Summary
Taxonomy
Eggplant latent viroid (ELVd) is the only species of the genus Elaviroid (family Avsunviroidae). All the viroids in the family Avsunviroidae contain hammerhead ribozymes in the strands of both polarities, and are considered to replicate in the chloroplasts of infected cells. This family includes two other genera: Avsunviroid and Pelamoviroid.
Physical properties
ELVd consists of a single‐stranded, circular, non‐coding RNA of 332–335 nucleotides that folds in a branched quasi‐rod‐like minimum free‐energy conformation. RNAs of complementary polarity exist in infected cells and are considered to be replication intermediates. Plus (+) polarity is assigned arbitrarily to the strand that accumulates at a higher concentration in infected tissues.
Host
To date, ELVd has only been shown to infect eggplant (Solanum melongena L.), the species in which it was discovered. A very narrow host range seems to be a common property in members of the family Avsunviroidae.
Symptoms
ELVd infections of eggplants are apparently symptomless.
Transmission
ELVd is transmitted mechanically and by seed.
Useful website
Keywords: chloroplast, eggplant, ribozyme, RNA replication, RNA ligation, RNA traffic, viroid
Introduction
Viroids are infectious agents of higher plants that consist exclusively of circular, highly base‐paired, non‐coding RNAs which, in the species known to date, range from 246 to 401 nucleotides (Ding, 2009; Flores et al., 2005; Palukaitis, 2014). Most viroids induce disease, although some apparently cause only symptomless infections. The more than 30 viroid species currently accepted by the International Committee on Taxonomy of Viruses (Owens et al., 2012) are classified into two families. Most currently known viroids belong to the family Pospiviroidae. They all contain a characteristic central conserved region (CCR) around the centre of their molecules, when folded in their minimum free‐energy conformations. In contrast, Avocado sunblotch viroid (ASBVd) (Symons, 1981), Peach latent mosaic viroid (PLMVd) (Flores et al., 2006; Hernández and Flores, 1992), Chrysanthemum chlorotic mottle viroid (CChMVd) (Navarro and Flores, 1997) and Eggplant latent viroid (ELVd) (Fadda et al., 2003) lack a CCR, but the strands of both polarities can form hammerhead ribozymes, which are active at self‐cleaving the RNAs. These four viroids are classified in the family Avsunviroidae (Flores et al., 2000). ASBVd is the type member of this family and is classified in the genus Avsunviroid. PLMVd and CChMVd are included in the genus Pelamoviroid, whereas ELVd is the sole species in the genus Elaviroid.
Viroids replicate through a rolling‐circle mechanism with RNA intermediates (Branch and Robertson, 1984). Viroids in the families Pospiviroidae and Avsunviroidae follow slightly different variations of this mechanism, called asymmetric and symmetric pathways, depending on the existence of one or two symmetrical rolling circles. Members of Pospiviroidae replicate through the asymmetric pathway in the nucleus of infected cells, whereas members of Avsunviroidae do so through the symmetric pathway in chloroplasts (Daròs et al., 2006). During viroid replication, host DNA‐dependent RNA polymerases transcribe multimeric viroid RNAs (Hutchins et al., 1985; Mühlbach and Sänger, 1979; Navarro et al., 2000; Spiesmacher et al., 1983). Some of these intermediates are cleaved to monomers, either autocatalytically through hammerhead ribozymes in the family Avsunviroidae (Hutchins et al., 1986) or by a host type‐III RNase in the family Pospiviroidae (Gas et al., 2007). Finally, monomers are circularized by host DNA ligase 1 (family Pospiviroidae) (Nohales et al., 2012a), or by a chloroplastic isoform of tRNA ligase (family Avsunviroidae) (Nohales et al., 2012b).
Much of our knowledge about viroid biology derives from research into Potato spindle tuber viroid (PSTVd) (Gross et al., 1978; Owens, 2007), the first viroid to be discovered (Diener, 1971, 2003) and the type member of the family Pospiviroidae. Research into this viroid family has also been facilitated by the many species within it. In contrast, research into the family Avsunviroidae has been made more difficult as it contains very few species and, in particular, because of the problematic (mostly ligneous) hosts. Viroids in the family Avsunviroidae show an extremely narrow host range, which is restricted basically to the species in which they were discovered (avocado, peach, chrysanthemum or eggplant), or to some related species (Fadda et al., 2003; Flores et al., 2000). Nevertheless, recent research has shown that ELVd is possibly the friendliest experimental system in the family Avsunviroidae, and substantial advances in RNA replication, processing and trafficking, and even some biotechnological applications, have resulted from the experimental work that has employed this viroid. It should be noted that, according to current knowledge, ELVd cannot be strictly considered as a pathogen because ELVd‐infected eggplants do not display noticeable symptoms. However, research on this viroid is aiding in our understanding of this family, which does include well‐defined pathogens. Moreover, the term ‘pathogen’ is not without controversy (Casadevall and Pirofski, 2014; Pirofski and Casadevall, 2012). For those researchers interested in viroid biology, or even in some basic RNA molecular biology aspects, who may consider adopting ELVd as one of their experimental systems, I have herein attempted to review all current knowledge on this viroid.
ELVd Discovery
In 1994, Fagoaga et al. published a survey of viroid and viroid‐like agents in a series of vegetable crop species typically grown in eastern and southern Spain, known to be easily infected with vector‐transmitted viruses (Fagoaga et al., 1994). Tissue samples (symptomatic and asymptomatic) were collected at Servicio de Plantas de Vivero (Valencia, Spain) and included 12 species in the families Fabaceae, Apiaceae, Brassicaceae, Cucurbitaceae, Solanaceae and Amaranthaceae. Some samples that corresponded to eggplant (Solanum melongena L.), squash (Cucurbita pepo L.) and tomato (Solanum lycopersicum L.) were shown to be infected with Cucumber mosaic virus satellite RNA. Interestingly, two of the 24 eggplant samples were shown to contain a viroid‐like RNA. This was found in the plants that belonged to the cultivar Sonja. Some other eggplant cultivars (Avan, Baluroi and Bonica) were also analysed in the survey, although with negative results with respect to this viroid‐like RNA (Fagoaga et al., 1994).
An analysis of the RNA preparations obtained from these two plants by sequential polyacrylamide gel electrophoresis (PAGE) and silver staining revealed a likely circular species that migrated similarly, but not exactly, as Chrysanthemum stunt viroid, and two variants (CVd‐Ia and CVd‐Ib) of Citrus bent leaf viroid. Nuclease treatment confirmed this species to be RNA. Fagoaga et al. used these RNA preparations to mechanically inoculate new eggplants. They observed that most inoculated plants accumulated a circular RNA, 2 months after inoculation, as revealed by sequential PAGE. However, no symptoms were distinguished in these infected plants. Remarkably, hybridization analyses with probes that represented the major viroid groups [ASBVd, Apple scar skin viroid, Citrus exocortis viroid (CEVd) and Hop stunt viroid] gave negative results. Eggplant circular RNA was also shown to poorly precipitate in 2 m LiCl. Taking together all of these results, Fagoaga et al. (1994) considered the pathogen to be a new viroid and tentatively designated it as Eggplant latent viroid.
ELVd Infectivity and Symptoms
ELVd RNA was mechanically inoculated into some typical hosts of known viroids (tomato, cucumber, chrysanthemum and citron) with negative results (Fagoaga et al., 1994). However, ELVd infectivity has been demonstrated in a series of tested eggplant cultivars, all of which remained symptomless (Fagoaga and Duran‐Vila, 2003). Weekly observations of the growing pattern, flower number and weight, and the number and appearance of fruits over 3 months, failed to reveal any symptoms in infected eggplants (Fadda et al., 2003). Figure 1 shows the leaves and flowers of mock‐inoculated and ELVd‐infected eggplants belonging to the cultivar Black Beauty. No differences can be observed 2 months post‐inoculation.
Figure 1.

Eggplant latent viroid (ELVd) induces apparently symptomless infections in eggplant. Leaves and flowers of mock‐inoculated (left) and ELVd‐infected (right) eggplants (cv. Black Beauty). Photograph was taken 2 months after inoculation.
ELVd has been shown to be mechanically transmitted by cutting tools with 55% efficiency and seed transmitted with an efficiency of approximately 20% (Fadda et al., 2003; Fagoaga and Duran‐Vila, 2003). Seed treatment with 1% sodium hypochlorite did not affect the seed transmission ratio, which indicates that this does not result from accidental seedling inoculation with the remains of contaminated tissues during the germination process.
ELVd Cloning and Sequencing
ELVd RNA preparations were obtained from mechanically inoculated eggplants that belonged to the cultivar Redonda Morada, and cDNAs were synthesized by reverse transcription‐polymerase chain reaction (RT‐PCR) by an approach that requires no prior sequence knowledge (Navarro et al., 1997). Sequences from a partial ELVd clone were used to design a pair of primers to amplify full‐length viroid cDNAs by RT‐PCR, which were cloned and sequenced. These sequences were used to design a second pair of primers in a different region of the molecule to amplify new full‐length cDNAs, which were cloned and sequenced again (Fadda et al., 2003). A total of 10 full‐length ELVd cDNAs were sequenced by Fadda et al. (2003). The sequence analysis led to three interesting conclusions. First, the sequences obtained with both pairs of primers were coherent with a circular RNA template, which confirmed the anticipated conclusion based on electrophoretic migration (Fagoaga et al., 1994). Second, only two of the 10 analysed full‐length sequences were identical, which denoted a viroid species with high sequence variability. More specifically, ELVd sequence variants were classified into four groups. Group I consisted of three variants of 335 nucleotides, which shared more than 98% sequence identity. Group II comprised three other variants of 332, 333 and 334 nucleotides, once again sharing more than 98% identity. Group III contained two variants of 335 nucleotides, which were 98% identical. The last sequence (334 nucleotides) was included in Group IV. Representative sequences in each group were selected and named ELVd‐1, ELVd‐2, ELVd‐3 and ELVd‐4, respectively, and were deposited in GenBank (accession numbers AJ536612, AJ536613, AJ536614 and AJ536615, respectively). Interestingly, the variants that belonged to the different groups displayed rather low identity, despite forming part of the same species. Thus, ELVd‐1 was only 88% identical to ELVd‐2, ELVd‐3 or ELVd‐4, ELVd‐2 was 91% identical to ELVd‐3 or ELVd‐4, and ELVd‐3 was 92% identical to ELVd‐4. All of these sequence variants had a guanine plus cytosine (G + C) content of 53%–54%. Third, and remarkably, all of these sequence variants contained the conserved nucleotide residues and double‐stranded elements that connect them to form hammerhead ribozyme structures in the strands of both polarities (Fadda et al., 2003).
ELVd‐2 was probably the most representative of the sequence variants obtained, because somehow it had intermediate properties. For this reason, ELVd‐2 was employed in almost all of the subsequent experiments. However, ELVd‐1 is the reference variant of the species (Owens et al., 2012). Figure 2 presents the sequence and minimum free‐energy conformation of ELVd‐2 obtained by the Mfold algorithm (Zuker, 2003). This is a quasi‐rod‐like structure with two bifurcations (upper left and upper right hairpins) at both ends. Most of the variability encountered in the other ELVd sequence variants was accommodated in this conformation as compensatory mutations or covariations, which suggested that this conformation, or something similar, most likely exists in vivo (Fadda et al., 2003). Giguère et al. (2014) experimentally determined the secondary structures of all the species in the family Avsunviroidae by a selective 2′‐hydroxyl acylation analysed by primer extension (SHAPE) approach. For ELVd, the analysis was performed with the National Center for Biotechnology Information (NCBI) reference sequence variant (NC_004728, which is ELVd‐1, AJ536612). Both the + and − strands were shown to fold in similar structures, composed of a relatively long rod‐like central domain, which ended with a three‐way junction on the left and three‐way (+ strand) or five‐way (– strand) junctions on the right. The presence of MgCl2 in the analysis did not allow significant differences to be detected, which suggests that ELVd, as for ASBVd, and unlike PLMVd and CChMVd, lacks a pseudoknot (Giguère et al., 2014).
Figure 2.
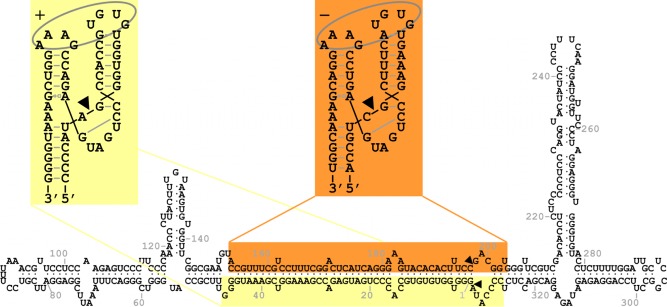
Sequence of Eggplant latent viroid (ELVd)−2 (AJ536613) folded in the secondary structure of minimum free energy predicted by Mfold. The domains of the + and − hammerhead ribozymes are highlighted on yellow and orange backgrounds, respectively. Ribozyme cleavage sites are indicated by arrowheads. The structures of the + and − ribozymes, represented in the Y‐shaped conformation supported by X‐ray crystallography and NMR data, are also shown. In these structures, black lines indicate the continuity of the nucleotide strands, black arrowheads indicate self‐cleavage sites, grey dashes and lines indicate base pairing, grey open squares next to open triangles indicate Hoogsteen/sugar edge base pairing and grey ellipses indicate tertiary interactions between loops.
The availability of cDNA clones facilitates ELVd detection in infected tissues. Figure 3 shows a screening of a series of eggplants by northern blot hybridization. RNA was purified from leaves of different plants and separated by denaturing PAGE (Fig. 3a). Then, RNAs were transferred to a membrane and ELVd RNAs were detected by hybridization with a radioactive probe of − polarity. Strong hybridization signals are detected in eggplants infected with ELVd (Fig. 3b). This analysis also shows that the monomeric circular and linear ELVd RNAs of + polarity are the forms reaching higher concentrations in infected tissues.
Figure 3.
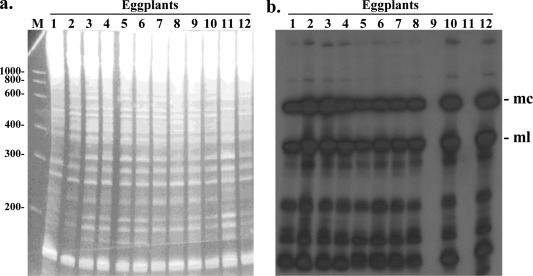
Detection of Eggplant latent viroid (ELVd) RNA by molecular hybridization. Analysis of eggplant RNA preparations by denaturing polyacrylamide gel electrophoresis (PAGE) followed by ethidium bromide staining (a), and northern blot hybridization using an ELVd radioactive probe of − polarity (b). The positions of the monomeric circular (mc) and monomeric linear (ml) ELVd forms are indicated. Additional hybridization bands correspond to oligomeric replication intermediates and degradation products. This analysis shows that samples 1–8, 10 and 12 are infected with ELVd, whereas 9 and 11 remain non‐infected. Lane M, RNA marker with the lengths of the different species indicated on the left in nucleotides.
ELVd Ribozymes
The molecular characterization of ELVd showed that both viroid strands contained hammerhead structures. Fadda et al. (2003) produced ELVd‐2 monomeric transcripts of both polarities, and observed that these transcripts spontaneously self‐cleaved during the transcription reactions and produced fragments whose size matched expectations according to the theoretical cleavage sites of both ribozymes. When they purified the uncleaved fractions of the monomeric transcripts and subjected them to an in vitro self‐cleavage reaction with absolutely no proteins, these transcripts self‐cleaved again, but only in the presence of 5 mm MgCl2. A primer extension analysis of the cleavage products confirmed the theoretical ribozyme self‐cleavage site (Fadda et al., 2003). ELVd‐2 hammerhead ribozymes are schematically represented in Fig. 2. They contain long helices I and stable helices III, similar to the observations in PLMVd and CChMVd, but in contrast with ASBVd ribozymes, which self‐cleave in dimeric molecules through the formation of double hammerhead structures (Davies et al., 1991; Forster et al., 1988). Both ELVd hammerhead structures resemble each other, which is quite common in ribozyme‐containing pathogenic RNAs, and suggests some kind of reverse‐complement duplication of the domain in their evolutionary origin.
The hammerhead ribozyme present in the ELVd strand of + polarity displayed the peculiarity that the cleavage site was preceded by the trinucleotide AUA, which is absent in other natural hammerhead structures. Carbonell et al. (2006) mutagenized this trinucleotide to GUA, GUC and AUC (absent in natural hammerheads), and assayed self‐cleavage in vitro at the low Mg2+ concentrations that exist in vivo. They found that all the mutant hammerhead structures had a higher self‐cleavage rate constant than the wild‐type. Interestingly, the hammerhead ribozyme with the AUC trinucleotide displayed a remarkably high self‐cleavage rate constant. In contrast, the ELVd + wild‐type ribozyme (AUA trinucleotide) showed efficient self‐cleavage during transcription compared with the mutants (GUA, GUC and AUC). These results suggest that natural hammerheads have been evolutionarily selected to function co‐transcriptionally. An efficient hammerhead ribozyme during transcription, with low efficiency during the self‐cleavage reaction after transcription, may reflect a strategy to cleave the replication intermediates co‐transcriptionally whilst protecting the viroid progeny.
ELVd Taxonomic Status
After the ELVd sequence had been determined, the species was assigned to the family Avsunviroidae (Fadda et al., 2003), given the presence of hammerhead ribozymes in both polarity strands, and because the CCR was lacking, which is characteristic of family Pospiviroidae members. The strict host range is also a property that is common in the family (Flores et al., 2000). However, ELVd properties were somewhat intermediate between those of ASBVd (the only species in the genus Avsunviroid) and those of PLMVd and CChMVd (which belong to the genus Pelamoviroid). The ELVd secondary structure was quasi‐rod‐like, similar to ASBVd. PLMVd and CChMVd had multi‐branched conformations, which included a kissing loop (Bussière et al., 2000; Giguère et al., 2014). However, the ELVd hammerhead structures had stable helices III, similar to those of the last two viroids, and unlike the ASBVd hammerhead, which self‐cleaved in dimeric molecules through the formation of double hammerhead structures (Davies et al., 1991). In addition, the ELVd G + C content was high, like those of PLMVd and CChMVd, and unlike ASBVd. As a result of these singular properties, Fadda et al. (2003) proposed the assignment of ELVd to a new genus in the family Avsunviroidae, and offered the name Elaviroid. This genus was later accepted by the International Committee on Taxonomy of Viruses, and ELVd remains its only species to date (Owens et al., 2012). The assignment of ELVd to the family Avsunviroidae also implied that this viroid may replicate and accumulate in the plastids of infected plants, as these are common properties of the other viroids in the family (Flores et al., 2000).
ELVd Replication
A northern blot hybridization analysis of the RNA preparations obtained from ELVd‐infected eggplants, using RNA probes of both polarities, showed that monomeric circular and linear ELVd molecules of both polarities accumulated in infected tissues (Fadda et al., 2003). Accumulation was not symmetrical and the RNAs of one of the polarities were more abundant. This result served to assign the + polarity to this more abundant strand. In any case, the presence of the monomeric circular ELVd RNA of – polarity, the hallmark of the symmetric pathway in the viroid rolling‐circle replication mechanism (Branch and Robertson, 1984), indicated that this viroid, like all other members of the family Avsunviroidae, followed this replication pathway (Fadda et al., 2003). In this pathway, the oligomeric RNAs of both polarities are cleaved to monomers by hammerhead ribozymes during a reaction which, in vivo, can be assisted by host proteins (Daròs and Flores, 2002). ELVd hammerhead ribozymes are very active in vitro (Fadda et al., 2003), and apparently also in vivo, because only very small amounts of dimeric ELVd RNAs have been detected in eggplant infected tissues (Martínez et al., 2009). In ASBVd, viroid RNAs have been proposed to be transcribed by a nuclear‐encoded RNA polymerase (NEP) of chloroplast localization (Navarro et al., 2000). The NEP orthologue in eggplant may transcribe ELVd RNA strands, which would subsequently self‐cleave through the activity of the embedded hammerhead ribozymes to yield monomeric RNAs with 5′‐hydroxyl and 2′,3′‐cyclic phosphodiester termini.
There has been some speculation over the years about the mechanism of circularization during replication in Avsunviroidae (Flores et al., 2005; Navarro et al., 2012). The monomeric linear intermediates with 5′‐hydroxyl and 2′,3′‐cyclic phosphodiester termini that result from hammerhead self‐cleavage can be circularized by a host RNA ligase. However, at that time, no such enzyme had been described in the chloroplast. An alternative proposal was an RNA autocatalytic reaction, which had been shown to occur in PLMVd self‐cleavage monomers in the presence of magnesium, which produces a non‐physiological 2′,5′‐phosphodiester bond (Côté and Perreault, 1997). Another proposal has been that the hammerhead ribozyme would mediate the reverse (ligation) reaction by starting from self‐cleavage monomers. This reaction, which produces the physiological 3′,5′‐phosphodiester bond, has been shown in vitro with some hammerheads, but with poor efficiency (Canny et al., 2007; Nelson et al., 2005). In this context, Molina‐Serrano et al. (2007) developed an experimental system that consisted of transplastomic lines of the green alga Chlamydomonas reinhardtii, which expressed viroid transcripts in the chloroplast under the control of the rbcL promoter and the psaB terminator. A northern blot analysis of the RNA preparations purified from C. reinhardtii cultures showed that, unlike the transcripts of a nuclear viroid (CEVd), the transcripts of the viroids in the family Avsunviroidae generally processed efficiently in C. reinhardtii chloroplasts. However, no RNA‐to‐RNA amplification of viroid RNA was detected. This experimental system was proposed to address intriguing questions about viroid RNA processing and, in particular, about the cellular factors involved in cleavage and ligation (Molina‐Serrano et al., 2007). It is worth noting that, of the different viroids (ASBVd, CChMVd, ELVd and CEVd) tested in this work, ELVd was the viroid whose transcripts processed more efficiently in the C. reinhardtii chloroplast with an excellent yield of circular molecules.
Using this experimental system, Martínez et al. (2009) carried out a mutational analysis of sequence and structural elements in the ELVd molecule involved in transcript processing in vivo in a chloroplastic context. A collection of insertion and deletion mutants suggested that the only domain involved in cleavage was that of the hammerhead ribozyme, but additional sequences were involved in ligation. More specifically, the results obtained with the two deletion mutants that cleaved efficiently, but showed defects in ligation, suggested that a quasi‐double‐stranded structure in the central part of the molecule, which contained the ligation site in an internal loop, would be involved in ligation (Martínez et al., 2009). Incidentally, the control inoculations of all of these mutants in the viroid natural host (eggplant) revealed that this viroid admits certain insertions into the terminal loop of the upper right hairpin with no loss of infectivity (Martínez et al., 2009). This raised the possibility of tagging the viroid during infection for tracking and affinity purification. We are currently investigating this possibility (E. Majer and J.‐A. Daròs, unpublished results). Finally, based on the ELVd circularization efficiency observed in C. reinhardtii chloroplasts, Martínez et al. (2009) concluded that a chloroplastic RNA ligase should be involved in the process.
Englert et al. (2007) demonstrated that Arabidopsis thaliana tRNA ligase, an enzyme involved in nuclear tRNA maturation conserved in all eukaryotes (Abelson et al., 1998), in addition to the nucleus, also localized in the cytoplasm and chloroplast. As this enzyme efficiently recognizes the 5′‐hydroxyl and 2′,3′‐cyclic phosphodiester RNA termini produced by hammerhead ribozymes, it was a good candidate to mediate chloroplastic viroid circularization. To prove this hypothesis, Nohales et al. (2012b) cloned the chloroplastic isoform of eggplant tRNA ligase. An analysis of the protein sequence with the ChloroP algorithm predicted an amino‐terminal transit peptide to the chloroplast for this protein. A recombinant version of the protein was expressed in Escherichia coli and purified. Reactions in vitro with different ELVd RNA transcripts demonstrated that this enzyme very efficiently circularized self‐cleavage monomeric ELVd RNA of + polarity, which contained 5′‐hydroxyl and 2′,3′‐cyclic phosphodiester ends. Interestingly, the enzyme did not circularize five other monomeric ELVd + RNAs that contained the same terminal groups, but opened at positions different from the ribozyme self‐cleavage site. The enzyme has also been shown to efficiently circularize the + and – self‐cleavage monomeric RNAs of the four members of the family Avsunviroidae (ASBVd, PLMVd, CChMVd and ELVd) (Nohales et al., 2012b). Finally, an in vivo assay, in which dimeric ELVd and Coconut cadang‐cadang viroid (Pospiviroidae) transcripts were transitorily expressed in Nicotiana benthamiana plants, whose endogenous tRNA ligase was silenced by virus‐induced gene silencing, supported the involvement of this enzyme in ELVd, but not Coconut cadang‐cadang viroid, circularization (Nohales et al., 2012b).
ELVd Traffic
The initial molecular characterization of ELVd implied that this viroid could replicate and accumulate in plastids, as do other members of the family Avsunviroidae. Indeed, our own observations of eggplant ELVd‐infected tissues by in situ hybridization with digoxigenin‐labelled RNA probes support the chloroplastic accumulation of this viroid (J. Marqués and J.‐A. Daròs, unpublished results). Exactly how members of the family Avsunviroidae reach plastids has been an intriguing question for a long time (Daròs et al., 2006), but Gómez and Pallás (2010b) published the fascinating observation that a green fluorescent protein (GFP), transitorily expressed in N. benthamiana tissues by Agrobacterium tumefaciens infiltration, was translated in the chloroplast when ELVd sequences were fused at the 5′ untranslated region (UTR) of messenger RNA (mRNA). More specifically, these authors inserted a chimeric ELVd sequence that consisted of a fragment of the minus strand of ELVd‐AJ536613 (position 54–267, note that the numbering of the minus strand goes backwards), followed by a fragment of the plus strand of ELVd‐AJ536613 (position 54–261, including two mutations), immediately upstream of the initiation codon of a fluorescent protein mRNA (see GenBank accession number HM136583). This chimeric ELVd sequence did not contain either an alternative AUG in frame with that of GFP or complete viroid ribozymes that may mediate mRNA cleavage. An analysis of the infiltrated tissues by laser scanning confocal microscopy showed GFP fluorescence in the chloroplasts, in contrast with the situation (nucleocytoplasmic GFP fluorescence) in the control constructs in which GFP mRNA with no ELVd insertion was expressed (Gómez and Pallás, 2010b). A western blot analysis of expressed GFP supported the fact that the ELVd chimeric sequence acted as a non‐coding RNA by mediating the transit of the whole mRNA to the chloroplast for GFP translation inside this organelle. Furthermore, ELVd‐containing GFP mRNA was detected by RT‐PCR in the chloroplasts isolated from agroinfiltrated tissues (Gómez and Pallás, 2010b). Figure 4 shows the green and red fluorescence of N. benthamiana chloroplasts in which a GFP mRNA, with a chimeric ELVd in the leader sequence, was transiently expressed.
Figure 4.
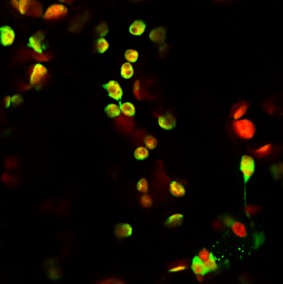
Laser scanning confocal microscopy image showing Nicotiana benthamiana chloroplasts exhibiting green fluorescent protein (GFP) fluorescence after infiltration with a construct that contains an Eggplant latent viroid (ELVd) chimeric sequence at the 5′ untranslated region (UTR), which facilitates GFP mRNA uptake by chloroplasts for translation. Image provided by G. G. Gómez [Instituto de Biología Molecular y Celular de Plantas (IBMCP), Valencia, Spain].
In a subsequent study, Gómez and Pallás (2010a) dissected the chimeric ELVd sequence capable of mediating GFP mRNA import into the chloroplast. They found that the 110‐nucleotide‐long central fragment of their chimeric sequence was sufficient. Interestingly, although the left 108‐nucleotide and right 112‐nucleotide fragments of the chimeric ELVd sequence were unable to mediate mRNA import into the chloroplast, each increased the efficiency of the process when fused to the central fragment. In another study, the same authors repeated the observation of GFP mRNA translation in chloroplasts by inserting a full‐length ELVd of + polarity (position 1–333 of AJ536613) into the 5′ UTR (Gómez and Pallás, 2012b). On the basis of all of these results, these authors proposed the existence of a novel plant signalling mechanism capable of regulating the selective import of nuclear transcripts into chloroplasts. As an alternative to the pathway mediated by chloroplastic transit peptides (Shi and Theg, 2013), the mRNAs of some proteins may contain the RNA elements recognized by the same machinery which mediates ELVd translocation into chloroplasts. Incidentally, members of Avsunviroidae may have evolved to take advantage of this pathway in order to achieve import into the chloroplast to start replication (Gómez and Pallás, 2010a, 2010b).
Gómez and Pallás (2012b) also applied an ingenious experimental system based on Potato virus X (PVX), which has been used previously to demonstrate the nuclear targeting of PSTVd (Abraitiene et al., 2008; Zhao et al., 2001), to analyse ELVd intracellular trafficking. This system consisted of a recombinant version of PVX, which expresses GFP under the control of a coat protein (CP) subgenomic promoter (viral CP was expressed downstream of the genome through a duplicated version of this same promoter). The GFP gene contains the intron IV2 derived from the potato (Solanum tuberosum L.) gene ST‐LS1, and fluorescence can only be detected if intron‐containing GFP mRNA traffics into the nucleus for appropriate splicing. Similar to the previous work conducted with PSTVd (Abraitiene et al., 2008; Zhao et al., 2001), Gómez and Pallás (2012b) inserted a full‐length monomeric ELVd‐AJ536613 (from position 1 to 333) in + polarity orientation into the IV2 intron and infected N. benthamiana plants with the transcripts of the resulting recombinant PVX clone. A few days after inoculation, GFP fluorescence was detected in systemically infected leaves. This indicated that ELVd RNA directs the cytoplasmic GFP transcript to the nucleus, where the intron is efficiently removed. Moreover, dissection of the ELVd sequence in three regions (left, from position 15 to 181; upper, from position 89 to 294; and right, from position 187 to 333) indicated that the left region of the ELVd molecule sufficed to mediate GFP mRNA trafficking into the nucleus (Gómez and Pallás, 2012b). This result added unexpected complexity to ELVd intracellular trafficking, which could be general for all Avsunviroidae. On entry to the host cell, ELVd would move from the cytoplasm to the nucleus to be subsequently delivered from the nucleus to chloroplasts, where this pathogenic RNA can efficiently replicate and accumulate (Gómez and Pallás, 2012a, 2012b).
Finally, at the tissue level, initial analyses have shown that ELVd is uniformly distributed in the leaves, stems and fruits (skin and pulp) of infected plants (Fadda et al., 2003).
ELVd‐Related Biotechnological Aspects
First, plastids are organelles that have attracted a great deal of attention in plant biotechnology. Given their prokaryotic evolutionary origin, rules to express recombinant proteins are rather particular and large accumulations have been achieved. Many important reactions of the plant primary and secondary metabolism also occur in these organelles. As plastids are scarce in pollen, plastid transgenes can be more contained. The unique ability of ELVd to mediate RNA import into the chloroplast (Gómez and Pallás, 2010b) may result in a use for this viroid as a valuable tool to transport RNAs into this organelle for translation into proteins of interest or for regulatory purposes. Second, two mutant versions of ELVd hammerhead ribozymes of + polarity, those that contain AUC and GUC trinucleotides instead of AUA preceding the self‐cleavage site, display a remarkably high self‐cleavage rate constant (Carbonell et al., 2006; Carbonell et al., 2011), and could be a good starting point to build efficient trans‐cleaving devices at the low Mg2+ concentrations that exist in vivo. Third, and finally, our recent research has shown that the co‐expression in E. coli of ELVd transcripts and the chloroplastic isoform of eggplant tRNA ligase leads to the accumulation of large amounts of the ELVd circular RNA. The insertion of an RNA of interest into certain particular positions of the ELVd molecule does not affect accumulation, which makes this approach an excellent system to over‐produce recombinant RNAs in E. coli cultures using ELVd as a scaffold (Daròs et al., 2014).
Acknowledgements
I thank Gustavo G. Gómez [Instituto de Biología Molecular y Celular de Plantas (IBMCP), Valencia, Spain] for Fig. 4 which illustrates ELVd trafficking into chloroplasts. This work was supported by grants AGL2013‐49919‐EXP and BIO2014‐54269‐R from the Spanish Ministerio de Economía y Competitividad (MINECO). The author declares no conflict of interest.
References
- Abelson, J. , Trotta, C.R. and Li, H. (1998) tRNA splicing. J. Biol. Chem. 273, 12685–1288. [DOI] [PubMed] [Google Scholar]
- Abraitiene, A. , Zhao, Y. and Hammond, R. (2008) Nuclear targeting by fragmentation of the potato spindle tuber viroid genome. Biochem. Biophys. Res. Commun. 368, 470–475. [DOI] [PubMed] [Google Scholar]
- Branch, A.D. and Robertson, H.D. (1984) A replication cycle for viroids and other small infectious RNAs. Science, 223, 450–455. [DOI] [PubMed] [Google Scholar]
- Bussière, F. , Ouellet, J. , Côté, F. , Lévesque, D. and Perreault, J.P. (2000) Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74, 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny, M.D. , Jucker, F.M. and Pardi, A. (2007) Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry, 46, 3826–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A. , De la Peña, M. , Flores, R. and Gago, S. (2006) Effects of the trinucleotide preceding the self‐cleavage site on eggplant latent viroid hammerheads: differences in co‐ and post‐transcriptional self‐cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res. 34, 5613–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A ., Flores, R. and Gago, S. (2011) Trans‐cleaving hammerhead ribozymes with tertirary stabilizing motifs: in vitro and in vivo activity against a structured viroid RNA. Nucleic Acids Res. 39, 2432–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall, A. and Pirofski, L.A. (2014) Ditch the term pathogen. Nature, 516, 165–166. [DOI] [PubMed] [Google Scholar]
- Côté, F. and Perreault, J.P. (1997) Peach latent mosaic viroid is locked by a 2′,5′‐phosphodiester bond produced by in vitro self‐ligation. J. Mol. Biol. 273, 533–543. [DOI] [PubMed] [Google Scholar]
- Daròs, J.A. and Flores, R. (2002) A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead‐mediated self‐cleavage. EMBO J. 21, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs, J.A. , Elena, S.F. and Flores, R. (2006) Viroids: an Ariadne's thread into the RNA labyrinth. EMBO Rep. 7, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs, J.A. , Aragonés, V. and Cordero, M.T. (2014) Recombinant RNA production. Patent EP14382177.5, PCT/EP2015/060912.
- Davies, C. , Sheldon, C.C. and Symons, R.H. (1991) Alternative hammerhead structures in the self‐cleavage of avocado sunblotch viroid RNAs. Nucleic Acids Res. 19, 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, T.O. (1971) Potato spindle tuber “virus” IV. A replicating, low molecular weight RNA. Virology, 45, 411–428. [DOI] [PubMed] [Google Scholar]
- Diener, T.O. (2003) Discovering viroids – a personal perspective. Nat. Rev. Microbiol. 1, 75–80. [DOI] [PubMed] [Google Scholar]
- Ding, B. (2009) The biology of viroid–host interactions. Annu. Rev. Phytopathol. 47, 105–131. [DOI] [PubMed] [Google Scholar]
- Englert, M. , Latz, A. , Becker, D. , Gimple, O. , Beier, H. and Akama, K. (2007) Plant pre‐tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie, 89, 1351–1365. [DOI] [PubMed] [Google Scholar]
- Fadda, Z. , Daròs, J.A. , Fagoaga, C. , Flores, R. and Duran‐Vila, N. (2003) Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J. Virol. 77, 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoaga, C. and Duran‐Vila, N. (2003) Eggplant latent In Viroids (Hadidi A., Flores R., Randless J.W. and Semancik J.S., eds), p. 333 Collingwood, Vic: CSIRO Publishing. [Google Scholar]
- Fagoaga, C. , Pina, J.A. and Duran‐Vila, N. (1994) Occurrence of small RNAs in severely diseased vegetable crops. Plant Dis. 78, 749–753. [Google Scholar]
- Flores, R. , Daròs, J.A. and Hernandez, C. (2000) Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 55, 271–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R. , Hernández, C. , Martínez de Alba, A.E. , Daròs, J.A. and Di Serio, F. (2005) Viroids and viroid–host interactions. Annu. Rev. Phytopathol. 43, 117–139. [DOI] [PubMed] [Google Scholar]
- Flores, R. , Delgado, S. , Rodio, M.E. , Ambrós, S. , Hernández, C. and Serio, F.D. (2006) Peach latent mosaic viroid: not so latent. Mol. Plant Pathol. 7, 209–221. [DOI] [PubMed] [Google Scholar]
- Forster, A.C. , Davies, C. , Sheldon, C.C. , Jeffries, A.C. and Symons, R.H. (1988) Self‐cleaving viroid and newt RNAs may only be active as dimers. Nature, 334, 265–267. [DOI] [PubMed] [Google Scholar]
- Gas, M.E. , Hernández, C. , Flores, R. and Daròs, J.A. (2007) Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 3, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère, T. , Adkar‐Purushothama, C.R. , Bolduc, F. and Perreault, J.P. (2014) Elucidation of the structures of all members of the Avsunviroidae family. Mol. Plant Pathol. 15, 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2010a) Can the import of mRNA into chloroplasts be mediated by a secondary structure of a small non‐coding RNA? Plant Signal. Behav. 5, 1517–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2010b) Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One, 5, e12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2012a) A pathogenic non coding RNA that replicates and accumulates in chloroplasts traffics to this organelle through a nuclear‐dependent step. Plant Signal. Behav. 7, 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2012b) Studies on subcellular compartmentalization of plant pathogenic noncoding RNAs give new insights into the intracellular RNA‐traffic mechanisms. Plant Physiol. 159, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, H.J. , Domdey, H. , Lossow, C. , Jank, P. , Raba, M. , Alberty, H. and Sänger, H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Hernández, C. and Flores, R. (1992) Plus and minus RNAs of peach latent mosaic viroid self‐cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA, 89, 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, C.J. , Keese, P. , Visvader, J.E. , Rathjen, P.D. , McInnes, J.L. and Symons, R.H. (1985) Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 4, 293–304. [DOI] [PubMed] [Google Scholar]
- Hutchins, C.J. , Rathjen, P.D. , Forster, A.C. and Symons, R.H. (1986) Self‐cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, F. , Marqués, J. , Salvador, M.L. and Daròs, J.A. (2009) Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J. Gen. Virol. 90, 3057–3065. [DOI] [PubMed] [Google Scholar]
- Molina‐Serrano, D. , Suay, L. , Salvador, M.L. , Flores, R. and Daròs, J.A. (2007) Processing of RNAs of the family Avsunviroidae in Chlamydomonas reinhardtii chloroplasts. J. Virol. 81, 4363–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbach, H.P. and Sänger, H.L. (1979) Viroid replication is inhibited by α‐amanitin. Nature, 278, 185–188. [DOI] [PubMed] [Google Scholar]
- Navarro, B. and Flores, R. (1997) Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self‐cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA, 94, 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, B. , Daròs, J.A. and Flores, R. (1996) A general strategy for cloning viroids and other small circular RNAs that uses minimal amounts of template and does not require prior knowledge of its sequence. J. Virol. Methods, 56, 59–66. [DOI] [PubMed] [Google Scholar]
- Navarro, B. , Gisel, A. , Rodio, M.E. , Delgado, S. , Flores, R. and Di Serio, F. (2012) Viroids: how to infect a host and cause disease without encoding proteins. Biochimie, 94, 1474–1480. [DOI] [PubMed] [Google Scholar]
- Navarro, J.A. , Vera, A. and Flores, R. (2000) A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology, 268, 218–225. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A. , Shepotinovskaya, I. and Uhlenbeck, O.C. (2005) Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry, 44, 14577–14585. [DOI] [PubMed] [Google Scholar]
- Nohales, M.A. , Flores, R. and Daròs, J.A. (2012a) Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA, 109, 13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales, M.A. , Molina‐Serrano, D. , Flores, R. and Daròs, J.A. (2012b) Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae . J. Virol. 86, 8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, R.A. (2007) Potato spindle tuber viroid: the simplicity paradox resolved? Mol. Plant Pathol. 8, 549–560. [DOI] [PubMed] [Google Scholar]
- Owens, R.A. , Flores, R. , Di Serio, F. , Li, S.F. , Pallás, V. , Randles, J.W. , Sano, T. and Vidalakis, G. (2012) Viroids In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J. eds), pp. 1221–1234. London: Elsevier, Academic Press. [Google Scholar]
- Palukaitis, P. (2014) What has been happening with viroids? Virus Genes, 49, 175–184. [DOI] [PubMed] [Google Scholar]
- Pirofski, L.A. and Casadevall, A. (2012) What is a pathogen? A question that begs the point. BMC Biol. 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L.X. and Theg, S.M. (2013) The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta, 1833, 314–331. [DOI] [PubMed] [Google Scholar]
- Spiesmacher, E. , Mühlbach, H.P. , Schnölzer, M. , Haas, B. and Sänger, H.L. (1983) Oligomeric forms of potato spindle tuber viroid (PSTV) and of its complementary RNA are present in nuclei isolated from viroid‐infected potato cells. Biosci. Rep. 3, 767–774. [DOI] [PubMed] [Google Scholar]
- Symons, R.H. (1981) Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res. 9, 6527–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Owens, R.A. and Hammond, R.W. (2001) Use of a vector based on Potato virus X in a whole plant assay to demonstrate nuclear targeting of Potato spindle tuber viroid . J. Gen. Virol. 82, 1491–1497. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]


