Summary
Oidium heveae, an obligate biotrophic pathogen of rubber trees (Hevea brasiliensis), causes significant yield losses of rubber worldwide. However, the molecular mechanisms underlying the interplay between O. heveae and rubber trees remain largely unknown. In this study, we isolated an O. heveae strain, named HN1106, from cultivated H. brasiliensis in Hainan, China. We found that O. heveae HN1106 triggers the hypersensitive response in a manner that depends on the effector‐triggered immunity proteins EDS1 (Enhanced Disease Susceptibility 1) and PAD4 (Phytoalexin Deficient 4) and on salicylic acid (SA) in the model plant Arabidopsis thaliana. However, SA‐independent resistance also appears to limit O. heveae infection of Arabidopsis, because the pathogen does not produce conidiospores on npr1 (nonexpressor of pr1), sid2 (SA induction deficient 2) and NahG plants, which show disruptions in SA signalling. Furthermore, we found that the callose synthase PMR4 (Powdery Mildew Resistant 4) prevents O. heveae HN1106 penetration into leaves in the early stages of infection. To elucidate the potential mechanism of resistance of Arabidopsis to O. heveae HN1106, we inoculated 47 different Arabidopsis accessions with the pathogen, and analysed the plant disease symptoms and O. heveae HN1106 hyphal growth and conidiospore formation on the leaves. We found that the accession Lag2‐2 showed significant susceptibility to O. heveae HN1106. Overall, this study provides a basis for future research aimed at combatting powdery mildew caused by O. heveae in rubber trees.
Keywords: Arabidopsis thaliana, enhanced disease susceptibility 1, Hevea brasiliensis, incompatible interaction, Oidium heveae, powdery mildew resistant 4, salicylic acid
Introduction
Powdery mildews are a group of parasitic ascomycete fungi that infect many plant species and result in significant crop yield losses worldwide. Outbreaks of powdery mildew disease of rubber trees (Hevea brasiliensis) in Malaysia, India, Brazil and Papua New Guinea (Beeley, 1933; Mitra and Mehta, 1938; Saranya et al., 2005) have significantly reduced rubber yields. Oidium heveae, the powdery mildew that infects rubber trees, usually grows in living host cells of young leaves or buds, resulting in defoliation of young leaves and discoloration and curling of the margins of old leaves. When an O. heveae conidiospore lands on a leaf, a primary germ tube is generated which produces an appressorium at the site at which the fungal pathogen attempts to breach the plant cell wall. Successful penetration leads to invagination of the host plasma membrane to form a lobe‐shaped structure, the haustorium. Subsequently, secondary hyphae form from the spore and new appressoria penetrate neighbouring cells and develop secondary haustoria (Saranya et al., 2005). Mycelia on the leaves are amphigenous and conidiospores develop from the vegetative hyphae, forming straight, cylindrical foot cells, followed by one to three additional cells. The first conidium that matures on a conidiospore is ellipsoidal with a rounded tip; secondary conidia are ellipsoidal‐cylindrical in shape (Saranya et al., 2005). At present, brimstone powder is often used to prevent outbreaks of powdery mildew on rubber trees. However, this substance is toxic to the environment. Therefore, there is an urgent need to elucidate the molecular profiles of O. heveae–host interactions, and subsequently to develop an effective strategy to cope with the disease.
To protect themselves against pathogen invasion, plants possess two layers of immune recognition. The first line of disease resistance, triggered by pathogen/microbe‐associated molecular patterns (PAMPs/MAMPs) (Boller and He, 2009; Zhou and Chai, 2008), is termed PAMP‐triggered immunity (PTI). For example, chitin present in the growing hyphae and conidia of the powdery mildew Golovinomyces cichoracearum functions as a PAMP that elicits PTI responses during infection of Arabidopsis thaliana (Ramonell et al., 2005). The second, effector‐triggered immunity (ETI), is activated by plant resistance (R) proteins on recognition of race‐specific effectors, and is usually associated with a hypersensitive response (HR), which involves rapid programmed host cell death at the infection site (Jones and Dangl, 2006). Two avirulence (AVR) proteins, AVRa10 and AVRk1, encoded by the powdery mildew Blumeria graminis f. sp. hordei, elicit host defences in resistant barley (Hordeum vulgare) varieties (Catanzariti et al., 2007; Ridout et al., 2006).
Several proteins play crucial roles in the activation of R proteins in Arabidopsis (Day et al., 2006; Hammond‐Kosack and Parker, 2003). For example, EDS1 (Enhanced Disease Susceptibility 1) is generally required for the activation of the Toll‐Interleukin1 Receptor (TIR)‐nucleotide binding (NB)‐leucine‐rich repeat (LRR) subclass of R proteins, such as resistance to Peronospora parasitica RPP(s) and resistance to Pseudomonas syringae (RPS) 4 (Aarts et al., 1998; Moreau et al., 2012; Parker et al., 1996; Rietz et al., 2011). NDR1 (Non‐Race‐Specific Disease Resistance 1) is largely required for ETI conditioned by coiled‐coil (CC)‐NB‐LRR R proteins, such as resistance to Pseudomonas syringae pv. maculicola (RPM) 1, RPS2 and RPS5 (Aarts et al., 1998; Century et al., 1997; Day et al., 2006; Moreau et al., 2012). In Arabidopsis, EDS1 interacts with two signalling partners, PAD4 (Phytoalexin Deficient 4) and SAG (Senescence‐Associated Gene) 101, to form soluble cytoplasmic and nuclear signalling complexes (Cui et al., 2015; Feys et al., 2001, 2005; Wagner et al., 2013). The Heat Shock Protein 90 (HSP90) co‐chaperones RAR1 (Required for Mla12 Resistance) and SGT1 (Suppressor of G‐Two Allele of Skp1) are required to stabilize some R proteins, and rar1 and sgt1 mutants often fail to induce R protein‐mediated resistance (Hubert et al., 2009).
R protein‐mediated resistance responses are usually accompanied by increased levels of salicylic acid (SA) in local and distant parts of the plant, as well as the up‐regulation of a large set of defence genes, including pathogenesis‐related (PR) genes (Gaffney et al., 1993; McDowell and Dangl, 2000; Ward et al., 1991). Arabidopsis mutants that are impaired in pathogen‐induced SA accumulation include eds1 (Falk et al., 1999), eds5 (Nawrath et al., 2002), sid2 (SA induction deficient 2) (Wildermuth et al., 2001) and pad4 (Jirage et al., 1999). In addition, mutants exist that are defective in SA responsiveness, for example npr1 (nonexpressor of pr1) (Cao et al., 1997). Transgenic Arabidopsis plants expressing NahG, a bacterial SA hydroxylase gene, are also unable to accumulate SA and show enhanced susceptibility to incompatible pathogens (Delaney et al., 1994). In Arabidopsis, SA and EDS1 are thought to function in a positive feedback loop, because SA up‐regulates EDS1 expression and EDS1 enhances SA production (Falk et al., 1999; Venugopal et al., 2009). EDS1 mediates resistance via both SA‐dependent and SA‐independent pathways (Bartsch et al., 2006).
In this study, we isolated the O. heveae strain HN1106 from the rubber species H. brasiliensis GT‐1. When infected by O. heveae HN1106, H. brasiliensis GT‐1 displays severe disease symptoms, such as irregular patches on the upper and lower surfaces of the leaves. Interestingly, we found that Arabidopsis displays a typical resistance response to O. heveae, characterized by cell death activation, hydrogen peroxide (H2O2) production, callose deposition and PR1 (Pathogenesis‐Related 1) gene expression. In addition, we found that the eds1 and pad4 mutants, but not the ndr1 mutant, are significantly compromised in resistance to O. heveae HN1106. These results collectively suggest that EDS1‐mediated ETI signalling is involved in the resistance to the pathogen O. heveae.
Results
Arabidopsis and O. heveae HN1106 exhibit an incompatible interaction
In this study, we isolated a powdery mildew strain from H. brasiliensis GT‐1 leaves displaying powdery mildew symptoms, and named it O. heveae HN1106. The ribosomal DNA internal transcribed spacer (ITS) sequence of O. heveae HN1106 was analysed and found to be identical to the Thailand O. heveae MUMH2602 isolate (Fig. S1, see Supporting Information). Furthermore, we generated a phylogenetic tree based on 11 powdery mildew ITS sequences, and showed that O. heveae is more closely related to Erysiphe cruciferarum (Fig. S2, see Supporting Information), a powdery mildew that infects Arabidopsis (Gollner et al., 2008), than it is to B. graminis f. sp. hordei DH14, a non‐adapted pathogen in Arabidopsis (Stein et al., 2006).
To test whether O. heveae HN1106 is pathogenic to Arabidopsis, we inoculated A. thaliana Col‐0 leaves and H. brasiliensis GT‐1 leaves (as a control) with the pathogen. We collected whole leaves every 3 h post‐inoculation (hpi) during the early stages of infection, and examined the leaves by light microscopy. Germ tubes arose from the ends of conidia on both Col‐0 and GT‐1 leaves by 3 hpi (Fig. 1A). The primary germ tubes produced appressoria on GT‐1 leaves by 3 hpi and on Arabidopsis leaves by 6 hpi (Fig. 1A). Secondary germ tubes often emerged from conidia on GT‐1 leaves by 6 hpi and on Arabidopsis by 24 hpi (Fig. 1A). By 24 hpi, the majority of the germ tubes had penetrated the epidermal cells on GT‐1 leaves, but a large proportion of the germ tubes had not completely penetrated the Arabidopsis cells (Fig. 2A). By 2 days post‐inoculation (dpi), the hyphae had elongated over 300 μm on H. brasiliensis, but only about 50 μm on Arabidopsis (Fig. 2B). By 4 dpi, fungal growth was apparent to the naked eye on H. brasiliensis leaves (Fig. 1C), and conidiospores had begun to develop perpendicular to the leaf surface (Fig. 1B). In the following days, fungal mycelia spread rapidly, adjacent patches of mycelia coalesced and mature conidia appeared at the ends of the conidiospores by 10 dpi on H. brasiliensis leaves (Fig. 1B, 2C). By contrast, chlorotic/necrotic flecks at the inoculation sites were apparent on Arabidopsis leaves by 10 dpi (Fig. 1C). Microscopy analysis showed that O. heveae HN1106 had a significantly lower penetration ratio on Arabidopsis than on H. brasiliensis, and had limited growth and failed to form a hyphal network and conidiospores on Arabidopsis (Fig. 1B). Thus, O. heveae HN1106 cannot complete its life cycle on Arabidopsis Col‐0. Cell death is a second line of defence that is induced when non‐host fungi successfully penetrate epidermal cells (Consonni et al., 2006). Here, we found that O. heveae HN1106 triggers chlorotic/necrotic symptoms on Arabidopsis, suggesting that the interaction between Arabidopsis and O. heveae HN1106 is incompatible, but not a non‐host interaction.
Figure 1.
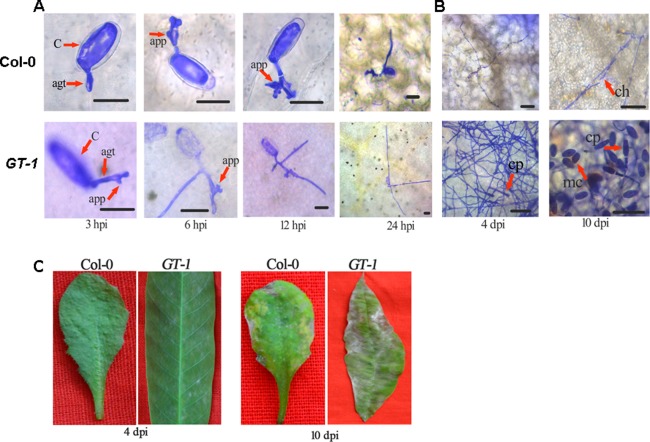
Light micrographs and symptoms of Arabidopsis Col‐0 and Hevea brasiliensis GT‐1 plants infected with Oidium heveae HN1106. Leaves of 4‐week‐old Col‐0 plants and H. brasiliensis GT‐1 leaves in the light‐green phase were inoculated with O. heveae HN1106. (A) Infected leaves stained with Coomassie brilliant blue were observed by microscopy at 3, 6, 12 and 24 h post‐inoculation (hpi). Bar, 20 μm. (B) Infected leaves stained with Coomassie brilliant blue were observed by microscopy at 4 and 10 dpi. Bar, 50 μm. (C) Symptoms were analysed at 4 and 10 dpi. agt, appressorial germ tube; app, appressorium; C, conidia; ch, collapsed hyphae cp, conidiospores; mc, mature conidia. These experiments were repeated twice with similar results.
Figure 2.
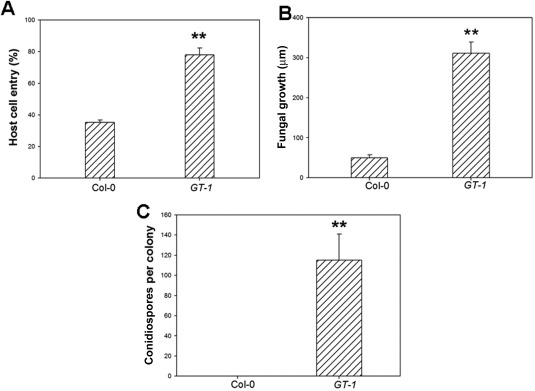
Quantitative analysis of Oidium heveae HN1106 infection on wild‐type Col‐0 and Hevea brasiliensis GT‐1 leaves. (A) Quantitative assessment of host cell entry rates. Data represent the mean ± standard deviation (SD) of three experiments, each based on five to seven leaves per plant line. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (B) Quantitative analysis of hyphal growth of O. heveae HN1106 at 48 h post‐inoculation (hpi). Leaves were stained with Coomassie brilliant blue at 48 hpi, and hyphal lengths per colony were measured from photographs using mie 3.1 software. At least 10 colonies were measured for hyphal length. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (C) The numbers of conidiospores per colony were counted at 10 days post‐inoculation (dpi). Data represent the mean ± SD of at least 20 fungal colonies per plant line. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). These experiments were repeated twice with similar results.
Arabidopsis resistance to O. heveae HN1106 requires EDS1 and PAD4, but not NDR1
Based on our observation that O. heveae HN1106 is an incompatible pathogen on Arabidopsis, we next determined whether the disease resistance against O. heveae HN1106 depends on some known ETI signalling components. We incubated wild‐type Arabidopsis (Col‐0), eds1, pad4, rar1 and ndr1 mutants with O. heveae HN1106, and evaluated the Arabidopsis resistance primarily based on conidiospore formation and plant disease symptoms. We observed a higher cell entry ratio (Fig. 3A) and significantly higher mycelium growth (Fig. 3B) on eds1, pad4 and rar1 mutants compared with Col‐0. Typical powdery mildew patches occurred on eds1 and pad4 mutants by 10 dpi (Fig. 4A). Whereas ndr1 mutants and Col‐0 plants developed severe chlorosis flecks, rar1 mutants exhibited relatively mild chlorosis (Fig. 4A). Many fungal conidiospores formed on eds1 and pad4 mutants, but none formed on Col‐0, ndr1 and rar1 plants (Fig. 3D, 4B). Furthermore, the O. heveae mature fungal spores produced on eds1 leaves successfully infected H. brasiliensis GT‐1 leaves (Fig. S3, see Supporting Information). The rar1 mutant supports the formation of a dense hyphal network (Fig. 4B). These results indicate that O. heveae can complete its life cycle on eds1 and pad4 plants, and that O. heveae‐triggered disease resistance partially requires RAR1. In addition, we found that cell death (Fig. 4C), H2O2 production (Fig. 4D), callose deposition (Fig. 3C) and PR1 expression (Fig. 3E) were strongly induced in Col‐0 and ndr1, and mildly in rar1, but weakly in eds1 and pad4 mutants. Taken together, these results indicate that EDS1, PAD4 and RAR1 are required for O. heveae HN1106‐triggered resistance responses.
Figure 3.
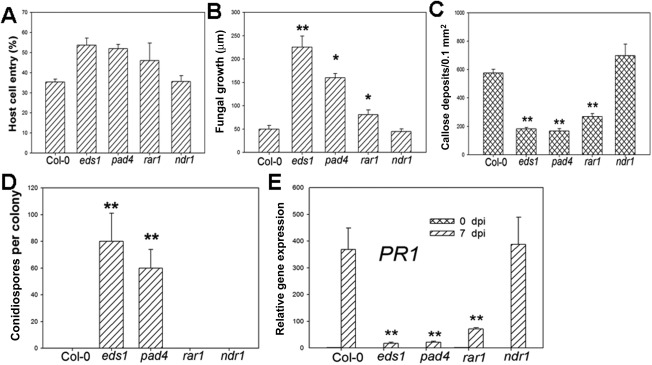
EDS1 (Enhanced Disease Susceptibility 1) and PAD4 (Phytoalexin Deficient 4) are required for Oidium heveae HN1106‐triggered disease resistance and defence responses in Arabidopsis. Four‐week‐old Arabidopsis wild‐type (WT) Col‐0 and mutants were inoculated with O. heveae HN1106. (A) Quantitative assessment of host cell entry rates. Data represent the mean ± standard deviation (SD) of three experiments, each based on five to seven leaves per plant line. (B) Quantitative analysis of hyphal growth of O. heveae HN1106 at 48 h post‐inoculation (hpi). Leaves were stained with Coomassie brilliant blue at 48 hpi, and hyphal lengths per colony were measured from photographs using mie 3.1 software. At least 10 colonies were measured for hyphal length. *Significant difference from Col‐0 (Student's t‐test, P < 0.05). **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (C) Average numbers of callose deposits per microscope field of 0.1 mm2 on Col‐0 and mutant leaves determined at 7 days post‐inoculation (dpi). **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (D) The numbers of conidiospores per colony were counted at 10 dpi. Data represent the mean ± SD of at least 20 fungal colonies per plant line. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (E) The abundance of PR1 (Pathogenesis‐Related 1) mRNA was determined at the indicated time points by quantitative polymerase chain reaction. Data represent the mean ± SD of three RNA replicates. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). These experiments were repeated twice with similar results.
Figure 4.
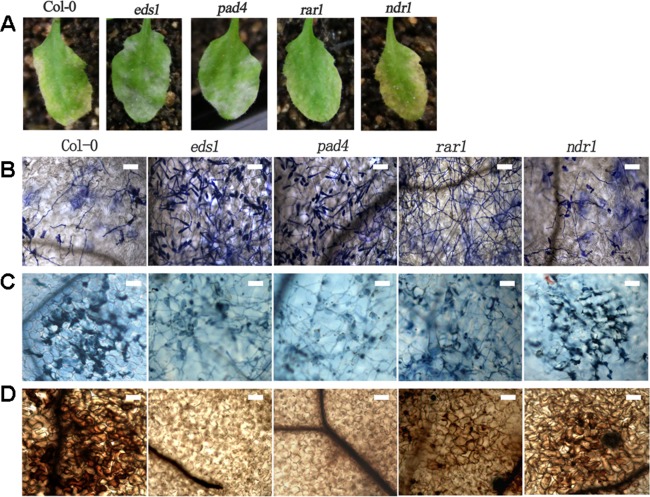
Symptoms and light micrographs of Arabidopsis Col‐0 and eds1 (enhanced disease susceptibility 1), pad4 (phytoalexin deficient 4), rar1 (required for Mla12 resistance) and ndr1 (non‐race‐specific disease resistance 1) mutants infected with Oidium heveae HN1106. Four‐week‐old Arabidopsis wild‐type (WT) Col‐0 and mutants were inoculated with O. heveae HN1106. (A) Symptoms were photographed at 10 days post‐inoculation (dpi). (B) Light microscopy images were taken after fungal structures had been stained with Coomassie brilliant blue at 10 dpi. Bar, 50 μm. (C) Infected leaves were stained with trypan blue at 10 dpi. Bar, 50 μm. (D) Infected leaves were stained with 3,3′‐diaminobenzidine (DAB) at 7 dpi. Bar, 50 μm. These experiments were repeated twice with similar results.
SA is required for resistance to O. heveae HN1106 in Arabidopsis
The SA‐dependent pathway is usually required for R protein‐mediated resistance in Arabidopsis (Bartsch et al., 2010; Rusterucci et al., 2001). To test whether O. heveae HN1106‐induced defence responses and pathogen resistance are SA dependent, we inoculated sid2, npr1, NahG and Col‐0 plants with O. heveae HN1106. Similar to rar1 mutants, the sid2, npr1 and NahG plants displayed higher penetration ratios (Fig. 5A) and fungal growth (Fig. 5B) than did Col‐0. By 10 dpi, O. heveae HN1106 had induced typical chlorosis symptoms on wild‐type Col‐0, but no symptoms on sid2, npr1 and NahG plants (Fig. 6A). The hyphal networks were slightly denser on sid2, npr1 and NahG leaves than on those of the wild‐type (Fig. 6B); however, conidiospores were not detected on sid2, npr1 and NahG leaves (Fig. 6B), indicating that O. heveae HN1106 cannot complete its life cycle on plants with compromised SA signalling. Consistent with the disease symptoms, the levels of cell death (Fig. 6C), H2O2 production (Fig. 6D), callose deposition (Fig. 5C) and PR1 gene expression (Fig. 5D) triggered by O. heveae HN1106 were significantly lower in sid2, npr1 and NahG plants than in the wild‐type. These results indicate that SA is required for the cell death response associated with O. heveae HN1106 infection, and contributes to the penetration and hyphal growth resistance against the pathogen.
Figure 5.
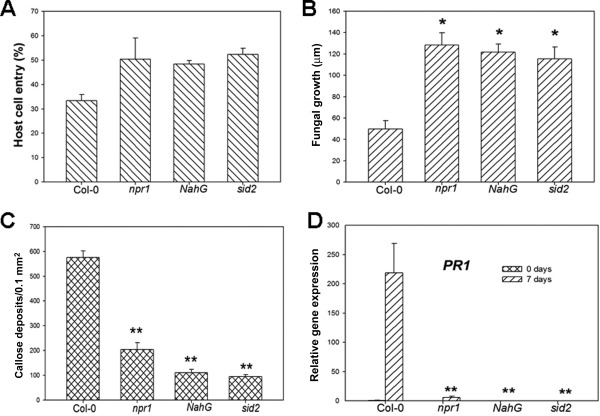
Salicylic acid (SA) contributes to the penetration and hyphal growth resistance and defence responses against Oidium heveae HN1106. Four‐week‐old Arabidopsis wild‐type Col‐0 and mutants were inoculated with O. heveae HN1106. (A) Quantitative assessment of host cell entry rates. Data represent the mean ± standard deviation (SD) of three experiments, each based on five to seven leaves per plant line. (B) Quantitative analysis of hyphal growth of O. heveae HN1106 at 48 h post‐inoculation (hpi). Leaves were stained with Coomassie brilliant blue at 48 hpi, and hyphal lengths per colony were measured from photographs using mie 3.1 software. At least 10 colonies were measured for hyphal length. *Significant difference from Col‐0 (Student's t‐test, P < 0.05). (C) Average numbers of callose deposits per microscopic field of 0.1 mm2 on Col‐0 and mutant leaves determined at 7 days post‐inoculation (dpi). **Significant difference from Col‐0 (Student's t‐test, P < 0.01). (D) The abundance of PR1 (Pathogenesis‐Related 1) mRNA was determined at the indicated time points by quantitative polymerase chain reaction. Data represent the mean ± SD of three RNA replicates. **Significant difference from Col‐0 (Student's t‐test, P < 0.01). These experiments were repeated twice with similar results.
Figure 6.
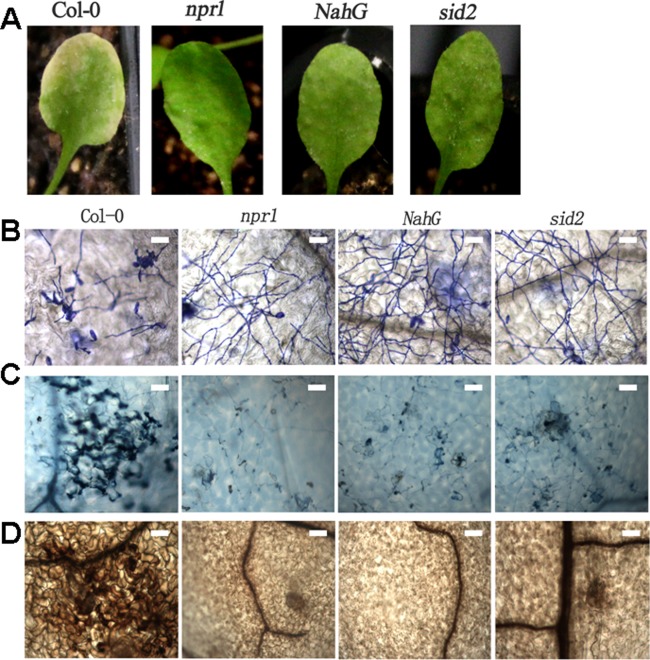
Salicylic acid (SA) is required for a cell death response against Oidium heveae HN1106 on Arabidopsis. Symptoms and light micrographs of Arabidopsis Col‐0 and npr1 (nonexpressor of pr1), NahG and sid2 (SA induction deficient 2) mutants infected with O. heveae HN1106. Four‐week‐old Arabidopsis wild‐type (WT) Col‐0 and mutants were inoculated with O. heveae HN1106. (A) Symptoms were photographed at 10 days post‐inoculation (dpi). (B) Light microscopy images were taken after fungal structures had been stained with Coomassie brilliant blue at 10 dpi. Bar, 50 μm. (C) Infected leaves were stained with trypan blue at 10 dpi. Bar, 50 μm. (D) Infected leaves were stained with 3,3′‐diaminobenzidine (DAB) at 7 dpi. Bar, 50 μm. These experiments were repeated twice with similar results.
Callose deposition contributes to penetration resistance to O. heveae HN1106 in Arabidopsis
Callose deposition has been reported to be involved in penetration resistance in incompatible interactions between Arabidopsis and powdery mildews (Nishimura et al., 2003). To test whether callose deposition is required for penetration resistance to O. heveae HN1106, the callose synthase mutant powdery mildew resistant 4 (pmr4) and Col‐0 were inoculated with O. heveae HN1106. At 1 dpi, the ratio of fungal entry was 72% on the pmr4 mutant (Fig. 7E), but 35% on Col‐0 (Fig. 7E), suggesting that callose deposition contributes to the penetration resistance to O. heveae HN1106. By 5 dpi, chlorosis symptoms were present in pmr4 mutants, but not in Col‐0 (Fig. 7A). This could be a result of the increased entry rate of O. heveae HN1106 in pmr4 mutants. Consistent with the phenotypes, O. heveae HN1106 triggers a higher level of cell death (Fig. 7B) and PR gene expression (Fig. 7C, D) in pmr4 than in Col‐0 at 5 dpi.
Figure 7.
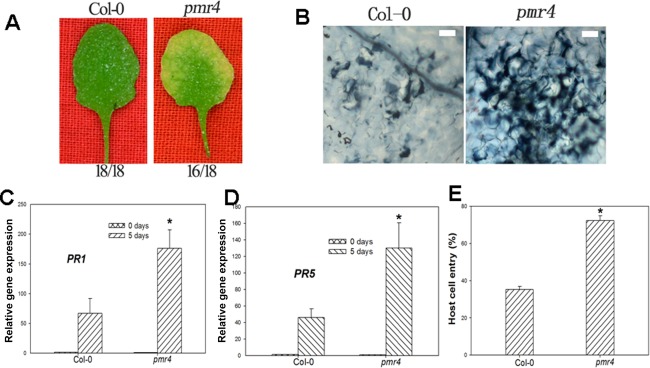
PMR4 (Powdery Mildew Resistant 4) contributes to the penetration resistance to Oidium heveae HN1106 in Arabidopsis. Four‐week‐old plants were inoculated with O. heveae HN1106. (A) The symptoms were examined for Col‐0 and the pmr4 mutant at 5 days post‐inoculation (dpi). The numbers below each panel indicate the number of leaves surveyed (denominator) and the number of leaves showing the phenotypes (numerator). (B) Infected leaves of Col‐0 and the pmr4 mutant were stained with trypan blue at 5 dpi. Bar, 50 μm. (C) The mRNA abundance of PR1 (Pathogenesis‐Related 1) was determined at 0 and 5 dpi. Data represent the mean ± standard deviation (SD) of three RNA replicates. *Significant difference from Col‐0 (Student's t‐test, P < 0.05). (D) The mRNA abundance of PR5 was determined at 0 and 5 dpi. Data represent the mean ± SD of three RNA replicates. *Significant difference from Col‐0 (Student's t‐test, P < 0.05). (E) Quantitative assessment of host cell entry rates. Data represent the mean ± SD of three experiments, each based on five to seven leaves per plant line. *Significant difference from Col‐0 (Student's t‐test, P < 0.05). These experiments were repeated twice with similar results.
The Arabidopsis accession Lag2‐2 is susceptible to O. heveae HN1106
Arabidopsis accessions exhibit natural genetic variation; although the Col‐0 accession is resistant to O. heveae HN1106, other accessions may be susceptible. To test this hypothesis, a total of 47 Arabidopsis accessions obtained from the Nottingham Arabidopsis Stock Centre (NASC) and Col‐0 as a control were challenged with O. heveae HN1106. We found that 37 Arabidopsis accessions, including Shigu‐2 (Fig. 8A, B) and Col‐0 (Fig. 8A, B), were resistant, showing characteristic chlorosis symptoms, cell death, limited fungal growth and the absence of conidiospores (Table 1). By contrast, nine Arabidopsis accessions, including HKT2‐4 (Fig. 8A, B), displayed reduced disease symptoms, such as a sparse hyphal network, no apparent cell death and no formation of conidiospores (Table 1). Only the Lag2‐2 accession showed typical powdery mildew symptoms, as indicated by strong hyphal growth and conidiospore formation (Fig. 8A, B). These results indicate that O. heveae HN1106 and Lag2‐2 exhibit a compatible interaction.
Figure 8.
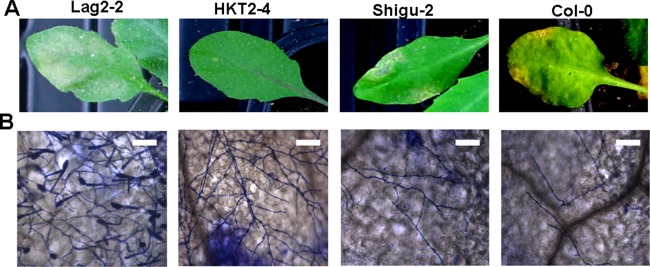
Ecotype Lag2‐2 is susceptible to Oidium heveae HN1106. Leaves from 4‐week‐old plants were inoculated with O. heveae HN1106. (A) The symptoms of Arabidopsis accessions Lag2‐2, HKT‐2, Shigu‐2 and Col‐0 infected with O. heveae HN1106 at 10 days post‐inoculation (dpi). (B) Light micrographs of O. heveae growth in the Lag2‐2, HKT‐2, Shigu‐2 and Col‐0 ecotypes at 10 dpi. Light microscopy images were taken after visualization of the fungal structures using Coomassie brilliant blue at 10 dpi. Bar, 50 μm. These experiments were repeated twice with similar results.
Table 1.
Infection phenotypes of 47 Arabidopsis thaliana accessions on challenge with Oidium heveae HN1106.
| Accession | NASC code | Chlorosis symptom | Cell death | Hyphal network | Conidiospores |
|---|---|---|---|---|---|
| Altenb‐2 | N76353 | + | + | – | – |
| Apost‐1 | N76368 | + | + | – | – |
| Nemrut‐1 | N76398 | + | + | – | – |
| Xan‐1 | N76387 | + | + | – | – |
| Shigu‐2 | N76374 | + | + | – | – |
| Slavi‐1 | N76419 | + | + | – | – |
| Ped‐0 | N76415 | + | + | – | – |
| Sha | N76382 | + | + | – | – |
| Petro‐1 | N76370 | + | + | – | – |
| Bak‐2 | N76392 | + | + | – | – |
| Don‐0 | N76411 | + | + | – | – |
| Lerik1‐3 | N76388 | + | + | – | – |
| Nie1‐2 | N76402 | + | + | – | – |
| Vie‐0 | N76418 | + | + | – | – |
| Lag2‐2 | N76390 | – | – | + | + |
| Tu‐Scha‐9 | N76401 | + | + | – | – |
| Kidr‐1 | N76376 | + | + | – | – |
| Vash‐1 | N76391 | + | + | – | – |
| Sij1 | N76379 | + | + | – | – |
| Dobra‐1 | N76369 | + | + | – | – |
| Angel‐1 | N76362 | + | + | – | – |
| Lago‐1 | N76367 | – | – | + | – |
| Star‐8 | N76400 | – | – | + | – |
| Agu‐1 | N76409 | + | + | – | – |
| Moran‐1 | N76363 | + | + | – | – |
| Yeg‐1 | N76394 | + | + | – | – |
| Rovero‐1 | N76351 | + | + | – | – |
| Kly1 | N76385 | + | + | – | – |
| Castelfed‐4 | N76355 | + | + | – | – |
| Shigu‐1 | N76375 | + | + | – | – |
| Tu‐Wa1‐2 | N76405 | + | + | – | – |
| Bak‐7 | N76393 | + | + | – | – |
| Vezzano‐2 | N76349 | – | – | + | – |
| Voeran‐1 | N76352 | – | – | + | – |
| Pra‐6 | N76416 | + | + | – | – |
| Istisu‐1 | N76389 | + | + | – | – |
| Bolin‐1 | N76373 | + | + | – | – |
| Lecho‐1 | N76371 | – | – | + | – |
| Da(1)‐12 | N76470 | – | – | + | – |
| Qui‐0 | N76417 | + | + | – | – |
| Monte‐1 | N76361 | – | – | + | – |
| Ciste‐1 | N76359 | – | – | + | – |
| HKT2‐4 | N76404 | – | – | + | – |
| Ler‐0 | N77020 | + | + | – | – |
| Kas‐1 | N22636 | + | + | – | – |
| Nossen | N3081 | + | + | – | – |
| Ws‐0 | N76303 | + | + | – | – |
NASC, Nottingham Arabidopsis Stock Centre.
+, chlorosis symptom or cell death similar to Col‐0, conidiospores formation and hyphal network formation. –, no chlorosis symptom, cell death, conidiospores or hyphal network formation.
To further determine whether the susceptibility of Lag2‐2 is specific to O. heveae strain HN1106, Arabidopsis Lag2‐2 plants were inoculated with two other O. heveae strains, HN1208 and YN7834, isolated in Hainan and YunNan, China, respectively. By 10 dpi, Lag2‐2 seemed to be equally susceptible to HN1208 and HN1106 infection (Fig. S4, see Supporting Information). However, the Arabidopsis accession was found to be resistant to YN7834, because YN7834 only developed an apparent hyphal network, but no conidiospores (Fig. S4), suggesting that the susceptibility of Lag2‐2 is specific to O. heveae strains HN1106 and HN1208.
Discussion
Powdery mildew is an important disease affecting rubber plantations worldwide. However, studies of the molecular mechanism underlying the interaction between H. brasiliensis and O. heveae have been hampered largely by the lack of genetic background and methodology for these species. The model plant A. thaliana has been widely used to investigate the molecular mechanisms underlying powdery mildew disease. Several isolates of powdery mildew, Golovinomyces cichoracearum (Adam and Somerville, 1996), G. cruciferarum (Koch and Slusarenko, 1990), G. orontii (Plotnikova et al., 1998) and Oidium neolycopersici (Xiao et al., 2001), can infect Arabidopsis. Microscopy analysis showed that O. heveae HN1106 exhibited similar pathogenicity on H. brasiliensis and Arabidopsis eds1 mutants. In addition, mature conidia developed from Arabidopsis eds1 mutant leaves can successfully infect H. brasiliensis, suggesting that studies of the interaction between Arabidopsis and O. heveae might further our understanding of powdery mildew disease in economically important rubber trees.
The interactions between plants and powdery mildew can be divided into non‐host, compatible and incompatible interactions. Non‐host resistance usually occurs in plant resistance against some non‐adapted powdery mildews. Post‐penetration resistance, mediated by EDS1 or PAD4, and penetration resistance, conferred by PENETRATION2 (PEN2) or PEN3, limit invasion, growth and asexual reproduction of the non‐adapted powdery mildew Erysiphe pisi on Col‐0 Arabidopsis (Stein et al., 2006). In contrast with non‐host interactions, penetration resistance conditioned by PEN2 or PEN3 does not appear to be required for compatible and incompatible interactions. In our study, O. heveae HN1106 displayed an entry ratio of about 35% on Col‐0 and about 55% on the eds1 mutant, which are significantly higher than that in non‐host interactions (10%), suggesting that the interaction between Arabidopsis and O. heveae HN1106 is not a non‐host interaction. Furthermore, O. heveae HN1106 can finish its life cycle and produce many conidiospores on eds1 or pad4 single mutants, indicating that post‐penetration resistance is sufficient to limit infection by O. heveae HN1106.
NBS‐LRR‐type R proteins that respond to powdery mildews have been characterized in many plant species, such as the Mla genes in barley (Shen et al., 2007), Ol genes in Solanum lycopersicum (tomato; Bai et al., 2005), Run1 in Vitis vinifera (grapevine; Barker et al., 2005) and Rpp1 in Rosa multiflora (rose; Linde et al., 2004). Golovinomyces cruciferarum is an incompatible pathogen on Arabidopsis ecotype Ms‐0. However, no canonical powdery mildew R genes have been identified in Arabidopsis to date. Thus, Arabidopsis might not have co‐evolved with G. cruciferarum long enough to select for canonical R genes. The RPW8 locus of Ms‐0 was found to be involved in disease resistance to G. cruciferarum (Xiao et al., 2003). RPW8 is an atypical R protein with a transmembrane (TT)‐CC domain, and mediates broad‐spectrum resistance against powdery mildews through EDS1, PAD4 and SA signalling (Xiao et al., 2001, 2005). Here, we found that Arabidopsis Col‐0 is a resistant host of O. heveae HN1106, and proposed that similar mechanisms might function in O. heveae HN1106–Col‐0 and G. cruciferarum–Ms‐0 interactions. Nevertheless, O. heveae HN1106 triggers disease resistance in Arabidopsis Col‐0 in an EDS1‐ and PAD4‐dependent manner, which is different from the compatible interaction between G. cruciferarum and Col‐0. Therefore, we speculate that EDS1‐mediated ETI signalling, which is usually activated by TIR‐NB‐LRR proteins, is responsible for O. heveae‐induced resistance in Arabidopsis. If this is indeed the case, canonical TIR‐NB‐LRR‐type R gene(s) should be present in Col‐0 in response to O. heveae infection. However, which TIR‐NB‐LRR protein(s) confers resistance to O. heveae HN1106 remains to be elucidated. CC‐NB‐LRR‐type R proteins are not likely to be involved in resistance to O. heveae, because NDR1 is not required for O. heveae HN1106‐induced disease resistance and defence responses.
In plant–microbe interactions, the full output of resistance conditioned by TIR‐NB‐LRR‐type R proteins requires both SA‐dependent and SA‐independent resistance pathways. In addition, both need the signalling nodes of EDS1 and PAD4. For example, the EDS1‐ and PAD4‐mediated Flavin‐Dependent Monooxygenase 1 (FMO 1) signalling pathway, which is activated by the TIR‐NB‐LRR‐type R proteins RPS4 and RPP2, represents an SA‐independent resistance pathway (Bartsch et al., 2006). In this study, although Arabidopsis lost the ability to trigger the cell death response to O. heveae HN1106 when SA signalling was disrupted, a modest resistance was still present, suggesting that SA‐independent pathways, together with SA signalling, are required for full resistance to O. heveae HN1106 in Arabidopsis. However, whether FMO 1 signalling is involved in resistance to O. heveae HN1106 remains to be determined.
In incompatible interactions between Arabidopsis and some powdery mildews, callose deposition is fully responsible for penetration resistance. However, the pmr4 mutant exhibited increased resistance to compatible powdery mildews, and this was caused by the hyperactivation of SA‐mediated defence responses in the pmr4 mutant (Nishimura et al., 2003). In this study, we found that O. heveae HN1106 exhibits an incompatible interaction with Arabidopsis, and induces chlorosis symptoms and defence responses more rapidly in pmr4 mutants than in Col‐0, presumably because of the significantly higher host cell entry rate in pmr4. ETI is often accompanied by callose deposition at infection sites, and callose synthesis is significantly compromised in mutants in which ETI is disrupted. Here, we found that eds1, pad4, rar1, npr1, NahG and sid2 mutants displayed considerably less callose deposition, which underlies the higher penetration ratio of O. heveae HN1106 in these mutants.
Arabidopsis Col‐0 is the resistant host for O. heveae HN1106. Thus, we postulated that a susceptible ecotype may exist in natural Arabidopsis accessions. We screened 47 Arabidopsis accessions for resistance to O. heveae HN1106 and identified significant genetic diversity. Most accessions (46 of 47) were resistant to O. heveae HN1106, and did not support conidiospore formation. Among the 46 resistant accessions, 37 accessions showed chlorotic/necrotic flecks with collapsed hyphae at inoculation sites. Another nine resistant accessions displayed observable hyphal networks with no apparent cell death, which is consistent with a previous study, which showed that powdery mildew resistance is not always associated with a rapid HR (Adam and Somerville, 1996), indicating that resistance mechanisms other than rapid necrosis arrest pathogen development. Only one accession, Lag2‐2, was susceptible to O. heveae HN1106. Lag2‐2 was further found to be susceptible to HN1208, but resistant to O. heveae YN7834. We propose that the R proteins involved in O. heveae HN1106 and HN1208 resistance may have been lost or may display sequence divergence from their counterparts in other accessions. Furthermore, we predict that the effector proteins secreted by O. heveae YN7834 are also probably different from those secreted by O. heveae HN1106 and HN1208.
Experimental Procedures
Arabidopsis and mutants
A full list of A. thaliana accessions used in this study is provided in Table 1. The common laboratory Arabidopsis plants used in this study included the wild‐type (Col‐0) and the following mutants: eds1‐2 (backcrossed multiple times into the Col‐0 background) (Aarts et al., 1998), pad4‐1 (Glazebrook et al., 1997), rar1‐20 (Tornero et al., 2002), ndr1‐1 (Century et al., 1997), sid2‐2 (Wildermuth et al., 2001), NahG (Lawton et al., 1995), npr1‐1 (Cao et al., 1997) and pmr4‐1 (Nishimura et al., 2003).
Powdery mildew infections
The O. heveae strain HN1106 was used throughout this study and was maintained in the leaves of the susceptible H. brasiliensis clone GT‐1. Actively growing fungal spores (at 12–15 dpi) were used as a source of inoculum. To ensure an even inoculation density, the plants were placed under a modified settling tower (diameter, 40 cm; height, 60 cm), which was covered with a nylon mesh. Conidia from three to five infected GT‐1 leaves were dusted on top of the tower, and the inoculated plants were kept under the settling tower for 1 h before being moved to the growth chamber (Adam and Somerville, 1996).
Phylogenetic tree creation
A phylogenetic tree was created based on 11 powdery mildew ITS sequences using mega 5.0. The analysis preferences were as follows: test of phylogeny, bootstrap method; number of bootstrap replications, 500; number of initial trees (random addition), 2; Mp search level, 1; and maximum number of trees retained, 100.
Host cell entry
Three inoculated leaves were harvested at 1 dpi and stained with Coomassie brilliant blue. The proportion of germinated fungal sporelings that developed secondary hyphae was assessed (minimum of 50 germinated sporelings/leaf evaluated). Fungal penetration success on each plant was quantified in at least three independent experiments.
Analysis of fungal growth
Three different plants per line were inoculated with a low density of powdery mildew spores (to avoid overlap of fungal colonies) and four leaves per plant were harvested at 48 hpi. At least 20 images of single colonies per line and time point were taken and analysed with mie 3.1 software (http://www.miesoftware.com/).
Quantification of conidiospores per colony
Eight inoculated leaves were harvested at 10 dpi and stained with Coomassie brilliant blue. Conidiospores were counted. This procedure was repeated two to four times.
DNA extraction and polymerase chain reaction (PCR) amplification
DNA was extracted from fungal spores and hyphae with 2 × CTAB (hexadecyltrimethylammonium bromide) buffer (Martin and Rygiewicz, 2005). The nuclear rDNA ITS region (644 bp), which included the 31‐bp fragment at the 3′ end of the 18S (small subunit) rRNA gene, 220‐bp fragment in the first ITS (ITS1), 154‐bp fragment in the complete 5.8S rRNA gene, 183‐bp fragment in the second ITS (ITS2) and 56‐bp fragment at the 5′ end of the 28S (large subunit) rRNA gene, was amplified using the primers ITS1 (5′‐TCCGTAGGTGAACCTGCGG‐3′) and ITS4 (5′‐TCCTCCGCTTATTGATATGC‐3′).
Callose deposition assay
The leaves of 4‐week‐old Arabidopsis plants were inoculated with O. heveae HN1106. Leaves were harvested at 7 dpi, cleared, stained with aniline blue (Hauck et al., 2003) and mounted in 50% glycerol. The leaves were examined with a fluorescence microscope under ultraviolet light. The number of callose deposits per microscopic field of 0.1 mm2 was calculated from six leaves using Image J software (http://www.uhnresearch.ca/wcif).
RNA isolation and real‐time reverse transcription (RT)‐PCR
The leaves of 4‐week‐old Arabidopsis plants were inoculated with O. heveae HN1106, and RNA was isolated at the indicated time points. Three to five micrograms of RNA were used for cDNA synthesis. mRNA was quantified by real‐time RT‐PCR using a SYBR Premix Ex Taq Kit (TaKaRa, Changping, Beijing, China). Arabidopsis ACTIN was used as the reference gene. Primers 5′‐TACGCAGAACAACTAAGAGG‐3′ and 5′‐TCGTTCACATAATTCCCACG‐3′ were used to amplify PR1, and primers 5′‐TGGTGGAAGCACAGAAGTTG‐3′ and 5′‐GATCCATGTTTGGCTCCTTC‐3′ were used to amplify ACTIN. Primers 5′‐GCACAGAGACACACACAAAA‐3′ and 5′‐TGTTCCTTAGAGTGAAGTCTG‐3′ were used to amplify PR5. The RT‐PCR conditions were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s and 72°C for 25 s. The expression level was normalized to that of the ACTIN control, and relative expression values were determined against uninfected samples or wild‐type Col‐0 using the comparative C t method.
Trypan blue staining
The leaves of 4‐week‐old Arabidopsis plants were inoculated with O. heveae HN1106, and leaves were harvested at 10 dpi. Fungal structures and dead plant cells were stained with trypan blue and cleared with chloral hydrate overnight at room temperature (Frye and Innes, 1998). Cleared leaves were mounted under coverslips in 50% glycerol and examined with a microscope.
H2O2 production assay
The leaves of 4‐week‐old Arabidopsis plants were inoculated with O. heveae HN1106, and leaves were collected at 7 dpi. H2O2 was detected by staining with 3,3′‐diaminobenzidine‐HCl, pH 3.8, for 8 h, followed by clearing in 95% ethanol overnight at room temperature (Xiao et al., 2003). Cleared leaves were mounted under coverslips in 50% glycerol and examined with a microscope.
Coomassie brilliant blue staining
Infected leaves were fixed and cleared in an ethanol solution containing 6.7% phenol, 6.7% lactic acid, 13.3% glycerol and 6.7% H2O. Fungal structures in cleared leaves were visualized by staining with an ethanolic solution containing 0.6% Coomassie brilliant blue (Lipka et al., 2005).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The internal transcribed spacer (ITS) sequence of Oidium heveae HN1106.
Fig. S2 Phylogenetic analysis of the Oidium heveae HN1106 internal transcribed spacer (ITS) sequence. A phylogenetic tree was created based on 11 powdery mildew ITS sequences using mega 5.0. The phylogeny reconstruction used the maximum parsimony method. The percentage bootstrap support (500 replications) is shown above the branches.
Fig. S3 Symptoms of Hevea brasiliensis GT‐1 infection with mature conidia on an Arabidopsis eds1 mutant. Light‐green phase H. brasiliensis GT‐1 leaves were inoculated with mature conidia from Arabidopsis eds1 (enhanced disease susceptibility 1) mutants, and symptoms were examined at 15 days post‐inoculation (dpi). These experiments were repeated twice with similar results.
Fig. S4 Ecotype Lag2‐2 is resistant to Oidium heveae YN7834. Leaves of 4‐week‐old plants were inoculated with O. heveae strains HN1106, HN1208, and YN7834. (A) The plants were photographed at 10 days post‐inoculation (dpi). (B) Light micrograph of O. heveae growth on ecotype Lag2‐2 at 10 dpi. Light microscopy images were taken after fungal structures had been stained with Coomassie brilliant blue at 10 dpi. Bar, 50 μm. These experiments were repeated twice with similar results.
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (No. 31201483, No. 31560296) and the Natural Science Foundation of Hainan Province (No. 314053, No. 314043).
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, L. and Somerville, S.C. (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . Plant J. 9, 341–356. [DOI] [PubMed] [Google Scholar]
- Bai, Y. , van der Hulst, R. , Bonnema, G. , Marcel, T.C. , Meijer‐Dekens, F. , Niks, R.E. and Lindhout, P. (2005) Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate‐dependent resistance via a different mechanism than recessive ol‐2. Mol. Plant–Microbe Interact. 18, 354–362. [DOI] [PubMed] [Google Scholar]
- Barker, C.L. , Donald, T. , Pauquet, J. , Ratnaparkhe, M.B. , Bouquet, A. , Adam‐Blondon, A.F. , Thomas, M.R. and Dry, I. (2005) Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theoretical and applied genetics, 111, 370–377. [DOI] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, M. , Bednarek, P. , Vivancos, P.D. , Schneider, B. , von Roepenack‐Lahaye, E. , Foyer, C.H. , Kombrink, E. , Scheel, D. and Parker, J.E. (2010) Accumulation of isochorismate‐derived 2,3‐dihydroxybenzoic 3‐O‐beta‐D‐xyloside in Arabidopsis resistance to pathogens and ageing of leaves. J. Biol. Chem. 285, 25 654–25 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeley, F. (1933) Oidium heveae: report on the 1933 outbreak of Hevea leaf mildew. J. Rubber Res. Inst. Malaysia, 5, 5–13. [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Glazebrook, J. , Clarke, J.D. , Volko, S. and Dong, X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. and Ellis, J.G. (2007) Avirulence proteins from haustoria‐forming pathogens. FEMS Microbiol. Lett. 269, 181–188. [DOI] [PubMed] [Google Scholar]
- Century, K.S. , Shapiro, A.D. , Repetti, P.P. , Dahlbeck, D. , Holub, E. and Staskawicz, B.J. (1997) NDR1, a pathogen‐induced component required for Arabidopsis disease resistance. Science, 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Consonni, C. , Humphry, M.E. , Hartmann, H.A. , Livaja, M. , Durner, J. , Westphal, L. , Vogel, J. , Lipka, V. , Kemmerling, B. , Schulze‐Lefert, P. , Somerville, S.C. and Panstruga, R. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Day, B. , Dahlbeck, D. and Staskawicz, B.J. (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis . Plant Cell, 18, 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Falk, A. , Feys, B.J. , Frost, L.N. , Jones, J.D. , Daniels, M.J. and Parker, J.E. (1999) EDS1, an essential component of R gene‐mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA, 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J. , Wiermer, M. , Bhat, R.A. , Moisan, L.J. , Medina‐Escobar, N. , Neu, C. , Cabral, A. and Parker, J.E. (2005) Arabidopsis SENESCENCE‐ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell, 17, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A. and Innes, R.W. (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell, 10, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Zook, M. , Mert, F. , Kagan, I. , Rogers, E.E. , Crute, I.R. , Holub, E.B. , Hammerschmidt, R. and Ausubel, F.M. (1997) Phytoalexin‐deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics, 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollner, K. , Schweizer, P. , Bai, Y. and Panstruga, R. (2008) Natural genetic resources of Arabidopsis thaliana reveal a high prevalence and unexpected phenotypic plasticity of RPW8‐mediated powdery mildew resistance. New Phytol. 177, 725–742. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, D.A. , He, Y. , McNulty, B.C. , Tornero, P. and Dangl, J.L. (2009) Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB‐LRR disease resistance protein regulation. Proc. Natl. Acad. Sci. USA, 106, 9556–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D. , Tootle, T.L. , Reuber, T.L. , Frost, L.N. , Feys, B.J. , Parker, J.E. , Ausubel, F.M. and Glazebrook, J. (1999) Arabidopsis thaliana PAD4 encodes a lipase‐like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA, 96, 13 583–13 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A.J. (1990) Fungal pathogens of Arabidopsis thaliana (L.) Heynh. Bot Helv, 100, 257–268. [Google Scholar]
- Lawton, K. , Weymann, K. , Friedrich, L. , Vernooij, B. , Uknes, S. and Ryals, J. (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant–Microbe Interact. 8, 863–870. [DOI] [PubMed] [Google Scholar]
- Linde, M. , Mattiesch, L. and Debener, T. (2004) Rpp1, a dominant gene providing race‐specific resistance to rose powdery mildew (Podosphaera pannosa): molecular mapping, SCAR development and confirmation of disease resistance data. Theor. Appl. Genet. (Theoretische und angewandte Genetik) 109, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Lipka, V. , Dittgen, J. , Bednarek, P. , Bhat, R. , Wiermer, M. , Stein, M. , Landtag, J. , Brandt, W. , Rosahl, S. , Scheel, D. , Llorente, F. , Molina, A. , Parker, J. , Somerville, S. and Schulze‐Lefert, P. (2005) Pre‐ and postinvasion defenses both contribute to nonhost resistance in Arabidopsis . Science, 310, 1180–1183. [DOI] [PubMed] [Google Scholar]
- Martin, K.J. and Rygiewicz, P.T. (2005) Fungal‐specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M. and Dangl, J.L. (2000) Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- Mitra, M. and Mehta, P.R. (1938) Some leaf diseases of Hevea brasiliensis new to India. Indian J. Agric. Sci. 8, 185–188. [Google Scholar]
- Moreau, M. , Degrave, A. , Vedel, R. , Bitton, F. , Patrit, O. , Renou, J.P. , Barny, M.A. and Fagard, M. (2012) EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora . Mol. Plant–Microbe Interact. 25, 421–430. [DOI] [PubMed] [Google Scholar]
- Nawrath, C. , Heck, S. , Parinthawong, N. and Metraux, J.P. (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell, 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T. , Stein, M. , Hou, B.H. , Vogel, J.P. , Edwards, H. and Somerville, S.C. (2003) Loss of a callose synthase results in salicylic acid‐dependent disease resistance. Science, 301, 969–972. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova, J.M. , Reuber, T.L. and Ausubel, F.M. (1998) Powdery mildew pathogenesis of Arabidopsis thaliana . Mycologia, 90, 1009–1016. [Google Scholar]
- Ramonell, K. , Berrocal‐Lobo, M. , Koh, S. , Wan, J. , Edwards, H. , Stacey, G. and Somerville, S. 2005. Loss‐of‐function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum . Plant Physiol. 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout, C.J. , Skamnioti, P. , Porritt, O. , Sacristan, S. , Jones, J.D. and Brown, J.K. 2006. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell, 18, 2402–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz, S. , Stamm, A. , Malonek, S. , Wagner, S. , Becker, D. , Medina‐Escobar, N. , Vlot, A.C. , Feys, B.J. , Niefind, K. and Parker, J.E. 2011. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Rusterucci, C. , Aviv, D.H. , Holt, B.F., 3rd , Dangl, J.L. and Parker, J.E. 2001. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis . Plant Cell, 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saranya, L. , Sawanee, K. , Edson, L. , Kon, W. , Baharuddin, S. , Yukio, S. and T., S. 2005. Molecular phylogenetic and morphological analyses of Oidium heveae, a powdery mildew of rubber tree. J. Mol. Evol. 46, 220–226. [Google Scholar]
- Shen, Q.H. , Saijo, Y. , Mauch, S. , Biskup, C. , Bieri, S. , Keller, B. , Seki, H. , Ulker, B. , Somssich, I.E. and Schulze‐Lefert, P. 2007. Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Stein, M. , Dittgen, J. , Sanchez‐Rodriguez, C. , Hou, B.H. , Molina, A. , Schulze‐Lefert, P. , Lipka, V. and Somerville, S. 2006. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell, 18, 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P. , Merritt, P. , Sadanandom, A. , Shirasu, K. , Innes, R.W. and Dangl, J.L. 2002. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell, 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal, S.C. , Jeong, R.D. , Mandal, M.K. , Zhu, S. , Chandra‐Shekara, A.C. , Xia, Y. , Hersh, M. , Stromberg, A.J. , Navarre, D. , Kachroo, A. and Kachroo, P. 2009. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene‐mediated signaling. PLoS Genetics, 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, S. , Stuttmann, J. , Rietz, S. , Guerois, R. , Brunstein, E. , Bautor, J. , Niefind, K. and Parker, J.E. 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe, 14, 619–630. [DOI] [PubMed] [Google Scholar]
- Ward, E.R. , Uknes, S.J. , Williams, S.C. , Dincher, S.S. , Wiederhold, D.L. , Alexander, D.C. , Ahl‐Goy, P. , Metraux, J.P. and Ryals, J.A. 1991. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell, 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Ellwood, S. , Calis, O. , Patrick, E. , Li, T. , Coleman, M. and Turner, J.G. 2001. Broad‐spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science, 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Brown, S. , Patrick, E. , Brearley, C. and Turner, J.G. 2003. Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid‐dependent amplification circuit is required for hypersensitive cell death. Plant Cell, 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Calis, O. , Patrick, E. , Zhang, G. , Charoenwattana, P. , Muskett, P. , Parker, J.E. and Turner, J.G. 2005. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis . Plant J. Cell Mol. Biol. 42, 95–110. [DOI] [PubMed] [Google Scholar]
- Zhou, J.M. and Chai, J. 2008. Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The internal transcribed spacer (ITS) sequence of Oidium heveae HN1106.
Fig. S2 Phylogenetic analysis of the Oidium heveae HN1106 internal transcribed spacer (ITS) sequence. A phylogenetic tree was created based on 11 powdery mildew ITS sequences using mega 5.0. The phylogeny reconstruction used the maximum parsimony method. The percentage bootstrap support (500 replications) is shown above the branches.
Fig. S3 Symptoms of Hevea brasiliensis GT‐1 infection with mature conidia on an Arabidopsis eds1 mutant. Light‐green phase H. brasiliensis GT‐1 leaves were inoculated with mature conidia from Arabidopsis eds1 (enhanced disease susceptibility 1) mutants, and symptoms were examined at 15 days post‐inoculation (dpi). These experiments were repeated twice with similar results.
Fig. S4 Ecotype Lag2‐2 is resistant to Oidium heveae YN7834. Leaves of 4‐week‐old plants were inoculated with O. heveae strains HN1106, HN1208, and YN7834. (A) The plants were photographed at 10 days post‐inoculation (dpi). (B) Light micrograph of O. heveae growth on ecotype Lag2‐2 at 10 dpi. Light microscopy images were taken after fungal structures had been stained with Coomassie brilliant blue at 10 dpi. Bar, 50 μm. These experiments were repeated twice with similar results.


