Summary
Xanthomonas axonopodis pv. manihotis (Xam) employs transcription activator‐like (TAL) effectors to promote bacterial growth and symptom formation during infection of cassava. TAL effectors are secreted via the bacterial type III secretion system into plant cells, where they are directed to the nucleus, bind DNA in plant promoters and activate the expression of downstream genes. The DNA‐binding activity of TAL effectors is carried out by a central domain which contains a series of repeat variable diresidues (RVDs) that dictate the sequence of bound nucleotides. TAL14Xam668 promotes virulence in Xam strain Xam668 and has been shown to activate multiple cassava genes. In this study, we used RNA sequencing to identify the full target repertoire of TAL14Xam668 in cassava, which includes over 50 genes. A subset of highly up‐regulated genes was tested for activation by TAL14CIO151 from Xam strain CIO151. Although TAL14CIO151 and TAL14Xam668 differ by only a single RVD, they display differential activation of gene targets. TAL14CIO151 complements the TAL14Xam668 mutant defect, implying that shared target genes are important for TAL14Xam668‐mediated disease susceptibility. Complementation with closely related TAL effectors is a novel approach to the narrowing down of biologically relevant susceptibility genes of TAL effectors with multiple targets. This study provides an example of how TAL effector target activation by two strains within a single species of Xanthomonas can be dramatically affected by a small change in RVD–nucleotide affinity at a single site, and reflects the parameters of RVD–nucleotide interaction determined using designer TAL effectors in transient systems.
Keywords: Manihot esculenta (cassava); susceptibility (S) gene; TAL14; transcription activator‐like effector (TAL effector), Xanthomonas axonopodis pv. manihotis (Xam)
Introduction
The transcription activator‐like (TAL) effectors of plant‐pathogenic Xanthomonas species promote bacterial growth and disease symptom formation in diverse plant hosts (Schornack et al., 2013). After translocation into plant cells via the bacterial type III secretion system, TAL effectors are targeted to the nucleus, where they bind specific DNA sequences in gene promoters and activate the expression of downstream genes (Boch and Bonas, 2010; Bogdanove et al., 2010). TAL effectors have a modular architecture consisting of an N‐terminal region with a type III secretion signal, a C‐terminal region with nuclear localization signals and an acidic activation domain, and a central repeat domain that mediates DNA binding. A TAL effector's specific function during disease progression is determined by the host gene(s) that it activates, making the DNA‐binding domain of particular interest, as it dictates where in the plant genome the TAL effector will bind. Each repeat mediates binding to a single consecutive nucleotide and the sequence of nucleotides bound by a TAL effector is termed the effector binding element (EBE).
The majority of polymorphism between repeats within a TAL effector DNA‐binding domain is at the 12th and 13th amino acids, termed the repeat variable diresidues (RVDs). By observing the correspondence of RVDs with specific nucleotides in known TAL effector–EBE pairs, the TAL effector–DNA binding ‘code’ was elucidated (Boch et al., 2009; Moscou and Bogdanove, 2009). On the basis of these association frequencies, the RVDs NG (asparagine–glycine, Asn–Gly), NI (asparagine–isoleucine, Asn–Ile) and HD (histidine–aspartic acid, His–Asp) have been shown to be highly specific for thymine (T), adenine (A) and cytosine (C), respectively, whereas NS (asparagine–serine, Asn–Ser) and NN (Asn–Asn) are less specific in their nucleotide preference (Boch et al., 2009; Moscou and Bogdanove, 2009). Crystal structures of TAL effectors bound to their DNA targets confirmed the one‐to‐one nature of RVD–nucleotide interaction and showed that the 13th amino acid of the repeat interacts directly with the DNA base, and is referred to as the base‐specifying residue (BSR) (de Lange et al., 2014; Deng et al., 2012; Mak et al., 2012). The majority of natural TAL effector EBEs are directly preceded by a thymine (T0) which is required for efficient TAL effector binding and gene activation (Boch et al., 2009; Doyle et al., 2013; Römer et al., 2009b, 2010).
The knowledge of the TAL effector–DNA binding code has made it possible to predict potential EBEs in a genome, albeit with a large number of false‐positive predictions, and has enabled the development of TAL effector‐based biotechnological applications, such as TAL effector nucleases (TALENs) (Bogdanove and Voytas, 2011; Carroll, 2014; Joung and Sander, 2013; Sun and Zhao, 2013). Driven by the need for more specific and efficient DNA targeting by TAL effector‐based biotechnologies, much has been clarified in recent years with regard to the parameters that influence TAL effector–DNA binding, such as RVD efficiency, RVD–nucleotide affinity, polarity effects and the vicinity of EBEs to core promoter elements (Cernadas et al., 2014; Cong et al., 2012; Grau et al., 2013; Meckler et al., 2013; Moore et al., 2014; Streubel et al., 2012). These studies have primarily been performed using highly expressed artificial TAL effector–EBE pairs assayed transiently for activation strength. The way in which RVD substitutions affecting TAL effector binding efficiency affect the function of natural TAL effectors in the context of their plant hosts is an open question.
Several TAL effector‐targeted host genes have been shown to promote disease susceptibility upon activation (Hutin et al., 2015). The pepper (Capsicum annuum) transcription factor UPA20, a target of the TAL effector AvrBs3 from Xanthomonas euvesicatoria (Xe), contributes to symptom formation by inducing cell hypertrophy (Kay et al., 2007). The sweet orange (Citrus sinensis) transcription factor CsLOB1, which is targeted by multiple TAL effectors from X. citri ssp. citri (Xcc), promotes pustule formation and in planta bacterial growth (Hu et al., 2014; Li et al., 2014). TAL2g of X. oryzae pv. oryzicola activates the predicted sulfate transporter OsSULTR3;6 to promote symptom formation and bacterial exudation from leaves of rice (Oryza sativa) (Cernadas et al., 2014). SWEET sugar transporters have been established as important virulence targets for TAL effectors of X. oryzae pv. oryzae (Xoo) and X. axonopodis pv. manihotis (Xam) during bacterial blight of rice and cassava (Manihot esculenta), respectively (Antony et al., 2010; Chen et al., 2010, 2012; Cohn et al., 2014; Yang et al., 2006).
Cassava bacterial blight (CBB), which is elicited by Xam, is a devastating disease of this staple food crop. Cassava is grown in tropical and subtropical areas of Africa, Asia and South America, and is an inexpensive source of dietary calories and nutrients for millions of people in these regions (Howeler et al., 2013). Outbreaks of CBB can cause extensive crop damage and few reports of CBB resistance exist (reviewed in López and Bernal, 2012; Lozano, 1986). The recent surge of next‐generation sequencing technologies has allowed sophisticated questions regarding the molecular interactions between Xam and its non‐model host cassava to be addressed (Arrieta‐Ortiz et al., 2013; Bart et al., 2012). In particular, TAL effectors have been shown to be important in promoting Xam virulence (Castiblanco et al., 2012; Cohn et al., 2014).
The highly virulent Xam strain Xam668 contains five TAL effectors, two of which, TAL20Xam668 and TAL14Xam668, have been shown to have virulence contributions (Cohn et al., 2014). TAL14Xam668 contributes to in planta bacterial growth and TAL20Xam668 promotes both bacterial growth and water‐soaking symptom development during CBB infection. RNA‐Sequencing (RNA‐Seq) has revealed a single target gene for TAL20Xam668, MeSWEET10a, a SWEET sugar transporter that has been shown to be responsible for the TAL20Xam668‐mediated susceptibility to CBB through the use of designer TAL effectors (dTALEs), which independently activate MeSWEET10a and complement the TAL20Xam668 mutant phenotype (Cohn et al., 2014). A search for TAL effector targets among the most highly up‐regulated genes during Xam infection of cassava leaves has revealed that TAL14Xam668 has multiple targets, which contrasts with TAL20Xam668's activation of a single gene.
For this study, we originally set out to identify the TAL14Xam668‐targeted gene responsible for the promotion of in planta bacterial growth. We anticipated the testing of roughly 10 candidate target genes using dTALEs for complementation of the TAL14Xam668 mutant phenotype. However, RNA‐Seq revealed that TAL14Xam668 is a promiscuous TAL effector with a collection of over 50 target genes. We found that TAL14CIO151, which differs from TAL14Xam668 by a single repeat, is able to activate only a small subset of the highly up‐regulated gene targets of TAL14Xam668. Nevertheless, TAL14CIO151 complements a TAL14Xam668 mutant strain, suggesting that the TAL effectors activate the same physiologically relevant host susceptibility (S) gene(s). By analysis of the predicted EBEs for TAL14Xam668 and TAL14CIO151 in the promoters of genes activated by one or both TAL14 proteins, we found that this difference in gene activation capability is consistent with known effects of differential RVD specificity and nucleotide affinity. This study reveals the dramatic impact of a single RVD difference on host targets in two naturally occurring TAL effectors within a single species of Xanthomonas, and demonstrates the use of highly similar TAL effectors as tools to rapidly eliminate false‐positive candidate S genes.
Results
TAL14Xam668 is predicted to target many cassava promoters
TAL14Xam668 has a large number of predicted target EBEs relative to the other TAL effectors of Xam668 (Cohn et al., 2014). Target predictions were generated by TALE‐NT (2.0) Target Finder, which identifies potential targets on the basis of the observed RVD–nucleotide association frequencies (i.e. RVD specificity), but does not take into account RVD efficiency or EBE location (Doyle et al., 2012). The observation that TAL14Xam668 has a relatively large number of predicted targets was confirmed when we ran Target Finder on the 1‐kb cassava promoterome, defined as 1 kb upstream of the annotated transcriptional start sites, with more stringent parameters than previously reported (Fig. 1a). We first hypothesized that the number of TAL14Xam668 targets was large, because its small size (low repeat number) makes it more likely to match a suitable nucleotide sequence at random than a TAL effector with more repeats. We compared the number of predicted targets for the Xam668 TAL effectors which have RVD numbers ranging from 13 to 22, and found that TAL14Xam668 has more targets than would be expected given that it differs in length from TAL13Xam668 and TAL15Xam668 by only one RVD (Fig. 1a).
Figure 1.
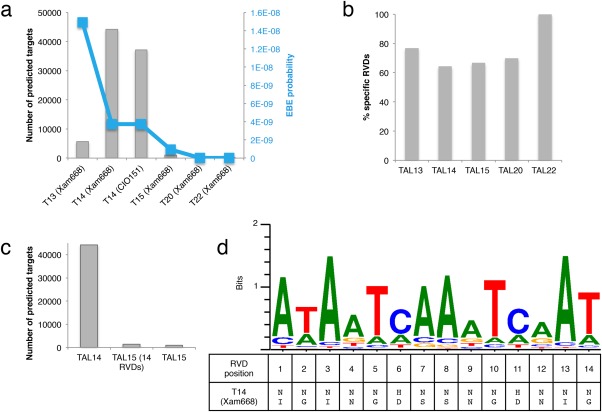
TAL14Xam668 is predicted to target many cassava promoters. (a) Target effector binding elements (EBEs) for the transcription activator‐like (TAL) effectors of Xam668 and TAL14CIO151 were predicted using Target Finder (TALE‐NT 2.0) in the 1‐kb cassava promoterome. The predicted target number is plotted (left y‐axis, bar graph) for each TAL effector. The number in each TAL effector name is the number of repeat variable diresidues (RVDs) present in the protein. To visualize the effect of RVD number on EBE prediction, we show the rate at which a correct EBE sequence would occur in the genome at random, where each RVD has a 25% chance of being aligned to its preferred nucleotide (0.25RVD#) (right y‐axis, line graph). (b) The percentage of specific RVDs (NI, NG, HD) for each Xam668 TAL effector is plotted for comparison. (c) EBEs for the first 14 RVDs of TAL15Xam668 predicted as in (a) and plotted alongside the number of predicted targets of TAL14Xam668 and TAL15Xam668 for comparison. (d) The consensus predicted EBE for the top 5000 predicted TAL14Xam668 EBEs in the 1‐kb cassava promoterome, together with the RVD sequence of TAL14Xam668.
We next speculated that TAL14Xam668 has a large number of predicted targets because it contains a higher proportion of RVDs with relaxed specificity. Of the RVDs present in Xam668 TAL effectors, HD, NG and NI are specific with a preference for binding C, T and A, respectively, whereas the RVDs NN and NS are less stringent in their nucleotide specificity (Moscou and Bogdanove, 2009). We calculated the percentage of specific RVDs (NI, NG, HD) present in the RVD sequence for each TAL effector to determine whether TAL14Xam668 is generally less specific. TAL14Xam668 has the lowest percentage of specific RVDs (64.3%, 9/14); however, this percentage does not differ greatly from that of TAL15Xam668, which has 66.7% (10/15) specific RVDs (Fig. 1b). Furthermore, the first 14 RVDs of TAL15Xam668 consist of the same percentage of specific RVDs as those of TAL14Xam668, yet maintain a much smaller number of predicted targets in the 1‐kb cassava promoterome (Fig. 1c). Both TAL14Xam668 and TAL15Xam668 begin with the RVDs NI‐NG‐NI which, together with a 5′ T (T0), prefer to bind the sequence TATA. Therefore, both TAL14Xam668 and TAL15Xam668 are likely to have predicted target EBEs that are anchored in a TATA‐box sequence (TATAWA), showing that predicted binding to a core promoter motif may contribute to TAL14Xam668's apparent target promiscuity, but is not the only explanation (Butler and Kadonaga, 2002). We conclude that TAL14Xam668 is predicted to bind a group of nucleotide sequences that are relatively common in the cassava promoterome compared with the groups of sequences predicted to be bound by the other TAL effectors of Xam668. The consensus sequence for the top 5000 predicted targets of TAL14Xam668 is displayed in Fig. 1d.
RNA‐Seq identifies targets of TAL14Xam668
The prediction of TAL effector target EBEs using only RVD–nucleotide association frequencies results in many false‐positive predictions, making transcriptomic approaches necessary to identify true TAL effector targets (Cohn et al., 2014; Grau et al., 2013). We conducted an RNA‐Seq experiment to identify the full repertoire of genes activated by TAL14Xam668 because of its large number of predicted targets and also because of the virulence defect seen in the TAL14Xam668 mutant strain (Cohn et al., 2014). Xe strain 85‐10, which does not cause disease on cassava and has no TAL effectors of its own, was used to deliver TAL14Xam668 to cassava leaf tissue independently of the other TAL effectors of Xam668 (Cohn et al., 2014). We compared genes that were up‐regulated in leaf tissue infiltrated with mock inoculation (10 mm MgCl2) versus Xam668, and those that were up‐regulated between Xe and Xe(TAL14) at a single time point (48 h post‐inoculation, hpi). The genes present in both comparisons were designated as potential targets of TAL14Xam668 (Fig. 2). We found a total of 52 genes up‐regulated in a TAL14Xam668‐dependent manner [log2(fold change) >1, fragments per kilobase transcript per million mapped reads (FPKM) difference >5] (Fig. 3). These target genes encode a range of proteins, including pectate lyases and other cell wall‐modifying enzymes, EamA‐like/MtN21 transporters, several proteases, glyceraldehyde‐3‐phosphate dehydrogenases, leucine‐rich repeat (LRR) kinases, and proteins of unknown function.
Figure 2.
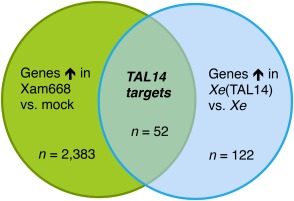
Venn diagram illustrating the RNA‐Sequencing (RNA‐Seq) experimental approach to identify TAL14Xam668 targeted genes. Cassava leaf tissue was infiltrated with strains at an optical density at 600 nm (OD600) of 0.5 and collected after 48 h. RNA‐Seq analysis identified genes up‐regulated [log2(fold change) >1 and fragments per kilobase transcript per million mapped reads (FPKM) difference >5] by both Xanthomonas axonopodis pv. manihotis (Xam) strain Xam668 compared with mock inoculation (10 mm MgCl2) and by X. euvesicatoria 85‐10 delivering TAL14Xam668 [Xe(TAL14)] versus wild‐type Xe as potential TAL14Xam668 targets. The total number of genes identified in each group is displayed.
Figure 3.
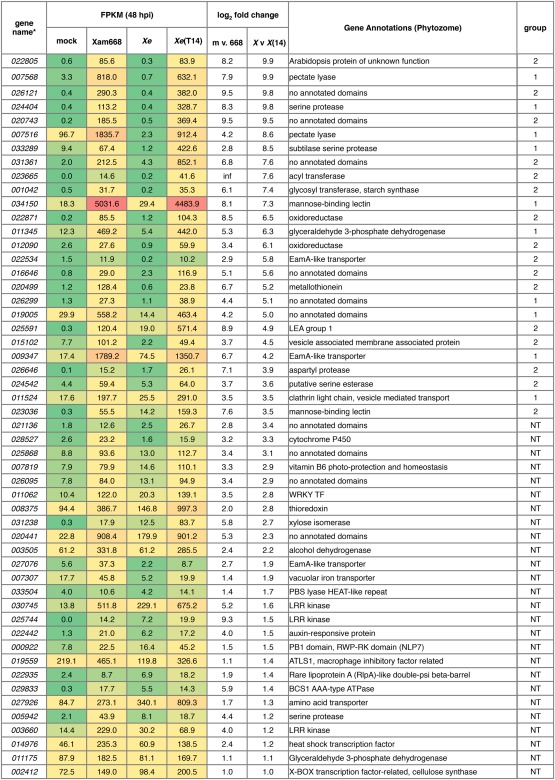
RNA‐Sequencing (RNA‐Seq) reveals 52 genes activated in a TAL14Xam68‐dependent manner. Genes activated by TAL14Xam668 are defined as showing log2(fold change) > 1 and fragments per kilobase transcript per million mapped reads (FPKM) difference > 5 in both the mock versus Xanthomonas axonopodis pv. manihotis (Xam) strain Xam668 and Xanthomonas euvesicatoria (Xe) versus Xe(T14) comparisons. For each cassava gene, FPKM values for mock, Xam668, Xe and Xe(T14) are listed and colour coded by value, together with the log2(fold change) between mock and Xam668 (m v 668), and between Xe and Xe(T14) [X v X(14)]. Genes are listed by decreasing values for the latter comparison. Phytozome gene annotations are listed, together with the assigned promoter group (NT, not tested). *Only gene numbers are shown; full gene names are cassava4.1_xxxxxx, where XXXXXX represents the unique 6‐digit gene number shown above.
We validated the TAL14Xam668‐dependent activation of the 26 most highly activated genes through semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) of cassava leaf tissue inoculated with wild‐type Xam668 and a mutant Xam668 strain missing the plasmid segment (ps) which encodes the TAL14Xam668 gene (Xam668ΔTAL14ps) (Figs 4a, S1a, b, see Supporting Information). Xam668ΔTAL14ps is a markerless strain that shows the same bacterial growth defect as the previously studied TAL14Xam668 knockout strain (Xam668ΔTAL14), and is fully complemented by the introduction of wild‐type TAL14Xam668 (Fig. 5a) (Cohn et al., 2014). Twenty‐five of the 26 tested genes are up‐regulated by TAL14Xam668 alone. As the activation of cassava4.1_009347 was induced by Xam668, but also by Xam668ΔTAL14ps, we tested for activation by the other Xam668 TAL effectors, and found that both TAL14Xam668 and TAL22Xam668 activate this gene (Fig. S2, see Supporting information).
Figure 4.
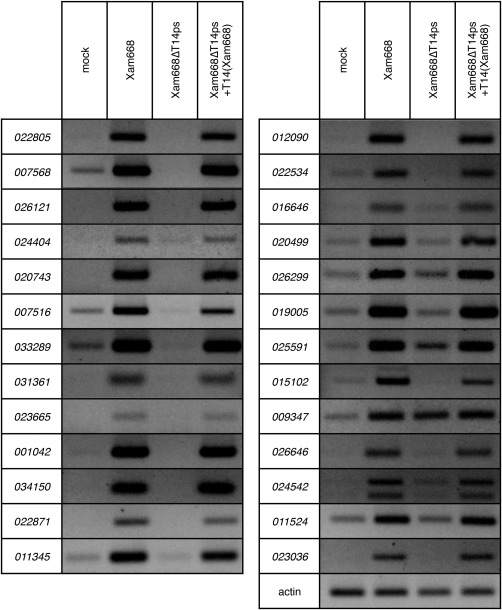
Confirmation of TAL14Xam668‐dependent target activation by semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR). The 26 most highly activated TAL14Xam668 targets were confirmed by semi‐quantitative RT‐PCR. Gene expression was measured at 48 h post‐inoculation (hpi) in leaf tissue inoculated with mock infiltration (10 mm MgCl2), Xam668, Xam668ΔTAL14ps and Xam668ΔTAL14ps(T14) strains at an optical density at 600 nm (OD600) of 0.5.
Figure 5.
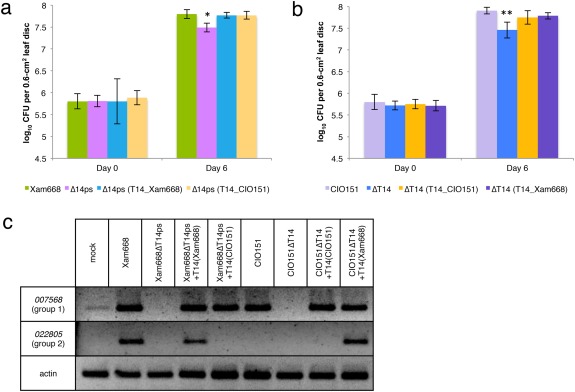
TAL14CIO151 complements Xam668ΔT14ps and TAL14Xam668 complements CIO151ΔT14 in bacterial growth assays. (a, b) Bacterial populations at leaf midvein inoculation points were measured at days 0 and 6. Data are represented as the mean colony‐forming units (CFU) per 0.6‐cm2 leaf disc encompassing the inoculation point (± standard deviation). *Significantly lower growth than Xam668, two‐tailed t‐test, P < 0.001. **Significantly lower growth than CIO151, two‐tailed t‐test, P < 0.005. CIO151ΔTAL14(TAL14Xam668) and CIO151ΔTAL14(TAL14CIO151) grow to higher levels than CIO151ΔTAL14 (P = 0.054 and 0.016, respectively). Growth assays were repeated four times with similar results. (c) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) of a group 1 gene (cassava4.1_007568) and a group 2 gene (cassava4.1_022805) is displayed to show that the TAL14 proteins retain their gene activation specificities in a non‐native context.
TAL14CIO151 from Xam strain CIO151 activates a subset of TAL14Xam668 targets
Xam strain CIO151 has two TAL effectors: TAL14CIO151 and TAL21CIO151 (Bart et al., 2012). Growth assays of CIO151 and a TAL14CIO151 knockout strain (CIO151ΔTAL14) showed that TAL14CIO151 promotes in planta bacterial growth (Figs 5b and S1c). TAL14Xam668 and TAL14CIO151 have identical amino acid sequences, except for three amino acid differences in the fifth DNA‐binding domain repeat (Fig. 6a) (Bart et al., 2012). The fifth RVD of TAL14Xam668 is NG which preferentially interacts with T, whereas the fifth RVD of TAL14CIO151 is NI which preferentially interacts with A. Similar to the observations for TAL14Xam668, TAL14CIO151 has an unexpectedly high number of predicted EBEs in the 1‐kb cassava promoterome (Fig. 1a). Using semi‐quantitative RT‐PCR, we tested the ability of TAL14CIO151 to activate the top targets of TAL14Xam668. Given the similarity between the two TAL14 proteins, we were surprised to find that TAL14CIO151 was only able to activate 10 of the 26 top TAL14Xam668 targets (Fig. 6b). We subsequently refer to the group of genes activated by both TAL14Xam668 and TAL14CIO151 as group 1, and to the group of genes activated by only TAL14Xam668 as group 2 (Figs 3 and 6b). Gene targets were tested for activation in the presence of 50 µm cycloheximide (CHX), an inhibitor of eukaryotic protein synthesis, to determine whether they were direct targets of the TAL14 proteins (Fig. S3, see Supporting information). All targets were directly activated, except the group 2 gene, cassava4.1_022534. Gene groups 1 and 2 were not distinguished by common function: group 1 contains two pectate lyases, two proteases and a mannose‐binding lectin, among others; group 2 contains an acyl transferase, a glycosyl transferase, two EamA‐like transporters and multiple proteins with no annotated domains, among others.
Figure 6.
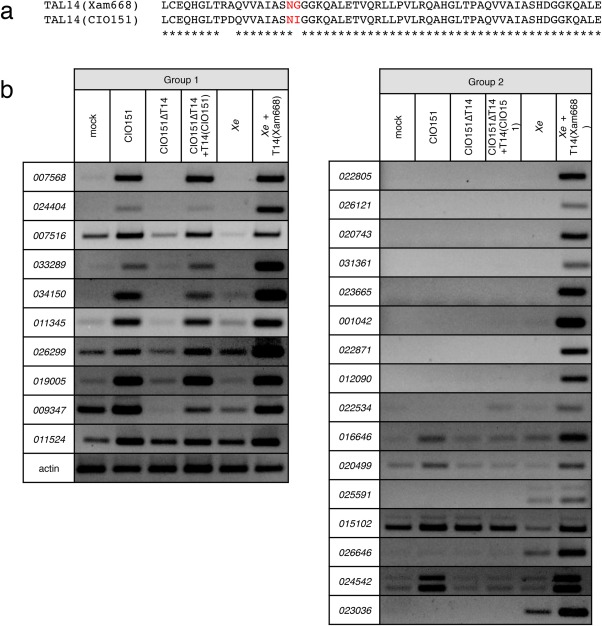
TAL14Xam668 and TAL14CIO151 differ by one repeat variable diresidue (RVD) and differentially activate host target genes. (a) Amino acid alignment of the fifth DNA‐binding domain repeat of TAL14Xam668 and TAL14CIO151. RVDs are coloured red. (b) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) of leaf tissue inoculated with mock inoculation (10 mm MgCl2), Xanthomonas axonopodis pv. manihotis (Xam) strain CIO151, CIO151ΔTAL14, CIO151ΔTAL14(TAL14CIO151), X. euvesicatoria (Xe) and Xe(TAL14Xam668) at an optical density at 600 nm (OD600) of 0.5, and collected at 48 h post‐inoculation (hpi), shows activation of genes by both TAL14Xam668 and TAL14CIO151 (group 1 genes), or by TAL14Xam668 only (group 2 genes).
To gain more information with regard to why TAL14CIO151 is able to activate some of the targets of TAL14Xam668 and not others, we decided to compare the consensus of the predicted EBEs for TAL14Xam668 and TAL14CIO151 in the directly targeted promoters of groups 1 and 2 using two EBE prediction programs: Target Finder (TALE‐NT 2.0) and TALgetter (Doyle et al., 2012; Grau et al., 2013). Target Finder takes into account only RVD–nucleotide association frequencies (i.e. binding specificity), whereas TALgetter takes into account RVD binding specificity, the positive contribution of binding efficiency (i.e. the strength of a matching RVD–nucleotide interaction) and the negative penalty for non‐matching RVD–nucleotide pairs. The gene promoters of groups 1 and 2 were defined as 300 bp upstream of the start codon, a region which has been shown to be enriched in TAL effector target sites (Grau et al., 2013).
Both the Target Finder and TALgetter analyses showed a greater occurrence of an A at position 5 in the group 1 consensus TAL14CIO151 EBE than in the group 2 consensus TAL14CIO151 EBE. This was in contrast with position 5 of the consensus TAL14Xam668 EBE for the group 1 and 2 promoters, which showed a roughly equal occurrence of an A or T at this position in both promoter groups (Figs 7 and S4, see Supporting Information). There was also a better consensus for a canonical TATA‐box (TATAWA) in the group 1 consensus TAL14CIO151 EBE versus group 2 (Butler and Kadonaga, 2002) (Figs 7 and S4). We compared the average normalized EBE scores for the TAL14Xam668 and TAL14CIO151 Target Finder‐predicted EBEs in the two promoter groups. The average normalized EBE scores were the same for TAL14Xam668 and TAL14CIO151 in the group 2 promoters (3.1), whereas, in the group 1 promoters, the average normalized score for the TAL14CIO151 EBEs (2.8) was lower (i.e. better) than the average normalized score for the TAL14Xam668 EBEs (3.0), indicating a stricter requirement for ideal alignment in promoters which are activated by TAL14CIO151.
Figure 7.
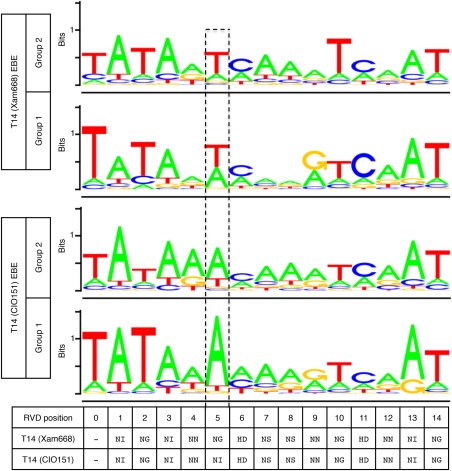
Consensus predicted TAL14CIO151 effector binding elements (EBEs) in group 1 promoters show a strict requirement for an A at position 5. TALgetter (1.0) was used to predict EBEs for TAL14Xam668 and TAL14CIO151 in group 1 and 2 promoters (Grau et al., 2013). The consensus EBE for each group is displayed with position 5 outlined by broken lines. The height of the consensus indicates the nucleotide conservation at that site (measured in bits), whereas the heights of the nucleotides within the consensus reflect their relative frequency. TAL14Xam668 and TAL14CIO151 repeat variable diresidue (RVD) sequences are displayed.
TAL14CIO151 complements Xam668ΔTAL14ps in growth assays
dTALEs that directly activate Xanthomonas TAL effector targets have been used to complement TAL effector mutant phenotypes and to attribute the promotion of disease susceptibility to the activation of specific genes (Cernadas et al., 2014; Cohn et al., 2014; Hu et al., 2014; Li et al., 2014; Morbitzer et al., 2010). dTALEs were made for eight of the highly up‐regulated previously published TAL14Xam668 targets and conjugated into Xam668ΔTAL14ps (Cohn et al., 2014). Of the dTALE targets, four are in group 1 (cassava4.1_007568, cassava4.1_007516, cassava4.1_034150 and cassava4.1_024404) and four are in group 2 (cassava4.1_031361, cassava4.1_026121, cassava4.1_026646 and cassava4.1_020499). Xam668ΔTAL14ps (dTALE) strains were then tested for target gene activation and complementation of the TAL14Xam668 mutant growth defect (Fig. S5, see Supporting information). No single tested target or target class promoted growth to the same level as complementation with TAL14Xam668. Given the level of variability inherent in growth assays, we were not able to detect any statistically significant partial complementation by the tested dTALEs. When RNA‐Seq revealed the large number of genes activated by TAL14Xam668, the dTALE approach became unfeasible in the timeframe of our study and we turned to TAL14CIO151 as a tool to narrow down candidate S genes that might be important for CBB susceptibility.
TAL14CIO151 was conjugated into Xam668ΔTAL14ps on a plasmid and the resulting strain was tested for complementation of the mutant bacterial growth defect. TAL14CIO151 promoted growth to wild‐type levels in Xam668ΔTAL14ps whilst maintaining its target specificity (Fig. 5a,c), indicating that, although TAL14CIO151 activates only a subset of TAL14Xam668‐activated genes, the two TAL effectors are functionally interchangeable. We found that TAL14Xam668 also complemented the growth defect seen in CIO151ΔTAL14 in a manner similar to TAL14CIO151 (Fig. 5b). This result indicates that one or more of the group 1 genes are important for susceptibility to CBB.
Discussion
TAL14Xam668 of Xam strain Xam668 promotes bacterial growth in the host plant cassava (Cohn et al., 2014). In this study, we used RNA‐Seq to identify the full repertoire of genes activated by TAL14Xam668 in the cassava genome. We tested the 26 most highly up‐regulated TAL14Xam668 targets for activation by TAL14CIO151 from Xam strain CIO151, whose RVD sequence differs from that of TAL14Xam668 at the fifth repeat in the DNA‐binding domain. The fifth RVD of TAL14Xam668 is NG, whereas the fifth RVD of TAL14CIO151 is NI. We found that TAL14CIO151 only activates a subset of the TAL14Xam668 targets because of the stricter binding requirement of NI to A, yet is able to complement the in planta growth defect of a Xam668 strain lacking TAL14Xam668, implying that shared targets of the two TAL14 variants are responsible for TAL14Xam668's contribution to virulence.
TAL14Xam668 and TAL14CIO151 are predicted to bind a large number of EBEs in the cassava promoterome when these EBEs are predicted on the basis of the original TAL effector–DNA binding code which assigns binding scores on the basis of RVD specificity (Doyle et al., 2012; Moscou and Bogdanove, 2009). The disproportionately large number of predicted targets for TAL14Xam668 and TAL14CIO151 cannot be explained by RVD number or non‐specific RVD content alone. By comparing EBE predictions among the Xam668 TAL effectors, we believe that the best explanation for the large number of predicted targets is that the TAL14 proteins are predicted to bind a group of nucleotide sequences that are relatively common in the cassava promoterome compared with the groups of sequences predicted to be bound by the other TAL effectors of Xam668. The consensus of the top 5000 predicted TAL14Xam668 targets reveals that TAL14Xam668 is predicted to target AT‐rich promoter sequences, and that the motif ‘TATA‐T‐AA‐T‐ ‐ AT’ may be a relatively common sequence in cassava promoters, where the dashes indicate variable sites.
The original TAL effector–DNA binding code revealed a level of degeneracy in RVD–nucleotide specificity (Boch et al., 2009; Moscou and Bogdanove, 2009). Given this ability for RVDs to occasionally bind imperfectly to mismatched nucleotides, it was surprising that TAL14CIO151 only activates 10 of the 26 tested targets of TAL14Xam668, despite the two proteins differing by only a single RVD. However, this result is supported by those of recent studies regarding the parameters that influence TAL effector binding, including RVD efficiency and nucleotide affinity, polarity effects, the distance of EBE to the transcriptional start site and the vicinity of EBEs to core promoter elements, such as the TATA‐box (Cernadas et al., 2014; Cong et al., 2012; Grau et al., 2013; Meckler et al., 2013; Moore et al., 2014; Pereira et al., 2014; Streubel et al., 2013).
We compared the consensus EBEs for TAL14Xam668 and TAL14CIO151 in the promoters of group 1 genes (which are activated by both TAL14Xam668 and TAL14CIO15) and group 2 genes (which are only activated by TAL14Xam668). Although the fifth RVD of TAL14Xam668 (NG) appears to tolerate binding to both T and A, the fifth RVD of TAL14CIO151 (NI) has a stricter requirement for binding A in order to activate gene expression. The RVDs NG and NI form relatively weak van der Waals’ interactions with their corresponding nucleotides and are considered weak in their binding efficiency, yet are still highly specific in their preference for T and A, respectively (Deng et al., 2012; Mak et al., 2012; Streubel et al., 2012). Interestingly, the specificity of NG for T comes from the glycine residue allowing space for the 5‐methyl group of thymine, and this space probably accommodates mismatched nucleotides, whereas the bulkier side chain of isoleucine does not (Deng et al., 2012; Mak et al., 2012). In addition, the relative affinity of NI and NG for their corresponding nucleotides is different, with NI having an affinity for A that is about three times the strength of the affinity of NG for T (Cong et al., 2012; Moore et al., 2014). The affinity of NI for A is 22 times its affinity for T, whereas the affinity of NG for T is only about 2.5 times its affinity for A (Cong et al., 2012; Moore et al., 2014). Therefore, TAL14CIO151 has an RVD at position 5 with a stronger preference for its corresponding nucleotide than does TAL14Xam668 and, consistent with our findings, is less likely to functionally bind mismatched nucleotides.
The RVD difference between TAL14Xam668 and TAL14CIO151 occurs towards the N‐terminus of the DNA‐binding domain (RVD5 of 14), where mismatches are less likely to be tolerated (Meckler et al., 2013; Römer et al., 2010). In addition, TAL14Xam668 and TAL14CIO151 both appear to prefer binding to a TATA‐rich sequence, with TAL14CIO151 showing a strong requirement for binding the canonical TATA‐box (TATAWA) if gene activation is to be achieved. Because a preference for binding a TATA‐box has been seen for multiple TAL effectors, it is widely speculated that EBE overlap with this core promoter motif has functional significance (Antony et al., 2010; Boch et al., 2009; Grau et al., 2013; Pereira et al., 2014; Römer et al., 2010). Several non‐mutually exclusive reasons why EBEs may overlap with the TATA‐box motif have been proposed. These include a requirement to bind regions of open chromatin, to cooperate with host factors for transcription initiation, to create a functional gene product by having the correct translational start site, and to bind in the ‘safe haven’ of the TATA‐box where sequence changes through mutation are unlikely because of the resulting negative effects on endogenous transcription by the host (Grau et al., 2013).
Although the TAL effector–DNA binding code is a powerful tool, many false‐positive predictions are made when using association frequencies alone to predict TAL effector targets (Cohn et al., 2014; Grau et al., 2013). We anticipated that most of our predicted TAL14Xam668 targets were likely false positives, prompting us to undertake a transcriptomic approach to identify true targets of TAL14Xam668. RNA‐Seq revealed that TAL14Xam668 activates 52 cassava genes. In contrast, TAL20Xam668 was shown to target only a single gene, MeSWEET10a, a sugar transporter that promotes bacterial growth and water‐soaking symptom development during the CBB disease process (Cohn et al., 2014). It is unclear whether TAL14Xam668 targets a conserved core promoter motif that activates many off‐target genes, but only one is truly required to promote disease, or whether TAL14Xam668 activates multiple genes whose collective over‐ or ectopic expression promotes disease. What we do know is that TAL14CIO151 complements the growth defect seen in Xam668ΔTAL14ps in a manner identical to TAL14Xam668, indicating that one or several of the target genes in group 1 are important for disease susceptibility. We point out that we only tested the 26 most highly up‐regulated genes, and so other group 1 genes may exist. For example, cassava4.1_014976 was predicted to be a virulence target of TALEXam1 (which has an RVD sequence identical to TAL14CIO151) based on gene expression analysis and EBE prediction, although its role in susceptibility was not experimentally validated (Muñoz‐Bodnar et al., 2014). This gene was also activated by TAL14Xam668 with a mock versus Xam668 log2(fold change) of 2.4, and was thus outside the group of targets tested here for TAL14CIO151 activation.
dTALEs are a useful tool for the implication of single TAL effector targets as S genes (Cernadas et al., 2014; Cohn et al., 2014; Hu et al., 2014; Li et al., 2014). Previously, it has been observed that when a TAL effector targets more than one gene, the activation of only one of the genes is sufficient to complement the TAL effector mutant phenotype. Of the two gene targets of Xoc Tal2g in rice, only dTALEs targeting OsSULTR3;6 complement the mutant phenotype, and, of the two gene targets of the TAL effector PthA4 in citrus, only dTALEs targeting CsLOB1 complement the mutant phenotype (Cernadas et al., 2014; Hu et al., 2014; Li et al., 2014). We tested eight of the highly up‐regulated targets that were identified in our previously published Xam TAL effector study for their role in the promotion of growth in the TAL14Xam668 mutant background, but did not identify a single S gene (Cohn et al., 2014).
Given the deduced role of group 1 genes in the promotion of CBB, we utilized the program Mapman to assign these genes to functional categories, and found that half of them are predicted to be involved in responses to biotic stress (Thimm et al., 2004). These include the proteases cassava4.1_033289 and cassava4.1_024404, the mannose‐binding lectin cassava4.1_034150, and the pectate lyases cassava4.1_007516 and cassava4.1_007568.
The pectate lyases are particularly interesting, given that plant pectin modification is seen during Xam infection of cassava (Boher et al., 1995). Xam‐infected xylem vessels were found to be occluded by pectinaceous material associated with tyloses, which are outgrowths of the xylem parenchyma that form blockages in xylem vessels as a defence against the spread of vascular pathogens (Yadeta and Thomma, 2013). Secretion of pectin during tylose formation is thought to plug spaces not occluded by the tylose itself (Rioux et al., 1998). The assumption that TAL effectors activate genes important for susceptibility would lead one to hypothesize that Xam is activating plant pectate lyases as a way to degrade these occlusive materials.
Interestingly, a dTALE activating the TAL effector‐targeted pectate lyases (dT_007568, 007516) expressed by Xam668ΔTAL14ps resulted in fewer colony‐forming units (CFU) at the site of midvein inoculation than Xam668ΔTAL14ps. It is possible that the activities of the pectate lyases actually have negative effects on Xam growth by aiding in the secretion of pectinaceous vessel‐occluding material. In this scenario, the pectate lyases would be similar to executor (E) genes, which are TAL effector‐activated genes that promote disease resistance (Gu et al., 2005; Römer et al., 2007; Schornack et al., 2013; Strauß et al., 2012; Tian et al., 2014). It would then follow that the negative effects of pectate lyase activation would be overcome by other targets of TAL14Xam668 which promote disease susceptibility.
Another group 1 target, the MtN21/EamA‐like transporter cassava4.1_009347, is of interest as it is targeted by TAL14CIO151, TAL14Xam668 and TAL22Xam668, and may therefore be a virulence hub (Hutin et al., 2015). MtN21/EamA‐like transporters have been associated with both amino acid and auxin transport (Denance et al., 2014). The Arabidopsis MtN21/EamA‐like transporter AtUMAMIT18/SlAR1 is thought to be involved in amino acid loading of the apoplasm and the xylem, and is primarily expressed in vascular tissues of source leaves (Ladwig et al., 2012). Similar to TAL20Xam668, which probably activates MeSWEET10a to promote the accumulation of a carbon source in the apoplasm at the site of bacterial infection, TAL14Xam668 may be activating cassava4.1_009347 to export amino acids into the apoplasm to be used as a bacterial nitrogen and carbon source.
Studies of RVD efficiency have been primarily performed using highly expressed artificial TAL effector–EBE pairs in transient reporter assays. In this study, we have shown that a single repeat difference in two TAL effectors within one species of Xanthomonas can have a dramatic effect on target activation that is consistent with the RVD–nucleotide binding parameters that have been determined through studies using artificial TAL effector–EBE pairs. TAL14 proteins from Xam strains Xam668 and CIO151 promote bacterial growth in the host plant cassava, making strategies of resistance in response to these proteins likely to be effective (Dangl et al., 2013). One resistance strategy against TAL effectors that activate known S genes is the targeted editing of the TAL effector EBE in the S gene promoter, so that it is no longer activated on infection (Li et al., 2012). The identification of biologically relevant TAL effector‐targeted S genes is made difficult when TAL effectors have multiple targets, as is the case with TAL14Xam668 (Wilkins et al., 2015). Although we demonstrated the use of TAL14CIO151 as a tool to narrow down the biologically relevant S gene targets of Xam, the identification of a single TAL14‐targeted S gene target awaits future study. In the absence of known S genes, a feasible resistance strategy is an executor (E) gene approach, in which a resistance‐triggering gene is engineered downstream from a TAL14‐targeted promoter, so that Xam is ‘tricked’ into activating resistance on infection (Boch et al., 2014; Hummel et al., 2012; Römer et al., 2009a; Schornack et al., 2013). The ideal E gene promoter would be activated by as many strains as possible. The results of this study show that, in order to determine whether an E gene construct will be effective against a group of TAL14‐containing Xam strains, we require a knowledge of not only whether they contain a TAL effector of the appropriate size, but also the TAL effector RVD sequence and binding capacity for the E gene promoter. Our collective findings on the TAL effectors of Xam suggest that stacking promoter mutations that abolish TAL20Xam668‐mediated MeSWEET10a activation with transformation of a TAL14Xam668/TAL20Xam668‐targeted E gene construct will generate cassava plants with increased durable resistance to CBB.
Experimental Procedures
TAL effector target prediction with TALE‐NT (2.0) Target Finder
For the prediction of targets of Xam668 TAL effectors, Target Finder searched the cassava promoterome (1 kb upstream of the annotated transcriptional start sites, cassava genome version 4.1) (Doyle et al., 2012; Prochnik et al., 2012). Only the forward DNA strand was searched and a 5′ T (T0) was required. Each TAL effector was assigned a best binding score given its RVD sequence, and sites within a three‐fold cut‐off of this score were considered as potential EBEs.
For the prediction of targets of TAL14Xam668 and TAL14CIO151 in group 1 and 2 target promoters, Target Finder searched 300 bp upstream of the gene start codons (cassava genome version 4.1). Only the forward DNA strand was searched and a 5′ T (T0) was required. Each TAL effector was assigned a best binding score given its RVD sequence, and sites within a 3.5‐fold cut‐off of this score were considered as potential EBEs.
TAL effector target prediction with TALgetter
TALgetter (version 1.0) predicted targets of TAL14Xam668 and TAL14CIO151 in group 1 and 2 target promoters 300 bp upstream of the gene start codons (cassava genome version 4.1) (Grau et al., 2013; Prochnik et al., 2012). Computation of P values was fine‐grained, and the maximum P value was set at 1e‐2. The TALgetter standard model was used for model training.
TAL effector mutant and complementation constructs
CIO151ΔTAL14 was generated through integration of a suicide vector into the TAL14CIO151 coding region, as described by Cohn et al. (2014). The loss of TAL14CIO151 in CIO151ΔTAL14 was confirmed by Western blot. A TAL14CIO151 complementation construct was generated as described for TAL14Xam668 (Cohn et al., 2014).
Xam668ΔTAL14ps was generated by cycling Xam668 on rifampicin (100 µg/mL) and screening for TAL effector loss by Southern blot. Visualization of plasmids by agarose gel electrophoresis revealed a loss of TAL14Xam668 ps. Xam668ΔTAL14ps was complemented with TAL14Xam668, as described by Cohn et al. (2014).
Plant inoculations and virulence assays
For midvein growth assays, 5‐µL drops of bacterial suspension at an optical density at 600 nm (OD600) of 0.2 were placed in 2‐mm holes made in leaf midveins of 2–4‐month‐old cassava plants (cultivar TMS60444). Approximately 0.6‐cm2 leaf discs were taken with inoculation points exactly in the middle and ground in 10 mm MgCl2 in a beadbeater. Serial dilutions were plated on the appropriate selection plus CHX to inhibit fungal growth. A detailed description of the midvein growth assay protocol is given in Cohn et al. (2015).
TAL14Xam668 RNA‐Seq
Plant inoculations for RNA‐Seq
Xam and Xe grown on nutrient yeast glycerol agar (NYGA) plates supplemented with rifampicin (100 μg/mL), or rifampicin and kanamycin (25 μg/mL) for Xe(TAL14Xam668), were re‐suspended in 10 mm MgCl2 at OD600 = 0.5. Abaxial nicks were made on leaves of cassava cultivar TMS60444 with a razor blade and culture was injected into the leaf via a 1‐mL needleless syringe. Mock infiltrations were performed with 10 mm MgCl2. Tissue was harvested and frozen at 48 hpi.
Library preparation
Total RNA from inoculated leaf tissue (30–50 mg) was extracted with the Spectrum Plant Total RNA Kit (Sigma‐Aldrich, St. Louis, MO, USA), with an on‐column DNaseI digestion step included. RNA quality was checked by Bioanalyzer [RNA pico chip, Functional Genomics Laboratory (FGL), UC Berkeley, CA, USA]. RNA‐Seq libraries were made using the TruSeq RNA sample preparation kit, v2, adapter set B (Illumina, San Diego, CA, USA), starting with 1 µg of total RNA. Library quality was assessed by Bioanalyzer (DNA 1000 chip, FGL). Quantification by quantitative PCR and pooling of samples were carried out at UC Berkeley's Genomics Sequencing Laboratory (GSL). Libraries were sequenced at GSL in a single lane of an Illumina HiSeq 2000, generating 100‐bp paired‐end reads.
Data analysis
TAL14Xam668 RNA‐Seq data analysis was carried out using the Galaxy platform (Blankenberg et al., 2001; Giardine et al., 2005; Goecks et al., 2010). Reads were trimmed using Trim Galore with default settings (Cutadapt version 1.2.1) (Martin, 2011). Data quality was assessed using FastQC:ReadQC (version 0.51). Trimmed reads were aligned to cassava reference genome AM560‐2 version 4.1 by Tophat2 (version 0.5; mean inner distance, 150; N mode; min intron, 45 bp; max intron, 5 kb; all other parameters set at default) (Prochnik et al., 2012; Trapnell et al., 2009). We allowed two mismatches per read to accommodate single nucleotide polymorphisms (SNPs) between our experimental cultivar TMS60444 and the sequenced cultivar AM560‐2. Transcript assembly was carried out by Cufflinks (version 0.0.5) using the reference annotation as a guide (Trapnell et al., 2010). Quartile normalization, bias correction and multi‐read correction were enabled. Cuffmerge (version 0.0.5) merged the cufflinks output from the various datasets (treatments) and differential gene expression analysis was carried out by Cuffdiff (version 0.0.5; false discovery rate, 0.05; enabled quartile normalization, bias correction and multi read correction) (Trapnell et al., 2012). Results were filtered for log2(fold change) >1.0 and FPKM difference >5 for genes activated by Xam668 versus mock infiltration, and by Xe(TAL14) versus Xe alone. Genes that were statistically significantly up‐regulated (P < 0.001) in at least one of the two comparisons were considered.
Semi‐quantitative RT‐PCR analysis
Semi‐quantitative RT‐PCR was carried out as described by Cohn et al. (2014). Primers and corresponding PCR cycle numbers are listed in Table S1 (see Supporting Information). For CHX treatments, bacterial infiltrations were performed with suspensions containing 50 µm CHX. Actin (gene ID cassava4.1_009807) was used as a control for all RT‐PCRs.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Xanthomonas axonopodis pv. manihotis (Xam) strain Xam668ΔTAL14ps is missing the TAL14Xam668 plasmid segment. (a) Western blot showing the absence of TAL14Xam668 protein expression (marked by an asterisk) in Xam668ΔTAL14ps (Xam668ΔT14ps), and restoration of TAL14Xam668 expression in the complemented line [Xam668ΔT14ps(T14)]. (b) Plasmid preparation of Xam668ΔTAL14ps (Xam668ΔT14ps) shows a downward shift in the middle TAL14Xam668‐containing plasmid (marked by an asterisk), indicating loss of the TAL14Xam668 plasmid segment. (c) Western blot showing the absence of TAL14CIO151 protein expression (marked by an asterisk) in Xam strain CIO151ΔTAL14 (CIO151ΔT14).
Fig. S2 TAL14Xam668 and TAL22Xam668 increase the expression of cassava4.1_009347. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) of cassava4.1_009347 expression at 48 h post‐inoculation (hpi) in leaf tissue inoculated at an optical density at 600 nm (OD600) of 0.5 with Xanthomonas euvesicatoria (Xe) 85‐10 delivering each of the five Xam668 transcription activator‐like (TAL) effectors.
Fig. S3 Group 1 and group 2 genes are directly activated by Xam TAL14 proteins. Xanthomonas axonopodis pv. manihotis (Xam) strains Xam668 and CIO151 and mock (10 mm MgCl2) were inoculated into cassava leaves in the presence of 50 µm cycloheximide (CHX) and harvested at 24 h post‐inoculation (hpi). Genes activated above background levels in the presence of CHX are direct targets of their corresponding TAL14 proteins. The polymerase chain reaction (PCR) cycle number for each gene primer set is listed in Table S1 (see Supporting Information). *30 PCR cycles were required for the visualization of cassava4.1_023665 transcripts at 24 hpi. Actin expression is shown for all samples as a loading control, and the expression of cassava4.1_009922 is shown as a control for the effect of CHX on secondary transcript accumulation. Genes that have been shown previously to be direct targets of TAL14Xam668 are referenced (Cohn et al., 2014).
Fig. S4 Consensus TAL14Xam668 and TAL14CIO151 effector binding elements (EBEs) predicted by TALE‐NT (2.0) Target Finder in group 1 and group 2 promoters. EBEs were predicted by TALE‐NT (2.0) Target Finder in the 300‐bp promoters of group 1 and group 2 genes by searching only the forward DNA strand and requiring a thymine (T) at position 0 (Doyle et al., 2012). EBEs with scores within 3.5‐fold of the best possible TAL14 binding score were considered. The consensus EBE for each group is displayed with position 5 outlined by broken lines. Consensus sequences were generated with Weblogo 3.4 (Crooks et al., 2004; Schneider & Stephens, 1990). The height of the consensus indicates the nucleotide conservation at that site (measured in bits), whereas the height of the nucleotides within the consensus reflects their relative frequency. TAL14Xam668 and TAL14CIO151 repeat variable diresidue (RVD) sequences are displayed.
Fig. S5 Designer transcription activator‐like effectors (dTALEs) do not complement the TAL14Xam668 mutant growth defect. (a) dTALEs activating TAL14Xam668 target genes cassava4.1_007568/007516, cassava4.1_034150, cassava4.1_031361, cassava4.1_026646, cassava4.1_026121, cassava4.1_020499 and cassava4.1_024404 were conjugated into Xam668ΔTAL14ps and tested for their ability to complement the growth defect seen in the TAL14Xam668 mutant strain. Bacterial populations at leaf midvein inoculation points were measured at days 1 and 6. Data are represented as the mean colony‐forming units (CFU) per 0.6‐cm2 leaf disc encompassing the inoculation point (± standard deviation). *Significantly higher growth than Xam668ΔTAL14ps, two‐tailed t‐test, P < 0.005. Growth assays were repeated twice with similar results. (b) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) shows that the dTALEs activate their intended target genes. For each gene listed in the left column, Xam668ΔT14ps + dTALE_X is the strain carrying the dTALE that activates that gene. (c) Actin expression is shown for each dTALE strain.
Table S1 Reverse transcription‐polymerase chain reaction (RT‐PCR) primers used in this study with corresponding PCR cycle number.
Acknowledgements
This work was funded by the Two Blades Foundation (BJS), National Science Foundation/Basic Research to Enable Agricultural Development (NSF/BREAD) (grant 0965418, BJS), NSF Graduate Research Fellowship (MC) and National Institutes of Health (NIH) Genetics Training Grant 2T32GM007127‐36A1 (MC), and used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. Work on TALE‐based custom DNA‐binding domains in the Lahaye laboratory is supported by grants from the Deutsche Forschungsgemeinschaft (LA1338/5‐1).
References
- Antony, G. , Zhou, J. , Huang, S. , Li, T. , Liu, B. , White, F. and Yang, B. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os‐11N3 . Plant Cell, 22, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta‐Ortiz, M.L. , Rodríguez‐R, L.M. , Pérez‐Quintero, Á.L. , Poulin, L. , Díaz, A.C. , Rojas, N.A. , Trujillo, C. , Benavides, M.R. , Bart, R. , Boch, J. , Boureau, T. , Darrasse, A. , David, P. , Dugé de Bernonville, T. , Fontanilla, P. , Gagnevin, L. , Guérin, F. , Jacques, M.A. , Lauber, E. , Lefeuvre, P. , Medina, C. , Medina, E. , Montenegro, N. , Bodnar, A.M. , Noël, L.D. , Quiñones, J.F.O. , Osorio, D. , Pardo, C. , Patil, P.B. , Poussier, S. , Pruvost, O. , Robéne‐Soustrade, I. , Ryan, R.P. , Tabima, J. , Morales, O.G.U. , Verniére, C. , Carrere, S. , Verdier, V. , Szurek, B. , Restrepo, S. , Lopez, C. , Koebnik, R. and Bernal, A. (2013) Genomic survey of pathogenicity determinants and VNTR markers in the cassava bacterial pathogen Xanthomonas axonopodis pv. manihotis strain CIO151. PLoS One, 8, e79704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart, R. , Cohn, M. , Kassen, A. , McCallum, E.J. , Shybut, M. , Petriello, A. , Krasileva, K. , Dahlbeck, D. , Medina, C. , Alicai, T. , Kumar, L. , Moreira, L.M. , Rodrigues Neto, J. , Verider, V. , Santana, M.A. , Kositcharoenkul, N. , Vanderschuren, H. , Gruissem, W. , Bernal, A. and Staskawicz, B. (2012) High‐throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc. Natl. Acad. Sci. USA, 109, E1972–E1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg, D. , Kuster, G.V. , Coraor, N. , Ananda, G. , Lazarus, R. , Mangan, M. , Nekrutenko, A. and Taylor, J. (2001) Galaxy: A Web‐Based Genome Analysis Tool for Experimentalists. Hoboken, NJ: John Wiley & Sons, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Bonas, U. and Lahaye, T. (2014) TAL effectors – pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. and Voytas, D.F. (2011) TAL effectors: customizable proteins for DNA targeting. Science, 333, 1843–1846. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Schornack, S. and Lahaye, T. (2010) TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Boher, B. , Kpemoua, K. , Nicole, M. , Luisetti, J. and Geiger, J.P. (1995) Ultrastructure of interactions between cassava and Xanthomonas campestris pv. manihotis: cytochemistry of cellulose and pectin degradation in a susceptible cultivar. Phytopathology, 85, 777–788. [Google Scholar]
- Butler, J.E.F. and Kadonaga, J.T. (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16, 2583–2592. [DOI] [PubMed] [Google Scholar]
- Carroll, D. (2014) Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. [DOI] [PubMed] [Google Scholar]
- Castiblanco, L.F. , Gil, J. , Rojas, A. , Osorio, D. , Gutiérrez, S. , Muñoz‐Bodnar, A. , Pérez‐Quintero, Á.L. , Koebnik, R. , Szurek, B. , López, C. , Restrepo, S. , Verdier, V. and Bernal, A.J. (2012) TALE1 from Xanthomonas axonopodis pv. manihotis acts as a transcriptional activator in plant cells and is important for pathogenicity in cassava plants. Mol. Plant Pathol. 14, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas, R.A. , Doyle, E.L. , Niño‐Liu, D.O. , Wilkins, K.E. , Bancroft, T. , Wang, L. , Schmidt, C.L. , Caldo, R. , Yang, B. , White, F.F. , Nettleton, D. , Wise, R.P. and Bogdanove, A.J. (2014) Code‐assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10, e1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐Q. , Hou, B.‐H. , Lalonde, S. , Takanaga, H. , Hartung, M.L. , Qu, X.‐Q. , Guo, W.‐J. , Kim, J.‐G. , Underwood, W. , Chaudhuri, B. , Chermak, D. , Antony, G. , White, F.F. , Somerville, S.C. , Mudgett, M. and Frommer, W.B. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature, 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐Q. , Qu, X.‐Q. , Hou, B.‐H. , Sosso, D. , Osorio, S. , Fernie, A.R. and Frommer, W.B. (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science, 335, 207–211. [DOI] [PubMed] [Google Scholar]
- Cohn, M. , Bart, R.S. , Shybut, M. , Dahlbeck, D. , Gomez, M. , Morbitzer, R. , Hou, B.‐H. , Frommer, W.B. , Lahaye, T. and Staskawicz, B.J. (2014) Xanthomonas axonopodis virulence is promoted by a transcription activator‐like effector‐mediated induction of a SWEET sugar transporter in cassava. Mol. Plant–Microbe Interact. 27, 1186–1198. [DOI] [PubMed] [Google Scholar]
- Cohn, M. , Shybut, M. , Dahlbeck, D. and Staskawicz, B.J. (2015) Assays to assess virulence of Xanthomonas axonopodis pv. manihotis on cassava. Bio‐Protocol, 5, e1522. [Google Scholar]
- Cong, L. , Zhou, R. , Kuo, Y.‐C. , Cunniff, M. and Zhang, F. (2012) Comprehensive interrogation of natural TALE DNA‐binding modules and transcriptional repressor domains. Nat. Commun. 3, 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E ., Hon, G ., Chandonia, J‐M. and Brenner, S.E . (2004) Weblogo: A sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, O. , Binder, A. and Lahaye, T. (2014) From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J. 78, 753–771. [DOI] [PubMed] [Google Scholar]
- Denance, N. , Szurek, B. and Noel, L.D. (2014) Emerging functions of nodulin‐like proteins in non‐nodulating plant species. Plant Cell Physiol. 55, 469–474. [DOI] [PubMed] [Google Scholar]
- Deng, D. , Yan, C. , Pan, X. , Mahfouz, M. , Wang, J. , Zhu, J.‐K. , Shi, Y. and Yan, N. (2012) Structural basis for sequence‐specific recognition of DNA by TAL effectors. Science, 335, 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, E.L. , Booher, N.J. , Standage, D.S. , Voytas, D.F. , Brendel, V.P. , VanDyk, J.K. and Bogdanove, A.J. (2012) TAL Effector‐Nucleotide Targeter (TALE‐NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, E.L. , Hummel, A.W. , Demorest, Z.L. , Starker, C.G. , Voytas, D.F. , Bradley, P. and Bogdanove, A.J. (2013) TAL effector specificity for base 0 of the DNA target is altered in a complex, effector‐ and assay‐dependent manner by substitutions for the tryptophan in cryptic repeat −1. PLoS One, 8, e82120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine, B. , Riemer, C. , Hardison, R.C. , Burhans, R. , Elnitski, L. , Shah, P. , Zhang, Y. , Blankenberg, D. , Albert, I. , Taylor, J. , Miller, W. , Kent, W.J. and Nekrutenko, A. (2005) Galaxy: a platform for interactive large‐scale genome analysis. Genome Res. 15, 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks, J. , Nekrutenko, A. and Taylor, J. (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau, J. , Wolf, A. , Reschke, M. , Bonas, U. , Posch, S. and Boch, J. (2013) Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput. Biol. 9, e1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, K. , Yang, B. , Tian, D. , Wu, L. , Wang, D. , Sreekala, C. , Yang, F. , Chu, Z. , Wang, G.‐L. , White, F.F. and Yin, Z. (2005) R gene expression induced by a type‐III effector triggers disease resistance in rice. Nature, 435, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Howeler, R. , Lutaladio, N. and Thomas, G. (2013) Save and Grow Cassava: A Guide to Sustainable Production Intensification, Food and Agriculture Organization of the United Nations (FAO), Rome: 1–24.
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. , Yang, B. , White, F.F. , Wang, N. and Jones, J.B. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111, E521–E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel, A.W. , Doyle, E.L. and Bogdanove, A.J. (2012) Addition of transcription activator‐like effector binding sites to a pathogen strain‐specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195, 883–893. [DOI] [PubMed] [Google Scholar]
- Hutin, M. , Pérez‐Quintero, Á.L. , López, C. and Szurek, B. (2015) MorTAL Kombat: the story of defense against TAL effectors through loss‐of‐susceptibility. Front. Plant Sci. 6, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, J.K. and Sander, J.D. (2013) TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Ladwig, F. , Stahl, M. , Ludewig, U. , Hirner, A.A. , Hammes, U.Z. , Stadler, R. , Harter, K. and Koch, W. (2012) Siliques are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol. 158, 1643–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Zou, L. , Ye, G. , Xiong, L. , Ji, Z. , Zakria, M. , Hong, N. , Wang, G. and Chen, G. (2014) A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Mol. Plant. 7, 912–915. [DOI] [PubMed] [Google Scholar]
- López, C. and Bernal, A.J. (2012) Cassava bacterial blight: using genomics for the elucidation and management of an old problem. Trop. Plant Biol. 5, 117–126. [Google Scholar]
- Lozano, J.C. (1986) Cassava bacterial blight: a manageable disease. Plant Dis. 70, 1089–1093. [Google Scholar]
- Mak, A.N.S. , Bradley, P. , Cernadas, R.A. , Bogdanove, A.J. and Stoddard, B.L. (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science, 335, 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet. J. 17, 10–12. [Google Scholar]
- Meckler, J.F. , Bhakta, M.S. , Kim, M.S. , Ovadia, R. , Habrian, C.H. , Zykovich, A. , Yu, A. , Lockwood, S.H. , Morbitzer, R. , Elsaesser, J. , Lahaye, T. , Segal, D.J. and Baldwin, E.P. (2013) Quantitative analysis of TALE–DNA interactions suggests polarity effects. Nucleic Acids Res. 41, 4118–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. , Chandrahas, A. and Bleris, L. (2014) Transcription activator‐like effectors: a toolkit for synthetic biology. ACS Synth. Biol. 3, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer, R. , Römer, P. , Boch, J. and Lahaye, T. (2010) Regulation of selected genome loci using de novo‐engineered transcription activator‐like effector (TALE)‐type transcription factors. Proc. Natl. Acad. Sci. USA, 107, 21617–21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009. ) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Bodnar, A. , Pérez‐Quintero, Á.L. , Gomez‐Cano, F. , Gil, J. , Michelmore, R. , Bernal, A. , Szurek, B. and López, C. (2014) RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non‐pathogenic strains of Xanthomonas axonopodis pv. manihotis . Plant Cell Rep. 33, 1901–1912. [DOI] [PubMed] [Google Scholar]
- Pereira, A.L. , Carazzolle, M.F. , Abe, V.Y. , de Oliveira, M.L. , Domingues, M.N. , Silva, J.C. , Cernadas, R.A. and Benedetti, C.E. (2014) Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics, 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik, S. , Marri, P.R. , Desany, B. , Rabinowicz, P.D. , Kodira, C. , Mohiuddin, M. , Rodriguez, F. , Fauquet, C. , Tohme, J. , Harkins, T. , Rokhsar, D.S. and Rounsley, S. (2012) The cassava genome: current progress, future directions. Trop. Plant Biol. 5, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux, D. , Nicole, M. , Simard, M. and Ouellette, G.B. (1998) Immunocytochemical evidence that secretion of pectin occurs during gel (Gum) and tylosis formation in trees. Phytopathology, 88, 494–505. [DOI] [PubMed] [Google Scholar]
- Römer, P. , Hahn, S. , Jordan, T. , Strauß, T. , Bonas, U. and Lahaye, T. (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 318, 645–648. [DOI] [PubMed] [Google Scholar]
- Römer, P. , Recht, S. and Lahaye, T. (2009a) A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA, 106, 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P. , Strauß, T. , Hahn, S. , Scholze, H. , Morbitzer, R. , Grau, J. , Bonas, U. and Lahaye, T. (2009b) Recognition of AvrBs3‐like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150, 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P. , Recht, S. , Strauß, T. , Elsaesser, J. , Schornack, S. , Boch, J. , Wang, S. and Lahaye, T. (2010) Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae . New Phytol. 187, 1048–1057. [DOI] [PubMed] [Google Scholar]
- Schneider, T.D ., Stephens, R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , Moscou, M.J. , Ward, E.R. and Horvath, D.M. (2013) Engineering plant disease resistance based on TAL effectors. Annu. Rev. Phytopathol. 51, 383–406. [DOI] [PubMed] [Google Scholar]
- Strauß, T. , van Poecke, R.M.P. , Strauß, A. , Römer, P. , Minsavage, G.V. , Singh, S. , Wolf, C. , Strauss, A. , Kim, S. , Lee, H. , Yeom, S. , Parniske, M. , Stall, R.E. , Jones, J.B. , Choi, D. , Prins, M. and Lahaye, T. (2012) RNA‐seq pinpoints a Xanthomonas TAL‐effector activated resistance gene in a large‐crop genome. Proc. Natl. Acad. Sci. USA, 109, 19480–19485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel, J. , Blücher, C. , Landgraf, A. and Boch, J. (2012) TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30, 593–595. [DOI] [PubMed] [Google Scholar]
- Streubel, J. , Pesce, C. , Hutin, M. , Koebnik, R. , Boch, J. and Szurek, B. (2013) Five phylogenetically close rice SWEET genes confer TAL effector‐mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200, 808–819. [DOI] [PubMed] [Google Scholar]
- Sun, N. and Zhao, H. (2013) Transcription activator‐like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 110, 1811–1821. [DOI] [PubMed] [Google Scholar]
- Thimm, O. , Bläsing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Krüger, P. , Selbig, J. , Müller, L.A. , Rhee, S.Y. and Stitt, M. (2004) Mapman: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tian, D. , Wang, J. , Zeng, X. , Gu, K. , Qiu, C. , Yang, X. , Zhou, Z. , Goh, M. , Luo, Y. , Murata‐Hori, M. , White, F.F. and Yin, Z. (2014) The rice TAL effector‐dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell, 26, 497–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. and Salzberg, S.L. (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. , Wold, B.J. and Pachter, L. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. , Pimentel, H. , Salzberg, S.L. , Rinn, J.L. and Pachter, L. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, K.E. , Booher, N.J. , Wang, L. and Bogdanove, A.J. (2015) TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 6, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta, K.A. and Thomma, B.P.H.J. (2013) The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Xanthomonas axonopodis pv. manihotis (Xam) strain Xam668ΔTAL14ps is missing the TAL14Xam668 plasmid segment. (a) Western blot showing the absence of TAL14Xam668 protein expression (marked by an asterisk) in Xam668ΔTAL14ps (Xam668ΔT14ps), and restoration of TAL14Xam668 expression in the complemented line [Xam668ΔT14ps(T14)]. (b) Plasmid preparation of Xam668ΔTAL14ps (Xam668ΔT14ps) shows a downward shift in the middle TAL14Xam668‐containing plasmid (marked by an asterisk), indicating loss of the TAL14Xam668 plasmid segment. (c) Western blot showing the absence of TAL14CIO151 protein expression (marked by an asterisk) in Xam strain CIO151ΔTAL14 (CIO151ΔT14).
Fig. S2 TAL14Xam668 and TAL22Xam668 increase the expression of cassava4.1_009347. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) of cassava4.1_009347 expression at 48 h post‐inoculation (hpi) in leaf tissue inoculated at an optical density at 600 nm (OD600) of 0.5 with Xanthomonas euvesicatoria (Xe) 85‐10 delivering each of the five Xam668 transcription activator‐like (TAL) effectors.
Fig. S3 Group 1 and group 2 genes are directly activated by Xam TAL14 proteins. Xanthomonas axonopodis pv. manihotis (Xam) strains Xam668 and CIO151 and mock (10 mm MgCl2) were inoculated into cassava leaves in the presence of 50 µm cycloheximide (CHX) and harvested at 24 h post‐inoculation (hpi). Genes activated above background levels in the presence of CHX are direct targets of their corresponding TAL14 proteins. The polymerase chain reaction (PCR) cycle number for each gene primer set is listed in Table S1 (see Supporting Information). *30 PCR cycles were required for the visualization of cassava4.1_023665 transcripts at 24 hpi. Actin expression is shown for all samples as a loading control, and the expression of cassava4.1_009922 is shown as a control for the effect of CHX on secondary transcript accumulation. Genes that have been shown previously to be direct targets of TAL14Xam668 are referenced (Cohn et al., 2014).
Fig. S4 Consensus TAL14Xam668 and TAL14CIO151 effector binding elements (EBEs) predicted by TALE‐NT (2.0) Target Finder in group 1 and group 2 promoters. EBEs were predicted by TALE‐NT (2.0) Target Finder in the 300‐bp promoters of group 1 and group 2 genes by searching only the forward DNA strand and requiring a thymine (T) at position 0 (Doyle et al., 2012). EBEs with scores within 3.5‐fold of the best possible TAL14 binding score were considered. The consensus EBE for each group is displayed with position 5 outlined by broken lines. Consensus sequences were generated with Weblogo 3.4 (Crooks et al., 2004; Schneider & Stephens, 1990). The height of the consensus indicates the nucleotide conservation at that site (measured in bits), whereas the height of the nucleotides within the consensus reflects their relative frequency. TAL14Xam668 and TAL14CIO151 repeat variable diresidue (RVD) sequences are displayed.
Fig. S5 Designer transcription activator‐like effectors (dTALEs) do not complement the TAL14Xam668 mutant growth defect. (a) dTALEs activating TAL14Xam668 target genes cassava4.1_007568/007516, cassava4.1_034150, cassava4.1_031361, cassava4.1_026646, cassava4.1_026121, cassava4.1_020499 and cassava4.1_024404 were conjugated into Xam668ΔTAL14ps and tested for their ability to complement the growth defect seen in the TAL14Xam668 mutant strain. Bacterial populations at leaf midvein inoculation points were measured at days 1 and 6. Data are represented as the mean colony‐forming units (CFU) per 0.6‐cm2 leaf disc encompassing the inoculation point (± standard deviation). *Significantly higher growth than Xam668ΔTAL14ps, two‐tailed t‐test, P < 0.005. Growth assays were repeated twice with similar results. (b) Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) shows that the dTALEs activate their intended target genes. For each gene listed in the left column, Xam668ΔT14ps + dTALE_X is the strain carrying the dTALE that activates that gene. (c) Actin expression is shown for each dTALE strain.
Table S1 Reverse transcription‐polymerase chain reaction (RT‐PCR) primers used in this study with corresponding PCR cycle number.


