colonization of the tomato leaf apoplast, C. fulvum secretes CfAvr4, a chitin‐binding effector protein that protects the cell wall of this fungus against hydrolysis by secreted host chitinases (van den Burg et al., 2006; Joosten et al., 1994). However, in the presence of the Cf‐4 immune receptor of tomato, CfAvr4 triggers an HR, which renders the pathogen avirulent (Joosten et al., 1994; Thomas et al., 1997). Recently, several orthologues of CfAvr4 have been identified from phylogenetically closely related species of Dothideomycete fungi that infect different hosts, including pine (D. septosporum; DsAvr4), banana (M. fijiensis; MfAvr4), celery (Ce. apii; CaAvr4), sugar beet (Ce. beticola; CbAvr4) and tobacco (Ce. nicotianae; CnAvr4) (Stergiopoulos et al., 2010; de Wit et al., 2012). All are expected to be functional orthologues of CfAvr4, because DsAvr4, MfAvr4, CaAvr4 and CbAvr4 bind chitin (Stergiopoulos et al., 2010; this study), whereas MfAvr4 has also been shown to provide protection against basic plant chitinases (Stergiopoulos et al., 2010). Notably, of the five CfAvr4 orthologues mentioned above, DsAvr4 and MfAvr4 also trigger a Cf‐4‐dependent HR, but CaAvr4 and CbAvr4 do not (Stergiopoulos et al., 2010; de Wit et al., 2012; this study). In the present study, we took advantage of this discrepancy by producing and phenotypically characterizing a series of chimeric and mutant proteins, in which specific protein regions or single amino acid residues, respectively, had been exchanged between CfAvr4 and CaAvr4 or DsAvr4 and CbAvr4, to identify a Pro residue conserved between CfAvr4 (Pro87), DsAvr4 (Pro102) and MfAvr4 (Pro83; Fig. 1), but absent from CaAvr4, CbAvr4 and CnAvr4, that is required to trigger a Cf‐4‐dependent HR. Indeed, when Pro87 of CfAvr4 and Pro102 of DsAvr4 were exchanged with their corresponding amino acid residue from CaAvr4 and CbAvr4, respectively [arginine (Arg) 83], both proteins (CfAvr4‐Pro87Arg and DsAvr4‐Pro102Arg) lost the ability to trigger a Cf‐4‐dependent HR. Conversely, when Arg83 of CaAvr4 and CbAvr4 was exchanged with Pro87 of CfAvr4 and Pro102 of DsAvr4, respectively, both proteins (CaAvr4‐Arg83Pro and CbAvr4‐Arg83Pro) gained the ability to trigger a Cf‐4‐dependent HR.
Although our results clearly indicate that Avr4 effectors require a conserved Pro residue to trigger a Cf‐4‐dependent HR, a mechanistic understanding as to how this residue promotes the interaction between these proteins and the Cf‐4 immune receptor is lacking. In the remaining discussion, we put forward three hypotheses for the role of the identified Pro residue in the promotion of this interaction. All three hypotheses hold substantial merit and will require additional experimental testing in order to be proven or disproven.
Hypothesis 1: the conserved Pro is an interfacial residue that interacts directly with Cf‐4 or a host protein (Avr4 virulence target) guarded by Cf‐4
Protein–protein interactions are crucial to the molecular communication between hosts and their pathogens. With regard to the recognition of pathogen effectors by host immune receptors, this interaction can be direct, with binding between the two (e.g. Jia et al., 2000), or indirect, with the immune receptor binding an effector‐modulated host protein (e.g. Rooney et al., 2005). It has yet to be experimentally proven whether Cf‐4 interacts directly or indirectly with CfAvr4, DsAvr4 and MfAvr4, but direct interaction is anticipated, given that both CfAvr4 and MfAvr4 are protective effectors with a fungal target, chitin, and that CfAvr4 probably lacks a virulence target in the host guarded by Cf‐4 (van den Burg et al., 2006; van Esse et al., 2007; Stergiopoulos et al., 2010). In support of a direct interaction, an analysis of CfAvr4 allelic variation across C. fulvum strains indicated a preference for sequence diversification (i.e. non‐synonymous mutations) over frame‐shift mutations or gene loss as a means of overcoming Cf‐4‐mediated resistance (Stergiopoulos et al., 2007). This type of sequence diversification is commonly associated with genes encoding effectors that interact directly with their corresponding immune receptors (e.g. Dodds et al., 2006). Of note, Pro87 of CfAvr4 is predicted to be surface exposed by three‐dimensional structural modelling (Fig. S1, see Supporting Information), suggesting that this residue may indeed be available for direct recognition by Cf‐4. Thus, with this in mind, it is possible that Cf‐4 interacts directly with Pro87 of CfAvr4, Pro102 of DsAvr4 and Pro83 of MfAvr4, together with one or more conserved and/or physicochemically similar amino acid residues shared between CfAvr4, DsAvr4, MfAvr4, CaAvr4 and CbAvr4, to form a strong interaction surface necessary for the elicitation of an HR. Interestingly, Pro87 is located close to the six amino acid residues of CfAvr4 putatively involved in chitin binding, which are also predicted to be surface exposed (van den Burg et al., 2004; Fig. S1). A detailed characterization of WT and mutant Avr4 effector tertiary structures, both alone and in a complex with Cf‐4, will now be required to determine whether these amino acid residues, the identified Pro residue, and/or other conserved and/or physicochemically similar amino acid residues do in fact form an interface between Avr4 effectors and Cf‐4.
Although current evidence points towards a direct interaction, it is also possible that the identified Pro residue is required for a function associated with the indirect recognition of CfAvr4, DsAvr4 and MfAvr4 by Cf‐4. However, any such function would be independent of chitin binding, because no causal relationship exists between the ability of Avr4 orthologues to bind chitin and their ability to trigger a Cf‐4‐dependent HR. As evidence of this, CfAvr4, DsAvr4 and MfAvr4 trigger an HR in Cf‐4–tomato and/or Cf‐4–N. benthamiana in the absence of chitin (van der Hoorn et al., 2000; Joosten et al., 1997; Stergiopoulos et al., 2010; this study). Consequently, the identified Pro residue would need to take part in an additional biological function, for example, in manipulating a host protein guarded by Cf‐4. However, this would imply that CaAvr4, CbAvr4 and CnAvr4, although core effectors based on chitin‐binding ability, are unable to perform this additional biological function in the absence of the identified Pro residue, or that their specific manipulation of the same host protein is not guarded by Cf‐4. Thus, the possibility that the identified Pro residue is required for a function associated with the indirect recognition of CfAvr4, DsAvr4 and MfAvr4 by Cf‐4 seems less likely.
Hypothesis 2: the conserved Pro residue dictates the position and/or orientation of other conserved and/or physicochemically similar amino acid residues shared between Avr4 effector proteins
Pro residues are typically important for determining the local structural conformation of proteins (MacArthur and Thornton, 1991). Therefore, it is also conceivable that the identified Pro residue provides a local backbone conformation that promotes the recruitment of multiple conserved and/or physicochemically similar amino acid residues shared between CfAvr4, DsAvr4 and MfAvr4 that are required to trigger a Cf‐4‐dependent HR. Given that a Cf‐4‐dependent HR is triggered by CaAvr4‐Arg83Pro and CbAvr4‐Arg83Pro, these residues would also be present in CaAvr4 and CbAvr4 (and, probably, CnAvr4). However, as CaAvr4, CbAvr4 and CnAvr4 lack the identified Pro residue, these residues would, according to this hypothesis, not be recruited in the conformation required to trigger a Cf‐4‐dependent HR. Certainly, validation of this hypothesis would again require a detailed characterization of protein tertiary structure, including that of WT and mutant Avr4 effectors, both alone and in a complex with Cf‐4. Such a detailed structural characterization would also provide an opportunity to assess whether Arg83 of CaAvr4, CbAvr4 and CnAvr4 prevents, for example, through its size and/or charge, the availability of conserved and/or physicochemically similar amino acid residues shared between all six Avr4 effectors that are required to trigger a Cf‐4‐dependent HR.
Hypothesis 3: the conserved Pro residue is required for the conformational stability of Avr4 effector proteins in the protease‐rich environment of the leaf apoplast
Another possibility is that the identified Pro residue is required for the conformational stability of CfAvr4, DsAvr4 and MfAvr4 in the protease‐rich leaf apoplast of tomato and/or N. benthamiana, and that this stability is, in turn, necessary for these proteins to trigger a Cf‐4‐dependent HR. However, this would imply that CaAvr4, CbAvr4 and CnAvr4 are unable to trigger a Cf‐4‐dependent HR in tomato and/or N. benthamiana as a result of their poor stability in the leaf apoplast. Likewise, this would also imply that CaAvr4‐Arg83Pro and CbAvr4‐Arg83Pro are able to trigger a Cf‐4‐dependent HR in N. benthamiana as a consequence of their enhanced stability in the leaf apoplast. Certainly, amino acid composition is known to be critically important to the stability of CfAvr4 in the leaf apoplast of tomato. This is best illustrated by natural isoforms of CfAvr4, which are produced by most known race 4 strains of C. fulvum that have evolved to overcome Cf‐4‐mediated resistance in tomato. The majority carry a single amino acid substitution that disrupts one of three disulfide bonds conserved as part of the CBM14 motif, specifically Cys40–70 (Cys70Tyr), Cys64–109 (Cys64Tyr and Cys109Tyr) and Cys86–101 (Cys86Phe) (Joosten et al., 1997; Stergiopoulos et al., 2007). Two additional isoforms carrying a Thr66Ile or Tyr67His amino acid substitution have also been identified (Joosten et al., 1997). Regardless of the substitution type, all exhibit decreased conformational stability in the leaf apoplast of tomato and, as a consequence, are rapidly degraded by host proteases, preventing their recognition by Cf‐4 (van den Burg et al., 2003; Joosten et al., 1997). Indeed, reduced conformational stability may provide an explanation as to why, in our study, CfAvr4‐CaCys5–Cys6, but not its equivalent chimera, DsAvr4‐CbCys5–Cys6, triggered a Cf‐4‐dependent HR of reduced intensity in N. benthamiana relative to its WT counterpart. In the same way, reduced conformational stability may also provide an explanation as to why a recently identified natural isoform of MfAvr4, Thr109Ile, failed to trigger a Cf‐4‐dependent HR in Cf‐4–N. benthamiana (Stergiopoulos et al., 2014), given that Thr109 is not conserved or physicochemically similar between MfAvr4, CfAvr4 and DsAvr4, and that this residue falls outside of the Cys6–Cys7 region identified in our study (Fig. 1). Unfortunately, in our study, none of the WT, chimeric or mutant Avr4 proteins could be detected in the leaf apoplast of N. benthamiana or following their incubation with apoplastic fluid of N. benthamiana or tomato. Consequently, it currently remains unclear whether the identified Pro residue plays a role in the stability of these proteins in planta.
Conclusions
In summary, we have identified a conserved Pro residue in CfAvr4 and DsAvr4 that is required to trigger a Cf‐4‐dependent HR. In doing so, we have provided the first step in deciphering how a Cf‐4‐dependent HR can be triggered by Avr4 orthologues from different Dothideomycete fungi. In addition, we have shown that DsAvr4, CaAvr4 and CbAvr4 bind chitin, providing further evidence that they, together with CfAvr4 and MfAvr4, are core effectors, i.e. effectors with a conserved biological function to promote the virulence of multiple plant‐pathogenic fungi on distantly related host species. Future studies will focus on providing a mechanistic understanding of how the identified Pro residue promotes the interaction between Avr4 effectors and the Cf‐4 immune receptor.
Experimental Procedures
Nucleotide sequences
GenBank (GB) or Joint Genome Institute (JGI) accession numbers for nucleotide sequences used in this study are as follows: CaAvr4 from Ce. apii, GU574326 (GB); CbAvr4 from Ce. beticola, GU574324 (GB); CfAvr4 from C. fulvum, X78829 (GB); DsAvr4 from D. septosporum, 36707 (JGI).
Protein sequence alignments
Protein sequence alignments were performed between CaAvr4 (GB accession number ADE28521), CbAvr4 (ADE28519), CfAvr4 (Q00363), CnAvr4 (ADE28520), DsAvr4 (EME41286) and MfAvr4 (XP_007926047) using Clustal Omega (Sievers et al., 2011).
Generation of ATTA expression vectors
ATTA expression vectors generated in this study are listed in Table S1 (see Supporting Information). A polymerase chain reaction (PCR) amplification scheme for the generation of these vectors is presented in Table S2 (see Supporting Information), and an associated primer list is provided in Table S3 (see Supporting Information). In brief, the cDNA sequence encoding a mature WT, chimeric or mutant Avr4 protein was fused downstream of the cDNA sequence encoding the Pathogenesis‐Related 1A (PR1A) signal peptide of Nicotiana tabacum (for targeted secretion into the plant apoplast; Hammond‐Kosack et al., 1995), and cloned into the binary vector pK2GW7 (Karimi et al., 2002) behind the Cauliflower mosaic virus (CaMV) 35S promoter. For this purpose, the cDNA sequences encoding PR1A‐DsAvr4, PR1A‐CfAvr4‐CaCys1–Cys4, PR1A‐CfAvr4‐CaCys5–Cys6, PR1A‐CfAvr4‐CaCys6–Cys7, PR1A‐CfAvr4‐CaCys6–Cys8, PR1A‐DsAvr4‐CbCys1–Cys4, PR1A‐DsAvr4‐CbCys4–Cys6 and PR1A‐DsAvr4‐CbCys6–Cys8, flanked by 5′ attB1 and 3′ attB2 sites for recombination into the GATEWAY entry vector pDONR207 (Invitrogen, Carlsbad, CA, USA), were artificially synthesized by Shanghai Shinegene Molecular Biotechnology (Shanghai, China) or Integrated DNA Technologies (Coralville, IA, USA). All other sequences were generated by PCR, with overlap extension PCR (for fusion of PCR amplicons) performed according to the protocol described by Mesarich et al. (2014). PCRs were set up in a final volume of 20 μL containing 20 ng plasmid DNA template, as well as 0.5 μm of forward and reverse primer, and were carried out using Phusion Flash High Fidelity PCR Master Mix (Finnzyme; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. In each case, a final primer pair was used to integrate 5′ attB1 and 3′ attB2 sites for recombination into pDONR207. PCR amplicons were purified using an Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Milwaukee, WI, USA) according to the manufacturer's instructions. Artificially synthesized and PCR‐generated sequences were recombined into pDONR207 using the GATEWAY cloning system (Invitrogen) according to the manufacturer's instructions, and transformed into Subcloning Efficiency DH5α Competent Cells of Escherichia coli (Invitrogen) using a standard heat shock protocol, with selection using 25 μg/mL gentamicin (Sigma‐Aldrich, St. Louis, MO, USA). Plasmid DNA was purified using a QIAprep Spin Miniprep Kit (Qiagen, Benelux BV, Venlo, the Netherlands), and sequence authenticity was confirmed by custom DNA sequencing at Macrogen Inc. (Amsterdam, the Netherlands) using the pDONR207_F and pDONR207_R primer pair. Sequences were recombined into the destination vector pK2GW7 using the GATEWAY cloning system, and transformed into E. coli, with selection using 100 μg/mL spectinomycin (Sigma‐Aldrich). Plasmid DNA was purified, and the authenticity of each ATTA expression vector was confirmed by a diagnostic digest using the NotI restriction enzyme (New England Biolabs, Ipswich, MA, USA). ATTA expression vectors were transformed into A. tumefaciens strain GV3101 by electroporation according to the protocol described by Takken et al. (2000).
Agrobacterium tumefaciens transient transformation assays
Transgenic N. benthamiana plants carrying the Cf‐4 immune receptor gene of tomato (Cf‐4–N. benthamiana), with expression controlled by the CaMV 35S promoter (Gabriels et al., 2006), were used for all ATTAs. Agroinfiltration of Cf‐4–N. benthamiana leaves was essentially performed as described by van der Hoorn et al. (2000) as well as Stergiopoulos et al. (2010). Briefly, to prepare A. tumefaciens transformants for leaf infiltration, each was inoculated into 3 mL of Luria–Bertani liquid medium containing 100 μg/mL of spectinomycin and 25 μg/mL of rifampicin (Sigma‐Aldrich), and incubated for 1.5–2 days at 28 °C and 200 r.p.m. From this liquid culture, 250 μL were used to seed 15 mL of yeast extract and beef liquid medium [0.5% (w/v) beef extract, 0.5% (w/v) bacteriological peptone, 0.5% (w/v) sucrose, 0.1% (w/v) yeast extract, 2 mm MgSO4] containing 20 mm acetosyringone, 100 μg/mL spectinomycin, 25 μg/mL rifampicin and 10 mm 2‐[N‐morpholino]ethanesulfonic acid (MES), and cultured overnight at 28 °C and 200 r.p.m. Cultures were pelleted by centrifugation at 4000 g for 8 min, and resuspended in 10 mL of minimal medium A liquid medium [2.0% (w/v) sucrose, 0.5% (w/v) Murashige and Skoog salts without vitamins, 0.195% (w/v) MES, pH 5.6] containing 200 μm acetosyringone to a final optical density at 600 nm (OD600) of 0.5. Cultures were incubated at room temperature (RT) for 3 h, and infiltrated into the abaxial side of leaves from 5‐week‐old Cf‐4–N. benthamiana plants using a 1‐mL needleless syringe. Inoculated plants were grown in a climate chamber at 70% relative humidity, with temperatures of 21 °C during the day and 19 °C during the night, and a light/dark regime of 12 h/12 h. HR induction was assessed at 7 days post‐infiltration.
Generation of P. pastoris expression vectors
cDNA sequences encoding WT and mutant Avr4 proteins, for the generation of P. pastoris expression vectors used in this study (Table S1), were amplified by PCR from the pK2GW7 ATTA expression vectors described above using the primers listed in Table S3. For this purpose, the forward and reverse primers introduced 5′ SmaI and 3′ EcoRI restriction sites, respectively, for directional cloning into the destination vector pPIC9‐His6 (van Esse et al., 2006; Rooney et al., 2005), and the forward primer also introduced a 24‐bp stretch of nucleotide sequence encoding an N‐terminal FLAG tag for the detection of protein by Western blotting. PCR amplicons were purified, cloned into pGEM‐T Easy (Promega, Madison, WI, USA) according to the manufacturer's instructions, and transformed into E. coli. Plasmid DNA was purified, and sequence authenticity was confirmed by custom DNA sequencing using the universal M13_F/M13_R primer pair. Insert DNA was isolated from pGEM‐T Easy using the SmaI and EcoRI restriction enzymes (New England Biolabs), and directionally ligated into the SmaI/EcoRI‐opened polylinker of pPIC9‐His6 using T4 DNA ligase (Promega) according to the manufacturer's instructions. Ligation products were transformed into E. coli, with selection using 100 μg/mL ampicillin (Sigma‐Aldrich), and plasmid DNA was isolated. Final sequence authenticity was confirmed by custom DNA sequencing using the universal AOX1_F/AOX1_R primer pair. Plasmid DNA was linearized with the SalI restriction enzyme (New England Biolabs), and 10 μg were transformed into the methylotrophic yeast P. pastoris (strain GS115; Invitrogen) by electroporation (Rooney et al., 2005). His+ Mut+ (methanol utilization plus) transformants of P. pastoris were selected according to the Pichia Expression Kit manual (Invitrogen).
Heterologous production of His6‐FLAG‐tagged WT and mutant Avr4 proteins by P. pastoris
Heterologous production of His6‐FLAG‐tagged mutant and WT Avr4 proteins was carried out as described by van den Burg et al. (2001). Briefly, for each P. pastoris transformant, a single colony was cultured in 10 mL of buffered glycerol‐complex liquid medium [1% (w/v) yeast extract, 2% (w/v) peptone, 1.34% (w/v) yeast nitrogen base (YNB) without amino acids, 1% (v/v) glycerol, 0.00004% (w/v) biotin, 100 mm potassium phosphate, pH 6.0) at 28 °C and 250 r.p.m. to an OD600 of 2.0. To induce expression of the heterologous gene, the culture was collected by centrifugation at 3000 g for 5 min, resuspended to an OD600 of 1.0 in buffered methanol‐complex liquid medium [1% (w/v) yeast extract, 2% (w/v) peptone, 1.34% (w/v) YNB without amino acids, 0.5% (v/v) methanol, 0.00004% (w/v) biotin, 100 mm potassium phosphate, pH 6.0), and incubated for 3 days as above, with 0.5% (v/v) methanol added every 24 h. Culture filtrate containing the protein of interest was collected at 1, 2 and 3 days post‐methanol induction, and stored at −20 °C until required.
Polysaccharide affinity precipitation assay and Western blotting
The ability of His6‐FLAG‐tagged WT and mutant Avr4 proteins to bind the insoluble polysaccharides crab‐shell chitin and chitosan (both from Sigma‐Aldrich) was investigated using an in vitro polysaccharide affinity precipitation assay, as described previously by Marshall et al. (2011). As a starting point, 3 mg of each polysaccharide were washed with 500 μL of 100 mm potassium phosphate buffer (pH 6.0) for 10 min at RT and 15 r.p.m., and collected by centrifugation at 13 000 g for 5 min. Polysaccharides were resuspended in 500 μL of unpurified cell‐free P. pastoris culture filtrate containing His6‐FLAG‐tagged WT or mutant Avr4 protein, incubated for 1 h at RT and 15 r.p.m., centrifuged as above, and the supernatant was discarded. Pelleted polysaccharides were washed three times as above, with collection by centrifugation at 13 000 g for 5 min after each washing step. Polysaccharides were resuspended in 200 μL Tricine–sodium dodecylsulfate–polyacrylamide gel electrophoresis (TSDS–PAGE) sample buffer [2% (w/v) SDS, 50 mm dithiothreitol, 10% (v/v) glycerol, 0.0005% (w/v) bromophenol blue, 50 mm tris(hydroxymethyl)aminomethane (Tris)‐HCl, pH 6.8], and the His6‐FLAG‐tagged protein was released by denaturation at 95 °C for 5 min. Polysaccharides were collected by centrifugation, and the supernatant (bound His6‐FLAG‐tagged protein fraction) was collected. Western blotting was performed as follows: 30–50 μL of bound His6‐FLAG‐tagged protein fraction was resolved by 16% TSDS–PAGE, and transferred to a polyvinylidene difluoride membrane (Bio‐Rad, Hercules, CA, USA) by electroblotting for 2 h at 200 mA. The membrane (blot) was blocked in Tris‐buffered saline–skimmed milk [TBS–SM; 50 mm Tris, pH 7.4, 150 mm NaCl; 3% (w/v) SM powder] for 2 h at RT with gentle agitation. Once blocked, the blot was incubated in TBS–SM containing primary (rabbit) monoclonal anti‐FLAG M2 antibody (diluted 1 : 2000; Sigma‐Aldrich) at 4 °C overnight with gentle agitation, washed three times with TBST [TBS with 0.1% (v/v) Tween 20] for 10 min, each with gentle agitation, and then incubated in TBS–SM containing secondary (mouse) enhanced chemiluminescence anti‐horseradish peroxidase‐conjugated antibody (diluted 1 : 2000; GE Healthcare) for 2 h at RT with gentle agitation. Finally, the blot was washed twice with TBST and once with TBS for 10 min, each with gentle agitation, and His6‐FLAG‐tagged proteins were detected using a SuperSignal West Pico or Femto Chemiluminescent Substrate Kit (Rockford, IL, USA) according to the manufacturer's instructions.
Structural modelling
A three‐dimensional model of the CfAvr4 protein structure was generated using the iterative threading assembly refinement (I‐TASSER) server (http://zhanglab.ccmb.med.umich.edu/I‐TASSER/) (Roy et al., 2010), and was assisted with CfAvr4 disulfide bond constraints (van den Burg et al., 2003). The model with the lowest confidence (C)‐score was selected for visualization using the PyMOL molecular graphics system (http://www.pymol.org).
Supporting information
Fig. S1 Pro87 of CfAvr4 is predicted to be surface exposed. A three‐dimensional structure of the mature CfAvr4 protein was predicted using the I‐TASSER server (Roy et al., 2010) with a confidence (C)‐score of −1.11. The molecular surface of the predicted structure is shown. Amino acid residues putatively involved in chitin binding (Asn93, Asp94, Asn95, Trp100, Asp102 and Tyr103; van den Burg et al., 2004) are coloured orange, whereas Pro87 is coloured yellow. The figure was prepared using PyMOL (http://www.pymol.org).
Table S1 Plasmid vectors used in this study.
Table S2 Polymerase chain reaction (PCR) amplification scheme for the generation of Agrobacterium tumefaciens transient transformation assay (ATTA) and Pichia pastoris protein expression vectors.
Table S3 Primers used in this study.
Acknowledgements
We are thankful to Dr Jérôme Collemare and Dr Matthieu Joosten for critically reviewing the manuscript, as well as to Dr Kee Sohn for providing input into the experimental design. Financial assistance for this research was provided by Wageningen University, the Royal Netherlands Academy of Arts and Sciences (KNAW), European Research Area–Plant Genomics (ERA‐PG), the Centre for BioSystems Genomics [CBSG; part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO)], Scion (New Zealand Forest Research) and the New Zealand Bio‐Protection Research Centre. The authors declare no conflicts of interest.
References
- Bolton, M.D. , van Esse, H.P. , Vossen, J.H. , de Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J.E. , van den Berg, G.C.M. , Borrás‐Hidalgo, O. , Dekker, H.L. , de Koster, C.G. , de Wit, P.J.G.M. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , de Wit, P.J. and Vervoort, J. (2001) Efficient 13C/15N double labeling of the avirulence protein AVR4 in a methanol‐utilizing strain (Mut+) of Pichia pastoris . J. Biomol. NMR, 20, 251–261. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Westerink, N. , Francoijs, K.J. , Roth, R. , Woestenenk, E. , Boeren, S. , de Wit, P.J. , Joosten, M.H. and Vervoort, J. (2003) Natural disulfide bond‐disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf‐4‐mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278, 27 340–27 346. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Spronk, C.A. , Boeren, S. , Kennedy, M.A. , Vissers, J. , Vuister, G.W. , de Wit, P.J. and Vervoort, J. (2004) Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein–protein interactions: the chitin‐binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin‐binding domain. J. Biol. Chem. 279, 16 786–16 796. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Harrison, S.J. , Joosten, M.H. , Vervoort, J. and de Wit, P.J. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant–Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Chang, T.‐C. and Stergiopoulos, I. (2015) Inter‐ and intra‐domain horizontal gene transfer, gain–loss asymmetry and positive selection mark the evolutionary history of the CBM14 family. FEBS J. in press. doi: 10.1111/febs.13256. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Teh, T. , Wang, C.‐I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H. , Thomma, B. , van't Klooster, J.W. and de Wit, P.J. (2006) Affinity‐tags are removed from Cladosporium fulvum effector proteins expressed in the tomato leaf apoplast. J. Exp. Bot. 57, 599–608. [DOI] [PubMed] [Google Scholar]
- van Esse, H. , Bolton, M.D. , Stergiopoulos, I. , de Wit, P.J. and Thomma, B. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant–Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- van Esse, H. , van't Klooster, J.W. , Bolton, M.D. , Yadeta, K.A. , van Baarlen, P. , Boeren, S. , Vervoort, J. , de Wit, P.J. and Thomma, B. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels, S.H. , Takken, F.L. , Vossen, J.H. , de Jong, C.F. , Liu, Q. , Turk, S.C. , Wachowski, L.K. , Peters, J. , Witsenboer, H.M. , de Wit, P.J. and Joosten, M.H. (2006) cDNA‐AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant–Microbe Interact. 19, 567–576. [DOI] [PubMed] [Google Scholar]
- Grover, A. (2012) Plant chitinases: genetic diversity and physiological roles. Crit. Rev. Plant Sci. 31, 57–73. [Google Scholar]
- Hammond‐Kosack, K.E. , Staskawicz, B.J. , Jones, J.D.G. and Baulcombe, D.C. (1995) Functional expression of a fungal avirulence gene from a modified potato virus X genome. Mol. Plant–Microbe Interact. 8, 181–185. [Google Scholar]
- Heath, M.C. (2000) Hypersensitive response‐related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A. , Roth, R. and de Wit, P.J. (2001) Identification of distinct specificity determinants in resistance protein Cf‐4 allows construction of a Cf‐9 mutant that confers recognition of avirulence protein Avr4. Plant Cell, 13, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Laurent, F. , Roth, R. and de Wit, P.J.G.M. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf‐9‐induced and Avr4/Cf‐4‐induced necrosis. Mol. Plant–Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , McAdams, S.A. , Bryan, G.T. , Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , Peter van Esse, H. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , van der Krol, S. , Shibuya, N. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H. , Vogelsang, R. , Cozijnsen, T.J. , Verberne, M.C. and de Wit, P.J. (1997) The biotrophic fungus Cladosporium fulvum circumvents Cf‐4‐mediated resistance by producing unstable AVR4 elicitors. Plant Cell, 9, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. , Cozijnsen, T.J. and de Wit, P.J.G.M. (1994) Host resistance to a fungal tomato pathogen lost by a single base‐pair change in an avirulence gene. Nature, 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kruijt, M. , de Kock, M.J. and de Wit, P.J. (2005) Receptor‐like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97. [DOI] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H.A.J. , van den Ackerveken, G.F.J.M. , van den Broek, H.W.J. and de Wit, P.J.G.M. (1997) The in planta‐produced extracellular proteins ECP1 and ECP2 of Cladosporium fulvum are virulence factors. Mol. Plant–Microbe Interact. 10, 725–734. [Google Scholar]
- MacArthur, M.W. and Thornton, J.M. (1991) Influence of proline residues on protein conformation. J. Mol. Biol. 218, 397–412. [DOI] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P. and Rudd, J.J. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesarich, C.H. , Griffiths, S.A. , van der Burgt, A. , Ökmen, B. , Beenen, H.G. , Etalo, D.W. , Joosten, M.H.A.J. and de Wit, P.J.G.M. (2014) Transcriptome sequencing uncovers the Avr5 avirulence gene of the tomato leaf mold pathogen Cladosporium fulvum . Mol. Plant–Microbe Interact. 27, 846–857. [DOI] [PubMed] [Google Scholar]
- Ökmen, B. , Etalo, D.W. , Joosten, M.H.A.J. , Bouwmeester, H.J. , de Vos, R.C.H. , Collemare, J. and de Wit, P.J.G.M. (2013) Detoxification of α‐tomatine by Cladosporium fulvum is required for full virulence on tomato. New Phytologist, 198, 1203–1214. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C. , van't Klooster, J.W. , van der Hoorn, R.A. , Joosten, M.H. , Jones, J.D. and de Wit, P.J. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. and Zhang, Y. (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seear, P.J. and Dixon, M.S. (2003) Variable leucine‐rich repeats of tomato disease resistance genes Cf‐2 and Cf‐5 determine specificity. Mol. Plant Pathol. 4, 199–202. [DOI] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. , McWilliam, H. , Remmert, M. , Söding, J. , Thompson, J.D. and Higgins, D.G. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , de Kock, M.J.D. , Lindhout, P. and de Wit, P.J.G.M. (2007) Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol. Plant–Microbe Interact. 20, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , van den Burg, H.A. , Ökmen, B. , Beenen, H.G. , van Liere, S. , Kema, G.H.J. and de Wit, P.J.G.M. (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA, 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Cordovez, V. , Ökmen, B. , Beenen, H.G. , Kema, G.H.J. and de Wit, P.J.G.M. (2014) Positive selection and intragenic recombination contribute to high allelic diversity in effector genes of Mycosphaerella fijiensis, causal agent of the black leaf streak disease of banana. Mol. Plant Pathol. 15, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken, F.L. , Luderer, R. , Gabriels, S.H. , Westerink, N. , Lu, R. , de Wit, P.J. and Joosten, M.H. (2000) A functional cloning strategy, based on a binary PVX‐expression vector, to isolate HR‐inducing cDNAs of plant pathogens. Plant J. 24, 275–283. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M. , Jones, D.A. , Parniske, M. , Harrison, K. , Balint‐Kurti, P.J. , Hatzixanthis, K. and Jones, J.D. (1997) Characterization of the tomato Cf‐4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf‐4 and Cf‐9. Plant Cell, 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , van Esse, H.P. , Crous, P.W. and de Wit, P.J.G.M. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. [DOI] [PubMed] [Google Scholar]
- de Wit, P.J. , Mehrabi, R. , van den Burg, H.A. and Stergiopoulos, I. (2009a) Fungal effector proteins: past, present and future. Mol. Plant Pathol. 10, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, P.J.G.M. and Kodde, E. (1981) Further characterization and cultivar‐specificity of glycoprotein elicitors from culture filtrates and cell walls of Cladosporium fulvum (syn. Fulvia fulva). Physiol. Plant Pathol. 18, 297–314. [Google Scholar]
- de Wit, P.J.G.M. and Roseboom, P.H.M. (1980) Isolation, partial characterization and specificity of glycoprotein elicitors from culture filtrates, mycelium and cell walls of Cladosporium fulvum (syn. Fulvia fulva). Physiol. Plant Pathol. 16, 391–408. [Google Scholar]
- de Wit, P.J.G.M. , Joosten, M.H.A.J. , Thomma, B.P.H.J. and Stergiopoulos, I. (2009b) Gene for gene models and beyond: the Cladosporium fulvum–tomato pathosystem In: The Mycota V: Plant Relationships, 2nd edn (Deising H.B., ed.), pp. 135–156. Berlin: Springer. [Google Scholar]
- de Wit, P.J.G.M. , van der Burgt, A. , Ökmen, B. , Stergiopoulos, I. , Abd‐Elsalam, K.A. , Aerts, A.L. , Bahkali, A.H. , Beenen, H.G. , Chettri, P. , Cox, M. , Datema, E. , de Vries, R. , Dhillon, B. , Ganley, A.R. , Griffiths, S.A. , Guo, Y. , Hamelin, R.C. , Henrissat, B. , Kabir, M.S. , Jashni, M.K. , Kema, G. , Klaubauf, S. , Lapidus, A. , Levasseur, A. , Lindquist, E. , Mehrabi, R. , Ohm, R.A. , Owen, T.J. , Salamov, A. , Schwelm, A. , Schijlen, E. , Sun, H. , van den Burg, H.A. , van Ham, R.C.H.J. , Zhang, S. , Goodwin, S.B. , Grigoriev, I.V. , Collemare, J. and Bradshaw, R.E. (2012) The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8, e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, B.B. , Thomas, C.M. , Smoker, M. , Grant, M. and Jones, J.D. (2001) Domain swapping and gene shuffling identify sequences required for induction of an Avr‐dependent hypersensitive response by the tomato Cf‐4 and Cf‐9 proteins. Plant Cell, 13, 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, B.B. , Heese, A. , Tomlinson‐Buhot, L. , Jones, D.A. , Pena, M. and Jones, J.D. (2009b) The major specificity‐determining amino acids of the tomato Cf‐9 disease resistance protein are at hypervariable solvent‐exposed positions in the central leucine‐rich repeats. Mol. Plant–Microbe Interact. 22, 1203–1213. [DOI] [PubMed] [Google Scholar]
- Wulff, B.B.H. , Chakrabarti, A. and Jones, D.A. (2009a) Recognitional specificity and evolution in the tomato–Cladosporium fulvum pathosystem. Mol. Plant–Microbe Interact. 22, 1191–1202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Pro87 of CfAvr4 is predicted to be surface exposed. A three‐dimensional structure of the mature CfAvr4 protein was predicted using the I‐TASSER server (Roy et al., 2010) with a confidence (C)‐score of −1.11. The molecular surface of the predicted structure is shown. Amino acid residues putatively involved in chitin binding (Asn93, Asp94, Asn95, Trp100, Asp102 and Tyr103; van den Burg et al., 2004) are coloured orange, whereas Pro87 is coloured yellow. The figure was prepared using PyMOL (http://www.pymol.org).
Table S1 Plasmid vectors used in this study.
Table S2 Polymerase chain reaction (PCR) amplification scheme for the generation of Agrobacterium tumefaciens transient transformation assay (ATTA) and Pichia pastoris protein expression vectors.
Table S3 Primers used in this study.


