Summary
HRT is a plant coiled‐coil, nucleotide‐binding and leucine‐rich repeat (CC‐NB‐LRR) disease resistance protein that triggers the hypersensitive response (HR) on recognition of Turnip crinkle virus (TCV) coat protein (CP). The molecular mechanism and significance of HR‐mediated cell death for TCV resistance have not been fully elucidated. To identify the genes involved in HRT/TCV CP‐mediated HR in Nicotiana benthamiana, we performed virus‐induced gene silencing (VIGS) of 459 expressed sequence tags (ESTs) of pathogen‐responsive Capsicum annuum genes. VIGS of CaBLP5, which encodes an endoplasmic reticulum (ER)‐associated immunoglobulin‐binding protein (BiP), silenced NbBiP4 and NbBiP5 and significantly reduced HRT‐mediated HR. The induction of ER stress‐responsive genes and the accumulation of ER‐targeted BiPs in response to HRT‐mediated HR suggest that ER is involved in HR in N. benthamiana. BiP4/5 silencing significantly down‐regulated HRT at the mRNA and protein levels, and affected SGT1 and HSP90 expression. Co‐expression of TCV CP in BiP4/5‐silenced plants completely abolished HRT induction. Transient expression of TCV CP alone induced selected ER stress‐responsive gene transcripts only in Tobacco rattle virus (TRV)‐infected plants, and most of these genes were induced by HRT/TCV CP, except for bZIP60, which was induced specifically in response to HRT/TCV CP. TCV CP‐mediated induction of ER stress‐responsive genes still occurred in BiP4/5‐silenced plants, but HRT/TCV CP‐mediated induction of these genes was defective. Tunicamycin, a chemical that inhibits protein N‐glycosylation, inhibited HRT‐mediated HR, suggesting that ER has a role in HR regulation. These results indicate that BiP and ER, which modulate pattern recognition receptors in innate immunity, also regulate R protein‐mediated resistance.
Keywords: BiP, ER stress, HR cell death, HRT, R protein, TCV CP
Introduction
Effector‐triggered immunity (ETI) is one of the sophisticated disease resistance mechanisms that plants have evolved to protect themselves against a wide range of pathogens. ETI is generally triggered when a plant resistance (R) protein directly or indirectly recognizes a corresponding pathogen effector protein (Gassmann and Bhattacharjee, 2012). ETI triggers diverse cellular responses, including an oxidative burst, the expression of defence‐related genes and the hypersensitive response (HR) (Hammond‐Kosack and Parker, 2003). HR triggers rapid and localized cell death at infection sites, and is a potent resistance response that limits pathogen growth in plants.
Many R proteins encoded by plant genomes belong to the nucleotide‐binding and leucine‐rich repeat (NB‐LRR) protein family. The NB‐LRR class of R proteins can be further categorized into two groups depending on whether they have a Toll/interleukin‐1 receptor (TIR) domain or a coiled‐coil (CC) domain at the N‐terminus (Collier and Moffett, 2009). Several important signalling components involved in R protein function have been identified and extensively characterized. R proteins with TIR or CC domains require EDS1 (enhanced disease susceptibility 1) or NDR1 (non‐race‐specific disease resistance 1), respectively, for downstream signal transduction (Aarts et al., 1998). Several co‐chaperone proteins, such as heat shock protein 90 (HSP90), SGT1 (suppressor of G2 allele of Skp1) and RAR1 (required for Mla12 resistance), are critical for the stabilization of R proteins in plant cells (Hubert et al., 2009; Shirasu, 2009). CRT/MORC1 (compromised recognition of TCV/Microchidia 1) was recently identified as a molecular modulator that regulates diverse R protein functions and mostly localizes to endomembrane systems in plants (Kang et al., 2010).
The HSP70 protein family contains a large group of well‐characterized chaperones. In Arabidopsis, cytosolic and nuclear heat shock cognate 70 (HSC70) proteins interact with SGT1 and regulate immune responses (Noel et al., 2007). Another subclass of HSP70s, the luminal binding proteins (BiPs; also known as Glucose Regulated Protein 78, GRP78), is targeted to the endoplasmic reticulum (ER) in yeast and mammals (Kohno et al., 1993; Lee, 1992). In yeast, BiP associates with nascent polypeptides and functions in the translocation and folding of newly synthesized proteins (Zimmermann et al., 2011). Plant BiPs function similarly to other eukaryotic BiPs; a tobacco BiP can complement a yeast temperature‐sensitive kar2 mutant (Denecke et al., 1991), and BiP overexpression in tobacco alleviates ER stress resulting from tunicamycin (Tm) treatment, an inhibitor of N‐linked protein glycosylation (Leborgne‐Castel et al., 1999). However, plants have multiple BiP genes, unlike other eukaryotes (Denecke et al., 1991; Kalinski et al., 1995; Wrobel et al., 1997). Although some studies have reported that plant BiPs are involved in environmental stress responses, the functional specificity and redundancy among plant BiP family members remain to be elucidated.
Plant BiPs are involved in a wide range of functions, including immune responses. AtBiP2 silencing in Arabidopsis attenuates NPR1 (nonexpressor of pathogenesis‐related genes 1)‐dependent secretion of PR1 (pathogenesis‐related 1) proteins, indicating that AtBiP2 is involved in the efficient induction of systemic acquired resistance (SAR) (Wang et al., 2005). BiP3 overexpression in rice reduces innate immunity mediated by receptor‐like kinase Xa21 (Park et al., 2010, 2014a), suggesting that this protein negatively regulates innate immunity. Yeast ERD2 (endoplasmic reticulum retention defective 2) homologues, which are ER luminal protein receptors, have a critical role in the regulation of HR induction in N. benthamiana; silencing of the tobacco homologues NtERD2a and NtERD2b in N. benthamiana increases HR induced by R proteins, such as tobacco N and tomato Cf9 and Pto (Xu et al., 2012). Recent work has shown that overexpression of tobacco BiP4 or BiP2 in soybean enhances non‐host HR triggered by Pseudomonas syringae pv. tomato, further confirming that plant BiP proteins have important roles in plant immunity (Carvalho et al., 2014).
The Arabidopsis R protein HRT, which confers disease resistance to Turnip crinkle virus (TCV), belongs to the HRT/RPP8 (hypersensitive response of TCV/recognition of Peronospora parasitica 8) protein family, whose members contain CC‐NB‐LRR domains (Cooley et al., 2000). On recognition of the TCV coat protein (CP), HRT triggers HR in inoculated leaves and local and systemic defence responses. A recessive allele, designated rrt, is required for HRT‐mediated HR and complete resistance to TCV infection (Cooley et al., 2000). The requirement for rrt in resistance can be bypassed by increasing the HRT levels via the exogenous application of salicylic acid (SA) or by transgenic HRT overexpression. A limited number of host proteins required for HRT/rrt‐mediated resistance, including EDS1, SID2 (salicylic acid induction‐deficient 2), PAD4 (protein arginine deiminase 4), EDS5 and CRT1/MORC1, have been identified in N. benthamiana transient expression studies and Arabidopsis mutant analyses (Chandra‐Shekara et al., 2004; Kang et al., 2008).
In this study, we used virus‐induced gene silencing (VIGS) to identify new signalling components associated with HRT/TCV CP‐mediated HR in N. benthamiana. We found that transient co‐expression of HRT and its elicitor TCV CP induced cell death in N. benthamiana. We used this system in conjunction with VIGS, and showed that the silencing of BiP4 and BiP5 genes, which encode ER‐resident HSP70 proteins of N. benthamiana, attenuates HRT‐mediated HR by down‐regulating HRT mRNA levels and protein expression. This work shows that ER stress responses play a role in HRT‐mediated HR induction.
Results
Agrobacterium‐mediated TCV CP and HRT co‐expression induces HR‐like cell death in N. benthamiana
On recognition of TCV CP, transgenic Arabidopsis overexpressing HRT undergoes HR‐like cell death (Cooley et al., 2000). To investigate whether the ectopic expression of HRT protein in conjunction with TCV CP can specifically induce HR in other plant species, we transiently expressed both proteins in N. benthamiana. When either HRT or TCV CP alone was constitutively expressed, no cell death was detected; however, when HRT or haemagglutinin (HA)‐tagged HRT was co‐expressed with TCV CP, HR‐like cell death was induced within 48 h after agroinfiltration (Figs 1A and S1, see Supporting Information). The extent of cell death was comparable with that in plants co‐expressing Pto and AvrPto, which are known to trigger cell death in N. benthamiana and were used as a positive control (Oh et al., 2010). The visual cell death phenotype was verified biochemically by quantitative measurements of ion leakage from infiltrated leaf discs, which occurs during plant cell death (Fig. 1B) (Tamagnone et al., 1998). To determine whether this specific recognition is conserved in N. benthamiana, we co‐expressed HRT and a mutant TCV CP (D4N), which is known to escape HRT recognition (Zhao et al., 2000). Unlike the effects of HRT/TCV CP, co‐expression of HRT/TCV CP (D4N) did not induce HR‐like cell death symptoms (Fig. 1A). Consistently, the ion leakage measurements indicated that there was no cell death response in N. benthamiana leaves expressing HRT alone or HRT with TCV CP (D4N) (Fig. 1B). These combined results indicate that HRT does not have autoactivation activity, but causes HR‐like cell death by specific recognition of TCV CP in N. benthamiana.
Figure 1.
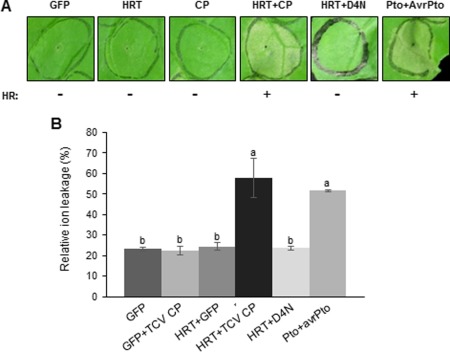
Co‐expression of HRT and Turnip crinkle virus coat protein (TCV CP) induced the hypersensitive response (HR) in Nicotiana benthamiana. (A) HR induction on leaves of N. benthamiana after transient expression of the indicated genes. Photographs were taken at 3 days post‐infiltration (dpi). GFP, green fluorescent protein; +, HR; −, no HR. (B) Electrolyte leakage assay. Measurements of ion conductivity were performed at 3 dpi as in (A). Error bars represent the standard deviation of duplicate measurements calculated from 40 leaf discs (diameter, 1 cm) per construct. Means with the same letter are not significantly different (t‐test, P < 0.05).
BiP4/5 silencing in N. benthamiana disrupts cell death triggered by the interaction between HRT and TCV CP
To identify signalling components that modulate HRT/TCV CP‐mediated HR in N. benthamiana, we performed Tobacco rattle virus (TRV)‐mediated VIGS of 459 expressed sequence tags (ESTs) from Capsicum annuum, which were induced in response to pathogen challenge (Lee et al., 2013). Silencing of one of these ESTs consistently reduced HR induction caused by HRT/TCV CP in N. benthamiana (Fig. 2A). This EST, which appeared to lack the native N‐terminal region, corresponds to a UniGene (cacn15824) with 34 ESTs. This gene was named CaBLP5 (Capsicum annum BiP5‐like protein; GenBank accession no. AGS42239) because it had 97% amino acid identity with tomato SlBiP5 protein, which is a luminal binding protein belonging to the HSP70 family of ER‐targeted proteins (Denecke et al., 1991). A domain search using InterProScan revealed that, like SlBiP5, the CaBLP5 protein contains an N‐terminal signal peptide (SP), an ATPase domain, a C‐terminal substrate binding domain and an ER retention motif (Denecke et al., 1991).
Figure 2.
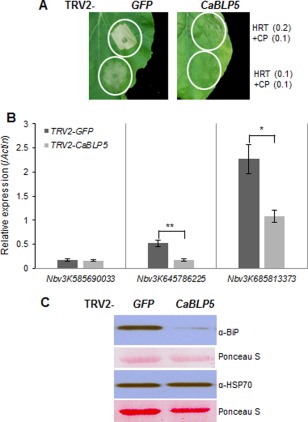
Silencing of the CaBLP5 homologue in Nicotiana benthamiana disrupts the HRT/Turnip crinkle virus coat protein (TCV CP)‐mediated hypersensitive response (HR). (A) HR phenotypes induced by co‐expression of HRT and TCV CP in TRV2‐GFP or TRV2‐CaBLP5‐infected plants (GFP, green fluorescent protein; TRV, Tobacco rattle virus). Samples were photographed at 3 days post‐infiltration (dpi). The numbers indicate Agrobacterium cell concentrations determined from the optical density at 600 nm (OD600). (B) Suppression of CaBLP5 homologous gene expression in GFP‐ or CaBLP5‐silenced N. benthamiana plants. Error bars indicate the standard error of three replicates, and asterisks indicate a significant difference from the corresponding control (Student's t‐test, *P < 0.05; **P < 0.005). (C) BiP protein levels in GFP‐ and CaBLP5‐silenced N. benthamiana leaves. Total protein samples were extracted after 21 days of infection. Samples were probed with the indicated antibodies for each protein.
The HSP70 family contains many genes, including BiP. Although HSP70 proteins are highly similar, their variable N‐ and C‐termini are responsible for their subcellular localization and intra‐ or intermolecular interactions (Lin et al., 2001). The GenBank database identified 26 proteins closely related to CaBLP5 from diverse plant species. Based on the phylogenetic analysis of these homologues, CaBLP5 was grouped together with tomato SlBiP5 and potato StBiP5‐L (Fig. S2, see Supporting Information).
To analyse the specificity of TRV2‐CaBLP5‐mediated gene silencing, we searched for CaBLP5 homologues by performing a blast search in the N. benthamiana_ transcriptome_v3_ unigenes 95 database (http://benth-web-pro-1.ucc.usyd.edu.au/blast/search.php). This analysis retrieved six NbBiP homologues, and their nucleotide sequences aligned with CaBLP5 (Table S1 and Fig. S3, see Supporting Information). A sequence of more than 21 nucleotides with 100% identity between the trigger and the target sequence is required to cause gene silencing during VIGS (Liu et al., 2002a). Therefore, we examined whether the six NbBiP sequences have a 21‐nucleotide sequence with 100% identity with TRV2‐CaBLP5 (Fig. S3). Consequently, we found that TRV2‐CaBLP5 contains a stretch of 21 nucleotides with 100% identity with Nbv3K645786225 and Nbv3K645789686, and multiple sites with more than 21‐nucleotide stretches that are identical with Nbv3K685813373. By contrast, the other three BiP homologues, Nbv3K585690033, Nbv3K585703505 and Nbv3K765636570, did not have a 21‐nucleotide stretch with 100% identity to TRV2‐CaBLP5 (Fig. S3). A phylogenetic analysis based on the amino acid sequence alignment of NbBiPs with CaBLP5 and the tobacco and Arabidopsis BiPs places Nbv3K685813373 and Nbv3K645786225 (Nbv3K645789686 amino acid sequence is identical to that of Nbv3K645786225) in the same clade with CaBLP5 (Fig. S4 and S5, see Supporting Information). Nbv3K685813373 grouped with NtBiP5 is named NbBiP5. This clone shares 93% identity with CaBLP5 at the nucleotide level (Table S1). The clone Nbv3K645786225 (identical to Nbv3K645789686), which shares 82% identity with CaBLP5 at the nucleotide level, was named NbBiP4 based on homology with other BiP4s (Figs S3 and S4, Table S1).
To confirm the specificity of VIGS in N. benthamiana, we assessed the transcript levels of three CaBLP5 homologues by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) using specific primers for each N. benthamiana BiP gene in GFP (green fluorescent protein, negative control for VIGS) or CaBLP5‐silenced plants after 3 weeks of TRV infection (Figs 2B, S3 and S4). The qRT‐PCR results showed that the CaBLP5 fragment targeted the NbBiP5 and NbBiP4 homologues (Nbv3K685813373 and Nbv3K645786225), whereas Nbv3K585690033 (encoding the Arabidopsis BiP2 homologue) expression was not reduced in TRV2‐CaBLP5‐silenced leaves relative to the TRV2‐GFP control (Figs 2B, S3 and S4). These results indicate that NbBiP4 and NbBiP5 are the targets of TRV2‐CaBLP5 gene silencing in N. benthamiana. To determine whether specific down‐regulation of BiP4/5 in silenced plants was reflected in encoded protein levels, we assessed ER‐resident BiP and cytosol‐resident HSP70 using two anti‐HSP70 antibodies that differentially detect these homologous proteins (Ko et al., 2007; Noel et al., 2007). As anticipated, the levels of ER‐resident BiP were greatly reduced, whereas cytosolic HSP70 levels were unchanged (Fig. 2C). These results further confirmed the specific down‐regulation of ER‐resident BiP proteins, but not cytosolic HSP70 proteins, by VIGS targeting of CaBLP5.
Co‐expression of HRT and TCV CP induced a strong ER stress response in N. benthamiana
There are no reports that HRT‐mediated HR is associated with ER stress responses, and so we assessed the possibility that HR suppression in BiP4/5‐silenced plants was caused by the pleiotropic effects of silencing. To this end, we monitored the expression of a set of ER stress‐responsive genes, including BiP4/5, in response to the expression of HRT and/or TCV CP in N. benthamiana leaves. We observed that transient HRT expression induced weak expression of ER stress‐responsive genes compared with the expression of the YFP (yellow fluorescent protein) control, whereas TCV CP expression did not increase the expression of ER stress‐responsive genes above that induced by the YFP control (Fig. 3A). However, we observed strong induction of ER stress‐responsive genes by co‐expression of HRT and TCV CP, indicating that the induction of ER stress‐responsive genes specifically responds to the interaction between HRT and TCV CP. We also observed that, although HRT and TCV CP expression was driven by the 35S cauliflower mosaic virus (CaMV) promoter, HRT transcripts were strongly induced by the co‐expression of TCV CP, whereas TCV CP transcripts were significantly reduced by the co‐expression of HRT (Fig. 3A). These results suggest that the interaction between HRT and TCV CP affects the post‐transcriptional regulation of their corresponding genes.
Figure 3.
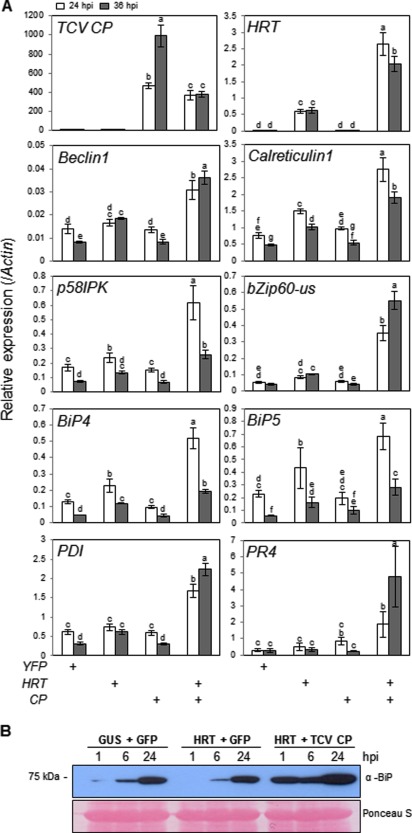
Interaction between HRT and Turnip crinkle virus coat protein (TCV CP) induced the endoplasmic reticulum (ER) stress response. (A) The transcriptional levels of ER stress‐responsive genes following the transient expression of TCV CP or HRT were assayed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The expression of Calreticulin1, Beclin1, BiP4/5 and PDI, as markers for ER quality control (QC), and bZIP60, p58IPK and PR4, as markers for ER stress, was analysed at 24 and 36 h post‐infiltration (hpi). Error bars indicate standard deviation (n = 3). Bars with the same letter are not significantly different (t‐test, P < 0.05). (B) BiP protein levels under the HRT‐mediated hypersensitive response (HR) in Nicotiana benthamiana leaves. Total protein was extracted from N. benthamiana leaves expressing the indicated proteins at the designated time points. BiP proteins were analysed by immunoblot analysis using an anti‐BiP antibody.
The induction of BiP protein expression was detected during HR progression (Fig. 3B). The expression of HRT/GFP and β‐glucuronidase (GUS)/GFP was used as a negative control for HR. Following agroinfiltration to express HRT/GFP or GUS/GFP, BiP levels gradually increased over the next 24 h (Fig. 3B). Transient co‐expression of HRT and TCV CP also increased BiP accumulation, but the induction occurred much earlier than that in the controls (Fig. 3B). These combined results suggest that ER stress responses are associated with the interaction of HRT and TCV CP, and that BiP is induced in response to HRT‐mediated cell death.
Effect of SGT1 and HSP90 in conjunction with BiP4/5 in HRT‐mediated HR
To study the interaction of BiP4/5 with HSP90 and SGT1, which are involved in R protein regulation as chaperone or co‐chaperone (Bhattarai et al., 2007; Hubert et al., 2003; Lu et al., 2003), we assessed HRT‐mediated HR in BiP4/5‐, SGT1‐ and HSP90‐silenced N. benthamiana plants (Figs 4 and S7, see Supporting Information). Rx‐mediated HR, which confers resistance to Potato virus X (PVX) via recognition of its CP (Bendahmane et al., 2002; Cooley et al., 2000), was used as an experimental control for SGT1 and HSP90 involvement in Rx/PVX‐CP‐mediated HR. HRT‐ and Rx‐mediated HR was visibly disrupted in CaBLP5‐, HSP90‐ and SGT1‐silenced leaves, whereas normal HR was induced in TRV2‐GFP and TRV2‐empty vector (EV) leaves (Fig. 4A). Reduced HR was confirmed by trypan blue staining and electrolyte leakage assay (Fig. 4B,C). Co‐infiltrated HRT/GFP, mutant PVX CP (PVX mCP; Bazzini et al., 2006) and Rx were used as HR‐negative controls. Similar to the results for HRT/TCV CP, Rx and PVX‐CP co‐expression also resulted in higher levels of BiP accumulation than those observed from Rx and PVX‐mCP co‐expression (Fig. S6, see Supporting Information). These results indicate that, in addition to SGT1 and HSP90 silencing, BiP4/5 silencing disrupts HRT‐ and Rx‐mediated HR in N. benthamiana.
Figure 4.
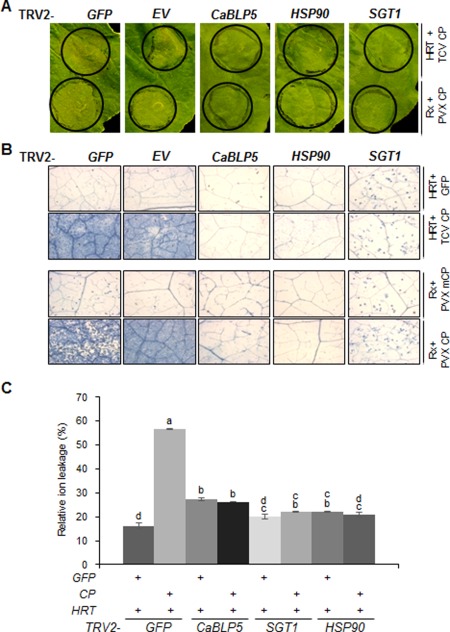
Silencing of CaBLP5 reduces the HRT‐ or Rx‐mediated hypersensitive response (HR) to a level similar to that observed after suppression of HSP90 and SGT1. (A) Visual HR phenotypes of the gene‐silenced plants after agroinfiltration to co‐express Turnip crinkle virus coat protein (TCV CP) and HRT, or Potato virus X (PVX) CP and Rx, a resistance protein. Black circles indicate the agroinfiltrated regions. Photographs were taken at 3 days post‐infiltration (dpi). (B) Trypan blue staining at 40 h post‐infiltration (hpi). Green fluorescent protein (GFP) and mutant type CP of PVX were employed as negative reference to TCV CP and PVX CP, respectively. After staining, the leaves were viewed under a light microscope to reveal necrotic plant cells. (C) Electrolyte leakage assay. Ion conductivity was measured at the indicated time points (hpi), and means ± standard deviation were calculated from 60 leaf discs (diameter, 1 cm) per construct; similar results were obtained in three independent experiments, and means with the same letter are not significantly different (t‐test, P < 0.05).
To confirm the gene‐specific down‐regulation of BiP4/5, HSP90 and SGT1 in plants subjected to VIGS, qRT‐PCR analyses were performed (Fig. 5A). Consistent with earlier results, HSP90 and SGT1 silencing affected the expression of each other, and the expression of BiP4/5 was down‐regulated by SGT1 or HSP90 silencing. However, HSP90 expression was not affected in the CaBLP5‐silenced plant, whereas BiP4/5 and SGT1 expression was strongly reduced. These results indicate that there is cross‐regulation among the three genes, although the interaction of BiP4/5 with SGT1 and HSP90 appears to differ. Expression of these genes in non‐silenced N. benthamiana leaves was observed as a reference.
Figure 5.
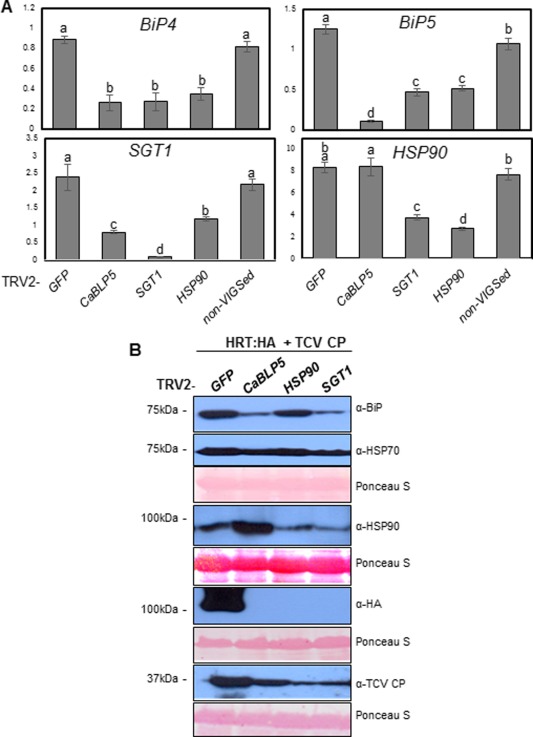
Silencing of BiP4/5, HSP90 or SGT1 in Nicotiana benthamiana reduced HRT protein expression. (A) Verification of CaBLP5, SGT1 and HSP90 gene silencing in N. benthamiana plants. The expression of CaBLP5, SGT1 and HSP90 genes in the silenced plants at 21 days post‐infiltration (dpi) was analysed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) using the specific primers shown in Table S2, and the values were normalized to the level of NbActin. Error bars represent standard deviation (n = 3). Means with the same letter are not significantly different (t‐test, P < 0.05). Normal N. benthamiana leaves (non‐virus‐induced gene silenced) were also used as a control. (B) Levels of BiP, HRT‐HA, TCV CP and HSP90 in BiP4/5‐silenced N. benthamiana leaves. Total proteins were isolated from leaves of the indicated gene‐silenced N. benthamiana, co‐infiltrated with haemagglutinin (HA)‐tagged HRT and Turnip crinkle virus coat protein (TCV CP). The protein extracts were analysed by immunoblotting with anti‐BiP, anti‐cytosol‐resident HSP70, anti‐HSP90, anti‐HA or anti‐TCV CP antibodies to detect the protein levels.
BiP5 is probably a chaperone. Therefore, we assessed whether BiP affects the accumulation of HRT protein. To determine the effect of BiP on the accumulation of HRT protein, we transiently expressed HA‐tagged HRT protein together with TCV CP in BiP4/5‐silenced N. benthamiana plants. We examined BiP protein accumulation in the silenced plants during co‐expression of HRT and TCV CP. Silencing of BiP4/5, SGT1 and HSP90 reduced BiP levels, whereas cytosolic HSP70 levels were not affected (Fig. 5). HSP90 accumulated at high levels in BiP4/5‐silenced plants, whereas HSP90 levels were reduced in SGT1‐silenced plants. These results raise the possibility that BiP, HSP90 and SGT1 functionally interact with different mechanisms during HRT‐mediated HR. Next, we measured HRT protein levels in gene‐silenced plant leaves using an anti‐HA antibody. Although HRT proteins were efficiently expressed in TRV‐GFP leaves, these proteins were not detected in TRV2‐CaBLP5, TRV2‐HSP90 and TRV2‐SGT1 leaves (Fig. 5B). Previous studies have shown that HSP90 and SGT1 regulate the stability/accumulation of various R proteins (Holt et al., 2005; Lu et al., 2003), which raises the possibility that they serve as co‐chaperone and chaperone for the assembly of an active R protein complex. Consistent with these previous reports, our results indicate that BiP4/5, HSP90 and SGT1 are important for HRT protein accumulation in N. benthamiana. TCV CP was detected in all silenced plants, although its accumulation was reduced in CaBLP5‐, HSP90‐ and SGT1‐silenced leaves.
BiP4/5, SGT1 and HSP90 modulate HRT expression
To study HRT transcriptional regulation and the effects of BiP4/5, HSP90 and SGT1 silencing on HRT‐mediated HR, we investigated the expression of BiP4/5, HSP90 and SGT1 in response to HRT and/or TCV CP (Fig. 6). We did not observe any cell death response in HRT/TCV CP co‐infiltrated leaves up to 24 h post‐infiltration (hpi); however, after 36–40 hpi, HR appeared to be initiated specifically on leaf spots in which HRT and TCV CP were co‐expressed. To analyse the gene expression patterns before and after HR was triggered, transcript levels were measured by qRT‐PCR at 24 or 41 hpi (Fig. 6). In the same samples, we also confirmed the expression of HRT and TCV CP by qRT‐PCR (Fig. 6).
Figure 6.
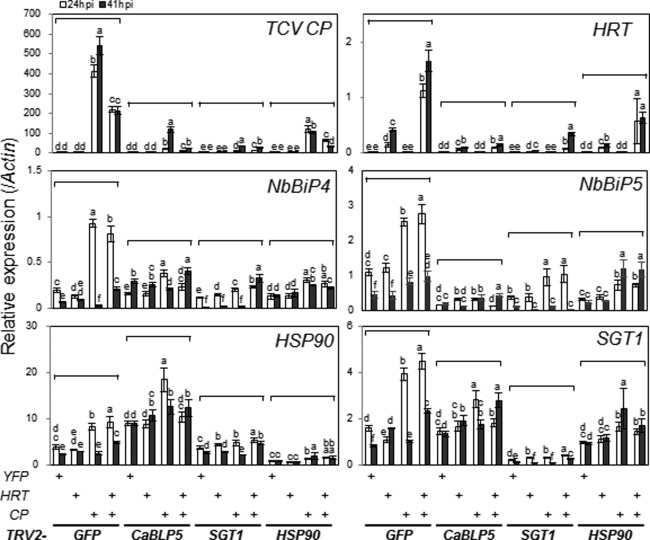
HRT and Turnip crinkle virus coat protein (TCV CP) were impeded at mRNA levels in BiP4/5‐silenced Nicotiana benthamiana. Total RNA was obtained from leaves of the gene‐silenced plants after expression of either HRT alone or in conjunction with TCV CP at 24 h post‐infiltration (hpi) (white bar) or 41 hpi (dark grey bar). The transcriptional levels of the specific genes were assayed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Error bars indicate standard deviation (n = 3). Bars with the same letter are not significantly different (t‐test, P < 0.05). Two additional biologically independent experiments yielded similar results.
First, we checked whether there were differences in gene expression in non‐silenced, TRV‐infected control leaves expressing HRT or TCV CP alone, and in leaves expressing HRT plus TCV CP (Fig. 6). We found significant differences in gene expression in response to TCV CP between GFP‐silenced control and non‐silenced N. benthamiana plants (Figs 3A and 6). In GFP‐silenced control plants, transient TCV CP expression induced higher expression of BiP4/5, SGT1 and HSP90 than in YFP‐expressing leaves (Fig. 6), although the TCV CP expression intensity and pattern were similar to those of non‐silenced plants. Significant induction of BiP4/5, SGT1, HSP90 and HRT was observed during HRT/TCV CP‐mediated HR in GFP‐silenced leaves. TCV CP‐mediated induction of BiP4/5, SGT1 and HSP90 was observed only in the 24‐hpi sample, whereas induction of these genes by HRT plus TCV CP was sustained until 41 hpi compared with the duration in the YFP‐expressing control.
In TRV2‐GFP plant leaves, HRT expression increased more than two‐fold during HRT‐mediated HR compared with the absence of HR (Fig. 6). However, HRT expression was most severely reduced in TRV2‐CaBLP5 leaves than in GFP‐silenced control, SGT1‐ and HSP90‐VIGSed leaves (Fig. 6). HRT expression in SGT1‐ and HSP90‐silenced leaves showed a similar pattern to that in GFP‐silenced leaves, although the overall expression was much less (Fig. 6). These results indicate that BiP4/5, HSP90 and SGT1 affect HRT expression at both the mRNA and protein levels (Figs 5B and 6). TCV CP expression was also strongly reduced in the BiP4/5‐, SGT1‐ and HSP90‐silenced plants, but the expression level did not differ significantly.
SGT1, BiP and HSP90 have been reported to be involved in Agrobacterium‐mediated gene transfer in N. benthamiana (Anand and Mysore, 2013; Park et al., 2014b); therefore, it is possible that low HRT expression in the silenced plants was caused by the low efficiency of transgene expression. To test this possibility, we searched for a gene that has similar function to that of BiP, HSP90 or SGT1, but does not affect HRT/TCV CP‐mediated HR. We found the RAR1 gene (Anand and Mysore, 2013), which functions as a chaperone for several R genes involved in Agrobacterium‐mediated gene transfer, but not in HRT‐mediated immune responses in Arabidopsis (Chandra‐Shekara et al., 2004). We observed strong HR by co‐expression of HRT/TCV CP in RAR1‐silenced plants, although the HRT expression level was lower than that in the control (Fig. S8, see Supporting Information). These results suggest that the efficiency of Agrobacterium‐mediated gene transfer is not the primary reason for HR suppression in BiP4/5‐, SGT1‐ and HSP90‐silenced plants.
Among the BiP4/5, SGT1 and HSP90 genes, BiP4/5 silencing affected the expression of HSP90 and SGT1. The induction of HSP90 transcripts in response to TCV CP still occurred in BiP4/5‐silenced plants relative to YFP‐expressing leaves, whereas the induction during HRT‐mediated HR was defective (Fig. 6). SGT1 induction in response to both TCV CP and HRT/TCV CP was suppressed in BiP4/5‐silenced plants. SGT1 silencing reduced basal BiP4/5 expression and abolished its induction by TCV CP and HRT/TCV CP; however, HSP90 expression only affected its induction by TCV CP and HRT/TCV CP in SGT1‐silenced plants. In HSP90‐silenced plants, SGT1 and BiP4/5 expression was slightly reduced, and their induction in response to TCV CP and HRT/TCV CP was strongly suppressed. Collectively, these results suggest that co‐chaperone SGT1 expression might be correlated with both the expression of BiP4/5 and HSP90 chaperones with different interactions, and BiP4/5 and HSP90 expression might be regulated independently, especially in HRT‐mediated immune responses.
BiP4/5 silencing alters the expression of stress‐related genes
We determined whether BiP4/5, SGT1 or HSP90 changes the expression of defence‐related and ER stress‐responsive genes in response to TCV CP and/or HRT. We assessed the expression of two HR marker genes [Hsr203J and HIN1 (harpin‐induced 1)], four basal defence‐related genes (PR1a, PR2, CYP71D20 and WRKY8) and six ER stress‐responsive genes [Beclin 1, Calreticulin 1, PDI (protein disulfide isomerase), p58IPK, bZiP60 and PR4] (Fig. 7). In cells transiently overexpressing YFP, selected ER stress‐responsive genes were up‐regulated in CaBLP5‐ and HSP90‐silenced plants relative to GFP‐silenced control plants (Fig. 7A). Beclin1, PDI and PR4 were up‐regulated in HSP90‐silenced plants, whereas the expression of selected ER stress‐responsive genes (except for PR4) in SGT1‐silenced plants was comparable with that in GFP‐silenced control plants (Fig. 7A). PR4 is down‐regulated by IRE1‐dependent mRNA decay under ER stress (Mishiba et al., 2013). Expression of TCV CP with or without HRT significantly up‐regulated the examined ER stress‐responsive genes, with the exception of bZiP60 (Fig. 7A). Expression of bZIP60 only increased in response to HRT and TCV CP co‐expression. Expression of ER stress‐responsive genes during HRT/TCV CP‐triggered HR was significantly reduced in response to BiP4/5 silencing, except for PDI, whereas induction of the same genes by TCV CP was not suppressed (Fig. 7A). By contrast, SGT1 silencing induced the genes with the same expression patterns, but with lower intensities. HSP90 silencing abolished the induction of all tested genes in response to TCV CP and HRT/TCV CP. These combined results suggest that BiP4/5, HSP90 and SGT1 are involved in HRT‐mediated HR via regulation of HRT, but have distinct relationships with ER stress responses.
Figure 7.
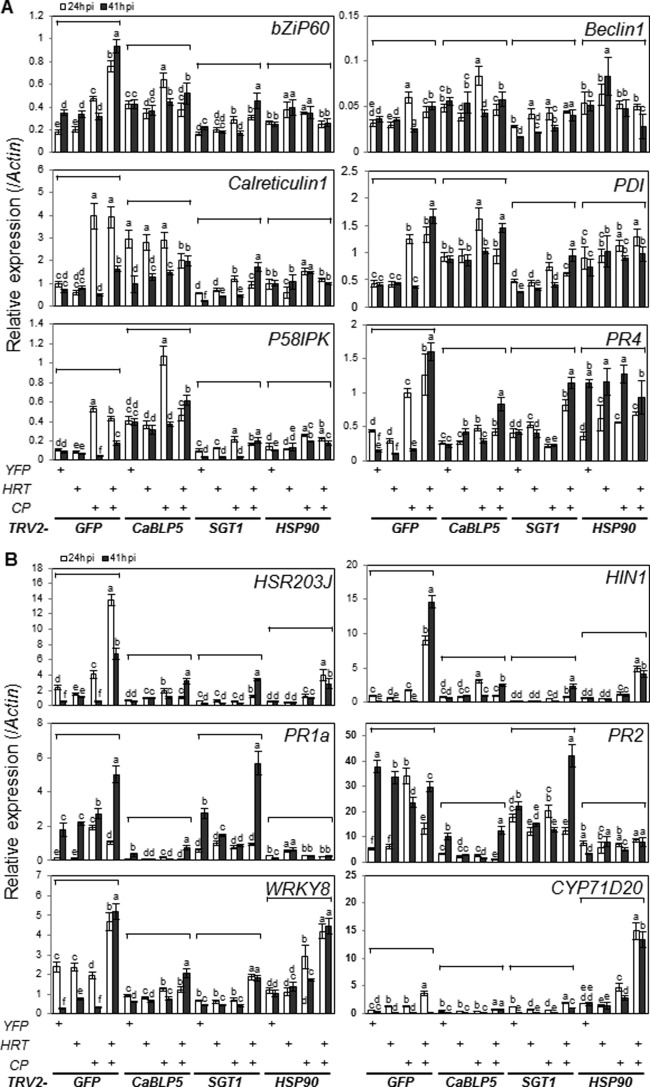
Silencing of BiP4/5 reduced defence or endoplasmic reticulum (ER) stress‐related gene expression in Nicotiana benthamiana. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assay was conducted as shown in Fig. 6, using primers specific to ER stress‐responsive genes (A) and defence‐related genes (B). Bars with the same letter are not significantly different (t‐test, P < 0.05). Two additional biologically independent experiments yielded similar results.
Previous work has shown that PR1a, HIN1 and Hsr203J are strongly induced during HR cell death (Heath, 2000). HRT and TCV CP co‐expression induced the expression of the HR marker genes PR1a, HIN1 and HSR203, and the basal defence genes CYP71D20 and WRKY8, but BiP4/5 silencing reduced the induction of all genes (Fig. 7B). The expression of most defence‐related genes was similarly reduced in HSP90‐ and SGT1‐silenced plants, with the exceptions of PR1a and PR2 in SGT1‐silenced plants, and WRKY8 and CYP71D20 in HSP90‐silenced plants (Fig. 7B). Taken together, these results indicate that silencing of BiP4/5, SGT1 and HSP90 triggers distinct ER stress responses and alters the expression patterns of defence‐related genes in response to TCV CP or HRT/TCV CP.
Tm treatment blocks HRT‐mediated HR in N. benthamiana
In mammals, BiPs are master regulators of signal transduction in the unfolded protein response (UPR) following ER stress (Zhang and Kaufman, 2004). In general, the accumulation of unfolded proteins in the ER triggers the UPR pathway, which results in elevated chaperone protein expression to maintain organellar homeostasis. Little is known about the effects of the UPR pathway on R‐mediated HR. Therefore, we examined the effect of ER stress on R‐mediated HR in N. benthamiana. We induced ER stress using Tm, which inhibits N‐glycosylation of proteins passing through the ER (Samali et al., 2010) and activates the UPR pathway in mammals and plants. Nicotiana benthamiana leaves were co‐infiltrated with HRT/TCV CP in the presence or absence of Tm. During cell death, plant cells accumulate fluorescent phenolic compounds in the cell wall, and the secondary metabolites of these compounds can be visualized by observing the fluorescence they emit under ultraviolet (UV) irradiation (Tang et al., 1999). Treatment with Tm strongly inhibited HRT‐mediated HR (Figs 8A and S9, see Supporting Information). Tm treatment increased BiP protein levels, as expected, but reduced HRT levels (Fig. 8B). These results indicate that HRT accumulation and stability are affected by ER stress and N‐glycosylation, and dysregulation reduces cell death induction.
Figure 8.
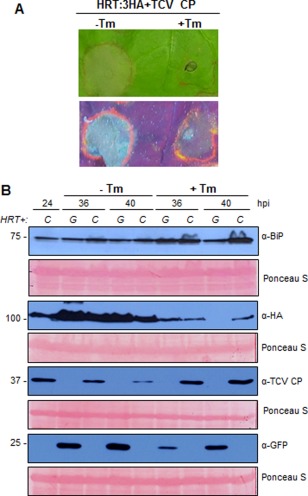
Tunicamycin (Tm) treatment blocked the HRT‐mediated hypersensitive response (HR) and HRT accumulation, but increased endoplasmic reticulum (ER)‐resident heat shock protein 70 (HSP70) levels. (A) Verification of HR initiation after Tm treatment. HRT was co‐expressed with Turnip crinkle virus coat protein (TCV CP) in leaves of Nicotiana benthamiana and Tm was infiltrated at a concentration of 10 µm. The images of the infiltrated leaves were photographed under white light or UV illumination at 3 days post‐infiltration (dpi). (B) Effects of Tm treatment on the accumulation of the indicated proteins. Total protein was obtained at 24, 36 and 40 h post‐infiltration (hpi) after expression of haemagglutinin (HA)‐tagged HRT with TCV CP (CP) (C) or green fluorescent protein (GFP) (G). Western blotting was performed with anti‐BiP, anti‐HA, anti‐TCV CP or anti‐GFP antibodies to detect the protein levels.
To confirm that Tm treatment triggered the UPR pathway, we measured BiP5 and bZiP60 expression by qRT‐PCR, and monitored the BiP expression in GFP‐overexpressing control plants. Expression of both genes increased as a function of Tm concentration (Fig. S10A, see Supporting Information), and BiP proteins accumulated in response to Tm treatment (Fig. S10B), confirming that Tm treatment triggers the UPR pathway. Cell death‐like damage was observed in Tm‐treated leaves at 5 dpi (data not shown).
Discussion
Many mammalian viruses target ER stress sensors and utilize the host UPR pathway to enhance the folding of viral proteins or to modulate immune responses. The significance of ER stress and the UPR pathway in plant–virus interactions has started to be appreciated (Ye et al., 2011, 2012; Zhang and Wang, 2012). However, it remains unclear how ER stress and the UPR pathway contribute to R protein‐mediated HR. In this study, our VIGS screen for signalling factors that modulate HRT‐mediated HR identified the ER‐resident HSP70s, BiP4 and BiP5 proteins. We demonstrated that the BiP4/5‐associated ER stress response has an important role in R protein‐mediated cell death, but that this response is dissimilar to the ER stress response induced by the N‐glycosylation inhibitor Tm.
Recent studies have reported that plant viral proteins interact with components of the UPR machinery to enhance pathogenesis. For example, a movement protein of PVX, TGB3, resides in the ER during PVX infection (Ye et al., 2011). This viral protein induces the expression of ER‐resident chaperones, including BiP, through the bZiP60 signalling pathway (Garcia‐Marcos et al., 2009; Ye et al., 2011). Silencing of bZiP60 suppresses PVX replication, and BiP overexpression inhibits the TGB3‐induced cell death response (Ye et al., 2012). Unlike TGB3, TCV CP did not induce the expression of any ER stress‐responsive genes; however, TCV CP overexpression induced several ER stress‐responsive genes in TRV‐GFP‐silenced control plants (Fig. 7A). TRV is a positive‐strand RNA virus with a bipartite genome. TRV can move systemically in many plants (Ratcliff et al., 2001). Several studies have identified a cysteine‐rich 16K protein of TRV as the main suppressor of RNA silencing (Martínez‐Priego et al., 2008; Reavy et al., 2004), which occurs as the result of the inhibition of the formation of the initial RNA silencing complex by interference with AGO (argonaute) proteins (Fernández‐Calvino et al., 2016). Previous work has reported the strong RNA‐silencing activity of TCV CP in N. benthamiana using an Agrobacterium‐mediated transient expression study (Qu et al., 2003). Therefore, the transient overexpression of TCV CP increased TRV accumulation probably via the inhibition of RNA silencing‐mediated plant basal defence responses, leading to the induction of ER stress responses and suggesting that this response may be proportional to the level of the virus. A few reports have explained the involvement of ER stress responses in plant immunity. The tobacco N gene, which confers resistance against Tobacco mosaic virus (TMV), interacts with multiple cytosolic and ER‐resident quality control (QC) components that are essential for N‐mediated HR and TMV resistance (Caplan et al., 2009). In cucumber, it has been reported that chemically induced UPR promotes SAR (Sticher and Metraux, 2000). Nagashima et al. (2014) have shown that exogenous SA treatment activates bZIP28, and consequently induces UPR gene expression in an IRE1‐dependent and NPR1‐independent manner. Chandra‐Shekara et al. (2004) have reported that a high SA level up‐regulates HRT transcription and HRT‐mediated resistance is required for SA‐dependent signal transduction (Chandra‐Shekara et al., 2004). The current study showed diminished HRT expression in BiP4/5‐, SGT1‐ and HSP90‐silenced leaves, and HRT/TCV CP‐specific bZIP60 induction. These results suggest that HRT expression is up‐regulated by ER stress response‐triggered transcription factors, such as bZIP60. Further analysis will be performed to explore this possibility by investigating HRT expression in ER stress‐related genetic mutants.
The HRT/TCV CP‐induced HR was disrupted in both BiP4/5‐silenced and Tm‐treated plants. The ER‐resident BiP protein levels responded in opposite ways to these two conditions, as follows: BiP protein levels were greatly reduced in BiP4/5‐silenced plants, whereas BiP proteins accumulated in Tm‐treated plants (Figs 5B and 8B). In both cases, ER‐QC was dysfunctional, which probably significantly reduced HRT protein accumulation. ER‐QC is crucial in plants for the abundance, quality and signalling of transmembrane immune receptors, including receptor‐like R proteins, such as Cf4 (Liebrand et al., 2012) and Ve1 (Liebrand et al., 2014), and pathogen‐associated molecular pattern receptors, such as EFR (elongation factor Tu receptor) and, to a lesser degree, FLS2 (flagellin‐sensitive 2) (Nekrasov et al., 2009). There are four BiP and three calreticulin genes in tomato. Among these, only calreticulin 3a silencing disrupted Cf4 protein function, whereas silencing of any BiP (except for BiP4) or calreticulin genes impaired Ve1‐mediated resistance (Liebrand et al., 2012). This indicates a differential requirement for ER‐QC in these two R gene‐mediated responses. Phylogenetic analysis revealed that NbBiP5 is very similar to NbBiP4 (Figs S2 and S4). Therefore, we compared NbBiP4 and NbBiP5 expression patterns, and found that they were largely similar (Fig. 3). Plant BiPs exhibit tissue‐specific expression and differential regulation in response to abiotic and biotic stresses through distinct signalling pathways (Moreno and Orellana, 2011; Tintor and Saijo, 2014). These lines of evidence suggest that different mechanisms can regulate the expression of even very close homologues, such as BiP4 and BiP5, in N. benthamiana. To investigate this issue further, we are currently engaged in a study to determine the specific requirement of ER‐QC components for HRT, such as BiP4, BiP5 and CRTs, using a specific VIGS approach to target individual ER‐QC components and yeast two‐hybrid experiments.
Although many studies have used VIGS to study plant–pathogen interactions, its use has several limitations (Senthil‐Kumar and Mysore, 2011). For instance, in studies designed to identify plant genes involved in antiviral mechanisms, silencing of the candidate plant gene can affect viral replication, thus making it hard to determine whether the phenotype is caused by the silencing of the plant gene or high virus proliferation (Burch‐Smith et al., 2004). To study further the biological significance of BiP4/5 in HRT‐mediated TCV disease resistance, we are generating transgenic N. benthamiana plants expressing HRT and studying the role of BiPs in Arabidopsis TCV resistance. Because TRV and TCV viruses have different properties (e.g. in replication strategy, proliferation, viral RNA levels and silencing suppressor), the effect of TRV‐mediated VIGS must be carefully evaluated to arrive at a full understanding of TCV resistance in N. benthamiana.
Experimental Procedures
Plant materials
The GFP‐expressing N. benthamiana line used for gene silencing experiments was kindly provided by Dr Dinesh Kumar (University of California‐Davis, CA, USA). For protein expression studies, leaves of 4‐week‐old N. benthamiana plants were subjected to agroinfiltration (Agrobacterium‐mediated transient transfection). Plants were grown in pots at 23°C in a growth chamber under a 16 h light/8 h dark cycle.
Plasmid construction for VIGS
To clone the N. benthamiana cDNA fragments corresponding to the HSP90 gene, we searched GenBank sequence information and designed specific primers. A 542‐bp cDNA fragment of N. benthamiana HSP90 (AY368904) was amplified using a pair of primers (5′‐TGACTGGGAGGAGCATTTG‐3′ and 5′‐GGTACCGCAGCAGTTCAGC‐3′). This cDNA was cloned into the T&A vector (Bioneer, Daejeon, South Korea). The cDNA sequence was confirmed by DNA sequencing, and it was cloned into the pTRV2 silencing vector.
VIGS and Agrobacterium‐mediated transient assays
VIGS and Agrobacterium‐mediated transient assays were performed according to the protocol described previously (Lee et al., 2013; Liu et al., 2002b) Each silencing experiment was repeated at least three times, and each experiment included at least three independent plants.
For transient expression assay, A. tumefaciens strain GV2260 containing the HRT, TCV CP, GFP, Rx, PVX‐mCP, PVX‐CP or HRT:3HA gene, driven by the 35S promoter, was cultured overnight and harvested by centrifugation. Cells were resuspended in infiltration medium [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.6, 10 mm MgCl2 and 200 μm acetosyringone] to an optical density at 600 nm (OD600) of 0.5 for HRT:3HA and 0.1 for the other genes, and then incubated for 2 h at room temperature. The inoculum was infiltrated into 4‐week‐old N. benthamiana or 3‐week‐old gene‐silenced N. benthamiana leaves with a 1‐mL needleless syringe. To observe the effect of Tm (Calbiochem, Los Angeles, CA, USA) on HR or protein status, drugs were prepared as 10 mm stocks in dimethylsulfoxide (DMSO) and stored at −20°C until use.
Cell death measurement by trypan blue staining or electrolyte leakage assay
Cell death was monitored by trypan blue staining according to a previously described method (Koch and Slusarenko, 1990). Samples were analysed with an Axiophot photomicroscope (Zeiss, Munich, Germany) under bright‐field optics.
The extent of cell death was measured quantitatively by monitoring electrolyte leakage. Ten leaf discs (diameter, 1 cm) were punched out of the infiltrated leaf tissue, and washed in distilled water for 30 min with gentle shaking to remove ions released by sampling‐related injury. Then, leaf discs were transferred to a plate containing 10 mL of distilled/deionized water, which was incubated in a growth chamber until 70 hpi. Conductivity measurements were taken at the indicated times using a NeoMet EC Meter EC‐470L conductivity meter (iSTE, Seoul, South Korea); three replicates were performed for each treatment.
qRT‐PCR analysis
Total RNAs were extracted from infiltrated leaf tissues using Trizol reagent (Molecular Research Center, Inc., Cincinnati, OH, USA), and cDNA synthesis was performed with MMLV reverse transcriptase (Invitrogen, Grand Island, NY, USA) using oligo(dT), according to the manufacturer's instructions. qRT‐PCR was performed as described by Liu et al. (2002b) with some modifications. Briefly, the PCR conditions were as follows: first denaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 20 s; annealing at a primer‐specific temperature for 30 s; elongation at 72°C for 20 s; and a melting curve consisting of 1°C steps from 65 to 95°C. Reactions were performed on a CFX Connect™ Real‐Time System (Bio‐Rad, Hercules, CA, USA). Relative expression levels of the indicated genes, normalized against the expression level of NbActin, were calculated via the comparative C(T) method. The primers used are listed in Table S2 (see Supporting Information).
Statistical analysis
Data were analysed using an analysis of variance (ANOVA) to evaluate the expression levels of the relative genes or the conductivity of ion leakage. Student's t‐test, or one‐way ANOVA, followed by Tukey's honestly significant difference (HSD) test, was used to compare the means of the treatments in each experiment. All statistical analyses were conducted using SPSS v. 18.
Immunoblot analysis
Total cellular protein was extracted from three infiltrated leaf discs using urea lysis buffer (8 m urea, 100 mm NaH2PO4, 10 mm Tris‐HCl, pH 8.0), and the extracts were separated by 10% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to a poly(vinylidene difluoride) (PVDF) membrane (Pierce, Rockford, IL, USA) using a wet‐transfer apparatus. The protein blots were incubated overnight in 5% (w/v) non‐fat dry milk in phosphate‐buffered saline (PBS; 10 mm Na2HPO4, 2 mm KH2PO4, 137 mm NaCl, pH 7.4) supplemented with 0.1% (v/v) Tween 20 (PBST) containing anti‐HA (Roche, Lewes, UK), anti‐TCV CP (Kang et al., 2010), anti‐BiP (SPA818), anti‐HSP70 (SPA817) (Stressgen Biotechnology, Victoria, BC, Canada), anti‐HSP90 and anti‐GFP (Santa Cruz Biotechnology, Dallas, TX, USA) antibodies.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Haemagglutinin (HA)‐tagged HRT is functional. Co‐expression of HRT‐HA and Turnip crinkle virus (TCV) coat protein (CP) induced the hypersensitive response (HR) in Nicotiana benthamiana. Numbers indicate concentration (optical density at 600 nm, OD600) of Agrobacterium cells. The images were photographed at 3 days post‐infiltration (dpi).
Fig. S2 Phylogenetic analysis of CaBLP5 (GenBank accession no. KC912859) and its homologues. The amino acid sequences of 26 CaBLP5 homologues were imported into MEGA 6 (Tamura et al., 2013) for multiple sequence alignment with ClustalW (Larkin et al., 2007). Phylogenetic analysis was performed using the neighbour‐joining (Saitou and Nei., 1987) and bootstrap methods. The bootstrap consensus tree was inferred from 500 replicates. The scale bar indicates the lengths of the branches (relative evolutionary distance). The protein sequences are deposited in GenBank under the following accession numbers: Solanum lycopersicum SlBiP5 (XP_004234985.1) and SlBiP (NP_001234636.1), Solanum tuberosum StBiP5‐L (XP_006350519.1) and StBiP‐L (XP_006343810.1), Nicotiana benthamiana NbBLP4 (ACK55195.1), Zea mays ZmBiP3 (NP_001105894.1) and ZmBiP2 (NP_001105893.1), Arabidopsis thaliana AtBiP2 (NP_851119.1) and AtBiP1 (NP_198206.1), Nicotiana tabacum NtBiP4 (Q03684.1), Cucumis sativus CsBiP5‐L (XP_004143862.1), Vitis vinifera VvBiP5 (XP_002263323.1) and VvBiP5‐L (XP_002276268.2), Nicotiana sylvestris NsBiP5 (XP_009773333.1), NsBiP4 (XP_009788736.1), NsBiP (XP_009802727.1) and NsBiP4‐L (XP_009770477.1), Nicotiana tomentosiformis NtoBiP5 (XP_009592769.1), NtoBiP4 (XP_009588550.1), NtoBiP (XP_009593820.1) and NtoBiP4‐L (XP_009619852.1), Ricinus communis RsHSP (XP_002518865.1), Cucumis sativus CsBiP5‐l (XP_004143862.1) and Glycine max GmBiP (XP_003525327.2), GmBiP isoform A (NP_001234941.1) and GmBiP isoform B (NP_001238736.1).
Fig. S3 Multiple sequence alignment of CaBLP5 and six Nicotiana benthamiana BiP genes with Clustal 2.1. Asterisks on the bottom line of the alignment indicate identical residues in a given sequence position. Within the aligned sequences, dashes indicate the gaps that were inserted to optimize the alignment. The CaBLP5 nucleotide sequences in red were used for the Tobacco rattle virus (TRV)2‐CaBLP5 construct. Residues underlined in red represent gene‐specific primer sequences used for the detection of gene expression in GFP‐ or CaBLP5‐silenced plants. Residues in turquoise indicate stretches of more than 21 nucleotides (Liu et al., 2002a) that are identical between TRV2‐CaBLP5 and the corresponding NbBiP homologues. It should be noted that TRV2‐CaBLP5 contains a stretch of 21 nucleotides with 100% identity to Nbv3K645786225 and Nbv3K645789686, and multiple 21‐nucleotide stretches with 100% identity to Nbv3K685813373, but no stretches of 21 nucleotides with perfect matches to other BiP homologues.
Fig. S4 Phylogenetic analysis of CaBLP5 and its tobacco and Arabidopsis homologues. The amino acid sequences of 11 CaBLP5 homologues were imported into MEGA 6 for multiple sequence alignment with ClustalW. Phylogenetic analysis was performed using the neighbour‐joining and bootstrap methods. The bootstrap consensus tree was inferred from 500 replicates. The scale bar indicates the lengths of the branches (relative evolutionary distance). GenBank accession numbers for the homologous proteins are given in the legend of Fig. S2.
Fig. S5 Putative amino acid sequence alignment of CaBLP5 (AGS42239) and its Nicotiana benthamiana homologues. The amino acid sequences of three CaBLP5 homologues were aligned with ClustalW. Asterisks on the bottom line of the alignment indicate identical residues in a given sequence position; single and double dots refer to highly and moderately conserved residues, respectively. Within the aligned sequences, dashes indicate gaps that were inserted to optimize the alignment.
Fig. S6 Induction of Rx‐mediated hypersensitive response (HR) resulted in an increase in the level of BiPs. Rx and mutant Potato virus X coat protein (PVX mCP) or wild‐type PVX CP (PVX CP) were co‐expressed in leaves of Nicotiana benthamiana. Total protein was extracted from N. benthamiana leaves expressing the indicated proteins at the designated time points and analysed by immunoblotting with anti‐BiP or anti‐cytosolic heat shock protein (HSP) 70 antibodies. Ponceau S staining of Rubisco was used as a loading control.
Fig. S7 Morphological phenotype of the gene‐silenced Nicotiana benthamiana plant. GFP‐ (green fluorescent protein) and PDS (phytoene desaturase)‐silenced plants were used as negative and positive controls for the virus‐induced gene silencing (VIGS) experiment. The silenced plants were photographed at 21 days post‐infiltration (dpi).
Fig. S8 Silencing of RAR1. (A) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis was performed to assess the efficiency of gene silencing. Primers directed to a specific gene or NbActin were used with equal amounts of cDNA from silenced plants. Numbers indicate PCR cycles. (B) HRT/coat protein (CP)‐mediated hypersensitive response (HR) induction in the leaves of RAR1‐silenced plants. (C) HRT transcriptional levels in GFP‐ and RAR1‐silenced plants. The values were normalized to the expression of NbActin. Error bars represent standard deviation (n = 3). Means with the same letter are not significantly different (t‐test, P < 0.001).
Fig. S9 Effect of tunicamycin (Tm) on cell death during co‐expression of HRT/Turnip crinkle virus coat protein (TCV CP) or Rx/Potato virus X coat protein (PVX CP). TM, an endoplasmic reticulum (ER) stress‐inducing chemical, was co‐infiltrated at the indicated concentrations, and infiltration buffer was used as a control. Photographs were taken at 3 days post‐infiltration (dpi).
Fig. S10 Triggering of the unfolded protein response (UPR) pathway by tunicamycin (Tm) treatment. (A) Expression of BiP5 and bZiP60 in Tobacco rattle virus2‐green fluorescent protein (TRV2‐GFP)‐infected plants by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Error bars present the standard error of three replicates. Asterisks indicate a significant difference from the corresponding control (Student's t‐test, **P < 0.005). Tm 10, 10 µm Tm; Tm 50, 50 µm Tm. (B) Effects of Tm treatment on the BiP protein level. Total protein was prepared at the indicated time points (h). BiP and GFP proteins were detected using anti‐BiP and anti‐GFP antibodies.
Table S1. Putative CaBLP5 homologues identified in Nicotiana benthamiana. CaBLP5 homologues were isolated using blast searches in the N. benthamiana_transcriptome_v3_ unigenes95 database (http://benth-web-pro-1.ucc.usyd.edu.au/blast/search.php). Six CaBLP5 homologues were retrieved, which have an e‐value of less than 10−9 and more than 80% identity with positive orientation.
Table S2. List of the oligonucleotide primers used in this study.
Acknowledgements
We thank Dr Dinesh Kumar for providing the pTRV1, pTRV2 and pTRV2:SGT1 constructs, Dr Daniel F. Klessig for providing the HA‐tagged HRT clone and antisera against TCV CP, and Dr David Baulcombe for providing the Rx, PVX‐mCP and PVX‐CP clones. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF‐2009‐0090805), and by the KRIBB initiative program to JMP. The research was partly funded by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ009762), Rural Development Administration to C‐SO and the Texas State University‐Faculty Standup Program to H‐GK.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, A. and Mysore, K.S. (2013) The role of RAR1 in Agrobacterium‐mediated plant transformation. Plant Signal. Behav. 8, e26784. doi: 10.4161/psb 26784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini, A.A. , Asurmendi, S. , Hopp, H.E. and Beachy, R.N. (2006) Tobacco mosaic virus (TMV) and potato virus X (PVX) coat proteins confer heterologous interference to PVX and TMV infection, respectively. J. Gen. Virol. 87, 1005–1012. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Farnham, G. , Moffett, P. and Baulcombe, D.C. (2002) Constitutive gain‐of‐function mutants in a nucleotide binding site‐leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Bhattarai, K.K. , Li, Q. , Liu, Y. , Dinesh‐Kumar, S.P. and Kaloshian, I. (2007) The MI‐1‐mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol, 144, 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Anderson, J.C. , Martin, G.B. and Dinesh‐Kumar, S.P. (2004) Applications and advantages of virus‐induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. [DOI] [PubMed] [Google Scholar]
- Caplan, J.L. , Zhu, X. , Mamillapalli, P. , Marathe, R. , Anandalakshmi, R. and Dinesh‐Kumar, S.P. (2009) Induced ER chaperones regulate a receptor‐like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe, 6, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, H.H. , Silva, P.A. , Mendes, G.C. , Brustolini, O.J. , Pimenta, M.R. , Gouveia, B.C. , Valente, M.A. , Ramos, H.J. , Soares‐Ramos, J.R. and Fontes, E.P. (2014) The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 164, 654–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra‐Shekara, A.C. , Navarre, D. , Kachroo, A. , Kang, H.G. , Klessig, D. and Kachroo, P. (2004) Signaling requirements and role of salicylic acid in HRT‐ and rrt‐mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 40, 647–659. [DOI] [PubMed] [Google Scholar]
- Collier, S.M. and Moffett, P. (2009) NB‐LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B. , Pathirana, S. , Wu, H.J. , Kachroo, P. and Klessig, D.F. (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell, 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke, J. , Goldman, M.H. , Demolder, J. , Seurinck, J. and Botterman, J. (1991) The tobacco luminal binding protein is encoded by a multigene family. Plant Cell, 3, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Calvino, L. , Martínez‐Priego, L. , Szabo, E.Z. , Guzmán‐Benito, I. , González, I. , Canto, T. , Lakatos, L. and Llave, C. (2016) Tobacco rattle virus 16K silencing suppressor binds ARGONAUTE 4 and inhibits formation of RNA silencing complexes. J. Gen. Virol. 97, 246–257. [DOI] [PubMed] [Google Scholar]
- Garcia‐Marcos, A. , Pacheco, R. , Martianez, J. , Gonzalez‐Jara, P. , Diaz‐Ruiz, J.R. and Tenllado, F. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant–Microbe Interact. 22, 1431–1444. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. and Bhattacharjee, S. (2012) Effector‐triggered immunity signaling: from gene‐for‐gene pathways to protein–protein interaction networks. Mol. Plant–Microbe Interact. 25, 862–868. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Hypersensitive response‐related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Holt, B.F. III , Belkhadir, Y. and Dangl, J.L. (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932. [DOI] [PubMed] [Google Scholar]
- Hubert, D.A. , Tornero, P. , Belkhadir, Y. , Krishna, P. , Takahashi, A. , Shirasu, K. and Dangl, J.L. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, D.A. , He, Y. , McNulty, B.C. , Tornero, P. and Dangl, J.L. (2009) Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB‐LRR disease resistance protein regulation. Proc. Natl. Acad. Sci. USA, 106, 9556–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski, A. , Rowley, D.L. , Loer, D.S. , Foley, C. , Buta, G. and Herman, E.M. (1995) Binding‐protein expression is subject to temporal, developmental and stress‐induced regulation in terminally differentiated soybean organs. Planta, 195, 611–621. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Kuhl, J.C. , Kachroo, P. and Klessig, D.F. (2008) CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe, 3, 48–57. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Oh, C.S. , Sato, M. , Katagiri, F. , Glazebrook, J. , Takahashi, H. , Kachroo, P. , Martin, G.B. and Klessig, D.F. (2010) Endosome‐associated CRT1 functions early in resistance gene‐mediated defense signaling in Arabidopsis and tobacco. Plant Cell, 22, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, C.B. , Woo, Y.M. , Lee, D.J. , Lee, M.C. and Kim, C.S. (2007) Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol Biochem. 45, 722–728. [DOI] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, K. , Normington, K. , Sambrook, J. , Gething, M.J. and Mori, K. (1993) The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leborgne‐Castel, N. , Jelitto‐Van Dooren, E.P. , Crofts, A.J. and Denecke, J. (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell, 11, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.S. (1992) Mammalian stress response: induction of the glucose‐regulated protein family. Curr. Opin. Cell Biol. 4, 267–273. [DOI] [PubMed] [Google Scholar]
- Lee, J.H. , Kim, Y.C. , Choi, D. and Park, J.M. (2013) Identification of novel pepper genes involved in Bax‐ or INF1‐mediated cell death responses by high‐throughput virus‐induced gene silencing. Int. J. Mol. Sci. 14, 22 782–22 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W. , Smit, P. , Abd‐El‐Haliem, A. , de Jonge, R. , Cordewener, J.H. , America, A.H. , Sklenar, J. , Jones, A.M. , Robatzek, S. , Thomma, B.P. , Tameling, W.I. and Joosten, M.H. (2012) Endoplasmic reticulum‐quality control chaperones facilitate the biogenesis of Cf receptor‐like proteins involved in pathogen resistance of tomato. Plant Physiol. 159, 1819–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W. , Kombrink, A. , Zhang, Z. , Sklenar, J. , Jones, A.M. , Robatzek, S. , Thomma, B.P. and Joosten, M.H . (2014) Chaperones of the endoplasmic reticulum are required for Ve1‐mediated resistance to Verticillium . Mol. Plant Pathol. 15, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.L. , Wang, J.S. , Liu, H.C. , Chen, R.W. , Meyer, Y. , Barakat, A. and Delseny, M . (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana . Cell Stress Chaperones, 6, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2002a) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. and Dinesh‐Kumar, S.P. (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D.C. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Priego, L. , Donaire, L. , Barajas, D. and Llave, C. (2008) Silencing suppressor activity of the Tobacco rattle virus‐encoded 16‐kDa protein and interference with endogenous small RNA‐guided regulatory pathways. Virology, 376, 346–356. [DOI] [PubMed] [Google Scholar]
- Mishiba, K. , Nagashima, Y. , Suzuki, E. , Hayashi, N. , Ogata, Y. , Shimada, Y. and Koizumi, N. (2013) Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA, 110, 5713–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A.A. and Orellana, A. (2011) The physiological role of the unfolded protein response in plants. Biol. Res. 44, 75–80. [DOI] [PubMed] [Google Scholar]
- Nagashima, Y. , Iwata, Y. , Ashida, M. , Mishiba, K. and Koizumi, N. (2014) Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55, 1772–1778. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Li, J. , Batoux, M. , Roux, M. , Chu, Z.H. , Lacombe, S. , Rougon, A. , Bittel, P. , Kiss‐Papp, M. , Chinchilla, D. , van Esse, H.P. , Jorda, L. , Schwessinger, B. , Nicaise, V. , Thomma, B.P. , Molina, A. , Jones, J.D. and Zipfel, C. (2009) Control of the pattern‐recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L.D. , Cagna, G. , Stuttmann, J. , Wirthmüller, L. , Betsuyaku, S. , Witte, C.P. , Bhat, R. , Pochon, N. , Colby, T. and Parker, J.E. (2007) Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell, 19, 4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.S. , Pedley, K.F. and Martin, G.B. (2010) Tomato 14‐3‐3 protein 7 positively regulates immunity‐associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK {alpha}. Plant Cell, 22, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.J. , Bart, R. , Chern, M. , Canlas, P.E. , Bai, W. and Ronald, P.C. (2010) Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21‐mediated innate immunity in rice. PLoS One, 5, e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.J. , Song, M.Y. , Kim, C.Y. , Jeon, J.S. and Ronald, P.C. (2014a) Rice BiP3 regulates immunity mediated by the PRRs XA3 and XA21 but not immunity mediated by the NB‐LRR protein, Pi5. Biochem. Biophys. Res. Commun. 448, 70–75. [DOI] [PubMed] [Google Scholar]
- Park, S.Y. , Yin, X. , Duan, K. , Gelvin, S.B. and Zhang, Z.J. (2014b) Heat shock protein 90.1 plays a role in Agrobacterium‐mediated plant transformation. Mol. Plant, 7, 1793–1796. [DOI] [PubMed] [Google Scholar]
- Qu, F. , Ren, T. and Morris, T.J. (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F. , Martin‐Hernandez, A.M. and Baulcombe, D.C. (2001) Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Reavy, B. , Dawson, S. , Canto, T. and MacFarlane, S.A. (2004) Heterologous expression of plant virus genes that suppress post‐transcriptional gene silencing results in suppression of RNA interference in Drosophila cells. BMC Biotechnol. 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali, A. , Fitzgerald, U. , Deegan, S. and Gupta, S. (2010) Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell Biol. 2010, p. 830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665. [DOI] [PubMed] [Google Scholar]
- Shirasu, K. (2009) The HSP90‐SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. [DOI] [PubMed] [Google Scholar]
- Sticher, L.A. and Metraux, J.P. (2000) Inhibitors of N‐linked glycosylation induce systemic acquired resistance in cucumber. Physiol. Mol. Plant Pathol. 56, 245–252. [Google Scholar]
- Tamagnone, L. , Merida, A. , Stacey, N. , Plaskitt, K. , Parr, A. , Chang, C.F. , Lynn, D. , Dow, J.M. , Roberts, K. and Martin, C. (1998) Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell, 10, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Xie, M. , Kim, Y.J. , Zhou, J. , Klessig, D.F. and Martin, G.B. (1999) Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell, 11, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor, N. and Saijo, Y. (2014) ER‐mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Front. Plant Sci. 5, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Weaver, N.D. , Kesarwani, M. and Dong, X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science, 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wrobel, R.L. , O'Brian, G.R. and Boston, R.S. (1997) Comparative analysis of BiP gene expression in maize endosperm. Gene, 204, 105–113. [DOI] [PubMed] [Google Scholar]
- Xu, G. , Li, S. , Xie, K. , Zhang, Q. , Wang, Y. , Tang, Y. , Liu, D. , Hong, Y. , He, C. and Liu, Y. (2012) Plant ERD2‐like proteins function as endoplasmic reticulum luminal protein receptors and participate in programmed cell death during innate immunity. Plant J. 72, 57–69. [DOI] [PubMed] [Google Scholar]
- Ye, C. , Dickman, M.B. , Whitham, S.A. , Payton, M. and Verchot, J. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, C.M. , Kelly, V. , Payton, M. , Dickman, M.B. and Verchot, J. (2012) SGT1 is induced by the potato virus X TGBp3 and enhances virus accumulation in Nicotiana benthamiana . Mol. Plant, 5, 1151–1153. [DOI] [PubMed] [Google Scholar]
- Zhang, K. and Kaufman, R.J. (2004) Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279, 25 935–25 938. [DOI] [PubMed] [Google Scholar]
- Zhang, L. and Wang, A. (2012) Virus‐induced ER stress and the unfolded protein response. Front. Plant Sci. 3, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , DelGrosso, L. , Yigit, E. , Dempsey, D.M.A. , Klessig, D.F. and Wobbe, K.K. (2000) The amino terminus of the coat protein of Turnip crinkle virus is the AVR factor recognized by resistant Arabidopsis. Mol. Plant–Microbe Interact. 13, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Zimmermann, R. , Eyrisch, S. , Ahmad, M. and Helms, V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta, 1808, 912–924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Haemagglutinin (HA)‐tagged HRT is functional. Co‐expression of HRT‐HA and Turnip crinkle virus (TCV) coat protein (CP) induced the hypersensitive response (HR) in Nicotiana benthamiana. Numbers indicate concentration (optical density at 600 nm, OD600) of Agrobacterium cells. The images were photographed at 3 days post‐infiltration (dpi).
Fig. S2 Phylogenetic analysis of CaBLP5 (GenBank accession no. KC912859) and its homologues. The amino acid sequences of 26 CaBLP5 homologues were imported into MEGA 6 (Tamura et al., 2013) for multiple sequence alignment with ClustalW (Larkin et al., 2007). Phylogenetic analysis was performed using the neighbour‐joining (Saitou and Nei., 1987) and bootstrap methods. The bootstrap consensus tree was inferred from 500 replicates. The scale bar indicates the lengths of the branches (relative evolutionary distance). The protein sequences are deposited in GenBank under the following accession numbers: Solanum lycopersicum SlBiP5 (XP_004234985.1) and SlBiP (NP_001234636.1), Solanum tuberosum StBiP5‐L (XP_006350519.1) and StBiP‐L (XP_006343810.1), Nicotiana benthamiana NbBLP4 (ACK55195.1), Zea mays ZmBiP3 (NP_001105894.1) and ZmBiP2 (NP_001105893.1), Arabidopsis thaliana AtBiP2 (NP_851119.1) and AtBiP1 (NP_198206.1), Nicotiana tabacum NtBiP4 (Q03684.1), Cucumis sativus CsBiP5‐L (XP_004143862.1), Vitis vinifera VvBiP5 (XP_002263323.1) and VvBiP5‐L (XP_002276268.2), Nicotiana sylvestris NsBiP5 (XP_009773333.1), NsBiP4 (XP_009788736.1), NsBiP (XP_009802727.1) and NsBiP4‐L (XP_009770477.1), Nicotiana tomentosiformis NtoBiP5 (XP_009592769.1), NtoBiP4 (XP_009588550.1), NtoBiP (XP_009593820.1) and NtoBiP4‐L (XP_009619852.1), Ricinus communis RsHSP (XP_002518865.1), Cucumis sativus CsBiP5‐l (XP_004143862.1) and Glycine max GmBiP (XP_003525327.2), GmBiP isoform A (NP_001234941.1) and GmBiP isoform B (NP_001238736.1).
Fig. S3 Multiple sequence alignment of CaBLP5 and six Nicotiana benthamiana BiP genes with Clustal 2.1. Asterisks on the bottom line of the alignment indicate identical residues in a given sequence position. Within the aligned sequences, dashes indicate the gaps that were inserted to optimize the alignment. The CaBLP5 nucleotide sequences in red were used for the Tobacco rattle virus (TRV)2‐CaBLP5 construct. Residues underlined in red represent gene‐specific primer sequences used for the detection of gene expression in GFP‐ or CaBLP5‐silenced plants. Residues in turquoise indicate stretches of more than 21 nucleotides (Liu et al., 2002a) that are identical between TRV2‐CaBLP5 and the corresponding NbBiP homologues. It should be noted that TRV2‐CaBLP5 contains a stretch of 21 nucleotides with 100% identity to Nbv3K645786225 and Nbv3K645789686, and multiple 21‐nucleotide stretches with 100% identity to Nbv3K685813373, but no stretches of 21 nucleotides with perfect matches to other BiP homologues.
Fig. S4 Phylogenetic analysis of CaBLP5 and its tobacco and Arabidopsis homologues. The amino acid sequences of 11 CaBLP5 homologues were imported into MEGA 6 for multiple sequence alignment with ClustalW. Phylogenetic analysis was performed using the neighbour‐joining and bootstrap methods. The bootstrap consensus tree was inferred from 500 replicates. The scale bar indicates the lengths of the branches (relative evolutionary distance). GenBank accession numbers for the homologous proteins are given in the legend of Fig. S2.
Fig. S5 Putative amino acid sequence alignment of CaBLP5 (AGS42239) and its Nicotiana benthamiana homologues. The amino acid sequences of three CaBLP5 homologues were aligned with ClustalW. Asterisks on the bottom line of the alignment indicate identical residues in a given sequence position; single and double dots refer to highly and moderately conserved residues, respectively. Within the aligned sequences, dashes indicate gaps that were inserted to optimize the alignment.
Fig. S6 Induction of Rx‐mediated hypersensitive response (HR) resulted in an increase in the level of BiPs. Rx and mutant Potato virus X coat protein (PVX mCP) or wild‐type PVX CP (PVX CP) were co‐expressed in leaves of Nicotiana benthamiana. Total protein was extracted from N. benthamiana leaves expressing the indicated proteins at the designated time points and analysed by immunoblotting with anti‐BiP or anti‐cytosolic heat shock protein (HSP) 70 antibodies. Ponceau S staining of Rubisco was used as a loading control.
Fig. S7 Morphological phenotype of the gene‐silenced Nicotiana benthamiana plant. GFP‐ (green fluorescent protein) and PDS (phytoene desaturase)‐silenced plants were used as negative and positive controls for the virus‐induced gene silencing (VIGS) experiment. The silenced plants were photographed at 21 days post‐infiltration (dpi).
Fig. S8 Silencing of RAR1. (A) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis was performed to assess the efficiency of gene silencing. Primers directed to a specific gene or NbActin were used with equal amounts of cDNA from silenced plants. Numbers indicate PCR cycles. (B) HRT/coat protein (CP)‐mediated hypersensitive response (HR) induction in the leaves of RAR1‐silenced plants. (C) HRT transcriptional levels in GFP‐ and RAR1‐silenced plants. The values were normalized to the expression of NbActin. Error bars represent standard deviation (n = 3). Means with the same letter are not significantly different (t‐test, P < 0.001).
Fig. S9 Effect of tunicamycin (Tm) on cell death during co‐expression of HRT/Turnip crinkle virus coat protein (TCV CP) or Rx/Potato virus X coat protein (PVX CP). TM, an endoplasmic reticulum (ER) stress‐inducing chemical, was co‐infiltrated at the indicated concentrations, and infiltration buffer was used as a control. Photographs were taken at 3 days post‐infiltration (dpi).
Fig. S10 Triggering of the unfolded protein response (UPR) pathway by tunicamycin (Tm) treatment. (A) Expression of BiP5 and bZiP60 in Tobacco rattle virus2‐green fluorescent protein (TRV2‐GFP)‐infected plants by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Error bars present the standard error of three replicates. Asterisks indicate a significant difference from the corresponding control (Student's t‐test, **P < 0.005). Tm 10, 10 µm Tm; Tm 50, 50 µm Tm. (B) Effects of Tm treatment on the BiP protein level. Total protein was prepared at the indicated time points (h). BiP and GFP proteins were detected using anti‐BiP and anti‐GFP antibodies.
Table S1. Putative CaBLP5 homologues identified in Nicotiana benthamiana. CaBLP5 homologues were isolated using blast searches in the N. benthamiana_transcriptome_v3_ unigenes95 database (http://benth-web-pro-1.ucc.usyd.edu.au/blast/search.php). Six CaBLP5 homologues were retrieved, which have an e‐value of less than 10−9 and more than 80% identity with positive orientation.
Table S2. List of the oligonucleotide primers used in this study.


