Summary
The mycovirus Fusarium graminearum virus 1 (FgV1) is associated with reduced virulence (hypovirulence) of Fusarium graminearum. Transcriptomic and proteomic expression profiling have shown that many F. graminearum genes are differentially expressed as a consequence of FgV1 infection. Several of these genes may be related to the maintenance of the virus life cycle. The host gene, FgHal2, which has a highly conserved 3′‐phosphoadenosine 5′‐phosphatase (PAP phosphatase‐like) domain or inositol monophosphatase (IMPase) superfamily domain, shows reduced expression in response to FgV1 infection. We generated targeted gene deletion and over‐expression mutants to clarify the possible function(s) of FgHal2 and its relationship to FgV1. The gene deletion mutant showed retarded growth, reduced aerial mycelia formation and reduced pigmentation, whereas over‐expression mutants were morphologically similar to the wild‐type (WT). Furthermore, compared with the WT, the gene deletion mutant produced fewer conidia and these showed abnormal morphology. The FgHal2 expression level was decreased by FgV1 infection at 120 h post‐inoculation (hpi), whereas the levels were nine‐fold greater for both the virus‐free and virus‐infected over‐expression mutant than for the WT. FgV1 RNA accumulation was decreased in the deletion mutant at 48, 72 and 120 hpi. FgV1 RNA accumulation in the over‐expression mutant was reduced relative to that of the WT at 48 and 120 hpi, but was similar to that of the WT at 72 hpi. The vertical transmission rate of FgV1 in the gene deletion mutant was low, suggesting that FgHal2 may be required for the maintenance of FgV1 in the host cell. Together, these results indicate that the putative 3′(2′),5′‐bisphosphate nucleotidase gene, FgHal2, has diverse biological functions in the host fungus and may affect the viral RNA accumulation and transmission of FgV1.
Keywords: 3′(2′), 5′‐bisphosphate nucleotidase, FgHal2, Fusarium graminearum, Fusarium graminearum virus 1, secondary metabolism, virus–host interaction
Introduction
Mycoviruses are common in plant‐pathogenic and other fungi and have genomes of double‐stranded RNA (dsRNA), single‐stranded RNA (ssRNA) or DNA (Xie and Jiang, 2014). Although many mycoviruses infect their fungal hosts asymptomatically, some viruses confer hypovirulence, i.e. they reduce the virulence of the pathogenic fungus (Pearson et al., 2009). From an agricultural perspective, mycovirus‐induced hypovirulence could contribute to the development of sustainable biological control (Ghabrial and Suzuki, 2009; Cho et al., 2013). Furthermore, the study of the interactions between a hypovirulence‐associated virus and its host provides an opportunity to characterize viral determinants for the virus disease cycle and to identify the fungal host genes involved in the interaction (Ghabrial and Suzuki, 2009).
Previous research has demonstrated that mycovirus infections alter the expression of a broad range of host genes and cause hypovirulence or phenotypic alterations in the fungal host (Allen and Nuss, 2004; Lee et al., 2014; McBride et al., 2013). Several host genes regulated by Cryphonectria hypovirus 1 strain EP713 (CHV1) infection have been identified in the plant‐pathogenic fungus Cryphonectria parasitica. For example, the expression of PRO1, which is required for female fertility, conidiation and the stable inheritance of CHV1 infection, is down‐regulated (Sun et al., 2009). CpBir, which is involved in fungal conidiation and virulence, is also significantly down‐regulated by CHV1 infection and affects the transmission of the hypovirus in C. parasitica (Gao et al., 2013). CHV1 infection also down‐regulates the expression of the tannic acid‐inducible, small heat shock protein Hsp24, and is associated with the fungal stress response and virulence (Baek et al., 2014).
Fusarium graminearum Schwabe [teleomorph: Gibberella zeae (Schwein.) Petch] is a homothallic ascomycetous fungus which causes Fusarium head blight (FHB) in small grain cereals and ear rot in maize (Desjardins and Proctor, 2007). In addition to reducing the yield of economically important crops, this plant‐pathogenic fungus produces mycotoxins, such as trichothecenes and zearalenone, in cereals, which are very harmful to humans and other animals (Goswami and Kistler, 2004). Recently, several mycoviruses have been detected and identified in Fusarium species, including Fusarium graminearum (Cho et al., 2013). Among Fusarium graminearum viruses, Fusarium graminearum virus 1 strain DK21 (currently named FgV1) has been associated with reduced host virulence, a reduced rate of mycelial growth, increased pigmentation and inhibition of mycotoxin production (Chu et al., 2002). FgV1 mainly occurs as the dsRNA form, but expresses its genome in a manner similar to that of positive‐strand potex‐like RNA viruses, and is phylogenetically similar to members of the genus Hypovirus based on the alignment of proteins containing RNA‐dependent RNA polymerase domains (Kwon et al., 2009b).
With the availability of well‐developed forward and reverse genetic tools, including an efficient DNA‐mediated transformation system and the availability of a completely sequenced genome, F. graminearum can be used as a model system to understand virus–host interactions (Güldener et al., 2006; Son et al., 2013). By using transcriptomic and proteomic approaches, we have determined previously that many fungal host genes are differentially expressed in response to FgV1 infection. These host genes are involved in cell signalling pathways, the stress response, post‐transcriptional gene silencing, differentiation and other processes (Cho et al., 2012; Kwon et al., 2009a; Lee et al., 2014). Researchers have proposed that some of these genes might function as host factors to maintain the life cycle of FgV1 (Kwon et al., 2009a; Son et al., 2013). For example, Hex1, a major protein of the Woronin body, plays a crucial role in maintaining the cellular integrity and pathogenicity of F. graminearum (Son et al., 2013). Moreover, the accumulation of FgV1 viral RNA depends on the HEX1 gene expression level, although the molecular mechanisms underlying Hex1 protein–FgV1 interactions are not clearly understood (Son et al., 2013). Other host genes in F. graminearum which may directly and/or indirectly affect the life cycle of FgVs, however, have not been identified.
This article concerns the characterization of one F. graminearum gene, FgHal2, which shows a reduced gene expression level (transcript and protein expression profiles) in response to FgV1 infection (Kwon et al., 2009a; Lee et al., 2014). FgHal2 is an orthologue of the yeast gene Hal2 (MET22) and encodes a hypothetical protein that has a conserved domain of 3′‐phosphoadenosine‐5′‐phosphate (pAp) phosphatase. Hal2 encodes 3′(2′),5′‐bisphosphate nucleotidase, which is involved in methionine biosynthesis in the sulfate assimilation pathway and which also affects salt tolerance in yeast (Gil‐Mascarell et al., 1999). Met22p removes the 3′ phosphate from pAp, thus producing adenosine monophosphate (AMP), and also hydrolyses 3′‐phosphoadenosine‐5′‐phosphosulfate (pApS) (Hudson and York, 2012). Met22p can suppress viral RNA recombination of Tomato bushy stunt virus (TBSV) in yeast and is related to the Xrn1p 5′–3′ ribonuclease, a known suppressor of viral RNA recombination (Jaag and Nagy, 2010). SAL1, one of the Arabidopsis orthologues of yeast Hal2, also has multiple functions. It catabolizes inositol 1,4,5‐trisphosphate (IP3) and pAp (Estavillo et al., 2011; Hudson and York, 2012), represses post‐transcriptional gene silencing by degrading the exoribonuclease inhibitor pAp, and functions in stress signalling and developmental processes in plants (Estavillo et al., 2011; Gy et al., 2007).
In this study, we provide evidence that the putative 3′(2′),5′‐bisphosphate nucleotidase gene, FgHal2, in F. graminearum is down‐regulated following FgV1 infection. We also investigate the possible function(s) of FgHal2 in F. graminearum using gene deletion and gene over‐expression mutants. We found that deletion of FgHal2 reduced conidiation, mycelial growth and the production of secondary metabolites. Moreover, deletion of FgHal2 decreased viral RNA accumulation and the vertical transmission of FgV1 via conidia. Together, these results indicate that F. graminearum can down‐regulate one of its major multifunctional genes, FgHal2, in response to FgV1 infection.
Results
Sequence analysis of FgHAL2
The genomic sequence of FGSG_09532, corresponding to the FgHAL2 gene, was determined at the Munich Information Centre of Protein Sequences (MIPS) and the Fusarium comparative database [Fusarium Comparative Sequencing Project, Broad Institute of Harvard and Massachusetts Institute of Technology (MIT) (http://www.broadinstitute.org)]. We aligned amino acid sequences of FgHal2 and FgHal2 orthologues using the GeneDoc program (http://www.nrbsc.org/gfx/genedoc/). The result of multiple amino acid sequence alignment indicated that the 3′‐phosphoadenosine 5′‐phosphatase (PAP phosphatase‐like) or inositol monophosphatase (IMPase) superfamily domain is highly conserved among all orthologues and FgHal2 (Fig. 1). The deduced amino acid sequence of FgHal2 in F. graminearum shares 45% sequence identity with the Saccharomyces cerevisiae Met22 protein. FgHal2 also shows high sequence identity with proteins in F. oxysporum (95%), F. verticillioides (95%), Neurospora crassa (76%) and Magnaporthe oryzae (78%). The F. graminearum chromosome has two other putative genes that contain a PAP phosphatase‐like domain: Fg07103 and Fg01708. The comparison of the deduced amino acid sequence of FgHal2 with those of Fg07103 and Fg01708 showed 42% and 35% sequence identity, respectively. When we conducted real‐time quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) to quantify the alteration in the expression level of Fg07103, Fg01708 and FgHal2 after FgV1 infection, only the FgHal2 gene was down‐regulated after virus infection; the transcription levels of Fg07103 and Fg01708 genes did not change significantly (Fig. S1, see Supporting Information).
Figure 1.
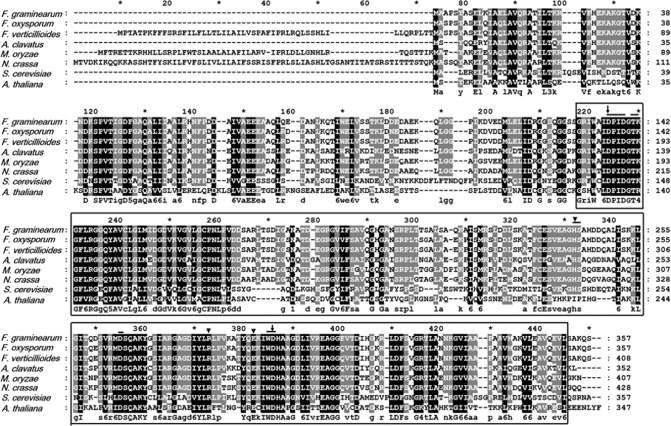
Amino acid sequence alignment of FgHal2 corresponding to the conserved 3′‐phosphoadenosine 5′‐phosphatase (PAP phosphatase‐like) domain of the predicted FgHal2 protein and orthologues. The positions of the amino acid sequences are indicated on the right. Species and GenBank accession numbers are as follows: Arabidopsis thaliana (NP201205.1); Aspergillus clavatus (XP001268064.1); Magnaporthe oryzae (XP003719499.1); Fusarium graminearum (XP389708.1); Fusarium oxysporum (EGU82355.1); Fusarium verticillioides (EWG42162); Neurospora crassa (EAA28448.2); Saccharomyces cerevisiae (NP014577.1) The conserved amino acids are shaded. The alignment was generated with GeneDoc software. The PAP phosphatase‐like domain [cd01517] is boxed; the conserved active site residues are indicated by a bar above the sequence; the substrate binding site residues are indicated by arrowheads; and the putative Li+/Na+ binding sites are indicated by black arrows.
Generation of mutants
To investigate the function of FgHal2 in F. graminearum, we generated gene deletion and gene over‐expression mutants using a homologous recombination strategy (Fig. 2A). For the construction of the gene deletion mutant, the 5′‐ and 3′‐flanking regions were amplified from wild‐type (WT) G. zeae PH‐1 genomic DNA by PCR and fused to the geneticin (gen) resistance cassette. The fused DNA construct was used to transform fungal protoplast, and gen‐resistant transformants were confirmed by Southern blot hybridization. For the construction of the complementation strain of FgHal2, the open reading frame (ORF) of FgHal2 and the green fluorescent protein (GFP) fusion construct under the control of the FgHal2 native promoter were inserted into the genome of the FgHal2 deletion mutant. We also generated an FgHal2 over‐expression strain, which contained the elongation factor 1α (EF1α) promoter of F. verticillioides upstream of the FgHal2 ORF. All transformed fungal mutants were confirmed by Southern blot analysis (Fig. 2B). qRT‐PCR of the virus‐free strains confirmed that the transcript level of FgHal2 was not detected at all in the deletion mutant, greatly increased in the over‐expression mutant, and similar in the WT and the complementation mutant (Fig. 2C). All fungal strains used in this study are listed in Table 1.
Figure 2.
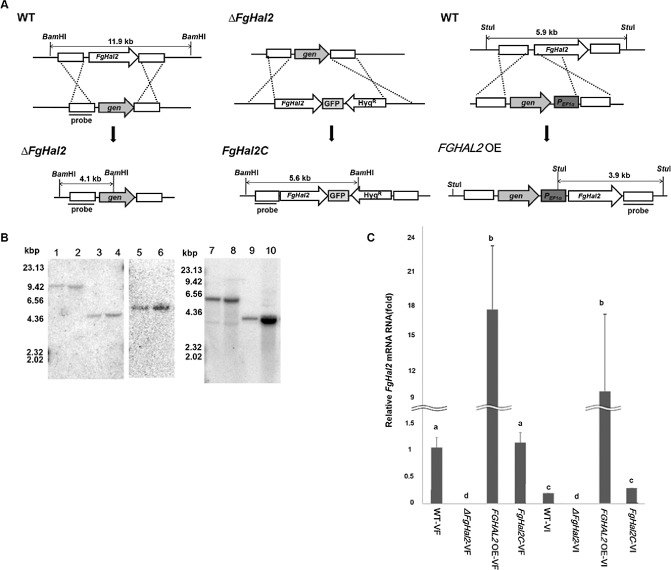
Generation of FgHal2 gene deletion, complementation and over‐expression mutants of Fusarium graminearum. (A) Schematic representation of the homologous gene recombination strategy used to generate the FgHal2 deletion mutants (left), FgHal2 complementation mutant (middle) and FgHal2 over‐expression mutant (right). The FgHal2 complementation mutant strain was fused with green fluorescent protein (GFP). The promoter was replaced with the elongation factor 1α (EF1α) promoter in the FGHAL2 OE strain. WT, F. graminearum wild‐type strain PH‐1; ΔFgHal2, FgHal2 gene deletion mutant; FgHal2C, FgHal2 complementation and GFP‐fused mutant; FGHAL2 OE, FgHal2 over‐expression mutant. (B) Southern blot hybridization of the F. graminearum mutant strains. The sizes of the DNA standards are indicated to the left of the blot. Lanes 1–6 indicate Southern blot images of BamHI‐digested genomic DNA of each strain. Lanes 1 and 2 represent the WT, lanes 3 and 4 represent ΔFgHal2, and lanes 5 and 6 represent the complementary strain (FgHal2C). A 32P‐labelled DNA fragment of the 5′ flanking region of the FgHal2 gene was used as a probe for Southern blot hybridization (lanes 1–6). Lanes 7–10 show Southern blot images of StuI‐digested genomic DNA of each strain. Lanes 7 and 8 represent the WT, and lanes 9 and 10 represent FGHAL2 OE. A 32 P‐labelled DNA fragment of the 3′ flanking region of the FgHal2 gene was used as a probe (lanes 7–10). In lanes 1–10, odd‐numbered lanes indicate virus‐free strains and even‐numbered lanes indicate virus‐infected strains. (C) Relative FgHal2 mRNA transcript levels in the WT and mutant strains. Relative transcript levels were normalized to elongation factor 1α and ubiquitin C‐terminal hydrolase. cDNAs were generated from total RNA extracts from mycelia harvested after 120 h of incubation. Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test. VF, virus‐free; VI, Fusarium graminearum virus 1 (FgV1)‐infected strains.
Table 1.
Fusarium graminearum strains used in this study
| Strain | Description | References |
|---|---|---|
| WT‐VF | Wild‐type F. graminearum (lineage 7), Gibberella zeae PH‐1 | Lee et al. (2014) |
| WT‐VI | Wild‐type PH‐1 infected with FgV1 | Lee et al. (2014) |
| ΔFgHal2‐VF | FgHal2 gene deletion mutant in WT‐VF genetic background | This study |
| ΔFgHal2‐VI | FgV1 introduced into ΔFgHal2‐VF by hyphal anastomosis | This study |
| FGHAL2 OE‐VF | FgHal2 gene over‐expression mutant in WT‐VF genetic background | This study |
| FGHAL2 OE‐VI | FgV1 introduced into FGHAL2 OE‐VF by hyphal anastomosis | This study |
| FgHal2C‐VF | FgHal2 gene complementation mutant in WT‐VF genetic background | This study |
| FgHal2C‐VI | FgV1 introduced into FgHal2C‐VF by hyphal anastomosis | This study |
Effect of FgV1 infection on FgHal2 gene expression level
To determine whether the expression level of FgHal2 mRNA differed among the F. graminearum strains, we introduced FgV1 into the virus‐free FgHal2 gene deletion strain, complementation strain and over‐expression strain through hyphal anastomosis. As indicated by qRT‐PCR, the expression level of FgHal2 mRNAs was significantly lower in the FgV1‐infected WT than in the virus‐free WT at 120 h post‐inoculation (hpi) (Fig. 2C). The FgHal2 transcript level was also significantly lower in the virus‐infected WT and the complemented strain than in the corresponding virus‐free strains when the strains were incubated for 48 and 72 h in complete medium (CM) liquid culture (data not shown). Previously, FgHal2 was found to be down‐regulated by FgV1 infection when two‐dimensional electrophoresis was used to determine the protein expression level of virus‐infected and virus‐free F. graminearum (Kwon et al., 2009a). This down‐regulation was consistently observed at the transcription level when qRT‐PCR was performed in the current study. These results demonstrate that FgV1 infection reduces the expression of FgHal2 in F. graminearum. FgHal2 transcript levels in over‐expression strains, however, were similar regardless of virus infection. The latter finding might be explained by the strong constitutive EF1α promoter that was inserted into the over‐expression strain.
Effects of FgHal2 deletion and over‐expression on vegetative growth and pigmentation
WT‐VF, FGHAL2 OE‐VF and FgHal2C‐VF showed similar morphologies on CM at 120 hpi, but the deletion of FgHal2 (ΔFgHal2‐VF) led to abnormal colony morphology, a slow growth rate, reduced aerial mycelium formation and reduced red pigmentation (Fig. 3). ΔFgHal2‐VF formed a hyphal mass that was pale yellow or white, whereas the WT, complementation strain and over‐expression strain formed a pink or red hyphal mass. Relative to the virus‐free strains, the virus‐infected strains had an irregular morphology, increased pigmentation and reduced aerial mycelia at 120 hpi on CM, as reported previously (Lee et al., 2014). Radial growth on CM was reduced for the deletion mutant whether it was virus‐free (ΔFgHal2‐VF) or virus‐infected (ΔFgHal2‐VI). The radial growth of FGHAL2 OE‐VF did not differ from that of WT‐VF, and the same was true for FGHAL2 OE‐VI and WT‐VI.
Figure 3.
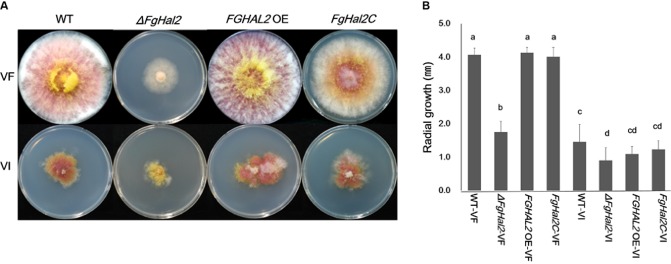
Colony morphology and mycelial growth of virus‐free and Fusarium graminearum virus 1 (FgV1)‐infected strains [wild‐type (WT) and FgHal2 gene deletion, complementation and over‐expression mutants] of F. graminearum. (A) Colony morphology on complete medium (CM) agar. Colonies were photographed after 5 days of incubation. (B) Radial growth after 5 days of incubation on CM agar. Means and standard deviations were calculated from three repeated experiments. Bars with different letters are significantly different (P < 0.05) based on Tukey's test, which was conducted using SPSS 12.0 (SPSS, Inc., Chicago, IL, USA).
As shown in Fig. 3, both the deletion of FgHal2 and FgV1 infection reduced the production of aerial mycelia. Because the aerial hyphae of many fungi have hydrophobic cell surfaces (Jiang et al., 2011), we hypothesized that deletion of FgHal2 or infection by FgV1 might reduce the hydrophobicity of the aerial hyphae. To measure the hydrophobicity of the hyphal surface, we applied 15 μL of water containing bromophenol blue to 3‐day‐old colonies of all virus‐free and virus‐infected strains (Fig. S2, see Supporting Information). The water formed spherical droplets on all virus‐free strains that produced abundant mycelia, including the WT, over‐expression strain and complementation strain. In contrast, water droplets were absorbed by the mycelia of the knockout mutant and virus‐infected WT and mutant strains. These results indicate that FgHal2, in addition to being involved in mycelial growth, is involved in the hydrophobicity of F. graminearum aerial hyphae. FgV1 infection also affected the hydrophobicity of the aerial mycelia, perhaps because FgV1 infection reduces the expression of FgHal2 (Fig. 2C).
Effects of FgHal2 deletion and over‐expression on conidiation and conidial morphology
To determine the effects of FgHal2 on conidiation and conidial morphology, we quantified the production of conidia by virus‐free and virus‐infected strains on carboxymethyl cellulose (CMC) medium (Fig. 4A). Deletion of FgHal2 dramatically reduced conidial production on CMC medium regardless of FgV1 infection. Conidial production did not differ significantly among the WT, complementation strain or over‐expression strain regardless of FgV1 infection (Fig. 4A). Our previous study also indicated that FgV1 infection did not affect conidial production (Lee et al., 2014).
Figure 4.
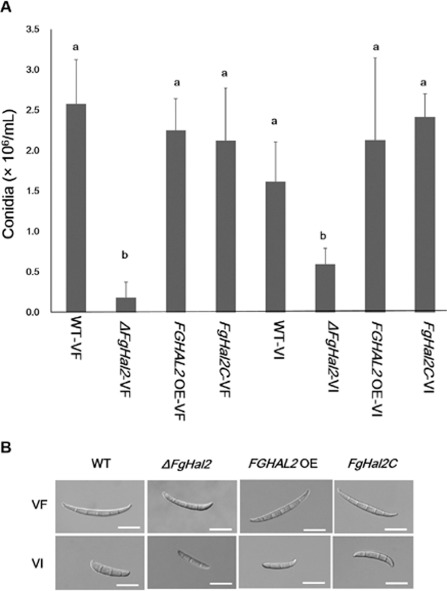
Conidial production by virus‐free and Fusarium graminearum virus 1 (FgV1)‐infected strains [wild‐type (WT) and FgHal2 gene deletion, complementation and over‐expression mutants] of F. graminearum. (A) Conidia production after 5 days on carboxymethyl cellulose (CMC). Values are means and standard deviations of three repeated experiments. Means with different letters are significantly different (P < 0.05) based on Tukey's test. (B) Conidial morphology. Representative conidia of virus‐free (VF) and virus‐infected (VI) strains were photographed with differential interference contrast optics. Bar, 20 μm.
We have shown previously that FgV1 infection affects conidium morphology, i.e. FgV1‐infected F. graminearum produce swollen and short conidia (Lee et al., 2014). In the current study, all virus‐infected strains also produced swollen and short conidia (Fig. 4B). With respect to virus‐free strains, ΔFgHal2‐VF produced shorter conidia than WT‐VF, FgHal2C‐VF or FGHAL2 OE‐VF (Fig. 4B). Overall, these results suggest that the FgHal2 gene is related to conidial production and development.
Effect of FgHal2 deletion and over‐expression on virulence
To determine the effect of FGHAL2 on F. graminearum virulence, we inoculated wheat heads with conidial suspensions of virus‐free and virus‐infected WT or mutant F. graminearum strains, including the deletion mutant ΔFgHal2. FHB symptoms appeared on wheat heads inoculated with the virus‐free WT (WT‐VF), virus‐free over‐expression strain (FGHAL2 OE‐VF) and virus‐free complementation strain (FgHal2C‐VF) (Fig. 5A). However, inoculation with ΔFgHal2‐VF did not result in bleaching of the head or even of the inoculated spikelet. Because the growth rate of ΔFgHal2‐VF was severely decreased relative to that of the WT, we monitored inoculated wheat heads for a prolonged period. Even at 21 days post‐inoculation, however, no symptoms were caused by ΔFgHal2‐VF. None of the virus‐infected strains caused visual symptoms on the wheat heads or inoculated spikelets (Fig. 5B).
Figure 5.
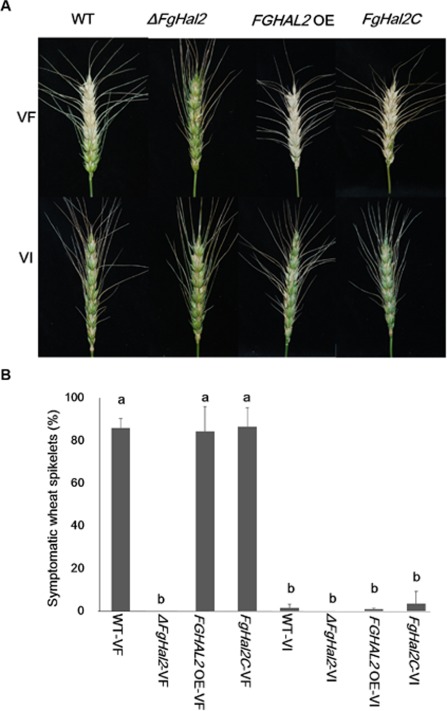
Virulence of virus‐free and Fusarium graminearum virus 1 (FgV1)‐infected F. graminearum strains [wild‐type (WT) and FgHal2 gene deletion, complementation and over‐expression mutants] on wheat. (A) Representative wheat heads 14 days after they were inoculated with conidia. (B) The percentage of symptomatic wheat spikelets 14 days after inoculation. Means and standard deviations were calculated from independent repeated experiments. Error bars indicate standard deviation. Bars with different letters are significantly different (P < 0.05).
Effect of FgHal2 deletion and over‐expression on vertical transmission
To assess the stability of the FgV1 virus in F. graminearum, we quantified the vertical transmission of FgV1 through the asexual spores of the WT and mutant strains (Table 2). Transmission to the first, second and third generations of conidia was 100% for the WT and complementation strain FgHal2C. Transmission for the over‐expression strain FGHAL2 OE was somewhat reduced in the first generation, but was 100% for the second and third generations. Transmission for the deletion strain ΔFgHal2 decreased substantially with each generation. These data indicate that deletion of the FgHal2 gene reduces the vertical transmission via conidia or reduces the maintenance of FgV1 in infected host cells.
Table 2.
Vertical transmission of Fusarium graminearum virus 1 (FgV1) between generations of F usarium graminearum strains
| Strain | Vertical transmission rate (%) | ||
|---|---|---|---|
| 1st generation | 2nd generation | 3rd generation | |
| WT‐VI | 100 | 100 | 100 |
| ΔFgHal2‐VI | 63 | 45 | 15 |
| FGHAL2 OE‐VI | 84 | 100 | 100 |
| FgHal2C‐VI | 100 | 100 | 100 |
Vertical transmission was measured as the percentage of FgV1‐positive isolates among the total number of single‐conidium isolates. Each value is based on ≥30 single‐conidium isolates.
Effect of FgHal2 deletion and over‐expression on the accumulation of FgV1 RNA
We performed qRT‐PCR to determine whether the deletion or over‐expression of FgHal2 affects viral RNA accumulation (Fig. 6). At 48 hpi, viral RNA levels were significantly greater in the WT than in the deletion mutant, over‐expression mutant or complemented mutant. At 72 hpi, viral RNA levels had significantly increased in all strains except the deletion mutant. At 120 hpi, viral RNA levels remained low in the deletion mutant, had decreased substantially in the over‐expression mutant and had dropped only moderately in the WT and complemented mutant. These results suggest that FgHal2 may affect FgV1 RNA accumulation either directly or indirectly.
Figure 6.
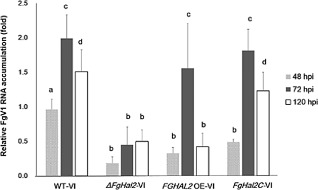
Accumulation of Fusarium graminearum virus 1 (FgV1) viral RNA in FgV1‐infected wild‐type (WT), ΔFgHal2, FgHal2C and FGHAL2 OE strains of F. graminearum. Quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) was used to quantify FgV1 viral RNA at 48, 72 and 120 h post‐inoculation (hpi). Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test.
Discussion
On the basis of computational sequence analysis, we assumed that FgHal2 encodes a 3′‐nucleotidase which is highly conserved in fungi. This nucleotidase is a member of the family of phosphatases that share the properties of sensitivity to Li+ and Na+ ions, metal dependence and substrate selectivity (Hudson and York, 2012). Only one gene encodes for the 3′‐nucleotidase in S. cerevisiae, but two or more genes code for this enzyme in other fungi and plants, including Arabidopsis. The expression of isoforms of pAp phosphatase enzymes may be differentially regulated during organ differentiation and stress responses, and such enzymes usually exhibit substrate stringency and sensitivity to toxic cations (Gil‐Mascarell et al., 1999; Hudson and York, 2012). Fusarium graminearum has three putative proteins with pAp phosphatase‐conserved domains. Interestingly, only FgHal2 is significantly down‐regulated by FgV1 infection. This result suggests that FgHal2 may be closely related to certain regulatory processes that are induced by FgV1 infection.
As mentioned earlier, Hal2 (Met22) has been identified and characterized in yeasts (Gläser et al., 1993; Vaupotič et al., 2007) and plants (Gil‐Mascarell et al., 1999). The function of this gene in plant‐pathogenic fungi, however, has not been determined. Because the inhibition of Hal2 causes PAP accumulation in the cell, it also reduces sulfotransferase activity, RNA‐processing enzymes and the sulfur assimilation pathway (Belles and Serrano, 1995; Gašparič et al., 2013). Accordingly, these effects impede growth. FRY1 in Arabidopsis is a homologue of MET22 in yeast, and the fry1 mutant generates fewer lateral roots and shows stunted growth, short hypocotyls, curly leaves and a retarded phase transition and leaf initiation (Chen and Xiong, 2010). In this study, deletion of the FgHal2 gene significantly reduced the growth of hyphae in and above the agar medium. Deletion of FgHal2 also reduced conidial production and resulted in abnormal conidial morphology. These results suggest that FgHal2 is involved in the regulation of F. graminearum growth and development. In addition, FgHal2 deletion reduced the hydrophobicity of the fungal colony, suggesting that FgHal2 contributes to cell wall integrity.
The colonies of ΔFgHal2‐VF and ΔFgHal2‐VI growing on CM agar showed a dramatic decrease in red pigmentation (Fig. 3A). This indicates that deletion of FgHal2 may affect secondary metabolism in F. graminearum. Fusarium graminearum produces diverse secondary metabolites, including the polyketide pigments aurofusarin (AUR) and rubrofusarin (Kim et al., 2006). To date, few genes have been characterized that participate in the biosynthesis of AUR (Geng et al., 2014). Gip2 is a putative positive regulator of the AUR biosynthetic gene cluster, and polyketide synthase 12 (Pks12) lies upstream of Gip1 and is responsible for changing a precursor to AUR (Geng et al., 2014). Furthermore, an osmosensor histidine kinase (FgOs1) and an osmoregulatory mitogen‐activated protein kinase (MAPK) pathway (consisting of FgOs4, FgOs5 and FgOs2) are associated with the regulation of AUR in F. graminearum (Ochiai et al., 2007). Among the genes related to secondary metabolite synthesis in F. graminearum, FgOs2, Gip2, Pks12 and AURj were not detected by qRT‐PCR in ΔFgHal2‐VF (Fig. S3, see Supporting Information). In addition, qRT‐PCR indicated that the transcript levels of FgOs2 and Pks12 were significantly different in FGHAL2 OE‐VI and WT‐VI than in WT‐VF (Fig. S3). These results support the inference that FgV1 infection and FgHal2 may regulate AUR biosynthesis so as to affect osmoregulation by the MAPK signalling pathway. Together, the morphological phenotype of ΔFgHal2‐VF and the alteration in the transcript levels of secondary metabolite synthesis genes in FGHAL2 OE and WT‐VI suggest that FgHal2 is involved in secondary metabolite production. For this reason, the suppression of FgHal2 in WT‐VI may be involved in the biological and physiological changes caused by FgV1 infection. Presumably, mycovirus infection and fungal secondary metabolism are related, sharing various regulatory pathways. Previous research has shown that CHV1‐EP713 hypovirus alters host G‐protein signal transduction (Dawe et al., 2004) and the Hog1‐homologue cpmk1 gene of the MAPK pathway of C. parasitica (Park et al., 2004). These signalling pathways are associated with the regulation of secondary metabolite synthesis. In this regard, the expression levels of genes in the osmotic stress signal transduction pathway (FgOs5 and FgOs2) and those related to polyketide synthase (Pks12, Gip4 and Gip5) were significantly different in the FgV1‐infected WT than in the virus‐free WT (Lee et al., 2014). It would be useful to determine how FgHal2 affects secondary metabolite synthesis pathways that may be related to FgV1 infection.
We used qRT‐PCR to assess FgV1 RNA accumulation in our F. graminearum strains at 48, 72 and 120 hpi. Because FgHal2 was repressed by FgV1 infection, we expected that the viral RNA accumulation level would be similar or higher in virus‐infected ΔFgHal2 than in the virus‐infected WT. However, we found that significantly less FgV1 RNA accumulated in ΔFgHal2‐VI than in WT‐VI. This suggests that the FgHal2 gene may also directly or indirectly affect virus RNA accumulation. The FgV1 RNA level in FGHAL2 OE‐VI was low relative to that in WT‐VI at 48 hpi, increased at 72 hpi and then returned to a low level at 120 hpi (Fig. 6). The FgHal2 expression level, however, was approximately nine‐fold greater in FGHAL2 OE‐VI than in WT‐VI. A possible explanation for this decrease in the FgV1 RNA level in FGHAL2 OE‐VI is that over‐expression of FgHal2 may help the fungus to overcome osmotic and other stresses, as demonstrated for the orthologue of FgHal2, Hal2 (Gašparič et al., 2013; Shinozaki and Yamaguchi‐Shinozaki, 1999). This result suggests that FgHal2 may affect the host defence response. If FgV1 infection is considered as a biological stress to F. graminearum, fungal stress‐responsive pathways or elements may also be affected by virus infection. As noted earlier, the small heat shock protein CpHsp24 contributes to stress adaptation, but not to hypoviral replication, in C. parasitica (Baek et al., 2014). Perhaps the reduced expression level of FgHal2 in response to virus infection does not protect the fungal host in some respects, but does generate unsuitable conditions for FgV1 persistence in host cells.
The FgV1 transmission experiment in the current study suggests that FgHal2 may help to maintain FgV1 infection. Relative to the vertical transmission in the FgV1‐infected WT, the vertical transmission in ΔFgHal2‐VI decreased gradually and was only 15% in the third generation (Table 2). This failure to maintain FgV1 was also observed during serial mycelial subcultures on agar. During repeated subculture on potato dextrose agar (PDA), WT‐VI, FgHal2C‐VI and FGHAL2 OE‐VI did not produce virus‐cured isolate under typical conditions. Unlike these strains, ΔFgHal2‐VI rapidly produced a colony with virus‐free and virus‐infected sectors (Fig. S4, see Supporting Information). These observations, which indicate that FgHal2 is necessary for the maintenance of FgV1 infection, may explain why F. graminearum down‐regulates FgHal2 expression after FgV1 infection. The C. parasitica pro1 gene, which encodes the Zn(II)2Cys6 transcription factor, is also down‐regulated in response to CHV1 infection (Sun et al., 2009). The hypovirus‐infected pro1‐disruption mutant was frequently recovered from hypovirus and produced colony coexisting the virus‐free sector and virus‐infected sectors on PDA. Like pro1, FgHal2 levels in WT were reduced, but not eliminated, by FgV1 infection. The decrease in vertical transmission and low stability after successive subcultures on agar for the FgHal2 deletion mutant indicate that FgV1 requires FgHal2 to maintain itself in the host. Therefore, the down‐regulation of the expression of the FgHal2 gene might be harmful for the fungal host, but this reduction is an inevitable consequence to provoke the persistence of the virus and to induce the defence response. However, the association between FgHal2 and the maintenance of FgV1 infection requires further investigation.
Taken together, our results suggest that F. graminearum requires the putative 3′(2′),5′‐bisphosphate nucleotidase gene FgHal2 for normal mycelial growth, conidial production, conidial maturation and secondary metabolite synthesis. When challenged by FgV1, however, F. graminearum represses FgHal2 expression, possibly to reduce viral RNA accumulation and to generate unstable conditions for the maintenance of FgV1.
Experimental Procedures
Fungal strains and media
The PH‐1 strain of F. graminearum was used as the WT in this study (Lee et al., 2014). Transgenic mutants were generated from the WT, and FgV1 was transferred to each strain by hyphal anastomosis with the PH‐1/FgV1 strain, as described previously (Lee et al., 2014). All strains used in this study are listed in Table 1. For extractions of total RNA and genomic DNA, strains were grown in 50 mL of liquid CM at 25 °C for 5 days on a rotary shaker at 150 rpm. The WT and mutant strains were stored in 15% (v/v) glycerol at −80 °C and were reactivated on CM agar medium.
Sequence analysis
Nucleotide sequence and amino acid sequences were obtained from the National Center for Biotechnology Information (NCBI) database and Fusarium comparative database (Fusarium Comparative Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org). Amino acid sequences were aligned using the MegAlign program in DNASTAR and GeneDoc programs (http://www.nrbsc.org/gfx/genedoc/). A conserved domain search and sequence similarity analysis were conducted with the NCBI blast program.
Construction of FgHAL2 gene deletion, over‐expression and complementation mutants
DNA constructs for targeted gene deletion, complementation and over‐expression were amplified by the double‐joint (DJ) PCR method, as described in a previous study, with slight modifications (Son et al., 2013). The 5′‐ and 3′‐flanking regions of the FgHal2 gene and a gen resistance cassette were amplified from F. graminearum PH‐1 and pII99, and fused by DJ PCR under the PCR conditions described previously (Lee S. et al., 2011b; Lin et al., 2011). The primer sets used in this study are listed in Table S1 (see Supporting Information). To complement the FgHal2 gene deletion mutant, the DNA fragment carrying the native promoter and the FgHal2 ORF was amplified with 9532 5F2/9532 5′ RC primers and then fused with the green fluorescent protein gene (gfp) and the hygromycin (hyg) resistance cassette, which was amplified from the pIGPAPA vector (Lin et al., 2011) by DJ PCR. To generate over‐expressing FgHal2 mutants, the 5′‐ and 3′‐ends of the FgHal2 region were amplified by the primer pairs 9532 5F/9532 5F Gen‐R and EF1 9532‐3F/9532 OE‐3R, respectively. The gen‐Pef1α construct, containing the gen resistance cassette and the EF1α promoter (Pef1α) from F. verticillioides, was amplified from pSKGEN (Lee S. et al., 2011b) with Gen_EF1 F/EF1 pro R primers. Amplicons were joined by DJ PCR as described above, and a final PCR product was generated using the nested primers. These three final DNA constructs were used for fungal transformation. Fungal protoplasts were transformed with polyethyleneglycol (PEG) as described previously (Son et al., 2013). Transformants were selected on PDA supplemented with 50 μg/mL of hyg or gen. After analysis of all transgenic strains using Southern blot (described below), FgV1 was introduced into the virus‐free transformant strains through hyphal anastomosis. Viral infection was confirmed by RT‐PCR using FgV1‐specific primer pairs.
DNA extraction and Southern blot hybridization
After fungal strains had been incubated for 5 days on CM liquid medium, mycelia were collected by filtration through Whatman 3MM filter paper (GE Healthcare, Uppsala, Sweden), washed with distilled water, pressed between paper towels to remove excess water and stored at −80 °C. Genomic DNA was extracted as described previously (Lee K.M. et al., 2011a). For the Southern blot hybridization of WT and transgenic mutants, 10 μg of genomic DNA were digested with the appropriate enzyme. Agarose gel electrophoresis, capillary blotting and hybridization were performed following standard protocols, as described previously (Lee K.M. et al., 2011a). Labelled probes were generated by standard reaction with DNA fragments in 20 mL of 10 mm Tris‐HCl (pH 7.5), 7 mm MgCl2, 0.1 mm dithiothreitol (DTT), 30 μCi [α‐32P]dCTP, 3 mm deoxynucleoside triphosphate (dNTP) mix, 10 pmol of random primers and 2 U Klenow fragment (TaKaRa Bio Inc., Ohtsu, Japan). Hybridized blots were exposed to phosphoimaging screens (BAS‐IP MS 2040, Fuji Photo Film Co., Tokyo, Japan) and analysed using a BAS‐2500 image analysis system (Fuji Photo Film Co).
Mycelial growth and conidiation test
The morphologies of the virus‐free and virus‐infected WT and transformed mutant strains were observed after the strains had grown for 5 days at 25 °C on PDA, CM and minimal medium (MM; 0.05% KCl, 0.2% NaNO3, 3% sucrose, 1% KH2PO4, 0.05% MgSO4·7H2O, 0.02% trace elements, 2% agar). The radial growth was measured on PDA after 3, 4 and 5 days at 25 °C. The experiment was repeated three times independently. To assess colony surface hydrophobicity, a 15‐μL droplet of dH2O coloured with bromophenol blue was added to the top of 4‐day‐old fungal colonies grown on CM agar; the colonies were photographed 10 min later.
For the assessment of conidial production, the virus‐free and FgV1‐infected strains were incubated for 3 days on CM liquid medium. Mycelia were collected by filtration through Whatman 3MM filter paper and washed twice with distilled water. The harvested mycelia were spread on yeast malt agar (YMA) and incubated for 48–72 h at 25 °C under near‐UV light (wavelength, 365 nm; HKiv Import and Export Co., Ltd, Xiamen, China). Conidia were collected in distilled water, filtered through cheesecloth and suspended in distilled water. A 10‐μL volume of this suspension (1 × 105 conidia/mL) was placed in 5 mL of CMC medium (1.5% CMC, 0.1% yeast extract, 0.05% MgSO4·H2O, 0.1% NH4NO3 and 0.1% KH2PO4) for 120 h at 25 °C on a rotary shaker (150 rpm). The conidia produced in the CMC culture were filtered through six layers of sterilized gauze, collected by centrifugation and counted using a haemocytometer. The experiment was repeated independently three times.
Microscopic observation of conidial and hyphal morphology
To assess conidial morphology and length, the conidia produced by each strain on YMA or CMC were harvested and examined with differential interference contrast (DIC) optics using a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany). The localization of GFP in the complementation strain was determined with a DE/Axio imager A1 microscope and a GFP filter (470/40 nm excitation, 495 nm dichroic, 525/50 nm emission).
Virulence assay on flowering wheat heads
A virulence test was performed on wheat cv. Jokyoung as described previously (Son et al., 2013). To prepare the conidial inoculum, mycelial plugs were incubated in CMC liquid culture for 5 days. To harvest conidia, the cultures were filtered through six layers of sterilized gauze and washed with distilled water; conidia in the filtrate were collected by centrifugation. A 10‐μL volume of the conidial suspension (1 × 105 conidia/mL) of each strain was injected into 10 replicate wheat head florets at early–mid anthesis. Virulence was assessed 14 days after inoculation by quantifying the percentage of spikelets with head blight symptoms. Statistical analysis was conducted with PASW statistics software 20.0 (IBM SPSS Inc., Armonk, NY, USA).
Total RNA extraction and gene expression level analysis
Total RNAs of the virus‐free or virus‐infected WT and mutant strains were extracted from the mycelia incubated on CM liquid for 48, 72 and 120 h. Total RNA was extracted and cDNA was synthesized as described previously (Lee et al., 2014). qRT‐PCR was performed on a CFX96 Real‐Time PCR System (Bio‐Rad, Hercules, CA, USA) using SsoFast™ EvaGreen® Supermix (Bio‐Rad) according to the manufacturer's instructions. After initial denaturation at 95 °C for 5 min, 40 cycles consisting of 5 s at 95 °C and 5 s at 58 °C were performed. Two endogenous reference genes, ubiquitin C‐terminal hydrolase (UBH, locus FGSG_01231; Kim and Yun, 2011) and EF1α (locus FGSG_08811), were used as internal controls to normalize qRT‐PCR results.
Vertical transmission
Vertical transmission of FgV1 by virus‐infected WT and mutant strains was measured on the basis of conidial sporulation and subculturing on PDA for three generations. Conidia were harvested from FgV1‐infected strains as described above, and 10–15 spores of this first generation were spread onto one PDA plate. At least 50 conidia of each strain were spread onto PDA plates per generation. The plates were cultured for 2–3 days to allow the spores to germinate. Germinated single spores were transferred to new PDA plates, each containing 30 independent germinates. These were incubated at 25 °C for an additional 5 days. The morphology and pigmentation of the colonies that developed were assessed, and each colony of this first generation was then assessed for the presence of FgV1 RNA using RT‐PCR, virus‐specific primers and treatment with DNase I and S1 nuclease (TaKaRa Bio Inc.). The colonies were then used to generate a second generation of conidia, and the process was repeated to generate and assess a third generation. The percentage of conidia that produced colonies containing FgV1 RNA was determined for each generation.
Supporting information
Fig. S1 (A) Amino acid sequence alignment of the 3′‐phosphoadenosine 5′‐phosphatase (PAP phosphatase‐like) domain in Fusarium gramineraum. The predicted FgHal2 protein, hypothetical protein FGSG_07103 and FGSG_01708 were used for this analysis. The positions of the amino acid sequences are indicated on the right. Protein names and GenBank accession numbers are as follows: Fusarium graminearum FGSG_09532 (XP389708.1); Fusarium graminearum FGSG_07103 (XP387279.1); Fusarium graminearum FGSG_01708 (XP381884.1). (B) Relative mRNA expression in the virus‐free and virus‐infected F. graminearum PH‐1 wild‐type (WT) strain. Relative transcript levels of fg07103, fg01708 and FgHal2 were normalized to elongation factor 1α and ubiquitin C‐terminal hydrolase. cDNAs were generated from total RNA extracts obtained after 120 h of incubation. Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test. VF, virus‐free; VI, virus‐infected.
Fig. S2 Effect of FgHal2 gene deletion on the hydrophobicity of the surface mycelium. The wild‐type (WT), FgHal2 gene deletion mutant and Fusarium graminearum virus 1 (FgV1)‐infected WT were grown on complete medium (CM) for 3 days. Each colony was treated with 15 μL of a 2.5% bromophenol blue solution. Colonies were photographed 10 min later. VF, virus‐free; VI, virus‐infected.
Fig. S3 Confirmation of the relative expression of genes related to secondary metabolite synthesis. (A) Relative mRNA expression in the FgHal2 over‐expression strain. Relative transcript levels of FgOs‐2 and Pks12 were normalized to elongation factor 1α (EF1α) and ubiquitin C‐terminal hydrolase (UBH). cDNAs were generated from total RNA extracts after 72 h of incubation. Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test. VF, virus‐free; VI, virus‐infected. (B) Semi‐quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) of FgOs‐2, Gip2, Pks12, AURj and FgHal2 genes using Fusarium graminearum strains. cDNAs were generated from total RNA extracts obtained after 72 h of incubation. After 25 cycles, amplified DNA was analysed by 1.5% agarose gel electrophoresis. The EF1α gene‐specific primer set was loaded as a control.
Fig. S4 The effects of subculturing on the morphology of virus‐infected Fusarium graminearum strains growing on potato dextrose agar (PDA). Single spores of all virus‐infected strains were inoculated on PDA containing appropriate antibiotics. After 5 days, a mycelial plug from each plate was transferred to fresh PDA. ΔFgHal2‐VI was transferred once more and photographed 5 days later.
Table S1 Oligonucleotide primers used in this study.
Acknowledgements
This research was supported in part by grants from the Center for Fungal Pathogenesis [grant no. 20110000959, funded by the Ministry of Education, Science and Technology (MEST)] and from the Next‐Generation BioGreen 21 Program (grant no. PJ00819801), Rural Development Administration, Republic of Korea. M. Son, K.‐M. Lee and J. Yu were supported by graduate research fellowships from MEST through the Brain Korea 21 Project.
References
- Allen, T.D. and Nuss, D.L. (2004) Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 78, 4145–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, J.‐H. , Park, J.‐A. , Kim, J.‐M. , Oh, J.‐M. , Park, S.‐M. and Kim, D.‐H. (2014) Functional analysis of a tannic‐acid‐inducible and hypoviral‐regulated small heat‐shock protein Hsp24 from the chestnut blight fungus Cryphonectria parasitica . Mol. Plant–Microbe Interact. 27, 56–65. [DOI] [PubMed] [Google Scholar]
- Belles, J.M. and Serrano, R. (1995) A salt‐sensitive 3′(2),5′‐bisphosphate nucleotidase involved in sulfate activation. Science, 267, 232–234. [DOI] [PubMed] [Google Scholar]
- Chen, H. and Xiong, L. (2010) The bifunctional abiotic stress signalling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 33, 2180–2190. [DOI] [PubMed] [Google Scholar]
- Cho, W.K. , Yu, J. , Lee, K.‐M. , Son, M. , Min, K. , Lee, Y.‐W. and Kim, K.‐H. (2012) Genome‐wide expression profiling shows transcriptional reprogramming in Fusarium graminearum by Fusarium graminearum virus 1‐DK21 infection. BMC Genomics, 13, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, W.K. , Lee, K.‐M. , Yu, J. , Son, M. and Kim, K.‐H. (2013) Insight into mycoviruses infecting Fusarium species. Adv. Virus Res. 86, 273–288. [DOI] [PubMed] [Google Scholar]
- Chu, Y.M. , Jeon, J.J. , Yea, S.J. , Kim, Y.H. , Yun, S.H. , Lee, Y.‐W. and Kim, K.‐H. (2002) Double‐stranded RNA mycovirus from Fusarium graminearum . Appl. Environ. Microbiol. 68, 2529–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, A.L. , Segers, G.C. , Allen, T.D. , McMains, V.C. and Nuss, D.L. (2004) Microarray analysis of Cryphonectria parasitica Gα‐and Gβγ‐signalling pathways reveals extensive modulation by hypovirus infection. Microbiology, 150, 4033–4043. [DOI] [PubMed] [Google Scholar]
- Desjardins, A. and Proctor, R. (2007) Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 119, 47–50. [DOI] [PubMed] [Google Scholar]
- Estavillo, G.M. , Crisp, P.A. , Pornsiriwong, W. , Wirtz, M. , Collinge, D. , Carrie, C. , Giraud, E. , Whelan, J. , David, P. , Javot, H. , Brearley, C. , Hell, R. , Marin, E. and Pogson, B.J. (2011) Evidence for a SAL1‐PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis . Plant Cell, 23, 3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, K. , Xiong, Q. , Xu, J. , Wang, K. and Wang, K. (2013) CpBir1 is required for conidiation, virulence and anti‐apoptotic effects and influences hypovirus transmission in Cryphonectria parasitica . Fungal Genet. Biol. 51, 60–71. [DOI] [PubMed] [Google Scholar]
- Gašparič, M.B. , Lenassi, M. , Gostinčar, C. , Rotter, A. , Plemenitaš, A. , Gunde‐Cimerman, N. , Gruden, K. and Žel, J. (2013) Insertion of a specific fungal 3′‐phosphoadenosine‐5′‐phosphatase motif into a plant homologue improves halotolerance and drought tolerance of plants. PLoS ONE, 8, e81872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, Z. , Zhu, W. , Su, H. , Zhao, Y. , Zhang, K.‐Q. and Yang, J. (2014) Recent advances in genes involved in secondary metabolite synthesis, hyphal development, energy metabolism and pathogenicity in Fusarium graminearum (teleomorph Gibberella zeae). Biotech. Adv. 32, 390–402. [DOI] [PubMed] [Google Scholar]
- Ghabrial, S.A. and Suzuki, N. (2009) Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384. [DOI] [PubMed] [Google Scholar]
- Gil‐Mascarell, R. , López‐Coronado, J.M. , Bellés, J.M. , Serrano, R. and Rodríguez, P. (1999) The Arabidopsis HAL2‐like gene family includes a novel sodium‐sensitive phosphatase. Plant J. 17, 373–383. [DOI] [PubMed] [Google Scholar]
- Gläser, H. , Thomas, D. , Gaxiola, R. , Montrichard, F. , Surdin‐Kerjan, Y. and Serrano, R. (1993) Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 12, 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Güldener, U. , Mannhaupt, G. , Münsterkötter, M. , Haase, D. , Oesterheld, M. , Stümpflen, V. , Mewes, H.W. and Adam, G. (2006) FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum . Nucleic Acids Res. 34, D456–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy, I. , Gasciolli, V. , Lauressergues, D. , Morel, J.‐B. , Gombert, J. , Proux, F. , Proux, C. , Vaucheret, H. and Mallory, A.C. (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell, 19, 3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, B.H. and York, J.D. (2012) Roles for nucleotide phosphatases in sulfate assimilation and skeletal disease. Adv. Biol. Regul. 52, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag, H.M. and Nagy, P.D. (2010) The combined effect of environmental and host factors on the emergence of viral RNA recombinants. PLoS Pathog. 6, e1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Liu, X. , Yin, Y. and Ma, Z. (2011) Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum . PLoS ONE, 6, e28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.‐K. and Yun, S.‐H. (2011) Evaluation of potential reference genes for quantitative RT‐PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol. J. 27, 301–309. [Google Scholar]
- Kim, J.‐E. , Jin, J. , Kim, H. , Kim, J.‐C. , Yun, S.‐H. and Lee, Y.‐W. (2006) GIP2, a putative transcription factor that regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae . Appl. Environ. Microbiol. 72, 1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S.J. , Cho, S.Y. , Lee, K.M. , Yu, J. , Son, M. and Kim, K.‐H. (2009a) Proteomic analysis of fungal host factors differentially expressed by Fusarium graminearum infected with Fusarium graminearum virus 1‐DK21. Virus Res. 144, 96–106. [DOI] [PubMed] [Google Scholar]
- Kwon, S.J. , Lim, W.S. , Park, S.H. , Park, M.R. and Kim, K.H. (2009b) Molecular characterization of a dsRNA mycovirus, Fusarium graminearum virus 1‐DK21, which is phylogenetically related to hypoviruses but has a genome organization and gene expression strategy resembling those of plant potex‐like viruses. Mol. Cells, 28, 73–74. [PubMed] [Google Scholar]
- Lee, K.M. , Yu, J. , Son, M. , Lee, Y.‐W. and Kim, K.‐H. (2011a) Transmission of Fusarium boothii mycovirus via protoplast fusion causes hypovirulence in other phytopathogenic fungi. PLoS ONE, 6, e21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.‐M. , Cho, W.K. , Yu, J. , Son, M. , Choi, H. , Min, K. , Lee, Y.‐W. and Kim, K.‐H. (2014) A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS ONE, 9, e100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Son, H. , Lee, J. , Min, K. , Choi, G.J. , Kim, J.‐C. and Lee, Y.‐W. (2011b) Functional analyses of two acetyl coenzyme A synthetases in the ascomycete Gibberella zeae . Eukaryot. Cell, 10, 1043–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Son, H. , Lee, J. , Min, K. , Choi, G.J. , Kim, J.‐C. and Lee, Y.‐W. (2011) A putative transcription factor MYT1 is required for female fertility in the ascomycete Gibberella zeae . PLoS ONE, 6, e25586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, R.C. , Boucher, N. , Park, D.S. , Turner, P.E. and Townsend, J.P. (2013) Yeast response to LA virus indicates coadapted global gene expression during mycoviral infection. FEMS Yeast Res. 13, 162–179. [DOI] [PubMed] [Google Scholar]
- Ochiai, N. , Tokai, T. , Nishiuchi, T. , Takahashi‐Ando, N. , Fujimura, M. and Kimura, M. (2007) Involvement of the osmosensor histidine kinase and osmotic stress‐activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum . Biochem. Biophys. Res. Commun. 363, 639–644. [DOI] [PubMed] [Google Scholar]
- Park, S.M. , Choi, E.S. , Kim, M.J. , Cha, B.J. , Yang, M.S. and Kim, D.H. (2004) Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus‐mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277. [DOI] [PubMed] [Google Scholar]
- Pearson, M.N. , Beever, R.E. , Boine, B. and Arthur, K. (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K. and Yamaguchi‐Shinozaki, K. (1999) Molecular Responses to Cold Drought, Heat, and Salt Stress in Higher Plants. Austin, TX: RG Landes Company. [Google Scholar]
- Son, M. , Lee, K.‐M. , Yu, J. , Kang, M. , Park, J.M. , Kwon, S.‐J. and Kim, K.‐H. (2013) The HEX1 gene of Fusarium graminearum is required for fungal asexual reproduction and pathogenesis and for efficient viral RNA accumulation of Fusarium graminearum virus 1. J. Virol. 87, 10 356–10 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Choi, G.H. and Nuss, D.L. (2009) Hypovirus‐responsive transcription factor gene pro1 of the chestnut blight fungus Cryphonectria parasitica is required for female fertility, asexual spore development, and stable maintenance of hypovirus infection. Eukaryot. Cell, 8, 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupotič, T. , Gunde‐Cimerman, N. and Plemenitaš, A. (2007) Novel 3′‐phosphoadenosine‐5′‐phosphatases from extremely halotolerant Hortaea werneckii reveal insight into molecular determinants of salt tolerance of black yeasts. Fungal Genet. Biol. 44, 1109–1122. [DOI] [PubMed] [Google Scholar]
- Xie, J. and Jiang, D. (2014) New insights into mycoviruses and exploration for biological control of crop fungal diseases. Annu. Rev. Phypopathol. 52, 45–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (A) Amino acid sequence alignment of the 3′‐phosphoadenosine 5′‐phosphatase (PAP phosphatase‐like) domain in Fusarium gramineraum. The predicted FgHal2 protein, hypothetical protein FGSG_07103 and FGSG_01708 were used for this analysis. The positions of the amino acid sequences are indicated on the right. Protein names and GenBank accession numbers are as follows: Fusarium graminearum FGSG_09532 (XP389708.1); Fusarium graminearum FGSG_07103 (XP387279.1); Fusarium graminearum FGSG_01708 (XP381884.1). (B) Relative mRNA expression in the virus‐free and virus‐infected F. graminearum PH‐1 wild‐type (WT) strain. Relative transcript levels of fg07103, fg01708 and FgHal2 were normalized to elongation factor 1α and ubiquitin C‐terminal hydrolase. cDNAs were generated from total RNA extracts obtained after 120 h of incubation. Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test. VF, virus‐free; VI, virus‐infected.
Fig. S2 Effect of FgHal2 gene deletion on the hydrophobicity of the surface mycelium. The wild‐type (WT), FgHal2 gene deletion mutant and Fusarium graminearum virus 1 (FgV1)‐infected WT were grown on complete medium (CM) for 3 days. Each colony was treated with 15 μL of a 2.5% bromophenol blue solution. Colonies were photographed 10 min later. VF, virus‐free; VI, virus‐infected.
Fig. S3 Confirmation of the relative expression of genes related to secondary metabolite synthesis. (A) Relative mRNA expression in the FgHal2 over‐expression strain. Relative transcript levels of FgOs‐2 and Pks12 were normalized to elongation factor 1α (EF1α) and ubiquitin C‐terminal hydrolase (UBH). cDNAs were generated from total RNA extracts after 72 h of incubation. Error bars indicate standard deviations. Values with different letters are significantly different (P < 0.05) based on Tukey's test. VF, virus‐free; VI, virus‐infected. (B) Semi‐quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) of FgOs‐2, Gip2, Pks12, AURj and FgHal2 genes using Fusarium graminearum strains. cDNAs were generated from total RNA extracts obtained after 72 h of incubation. After 25 cycles, amplified DNA was analysed by 1.5% agarose gel electrophoresis. The EF1α gene‐specific primer set was loaded as a control.
Fig. S4 The effects of subculturing on the morphology of virus‐infected Fusarium graminearum strains growing on potato dextrose agar (PDA). Single spores of all virus‐infected strains were inoculated on PDA containing appropriate antibiotics. After 5 days, a mycelial plug from each plate was transferred to fresh PDA. ΔFgHal2‐VI was transferred once more and photographed 5 days later.
Table S1 Oligonucleotide primers used in this study.


