Summary
Comparison of Arabidopsis thaliana (Arabidopsis) gene expression induced by Myzus persicae (green peach aphid) feeding, aphid saliva infiltration and abscisic acid (ABA) treatment showed a significant positive correlation. In particular, ABA‐regulated genes are over‐represented among genes that are induced by M. persicae saliva infiltration into Arabidopsis leaves. This suggests that the induction of ABA‐related gene expression could be an important component of the Arabidopsis–aphid interaction. Consistent with this hypothesis, M. persicae populations induced ABA production in wild‐type plants. Furthermore, aphid populations were smaller on Arabidopsis aba1‐1 mutants, which cannot synthesize ABA, and showed a significant preference for wild‐type plants compared with the mutant. Total free amino acids, which play an important role in aphid nutrition, were not altered in the aba1‐1 mutant line, but the levels of isoleucine (Ile) and tryptophan (Trp) were differentially affected by aphids in wild‐type and mutant plants. Recently, indole glucosinolates have been shown to promote aphid resistance in Arabidopsis. In this study, 4‐methoxyindol‐3‐ylmethylglucosinolate was more abundant in the aba1‐1 mutant than in wild‐type Arabidopsis, suggesting that the induction of ABA signals that decrease the accumulation of defence compounds may be beneficial for aphids.
Keywords: abscisic acid, amino acids, aphid choice, Arabidopsis, glucosinolates, Myzus persicae
Introduction
As a result of their capacity for extremely rapid population growth and their specialized feeding mode that causes the depletion of photoassimilates, aphids can have a large negative impact on their host plants (Goggin, 2007). Wingless females reproduce clonally on herbaceous annual hosts in the summer and winged adults move to perennial plants in the autumn, where they produce eggs that survive the winter. As a result of this alternating life cycle, aphids are well suited to colonize plants grown in monoculture, making them some of the most serious agricultural pests. The negative effects of aphid infestations are further compounded by their ability to transmit damaging plant viruses (Goggin, 2007; Walling, 2008).
Unlike chewing herbivores, aphids and other phloem‐feeding insects cause minimal mechanical damage to the plant. Thus, it is thought that most of the plant responses elicited by aphids are caused by the recognition of proteins and/or small molecules in the aphid saliva, physiological changes in phloem sieve elements and the metabolic stress that is induced by aphid colonization (Hogenhout & Bos, 2011; Smith and Boyko, 2007; Walling, 2008). However, the molecular mechanisms that control these responses and the actual effectors of plant defence against aphids are not well understood. Several recent publications have described basal defence in compatible interactions (De Vos et al., 2005; Dubey et al., 2013; Kerchev et al., 2013; Kuśnierczyk et al., 2008; Li et al., 2008; Studham and MacIntosh, 2013), and others have looked at resistant interactions (Li et al., 2008; Smith et al., 2010; Studham and MacIntosh, 2013). However, it remains to be determined which parts of the observed plant responses are actually defence mechanisms rather than merely consequences of the stress associated with aphid colonization, and whether aphids can elicit changes that make plants better hosts through metabolic hijacking and repression of defence pathways (Walling, 2008).
There is considerable evidence that secondary metabolites, including glucosinolates, hydroxamic acids, alkaloids and terpenes, can have a negative impact on aphid growth (De Vos et al., 2007; Goggin, 2007). In the Brassicaceae, the role of glucosinolates in the interaction with aphids is well documented. Glucosinolate accumulation is regulated by the salicylic acid (SA), ethylene (ET) and jasmonic acid (JA) signalling pathways (Mewis et al., 2005) and, although total glucosinolate levels do not change after aphid colonization and do not correlate with aphid performance, the level of at least one indolic glucosinolate, 4‐methoxyindol‐3‐ylmethylglucosinolate (4MI3M), increases in response to aphid feeding and has negative effects on aphid performance (De Vos and Jander, 2009; Kim and Jander, 2007; Kim et al., 2008; Pfalz et al., 2009). Unlike glucosinolate effects on chewing insects, the effect of 4MI3M on aphids does not depend on the cleavage by plant myrosinases (Barth and Jander, 2006).
It has been proposed that the colonization strategy of phloem feeders involves the suppression of effective plant defences. Microarray analysis of the Arabidopsis response to Bemisia tabaci (silverleaf whitefly) showed that SA is a major component of the response to these insects. However, B. tabaci performs poorly on npr1 plants that are deficient in SA signalling (Zarate et al., 2007). Moreover, although JA signalling is not evident in transcript profiling experiments, aphids reproduce better on Arabidopsis coi1 mutants that are insensitive to JA (Ellis et al., 2002; Mewis et al., 2006), and JA treatments of Arabidopsis, sorghum and Medicago truncatula plants retard aphid population expansion (Gao et al., 2007; Walling, 2008; Zhu‐Salzman et al., 2004). As a result of these observations, it was hypothesized that aphids induce a decoy response, possibly through an SA‐dependent pathway, that suppresses effective JA‐dependent defences (Goggin, 2007; Walling, 2008).
Another plant hormone that could interfere with SA and JA signalling pathways and affect defences is abscisic acid (ABA). Transcriptome analyses of several plant–aphid interactions suggest that ABA is part of a common response to aphid feeding (Kerchev et al., 2013; Park et al., 2006; Smith et al., 2010; Studham and MacIntosh, 2013; Zhu‐Salzman et al., 2004). It has been suggested that ABA signalling is induced in response to water stress associated with aphid colonization (Zhu‐Salzman et al., 2004). Alternatively, aphids could induce the ABA pathway to exploit phytohormone antagonism that suppresses effective defences (Anderson et al., 2004; De Torres Zabala et al., 2009; Fan et al., 2009). For example, ABA negatively affects the Arabidopsis response to Flg22, an elicitor derived from bacterial flagellin, completely suppressing the Flg22‐induced callose response and reducing the expression of Flg22‐induced defence genes, probably by antagonizing an ET‐dependent defence response (Clay et al., 2009). Similarly, ABA negatively regulates the synthesis of capsidiol, the major wild tobacco sesquiterpenoid phytoalexin, in plants treated with fungal elicitors or challenged with Botrytis cinerea (Mialoundama et al., 2009).
The role of ABA in plant–aphid interactions is not yet well defined. Kerchev et al. (2013) analysed the transcriptome changes associated with aphid feeding in Arabidopsis and found that ABA‐responsive transcripts were altered by aphid feeding. Some transcripts normally induced by ABA were also induced during the early stages of aphid feeding; however, other ABA‐induced transcripts were repressed in response to Myzus persicae. Genetic analyses showed that aphid fecundity was decreased on the leaves of mutants that were defective in either ABA synthesis (aba2) or the negative regulation (abi1) of the ABA signalling pathways, suggesting that ABA and the ABI1 signalling pathway favour lower aphid resistance (Kerchev et al., 2013). A microarray analysis of transcriptome changes of susceptible soybean plants in response to aphid colonization showed that the suppression of JA responses during the later stages of aphid colonization correlates with an important increase in the expression of ABA biosynthetic and signalling transcripts. These findings led to the hypothesis that aphid effectors induce an ABA response that blocks effective plant defences (Studham and MacIntosh, 2013).
Here, we describe experiments to investigate further the role of ABA in Arabidopsis–M. persicae interactions. We found that aphid saliva elicits an ABA response. Choice experiments showed that aphids preferred wild‐type plants to plants deficient in ABA biosynthesis, and aphid population growth was compromised on an ABA‐deficient mutant. Aphid‐induced changes in amino acid levels and glucosinolate accumulation are affected in aba1‐1 mutants, which could explain the observed changes in aphid behaviour and performance. Thus, our results suggest that aphid feeding can elicit ABA signals that could interfere with effective Arabidopsis defences.
Results
Aphid saliva triggers an ABA‐dependent response in Arabidopsis
We hypothesized that aphids induce ABA‐dependent responses that block effective plant defences. Alternatively, ABA signalling could be a response to the stress produced in the plant by phloem‐feeding aphids, particularly changes in water balance. In previous research, we showed that aphid saliva contains effectors able to trigger a plant response, independent of direct aphid feeding (De Vos and Jander, 2009). DNA microarray experiments identified 125 transcripts that are differentially regulated in Columbia‐0 (Col‐0) Arabidopsis infiltrated with aphid saliva, using mock‐treated plants as control. To determine whether ABA could be involved in this transcriptional response, we extracted information on the ABA‐mediated regulation of these genes from public databases. We found that 63% of the genes that were differentially regulated by aphid saliva infiltration also respond to ABA treatments, significantly more than the 21% expected by chance (χ2 test, P < 0.0001; Fig. 1 and File S1). In most cases, the direction of the regulation by saliva was the same as the direction of the regulation by ABA; 57% of the genes up‐regulated by aphid saliva were also up‐regulated by ABA (χ2 test, P < 0.0001; Fig. 1), whereas 41% of the genes down‐regulated by saliva were also repressed by the hormone (χ2 test, P = 0.0007; Fig. 1). The set of genes regulated by saliva include traditional ABA‐response markers, such as the caleosin RD20 (Takahashi et al., 2000), and genes encoding the late embryogenesis abundant proteins cor47 and LEA5/SAG21 (Gilmour et al., 1992; Hundertmark and Hincha, 2008) (AT2G33380, AT1G20440 and AT4G02380, respectively). Overall, the Col‐0 transcriptional response to aphid saliva correlated better with ABA regulation than with regulation by aphid feeding. Although aphid saliva and aphid feeding responses are also significantly correlated (see De Vos and Jander, 2009), the direction of regulation is more similar between aphid saliva and ABA (51% regulated in the same direction and 11% in the opposite direction) than between aphid saliva and aphid feeding (49% in the same direction and 21% in the opposite direction). Thus, our analysis suggests that aphid saliva infiltration can trigger an ABA‐dependent response in Arabidopsis, without the need for actual aphid feeding and the associated potential water stress.
Figure 1.
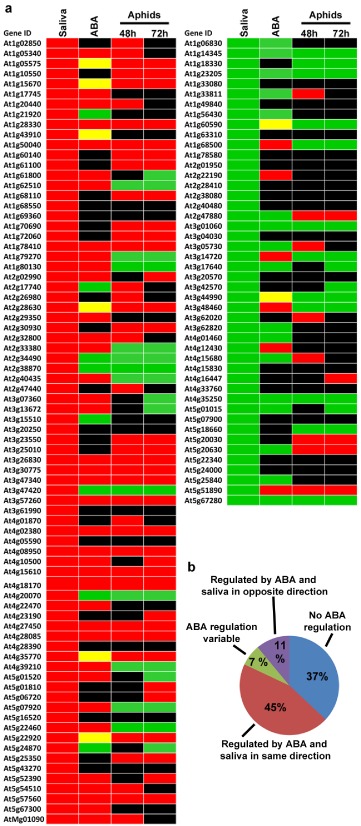
Comparison of the effects of aphid saliva infiltration, abscisic acid (ABA) treatments and aphid feeding on the Arabidopsis transcriptome. (a) Diagram showing all the differentially expressed genes, either induced (red) or repressed (green), in response to Myzus persicae saliva in Arabidopsis, and the regulation of these genes by ABA treatment and aphid feeding. Different ABA treatments are summarized in one column. Results for aphid feeding for 48 and 72 h are shown individually. Yellow boxes indicate variable expression in response to ABA in different reports. Black boxes indicate no effect of the particular treatment on gene expression. (b) Summary of the results from the comparison of aphid saliva treatment versus ABA treatment presented in (a).
We also determined the effect of aphid colonization on endogenous ABA levels. Aphids were allowed to feed on 3‐week‐old Col‐0 Arabidopsis plants for 72 h before collecting leaf tissue for analysis. Aphid feeding caused a four‐fold increase in the level of ABA (Fig. 2), suggesting that this hormone is indeed involved in the response to the insect in Arabidopsis.
Figure 2.

Aphid feeding increases Arabidopsis abscisic acid (ABA) content. Myzus persicae were caged on individual Arabidopsis leaves for 3 days before leaves were harvested for ABA analysis. Control leaves received empty cages. Mean ± standard error (SE) of n = 6 (control) and n = 5 (M. persicae). *P < 0.05, t‐test.
Aphids are deterred from and have compromised performance on ABA‐deficient plants
ABA signalling could be part of an effective defence response or could control a decoy response that results in the suppression of effective defences. To test the role of ABA in the Arabidopsis response to M. persicae, we compared aphid population growth on abi2 and aba1‐1 mutants with that on wild‐type Landsberg erecta (Ler) plants. The abi2 mutant is insensitive to ABA and shows deficiency in a subset of ABA‐regulated responses (Hubbard et al., 2010; Koornneef et al., 1984; Rubio et al., 2009). The aba1‐1 mutation affects a zeaxanthin epoxidase gene that functions in the first step of the biosynthesis of ABA, and thus mutant plants cannot synthesize the hormone and lack all ABA‐dependent responses (Koornneef et al., 1982; Marin et al., 1996). In this no‐choice experiment, Arabidopsis plants were infested with 10 aphids per plant, and the number of aphids was evaluated after 1 week. Aphid population growth was the same on the wild‐type and abi2 mutant. However, aphid numbers were reduced by about 30% on the aba1‐1 mutant (Fig. 3), suggesting that ABA synthesis is necessary for optimal aphid colonization in Arabidopsis and that, without ABA, the plant could display an increased level of defence.
Figure 3.

Aphid performance on abscisic acid (ABA) mutants. Four‐week‐old wild‐type and mutant Arabidopsis plants were infested with 10 aphids (adult apterae) and the number of aphids per plant was quantified after 1 week. Statistical analysis was performed using a t‐test assuming unequal variance. Results are the average of eight independent experiments, with at least 12 plants per treatment in each experiment. Error bars indicate standard error (SE).
Next, we tested whether aphids have a preference for wild‐type Arabidopsis or ABA biosynthetic mutants. One wild‐type and one aba1‐1 mutant plant were grown in the same pot. One plant of each pair was infested with 10 aphids. Aphids were allowed to move freely between plants in each pot (Fig. S1a, see Supporting Information). After 1 week, aphids were quantified on each plant. Control experiments consisted of two plants of the same genotype in the pot. In general, aphids remained on the plant on which they had been deposited. The movement from wild‐type to wild‐type and from aba1‐1 to aba1‐1 was similar, with about 15% of the individuals found on the plant not originally infested after 1 week. However, less than 4% of aphids moved from wild‐type to aba1‐1, whereas about 40% moved from aba1‐1 to wild‐type plants (Fig. 4). This experiment suggested that aphids have a preference for wild‐type plants that can produce ABA. However, as aphids were quantified after 1 week, differential reproduction on each plant line (as shown in Fig. 3) could have affected these results.
Figure 4.
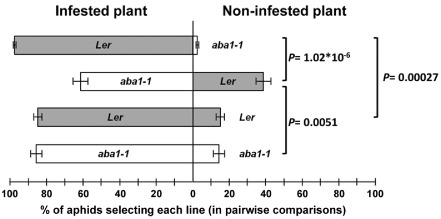
Aphids choose between wild‐type and aba1‐1 Arabidopsis plants. Wild‐type and aba1‐1 mutant plants were grown in the same pot. When plants were 4 weeks old, one plant of each pair was infested with 10 aphids. Aphids were allowed to move freely between plants in each pot. Controls had two plants of the same genotype. Aphids on each plant were quantified after 1 week of infestation. The number of aphids on the originally infested plant is shown to the left and the number of aphids on the originally non‐infested plant is shown to the right. Statistical analysis was performed using paired t‐tests. Results are the average of three independent experiments. Error bars indicate standard error (SE).
Thus, we used an additional choice test to evaluate aphid preference for wild‐type or ABA‐deficient mutants. Wild‐type and ABA‐deficient mutant plants were grown in different pots that were arranged as shown in Fig. S1b. Thirty aphids were placed on filter paper connected to the pots, at the same distance from each plant. Aphids were allowed to move freely among plants in each arrangement. After 48 h, no aphids were found on the paper, and the number of aphids on each plant was quantified. In this case, only adult apterae were quantified to eliminate any differences caused by differential reproduction on wild‐type or mutant plants. Again, aphids showed a significant preference for wild‐type plants (Fig. 5). The aba1‐1 mutant used in our study is in the Ler genetic background (Koornneef et al., 1982). To determine whether this preference effect was general or ecotype dependent, we also tested aphid preference using wild‐type Col‐0 and the aba2‐1 biosynthetic mutant in the same genetic background (Léon‐Kloosterziel et al., 1996). ABA2 encodes a cytosolic short‐chain dehydrogenase/reductase involved in the conversion of xanthoxin to ABA‐aldehyde during ABA biosynthesis, and mutants have significant reduction in ABA levels (González‐Guzmán et al., 2002). Again, aphids showed a significant preference for wild‐type plants (Fig. 5), indicating that the increase in aphid deterrence is not ecotype specific, and that mutations in two different ABA biosynthetic steps produce the same effect.
Figure 5.
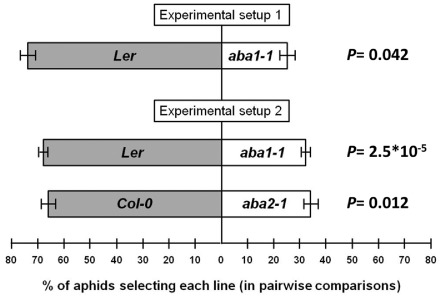
Aphid choice is not ecotype dependent. Wild‐type and aba mutant plants were grown in different pots. When plants were 4 weeks old, pots were arranged as indicated for setup 1 or setup 2 (see Fig. S1). Thirty aphids were placed on filter paper connected to the pots, at the same distance from each plant. Aphids were allowed to move freely among plants in each arrangement. The number of aphids on each plant was determined after 48 h. Only adult apterae were quantified to eliminate any difference caused by variable reproduction on wild‐type or mutant plants. Statistical analysis was performed using paired t‐tests. Results are the average of five independent experiments. Error bars indicate standard error (SE).
Free amino acids and indole glucosinolate levels are altered in the aba1‐1 mutant
The reduced aphid performance observed on the aba1‐1 mutant could be caused by changes in the levels of toxic compounds that have an antibiosis effect on the insects. Alternatively, it could be caused by a change in the nutritional composition of the plant. Nitrogen availability, mainly in the form of free amino acids, is a limiting factor on aphid performance (Dixon, 1998), and ABA controls the increase in proline concentration observed in response to drought in plants (Szabados and Savouré, 2010). Moreover, aphid feeding induced changes in the abundance of free amino acids in plants (Chiozza et al., 2010). Thus, it is reasonable to expect that aphid‐triggered ABA signals could alter the availability of amino acids and increase the nutritional value of the plant. Consequently, we analysed aphid‐elicited free amino acid changes in leaves of wild‐type and aba1‐1 mutant plants, after 7 days of aphid colonization. Aphids did not elicit a significant change in total amino acid levels (not shown). However, a significant decrease in the levels of isoleucine (Ile) and tryptophan (Trp) was observed in aphid‐infested wild‐type Ler plants (Fig. 6). This change was not observed in the aba1‐1 mutant.
Figure 6.
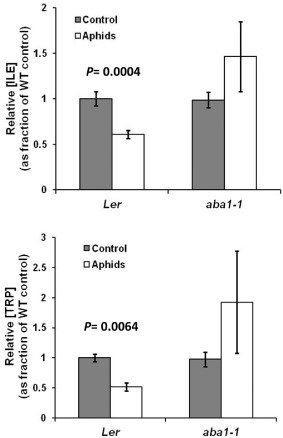
Free amino acid levels in wild‐type and aba1‐1 plants. Four‐week‐old wild‐type Ler and aba1‐1 mutant plants were mock treated or infested with aphids. After 1 week of infestation, aphids were removed, leaves were collected and amino acids were analysed by gas chromatography‐flame ionization detection (GC‐FID). Only amino acids for which significant differences were found in response to aphid feeding are shown. Statistical analysis was performed using a t‐test assuming unequal variance. Only wild‐type plants showed significant differences in response to aphids. Results are the average of two independent experiments with four replicates in each experiment. Error bars indicate standard error (SE). ILE, isoleucine; TRP, tryptophan; WT, wild‐type.
As nutritional factors did not seem to be the cause of poor performance on the aba1‐1 mutant, we also analysed the level of glucosinolate accumulation in wild‐type and aba1‐1 mutant plants under control or aphid‐infested conditions. During the aphid performance experiments, we noticed that the insects preferred to feed from the upper flower stalks and flowers, rather than the leaves of wild‐type plants. However, this behaviour was less evident on the aba1‐1 mutant [ratio of apical versus leaf aphids for wild‐type was 2.9 ± 0.68 (mean ± SE) and for aba1‐1 was 1.63 ± 0.19 (mean ± SE), P = 0.037, z‐test]. Thus, glucosinolate analysis was performed on the lower portion of the stem and on the flowers of infested and mock‐treated plants. Although most glucosinolates were not significantly different between wild‐type and mutant plants (not shown), we observed significantly elevated levels of indol‐3‐ylmethylglucosinolate (I3M) and 4MI3M in the aba1‐1 mutant irrespective of the presence or absence of aphids (Fig. 7). The highest levels of both glucosinolates were found in the stems of aba1‐1 aphid‐infested plants.
Figure 7.
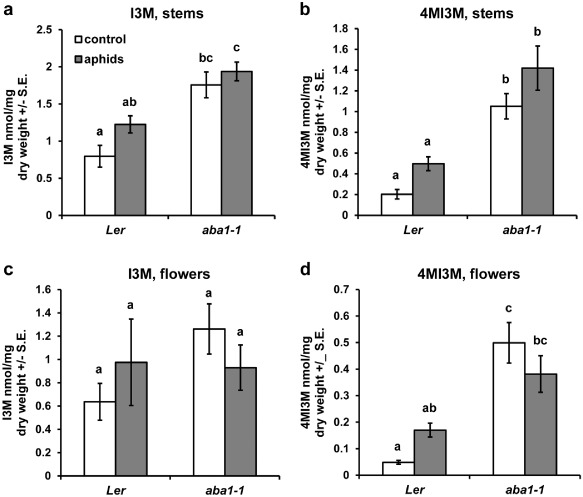
Glucosinolate content in wild‐type and aba1‐1 plants, with and without aphids. Four‐week‐old wild‐type and aba1‐1 mutant plants were mock treated or infested with aphids. After 1 week of infestation, aphids were removed and stems and flowers were collected for glucosinolate analysis by high‐performance liquid chromatography (HPLC). Only glucosinolates with significant changes are shown. (a) I3M in stems. (b) 4MI3M in stems. (c) I3M in flowers. (d) 4MI3M in flowers. I3M, indol‐3‐ylmethylglucosinolate; 4MI3M, 4‐methoxyindol‐3‐ylmethylglucosinolate. Mean ± standard error (SE) of n = 10. Different letters indicate significant differences, P < 0.05, Tukey–Kramer honestly significant difference (HSD) test.
Discussion
In this work, we have shown that M. persicae saliva infiltration regulates a set of Arabidopsis genes in a manner similar to ABA treatment, and that aphid feeding increases the level of ABA in Arabidopsis leaves. Aphids also performed better on wild‐type plants than on ABA‐deficient mutants, and the insects preferred wild‐type over mutant plants in choice experiments. These results show that ABA accumulation is beneficial to aphids, and could support the hypothesis that aphid feeding elicits an ABA‐dependent decoy response that could interfere with effective defences and make plants more suitable for aphid colonization.
Increased aphid susceptibility could be partly caused by a negative effect of ABA on indole glucosinolate production. Two glucosinolates, I3M and 4MI3M, show at least two‐fold higher levels in the ABA‐deficient mutant. The antibiosis and antixenosis effects of indole glucosinolates against M. persicae have been well established. The addition of 4MI3M to artificial diets or to petioles of Arabidopsis mutants deficient in indole glucosinolate synthesis showed that this compound has a potent repelling effect on the insects (Kim and Jander, 2007). This is consistent with results from choice experiments showing that M. persicae prefers wild‐type plants to ABA‐deficient mutants that accumulate higher levels of glucosinolates.
It has also been reported that the M. persicae population size is smaller on Arabidopsis mutants that accumulate elevated levels of indole glucosinolates than on wild‐type plants (Kim et al., 2008), and aphid reproduction is elevated on the cyp81F2 Arabidopsis mutant that cannot produce 4MI3M (De Vos and Jander, 2009; Pfalz et al., 2009). Analyses of M. persicae honeydew identified indole glucosinolate breakdown products, which increased when M. persicae was fed on atr1D mutant plants that have elevated indole glucosinolates, but were absent when M. persicae was fed on cyp79B2 cyp79B3 double mutants that lack indole glucosinolates (Kim et al., 2008). Experiments with myrosinase‐deficient tgg1 tgg2 double mutants indicated that the breakdown is independent of these plant enzymes (Kim et al., 2008). Breakdown products of I3M can react with ascorbate, glutathione and cysteine to produce diindolylmethylcysteines and other conjugates that have antifeedant effects on M. persicae when added to artificial diets (Kim et al., 2008). Thus, an increase in indole glucosinolates in the ABA‐deficient plants could also explain the decrease in population growth observed in our experiments.
Unlike in previous experiments (Kim and Jander, 2007; Kim et al., 2008; Pfalz et al., 2009), we did not observe a statistically significant increase in 4MI3M levels in wild‐type or mutant plants in response to aphid feeding. Several factors could have resulted in this difference. Previous experiments analysed indole glucosinolate levels in leaves, whereas our experiment analysed these compounds in stems and flowers. In addition, several of the previous experiments were performed with caged aphids, and we allowed aphids to move freely from the leaves to the tissues analysed; thus, it is possible that the higher variability could be the result of differences among plants or the time of aphid feeding on stems or leaves.
The role of ABA in M. persicae–Arabidopsis interactions has not been characterized extensively. A recent screen of mutants in the ABA pathway showed that aphids perform better on plants that are able to produce ABA (Kerchev et al., 2013), consistent with the results presented here. The same report showed that aphid resistance is also increased in the abi1‐1 mutant. Remarkably, we found that aphid performance is not compromised on the abi2 mutant. Both ABI1 and ABI2 encode for protein phosphatases in the PP2C family that negatively regulate ABA signalling through interaction with ABA receptors (Hubbard et al., 2010); however, both the abi1‐1 mutant used by Kerchev et al. (2013) and the abi2 mutant used in our experiments cause ABA insensitivity (Leung et al., 1997; Rodriguez et al., 1998). Together with a third phosphatase, PP2CA, these enzymes fine‐tune the response to ABA through partially overlapping pathways. Thus, the ABA effect on M. persicae performance seems to be mediated through an ABI1 signalling pathway and independent of ABI2. Kerchev et al. (2013) also found that specific components of the ABA signalling pathway, specifically ABI4, may mediate increased resistance to aphid infestation, in an ABA‐independent fashion.
A previous study (Nalam et al., 2012) suggested that the inability to acquire sufficient water is detrimental to M. persicae and decreases its ability to colonize Arabidopsis. Thus, it could be possible that the decreased performance on ABA‐deficient mutants observed in our experiments is a result of the proclivity of these mutants to lose water, which would then be less available for aphids colonizing the plant. However, several observations argue against this possibility. First, aphids performed well on the abi2 mutant, which shows drought sensitivity similar to other ABA mutants. Second, it has been shown recently that M. persicae performs better on Arabidopsis plants subjected to drought conditions (Khan et al., 2010; Mewis et al., 2012; Tariq et al., 2012), even though these plants have lower water content than non‐stressed plants (Tariq et al., 2012). This increase in aphid numbers was correlated with a decrease in indole glucosinolate levels, including 4MI3M, triggered by the drought treatment (Khan et al., 2010; Mewis et al., 2012). ABA is one of the main signalling pathways triggered by drought (Shinozaki and Yamaguchi‐Shinozaki, 2007), suggesting that ABA could mediate the drought effects that result in increased aphid performance, and is consistent with the results presented here.
The mechanism by which indole glucosinolate production and possibly other defence responses are suppressed by ABA during aphid colonization is not known. However, we observed a reduction in the levels of free Ile and Trp in wild‐type plants in response to green peach aphid feeding. Trp is a metabolic precursor for indole glucosinolates (Halkier & Gershenzon, 2006). Thus, a reduction in the level of this amino acid could slow down the production of I3M and 4MI3M. Ile is necessary for the formation of the JA–Ile conjugate which is the active form of the hormone and is able to interact with the jasmonate receptor to mediate intracellular signalling (Staswick and Tiryaki, 2004; Thines et al., 2007). In this case too, the decrease in free amino acid availability could have a negative effect on the formation of the conjugate. Alternatively, the possibility that the reduction in Trp and Ile is caused by an elevated consumption by the glucosinolate and JA pathways, respectively, cannot be disregarded, even if it seems unlikely on the basis of our results.
It has also been reported that the expression of genes encoding several enzymes involved in the biosynthesis and targeted delivery of Trp‐derived secondary metabolites is up‐regulated in the ABA biosynthetic mutant aba1‐6, including CYP81F2 (Sánchez‐Vallet et al., 2012). The CYP81F2 gene encodes a cytochrome P450 monooxygenase that is essential for the synthesis of 4MI3M (Bednarek et al., 2009). In addition, ABA treatment inhibits the Flg22‐induced expression of CYP81F2 (Clay et al., 2009). These results are consistent with the 4MI3M increase in aba1‐1 observed in our experiments (Fig. 7), and could indicate that ABA has an effect on glucosinolate synthesis at more than one level, reducing the availability of the precursor and the expression of genes involved in the synthesis and delivery of these secondary metabolites, which results in plants that are a better substrate for aphids.
It is clear that ABA can have an important role in the fine tuning of defence responses in many plant systems (Anderson et al., 2004; De Torres Zabala et al., 2009; Fan et al., 2009; Lackman et al., 2011; Nahar et al., 2012), and the generally negative effect of this signal on the main defence responses could be important for controlling the length and intensity of these responses to avoid excessive fitness costs associated with the deployment of defences in plants. Thus, aphids may have evolved the ability to exploit this regulatory node through the production of salivary effectors that are able to trigger an ABA response that antagonizes effective defences.
Experimental Procedures
Microarray comparisons
The list of differentially expressed genes from plants treated with aphid salivary secretions and the data on the regulation of these genes by aphid feeding were obtained from De Vos & Jander (2009) and De Vos et al. (2005). Data on ABA regulation of each gene were extracted from different microarray datasets: microarray experiments ME00333, ME00334, ME00335, ME00351 and ME00352 from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) database, and five additional experiments (Hoth et al., 2002; Li et al., 2006; Seki et al., 2002; Suzuki et al., 2003; Xin et al., 2005) using the TRABAS database and web‐based tools (Choudhury & Lahiri, 2008).
Plant material and plant growth
Seeds from the Arabidopsis thaliana ecotypes Col‐0 and Ler, and the mutant lines aba2‐1, were obtained from the Arabidopsis Biological Resource Center (ABRC), Ohio State University, Columbus, OH, USA. The aba1‐1 and abi2 seeds were kindly provided by Dr Michael Thomashow (Michigan State University, East Lansing, MI, USA). Plants were grown on soil (Lc01 Sunshine Mix, Sun Gro, Agawam, MA, USA) in growth chambers under 16 h of light (intensity of ∼150 μmol/m2/s) in 60% relative humidity at 21 °C.
Aphid treatments
A laboratory‐maintained M. persicae colony, originally provided by Dr Bryony Bonning (Iowa State University, Ames, IA, USA) and maintained on Col‐0 plants, was used as the source of insects for all the experiments. Aphid performance was analysed on 4‐week‐old Arabidopsis plants in a growth chamber under the same growth conditions as those used to maintain plants, by placing 10 adult apterae on one rosette leaf of each plant grown in individual pots. Ten plants were used for each genotype in each experiment, and five independent experiments were carried out. Pots were placed on 5‐cm‐tall bases that were, in turn, placed in a water‐filled tray to impede aphid movement from pot to pot. Aphids were allowed to colonize each plant for 7 days, and were then counted.
In the first aphid choice experiment, two plants, one of each of the two genotypes to be tested, were grown in the same pot and the rest of the setup was as described for aphid performance analysis. Ten adult wingless aphids were deposited on a rosette leaf of one of the plants, and the number of aphids on each plant was analysed after 1 week. Ten plant pairs were analysed for each comparison in each experiment, and three independent experiments were carried out. Results are expressed as a percentage of the total number of aphids on each plant relative to the total number of aphids on each pot. For the second choice experiments, four plants of an individual genotype were grown in one pot. When the plants were 4 weeks old, an 8 × 8‐cm2 piece of filter paper was used to connect either four pots (two for each genotype to be compared, setup 1) or two pots (one for each genotype to be compared, setup 2), and 30 adult wingless aphids were deposited on the filter paper. Aphids were allowed to move freely between the connected pots for 48 h and the number of adult aphids in each pot was determined. Only adult apterae were quantified to eliminate any difference caused by the differential reproduction on wild‐type or mutant plants. Results are expressed as a percentage of the number of aphids found on each plant genotype relative to the total number of aphids at the beginning of the experiment.
ABA analysis
For ABA analysis, 15 aphids were caged on individual leaves of 3‐week‐old Col‐0 plants. Control plants received cages without aphids. Leaves were harvested 72 h later, weighed, placed in tubes containing two 3‐mm steel balls, flash frozen in liquid nitrogen and stored at −80 °C until further use. Samples were homogenized with a paint shaker (model 5G‐HD, Harbil, Wheeling, IL, USA) for 45 s without thawing. One millilitre of extraction buffer (isopropanol–H2O–HCl, 2:1:0.005) was added to each sample with D6‐ABA (CDN Isotopes, Point‐Claire, QC, Canada) as an internal standard. Samples were extracted with dichloromethane and the solvent was allowed to evaporate. Following evaporation, samples were dissolved in 200 μL of methanol and 15 μL were analysed using a triple‐quadrupole liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) system (Quantum Access; Thermo Scientific, Waltham, MA, USA). Analytes were separated on a C18 reversed phase high‐performance liquid chromatography (HPLC) column (Gemini‐NX, 3 μm, 150 mm × 2.00 mm; Phenomenex, Torrance, CA, USA) using a gradient of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) at a flow rate of 300 μL/min. The initial condition of 10% B was kept for 2 min and increased to 100% solvent B at 20 min. Phytohormones were analysed by negative electrospray ionization, collision‐induced dissociation and selected reaction monitoring of compound‐specific parent/product ion transitions.
Amino acid and glucosinolate analyses
Wild‐type and aba1‐1 mutant Arabidopsis plants were mock treated or infested with 30 aphids (mixed instars) 4 weeks after germination. After 1 week of aphid infestation, tissue was collected for amino acid and glucosinolate analyses. First, aphids were carefully removed from the plant using a small brush. Mock‐treated plants were also brushed for a similar amount of time. For amino acid analysis, 50 mg of leaf tissue were collected and ground using liquid nitrogen. Amino acid quantification was carried out using the EZ:faast kit for free amino acids analysis (Phenomenex, KGO‐7165) by gas chromatography‐flame ionization detection (GC‐FID), as described previously (Chiozza et al., 2010). We were able to detect 24 amino acids: α‐aminoadipic acid, α‐aminobutyric acid, alanine, asparagine, aspartic acid, β‐aminoisobutyric acid, glutamine, glutamic acid, glycine, histidine, 4‐hydroxyproline, leucine, lysine, methionine, ornithine, phenylalanine, proline, sarcosine, serine, Trp, valine, threonine, Ile and tyrosine. For glucosinolate analysis, stems (the lower 5 cm) and flowers were collected from each treatment after aphid removal, and frozen in liquid nitrogen. HPLC analysis was performed as described previously (Kim and Jander, 2007). We were able to detect the following glucosinolates: 3‐hydroxypropylglucosinolate, 3‐methylsulfinylpropylglucoinolate, 4‐methylsulfinylbutylglucosinolate, 3‐methylthiopropylglucosinolate, 7‐methylsulfinylheptylglucosinolate, 8‐methylsulfinyloctylglucosinolate, I3M, 4MI3M, 1‐methoxyindol‐3‐ylmethylglucosinolate and 8‐methylthiooctylglucosinolate.
Supporting information
Fig. S1 Setup for choice experiments. (a) Aphid choice experiment 1. Wild‐type and aba1‐1 mutant plants were grown in the same pot. When plants were 4 weeks old, one plant of each pair was infested with 10 aphids. Aphids were allowed to move freely between plants in each pot. Controls had two plants of the same genotype. (b) Aphid choice experiment 2. Wild‐type and aba1‐1 or aba2‐1 mutant plants were grown in different pots. When plants were 4 weeks old, pots were arranged as indicated for setup 1 or setup 2. Thirty aphids were placed on filter paper connected to the pots, at the same distance from each plant. Aphids were allowed to move freely among plants in each arrangement.
File S1 Expression data for Arabidopsis transcripts differentially regulated by infiltration with aphid saliva.
Acknowledgements
This work was supported in part by the Iowa Soybean Association and the soybean checkoff through grants to GCM, United States Department of Agriculture award 2013‐03265 to CLC and US National Science Foundation award IOS‐1121788 to GJ. The authors declare that there are no conflicts of interest.
References
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, C. and Jander, G. (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46, 549–562. [DOI] [PubMed] [Google Scholar]
- Bednarek, P. , Piślewska‐Bednarek, M. , Svatoš, A. , Schneider, B. , Doubský, J. , Mansurova, M. , Humphry, M. , Consonni, C. , Panstruga, R. , Sanchez‐Vallet, A. , Molina, A. and Schulze‐Lefert, P. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad‐spectrum antifungal defense. Science, 323, 101–106. [DOI] [PubMed] [Google Scholar]
- Chiozza, M.V. , O'Neal, M.E. and MacIntosh, G.C. (2010) Constitutive and induced differential accumulation of amino acid in leaves of susceptible and resistant soybean plants in response to the soybean aphid (Hemiptera: Aphididae). Environ. Entomol. 39, 856–864. [DOI] [PubMed] [Google Scholar]
- Choudhury, A. and Lahiri, A. 2008) TRABAS: a database for transcription regulation by ABA signaling. In Silico Biol. 8, 511–516. [PubMed] [Google Scholar]
- Clay, N.K. , Adio, A.M. , Denoux, C. , Jander, G. and Ausubel, F.M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Sci. Signal. 323, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Torres Zabala, M. , Bennett, M.H. , Truman, W.H. and Grant, M.R. (2009) Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. [DOI] [PubMed] [Google Scholar]
- De Vos, M. and Jander, G. (2009) Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana . Plant Cell Environ. 32, 1548–1560. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M.P. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , Buchala, A.J. , Métraux, J.P. , Van Loon, L.C. , Dicke, M. and Pieterse, C.M.J. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant–Microbe Interact. 18, 923–937. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Kim, J.H. and Jander, G. (2007) Biochemistry and molecular biology of Arabidopsis–aphid interactions. Bioessays, 29, 871–883. [DOI] [PubMed] [Google Scholar]
- Dixon, A.F.G. (1998) Aphid Ecology. London: Chapman. [Google Scholar]
- Dubey, N.K. , Goel, R. , Ranjan, A. , Idris, A. , Singh, S.K. , Bag, S. , Chandrashekar, K. , Pandey, K.D. , Singh, P.K. and Sawant, S. (2013) Comparative transcriptome analysis of Gossypium hirsutum L. in response to sap sucking insects: aphid and whitefly. BMC Genomics, 14, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C. , Karafyllidis, L. and Turner, J.G. (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae . Mol. Plant–Microbe Interact. 15, 1025–1030. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Hill, L. , Crooks, C. , Doerner, P. and Lamb, C. (2009) Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L.‐L. , Anderson, J.P. , Klingler, J.P. , Nair, R.M. , Edwards, O.R. and Singh, K.B. (2007) Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago truncatula . Mol. Plant–Microbe Interact. 20, 82–93. [DOI] [PubMed] [Google Scholar]
- Gilmour, S. , Artus, N. and Thomashow, M. (1992) cDNA sequence analysis and expression of two cold‐regulated genes of Arabidopsis thaliana . Plant Mol. Biol. 18, 13–21. [DOI] [PubMed] [Google Scholar]
- Goggin, F.L. (2007) Plant–aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant Biol. 10, 399–408. [DOI] [PubMed] [Google Scholar]
- González‐Guzmán, M. , Apostolova, N. , Bellés, J.M. , Barrero, J.M. , Piqueras, P. , Ponce, M.R. , Micol, J.L. , Serrano, R. and Rodríguez, P.L. (2002) The short‐chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell, 14, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier, B.A. and Gershenzon, J. (2006) Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. [DOI] [PubMed] [Google Scholar]
- Hogenhout, S.A. and Bos, J.I.B. (2011) Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 14, 422–428. [DOI] [PubMed] [Google Scholar]
- Hoth, S. , Morgante, M. , Sanchez, J.‐P. , Hanafey, M.K. , Tingey, S.V. and Chua, N.‐H. (2002) Genome‐wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1‐1 mutant. J. Cell Sci. 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hubbard, K.E. , Nishimura, N. , Hitomi, K. , Getzoff, E.D. and Schroeder, J.I. (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes & Development 24, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark, M. and Hincha, D. (2008) LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana . BMC Genomics, 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev, P.I. , Karpińska, B. , Morris, J.A. , Hussain, A. , Verrall, S.R. , Hedley, P.E. , Fenton, B. , Foyer, C.H. and Hancock, R.D. (2013) Vitamin C and the abscisic acid‐insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis . Antioxid. Redox Signal. 18, 2091–2105. [DOI] [PubMed] [Google Scholar]
- Khan, M.A.M. , Ulrichs, C. and Mewis, I. (2010) Influence of water stress on the glucosinolate profile of Brassica oleracea var. italica and the performance of Brevicoryne brassicae and Myzus persicae . Entomol. Exp. Appl. 137, 229–236. [Google Scholar]
- Kim, J.H. and Jander, G. (2007) Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49, 1008–1019. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Lee, B.W. , Schroeder, F.C. and Jander, G. (2008) Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J. 54, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. , Jorna, M.L. , Brinkhorst, D.L.C. and Karssen, C.M. (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non‐germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. , Reuling, G. and Karssen, C.M. (1984) The isolation and characterization of abscisic acid‐insensitive mutants of Arabidopsis thaliana . Physiol. Plant. 61, 377–383. [Google Scholar]
- Kuśnierczyk, A. , Winge, P.E.R. , Jørstad, T.S. , Troczyńska, J. , Rossiter, J.T. and Bones, A.M. (2008) Towards global understanding of plant defence against aphids – timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115. [DOI] [PubMed] [Google Scholar]
- Lackman, P. , González‐Guzmán, M. , Tilleman, S. , Carqueijeiro, I. , Pérez, A.C. , Moses, T. , Seo, M. , Kanno, Y. , Häkkinen, S.T. , Van Montagu, M.C.E. , Thevelein, J.M. , Maaheimo, H. , Oksman‐Caldentey, K.M. , Rodriguez, P.L. , Rischer, H. and Goossens, A. (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci 108, 5891–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon‐Kloosterziel, K.M. , Gil, M.A. , Ruijs, G.J. , Jacobsen, S.E. , Olszewski, N.E. , Schwartz, S.H. , Zeevaart, J.A.D. and Koornneef, M. (1996) Isolation and characterization of abscisic acid‐deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Leung, J. , Merlot, S. and Giraudat, J. (1997) The Arabidopsis ABSCISIC ACID‐INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell, 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Lee, K.K. , Walsh, S. , Smith, C. , Hadingham, S. , Sorefan, K. , Cawley, G. and Bevan, M.W. (2006) Establishing glucose‐ and ABA‐regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 16, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zou, J. , Li, M. , Bilgin, D.D. , Vodkin, L.O. , Hartman, G.L. and Clough, S.J. (2008) Soybean defense responses to the soybean aphid. New Phytol. 179, 185–195. [DOI] [PubMed] [Google Scholar]
- Marin, E. , Nussaume, L. , Quesada, A. , Gonneau, M. , Sotta, B. , Hugueney, P. , Frey, A. and Marion‐Poll, A. (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana . EMBO J. 15, 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Mewis, I. , Appel, H.M. , Hom, A. , Raina, R. and Schultz, J.C. (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem‐feeding and chewing insects. Plant Physiol. 138, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis, I. , Tokuhisa, J.G. , Schultz, J.C. , Appel, H.M. , Ulrichs, C. and Gershenzon, J. (2006) Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry, 67, 2450–2462. [DOI] [PubMed] [Google Scholar]
- Mewis, I. , Khan, M.A.M. , Glawischnig, E. , Schreiner, M. and Ulrichs, C. (2012) Water stress and aphid feeding differentially influence metabolite composition in Arabidopsis thaliana (L.). PLoS ONE, 7, e48661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mialoundama, A.S. , Heintz, D. , Debayle, D. , Rahier, A. , Camara, B. and Bouvier, F. (2009) Abscisic acid negatively regulates elicitor‐induced synthesis of capsidiol in wild tobacco. Plant Physiol. 150, 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , Nzogela, Y.B. and Gheysen, G. (2012) Abscisic acid interacts antagonistically with classical defense pathways in rice–migratory nematode interaction. New Phytol. 196, 901–913. [DOI] [PubMed] [Google Scholar]
- Nalam, V.J. , Keeretaweep, J. , Sarowar, S. and Shah, J. (2012) Root‐derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell, 24, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.‐J. , Huang, Y. and Ayoubi, P. (2006) Identification of expression profiles of sorghum genes in response to greenbug phloem‐feeding using cDNA subtraction and microarray analysis. Planta, 223, 932–947. [DOI] [PubMed] [Google Scholar]
- Pfalz, M. , Vogel, H. and Kroymann, J. (2009) The gene controlling the Indole Glucosinolate Modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis . Plant Cell, 21, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P.L. , Benning, G. and Grill, E. (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421, 185–190. [DOI] [PubMed] [Google Scholar]
- Rubio, S. , Rodrigues, A. , Saez, A. , Dizon, M.B. , Galle, A. , Kim, T.‐H. , Santiago, J. , Flexas, J. , Schroeder, J.I. and Rodriguez, P.L. (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , López, G. , Ramos, B. , Delgado‐Cerezo, M. , Riviere, M. , Llorente, F. , Fernández, P.V. , Miedes, E. , Estevez, J.M. , Grant, M. and Molina, A. (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant Physiol. 160, 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M. , Narusaka, M. , Ishida, J. , Nanjo, T. , Fujita, M. , Oono, Y. , Kamiya, A. , Nakajima, M. , Enju, A. , Sakurai, T. , Satou, M. , Akiyama, K. , Taji, T. , Yamaguchi‐Shinozaki, K. , Carninci, P. , Kawai, J. , Hayashizaki, Y. and Shinozaki, K. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high‐salinity stresses using a full‐length cDNA microarray. Plant J. 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2007) Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Smith, C.M. and Boyko, E.V. (2007) The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol. Exp. Appl. 122, 1–16. [Google Scholar]
- Smith, C.M. , Liu, X. , Wang, L.J. , Liu, X. , Chen, M.‐S. , Starkey, S. and Bai, J. (2010) Aphid feeding activates expression of a transcriptome of oxylipin‐based defense signals in wheat involved in resistance to herbivory. J. Chem. Ecol. 36, 260–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. and Tiryaki, I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studham, M.E. and MacIntosh, G.C. (2013) Multiple phytohormone signals control the transcriptional response to soybean aphid infestation in susceptible and resistant soybean plants. Mol. Plant–Microbe Interact. 26, 116–129. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. , Ketterling, M.G. , Li, Q.‐B. and McCarty, D.R. (2003) Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132, 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados, L. and Savouré, A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Takahashi, S. , Katagiri, T. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2000) An Arabidopsis gene encoding a Ca2+‐binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 41, 898–903. [DOI] [PubMed] [Google Scholar]
- Tariq, M. , Wright, D.J. , Rossiter, J.T. and Staley, J.T. (2012) Aphids in a changing world: testing the plant stress, plant vigour and pulsed stress hypotheses. Agric. Forest Entomol. 14, 177–185. [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Walling, L.L. (2008) Avoiding effective defenses: strategies employed by phloem‐feeding insects. Plant Physiol. 146, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z. , Zhao, Y. and Zheng, Z.‐L. (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 139, 1350–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate, S.I. , Kempema, L.A. and Walling, L.L. (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu‐Salzman, K. , Salzman, R.A. , Ahn, J.‐E. and Koiwa, H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem‐feeding aphid. Plant Physiol. 134, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Setup for choice experiments. (a) Aphid choice experiment 1. Wild‐type and aba1‐1 mutant plants were grown in the same pot. When plants were 4 weeks old, one plant of each pair was infested with 10 aphids. Aphids were allowed to move freely between plants in each pot. Controls had two plants of the same genotype. (b) Aphid choice experiment 2. Wild‐type and aba1‐1 or aba2‐1 mutant plants were grown in different pots. When plants were 4 weeks old, pots were arranged as indicated for setup 1 or setup 2. Thirty aphids were placed on filter paper connected to the pots, at the same distance from each plant. Aphids were allowed to move freely among plants in each arrangement.
File S1 Expression data for Arabidopsis transcripts differentially regulated by infiltration with aphid saliva.


