Summary
Several studies have reported effects of the plant phenolic acids cinnamic acid (CA) and salicylic acid (SA) on the virulence of soft rot enterobacteria. However, the mechanisms involved in these processes are not yet fully understood. Here, we investigated whether CA and SA interfere with the quorum sensing (QS) system of two P ectobacterium species, P . aroidearum and P . carotovorum ssp. brasiliense, which are known to produce N‐acyl‐homoserine lactone (AHL) QS signals. Our results clearly indicate that both phenolic compounds affect the QS machinery of the two species, consequently altering the expression of bacterial virulence factors. Although, in control treatments, the expression of QS‐related genes increased over time, the exposure of bacteria to non‐lethal concentrations of CA or SA inhibited the expression of QS genes, including expI, exp R, PC 1_1442 (lux R transcriptional regulator) and luxS (a component of the AI‐2 system). Other virulence genes known to be regulated by the QS system, such as pec S, pel, peh and yhe O, were also down‐regulated relative to the control. In agreement with the low levels of expression of exp I and exp R, CA and SA also reduced the level of the AHL signal. The effects of CA and SA on AHL signalling were confirmed in compensation assays, in which exogenous application of N‐(β‐ketocaproyl)‐l‐homoserine lactone (eAHL) led to the recovery of the reduction in virulence caused by the two phenolic acids. Collectively, the results of gene expression studies, bioluminescence assays, virulence assays and compensation assays with eAHL clearly support a mechanism by which CA and SA interfere with Pectobacterium virulence via the QS machinery.
Keywords: bacterial virulence, cinnamic acid, Pectobacterium, quorum sensing, salicylic acid
Introduction
Pectobacterium spp., which were formerly considered to be members of the genus Erwinia, are plant pathogens of the Enterobacteriaceae family that cause soft rot on many types of fruit, ornamentals and vegetables (Ma et al., 2007; Park et al., 2012; Toth et al., 2003, 2011). Like other soft rot enterobacteria, Pectobacterium induces characteristic soft rot decay through the disruption of host cell integrity, which is promoted by a variety of plant cell wall‐degrading enzymes (PCWDEs) secreted by the bacterium (Davidsson et al., 2013; Toth and Birch, 2005). The synthesis of PCWDEs in Pectobacterium and other soft rot enterobacteria is mediated by quorum sensing (QS) (Barnard and Salmond, 2007; de Kievit and Iglewski, 2000; Pirhonen et al., 1993).
QS is a cell‐to‐cell communication system that allows bacteria to monitor the environment and to modulate gene expression according to population density (Fuqua et al., 1994). It is largely governed by the production, secretion, detection and response to freely diffusible chemical signal molecules, called auto‐inducers. Of these molecules, the most studied are the N‐acyl‐homoserine lactones (AHLs) (Fuqua et al., 2001; Waters and Bassler, 2005). QS plays an essential role in the virulence of pathogenic bacteria (Antunes et al., 2010; Deep et al., 2011; Rutherford and Bassler, 2012). Among soft rot enterobacteria species, the QS systems of Pectobacterium atrosepticum, Pectobacterium carotovorum and Dickeya spp. have been the subject of several studies (Crépin et al., 2012; Faure and Dessaux, 2007; Liu et al., 2008; Nasser et al., 1998).
Pectobacteria utilize 3‐oxohexanoyl‐l‐homoserine lactone (3‐oxo‐C6‐AHL) and 3‐oxooctanoyl‐l‐homoserine lactone (3‐oxo‐C8‐AHL) as QS signalling molecules (Jafra et al., 2006; Pollumaa et al., 2012). These molecules are synthesized by ExpI and detected by the receptor protein ExpR (Barnard and Salmond, 2007; Engebrecht and Silverman, 1984; Liu et al., 2008). ExpR is a transcriptional regulator that undergoes a conformational change, which subsequently decreases the expression of the rsmA gene. The products of this gene are negative regulators of the genes involved in the synthesis of PCWDEs (Barnard and Salmond, 2007; Barnard et al., 2007; Pollumaa et al., 2012; Toth et al., 2003). Therefore, the repression of rsmA ensures high levels of expression of PCWDEs, as well as type III secretion and other virulence factors (Burr et al., 2006; Cui et al., 2005; Smadja et al., 2004). Collectively, AHL‐mediated effects switch bacteria into a brute‐force mode to promote the rotting of the host tissue (Liu et al., 2008).
The recognition of the crucial role of QS in bacterial virulence has made QS an attractive target for new control strategies against bacterial infections. QS inhibitors have been shown to successfully reduce or attenuate the virulence of Pseudomonas aeruginosa (Hentzer et al., 2003; O'Loughlin et al., 2013), Vibrio harveyi (Manefield et al., 2000), Staphylococcus aureus (Murray et al., 2014) and P. carotovorum (Manefield et al., 2001). Moreover, the expression of heterologous AHL lactonase and co‐inoculation with bacteria producing this or other AHL‐degrading enzymes have been shown to be effective, novel biocontrol strategies for the management of P. carotovorum and Erwinia amylovora (Dong et al., 2001; Molina et al., 2003, 2005).
The development of QS interference strategies is becoming an increasingly important approach for the control of bacterial infections (Defoirdt et al., 2013; Galloway et al., 2012; Helman and Chernin, 2015; Kalia, 2013). Some plant phenolic compounds, such as coumaric acid and salicylic acid (SA), have been shown to interfere with the QS system of the plant pathogens Agrobacterium tumefaciens and P. carotovorum (Bodini et al., 2009; Lagonenko et al., 2013; Yuan et al., 2007). The elucidation of the mechanisms underlying the action of these compounds may pave the way to the development of effective measures for the control of soft rot pectobacteria.
Lagonenko et al. (2013) showed that SA inhibits the biofilm formation and motility of P. carotovorum and Pseudomonas syringae pv. syringae. Moreover, SA has been found to reduce infection in tissue‐cultured potato infected by Dickeya solani (Czajkowski et al., 2015). In agreement with these findings, we have shown recently that non‐lethal concentrations of several plant phenolic compounds, including SA and cinnamic acid (CA), affect several virulence‐associated traits and reduce the virulence of several Pectobacterium strains in different hosts (Joshi et al., 2015). To unravel the mechanism by which plant secondary metabolites, such as CA and SA, affect soft rot bacterial virulence, we investigated the effect of these compounds on the expression patterns of selected virulence‐related genes in Pectobacterium aroidearum and P. carotovorum ssp. brasiliense. The virulence genes were categorized as follows: (i) QS system (AI‐1 and AI‐2) genes; (ii) QS‐regulated genes; and (iii) other genes (related to membrane transporters, type III secretion, flagella and motility). Next, we attempted to elucidate the effect of CA and SA on AHL production using reporter bacteria that detect QS signals. Finally, bacterial infection assays were performed in the presence and absence of CA and SA to evaluate the effects of these compounds on virulence. Virulence assays were also performed to assess the ability of exogenous application of QS signal molecules to compensate for the impaired virulence caused by the phenolic compounds.
Results
Expression of QS genes in the presence of phenolic compounds
We have shown previously that several plant phenolic compounds, including CA and SA, influence virulence‐associated traits and reduce the virulence of several Pectobacterium species on different hosts when applied at non‐lethal concentrations (Joshi et al., 2015). To further investigate the mechanism by which plant phenolic compounds influence bacterial virulence factors, we assessed the effects of CA and SA on the expression of selected genes of two bacterial strains representing two Pectobacterium species: P. aroidearum PC1 and P. carotovorum ssp. brasiliense Pcb1692. Exponentially growing bacterial cells of these strains were used to inoculate fresh Lysogeny‐Broth (LB) with or without non‐lethal concentrations of CA or SA (0.25 and 0.21 mg/mL, respectively) at an initial bacterial concentration of 107 colony‐forming units (CFU)/mL. Gene expression levels in the presence of the compounds were measured by quantitative reverse transcription polymerase chain reaction (qRT‐PCR) at three time points that corresponded to different bacterial growth phases: acclimatization [1 h after inoculation (hai)], exponential phase (8 hai) and stationary phase (24 hai), as described in Experimental Procedures. At each of the mentioned time points, bacterial cell growth was evaluated by serial dilution plating to verify that bacterial concentrations of SA‐/CA‐treated and control cultures were at similar levels (Fig. S1, see Supporting Information).
Under these conditions, in the two strains, both CA and SA affected the expression of expI/expR QS system genes (Fig. 1) and the expression patterns of several genes whose expression is mediated by this QS system (Fig. 2). In control treatments, the relative expression of expI and expR increased with time and, after 24 h of growth, the expression of these genes was increased significantly (P = 0.05). At this time point, we observed an increase of about 2.5‐ and four‐fold for expI in PC1 and Pcb1692, respectively, and an increase of about 10‐ and four‐fold for expR in strains PC1 and Pcb1692, respectively, relative to the expression of these genes measured after 1 h of growth (Fig. 1). In contrast, during the same time period, bacterial cultures of both strains grown in the presence of CA and SA did not display such a time‐dependent increase in the expression of these genes, which was significantly lower than that of the untreated bacteria.
Figure 1.
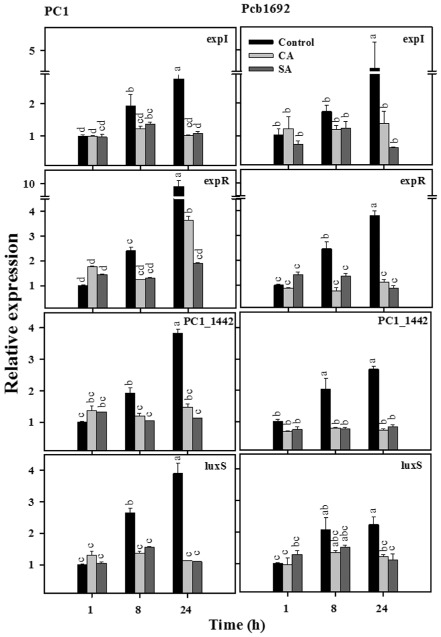
Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on transcript levels of quorum sensing (QS) system genes in P ectobacterium aroidearum PC1 and P ectobacterium carotovorum ssp. brasiliense Pcb1692. The transcript levels of QS (AI‐1 system) genes [exp I, exp R , PC 1_1442 (lux R transcription regulator)] and a QS (AI‐2 system) gene (lux S) were determined by quantitative reverse transcription polymerase chain reaction (qRT‐PCR) of cDNA samples prepared from RNA extracts of cultures grown in Lysogeny‐Broth (LB) (28 °C, continuous shaking at 150 rpm) with or without the phenolic acids. The data represent the results from one representative experiment, out of three with similar results. Means ± standard errors (SE) of the relative expression of each gene (three replicates per treatment) are shown. Different letters indicate significant differences (P = 0.05) among treatments for each time point in each gene/strain combination.
Figure 2.
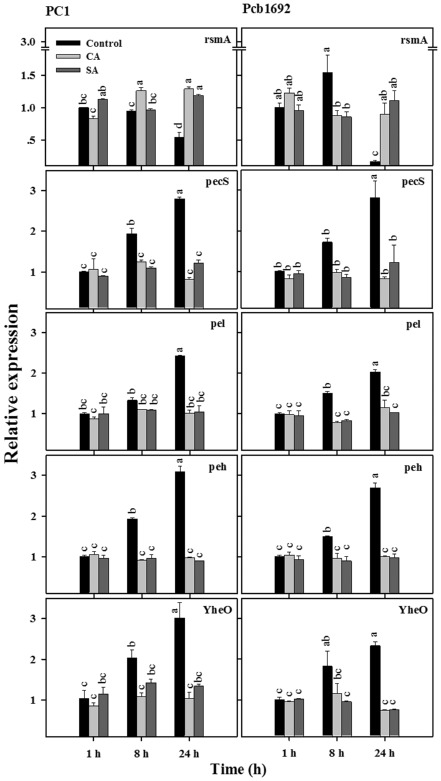
Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on transcript levels of quorum sensing‐controlled genes in P ectobacterium aroidearum PC1 and P ectobacterium carotovorum ssp. brasiliense Pcb1692. The transcript levels of rsmA, pec S, pel, peh and yhe O in DNA‐free RNA prepared from the bacterial strains grown in Lysogeny‐Broth (LB) medium (28 °C, continuous shaking at 150 rpm) with or without CA and SA were determined by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). Means ± standard errors (SE) of relative expression of each gene (three replicates per treatment) are shown. Different letters indicate significant differences (P = 0.05) among treatments for each time point in each gene/strain combination.
The expression levels of PC1_1442 (luxR transcription regulator belonging to the AI‐1 system) also increased significantly (P = 0.05) with time under control conditions in both strains, whereas exposure to the phenolic compounds did not allow for an increase in PC_1442 expression over time (Fig. 1). Interestingly, similar results were also observed for the expression of the luxS gene, which is part of the AI‐2 system (Fig. 1).
Overall, under control conditions, a significant increase (P = 0.05) in the relative expression of all of the tested QS system genes was observed following 8 h of growth, whereas no such increase in the relative expression of these genes was observed following exposure to CA and SA (Fig. 1).
Expression of QS‐regulated genes in the presence of phenolic compounds
To further characterize the response of the Pectobacterium strains to CA and SA, we also analysed the expression of genes contributing to production of PCWDEs: pecS, pel, peh and yheO, and their negative regulator rsmA (Fig. 2). These genes are located downstream of the QS cascade and are considered to be QS dependent (Barnard and Salmond, 2007; Barnard et al., 2007; Kõiv et al., 2013; Liu et al., 2008; Smith et al., 2006). The relative expression of yheO increased significantly (P = 0.05), in a gradual manner, during the experiments, reaching a maximum at 24 h in the control cultures. A pattern of increased expression over time was also observed for pecS, pel and peh. In contrast, the expression of these genes in the CA‐ and SA‐treated cultures did not change substantially with time under the experimental conditions, with significantly lower expression levels (P = 0.05) observed in these cultures relative to controls after 24 h (Fig. 2).
In contrast with pecS, pel, peh and yheO, expression levels of the post‐transcriptional master regulator of PCWDEs, rsmA, decreased significantly (P = 0.05) at the stationary phase (24 h) in the controls of both strains (Fig. 2). Exposure to the phenolic compounds did not alter significantly rsmA expression over time in either strain. In contrast with the untreated cells, for which a significant (P = 0.05) reduction in the expression of rsmA was observed at 24 h relative to 8 h, CA‐ and SA‐treated cells did not exhibit this reduction in the relative expression of this gene. Moreover, relative rsmA expression values at 24 h were significantly (P = 0.05) higher in the CA‐ and SA‐treated cells relative to the controls, with the exception of the SA treatment in PC1, in which a higher level of expression was observed in the control.
Expression of other virulence genes in the presence of phenolic compounds
In this study, we also assessed the effects of CA and SA on the relative expression of genes that have been associated with virulence, but whose expression is not necessarily associated with QS (Table 1). This group of genes included efflux pump genes (acrD and PC1_3330), a membrane transporter gene (nssA), hypersensitive response genes (hrpN and hrpL) and a stress‐related gene (mtlD). Overall, in contrast with the effects observed among QS and QS‐regulated genes, the expression of most of the above genes was not affected by exposure to CA or SA over time (Fig. 3). Moreover, QS‐independent transcription activators (hor and hrpS), which were also tested, were not affected by phenolics treatment (Fig. S2, see Supporting Information).
Table 1.
Genes assessed in this study
| Genes/gene ID | Description of function | References | |
|---|---|---|---|
| QS (AI‐1 system) |
expI
(8135146) |
QS signal generator; acyl‐homoserine lactone synthase | Nasser et al. (1998) |
|
expR
(8135145) |
QS transcriptional regulator | Nasser et al. (1998) | |
|
PC1_1442
(8132381) |
Transcriptional regulator of LuxR‐like proteins; alignment score showing 71% similarity with expR | NCBI | |
| QS (AI‐2) |
luxS
(8134173) |
S‐Ribosylhomocysteinase; QS autoinducer‐2 production | Schauder et al. (2001); Taga et al. (2001) |
| Genes regulated by QS |
rsmA
(8134182) |
Controls the production of PCWDEs, carbon storage and cell motility | Broberg et al. (2014); Kõiv et al. (2013) |
|
pecS
(8133198) |
Transcriptional regulator that controls the production of various virulence factors including pectinase and cellulase. | Hommais et al. (2008); Praillet et al. (1996); Reverchon et al. (1994) | |
|
pel
(8133114) |
Secretion of pectate lyase | NCBI | |
|
peh
(8131918) |
Secretion of polygalacturonase | NCBI | |
|
PC1_2249
(yheO) (8133193) |
yheO domain‐containing protein; DNA‐binding protein controlling the production of pectolytic enzymes | Lee et al. (2013) | |
| Other genes |
acrD
(8134910) |
Multidrug efflux pump | NCBI |
|
nssA
(8130937) |
Putative sodium/sulfate symporter or membrane transporter | NCBI | |
|
mtlD
(2885531) |
Mannitol dehydrogenase; protects against multiple types of stress (high salt and oxidative stress) | Hema et al. (2014) | |
|
hrpN
(8133133) |
Hypersensitive response gene | Jang et al. (2006) | |
|
hrpL
(8133148) |
Controls expression of hrp genes, including hrpN; elicits hypersensitive response | Wei and Beer (1995) | |
|
PC1_3330
(8134303) |
Efflux pump membrane protein; multidrug resistance protein | NCBI |
Gene IDs mentioned in this table were taken from GenBank (National Center for Biotechnology Information, NCBI).
PCWDEs, plant cell wall‐degrading enzymes; QS, quorum sensing.
Figure 3.
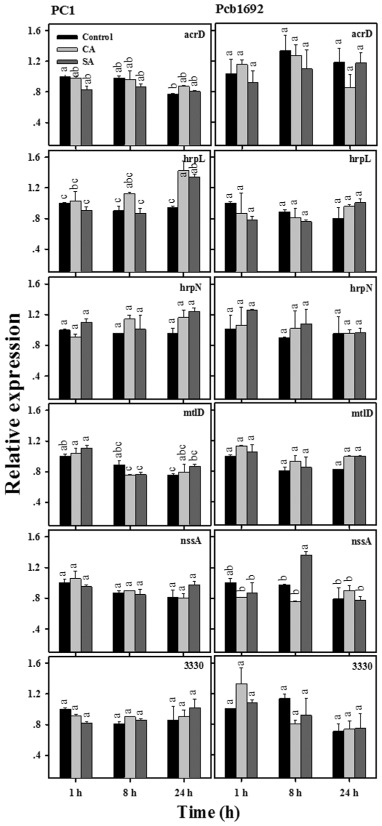
Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on transcript levels of other genes (associated with transporters, efflux pumps and multidrug resistance) in P ectobacterium aroidearum PC1 and P ectobacterium carotovorum ssp. brasiliense Pcb1692. The transcript levels of acr D, hrp L, hrp N, mtl D, nss A and PC 1_3330 in DNA‐free RNA prepared from the strains grown in Lysogeny‐Broth (LB) medium (28 °C, continuous shaking at 150 rpm) with or without CA and SA were determined by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). Means ± standard errors (SE) of relative expression of each gene (three replicates per treatment) are shown. Different letters indicate significant differences (P = 0.05) among treatments for each time point in each gene/strain combination.
Effects of CA and SA on the production of QS signalling molecules
AHLs are known signalling molecules in QS systems that control the synthesis of PCWDEs in Pectobacterium spp. and other soft rot bacteria (de Kievit and Iglewski, 2000), among other processes. To gain further insight into the effects of CA and SA on QS and on the virulence of Pectobacterium, we assessed how these compounds influence the production of AHLs, using bacterial reporter strains. The first assay was based on the reporter bacterium Chromobacterium violaceum CV026. In the presence of external AHLs, this strain produces the purple pigment violacein (McClean et al., 1997). Figure 4A clearly shows that PC1 and Pcb1692 cells pretreated with CA or SA induced lower purple coloration in C. violaceum CV026. The effect was also studied quantitatively using an Escherichia coli strain carrying the bioluminescence QS reporter plasmid pSB401. Supernatants of PC1 and Pcb1692 cultures grown with or without (control) exposure to CA or SA were compared for their ability to induce bioluminescence in the reporter strain. Supernatants of cell cultures that were exposed to both SA and CA induced lower bioluminescence levels, as compared with the bioluminescence induced by supernatants of control cells (Fig. 4B).
Figure 4.
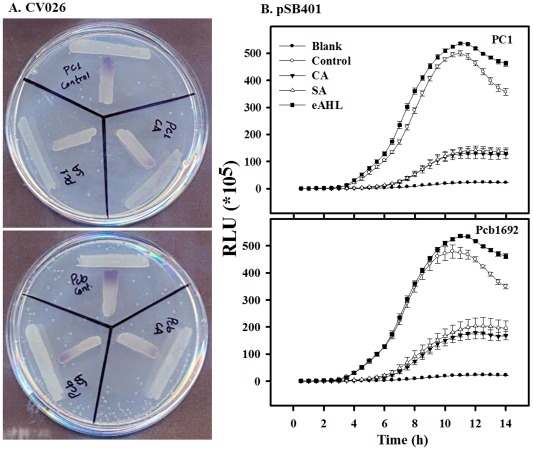
Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on the production of quorum sensing (QS) signalling molecules by P ectobacterium aroidearum PC1 and P ectobacterium carotovorum ssp. brasiliense Pcb1692. (A) Purple colour exhibited by CV026, as response to N‐acyl‐homoserine lactones (AHLs) produced by PC1 (top) or Pcb1692 (bottom) that were grown with (control) and without non‐lethal concentrations of CA or SA. (B) Intensity of luminescence produced by E scherichia coli pSB401 induced by supernatants of strains PC1 (top) and Pcb1692 (bottom) grown with or without non‐lethal concentrations of CA or SA. Luminescence (250 ms) and absorbance (600 nm) were measured every 30 min for 14 h, and the relative luminescence (RLU = LU/OD 600 nm) was calculated. Blank was E . coli pSB401 grown in the absence of supernatants of Pectobacterium strains, and also E . coli pSB401 was supplemented with exogenous N‐(β‐ketocaproyl)‐l‐homoserine lactone (eAHL) at 100 nm. Each data point represents the mean ± standard error (SE) of eight replicates per treatment of one experiment, representative of two independent experiments with similar results. OD, optical density; LU, light units; RLU, relative light units.
Restoration of virulence of CA‐ or SA‐treated cells by the addition of exogenous AHL
We have reported recently that CA, SA and other plant phenolics significantly reduce the virulence of Pectobacterium strains on Zantedeschia aethiopica (calla lily) and Solanum tuberosum (potato) (Joshi et al., 2015). To determine whether the impairment of the virulence in Pectobacterium by CA and SA is associated with the direct interference of these compounds with the QS system, we assessed whether exogenous application of AHL can restore wild‐type levels of virulence in PC1 and Pcb1692 cells pretreated with CA or SA. Bacterial cultures of strains PC1 and Pcb1692 were grown in the absence (controls) or presence of non‐lethal concentrations of CA or SA. Next, each culture was divided into two groups: in one group, N‐(β‐ketocaproyl)‐l‐homoserine lactone (eAHL) was added at a final concentration of 100 nm and the other group served as a no‐eAHL control. Bacterial cultures were used in virulence assays on calla lily leaf discs and potato tubers as described in Experimental Procedures.
Untreated bacterial cultures (controls) produced characteristic tissue necrosis in both hosts (Fig. 5), typical of the development of soft rot and indicative of extensive exoenzyme‐mediated tissue damage. The addition of eAHL to the control cultures did not increase disease severity in either host, relative to the control (data not shown). As expected from previous studies (Joshi et al., 2015), CA‐ and SA‐treated bacterial cultures failed to induce disease symptoms (Fig. 5). However, exogenous application of AHL to these cultures successfully restored their virulence on both hosts (Fig. 5). In some of the treatments, exogenous application of AHL to CA‐ or SA‐treated cells fully complemented their virulence (i.e. CA‐treated cultures of PC1 and both CA‐ and SA‐treated cultures of Pcb1692 in potato). In other cases, a partial restoration of virulence relative to untreated controls was observed. This was the case for both strains on calla lily. Importantly, however, in all cases, disease severity induced by CA/eAHL‐ and SA/eAHL‐treated cultures was significantly higher (P = 0.05) than that induced by CA and SA treatments (Fig. 5). Overall, these results support the hypothesis that SA and CA affect bacterial virulence, at least in part, by impairing the QS machinery, as also evidenced by gene expression and AHL reporter assays.
Figure 5.
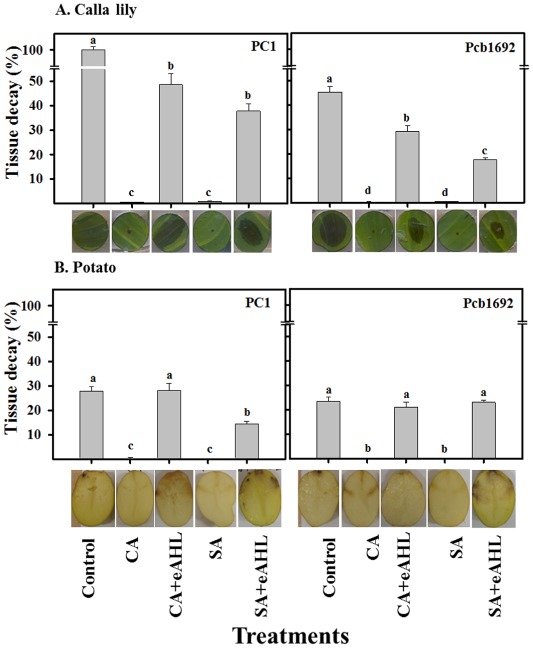
Effects of cinnamic acid (CA; 0.74 mg/mL), salicylic acid (SA; 0.42 mg/mL) and N‐(β‐ketocaproyl)‐l‐homoserine lactone (eAHL) on the virulence of P ectobacterium aroidearum PC1 and P ectobacterium carotovorum ssp. brasiliense Pcb1692 on calla lily (A) and potato (B). Bacterial strains were exposed to non‐lethal concentrations of CA and SA before they were used to inoculate the plant tissue. Parts of the CA‐ and SA‐treated cultures were supplemented with 100 nm of eAHL before inoculation. Calla lily leaf discs and potato tubers were injected with 10 μL of bacterial suspension [106 colony‐forming units (CFU)] and incubated at 28 °C. Virulence was determined as the percentage of decayed tissue 15 and 48 h after the inoculation of the calla lily leaf discs and potato tubers, respectively, relative to the decay induced by untreated bacterial cultures (control). Data represent the means ± standard errors (SE) of two independent experiments with 10 replicates for calla lily and four replicates for potato in each experiment. Treatments that are not labelled with the same letter in each panel are significantly (P = 0.05) different. Representative photographs of infected discs of calla lily and potato tubers are shown for each treatment.
Discussion
Plant colonization by Pectobacterium spp. is a very complex process in which a variety of factors, including the secretion and activity of PCWDEs, are involved. Several studies have suggested that, in soft rot enterobacteria, PCWDE activity is strictly under the control of the QS system (Barnard and Salmond, 2007; Barnard et al., 2007; Jones et al., 1993; Liu et al., 2008). The best‐studied QS systems are dependent on AHL signalling molecules which, at high cell densities, bind to receptor proteins belonging to the LuxR family of transcriptional regulators, resulting in altered gene expression (Jones et al., 1993). In soft rot enterobacteria, these systems have been reported to regulate not only PCWDEs, but other virulence factors, including the virulence regulators expR, rsmA and virR (Liu et al., 2008).
There is increasing evidence indicating that pectobacteria attack the cell wall and simultaneously target host defence mechanisms (Liu et al., 2008). However, plants respond to pathogen attack by the activation of defence responses. In this regard, small plant molecules, such as phenolic acids, are known to play a role in the elicitation of plant defence responses and also contribute to plant defence by exerting direct antimicrobial activity or by targeting virulence factors of the pathogen (Daglia, 2012; Matern and Kneusel, 1988). Indeed, some plant phenolic compounds have been reported to directly affect the production of virulence factors by P. carotovorum and Ps. syringae pv. syringae (Lagonenko et al., 2013; Sova, 2012). For example, SA has been shown to reduce the production of AHL in Ps. aeruginosa and P. carotovorum, and also to interfere with the QS system of A. tumefaciens (Chang et al., 2014; Lagonenko et al., 2013; Yuan et al., 2007). Furthermore, o‐coumaric acid and t‐cinnamic acid enhance the expression of type III secretion system (T3SS) genes in Dickeya dadantii 3937 (Yang et al., 2008). In contrast, p‐coumaric acid has been shown to repress the T3SS genes of the same pathogen, indicating that different phenolics, including different isomers of certain phenolic acids, can induce or repress the expression of the same set of genes in a given pathogen (Li et al., 2009; 2015). Phenolic acids are also known to regulate the infection efficiency during the exchange of signals between legumes and rhizobia (Mandal et al., 2010). However, despite the known effects of phenolic compounds on bacterial virulence, the mechanisms of action behind them are, as yet, unresolved (Chow et al., 2011; Mirzoeva et al., 1997; Prithiviraj et al., 2005).
Recently, we have shown that non‐lethal concentrations of various phenolic compounds affect PCWDE activity, biofilm formation and the motility of several Pectobacterium species, as well as their virulence in interactions with various host plants (Joshi et al., 2015). In the present study, we investigated the effects of two phenolic acids, CA and SA, on the expression of several virulence‐associated genes in pectobacteria. As a result of the demonstrated role of QS in PCWDE activity in soft rot enterobacteria, our efforts were focused on QS genes and on genes that are known to be regulated by the QS machinery in these bacteria. Two strains that are considered to be type strains of two Pectobacterium species, PC1 (P. aroidearum) and Pcb1692 (P. carotovorum ssp. brasiliense), were used in this study (Nabhan et al., 2013; Onkendi and Moleleki, 2014). Pathogens belonging to these species are able to infect monocot and dicot hosts of agricultural importance, especially under warm conditions (Nabhan et al., 2012, 2013; Onkendi and Moleleki, 2014).
In the present study, we explored the direct effects of CA and SA on the expression of virulence genes in Pectobacterium, distinct from their recognized role as bactericidal compounds (Daglia, 2012), or as signal molecules involved in plant defence, a well‐known role of SA (Zhang et al., 2010). The direct mechanism of action of such compounds on pectobacteria has hardly been studied, and the accumulation of information with regard to this subject from other soft rot enterobacterial models points to the importance of the examination of this mechanism. On the basis of previous studies, several virulence genes reported to be involved in Pectobacterium virulence were selected and their expression over time was analysed quantitatively using qRT‐PCR with and without exposure to the phenolic acids. As QS is a cell density‐dependent phenomenon, we followed the bacterial densities in treated and untreated cultures under the experimental conditions, with the results showing no differences between treatments in this parameter. Moreover, bacterial levels in all treatments considerably exceeded the threshold level required for the production of QS signalling molecules by Pectobacterium (Liu et al., 2008). We also studied the transcriptional patterns of QS system (AI‐1 and AI‐2) genes, QS‐regulated genes and other genes, mainly coding for membrane transporters and efflux pumps.
The most significant effects of the two phenolic acids were observed for the QS system genes expI, expR and PC1_1442, which are part of the AI‐1 QS system, and luxS, which is involved in the AI‐2 QS system. expI and expR define the AI‐1 QS system and are involved in a two‐component signal transduction system in Pectobacterium (Marchler‐Bauer et al., 2013; Pollumaa et al., 2012). The increased expression of QS system genes from the exponential growth phase and onwards was observed in both P. aroidearum PC1 and P. carotovorum ssp. brasiliense Pcb1692. Similar observations have been reported in several studies, where the efficient performance of the QS system was observed under high cell densities, represented by higher levels of AHL in Photobacterium fischeri and Ps. aeruginosa (Nealson et al., 1970; Wagner et al., 2003). However, neither of the Pectobacterium spp. displayed such an increase in QS system genes in the presence of CA or SA, although bacterial densities were similar to those of untreated controls, indicating that CA and CA directly repress the QS system. Our findings are also in line with those of Bandara et al. (2006) and Yang et al. (2009), who claimed that SA is a potential QS inhibitor in Ps. aeruginosa.
The second group of genes included a negative regulator of PCWDEs (rsmA) and four genes (pecS, pel, peh and yheO) that are involved in synthesis of PCWDEs identified in previous studies (Table 1). The regulator displayed a different expression pattern, with the lowest level of expression observed at a later stage of growth, when expression levels of QS system genes and genes contributing to the secretion of PCWDEs were at their peak. In addition, rsmB (small regulatory RNA of Rsm system) displayed a high level of expression, whereas rsmC (post‐transcriptional regulator of Rsm system), similar to rsmA, showed a low level of expression (data not shown). This observation fits the existing QS model already defined in P. atrosepticum, according to which rsmA negatively regulates the secretion of PCWDE (Chatterjee et al., 1995; Mole et al., 2007; Mukherjee et al., 1996; Pollumaa et al., 2012). These findings are in line with those of Cui et al. (2005), indicating that the availability of AHL (synthesized by ExpI) engages ExpR, thus making it unavailable for its target (rsmA promoter) and resulting in decreased rsmA production. The smaller amount of rsmA followed by higher levels of pecS, pel, peh and yheO expression in control treatment at the last stage of growth is typical of negative regulation of rsmA, which accounts for synthesis of virulence factors, such as PCWDEs. (Table 1; Charkowski et al., 2012). Exposure of the bacteria to CA or SA did not allow the normal expression pattern of the regulator rsmA or of the PCWDE genes pecS, pel, peh and YheO. Furthermore, the inhibition of any increase in the expression of QS genes might have contributed to the down‐regulation of virulence genes (such as pecS, pel, peh and yheO), downstream of the QS apparatus, and consequently the impairment of bacterial virulence (Chatterjee et al., 2005; Cui et al., 2005). No response of genes related to membrane transport was observed following the exposure to CA or SA, suggesting that efflux pumps and type III secretion were not part of the inhibitory effect of the compounds.
The inhibitory effects of CA and SA on the expression of QS‐related genes made it reasonable to assume that these compounds might affect directly the accumulation of QS signalling molecules (AHL in Pectobacterium). This hypothesis was confirmed employing two commonly used reporter assays for the presence of AHL molecules (Krishnan et al., 2012; McClean et al., 1997). Our results clearly demonstrate that treatment of PC1 and Pcb1692 with CA and SA reduces the production/accumulation of AHL.
To further clarify the results, virulence assays were conducted on two hosts representing two taxonomic plant groups, namely the monocot host calla lily and the dicot host potato. According to our hypothesis, if exposure to SA or CA actually affects virulence via QS signalling, the exogenous application of QS molecules (eAHL) might compensate for the deficiency of AHL molecules, leading to the recovery of virulence. As expected from our previous findings (Joshi et al., 2015), exposure of both Pectobacterium strains to CA and SA impaired their virulence. However, the supply of eAHL to the CA‐ or SA‐treated bacteria prior to infection restored bacterial virulence, fully in potato tubers and partially in calla lily leaves. This differential recovery might be a result of the nature of the tissue, where potato is a storage organ, rich in starch, whereas the calla lily leaf is a less favourable target. The time of exposure to the bacteria was also different, 48 h for potato and 15 h for calla lily, as no remarkable disease symptoms were developed in potato after 15 h. Nevertheless, this observation indicates that CA and SA interfere directly with the QS machinery of pectobacteria. Also, it indicates that, during plant infection, the accumulation of the AHL signal is sufficient for efficient PCWDE production, even in the absence of the auto‐inducer AI‐2, which, according to gene expression analyses, is also suppressed by CA or SA. In agreement with this notion, Laasik et al. (2006) observed that an AI‐2 mutant of P. carotovorum does not substantially differ from the wild‐type in PCWDE activity.
To conclude, the results of this work strongly support the hypothesized existence of a mechanism by which the plant phenolic acids CA and SA act through the QS machinery. CA and SA reduce the virulence of Pectobacterium spp., apparently by the inhibition of AHL production/accumulation and, ultimately, the production and secretion of PCWDEs. Further work is required to determine what is/are the exact site(s) of action of these compounds. The understanding of such aspects could hold potential for the future development of control measures against soft rot bacteria, as well as other plant pathogenic bacterial species that rely on AHL‐mediated QS systems for their fitness and virulence.
Experimental Procedures
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 2. PC1, Pcb1692 and CV026 were cultivated at 28 °C, whereas E. coli pSB401 was cultivated at 37 °C. All strains were grown in LB medium (Difco Laboratories, Detroit, MI, USA) under continuous shaking (150 rpm) in a TU‐400 incubator shaker (MRC, Holon, Israel). Murashige and Skoog minimal medium (MS; Duchefa, Haarlem, the Netherlands) was used for plant inoculation assays.
Table 2.
Strains used in this study
| Strain | Description | Reference/source |
|---|---|---|
| Pectobacterium aroidearum PC1 | Monocot strain isolated from Ornithogalum dubium, NCBI Accession no. PRJNA31289 | Yishay et al. (2008) |
| Pectobacterium carotovorum ssp. brasiliense Pcb1692 | Dicot strain isolated from Solanum tuberosum, NCBI Accession no. PRJNA31121 | Yishay et al. (2008) |
| Chromobacterim violaceum CV026 | Mini‐Tn5 mutant derived from C. violaceum ATCC 31532 HgR, cvil::Tn5 xylE, KanR, plus spontaneous StrR. AHL (C4–C8) biosensor, produces violacein (pigment) only in the presence of eAHL | McClean et al. (1997) |
| Escherichia coli pSB401 | luxRluxl′ (Photobacterium fischeri [ATCC 7744])::luxCDABE (Photorhabdus luminescens [ATCC 29999]) fusion; pACYC184‐derived, TetR, AHL bioluminescent biosensor | Winson et al. (1998) |
AHL, N‐acyl‐homoserine lactone; eAHL, N‐(β‐ketocaproyl)‐l‐homoserine lactone; HgR, hygromycin‐resistant; KanR, kanamycin‐resistant; NCBI, National Center for Biotechnology Information; StrR, streptomycin‐resistant; TetR, tetracycline‐resistant.
RNA extraction and cDNA preparation
Pectobacterium strains were grown overnight at 28 °C in LB medium under continuous shaking. Then, 20–40 μL from these cultures were transferred to 10 mL of fresh LB containing non‐lethal concentrations of cinnamic acid (CA, 0.25 mg/mL) or SA (0.21 mg/mL), and brought to a concentration of 107 CFU/mL. Controls were treated in the same manner, but with the absence of CA or SA. The fresh cultures were then grown at 28 °C under continuous shaking. Two‐millilitre samples were collected at 1, 8 and 24 h (corresponding to lag, log and stationary phases, respectively; Fig. S3a, see Supporting Information) for RNA extraction. RNA was extracted using the EZ‐RNA II kit (Biological Industries, Kibbutz Beit Haemek, Israel) following the manufacturer's instructions. Extracted RNA was employed to prepare cDNA using a cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA). A minimum of 1 μg of RNA was used with a total reaction volume of 20 μL. The cDNA reverse transcription reaction was performed using a programmable thermal controller (MJ Research, St. Bruno, QC, Canada) programmed to one cycle at 42 °C for 30 min, followed by inactivation at 95 °C for 2 min, after which the cDNA was stored at −20 °C for future use.
Quantification of mRNA by qRT‐PCR
Virulence genes from P. aroidearum and P. carotovorum ssp. brasiliense were divided into three different categories, as described in Table 1: QS genes, QS‐regulated genes and others. These two bacterial strains were found to contain a similar gene repertoire for the QS and Rsm systems (with high sequence similarity, >90%) as established in P. atrosepticum. Primers employed for mRNA reverse transcription were designed using the National Center for Biotechnology Information (NCBI) primer blast software (http://www.ncbi.nlm.nih.gov/tools/primer‐blast/), and are detailed in Table S1 (see Supporting Information). The primers were designed to generate 100–120‐bp‐sized amplicons and primer melting temperatures were designed for 60 °C, with a melting temperature difference of less than 5 °C for each primer pair. Primer sequences were subjected to blast analysis (using NCBI blast software) against the database for the genus Pectobacterium to eliminate the likelihood of non‐specific binding. qRT‐PCR mixtures contained 3.4 μL (17 ng) of cDNA, 5 μL of fast Syber green master mix (Applied Biosystems) and 0.8 μL (5 μm) of each forward and reverse primer. Reactions were performed using a Step One Plus Real‐Time PCR system (Applied Biosystems) with the following cycling parameters: holding stage, 95 °C for 20 s; cycling stage for 40 cycles of 95 °C for 3 s and 60 °C for 30 s; and melting curve stage, 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. The data were analysed by the comparative C T (ΔΔC T) method, with expression normalized to the expression of the reference gene recA, as described by Takle et al. (2007).
Qualitative assays for the detection of AHL molecules
Strain CV026 is a mini‐Tn5 mutant of C. violaceum in which the purple pigment violacein is induced in the presence of AHL compounds with N‐acyl C4–C8 side chains (McClean et al., 1997). This assay was performed as a qualitative tool to assess the effect of CA and SA on the production of AHLs in Pectobacterium strains. PC1 and Pcb1692 were grown in LB overnight as described above. Cultures were then centrifuged (7000 g, 5 min, at 28 °C) and bacterial pellets were resuspended in fresh LB supplemented or not with non‐lethal concentrations of CA or SA, and incubated for 8 h at 28 °C. The reporter strain CV026 was grown in fresh LB medium supplemented with kanamycin (10 μg/mL). Then, CV026 and Pectobacterium strains were spread perpendicularly (in a T‐shape) on LB plates, with the reporter strain being spread a few millimetres away from the tested bacteria. The plates were incubated overnight at 28 °C and the intensity of the purple colour exhibited by the reporter strain was then assessed.
Quantitative assay for AHL molecules using a bioluminescence‐based assay
Escherichia coli pSB401 is a bioluminescence‐based QS biosensor, which was generated on the background of E. coli strain JM109. This strain carries the plasmid pSB401, which possesses the luxRI′:luxCDABE bioluminescent reporter gene fusion (Winson et al., 1998). This system can detect AHLs with acyl chains ranging from six to eight carbons in length (C6–C8 AHLs; Middleton et al., 2002). This strain was used to quantitatively assess the secretion of AHL molecules by Pectobacterium strains PC1 and Pcb1692 in the presence of CA and SA. Bacteria were grown overnight (PC1 and Pcb1692 at 28 °C and E. coli pSB401 at 37 °C) in liquid LB medium under continuous shaking (150 rpm), centrifuged (7000 g, 5 min, at 28 °C) and the supernatants were removed. Bacterial pellets of Pectobacterium strains were suspended in fresh LB supplemented (or not) with non‐lethal concentrations of CA (0.25 mg/mL) or SA (0.21 mg/mL), and incubated for 8 h at 28 °C, whereas psB401 was suspended in fresh LB with tetracycline (10 μg/mL) and incubated at 37 °C. Control and treated suspensions of strains PC1 and Pcb1692 were then centrifuged (7000 g, 5 min, at 28 °C) and 10 μL of supernatant were mixed with 190 μL of 5 × 106 CFU/mL E. coli pSB401 in fresh LB medium in 96‐well microtitre plates. The supernatant used (10 μL) contained CA and SA, diluted 20‐fold, to a final volume of 200 μL of fresh LB. As the concentration used for the Pectobacterium strains did not affect the growth of psB401 (Fig. S3b, see Supporting Information), the effect of the diluted phenolics concentration on the reporter strain could be neglected. Two hundred microlitres of the reporter strain were used as a blank for the experiment and 100 nm of synthetic eAHL (Sigma, St. Louis, MO, USA) was added to the reporter strain as a positive control. The plates were incubated at 37 °C for 15 h in an Enspire 2300 multilabel reader (Perkin‐Elmer, Santa Clara, CA, USA). Bioluminescence and optical density (OD) were automatically and simultaneously determined every 30 min at 250 ms and 600 nm, respectively. Bioluminescence was calculated as the number of relative light units (RLU) per unit of optical density at 600 nm, which accounted for the influence of the different treatments on total bioluminescence, as described by Winzer et al. (2000).
AHL virulence compensation assays
Virulence was measured by the assessment of symptom severity in two plants, Zantedeschia aethiopica (calla lily) and Solanum tuberosum (potato) ‘Lady Rosetta’, as described by Yishay et al. (2008). To explore the effect of eAHL, higher concentrations of CA and SA (0.74 mg/mL and 0.42 mg/mL, respectively) were used in these experiments. These concentrations did not lead to the development of visible symptoms within 12 h of infection and were found to reduce the growth of the strains by 50% (Joshi et al., 2015). The experiment was performed on fully expanded young leaves of calla lily and small (about 25–50 g) potato tubers that were surface sterilized by soaking in 0.4% sodium hypochlorite for 20 min, and then washed twice with sterilized distilled water. In the case of calla lily, leaf discs (20 mm in diameter) were excised from disinfected leaves and transferred to Petri dishes containing MS medium. Whole disinfected potato tubers were used for infection assays. Bacterial strains were grown overnight in LB liquid medium at 28 °C with continuous shaking and diluted to 108 CFU/mL (OD600 = 0.1) in sterile double‐distilled water containing CA or SA alone or in combination with 100 nm eAHL. The bacterial suspensions were shaken (150 rpm) in a TU‐400 incubator shaker for 2 h at 28 °C before inoculation. Leaf discs and potato tubers were then pierced at the centre with a sterile tip and inoculated with 10 μL of bacterial suspension (106 CFU). The inoculated plant material was incubated at 28 °C. For calla lily, the disease severity was expressed as the percentage of decayed tissue relative to the total area of the disc, and was recorded 15 h after inoculation. For potato tubers, the disease severity was expressed as the percentage of rotten tissue, which was determined by weighing decayed tissues after 48 h of inoculation. Two independent experiments were carried out, with 10 and four replicates for each treatment, for the leaf disc and tuber assays, respectively.
Data analysis
Data were analysed by one‐way analysis of variance (ANOVA) using JMP software (Version 5, Medmenham, Buckinghamshire, UK). When ANOVA indicated a significant difference (P < 0.05), a Tukey–Kramer multiple comparison test was performed. Graphs were generated with Sigma Plot Version 10.0 (Systat Software, San Jose, CA, USA).
Supporting information
Fig. S1 Effects of cinnamic acid (CA) and salicylic acid (SA) on the growth of Pectobacterium aroidearum PC1 and Pectobacterium carotovorum ssp. brasiliense Pcb1692. The strains were grown in liquid Lysogeny‐Broth (LB) and LB with non‐lethal concentrations of CA or SA (0.25 mg/mL and 0.21 mg/mL, respectively). After 1, 8 and 24 h of growth, the bacterial cultures were serially diluted and plated on LB agar plates. The numbers of colonies were counted and CFU (colony forming units)/mL are expressed on a logarithmic scale.
Fig. S2 Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on the transcript levels of hor and hrpS. These genes are transcriptional regulators, which were not affected by the supplied phenolic compounds.
Fig. S3 (a) Growth curve of Pectobacterium strains (P. aroidearum PC1 and P. carotovorum ssp. brasiliense Pcb1692) in the control and in the presence of non‐lethal concentrations of cinnamic acid (CA) and salicylic acid (SA). Bacteria were grown at 28 °C under continuous shaking for 24 h under control and treatment conditions, and growth was assessed by measuring the absorbance (optical density, O.D.) at 600 nm every hour. The data represent means ± standard errors with at least five replicates for each experiment. (b) Growth curves of psB401 in control and in the presence of non‐lethal concentrations of CA and SA. Bacteria were grown at 37 °C under continuous shaking with measurements taken every 30 min for 24 h.
Table S1 Primers used for real‐time polymerase chain reaction (PCR).
Acknowledgements
We thank Dr Leonid Chernin and Dr Yael Helman for providing quorum sensing reporter strains (Chromobacterium violaceum CV026 and Escherichia coli pSB401), and Dr Leonid Chernin for critical reading of the manuscript.
References
- Antunes, L.C. , Ferreira, R.B. , Buckner, M.M. and Finlay, B.B. (2010) Quorum sensing in bacterial virulence. Microbiology, 156, 2271–2282. [DOI] [PubMed] [Google Scholar]
- Bandara, M.B. , Zhu, H. , Sankaridurg, P.R. and Willcox, M.D. (2006) Salicylic acid reduces the production of several potential virulence factors of Pseudomonas aeruginosa associated with microbial keratitis. Invest. Ophthalmol. Vis. Sci. 47, 4453–4460. [DOI] [PubMed] [Google Scholar]
- Barnard, A.L. and Salmond, G.C. (2007) Quorum sensing in Erwinia species. Annal. Bioanal. Chem. 387, 415–423. [DOI] [PubMed] [Google Scholar]
- Barnard, A.M. , Bowden, S.D. , Burr, T. , Coulthurst, S.J. , Monson, R.E. and Salmond, G.P. (2007) Quorum sensing, virulence and secondary metabolite production in plant soft‐rotting bacteria. Philos. Trans. R. Soc. London, B: Biol. Sci. 362, 1165–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodini, S.F. , Manfredini, S. , Epp, M. , Valentini, S. and Santori, F. (2009) Quorum sensing inhibition activity of garlic extract and p‐coumaric acid. Lett. Appl. Microbiol. 49, 551–555. [DOI] [PubMed] [Google Scholar]
- Broberg, M. , Lee, G.W. , Nykyri, J. , Lee, Y.H. , Pirhonen, M. and Palva, E.T. (2014) The global response regulator ExpA controls virulence gene expression through RsmA‐mediated and RsmA‐independent pathways in Pectobacterium wasabiae SCC3193. Appl. Environ. Microbiol. 80, 1972–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, T. , Barnard, A.M.L. , Corbett, M.J. , Pemberton, C.L. , Simpson, N.J.L. and Salmond, G.P.C. (2006) Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59, 113–125. [DOI] [PubMed] [Google Scholar]
- Chang, C.‐Y. , Krishnan, T. , Wang, H. , Chen, Y. , Yin, W.‐F. , Chong, Y.‐M. , Tan, L.Y. , Chong, T.M. and Chan, K.‐G. (2014) Non‐antibiotic quorum sensing inhibitors acting against N‐acyl homoserine lactone synthase as druggable target. Sci. Rep. 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. , Hugouvieux‐Cotte‐Pattat, N. , Solanilla, E.L. , Low, D. , Moleleki, L. , Pirhonrn, M. , Pitman, A. , Perna, N. , Reverchon, S. , Palenzuela, P.R. , Francisco, M.S. , Toth, I. , Tsuyumu, S. , Waals, J.D. , Wolf, J.V.D. , Gijsegem, F.V. , Yang, C.‐H. and Yedidia, I. (2012) The role of secretion systems and small molecules in soft‐rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A. , Cui, Y. , Liu, Y. , Dumenyo, C.K. and Chatterjee, A.K. (1995) Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density‐sensing signal, N‐(3‐oxohexanoyl)‐L‐homoserine lactone. Appl. Environ. Microbiol. 61, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A. , Cui, Y. , Hasegawa, H. , Leigh, N. , Dixit, V. and Chatterjee, A.K. (2005) Comparative analysis of two classes of quorum‐sensing signaling systems that control production of extracellular proteins and secondary metabolites in Erwinia carotovora subspecies. J. Bacteriol. 187, 8026–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, S. , Gu, K. , Jiang, L. and Nassour, A. (2011) Salicylic acid affects swimming, twitching and swarming motility in Pseudomonas aeruginosa, resulting in decreased biofilm formation. J. Exp. Microbiol. Immunol. 15, 22–29. [Google Scholar]
- Crépin, A. , Barbey, C. , Beury‐Cirou, A. , Hélias, V. , Taupin, L. , Reverchon, S. , Nasser, W. , Faure, D. , Dufour, A. , Orange, N. , Feuilloley, M. , Heurlier, K. , Burini, J.‐F. and Latour, X. (2012) Quorum sensing signaling molecules produced by reference and emerging soft‐rot bacteria (Dickeya and Pectobacterium spp. PLoS ONE, 7, e35176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. , Dixit, V. , Leigh, N. and Chatterjee, A.K. (2005) ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA‐binding protein. J. Bacteriol. 187, 4792–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski, R. , van der Wolf, J. , Krolicka, A. , Ozymko, Z. , Narajczyk, M. , Kaczynska, N. and Lojkowska, E. (2015) Salicylic acid can reduce infection symptoms caused by Dickeya solani in tissue culture grown potato (Solanum tuberosum L.) plants. Eur. J. Plant Pathol. 141, 545–558. [Google Scholar]
- Daglia, M. (2012) Polyphenols as antimicrobial agents. Curr. Opin. Biotechnol. 23, 174–181. [DOI] [PubMed] [Google Scholar]
- Davidsson, P.R.R. , Kariola, T. , Niemi, O. and Palva, T. (2013) Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deep, A. , Chaudhary, U. and Gupta, V. (2011) Quorum sensing and bacterial pathogenicity: from molecules to disease. J. Lab. Physicians, 3, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt, T. , Brackman, G. and Coenye, T. (2013) Quorum sensing inhibitors: how strong is the evidence? Trends Microbiol. 21, 619–624. [DOI] [PubMed] [Google Scholar]
- Dong, Y.‐H. , Wang, L.‐H. , Xu, J.‐L. , Zhang, H.‐B. , Zhang, X.‐F. and Zhang, L.‐H. (2001) Quenching quorum‐sensing‐dependent bacterial infection by an N‐acyl homoserine lactonase. Nature, 411, 813–817. [DOI] [PubMed] [Google Scholar]
- Engebrecht, J. and Silverman, M. (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA, 81, 4154–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, D. and Dessaux, Y. (2007) Quorum sensing as a target for developing control strategies for the plant pathogen Pectobacterium . Eur. J. Plant Pathol. 119, 353–365. [Google Scholar]
- Fuqua, C. , Parsek, M.R. and Greenberg, E.P. (2001) Regulation of gene expression by cell‐to‐cell communication: acyl‐homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468. [DOI] [PubMed] [Google Scholar]
- Fuqua, W.C. , Winans, S.C. and Greenberg, E.P. (1994) Quorum sensing in bacteria: the LuxR‐LuxI family of cell density‐responsive transcriptional regulators. J. Bacteriol. 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway, W.R. , Hodgkinson, J.T. , Bowden, S. , Welch, M. and Spring, D.R. (2012) Applications of small molecule activators and inhibitors of quorum sensing in Gram‐negative bacteria. Trends Microbiol. 20, 449–458. [DOI] [PubMed] [Google Scholar]
- Helman, Y. and Chernin, L. (2015) Silencing the mob: disrupting quorum sensing as a means to fight plant disease. Mol. Plant Pathol. 16, 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema, R. , Vemanna, R.S. , Sreeramulu, S. , Reddy, C.P. , Senthil‐Kumar, M. and Udayakumar, M. (2014) Stable expression of mtlD gene imparts multiple stress tolerance in finger millet. PLoS ONE, 9, e99110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer, M. , Wu, H. , Andersen, J.B. , Riedel, K. , Rasmussen, T.B. , Bagge, N. , Kumar, N. , Schembri, M.A. , Song, Z. , Kistoffersen, P. , Manefield, M. , Costerton, J.W. , Molin, S. , Eberl, L. , Steinberg, P. , Kjellebreg, S. , Hoiby, N. and Givskov, M. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais, F. , Oger‐Desfeux, C. , Van Gijsegem, F. , Castang, S. , Ligori, S. , Expert, D. , Nasser, W. and Reverchon, S. (2008) PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J. Bacteriol. 190, 7508–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafra, S. , Jalink, H. , Van Der Schoor, R. and Van Der Wolf, J.M. (2006) Pectobacterium carotovorum subsp. carotovorum strains show diversity in production of and response to N‐acyl homoserine lactones. J. Phytopathol. 154, 729–739. [Google Scholar]
- Jang, Y.‐S. , Sohn, S.‐I. and Wang, M.‐H. (2006) The hrpN gene of Erwinia amylovora stimulates tobacco growth and enhances resistance to Botrytis cinerea . Planta, 223, 449–456. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Yu, B. , Bainton, N.J. , Birdsall, M. , Bycroft, B.W. , Chhabra, S.R. , Cox, A.J. , Golby, P. , Reeves, P.J. and Stephens, S. (1993) The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa . EMBO J. 12, 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, J.R. , Burdman, S. , Lipsky, A. and Yedidia, I. (2015) Effects of plant antimicrobial phenolic compounds on virulence of the genus Pectobacterium . Res. Microbiol. 166, 535–545. doi: 10.1016/j.resmic.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Kalia, V.C. (2013) Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. [DOI] [PubMed] [Google Scholar]
- de Kievit, T.R. and Iglewski, B.H. (2000) Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõiv, V. , Andresen, L. , Broberg, M. , Frolova, J. , Somervuo, P. , Auvinen, P. , Pirhonen, M. , Tenson, T. and Mae, A. (2013) Lack of RsmA‐mediated control results in constant hypervirulence, cell elongation, and hyperflagellation in Pectobacterium wasabiae . PLoS ONE, 8, e54248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, T. , Yin, W.F. and Chan, K.G. (2012) Inhibition of quorum sensing‐controlled virulence factor production in Pseudomonas aeruginosa PAO1 by Ayurveda spice clove (Syzygium aromaticum) bud extract. Sensors, 12, 4016–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laasik, E. , Andresen, L. and Mäe, A. (2006) Type II quorum sensing regulates virulence in Erwinia carotovora ssp. carotovora . FEMS Microbiol. Lett. 258, 227–234. [DOI] [PubMed] [Google Scholar]
- Lagonenko, L. , Lagonenko, A. and Evtushenkov, A. (2013) Impact of salicylic acid on biofilm formation by plant pathogenic bacteria. J. Biol. Earth Sci. 3, B176–B181. [Google Scholar]
- Lee, D.H. , Lim, J.A. , Lee, J. , Roh, E. , Jung, K. , Choi, M. , Oh, C. , Ryu, S. , Yun, J. and Heu, S. (2013) Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology, 159, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Peng, Q. , Selimi, D. , Wang, Q. , Charkowski, A.O. , Chen, X. and Yang, C.‐H. (2009) The plant phenolic compound p‐coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microbiol. 75, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Hutchins, W. , Wu, X. , Liang, C. , Zhang, C. , Yuan, X. , Khokhani, D. , Chen, X. , Che, Y. , Wang, Q. and Yang, C.‐H. (2015) Derivative of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two‐component signal transduction and Rsm systems. Mol. Plant Pathol. 16, 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Coulthurst, S.J. , Pritchard, L. , Hedley, P.E. , Ravensdale, M. , Humphris, S. , Burr, T. , Takle, G. , Brurberg, M.‐B. , Brich, P.R.J. , Salmond, G.P.C. and Toth, I.K. (2008) Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum . PLoS Path. 4, e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Hibbing, M.E. , Kim, H.‐S. , Reedy, R.M. , Yedidia, I. , Breuer, J. , Breuer, J. , Glasner, J.D. , Perna, N.T. , Kelman, A. and Charkowski, A.O. (2007) Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya . Phytopathology, 97, 1150–1163. [DOI] [PubMed] [Google Scholar]
- Mandal, S.M. , Chakraborty, D. and Dey, S. (2010) Phenolic acids act as signaling molecules in plant–microbe symbioses. Plant Signal. Behav. 5, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield, M. , Harris, L. , Rice, S.A. , de Nys, R. and Kjelleberg, S. (2000) Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 66, 2079–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield, M. , Welch, M. , Givskov, M. , Salmond, G.P. and Kjelleberg, S. (2001) Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora . FEMS Microbiol. Lett. 205, 131–138. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Zheng, C. , Chitsaz, F. , Derbyshire, M.K. , Geer, L.Y. , Geer, R.C. , Gonzales, N.R. , Gwadz, M. , Hurwitz, D.I. , Lanczcki, C.J. , Lu, F. , Marchler, G.H. , Song, J.S. , Thanki, N. , Yamashita, R.A. , Zhang, D. and Bryant, S.H. (2013) CDD: conserved domains and protein three‐dimensional structure. Nucleic Acid Res. 41, 348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern, U. and Kneusel, R. (1988) Phenolic compounds in plant disease resistance. Phytoparasitica, 16, 153–170. [Google Scholar]
- McClean, K.H. , Winson, M.K. , Fish, L. , Taylor, A. , Chhabra, S.R. , Camara, M. , Daykin, M. , Lamb, J.H. , Swift, S. , Bycroft, B.W. , Stewart, G.S. and Williams, P. (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N‐acylhomoserine lactones. Microbiology, 143, 3703–3711. [DOI] [PubMed] [Google Scholar]
- Middleton, B. , Rodgers, H.C. , Cámara, M. , Knox, A.J. , Williams, P. and Hardman, A. (2002) Direct detection of N‐acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol. Lett. 207, 1–7. [DOI] [PubMed] [Google Scholar]
- Mirzoeva, O.K. , Grishanin, R.N. and Calder, P.C. (1997) Antimicrobial action of propolis and some of its components: the effects on growth, membrane potential and motility of bacteria. Microbiol. Res. 152, 239–246. [DOI] [PubMed] [Google Scholar]
- Mole, B.M. , Baltrus, D.A. , Dangl, J.L. and Grant, S.R. (2007) Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15, 363–371. [DOI] [PubMed] [Google Scholar]
- Molina, L. , Constantinescu, F. , Michel, L. , Reimmann, C. , Duffy, B. and Défago, G. (2003) Degradation of pathogen quorum‐sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 45, 71–81. [DOI] [PubMed] [Google Scholar]
- Molina, L. , Rezzonico, F. , Defago, G. and Duffy, B. (2005) Autoinduction in Erwinia amylovora: evidence of an acyl‐homoserine lactone signal in the fire blight pathogen. J. Bacteriol. 187, 3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, A. , Cui, Y. , Liu, Y. , Dumenyo, C.K. and Chatterjee, A.K. (1996) Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology, 142, 427–434. [DOI] [PubMed] [Google Scholar]
- Murray, E.J. , Crowley, R.C. , Truman, A. , Clarke, S.R. , Cottam, J.A. , Jadhav, G.P. , Steele, V.R. , O'Shea, P. , Lindholm, C. , Cockayne, A. , Chhabra, S.R. , Chan, W.C. and Williams, P. (2014) Targeting Staphylococcus aureus quorum sensing with nonpeptidic small molecule inhibitors. J. Med. Chem. 57, 2813–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan, S. , De Boer, S.H. , Maiss, E. and Wydra, K. (2012) Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J. Appl. Microbiol. 113, 904–913. [DOI] [PubMed] [Google Scholar]
- Nabhan, S. , De Boer, S.H. , Maiss, E. and Wydra, K. (2013) Pectobacterium aroidearum sp. nov., a soft rot pathogen with preference for monocotyledonous plants. Int. J. Syst. Evol. Microbiol. 63, 2520–2525. [DOI] [PubMed] [Google Scholar]
- Nasser, W. , Bouillant, M.L. , Salmond, G. and Reverchon, S. (1998) Characterization of the Erwinia chrysanthemi expI‐expR locus directing the synthesis of two N‐acyl‐homoserine lactone signal molecules. Mol. Microbiol. 29, 1391–1405. [DOI] [PubMed] [Google Scholar]
- Nealson, K.H. , Platt, T. and Hastings, J.W. (1970) Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loughlin, C.T. , Miller, L.C. , Siryaporn, A. , Drescher, K. , Semmelhack, M.F. and Bassler, B.L. (2013) A quorum‐sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA, 110, 17 981–17 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onkendi, E. and Moleleki, L. (2014) Characterization of Pectobacterium carotovorum subsp. carotovorum and brasiliense from diseased potatoes in Kenya. Eur. J. Plant Pathol. 139, 557–566. [DOI] [PubMed] [Google Scholar]
- Park, T.‐H. , Choi, B.‐S. , Choi, A.‐Y. , Choi, I.‐Y. , Heu, S. and Park, B.‐S. (2012) Genome sequence of Pectobacterium carotovorum subsp. carotovorum strain PCC21, a pathogen causing soft rot in Chinese cabbage. J. Bacteriol. 194, 6345–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhonen, M. , Flego, D. , Heikinheimo, R. and Palva, E.T. (1993) A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora . EMBO J. 12, 2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollumaa, L. , Alamae, T. and Mae, A. (2012) Quorum sensing and expression of virulence in pectobacteria. Sensors, 12, 3327–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praillet, T. , Nasser, W. , Robert‐Baudouy, J. and Reverchon, S. (1996) Purification and functional characterization of PecS, a regulator of virulence‐factor synthesis in Erwinia chrysanthemi . Mol. Microbiol. 20, 391–402. [DOI] [PubMed] [Google Scholar]
- Prithiviraj, B. , Bais, H.P. , Weir, T. , Suresh, B. , Najarro, E.H. , Dayakar, B.V. , Schweizer, H.P. and Vivanco, J.M. (2005) Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans . Infect. Immun. 73, 5319–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon, S. , Nasser, W. and Robert‐Baudouy, J. (1994) pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi . Mol. Microbiol. 11, 1127–1139. [DOI] [PubMed] [Google Scholar]
- Rutherford, S.T. and Bassler, B.L. (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder, S. , Shokat, K. , Surette, M.G. and Bassler, B.L. (2001) The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum‐sensing signal molecule. Mol. Microbiol. 41, 463–476. [DOI] [PubMed] [Google Scholar]
- Smadja, B. , Latour, X. , Faure, D. , Chevalier, S. , Dessaux, Y. and Orange, N. (2004) Involvement of N‐acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol. Plant–Microbe Interact. 17, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Smith, D. , Wang, J.H. , Swatton, J.E. , Davenport, P. , Price, B. , Mikkelsen, H. , Stickland, H. , Nishikawa, K. , Gardiol, V. , Spring, D.R. and Welch, M. (2006) Variations on a theme: diverse N‐acyl homoserine lactone‐mediated quorum sensing mechanisms in gram‐negative bacteria. Prog. Surf. Sci. 89, 167–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova, M. (2012) Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini Rev. Med. Chem. 12, 749–767. [DOI] [PubMed] [Google Scholar]
- Taga, M.E. , Semmelhack, J.L. and Bassler, B.L. (2001) The LuxS‐dependent autoinducer AI‐2 controls the expression of an ABC transporter that functions in AI‐2 uptake in Salmonella typhimurium . Mol. Microbiol. 42, 777–793. [DOI] [PubMed] [Google Scholar]
- Takle, G.W. , Toth, I.K. and Brurberg, M.B. (2007) Evaluation of reference genes for real‐time RT‐PCR expression studies in the plant pathogen Pectobacterium atrosepticum . BMC Plant Biol. 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, I.K. and Birch, P.R.J. (2005) Rotting softly and stealthily. Curr. Opin. Plant Biol. 8, 424–429. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , Bell, K.S. , Holeva, M.C. and Birch, P.R.J. (2003) Soft rot Erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , van der Wolf, J.M. , Saddler, G. , Lojkowska, E. , Hélias, V. , Pirhonen, M. , Tsror (Lahkim), L. and Elphinstone, J.G. (2011) Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 60, 385–399. [Google Scholar]
- Wagner, V.E. , Bushnell, D. , Passador, L. , Brooks, A.I. and Iglewski, B.H. (2003) Microarray analysis of Pseudomonas aeruginosa quorum‐sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, C.M. and Bassler, B.L. (2005) Quorum sensing: cell‐to‐cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. [DOI] [PubMed] [Google Scholar]
- Wei, Z.M. and Beer, S.V. (1995) hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J. Bacteriol. 177, 6201–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson, M.K. , Swift, S. , Fish, L. , Throup, J.P. , Jørgensen, F. , Chhabra, S.R. , Bycroft, B.W. , Williams, P. and Stewart, G.S.A.B. (1998) Construction and analysis of luxCDABE‐based plasmid sensors for investigating N‐acyl homoserine lactone‐mediated quorum sensing. FEMS Microbiol. Lett. 163, 185–192. [DOI] [PubMed] [Google Scholar]
- Winzer, K. , Falconer, C. , Garber, N.C. , Diggle, S.P. , Camara, M. and Williams, P. (2000) The Pseudomonas aeruginosa lectins PA‐IL and PA‐IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182, 6401–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Rybtke, M.T. , Jakobsen, T.H. , Hentzer, M. , Bjarnsholt, T. , Givskov, M. and Tolker‐Nielsen, T. (2009) Computer‐aided identification of recognized drugs as Pseudomonas aeruginosa quorum‐sensing inhibitors. Antimicrob. Agents Chemother. 53, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Peng, Q. , San Francisco, M. , Wang, Y. , Zeng, Q. and Yang, C.H. (2008) Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE, 3, e2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yishay, M. , Burdman, S. , Valverde, A. , Luzzatto, T. , Ophir, R. and Yedidia, I. (2008) Differential pathogenicity and genetic diversity among Pectobacterium carotovorum ssp. carotovorum isolates from monocot and dicot hosts support early genomic divergence within this taxon. Environ. Microbiol. 10, 2746–2759. [DOI] [PubMed] [Google Scholar]
- Yuan, Z.C. , Edlind, M.P. , Liu, P. , Saenkham, P. , Banta, L.M. , Wise, A.A. , Ronzone, E. , Binns, A.N. , Kerr, K. and Nester, E.U. (2007) The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone‐quenching genes in Agrobacterium . Proc. Natl. Acad. Sci. USA, 104, 11790–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Xu, S. , Ding, P. , Wang, D. , Cheng, Y.T. , He, J. , Gao, M. , Xu, F. , Li, Y. , Zhu, Z. , Li, X. and Zhang, Y. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant‐specific family of transcription factors. Proc. Natl. Acad. Sci. USA, 107, 18220–18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Effects of cinnamic acid (CA) and salicylic acid (SA) on the growth of Pectobacterium aroidearum PC1 and Pectobacterium carotovorum ssp. brasiliense Pcb1692. The strains were grown in liquid Lysogeny‐Broth (LB) and LB with non‐lethal concentrations of CA or SA (0.25 mg/mL and 0.21 mg/mL, respectively). After 1, 8 and 24 h of growth, the bacterial cultures were serially diluted and plated on LB agar plates. The numbers of colonies were counted and CFU (colony forming units)/mL are expressed on a logarithmic scale.
Fig. S2 Effects of cinnamic acid (CA; 0.25 mg/mL) and salicylic acid (SA; 0.21 mg/mL) on the transcript levels of hor and hrpS. These genes are transcriptional regulators, which were not affected by the supplied phenolic compounds.
Fig. S3 (a) Growth curve of Pectobacterium strains (P. aroidearum PC1 and P. carotovorum ssp. brasiliense Pcb1692) in the control and in the presence of non‐lethal concentrations of cinnamic acid (CA) and salicylic acid (SA). Bacteria were grown at 28 °C under continuous shaking for 24 h under control and treatment conditions, and growth was assessed by measuring the absorbance (optical density, O.D.) at 600 nm every hour. The data represent means ± standard errors with at least five replicates for each experiment. (b) Growth curves of psB401 in control and in the presence of non‐lethal concentrations of CA and SA. Bacteria were grown at 37 °C under continuous shaking with measurements taken every 30 min for 24 h.
Table S1 Primers used for real‐time polymerase chain reaction (PCR).


