Summary
To ensure the optimal infectivity on contact with host cells, pathogenic Pseudomonas syringae has evolved a complex mechanism to control the expression and construction of the functional type III secretion system (T3SS) that serves as a dominant pathogenicity factor. In this study, we showed that the hrpF gene of P. syringae pv. averrhoi, which is located upstream of hrpG, encodes a T3SS‐dependent secreted/translocated protein. Mutation of hrpF leads to the loss of bacterial ability on elicitation of disease symptoms in the host and a hypersensitive response in non‐host plants, and the secretion or translocation of the tested T3SS substrates into the bacterial milieu or plant cells. Moreover, overexpression of hrpF in the wild‐type results in delayed HR and reduced t3ss expression. The results of protein–protein interactions demonstrate that HrpF interacts directly with HrpG and HrpA in vitro and in vivo, and protein stability assays reveal that HrpF assists HrpA stability in the bacterial cytoplasm, which is reduced by a single amino acid substitution at the 67th lysine residue of HrpF with alanine. Taken together, the data presented here suggest that HrpF has two roles in the assembly of a functional T3SS: one by acting as a negative regulator, possibly involved in the HrpSVG regulation circuit via binding to HrpG, and the other by stabilizing HrpA in the bacterial cytoplasm via HrpF–HrpA interaction prior to the secretion and formation of Hrp pilus on the bacterial surface.
Keywords: hrpF, Pseudomonas syringae, T3SS, t3ss regulation
Introduction
Pseudomonas syringae, a plant‐pathogenic bacterium that elicits necrotic spots and a hypersensitive response (HR) on the foliage of host and non‐host plants, respectively, can be classified into numerous pathovars based on their specific interactions with different plant species (Hirano and Upper, 2000). The ability of P. syringae to infect its host plants depends on a cluster of genes, the hrp (hypersensitive response and pathogenicity)/hrc (hypersensitive response and conserved) genes, residing in a pathogenicity island (PAI) known as the Hrp PAI (Alfano et al., 2000). The hrp/hrc genes coding for the type III secretion system (T3SS) are conserved in many Gram‐negative plant‐ and animal‐pathogenic bacteria (Galan and Collmer, 1999). In plant pathogens, the T3SS is a syringe‐like structure and a protruding Hrp pilus that functions as a conduit to deliver virulence proteins, also known as type III effectors (T3Es), into host cells (Wei et al., 2000). Under suitable conditions, the transcription of P. syringae t3ss is induced by multiple proteins, including HrpR, HrpS and HrpL, and the mode of regulation is regarded as group 1 Hrp T3SS (Alfano and Collmer, 1997), in which HrpL acts as an alternative sigma factor that recognizes the consensus ‘hrp box’ in the promoter regions of hrp operons (Fouts et al., 2002; Xiao et al., 1994). The transcription of hrpL is induced by RpoN (σ54)‐RNA polymerase holoenzyme (Hendrickson et al., 2000) and the heterodimeric enhancer‐binding proteins HrpR and HrpS (Hutcheson et al., 2001; Jovanovic et al., 2011). A mutation in the Hrp pilin‐encoding hrpA gene of P. syringae pv. tomato (Pto) DC3000 has been reported to severely reduce the expression of hrpRS, hrpL and t3ss genes through an unknown mechanism (Wei et al., 2000), suggesting that HrpA may have a positive regulatory effect on t3ss expression. In addition to the positive regulatory network, negative regulations involving Lon‐HrpR (Bretz et al., 2002) and HrpG–HrpV–HrpS (Wei et al., 2005) have been reported to fine tune t3ss expression under different environmental conditions. The negative regulation is analogous to that found in the opportunistic animal pathogen Pseudomonas aeruginosa, whose t3ss is regulated by ExsADCE proteins (Dasgupta et al., 2004; Frank and Iglewski, 1991; McCaw et al., 2002; Thibault et al., 2009). The involvement of HrpS, HrpV and HrpG in the regulation of t3ss expression is characteristically equivalent to ExsA (a transcriptional activator), ExsD (an anti‐activator) and ExsC (an anti‐anti‐activator), respectively (Buttner, 2012; Dasgupta et al., 2004; Frank and Iglewski, 1991; McCaw et al., 2002). However, to date, no P. syringae Hrp protein equivalent to P. aeruginosa ExsE has been identified.
The most prominent T3SS structure extending from bacterial surfaces is the needle filament of animal pathogens or the pilus of plant pathogens, which is polymerized by one small protein (e.g. PrgI in Salmonella sp., MxiH in Shigella sp., PscF in P. aeruginosa, YscF in Yersinia pestis and HrpA in P. syringae) (Blocker et al., 2001; Hoiczyk and Blobel, 2001; Kimbrough and Miller, 2000; Pastor et al., 2005; Roine et al., 1997b). The T3SS needle‐assembling protein of animal pathogens employs molecular chaperones, e.g. EscE‐EscG of enteropathogenic Escherichia coli (Sal‐Man et al., 2013) and PscE‐PscG cochaperones of P. aeruginosa (Quinaud et al., 2005), as a general strategy to maintain the monomeric proteins in the cytoplasm (Quinaud et al., 2007). The chaperone function for the prevention of polymerization of pilus protein prior to secretion might also exist in plant‐pathogenic P. syringae or Xanthomonas vesicatoria; however, the identities of the pilus chaperones remain unknown.
A small secreted HrpF protein, encoded by the first gene adjacent to hrpG in the hrpC operon, has been demonstrated previously to be a pathogenicity factor in P. syringae pv. syringae 61 (Psy61) (Deng et al., 1998; Ramos et al., 2007), but its biochemical role in T3SS remains unclear. In this study, the coding sequence of hrpF was deleted in P. syringae pv. averrhoi strain HL1 (PavHL1) (a pathogen of starfruit; Wei et al., 2012) to produce the unmarked mutant strain HL1‐N1589. This mutant strain lost the ability to elicit HR in non‐host plants and to multiply in host plants, as described for phenotypes of the Psy61 hrpF mutant (Deng et al., 1998). We further analysed HrpF function in t3ss expression and its involvement in T3E secretion by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) and assays of protein secretion and protein–protein interactions, and found that HrpF negatively regulates t3ss expression via binding to HrpG and is essential for T3SS function by preventing HrpA pilin from degradation in the bacterial cytoplasm. In addition, the biochemical activities of HrpF on HrpA were severely compromised by the substitution of the 67th lysine residue with alanine (Ala).
Results
hrpF mutant of PavHL1 cannot elicit the HR on tobacco or cause disease on host plants
PavHL1, the causal agent of bacterial spot disease of carambola in Taiwan (Wei et al., 2012), contains a functional T3SS encoded by the hrp/hrc gene cluster (GenBank accession number KJ534576), whose gene organization and nucleotide sequences are similar to those of the gene clusters reported in other P. syringae pathovars (Alfano et al., 2000; Buell et al., 2003; Feil et al., 2005; Joardar et al., 2005). Comparison of the amino acid sequence revealed that HrpFPav is 80%–100% identical to HrpF of several P. syringae pathovars, but shares only 38% identity with HrpF of Pto (Fig. S1A, see Supporting Information). In this study, two homologous alleles of hrpF, one from PavHL1 (hrpF Pav) and one from PtoDC3000 (hrpF Pto), were chosen for biological analyses. The virulence functions of HrpF in PavHL1 and PtoDC3000 were first characterized by creating unmarked deletions of hrpF, followed by the infiltration of the mutant strains into plants for bioassays. As shown in Figs 1A–D and S1B,C, both mutants failed to elicit disease symptoms in host plants and the HR in non‐host tobacco plants, and their multiplication in host plants was severely reduced in comparison with the wild‐type (WT) strains. The starfruit leaves sprayed with PavHL1 and hrpF‐complementing strain showed necrotic spots with surrounding chlorotic haloes (Fig. 1A) at 7 dpi (days post‐inoculation). Interestingly, although hrpF orthologues from the pathovars averrhoi and tomato display low identity, heterologous expression of HrpFPto in the Pav hrpF mutant HL1‐N1589 partially restored the ability of the hrpF Pav mutant to multiply in starfruit leaves (Fig. 1B) and to trigger ion leakage at 7 hpi (hours post‐inoculation) (Fig. 1D) and HR at 24 hpi in tobacco leaves (Fig. 1C). Pav HrpF could also partially complement the HR phenotype of the Pto hrpF mutant (see Fig. S1C), suggesting that the functions of HrpF are conserved among different P. syringae pathovars. We tested whether the hrpF mutant HL1‐N1589 is defective in T3SS function by growing bacteria in hrp‐inducing medium HrpMM (Hrp minimal medium; Huynh et al., 1989) and then assaying T3E secretion using an immunoblotting method. Compared with the WT PavHL1, HopAK1, HrpZ and HrpA were not detectable in the supernatant fractions of HL1‐N1589 or the T3SS‐defective hrcC mutant, indicating that the mutation of hrpF Pav abolished the T3SS function to secrete proteins (Fig. 1E). In addition, the amount of HrpA in the cell pellet fraction of the hrpF mutant was consistently lower than that in the WT and hrcC mutant.
Figure 1.
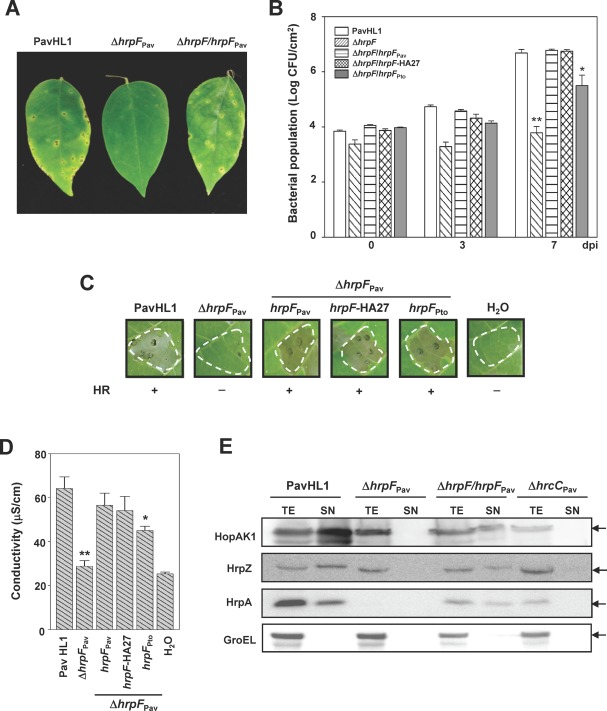
Phenotypes of the hrpF mutant strains derived from Pseudomonas syringae pv. averrhoi HL1 (PavHL1). (A) Necrotic spot symptoms on host plants caused by PavHL1, hrpF mutant (ΔhrpF Pav) and the complemented strain (ΔhrpF/hrpF Pav). (B) Bacterial growth in starfruit leaves. Each bar represents the mean and standard deviation of triplicate samples. CFU, colony‐forming units; dpi, days post‐inoculation. Hypersensitive response (HR) elicitation (C) and conductivity assays (D) demonstrate incompatible reactions elicited by the test strains. +, HR positive; –, HR negative. The mean values of bacterial populations and conductivity between wild‐type and test strains were significantly different according to Student's t‐test (*P < 0.05; **P < 0.01). (E) Immunoblot analysis of type III effector (T3E) secretion in the test strains using antisera against HopAK1, HrpZ, HrpA and GroEL. Bacterial cultures in Hrp minimal medium (HrpMM) at 6 h post‐inoculation (hpi) were separated into culture supernatant (SN) and total cell extract (TE) according to Experimental procedures. GroEL was detected as a reference for cellular integrity. PavHL1, wild‐type harbouring pBBR1MCS‐5; ΔhrpF Pav, HL1‐N1589; ΔhrpF/hrpF Pav, HL1‐N1589 harbouring pNCHU1591; ΔhrpF/hrpF‐HA27, HL1‐1589 harbouring pNCHU1810; ΔhrpF/hrpF Pto, HL1‐N1589 harbouring pNCHU1809; ΔhrcC Pav, PavHL1‐N1033.
The calmodulin‐dependent adenylate cyclase (Cya) reporter system (Schechter et al., 2004) was used to analyse the translocation of P. syringae T3Es into plant cells. The results shown in Table 1 confirm that the mutation in hrpF abolishes the translocation of T3Es [e.g. AvrPto1‐cCya in Psy61 (Ramos et al., 2007) and HopI1‐cCya in Pav (this study)], and HrpF‐cCya is translocated into plant cells in a T3SS‐dependent manner. Taken together, it can be concluded that Pav HrpF is involved in T3SS function by assisting the secretion and translocation of T3Es, and HrpF itself can also be translocated into plant cells.
Table 1.
Translocation of HrpF‐cCya or HopI1‐cCya into Nicotiana tabacum cells by Pseudomonas syringae pv. averrhoi.
| P. syringae pv. averrhoi | Genotype | Assay target* | Translocation † (pmol cAMP/μg protein) |
|---|---|---|---|
| HL1 | Wild‐type | HrpF‐cCya (pNCHU1947) | 32.22 ± 14.19 |
| HopI1‐cCya (pNCHU1881) | 355.18 ± 155.7 | ||
| HL1‐N1033 | ΔhrcC::nptII | HrpF‐cCya (pNCHU1947) | 1.37 ± 0.23 |
| HopI1‐cCya (pNCHU1881) | 1.84 ± 1.9 | ||
| HL1‐N1589 | ΔhrpF | HrpF‐cCya (pNCHU1947) | 2.47 ± 1.16 |
| HopI1‐cCya (pNCHU1881) | 2.33 ± 0.95 | ||
| HL1‐N1589/pNCHU1591 | ΔhrpF/hrpF | HopI1‐cCya (pNCHU1938) | 29.07 ± 14.31 |
| HL1‐N1589/pNCHU1808 | ΔhrpF/hrpF K67A | HopI1‐cCya (pNCHU1938) | 2.16 ± 0.13 |
| HL1/pBBR1MCS‐5 | Wild‐type/EV ‡ | HopI1‐cCya (pNCHU1938) | 354.07 ± 81.55 |
| HL1/pNCHU1591 | Wild‐type/hrpF | HopI1‐cCya (pNCHU1938) | 22.46 ± 16.76 |
| HL1/pNCHU1593 | Wild‐type/hrpG | HopI1‐cCya (pNCHU1938) | 332.92 ± 55.01 |
| HL1/pNCHU1866 | Wild‐type/hrpFG | HopI1‐cCya (pNCHU1938) | 101.40 ± 12.79 |
*Bacterial strains carried pNCHU1947, pNCHU1881 or pNCHU1938 (Table S1) which expresses HrpF‐cCya or HopI1‐cCya from a lac promoter of the plasmid.
†Levels of cyclic adenosine monophosphate (cAMP) in the leaf samples of Nicotiana tabacum were determined at 6 h post‐infiltration with bacterial inoculum at an optical density at 600 nm (OD600) of 0.3, and the values represent the mean and standard deviation (n = 3) in each treatment.
‡EV, empty vector.
hrpF mutant HL1‐N1589 expressing hemagglutinin (HA)‐tagged HrpFPav (HrpF‐HA27) restores WT‐like phenotypes
In order to analyse the functions of HrpF, the plasmid pNCHU1810 was constructed to express HA‐tagged HrpF from the lacZ promoter. The HA tag was inserted between the 27th glutamic acid (Glu) and 28th glycine (Gly) residues (E27‐GYPYDVPDYAG‐G28) to produce a HrpF‐HA27 fusion protein, because the tag‐fused HrpF proteins at the C‐terminus (e.g. HrpF‐cHA, HrpF‐cFLAG or HrpF‐cCya) failed to complement hrpF mutant phenotypes (data not shown). According to the predicted protein secondary structure at the Phyre Server (Kelley and Sternberg, 2009), the E27 and G28 residues reside in a loop region connecting two α‐helices; therefore, the structural integrity and function of HrpF are probably maintained on insertion of the HA epitope. Inoculation assays were performed to test whether HrpF‐HA27 was functional, and the results showed that HrpF‐HA27 fully complemented the mutant phenotypes of HL1‐N1589 in bacterial multiplication in starfruit leaves (Fig. 1B) and triggered the HR and ion leakage in tobacco leaves (Fig. 1C,D). In addition, HrpF‐HA27 was secreted to the culture supernatant via T3SS (Fig. 5D) by culturing the HrpF‐HA27‐expressing strain HL1‐N1589 (pNCHU1810) in HrpMM. Taken together, HrpF‐HA27 is functionally equivalent to the native HrpF protein.
Figure 5.
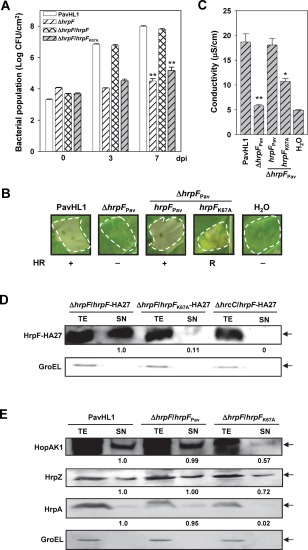
The effects of in‐trans expression of HrpFK67A on plant responses and type III effector (T3E) secretion. (A) Bacterial growth in starfruit leaves. (B) Elicitation of the hypersensitive response (HR) on tobacco leaves: +, HR positive; −, HR negative; R, reduced HR. (C) Conductivity on tobacco leaves. PavHL1, Pseudomonas syringae pv. averrhoi HL1 (pBBR1MCS‐5); ΔhrpF Pav, HL1‐N1589 (pBBR1MCS‐5); ΔhrpF/hrpF Pav, HL1‐N1589 (pNCHU1591); ΔhrpF/hrpF K67A, HL1‐N1589 (pNCHU1808). The mean values of the bacterial population (A) and conductivity (C) between the wild‐type and individual strain were significantly different according to Student's t‐test (*P < 0.05; **P < 0.01). (D) Differential secretion of HrpF‐HA27 and HrpFK67A‐HA27 in the Pav hrpF mutant was detected by immunoblotting against anti‐haemagglutinin (anti‐HA) antibody and expressed as the fold change in the banding intensity using HrpF‐HA27‐expressing HL1‐1589 as the denominator. (E) The secretion of HopAK1, HrpZ and HrpA in PavHL1 and HL1‐1589 harbouring hrpF or hrpF K67A was detected by immunoblotting. Differential secretion of T3Es was indicated by the fold change using PavHL1 as the denominator. In (D) and (E), GroEL was detected as a marker of cellular integrity. SN, culture supernatant; TE, total cellular extract.
Overexpression of HrpFPav in PavHL1 reduces the level of HR elicitation, and co‐expression of HrpFG counteracts the negative effect
Because the delayed HR phenotype was consistently observed when the hrpF complementing strain HL1‐N1589 (pNCHU1591) was inoculated into tobacco leaves at 106 colony‐forming units (CFU)/mL (data not shown), the negative effect of HrpF on T3SS function was tested by HR assays using serially titrated suspensions of the WT PavHL1 overexpressing hrpF as inocula. On infiltration of bacteria at a concentration of 2 × 106 CFU/mL into tobacco leaves, HL1 expressing lac promoter‐driven HrpF or HrpF‐HA27, in comparison with HL1 and HL1 expressing hrpG, displayed lower levels of ion leakage and delayed HR at 24 hpi. This phenomenon could be counteracted by the co‐expression of hrpFG (Fig. 2A), whose expression was driven by the same lac promoter. There were no discernible differences between the above tested strains in the HR when the inoculum was higher than 2 × 106 CFU/mL (Fig. 2A, left panel). The negative effect on HR elicitation was further confirmed by the overexpression of hrpF‐HA27 driven by a stronger tac promoter. The inoculation results showed that HL1 expressing the tac‐driven HrpF‐HA27 (pNCHU2164; Table S1, see Supporting Information) had a stronger negative effect than the lac‐driven expression of HrpF‐HA27 (pNCHU1810; Table S1) on HR elicitation by suppressing HR at 6 × 106 CFU/mL (Fig. S2 see Supporting Information), suggesting that the cumulative amount of HrpF in HL1 cells is negatively correlated with the ability to elicit HR, and the negative effect of HrpF on HR elicitation can be counteracted by the co‐expression of HrpG.
Figure 2.
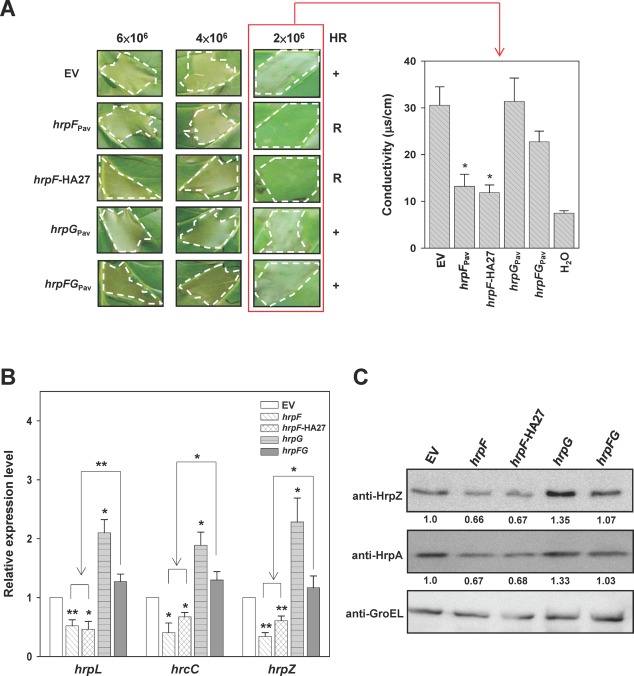
Overexpression of HrpF showed negative effects on the hypersensitive response and Hrp expression. (A) Tobacco leaves were infiltrated at the indicated concentrations with Pseudomonas syringae pv. averrhoi HL1 (PavHL1) harbouring pBBR1MCS‐5 (EV), pNCHU1591 (hrpF Pav), pNCHU1810 (hrpF‐HA27), pNCHU1593 (hrpG Pav) or pNCHU1866 (hrpFG Pav), and photographed at 24 h post‐inoculation (hpi) for the hypersensitive response (HR, left panel). The ionic conductivity was assayed at 24 hpi (right panel) with the indicated inocula for quantitative analysis of cell death. +, wild‐type HR; R, reduced HR. The conductivity values were significantly different between PavHL1 (EV) and the strains overexpressing hrpF Pav or hrpF‐HA27 according to Student's t‐test (*P < 0.05). (B) Relative transcription of hrpL, hrcC and hrpZ in PavHL1 overexpressing hrpF Pav, hrpF‐HA27, hrpG Pav and hrpFG Pav at 6 hpi. The 16S rRNA was quantified and used for normalization by the comparative CT (cycle threshold) method, and the relative expression level was expressed as the fold change using the quantitative polymerase chain reaction (qPCR) result of PavHL1 carrying the empty vector as the denominator and those of PavHL1 expressing the target gene as the numerators. The error bars denote standard deviation (n = 3). Statistical analysis by Student's t‐test (*P < 0.05; **P < 0.01) was applied to compare the fold change of transcription between PavHL1 carrying the empty vector (EV) and various hrp‐expressing plasmids. (C) The production of HrpZ and HrpA in PavHL1 overexpressing hrpF Pav, hrpF‐HA27, hrpG Pav and hrpFG Pav was determined by immunoblot analysis at 6 hpi. GroEL was detected as a protein loading control, and the relative translations of HrpZ and HrpA in PavHL1‐derived strains are shown as fold changes in the banding intensity using the wild‐type as the denominator.
HrpF interacts with HrpG and HrpA
The HrpF–HrpG and HrpF–HrpA interactions were examined on the basis of two findings: (i) the co‐expression of HrpF and HrpG in the WT PavHL1 relieved the phenotypes, i.e. the suppression of HR elicitation by overexpression of HrpF only (Fig. 2A); and (ii) the cumulative HrpA protein in the whole cell fraction of the hrpF mutant was very low (Fig. 1E). A BacterioMatch II Two‐Hybrid System (B2H) (Stratagene, Santa Clara, CA, USA) was used to determine protein interactions in vivo. The Escherichia coli transformants harbouring pTRG‐HrpG and pBT bait vector showed about 95% interaction activity (data not shown), suggesting that the chimeric HrpG‐RNA polymerase alone can activate the expression of the reporter cassette. Therefore, pTRG‐HrpG was not included in this assay. As shown in Fig. 3A, compared with the positive control strain harbouring pBT‐LGF2 and pTRG‐Gal11 as 100% interaction activity, the transformants co‐expressing pTRG‐HrpF with pBT‐HrpG or pBT‐HrpA and the pairs of pTRG‐HrpA with pBT‐HrpF or pBT‐HrpA exhibited 74.4 ± 2.1%, 75.6 ± 7.0%, 83.3 ± 6.4% and 70.6 ± 4.5% interaction activities, respectively. Because HrpA is the major component of Hrp pilus (Li et al., 2002; Roine et al., 1997b), the interaction of HrpA fusion proteins expressed from pTRG‐HrpA and pBT‐HrpA was also tested to show that HrpA indeed binds to itself in B2H. The interaction activity (c. 30%) of co‐expression of pTRG‐HrpF and pBT‐HrpF was not significantly greater than that of co‐expression of pTRG‐HrpF and pBT, indicating that HrpF does not interact with itself in B2H. The results reveal that HrpF interacts with HrpG and HrpA, but does not self‐associate, in vivo.
Figure 3.
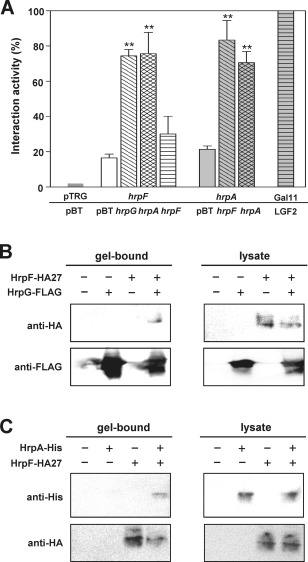
HrpF interacts with HrpG and HrpA in bacterial two‐hybrid and immunoprecipitation assays. (A) In vivo protein interactions were tested by bacterial two‐hybrid assays. Statistical differences were assessed by Student's t‐test (**P < 0.01) on the interaction between pBT vector and each pBT‐expressing hrp gene when pTRG‐hrpF or pTRG‐hrpA was used as the bait. (B, C) Immunoblots of immunoprecipitated proteins probed with monoclonal anti‐haemagglutinin (anti‐HA) (B) and anti‐histidine (anti‐His) (C) antibodies to show in vitro protein interactions. Escherichia coli lysates expressing HrpG‐FLAG, HrpA‐His, HrpF‐HA27 (indicated by ‘+’) or vector alone (indicated by ‘−’) were mixed with anti‐FLAG M2 (B) or anti‐HA (C) agarose beads for immunoprecipitation, as described in Experimental procedures. Gel‐bound, immunoprecipitated proteins; lysate, total proteins.
Next, an immunoprecipitation assay using anti‐FLAG (binding with HrpG‐FLAG) or anti‐HA (binding with HrpF‐HA27) affinity gel was applied to detect protein interactions in vitro. The FLAG‐ or HA agarose‐bound proteins were analysed by immunoblotting using antibodies against HA or histidine (His) epitope, respectively. The results showed that HrpF‐HA27 can be detected in the FLAG‐gel‐bound fraction (Fig. 3B) and HrpA‐His (encoded by pNCHU1944, see Table S1 for plasmid descriptions) in the HA‐bound fraction (Fig. 3C), revealing that HrpF binds to HrpG and HrpA in vitro. By contrast, HrpA‐His and HrpG‐FLAG could not be co‐precipitated with the anti‐FLAG or Ni‐NTA resin (Fig. S3, see Supporting Information), suggesting that HrpA protein does not interact with HrpG protein in vitro.
HrpF negatively regulates the expression of t3ss
The results shown in Figs 2A and 3 indicate that HrpF may be involved in the negative feedback loop of t3ss expression via interaction with HrpG; therefore, we measured t3ss transcription in the hrpF mutant and WT overexpressing hrpF by qRT‐PCR. For comparison of the hrp/hrc expression level between the WT and its derivative strains, the plasmids (Table S1) overexpressing hrp genes of interest driven by the lac promoter in the WT were consistently used in the following experiments. The transcription of four hrp/hrc genes, i.e. hrpL, hrcC, hrpA and hrpZ, which reside in different hrp operons and encode different components of T3SS, was monitored. After normalization with the 16S rRNA transcript, the relative quantities of the hrp genes were calculated as described in Experimental procedures. As shown in Fig. S4A (see Supporting Information), the levels of hrpL, hrcC, hrpZ and hrpA transcripts in WT increased gradually with increasing incubation time, and the transcription patterns of hrpA and hrpZ residing in the same transcription unit (Huang et al., 1995) were the same (Fig. 4A). Similarly, the immunoblot results showed that HrpZ production in the hrpF mutant was about 1.5‐fold that of the amount in WT, whereas the cumulative HrpA in the hrpF mutant was not positively correlated with the transcription level. At 6 hpi, the detectable HrpA in the hrpF mutant was about 3% of that produced in WT, suggesting that HrpA is post‐translationally processed in the hrpF mutant (Fig. 4B). The fold changes of hrpL, hrcC, hrpZ and hrpA transcripts in the hrpF mutant strain HL1‐N1589 compared with WT were greater than unity (Fig. 4A), and HL1‐N1589 showed the maximum transcription of these four genes at 3 hpi (Fig. 4A), whereas these transcripts reached the highest level at 6 hpi in the PtoDC3000 hrpF mutant (Fig. S4B, top panel), indicating that HrpF has a negative regulatory function in early t3ss expression. This result was different from that for the nptII‐marked hrpF mutant of Psy61, which showed reduced t3ss expression (Fig. S4B, bottom panel) (Ramos et al., 2007). The opposite patterns of t3ss expression in the marked and unmarked hrpF mutants are most probably a result of the nptII promoter‐driven, constitutive expression of the downstream negative regulator hrpV (Wei et al., 2005). Moreover, compared with WT, in‐trans expression of hrpF or hrpF‐HA27 in WT PavHL1 reduced the transcription of the three hrp/hrc genes at 6 hpi (Fig. 2B). The fold changes of the three hrp/hrc and hrpF genes in WT ecotopically expressing hrpG were between two and four (Figs 2B and S4C), whereas co‐expressed hrpFG genes counteracted the positive regulatory effect of HrpG on the transcription of the three hrp/hrc genes, reducing the fold changes to close to unity (Fig. 2B), indicating that HrpF and HrpG show inverse effects on t3ss expression. Overexpression of either hrpF or hrpG, which are organized in the same transcription unit (Huang et al., 1995), may disrupt the stoichiometry of interacting proteins, leading to the change in t3ss expression. Immunoblot analyses using sera against HrpZ and HrpA and effector translocation assays using the Cya reporter system were also conducted to monitor T3SS expression and function. As shown in Fig. 2C, the cumulative levels of HrpZ and HrpA in WT harbouring the plasmids containing hrpF, hrpG or hrpFG were consistent with the qPCR results (Fig. 2B), and the expression patterns were positively correlated with the results of the HR phenotype and ionic conductivity (Fig. 2A). Cya translocation assay revealed that, in WT overexpressing hrpF, T3SS was functional and could secrete and translocate T3Es, but the translocated HopI1 was reduced to c. 6% relative to WT (Table 1). Taken together, the results reveal that the function of HrpF is characteristically equivalent to a negative regulator, and the presence of HrpG counteracts the negative effect of HrpF.
Figure 4.
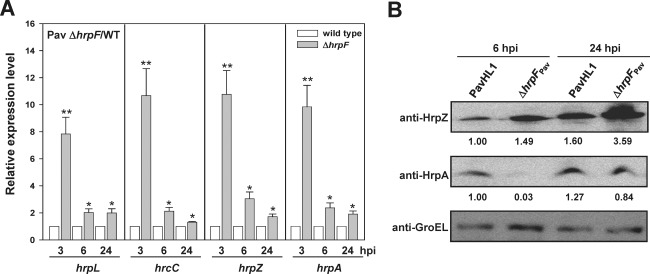
Expression of hrp genes was elevated in the hrpF mutant. (A) Relative transcription of hrpL, hrcC, hrpZ and hrpA in the hrpF mutant and wild‐type. Transcription of each hrp gene was normalized by the 16S rRNA transcript as described in Experimental procedures, and the relative expression levels of the hrp genes were determined at 3, 6 and 24 h post‐inoculation (hpi) using the quantitative polymerase chain reaction (qPCR) results of the wild‐type as the denominator. The relative expression levels of the hrpL, hrcC, hrpZ and hrpA transcripts were significantly different between the wild‐type and hrpF mutant at 3 hpi. The error bars denote standard deviation (n = 3). Statistical analysis by Student's t‐test (*P < 0.05; **P < 0.01) was applied to compare the fold change of transcription between the wild‐type and hrpF mutant. (B) Immunoblot analysis of HrpZ and HrpA at 6 and 24 hpi in the wild‐type and hrpF mutant. GroEL served as a protein loading control, and the relative production of HrpZ and HrpA is shown as the fold change in the banding intensity using the wild‐type at 6 hpi as the denominator.
A single amino acid substitution (K67A) at the C‐terminus of HrpFPav affects the virulence and secretion/translocation of T3Es
As it was found that epitope tagging at the C‐terminus of HrpF did not complement the hrp‐minus phenotypes of the hrpF mutant (data not shown), the importance of the HrpF C‐terminus in T3SS function was further analysed. The C‐terminal three conserved amino acids of HrpF (denoted by asterisks in Fig. S1A) were replaced with Ala to produce HrpFK67A, HrpFD71A and HrpFQ74A. These HrpF variants were expressed in HL1‐N1589 for complementation assays. The alleles encoding HrpFD71A and HrpFQ74A in HL1‐N1589 restored bacterial multiplication to the WT level in starfruit leaves (data not shown), whereas the population of HL1‐N1589 harbouring pNCHU1808 (hrpF K67A) only reached 5.2 logs, not significantly different from the 4.9 logs of the hrpF mutant (CFU/cm2 at 7 dpi) (Fig. 5A). By comparison with WT and HL1‐N1589 carrying the native hrpF, the hrpF mutant harbouring HrpFK67A also reduced the ability to induce the HR (Fig. 5B) and apoplastic ion leakage at 7 hpi (Fig. 5C) on tobacco leaves.
As HrpF is a secreted protein, we further investigated whether HrpFK67A is secreted into the bacterial milieu via T3SS. HL1‐N1589 harbouring pNCHU1810 (HrpF‐HA27) or pNCHU1945 (HrpFK67A‐HA27, HrpFK67A was internally tagged with HA as described for the preparation of HrpF‐HA27) was cultured in HrpMM and subjected to a protein secretion assay using an immunoblotting method. The immunoblots showed that HrpFK67A‐HA27 can also be secreted to the culture supernatant, but the amount of HrpFK67A‐HA27 in the supernatant fraction is much lower than that of HrpF‐HA27 (Fig. 5D), suggesting that the 67th lysine residue of HrpF is critical for its own secretion. Furthermore, the secretion of T3E substrates in HL1‐N1589 harbouring hrpF K67A was quantitatively monitored. As shown in Fig. 5E, the immunoblot revealed that the amounts of secreted HopAK1 and HrpZ in HL1‐N1589 harbouring hrpF K76A were lower than those in WT and WT expressing the native HrpF, and the secretion of HrpA in the complementing strain expressing hrpF K76A was severely reduced to approximately 2% of the amount secreted by WT. The efficiency of the HopI1‐cCya translocation was also greatly reduced in HL‐N1589 harbouring hrpF K67A (Table 1), indicating that the 67th lysine residue of HrpF is important for its function in assisting the secretion and translocation of T3E substrates.
HrpF is required for HrpA stability in the bacterial cytoplasm
As shown by the results illustrated in Figs 1E, 4B and 5E, the cumulative amount of HrpA in the cell pellets was severely reduced in the hrpF mutant, and the plasmid‐borne hrpF K67A cannot fully complement the mutant phenotypes. Therefore, the involvement of the 67th lysine residue of HrpF in T3SS function was further analysed by testing the interactions between HrpFK67A‐HA27 and HrpA. The immunoprecipitation assay (Fig. 6A) showed that the amount of HrpA pulled down by HrpFK67A‐HA27 was about 35% of that by HrpF‐HA27. Far western analysis confirmed that HrpF‐HA27, but not HrpFK67A‐HA27 (barely dectectable), binds to denatured HrpA (Fig. 6B), indicating that the 67th lysine residue of HrpF is critical for its interaction with HrpA. The biochemical activity of HrpF on HrpA was tested by the culture of PavHL1 and its derivatives harbouring pNCHU2046 (co‐expressing HrpA and HrpZ from the constitutive lac promoter on the vector) in HrpMM to induce t3ss expression. The translation of T3SS was stopped by the addition of 100 ppm of chloramphenicol to the culture medium at 2 hpi. Total cell cultures containing culture medium and bacterial cells were harvested at 0, 2 and 4 h post‐chloramphenicol treatment for immunoblot analyses using sera against HrpA and HrpZ. As shown in Fig. 6C, the level of HrpZ remained constant in PavHL1 and its derivatives at different time points, whereas the cumulative amount of HrpA decreased significantly in the hrpF mutant on chloramphenicol treatment, which can be greatly improved by complementing the mutant strain with a plasmid‐borne hrpF, or moderately by hrpF K67A. The data, together with the protein–protein interaction assays in Fig. 3, indicate that HrpF binds to HrpA to prevent HrpA degradation, and the 67th lysine residue of HrpF is important for its binding to HrpA and the maintenance of HrpA stability. In comparison with HrpF‐HA27, the binding of HrpFK67A‐HA27 to HrpG‐FLAG was reduced to approximately 70% (Fig. S5A, see Supporting Information). Therefore, the effect of HrpFK67A on t3ss expression was measured by in‐trans expression of lac promoter‐driven HrpFK67A in PavHL1 to show that there were no apparent differences between HrpF‐ and HrpFK67A‐expressing strains in HR elicitation (see Fig. S5B,C) or HrpZ/HrpA production in the cytoplasm (see Fig. S5D). Hence, the 67th lysine residue of HrpF is important for binding with HrpA, but has little effect on t3ss expression.
Figure 6.
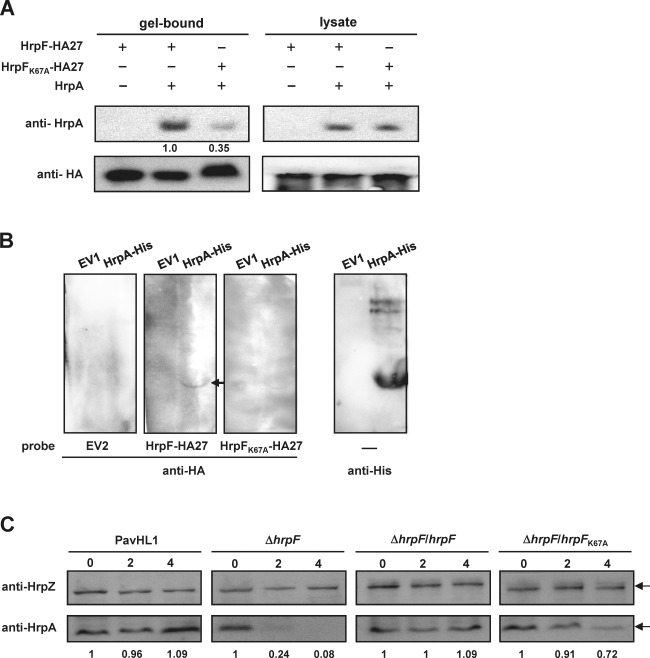
HrpF stabilizes HrpA, and the 67th lysine residue of HrpF is critical for its interaction with HrpA. (A) Immunoblots of immunoprecipitated proteins probed with anti‐HrpA (top) and anti‐haemagglutinin (anti‐HA) (bottom) antibodies to show in vitro protein interactions. Escherichia coli lysates expressing HrpF‐HA27 (pNCHU1867), HrpFK67A‐HA27 (pNCHU1868), HrpA (pNCHU1869) (‘+’) or vector alone (‘−’) were mixed with anti‐HA agarose beads for immunoprecipitation and detected by anti‐HrpA serum as described in Experimental procedures. Gel‐bound, immunoprecipitated proteins; lysate, total proteins as a control to detect the expression of recombinant proteins. (B) Far western analysis shows that HrpF‐HA27 and HrpFK67A‐HA27 bind to HrpA‐His in vitro. Escherichia coli‐expressed HrpA‐His (pNCHU1944) or EV1 (pETite) was probed with bacterial lysates expressing EV2 (pET29a), HrpF‐HA27 (pNCHU1867) or HrpFK67A‐HA27 (pNCHU1868) as described in Experimental procedures. HrpA‐His‐bound HrpF‐HA was further detected with HA monoclonal antibody. (C) Immunodetection of HrpA stability in the Pseudomonas syringae pv. averrhoi (Pav) hrpF mutant. PavHL1, wild‐type harbouring pRK415 and pNCHU2046; ΔhrpF, HL1‐N1589 harbouring pRK415 and pNCHU2046; ΔhrpF/hrpF, HL1‐N1589 harbouring pNCHU1590 and pNCHU2046, ΔhrpF/hrpF K67A, HL1‐N1589 harbouring pNCHU2068 and pNCHU2046. Total cell cultures were harvested at 0, 2 and 4 h post‐chloramphenicol treatment for immunodetection with anti‐HrpZ and anti‐HrpA sera. The stability of HrpA is shown by the fold changes in HrpA banding intensity, which was first normalized by the HrpZ bands and calculated using the value at 0 h post‐chloramphenicol treatment as the denominator.
Discussion
The hrp/hrc genes coding for T3SS in plant bacterial pathogens are organized into several operons and reside in a PAI that is located on a chromosome or a large plasmid of the pathogenic bacteria (Arnold et al., 2003). In this study, we characterized the role of Pav HrpF, whose coding gene is immediately upstream of hrpG, in T3SS function. The results shown here demonstrate that: (i) the hrpF mutant displays hrp‐minus phenotypes and defects in the secretion/translocation of T3Es, indicating that Pav HrpF is essential for T3SS function; (ii) the expression of t3ss is increased in the unmarked hrpF mutants of PavHL1 and PtoDC3000, and ectopic expression of hrpF in WT PavHL1 reduces t3ss expression, whereas co‐expression of HrpFG from a plasmid derepresses the negative impact of high‐dosage HrpF on t3ss transcription, suggesting that HrpF acts together with HrpG to modulate t3ss expression; (iii) HrpF binds to HrpA to prevent HrpA degradation in the bacterial cytosol; and (iv) the 67th lysine of HrpF is a critical residue for HrpF–HrpA interaction.
In the early stage of P. syringae–plant interactions, a functional T3SS is essential for the suppression of plant defence and subsequent bacterial colonization (Abramovitch and Martin, 2004; Abramovitch et al., 2006). hrpF mutant strains of P. syringae fail to elicit disease symptoms or programmed cell death in their host or non‐host plant species, and the secretion or translocation of T3SS substrates is abolished under t3ss‐inducing conditions (Deng et al., 1998; Figs 1 and S1). Transmission electron micrographs also showed the lack of the T3SS pilus structure on the surface of the hrpF mutant (Fig. S6, see Supporting Information). These results indicate that HrpF is essential for T3SS function. HrpF is a small, acidic protein (pI values ranging from 3.8 to 4.6 among HrpF homologues), and is conserved in all P. syringae pathovars, but shares low sequence similarity with other proteins, e.g. HrpA pilin protein (Deng et al., 1998). The threading protein structure at the Phyre Protein Fold Recognition Server (http://www.sbg.bio.ic.ac.uk/~phyre/index.cgi) (Kelley and Sternberg, 2009) predicted that HrpF shares low structural similarity with the T3SS needle proteins of Shigella flexneri MxiH (E‐value: 8.7), Y. pestis YscF (E‐value: 20) and Salmonella enterica PrgI (E‐value: 23) in the region rich in α‐helical coiled‐coil structures. The coiled‐coil structure is a versatile protein domain commonly found in molecular chaperones and proteins with various biological functions (Lupas and Gruber, 2005; Mason and Arndt, 2004), and polymerization of the T3SS needle filaments is also dependent on the coiled‐coil interactions between monomeric needle proteins (Blocker et al., 2008). Prior to secretion, the prevention of premature polymerization of the needle proteins is accomplished by T3SS chaperone functions (Cornelis, 2006). Many T3SS needle proteins, including P. aeruginosa PscF, Y. pestis YscF and Aeromonas hydrophilia AscF, have been found to employ a bimolecular chaperone, i.e. PscE–PscG (Quinaud et al., 2005), YscE–YscG (Sun et al., 2008) and AscE–AscG (Chatterjee et al., 2011), for the maintenance of the monomeric state and protein stability within the bacterial cytoplasm. The pscE‐ and pscG‐deleted strains of P. aeruginosa cannot accumulate PscF protein in the bacterial cytoplasm or secrete T3Es into the bacterial milieu under Ca2+‐depleted conditions, nor can they induce T3SS‐mediated cytotoxicity in a macrophage cell line, indicating that the needle chaperones are absolutely required for T3SS function (Quinaud et al., 2005). In this study, the biochemical assays of protein–protein interactions and HrpA stability suggest that HrpF may function like a T3SS pilus stabilizer to prevent HrpA degradation in the bacterial cytoplasm, which is important in the early t3ss‐inducing phase (e.g. 6 hpi in Fig. 4B) to facilitate the assembly of the Hrp pilus. Thus, bacteria lacking HrpF cannot form the Hrp pilus, nor can they secrete or translocate T3Es to the bacterial milieu or plant cells, resulting in the accumulation of HrpA in the bacterial cytosol in the late t3ss‐inducing phase (e.g. 24 hpi in Fig. 4B). Interestingly, differential secretome analysis of PtoDC3000 (Schumacher et al., 2014) revealed that extracellular HrpA was exclusively processed at the N‐terminus at 6 h post‐t3ss induction, and the secreted form was switched to full‐length HrpA at 48 hpi. Similar results, showing that the extracellular HrpA contains both full‐length and N‐terminally truncated forms at 2 dpi, have been reported by Roine et al. (1997a). Considering the interaction between HrpA and HrpF in the bacterial cytoplasm, HrpF may play a role in the temporal regulation of HrpA cleavage by direct binding. In the early stage of t3ss expression, HrpA bound with HrpF remains intact in the bacterial cytoplasm; on completion of the T3SS, HrpA is dissociated from HrpF and processed at or after secretion by an unidentified protease, and assembles into the Hrp pilus to facilitate the subsequent secretion and translocation of HrpF and T3Es. In later stages, HrpA, HrpF and T3Es may be secreted together into the bacterial milieu in vitro, resulting in the high abundance of HrpA in the culture supernatant and the alteration of the truncated form to the intact form. As HrpF does not contain any protease domain, the role of HrpF in the post‐translational cleavage of HrpA remains to be addressed.
On induction, a negative feedback regulatory circuit acts together with the positive feed‐forward loop to create an oscillation motif in the t3ss regulatory network. In P. syringae, HrpV negatively regulates t3ss expression via binding to the positive regulator HrpS (Buttner, 2012; Jovanovic et al., 2014; Preston et al., 1998; Tang et al., 2006), whereas HrpG acts as a suppressor of HrpV to release HrpS for the up‐regulatory state (Jovanovic et al., 2011; Wei et al., 2005). So far, this regulatory mode has only been found in the group 1 Hrp T3SSs (Wei et al., 2005). HrpG and HrpV of P. syringae and Erwinia amylovora share 32% and 40.8% sequence similarity, respectively, and the two proteins have been found to form a stable heterodimer in E. amylovora (Gazi et al., 2015). However, the function of HrpF in E. amylovora has not been studied. Although there is no apparent sequence similarity in the t3ss regulatory proteins of P. aeruginosa and P. syringae, the HrpSVG negative feedback regulatory system shares common features with the ExsADCE system of P. aeruginosa, in which the ExsA‐dependent t3ss activation is coupled with ExsE secretion (Buttner, 2012; Rietsch et al., 2005; Urbanowski et al., 2005). The results presented in this study reveal that the expression of the hrp regulon is transiently induced in the hrpF mutant and repressed in WT overexpressing the hrpF gene, and the negative effect of ectopically expressed HrpF on t3ss expression can be suppressed by the co‐expression of hrpF and hrpG genes. Moreover, the chaperone‐like HrpG interacts directly with HrpV (Wei et al., 2005) and HrpF (Fig. 3), suggesting that HrpF might act as the ExsE counterpart in P. syringae.
Although we cannot rule out the possibility that in‐trans expression of HrpF leads to the accumulation of misfolded proteins as inclusion bodies in the cytoplasm, circumstantial evidence obtained from the ecotopic expression of HrpF‐HA27 in the hrpF mutant (Fig. 5D,E) shows normal secretion of Hrp outer proteins (HrpF‐HA27, HopAK1, HrpA and HrpZ), indicating that T3SS is functional, and the negative effect of overexpressed HrpF on the elicitation of HR is probably caused by the suppression of t3ss. Alternatively, the negative effect of HrpF on t3ss expression may result partially from its interaction with HrpA, which has been found to be involved in the positive regulation of t3ss via an unknown mechanism (Wei et al., 2000). As the truncated and intact HrpA can be secreted in a temporal manner (Schumacher et al., 2014), it can be postulated that the truncated HrpA forms the T3SS pilus, whereas the intact HrpA may act as a secretion‐competent regulator that activates t3ss in the cytoplasm, and its positive effect on t3ss expression can be reduced on secretion. Nevertheless, the positive regulation of HrpA is probably monitored by its chaperone‐like protein HrpF, and the mutation of hrpF reduces the stability of HrpA, which greatly affects the construction of the T3SS conduit, leading to the transient activation of t3ss expression [Figs 4A and S4B (top panel)]. The actual role of HrpA in t3ss expression needs to be investigated. This hypothetical t3ss regulatory mechanism involving HrpA–HrpF–HrpG–HrpV–HrpS in P. syringae is not found in the other t3ss regulatory systems; however, the model describes how HrpA regulates t3ss expression without direct binding to any known regulatory components (Wei et al., 2000).
A comparison of HrpF orthologues from P. syringae pathovars reveals that HrpFPav and HrpFPto share 62% similarity in amino acid sequences (Fig. S1A). The three amino acid residues at the C‐terminus, K67, D71 and Q74, which are conserved in all HrpF orthologues, were selected for preliminary characterization. Single amino acid substitution of the three residues with Ala showed that HrpF harbouring the K67A substitution only partially restored the ability of the Pav hrpF mutant HL1‐N1589 to elicit plant responses in planta (Fig. 5B,C). In vitro secretion assays of three representative Hrp outer proteins, i.e. HopAK1, HrpZ and HrpA, showed that HrpFK67A had a minor effect on the secretion of HopAK1 and HrpZ; however, the cumulative HrpA levels in both the cell‐bound and secreted fractions were low (Fig. 5E), revealing that the 67th lysine residue is critical for HrpA stability (Fig. 6C). The 67th lysine residue of HrpF is a negatively charged amino acid and may be involved in protein–protein interactions with other Hrp proteins. In vitro immunoprecipitation assays and far western analysis revealed that single amino acid substitution attenuates the binding of HrpFK67A to HrpA (Fig. 6A,B). The secretion of HrpZ and HopAK1 is dependent on the HrpA pilus (Jin and He, 2001); therefore, we predict that bacteria expressing the variant HrpFK67A can transiently assemble a functional T3SS to aid in T3E secretion, thus partially compensating for the HrpF function in HR elicitation (Fig. 5B,C), but not bacterial growth (Fig. 5A).
In conclusion, we found that HrpF has a chaperone‐like activity for the stabilization of HrpA in the bacterial cytoplasm, and is a secreted t3ss regulator in plant‐pathogenic P. syringae. t3ss regulation in P. syringae involves a complex protein–protein coupling interaction, which presumably builds up an oscillating regulatory circuit by sensing small changes in the cellular concentrations of the key regulators to quickly respond to the surroundings (Tian et al., 2009). Based on the results of this study, a hypothetical model is proposed, in which small amounts of HrpR and HrpS at the early stages of t3ss expression result in a low concentration of type III secretion proteins, and the negative regulator HrpV binds to HrpS to suppress t3ss expression. Meanwhile, HrpG interacts with HrpF, and HrpF binds to and stabilizes HrpA in the cytoplasm to form a stable protein complex. On contact with plant cells, the assembly of T3SS on the cell envelop facilitates the secretion of HrpA and HrpF, shifting the equilibrium of protein interaction from HrpF–HrpG to HrpG–HrpV to release HrpS for subsequent up‐regulation of t3ss. The binding affinity between these proteins is unclear and worthy of further investigation. The findings in this study denote the stealthy life style of pathogenic P. syringae, which uses interlinked regulatory networks to up‐ and down‐regulate t3ss expression and to modulate pilus protein stability prior to the assembly of extracellular filaments.
Experimental Procedures
Bacterial strains, plasmids, culture conditions and DNA manipulations
The bacterial strains, plasmids and primers used in this study are described in Tables S1 and S2 (see Supporting Information). The cultural conditions of E. coli and P. syringae, and DNA manipulation, are described in Methods S1 (see Supporting Information).
Generation of non‐polar, unmarked mutants of HL1‐N1589 and DC3000‐N1865, and non‐polar nptII insertion of hrcC mutant (HL1‐N1033) and complementation
The unmarked mutation of hrpF in PavHL1 was constructed using the procedures described by Wei et al. (2007). To generate the unmarked hrpF‐deleted mutant (HL1‐N1589) of PavHL1, the flanking fragments of hrpF were cloned into the suicide vector pK18mobsacB (Schafer et al., 1994) to produce the respective pNCHU1589 (details in Methods S1), followed by transformant selection and counter‐selection for mutants, as described previously (Wei et al., 2007). For the generation of a non‐polar mutation of hrcC in PavHL1, the partial sequence of hrcC was replaced with an nptII marker (Alfano et al., 1996) (Table S1 and Methods S1). For the expression of an HA‐tagged HrpF, a chimeric hrpF Pav‐HA gene, coding for internally tagged HA between the 27th and 28th codons of HrpFPav, was generated by crossover PCR (Sukdeo and Charles, 2003) with primers prhrpF‐1F/prhrpFnr‐HA and prhrpFcf‐HA/prhrpF‐2R to produce pNCHU1810, and the Pav hrpF K67A allele was synthesized by PCR using the primers prhrpF‐1F/prhrpFK67A‐R to generate pNCHU1808 (Table S1 and S2). These recombinant plasmids were conjugated into hrpF Pav mutant HL1‐N1589 for virulence assays on host plants and HR elicitation and conductivity on non‐host plants.
Plant bioassays
Tobacco, tomato and starfruit plants were grown under glasshouse conditions as described in Methods S1. For disease symptom development in the leaves of starfruit and tomato, the bacterial suspension was adjusted to a cell density of 1 × 108 CFU/mL for spray or dipping inoculation, and inoculated plants were kept humid by wrapping in plastic bags at 28–30 ºC. For bacterial multiplication assays, a cell density of 1 × 105 CFU/mL was infiltrated into the leaves of starfruit and tomato, and three leaf discs (6 mm in diameter) were harvested from three infiltrated leaves for bacterial enumeration, as described previously (Wei et al., 2007, 2012). Tobacco leaves were infiltrated with bacteria at 1 × 108 CFU/mL or by serial titration for HR elicitation and ion leakage experiments, and the ionic conductivity was measured as an indicator of membrane damage during the onset of HR (Goodman, 1968) at 7 or 24 hpi by a conductivity meter (Radiometer analytical IONcheck 30, Lyon, France), as described by Wei et al. (2012).
Immunodetection of Hrc/Hrp proteins
After induction of hrp/hrc expression in HrpMM (Huynh et al., 1989) for 6 h at 22 ºC, extracellular and cytoplasmic proteins of cultured bacteria were separated and collected by centrifugation. The total protein concentration of the cell lysates and supernatants was quantified, and equal amounts of the total proteins were subjected to immunodetection (details in Methods S1). The intensity of chemiluminescence shown in immunoblots was quantified by an LAS‐4000mini and analysed by Multi Gauge 3.0 software (Fujifilm Co., Tokyo, Japan).
Isolation of total bacterial RNA and quantitative determination of t3ss expression
The High Pure RNA Isolation Kit (Roche, Mannheim, Germany) was used to prepare total RNA from PavHL1 and its derivatives. Bacterial t3ss expression was induced in HrpMM for 3, 6 and 24 h at 22 ºC. Bacterial cells were collected, and the extracted RNAs were subjected to qPCR experiments as described in Methods S1. For the detection of the transcripts of hrpL, hrcC, hrpZ and hrpA in PavHL1 and its derivative strains, the following primer pairs (Table S2), i.e. prhrpL‐3F/4R, prhrcC‐1F/2R, prhrpZ‐5F/6R and prhrpA‐17F/18R, respectively, specific for PavHL1, were employed. The amplification of the 16S rRNA gene using the primers pr16S‐3F/4R (Table S2) was applied to normalize the relative expression of t3ss by the comparative CT (cycle threshold) method using the formula 2–ΔΔCT (Livak and Schmittgen, 2001).
Cya translocation reporter assays
PavHL1, hrcC mutant HL1‐N1033 and hrpF mutant HL1‐N1589, carrying the plasmids expressing HrpF‐cCya or HopI1‐cCya fusion proteins, were employed for Cya activity assays in tobacco plants at 6 h after infiltration with bacterial concentrations at an optical density at 600 nm (OD600) of 0.3 using the procedures described by Wei et al. (2005).
Assays to determine protein–protein interactions of HrpF with HrpG and HrpA
Bacterial two‐hybrid assay
The assay was performed using a BacterioMatch® II Two‐Hybrid System Library Construction Kit (Stratagene). The coding sequences of hrpF Pav, hrpG Pav and hrpA Pav were translationally fused to reporter genes in the target vector pTRG or bait vector pBT. The derivatives of pTRG and pBT (corresponding plasmids described in Table S1) were simultaneously transformed into E. coli XL1 blue MRF′ Km strain carrying HIS3 and aadA reporter genes. Protein–protein interaction was selected for the growth of transformants harbouring the bait and target plasmids on a selective medium containing 5 mm 3‐amino‐1,2,4‐triazole (3‐AT). The strength of the interaction was calculated (%) by comparing the number of bacterial colonies grown on the selective medium with those grown on the non‐selective medium that does not contain 3‐AT, as described by the manufacturer.
Immunoprecipitation assay with an anti‐HA agarose or anti‐FLAG agarose
The recombinant proteins were induced and collected from E. coli C41 (DE3) carrying NCHU1867 (pET29a::hrpF Pav ‐HA27) or pNCHU1868 (pET29a::hrpF K67A ‐HA27), or BL21 (DE3) carrying pNCHU1869 (pET29a:: hrpA Pav), pNCHU1944 (pETite::hrpA Pav‐His) (Table S1) or pNCHU668 (pT7‐7::hrpG Psy‐FLAG) (Wei et al., 2005), as described in Methods S1. HrpG‐FLAG protein was filtered through a 0.45‐μm membrane (Millipore, Bedford, MA, USA) to remove inclusion bodies, and the concentration of total proteins was adjusted to 1 mg/mL. To pull down FLAG‐bound HrpF‐HA27, HA‐bound HrpA protein or HA‐bound HrpA‐His protein, the anti‐FLAG M2 agarose (Sigma‐Aldrich, St. Louis, MO, USA) or polyclonal anti‐HA agarose (Immunology Consultants Laboratory, Inc., Newberg, OR, USA) was prepared according to the manufacturer's instructions. The gel‐bound proteins were eluted and subjected to immunoblotting analysis as described in Methods S1.
Far western blotting analysis
Escherichia coli C41 (DE3) carrying pNCHU1867 (pET29a:: hrpF Pav ‐HA27) or pNCHU1868 (pET29a::hrpF K67A ‐ HA27), and E. coli BL21 (DE3) harbouring pET29a, pETite or pNCHU1944 (pETite::hrpA Pav‐His), were used to express the target proteins (see Methods S1 for details). The HrpF‐HA27 or HrpFK67A‐HA27 protein extract served as the protein probe for the detection of the Ni‐NTA resin‐purified HrpA‐His protein, which was prepared as described previously (Steen et al., 1986; Studier et al., 1990). HrpA‐His protein and control proteins expressed from the empty vector were separated by 15% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to poly(vinylidene difluoride) (PVDF) membrane (Millipore). The membranes were probed with the sonicated cell lysate containing HrpF‐HA27 or HrpFK67A‐HA27, as reported previously (Wei et al., 2005). Immunodetection was performed using monoclonal anti‐HA antibody (Sigma).
HrpA stability analysis
For the analysis of HrpA stability, pNCHU2046 (pBBR1MCS‐2::hrpAZ Pav) constitutively expressing hrpAZ was transformed into PavHL1 (WT), HL1‐N1589 (hrpF mutant) and HL1‐N1589 harbouring pNCHU1590 (pRK415::hrpF) or pNCHU2068 (pRK415::hrpF K67A). The transformants were cultured in King's medium B (KB) broth to an OD600 of 0.3, changed to HrpMM and incubated for 2 h to induce t3ss expression. To stop translation, chloramphenicol was added at a final concentration of 100 ppm, and the total cell cultures (culture medium plus cell mass) were harvested at 0, 2 and 4 h post‐chloramphenicol treatment, sonicated and precipitated by 5% cold trichloroacetic acid (TCA) for immunodetection as described above.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phenotypes of the hrpF mutant strains derived from Pseudomonas syringae pv. tomato DC3000.
Fig. S2 The hypersensitive response (HR) on tobacco leaves elicited by PavHL1 and its derived strains.
Fig. S3 Immunoprecipitation assays of HrpA and HrpG.
Fig. S4 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analyses of hrp transcripts in Pseudomonas syringae strains grown in Hrp minimal medium (HrpMM).
Fig. S5 The effects of HrpFK67A on t3ss expression.
Fig. S6 Immunogold labelling of Hrp pilus on bacterial surfaces.
Table S1 List of bacterial strains and plasmids used in this study.
Table S2 Primers used in this study.
Methods S1 Experimental procedures.
Acknowledgements
We thank Pao‐Sheng Hou, Ming‐Lung Cheng and Cheng‐Hsien Kuo for construction of the hrcC mutant of PavHL1 (PavHL1‐N1033), plasmid pNCHU1161 and pNCHU1645, respectively. This work was supported by NSC (National Science Council, Taiwan) grants NSC96‐2317‐B‐005‐005, NSC96‐2752‐B‐005‐003‐APE and NSC102‐2911‐I‐005‐301.
References
- Abramovitch, R.B. and Martin, G.B. (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7, 356–364. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.B. (2006) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. , Bauer, D.W. , Milos, T.M. and Collmer, A. (1996) Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non‐polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19, 715–728. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. , Charkowski, A.O. , Deng, W.L. , Badel, J.L. , Petnicki‐Ocwieja, T. , van Dijk, K. and Collmer, A. (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA, 97, 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, D.L. , Pitman, A. and Jackson, R.W. (2003) Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4, 407–420. [DOI] [PubMed] [Google Scholar]
- Blocker, A. , Jouihri, N. , Larquet, E. , Gounon, P. , Ebel, F. , Parsot, C. , Sansonetti, P. and Allaoui, A. (2001) Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39, 652–663. [DOI] [PubMed] [Google Scholar]
- Blocker, A.J. , Deane, J.E. , Veenendaal, A.K. , Roversi, P. , Hodgkinson, J.L. , Johnson, S. and Lea, S.M. (2008) What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. USA, 105, 6507–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz, J. , Losada, L. , Lisboa, K. and Hutcheson, S.W. (2002) Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae . Mol. Microbiol. 45, 397–409. [DOI] [PubMed] [Google Scholar]
- Buell, C.R. , Joardar, V. , Lindeberg, M. , Selengut, J. , Paulsen, I.T. , Gwinn, M.L. , Dodson, R.J. , Deboy, R.T. , Durkin, A.S. , Kolonay, J.F. , Madupu, R. , Daugherty, S. , Brinkac, L. , Beanan, M.J. , Haft, D.H. , Nelson, W.C. , Davidsen, T. , Zafar, N. , Zhou, L. , Liu, J. , Yuan, Q. , Khouri, H. , Fedorova, N. , Tran, B. , Russell, D. , Berry, K. , Utterback, T. , Van Aken, S.E. , Feldblyum, T.V. , D'Ascenzo, M. , Deng, W.L. , Ramos, A.R. , Alfano, J.R. , Cartinhour, S. , Chatterjee, A.K. , Delaney, T.P. , Lazarowitz, S.G. , Martin, G.B. , Schneider, D.J. , Tang, X. , Bender, C.L. , White, O. , Fraser, C.M. and Collmer, A. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 100, 10 181–10 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, C. , Kumar, S. , Chakraborty, S. , Tan, Y.W. , Leung, K.Y. , Sivaraman, J. and Mok, Y.K. (2011) Crystal structure of the heteromolecular chaperone, AscE‐AscG, from the type III secretion system in Aeromonas hydrophila . PLoS One, 6, e19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis, G.R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. [DOI] [PubMed] [Google Scholar]
- Dasgupta, N. , Lykken, G.L. , Wolfgang, M.C. and Yahr, T.L. (2004) A novel anti‐anti‐activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53, 297–308. [DOI] [PubMed] [Google Scholar]
- Deng, W.L. , Preston, G. , Collmer, A. , Chang, C.J. and Huang, H.C. (1998) Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 180, 4523–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil, H. , Feil, W.S. , Chain, P. , Larimer, F. , DiBartolo, G. , Copeland, A. , Lykidis, A. , Trong, S. , Nolan, M. , Goltsman, E. , Thiel, J. , Malfatti, S. , Loper, J.E. , Lapidus, A. , Detter, J.C. , Land, M. , Richardson, P.M. , Kyrpides, N.C. , Ivanova, N. and Lindow, S.E. (2005) Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 102, 11 064–11 069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts, D.E. , Abramovitch, R.B. , Alfano, J.R. , Baldo, A.M. , Buell, C.R. , Cartinhour, S. , Chatterjee, A.K. , D'Ascenzo, M. , Gwinn, M.L. , Lazarowitz, S.G. , Lin, N.C. , Martin, G.B. , Rehm, A.H. , Schneider, D.J. , van Dijk, K. , Tang, X. and Collmer, A. (2002) Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA, 99, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.W. and Iglewski, B.H. (1991) Cloning and sequence analysis of a trans‐regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa . J. Bacteriol. 173, 6460–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.E. and Collmer, A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Gazi, A.D. , Charova, S. , Aivaliotis, M. , Panopoulos, N.J. and Kokkinidis, M. (2015) HrpG and HrpV proteins from the Type III secretion system of Erwinia amylovora form a stable heterodimer. FEMS Microbiol. Lett. 362, 1–8. [DOI] [PubMed] [Google Scholar]
- Goodman, R.N. (1968) The hypersensitive reaction in tobacco: a reflection of changes in host cell permeability. Phytopathology, 58, 872–873. [Google Scholar]
- Hendrickson, E.L. , Guevera, P. and Ausubel, F.M. (2000) The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J. Bacteriol. 182, 3508–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S.S. and Upper, C.D. (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64, 624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk, E. and Blobel, G. (2001) Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. USA, 98, 4669–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.C. , Lin, R.H. , Chang, C.J. , Collmer, A. and Deng, W.L. (1995) The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol. Plant–Microbe Interact. 8, 733–746. [DOI] [PubMed] [Google Scholar]
- Hutcheson, S.W. , Bretz, J. , Sussan, T. , Jin, S. and Pak, K. (2001) Enhancer‐binding proteins HrpR and HrpS interact to regulate hrp‐encoded type III protein secretion in Pseudomonas syringae strains. J. Bacteriol. 183, 5589–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, T.V. , Dahlbeck, D. and Staskawicz, B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Jin, Q. and He, S.Y. (2001) Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae . Science, 294, 2556–2558. [DOI] [PubMed] [Google Scholar]
- Joardar, V. , Lindeberg, M. , Jackson, R.W. , Selengut, J. , Dodson, R. , Brinkac, L.M. , Daugherty, S.C. , Deboy, R. , Durkin, A.S. , Giglio, M.G. , Madupu, R. , Nelson, W.C. , Rosovitz, M.J. , Sullivan, S. , Crabtree, J. , Creasy, T. , Davidsen, T. , Haft, D.H. , Zafar, N. , Zhou, L. , Halpin, R. , Holley, T. , Khouri, H. , Feldblyum, T. , White, O. , Fraser, C.M. , Chatterjee, A.K. , Cartinhour, S. , Schneider, D.J. , Mansfield, J. , Collmer, A. and Buell, C.R. (2005) Whole‐genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187, 6488–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic, M. , James, E.H. , Burrows, P.C. , Rego, F.G. , Buck, M. and Schumacher, J. (2011) Regulation of the co‐evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat. Commun. 2, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic, M. , Lawton, E. , Schumacher, J. and Buck, M. (2014) Interplay among Pseudomonas syringae HrpR, HrpS and HrpV proteins for regulation of the type III secretion system. FEMS Microbiol. Lett. 356, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kimbrough, T.G. and Miller, S.I. (2000) Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA, 97, 11 008–11 013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.M. , Brown, I. , Mansfield, J. , Stevens, C. , Boureau, T. , Romantschuk, M. and Taira, S. (2002) The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 21, 1909–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lupas, A.N. and Gruber, M. (2005) The structure of alpha‐helical coiled coils. Adv. Protein Chem. 70, 37–78. [DOI] [PubMed] [Google Scholar]
- Mason, J.M. and Arndt, K.M. (2004) Coiled coil domains: stability, specificity, and biological implications. ChemBioChem, 5, 170–176. [DOI] [PubMed] [Google Scholar]
- McCaw, M.L. , Lykken, G.L. , Singh, P.K. and Yahr, T.L. (2002) ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 46, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Pastor, A. , Chabert, J. , Louwagie, M. , Garin, J. and Attree, I. (2005) PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 253, 95–101. [DOI] [PubMed] [Google Scholar]
- Preston, G. , Deng, W.L. , Huang, H.C. and Collmer, A. (1998) Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180, 4532–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinaud, M. , Chabert, J. , Faudry, E. , Neumann, E. , Lemaire, D. , Pastor, A. , Elsen, S. , Dessen, A. and Attree, I. (2005) The PscE‐PscF‐PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa . J. Biol. Chem. 280, 36 293–36 300. [DOI] [PubMed] [Google Scholar]
- Quinaud, M. , Ple, S. , Job, V. , Contreras‐Martel, C. , Simorre, J.P. , Attree, I. and Dessen, A. (2007) Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. USA, 104, 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, A.R. , Morello, J.E. , Ravindran, S. , Deng, W.L. , Huang, H.C. and Collmer, A. (2007) Identification of Pseudomonas syringae pv. syringae 61 type III secretion system Hrp proteins that can travel the type III pathway and contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189, 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch, A. , Vallet‐Gely, I. , Dove, S.L. and Mekalanos, J.J. (2005) ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa . Proc. Natl. Acad. Sci. USA, 102, 8006–8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine, E. , Saarinen, J. , Kalkkinen, N. and Romantschuk, M. (1997a) Purified HrpA of Pseudomonas syringae pv. tomato DC3000 reassembles into pili. FEMS Lett. 417, 168–172. [DOI] [PubMed] [Google Scholar]
- Roine, E. , Wei, W. , Yuan, J. , Nurmiaho‐Lassila, E.L. , Kalkkinen, N. , Romantschuk, M. and He, S.Y. (1997b) Hrp pilus: an hrp‐dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 94, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sal‐Man, N. , Setiaputra, D. , Scholz, R. , Deng, W. , Yu, A.C. , Strynadka, N.C. and Finlay, B.B. (2013) EscE and EscG are cochaperones for the type III needle protein EscF of enteropathogenic Escherichia coli . J. Bacteriol. 195, 2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, A. , Tauch, A. , Jager, W. , Kalinowski, J. , Thierbach, G. and Puhler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Schechter, L.M. , Roberts, K.A. , Jamir, Y. , Alfano, J.R. and Collmer, A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a cya translocation reporter. J. Bacteriol. 186, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J. , Waite, C.J. , Bennett, M.H. , Perez, M.F. , Shethi, K. and Buck, M. (2014) Differential secretome analysis of Pseudomonas syringae pv. tomato using gel‐free MS proteomics. Front. Plant Sci. 5, 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen, R. , Dahlberg, A.E. , Lade, B.N. , Studier, F.W. and Dunn, J.J. (1986) T7 RNA polymerase directed expression of the Escherichia coli rrnB operon. EMBO J. 5, 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, W.F. , Rosenberg, A.H. , Dunn, J.J. and Dubendorff, J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Method Enzymol. 185, 60–89. [DOI] [PubMed] [Google Scholar]
- Sukdeo, N. and Charles, T.C. (2003) Application of crossover‐PCR‐mediated deletion‐insertion mutagenesis to analysis of the bdhA‐xdhA2‐xdhB2 mixed‐function operon of Sinorhizobium meliloti . Arch. Microbiol. 179, 301–304. [DOI] [PubMed] [Google Scholar]
- Sun, P. , Tropea, J.E. , Austin, B.P. , Cherry, S. and Waugh, D.S. (2008) Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Xiao, Y. and Zhou, J.M. (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant–Microbe Interact. 19, 1159–1166. [DOI] [PubMed] [Google Scholar]
- Thibault, J. , Faudry, E. , Ebel, C. , Attree, I. and Elsen, S. (2009) Anti‐activator ExsD forms a 1 : 1 complex with ExsA to inhibit transcription of type III secretion operons. J. Biol. Chem. 284, 15 762–15 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X.J. , Zhang, X.P. , Liu, F. and Wang, W. (2009) Interlinking positive and negative feedback loops creates a tunable motif in gene regulatory networks. Phys. Rev. E 80, 011926. [DOI] [PubMed] [Google Scholar]
- Urbanowski, M.L. , Lykken, G.L. and Yahr, T.L. (2005) A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. USA, 102, 9930–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.F. , Deng, W.L. and Huang, H.C. (2005) A chaperone‐like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 57, 520–536. 15978082 [Google Scholar]
- Wei, C.F. , Kvitko, B.H. , Shimizu, R. , Crabill, E. , Alfano, J.R. , Lin, N.C. , Martin, G.B. , Huang, H.C. and Collmer, A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1‐1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 51, 32–46. [DOI] [PubMed] [Google Scholar]
- Wei, C.F. , Hsu, S.T. , Deng, W.L. , Wen, Y.D. and Huang, H.C. (2012) Plant innate immunity induced by flagellin suppresses the hypersensitive response in non‐host plants elicited by Pseudomonas syringae pv. averrhoi . PLoS One, 7, e41056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. , Plovanich‐Jones, A. , Deng, W.L. , Jin, Q.L. , Collmer, A. , Huang, H.C. and He, S.Y. (2000) The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato . Proc. Natl. Acad. Sci. USA, 97, 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Heu, S. , Yi, J. , Lu, Y. and Hutcheson, S.W. (1994) Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phenotypes of the hrpF mutant strains derived from Pseudomonas syringae pv. tomato DC3000.
Fig. S2 The hypersensitive response (HR) on tobacco leaves elicited by PavHL1 and its derived strains.
Fig. S3 Immunoprecipitation assays of HrpA and HrpG.
Fig. S4 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analyses of hrp transcripts in Pseudomonas syringae strains grown in Hrp minimal medium (HrpMM).
Fig. S5 The effects of HrpFK67A on t3ss expression.
Fig. S6 Immunogold labelling of Hrp pilus on bacterial surfaces.
Table S1 List of bacterial strains and plasmids used in this study.
Table S2 Primers used in this study.
Methods S1 Experimental procedures.


