Summary
Deoxynivalenol (DON), a trichothecene mycotoxin produced by Fusarium graminearum, is harmful to humans and animals. Because different nitrogen sources are known to have opposite effects on DON production, in this study, we characterized the regulatory mechanisms of the AREA transcription factor in trichothecene biosynthesis. The ΔareA mutant showed significantly reduced vegetative growth and DON production in cultures inoculated with hyphae. Suppression of TRI gene expression and DON production by ammonium were diminished in the ΔareA mutant. The deletion of AREA also affected the stimulatory effects of arginine on DON biosynthesis. The AreA‐green fluorescent protein (GFP) fusion complemented the ΔareA mutant, and its localization to the nucleus was enhanced under nitrogen starvation conditions. Site‐directed mutagenesis showed that the conserved predicted protein kinase A (PKA) phosphorylation site S874 was important for AreA function, indicating that AreA may be a downstream target of the cyclic adenosine monophosphate (cAMP)‐PKA pathway, which is known to regulate DON production. We also showed that AreA interacted with Tri10 in co‐immunoprecipitation assays. The interaction of AreA with Tri10 is probably related to its role in the regulation of TRI gene expression. Interestingly, the ΔareA mutant showed significantly reduced PKA activity and expression of all three predicted ammonium permease (MEP) genes, in particular MEP1, under low ammonium conditions. Taken together, our results show that AREA is involved in the regulation of DON production by ammonium suppression and the cAMP‐PKA pathway. The AreA transcription factor may interact with Tri10 and control the expression and up‐regulation of MEP genes.
Keywords: ammonium suppression, DON production, Gibberella zeae, nitrogen metabolism, TRI6 expression
Introduction
Fusarium head blight (FHB) or scab is one of the most important diseases of wheat and barley. Fusarium graminearum (teleomorph Gibberella zeae) is a major causal agent of FHB in North America and other parts of the world (Bai and Shaner, 2004; Goswami and Kistler, 2004). In addition to causing severe yield losses, this pathogen produces mycotoxins, such as deoxynivalenol (DON) and zearalenone, which are harmful to humans and animals (Desjardins et al., 2000). As a trichothecene, DON is a potent inhibitor of protein synthesis in eukaryotic organisms and is also toxic to plant cells (Maier et al., 2006). Indeed, DON production was the first and best studied virulence factor in F. graminearum (Desjardins et al., 2000; Proctor et al., 1997).
In the past decade, the genes involved in trichothecene biosynthesis have been well characterized (Desjardins et al., 2000). In F. graminearum, most of the TRI genes are in the core TRI cluster, including the trichodiene synthase gene TRI5 and two transcription factor genes TRI6 and TRI10. TRI101 and TRI1 are located on separate chromosomal regions (Alexander et al., 2009; Brown et al., 2004; Gale et al., 2005; Kimura et al., 2003). Tri5 catabolizes the cyclization of farnesyl pyrophosphate (FPP), the first step of the trichothecene biosynthesis pathway. Tri6 is a C2H2 zinc finger protein that plays a more critical role than Tri10 in DON biosynthesis and plant infection, but both are important for the regulation of TRI gene expression (Nasmith et al., 2011; Seong et al., 2009). Tri6 and Tri10 also regulate the expression of the genes involved in the isoprenoid pathway, which is responsible for the biosynthesis of FPP, a precursor for ergosterol and trichothecene biosynthesis (Seong et al., 2009).
In the past decade, various environmental and physiological conditions have been shown to affect DON synthesis in F. graminearum, including different nitrogen sources, pH, reactive oxygen species (ROS) and fungicide treatments (Audenaert et al., 2010; Gardiner et al., 2009a, 2009b; Merhej et al., 2011; Montibus et al., 2013; Ochiai et al., 2007). Similar to its inhibition of secondary metabolism in other fungi, ammonium, a preferred nitrogen source for most fungi, suppresses DON production in F. graminearum (Pestka et al., 1985). However, several nitrogen sources, such as arginine, are known to induce DON biosynthesis (Gardiner et al., 2009a). Similar to Gln3 or Gat1 in Saccharomyces cerevisiae (Coffman et al., 1996; Minehart and Magasanik, 1991), NIT‐2 and areA serve as the global regulators of nitrogen metabolism in Neurospora crassa and Aspergillus nidulans, respectively (Caddick et al., 1986; Fu and Marzluf, 1990). AreA orthologues are also known to function as positive regulators of the genes required for the utilization of secondary nitrogen sources in several other filamentous fungi (Christensen et al., 1998; Divon et al., 2006; Kim and Woloshuk, 2008; Mihlan et al., 2003). In F. graminearum, deletion of AREA, the orthologue of areA, resulted in defects in plant infection and trichothecene biosynthesis, but zearalenone production was not affected (Giese et al., 2013; Min et al., 2012).
Because of the importance of nitrogen metabolism in inducing DON production, in this study, we further characterized the regulatory role of the AreA transcription factor. Deletion of AREA led to a decrease in the suppression of TRI gene expression and in DON production by ammonium. The putative nuclear localization signal (NLS) sequence NLS3 and a conserved protein kinase A (PKA) phosphorylation site S874 were found to be important for AreA function. We found that the ΔareA mutant showed significantly reduced PKA activities, Gpmk1 phosphorylation and expression of all three predicted ammonium permease (MEP) genes in F. graminearum. Furthermore, we showed that AreA interacted with Tri10 in co‐immunoprecipitation assays. The interaction of AreA with Tri10 may be important for its regulation of DON biosynthesis by ammonium sensing in F. graminearum.
Results
Ammonium inhibits AREA expression and DON production
In filamentous fungi, ammonium is inhibitory to the use of secondary nitrogen sources, which is regulated by AreA and its orthologues (Bolton and Thomma, 2008). To determine the effect of ammonium on AREA (FGSG_08634) expression, we isolated RNA from vegetative hyphae of the wild‐type strain PH‐1 (Table 1), which were harvested from 2‐day‐old complete medium (CM) cultures and further incubated in medium with 50 mm nitrate or ammonium as the nitrogen source for 1 h. When assayed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), the expression level of AREA in ammonium samples was 20‐fold lower than that in nitrate samples (Fig. 1A), suggesting that AREA expression was up‐regulated in the medium with 50 mm nitrate as the sole nitrogen source compared with the medium with 50 mm ammonium.
Table 1.
Wild‐type and mutant strains of Fusarium graminearum used in this study
| Name | Brief description | Reference |
|---|---|---|
| PH‐1 | Wild‐type | Cuomo et al. (2007) |
| RH12 | ΔareA deletion mutant of PH‐1 | This study |
| AC6 | ΔareA/AREA‐GFP transformant of mutant RH12 | This study |
| RL10 | ΔareA/AREA ΔNLS1‐GFP transformant of mutant RH12 | This study |
| RL11 | ΔareA/AREA ΔNLS1‐GFP transformant of mutant RH12 | This study |
| RL12 | ΔareA/AREA ΔNLS2‐GFP transformant of mutant RH12 | This study |
| RL13 | ΔareA/AREA ΔNLS2‐GFP transformant of mutant RH12 | This study |
| RE8 | ΔareA/AREA ΔNLS3‐GFP transformant of mutant RH12 | This study |
| RE15 | ΔareA/AREA ΔNLS3‐GFP transformant of mutant RH12 | This study |
| RP2 | ΔareA/AREA ΔS874‐GFP (PKA site) transformant of RH12 | This study |
| RP4 | ΔareA/AREA ΔS874‐GFP (PKA site) transformant of RH12 | This study |
| RM3 | ΔareA/AREA ΔS657‐S658‐GFP (MAPK site) transformant of RH12 | This study |
| RM6 | ΔareA/AREA ΔS657‐S658‐GFP (MAPK site) transformant of RH12 | This study |
| RPA6 | ΔareA/AREA S874A‐GFP (PKA site) transformant of RH12 | This study |
| RPA8 | ΔareA/AREA S874A‐GFP (PKA site) transformant of RH12 | This study |
| AT61 | PRP27‐AREA‐3 × FLAG and TRI6‐GFP transformant of PH‐1 | This study |
| AT63 | PRP27‐AREA‐3 × FLAG and TRI6‐GFP transformant of PH‐1 | This study |
| AT02 | PRP27‐AREA‐3 × FLAG and TRI10‐GFP transformant of PH‐1 | This study |
| PH1Δtri6 | Δtri6 deletion mutant of PH‐1 | Seong et al. (2009) |
| PH1Δtri10 | Δtri10 deletion mutant of PH‐1 | Seong et al. (2009) |
| 6N5 | Δtri6/TRI6 complementation transformant of mutant RH12 | This study |
Figure 1.
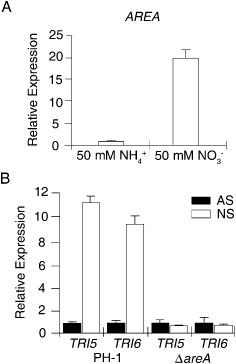
AREA expression and its role in the regulation of the TRI gene. (A) Relative expression level of AREA in complete medium (CM) cultures with 50 mm ammonium (NH4+; arbitrarily set to unity) or nitrate (NO3 −) as the sole nitrogen source. (B) The expression level of TRI5 and TRI6 in cultures of the wild‐type strain PH‐1 and ΔareA mutant grown under nitrogen starvation (NS) or ammonium suppression (AS; arbitrarily set to unity) conditions.
Because ammonium is known to inhibit secondary metabolism in fungi (Pestka et al., 1985), we added 50 mm ammonium to 21‐day‐old rice grain cultures of PH‐1. DON production was assayed after incubation for another 5 days. In comparison with the control (no ammonium added), ammonium treatment reduced DON production 2.6‐fold (Table 2). When assayed in RNA samples isolated from vegetative hyphae of PH‐1 cultured in medium with 1 mm nitrate (nitrogen starvation, NS) or 50 mm ammonium (ammonium suppression, AS) as the sole nitrogen source for 6 h, the expression levels of TRI5 and TRI6 were 11.4‐ and 9.4‐fold higher, respectively, under NS than AS conditions (Fig. 1B), confirming the inhibitory role of ammonium on trichothecene synthesis.
Table 2.
Growth rate, conidiation and deoxynivalenol (DON) production in the Δare A mutant
| Strain | Growth rate (mm/day)b | Conidiation (×105/mL)c | DON/Ergd | |
|---|---|---|---|---|
| Control | 50 mm NH4 + | |||
| PH‐1 (WT) | 6.9 ± 0.1A a | 10.5 ± 1.1A | 0.13 ± 0.03A | 0.05 ± 0.01A |
| RH12 (ΔareA) | 5.8 ± 0.0B | 1.9 ± 0.2B | 0.02 ± 0.01B | 0.25 ± 0.09B |
| AC6 (ΔareA/AREA) | 7.0 ± 0.1A | 10.2 ± 1.8A | 0.11 ± 0.05A | 0.06 ± 0.02A |
WT, wild‐type.
Data from three replicates were analysed with the protected Fisher's least‐significant difference (LSD) test. The same letter indicates that there is no significant difference (P = 0.05).
Growth rate was assayed with 3‐day‐old complete medium (CM) agar cultures.
Conidiation was measured with 5‐day‐old CarboxyMethylCellulose (CMC) cultures.
DON/ergosterol (Erg) ratio was assayed with 3‐week‐old rice grain cultures with or without 50 mm ammonium phosphate added at day 5. Erg was measured to quantify fungal biomass.
Inhibitory effect of NH4+ on DON production and utilization of secondary nitrogen sources requires AREA
We generated the AREA gene replacement mutant of the wild‐type strain PH‐1 (Table 1) by the split‐marker approach. Similar to the ΔareA mutant of strain GZ03639 (Min et al., 2012), the ΔareA mutant of PH‐1 showed a significantly reduced growth rate (Fig. S1, see Supporting Information), virulence (Fig. S2, see Supporting Information), conidiation and DON production (Table 2). In the absence of other nitrogen sources, high concentrations of ammonium and glutamine, but not nitrate, partially recovered the growth defects of the ΔareA mutant (Fig. 2A; Table 3). Furthermore, we found that the growth rate of ΔareA increased with an increase in concentration of ammonium from 5 to 50 mm (Fig. 2A; Table 3).
Figure 2.
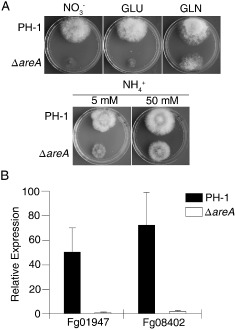
Defects of the ΔareA mutant in response to different nitrogen sources. (A) Cultures of the wild‐type strain PH‐1 and ΔareA mutant grown on complete medium (CM) with 50 mm sodium nitrate (NO3 –), glutamate (GLU) or glutamine (GLN) and 5 or 50 mm ammonium phosphate (NH4 +) as the nitrogen source. (B) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assays of the nitrate (FGSG_01947) and nitrite (FGSG_08402) reductase genes in PH‐1 and ΔareA mutant grown under nitrogen starvation (NS) and ammonium suppression (AS; arbitrarily set to 1) conditions.
Table 3.
Growth rates of PH‐1 and Δare A mutant in cultures with different nitrogen sources and different concentrations of ammonium
| Strain | Growth rate (mm/day)b | |||||
|---|---|---|---|---|---|---|
| NH4 + | NO3 − | Glutamine | Glutamate | |||
| 1 mm | 10 mm | 100 mm | ||||
| PH‐1 (WT) | 3.5 ± 0.1A a | 3.5 ± 0.1A | 4.1 ± 0.1A | 3.9 ± 0.1A | 4.4 ± 0.2A | 3.8 ± 0.1A |
| RH12 (ΔareA) | 2.0 ± 0.0B | 2.0 ± 0.1B | 3.4 ± 0.2B | 1.8 ± 0.1B | 2.7 ± 0.1B | 1.3 ± 0.1B |
WT, wild‐type.
Data from three replicates were analysed with the protected Fisher's least‐significant difference (LSD) test. Different letters are used to mark statistically significant differences (P = 0.05).
Growth rates on modified complete medium (CM) with different nitrogen sources or different concentrations of ammonium were measured after incubation for 3 days.
In rice grain cultures, the addition of ammonium reduced DON production in the wild‐type (Table 2), which is consistent with the inhibitory effect of ammonium on secondary metabolism (Teichert et al., 2008). However, the ΔareA mutant showed more than an 11.4‐fold increase in DON biosynthesis in cultures supplemented with 50 mm ammonium compared with regular rice grain cultures (Table 2). These results suggest that, in addition to the stimulatory effect of ammonium on growth, deletion of AREA may release its inhibitory effect on DON biosynthesis. To test this hypothesis, we assayed the expression levels of TRI5 and TRI6 by qRT‐PCR. In the ΔareA mutant, TRI5 and TRI6 expression was not significantly changed (less than 1.4‐fold) in AS cultures relative to NS cultures (Fig. 1B). Therefore, AREA is important for the suppression of TRI5 and TRI6 expression by ammonium.
Because AreA orthologues are important for the utilization of secondary nitrogen sources, we also assayed the expression of the nitrate reductase (FGSG_01947) and nitrite reductase (FGSG_08402) genes in vegetative hyphae of PH‐1 and the ΔareA mutant incubated under NS or AS conditions for 6 h. In the wild‐type strain, the expression levels of FGSG_01947 and FGSG_08402 increased 49.4‐ and 74.6‐fold, respectively, in NS cultures relative to AS cultures (Fig. 2B). However, NS failed to induce the expression of the nitrate and nitrite reductase genes in the ΔareA mutant (Fig. 2B).
Induction of DON biosynthesis by arginine is affected by AREA deletion
Because arginine can stimulate DON production, we assayed TRI gene expression in liquid cultures with 5 mm nitrate or arginine as described by Gardiner et al. (2009a). In PH‐1, the expression levels of TRI5, TRI6 and TRI10 increased 79.3‐, 205.9‐ and 42.3‐fold, respectively, in arginine cultures relative to nitrate cultures (Fig. 3A). In the ΔareA mutant RH12, the expression levels of TRI5, TRI6 and TRI10 were induced by arginine to 5.1‐, 12.7‐ and 2.6‐fold, respectively (Fig. 3A). DON production in arginine cultures of PH‐1 and mutant RH12 was also 379.3‐ and 96.1‐fold higher, respectively, than in nitrate cultures (Table 4). These results indicate that the ΔareA mutant is still induced by arginine for TRI gene expression and DON production, although to a much lesser degree in comparison with the wild‐type. Therefore, AREA is important, but not essential, for arginine‐induced DON biosynthesis.
Figure 3.
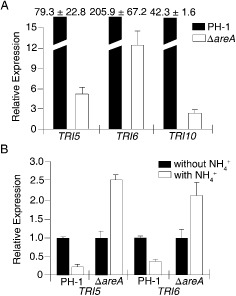
Stimulation of deoxynivalenol (DON) production by arginine is affected by AREA. (A) The expression levels of TRI5, TRI6 and TRI10 in the wild‐type strain PH‐1 and ΔareA mutant in liquid DON‐inducing medium with 5 mm nitrate or arginine. For each gene, the expression level in nitrate cultures was arbitrarily set to unity. (B) The expression levels of TRI5 and TRI6 in DON‐inducing liquid cultures (with arginine) of PH‐1 and ΔareA mutant with 0 or 50 mm ammonium added at the fourth day. Mean and standard deviations were calculated with results from three independent biological replicates.
Table 4.
Deoxynivalenol (DON) production in liquid cultures of the wild‐type (WT) and Δare A mutant
| Strain | DON production (ppm)b | |
|---|---|---|
| NaNO3 | Arginine | |
| PH‐1 (WT) | 6.2 ± 2.1A a | 2348.2 ± 156.8A |
| RH12 (ΔareA) | 1.7 ± 0.4B | 164.6 ± 32.5B |
Data from three replicates were analysed with the protected Fisher's least‐significant difference (LSD) test. Different letters are used to mark statistically significant differences (P = 0.05).
DON production in liquid medium with 5 mm NaNO3 or arginine as the nitrogen source was assayed as described by Gardiner et al. (2009a).
To test whether it can suppress the stimulatory effect of arginine on DON biosynthesis, ammonium was added to 4‐day‐old DON‐inducing cultures to a final concentration of 50 mm. After incubation for another 3 days, the expression level of TRI5 and TRI6 in the wild‐type strain PH‐1 decreased 4.0‐ and 2.7‐fold, respectively, in ammonium‐treated samples compared with the controls (Fig. 3B). However, ammonium failed to inhibit the expression of these TRI genes in the ΔareA mutant. In contrast, TRI5 and TRI6 expression was increased 2.6‐ and 2.1‐fold in ammonium‐treated samples (Fig. 3B), indicating that AREA is important for the inhibitory effects of ammonium on DON biosynthesis induced by arginine.
Localization of AreA‐green fluorescent protein (GFP) fusion to the nucleus is enhanced under NS conditions
For complementation assays, AREA‐GFP fusion constructs were generated and introduced into the ΔareA deletion mutant RH12. The resulting ΔareA/AREA‐GFP transformant AC6 (Table 1) was rescued in all the defects of ΔareA, including growth and plant infection (Figs S1 and S2). In transformant AC6, weak GFP signals were observed in the nucleus in conidia, germ tubes and vegetative hyphae. To determine the effects of nitrogen sources on AreA‐GFP, conidia (Fig. 4A) and hyphae (Fig. 4B) of the ΔareA/AREA‐GFP transformant were cultured under NS conditions for 1 h. In both conidia and hyphae, GFP signals in the nucleus were stronger under NS treatment conditions (Fig. 4), indicating that NS increased the expression and nuclear localization of AreA‐GFP fusion proteins.
Figure 4.
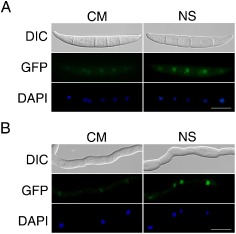
Expression and subcellular localization of AreA‐GFP fusion proteins. (A) Conidia of the ΔareA/ AREA‐GFP transformant AC6 were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) and examined by differential interference contrast (DIC) or epifluorescence microscopy with or without incubation for 1 h under nitrogen starvation (NS) conditions. (B) Germlings of transformant AC6 were stained with DAPI and examined by DIC or epifluorescence microscopy with or without incubation for 1 h under NS conditions. CM, complete medium; GFP, green fluorescent protein. Bar, 10 μm.
Functional characterization of putative NLS sequences of AreA proteins
The PIKSRKE (NLS1), RKRP (NLS2) and LHGVVRPLSL (NLS3) sequences of AreA are three putative NLS sequences (Fig. 5A) that are conserved in its orthologues from Fusarium verticillioides, Fusarium oxysporum, Magnaporthe oryzae, N. crassa and A. nidulans (Hunter et al., 2014). To determine their functions in the localization of AreA to the nucleus, we generated the AREA ΔNLS1‐GFP, AREA ΔNLS2‐GFP and AREA ΔNLS3‐GFP constructs and transformed them into the ΔareA mutant RH12. The resulting ΔareA/AREA ΔNLS1‐GFP and ΔareA/AREA ΔNLS2‐GFP transformants (Table 1) showed the wild‐type growth rate and produced normal colonies (Fig. 5B). GFP signals were mainly observed in the nucleus in these transformants under different growth conditions, indicating that deletion of NLS1 or NLS2 had no effect on the function and nuclear localization of AreA proteins (Fig. S3, see Supporting Information).
Figure 5.
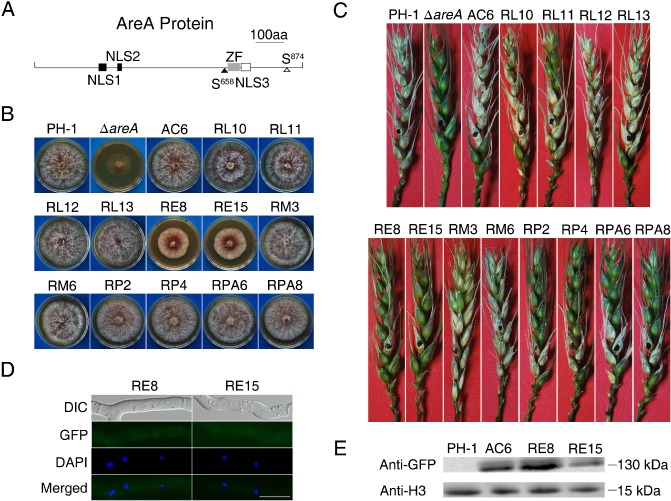
Site‐directed mutagenesis analysis with the AREA gene. (A) The AreA protein is predicted to have three conserved nuclear localization signal (NLS) sequences (NLS1, 238–244; NLS2, 287–290; NLS3, 716–725), one consensus protein kinase A (PKA) (S874) and one mitogen‐activated protein kinase (MAPK) (S658) phosphorylation site, and one zinc figure domain (ZF, 686–710). (B) Four‐day‐old potato dextrose agar (PDA) cultures of the wild‐type strain PH‐1, ΔareA mutant, ΔareA/ AREAΔNLS1‐GFP (RL10 and RL11), ΔareA/ AREAΔNLS2‐GFP (RL12 and RL13), ΔareA/ AREAΔNLS3‐GFP (RE8 and RE15), ΔareA/ AREAΔS874‐GFP (RP2 and RP4), ΔareA/ AREAΔS657 ‐658‐GFP (RM3 and RM6) and ΔareA/ AREAS874A‐GFP (RPA6 and RPA8) transformants, and complementation strain (AC6). (C) Wheat heads inoculated with the same set of strains were examined for scab symptoms at 14 days post‐inoculation (dpi). (D) Germlings of transformants RE8 and RE15 were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) and examined by differential interference contrast (DIC) or epifluorescence microscopy. GFP, green fluorescent protein. Bar, 10 μm. (E) Western blot analysis with total proteins isolated from vegetative hyphae of PH‐1 and ΔareA/ AREAΔNLS3‐GFP transformants RE8 and RE15. The AreA‐GFP band was detected with the anti‐GFP antibody. Detection with an anti‐H3 antibody was used as the control to show that similar amount of proteins were loaded in each lane.
However, the ΔareA/AREA ΔNLS3‐GFP transformants RE8 and RE15 showed partially recovered growth rates (Fig. 5B), although plant infection was still defective (Fig. 5C). When examined under an epifluorescence microscope, we failed to observe GFP signals in the nucleus in conidia or hyphae of these transformants, even under NS conditions (Fig. 5D). On Western blots of proteins isolated from transformants RE8 and RE15, a 130‐kDa band was detected with an anti‐GFP antibody (Fig. 5E). When assayed as described by Gardiner et al. (2009a), the AREA ΔNLS1 and AREA ΔNLS2 transformants were normal, but the AREA ΔNLS3 transformants RE8 and RE15 were only partially recovered in DON production in liquid cultures (Table S2, see Supporting Information). These results indicate that AreAΔNLS3‐GFP fusion proteins were expressed, but not localized, to the nucleus. Therefore, this bipartite NLS sequence must be essential for the localization of AreA to the nucleus and its function during plant infection and DON production in F. graminearum.
S874 is a putative PKA phosphorylation site important for AREA function
Sequence analysis revealed that the AreA protein has one putative consensus mitogen‐activated protein kinase (MAPK) phosphorylation site (PSS658P) and two putative optimal PKA phosphorylation sites (S32 and S874) (Fig. 5A). However, only the S658 and S874 residues are well conserved in AREA orthologues from M. oryzae, A. nidulans and N. crassa. To determine the role of these MAPK and PKA phosphorylation sites, we generated the AREA ΔS657‐658‐GFP and AREA ΔS874‐GFP constructs and transformed them into the ΔareA deletion mutant RH12. The ΔareA/AREA ΔS874 (RP2 and RP4) and ΔareA/AREA ΔS657‐658 (RM3 and RM6) transformants showed similar growth rates to PH‐1 (Fig. 5B). In infection assays with flowering wheat heads, transformants RM3 and RM6 were as virulent as the wild‐type and the complementation transformant. However, transformants RP2 and RP4 were more virulent than the ΔareA mutant, but not as virulent as the wild‐type (Fig. 5C), suggesting that AREA ΔS874 only partially complemented the defects of ΔareA in plant infection.
To further confirm that S874 is important for AreA function during plant infection, we generated the AREA S874A‐GFP construct and transformed it into the ΔareA deletion mutant RH12. Similar to the AREA ΔS874 transformants, the ΔareA/AREA S874A‐GFP transformants RPA6 and RPA8 showed fully recovered growth rates (Fig. 5B), but only partially rescued virulence in infection assays with flowering wheat heads (Fig. 5C). Nevertheless, the AREA ΔS874 and AREA S874A transformants were similar to the wild‐type and complementation transformant with regard to DON production (Table S2). In these transformants, GFP signals were also mainly observed in the nucleus, but localization of the GFP fusion proteins was not increased by NS (data not shown). These results indicate that S874 is not important for the expression and subcellular localization of AreA, but is important for its function during plant infection.
Expression of ammonium permease genes is regulated by AreA
AreA orthologues are known to regulate the expression of ammonium permease genes in A. nidulans (Monahan et al., 2006) and S. cerevisiae (Magasanik and Kaiser, 2002). In F. graminearum, all three predicted ammonium permease genes, FGSG_02094 (MEP1), FGSG_00620 (MEP2) and FGSG_00529 (MEP3), have multiple GATA sequences as putative AreA‐binding sites in their promoter regions (Fig. S4, see Supporting Information). When assayed by qRT‐PCR, the expression levels of MEP1, MEP2 and MEP3 were significantly reduced in the ΔareA mutant compared with the wild‐type in low‐concentration ammonium (0.5 mm) cultures (Fig. 6A). In comparison with PH‐1, the expression levels of MEP1, MEP2 and MEP3 were reduced 2275‐, 134‐ and 904‐fold in the ΔareA mutant in low‐concentration ammonium (0.5 mm) cultures (Fig. 6A). Even in high‐concentration ammonium (50 mm) cultures, the expression levels of MEP1, MEP2 and MEP3 were reduced 2.2‐, 3.1‐ and 23.5‐fold, respectively, in the ΔareA mutant compared with PH‐1 (Fig. 6A).
Figure 6.
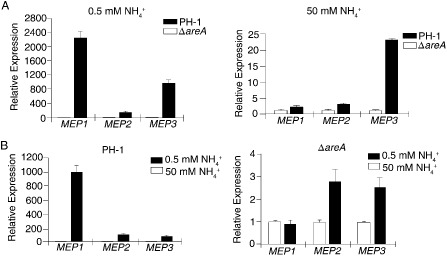
Assays for the expression levels of three ammonium permease genes by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). (A) The expression levels of MEP1, MEP2 and MEP3 were compared between the wild‐type strain PH‐1 and ΔareA mutant, which were cultured in medium with 0.5 mm (low) or 50 mm (high) ammonium. For each gene, the relative expression level in the ΔareA mutant was arbitrarily set to unity. (B) The expression level of each MEP gene in cultures of PH‐1 or ΔareA mutant with 0.5 mm ammonium was compared with that of cultures with 50 mm ammonium (arbitrarily set to unity). Mean and standard deviation were calculated with data from three independent biological replicates.
In the wild‐type, the expression levels of MEP1, MEP2 and MEP3 were 1003.3‐, 120.2‐ and 97.3‐fold higher, respectively, in low‐concentration ammonium cultures than in high‐concentration ammonium cultures, suggesting that MEP1 was the MEP gene with the most significant up‐regulation by low‐concentration ammonium conditions. In the ΔareA mutant, MEP1 expression was not affected by high or low concentrations of ammonium (Fig. 6B). The expression levels of MEP2 and MEP3 were increased 2.8‐ and 2.5‐fold under low‐concentration relative to high‐concentration ammonium conditions (Fig. 6B). Therefore, deletion of AREA also affected the up‐regulation of MEP2 and MEP3, although to a much lesser degree than MEP1, by low‐concentration ammonium conditions.
PKA activities and Gpmk1 phosphorylation are reduced in the ΔareA mutant
It has been shown that both the cyclic adenosine monophosphate (cAMP)‐PKA and MAPK pathways function downstream from Ras signalling in fungi (Li et al., 2012; Zhao and Xu, 2007), and Ras proteins may interact with the C‐terminal region of MEP2 in S. cerevisiae (Lorenz and Heitman, 1998) and Candida albicans (Biswas and Morschhauser, 2005). It is possible that the MEP–RAS association may activate the PKA or MAPK pathway, which, in turn, activates the downstream AreA transcription factor in F. graminearum. To test this hypothesis, we assayed the phosphorylation level of Gpmk1. When detected with an anti‐TpEY antibody, the ΔareA mutant showed a significantly reduced phosphorylation level of the Gpmk1 MAPK, but no obvious changes in Mgv1 activation (Fig. 7A). We also assayed PKA activities with proteins isolated from vegetative hyphae of yeast extract peptone dextrose (YEPD) cultures as described by Adachi and Hamer (1998). In comparison with PH‐1, the ΔareA mutant showed significantly reduced PKA activities (Fig. 7B). These data indicate that deletion of AREA may affect the cAMP‐PKA pathway and the activation of Gpmk1 MAPK in F. graminearum.
Figure 7.
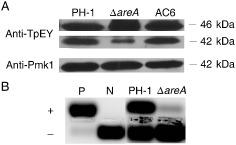
Mitogen‐activated protein kinase (MAPK) phosphorylation and protein kinase A (PKA) activity assays. (A) Assays for the activation of Mgv1 and Gpmk1 MAPKs. Total proteins were isolated from vegetative hyphae of the wild‐type PH‐1, ΔareA mutant and complementation strain (AC6). The phosphorylation of Mgv1 (46 kDa) and Gpmk1 (42 kDa) was detected with the anti‐TpEY antibody. The expression level of Gpmk1 was detected with the anti‐Pmk1 antibody. (B) PKA activities were assayed with proteins isolated from hyphae of PH‐1 and ΔareA mutant using the PepTag A1 PKA substrate peptide. Whereas phosphorylated peptides migrated towards the anode (+), unphosphorylated peptides migrated towards the cathode (–) on a 0.8% agarose gel. N, non‐phosphorylated sample control; P, phosphorylated sample control.
AreA interacts with Tri10 in vivo
One possibility for the regulation of DON production by AreA is that it may interact with Tri6 or Tri10, two key transcription factors in the TRI cluster. To test this hypothesis, we first attempted to study their interactions by yeast two‐hybrid assays, but Tri6, Tri10 and AreA all showed strong self‐activation activities. Therefore, we generated the PRP27‐TRI6‐GFP and PRP27‐TRI10‐GFP constructs and co‐transformed them into PH‐1 with PRP27‐AREA‐3 × FLAG. Because GFP signals were not observed in transformants expressing the TRI6‐GFP and TRI10‐GFP constructs that complemented the Δtri6 and Δtri10 deletion mutants (Seong et al., 2009), we used the RP27 promoter, a strong, constitutive promoter, to overexpress these two genes. Unfortunately, we detected the AreA‐3 × FLAG band, but failed to detect the Tri6‐GFP band, in the transformant AT61 (Fig. 8), which was confirmed by PCR analysis to contain the PRP27‐TRI6‐GFP and AREA‐3 × FLAG fusion constructs. Therefore, it is impossible to assay the interaction between AreA and Tri6.
Figure 8.
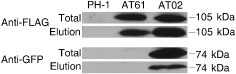
Co‐immunoprecipitation assays for the interactions between Tri10 and AreA. Total proteins (Total) isolated from vegetative hyphae of PH‐1, transformant AT61 (expressing the AREA‐3 × FLAG and TRI6‐GFP constructs) and transformant AT02 (expressing the AREA‐3 × FLAG and TRI10‐GFP constructs) and proteins eluted (Elution) from anti‐FLAG M2 beads were separated on 10% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel. After transfer to nitrocellulose membranes, the presence of AreA‐3 × FLAG, Tri10‐GFP or Tri6‐GFP fusion proteins was detected with the anti‐FLAG and anti‐GFP antibodies. Whereas the Tri6‐GFP band was not detectable in transformant AT61, the 74‐kDa Tri10‐GFP band was detected in total proteins and proteins eluted from anti‐FLAG beads in transformant AT02. GFP, green fluorescent protein.
In total proteins isolated from the PRP27‐TRI10‐GFP AREA‐3 × FLAG transformant AT02, both the 105‐kDa AreA‐3 × FLAG band and the 74‐kDa Tri10‐GFP band were detected with the anti‐FLAG and anti‐GFP antibodies, respectively (Fig. 8). The same antibodies failed to detect these two bands in proteins isolated from the wild‐type strain PH‐1 (Fig. 8). In proteins eluted from anti‐FLAG beads, both AreA‐3 × FLAG and Tri10‐GFP fusion proteins were detected in transformant AT02 (Fig. 8) over three independent replicates. Under the same conditions, only the AreA‐3 × FLAG band was detected in proteins eluted from anti‐FLAG beads for transformant AT61. These data indicate that AreA interacts with Tri10 in F. graminearum, which may be important for the function of AreA in the regulation of DON biosynthesis.
Discussion
AreA orthologues are well conserved in filamentous ascomycetes for the regulation of the utilization of secondary nitrogen sources (Tudzynski, 2014). In F. graminearum, AREA is required for the induced expression of the nitrate and nitrite reductase genes when nitrate is the sole nitrogen source, which is similar to FNR1 in F. oxysporum and AREA orthologues in other fungi (Divon et al., 2006; Feng and Marzluf, 1998). The ΔareA mutant is defective in the utilization of nitrate or glutamate as the sole nitrogen source and shows limited growth, which is similar to the areA mutant of A. oryzae (Christensen et al., 1998).
Because of its importance in secondary metabolism, we had started to functionally characterize the AreA orthologue before the publication on AreA in a different isolate of F. graminearum (Min et al., 2012). Similar to the earlier report (Min et al., 2012), we found that the ΔareA mutant was defective in growth and conidiation. Although at a significantly reduced level, the ΔareA mutant still produced DON in rice grain cultures or DON‐inducing liquid cultures when hyphae were used as the inoculum. It also showed reduced suppression of DON production and reduced expression of TRI5 and TRI6 by ammonium. These results indicate that AREA is important, but not essential, for DON biosynthesis and the suppression of DON biosynthesis by ammonium. As a result of its defects in hyphal growth, the ΔareA mutant was defective in DON production in DON‐inducing liquid medium inoculated with conidia, which is similar to the results reported by Min et al. (2012).
Although AreA orthologues are known to be important for AS of secondary metabolism, such as fumonisin biosynthesis in F. verticillioides (Kim and Woloshuk, 2008) and gibberellin synthesis in Gibberella fujikuroi (Mihlan et al., 2003; Teichert et al., 2004), the underlying mechanisms are not well characterized. In this study, we showed that AreA interacted with Tri10 in co‐immunoprecipitation assays. Tri10 and Tri6 are two transcription factors in the TRI gene cluster important for the regulation of TRI gene expression and DON biosynthesis (Seong et al., 2009). It is possible that the interaction of Tri10 with AreA is important for the induction of TRI gene expression. The suppression of AREA expression by ammonium may be directly responsible for its inhibitory effects on DON production.
In G. fujikuroi, the AreA orthologue appears to mediate gibberellin (GA) production by binding to the promoters of the GA biosynthesis genes (Mihlan et al., 2003; Teichert et al., 2004; Tudzynski et al., 1999). The promoter sequences of TRI6, TRI10 and other TRI genes all have multiple 5′‐HGATAR‐3′ sequences that may function as putative AreA‐binding sites (Ravagnani et al., 1997; Scazzocchio, 2000). It is possible that AreA may directly regulate the expression of some of these TRI genes. However, the HGATAR sequence occurs randomly at least once in every 256 bp (both directions). The promoter of TRI6 alone has six putative AreA‐binding sites. Therefore, it is difficult to predict and functionally identify which is the actual AreA‐binding site in the TRI genes of F. graminearum. In the future, it will be important to conduct chromatin immunoprecipitation sequencing (ChIP‐seq) analysis to identify promoter sequences of TRI genes binding to AreA in F. graminearum.
In F. graminearum, the expression of AREA was repressed by ammonium and up‐regulated when 50 mm nitrate was the sole nitrogen source. In the AREA‐GFP transformant, AreA‐GFP constitutively localized to the nucleus, although GFP signals in the nucleus were enhanced when nitrate was the sole nitrogen source, which is consistent with its role in the positive regulation of nitrate assimilation genes. In A. nidulans, AreA is located primarily in the cytoplasm when ammonium is present, but is accumulated in the nucleus in response to NS (Todd et al., 2005). In S. cerevisiae, Gln3 localizes to the nucleus to positively regulate the expression of nitrogen catabolite repression (NCR)‐sensitive genes (Beck and Hall, 1999). Therefore, the subcellular localization of AreA‐GFP is slightly different from its orthologues in A. nidulans and S. cerevisiae.
NLS1 and NLS2 are two well‐conserved NLS sequences in AreA and its orthologues. However, deletion of either NLS1 or NLS2 of AreA had no obvious effect on its localization and function in F. graminearum. In A. nidulans, mutations at these two NLS sites also had no effect on the subcellular localization of AreA (Hunter et al., 2014). However, AreA has a non‐canonical arginine‐based bipartite NLS sequence that is important for its nuclear localization. This NLS sequence is conserved in most fungi, including F. graminearum. To avoid interfering with the nearby zinc finger DNA‐binding domain, only part of this NLS sequence was deleted in the AREA ΔNLS3 allele. Our data showed that AreAΔNLS3‐GFP proteins were detected by Western blot analysis, but no GFP signals were observed in the nucleus. Therefore, NLS3 must be the NLS sequence that is responsible for the subcellular localization of AreA in F. graminearum. In A. nidulans, mutations in the four R residues in this NLS also significantly reduced the localization of AreA to the nucleus (Hunter et al., 2014). In F. graminearum, no detectable GFP signals were observed in the AREA ΔNLS3‐GFP transformant, even under NS conditions.
The AreA protein has a putative PKA phosphorylation site that is well conserved among its orthologues from filamentous ascomycetes. Site‐directed mutagenesis showed that this PKA phosphorylation site was important for the increased localization of AreA‐GFP to the nucleus and full virulence, suggesting the importance of this consensus PKA site. In S. cerevisiae, phosphorylation of Gln3 is dependent on nitrogen sources in the medium. In the presence of primary nitrogen sources, Gln3 is phosphorylated by the Tor1 and Tor2 TOR kinases (Beck and Hall, 1999; Bertram et al., 2000). The Snf1 and Npr1 kinases are also probably involved in Gln3 regulation (Bertram et al., 2002; Cox et al., 2002; Crespo et al., 2004). In A. nidulans, the regulation of AreA phosphorylation is different from that in S. cerevisiae (Todd et al., 2005), and it is not known to involve PKA. Direct phosphorylation of AreA orthologues by PKA has also not been reported in other fungi. Therefore, further studies are necessary to determine whether S874 is phosphorylated by Cpk1 or Cpk2, although it is a conserved, consensus PKA phosphorylation site based on bioinformatics analysis. Cpk1 and Cpk2 are two catalytic subunits of PKA that are essential for DON production and plant infection in F. graminearum (Hu et al., 2014).
In S. cerevisiae, the Gln3 and Nil1 GATA factors mediate NCR. They control the expression of three ammonium permease genes, named MEP1, MEP2 and MEP3 (Marini et al., 1997). In A. nidulans, the expression of all three AMT/MEP is also regulated by AreA under different nitrogen conditions. The AreA/Nit‐2 global nitrogen regulator binds specifically to GATA sequences in the promoters of its target genes to regulate their expression. In this study, we found that all three predicted F. graminearum MEP genes have several GATA sequences in the promoter regions, and their expression was significantly reduced in the ΔareA mutant, particularly MEP1, under low‐concentration ammonium conditions. Interestingly, the ΔareA mutant showed reduced PKA activity and Gpmk1 phosphorylation. Both the cAMP‐PKA and Gpmk1 MAPK pathways are known to be important for plant infection, sexual reproduction and DON production in F. graminearum (Hu et al., 2014; Jenczmionka et al., 2003), which may be related to some of the phenotypes of the ΔareA mutant. RAS proteins are known to function upstream from the cAMP signalling and MAPK pathways in S. cerevisiae, M. oryzae and other fungi (Li et al., 2012; Zhao and Xu, 2007). In C. albicans, the C‐terminal region of Mep2 has been implicated in association with Ras proteins. In F. graminearum, deletion of AREA significantly reduced the expression of all three MEP genes, which may, in turn, affect RAS signalling and the downstream cAMP‐PKA and MAPK pathways. Therefore, it will be important to determine the relationship between MEPs and RAS signalling in F. graminearum in the future.
Experimental Procedures
Strains and culture conditions
The wild‐type strain PH‐1 (Cuomo et al., 2007) and mutants of F. graminearum generated in this study were routinely cultured at 25 °C and preserved in 15% glycerol at −80 °C. The split‐marker approach was used to generate the gene replacement constructs for the AREA gene (Fig. S1). Putative knockout mutants were identified by PCR and further confirmed by Southern blot hybridizations. Cultures grown on CM, potato dextrose agar (PDA) and minimal medium (MM) were used for measurement of the growth rate or colony morphology (Hou et al., 2002). Conidiation was assayed in 5‐day‐old liquid CarboxyMethylCellulose (CMC) cultures as described previously (Ding et al., 2009; Zhou et al., 2010). Protoplasts were prepared and used for transformation as described previously (Hou et al., 2002). Hygromycin B (CalBiochem, La Jolla, CA, USA) and geneticin (Sigma‐Aldrich, St. Louis, MO, USA) were added to final concentrations of 300 and 400 μg/mL, respectively, to the regeneration medium for transformant selection.
Generation of the AreA‐GFP fusion construct and transformant
For complementation assays, a 4.3‐kb fragment containing the entire AREA gene and 1.5‐kb promoter region was amplified and cloned into pFL2 (Zhou et al., 2011a) by the yeast in vivo homologous recombination approach as described previously (Bruno et al., 2004). The resulting AreA‐GFP construct was transformed into the areA deletion mutant RH12. Transformants expressing the AreA‐GFP construct were verified by PCR. GFP signals in conidia and germlings were observed with an Olympus BX51 epifluorescence microscope (Olympus Corporation, Tokyo, Japan).
Generation of the AREAΔNLS1‐, AREAΔNLS2‐, AREAΔNLS3‐, AREAΔ657–658‐, AREAΔ874‐ and AREAS874A‐GFP fusion constructs and transformants
To generate the AREA ΔNLS1‐GFP fusion construct by the yeast gap repair approach (Bruno et al., 2004), PCR products amplified with the primer pairs AREA‐COMP‐F/NLS1‐R and NLS1‐F/AREA‐COMP‐R were co‐transformed into the yeast strain XK1‐25 with XhoI‐digested plasmid pFL2 (Zhou et al., 2011a). Plasmids rescued from the Trp+ yeast transformants were then sequenced to confirm the AREA ΔNLS1‐GFP construct. Similar strategies were used to generate the AREA ΔNLS2‐, AREA ΔNLS3‐, AREA Δ657–658‐, AREA Δ874‐ and AREA S874A‐GFP fusion constructs by yeast gap repair (Bruno et al., 2004) with the primers listed in Table S1 (see Supporting Information). All these mutant alleles of AREA were confirmed by sequencing analysis and transformed into the ΔareA deletion mutant RH12. The resulting transformants were screened by PCR to confirm the presence of transforming constructs and examined for GFP signals by epifluorescence microscopy.
Sexual reproduction assays
To assay defects in sexual reproduction with the areA mutant and complementation strains, aerial hyphae of 7‐day‐old carrot agar cultures were pressed down with sterile 0.1% Tween 20 and incubated at 25 °C as described previously (Li et al., 2011; Luo et al., 2014). Perithecium formation, cirrhi production and the formation of asci and ascospores were examined 2–3 weeks after induction (Zheng et al., 2013).
Plant infection and DON production assays
Conidia freshly harvested from CMC cultures were re‐suspended to 105 spores/mL in sterile water and used for plant infection assays. Flowering wheat heads of cultivars Norm and Xiaoyan22 were inoculated with 10 μL of conidium suspensions at the fifth spikelet from the base of the inflorescence (Gale et al., 2007). Spikelets with typical head blight symptoms on each inoculated head were counted at 14 days post‐inoculation (dpi). Results from three biological replicates with over 10 inoculated wheat heads each were used to estimate the disease index (Chen et al., 2014; Hou et al., 2002). Stalks of 8‐week‐old corn plants of cv. Pioneer 2375 were inoculated with tooth‐picks dipped in conidium suspensions and examined for stalk rot symptoms as described previously (Zhou et al., 2010). For rice grain cultures (Seo et al., 1996), DON and ergosterol production were assayed as described previously (Bluhm et al., 2007; Seong et al., 2009). For each strain, DON production was also assayed with liquid‐inducing cultures containing 5 mm NaNO3 or arginine (Gardiner et al., 2009a).
qRT‐PCR analysis
RNA was isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) from vegetative hyphae harvested from 24 h YEPD cultures. For qRT‐PCR analysis, first‐strand cDNA was synthesized with the Fermentas 1st cDNA synthesis kit (Hanover, MD, USA) following the instructions provided by the manufacturer. The β‐tubulin gene FGSG_06611 of F. graminearum was used as the internal control (Bluhm et al., 2007). Relative expression levels of each gene were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001). Data from three biological replicates were used to calculate the mean and standard deviation.
Yeast two‐hybrid assays
The Matchmaker yeast two‐hybrid system (Clontech, Mountain View, CA, USA) was used to assay the interactions of AreA with Tir6 and Tri10. The open reading frame (ORF) of the AREA gene was amplified from first‐strand cDNA of PH‐1 and cloned into pGBK7 (Clontech) as the bait construct. For the TRI6 and TRI10 genes, their ORFs were cloned into pGADT7 as the prey constructs. The resulting bait and prey vectors were transformed into the yeast strain AH109 (Clontech). Growth on synthetic dropout medium lacking tryptophan, leucine, and histidine (SD‐Trp‐Leu‐His) medium and galactosidase activity were assayed as described previously (Zhou et al., 2011b). The positive and negative controls were provided in the Matchmaker Library Construction and Screening Kits (Clontech).
Co‐immunoprecipitation assays
The TRI6 and TRI10 genes were amplified and cloned into pDL2 by the yeast gap repair approach (Bruno et al., 2004) to generate the GFP fusion constructs. Similar approaches were used to generate the 3 × FLAG fusion constructs for the AREA gene with the pFL7 vector (Zhou et al., 2011a). The resulting fusion constructs were verified by sequencing analysis and transformed in pairs into protoplasts of PH‐1. Transformants expressing both GFP and FLAG fusion constructs were confirmed by Western blot analysis. For co‐immunoprecipitation assays, total proteins were isolated (Bruno et al., 2004) and incubated with the anti‐FLAG M2 beads as described previously (Liu et al., 2011). Western blots of proteins eluted from anti‐FLAG beads were detected with monoclonal anti‐GFP and anti‐FLAG antibodies (Roche, Indianapolis, IN, USA).
Assays for Threonine‐Glutamine‐Tyrosine (TEY) phosphorylation and PKA activities
Total proteins were isolated from hyphae harvested from 48‐h YEPD cultures and separated on 12.5% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels (Bruno et al., 2004; Ding et al., 2010). After transfer onto nitrocellulose membranes, the phosphorylation of Mgv1 and Gpmk1 was detected with the PhophoPlus p44/42 antibody kit (Cell Signaling Technology, Danvers, MA, USA) as described previously (Liu et al., 2011). The expression level of Gpmk1 was detected with the anti‐Pmk1 antibody generated in a previous study (Bruno et al., 2004). PKA activities were assayed with the PepTag nonradioactive PKA assay kit (Promega, Madison, WI, USA) as described previously (Adachi and Hamer, 1998; Nishimura et al., 2003).
Supporting information
Fig. S1 The AREA gene replacement construct and mutant. (A) Generation of the AREA gene replacement construct. 1F, 2R, 3F and 4R are primers used to amplify the flanking sequences. hph, hygromycin phosphotransferase gene. (B) Southern blots of the wild‐type strain PH‐1 and ΔareA transformants (M1, M2 and M3) were hybridized with probe 1 amplified with primers AREA/5F and AREA/6R and probe 2 amplified with H852 and H850. (C) Three‐day‐old complete medium (CM) and minimal medium (MM) cultures of PH‐1, ΔareA mutant RH12 and complementation strain (AC6). (D) Conidia of the same set of strains incubated in yeast extract peptone dextrose (YEPD) for 6 h. Bar, 20 μm.
Fig. S2 Defects of the ΔareA mutant in plant infection and self‐crosses. (A) Flowering wheat heads were drop inoculated with conidia from the wild‐type strain PH‐1, ΔareA mutant and complementation strain (AC6). Black dots mark the inoculated spikelets. (B) Corn stalks were inoculated with the same set of strains. Photographs were taken at 14 days post‐inoculation (dpi). (C) Perithecia and cirrhi produced by PH‐1, ΔareA mutant and AC6. Bar, 1 mm. (D) Two‐week‐old perithecia were cracked open and examined for asci and ascospores with the same set of strains. Bar, 50 μm.
Fig. S3 Functional characterization of two putative nuclear localization signal (NLS) sequences. Germlings of the ΔareA/AREA ΔNLS1‐GFP transformants (RL10 and RL11) and ΔareA/AREA ΔNLS2‐GFP transformants (RL12 and RL13) were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) and examined by differential interference contrast (DIC) and epifluorescence microscopy. GFP, green fluorescent protein. Bar, 10 μm.
Fig. S4 Putative AreA‐binding sites in promoter region (within 1 kb) of the three MEP genes. White triangle, H (A, C or T)/GATA/R (A or G); black triangle, Y (T, G or A)/TATC/D (T or C).
Table S1 Polymerase chain reaction (PCR) primers used in this study.
Table S2 Deoxynivalenol (DON) production in PH‐1 and its transformants expressing different mutant alleles of AREA.
Acknowledgements
We thank Dr Huiquan Liu for fruitful discussions and assistance with phylogenetic analysis. We also thank Mr Shijie Zhang for assistance with microscopic examinations. This work was supported by the National Major Project of Breeding for New Transgenic Organisms (2012ZX08009003), the Nature Science Foundation of China (No. 31271989) and the Program for New Century Excellent Talents in University. The authors have no conflicts of interest to declare.
References
- Adachi, K. and Hamer, J.E. (1998) Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea . Plant Cell, 10, 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, N.J. , Proctor, R.H. and McCormick, S.P. (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 28, 198–215. [Google Scholar]
- Audenaert, K. , Callewaert, E. , Hofte, M. , De Saeger, S. and Haesaert, G. (2010) Hydrogen peroxide induced by the fungicide prothioconazole triggers deoxynivalenol (DON) production by Fusarium graminearum . BMC Microbiol. 10, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, G.H. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. [DOI] [PubMed] [Google Scholar]
- Beck, T. and Hall, M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient‐regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Bertram, P.G. , Choi, J.H. , Carvalho, J. , Ai, W.D. , Zeng, C.B. , Chan, T.F. and Zheng, X.F. (2000) Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275, 35 727–35 733. [DOI] [PubMed] [Google Scholar]
- Bertram, P.G. , Choi, J.H. , Carvalho, J. , Chan, T.F. , Ai, W. and Zheng, X.F. (2002) Convergence of TOR‐nitrogen and Snf1‐glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22, 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, K. and Morschhauser, J. (2005) The Mep2p ammonium permease controls nitrogen starvation‐induced filamentous growth in Candida albicans . Mol. Microbiol. 56, 649–669. [DOI] [PubMed] [Google Scholar]
- Bluhm, B.H. , Zhao, X. , Flaherty, J.E. , Xu, J.R. and Dunkle, L.D. (2007) RAS2 regulates growth and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 20, 627–636. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. and Thomma, B.P.H.J. (2008) The complexity of nitrogen metabolism and nitrogen‐regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72, 104–110. [Google Scholar]
- Brown, D.W. , Dyer, R.B. , McCormick, S.P. , Kendra, D.F. and Plattner, R.D. (2004) Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 41, 454–462. [DOI] [PubMed] [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J.R. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryotic Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick, M.X. , Arst, H.N. Jr , Taylor, L.H. , Johnson, R.I. and Brownlee, A.G. (1986) Cloning of the regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans . EMBO J. 5, 1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Wang, Y. , Zhou, X. , Wang, Y. and Xu, J.R. (2014) The Sch9 kinase regulates conidium size, stress responses, and pathogenesis in Fusarium graminearum . PLoS ONE, 9, e105811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, T. , Hynes, M.J. and Davis, M.A. (1998) Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl. Environ. Microbiol. 64, 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman, J.A. , Rai, R. , Cunningham, T. , Svetlov, V. and Cooper, T.G. (1996) Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen‐catabolic genes in Saccharomyces cerevisiae . Mol. Cell. Biol. 16, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, K.H. , Tate, J.J. and Cooper, T.G. (2002) Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae . J. Biol. Chem. 277, 37 559–37 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, J.L. , Helliwell, S.B. , Wiederkehr, C. , Demougin, P. , Fowler, B. , Primig, M. and Hall, M.N. (2004) NPR1 kinase and RSP5‐BUL1/2 ubiquitin ligase control GLN3‐dependent transcription in Saccharomyces cerevisiae . J. Biol. Chem. 279, 37 512–37 517. [DOI] [PubMed] [Google Scholar]
- Cuomo, C.A. , Guldener, U. , Xu, J.R. , Trail, F. , Turgeon, B.G. , Di Pietro, A. , Walton, J.D. , Ma, L.J. , Baker, S.E. , Rep, M. , Adam, G. , Antoniw, J. , Baldwin, T. , Calvo, S. , Chang, Y.L. , Decaprio, D. , Gale, L.R. , Gnerre, S. , Goswami, R.S. , Hammond‐Kosack, K. , Harris, L.J. , Hilburn, K. , Kennell, J.C. , Kroken, S. , Magnuson, J.K. , Mannhaupt, G. , Mauceli, E. , Mewes, H.W. , Mitterbauer, R. , Muehlbauer, G. , Münsterkötter, M. , Nelson, D. , O'donnell, K. , Ouellet, T. , Qi, W. , Quesneville, H. , Roncero, M.I. , Seong, K.Y. , Tetko, I.V. , Urban, M. , Waalwijk, C. , Ward, T.J. , Yao, J. , Birren, B.W. , Kistler, H.C. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317, 1400–1402. [DOI] [PubMed] [Google Scholar]
- Desjardins, A.E. , Bai, G.H. , Plattner, R.D. and Proctor, R.H. (2000) Analysis of aberrant virulence of Gibberella zeae following transformation‐mediated complementation of a trichothecene‐deficient (Tri5) mutant. Microbiology, 146, 2059–2068. [DOI] [PubMed] [Google Scholar]
- Ding, S. , Mehrabi, R. , Koten, C. , Kang, Z. , Wei, Y. , Seong, K. , Kistler, H.C. and Xu, J.R. (2009) Transducin beta‐like gene FTL1 is essential for pathogenesis in Fusarium graminearum . Eukaryotic Cell, 8, 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.L. , Liu, W. , Iliuk, A. , Ribot, C. , Vallet, J. , Tao, A. , Wang, Y. , Lebrun, M.H. , Xu, J.R. (2010) The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae . Plant Cell, 22, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divon, H.H. , Ziv, C. , Davydov, O. , Yarden, O. and Fluhr, R. (2006) The global nitrogen regulator, FNR1, regulates fungal nutrition‐genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 7, 485–497. [DOI] [PubMed] [Google Scholar]
- Feng, B. and Marzluf, G.A. (1998) Interaction between major nitrogen regulatory protein NIT2 and pathway‐specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa . Mol. Cell. Biol. 18, 3983–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.H. and Marzluf, G.A. (1990) nit‐2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA‐binding domain. Mol. Cell. Biol. 10, 1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, L.R. , Bryant, J.D. , Calvo, S. , Giese, H. , Katan, T. , O'Donnell, K. , Suga, H. , Taga, M. , Usgaard, T.R. , Ward, T.J. and Kistler, H.C. (2005) Chromosome complement of the fungal plant pathogen Fusarium graminearum based on genetic and physical mapping and cytological observations. Genetics, 171, 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, L.R. , Ward, T.J. , Balmas, V. and Kistler, H.C. (2007) Population subdivision of Fusarium graminearum sensu stricto in the upper midwestern United States. Phytopathology, 97, 1434–1439. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009a) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 46, 604–613. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Osborne, S. , Kazan, K. and Manners, J.M. (2009b) Low pH regulates the production of deoxynivalenol by Fusarium graminearum . Microbiology, 155, 3149–3156. [DOI] [PubMed] [Google Scholar]
- Giese, H. , Sondergaard, T.E. and Sorensen, J.L. (2013) The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 117, 814–821. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Hou, Z. , Xue, C. , Peng, Y. , Katan, T. , Kistler, H.C. and Xu, J.R. (2002) A mitogen‐activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant–Microbe Interact. 15, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Zhou, X. , Gu, X. , Cao, S. , Wang, C. and Xu, J.R. (2014) The cAMP‐PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 27, 557–566. [DOI] [PubMed] [Google Scholar]
- Hunter, C.C. , Siebert, K.S. , Downes, D.J. , Wong, K.H. , Kreutzberger, S.D. , Fraser, J.A. , Clarke, D.F. , Hynes, M.J. , Davis, M.A. and Todd, R.B. (2014) Multiple nuclear localization signals mediate nuclear localization of the GATA transcription factor AreA. Eukaryotic Cell, 13, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenczmionka, N.J. , Maier, F.J. , Losch, A.P. and Schafer, W. (2003) Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head‐blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43, 87–95. [DOI] [PubMed] [Google Scholar]
- Kim, H. and Woloshuk, C.P. (2008) Role of AREA, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in Fusarium verticillioides . Fungal Genet. Biol. 45, 947–953. [DOI] [PubMed] [Google Scholar]
- Kimura, M. , Tokai, T. , O'Donnell, K. , Ward, T.J. , Fujimura, M. , Hamamoto, H. , Shibata, T. and Yamaguchi, I. (2003) The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non‐essential genes. FEBS Lett. 539, 105–110. [DOI] [PubMed] [Google Scholar]
- Li, G. , Zhou, X. and Xu, J.R. (2012) Genetic control of infection‐related development in Magnaporthe oryzae . Curr. Opin. Microbiol. 15, 678–684. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, C. , Liu, W. , Wang, G. , Kang, Z. , Kistler, H.C. and Xu, J.R. (2011) The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 487–496. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Zhou, X. , Li, G. , Li, L. , Kong, L. , Wang, C. , Zhang, H. and Xu, J.R. (2011) Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7, e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C. and Heitman, J. (1998) The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae . EMBO J. 17, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Zhang, H. , Qi, L. , Zhang, S. , Zhou, X. , Zhang, Y. and Xu, J.R. (2014) FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta‐tubulins in Fusarium graminearum . New Phytol. 204, 943–954. [DOI] [PubMed] [Google Scholar]
- Magasanik, B. and Kaiser, C.A. (2002) Nitrogen regulation in Saccharomyces cerevisiae . Gene, 290, 1–18. [DOI] [PubMed] [Google Scholar]
- Maier, F.J. , Miedaner, T. , Hadeler, B. , Felk, A. , Salomon, S. , Lemmens, M. , Kassner, H. and Schäfer, W. (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. [DOI] [PubMed] [Google Scholar]
- Marini, A.M. , Soussi‐Boudekou, S. , Vissers, S. and Andre, B. (1997) A family of ammonium transporters in Saccharomyces cerevisiae . Mol. Cell. Biol. 17, 4282–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej, J. , Richard‐Forget, F. and Barreau, C. (2011) The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum . Fungal Genet. Biol. 48, 275–284. [DOI] [PubMed] [Google Scholar]
- Mihlan, M. , Homann, V. , Liu, T.W. and Tudzynski, B. (2003) AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol. Microbiol. 47, 975–991. [DOI] [PubMed] [Google Scholar]
- Min, K. , Shin, Y. , Son, H. , Lee, J. , Kim, J.C. , Choi, G.J. and Lee, Y.W. (2012) Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae . FEMS Microbiol. Lett. 334, 66–73. [DOI] [PubMed] [Google Scholar]
- Minehart, P.L. and Magasanik, B. (1991) Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA‐binding domain. Mol. Cell. Biol. 11, 6216–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan, B.J. , Askin, M.C. , Hynes, M.J. and Davis, M.A. (2006) Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryotic Cell, 5, 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montibus, M. , Ducos, C. , Bonnin‐Verdal, M.N. , Bormann, J. , Ponts, N. , Richard‐Forget, F. and Barreau, C. (2013) The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum . PLoS ONE, 8, e83377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmith, C.G. , Walkowiak, S. , Wang, L. , Leung, W.W.Y. , Gong, Y.C. , Johnston, A. , Harris, L.J. , Guttman, D.S. and Subramaniam, R. (2011) Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum . PLoS Pathog. 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M. , Park, G. and Xu, J.R. (2003) The G‐beta subunit MGB1 is involved in regulating multiple steps of infection‐related morphogenesis in Magnaporthe grisea . Mol. Microbiol. 50, 231–243. [DOI] [PubMed] [Google Scholar]
- Ochiai, N. , Tokai, T. , Takahashi‐Ando, N. , Fujimura, M. and Kimura, M. (2007) Genetically engineered Fusarium as a tool to evaluate the effects of environmental factors on initiation of trichothecene biosynthesis. FEMS Microbiol. Lett. 275, 53–61. [DOI] [PubMed] [Google Scholar]
- Pestka, J.J. , el‐Bahrawy, A. and Hart, L.P. (1985) Deoxynivalenol and 15‐monoacetyl deoxynivalenol production by Fusarium graminearum R6576 in liquid media. Mycopathologia, 91, 23–28. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Hohn, T.M. and McCormick, S.P. (1997) ) Restoration of wild‐type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology, 143, 2583–2591. [DOI] [PubMed] [Google Scholar]
- Ravagnani, A. , Gorfinkiel, L. , Langdon, T. , Diallinas, G. , Adjadj, E. , Demais, S. , Gorton, D. , Arst, H.N. Jr and Scazzocchio, C. (1997) Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter‐specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16, 3974–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio, C. (2000) The fungal GATA factors. Curr. Opin. Microbiol. 3, 126–131. [DOI] [PubMed] [Google Scholar]
- Seo, J.A. , Kim, J.C. , Lee, D.H. and Lee, Y.W. (1996) Variation in 8‐ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia, 134, 31–37. [DOI] [PubMed] [Google Scholar]
- Seong, K.Y. , Pasquali, M. , Zhou, X. , Song, J. , Hilburn, K. , McCormick, S. , Dong, Y. , Xu, J.R. and Kistler, H.C. (2009) Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. [DOI] [PubMed] [Google Scholar]
- Teichert, S. , Schonig, B. , Richter, S. and Tudzynski, B. (2004) Deletion of the Gibberella fujikuroi glutamine synthetase gene has significant impact on transcriptional control of primary and secondary metabolism. Mol. Microbiol. 53, 1661–1675. [DOI] [PubMed] [Google Scholar]
- Teichert, S. , Rutherford, J.C. , Wottawa, M. , Heitman, J. and Tudzynski, B. (2008) Impact of ammonium permeases mepA, mepB, and mepC on nitrogen‐regulated secondary metabolism in Fusarium fujikuroi . Eukaryotic Cell, 7, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, R.B. , Fraser, J.A. , Wong, K.H. , Davis, M.A. and Hynes, M.J. (2005) Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryotic Cell, 4, 1646–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski, B. (2014) Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 5, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski, B. , Homann, V. , Feng, B. and Marzluf, G.A. (1999) Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi . Mol. Gen. Genet. 261, 106–114. [DOI] [PubMed] [Google Scholar]
- Zhao, X. and Xu, J.R. (2007) A highly conserved MAPK‐docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea . Mol. Microbiol. 63, 881–894. [DOI] [PubMed] [Google Scholar]
- Zheng, Q. , Hou, R. , Zhang, J., Ma, J. , Wu, Z. , Wang, G. , Wang, C. and Xu, J.R. (2013) The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum . PLoS ONE, 8, e66980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Li, G. and Xu, J.R. (2011a) Efficient approaches for generating GFP fusion and epitope‐tagging constructs in filamentous fungi. Methods Mol. Biol. 722, 199–212. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Liu, W. , Wang, C. , Xu, Q. , Wang, Y. , Ding, S. and Xu, J.R. (2011b) A MADS‐box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae . Mol. Microbiol. 80, 33–53. [DOI] [PubMed] [Google Scholar]
- Zhou, X.Y. , Heyer, C. , Choi, Y.E. , Mehrabi, R. and Xu, J.R. (2010) The CID1 cyclin C‐like gene is important for plant infection in Fusarium graminearum . Fungal Genet. Biol. 47, 143–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The AREA gene replacement construct and mutant. (A) Generation of the AREA gene replacement construct. 1F, 2R, 3F and 4R are primers used to amplify the flanking sequences. hph, hygromycin phosphotransferase gene. (B) Southern blots of the wild‐type strain PH‐1 and ΔareA transformants (M1, M2 and M3) were hybridized with probe 1 amplified with primers AREA/5F and AREA/6R and probe 2 amplified with H852 and H850. (C) Three‐day‐old complete medium (CM) and minimal medium (MM) cultures of PH‐1, ΔareA mutant RH12 and complementation strain (AC6). (D) Conidia of the same set of strains incubated in yeast extract peptone dextrose (YEPD) for 6 h. Bar, 20 μm.
Fig. S2 Defects of the ΔareA mutant in plant infection and self‐crosses. (A) Flowering wheat heads were drop inoculated with conidia from the wild‐type strain PH‐1, ΔareA mutant and complementation strain (AC6). Black dots mark the inoculated spikelets. (B) Corn stalks were inoculated with the same set of strains. Photographs were taken at 14 days post‐inoculation (dpi). (C) Perithecia and cirrhi produced by PH‐1, ΔareA mutant and AC6. Bar, 1 mm. (D) Two‐week‐old perithecia were cracked open and examined for asci and ascospores with the same set of strains. Bar, 50 μm.
Fig. S3 Functional characterization of two putative nuclear localization signal (NLS) sequences. Germlings of the ΔareA/AREA ΔNLS1‐GFP transformants (RL10 and RL11) and ΔareA/AREA ΔNLS2‐GFP transformants (RL12 and RL13) were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) and examined by differential interference contrast (DIC) and epifluorescence microscopy. GFP, green fluorescent protein. Bar, 10 μm.
Fig. S4 Putative AreA‐binding sites in promoter region (within 1 kb) of the three MEP genes. White triangle, H (A, C or T)/GATA/R (A or G); black triangle, Y (T, G or A)/TATC/D (T or C).
Table S1 Polymerase chain reaction (PCR) primers used in this study.
Table S2 Deoxynivalenol (DON) production in PH‐1 and its transformants expressing different mutant alleles of AREA.


