Summary
The circadian clock is the internal time‐keeping machinery in higher organisms. Cross‐talk between the circadian clock and a diverse range of physiological processes in plants, including stress acclimatization, hormone signalling, photomorphogenesis and defence signalling, is currently being explored. Recent studies on circadian clock genes and genes involved in defence signalling have indicated a possible reciprocal interaction between the two. It has been proposed that the circadian clock shapes the outcome of plant–pathogen interactions. In this review, we highlight the studies carried out so far on two model plant pathogens, namely Pseudomonas syringae and Hyaloperonospora arabidopsidis, and the involvement of the circadian clock in gating effector‐triggered immunity and pathogen‐associated molecular pattern‐triggered immunity. We focus on how the circadian clock gates the expression of various stress‐related transcripts in a prolific manner to enhance plant fitness. An understanding of this dynamic relationship between clock and stress will open up new avenues in the understanding of endogenous mechanisms of defence signalling in plants.
Keywords: biotic stress, circadian clock, defence signalling, plant immunity
Introduction
The biological processes of an organism change greatly during the day and night, and continuous entrainment by environmental factors results in the establishment of a biological rhythm. An endogenous biological rhythm with a period of approximately 24 h is known as the circadian rhythm (McClung, 2001). These circadian rhythms persist even in the absence of external cues for a definite time period, known as the free running time (Jones, 2009). The circadian rhythms are maintained by a well‐defined molecular machinery—the circadian clock. The circadian clock temporally coordinates biological processes with diurnal environmental changes, thus enhancing overall fitness. It is well documented that a diverse range of organisms, including cyanobacteria, algae, fungi, plants, flies, birds and mammals, possess their own circadian clock (Bell‐Pedersen et al., 2005). Plants, being sessile in nature, are dependent on this endogenous clock machinery to acclimatize themselves with environmental variations and to prepare for future changes. Many of the plant physiological and developmental processes are regulated by this endogenous regulator (Yakir et al., 2007). Since 1985, when the molecular study of the plant circadian clock began, the clock network has widened, and we are still trying to delineate it (Kloppstech, 1985; McClung, 2014). It has been documented that several abiotic factors, such as light and temperature, provide an input signal to the clock; in turn, the clock provides various output signals that influence the downstream processes (Kinmonth‐Schultz et al., 2013). In the model plant, Arabidopsis thaliana, the circadian clock framework is made up of various transcriptional–translational feedback loops (TTFLs). The circadian clock components constitute DNA‐binding transcription factors. Two MYB domain‐containing transcription factors, namely Circadian Clock Associated 1 (CCA1) and Late elongated HYpocotyl (LHY), and a pseudo‐response regulator (PRR), Timing Of CAB2 expression 1 (TOC1), were initially attributed as core clock components, as mutations in these genes showed severe alterations in phase, amplitude and period of the circadian clock (McClung, 2006). These three components together form a negative feedback loop, thus allowing their phase of the day‐specific expression. Negative feedback loops are formed by the majority of core clock proteins, where they inhibit their own expression at the transcriptional or translational level (Harmer, 2009). CCA1 and its close homologue LHY are morning‐expressed genes. CCA1/LHY bind to evening element (EE) present in the promoter region of TOC1, an evening‐expressed gene, and repress its expression (Alabadí et al., 2001). In turn, TOC1 directly represses the expression of CCA1, thus closing the loop (Gendron et al., 2012). The EE is a nine‐nucleotide ‘AAAATATCT’ conserved motif and the target site for the binding of several clock proteins, including CCA1, LHY and REVEILLE (RVE). Advancements in the biology of the circadian clock during the last decade have revealed several other core components, namely PRRs, LUX Arrhythmo (LUX), Early FLowering 3 and 4 (ELF3, 4) and the RVE group of transcription factors. These components form different feedback loops interconnected with the CCA1–LHY–TOC1 network (Fig. 1). The circadian clock network is more complex and robust in plants relative to other eukaryotes. This network is tightly regulated at the post‐transcriptional, post‐translational and epigenetic levels (Cui et al., 2013; Malapeira and Mas, 2013). The circadian clock model in higher plants is still evolving and is under developmental regulation (McClung, 2014).
Figure 1.
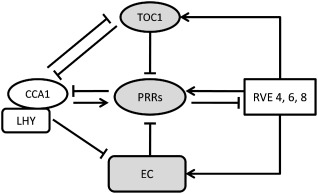
A simplified schematic representation of transcriptional regulatory networks of the Arabidopsis thaliana circadian clock. Arrows indicate activation and perpendicular bars represent repression. Circadian Clock Associated 1/Late elongated HYpocotyl (CCA1/LHY) represses the expression of Timing Of CAB2 expression 1 (TOC1) and evening‐expressed Early FLowering 3 and 4 (ELF4) and LUX Arrhythmo (LUX) via binding to evening element (EE). CCA1 also activates the transcription of PRR9 and PRR5. PRRs are transcriptional repressors and RVEs (REVEILLE) function as transcriptional activators. TOC1, an evening‐expressed component, represses the expression of other PRR's. EC, evening complex (ELF3, ELF4 and LUX together form an EC); PRRs, pseudo‐response regulators. Adapted from Hsu and Harmer (2014).
The application of genome‐wide ‘omics’ approaches has helped the scientific community to comprehend the clock‐related output pathways. A genome‐scale, time‐course expression profiling of A. thaliana revealed that about one‐third of global transcripts were circadian regulated (Covington et al., 2008). These large‐scale datasets provided a glance at the diversity of the circadian clock outputs. The clock output pathways vary from plant diurnal developmental processes, such as cotyledon and leaf movement (Engelmann and Johnsson, 1998), to biochemical and metabolic processes (Harmer et al., 2000), including acclimatization to abiotic and biotic stresses (Roden and Ingle, 2009; Sanchez et al., 2011) and hormone signalling (Robertson et al., 2009).
Roles of light and temperature in the modulation of plant immunity have been elucidated extensively (Genoud et al., 2002; Hua, 2013; Zeier et al., 2004); nevertheless, the role of the clock in defence signalling was under shade (McClung, 2011; Molina et al., 1997). Evidence from animal sciences has validated that the circadian clock plays an indispensable role in regulating the immune system (Scheiermann et al., 2013). The clock mutants of Drosophila display enhanced susceptibility to pathogen attack and a lower survival rate (Kuo et al., 2010). Interaction of the clock and immune system is known to be bidirectional in animal systems (Duhart et al., 2013). This reciprocal nature of clock–immune system interaction is now also evident in plants (Zhang et al., 2013). Studies hitherto have been based on two core clock components of the model system A. thaliana: CCA1 and LHY. These components were examined for their involvement in plant defence signalling towards anticipating pathogen attack (Wang et al., 2011; Zhang et al., 2013). Several lines of evidence have supported the involvement of the circadian clock in the enhancement of organismal adaptation towards predictable changes in nature (Bhardwaj et al., 2011; Goodspeed et al., 2012, 2013a, b; Roden and Ingle, 2009). How this interaction is beneficial for plants is discussed below.
Possible Interactions and Circadian Gating of Defence Genes
The circadian clock limits a physiological or biochemical process to a specific portion of the circadian cycle; this is known as circadian gating (Yakir et al., 2007). The circadian clock gates several signalling events to effectively employ resources. For instance, the expression of cold‐responsive genes is higher in the early morning and late evening, when the temperature drops to a sudden low level (Bieniawska et al., 2008). Likewise, the expression of defence‐related genes is required to orchestrate with the peak time of pathogen attack during a subjective day. Extensive data, gathered over the past few years, have provided unambiguous evidence supporting the involvement of the circadian clock in the defence response by plants (Bhardwaj et al., 2011; Wang et al., 2011; Zhang et al., 2013), and the studies indicate that the circadian clock is involved in the modulation of plant immunity.
The plant immune system can broadly be characterized at four levels. The presence of various physical barriers to restrict the entry of the pathogen to the inner core of plant tissues provides the first layer of defence (Campbell et al., 2012). Second is pathogen‐associated molecular pattern (PAMP)‐triggered immunity, or PTI, which involves the identification of various conserved PAMPs via pattern recognition receptors present at the plant cell surface, thus activating downstream defence signalling. This results in a cascade of events, including stomatal closure, the production of reactive oxygen species and callose deposition in the cell wall (Ingle et al., 2006; Nürnberger et al., 2004), to prevent the entry of bacterial pathogens or effectors secreted by pathogens. PTI serves as a basal defence system and offers a primary shield on pathogen attack. Some pathogens have evolved to evade PTI and secrete virulence effector molecules inside the host cell to suppress the signalling events induced by PTI and to develop effector‐triggered susceptibility (ETS) in the host (Jones and Dangl, 2006). Bacterial pathogens inject these effectors inside the host cell with the help of a well‐defined type III secretion system (T3SS), whereas oomycete pathogens secrete these effectors via the formation of invasion hyphae or haustoria (Büttner and He, 2009; Petre and Kamoun, 2014). A third level of the plant immune system has evolved to combat ETS, and is known as effector‐triggered immunity, or ETI. Specific resistance (R) proteins recognize cognate effectors and trigger a stronger defence response leading to, not strictly, programmed cell death (PCD) (Chisholm et al., 2006; Jones and Dangl, 2006). This basal and R‐mediated defence response sometimes induces a higher echelon of defence response, known as systemic acquired resistance (SAR) (Ryals et al., 1996). SAR is the fourth level of plant defence, producing whole‐plant resistance to a broad range of pathogens. The resistance is systemic in the sense that a local encounter leads to the activation of resistance to the rest of the plant organs via intra‐plant communication. SAR usually involves the accumulation of salicylic acid (SA) and leads to the induction of various SA‐mediated signalling events (Fu and Dong, 2013; Gaffney et al., 1993).
During PTI, PAMP perception by receptors at the cell surface initiates a mitogen‐activated protein kinase (MAPK) signalling cascade, resulting in the downstream activation of various physiological processes. Bhardwaj et al. (2011) analysed, in silico, the public microarray repository of the model system Arabidopsis and demonstrated that a leucine‐rich repeat receptor kinase, encoding a flagellin (bacterial PAMP) receptor FLS2 (FLagellin‐Sensitive 2), and downstream MAPK signalling components (MKK4/5–MAPK3/6–WRKY22 module) are circadian regulated, as they show peak expression in the subjective morning during a circadian cycle. This concurs with stomatal opening in the morning, when stomata are initially unable to prevent bacterial invasion. This implies that the basal defence level should be sufficiently adequate to counter pathogen attack whenever the physical barriers, i.e. stomata, are unable to prevent pathogen entry.
One of the earliest studies to provide a clue on clock–defence connection was of a pathogen‐responsive gene coding for glycine‐rich protein (GRP) in barley (Molina et al., 1997). Two GRPs (Hvgrp2 and Hvgrp3) were shown to be rapidly induced on attack by fungal pathogens. The expression level of Hvgrp3, an RNA‐binding GRP, was variable during different time courses in a light/dark cycle of 16 h/8 h. In Arabidopsis, a member of the GRP protein family, AtGRP7, is involved in the regulation of stomatal movement, stress responsiveness, floral transition and the circadian clock (Mangeon et al., 2010; Streitner et al., 2008; Zhang et al., 2013). GRP7 is also associated with PTI in plants. The evidence indicates the direct binding of GRP7 with several pattern recognition receptors encoding transcripts in response to pathogen attack. GRP7 is also one of the targets of a pathogen effector that inhibits its binding with pattern recognition receptor transcripts and results in the suppression of PTI (Nicaise et al., 2013). Although the expression of AtGRP7 is circadian regulated, it cannot affect the expression of core clock genes, and hence is designated as a ‘slave oscillator’ (Heintzen et al., 1997). A slave oscillator has a circadian output, but is dependent on a master or core oscillator for its function. AtGRP7 acts downstream of a core oscillator CCA1 via a putative interaction with an EE present in its promoter region.
Promoters enriched in EE could be hypothesized to act downstream of clock genes. Subsequently, to identify the set of defence‐related genes regulated by the circadian clock, we performed in silico analysis of Arabidopsis circadian‐regulated genes. A list of the circadian‐regulated genes was obtained from supporting information submitted with Covington et al. (2008). We further refined the list by selecting only those circadian‐regulated genes that were redundant in two independent experiments of Covington et al. (2008). The online web server Plant Gene Set Enrichment Analysis Toolkit (http://structuralbiology.cau.edu.cn/PlantGSEA/) was used for gene set enrichment analysis of the circadian‐regulated genes. Initially, 1595 circadian‐regulated genes were requested for analysis. After analysis of the enriched gene set, we identified 185 biotic stress‐ and defence‐related genes involved in various signalling pathways. We further screened their promoter sequences for the presence of EE of the CCA1‐binding site (CBS). We used the Athena program (http://www.bioinformatics2.wsu.edu/cgi‐bin/Athena/cgi/home.pl) to analyse the 1000‐bp upstream region up to the adjacent gene of these 185 genes (cut‐off P < 10−4). Of these 185 genes, 75 were identified to be significantly enriched with either EE (26) or CBS (42), or both (7) (Table 1). This analysis identified a number of putative defence‐related genes that could be the direct targets of CCA1/LHY and might be circadian regulated. The list includes genes involved in various signalling pathways which are triggered to anticipate pathogen attack. There is a possibility that some of the genes might act as slave oscillators that work on the framework in a similar manner to AtGRP7, although a more robust biochemical analysis is needed to support this hypothesis.
Table 1.
List of Arabidopsis defence‐related genes enriched with clock regulatory elements, identified from Covington et al. (2008)
| Locus | Gene name | Description | Clock element |
|---|---|---|---|
| At1g10760 | SEX1 (Starch Excess 1) | Encodes an α‐glucan, water dikinase | EE |
| At1g11260 | STP1 (Sugar Transporter 1) | H+/hexose co‐transporter | CBS |
| At1g18570 | MYB51 | Involved in indoleglucosinolate biosynthesis | EE |
| At1g20020 | LFNR2 | Encodes a leaf‐type ferredoxin:NADP(H) oxidoreductase | CBS |
| At1g20620 | CAT3 (Catalase 3) | Catalyses the breakdown of hydrogen peroxide (H2O2) | EE |
| At1g22410 | Class‐II DAHP synthetase | 3‐Deoxy‐7‐phosphoheptulonate synthase activity | CBS |
| At1g24100 | UGT74B1 | Involved in glucosinolate biosynthesis | CBS |
| At1g33590 | Leucine‐rich repeat (LRR) family protein | Signal transduction, defence response | CBS |
| At1g33600 | Leucine‐rich repeat (LRR) family protein | Signal transduction | CBS |
| At1g33970 | P‐loop‐containing NTP hydrolases | GTP binding | EE |
| At1g37130 | ATNR2 (Nitrate Reductase 2) | Encodes nitrate reductase structural gene | CBS |
| At1g53230 | TCP3 | Involved in leaf differentiation | CBS |
| At1g53320 | TLP7 (Tubby Like Protein 7) | Phosphoric diester hydrolase activity | CBS |
| At1g55210 | Disease resistance‐responsive protein | Defence response | CBS |
| At1g59870 | PDR8 (Pleiotropic Drug Resistance 8) | ATP‐binding cassette transporter | CBS |
| At1g61740 | Sulphite exporter TauE/SafE family protein | Regulation of plant‐type hypersensitive response | CBS |
| At1g64780 | AMT 1;2 (Ammonium Transporter 1;2) | Act as a high‐affinity transporter | CBS |
| At1g66760 | MATE efflux family protein | Antiporter activity, drug transmembrane transport | CBS |
| At1g66980 | SNC4 (Suppressor Of Npr1‐1 Constitutive 4) | An atypical receptor‐like kinase | CBS |
| At1g74710 | ICS1 (Isochorismate Synthase 1) | Mutants fail to accumulate salicylic acid | CBS |
| At1g75800 | PR thaumatin superfamily protein | Pathogenesis‐related protein | CBS |
| At1g78600 | LZF1 (Light‐Regulated Zinc Finger protein 1) | Negative regulation of defence response | CBS, EE |
| At2g02100 | PDF2.2 (Plant Defensin 2.2) | Predicted to encode a PR (pathogenesis‐related) protein | EE |
| At2g15890 | MEE14 (Maternal Effect Embryo Arrest 14) | Defence response to fungus | EE |
| At2g18290 | APC10 (Anaphase Promoting Complex 10) | Ubiquitin‐dependent protein catabolic process | CBS |
| At2g21660 | ATGRP7 (Glycine‐rich protein 7), CCR2 | Slave oscillator, promotes stomatal opening | EE |
| At2g22240 | MIPS2 (Inositol 3‐Phosphate Synthase 2) | Response to phosphate starvation | EE |
| At2g22540 | AGL22 (Agamous‐Like 22) | Nuclear protein that acts as a floral repressor | EE |
| At2g24550 | Unknown | Regulation of plant‐type hypersensitive response | CBS |
| At2g26170 | CYP711A1 (Cytochrome P450 Family) | Involved in flavonoid pathway | CBS |
| At2g29630 | PY (Pyrimidine Requiring) | Involved in thiamine biosynthesis | EE |
| At2g29650 | PHT4;1 (PHOSPHATE TRANSPORTER 4;1) | Inorganic phosphate transporter | CBS |
| At2g34690 | ACD11 (ACCELERATED CELL DEATH 11) | Transports the glycolipid precursor sphingosine | CBS |
| At2g37040 | PAL1 (Phenylalanine ammonia‐lyase) | l‐Phenylalanine catabolic process | CBS |
| At2g37220 | A chloroplast RNA‐binding protein | Involved in innate immune response | CBS,EE |
| At2g40460 | Major facilitator superfamily protein | Involved in oligopeptide transport | CBS |
| At2g41430 | LSR1 (Light Stress‐Regulated 1) | Hydrophilic protein lacking cysteine residues | EE |
| At2g42530 | COR15b (Cold Regulated 15B) | Response to cold and defence to fungus | CBS,EE |
| At2g46450 | CNGC12 (Cyclic Nucleotide‐Gated Channel) | Cation channel activity, RESPONSE TO FUNGUS | CBS |
| At3g05800 | AIF1 (AtBS1 Interacting Factor 1) | Brassinosteroid‐mediated signalling pathway | CBS,EE |
| At3g12580 | HSP70 (Heat Shock Protein 70) | Response to heat, ER stress, bacteria | CBS |
| At3g17020 | Adenine nucleotide α hydrolases‐like | Response to cold, molecule of fungal origin | EE |
| At3g21690 | MATE efflux family protein | Antiporter activity | EE |
| At3g27210 | Unknown protein | N‐terminal protein myristoylation | CBS |
| At3g46530 | RPP13 (Recognition of P. parasitica 13) | A nucleotide‐binding site leucine‐rich repeat (NBS‐LRR)‐type R protein | CBS |
| At3g58750 | CSY2 (CITRATE SYNTHASE 2) | Peroxisomal citrate synthase | EE |
| At4g16950 | RPP5 (Recognition of P. parasitica 5) | Putative TIR‐NB‐LRR receptors | EE |
| At4g16990 | RLM3 (Resistance To Leptosphaeria mac. 3) | Toll‐Interleukin receptor, ATP binding | CBS |
| At4g26850 | VTC2 (Vitamin C Defective 2) | GDP‐d‐glucose phosphorylase activity | CBS |
| At4g30650 | Low temperature and salt‐responsive protein | Defence response to fungus | EE |
| At4g32770 | SDX1 (Sucrose Export Defective 1) | Tocopherolcyclase involved in tocopherol synthesis | CBS |
| At4g33300 | ADR1 like 1 | Nucleotide‐binding leucine‐rich repeat (NB‐LRR) | CBS |
| At4g34590 | BZIP11 | Encodes a basic domain leucine zipper (bZip) | CBS |
| At4g37870 | PEPCK1 | Encodes a phosphoenolpyruvatecarboxykinase | CBS |
| At4g39980 | DHS1 (Phosphate Synthase 1) | Aromatic amino acid family biosynthetic process | CBS,EE |
| At5g02500 | HSP70‐1 (Heat Shock Protein 70‐1) | Member of heat shock protein 70 family | EE |
| At5g08330 | TCP11 (TCP Domain Protein 11) | Negative regulation of sequence‐specific DNA binding | CBS |
| At5g11670 | NADP‐ME2 (NADP‐Malic Enzyme 2) | Involved in malate metabolism | CBS |
| At5g15410 | DND1 (Defence No Death 1) | Mutated cyclic nucleotide‐gated cation channel | EE |
| At5g20630 | GLP3 (Germin‐Like Protein 3) | Cellular cation homeostasis | CBS,EE |
| At5g24530 | DMR6 (Downy Mildew Resistant 6) | Encodes a putative 2OG‐Fe(II) oxygenase | CBS |
| At5g35735 | Auxin‐responsive family protein | Negative regulation of defence response | CBS |
| At5g36910 | THIONIN 2.2 | Encodes a thionin, toxin receptor binding | EE |
| At5g40170 | RLP54 (Receptor Like Protein 54) | Kinase activity | EE |
| At5g42650 | CYTOCHROME P450 74A (CYP74A) | Allene oxide synthase | EE |
| At5g42810 | Inositol‐pentakisphosphate 2‐kinase 1 | Involved in the biosynthesis of phytic acid | CBS |
| At5g47220 | ERF2 (Ethylene Response Factor‐2) | Ethylene‐mediated signalling pathway | CBS |
| At5g48590 | DUF760 (Unknown function) | Starch biosynthetic process, systemic acquired resistance | CBS |
| At5g49450 | BZIP1 (Basic Leucine‐Zipper 1) | Positive regulator of plant tolerance to abiotic stresses | CBS |
| At5g51770 | Serine/threonine kinase family protein | Kinase activity, systemic acquired resistance | CBS |
| At5g51820 | PGM1 (Phosphoglucomutase 1) | Involved in controlling photosynthetic carbon flow | EE |
| At5g56280 | CSN6A | Subunit 6 of COP9 signalosome complex | CBS |
| At5g57110 | Autoinhibited Calcium ATPase, Isoform 8 | ATP biosynthetic process | CBS |
| At5g63780 | SHA1 (Shoot Apical Meristem Arrest 1) | Putative E3 ligase | CBS,EE |
| At5g66570 | OEE33 (Oxygen Evolving Complex 33) | An extrinsic subunit of photosystem II | CBS |
CBS, Circadian Clock Associated 1 (CCA1)‐binding site; EE, evening element.
A firm connection between the two diverse physiological processes clock and defence signalling has been demonstrated by Wang et al. (2011). A study of the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa), with regard to the delineation of the molecular basis of R gene‐mediated defence, revealed that R gene‐mediated resistance shares some common components with the basal defence mechanism. A leucine‐rich repeat receptor‐like kinase (LRR‐RLK, At1g35710) was identified as a connecting link between the two. Plants defective in Atg35710 were insensitive to a microbe‐associated molecular pattern (MAMP), EF‐Tu (elf18), and were highly susceptible to Hpa Emwa1 infection, demonstrating the functional inactivation of both R gene‐mediated and basal defence. The analysis of Arabidopsis mutants defective in R gene‐mediated defence (rrp4), to avirulent Hpa isolate Emwa1 (recognized by R protein RRP4), resulted in the identification of a unique set of 22 defence‐related genes critically involved in R gene‐mediated plant defence in response to Hpa. Many of these genes were rhythmic in terms of expression. Promoters of these 22 genes were also enriched with either EE and/or CBS, implicating their circadian regulation via CCA1/LHY.
The circadian clock therefore seems to modulate the outcome of plant–pathogen interactions via its core components. The precise mechanism is largely unknown. Several lines of evidence have demonstrated that the interaction of clock and immunity in plants is reciprocal. In other words, the pathogen attack may result in the modulation of circadian clock output and vice versa. An alteration in the rhythmicity of CCA1 expression was observed in plants on infection with Pseudomonas syringae and Hpa Emwa1. Even a 22‐amino‐acid synthetic peptide flg22 (mimics bacterial PAMP) could induce basal defence in plants with an altered rhythm and shortened period of CCA1 expression (Zhang et al., 2013). However, in the case of Hpa infection, the expression of another clock gene LHY remained unchanged (Zhang et al., 2013), implying different modes of regulation of the plant defence response via these core oscillators.
Timing of Plant–Pathogen Interaction
Various stages in the life cycle of an oomycete pathogen, starting from sporulation, spore dissemination with successive spore germination, formation of hyphae and establishment of primary haustoria, occur at a specific time in a subjective day (Donofrio and Delaney, 2001). Sporulation occurs mainly at night and spores are disseminated at dawn, soon after the clearance of dew (Slusarenko and Schlaich, 2003). The formation of penetration hyphae occurs approximately 12 h after spore germination and the development of primary haustoria starts approximately 24 h after spore germination (Donofrio and Delaney, 2001). Studies carried out on avirulent Hpa Emwa1 provided further evidence for a role of circadian component CCA1 (Wang et al., 2011) in defence signalling. The clock‐defective mutant of Arabidopsis, cca1, was more susceptible towards pathogen attack relative to the wild‐type, whereas CCA1‐overexpressing lines displayed enhanced resistance (Wang et al., 2011), as indicated by the extent of pathogen growth and spread. Defence genes show peak expression at midnight and at dawn, the time periods concurrent with the time of sporulation and spore dissemination, respectively. Similarly, plants infected at dusk were more susceptible to pathogen attack relative to those infected at dawn. Collectively, the data indicate that the circadian clock temporally coordinates the expression of defence‐related genes corresponding to the predictable time of pathogen attack.
In an analysis, wild‐type Arabidopsis plants and lines defective in RRP4 were infected with Hpa Emwa1, and the expression level of a cluster of defence genes, involved in R gene‐mediated PCD, was measured. In the first case, without a functional RRP4, the genes involved in basal defence showed a peak expression at 6, 16 and 24 h time points of pathogen attack, strengthening the plant defence against three major cyclic events of Hpa infection, namely spore germination, hyphal penetration and the formation of primary haustoria, respectively (Wang et al., 2011). Contrastingly, in the case of wild‐type infected plants that carry a functional RRP4, the peak expression of the defence genes at 6 h was abolished. Instead, prolonged expression of these genes was observed with peaks at 16 and 24 h. Based on the above observations, it could be hypothesized that early signalling events in the host, on encountering a pathogen, modulate the circadian clock and, in turn, the circadian clock gates the downstream defence signalling events. However, further investigation is required to delineate this complex signalling network and to answer how clock genes and RRP4 interact to shape the outcome of this interaction.
Studies on P. syringae–A. thaliana interaction have indicated that the extent of host susceptibility is dependent on the time of infection during a diurnal or circadian cycle. Initial studies on the effect of the light/dark (LD) cycle on the severity of pathogen attack indicated that plants infected during the light period are less susceptible than those infected in the dark. However, this was alleged to be caused by the sole effect of light conditions (Griebel and Zeier, 2008). Later experiments performed under constant light conditions provided the same results (Bhardwaj et al., 2011), indicating a role for the circadian clock. Circadian rhythms are endogenous and sustained for a free running period of approximately 24 h under constant conditions. Therefore, the constant conditions are best to study the outputs of the circadian clock. In order to understand the role of the circadian clock in the modulation of plant immunity against biotrophic pathogens, plants were infected with the bacterial pathogen P. syringae pv. tomato DC3000 (Pst DC3000) at specific time intervals and, later, the growth and spread of the pathogen were observed (Bhardwaj et al., 2011; Zhang et al., 2013). The results of two independent studies, carried out under constant and variable light conditions, provided an insight into the preferred time of bacterial pathogen–plant interaction. In one study, Arabidopsis leaves were infiltrated with bacterial solution at definite time intervals to infect plants. The wild‐type displayed greatest susceptibility when infected at subjective midnight (Bhardwaj et al., 2011). In another study, where spray infection was preferred to infect plants, the greatest susceptibility was observed in plants infected in the subjective morning (Zhang et al., 2013). Different modes of infection resulted in different outcomes. Spray infection may mimic the mode of bacterial infection in nature; therefore, in plants subjected to spray infection, stomata provide a barrier to restrict pathogen entry. In contrast, for bacterial infiltration, the extent of infection largely depends on the strength of the basal level of defence of the host. Variability in the degree of basal defence extensively correlates with stomatal movement. As a result, when stomata remain closed during the night, the intensity of basal defence remains lower. This temporal variation in the basal level of defence provides a clue that helps to confirm the phenomenon of clock modulation of the immune system.
Together, the above data indicate that the outcome of the interaction largely depends on light and the circadian clock. CCA1, in conjunction with LHY (as shown by Zhang et al., 2013), acts as a major link to establish the circadian gating of plant immunity and is also involved in the modulation of plant defence. Double mutants (cca1‐lhy) displayed severe alteration in rhythmicity of defence genes and were comparatively more susceptible towards pathogen attack than the single mutants. The extent of susceptibility was quantified as bacterial growth and the number of sporangiophores per cotyledon in the case of oomycete pathogens (Hpa). A two‐ to five‐fold increase in sporangiophore count and a significantly higher bacterial growth were observed in double mutants (Zhang et al., 2013). This suggests that both of the core clock components (CCA1 and LHY) act synergistically to modulate defence signalling in plants. In addition, two further clock components, namely ELF3 and ZTL‐4 (ZEITLUPE‐4), have also been reported to modulate plant immunity (Wang et al., 2011; Zhang et al., 2013).
Plant Growth Regulators, Defence and Clock
Plant hormones are known to be an integral part of the plant immune system (Robert‐Seilaniantz et al., 2011). Plant growth regulators involved in defence signalling mainly include SA, jasmonic acid (JA), auxin and ethylene. SA is a plant hormone involved in several different disease resistance mechanisms (Vlot et al., 2009). SAR in plants is associated with higher levels of SA accumulation. SA plays a central role in the establishment of SAR (Gaffney et al., 1993). SA accumulation in plants has been shown to be rhythmic, with a peak accumulation at subjective night (from 25 to 35 ng/g fresh weight) (Goodspeed et al., 2012). The time of peak SA accumulation is a few hours before the time of Hpa infection, hence preparing the plant for a predictable pathogen encounter. However, it is unknown how this small increase in SA level strengthens the plant defence against biotrophic pathogens (Goodspeed et al., 2012). SA biosynthesis is also regulated by the circadian clock. A clock component CHE (CCA1 HIKING EXPEDITION) has been shown to bind with the promoter element of ICS1 (ISOCHORISMATE SYNTHASE 1), a major gene involved in SA biosynthesis, and positively regulates its expression. Another gene NPR1 (NONEXPRESSER OF PR1) has been established as a connecting link between SA‐mediated defence signalling and the circadian clock (Zhou, 2014). Although SA accumulation is modulated by the circadian clock, clock‐mediated modulation of plant immunity by CCA1/LHY has been shown to be independent of SA (Zhang et al., 2013). This has been demonstrated by experiments carried out with A. thaliana defective in the ACD6 (accelerated cell death 6‐1) gene. The mutant line, acd6‐1, displays constitutive defence against the pathogen and accumulates higher levels of SA (Rate et al., 1999). Therefore, to analyse whether the higher level of SA accumulation complements the loss of disease resistance in clock‐defective mutants, the clock‐defective lines cca1 and lhy were crossed with acd6‐1 to generate double (acd6‐1‐cca1, acd6‐1‐lhy) and triple (acd6‐1‐cca1‐lhy) mutants. The double and triple mutants displayed a similar dwarf phenotype to acd6‐1 and a similar SA level, but plants were unsuccessful in recovering the loss of disease resistance because of the absence of a functional circadian clock (Zhang et al., 2013). These results indicate that an SA‐independent mechanism also exists in plant cells that links the circadian clock to disease resistance. A hormonal cross‐talk, activated in response to inputs from pathogens, results in a pathogen‐specific defence signalling cascade. Another plant growth regulator, JA, functions as an agonist to SA (Niki et al., 1998). JA is synthesized on pathogen perception and mediates defence signalling to encounter pathogen attack. JA is involved in the production of anti‐herbivore metabolites that protect plants against herbivore attack. JA is also involved in defence signalling against necrotrophic fungal pathogens (Antico et al., 2012). A clock component TIC (TIme for Coffee) function as a intermediate between JA and circadian clock signaling (Shin et al., 2012). JA accumulation peaks at the subjective day and is known to be circadian regulated (Goodspeed et al., 2012). The peak accumulation of JA corresponds to the peak time of herbivore attack. Similarly, a third plant hormone, ethylene, is also known to be involved in biotic stress signalling (Abeles et al., 1992; Thain et al., 2004). Ethylene plays a dual role in plant defence signalling. In some cases, ethylene is used by pathogens as a virulence factor and promotes pathogenesis, whereas, in other cases, it aids in stress alleviation (Van der Ent and Pieterse, 2012; van Loon et al., 2006). Broadly, the ethylene‐regulated defence responses depend on the outcome of interactions between multiple signals. Ethylene biosynthesis is also known to be circadian regulated (Thain et al., 2004). Another plant growth regulator, auxin, involved in stomatal movement, fluctuates in a circadian manner (Robertson et al., 2009). Auxin also regulates the clock as an input (Covington and Harmer, 2007).
Perspectives
The plant circadian clock is known to perform a wide variety of functions. Earlier, only abiotic factors, mainly light and temperature, were attributed as input signals to the clock. Recent advancements in clock–defence signalling in plants have indicated that biotic factors, such as pathogens, might also act as input signals to the circadian clock (Fig. 2). Consistent host–pathogen interactions in nature set down a circadian rhythm in the expression of related genes in both host and pathogen. This rhythmicity makes organisms more adaptive and comparatively fitter to encounter these routine interactions. As the defence response in plants is an energy‐consuming process, an appropriate synchronization of defence signalling events with other physiological processes is essential for the judicious utilization of energy resources. Therefore, according to the peak time of infection, the plant circadian rhythm orchestrates defence signals in such a way that it should be able to anticipate attacks in an enhanced manner. Studies carried out so far have not provided clear‐cut evidence to support CCA1‐mediated regulation of defence genes. As defence genes are not the only targets of CCA1 and a large proportion of global transcripts are speculated to be under the regulation of core clock components, a comprehensive biochemical and mechanistic approach is required to establish this cross‐talk. It is notable that, to determine the phenotypic outcome of the circadian clock, a comparative study under both light/dark and a constant free‐running period should be performed simultaneously. A time‐course genome‐wide profile of plant–pathogen interactions under different light conditions [constant light (LL), light/dark (LD) and constant dark (DD)] could help to identify those candidate defence genes which are solely regulated by the circadian clock and not light conditions. However, studies on clock–defence signalling hitherto have not covered these aspects. Abiotic factors, especially light and temperature, provide an input to the circadian clock via various receptors, such as phytochromes, that help to transmit signals from stimuli to clock, whereas, in the case of biotic factors, the connecting link between pathogen receptors and clock components is largely unknown (Fig. 2).
Figure 2.
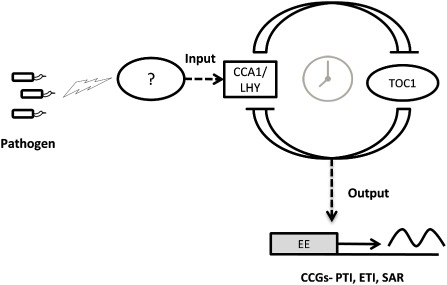
A model representing circadian clock–defence cross‐talk. Pathogen elicitors provide an input to the circadian clock via an unknown intermediate (depicted by a question mark). The circadian clock thus modulates the expression of various defence‐related circadian‐controlled genes (CCGs) via a putative binding of clock components to the promoter region of CCGs. CCA1/LHY, Circadian Clock Associated 1/Late elongated HYpocotyl; EE, evening element; ETI, effector‐triggered immunity; PTI, pathogen‐associated molecular pattern (PAMP)‐triggered immunity; SAR, systemic acquired resistance; TOC1, Timing Of CAB2 expression 1.
So far, all studies have been restricted to the model system Arabidopsis, and so it would be interesting to explore other plant species as well to identify the conservative nature of clock–defence cross‐talk. The circadian clock connections are vast and it is difficult to connect the loops without mathematical modelling. The mining of meaningful information from a large set of genome‐wide data could be easier if we used systems biological approaches coupled with biochemical analysis. A model built after utilizing the systems approach could help us to predict the phenotypic changes on modification of one of the components of this network. A delineated network might provide us with better candidate genes for crop improvement. Clock–defence cross‐talk is in its preliminary stage of research and several questions need to be addressed. The circadian clock of individual plant species is modulated in accordance with its surroundings. Thus, it would be interesting to identify the degree of conservation of this cross‐talk across a wider range of species from different geographical backgrounds. The identification of a connecting link between defence signals and the circadian clock is a major challenge. It is also important to define how hormonal signalling affects this cross‐talk. The combinatorial effect of major plant hormones on the outcome of plant pathogen interaction is difficult to understand. An integrative study of plant growth regulators, the circadian clock and defence signalling could demarcate the role of the above components in the establishment of disease resistance in plants.
Acknowledgements
We would like to thank Dr Laura Roden (University of Cape Town, South Africa) for useful suggestions and critical reading of the manuscript. We thank anonymous reviewers for their meaningful suggestions.
References
- Abeles, F.B. , Morgan, P.W. and Saltveit, M.E. Jr (1992) Ethylene in Plant Biology. San Diego, California: Academic Press, Inc. [Google Scholar]
- Alabadí, D. , Oyama, T. , Yanovsky, M.J. , Harmon, F.G. , Mas, P. and Kay, S.A. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science, 293, 880–883. [DOI] [PubMed] [Google Scholar]
- Antico, C.J. , Colon, C. , Banks, T. and Ramonell, K.M. (2012) Insights into the role of jasmonic acid‐mediated defenses against necrotrophic and biotrophic fungal pathogens. Front. Biol. 7, 48–56. [Google Scholar]
- Bell‐Pedersen, D. , Cassone, V.M. , Earnest, D.J. , Golden, S.S. , Hardin, P.E. , Thomas, T.L. and Zoran, M.J. (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. , Meier, S. , Petersen, L.N. , Ingle, R.A. and Roden, L.C. (2011) Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE, 6, e26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska, Z. , Espinoza, C. , Schlereth, A. , Sulpice, R. , Hincha, D.K. and Hannah, M.A. (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold‐responsive transcriptome. Plant Physiol. 147, 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, C.L. , Huang, J. and Payne, G.A. (2012) Defense at the perimeter: the outer walls and the gates. Plant Dis. Adv. Treat. 5, 103–120. [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Covington, M.F. and Harmer, S.L. (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS biol. 5, e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington, M.F. , Maloof, J.N. , Straume, M. , Kay, S.A. and Harmer, S.L. (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X. , Lu, F. , Li, Y. , Xue, Y. , Kang, Y. , Zhang, S. , Qui, Q. , Cui, X. , Zheng, S. , Liu, B. , Xu, X. and Cao, X. (2013) Ubiquitin‐specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donofrio, N.M. and Delaney, T.P. (2001) Abnormal callose response phenotype and hypersusceptibility to Peronospora parasitica in defense‐compromised Arabidopsis nim1‐1 and salicylate hydroxylase‐expressing plants. Mol. Plant–Microbe Interact. 14, 439–450. [DOI] [PubMed] [Google Scholar]
- Duhart, J.M. , Leone, M.J. , Paladino, N. , Evans, J.A. , Castanon‐Cervantes, O. , Davidson, A.J. and Golombek, D.A. (2013) Suprachiasmatic astrocytes modulate the circadian clock in response to TNF‐α. J. Immunol. 191, 4656–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, W. and Johnsson, A. (1998) Rhythms in organ movement In: Biological Rhythms and Photoperiodism in Plants (Lumsden P.J. and Millar A.J., eds), pp. 35–50. Oxford: BIOS Scientific Publishers. [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gendron, J.M. , Pruneda‐Paz, J.L. , Doherty, C.J. , Gross, A.M. , Kang, S.E. and Kay, S.A. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA‐binding transcription factor. Proc. Natl. Acad. Sci. USA, 109, 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud, T. , Buchala, A.J. , Chua, N.H. and Métraux, J.P. (2002) Phytochrome signalling modulates the SA‐perceptive pathway in Arabidopsis. Plant J. 31, 87–95. [DOI] [PubMed] [Google Scholar]
- Goodspeed, D. , Chehab, E.W. , Min‐Venditti, A. , Braam, J. and Covington, M.F. (2012) Arabidopsis synchronizes jasmonate‐mediated defence with insect circadian behavior. Proc. Natl. Acad. Sci. USA, 109, 4674–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed, D. , Chehab, E.W. , Covington, M.F. and Braam, J. (2013a) Circadian control of jasmonates and salicylates: the clock role in plant defense. Plant Sign. Behav. 8, e23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed, D. , Liu, J.D. , Chehab, E.W. , Sheng, Z. , Francisco, M. , Kliebenstein, D.J. and Braam, J. (2013b) Postharvest circadian entrainment enhances crop pest resistance and phytochemical cycling. Curr. Biol. 23, 1235–1241. [DOI] [PubMed] [Google Scholar]
- Griebel, T. and Zeier, J. (2008) Light regulation and daytime dependency of inducible plant defences in Arabidopsis: phytochrome signalling controls systemic acquired resistance rather than local defence. Plant Physiol. 147, 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L. (2009) The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L. , Hogenesch, J.B. , Straume, M. , Chang, H.S. , Han, B. , Zhu, T. , Wang, X. , Kreps, J.A. and Kay, S.A. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science, 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Heintzen, C. , Nater, M. , Apel, K. and Staiger, D. (1997) AtGRP7, a nuclear RNA‐binding protein as a component of a circadian‐regulated negative feedback loop in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.Y. and Harmer, S.L. (2014) Wheels within wheels: the plant circadian system. Trends plant sci. 19, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J. (2013) Modulation of plant immunity by light, circadian rhythm, and temperature. Curr. Opin. Plant Biol. 16, 406–413. [DOI] [PubMed] [Google Scholar]
- Ingle, R.A. , Carstens, M. and Denby, K.J. (2006) PAMP recognition and the plant–pathogen arms race. Bioessays, 28, 880–889. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, M.A. (2009) Entrainment of the Arabidopsis circadian clock. J. Plant Biol. 52, 202–209. [Google Scholar]
- Kinmonth‐Schultz, H.A. , Golembeski, G.S. and Imaizumi, T. (2013) Circadian clock‐regulated physiological outputs: dynamic responses in nature. Semin. Cell Dev. Biol. 24, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppstech, K. (1985) Diurnal and circadian rhythmicity in the expression of light‐induced nuclear messenger RNAs. Planta, 165, 502–506. [DOI] [PubMed] [Google Scholar]
- Kuo, T.H. , Pike, D.H. , Beizaeipour, Z. and Williams, J.A. (2010) Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFκB Relish. BMC Neurosci. 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Geraats, B.P. and Linthorst, H.J. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. [DOI] [PubMed] [Google Scholar]
- Malapeira, J. and Mas, P. (2013) A chromatin‐dependent mechanism regulates gene expression at the core of the Arabidopsis circadian clock. Plant Sign. Behav. 8, e24079–e24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeon, A. , Junqueira, R.M. and Sachetto‐Martins, G. (2010) Functional diversity of the plant glycine‐rich proteins superfamily. Plant Signal. Behav. 5, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C.R. (2001) Circadian rhythms in plants. Annu. Rev. Plant Biol. 52, 139–162. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2006) Plant circadian rhythms. Plant Cell, 18, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C.R. (2011) The genetics of plant clocks. Adv. Genet. 74, 105–139. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2014) Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, A. , Mena, M. , Carbonero, P. and García‐Olmedo, F. (1997) Differential expression of pathogen‐responsive genes encoding two types of glycine‐rich proteins in barley. Plant Mol. Biol. 33, 803–810. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Joe, A. , Jeong, B.R. , Korneli, C. , Boutrot, F. , Westedt, I. , Staiger, D. , Alfano, J.R. and Zipfel, C. (2013) Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 32, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki, T. , Mitsuhara, I. , Seo, S. , Ohtsubo, N. and Ohashi, Y. (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis‐related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39, 500–507. [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Petre, B. and Kamoun, S. (2014) How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12, e1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N. , Cuenca, J.V. , Bowman, G.R. , Guttman, D.S. and Greenberg, J.T. (1999) The gain‐of‐function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell, 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Robertson, F.C. , Skeffington, A.W. , Gardner, M.J. and Webb, A.A. (2009) Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 69, 419–427. [DOI] [PubMed] [Google Scholar]
- Roden, L.C. and Ingle, R.A. (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. Plant Cell, 21, 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, A. , Shin, J. and Davis, S.J. (2011) Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann, C. , Kunisaki, Y. and Frenette, P.S. (2013) Circadian control of the immune system. Nat. Rev. Immunol. 13, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J. , Heidrich, K. , Sanchez‐Villarreal, A. , Parker, J.E. and Davis, S.J. (2012) TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time‐of‐day regulation of jasmonate signaling in Arabidopsis Plant Cell. 24, 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarenko, A.J. and Schlaich, N.L. (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Pathol. 4, 159–170. [DOI] [PubMed] [Google Scholar]
- Streitner, C. , Danisman, S. , Wehrle, F. , Schöning, J.C. , Alfano, J.R. and Staiger, D. (2008) The small glycine‐rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana . Plant J. 56, 239–250. [DOI] [PubMed] [Google Scholar]
- Thain, S.C. , Vandenbussche, F. , Laarhoven, L.J. , Dowson‐Day, M.J. , Wang, Z.Y. , Tobin, E.M. and Van Der Straeten, D. (2004) Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 136, 3751–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent, S. and Pieterse, C.M. (2012) Ethylene: multi‐tasker in plant–attacker interactions. Annu. Plant Rev. 44, 343–378. [Google Scholar]
- Vlot, A.C. , Dempsey, D.M.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Barnaby, J.Y. , Tada, Y. , Li, H. , Tör, M. , Caldelari, D. , Lee, D.‐U. , Fu, X. and Dong, X. (2011) Timing of plant immune responses by a central circadian regulator. Nature, 470, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir, E. , Hilman, D. , Harir, Y. and Green, R.M. (2007) Regulation of output from the plant circadian clock. FEBS J. 274, 335–345. [DOI] [PubMed] [Google Scholar]
- Zeier, J. , Delledonne, M. , Mishina, T. , Severi, E. , Sonoda, M. and Lamb, C. (2004) Genetic elucidation of nitric oxide signalling in incompatible plant–pathogen interactions. Plant Physiol. 136, 2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Xie, Q. , Anderson, R.G. , Ng, G. , Seitz, N.C. , Peterson, T. , McClung, C.R. , McDowell, J.M. , Kong, D. , Kwak, J.M. and Lu, H. (2013) Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9, e1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M. (2014) The molecular interplay between the circadian clock and the plant immune signal, salicylic acid. PhD Thesis, Duke University. Available at http://hdl.handle.net/10161/8677.


