Summary
To ensure a successful long‐term infection cycle, begomoviruses must restrain their destructive effect on host cells and prevent drastic plant responses, at least in the early stages of infection. The monopartite begomovirus Tomato yellow leaf curl virus (TYLCV) does not induce a hypersensitive response and cell death on whitefly‐mediated infection of virus‐susceptible tomato plants until diseased tomatoes become senescent. The way in which begomoviruses evade plant defences and interfere with cell death pathways is still poorly understood. We show that the chaperone HSP90 (heat shock protein 90) and its co‐chaperone SGT1 (suppressor of the G2 allele of Skp1) are involved in the establishment of TYLCV infection. Inactivation of HSP90, as well as silencing of the Hsp90 and Sgt1 genes, leads to the accumulation of damaged ubiquitinated proteins and to a cell death phenotype. These effects are relieved under TYLCV infection. HSP90‐dependent inactivation of 26S proteasome degradation and the transcriptional activation of the heat shock transcription factors HsfA2 and HsfB1 and of the downstream genes Hsp17 and Apx1/2 are suppressed in TYLCV‐infected tomatoes. Following suppression of the plant stress response, TYLCV can replicate and accumulate in a permissive environment.
Keywords: cell death, geminivirus, heat stress transcription factor, HSP90
Introduction
Plants must be prepared to respond quickly and efficiently to abiotic and biotic stresses or to a combination of these stresses. These responses are complex and involve numerous pathways that are either antagonistic or synergistic, and are controlled by hormones, transcription factors, kinases and reactive oxygen species (ROS) (recently reviewed by Ben Rejeb et al., 2014; Suzuki et al., 2014).
To mitigate the effects of both biotic and abiotic stresses, the abundance of key stress proteins, such as transcription factors and chaperones (Baniwal et al., 2004; Verchot, 2012), is modulated by the regulation of assembly, aggregation and ubiquitin‐labelled degradation of misfolded proteins via the ubiquitin–proteasome system (UPS) (Lyzenga and Stone, 2011; Sherman and Goldberg, 2001). The 26S proteasome is a large ATP‐dependent proteolytic machine consisting of a barrel‐shaped proteolytic core complex (the 20S proteasome) capped at one or both ends by 19S regulatory complexes, which recognize ubiquitinated proteins (UbqPs) (Voges et al., 1999). In plants, the disruption of the 26S proteasome activates cell death (CD), accompanied by the production of ROS and followed by the release of H2O2 (Kim et al., 2003). The UPS functions not only in protein quality control, but also influences gene expression through the degradation of transcription factors (Goldberg, 2003).
Heat shock transcription factors (HSFs) are involved in the regulation of tolerance to biotic and abiotic stresses (reviewed by Wang et al., 2014b). In avian cells, it was shown that the inhibition of the 26S proteasome induced the expression of HSFs and, consequently, of the key heat shock proteins (HSPs) HSP70 and HSP90, even in the absence of heat stress (Kawazoe et al., 1998). In Arabidopsis, the inhibition of the 26S proteasome enhances the expression of HSFA2 and of the HSFA2‐regulated genes (Nishizawa‐Yokoi et al., 2010).
The key cellular chaperone HSP90 plays an essential role in the interaction, assembly and maintenance of the 26S proteasome. Functional loss of HSP90 on incubation with geldanamycin (GDA) causes the dissociation of the 26S proteasome and a significant decrease in its peptidase activity (Imai et al., 2003; Nishizawa‐Yokoi et al., 2010).
Plants have developed multiple layers of defence that recognize pathogens and rapidly respond before they cause severe damage. These surveillance systems, called innate immunity, are linked to specific pre‐programmed defence responses. However, pathogens have developed countermeasures that are able to suppress basal resistance in certain plant species. In this case, to restrain the spread of the pathogen, plants may respond by the induction of a hypersensitive response (HR) and CD (Freeman and Beattie, 2008). The signalling for controlled plant CD and its regulation has been the object of intensive research (Dickman and de Figueiredo, 2013; Lam, 2004). Plant infection with a bacterium, such as Pseudomonas syringae, or a fungus, such as Sclerotinia sclerotiorum, is followed by a conspicuous HR and CD on the inoculated leaves after 1–2 days (Kabbage et al., 2013). Infection of Nicotiana spp. with an RNA virus, such as Tobacco mosaic virus (TMV), also causes an HR similar to that induced by bacteria and fungi (Mandadi and Scholthof, 2013). By comparison, begomoviruses, such as Tomato yellow leaf curl virus (TYLCV), induce HR/CD in some plants (e.g. Nicotiana benthamiana) (van Wezel et al., 2002), but not in others (e.g. tomatoes). HR/CD starts to be observed in TYLCV‐infected tomato at the late stages of infection [50–60 days after the onset of infection (dpi)], when plants become senescent (Gorovits et al., 2007; Gorovits and Czosnek, 2007; Moshe et al., 2012).
TYLCV is a begomovirus (genus Begomovirus, family Geminiviridae) transmitted by the whitefly Bemisia tabaci (Czosnek, 2007). The role of the genes encoded by begomoviruses has been reviewed recently (Hanley‐Bowdoin et al., 2013). The virion‐sense strand comprises two genes, V1 and V2, whereas the complementary‐sense strand comprises four genes, C1–C4 (Czosnek, 2007). V1 encodes the coat protein (CP) that functions as a nuclear shuttle protein and mediates vector transmission (Priyadarshini et al., 2011). V2 encodes a protein involved in virus movement (Priyadarshini et al., 2011), in the suppression of post‐transcriptional gene silencing (PTGS) (Zhang et al., 2012) and in the suppression of methylation‐mediated transcriptional gene silencing (TGS) (Wang et al., 2014a). C1 encodes a replication‐associated protein (Rep) which initiates viral replication (Hanley‐Bowdoin et al., 2000). C2 is the transcriptional activator protein (TrAP), which interferes with TGS and PTGS (Trinks et al., 2005). C3 encodes the replication enhancer protein (REn) involved in viral replication (Settlage et al., 2005). C4 counteracts PTGS (Zhang et al., 2012).
TYLCV, like many other begomoviruses, is restricted to phloem‐associated cells and accumulates rather slowly in the plant (Ber et al., 1990). As geminiviruses do not encode their own DNA polymerases, they utilize the host nuclear DNA replication machinery and reprogram the cell cycle of mature plant cells to create a permissive environment for viral replication (reviewed by Hanley‐Bowdoin et al., 2013). Among other events, begomoviral proteins, such as Rep, bind to several host factors to ensure replication. Begomoviruses reprogram the expression of many plant genes related to DNA replication and methylation, stress response pathways, hormone‐inducible regulation of gene expression, tissue differentiation and resistance to infection (Baliji et al., 2010; Chen et al., 2013; Miozzi et al., 2014; Sade et al., 2013). To ensure a successful long‐term infection cycle, begomoviruses must restrain their destructive effect on the host cells, at least in the early stages of infection. Hence, these viruses have devised means to delay or prevent CD by interfering with the plant CD pathways. Begomoviruses encode proteins that promote CD, whereas others prevent it. For instance, the BV1 gene of the bipartite Tomato leaf curl New Delhi virus (ToLCNDV) encodes a nuclear shuttle protein that induces HR in N. tabacum and tomatoes when expressed under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. However, as HR is not evident in plants infected with ToLCNDV, the analysis of all ToLCNDV genes revealed that C2/TrAP was the factor mediating the anti‐HR effect (Hussain et al., 2007). Similarly, the V2 protein of the monopartite begomoviruses Papaya leaf curl virus (PaLCuV) and Cotton leaf curl Kokhran virus (CLCuKoV) induces CD when expressed in N. benthamiana and N. tabacum under the control of the CaMV 35S promoter, whereas the expression of the protein encoded by the C2 gene suppresses the CD elicited by the V2 protein (Mubin et al., 2010).
The way in which geminiviruses suppress CD is poorly understood. We describe the suppression effect on CD, induced by the inhibition of HSP90 and its co‐chaperone SGT1 (suppressor of the G2 allele of Skp1), in tomato plants infected by TYLCV. The levels of HSP90‐dependent UPS inactivation were reduced in the presence of TYLCV. In parallel, the activation of HSFA2 itself, HSFA2‐regulated gene expression, such as Hsp17 encoding small HSP17 and Apx1/2 encoding ascorbate peroxidases 1 and 2 (Nishizawa‐Yokoi et al., 2010), and the other heat shock transcription factor HSFB1 was sufficiently impaired in virus‐infected cells. As a result of the weakening of the plant stress response, TYLCV can accumulate and cause disease in susceptible tomato plants.
Results
Silencing of tomato Hsp90/Sgt1 genes induced CD, which was alleviated in TYLCV‐infected tomatoes
TYLCV causes only a weak HR and almost no CD in susceptible and resistant tomatoes (Eybishtz et al., 2010; Gorovits et al., 2007; Moshe et al., 2012). One of the well‐known regulators of plant cellular survival and CD is the chaperone HSP90; silencing of the Hsp90 gene in N. benthamiana plants induced CD (Liu et al., 2004). The HSP90 co‐chaperone SGT1 positively regulated CD during pathogen infection (Wang et al., 2010).
To understand the role of TYLCV in preventing HR/CD, the Hsp90‐1 and Sgt1 genes were silenced using the TRV‐based VIGS system. All leaves of tomato plants at the four to six true leaf stage (4 weeks after sowing) were agroinfiltrated with the Hsp90‐1 and Sgt1 silencing constructs (coined TRV‐Hsp90 and TRV‐Sgt1, respectively), and with an empty TRV vector (TRV‐0), 30 plants for each treatment. Five days after agroinfiltration, 15 plants from each treatment were infected with TYLCV using viruliferous whiteflies. The levels of Hsp90‐1 and Sgt1 transcripts were estimated by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) immediately before and 5 days after TRV introduction, at the time at which one‐half of the plants were inoculated with TYLCV using viruliferous whiteflies. The levels of Hsp90‐1 and Sgt1 transcripts were estimated 7 and 15 days thereafter in uninfected (o:TRV‐Hsp90, o:TRV‐Sgt1, o:TRV‐0) and TYLCV‐infected (i:TRV‐Hsp90, i:TRV‐Sgt1, i:TRV‐0) plants.
Figure 1A shows the silencing patterns of Hsp90 and Sgt1 with time. Inoculation of empty TRV (TRV‐0) had no effect on the expression of Hsp90 and Sgt1. The expression of Hsp90 and Sgt1 was reduced by 35%–40%, 5 days after silencing of these genes; the silencing effect remained stable for the next 15 days. Although 5 days after the onset of TRV‐mediated silencing, the extent of RNA silencing was not optimal (Eybishtz et al., 2009), it was sufficient to significantly deplete the amount of target gene transcript; indeed, VIGS still developed during TYLCV infection. TYLCV infection did not significantly change the levels of silencing of Hsp90 and Sgt1, as shown previously for other tomato genes (Eybishtz et al., 2009, 2010; Sade et al., 2012).
Figure 1.
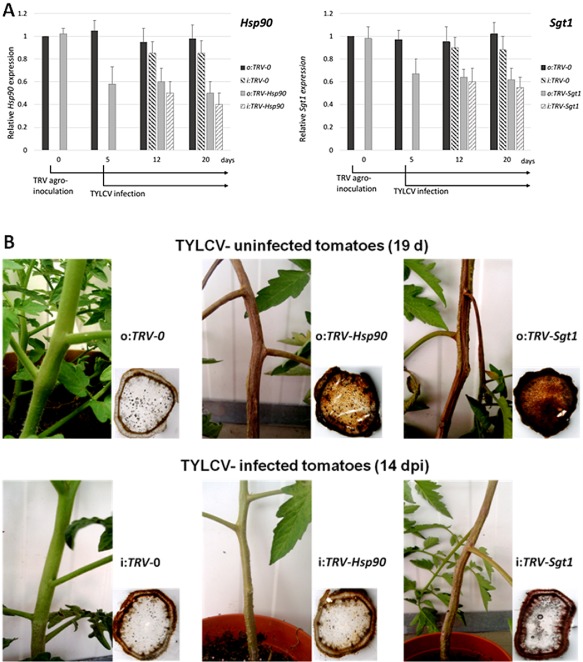
Silencing of tomato Hsp90/Sgt1 genes induces cell death (CD), which is alleviated in Tomato yellow leaf curl virus (TYLCV)‐infected plants. (A) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR)‐based expression analyses of Hsp90 (left panel) and Sgt1 (right panel) in tomato leaves agro‐infiltrated with the Tobacco rattle virus (TRV) vector alone (TRV‐0), and with the TRV Hsp90 and Sgt1 silencing constructs (TRV‐Hsp90 and TRV‐Sgt1), uninfected (o) and infected (i) with TYLCV. In TYLCV‐uninfected plants, qRT‐PCR was performed immediately before TRV inoculation (0), and 5, 12 and 20 days thereafter. Another set of tomato plants was infected with TYLCV 5 days after TRV inoculation; qRT‐PCR was performed 7 and 15 days (7, 15 dpi) thereafter (12 and 20 days after TRV inoculation). The relative expression of each gene was calculated in relation to TRV‐0 plants before TRV inoculation. The results were normalized using β‐actin as an internal marker. Bars represent the average and standard deviation of the relative expression from three technical repeats of three independent biological repeats. (B) Top panel: stems of tomato plants 19 days after agroinfiltration with TRV‐0, and with TRV‐Hsp90 and TRV‐Sgt1, not infected with TYLCV; bottom panel: stems of tomato plants 14 days after infection with TYLCV/19 days after agroinfiltration with TRV‐0, and with TRV‐Hsp90 and TRV‐Sgt1. 3,3′‐Diaminobenzidine (DAB) staining was performed on stem cross‐sections.
The growth of Hsp90‐ and Sgt1‐silenced tomatoes (o:TRV‐Hsp90, o:TRV‐Sgt1) started to deteriorate 2 weeks after inoculation with the silencing constructs. During the following week, the stems became necrotic (Fig. 1B), and the plants collapsed 27–28 days after inoculation with the silencing constructs. Silenced tomato plants of the same age, but infected with TYLCV (i:TRV‐Hsp90, i:TRV‐Sgt1), showed a much weaker necrosis on their stems (Fig. 1B), and were still alive and growing 27–28 days after the onset of silencing. Tomato plants treated with the TRV vector that did not carry any additional sequences (o:TRV‐0, i:TRV‐0) did not show necrosis on the stem, even after prolonged TYLCV infection (Fig. 1B).
CD symptoms in living tissues were visualized with 3,3′‐diaminobenzidine (DAB) (Fig. 1B). In the dying tissues, DAB was oxidized by the released H2O2 and appeared as a brown stain (Alvarez et al., 1998; Gorovits et al., 2007). TRV‐inoculated tomato stems (o:TRV‐0, i:TRV‐0) showed a ring of brownish colour of similar intensity, irrespective of TYLCV infection (shown in Fig. 1B at 14 dpi). Hsp90‐ and Sgt1‐silenced tomatoes (o:TRV‐Hsp90, o:TRV‐Sgt1) presented an intense brown stain all over the stem section. The stain was much less intense on TYLCV infection of silenced plants (i:TRV‐Hsp90, i:TRV‐Sgt1). The results indicate that TYLCV infection markedly alleviates the stress response experienced by tomato plants with silenced Hsp90 and Sgt1 genes (Fig. 1B).
Impairment of HSP90 and silencing of Hsp90/Sgt1‐enhanced protein ubiquitination; less enhancement in TYLCV‐infected plants
UPS is involved in the control of CD in plants (Lee et al., 2011; Li et al., 2012; Yang et al., 2006; Zeng et al., 2004). HSP90 has been shown to play a role in the assembly and maintenance of the 26S proteasome, and, consequently, in plant UPS activity. The accumulation of polyubiquitinated proteins in plant cells, induced by the inhibition of HSP90, has been attributed to an impairment of 26S proteasome function (Nishizawa‐Yokoi et al., 2010). Similar effects have been obtained in tomato plants treated with GDA, an inhibitor of HSP90 activity (Hahn et al., 2011).
As it is difficult to treat whole plants with GDA (infiltration or injection caused necrosis and CD, and the results were not reproducible), we used young detached leaves from TYLCV‐infected (at 21 dpi) and non‐infected tomato plants of the same age. The leaves were incubated with GDA (0.35 μm) for 24 h and total proteins were analysed by Western blot with antibodies raised against UbqPs and against the 26S proteasome 20S subunit (20S); an antibody against a chloroplast protein (OE33) served as an unrelated control (Fig. 2). The results were compared with mock‐treated plants (0) and with uninfected and TYLCV‐infected Hsp90‐ and Sgt1‐silenced plants (o:TRV‐Hsp90, o:TRV‐Sgt1 and i:TRV‐Hsp90, i:TRV‐Sgt1). The degree of polyubiquitination was estimated using protein slot blots (Fig. S1, see Supporting Information). Proteins extracted from GDA‐treated leaves were highly polyubiquitinated; the intensity of polyubiquitination was 2.5‐fold higher than that in mock‐treated leaves (0), and decreased by half in GDA‐treated, TYLCV‐infected leaves (22 and S1). The level of polyubiquitinated proteins in leaves of Hsp90‐ and Sgt1‐silenced tomatoes was also 2.4–2.8‐fold higher than in non‐silenced leaves; it decreased by 30%–40% in TYLCV‐infected plants (Figs 2 and S1). Furthermore, the release of the 20S proteasome subunit caused by the inhibition of HSP90, a marker of UPS inactivation (Yamano et al., 2008), was less pronounced in the presence of TYLCV (Fig. 2). It should be mentioned that TYLCV infection by itself, in the absence of HSP90 inhibition, did not notably affect the patterns of polyubiquitinated proteins and the abundance of the 20S proteasome.
Figure 2.
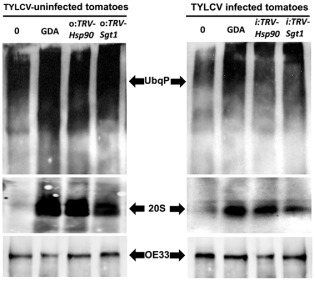
Inhibition of heat shock protein 90 (HSP90) and silencing of Hsp90/Sgt1 inactivate the ubiquitin–proteasome system (UPS); Tomato yellow leaf curl virus (TYLCV) infection down‐regulates proteasome inactivation. Western blot was applied to immunodetect ubiquitinated proteins (UbqP), the 20S core subunit of the 26S proteasome (20S) and OE33 chloroplast protein (OE33) as unrelated control in proteins extracted from tomato leaves [uninfected and TYLCV infected (21 days post‐infection, dpi)]. Protein patterns of detached leaves, treated with dimethylsulfoxide (DMSO) (0) and geldanamycin (GDA), were compared with proteins extracted from Hsp90‐ and Sgt1‐silenced tomatoes (TRV‐Hsp90 and TRV‐Sgt1).
Inhibition of HSP90 increased the accumulation of HSFA2 in heat‐treated tomato leaves and was mitigated in TYLCV‐infected tissues
UPS inactivation and the accumulation of polyubiquitinated proteins have been shown to trigger the induction of stress‐related transcription factors (HSFs), including HSFA2 (Yamada and Nishimura, 2008). However, plant HSP90 functions as a repressor of HSFs (Nishizawa‐Yokoi et al., 2010). Therefore, inactivation of HSP90 increased the expression of HSFA2 and, consequently, the expression of HSFA2‐regulated genes, as shown in Arabidopsis thaliana cell cultures (Nishizawa‐Yokoi et al., 2010).
We compared HSFA2 levels in protein extracts of detached tomato leaves on HSP90 inhibition in TYLCV‐infected and uninfected tomatoes (Fig. 3). The chloroplast protein OE33 detected with a specific antibody served as an unrelated control. In dimethylsulfoxide (DMSO)‐treated samples, considered as untreated controls, the levels of HSFA2 were low. As the expression of the HsfA2 gene increased under heat stress (Busch et al., 2005; Miller and Mittler, 2006; Nishizawa et al., 2006), we followed the fate of HSFA2 in detached tomato leaflets incubated at 42 °C for 2 h. The amount of HSFA2 greatly increased on heat treatment of the leaflets. It increased even more when the leaflets were treated with the HSP90 inhibitor GDA (0.35 μm) for 24 h prior to heat stress (Fig. 3A). The effect of short‐ and long‐term TYLCV infection (14 and 28 dpi) on HSFA2 amounts was investigated in tomatoes. At both stages of viral infection, the increase in HSFA2 levels on heat shock and GDA treatment was significantly mitigated in infected tissues (Fig. 3A).
Figure 3.
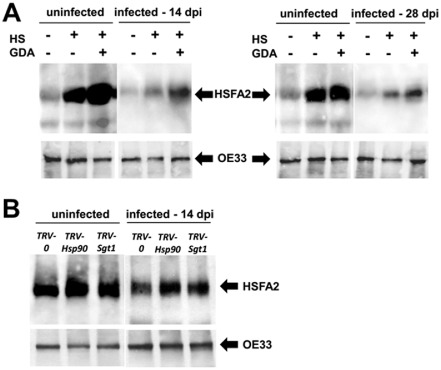
Impairment of heat shock protein 90 (HSP90) function increases the accumulation of HSFA2 in heat‐treated tomatoes; Tomato yellow leaf curl virus (TYLCV) infection mitigates this increase. Immunodetection of the transcription factor HSFA2 in leaf protein extracts following heat shock (HS) and TYLCV infection [uninfected and at 14 and 28 days post‐infection (dpi)]. The OE33 chloroplast protein (OE33) was used as unrelated control. (A) Detached leaves were treated with dimethylsulfoxide (DMSO) (GDA–) and with the HSP90 inhibitor geldanamycin (GDA+) and were subjected to heat shock (HS+) or incubated at room temperature (HS–) for 2 h. (B) Leaves of Hsp90‐ and Sgt‐silenced plants and Tobacco rattle virus (TRV)‐inoculated plants (TRV‐Hsp90 and TRV‐Sgt1; TRV‐0: empty vector) were subjected to HS.
Hsp90 and Sgt1 silencing increased HSFA2 amounts and HsfA2 expression; this increase was inhibited in TYLCV‐infected tissues
The levels of HSFA2 protein and HsfA2 gene expression were followed in tomato plants with silenced Hsp90 and Sgt1 genes. Detached leaves of tomatoes inoculated with empty TRV (TRV‐0) and the Hsp90‐1/Sgt1 silencing constructs (TRV‐Hsp90, TRV‐Sgt1), uninfected and infected with TYLCV (14 dpi), were subjected to heat shock (42 °C for 2 h). The silenced tomatoes showed increased levels of HSFA2 protein compared with TRV‐0 plants, whereas, in TYLCV‐infected plants, the increase was less pronounced (Fig. 3B).
The immunodetection of HSFA2 was paralleled with qRT‐PCR analyses to estimate the changes in HsfA2 transcription levels accompanying TYLCV infection (at 14 dpi). The expression levels of the other stress transcription factor HsfB1 and of the HSF‐regulated genes Hsp17 and Apx1/2 were also estimated (Fig. 4). TYLCV infection did not suppress the efficiency of Hsp90‐1 and Sgt1 gene silencing (see Fig. 1). Compared with uninfected plants, a decrease in the expression of Hsf and the HSF‐regulated genes (two‐ to six‐fold decrease for HsfA2, two‐ to seven‐fold for HsfB1, two‐ to nine‐fold for Hsp17 and two‐fold for Apx1/2) was detected in infected tomatoes (Fig. 4).
Figure 4.
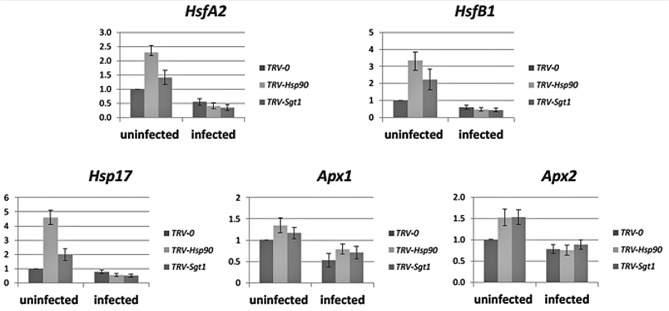
Silencing of the tomato Hsp90 and Sgt1 genes induces the accumulation of HsfA2 transcripts; Tomato yellow leaf curl virus (TYLCV) infection mitigates the accumulation of HsfA2 and the activation of HsfA2‐dependent gene expression. A quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) approach was used to analyse gene expression in uninfected and TYLCV‐infected (14 days post‐infection, dpi) tomatoes with Hsp90‐ and Sgt1‐silenced genes (TRV‐Hsp90 and TRV‐Sgt1); control plants were inoculated with the Tobacco rattle virus (TRV) vector alone (TRV‐0). For heat shock, the leaves were incubated at 42 °C for 2 h. The relative expression of each gene was calculated in relation to the TRV‐0 TYLCV‐uninfected sample. The results were normalized using the β‐actin gene as an internal marker. Bars represent the average and standard deviation of the relative expression from five independent biological repeats.
HSP90 inactivation resulted in increased TYLCV accumulation and CP aggregation
We monitored the accumulation of TYLCV DNA on HSP90 inhibition. Detached young leaflets of TYLCV‐infected tomatoes (21 dpi, infected at the four‐ to six‐leaf stage) were incubated for 24 h with GDA (0.25 and 0.35 μm) to inhibit HSP90; TYLCV DNA amounts were estimated by qPCR immediately thereafter. HSP90 inhibition led to increased levels of viral DNA (Fig. 5A). Similarly, TYLCV DNA amounts, measured at 10, 20 and 30 dpi, were higher in Hsp90‐ and Sgt1‐silenced tomatoes than in TRV‐inoculated control plants (Fig. 5B). The TYLCV CP was detected in GDA‐treated as well as in Hsp90/Sgt1‐silenced tomatoes using a specific antibody. The increase in CP amounts in these plants (Fig. 5C) was in agreement with the enhanced amounts of viral DNA (Fig. 5A,B). The increase in TYLCV DNA amounts in GDA‐treated plants provided indirect evidence of HSP90 inactivation.
Figure 5.
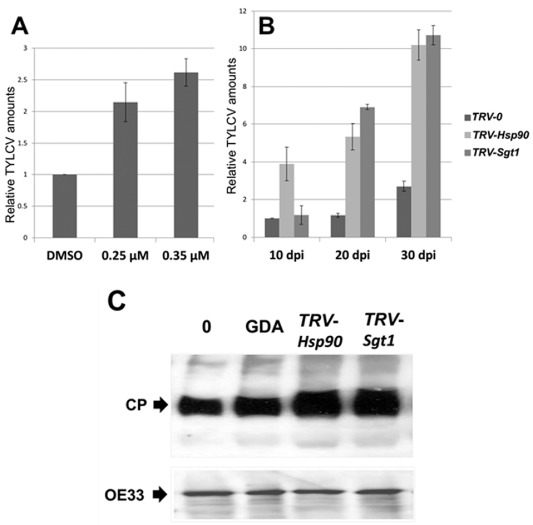
Heat shock protein 90 (HSP90) inactivation results in enhanced accumulation of Tomato yellow leaf curl virus (TYLCV). (A) Quantitative polymerase chain reaction (qPCR) estimation of TYLCV DNA amounts. The results were normalized using the tomato Expressed gene as an internal DNA marker. Bars represent the average and standard deviation of the relative expression from five independent biological repeats. Detached leaflets of TYLCV‐infected tomatoes (21 days post‐infection, dpi) were incubated for 24 h with 0.25 and 0.35 μm geldanamycin (GDA) to inhibit HSP90 [dimethylsulfoxide (DMSO) was used as the control without GDA]; TYLCV DNA amounts were estimated immediately thereafter. (B) Accumulation of TYLCV with time, estimated after 10, 20 and 30 dpi in tomatoes in which Hsp90 and Sgt1 had been silenced (TRV‐Hsp90 and TRV‐Sgt1); plants inoculated with the Tobacco rattle virus (TRV) vector alone (TRV‐0) served as control. (C) Immunodetection of TYLCV coat protein (CP) at 30 dpi in untreated tomato plants (0), in GDA‐inhibited HSP90 (GDA) plants and in tomatoes in which the Hsp90 and Sgt1 genes had been silenced (TRV‐Hsp90 and TRV‐Sgt1); the OE33 chloroplast protein (OE33) was used as unrelated control.
We have shown previously that the progress of TYLCV infection is accompanied by the accumulation of CP aggregates of increasing size in TYLCV‐infected tomato tissues (Gorovits et al., 2013a). Soluble and insoluble leaf proteins from TYLCV‐infected (21 dpi) Hsp90/Sgt1‐silenced and TRV‐0 tomato plants were resolved by ultracentrifugation on sucrose gradients (10%–50%) in non‐denaturing conditions (Fig. 6), a method allowing the separation of virus‐induced aggregates according to their size (Asurmendi et al., 2007; Bendahmane et al., 2007; Gorovits et al., 2013a, b). HSP70 patterns were used as protein control. TYLCV CP was not detected in TRV‐0‐uninfected tomatoes. The distribution of CP in gradients of TYLCV‐infected TRV‐0 tomatoes was similar to that of plants without TRV of similar age (Gorovits et al., 2013a), with most CP present in fractions containing mid‐size to large aggregates (fractions 8–10) (Fig. 6). In gradients of Hsp90/Sgt1‐silenced tomatoes, the abundance of CP increased; this increase was seen mostly in fractions containing large aggregates (9–10). Therefore, HSP90‐dependent UPS inactivation leads not only to an increase in the amounts of TYLCV DNA and CP, but also to an alteration in the degree of CP aggregation.
Figure 6.
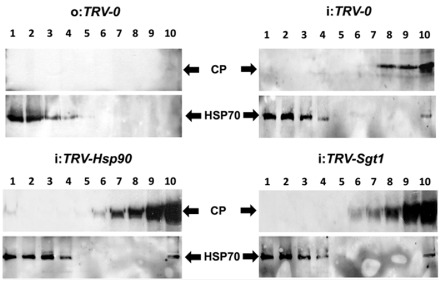
Heat shock protein 90 (HSP90) inactivation enhances the aggregation of Tomato yellow leaf curl virus (TYLCV) coat protein (CP). Immunodetection of TYLCV CP in tomato leaves (at 21 days post‐infection, dpi) distributed in a linear 10%–50% sucrose gradient; gradients were divided into 10 fractions, 1 (top) to 10 (bottom), and aliquots were subjected to sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). HSP70 was used as a marker of protein loading changes. No CP signal was detected in uninfected Tobacco rattle virus (TRV)‐inoculated tomatoes (o:TRV‐0); aggregated CP patterns were changed in Hsp90 (i:TRV‐Hsp90)‐ and Sgt1 (i:TRV‐Sgt1)‐silenced versus TRV‐inoculated (i:TRV‐0) plants.
HSP90 inactivation resulted in a size increase of the V2:GFP (V2:green fluorescent protein) aggregates
HSP90/UPS activities regulate not only the aggregation of TYLCV CP, but also of V2, the other gene product encoded on the TYLCV virion‐sense single‐stranded DNA. We have demonstrated that the state of V2 aggregation is dependent on the efficiency of 26S proteasome degradation: inhibition of the 26S proteasome results in an increase in size of the V2:GFP aggregates (Moshe et al., 2015). Hence, we wanted to determine whether HSP90 activity also influenced V2 aggregation that could be visualized under the fluorescence microscope. Nicotiana benthamiana leaves were infiltrated with the V2:GFP fusion. Transient expression of V2:GFP allowed the detection of fluorescent aggregates in the cytoplasm in contrast with the expression of GFP alone (Fig. 7A). Nicotiana benthamiana leaves expressing V2:GFP were treated with GDA to inhibit HSP90. The treatment caused a two‐fold increase in the size of the V2:GFP aggregates, on average, including large aggregates not seen in the control (Fig. 7B). When HSP90 and the 26S proteasome were inhibited simultaneously by treating leaves with GDA and MG132 (a cell‐permeable proteasome inhibitor), respectively, the effect of the double treatment on the size of the V2:GFP aggregates was the same as for HSP90 inhibition alone, confirming that HSP90 involvement in the aggregation/degradation of V2 was interconnected with UPS activity. Therefore, at least two TYLCV proteins may be degraded by the cellular HSP90‐dependent degradation machinery.
Figure 7.
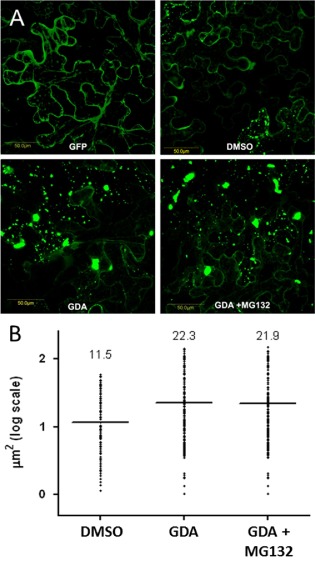
Heat shock protein 90 (HSP90) inactivation results in an increase in size of V2:GFP aggregates. (A) Expression of fusion V2:GFP in epidermal cells of Nicotiana benthamiana leaves infiltrated with Agrobacterium tumefaciens carrying the V2:GFP expression plasmid as observed with a laser confocal microscope. GFP, green fluorescent protein expression in leaves; DMSO, leaves infiltrated with dimethylsulfoxide as a control; GDA, leaves treated with geldanamycin to inhibit HSP90 activity; GDA + MG132, leaves treated with GDA and MG132 to inhibit HSP90 and 26S proteasome activities. Bar, 50 μm. (B) Sizes of aggregates. The microscope fields obtained in (A) were analysed with ImageJ; bars and numbers indicate the mean sizes of aggregates as measured for leaf areas of 50 mm2 in five independent experiments for each treatment.
Discussion
Plants have developed a variety of anti‐viral resistance mechanisms, either ready to meet incoming pathogens or induced by them, that are designed to minimize the damage caused by viral infection whilst quickly clearing the invading pathogen. CD is a commonly employed immune defence against viral infection, and many viruses potently induce or suppress CD during infection (Komatsu et al., 2010). In this hostile environment, begomoviruses prevent or counteract the plant defences, replicate in the phloem‐associated cells, and move to other cells and sites of the vascular system where they are acquired by their insect vectors (Hanley‐Bowdoin et al., 2013 and references cited therein). Geminiviruses probably suppress a range of host defence responses, which may include those that trigger CD.
A begomovirus, such as TYLCV, does not induce a macroscopic HR and CD in tomato leaves and stems until the very late stages of infection (50–60 dpi), at a time at which diseased tissues are senescent (Gorovits et al., 2007; Gorovits and Czosnek, 2007; Moshe et al., 2012). In comparison, the fungus S. sclerotiorum produces a CD‐related oxidative burst only a few hours after inoculation in the same tomato genotypes (Gorovits et al., 2007). In our hands, the only instance in which TYLCV infection caused CD was in a TYLCV‐resistant genotype in which the sucrose transporter gene LeHT1 and a Lipocalin‐like gene (two genes shown to be involved in TYLCV resistance) were silenced, but not in the silenced susceptible tomato near‐isogenic line (Eybishtz et al., 2010; Sade et al., 2012). We interpreted these observations as evidence that TYLCV induces the CD defence response only after the natural resistance to the virus has been significantly impaired.
We do not know how TYLCV suppresses the death of infected host plant cells or the cascade of events involved in this process, although many host genes associated with CD are up‐regulated (Ascencio‐Ibanez et al., 2008), indicating that geminiviruses effectively counteract the activation of CD pathways during infection. In the current study, we have shown that TYLCV suppresses CD induced by the inhibition of HSP90. Furthermore, the virus impairs the signal transduction pathways leading to CD, such as the UPS‐ and HSF‐regulated transcription of essential cellular stress genes. Plant HSP90 has been shown to play a key role in stress responses and resistance to viruses, nematodes and insects (reviewed by Xu et al., 2012). The inhibition of HSP90 activities, including the activities of the HSP90 co‐chaperone SGT1, leads to the appearance of CD signs in Arabidopsis and N. benthamiana (Liu et al., 2004; Wang et al., 2010). Silencing of Hsp90/Sgt1 genes in tomato plants is not an exception; obvious HR symptoms were detected by DAB staining in stem cross‐sections of silenced tomato (Fig. 1B). The effect of Hsp90/Sgt1 gene silencing was mitigated by TYLCV infection: instead of becoming even more diseased, the infected plants showed a dramatic reduction in the magnitude of CD. However, it should be noted that TYLCV infection does not suppress the silencing of Hsp90 and Hsgt1 (Fig. 1A). Therefore, TYLCV does not induce HR and CD in the infected tomatoes by itself, but is able to suppress CD, induced by the other stresses, such as inactivation of HSP90 signalling.
HSP90 and SGT1 are involved in the ubiquitination processes of defence response regulators, subsequently degraded by the 26S proteasome, in plants infected by RNA viruses (Seo et al., 2008). The UPS degradation pathway plays a role in R protein‐mediated defence, in the degradation of viral movement proteins and in the activity of viral RNA silencing suppressors (reviewed by Citovsky et al., 2009). An increase in the level of ubiquitinated proteins has been observed in plants with inhibited HSP90 and UPS activities (Nishizawa‐Yokoi et al., 2010). The inhibition of the 26S proteasome, followed by the accumulation of polyubiquitinated proteins, stimulates the expression of heat‐inducible genes in the cells of plants and mammals (Kim et al., 1999; Kurepa et al., 2008; Pena et al., 2007).
We have found that TYLCV infection of tomato plants by itself does not lead to significant changes in the pattern of polyubiquitinated proteins, but reduces the levels of protein ubiquitination and UPS inactivation caused by HSP90 inhibition (Fig. 2). Geminiviruses interact with the UPS pathways and alter them to achieve full infection. The Tomato yellow leaf curl Sardinia virus (TYLCSV) C2 interacts with the CSN5 subunit of the COP9 signalosome (CSN) complex in N. benthamiana, interfering with the derubylation activity of CSN. Consequently, the inactive CSN is unable to bind its target, a cullin–RING E3 ubiquitin–ligase enzyme complex, compromising the host cell polyubiquitination activity (Lozano‐Durán et al., 2011a, b). Impaired CSN function also suppresses jasmonic acid (JA)‐mediated defence responses and several other host processes in infected plants. The TYLCSV Rep protein interacts with the E2 small ubiquitin‐like modifier enzyme (SUMO)‐conjugating enzyme 1 (SCE1) (Castillo et al., 2004). Silencing of SCE1 or altering the Rep–SCE1 interaction reduces the accumulation of viral DNA, suggesting that the interaction is required for viral replication (Castillo et al., 2004). Transient Rep expression modifies the sumoylation state of selected host proteins that may play a role in viral replication (Sánchez‐Durán et al., 2011). Some monopartite begomoviruses, but not TYLCV, have been associated with betasatellites (Briddon et al., 2003). The betasatellite‐encoded protein βC1 binds to the tomato UPS element, the E2 enzyme ubiquitin‐conjugating enzyme 3 (UBC3), reducing the global accumulation of polyubiquitinated proteins and causing strong symptoms (Eini et al., 2009). Therefore, the capacity of geminiviruses to hijack the ubiquitination complexes represents a powerful strategy for the modulation of host function.
An active Hsp90 gene is required for normal plant development (Liu et al., 2004). One of the main regulators of HSP expression is the transcription factor HSFA2, which accumulates to high levels during heat stress and recovery (Baniwal et al., 2004; von Koskull‐Döring et al., 2007; Scharf et al., 2012), and whose expression is significantly increased under light and heat stress in Arabidopsis (Nishizawa et al., 2006). Indeed, the changes caused by high‐temperature stress involve the reprogramming of signal transduction components, transcription factors and proteins associated with the metabolism of ROS (reviewed by Grover et al., 2013). The involvement of HSP90 in the regulation of signal transduction via HSFA2 activation has been demonstrated in several plant species, including tomatoes (Hahn et al., 2011). In our hands, the levels of HSFA2 were rather low in leaves of uninfected tomato plants. The amounts of HSFA2 greatly increased on heat stress in uninfected tissues, and much less in TYLCV‐infected leaves (Fig. 3A). The inhibition of HSP90 activity caused an additional increase in HSFA2 expression (Figs 3 and 4). Subsequent TYLCV infection reduced HSFA2 levels, as well as the expression levels of HsfB1, Hsp17, Apx1 and Apx2 (Fig. 4).
TYLCV can suppress HSFA2 signalling, which is in charge of the key cellular stress response. These findings are in line with our previous results, which demonstrated that TYLCV does not induce, but rather reduces, the expression of several key stress response proteins, such as pathogenesis‐related proteins (PRs), mitogen‐activated protein kinases (MAPKs), proteases and various cellular HSPs (Gorovits et al., 2007; Moshe et al., 2012). TYLCV seems to moderate various plant stress responses to ensure an environment favourable to its multiplication. Indeed, in infected GDA‐treated and Hsp90/Sgt1‐silenced tomatoes, the amounts of TYLCV DNA were several times higher than in plants with intact HSP90‐dependent signalling (Fig. 5A,B).
It has already been shown that TYLCV invasion of susceptible tomato plants is accompanied by the accumulation of large quantities of CP and by increasing amounts of the large CP aggregates/inclusion bodies (Gorovits et al., 2013a, b). It has also been suggested that CP degradation is mediated by UPS in large aggregates (Gorovits et al., 2014). Hence, it is likely that the HSP90‐dependent reduction of UPS activity, and therefore of CP degradation, causes the accumulation of CP in tomato leaves (Fig. 5C). Furthermore, the accumulated CP is concentrated in large aggregates in tomato plants with silenced Hsp90/Sgt1 genes (Fig. 6). The inhibition of HSP90 function not only induces the aggregation of TYLCV CP, but also that of TYLCV V2. This can be best visualized in N. benthamiana infiltrated with the fusion protein V2:GFP, where the inhibition of HSP90 by GDA induces an increase in the size of V2 aggregates (Fig. 7). Leaves of N. benthamiana transiently expressing a CP:GFP show that the fluorescent CP accumulates only in the nuclei, independent of HSP90 inhibition (not shown). Taken together, the results indicate that treatments favouring the formation of large CP (and V2) aggregates enhance the accumulation of TYLCV (Figs 5, 6, 7).
HSP90‐dependent impairment of UPS not only increases the amounts of viral proteins, but also promotes their aggregation. The re‐localization of the key cellular chaperone HSP70 (Gorovits et al., 2013b) and the finding that 26S proteasome degradation is protein quality control associated in large TYLCV aggregates (Gorovits et al., 2014) point to a possible association of the systems with viral aggregates. The recruitment of chaperones by TYLCV may be required to reduce the deleterious effects of misfolded proteins and, consequently, the suppression of plant CD. UPS has previously been shown to promote geminiviral infection by the development of complexes between the viral Rep and the plant SUMO SCE (Sánchez‐Durán et al., 2011). The suggestion that nuclear aggregates, containing molecular chaperones and proteasome, can prevent the induction of CD (Woulfe, 2007) was confirmed by the identification of a proteolytic activity in nuclear foci (Iwata et al., 2005; Rockel et al., 2005). Mammalian viruses, such as Herpes simplex virus (HSV‐1), may protect themselves by inducing the formation of nuclear protein quality control centres to remodel or degrade potentially toxic aberrant nuclear proteins with the 26S proteasome, which otherwise would interfere with virus multiplication (Livingston et al., 2009). The appearance of TYLCV‐induced large aggregates, mostly pronounced in nuclei, is a feature of a successful TYLCV infection (Gorovits et al., 2013a).
In the current study, we have described the suppression effect of TYLCV infection on CD in tomato plants, induced by the inactivation of HSP90 and its co‐chaperone SGT1. Furthermore, the virus impairs the signal transduction pathways leading to CD, such as UPS‐ and HSF‐regulated transcription of essential cellular stress genes. The key processes described here are summarized in Fig. 8. Future studies will aim to advance the knowledge of the mechanisms by which plant virus TYLCV is able to restrain host cellular responses for its benefit and successful multiplication.
Figure 8.
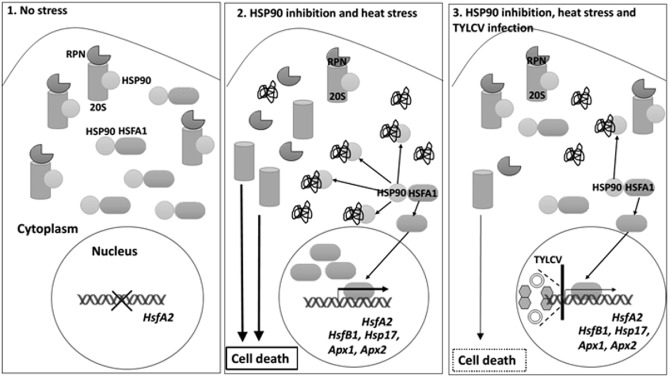
Summary of the key processes of stress response regulation by heat shock protein 90 (HSP90) in tomato plants, and down‐regulation by Tomato yellow leaf curl virus (TYLCV) infection. 1. In the absence of any stress, HSP90 binds HSFA1, one of the key regulators of the plant stress response, keeping it inactive; HSP90's substrate 26S proteasome (RPN regulatory subunits and the 20S catalytic subunit) is active. Therefore, there is no ubiquitin–proteasome system (UPS) inactivation, and consequently no cell death (CD) and no activation of HsfA2 signalling. 2. In heat‐stressed Hsp90‐silenced tomato plants, there is a massive appearance of damaged proteins, which sequester HSP90, causing the release of free HSFA1, which activates HsfA2 and downstream gene expression. The inhibition of HSP90 function also results in a dissociation of the 26S proteasome and a decrease in its peptidase activity. Inhibition of the 26S proteasome is accompanied by CD. 3. TYLCV infection (represented as virions and viral DNA) suppresses HSP90‐dependent 26S proteasome inactivation, CD and HSFA2 signal transduction pathways. Furthermore, loss of HSP90 function effects positively TYLCV accumulation: there are higher levels of viral DNA and CP, and the enrichment of CP/V2 in large aggregates, markers of a successful TYLCV infection.
Experimental Procedures
Sources of virus, insects and plants
TYLCV was maintained in tomato plants (cv. Daniella) by whitefly‐mediated inoculation. Whiteflies (B. tabaci B biotype) were reared on cotton plants grown in insect‐proof cages at 26 °C, as described by Zeidan and Czosnek (1991). All plants were grown in a temperature‐controlled glasshouse under standard rearing conditions. Tomato plants at their three to five true leaf stage were caged with viruliferous whiteflies (about 50 insects per plant at the onset of infection) for the duration of the experiments. Whiteflies were discarded before tissue sampling.
Gene silencing using a virus‐induced gene silencing (VIGS) vector
The silencing Tobacco rattle virus (TRV) vector was licensed from PBL (Norwich, UK). For Hsp90‐1 gene silencing, a fragment of 404 bp was amplified with the primers sense (5′‐AATTTATGGAGGCCATTGCT‐3′) and antisense (5′‐TTCATCAACCTCCTCTACCTTT‐3′) using tomato cDNA as template. For silencing of the Sgt1 gene, a fragment of 479 bp was amplified with the primers sense (5′‐ATGGCGTCCGATCTGG‐3′) and complementary sense (5′‐TTGACAGTTGGTTGAGCATCT‐3′), as described previously (Liu et al., 2002a). Both fragments were T/A cloned into the TOPO vector (Invitrogen, Grand Island, NY, USA), excised from the vector using EcoRI and XhoI and ligated to TRVII using the same enzymes. The plasmids were introduced by electroporation into Agrobacterium tumefaciens C58. The cells were grown at 28 °C on Luria–Bertani (LB) medium containing kanamycin and rifampicin (50 μg/mL each), as described previously (Liu et al., 2002b). All the leaves of tomato seedlings at the four to six true leaf stage (4 weeks after seeding) were infiltrated with a mixture of agrobacteria containing TRVI and TRVII‐Hsp90, TRVI and TRVII‐Sgt1, and TRVI and TRVII alone as control, using a needleless 1‐mL syringe. Five days later, the plants were caged with viruliferous whiteflies (about 30 insects per plant). Each silencing experiment was repeated at least three times.
Inhibition of HSP90 with GDA and of 26S proteasome with MG132
GDA (Enzo Life Science, Farmingdale, NY, USA) and carbobenzoxy‐Leu‐Leu‐leucinal (MG132, Calbiochem, Darmstadt, Germany) were dissolved in DMSO to prepare stock solutions of 250 μm. For in planta treatments, detached leaflets from infected tomato plants were placed for 24 h in micro‐tubes containing 0.25 or 0.35 μm GDA (two to three leaflets per tube, the tips of the petioles soaking in the solution). The same volume of water in DMSO was used as a control. For the treatment of N. benthamiana leaves transiently expressing V2:GFP, freshly prepared solutions of 0.25 μm GDA (an HSP90 inhibitor, Nakarai Tesque Inc., Kyoto, Japan) or 50 μm MG132 (a 26S proteasome inhibitor, APExBIO, Houston, TX, USA) plus 0.25 μm GDA were syringe infiltrated into the abaxial leaf side, 24 h after agroinfiltration. Leaves were examined under the microscope 15–20 h after inhibitor treatment. Control infiltrations were performed using the same final concentration of DMSO.
Extraction of proteins from tomato leaves
For analyses by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), proteins were extracted from minced leaves (pooled from three plants), frozen in liquid nitrogen and drill homogenized in a standard PAGE loading buffer supplemented with 2% SDS. Samples containing 50 μg protein each were subjected to 12% SDS‐PAGE.
Detection of H2O2
H2O2 accumulation was detected by endogenous peroxidase‐dependent in situ histochemical staining with DAB (Sigma, St. Louis, MO, USA), as described previously (Gorovits et al., 2007). Samples were mounted on microscope glass slides in 60% glycerol. H2O2 was detectable as a reddish brown precipitate. The intensity and pattern of DAB staining were assessed under an Olympus (Tokyo, Japan) SZET binocular coupled with a Nikon (Tokyo, Japan) LCD camera DXM1200 and photographed.
Sucrose density gradient analyses
Extracts of native proteins from 500 mg of tomato leaves, prepared as described by Gorovits et al. (2013a), were layered on 10‐mL linear 10%–50% sucrose gradients. After 20 h of centrifugation at 104 000 g at 4 °C [Beckman (Pasadena, CA, USA) SW27 rotor], the content of each gradient tube was divided into 10 fractions, as described previously (Asurmendi et al., 2007; Bendahmane et al., 2007; Gorovits et al., 2013a, b). Aliquots of 50 μL were subjected to 12% SDS‐PAGE and analysed by Western blot. Gradients from at least five experiments were analysed.
Immunodetection of viral and plant proteins
Western blotting was performed as described previously (Gorovits et al., 2013a, b) using antibodies against the TYLCV CP (Gorovits et al., 2013a, 2013b), HSP70 and ubiquitin (Agrisera, Vännäs, Sweden), proteasome 20S alpha‐beta (Abcam, Cambridge, UK), HSFA2 (a gift from Professor K. D. Scharf, Goethe University, Frankfurt‐am‐Main, Germany) and OE33 (a gift from Professor Z. Adam, The Hebrew University of Jerusalem, Rehovot, Israel). Incubation with the antibody was followed by ECL detection (Amersham, Amersham, UK). Each immunodetection was repeated at least three times for each set of plants (pooled tissues from three plants). Polyubiquitinated proteins were quantified using slot blots: 2 μg of leaf proteins per slot were subjected to detection with a specific antiserum; the relative density of the bands was calculated using ImageJ software (free download: http://imagej.nih.gov/ij/). The uninfected untreated sample was used as a standard of the relative amount set to unity.
DNA, RNA extractions and qPCR/qRT‐PCR
DNA was prepared from 100 mg of leaf tissue, as described previously (Gorovits et al., 2013a). For qPCR, 50 ng of DNA was used in triplicate for each DNA sample. RNA was prepared from 100 mg of leaf tissue using the Tri‐Reagent method (Sigma‐Aldrich, Grand Island, NY, USA); cDNA was prepared using the EZ‐first‐strand cDNA synthesis kit (Biological Industries, Beit HaEmek, Israel) according to the manufacturer. cDNA was used for qRT‐PCR in triplicate for each cDNA sample.
DNA and cDNA were subjected to qPCR and qRT‐PCR, respectively, in the presence of SYBR Green I (Takara, Kyoto, Japan), using a Corbett (Sydney, Australia) Research Rotor‐Gene 6000 cycler. The reaction was as follows: 30 s at 94 °C, followed by 40 cycles consisting of 10 s at 94 °C, 30 s at 59 °C and 20 s at 72 °C. The primers used to amplify a 182‐bp fragment of TYLCV (X15656) were sense (5′‐TCTGTTCAAGGATTTCGTTG‐3′) and complementary sense (5′‐GCTGTCGAAGTTCAGCCTTC‐3′). As an internal reference, a 183‐bp fragment of the tomato ‘Expressed’ housekeeping gene (SGN‐U346908) was amplified using the sense (5′‐CTAAGAACGCTGGACCTAATG‐3′) and complementary sense (5′‐TGGGTGTGCCTTTCTGAATG‐3′) primers. For the tomato β‐actin gene (TC178617), a 180‐bp fragment was amplified using the sense (5′‐GGAAAAGCTTGCCTATGTGG‐3′) and complementary sense (5′‐CCTGCAGCTTCCATACCAAT‐3′) primers. For tomato HsfA2 (CAA47870), a 66‐bp fragment was amplified using the sense (5′‐ACCTTGTGGATCAGCTTGGTTTCC‐3′) and complementary sense (5′‐AATAGTGGAGGAGGCCAGAGGAAC‐3′) primers. For tomato HsfB1 (CAA39034), a 74‐bp fragment was amplified using the sense (5′‐GGTGCAGGCGAAGAAACAATGC‐3′) and complementary sense (5′‐TCATATCGGGTGCAACCTTCACG‐3′) primers. For tomato Hsp17‐C1 (AJ225046), a 77‐bp fragment was amplified using the sense (5′‐ACTTGGCATCGTGTGGAACG‐3′) and complementary sense (5′‐TGATCCATCTTTGCGTTCTCTGG‐3′) primers. For tomato Apx1 (DQ099420), a 71‐bp fragment was amplified using the sense (5′‐ACGATGATATTGTGACACTCTTCCA‐3′) and complementary sense (5′‐AAGCGATGAAACCACAAAAACA‐3′) primers. For tomato Apx2 (DQ099421), a 190‐bp fragment was amplified using the sense (5′‐TGGGAGGGTGGTGACATATTTT‐3′) and complementary sense (5′‐TTGAAGTGCATAACTTCCCATCTTT‐3′) primers. For the Hsp90‐1 transcript (SGN‐U312354), a fragment of 77 bp was amplified using the sense (5′‐TGCGTTCTTGTATGGAAGTCTGC‐3′) and antisense (5′‐TGGACCACTTAGTCACGACCAATC‐3′) primers. For the Sgt1 transcript (SGN‐U596737), a fragment of 284 bp was amplified using the sense (5′‐CTGTAGTGCAGAGGCCTAATGT‐3′) and antisense (5′‐ GCTTCCTTCGACCTTCTTTG‐3′) primers.
Transient expression of V2:GFP in N. benthamiana
Agrobacterium tumefaciens transformed with pBIN19‐GFP or pBIN19‐V2:GFP, provided by Dr Gafni, Volcani Center, Bet‐Dagan, Israel (Glick et al., 2008), was used to infiltrate N. benthamiana leaves. The bacteria were grown for 48 h at 28 °C in LB medium containing 50 μg/mL kanamycin. The culture was centrifuged at 4000 g for 8 min, and the pellet was re‐suspended in water to a final optical density at 600 nm (OD600) of 0.6. Young leaves of 3‐week‐old plants were infiltrated with the suspension using a needleless syringe. The V2:GFP fusion and GFP proteins were visible under the fluorescence microscope 48 h after infiltration.
Measurement of number and size of aggregates
To determinate the cross‐sectional areas of aggregates, images were imported into ImageJ (free download: http://imagej.en.softonic.com/). The number of aggregates per area and their size were estimated using the ‘threshold’ and ‘particle analyzer’ functions of the program.
Supporting information
Fig. S1 A comparison of protein ubiquitination levels in tomato leaves of Tomato yellow leaf curl virus (TYLCV)‐infected and ‐uninfected tomatoes. Each lane of the slot blot contains 2 μg of total protein. The anti‐ubiquitin antiserum detected ubiquitinated protein (UbqP) levels in untreated leaves (0), those treated with geldanamycin (GDA) and in those silenced in Hsp90 and Sgt1 genes (TRV‐Hsp90 and TRV‐Sgt1) versus plants containing the Tobacco rattle virus (TRV) vector (TRV‐0). Leaf samples were taken from TYLCV‐uninfected and ‐infected (21 days post‐infection) tomatoes. The relative density of the bands was calculated using ImageJ software. The uninfected untreated sample was used as a standard with its relative density set to unity.
Acknowledgements
We thank Eduard Belausov (Institute of Plant Sciences, Volcani Center, Bet Dagan, Israel) for excellent technical assistance in fluorescent confocal microscopy studies and Klaus‐Dieter Scharf (Goethe University, Frankfurt‐am‐Main, Germany) for the anti‐HSFA2 antibody. This research was supported by a grant from the US Agency for International Development, Middle East Research and Cooperation (MERC) program (GEG‐G‐00‐02‐00003‐00).
References
- Alvarez, M.E. , Pennell, R.I. , Meijer, P.J. , Ishikawa, A. , Dixon, R.A. and Lamb, C. (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell, 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Ascencio‐Ibanez, J.T. , Sozzani, R. , Lee, T.J. , Chu, T.M. , Wolfinger, R.D. , Cella, R. and Hanley‐Bowdoin, L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asurmendi, S. , Berg, R.H. , Smith, T.J. , Bendahmane, M. and Beachy, R.N. (2007) Aggregation of TMV CP plays a role in CP functions and in coat‐protein‐mediated resistance. Virology, 366, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliji, S. , Lacatus, G. and Sunter, G. (2010) The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin‐responsive genes. Virology, 402, 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal, S.K. , Bharti, K. , Chan, K.Y. , Fauth, M. , Ganguli, A. , Kotak, S. , Mishra, S.K. , Nover, L. , Port, M. , Scharf, K.D. , Tripp, J. , Weber, C. , Zielinski, D. and von Koskull‐Döring, P. (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. [DOI] [PubMed] [Google Scholar]
- Ben Rejeb, I. , Pastor, V. and Mauch‐Mani, B. (2014) Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants, 3, 458–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, M. , Chen, I. , Asurmendi, S. , Bazzini, A.A. , Szecsi, J. and Beachy, R. (2007) Coat protein‐mediated resistance to TMV infection of Nicotiana tabacum involves multiple modes of interference by coat protein. Virology, 366, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ber, R. , Navot, N. , Zamir, D. , Antignus, Y. , Cohen, S. and Czosnek, H. (1990) Infection of tomato by the tomato yellow leaf curl virus: susceptibility to infection, symptom development and accumulation of viral DNA. Arch. Virol. 112, 169–180. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. , Bull, S.E. , Amin, I. , Idris, A.M. , Mansoor, S. , Bedford, I.D. , Dhawan, P. , Narayan Rishi, N. , Siwatch, S.S. , Abdel‐Salam, A.M. , Brown, J.K. , Zafar, Y. and Markham, P.G. (2003) Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology, 312, 106–121. [DOI] [PubMed] [Google Scholar]
- Busch, W. , Wunderlich, M. and Schöffl, F. (2005) Identification of novel heat shock factor‐dependent genes and biochemical pathways in Arabidopsis thaliana . Plant J. 41, 1–14. [DOI] [PubMed] [Google Scholar]
- Castillo, A.G. , Kong, L.J. , Hanley‐Bowdoin, L. and Bejarano, E.R. (2004) Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78, 2758–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Lv, Y. , Zhao, T. , Li, N. , Yang, Y. , Yu, W. , He, X. , Liu, T. and Zhang, B. (2013) Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by Tomato yellow leaf curl virus . PLoS ONE, 8, e80816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V. , Zaltsman, A. , Kozlovsky, S.V. , Gafni, Y. and Krichevsky, A. (2009) Proteasomal degradation in plant–pathogen interactions. Semin. Cell Dev. Biol. 20, 1048–1054. [DOI] [PubMed] [Google Scholar]
- Czosnek, H. (2007) Tomato yellow leaf curl virus disease In: Management, Molecular Biology, Breeding for Resistance (Czosnek H., ed.), p. 440 Dordrecht: Springer. [Google Scholar]
- Dickman, M.B. and de Figueiredo, P. (2013) Death be not proud—cell death control in plant fungal interactions. PLoS Pathog. 9, e1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eini, O. , Dogra, S. , Selth, L.A. , Dry, I.B. , Randles, J.W. and Rezaian, M.A. (2009) Interaction with a host ubiquitin‐conjugating enzyme is required for the pathogenicity of a geminiviral DNA β satellite. Mol. Plant–Microbe Interact. 22, 737–746. [DOI] [PubMed] [Google Scholar]
- Eybishtz, A. , Peretz, Y. , Sade, D. , Akad, F. and Czosnek, H. (2009) Silencing of a single gene in tomato plants resistant to Tomato yellow leaf curl virus renders them susceptible to the virus. Plant Mol. Biol. 71, 157–171. [DOI] [PubMed] [Google Scholar]
- Eybishtz, A. , Peretz, Y. , Sade, D. , Gorovits, R. and Czosnek, H. (2010) Tomato yellow leaf curl virus (TYLCV) infection of a resistant tomato line with a silenced sucrose transporter gene LeHT1 results in inhibition of growth, enhanced virus spread and necrosis. Planta, 231, 537–548. [DOI] [PubMed] [Google Scholar]
- Freeman, B.C. and Beattie, G.A. (2008) An overview of plant defenses against pathogens and herbivores. Plant Health Instructor. doi: 10.1094/PHI-I-2008-0226-01. [DOI] [Google Scholar]
- Glick, E. , Zrachya, A. , Levy, Y. , Mett, A. , Gidoni, D. , Belausov, E. , Citovsky, V. and Gafni, Y. (2008) Interaction with host SGS3 is required for suppression of RNA silencing by Tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA, 105, 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, A.L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature, 426, 895–899. [DOI] [PubMed] [Google Scholar]
- Gorovits, R. and Czosnek, H. (2007) Biotic and abiotic stress responses in breeding tomato lines resistant and susceptible to Tomato yellow leaf curl virus In: Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology and Breeding for Resistance (Czosnek H., ed.), pp. 223–237. Dordrecht: Springer. [Google Scholar]
- Gorovits, R. , Akad, F. , Beery, H. , Vidavsky, F. , Mahadav, A. and Czosnek, H. (2007) Expression of stress‐response proteins upon whitefly‐mediated inoculation of Tomato yellow leaf curl virus (TYLCV) in susceptible and resistant tomato plants. Mol. Plant–Microbe Interact. 20, 1376–1383. [DOI] [PubMed] [Google Scholar]
- Gorovits, R. , Moshe, A. , Kolot, M. , Sobol, I. and Czosnek, H. (2013a) Progressive aggregation of Tomato yellow leaf curl virus coat protein in systemically infected tomato plants, susceptible and resistant to the virus. Virus Res. 171, 33–43. [DOI] [PubMed] [Google Scholar]
- Gorovits, R. , Moshe, A. , Ghanim, M. and Czosnek, H. (2013b) Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS ONE, 8, e70280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits, R. , Moshe, A. , Ghanim, M. and Czosnek, H. (2014) Degradation mechanisms of the Tomato yellow leaf curl virus coat protein following inoculation of tomato plants by the whitefly Bemisia tabaci . Pest Manag. Sci. 70, 1632–1639. [DOI] [PubMed] [Google Scholar]
- Grover, A. , Mittal, D. , Negi, M. and Lavania, D. (2013) Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci. 205, 38–47. [DOI] [PubMed] [Google Scholar]
- Hahn, A. , Bublak, D. , Schleiff, E. and Scharf, K.D. (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell, 23, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley‐Bowdoin, L. , Settlage, S.B. , Orozco, B.M. , Nagar, S. and Robertson, D. (2000) Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35, 105–140. [PubMed] [Google Scholar]
- Hanley‐Bowdoin, L. , Bejarano, E.R. , Robertson, D. and Mansoor, S. (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Hussain, M. , Mansoor, S. , Iram, S. , Zafar, Y. and Briddon, R.W. (2007) The hypersensitive response to Tomato leaf curl New Delhi virus nuclear shuttle protein is inhibited by transcriptional activator protein. Mol. Plant–Microbe Interact. 20, 1581–1588. [DOI] [PubMed] [Google Scholar]
- Imai, J. , Maruya, M. , Yashiroda, H. , Yahara, I. and Tanaka, K. (2003) The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22, 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, A. , Christianson, J.C. , Bucci, M. , Ellerby, L.M. , Nukina, N. , Forno, L.S. and Kopito, R.R. (2005) Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. USA, 102, 13 135–13 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe, Y. , Nakai, A. , Tanabe, M. and Nagata, K. (1998) Proteasome inhibition leads to the activation of all members of the heat‐shock‐factor family. Eur. J. Biochem. 255, 356–362. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Kim, S.H. and Li, G.C. (1999) Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem. Biophys. Res. Commun. 254, 264–268. [DOI] [PubMed] [Google Scholar]
- Kim, M. , Ahn, J.W. , Jin, U.H. , Choi, D. , Paek, K.H. and Pai, H.S. (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J. Biol. Chem. 278, 19 406–19 415. [DOI] [PubMed] [Google Scholar]
- Komatsu, K. , Hashimoto, M. , Ozeki, J. , Yamaji, Y. , Maejima, K. , Senshu, H. , Himeno, M. , Okano, Y. , Kagiwada, S. and Namba, S. (2010) Viral‐induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol. Plant–Microbe Interact. 23, 283–293. [DOI] [PubMed] [Google Scholar]
- von Koskull‐Döring, P. , Scharf, K.D. and Nover, L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. [DOI] [PubMed] [Google Scholar]
- Kurepa, J. , Toh‐E, A. and Smalle, J.A. (2008) 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53, 102–114. [DOI] [PubMed] [Google Scholar]
- Lam, E. (2004) Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5, 305–315. [DOI] [PubMed] [Google Scholar]
- Lee, D.H. , Choi, H.W. and Hwang, B.K. (2011) The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid‐dependent defense response. Plant Physiol. 156, 2011–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Ahn, I.P. , Ning, Y. , Park, C.H. , Zeng, L. , Whitehill, J.G. , Lu, H. , Zhao, Q. , Ding, B. , Xie, Q. , Zhou, J.M. , Dai, L. and Wang, G.L. (2012) The U‐Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2002a) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Serino, G. , Deng, X.W. and Dinesh‐Kumar, S.P. (2002b) Role of SCF ubiquitin‐ligase and the COP9 signalosome in the N gene‐mediated resistance response to Tobacco mosaic virus. Plant Cell, 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Burch‐Smith, T. , Schiff, M. , Feng, S. and Dinesh‐Kumar, S.P. (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. [DOI] [PubMed] [Google Scholar]
- Livingston, C.M. , Ifrim, M.F. , Cowan, A.E. and Weller, S.K. (2009) Virus‐Induced Chaperone‐Enriched (VICE) domains function as nuclear protein quality control centers during HSV‐1 infection. PLoS Pathog. 5, e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Durán, R. , Rosas‐Díaz, T. , Gusmaroli, G. , Luna, A.P. , Taconnat, L. , Deng, X.W. and Bejarano, E.R. (2011a) Geminiviruses subvert ubiquitination by altering CSN‐mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana . Plant Cell, 23, 1014–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Durán, R. , Rosas‐Díaz, T. , Luna, A.P. and Bejarano, E.R. (2011b) Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE, 6, e22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga, W.J. and Stone, S.L. (2011) Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616. [DOI] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.‐B. (2013) Plant immune responses against viruses: how does a virus cause disease. Plant Cell, 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G. and Mittler, R. (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 98, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzi, L. , Napoli, C. , Sardo, L. and Accotto, G.P. (2014) Transcriptomics of the interaction between the monopartite phloem‐limited geminivirus Tomato yellow leaf curl Sardinia virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS ONE, 9, e89951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe, A. , Pfannstiel, J. , Brotman, Y. , Kolot, M. , Sobol, I. , Czosnek, H. and Gorovits, R. (2012) Stress responses to Tomato yellow leaf curl virus (TYLCV) infection of resistant and susceptible tomato plants are different. Metabolomics, S1, 006. doi: 10.4172/2153-0769.S1-006. [DOI] [Google Scholar]
- Moshe, A. , Belausov, E. , Niehl, A. , Heinlein, M. , Czosnek, H. and Gorovits, R. (2015) The Tomato yellow leaf curl virus V2 protein forms aggregates depending on the cytoskeleton integrity and binds viral genomic DNA. Sci. Rep. 5, 9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubin, M. , Amin, I. , Amrao, L. , Briddon, R.W. and Mansoor, S. (2010) The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 11, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa, A. , Yabuta, Y. , Yoshida, E. , Maruta, T. , Yoshimura, K. and Shigeoka, S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. [DOI] [PubMed] [Google Scholar]
- Nishizawa‐Yokoi, A. , Tainaka, H. , Yoshida, E. , Tamoi, M. , Yabuta, Y. and Shigeoka, S. (2010) The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 51, 486–496. [DOI] [PubMed] [Google Scholar]
- Pena, L.B. , Pasquini, L.A. , Tomaro, M.L. and Gallego, S.M. (2007) 20S proteasome and accumulation of oxidized and ubiquitinated proteins in maize leaves subjected to cadmium stress. Phytochemistry, 68, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Priyadarshini, P. , Ambika, C.G. , Tippeswamy, M.V. and Savithri, H.S. (2011) Functional characterization of coat protein and V2 involved in cell to cell movement of cotton leaf curl Kokhran virus‐Dabawali. PLoS ONE, 6, e26929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel, T.D. , Stuhlmann, D. and von Mikecz, A. (2005) Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 118, 5231–5242. [DOI] [PubMed] [Google Scholar]
- Sade, D. , Eybishtz, A. , Gorovits, R. , Sobol, I. and Czosnek, H. (2012) A developmentally regulated lipocalin‐like gene is overexpressed in Tomato yellow leaf curl virus‐resistant tomato plants upon virus inoculation, and its silencing abolishes resistance. Plant Mol. Biol. 80, 273–287. [DOI] [PubMed] [Google Scholar]
- Sade, D. , Brotman, Y. , Eybishtz, A. , Cuadros‐Inostroza, A. , Fernie, A.R. , Willmitzer, L. and Czosnek, H. (2013) Involvement of the hexose transporter gene LeHT1 and of sugars in resistance of tomato to Tomato yellow leaf curl virus . Mol. Plant 5, 1707–1710. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Durán, M.A. , Dallas, M.B. , Ascencio‐Ibañez, J.T. , Reyes, M.I. , Arroyo‐Mateos, M. , Ruiz‐Albert, J. , Hanley‐Bowdoin, L. and Bejarano, E.R. (2011) Interaction between geminivirus replication protein and the SUMO‐conjugating enzyme is required for viral infection. J. Virol. 85, 9789–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, K.D. , Berberich, T. , Ebersberger, I. and Nover, L. (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta, 1819, 104–119. [DOI] [PubMed] [Google Scholar]
- Seo, Y.S. , Lee, S.K. , Song, M.Y. , Suh, J.P. , Hahn, T.R. , Ronald, P. and Jeon, J.S. (2008) The HSP90–SGT1–RAR1 molecular chaperone complex: a core modulator in plant immunity. J. Plant Biol. 51, 1–10. [Google Scholar]
- Settlage, S.B. , See, R.G. and Hanley‐Bowdoin, L. (2005) Geminivirus C3 protein: replication enhancement and protein interactions. J. Virol. 79, 9885–9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, M.Y. and Goldberg, A.L. (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron, 29, 15–32. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Rivero, R.M. , Shulaev, V. , Blumwald, E. and Mittler, R. (2014) Abiotic and biotic stress combinations. New Phytol. 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Trinks, D. , Rajeswaran, R. , Shivaprasad, P.V. , Akbergenov, R. , Oakeley, E.J. , Veluthambi, K. , Hohn, T. and Pooggin, M.M. (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79, 2517–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. (2012) Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 3, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges, D. , Zwickl, P. and Baumeister, W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Li, F. , Huang, C. , Yang, X. , Qian, Y. , Xie, Y. and Zhou, X. (2014a) V2 of Tomato yellow leaf curl virus can suppress methylation‐mediated plant transcriptional gene silencing. J. Gen. Virol. 95, 225–230. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Sun, N. , Deng, T. , Zhang, L. and Zuo, K. (2014b) Genome‐wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genomics, 15, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Uppalapati, S.R. , Zhu, X. , Dinesh‐Kumar, S.P. and Mysore, K.S. (2010) SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant–pathogen interactions. Mol. Plant Pathol. 11, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel, R. , Dong, X. , Blake, P. , Stanley, J. and Hong, Y. (2002) Differential roles of geminivirus Rep and AC4 (C4) in the induction of necrosis in Nicotiana benthamiana . Mol. Plant Pathol. 3, 461–471. [DOI] [PubMed] [Google Scholar]
- Woulfe, J.M. (2007) Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 33, 2–42. [DOI] [PubMed] [Google Scholar]
- Xu, Z.S. , Li, Z.Y. , Chen, Y. , Chen, M. , Li, L.C. and Ma, Y.Z. (2012) Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 13, 15 706–15 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. and Nishimura, M. (2008) Cytosolic heat shock protein 90 regulates heat shock transcription factor in Arabidopsis thaliana . Plant Signal. Behav. 3, 660–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano, T. , Mizukami, S. , Murata, S. , Chiba, T. , Tanaka, K. and Udono, H. (2008) Hsp90‐mediated assembly of the 26S proteasome is involved in major histocompatibility complex class I antigen processing. J. Biol. Chem. 283, 28 060–28 065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.W. , González‐Lamothe, R. , Ewan, R.A. , Rowland, O. , Yoshioka, H. , Shenton, M. , Ye, H. , O'Donnell, E. , Jones, J.D. and Sadanandom, A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U‐BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell, 18, 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan, M. and Czosnek, H. (1991) Acquisition of tomato yellow leaf curl virus by the whitefly Bemisia tabaci . J. Gen. Virol. 72, 2607–2614. [DOI] [PubMed] [Google Scholar]
- Zeng, L.R. , Qu, S. , Bordeos, A. , Yang, C. , Baraoidan, M. , Yan, H. , Xie, Q. , Nahm, B.H. , Leung, H. and Wang, G.L. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U‐box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Dong, J. , Xu, Y. and Wu, J. (2012) V2 protein encoded by tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 163, 51–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 A comparison of protein ubiquitination levels in tomato leaves of Tomato yellow leaf curl virus (TYLCV)‐infected and ‐uninfected tomatoes. Each lane of the slot blot contains 2 μg of total protein. The anti‐ubiquitin antiserum detected ubiquitinated protein (UbqP) levels in untreated leaves (0), those treated with geldanamycin (GDA) and in those silenced in Hsp90 and Sgt1 genes (TRV‐Hsp90 and TRV‐Sgt1) versus plants containing the Tobacco rattle virus (TRV) vector (TRV‐0). Leaf samples were taken from TYLCV‐uninfected and ‐infected (21 days post‐infection) tomatoes. The relative density of the bands was calculated using ImageJ software. The uninfected untreated sample was used as a standard with its relative density set to unity.


