Summary
Root‐knot nematodes are sedentary biotrophic endoparasites that maintain a complex interaction with their host plants. Nematode effector proteins are synthesized in the oesophageal glands of nematodes and secreted into plant tissue through a needle‐like stylet. Effectors characterized to date have been shown to mediate processes essential for nematode pathogenesis. To gain an insight into their site of action and putative function, the subcellular localization of 13 previously isolated Meloidogyne incognita effectors was determined. Translational fusions were created between effectors and EGFP‐GUS (enhanced green fluorescent protein‐β‐glucuronidase) reporter genes, which were transiently expressed in tobacco leaf cells. The majority of effectors localized to the cytoplasm, with one effector, 7H08, imported into the nuclei of plant cells. Deletion analysis revealed that the nuclear localization of 7H08 was mediated by two novel independent nuclear localization domains. As a result of the nuclear localization of the effector, 7H08 was tested for the ability to activate gene transcription. 7H08 was found to activate the expression of reporter genes in both yeast and plant systems. This is the first report of a plant‐parasitic nematode effector with transcriptional activation activity.
Keywords: effector, interaction, Meloidogyne incognita, NLS, nuclear import, root‐knot nematode, transcriptional activation activity
Introduction
The sedentary endoparasitic root‐knot nematodes of the genus Meloidogyne are amongst the most economically important pathogens in agriculture with a host range that includes high‐value agronomic crops, such as coffee, cotton, peanuts, potato, soybean and tomato (Trudgill and Blok, 2001). Root‐knot nematodes utilize plant‐derived nutrients to support their own development. The plant‐derived nutrients are obtained through a specialized feeding site, established in the vascular parenchyma of host root cells (Kyndt et al., 2013). Interaction with the host plant begins when second‐stage juveniles (J2s), the infective stage of the root‐knot nematode life cycle, penetrate the root. J2s move intercellularly towards the root tip and, following reorientation, migrate towards the differentiating vascular cylinder. Four to eight root cells are transformed into giant cells, which collectively make up the nematode feeding site. Root‐knot nematodes maintain a biotrophic interaction with the host plant for the 3–6‐week duration of the nematode life cycle (Gheysen and Mitchum, 2011).
Successful pathogenesis by root‐knot nematodes is dependent on the ability of nematodes to migrate through host roots, to suppress host defence responses and to both form and maintain giant cells (Kyndt et al., 2013). This modulation of a diverse range of plant processes during nematode parasitism is hypothesized to be mediated by nematode‐secreted effector proteins. Effectors have been defined previously as ‘all pathogen proteins and small molecules that alter host cell structure and function’ (Hogenhout et al., 2009). The nematode effectors characterized to date are involved in plant cell wall degradation and modification, suppression of host defences, peptide mimicry and the manipulation of plant signalling pathways, such as those involving ubiquitination and phytohormones (Haegeman et al., 2012; Hewezi and Baum, 2013; Mitchum et al., 2013).
Effectors are synthesized in three specialized oesophageal glands in the nematode (Davis et al., 2008; Hussey, 1989; Rosso et al., 2011). The presence of an N‐terminal signal peptide directs the effector protein from the nuclear region of oesophageal gland cells to the secretory pathway, where the signal peptide is cleaved. Effectors are subsequently secreted into plant tissues through the stylet, a protractible, needle‐like structure in the nematode's head (Davis et al., 2008; Hussey, 1989). Immunocytological analyses have revealed that effectors are secreted along the nematode migratory path (Doyle and Lambert, 2002; Vieira et al., 2011; Wang et al., 1999), the plant apoplasm (Jaouannet et al., 2012; Vieira et al., 2011) and directly into the cytoplasm of host cells (Wang et al., 2010). Although the determination of the localization of nematode secretions is important, the ascertainment of the site of effector action is of greater interest in terms of the functional characterization of effectors. A number of nematode effector proteins contain nuclear localization signals (NLSs) (Elling et al., 2007; Jaouannet et al., 2012; Jones et al., 2009; Lin et al., 2013). Previous studies determining the subcellular localization of NLS‐containing nematode effectors have revealed cytoplasmic, nuclear and nucleolar localization (Elling et al., 2007; Jaouannet et al., 2012, 2013; Jones et al., 2009; Lin et al., 2013), demonstrating the importance of testing in silico predictions in planta. In other plant–pathogen systems, nuclear imported effectors have been shown to play a vital role in mediating the parasitism process (Rivas and Genin, 2011). Evidence is emerging that the nuclear compartment is a central target for the effector‐mediated suppression of host defence responses (Mukhtar et al., 2011; Rivas, 2012). Moreover, the formation of nematode feeding sites is associated with comprehensive changes in host gene transcription (Barcala et al., 2012; Damiani et al., 2012; Ithal et al., 2007; Jammes et al., 2005; Portillo et al., 2009, 2013). The objectives of this study were: (i) to determine the subcellular localization of representative Meloidogyne incognita effectors in planta; and (ii) to analyse whether or not nuclear imported M. incognita effectors have transcriptional activation activity in yeast and in plants.
Results
Sequence analysis of M. incognita effectors
Putative M. incognita effectors have been identified previously (Huang et al., 2003, 2004). Thirteen representative effectors were selected for analysis in this study (Table 1). Protein sequence similarity searches using blastp (Altschul et al., 1990) revealed that 12 of the effectors were novel, whereas one effector, 30G11, was homologous to an acid phosphatase (AFQ55439.1) from the cyst nematode Heterodera avenae (blastp score 467, 53% identity).
Table 1.
Prediction of subcellular localization of Meloidogyne incognita effectors
| Effector | psortii prediction | wolf psort prediction |
|---|---|---|
| 6F06 | Nuclear, 91.4% | 10 nucleus, 2 peroxisome |
| 2E07 | Cytoplasmic, 43.5% | 12 extracellular, 1 chloroplast |
| 7A01 | Cytoplasmic, 43.5% | 4 chloroplast, 3 cytoplasmic, 3 plasma membrane, 2 vacuolar, 1 nucleus |
| 7E12 | Nuclear, 69.6% | 6 extracellular, 4 chloroplast, 2 cytoplasmic, 1 vacuolar |
| 30G11 | Cytoplasmic, 26.1% | 5 cytoplasmic, 3 Golgi, 2 nucleus, 1 chloroplast, 1 mitochondria, 1 endoplasmic reticulum |
| 5C03B | Nuclear, 78.3% | 8 nucleus, 3 extracellular, 2 chloroplast |
| 2G02 | Nuclear, 73.9% | 10 nucleus, 7 cytoplasmic |
| 8H11 | Nuclear, 78.3% | 5 nucleus, 4 chloroplast, 3 mitochondria, 1 cytoplasmic |
| 1D08B | Nuclear, 60.9% | 7 extracellular, 4 nucleus, 2 chloroplast |
| 6G07 | Nuclear, 73.9% | 6 nucleus, 4 chloroplast, 4 cytoplasmic, 1 extracellular |
| 17H02 | Nuclear, 73.9% | 4 chloroplast, 3 cytoplasmic, 3 extracellular, 2 nucleus |
| 31H06 | Cytoplasmic, 60.9% | N/A* |
| 7H08 | Nuclear, 56.5% | 11 nucleus, 7 cytoplasmic, 1 plasma membrane |
*Sequence was too short for wolf psort.
The predicted subcellular localization of the 13 effectors was determined by psortii (Nakai and Horton, 1999) and wolf psort (Horton et al., 2007) with the signal peptide sequence, as predicted by SignalP, removed (Table 1). Nine of the 13 effectors were predicted by psortii to localize to the nucleus; only two effectors, 7H08 and 2G02, were predicted to contain putative monopartite NLSs, based on sequence similarity to the simian virus 40 (SV40) large T‐antigen NLS (Kalderon et al., 1984a, 1984b). The four remaining effectors were predicted to be cytoplasmic. For six of the effectors, the psortii prediction of nuclear localization was supported by wolf psort. 30G11 was predicted to be cytoplasmic by wolf psort analysis; the remaining effectors were predicted to accumulate in other compartments (Table 1).
Subcellular localizations of effector fusion proteins in planta
To test the predicted subcellular localization patterns in planta, effectors were transiently expressed in tobacco leaf epidermal cells. In order to prevent effectors from passively diffusing into the nucleus (Grebenok et al., 1997; Haasen et al., 1999), effectors (without the signal peptide) were fused with the β‐glucuronidase (GUS) and enhanced green fluorescent protein (EGFP) reporter genes (Fig. 1a). Confocal microscopy imaging revealed that GUS‐EGFP fusion proteins were restricted to the cytoplasm (Fig. 1b,c). Free EGFP was present in both the nucleus and cytoplasm (Fig. 1d).
Figure 1.
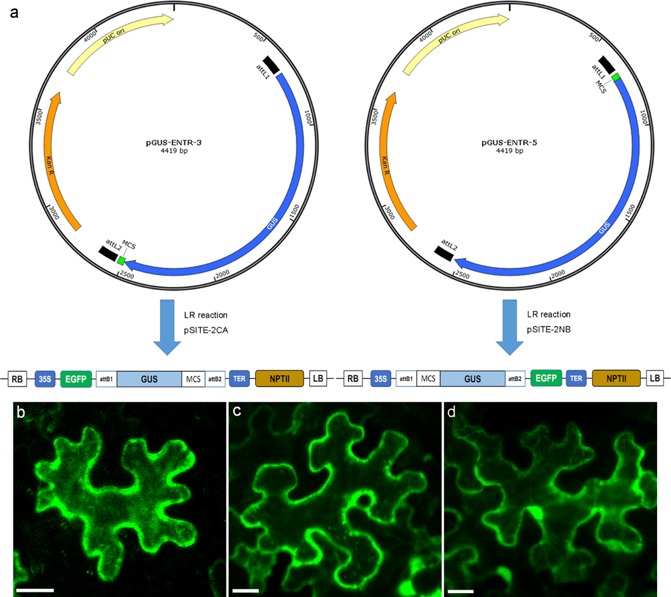
Construction of pGUS‐ENTR vectors for GUS‐EGFP (β‐glucuronidase‐enhanced green fluorescent protein) fusions. (a) To prevent the passive diffusion of effectors into the nucleus, effector cDNA was fused to the GUS gene in pGUS‐ENTR‐3 and pGUS‐ENTR‐5 vectors prior to fusion with the EGFP gene in pSITE‐2CA and pSITE‐2NB plasmids. (b) Transiently expressed EGFP‐GUS fusions and (c) GUS‐EGFP fusions in tobacco leaf cells localized to the cytoplasm. (d) Free EGFP was observed in both the cytoplasm and nucleus. Scale bar, 20 μm.
Although nine of the effectors were predicted to localize to the nucleus, 12 of the 13 selected effector proteins showed a cytoplasmic localization (Fig. 2a–l) and were observed in the cytoplasm and cytoplasmic strands. Among the selected effectors, only one showed specific nuclear localization in tobacco leaf cells (Fig. 2m). Strong fluorescence was observed in the nucleus of tobacco cells transformed with the 7H08 fusion protein, with no EGFP expression observed in the cytoplasm (Fig. 2m). The nuclear localization of 7H08 was confirmed by counterstaining with the nucleic acid stain 4′,6‐diamidino‐2‐phenylindole (DAPI) (Fig. 2n,o). 7H08 was predicted to contain a monopartite NLS. Monopartite NLSs were also predicted to occur in 2G02; however, 2G02 showed a cytoplasmic localization (Fig. 2g). No differences in effector localization patterns were observed when effector proteins were fused to the N‐terminus of EGFP in comparison with C‐terminal fusions (data not shown).
Figure 2.
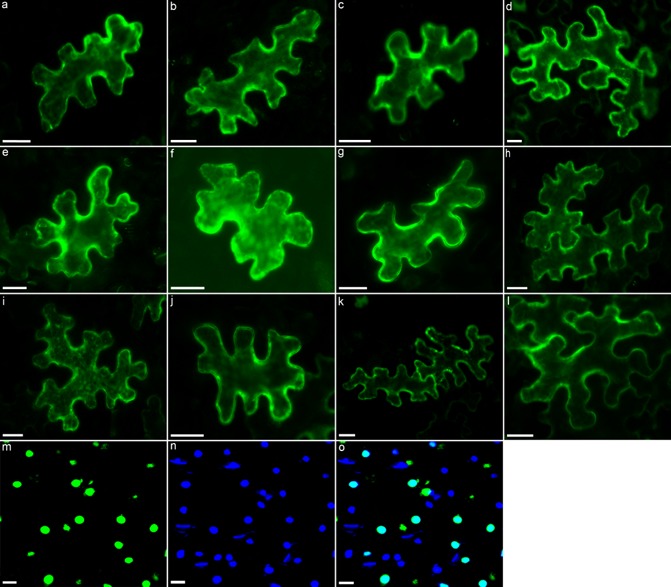
Subcellular localization of transiently expressed EGFP‐GUS (β‐glucuronidase‐enhanced green fluorescent protein)‐effector fusions in tobacco leaves. The subcellular compartments targeted by the following Meloidogyne incognita effectors were determined: (a) 6F06; (b) 2E07; (c) 7A01; (d) 7E12; (e) 30G11; (f) 5C03B; (g) 2G02; (h) 8H11; (i) 1D08D; (j) 6G07; (k) 17H02; (l) 31H06; (m) 7H08. (n) To confirm the nuclear localization of 7H08, leaves were counterstained with nucleic acid stain 4′,6‐diamidino‐2‐phenylindole (DAPI) and the image was merged with the subcellular localization of 7H08 (o). Scale bar, 20 μm.
Testing the functionality of predicted NLSs
The cytoplasmically localized 2G02 was predicted to contain two monopartite NLSs (62PKKCKVC68 and 189KRKK192). To test the functionality of the predicted NLSs, two short protein fragments containing each of the predicted NLSs (designated pNLS1 and pNLS2) were generated and fused to EGFP‐GUS. Both pNLS1 and pNLS2 fusion proteins localized to the cytoplasm in tobacco leaf cells (Fig. 3a,b), indicating that the predicted NLSs are not functional in planta. EGFP‐GUS fused to Arabidopsis histone 2B served as a positive control for nuclear import (Fig. 3c).
Figure 3.
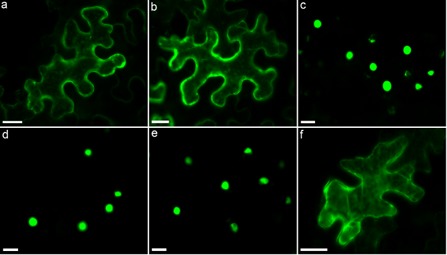
Determination of the functionality of predicted nuclear localization signals (NLSs). (a, b) Expression of two 2G02 protein fragments, each containing a predicted monopartite NLS, localized to the cytoplasm. (c) EGFP‐GUS (β‐glucuronidase‐enhanced green fluorescent protein) fused to Arabidopsis histone 2B (At1G07790) was used as a positive control for nuclear import. (d) Following the mutation of three lysine residues in the predicted monopartite NLS of 7H08 to alanine residues, nuclear localization of the effector was still observed. (e) 7H08 also localized to the nucleus following deletion of the predicted NLS. (f) Expression of a 7H08 protein fragment containing the predicted monopartite NLS localized to the cytoplasm.
Transiently expressed 7H08 fusion proteins localized to the nucleus in tobacco leaf cells. To determine whether or not the predicted monopartite NLS of 7H08 was responsible for nuclear localization, a 7H08 fusion protein was constructed in which three positively charged lysine residues in the predicted NLS motif were mutated to alanine residues (179PYGTKKK 185 to 179PYGTAAA 185). Nuclear localization was still observed for the mutated 7H08 fusion protein (Fig. 3d). Furthermore, deletion of the predicted NLS failed to alter the subcellular localization of 7H08 (Fig. 3e). In addition, the functionality of the predicted NLS was tested by fusing a protein fragment of 7H08 containing the predicted NLS to EGFP‐GUS. The resulting fusion product localized to the cytoplasm of tobacco leaf cells (Fig. 3f). Taken together, these experiments indicate that the predicted NLS in 7H08 was not responsible for nuclear import.
Identification of nuclear localization domains in 7H08
In order to identify the protein region(s) responsible for the nuclear localization of 7H08, a series of deletion variants was created. Initially, 7H08 was divided into three protein fragments: two 140‐amino‐acid fragments that contained the N‐terminus (7H08 23–162) and C‐terminus (7H08 161–300) of 7H08 and a 61‐amino‐acid fragment of 7H08 that lies between the terminal regions (7H08 132–192). The N‐terminal fragment begins at amino acid residue 23, as amino acids 1–22 encode the cleaved signal peptide. Protein fragments were used to construct EGFP‐GUS fusion proteins and transiently expressed in tobacco leaf cells. Nuclear localization was observed for both the N‐ and C‐terminal protein fragments (Fig. 4a). To gain further insight into the protein regions responsible for 7H08 nuclear localization, a deletion series of both the N‐terminal and C‐terminal fragments was created (Fig. 4b,c). All three deletion variants of the N‐terminal region localized to the cytoplasm (Fig. 4b), suggesting that the protein region responsible for nuclear import occurs between amino acids 23 and 57. Expression of the 7H08 23–57 protein fragment fused with EGFP‐GUS was localized to the nucleus (Fig. 4d), confirming the importance of this region for nuclear import. A deletion series was also created for the C‐terminal fragment. The 7H08 195–300 protein fragment was localized to the nucleus (Fig. 4c). Conversely, the protein fragments of 7H08 240–300 and 7H08 265–300 were both localized to the cytoplasm (Fig. 4c). This indicated that the region responsible for nuclear import in the C‐terminus occurs between amino acids 195 and 240. This was confirmed by the nuclear localization of the 7H08 195–240 protein fragment (Fig. 4d). As the regions identified in the N‐ and C‐terminus do not resemble classical monopartite or bipartite NLSs, they were termed nuclear localization domains (NLD1 and NLD2, respectively). In order to confirm the function of NLD1 and NLD2, a mutated 7H08 fusion protein was created in which both NLDs were deleted. The NLD deletion fusion protein was localized to the cytoplasm (Fig. 4d). This confirms that 7H08 contains two independent NLDs, which result in nuclear import of the effector.
Figure 4.
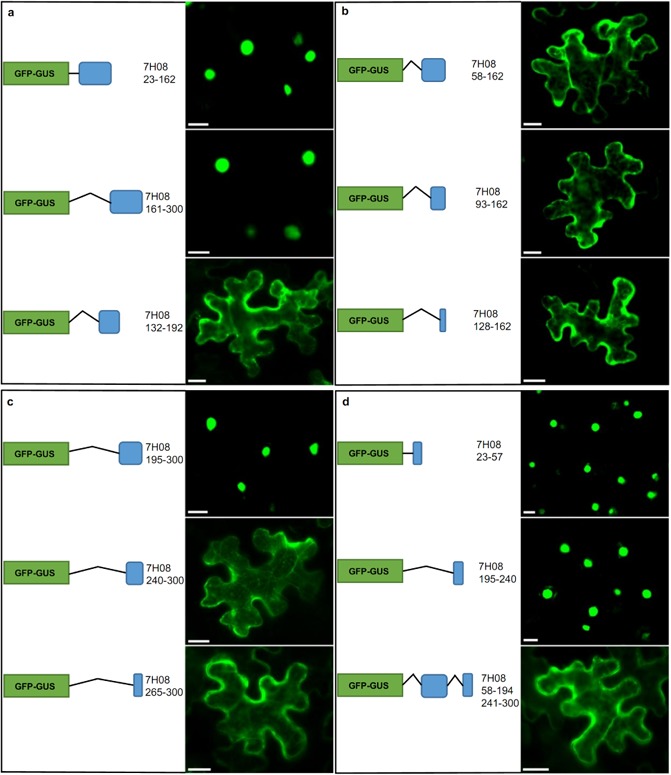
Determination of the regions of 7H08 responsible for nuclear import. (a) The 7H08 protein, minus the signal peptide, was divided into N‐terminal (7H08 23–162), C‐terminal (7H08 161–300) and middle region (7H08 132–192) protein fragments. Protein fragments were fused to EGFP‐GUS (β‐glucuronidase‐enhanced green fluorescent protein) and the subcellular localization was determined in tobacco leaf cells. (b) The N‐terminus of 7H08 was further divided into three subfragments. (c) Three subfragments were also generated from the C‐terminus of 7H08. (d) The two experimentally determined nuclear localization domains (7H08 23–57 and 7H08 195–240) were fused to EGFP‐GUS and expressed in tobacco. Deletion of the nuclear localization domains resulted in the cytoplasmic localization of 7H08. Scale bar, 20 μm.
Testing the transcriptional activation activity of 7H08 in yeast
As a result of the nuclear localization of 7H08, the ability of 7H08 to transcriptionally activate gene expression was tested using a yeast reporter system. Two yeast reporters were utilized: the ability of yeast colonies to grow on histidine minimal medium (SC/–TH), which indicates expression of the HIS3 reporter gene, and expression of MEL‐1, which results in the blue coloration of yeast colonies in the presence of the chromogenic substrate 5‐bromo‐4‐chloro‐3‐indolyl‐α‐d‐galactopyranoside (X‐α‐Gal). Transformation of yeast cells with a plasmid containing only the GAL4 DNA‐binding domain (GAL4BD) failed to activate either of the reporters (Fig. 5). Fusion of 7H08 (7H08 23–300) with GAL4BD activated both reporter systems in yeast cells, indicating that 7H08 is able to activate gene expression in yeast. To determine the protein regions responsible for the activation of gene expression, 7H08 was divided into N‐terminal (7H08 23–162) and C‐terminal (7H08 161–300) fragments and GAL4BD fusion proteins were generated. The N‐terminal fragment failed to activate reporters, whereas both reporters were activated by the C‐terminal fragment fusion (Fig. 5). Further deletions of the C‐terminus of 7H08 were created (7H08 161–240 and 7H08 220–300) with only the GAL4BD‐7H08 220–300 fusion protein able to activate gene expression (Fig. 5). Two subfragments were created from the 220–300‐amino‐acid region, both of which were able to activate reporter gene expression. This demonstrates that 7H08 is able to activate gene expression when recruited to the promoter region of reporter genes by GAL4BD.
Figure 5.
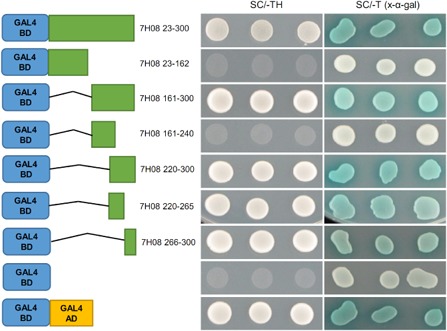
Testing of the transcriptional activation activity of 7H08 in yeast. The 7H08 protein was fused with the GAL4 DNA‐binding domain (GAL4BD) and expressed in yeast cells. The ability of yeast colonies to grow on synthetic medium minus tryptophan and histidine (SC/–TH) indicates expression of the HIS3 reporter gene. Expression of the MEL1 reporter gene was determined by the blue coloration of yeast colonies on SC/–T medium with the chromogenic substrate 5‐bromo‐4‐chloro‐3‐indolyl‐α‐d‐galactopyranoside (X‐α‐Gal). A series of deletion variants was created to localize the 7H08 region responsible for transcriptional activation activity.
Testing the transcriptional activation activity of 7H08 in planta
To determine whether or not 7H08 also had transcriptional activation activity in planta, an Agrobacterium tumefaciens‐mediated transient assay system using the luciferase (LUC) reporter gene was employed. 7H08 was fused with GAL4BD (Fig. 6a). The resulting construct was transformed into A. tumefaciens and transiently expressed in tobacco leaf cells, together with the LUC reporter gene, under the control of the GAL4 upstream activation sequence (GAL4UAS) (Yamamoto et al., 2001). To eliminate the expression of LUC in A. tumefaciens, an intron was introduced into the LUC gene. To monitor the transformation efficiency, a reporter under the direction of a constitutive promoter (CaMV35S‐GUSint) was co‐transformed and used as an internal control to normalize LUC expression (Fig. 6a). To test the suitability of the system, GAL4BD was fused to the transactivation domain of the herpes simplex virus VP16 protein (GAL4BD‐VP16). Transient expression of GAL4BD‐VP16 with the reporter construct resulted in significantly higher (P < 0.01) LUC activity in comparison with transient expression of GAL4BD only (Fig. 6b). Transient expression of GAL4BD‐7H08 23–300 increased significantly the LUC/GUS ratio compared with the GAL4BD negative control (P < 0.01) (Fig. 6c). This demonstrates that 7H08 is able to transcriptionally activate gene expression in planta. The 7H08 protein was divided into the same protein fragments as used to test transactivation activity in yeast and transiently expressed in tobacco. Generally, the protein fragments that were able to activate reporter systems in yeast were also able to significantly activate the LUC reporter in tobacco compared with the GAL4BD negative control (P < 0.01) (Fig. 6c). However, when the 220–300‐amino‐acid fragment, which strongly activated the LUC reporter in yeast, was divided into two subfragments, transactivation activity was no longer observed in planta. All effector fusion proteins were expressed at the expected sizes in the leaves of tobacco cells, as evidenced by western blotting (Fig. 6d).
Figure 6.
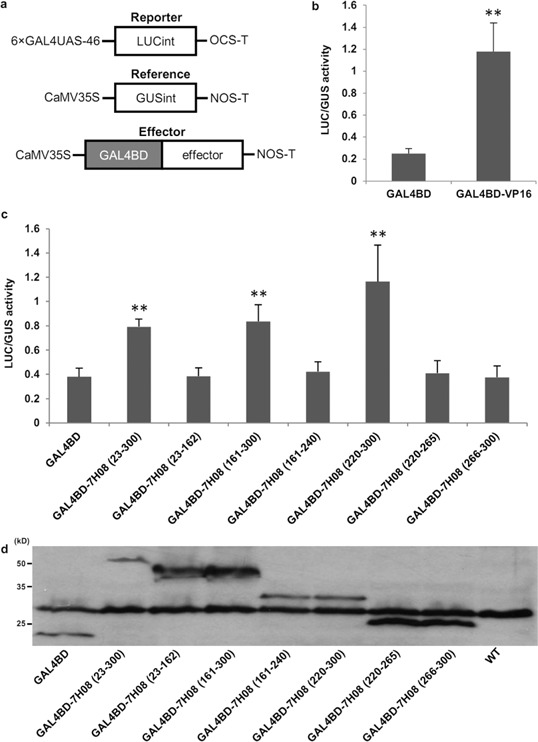
Testing of the transcriptional activation activity of 7H08 in plant cells. (a) Schematic diagram of reporter, reference and effector plasmids used in the transient assay. 7H08 was fused to the GAL4 DNA‐binding domain (GAL4BD), whereas the luciferase (LUC) reporter gene was fused to the GAL4 upstream activating sequence (GAL4UAS). (b) Fusion of the transactivation domain of the herpes simplex virus VP16 protein to GAL4BD (GAL4BD‐VP16) resulted in high LUC activity. Data represent the means ± standard deviation from four independent leaf samples. The experiment was replicated three times, and representative results are shown. **P < 0.01. (c) A series of deletion variants of 7H08 was fused to GAL4 and expressed in tobacco leaves to determine the regions of 7H08 able to activate expression of the LUC reporter gene in planta. Data represent means ± standard deviation from four independent leaf samples. The experiment was replicated three times, and representative results are shown. **P < 0.01. (d) Western blot analysis of GAL4BD and GAL4BD‐7H08 deletion series. The anti‐GAL4BD antibody was used to detect the accumulation of GAL4BD‐fused proteins in the leaves of tobacco plants.
Discussion
Plant‐parasitic nematode effectors modulate a diverse range of plant processes (Mitchum et al., 2013). The identification of the subcellular compartments targeted by nematode effectors can provide an insight into effector function. In silico analysis predicted that nine of the 13 selected effectors would localize to the nucleus. However, transient expression of translational fusions between effector cDNA (minus the signal peptide encoding region) and EGFP and GUS genes in tobacco leaf cells revealed that only one effector, 7H08, was localized to the nucleus. A series of deletion variants revealed that 7H08 contains two independent NLDs, which mediate nuclear import. Furthermore, 7H08 was capable of facilitating transcriptional activation in both yeast and plant systems.
7H08 was found to contain two NLDs, one in the N‐terminus and one in the C‐terminus. The NLDs of 7H08 did not contain stretches of basic amino acids, which are typical of classical NLSs (Hicks and Raikhel, 1995); no significant sequence similarities were found between NLD1 and NLD2. The nuclear localization of plant pathogen effectors that do not contain classical NLSs was also observed in the bacterial pathogen Pantoea agglomerans (Nissan et al., 2006; Weinthal et al., 2011). Similar to 7H08, the P. agglomerans effectors, HsvG and HsvB contained two domains that conferred nuclear localization, one in each protein terminus (Weinthal et al., 2011). The nuclear import of proteins that do not possess classical NLSs has been reported previously in other systems (Lin and Yen, 2006; Lindert et al., 2009; Siomi and Dreyfuss, 1995; Van Dusen et al., 2010). In these proteins, the region responsible for nuclear import was longer than classical NLSs (>25 amino acids) and, unlike classical NLSs, was not rich in basic amino acid residues.
7H08 is the first reported nuclear‐localized plant‐parasitic nematode effector to contain novel NLDs. The nuclear‐localized Mi‐EEF1 and MjNULG1a effectors, secreted by M. incognita and M. javanica, respectively, both contained classical NLSs (Jaouannet et al., 2012; Lin et al., 2013). In addition, several cyst nematode effectors containing predicted NLSs were nuclear imported (Elling et al., 2007; Jones et al., 2009). The nuclear localization of effectors containing classical NLSs has been reported in bacterial, fungal and oomycete phytopathogens (Rivas and Genin, 2011). In addition to NLS‐dependent nuclear import, evidence suggests that nematode effectors are able to translocate to the nucleus following modification within the host cytoplasm. The cyst nematode‐secreted HsUbiI, a putative ubiquitin extension protein, was predicted to localize to the cytoplasm. However, in host cells, the cleavable C‐terminal domain localized to the nucleolus, where it was hypothesized to be involved in ribosome synthesis (Tytgat et al., 2004). In addition, the cytoplasmically localized effector Hs10A07 translocated to the nucleus following interaction with an Arabidopsis protein kinase, indicating phosphorylation of the effector (Hewezi and Baum, 2013). This suggests that post‐translational modifications of effectors have an effect on their subcellular localization in plant cells, which may, in part, explain the discrepancy between the prediction of subcellular localizations by psortii and wolf psort and their actual localization in vivo. Discrepancies between computer‐predicted and experimentally observed subcellular localizations of proteins in the same range as found here also occurred in other organisms, including cyst nematodes and plants (Elling et al., 2007; Heazlewood et al., 2005; Jones et al., 2009; Koroleva et al., 2005; Sprenger et al., 2006; Tanaka et al., 2004). Both psortii and wolf psort take advantage of algorithms that facilitate the detection of known sorting signals or functional motifs in query protein sequences to estimate the likelihood of specific subcellular localizations (Horton et al., 2007; Nakai and Horton, 1999). Thus, it is possible that protein sequences that are similar to known sorting signals, for example short stretches of basic amino acids, a hallmark of NLSs, are present in the query protein but are not functional in planta, which could explain the relatively low level of accuracy of computer‐based predictions.
In addition to exhibiting nuclear localization, 7H08 from M. incognita was shown to possess transactivation activity in both yeast and plants. In yeast, two regions (7H08 220–265 and 7H08 266–300) were found to activate reporter expression, whereas, in the plant system, a larger region of 7H08 (220–300) was necessary. The different lengths of 7H08 required to activate gene expression in the two systems may be a result of differences in the transcriptional machinery of lower and higher eukaryotes (Escher et al., 2000; Remacle et al., 1997). The region of 7H08 found to confer transcriptional activation activity in planta did not possess any of the features that typify classical transcription activation domains, such as acidic, proline‐rich and glutamine‐rich regions (Triezenberg, 1995).
Prior to this study, no plant‐parasitic nematode effectors with transcriptional activation activity had been reported. Effectors with transcriptional activation activity have been most extensively characterized in Xanthomonas spp. Bacterial type III effectors belonging to the AvrBs3 family are the largest class of proteins known as transcription activator‐like (TAL) effectors (Kay and Bonas, 2009), which function as transcriptional activation activators in the plant nucleus. It has been demonstrated that AvrBs3 contains a novel modular DNA‐binding domain, which binds to a DNA‐binding motif in the promoter region of certain plant genes. AvrBs3 effectors also contain an acidic domain, which is involved in the activation of transcription (Kay et al., 2007, 2009). No homology was found between the acidic activation domain of AvrBs3 and the transactivation region of 7H08. The effector HsvG from P. agglomerans which, like 7H08, does not display homology to classical transactivation domains, has been demonstrated to activate transcription in yeast (Nissan et al., 2012). Furthermore, HsvG was found to bind to the promoter of a plant gene that harbours characteristic motifs of a eukaryotic transcription factor (Nissan et al., 2012). Transactivation domains in transcription activators play a fundamental role in the assemby and initiation of the cellular transcription machinery. It is generally believed that transactivation domains function by providing interfaces for protein interactions and binding to general transcription factors, co‐activators, mediators or chromatin‐modifying enzymes. Transactivation domains have been classified on the basis of the enrichment of certain types of amino acids, such as acidic amino acids, glutamine or proline, but they generally share little sequence or structural homology and normally lack folded structures (Lewis and Reinberg, 2003; Triezenberg, 1995; Uesugi and Verdine, 1999). Recent studies have suggested that intrinsically disordered regions (IDRs), which lack a fixed tertiary structure, are particularly important in transcriptional regulation (Kragelund et al., 2012; Uversky, 2011). A study showed that 83%–94% of transcription factors contain extended IDRs and that the degree of disorder is much higher in transactivation domains than in DNA‐binding domains (Liu et al., 2006). The extreme flexibility of IDRs allows transactivation domains to bind to different partners in transient interactions with high specificity and low affinity (Hilser and Thompson, 2011; Oldfield et al., 2008). A preliminary search of the transactivation region of 7H08 (220–300) for IDRs revealed two putative IDRs, located at amino acids 220–245 and 254–300, respectively. Hence, it is feasible that the transactivation region of 7H08 activates gene expression by interacting with and recruiting other components of the transcription machinery through its IDRs.
Transcriptome analyses have revealed that nematode parasitism results in extensive changes in plant gene expression across all plant processes, including a massive down‐regulation of genes involved in the plant defence response (Barcala et al., 2012; Damiani et al., 2012; Ithal et al., 2007; Jammes et al., 2005; Portillo et al., 2009, 2013). This poses the question of how a limited number of effector proteins can cause such extensive modifications to host gene expression. Rather than targeting individual genes, a previous study has demonstrated that effectors are likely to have converged to target a relatively small number of highly interconnected ‘hubs’ in the plant signalling network, such as those that control the plant immune system (Mukhtar et al., 2011). Although the processes by which nematode effector proteins modulate host gene expression are yet to be elucidated, the discovery of a M. incognita effector with transcriptional activation activity represents an important step in understanding the interaction between nematode effectors and host transcriptional networks.
Experimental Procedures
Sequence analyses
Representative effector genes were chosen from a pool of previously identified M. incognita effectors (Huang et al., 2003, 2004). Effector protein sequences were compared against the National Center for Biotechnology Information (NCBI) non‐redundant protein database using blastp set at default parameters. Signal peptides were predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP‐3.0/). The subcellular localization of effectors was predicted using psortii (http://psort.hgc.jp/form2.html) and wolf psort (http://wolfpsort.org/). IDRs were predicted using the program disembl 1.5 (http://dis.embl.de/) at default settings (Linding et al., 2003).
Construction of vectors for subcellular localization
To create the pGUS‐ENTR‐3 and pGUS‐ENTR‐5 vectors, the cloning vector pENTR/D‐TOPO (Life Technologies, Grand Island, NY, USA) was modified. To construct pGUS‐ENTR‐3, the GUS coding sequence and multiple cloning site (MCS), with a stop codon (5′‐CATATGGAATTCCCGGGGATCCGTCGACCTTAA‐3′), were amplified by polymerase chain reaction (PCR) from the binary vector pBI101. The GUS gene and MCS were directionally cloned into the pENTR/D‐TOPO vector. This created the pGUS‐ENTR‐3 vector in which MCS was fused to the 3′ end of the GUS gene, with both the GUS gene and MCS flanked by attL1 and attL2 Gateway cloning sequences. pGUS‐ENTR‐5 vector was constructed in a similar manner, but MCS, without a stop codon (5′‐CATATGGAATTCCCGGGGATCCGTCGACCT‐3′), was fused to the 5′ end of the GUS gene (Fig. 1a). Coding sequences of effectors, without the signal peptide encoding region (as predicted by SignalP 3.0), were amplified from full‐length cDNA clones (Huang et al., 2003, 2004). All PCR amplifications were performed with Phusion High‐Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) according to the manufacturer's instructions. Effector cDNA sequences were cloned into MCS of both the pGUS‐ENTR‐3 and pGUS‐ENTR‐5 vectors using EcoRI and BamHI restriction sites (NdeI/BamHI for 30G11 and 2G02). The resulting plasmids harboured the M. incognita effector gene fused with the GUS gene, flanked by attL1 and attL2 sequences. An LR reaction was performed to clone the GUS‐effector fusion from pGUS‐ENTR‐3 into the EGFP fusion binary vector pSITE‐2CA (EGFP‐GUS‐effector) (Chakrabarty et al., 2007) or effector‐GUS fusion from pGUS‐ENTR‐5 into pSITE‐2NB (effector‐GUS‐EGFP) (Chakrabarty et al., 2007). Clones were sequenced at Elim Biopharmaceuticals (Hayward, CA, USA) to confirm sequence identity. The vector maps shown in Fig. 1 were drawn using SnapGene Viewer software (http://www.snapgene.com/). Primers are detailed in Table S1 (see Supporting Information).
Testing of predicted NLSs
To test the function of the two predicted NLSs (pNLS) in 2G02, two regions of coding sequences that contained the predicted NLS sites were amplified by PCR using the primer pairs 2G02 pNLS1‐FP/2G02 pNLS1‐RP and 2G02 pNLS2‐FP/2G02 pNLS2‐RP, respectively. Regions were cloned as described above to generate EGFP‐GUS‐2G02 pNLS1 fusion and EGFP‐GUS‐2G02 pNLS2 fusion constructs. The cDNA of the Arabidopsis histone 2B gene (AT1G07790) was cloned as described above to generate an EGFP‐GUS‐AtH2B fusion, which served as positive nuclear import control. The predicted NLS in 7H08 was mutated by PCR, through the generation of overlapping PCR fragments. Three positively charged lysine residues in the predicted NLS motif were mutated to alanine residues (179PYGTKKK 185 to 179PYGTAAA 185). The first and second fragments were amplified using primer pairs 7H08‐pNLSm‐FP1/7H08‐pNLSm‐RP1 and 7H08‐pNLSm‐FP2/7H08‐pNLSm‐RP2. A third PCR fused both fragments using primer pair 7H08‐pNLSm‐FP1/7H08‐pNLSm‐RP2. The final PCR product 7H08‐pNLSmut was cloned into the pGUS‐ENTR‐3 vector and subsequently into pSITE‐2CA. Similarly, to delete the predicted NLS in 7H08, two overlapping PCR fragments were generated. The first and second fragments were amplified using primer pairs 7H08‐pNLSdel‐FP1/7H08‐pNLSdel‐RP1 and 7H08‐pNLSdel‐FP2/7H08‐pNLSdel‐RP2. A third PCR fused both fragments using primer pair 7H08‐pNLSdel‐FP1/7H08‐pNLSdel‐RP2. The final PCR product 7H08‐pNLSdel was cloned into the pGUS‐ENTR‐3 vector and subsequently into pSITE‐2CA. Furthermore, to test the nuclear targeting function of the predicted NLS of 7H08, a region containing the NLS and short flanking sequences at either side of the NLS was amplified by PCR using the primer pair 7H08‐pNLS‐FP/7H08‐pNLS‐RP. The resulting sequence was cloned as described above to generate EGFP‐GUS‐7H08‐pNLS. All primers are detailed in Table S2 (see Supporting Information). Clones were sequenced at Elim Biopharmaceuticals to confirm sequence identity.
Identification of NLDs in 7H08
To determine the protein domains responsible for 7H08 nuclear localization, different regions of 7H08 were amplified by PCR using specific primers. PCR products were cloned into the pGUS‐ENTR‐3 vector and subsequently into pSITE‐2CA. In order to delete the two experimentally confirmed NLDs in 7H08, two sets of primer pairs (7H08 58–194, 241–300‐FP1/7H08 58–194, 241–300‐RP1 and 7H08 58–194, 241–300‐FP2/7H08 58–194, 241–300‐RP2) were used to generate 7H08 NLDdel. The PCR product was cloned into the pGUS‐ENTR‐3 vector and subsequently into pSITE‐2CA. Primers are detailed in Table S2. Clones were sequenced at Elim Biopharmaceuticals.
Agrobacterium tumefaciens‐mediated transient expression of effector fusion constructs in tobacco leaves
Effector fusion constructs were introduced into A. tumefaciens GV3101 using the freeze–thaw method (Weigel and Glazebrook, 2006). Suspensions at a final optical density at 600 nm (OD600) of 0.6 in infiltration buffer (10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6, 150 μm acetosyringone) were infiltrated into the leaves (abaxial side) of 4‐week‐old Nicotiana benthamiana plants (Sparkes et al., 2006). EGFP‐GUS‐effector constructs were co‐infiltrated with HcPro, the suppressor of post‐transcriptional gene silencing of Potato virus Y (Brigneti et al., 1998), at a 1:1 ratio. For each construct, three to four leaves of two individual tobacco plants were infiltrated. Constructs with EGFP only and GUS/EGFP served as controls for subcellular localization (Fig. 1b–d). EGFP‐GUS fused to Arabidopsis histone 2B (At1G07790) served as an additional positive control for nuclear import (Fig. 3c). Leaf samples were harvested at 48 h post‐inoculation for EGFP detection. Infiltrated leaf sections were mounted in water and the EGFP fluorescence was visualized at an excitation wavelength of 488 nm, with emission collected between 510 and 550 nm on a LSM 510 META inverted confocal microscope (Carl Zeiss, Jena, Germany). To confirm the localization of plant nuclei, leaf sections were counterstained with DAPI. Tobacco leaves were infiltrated with DAPI solution (1 μg/mL) for 5 min at room temperature. Images of DAPI staining were collected at excitation/emission wavelengths of 350/470 nm on a LSM 510 META confocal microscope (Carl Zeiss).
Yeast transcriptional activation assay
Transactivation activity by 7H08 was determined in yeast using a system based on the Matchmaker Gold Yeast Two‐Hybrid Kit (Clontech, Mountain View, CA, USA). A series of 7H08 deletion variants was created by PCR (Table S3, see Supporting Information) and cloned into pGBKT7, in which the effector fragment was fused, in frame, with GAL4BD. Clones were sequenced at Elim Biopharmaceuticals to confirm sequence identity. Constructs were introduced into Saccharomyces cerevisiae strain AH109 using the Yeastmaker Yeast Transformation System 2 Kit (Clontech) and positive transformants were selected on synthetic medium without tryptophan (SC/–T). Three independent yeast colonies for each construct were suspended in sterile water and dotted onto synthetic medium without histidine or tryptophan (SC/–TH) to test for activation of the HIS3 reporter gene. Yeast colonies were also dotted onto SC/–T medium with the chromogenic substrate X‐α‐Gal (4 mg/mL). Expression of the MEL1 reporter gene causes yeast colonies to turn blue in the presence of X‐α‐Gal. All experiments were replicated three times and are based on three independent transformations. GAL4BD and GAL4BD‐GAL4AD served as negative and positive controls, respectively.
Plant transcriptional activation assay
To determine the transcriptional activation activity of 7H08 in plant cells, a previously described UAS‐LUC reporter system (Chern et al., 2012) was utilized. This assay determined the ability of 7H08 protein fragments to activate expression of the reporter gene LUC in tobacco leaf cells. To prevent A. tumefaciens from expressing the LUC gene, the UAS‐LUC/SK vector was modified by replacing the LUC gene with a LUC gene containing an intron. Specifically, an intron from pCAMBIA1301 was amplified by PCR (forward primer, 5′‐CGATGGATCCATGGTAGATCTGAGGGTAAATTTC‐3′; reverse primer, 5′‐ATCGCTGCAGACTGAATGCCCACAGGCCGTC‐3′) and cloned into the BamHI/PstI sites of pBluescript II SK+. LUC cDNA was amplified (forward primer, 5′‐ATCGCTGCAGATGGAAGACGCCAAAAACATAAAG‐3′; reverse primer, 5′‐CGATAAGCTTTTACAATTTGGACTTTCCGCC‐3′) from the UAS‐LUC/SK vector and cloned downstream of the intron in pBluescript II SK+ vector to fuse LUC with the intron (LUCint). The sequence of LUCint was amplified and used to replace the LUC gene in the UAS‐LUC/SK plasmid. The entire UAS‐LUCint cassette was transferred into the pDONR221 Gateway vector using BP reaction (Life Technologies). An LR reaction was performed to clone the UAS‐LUCint cassette into the binary vector pEarleygate 301 (Earley et al., 2006) for expression in plants (Fig. 6a). The binary vector pCAMBIA1301, containing a Cauliflower mosaic virus (CaMV) 35S‐driven and intron‐containing GUS reporter gene, was used as reference plasmid to determine the efficiency of tobacco cell transformation. To create constructs containing 7H08 and 7H08 deletions fused with GAL4BD, sequences of GAL4BD‐7H08 or 7H08 deletions were amplified by PCR from the corresponding constructs used in the yeast assay and cloned into pDONR221 (primers detailed in Table S3), and the resulting constructs were cloned into pSITE‐2NB for expression in plants. Clones were sequenced at Elim Biopharmaceuticals to confirm sequence identity. All three constructs were individually transformed into A. tumefaciens, as described above. Suspensions of A. tumefaciens containing the reporter construct, reference construct, GAL4BD‐7H08 deletions construct and HcPro at a final OD600 of 0.5 were infiltrated into N. benthamiana leaves as described above. Infiltrated leaf samples were collected in liquid nitrogen at 48 h post‐infiltration. Ground tissue samples were resuspended in extraction buffer [100 mm potassium phosphate, pH 7.8, 1 mm ethylenediaminetetraacetic acid (EDTA), 10 mm dithiothreitol (DTT), 5% glycerol, phenylmethylsulphonyl fluoride (PMSF, 17 μg/mL)], centrifuged at 15 700 g at 4 °C for 5 min, and the supernatant was used to determine GUS and LUC activity. LUC and GUS activity were determined as described in Chern et al. (2012). Briefly, LUC activity was measured using the Luciferase Assay System (Promega, Madison, WI, USA) with a VICTOR X5 multilabel plate reader (Perkin‐Elmer, Waltham, MA, USA) according to the manufacturer's instructions. GUS activity was assayed using the fluorometric substrate 4‐methylumbelliferone with a Fluoromax‐4 fluorescence spectrophotometer (Horiba Jobin‐Yvon, Edison, NJ, USA) according to the manufacturer's instructions and Jefferson et al. (1987). The relative reporter gene activity was calculated by LUC activity divided by GUS activity. A Student's t‐test was performed using SAS version 9.2 to determine statistically significant differences in LUC/GUS relative activity between GAL4BD control and GAL4BD‐effector. Four leaves per individual plant were inoculated per experiment. The experiment was replicated three times. Although the same relative activity pattern was observed in each experiment, statistical analysis revealed that data from each of the three independent experiments could not be combined for analysis. Individual replicates were analysed independently and representative results are shown.
Western blot analysis of protein expression in tobacco leaves
Total protein was extracted from leaf samples of N. benthamiana using protein extraction buffer (25 mm Tris‐HCl, pH 8.0, 2 mm EDTA, 150 mm NaCl, 1 mm DTT) and Protease Inhibitor Cocktail for plant cell extract (Sigma‐Aldrich, St. Louis, MO, USA). The reporter construct expressing GUS under the control of the CaMV35S promoter was used as indicator of transformation efficiency of tobacco leaf cells by A. tumefaciens. The amount of protein loaded was normalized to GUS activity to ensure equal loading of protein from transformed tobacco cells. Equal amounts of protein samples were separated on sodium dodecylsulphate‐polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Following blocking with 5% non‐fat milk in TBST solution (50 mm Tris‐HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) for 1 h at room temperature, the membrane was incubated with a 1:250 dilution of the anti‐GAL4‐DBD primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 °C overnight. After washing with TBST solution, the membrane was incubated with horseradish peroxidase‐conjugated anti‐mouse secondary antibody (1:4000 dilution) (Thermo Scientific, Rockford, IL, USA) for 1 h at room temperature. The antigen–antibody complex was detected using a BM Chemiluminescence Western Blot Kit (Roche Applied Sciences, Indianapolis, IN, USA). Western blots were replicated four times using protein extracts from two independent isolations.
Supporting information
Table S1 Details of primers used in the construction of vectors fusing effector cDNA with β‐glucuronidase (GUS) and enhanced green fluorescent protein (EGFP).
Table S2 Details of primers used in the analysis of 7H08 and 2G02 nuclear localization signal (NLS) domains.
Table S3 Details of primers used in the determination of the transcriptional activation activity of 7H08 in yeast and plants.
Acknowledgements
The authors thank Dr Richard Hussey (University of Georgia) for providing the cDNA clones of M. incognita effectors and Dr Pamela Ronald (UC Davis) for sharing the UAS‐LUC/SK and pCAMBIA1301 vectors. Dr Michael Goodin (University of Kentucky) provided the pSITE‐2CA and pSITE‐2NB vectors. We thank Dr Michael Konkel for technical support with the luciferase activity assay, Dr Helmut Kirchhoff for assistance with the GUS assay, and the Franceschi Microscopy and Imaging Center (Washington State University) for assistance with confocal microscopy. We thank Drs Linda Thomashow and Lee Hadwiger for critical reading of the manuscript. This study was funded by grants from the Washington State Department of Agriculture, Washington State Potato Commission, Idaho Potato Commission and Washington Grain Commission to A.A.E. PPNS no. 0648, Department of Plant Pathology, College of Agricultural, Human, and Natural Resource Sciences, Agricultural Research Center, Hatch Project No. WNP00744, Washington State University, Pullman, WA 99164‐6430, USA.
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Barcala, M. , Fenoll, C. and Escobar, C. (2012) Laser microdissection of cells and isolation of high‐quality RNA after cryosectioning. Methods Mol. Biol. 883, 87–95. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chakrabarty, R. , Banerjee, R. , Chung, S.M. , Farman, M. , Citovsky, V. , Hogenhout, S.A. , Tzfira, T. and Goodin, M. (2007) PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana–virus interactions. Mol. Plant–Microbe Interact. 20, 740–750. [DOI] [PubMed] [Google Scholar]
- Chern, M. , Bai, W. , Sze‐To, W.H. , Canlas, P.E. , Bartley, L.E. and Ronald, P.C. (2012) A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1‐mediated transcriptional activation. Plant Methods, 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani, I. , Baldacci‐Cresp, F. , Hopkins, J. , Andrio, E. , Balzergue, S. , Lecomte, P. , Puppo, A. , Abad, P. , Favery, B. and Herouart, D. (2012) Plant genes involved in harbouring symbiotic rhizobia or pathogenic nematodes. New Phytol. 194, 511–522. [DOI] [PubMed] [Google Scholar]
- Davis, E.L. , Hussey, R.S. , Mitchum, M.G. and Baum, T.J. (2008) Parasitism proteins in nematode–plant interactions. Curr. Opin. Plant Biol. 11, 360–366. [DOI] [PubMed] [Google Scholar]
- Doyle, E.A. and Lambert, K.N. (2002) Cloning and characterization of an esophageal‐gland‐specific pectate lyase from the root‐knot nematode Meloidogyne javanica . Mol. Plant–Microbe Interact. 15, 549–556. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elling, A.A. , Davis, E.L. , Hussey, R.S. and Baum, T.J. (2007) Active uptake of cyst nematode parasitism proteins into the plant cell nucleus. Int. J. Parasitol. 37, 1269–1279. [DOI] [PubMed] [Google Scholar]
- Escher, D. , Bodmer‐Glavas, M. , Barberis, A. and Schaffner, W. (2000) Conservation of glutamine‐rich transactivation function between yeast and humans. Mol. Cell. Biol. 20, 2774–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen, G. and Mitchum, M.G. (2011) How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 14, 415–421. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J. , Pierson, E. , Lambert, G.M. , Gong, F.C. , Afonso, C.L. , Haldeman‐Cahill, R. , Carrington, J.C. and Galbraith, D.W. (1997) Green‐fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11, 573–586. [DOI] [PubMed] [Google Scholar]
- Haasen, D. , Kohler, C. , Neuhaus, G. and Merkle, T. (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine‐rich nuclear export signals in Arabidopsis thaliana . Plant J. 20, 695–705. [DOI] [PubMed] [Google Scholar]
- Haegeman, A. , Mantelin, S. , Jones, J.T. and Gheysen, G. (2012) Functional roles of effectors of plant‐parasitic nematodes. Gene, 492, 19–31. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L. , Tonti‐Filippini, J. , Verboom, R.E. and Millar, A.H. (2005) Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis . Plant Physiol. 139, 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi, T. and Baum, T.J. (2013) Manipulation of plant cells by cyst and root‐knot nematode effectors. Mol. Plant–Microbe Interact. 26, 9–16. [DOI] [PubMed] [Google Scholar]
- Hicks, G.R. and Raikhel, N.V. (1995) Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11, 155–188. [DOI] [PubMed] [Google Scholar]
- Hilser, V.J. and Thompson, E.B. (2011) Structural dynamics, intrinsic disorder, and allostery in nuclear receptors as transcription factors. J. Biol. Chem. 286, 39 675–39 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A. , Terauchi, R. and Kamoun, S. (2009) Emerging concepts in effector biology of plant‐associated organisms. Mol. Plant–Microbe Interact. 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. , Gao, B. , Maier, T. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2003) A profile of putative parasitism genes expressed in the esophageal gland cells of the root‐knot nematode Meloidogyne incognita . Mol. Plant–Microbe Interact. 16, 376–381. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Dong, R. , Maier, T. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2004) Use of solid‐phase subtractive hybridization for the identification of parasitism gene candidates from the root‐knot nematode Meloidogyne incognita . Mol. Plant Pathol. 5, 217–222. [DOI] [PubMed] [Google Scholar]
- Hussey, R.S. (1989) Disease‐inducing secretions of plant‐parasitic nematodes. Annu. Rev. Phytopathol. 27, 123–141. [Google Scholar]
- Ithal, N. , Recknor, J. , Nettleton, D. , Maier, T. , Baum, T.J. and Mitchum, M.G. (2007) Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant–Microbe Interact. 20, 510–525. [DOI] [PubMed] [Google Scholar]
- Jammes, F. , Lecomte, P. , de Almeida‐Engler, J. , Bitton, F. , Martin‐Magniette, M.L. , Renou, J.P. , Abad, P. and Favery, B. (2005) Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis . Plant J. 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jaouannet, M. , Perfus‐Barbeoch, L. , Deleury, E. , Magliano, M. , Engler, G. , Vieira, P. , Danchin, E.G. , Da Rocha, M. , Coquillard, P. , Abad, P. and Rosso, M.N. (2012) A root‐knot nematode‐secreted protein is injected into giant cells and targeted to the nuclei. New Phytol. 194, 924–931. [DOI] [PubMed] [Google Scholar]
- Jaouannet, M. , Magliano, M. , Arguel, M.J. , Gourgues, M. , Evangelisti, E. , Abad, P. and Rosso, M.N. (2013) The root‐knot nematode calreticulin Mi‐CRT is a key effector in plant defense suppression. Mol. Plant–Microbe Interact. 26, 97–105. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.T. , Kumar, A. , Pylypenko, L.A. , Thirugnanasambandam, A. , Castelli, L. , Chapman, S. , Cock, P.J. , Grenier, E. , Lilley, C.J. , Phillips, M.S. and Blok, V.C. (2009) Identification and functional characterization of effectors in expressed sequence tags from various life cycle stages of the potato cyst nematode Globodera pallida . Mol. Plant Pathol. 10, 815–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D. , Richardson, W.D. , Markham, A.F. and Smith, A.E. (1984a) Sequence requirements for nuclear location of simian virus 40 large‐T antigen. Nature, 311, 33–38. [DOI] [PubMed] [Google Scholar]
- Kalderon, D. , Roberts, B.L. , Richardson, W.D. and Smith, A.E. (1984b) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Wieduwild, R. and Bonas, U. (2009) Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Δrep16. Plant J. 59, 859–871. [DOI] [PubMed] [Google Scholar]
- Koroleva, O.A. , Tomlinson, M.L. , Leader, D. , Shaw, P. and Doonan, J.H. (2005) High‐throughput protein localization in Arabidopsis using Agrobacterium‐mediated transient expression of GFP‐ORF fusions. Plant J. 41, 162–174. [DOI] [PubMed] [Google Scholar]
- Kragelund, B.B. , Jensen, M.K. and Skriver, K. (2012) Order by disorder in plant signaling. Trends Plant Sci. 17, 625–632. [DOI] [PubMed] [Google Scholar]
- Kyndt, T. , Vieira, P. , Gheysen, G. and de Almeida‐Engler, J. (2013) Nematode feeding sites: unique organs in plant roots. Planta, 238, 807–818. [DOI] [PubMed] [Google Scholar]
- Lewis, B.A. and Reinberg, D. (2003) The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116, 3667–3675. [DOI] [PubMed] [Google Scholar]
- Lin, B. , Zhuo, K. , Wu, P. , Cui, R. , Zhang, L.H. and Liao, J. (2013) A novel effector protein, MJ‐NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. Plant–Microbe Interact. 26, 55–66. [DOI] [PubMed] [Google Scholar]
- Lin, Y.T. and Yen, P.H. (2006) A novel nucleocytoplasmic shutting sequence of DAZAP1, a testis‐abundant RNA‐binding protein. RNA, 12, 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert, U. , Cramer, M. , Meuli, M. , Georgiev, O. and Schaffner, W. (2009) Metal‐responsive transcription factor 1 (MTF‐1) activity is regulated by a nonconventional nuclear localization signal and a metal‐responsive transactivation domain. Mol. Cell. Biol. 29, 6283–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding, R. , Jensen, L.J. , Diella, F. , Bork, P. , Gibson, T.J. and Russell, R.B. (2003) Protein disorder prediction: implications for structural proteomics. Structure, 11, 1453–1459. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Perumal, N.B. , Oldfield, C.J. , Su, E.W. , Uversky, V.N. and Dunker, A.K. (2006) Intrinsic disorder in transcription factors. Biochemistry, 45, 6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G. , Hussey, R.S. , Baum, T.J. , Wang, X. , Elling, A.A. , Wubben, M. and Davis, E.L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S. , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. , European Union Effectoromics, C. , Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J.L. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K. and Horton, P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Nissan, G. , Manulis‐Sasson, S. , Weinthal, D. , Mor, H. , Sessa, G. and Barash, I. (2006) The type III effectors HsvG and HsvB of gall‐forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol. Microbiol. 61, 1118–1131. [DOI] [PubMed] [Google Scholar]
- Nissan, G. , Manulis‐Sasson, S. , Chalupowicz, L. , Teper, D. , Yeheskel, A. , Pasmanik‐Chor, M. , Sessa, G. and Barash, I. (2012) The type III effector HsvG of the gall‐forming Pantoea agglomerans mediates expression of the host gene HSVGT. Mol. Plant–Microbe Interact. 25, 231–240. [DOI] [PubMed] [Google Scholar]
- Oldfield, C.J. , Meng, J. , Yang, J.Y. , Yang, M.Q. , Uversky, V.N. and Dunker, A.K. (2008) Flexible nets: disorder and induced fit in the associations of p53 and 14‐3‐3 with their partners. BMC Genomics, 9 (Suppl. 1), S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo, M. , Lindsey, K. , Casson, S. , Garcia‐Casado, G. , Solano, R. , Fenoll, C. and Escobar, C. (2009) Isolation of RNA from laser‐capture‐microdissected giant cells at early differentiation stages suitable for differential transcriptome analysis. Mol. Plant Pathol. 10, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo, M. , Cabrera, J. , Lindsey, K. , Topping, J. , Andres, M.F. , Emiliozzi, M. , Oliveros, J.C. , Garcia‐Casado, G. , Solano, R. , Koltai, H. , Resnick, N. , Fenoll, C. and Escobar, C. (2013) Distinct and conserved transcriptomic changes during nematode‐induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. [DOI] [PubMed] [Google Scholar]
- Remacle, J.E. , Albrecht, G. , Brys, R. , Braus, G.H. and Huylebroeck, D. (1997) Three classes of mammalian transcription activation domain stimulate transcription in Schizosaccharomyces pombe . EMBO J. 16, 5722–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas, S. (2012) Nuclear dynamics during plant innate immunity. Plant Physiol. 158, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas, S. and Genin, S. (2011) A plethora of virulence strategies hidden behind nuclear targeting of microbial effectors. Front. Plant Sci. 2, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.N. , Hussey, R.S. , Davis, E.L. , Smant, G. , Baum, T.J. , Abad, P. and Mitchum, M.G. (2011) Nematode effector proteins: targets and functions in plant parasitism In: Effectors in Plant–Microbe Interactions (Martin F. and Kamoun S., eds), pp. 327–354. Oxford: Wiley‐Blackwell. [Google Scholar]
- Siomi, H. and Dreyfuss, G. (1995) A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I.A. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sprenger, J. , Fink, J.L. and Teasdale, R.D. (2006) Evaluation and comparison of mammalian subcellular localization prediction methods. BMC Bionformatics, 7 (Suppl. 5), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, N. , Fujita, M. , Handa, H. , Murayama, S. , Uemura, M. , Kawamura, Y. , Mitsui, T. , Mikami, S. , Tozawa, Y. , Yoshinaga, T. and Komatsu, S. (2004) Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Mol. Gen. Genomics, 271, 566–576. [DOI] [PubMed] [Google Scholar]
- Triezenberg, S.J. (1995) Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5, 190–196. [DOI] [PubMed] [Google Scholar]
- Trudgill, D.L. and Blok, V.C. (2001) Apomictic, polyphagous root‐knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39, 53–77. [DOI] [PubMed] [Google Scholar]
- Tytgat, T. , Vanholme, B. , De Meutter, J. , Claeys, M. , Couvreur, M. , Vanhoutte, I. , Gheysen, G. , Van Criekinge, W. , Borgonie, G. , Coomans, A. and Gheysen, G. (2004) A new class of ubiquitin extension proteins secreted by the dorsal pharyngeal gland in plant parasitic cyst nematodes. Mol. Plant–Microbe Interact. 17, 846–852. [DOI] [PubMed] [Google Scholar]
- Uesugi, M. and Verdine, G.L. (1999) The α‐helical FXXΦΦ motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA, 96, 14 801–14 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky, V.N. (2011) Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 43, 1090–1103. [DOI] [PubMed] [Google Scholar]
- Van Dusen, C.M. , Yee, L. , McNally, L.M. and McNally, M.T. (2010) A glycine‐rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell. Biol. 30, 2552–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, P. , Danchin, E.G. , Neveu, C. , Crozat, C. , Jaubert, S. , Hussey, R.S. , Engler, G. , Abad, P. , de Almeida‐Engler, J. , Castagnone‐Sereno, P. and Rosso, M.N. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Lee, C. , Replogle, A. , Joshi, S. , Korkin, D. , Hussey, R.S. , Baum, T.J. , Davis, E.L. , Wang, X. and Mitchum, M.G. (2010) Dual roles for the variable domain in protein trafficking and host‐specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 187, 1003–1017. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Meyers, D. , Yan, Y. , Baum, T.J. , Smant, G. , Hussey, R.S. and Davis, E.L. (1999) In planta localization of a beta‐1,4‐endoglucanase secreted by Heterodera glycines . Mol. Plant–Microbe Interact. 12, 64–67. [DOI] [PubMed] [Google Scholar]
- Weigel, D. and Glazebrook, J. (2006) Transformation of Agrobacterium using the freeze–thaw method. CSH Protoc. doi: 10.1101/pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Weinthal, D.M. , Barash, I. , Tzfira, T. , Gaba, V. , Teper, D. , Sessa, G. and Manulis‐Sasson, S. (2011) Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall‐forming bacterium Pantoea agglomerans . Microbiology, 157, 1500–1508. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y.Y. , Deng, X. and Matsui, M. (2001) Cip4, a new COP1 target, is a nucleus‐localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell, 13, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Details of primers used in the construction of vectors fusing effector cDNA with β‐glucuronidase (GUS) and enhanced green fluorescent protein (EGFP).
Table S2 Details of primers used in the analysis of 7H08 and 2G02 nuclear localization signal (NLS) domains.
Table S3 Details of primers used in the determination of the transcriptional activation activity of 7H08 in yeast and plants.


