Summary
Many plant‐ and animal‐pathogenic Gram‐negative bacteria employ the type III secretion system (T3SS) to translocate effector proteins from bacterial cells into the cytosol of eukaryotic host cells. The effector translocation occurs through an integral component of T3SS, the channel‐like translocon, assembled by hydrophilic and hydrophobic proteinaceous translocators in a two‐step process. In the first, hydrophilic translocators localize to the tip of a proteinaceous needle in animal pathogens, or a proteinaceous pilus in plant pathogens, and associate with hydrophobic translocators, which insert into host plasma membranes in the second step. However, the pilus needs to penetrate plant cell walls in advance. All hydrophilic translocators so far identified in plant pathogens are characteristic of harpins: T3SS accessory proteins containing a unitary hydrophilic domain or an additional enzymatic domain. Two‐domain harpins carrying a pectate lyase domain potentially target plant cell walls and facilitate the penetration of the pectin‐rich middle lamella by the bacterial pilus. One‐domain harpins target plant plasma membranes and may play a crucial role in translocon assembly, which may also involve contrapuntal associations of hydrophobic translocators. In all cases, sensory components in the target plasma membrane are indispensable for the membrane recognition of translocators and the functionality of the translocon. The conjectural sensors point to membrane lipids and proteins, and a phosphatidic acid and an aquaporin are able to interact with selected harpin‐type translocators. Interactions between translocators and their sensors at the target plasma membrane are assumed to be critical for translocon assembly.
Keywords: harpins, plasma membrane, sensor, translocators, type III translocon
Introduction
A variety of Gram‐negative bacteria which are pathogens of plants (Alfano and Collmer, 2004; White et al., 2009) or animals (Chatterjee et al., 2013; Mueller et al., 2008) utilize the type III secretion system (T3SS) to transport proteins into the surrounding milieu or directly into the cytosol of eukaryotic host cells. The T3SS deploys a proteinaceous needle complex in animal pathogens (Kubori et al., 1998; Deane et al., 2006) and a proteinaceous pilus complex in plant pathogens (Jin and He, 2001) as an essential pathway for protein secretion and translocation. In the needle or pilus complex, an integral apparatus that governs protein translocation is called the type III translocon, which consists of T3SS accessory (structural) proteins, namely type III translocators, in association with sensory components potentially present in the target plasma membrane (PM) of host cells (Büttner, 2012; Matteï et al., 2011; Mueller et al., 2008). Translocated bacterial proteins serve as either virulence effectors to cause diseases or defence activators to confer disease resistance in eukaryotic hosts (Galán and Collmer, 1999). In particular, virulence effectors employ different mechanisms, avoiding or defeating the innate immunity and manipulating the transcriptomes of plants (Alfano and Collmer, 2004; Bogdanove and Voytas, 2011; Lindeberg et al., 2012; Scholze and Boch, 2010; White et al., 2009; White and Yang, 2009) and animals (Bliska et al., 2013; Plano and Schesser, 2013), respectively, to secure the bacterial ability to multiply and cause diseases in hosts (Galán and Collmer, 1999).
In the last 10 years, significant progress has been acquired in demonstrating T3SS components and architectures in animal (Büttner, 2012; Chatterjee et al., 2013; Matteï et al., 2011; Mueller et al., 2008) and plant (Alfano and Collmer, 2004; Choi et al., 2013; Kay and Bonas, 2009) pathogens. The essential compositions of the type III translocon and the characteristics of the translocators have been elucidated extensively for animal pathogens and have also attracted increasing attention from the plant bacteriology community. The accumulated knowledge has led to new research goals directed towards the determination of the accurate assembly of the translocon and the characterization of recognition sensors of the translocators putatively present in the target PM. This review briefly discusses T3SS organization and then focuses on the biochemical and pathological characteristics of the translocators from the pathogens of plants compared with animals, models of translocator associations to form the translocon, potential sensors of the translocators in the target PM, especially from plants, and possible interactions between the translocators and their sensors to facilitate effector translocation.
General Architecture of T3SS
In all bacteria studied to date, the T3SS is exquisitely yet similarly assembled as a nanomachine by refined associations of more than 20 structural proteins that are either diverse or conserved among different bacterial species (Abrusci et al., 2014; Bogdanove et al., 1996; Chatterjee et al., 2013; Matteï et al., 2011; Plano and Schesser, 2013). In every bacterial species, different structural proteins are produced hierarchically to build up the T3SS machinery as two associated parts: a membrane‐traversing basal body plus an extracellular outgrowth: a needle complex in animal pathogens and a pilus complex in plant pathogens (Fig. 1). The basal body spans bacterial inner (IMs) and outer (OMs) membranes, comprises ring structures that are presumably connected by a periplasmic rod (P‐rod) across the peptidoglycan mesh and associates with the needle or pilus complex. The needle or pilus elongates outwards from the bacterial surface and reaches the surface of host cells when the bacterium contacts with host cells (Abrusci et al., 2014; Matteï et al., 2011; Mueller et al., 2008).
Figure 1.
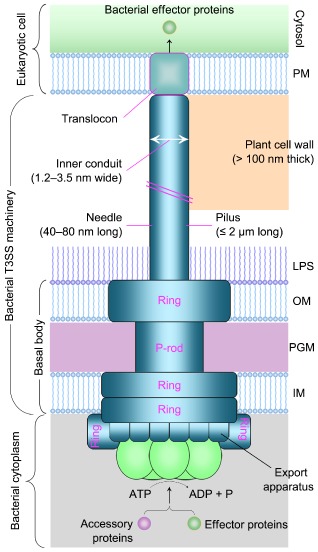
Schematic diagram illustrating type III secretion system (T3SS) architectures of animal‐ and plant‐pathogenic Gram‐negative bacteria. The basal body of the T3SS spans the bacterial inner membrane (IM) and outer membrane (OM), and comprises ring structures that are presumably connected by a periplasmic rod (P‐rod) across the peptidoglycan mesh (PGM) (Büttner 2012). The basal body associates with an extracellular needle (animal pathogens) or pilus (plant pathogens) appendage, which elongates across an OM peripheral layer containing lipopolysaccharide (LPS) and forms a channel‐like translocon that inserts into the host plasma membrane (PM). The energy for the docking and unfolding of T3SS substrates, including effector proteins and T3SS‐accessory extracellular proteins, is probably provided by a cytoplasmic ATPase associated with the T3SS (Büttner 2012). The width and length of the bacterial needle or pilus, as well as the thickness of the plant cell wall, are indicated in the diagram, and in the text these parameters are correlated with key steps of the needle or pilus growth towards the translocon assembly into the target PM of animal or plant cells.
In essence, the extracellular needle or pilus appendage comprises an approximately 1.2–3.5‐nm inner channel (Fig. 1), which hypothetically functions as a pathway for the secretion of unfolded/semi‐unfolded proteins through a trafficking route all the way from the bacterial cytoplasm to the bacteria–host interface (Jin and He, 2001; Büttner and Bonas, 2002; Kubori et al., 1998; Schraidt et al., 2010). The subsequent translocation of type III effectors from the tip of the needle or pilus appendage into the cytosol of host cells is facilitated by the channel‐like type III translocon situated at the distal end of the needle or pilus complex. The translocon extends to penetrate the target PM (Daniell et al., 2001; Faudry et al., 2006; Neyt and Cornelis, 1999; Ryndak et al., 2005; Schoehn et al., 2003) and has an opening inwards to the cytosol of host cells (Ide et al., 2001), allowing for direct injection of type III effectors from bacteria into host cells (Alfano and Collmer, 2004; Chatterjee et al., 2013; Kay and Bonas, 2009; Matteï et al., 2011).
As a prerequisite for translocon assembly at the bacteria–host interface, the distal end of the bacterial needle and pilus must reach to the outer plane of host cell PMs (Fig. 1). This requires a common mechanism by which the P‐rod of the T3SS basal body traverses the bacterial periplasm and, by this step, the basal body is able to associate with the OM‐located ring structure, which is connected outwards to the needle or pilus complex. Owing to the structural difference between plant and animal cells, the bacterial pilus needs to overcome an additional physical barrier, the plant cell wall (CW), which is at least 100 nm thick under most circumstances (Rose, 2003). Perhaps as a result of this particularity, the pilus elongating up to 2 μm is maximally 25‐fold longer than the needle extending, at most, to 80 nm in length (Blocker et al., 2001; Jin and He, 2001; Journet et al., 2003). Penetrations of bacterial periplasmic layers and plant CWs accordingly represent the characteristic similarity and difference between plant‐ and animal‐pathogenic bacteria in their T3SS organization towards the translocon assembly.
Penetration of Bacterial Periplasm by T3SS Basal Body
The periplasm of Gram‐negative bacteria comprises a peptidoglycan mesh (Fig. 1) with natural pores of approximately 2 nm in radius (Demchick and Koch, 1996). As argued circumstantially by Matteï et al. (2011), such natural pores are too narrow to accommodate the P‐rod, which is much wider than 3.5 nm (Fig. 1). Thus, bacteria need to deploy a mechanism for their T3SS machinery to traverse the periplasmic layer. This mechanism has been characterized as the production of lytic transglycosylases (LTs), which are assumed to locally degrade the peptidoglycan substrate (Mushegian et al., 1996; Ward et al., 2002) and to enlarge pores in the periplasmic mesh to accommodate the T3SS machinery (Koraimann, 2003; Oh et al., 2007; Wang et al., 2008; Zahrl et al., 2005).
LTs typically recognize a single motif with an invariant glutamate in the substrate and catalyse the cleavage of the β‐1,4‐glycosidic bond between N‐acetylglucosamine and N‐acetylmuramic acid in peptidoglycan, producing the terminal 1,6‐anhydromuramyl cleavage product (Boch et al., 2002; Mushegian et al., 1996). This catalytic activity can broaden natural pores in the peptidoglycan mesh, allowing the T3SS machinery to traverse the periplasmic barrier in animal‐pathogenic bacteria (Demchick and Koch, 1996; Zahrl et al., 2005) and possibly also in plant pathogens (Boch et al., 2002; Oh et al., 2007; Wang et al., 2008; Ward et al., 2002). In agreement with this hypothesis, the hypersensitive response and pathogenicity (Hrp) in non‐host and host plants, respectively, are regarded as the translocation‐associated phenotype of bacteria, and Hrp‐associated (Hpa) protein Hpa2 from Xanthomonas oryzae pv. oryzae (Xoo) and X. oryzae pv. oryzicola (Xoc), the pathogens that cause bacterial blight and bacterial streak of rice (Oryza sativa), respectively, has been characterized to be an LT enzyme with the ability to degrade the bacterial envelope (Zhang et al., 2008). The LT orthologue HpaH from X. campestris pv. vesicatoria (Xcv), the bacterial spot pathogen of pepper (Noël et al., 2002), may also be enzymatically active, because HpaH is almost identical in amino acid sequence to Hpa2, contributes to virulence and T3SS assembly, and promotes the secretion and translocation of effector proteins (Büttner et al., 2007). In Xcv, moreover, the function of HpaH is regulated by HrpB1 and HrpB2 in the Hrp peninsula (Büttner et al., 2007; Rossier et al., 2000). HrpB1 localizes to the bacterial periplasm and physically interacts not only with peptidoglycan but also with HrpB2 and several other T3SS components (Hausner et al., 2013), suggesting that these proteins are part of a periplasmic substructure of the T3SS machinery related to the potential role of HpaH in degrading the peptidoglycan substrate.
Predicted LTs from plant‐pathogenic bacteria (Boch et al., 2002; Oh et al., 2007; Wang et al., 2008; Ward et al., 2002) perform in a similar manner to their orthologues in animal pathogens, such as IpgB and IpgF from the animal diarrhoea pathogen Shigella spp. (Allaoui et al., 1993; Zahrl et al., 2005), with regard to the positive effect on T3SS and/or virulence. However, single LTs do not contribute significantly to T3SS and virulence, both of which may require cooperative functions of the LTs. For example, Pseudomonas syringae pv. tomato (Pst) produces different effectors, namely Hrp proteins or Hrp‐out proteins (Hops), to cause bacterial speck in host plants, such as tomato (Solanum lycopersicum). The pathogen deploys three LTs (HrpH, HopP1 and HopAJ1) under co‐regulation with the T3SS regulon HrpJ, which controls effector translocation by regulating the secretion of the corresponding translocators (Crabill et al., 2012; Fu et al., 2006). These LTs function collaboratively and contribute to the translocation of effector HopA1 and its virulence to the plant (Oh et al., 2007). HrpH and HopP1, but not HopAJ1, are T3SS substrates, and are themselves secreted (Oh et al., 2007), possibly to prevent further peptidoglycan degradation after the secretion machinery has traversed the periplasmic barrier (Büttner, 2012).
In addition, HopP1 is similar to a distinct group of T3SS‐secreted, effector‐helper, non‐effector proteins, collectively designated as harpins, with respect to the properties of lacking cysteine (except for Hpa1 from bacteria in the Xanthomonas genus; Zhu et al., 2000) and eliciting the hypersensitive response (HR) in leaves of tobacco (Nicotiana tabacum) (Kvitko et al., 2007), a non‐host plant of Pst and many other bacteria. Harpins are basically distinct from other T3SS‐secreted proteins, especially virulence effectors (He, 1998; Kim and Beer, 2000). A fundamental distinction is that harpins function outside plant cells; by contrast, to perform their pathological roles, effectors must translocate into the cytosol of plant cells (Alfano and Collmer, 2004; Galán and Collmer, 1999; He, 1998). Multiple effects of harpins elucidated in the last 23 years (since the first report by Wei et al., 1992) and the newly appreciated function of some harpins in effector translocation have been summarized in a recent review (Choi et al., 2013). This review fully discusses the structural, biochemical and functional features of harpins from different bacterial species. Below, we focus on the structure–function relationship of tested harpins that affects bacterial pilus growth towards PMs of plant cells.
Potential Roles of Two‐Domain Harpins in Bacterial Pilus Penetration of the Plant CW Barrier
To contact with the outer plane of plant PMs, the bacterial pilus must traverse at least 100‐nm‐wide plant CWs (Fig. 1). This process perhaps requires the aid of particular T3SS accessory proteins that have activities of CW‐degrading enzymes. Candidates have been found in so‐called two‐domain harpins, which have been defined, in contrast with one‐domain harpins, to emphasize the obvious structural difference related to pilus growth towards the target PM, but not to preclude other functional domains (Kvitko et al., 2007), such as predicted α‐helical and glycine‐rich motifs (Ji et al., 2011; Kvitko et al., 2007; Liu et al., 2006; Meyer et al., 2006; Wang et al., 2007, 2008). One‐domain harpins refer to early defined conventional type III accessory proteins that are characteristically acidic, hydrophilic and glycine‐rich, with the dual role of inducing HR in non‐host plants and/or contributing to virulence on hosts of the bacteria (He et al., 1993; Wei et al., 1992). Two‐domain harpins are hybrids of the corresponding canonical proteins, and each hybrid contains a residue‐indexed shortened version of a particular harpin at the nitroxyl (N) terminus and a carboxyl‐terminal (C‐terminal) enzymatic domain characteristic of either LT enzymes, which have been implicated in the degradation of the bacterial periplasmic layer (Kvitko et al., 2007; Zhang et al., 2008), or pectate lyase (PL) enzymes, which may catalyse the decomposition of plant CWs (Charkowski et al., 1998; Kim and Beer, 1998). So far, nine one‐domain harpins (HrpN, HrpZ1, HrpZ, HrpZj, Hpa1, HpaG, XopA, HreX and HrpXm) and six two‐domain harpins (HrpW, HopAK1, HrpW1, HopP1, PopW and HrpWk) have been identified in plant‐pathogenic bacteria and a rhizobium (Choi et al., 2013). The two classes of harpin are basically different with regard to potential targets in the bacterial envelope or plant CWs.
Although LT‐type two‐domain harpins have been proposed to act on the peptidoglycan mesh to facilitate the penetration of the bacterial periplasm by the T3SS basal body, as discussed above, PL‐containing two‐domain harpins have been found to target the compositions of plant CWs and are thought to be essential for the bacterial pilus to traverse the plant CW barrier in the pectin‐rich middle lamella. Early characterized two‐domain harpins were HrpW1 from Pst (Charkowski et al., 1998) and HrpW from Erwinia amylovora (Ea), the pathogen which causes fire blight of rosaceous plants (Kim and Beer, 1998). Ea HrpW contains 447 amino acids and comprises two domains connected by an intermediate sequence with abundant proline and serine residues. The N‐terminal domain is similar to the partial sequence of HrpN, especially with regard to glycine‐rich repeats, and the C‐terminal domain is characteristic of polysaccharide lyase family 3 (PL3) PLs (Kim and Beer, 1998). Similar dual domains have been found in Pst HrpW1 (Charkowski et al., 1998; Kvitko et al., 2007). At 424 amino acids in size, HrpW1 appears to be a canonical protein hybrid, in which the N‐terminal 1–188 region is equivalent to a residue‐indexed HrpZ1 and the C‐terminal fragment sequence is similar to PL3 PLs. The PL3 fragment truncated from HrpW1 is able to bind with calcium pectate, a major constituent of plant CWs, suggesting that the target of HrpW1 is in the plant CW (Charkowski et al., 1998).
However, neither the truncated PL3 fragment nor the full‐length HrpW protein is able to degrade pectate substrates (Charkowski et al., 1998; Kim and Beer, 1998; Li et al., 2010). By contrast, the HrpW orthologue of Rhizobium etli, called HrpWk (Choi et al., 2013), exhibits pectinolytic activity, which, however, is eliminated by replacing tryptophan‐192 with alanine (Fauvart et al., 2009). The alanine residue in HrpW1 is equivalent to tryptophan‐192 in HrpWk, whereas the tryptophan residue is conserved in all active PL enzymes, but not in HrpW and HrpW1 (Fauvart et al., 2009), suggesting that HrpW and HrpW1 are destined to be inactive. Presumably, the loss of enzymatic activity is a congenital strategy for the bacterial pilus machinery to translocate virulence effectors in a manner of friendship with plants, or without breakage of plant cell structures. If this hypothesis is true, HrpW or HrpW1 binding to, instead of degrading, the middle lamella is sufficient for this thin, soft, cementing layer to be penetrated by the pilus. Alternatively, pectate degradation may be provided by a different two‐domain harpin from the same bacteria, such as HopAK1, under the particular circumstance (Kvitko et al., 2007) in which HrpW1 binding to the middle lamella serves as a platform for HopAK1 to locally degrade the substrate. Such an ‘arena‐player’ mode is commonly used by hydrophilic translocators (Fig. 2A‐a), namely needle tip proteins (Chatterjee et al., 2011; Johnson et al., 2007; Mueller et al., 2005), of animal‐pathogenic bacteria to assist their hydrophobic counterparts to burrow into the target PM during type III translocon assembly (Büttner, 2012; Mueller et al., 2008).
Figure 2.
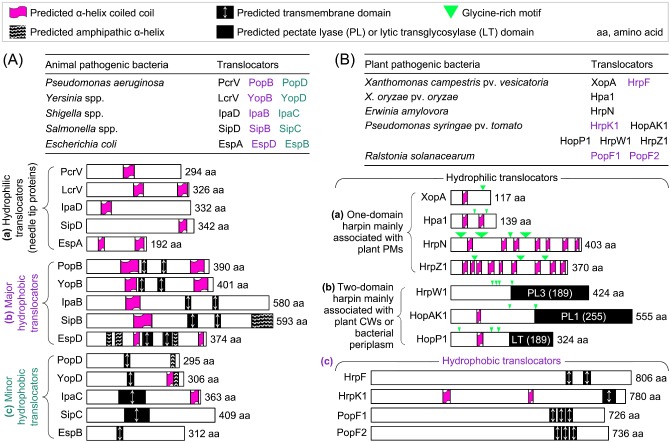
Schematic analysis of type III translocators from animal‐ and plant‐pathogenic Gram‐negative bacteria. The predicted characteristic domains are indicated by the symbols described in the figure and, in (A) and (B), they are located pro rata in the graphic protein sequences. Predicted α‐helical regions form a coiled coil, called an α‐helix coiled coil, or contain hydrophobic residues, designated as an amphipathic α‐helix. Additional information is to be stated below in (A) and (B). In both panels, translocators that belong to the same category are shown together in subpanels indicated by lowercase letters in parentheses. (A) Overview of translocators from animal pathogens. The depicted features of the proteins are constant in the literature, except for EspB, EspD and PcrV. For EspB, only the predicted transmembrane (TM) domain is shown, and the predicted 11 α‐helical regions, which vary in length and are located throughout the protein sequence (Hamada et al., 2005), are neglected. For PcrV and EspD, the number of amino acid residues shown here is the same as in the original research papers (Delahay and Frankel, 2002; Goure et al., 2004), but different from an in‐depth review article (Matteï et al., 2011). (B) Overview of translocators from plant pathogens. Hpa1 is a candidate for the translocator of the TAL effector PthXo1 based on studies shown in Fig. 4, whereas the other translocators have been studied previously (Choi et al., 2013). Predicted TM domains present in the hydrophobic translocators (Büttner et al., 2002), the enzymatic domains in two‐domain harpins (Kvitko et al., 2007) and predicted α‐helices present in HrpN, HrpZ1 and Hpa1 (Choi et al., 2013) are situated according to the literature. The predicted α‐helices present in other harpins were not stated in the literature and were determined by analysis with the online SIB (Swiss Institute of Bioinformatics) ExPASy program (http://www.ch.embnet.org/software/COILS_form.html).
Roles of One‐Domain Harpins in Pilus Targeting to Plant PM
Owing to their hydrophilicity, one‐domain harpins of plant‐pathogenic bacteria (Fig. 2B‐a) represent analogues of needle tip proteins from animal pathogens (Fig. 2A‐a) and are likely to locate at the tip of the pilus with direct access to the outer plane of plant PMs. Similar to the spatially contrapuntal situation of needle tip proteins of animal pathogens at the interface between the distal end of the needle and the targeted PM of host animal cells, one‐domain harpins of plant pathogens can localize to plant PMs or membranes of other eukaryotic cells, such as oocytes of Xenopus laevis, an African clawed frog frequently used in protein localization experiments (Maurel, 2007; Racape et al., 2005). Direct evidence has been obtained for Ralstonia solanacearum PopA1 binding to X. laevis oocytes (Racape et al., 2005), and for Ea HrpN (Oh and Beer, 2007) and Xoo Hpa1 (Sang et al., 2012) expressed in Arabidopsis thaliana. Direct evidence has also been found for HrpZ from P. syringae pv. phaseolicola, and this harpin binds to the PM of tobacco cells (Lee et al., 2001a). In addition, isolated or synthetic liposomes and/or synthetic planar lipid bilayers bind HreX from X. campestris pv. pelargonii (Engelhardt et al., 2009) and HrpZ orthologues of Pseudomonas spp. (Engelhardt et al., 2009; Lee et al., 2001b). Targeting to the plant PM may allow HrpN (Bocsanczy et al., 2008), HrpZ1 (Kvitko et al., 2007; Petnicki‐Ocwieja et al., 2005), Hpa1 (Sang et al., 2012) and XopA (Büttner et al., 2006; Noël et al. 2002) to integrate into the type II translocon assembly and facilitate pore formation in the target PM.
Hydrophilic Translocators of Plant‐Pathogenic Bacteria
With respect to the role in effector translocation, needle tip proteins are also called hydrophilic translocators (Fig. 2A‐a), which depend on a unitary motif, the predicted α‐helical coiled coil (Espina et al., 2006a, 2006b; Markham et al., 2008), to oligomerize into the tip of the needle complex (Yip et al., 2005). To date, three one‐domain harpins (HrpN, HrpZ1 and XopA) have been implicated in effector translocation from plant‐pathogenic bacteria (Fig. 2B‐a). These harpins resemble the orthologues of animal pathogens in their structural characteristics (Fig. 2B‐a compared with Fig. 2A‐a), especially with regard to the presence of predicted α‐helical coiled‐coil motifs (Ji et al., 2011; Kvitko et al., 2007; Liu et al., 2006; Wang et al., 2007, 2008). The three harpins have been demonstrated to play substantial roles in supporting the virulence and/or translocation of different effectors. XopA of Xcv is implicated in the translocation of type III effectors, as the Xcv xopa mutant incurs partial impairment in the ability to induce plant responses, but does not affect effector secretion (Büttner et al., 2006; Noël et al. 2002). Hence, XopA is regarded as a hydrophilic translocator candidate, but its direct role in effector translocation has not been determined. By contrast, HrpN of Ea has been convincingly elucidated to be an essential translocator for effector DspA/E of the bacterium (Bocsanczy et al., 2008).
Two‐domain harpins have also been implicated in effector translocation. It is quite interesting that Pst produces four harpins, one‐domain HrpZ1 and two‐domain HrpW1, HopP1 and HopAk1 (Fig. 2B‐b), which all affect effector translocation and virulence or HR induction (Kvitko et al., 2007). These harpins synergize hydrophobic translocator HrpK1 (Petnicki‐Ocwieja et al., 2005) on the basis of the following findings: (i) the quadruple harpin gene polymutant carrying the hrpK1 gene displays moderate reductions in AvrPto1 translocation and the ability to elicit HR or cause disease; (ii) these events are strongly reduced in the quadruple polymutant if hrpK1 is knocked out; and (iii) the quinary polymutant, in which hrpK1 and the four harpin genes are concurrently knocked out, can be complemented by any of hrpZ1, hrpW1 and hopAK1, but not hopP1 (Kvitko et al., 2007). Thus, HrpZ1, HrpW1 and HopAK1 are functionally redundant with HrpK1, but HopP1 is not, to mediate AvrPto1 translocation. However, HopP1 plays a role in the translocation of a different effector called HopA1 (Oh et al., 2007). This difference is attributable to the T3SS regulon HrpJ, which controls effector translocation by regulating the secretion of the corresponding translocators (Crabill et al., 2012; Fu et al., 2006). Indeed, HrpJ regulates the secretion of HrpZ1, HrpW1 and HopAK1, but not HopP1 (Crabill et al., 2012). These analyses indicate that secretory regulation received by translocators is important for their roles in effector translocation. These analyses also suggest that a particular translocator may be responsible for the translocation of some, but not all, of the effectors from a bacterium.
Hydrophobic Translocators of Plant‐Pathogenic Bacteria
So far, four hydrophobic translocators have been identified in plant‐pathogenic bacteria (Fig. 2B‐c), representing a very small number of potential translocators compared with the repertoire of animal pathogens (Fig. 2A‐bc). Moreover, each species of animal‐pathogenic bacterium possesses a major hydrophobic translocator that carries two predicted transmembrane (TM) domains (Fig. 2A‐a) and a minor one that contains a single predicted TM domain (Fig. 2A‐b). Predicted TM domains guide the hydrophobic translocators into the target PM to create a translocon pore (Faudry et al., 2006; Hakansson et al., 1996; Ryndak et al., 2005). In addition to TM, predicted α‐helix coiled‐coil and/or amphilipathic α‐helix motifs (Fig. 2A) are also present in most hydrophobic translocators (Derewenda et al., 2004; Espina et al., 2007; Johnson et al., 2007). Amphilipathic α‐helix and α‐helix coiled‐coil motifs allow for the dimerization and oligomerization of proteins, respectively (Twyman, 1998), and sometimes dimerization induces oligomerization by the same protein (Delahay and Frankel, 2002). In contrast with these extensive studies on translocators of animal pathogens, the current knowledge is far from sufficient to allow for a consideration of whether or not the attributive ‘major’ and ‘minor’ are pertinent to define the hydrophobic translocators of plant pathogens.
The first type III hydrophobic translocator discovered in plant‐pathogenic bacteria was HrpF of Xcv (Büttner et al., 2002). This protein locates within the HrpF pathogenicity peninsula conserved in different species of the Xanthomonas genus (Sugio et al., 2005) and contains two predicted TM domains (Büttner et al., 2002; Huguet and Bonas, 1997; Fig. 2B‐c). The role of Xcv HrpF as a translocator has been well demonstrated by Büttner et al. (2002). They elucidated that the C‐terminal region of the HrpF sequence plays an essential role in supporting the translocation of effector AvrBs3, and that the N‐terminal region contains a signal for secretion, but not for translocation. This suggests that secretion and translocation are sequential, but independent, processes under the control of different functional domains in the translocator. The same workers further showed that HrpF was able to bind with artificial lipids formed in silicon beads to induce altered electrical current fluxes, indicative of pore formation, in a synthetic planar lipid bilayer system. Quite interestingly, the C‐terminus required for lipid binding and effector translocation is dispensable for pore formation in the bionic membrane. This discrepancy may be caused by the higher sensitivity of the planar bilayer assay relative to the lipid‐binding measurement for the detection of the activity provided by the region towards the N‐terminus.
With a similarity (E value = 6e – 14) to Xcv HrpF, HrpK secreted by Pst contains two predicted α‐helical coiled‐coil motifs in the middle and a single predicted TM domain at the C‐terminal region of the protein sequence (Fig. 2B‐c). The predicted TM domain is required for the translocation of HrpK itself into plant cells and this translocation is subject to T3SS. Furthermore, HrpK is a putative type III translocator, as it is required for type III effector HopA1 to induce HR in the non‐host plant tobacco and to cause leaf speck in the host plants tomato and Arabidopsis (Petnicki‐Ocwieja et al., 2005).
In addition to HrpF and HrpK, PopF1 and PopF2 secreted by Ralstonia (previous Pseudomonas) solanacearum, a soil‐borne pathogen that causes bacterial wilt disease in more than 200 plant species (Arlat et al., 1994; Mansfield et al., 2012; Meyer et al., 2006), are also characteristic of hydrophobic translocators (Fig. 2B‐c). According to Meyer et al. (2006), PopF1 and PopF2 share significant homology with one another, with 81% identity and 86% similarity, and exhibit some degree of similarity to HrpF from Xanthomonas spp. and YopB from Yersinia spp. at the C‐terminal 200‐amino‐acid region, which contains the predicted TM domains (Fig. 2B‐c). Although PopF1 contributes more than PopF2 to virulence on tomato and HR induction in tobacco, both PopFs act similarly to mediate the translocation of effector AvrA. Both PopFs are partially exchangeable with Xcv HrpF, as HrpF can compensate for part of the compromised AvrA translocation in the popF1 popF2 double mutant. This finding and the similarity of PopFs and HrpFs suggest that translocators are functionally conserved in bacteria of the genera Ralstonia and Xanthomonas (Büttner, 2012; Meyer et al., 2006).
Models of Type III Translocon Assembly
As shown in Fig. 2A, every species of animal‐pathogenic bacterium possesses one hydrophilic and two hydrophobic translocators, each forming homogeneous oligomers before participating in translocon assembly (Chatterjee et al., 2013). Matteï et al. (2011) proposed two distinct models of type III translocon assembly by animal pathogens. One involves a hetero‐oligomer of two hydrophobic translocators, spontaneously contacting the hydrophilic counterpart (Fig. 3A). The other is called the ‘three‐tiered ring’ model: two hydrophobic translocators exist in oligomeric form, with the major partner inserting stably into the target PM, whereas the minor translocator, situated at the needle tip, serves as the link with the hydrophilic counterpart (Fig. 3B). Although most biochemical results point to the second model (Büttner, 2012; Faudry et al., 2006; Goure et al., 2004), the possibility of translocon assembly as a hetero‐oligomer cannot be ruled out at present, as argued by Matteï et al. (2011).
Figure 3.
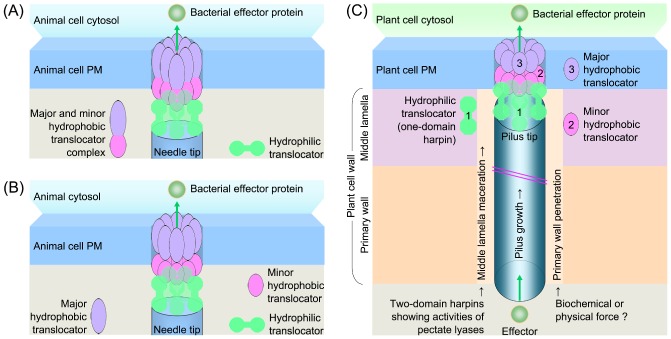
Models of the translocon assembly by bacteria that are pathogens of animals (A, B) and plants (C). (A, B) Both diagrams were created according to previous definitions (Matteï et al., 2011). (C) The diagram was created according to previous studies, which have been partially discussed in a recent review on harpins (Choi et al., 2013). The numbers indicate the potential location of translocator complexes. In (A)–(C), the number of a protein complex, five and eight in hydrophilic and hydrophobic translocators, respectively, is shown based on previous studies partially summarized in several review articles (Büttner, 2012; Chatterjee et al., 2013; Matteï et al., 2011; Mueller et al., 2008).
Known translocators of plant‐pathogenic bacteria diversify largely in terms of biochemistry (Fig. 2B), indicating the complexity of the process of translocon assembly, which may involve two steps (Fig. 3C). First, the pilus penetrates the plant CW with the aid of PL‐type two‐domain harpins. Additional CW‐degrading enzymes (Yang et al., 2008) or physical force only is required for pilus penetration of the primary wall, a thin, flexible and extensible layer formed by growing cells in young tissues that favours bacterial infection. Second, one‐domain harpins serving as hydrophilic translocators may assist the hydrophobic counterparts to burrow into the target PM. A similar ‘two‐step model’ has been proposed by Kvitko et al. (2007). They also presented a ‘consortium model’: the pilus machinery has an intrinsic ability to breach the plant CW, whereas harpins and HrpF homologues function as a consortium for effector translocation. The intrinsic CW‐breaching mechanism may be provided in the manner stated above, i.e. direct pilus penetration of the CW after binding of an inactive two‐domain harpin, or by cooperation of two‐domain harpin partners in the ‘arena‐player’ mode. The ‘consortium’ is, indeed, a translocon inserted into the target PM (Büttner, 2012; Mueller et al., 2008). In addition to the translocators, LTs may also participate in translocon assembly. For example, X. oryzae Hpa2 has been shown to degrade bacterial cell envelopes (Zhang et al., 2008), to target plant PMs and to interact with the hydrophobic translocator HrpF (Li et al., 2011).
Translocon assembly by a plant‐pathogenic bacterium probably resembles the ‘three‐tiered ring’ architecture with modifications (Fig. 3C): (i) the two assumed hydrophobic translocators simultaneously burrow into the target PM; and (ii) the harpin‐type hydrophilic translocator commits a dual association with the target PM at one side and with the minor hydrophobic counterpart at the other. This hypothesis is in agreement with certain discoveries: (i) HrpN and Hpa1 can bind to the outer surfaces of plant PMs (Oh and Beer, 2007; Sang et al., 2012); (ii) Hpa2 and HrpZ1 can bind to plant PMs or with the planar bilayer to create ion‐conducting pores (Lee et al., 2001a; Li et al., 2011); (iii) Hpa2 can interact with HrpF inside plant cells (Li et al., 2011); and (iv) Hpa1 and HrpZ1 oligomerize via the α‐helical coiled‐coil motif (Haapalainen et al., 2011; Ji et al., 2011). In addition, four translocators lack an α‐helix (Fig. 2B) and may oligomerize through hydrogen bonds (Twyman, 1998).
The possibility of dual association relies on multiple motifs in harpin sequences (Fig. 2B). The C‐terminal region containing the first three of nine predicted α‐helices determines HrpZ1 oligomerization and its ability to create pores in the bionic membrane (Haapalainen et al., 2011). Although the N‐terminus containing the first of two α‐helices is essential for Hpa1 to oligomerize and to induce plant defences and growth enhancement (Ji et al., 2011; Li et al., 2014b; Wang et al., 2007), the last two effects are inhibited by the glycine‐rich motif (Liu et al., 2006). Thus, the glycine‐rich motif is adverse to plants, but may be propitious to bacteria. Both motifs locate with a distance in the Hpa1 sequence and may function independently, enabling the translocator to associate with the pilus tip and the target PM or the assumed hydrophobic counterpart, such as HrpF (Li et al., 2011). Further studies are needed to determine the number and characters of translocators and their oligomerization into the translocon, as proposed in Fig. 3C, which differs from the models shown in Fig. 3A, B with regard to the particular translocators that associate with the postulated sensors in the target PM.
Potential PM Sensors for Type III Translocators
As discussed in detail in several recent reviews (Büttner, 2012; Cortes et al., 2014; Matteï et al., 2011), the recognition of type III translocators by postulated sensors in the target PM is critical for the creation and functionality of the translocon pore. Membrane lipids are curently known common sensors of translocators from animal and plant pathogens. Phospholipids are major components of cell membranes and are essential for the formation of the translocon pore in animal cells (Matteï et al., 2011). Cholesterol is a high‐density lipoprotein involved in cellular signalling and is implicated in IpB binding to the target PM and in the bilayer binding of PopB and PopD (Cortes et al., 2014). In plants, phosphatidic acid is implicated in the sensing of HrpZ1. HrpZ1 interacts with this compound and requires it to create pores in artificial lipid vesicles and in vesicles prepared from PMs of Arabidopsis cells (Haapalainen et al., 2011). It is possible that the plant PM‐integral phosphatidic acid may serve as a receptor of the HrpZ1 translocator.
In addition to membrane lipids, PM‐integral proteins are also implicated in plant recognition of harpin‐type translocators. Oh and Beer (2007) identified a HrpN‐interacting protein from apple (Malus sylvestris), HIPM (HrpN‐interacting protein from Malus), and found an orthologue in Arabidopsis, AtHIPM. Both HIPMs contain an N‐terminal signal peptide and a central TM domain, and associate with PMs of plant cells. Thus, HIPM and AtHIPM are membrane‐integral with a stretch to the cellular periphery in contact with the HrpN translocator.
One further potential PM sensor is an isoform of PM intrinsic proteins (PIPs), which constitute a unique aquaporin (AQP) family in plants (Abascal et al., 2014; Bansal and Sankararamakrishnan, 2007). Rice OsPIP1;3 interacts with the canonical form of Hpa1, but not the mutant version (Fig. 4A) generated by removal of the N‐terminal region, which contains the first of two α‐helical coiled coils (Fig. 2B) and is essential for the oligomerization of Hpa1 and its bioactivity in plants (Ji et al., 2011; Li et al. 2014a, b; Wang et al., 2008). Hpa1 contributes to the virulence of Xoo strain PXO99A on the rice variety Nipponbare, as the severity of bacterial blight can be markedly reduced when Hpa1 is deleted (Fig. 4B). The virulence is provided mainly by the effector PthXo1 (Yang et al., 2006; Yang and White, 2004), the deletion of which causes a substantial alleviation of blight symptoms (Fig. 4B). Single Hpa1 or PthXo1 protein deletion and double deletion result in similarly alleviated blight symptoms (Fig. 4B). Because virulence is executed only after injection of the effector into rice cells (Gürlebeck et al., 2006; White et al., 2009), these analyses provide a basis for further studies to determine the possible effects of OsPIP1;3 and Hpa1 on the virulence and translocation of PthXo1.
Figure 4.
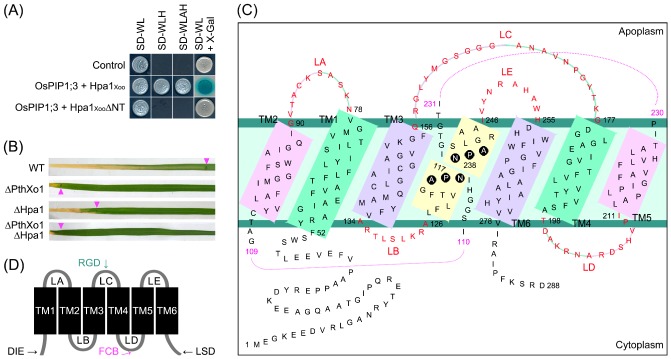
Xanthomonas oryzae pv. oryzae (Xoo) Hpa1 interacts with rice aquaporin OsPIP1;3 (A) and affects the virulence of Xoo effector PthXo1 on rice variety Nipponbare (B). The aquaporin structural complex is sketched according to the current model (C) which allows for the characterization of protein‐interacting motifs (D). (A) The OsPIP1;3–Hpa1 interaction analysed with a split ubiquitin‐based yeast two‐hybrid system (Dualsystems Biotech., Zurich, Switzerland). This assay was performed on the control protein combination (control) provided by the system manufacturer and the combination of OsPIP1;3 with Hpa1Xoo or Hpa1XooΔNT, a Hpa1Xoo mutant version generated by deleting the N‐terminal region, which contains 53 amino acids and the first of two predicted α‐helices (Li et al., 2014b). Three types of synthetic dropout (SD) amino acid nutrient medium were used in the screening of yeast hybrids. SD‐WL medium allows the growth of yeast cells irrespective of protein interactions. Yeast cells are able to grow on both SD‐WLH and SD‐WLAH media only when an interaction of the tested proteins occurs. The interaction can also be detected by the X‐Gal assay of colonies grown on SD‐WL (Möckli and Auerbach, 2004). (B) Bacterial blight symptoms in Nipponbare leaves photographed at 14 days after inoculation with the bacterial strain POX99A and its mutants. The symbol Δ prefixed to a protein name indicates that the protein has been deleted from POX99A by the method described previously (Song and Yang, 2010). Purple triangles indicate the end of bacterial blight lesions. WT, wild‐type. (C) Topological structure of OsPIP1;3 (accession number Q9SXF8.2) analysed according to a previously established model (Chaumont et al., 2001). Six transmembrane (TM) domains, TM1–TM6, are located in the plasma membrane (PM) in an alternate manner and are linked by five connecting loops, LA–LE. NPA is highlighted with black circles as it forms the sandglass‐shaped filter that decides substrate selectivity. Numbers indicate the positions of NPA turning points or actual adjacent residues, but plotted separately to yield space. (D) Protein‐interacting motifs or regions found in different aquaporins. DIE, maize (Zea mays) ZmPIP2 diacidic DIE motif (4−6 Asp‐Ile‐Glu); FCB, filensin‐ and calmodulin‐interacting region in lens AQP0; LSD, human AQP1 and AQP4 regions binding to the light‐sensing protein Killer Red; RGD, integrin β1‐binding RGD motif (Arg‐Gly‐Asp) in renal AQP2 (You et al., 2013).
AQP interacting with non‐AQP proteins is a recently appreciated mechanism that governs the physiological roles of distinct AQPs (Qin and Boron, 2013; Sjöhamn and Hedfalk, 2014) on the basis of the topological structures conserved in all AQPs, including OsPIP1;3. In the current topological model (Gomes et al., 2009; Maurel, 2007), OsPIP1;3 is characterized by six α‐helical TM domains (TM1–TM6) that are tilted along the plane of PM and linked one to the other by five connecting loops (LA–LE) (Fig. 4C). LB, LD and both termini are located in the cell cytosol and potentially bind to cytosolic substrates, similarly to lens AQP0 binding with filensin through its C‐terminus (Hu et al., 2012; Lindsey Rose et al., 2006; Sjöhamn and Hedfalk, 2014; Fig. 4D). By contrast, LA, LC and LE face the apoplasm and have the opportunity to contact with apoplastic substrates, similarly to renal AQP2 LE binding to integrin β1 (Jin et al., 2012). Presumably, PIPs depend on the extracellular LA, LC and LE to sense environmental cues (Chen et al., 2013; Maurel et al., 2008) and biotic signals (Ruiz‐Lozano et al., 2009; You et al., 2013), and thus extend their physiological roles beyond substrate transport. The experimental performances of wild‐type and mutant versions of OsPIP1;3, Hpa1 and PthXo1 (Fig. 4A, B) suggest that OsPIP1;3 potentially interacts with Hpa1 to facilitate PthXo1 translocation from Xoo cytoplasm into the cytosol of rice cells.
Conclusions
As an integral element of T3SS (Fig. 1), the type III proteinaceous translocon is believed to be assembled by refined associations of one hydrophilic and two hydrophobic translocators identified in animal‐pathogenic bacteria, or similar proteins from plant pathogens (Fig. 2). In a particular bacterium, the translocator counterparts may rely on the predicted α‐helix motifs to oligomerize into the translocon in a hierarchical and contrapuntal manner (Fig. 2). The translocon organization presumably follows distinct models (Fig. 3), which similarly comprise the essential role of translocator sensors potentially present in the target PM of animal or plant cells (Fig. 4). The aims of future studies will be to determine the accurate contrapuntal location of each translocator at the bacterium–host interface and to generate a complete atomic model of translocon assembly (Taufik et al., 2013; Wager et al., 2013).
This general perspective remains a great challenge for studies on plant‐pathogenic bacteria because their type III translocon and translocators are not as well understood as those of animal pathogens. Numerous studies are needed to address many basic questions regarding: (i) the accurate mechanisms by which the pilus machinery traverses plant CWs; (ii) the composition of the translocon and the number of translocators, especially hydrophilic counterparts other than harpins; (iii) the hydrophobic counterparts of the hydrophilic translocators; (iv) the oligomerization of each translocator; (v) the location of each translocator at the bacterium–plant interface; and (vi) the physical interaction of translocator counterparts into the translocon assembly.
Future studies to demonstrate how the translocators and putative PM sensors interact to facilitate effector translocation will also be a promising field of research, especially for the emerging rice–Xoo interaction system that possibly involves functional relationships among OsPIP1;3, Hpa1 and PthXo1 (Fig. 4). The OsPIP1;3–Hpa1 interaction assumed to occur at the target PM probably leads to a channel‐like pore which resembles the so‐called translocon pore. Critical studies are required to elucidate: (i) the structural basis for OsPIP1;3–Hpa1 interaction; (ii) the topological structure of the channel‐like pore at the target PM following molecular interaction; and (iii) the rice cell cytosol‐directional opening of the translocon in relation to the dynamic translocation of Xoo effectors, PthXo1 and possibly other proteins into the cytosol of rice cells. Similar studies are also needed to elucidate the translocon assembly in other bacterial pathosystems in addition to the rice–Xoo interaction.
Acknowledgements
We thank Bing Yang (Iowa State University, Ames, IA, USA) for the gift of vector pZWpthXo1, and Liping Han (graduate student in the laboratory) for the inoculation experiment. This study was supported by the National Key Basic Research Program of China (973 plan 2012CB114003) and Natural Science Foundation of China (31171830 and 31272072).
References
- Abascal, F. , Irisarri, I. and Zardoya, R. (2014) Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta, 1840, 1468–1481. [DOI] [PubMed] [Google Scholar]
- Abrusci, P. , McDowell, M.A. , Lea, S.M. and Johnson, S. (2014) Building a secreting nanomachine: a structural overview of the T3SS. Curr. Opin. Struct. Biol. 25C, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Allaoui, A. , Menard, R. , Sansonetti, P.J. and Parsot, C. (1993) Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61, 1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlat, M. , Van Gijsegem, F. , Huet, J.C. , Pernollet, J.C. and Boucher, C.A. (1994) PopA1, a protein which induces a hypersensitivity‐like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum . EMBO J. 13, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, A. and Sankararamakrishnan, R. (2007) Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Struct. Biol. 7, 27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska, J.B. , Wang, X. , Viboud, G.I. and Brodsky, I.E. (2013) Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell. Microbiol. 15, 1622–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker, A. , Jouihri, N. , Larquet, E. , Gounon, P. , Ebel, F. , Parsot, C. , Sansonetti, P. and Allaoui, A. (2001) Structure and composition of the Shigella flexneri ‘needle complex’, a part of its type III secreton. Mol. Microbiol. 39, 652–663. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Joarda, V. , Gao, L. , Robertson, T.L. , Lim, M. and Kunkel, B.N. (2002) Identification of Pseudomonas syringae genes induced during infection of Arabidopsis thaliana . Mol. Microbiol. 44, 73–88. [DOI] [PubMed] [Google Scholar]
- Bocsanczy, A.M. , Nissinen, R.M. , Oh, C.S. and Beer, S.V. (2008) HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. and Voytas, D.F. (2011) TAL effectors: customizable proteins for DNA targeting. Science, 333, 1843–1846. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Beer, S.V. , Bonas, U. , Boucher, C.A. , Collmer, A. , Coplin, D.L. , Cornelis, G.R. , Huang, H.C. , Hutcheson, S.W. , Panopoulos, N.J. and Van Gijsegem, F. (1996) Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20, 681–683. [DOI] [PubMed] [Google Scholar]
- Büttner, D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2002) Port of entry—the type III secretion translocon. Trends Microbiol. 10, 186–192. [DOI] [PubMed] [Google Scholar]
- Büttner, D. , Nennstiel, D. , Klüsener, B. and Bonas, U. (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. , Lorenz, C. , Weber, E. and Bonas, U. (2006) Targeting of two effector protein classes to the type III secretion system by a HpaC‐ and HpaB‐dependent protein complex from Xanthomonas campestris pv. vesicatoria . Mol. Microbiol. 59, 513–527. [DOI] [PubMed] [Google Scholar]
- Büttner, D. , Noel, L. , Stuttmann, J. and Bonas, U. (2007) Characterization of the non‐conserved hpaB‐hrpF region in the hrp pathogenicity island from Xanthomonas campestris pv. vesicatoria . Mol. Plant–Microbe Interact. 20, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Charkowski, A.O. , Alfano, J.R. , Preston, G. , Yuan, J. , He, S.Y. and Collmer, A. (1998) The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180, 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , Zhong, D. , Nordhues, B.A. , Battaile, K.P. , Lovell, S. and De Guzman, R.N. (2011) The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 20, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , Chaudhury, S. , McShan, A.C. , Kaur, K. and De Guzman, R.N. (2013) Structure and biophysics of type III secretion in bacteria. Biochemistry, 52, 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont, F. , Barrieu, F. , Wojcik, E. , Chrispeels, M.J. and Jung, R. (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Yin, X. , Wang, L. , Tian, J. , Yang, R. , Liu, D. , Yu, Z. , Ma, N. and Gao, J. (2013) Involvement of rose aquaporin RhPIP1;1 in ethylene‐regulated petal expansion through interaction with RhPIP2;1. Plant Mol. Biol. 83, 219–233. [DOI] [PubMed] [Google Scholar]
- Choi, M.S. , Kim, W. , Lee, C. and Oh, C.S. (2013) Harpins, multifunctional proteins secreted by gram‐negative plant‐pathogenic bacteria. Mol. Plant–Microbe Interact. 26, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Cortes, V.A. , Busso, D. , Maiz, A. , Arteaga, A. , Nervi, F. and Rigotti, A. (2014) Physiological and pathological implications of cholesterol. Front. Biosci. 19, 416–428. [DOI] [PubMed] [Google Scholar]
- Crabill, E. , Karpisek, A. and Alfano, J.R. (2012) The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol. Microbiol. 85, 225–238. [DOI] [PubMed] [Google Scholar]
- Daniell, S.J. , Takahashi, N. , Wilson, R. , Friedberg, D. , Rosenshine, I. , Booy, F.P. , Shaw, R.K. , Knutton, S. , Frankel, G. and Aizawa, S. (2001) The filamentous type III secretion translocon of enteropathogenic Escherichia coli . Cell. Microbiol. 3, 865–871. [DOI] [PubMed] [Google Scholar]
- Deane, J.E. , Roversi, P. , Cordes, F.S. , Johnson, S. , Kenjale, R. , Daniell, S. , Booy, F. , Picking, W.D. , Picking, W.L. , Blocker, A.J. and Lea, S.M. (2006) Molecular model of a type III secretion system needle: implications for host‐cell sensing. Proc. Natl. Acad. Sci. USA, 103, 12 529–12 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahay, R.M. and Frankel, G. (2002) Coiled‐coil proteins associated with type III secretion systems: a versatile domain revisited. Mol. Microbiol. 45, 905–916. [DOI] [PubMed] [Google Scholar]
- Demchick, P. and Koch, A.L. (1996) The permeability of the wall fabric of Escherichia coli and Bacillus subtilis . J. Bacteriol. 178, 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda, U. , Mateja, A. , Devedjiev, Y. , Routzahn, K.M. , Evdokimov, A.G. , Derewenda, Z.S. and Waugh, D.S. (2004) The structure of Yersinia pestis V‐antigen, an essential virulence factor and mediator of immunity against plague. Structure, 12, 301–306. [DOI] [PubMed] [Google Scholar]
- Engelhardt, S. , Lee, J. , Gabler, Y. , Kemmerling, B. , Haapalainen, M.L. , Li, C.M. , Wei, Z. , Keller, H. , Joosten, M. , Taira, S. and Nurnberger, T. (2009) Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion‐conducting pore formation and activation of plant immunity. Plant J. 57, 706–717. [DOI] [PubMed] [Google Scholar]
- Espina, M. , Ausar, S.F. , Middaugh, C.R. , Picking, W.D. and Picking, W.L. (2006a) Spectroscopic and calorimetric analyses of invasion plasmid antigen D (IpaD) from Shigella flexneri reveal the presence of two structural domains. Biochemistry, 45, 9219–9227. [DOI] [PubMed] [Google Scholar]
- Espina, M. , Olive, A.J. , Kenjale, R. , Moore, D.S. , Ausar, S.F. , Kaminski, R.W. , Oaks, E.V. , Middaugh, C.R. , Picking, W.D. and Picking, W.L. (2006b) IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri . Infect. Immun. 74, 4391–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina, M. , Ausar, S.F. , Middaugh, C.R. , Baxter, M.A. , Picking, W.D. and Picking, W.L. (2007) Conformational stability and differential structural analysis of LcrV, PcrV, BipD, and SipD from type III secretion systems. Protein Sci. 16, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faudry, E. , Vernier, G. , Neumann, E. , Forge, V. and Attree, I. (2006) Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa . Biochemistry, 45, 8117–8123. [DOI] [PubMed] [Google Scholar]
- Fauvart, M. , Verstraeten, N. , Dombrecht, B. , Venmans, R. , Beullens, S. , Heusdens, C. and Michiels, J. (2009) Rhizobium etli HrpW is a pectin‐degrading enzyme and differs from phytopathogenic homologues in enzymically crucial tryptophan and glycine residues. Microbiology, 155, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. , Guo, M. and Alfano, J.R. (2006) Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors and secretion of the HrpZ1 harpin. J. Bacteriol. 188, 6060–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán, J.E. and Collmer, A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Gomes, D. , Agasse, A. , Thiébaud, P. , Delrot, S. , Gerós, H. and Chaumont, F. (2009) Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim. Biophys. Acta, 1788, 1213–1228. [DOI] [PubMed] [Google Scholar]
- Goure, J. , Pastor, A. , Faudry, E. , Chabert, J. , Dessen, A. and Attree, I. (2004) The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72, 4741–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Haapalainen, M. , Engelhardt, S. , Kufner, I. , Li, C.M. , Nurnberger, T. , Lee, J. , Romantschuk, M. and Taira, S. (2011) Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol. Plant Pathol. 12, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson, S. , Schesser, K. , Persson, C. , Gaylov, E.E. , Rosqvist, R. , Homble, F. and Wolf‐Watz, H. (1996) The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact‐dependent membrane disrupting activity. EMBO J. 15, 5812–5823. [PMC free article] [PubMed] [Google Scholar]
- Hamada, D. , Kato, T. , Ikegami, T. , Suzuki, K.N. , Hayashi, M. , Murooka, Y. , Honda, T. and Yanagihara, I. (2005) EspB from enterohaemorrhagic Escherichia coli is a natively partially folded protein. FEBS J. 272, 756–768. [DOI] [PubMed] [Google Scholar]
- Hausner, J. , Hartmann, N. , Lorenz, C. and Büttner, D. (2013) The periplasmic HrpB1 protein from Xanthomonas spp. binds to peptidoglycan and to components of the type III secretion system. Appl. Environ. Microbiol. 79, 6312–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S.Y. (1998) Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36, 363–392. [DOI] [PubMed] [Google Scholar]
- He, S.Y. , Huang, H.C. and Collmer, A. (1993) Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell, 73, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Wang, B. , Qi, Y. and Lin, H. (2012) The Arg233Lys AQP0 mutation disturbs aquaporin 0–calmodulin interaction causing polymorphic congenital cataract. PLoS ONE, 7, e37637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet, E. and Bonas, U. (1997) hrpF of Xanthomonas campestris pv. vesicatoria encodes an 87‐kDa protein with homology to NoIX of Rhizobium fredii . Mol. Plant–Microbe Interact. 10, 488–498. [DOI] [PubMed] [Google Scholar]
- Ide, T. , Laarmann, S. , Greune, L. , Schillers, H. , Oberleithner, H. and Schmidt, M.A. (2001) Characterization of translocation pores inserted into plasma membranes by type III‐secreted Esp proteins of enteropathogenic Escherichia coli . Cell. Microbiol. 3, 669–679. [DOI] [PubMed] [Google Scholar]
- Ji, Z.L. , Song, C.F. , Lu, X.Z. and Wang, J.S. (2011) Two coiled‐coil regions of Xanthomonas oryzae pv. oryzae harpin differ in oligomerization and hypersensitive response induction. Amino Acids, 40, 381–392. [DOI] [PubMed] [Google Scholar]
- Jin, M. , Berrout, J. , Chen, L. and O'Neil, R.G. (2012) Hypotonicity‐induced TRPV4 function in renal collecting duct cells: modulation by progressive cross‐talk with Ca2+‐activated K+ channels. Cell Calcium, 51, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q. and He, S.Y. (2001) Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae . Science, 294, 2556–2558. [DOI] [PubMed] [Google Scholar]
- Johnson, S. , Roversi, P. , Espina, M. , Olive, A. , Deane, J.E. , Birket, S. , Field, T. , Picking, W.D. , Blocker, A.J. , Galyov, E.E. , Picking, W.L. and Lea, S.M. (2007) Self‐chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 282, 4035–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet, L. , Agrain, C. , Broz, P. and Cornelis, G.R. (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science, 302, 1757–1760. [DOI] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Kim, J.F. and Beer, S.V. (1998) HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.F. and Beer, S.V. (2000) hrp genes and harpins of Erwinia amylovora: a decade of discovery In: Fire Blight and its Causative Agent, Erwinia amylovora (Vanneste J.L., ed.), pp. 141–162. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Koraimann, G. (2003) Lytic transglycosylases in macromolecular transport systems of Gram‐negative bacteria. Cell. Mol. Life Sci. 60, 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori, T. , Matsushima, Y. , Nakamura, D. , Uralil, J. , Lara‐Tejero, M. , Sukhan, A. , Galán, J.E. and Aizawa, S.I. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science, 280, 602–605. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Ramos, A.R. , Morello, J.E. , Oh, H.S. and Collmer, A. (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189, 8059–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Klessig, D.F. and Nurnberger, T. (2001a) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis‐related gene HIN1 independent of extracellular calcium but dependent on mitogen‐activated protein kinase activity. Plant Cell, 13, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Klusener, B. , Tsiamis, G. , Stevens, C. , Neyt, C. , Tampakaki, A.P. , Panopoulos, N.J. , Noller, J. , Weiler, E.W. , Cornelis, G.R. et al. (2001b) HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion‐conducting pore in vitro. Proc. Natl. Acad. Sci. USA, 98, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.G. , Liu, H.X. , Cao, J. , Chen, L.F. , Gu, C. , Allen, C. and Guo, J.H. (2010) PopW of Ralstonia solanacearum, a new two‐domain harpin targeting the plant cell wall. Mol. Plant Pathol. 11, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.J. , Han, B. , Xu, M.Y. , Han, L.P. , Zhao, Y.Y. , Liu, Z.L. , Dong, H.S. and Zhang, C.L. (2014a) Plant growth enhancement and associated physiological responses are coregulated by ethylene and gibberellin following treatment with harpin protein Hpa1. Planta, 239, 831–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.J. , Zhao, Y.Y. , You, Z.Z. , Dong, H.S. and Zhang, C.L. (2014b) Harpin Hpa1 needs nitroxyl terminus to promote vegetative growth and leaf photosynthesis in Arabidopsis . J. Biosci. 39, 127–137. [DOI] [PubMed] [Google Scholar]
- Li, Y.R. , Che, Y.Z. , Zou, H.S. , Cui, Y.P. , Guo, W. , Zou, L.F. , Biddle, E.M. , Yang, C.H. and Chen, G.Y. (2011) Hpa2 required by HrpF to translocate Xanthomonas oryzae transcriptional activator‐like effectors into rice for pathogenicity. Appl. Environ. Microbiol. 77, 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. [DOI] [PubMed] [Google Scholar]
- Lindsey Rose, K.M. , Gourdie, R.G. , Prescott, A.R. , Quinlan, R.A. , Crouch, R.K. and Schey, K.L. (2006) The C terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest. Ophthalmol. Vis. Sci. 47, 1562–1570. [DOI] [PubMed] [Google Scholar]
- Liu, F.Q. , Liu, H.X. , Jia, Q. , Guo, X.J. , Zhang, S.J. , Wu, X. , Song, F. and Dong, H.S. (2006) The internal glycine‐rich motif and cysteine suppress several effects of the HpaGXooc protein in plants. Phytopathology, 96, 1052–1059. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham, A.P. , Birket, S.E. , Picking, W.D. , Picking, W.L. and Middaugh, C.R. (2008) pH sensitivity of type III secretion system tip proteins. Proteins, 71, 1830–1842. [DOI] [PubMed] [Google Scholar]
- Matteï, P.J. , Faudry, E. , Job, V. , Izoré, T. , Attree, I. and Dessen, A. (2011) Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 278, 414–426. [DOI] [PubMed] [Google Scholar]
- Maurel, C. (2007) Plant aquaporins: novel functions and regulation properties. FEBS Lett. 581, 2227–2236. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Verdoucq, L. , Luu, D.T. and Santoni, V. (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. [DOI] [PubMed] [Google Scholar]
- Meyer, D. , Cunnac, S. , Guéneron, M. , Declercq, C. , Van Gijsegem, F. , Lauber, E. , Boucher, C. and Arlat, M. (2006) PopF1 and PopF2, two proteins secreted by the type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 188, 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckli, N. and Auerbach, D. (2004) Quantitative β‐galactosidase assay suitable for high‐throughput applications in the yeast two‐hybrid system. Biotechniques, 36, 872–876. [DOI] [PubMed] [Google Scholar]
- Mueller, C.A. , Broz, P. , Muller, S.A. , Ringler, P. , Erne‐Brand, F. , Sorg, I. , Kuhn, M. , Engel, A. and Cornelis, G.R. (2005) The V‐antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science, 310, 674–676. [DOI] [PubMed] [Google Scholar]
- Mueller, C.A. , Broz, P. and Cornelis, G.R. (2008) The type III secretion system tip complex and translocon. Mol. Microbiol. 68, 1085–1095. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R. , Fullner, K.J. , Koonin, E.V. and Nester, E.W. (1996) A family of lysozyme‐like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. USA, 93, 7321–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyt, C. and Cornelis, G.R. (1999) Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33, 971–981. [DOI] [PubMed] [Google Scholar]
- Noël, L. , Thieme, F. , Nennstiel, D. and Bonas, U. (2002) Two novel type III‐secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the Hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.S. and Beer, S.V. (2007) AtHIPM, an ortholog of the apple HrpN‐interacting protein, is a negative regulator of plant growth and mediates the growth‐enhancing effect of HrpN in Arabidopsis . Plant Physiol. 145, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H.S. , Kvitko, B.H. , Morello, J.E. and Collmer, A. (2007) ) Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189, 8277–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki‐Ocwieja, T. , van Dijk, K. and Alfano, J.R. (2005) The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187, 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano, G.V. and Schesser, K. (2013) The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol. Res. 57, 237–245. [DOI] [PubMed] [Google Scholar]
- Qin, X. and Boron, W.F. (2013) Mutation of a single amino acid converts the human water channel aquaporin 5 into an anion channel. Am. J. Physiol. Cell Physiol. 305, C663–C672. [DOI] [PubMed] [Google Scholar]
- Racape, J. , Belbahri, L. , Engelhardt, S. , Lacombe, B. , Lee, J. , Lochman, J. , Marais, A. , Nicole, M. , Nurnberger, T. , Parlange, F. , Puverel, S. and Keller, H. (2005) Ca2+‐dependent lipid binding and membrane integration of PopA, a harpin‐like elicitor of the hypersensitive response in tobacco. Mol. Microbiol. 58, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C. (2003) The Plant Cell Wall. Oxford: Blackwell Publishing Ltd. [Google Scholar]
- Rossier, O. , Van den Ackerveken, G. and Bonas, U. (2000) HrpB2 and HrpF from Xanthomonas are type III‐secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38, 828–838. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Lozano, J.M. , del Mar Alguacil, M. , Bárzana, G. , Vernieri, P. and Aroca, R. (2009) Exogenous ABA accentuates the differences in root hydraulic properties between mycorrhizal and non mycorrhizal maize plants through regulation of PIP aquaporins. Plant Mol. Biol. 70, 565–579. [DOI] [PubMed] [Google Scholar]
- Ryndak, M.B. , Chung, H. , London, E. and Bliska, J.B. (2005) Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect. Immun. 73, 2433–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, S. , Li, X. , Gao, R. , You, Z. , Lü, B. , Liu, P. , Ma, Q. and Dong, H. (2012) Apoplastic and cytoplasmic location of harpin protein Hpa1Xoo plays different roles in H2O2 generation and pathogen resistance in Arabidopsis . Plant Mol. Biol. 79, 375–391. [DOI] [PubMed] [Google Scholar]
- Schoehn, G. , Di Guilmi, A.M. , Lemaire, D. , Attree, I. , Weissenhorn, W. and Dessen, A. (2003) Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas . EMBO J. 22, 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze, H. and Boch, J. (2010) TAL effector‐DNA specificity. Virulence, 1, 428–432. [DOI] [PubMed] [Google Scholar]
- Schraidt, O. , Lefebre, M.D. , Brunner, M.J. , Schmied, W.H. , Schmidt, A. , Radics, J. , Mechtler, K. , Galán, J.E. and Marlovits, T.C. (2010) Topology and organization of the Salmonella typhimurium type III secretion needle complex components. Plos Pathog. 6, e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöhamn, J. and Hedfalk, K. (2014) Unraveling aquaporin interaction partners. Biochim. Biophys. Acta, 1840, 1614–1623. [DOI] [PubMed] [Google Scholar]
- Song, C. and Yang, B. (2010) Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 23, 893–902. [DOI] [PubMed] [Google Scholar]
- Sugio, A. , Yang, B. and White, F.F. (2005) Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 18, 546–554. [DOI] [PubMed] [Google Scholar]
- Taufik, I. , Kedrov, A. , Exterkate, M. and Driessen, A.J. (2013) Monitoring the activity of single translocons. J. Mol. Biol. 425, 4145–4153. [DOI] [PubMed] [Google Scholar]
- Twyman, R.M. (1998) Advanced Molecular Biology. New York: BIOS Scientific Publishers Limited. [Google Scholar]
- Wager, B. , Faudry, E. , Wills, T. , Attree, I. and Delcour, A.H. (2013) Current fluctuation analysis of the PopB and PopD translocon components of the Pseudomonas aeruginosa type III secretion system. Biophys. J. 104, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Y. , Li, M. , Zhang, J.H. , Zhang, Y. , Zhang, G.Y. and Wang, J.S. (2007) Identification of a key functional region in harpins from Xanthomonas that suppresses protein aggregation and mediates harpin expression in E. coli . Mol. Biol. Rep. 34, 189–198. [DOI] [PubMed] [Google Scholar]
- Wang, X.Y. , Song, C.F. , Miao, W.G. , Ji, Z.L. , Wang, X.B. , Zhang, Y. , Zhang, J.H. , Hu, J.S. , Borth, W. and Wang, J.S. (2008) Mutations in the N‐terminal coding region of the harpin protein Hpa1 from Xanthomonas oryzae cause loss of hypersensitive reaction induction in tobacco. Appl. Microbiol. Biotechnol. 81, 359–369. [DOI] [PubMed] [Google Scholar]
- Ward, D.V. , Draper, O. , Zupan, J.R. and Zambryski, P.C. (2002) Peptide linkage mapping of the Agrobacterium tumefaciens vir‐encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA, 99, 11 493–11 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z.M. , Laby, R.J. , Zumoff, C.H. , Bauer, D.W. , He, S.Y. , Collmer, A. and Beer, S.V. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora . Science, 257, 85–88. [DOI] [PubMed] [Google Scholar]
- White, F.F. and Yang, B. (2009) Host and pathogen factors controlling the rice–Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. and White, F.F. (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant–Microbe Interact. 17, 1192–1200. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Peng, Q. , Zhang, Q. , Yi, X. , Choi, C.J. , Reedy, R.M. , Charkowski, A.O. and Yang, C.H. (2008) Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant–Microbe Interact. 21, 133–142. [DOI] [PubMed] [Google Scholar]
- Yip, C.K. , Finlay, B.B. and Strynadka, N.C. (2005) Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12, 75–81. [DOI] [PubMed] [Google Scholar]
- You, Z.Z. , Gao, R. , Tian, S. and Dong, H.S. (2013) Plant aquaporins: structure meets function as associating with sensing of Xanthomonas oryzae Hpa1 and subsequent signal transduction. Acta Phytopathol. Sin. 43, 232–248. [Google Scholar]
- Zahrl, D. , Wagner, M. , Bischof, K. , Bayer, M. , Zavecz, B. , Beranek, A. , Ruckenstuhl, C. , Zarfel, G.E. and Koraimann, G. (2005) Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology, 151, 3455–3467. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Wang, X. , Zhang, Y. , Zhang, G. and Wang, J. (2008) A conserved Hpa2 protein has lytic activity against the bacterial cell wall in phytopathogenic Xanthomonas oryzae . Appl. Microbiol. Biotechnol. 79, 605–616. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , MaGbanua, M.M. and White, F.F. (2000) Identification of two novel hrp‐associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]


