Summary
Several plant viruses encode movement proteins (MPs) classified in the 30K superfamily. Despite a great functional diversity, alignment analysis of MP sequences belonging to the 30K superfamily revealed the presence of a central core region, including amino acids potentially critical for MP structure and functionality. We performed alanine‐scanning mutagenesis of the Ourmia melon virus (OuMV) MP, and studied the effects of amino acid substitutions on MP properties and virus infection. We identified five OuMV mutants that were impaired in systemic infection in Nicotiana benthamiana and Arabidopsis thaliana, and two mutants showing necrosis and pronounced mosaic symptoms, respectively, in N. benthamiana. Green fluorescent protein fusion constructs (GFP:MP) of movement‐defective MP alleles failed to localize in distinct foci at the cell wall, whereas a GFP fusion with wild‐type MP (GFP:MPwt) mainly co‐localized with plasmodesmata and accumulated at the periphery of epidermal cells. The movement‐defective mutants also failed to produce tubular protrusions in protoplasts isolated from infected leaves, suggesting a link between tubule formation and the ability of OuMV to move. In addition to providing data to support the importance of specific amino acids for OuMV MP functionality, we predict that these conserved residues might be critical for the correct folding and/or function of the MP of other viral species in the 30K superfamily.
Keywords: 30K superfamily, movement protein, Ourmia melon virus, Ourmiavirus, plasmodesmata, tubule
Introduction
Successful infection of a plant host by a virus derives from its ability to spread from the initial infection site to distant tissues and organs. This process involves various steps in different cellular compartments: intracellularly from the replication site to the cell periphery, through the cell wall to the adjacent cell, across tissues to the vascular system (usually phloem sieve tubes) and through the conductive elements to distant host tissues, where the virus can initiate systemic infection (Harries and Nelson, 2008; Hull, 2014; Schoelz et al., 2011; Tilsner et al., 2014; Ueki and Citovsky, 2007). In all the intercellular stages, viruses must traverse a substantial barrier that surrounds the host cells: the plant cell wall. For this purpose, plant viruses have evolved a special class of proteins, called movement proteins (MPs), which facilitate the cell‐to‐cell transport of viral ribonucleoprotein complexes or viral particles to and through the plasmodesmata, thus enabling viral cell‐to‐cell and systemic spread (Benitez‐Alfonso et al., 2010; Carrington et al., 1996; Lucas, 2006; Scholthof, 2005; Taliansky et al., 2008). Attractive theories have been formulated to explain the origin and evolution of plant viral proteins involved in movement (Lucas, 2006; Melcher, 2000; Mushegian and Elena, 2015; Scholthof, 2005; Tilsner et al., 2014). Although MPs and viral movement can generally be categorized into subgroups sharing some mechanistic or molecular properties, many exceptions and differences exist in the details of movement processes in different virus–host systems (Hull, 2014), and the currently accepted MP classification has shown some limitations in describing the fine tuning accomplished by each virus in its host to move from cell to cell and long distance.
Ourmia melon virus (OuMV) was first reported in the late 1980s on melon crops in the Ourmia district of Iran (Lisa et al., 1988). The virus is the type species of the genus Ourmiavirus, which includes two other species: Epirus cherry virus (EpCV) and Cassava virus C (CaVC) (Rastgou et al., 2009). In addition to Cucurbitaceae, in experimental conditions, OuMV can be mechanically transmitted to a wide range of dicotyledonous plants (more than 30 species in 14 families), including many Nicotiana species, Capsicum spp. and Lycopersicum spp. (Lisa et al., 1988). Infection of the model plant Arabidopsis thaliana has been demonstrated recently (Rossi et al., 2015). The natural spread of the virus in several regions in Iran has been reported (Gholamalizadeh et al., 2008), but no biological vector has been implicated to date in natural plant‐to‐plant transmission (Rastgou et al., 2011).
OuMV virion morphology is unique, with bacilliform particles of 18 nm in diameter, showing apparently hemi‐icosahedral ends and cylindrical bodies of three different lengths (Rastgou et al., 2009). The virus has a positive single‐strand RNA tripartite genome. Each genomic RNA is mono‐cistronic: the largest RNA segment (RNA‐1) encodes an RNA‐dependent RNA polymerase (RdRp), RNA‐2 encodes an MP and RNA‐3 encodes the 22‐kDa coat protein (CP) (Rastgou et al., 2009). With only three virus‐encoded proteins, ourmiaviruses are arguably amongst the simplest, non‐defective viruses able to complete a full infection cycle.
In addition to their unique morphology, ourmiaviruses are in a peculiar phylogenetic position amongst plant viruses. Phylogenetic analysis has shown that their RdRp is distantly related to viruses in the family Narnaviridae (Rastgou et al., 2009), whose members mainly infect fungi, including the yeast Saccharomyces cerevisiae (Hillman and Cai, 2013). Notably, on the basis of RdRp homology, fungal Narnaviridae and plant ourmiaviruses appear to be the sole descendants of Leviviridae, the only known group of bacteriophages with a positive RNA genome (Koonin et al., 2015). Phylogenetic analysis of the OuMV MP protein showed distant similarity with plant‐infecting viruses in the Tombusvirus and Aureusvirus genera (Rastgou et al., 2009). The origin of the CP remains uncertain, even though limited similarity has been found with the CPs of plant and mammalian viruses (Rastgou et al., 2009). These data suggest that ourmiaviruses might have evolved by reassortment between a narnavirus‐like element from a plant‐pathogenic fungus and a tombusvirus (Koonin et al., 2015; Rastgou et al., 2009). In this context, the study of the OuMV MP is of interest to elucidate the origin of the Ourmiavirus lineage, and the evolution/diversification of MPs.
The OuMV MP belongs to the 30K MP superfamily, which includes viruses belonging to more than 15 genera, with numerous RNA viruses and also the DNA‐geminiviruses (Melcher, 1990, 2000; Mushegian and Elena, 2015; Mushegian and Koonin, 1993). Species in this superfamily, despite phylogenetic‐related MPs, use different mechanisms for cell‐to‐cell movement (Carrington et al., 1996; Melcher, 2000). A previous analysis of sequence and structural similarities of the MPs of 30K superfamily members (including OuMV) showed the presence of a central core region conserved amongst all MPs, consisting of seven predicted β‐strands (Melcher, 2000; Mushegian and Elena, 2015). Amino acids in this region are conserved in several 30K superfamily species, and have been predicted to induce main kinks and turns in the MP structure (Melcher, 2000; Mushegian and Elena, 2015). Attempts to predict the three‐dimensional structure of 30K superfamily MPs have been inconclusive so far, and their crystal structure is still missing. In this context, multiple alignments are a valuable tool to identify amino acids with potential functional significance, and site‐directed mutagenesis can be used to study the properties of the MPs and to verify the effect of specific mutations on various aspects of virus infection (Mushegian and Elena, 2015).
In this work, we generated a collection of OuMV MP mutants to identify conserved amino acids affecting MP function in two hosts, Nicotiana benthamiana and A. thaliana, using alanine‐scanning mutagenesis. The application of this technique to target amino acids of viral MPs was first reported for Red clover necrotic mosaic virus (Giesman‐Cookmeyer and Lommel, 1993) and, since then, it has been used extensively for the molecular characterization of the MPs of several viral species (Bertens et al., 2000; Chu et al., 1999; Huang et al., 2001). We selected for mutagenesis: (i) amino acids located in the 30K MP core region and conserved among OuMV and phylogenetically related species, and thus expected to potentially influence the correct folding of the core 30K domain; and (ii) solvent‐exposed amino acids, possibly responsible for MP self‐interaction, or for interactions with other viral proteins and/or host proteins. We were able to identify five OuMV mutants that were impaired in local and systemic infection in both hosts, and two mutants showing increased symptom severity in N. benthamiana. Green fluorescent protein fusion constructs (GFP:MP) of movement‐defective MP alleles failed to localize in distinct foci at the cell periphery or to induce the formation of tubular protrusions in protoplasts, suggesting a link between tubule formation and the ability of the virus to move. Given the conservation of the same amino acid residues in several viral species in the 30K superfamily, our work provides interesting target sites for further investigation of MP functionality in a wide range of plant viruses.
Results
Generation of 11 mutant MP constructs
The sequence alignment of MPs of ourmiaviruses, tombusviruses and aureusviruses showed the presence of conserved amino acids, mostly in the 30K superfamily core region (between amino acids 63 and 206 in the OuMV MP) (Fig. 1). Among them, topology prediction identified residues in positions 98, 149‐150‐151 and 202 as part of the protein helices possibly associated with cell membranes (Fig. S1, see Supporting Information). We selected the above five amino acid sites for mutagenesis. In addition, we targeted amino acids 97, 137 and 169, because they were also located in the central core region, and were conserved among all of the viral species considered in the phylogenetic analysis (Fig. 1). Five additional amino acids (positions 215, 251, 252, 253, 282), mainly located in the C‐terminal region, were selected for mutagenesis because they were predicted to be surface exposed. We generated 11 mutant clones, encoding for single (pGC‐MP98, pGC‐MP137, pGC‐MP150, pGC‐MP169, pGC‐MP202, pGC‐MP252, pGC‐MP282), double (pGC‐MP97, pGC‐MP215) or triple (pGC‐MP149, pGC‐MP253) amino acid substitutions (Table 1), which were used in the following experiments in comparison with the wild‐type MP (MPwt) construct.
Figure 1.

Alignment of the movement proteins (MPs) of ourmia‐, aureus‐ and tombusviruses using the PROMALDS3D program. Each sequence is coloured according to PSIPRED secondary structure predictions (red, α‐helix; blue, β‐strand). Consensus amino acid symbols are as follows: aliphatic, l; aromatic, @; hydrophobic, h; alcohol, o; polar residues, p; tiny, t; small, s; bulky residues, b; positively charged, +; negatively charged, –; charged, c. Conserved amino acids are in bold and capital letters. The last line shows the consensus amino acid sequence (Consensus_aa). Yellow shows the amino acids that were selected for alanine‐scanning site‐directed mutagenesis of Ourmia melon virus MP. Viral species: AMCV, Artichoke mottled crinkle virus; CBLV, Cucumber Bulgarian virus; CLSV, Cucumber leaf spot virus; CsVC, Cassava Virus C' CymRSV, Cymbidium ringspot virus EpCV, Epirus cherry virus; JCSMV, Johnson grass chlorotic stripe mosaic virus; MNSV, Maize necrotic streak virus; MWLMV, Maize white line mosaic virus; OuMV, Ourmia melon virus; PoLV, Pothos latent virus.
Table 1.
Genotype of the Ourmia melon virus movement protein (MP) mutants, and summary of the infectivity assays in Nicotiana benthamiana and Arabidopsis thaliana.
| Mutant clone | Amino acid substitution(s) position | Amino acid substitution(s) | Feature* | Systemic symptoms in Nicotiana benthamiana | Systemic symptoms in Arabidopsis thaliana |
|---|---|---|---|---|---|
| pGC‐MP97 | 97–98 | PH/AA | a,b,c | − | − |
| pGC‐MP98 | 98 | H/A | a,c | − | − |
| pGC‐MP137 | 137 | G/A | a,b | + | + |
| pGC‐MP149 | 149,150,151 | CLP/AAA | a | − | − |
| pGC‐MP150 | 150 | L/A | a | − | |
| pGC‐MP169 | 169 | P/A | a,b | − | − |
| pGC‐MP202 | 202 | P/A | a | + | + |
| pGC‐MP215 | 215,216 | QP/AA | c | + | + |
| pGC‐MP252 | 252 | E/A | c | + | + |
| pGC‐MP253 | 252,252,253 | EEE/AAA | c | + | + |
| pGC‐MP282 | 282 | R/A | c | + | + |
*a, amino acid conserved among Ourmiavirus–Aureusvirus species; b, amino acid conserved among Ourmiavirus–Tombusvirus species; c, solvent‐exposed predicted amino acid.
Amino acid substitutions can affect virus movement and symptom development
Starting at 6 days post‐inoculation (dpi), systemic viral symptoms were evident on un‐inoculated leaves from plants infiltrated with the MPwt construct, and similar symptoms were observed for the mutants pGC‐MP215, pGC‐MP252, pGC‐MP253 and pGC‐MP282 (Fig. 2A shows the symptoms at 10 dpi). At the same time point, mutant pGC‐MP202 showed increased symptom severity, with evident leaf curling and necrotic spots, whereas strong systemic mosaic was caused by pGC‐MP137 (Fig. 2A, B). No obvious symptoms were observed for constructs pGC‐MP97, pGC‐MP98, pGC‐MP149, pGC‐MP150 and pGC‐MP169 (Fig. 2A). This remained true for the entire period of observation (21 dpi). We also tested for possible complementation of defective mutants by co‐inoculations of constructs pGC‐MP98/pGC‐MP150, pGC‐MP98/pGC‐MP169 or pGC‐MP150/pGC‐MP169, but did not observe systemic infection (not shown).
Figure 2.
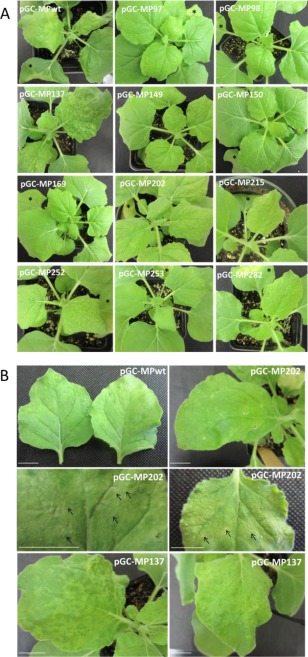
Characteristic symptoms on Nicotiana benthamiana systemic leaves after agroinfiltration with a bacterial mixture containing pGC‐RNA1, pGC‐RNA3 and (alternatively) wild‐type (WT) or mutant pGC‐MP constructs at 10 days post‐inoculation (dpi). (A) Symptoms induced by WT and the indicated movement protein (MP) mutants. (B) Close‐up view of systemic leaves from plants infiltrated with pGC‐MPwt, pGC‐MP202 and pGC‐MP137. Note the lack of symptoms for constructs pGC‐MP97, pGC‐MP98, pGC‐MP149, pGC‐MP150 and pGC‐MP169, the necrotic spots and leaf deformation for pGC‐MP202, and the strong mosaic for pGC‐MP137. Plants were agroinfiltrated with a bacterial (Agrobacterium tumefaciens strain C58C1) mixture diluted in agroinfiltration buffer at an optical density of 0.5 at 600 nm. Agroinfiltrated leaves are indicated by an asterisk when visible in the photograph. Scale bars, 1 cm.
Sequence analysis of reverse transcription‐polymerase chain reaction (RT‐PCR) products obtained from RNA extracted from local leaves confirmed the presence of the expected MP mutation(s) in all cases (data not shown), and the accumulation of CP and MP in infiltrated areas was detected for all of the mutants (Fig. 3A), demonstrating that the MPs carrying substitutions are not affected in overall stability or expression. The presence of viral RNA from the mutant MPs was also tested by RT‐PCR on the systemic tissues: sequence analysis of RT‐PCR products obtained from the mutants competent for systemic infections confirmed the presence of only the desired mutation(s), whereas no amplification product was observed for any of the five movement‐defective mutants (data not shown). Accordingly, Western blot analysis of protein extracts from systemic N. benthamiana leaves confirmed the accumulation of MP and CP by the clones causing systemic symptoms, whereas no accumulation was observed for the five movement‐defective constructs (Fig. 3B). To check whether the mutants impaired in systemic movement were also impaired in cell‐to‐cell movement, we mechanically inoculated the movement‐defective constructs on new N. benthamiana plants using, as source for inoculum, sap prepared from agroinfiltrated tissue (where all the mutants accumulated equally abundant MPs and CPs, as a result of the saturation provided by the agroinfiltration, Fig. 3A), and monitored MP and CP accumulation. For all movement‐defective constructs, neither infection foci (data not shown) nor CP/MP protein accumulation (Fig. 3C) could be detected for the entire period of observation (21 days). In contrast, infection foci and CP/MP protein accumulation were detected when pGC‐MPwt and pGC‐MP253 (as an infectious mutant control) were mechanically re‐inoculated in the same way, suggesting that the substitution of amino acids 97, 98, 149, 150 and 169 also abolished cell‐to‐cell movement, not allowing any expansion of the foci of infection from the initially infected cells.
Figure 3.
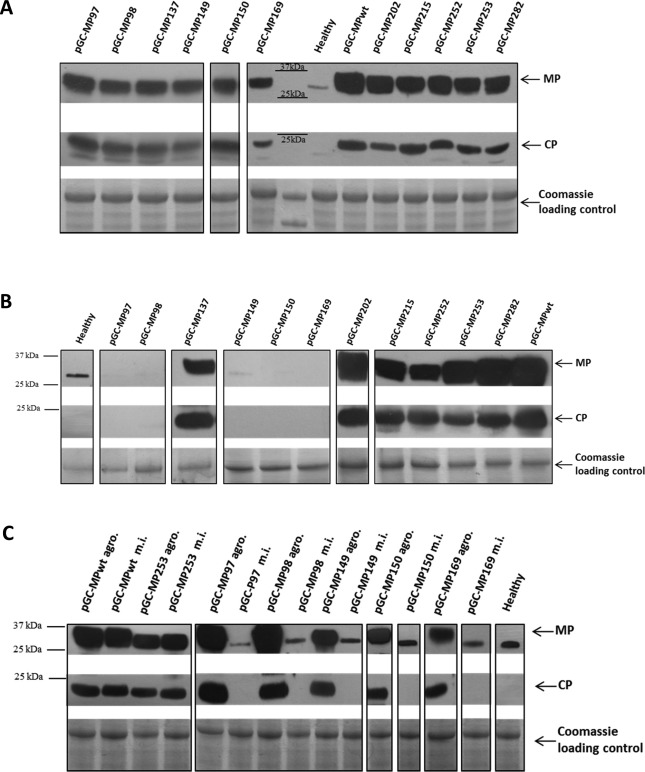
Ourmia melon virus movement protein (MP) and coat protein (CP) accumulation and total protein extracts in Nicotiana benthamiana leaves by Western blot and Coomassie staining. (A) Agroinfiltrated leaves. (B) Upper un‐inoculated leaves from agroinfiltrated plants. (C) Mechanically inoculated (m.i.) N. benthamiana leaves and their source of inoculum [agroinfiltrated leaves (‘agro.’) as in A] consisting of sap prepared from healthy plants or from leaves agroinfiltrated with a bacterial mixture containing pGC‐RNA1, pGC‐RNA3 and pGC‐MPwt, or mutants pGC‐MP253, pGC‐MP97, pGC‐MP98, pGC‐MP149, pGC‐MP150 or pGC‐MP169. Bands at c. 30 and 23 kDa show MP and CP, respectively. Note that a non‐specific band below MP is occasionally visible, probably as a result of cross‐reaction of the MP antibody with plant endogenous proteins, as shown in the healthy control samples.
Agroinfiltration of A. thaliana with the pGC‐MPwt construct resulted in visible yellowing and necrosis of older leaves by 15 dpi, which extended to the whole plant by the following week (Fig. 4A, photographs were taken at 18 dpi). Systemic symptoms were always observed for the mutants that infected N. benthamiana (Fig. 4A), and CP accumulation in systemic leaves was confirmed by enzyme‐linked immunosorbent assay (ELISA). In contrast, neither symptoms (Fig. 4A) nor CP accumulation were observed for the same five MP mutants that were impaired for movement in N. benthamiana (by ELISA, zero plants were positive from the five plants agroinfiltrated for each mutant). When we performed mechanical inoculation using sap from locally agroinfiltrated N. benthamiana tissue, chlorosis, necrosis and plant stunting were observed for pGC‐MPwt, and for the six infectious constructs (pGC‐MP137, pGC‐MP202, pGC‐MP215, pGC‐MP252, pGC‐MP253 and pGC‐MP282), but not for the five movement‐defective mutants (Fig. 4B). Accordingly, CP accumulation could not be detected in the upper leaves of A. thaliana inoculated with the five movement‐defective mutants, as seen by ELISA (by ELISA, zero plants were positive from the five plants inoculated for each mutant). No increased symptom severity was observed for pGC‐MP202 or pGC‐MP137, probably as a result of the already very strong symptoms developed by the wild‐type OuMV in this host, compared with N. benthamiana.
Figure 4.

Characteristic symptoms on Arabidopsis thaliana Col‐0. (A) Symptoms following agroinfiltration of bacterial mixtures containing pGC‐RNA1, pGC‐RNA3 and each of the movement protein (MP) constructs. (B) Symptoms following mechanical inoculation of wild‐type or mutant constructs, using sap obtained from Nicotiana benthamiana agroinfiltrated plants. Plants inoculated with MP constructs pGC‐MP97, pGC‐MP98, pGC‐MP149, pGC‐MP150 and pGC‐MP169 showed no symptoms, whereas yellowing and necrosis were observed for the other constructs. Photographs were taken at 18 days post‐inoculation (dpi) (A) and 10 dpi (B).
MPwt co‐localizes at the cell wall with plasmodesmata
Previous work has shown the association of GFP:MP with the cell wall in a punctate pattern in N. benthamiana (Crivelli et al., 2011), but no co‐localization with cell wall or plasmodesmata markers was investigated. Our GFP:MPwt localized as foci at the cell periphery of both N. benthamiana (as observed previously by Crivelli et al., 2011) and A. thaliana epidermal cells (Fig. 5A, B). The accumulation of GFP:MPwt foci between the borders of adjacent cells was also evident (Fig. 5A, top right panel), and three‐dimensional reconstructions of images from the infected cells showed the abundant presence of fluorescent spots at the cell periphery (Movies S1 and S2, see Supporting Information). Staining of leaf sections with propidium iodide, which stains pectin in the cell wall (Rounds et al., 2011), confirmed the co‐localization of GFP:MPwt with the cell wall (Fig. 6A). Following plasmolysis, MP fluorescent spots were observed on the plasma membrane as it retracted from the cell wall, but some fluorescent spots still co‐localized with the cell wall, apparently at the extremity of Hechtian strands (Fig. 6B), thin membranous structures that anchor the plasma membrane to the cell wall (Laporte et al., 2003). Staining of callose‐rich neck regions of plasmodesmata with aniline blue fluorochrome, which stains callose (Stone et al., 1984), showed the co‐localization of GFP:MP with callose deposits (Fig. 6C). Co‐localization was unambiguously evident in the plot showing the pixel fluorescence intensity of GFP:MP and aniline blue stain for a subset of foci (Fig. 7A), where the positions of the peaks of the two fluorescent markers clearly corresponded (Fig. 7B), indicating that MP co‐localizes with or is located in proximity to the plasmodesmata.
Figure 5.
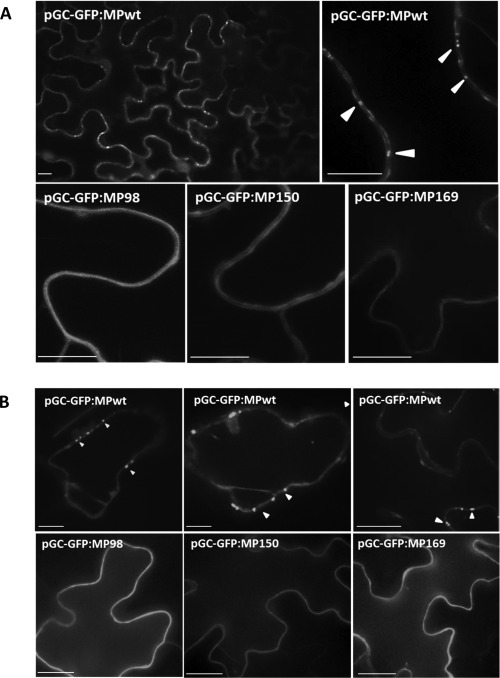
Green fluorescent protein fusion constructs (GFP:MP) of the mutants do not accumulate in punctate patterns at the cell periphery. Subcellular localization of GFP:MP was monitored in (A) Nicotiana benthamiana and (B) Arabidopsis thaliana leaf epidermal cells at 3 days post‐inoculation (dpi) following agroinfiltration of pGC‐RNA1, pGC‐RNA3 and the desired MP mutant construct (pGC‐GFP:MP98, pGC‐GFP:MP150 or pGC‐GFP:MP169) or pGC‐GFP:MPwt. For each genotype, about 100 cells were imaged. Arrowheads indicate MP localization. Scale bars, 5 µm.
Figure 6.
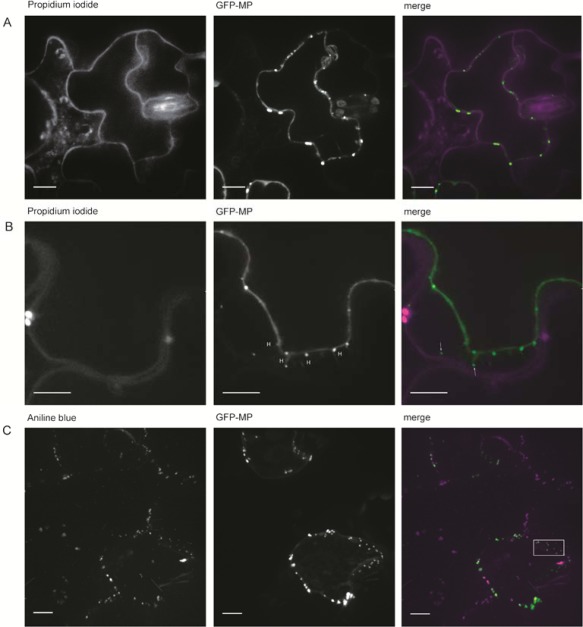
Ourmia melon virus GFP:MPwt (green fluorescent protein fusion with wild‐type movement protein) co‐localizes at the cell wall, on the cell membrane of plasmolysed cells and with plasmodesmata. The fluorescence was monitored on sections from Arabidopsis thaliana (A, C) and Nicotiana benthamiana (B) leaves, stained with (A) propidium iodide (which stains pectin in the cell wall) in Murashige and Skoog (MS) medium, (B) propidium iodide in 1 m mannitol solution, and (C) aniline blue fluorochrome (which stains callose) in MS medium (left panels). Middle panels show GFP:MP foci. Right panels are merged images of the two channels. The box in the right panel in (C) shows a subset of fluorescent dots used for the generation of the plots shown in Fig. 7. H, Hechtian strands. The images were taken at 3 days post‐inoculation (dpi). Scale bars, 5 µm.
Figure 7.
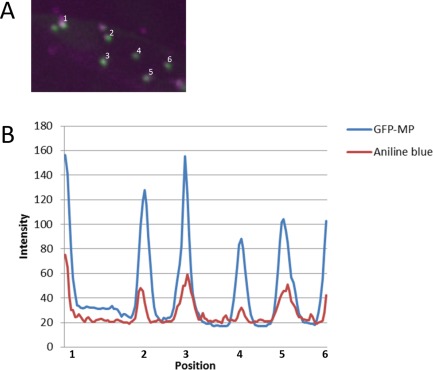
Co‐localization of GFP:MPwt (green fluorescent protein fusion with wild‐type movement protein) and aniline blue‐stained callose at the plasmodesmata sites. (A) Closer view of a subset of dots illustrated in Fig. 6C, showing overlapping (white colour) of GFP:MP (green pseudocolour) and aniline blue stain (magenta pseudocolour). (B) Plot showing pixel fluorescence intensity for the six spots shown in (A). The profile shows the co‐localization of the GFP:MPwt fusion and the aniline blue‐stained callose deposits.
GFP:MP mutants impaired in movement do not accumulate in punctate patterns at the periphery of N. benthamiana and A. thaliana epidermal cells
To investigate whether the absence of movement of mutants pGC‐MP98, pGC‐MP150 and pGC‐MP169 (selected from the five movement‐defective cDNA constructs because they encoded a single amino acid substitution in the MP) was associated with the differential localization of MP, we generated GFP:MP mutant fusion constructs. For all these constructs, the fluorescence signal was distributed evenly at the cell border (as observed for a free GFP‐negative control construct by Crivelli et al., 2011), and was not localized in foci as in GFP:MPwt, in either N. benthamiana (Fig. 5A) or A. thaliana (Fig. 5B).
GFP:MP mutants impaired in movement do not form protruding tubules in protoplasts
We could not observe protruding tubules from protoplasts obtained from leaves agroinfiltrated with the GFP:MP mutant constructs pGC‐GFP:MP98, pGC‐GFP:MP150 or pGC‐GFP:MP169 (each infiltrated together with Agrobacterium carrying pGC‐RNA1 and pGC‐RNA3 constructs) in N. benthamiana or A. thaliana (Fig. 8). By contrast, intense labelling of tubular structures or punctate extensions protruding from the plasma membrane into the extracellular space was evident for the GFP:MPwt construct, in both N. benthamiana (as observed previously by Crivelli et al., 2011) and A. thaliana (Fig. 8).
Figure 8.
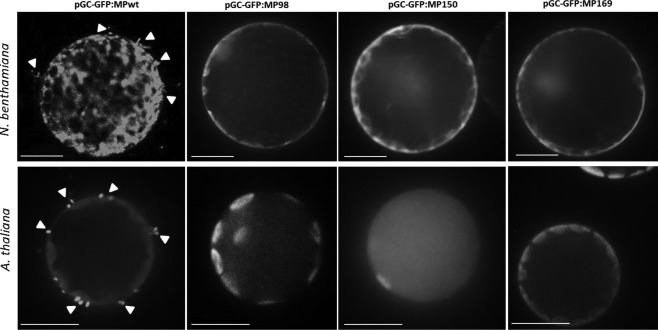
Movement‐defective Ourmia melon virus (OuMV) constructs do not form protruding tubular structures in protoplasts. Protoplasts were prepared from Nicotiana benthamiana and Arabidopsis thaliana leaves agroinfiltrated with pGC‐RNA1, pGC‐RNA3 and (alternatively) pGC‐GFP:MPwt, pGC‐GFP:MP98, pGC‐GFP:MP150 or pGC‐GFP:MP169. Fluorescence was assayed by confocal microscopy. Protoplasts were prepared at 3 days post‐inoculation (dpi). For each host–OuMV combination, at least 100 protoplasts were assayed. Scale bars, 10 µm.
Cell‐to‐cell movement is affected in MP mutants impaired in tubule formation
Following the inoculation of a dilution series of the Agrobacterium suspensions to obtain single transfected cells, we observed that the fluorescence of GFP:MP mutants impaired in tubule formation remained confined to the initially infected cells, as observed for the pBin:GFP negative control, whereas the fluorescence of GFP:MPwt in single cells was associated with a signal in one or more of the surrounding cells (Fig. 9). Single‐cell transfection was obtained with the 10−3 dilution; with further dilutions of all construct mixtures, including that with GFP:MPwt, no fluorescence was visible. These data support our hypothesis that the MP amino acids necessary for systemic movement are also involved in local movement.
Figure 9.
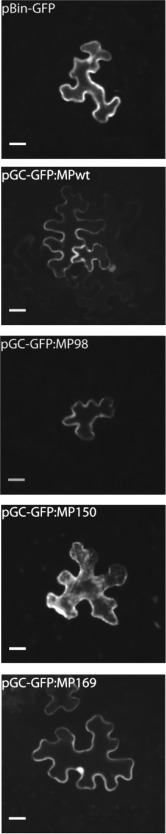
Green fluorescent protein fusion constructs (GFP:MP) of the mutants are impaired in cell‐to‐cell movement. Fully developed Nicotiana benthamiana leaves were agroinfiltrated with pGC‐RNA1, pGC‐RNA3 and the selected pGC‐GFP:MP construct in 1 : 10 serial dilutions of Agrobacterium suspensions grown to an optical density of 0.4 at 600 nm. Single‐cell transfection was obtained with the 10−3 dilution. Fluorescence of the GFP:MPwt fusion (GFP fusion with wild‐type movement protein) was observed in the initially infected cell and in one or more of the surrounding cells, whereas it remained limited to the transfected cell for mutants GFP:MP98, GFP:MP150 and GFP:MP169, as observed for the pBin:GFP negative control. With further dilutions of all construct mixtures, including that with GFP:MPwt, no fluorescence was visible. For each genotype, about 20 cells were imaged. Scale bars, 20 µm.
WT and mutant MPs are detected in the P30 membrane‐enriched fraction
A previous analysis has shown preferential accumulation of MPwt in membrane‐enriched P30 fractions (Crivelli et al., 2011). In this study, MP98, MP150 and MP169, as well as MPwt and MP253 (as an infectious mutant control), from 3‐dpi agroinfiltrated tissues and processed by differential centrifugation, were detected only in the P30 membrane‐enriched fraction and not in the S30 soluble fraction (Fig. 10A). It should be noted that the band visible in the S30 fraction had a smaller size than that expected for the MP protein, and corresponded to the band visible in the healthy samples, probably as a result of cross‐reactivity of the MP antibody with plant endogenous proteins.
Figure 10.
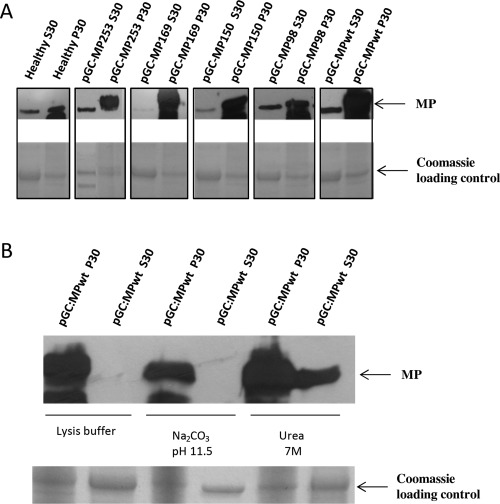
Ourmia melon virus wild‐type movement protein (MP) and selected MP mutants associate with biological membranes. (A) Pellet (P30) and soluble (S30) fractions were obtained by centrifugation of leaf extracts at 30 000 g, and analysed by Western blot using an anti‐MP antibody. (B) To analyse MP solubility properties, the P30 fractions were washed with different solutions. Pellet (P) and soluble (S) fractions were obtained by centrifugation at 30 000 g and analysed by Western blot. Localization was analysed on agroinfiltrated leaves at 3 dpi. Note that the lower band visible in the S30 fractions corresponds to the band visible in the healthy samples, probably as a result of cross‐reaction of the antibodies with host proteins.
To further characterize the nature of the association of the OuMV MP with cell membranes, the P30 fractions were treated with solubilizing agents. Treatment with Na2CO3 (pH 11.5), which has been reported to open the microsomes and thereby release the protein trapped in the luminal fraction (Fujiki et al., 1982), did not release the MP from the P30 pellet, as observed for the lysis buffer‐treated control (Fig. 10B). Treatment with 7 m urea, which should release all polypeptides from the membranes, with the exception of the integral membrane proteins, resulted in a 1 : 0.7 distribution of the MP between the detergent and aqueous phases (Fig. 10B), showing that this treatment can partially solubilize the MP.
Discussion
Viral MPs are critical for the establishment of successful viral infections, but relatively little is known about their structural and functional characteristics, and their interaction with host factors. MPs of different plant virus groups or even closely related genera often show low sequence similarity, and closely related viral species can employ different strategies for successful infection of their hosts (Hull, 2014). For example, the OuMV MP is able to form tubular structures, and is phylogenetically related to the MP of aureusviruses and tombusviruses, groups of plant viruses devoid of tubule formation (Rastgou et al., 2009). The OuMV MP belongs to the 30K superfamily, a group of proteins that share various degrees of sequence similarity to the MP of the founding member: Tobacco mosaic virus (TMV) (Melcher, 2000; Mushegian and Elena, 2015). Some functional and biological properties of the OuMV MP have been described previously in N. benthamiana (Crivelli et al., 2011; Rastgou et al., 2009). To further investigate the contribution of specific amino acids to MP function(s) and cellular localization, we generated 11 MP mutants, and studied their behaviour in two hosts, N. benthamiana and A. thaliana. We found five mutants that were unable to systemically infect either host. All carried mutations in the conserved MP core region, which includes seven predicted β‐strands (between amino acids 57 and 181) involved in protein folding (Melcher 2000; Mushegian and Elena 2015). Sequence alignment showed that three amino acids (amino acids 98, 149 and 150) of functionally defective MPs were conserved among Ourmiavirus and Aureusvirus species, and that amino acids 97 and 169 were also conserved in Tombusvirus species (Fig. 1). In this context, it would be interesting to investigate the effect of the same amino acid mutations on the systemic movement of other Ourmiavirus species, and of species phylogenetically related to ourmiaviruses.
In addition, residues 97, 98 and 150 are also conserved among distant members in the 30K superfamily (Mushegian and Elena, 2015). For instance, residue 97 (P, proline) is also found, among others, in Tomato torrado virus (ToTV, Torradovirus), Actinidia virus B (Vitivirus), Carrot mottle virus (CMotV, Umbravirus), Cycas necrotic stunt virus (Nepovirus), Alfalfa mosaic virus (Alfamovirus), Tomato spotted wilt virus (TSWV, Tospovirus) and Citrus psorosis virus (Ophiovirus) (Mushegian and Elena, 2015). Residue 98 (H, histidine) is conserved in ToTV, and in Actinidia virus B, CMotV and Cherry rasp leaf virus (Cheravirus). Residue 150 (L, leucine) is conserved in Cycas necrotic stunt virus (Nepovirus), Red clover necrotic mosaic virus (Dianthovirus), Rice stripe virus (Tenuivirus) and Lettuce necrotic yellows virus (Cytorhabdovirus) (Mushegian and Elena, 2015). The conservation of amino acids in viral species belonging to different taxa, and showing a differential genome structure and organization, suggests their importance for MP functionality across genera.
In this study, we also showed that the MP of a subset of OuMV mutants defective in systemic movement does not support movement to adjacent cells. These results suggest that the absence of systemic movement in this case could be a result of impairment in cell‐to‐cell movement. Mutants not able to systemically infect the host, but competent for cell‐to‐cell movement, were not present in our collection, and thus we cannot speculate on the involvement of distinct functional domains in the two types of movement, as described previously for the CP of Tobacco etch virus (Dolja et al., 1994, 1995), the MP of Red clover necrotic mosaic virus (Wang et al., 1998) and the MP of TSWV (Li et al., 2009).
In addition to being key players for the translocation of viruses among cells and through the plant, viral MPs play a significant role in host susceptibility and symptom development (Amari et al., 2012; García and Pallás, 2015; Pallas and García, 2011) , as shown for Cucumber mosaic virus (Choi et al., 2005), Brome mosaic virus (Rao and Grantham, 1995) , Turnip yellow mosaic virus (Tsai and Dreher, 1993) and TSWV (Li et al., 2009). In the study by Li et al. (2009), alanine substitution of five E (glutamic acid) residues in the C‐terminal region resulted in altered symptomatology and an inability to systemically infect the host. Three of the five E residues were substituted in our mutant pGC‐MP253, which induced symptoms indistinguishable from those of the pGC‐MPwt construct, suggesting that substitution of all five amino acids in this position is necessary to influence symptoms and the ability to infect the host, or that substitutions in this region do not affect OuMV‐induced symptomatology. In this study, we identified two single amino acid MP substitutions that affect symptom severity during OuMV infection. Alanine substitution of amino acid 202 (P) induced foliar deformation and necrosis in N. benthamiana (Fig. 2B) during active viral infection (infected with OuMV pGC‐RNA1, pGC‐RNA2 and pGC‐RNA3). Alanine substitution of amino acid 137 (G, glycine) also affected symptom severity, resulting in strong mosaic patterns on upper leaves following agroinfiltration. Such symptoms also developed after mechanical re‐inoculation onto new plants (data not shown), thus excluding an Agrobacterium/agroinfiltration effect. Possibly, altered interactions between OuMV MP and host component(s) might be related to the different symptom severity, as proposed previously for other virus–host interactions (Li et al., 2009).
At least two mechanisms have been proposed for viral movement: in one, typified by TMV, MP‐mediated gating of plasmodesmal connections is required to allow the passage of ribonucleoprotein complexes; in another, typified by comoviruses, the MP forms tubules that serve as conduits for the movement of viral particles between cells (Hull, 2014; Ritzenthaler and Hofmann, 2007). Based on the composition of viral nucleic acid–protein complexes that spread in the host, plant viruses can be classified into three categories: in type 1, CP is dispensable for movement and is not part of the moving complex; in type 2, CP is actively involved by probably protecting the viral genome; in type 3, CP is required as the viruses move as particles (Scholthof, 2005). Based on this classification, OuMV might belong to multiple categories. Indeed, OuMV moves from cell to cell as intact virus particles, as supported by electron microscopy observations and genetic analysis of MP localization (Crivelli et al., 2011; Rastgou et al., 2009). However, its CP is not strictly necessary for cell‐to‐cell or long‐distance movement (at least in N. benthamiana), as mixed inoculation of RNA‐1 and RNA‐2 results in localized infection in both the inoculated and upper un‐inoculated leaves, with infection failing to fully develop in the upper leaves (Crivelli et al., 2011). Interestingly, OuMV MP, which can induce tubules in protoplasts (this study and Crivelli et al., 2011), showed the closest similarity to aureusviruses (Rastgou et al., 2009), which do not form tubules. Taken together, these data indicate that OuMV is probably a tubule‐forming virus that does not strictly require CP for movement. It should be noted that tubule formation does not necessarily indicate dependence on tubules for movement: as an example, CMV MP can form tubules, but these structures are not necessary for viral cell‐to‐cell movement (Canto and Palukaitis, 1999). In this work, we showed that the GFP:MP from constructs pGC:GFP‐MP98, pGC:GFP‐MP150 and pGC:GFP‐MP169, which were impaired in both local and systemic movement, did not aggregate in foci at the cell borders of infected leaves, and did not form tubular protrusions in protoplasts derived from these leaves. Conversely, abundant foci in close proximity to (or at) the plasmodesmata and tubule formation were evident for GFP:MPwt, suggesting that OuMV crosses the cell wall through tubules that pass through highly modified plasmodesmata, as proposed for Cowpea mosaic virus (Bertens et al., 2003, 2000), Alfalfa mosaic virus (Sánchez‐Navarro and Bol, 2001) and TSWV (Leastro et al., 2015; Li et al., 2009). Despite the fact that foci at the cell periphery were not evident for the movement‐defective constructs, membrane fractionation assays showed that mutant MPs impaired in localization and movement were localized in the membrane‐enriched fractions, as observed for MPwt (Fig. 10A). We hypothesize that these mutant MPs are still able to reach the plasma membrane, but then cannot co‐localize with the plasmodesmata, self‐assemble or interact with host factors to generate tubules. The possible inability to interact with other viral proteins is not expected to play a role, as Crivelli et al. (2011) described that GFP:MP alone is able to form tubules in protoplasts prepared from agroinfiltrated tissues, showing that interaction with other viral proteins is not necessary to generate such structures. Given that amino acids 98, 150 and 169 are conserved in other 30k MPs (Mushegian and Elena, 2015), it would be interesting to test whether mutation of the same amino acid residue(s) is related to the ability to form tubules or to co‐localize with plasmodesmata in other members of the 30K superfamily.
The association of OuMV MP with plasmodesmata was further supported by plasmolysis experiments, which provided evidence for the presence of GFP:MP in punctate form on the cell wall, at the extremity of Hechtian strands, following retraction of the protoplast. Most Hechtian strands have been reported to be intimately linked to plasmodesmata (Oparka et al., 1994), further supporting a co‐localization with OuMV MP. The involvement of plasmodesmata in viral infection has long been accepted (Boevink and Oparka, 2005; Fernández‐Calvino et al., 2011; Lucas and Gilbertson, 1994; Nelson, 2005); however, de novo tubule formation might also play a role in OuMV movement. MP foci that did not co‐localize with plasmodesmata were occasionally apparent by confocal microscopy, but the resolution limit of this approach, or possible ineffective staining, did not allow us to explore this aspect in detail.
The membrane topology analysis revealed that OuMV MP possibly exists in two forms in the host cells. Indeed, chemical treatments used to remove the MP from the P30 membrane fractions revealed that, following washing with 7 m urea, a proportion of the protein was found in the soluble fraction. These results indicate that OuMV may exist as an integral or strictly attached membrane form and a more hydrophilic form, a model that could be consistent with the presence of tubules through the plasmodesmata as well as in the cell cytoplasm (Rastgou et al., 2009). Alternatively, the MP aggregates as tubules may not be soluble in sodium carbonate because of their compact nature, whereas they could be partially solubilized by urea treatment. A similar membrane topology behaviour was observed for the MP of four tospoviruses treated in the same conditions, and a tight association of these MPs to the biological membranes, without being typical integral membrane proteins, was proposed (Leastro et al., 2015). The MP of another tospovirus, Groundnut bud necrosis virus (GBDV), has been suggested to be membrane associated in vivo, and behaves as an integral membrane protein (Singh et al., 2014). An integral association with membrane structures has also been proposed for Alfalfa mosaic virus (Huang and Zhang, 1999), whereas a membrane‐associated pattern, without being an integral membrane protein, has been observed for other members of the 30K superfamily, such as TMV (Peiró et al., 2014) and Prunus necrotic ringspot virus (Martínez‐Gil et al., 2009).
Overall, our results demonstrate that amino acid substitutions in the OuMV MP determine drastic biological consequences in the viral cycle and host response. The major effects were observed by mutation of amino acids localized in the MP core region and conserved among species in the Ourmiavirus genus and in other members of the 30K superfamily, whereas mutations of charged amino acids in the C‐terminal region did not affect the ability of OuMV to move. We speculate that substitutions of the same key amino acids in other viral species could be equally important for movement and could induce phenotypic effects in multiple host species.
Experimental Procedures
Plant material and inoculations
Nicotiana benthamiana and A. thaliana Col‐0 plants were grown in a growth chamber set at 25 ± 1 ºC, with 16 h light and 8 h dark conditions, and inoculated 3–4 weeks after sowing. Mechanical inoculations were performed using sap obtained from infected, fresh N. benthamiana tissue ground in 0.05 m phosphate buffer, supplemented with carborundum and silica powders. Agroinfiltrations were performed using a syringe to infiltrate the bacterial mixtures prepared as described below.
Computational analysis for targeted amino acid selection
The alignment of MP sequences from viral species in the Ourmiavirus, Tombusvirus and Aureusvirus genera was performed using PROMALS3D (Pei and Grishin, 2014), which generates multiple sequence alignments and annotates secondary structural features through consensus predictions using the PSIPRED algorithm (Jones, 1999). Viral species and accession numbers used in the analysis were as follows: CaVC (205321012), EpCV (194351528), Maize necrotic streak virus (MNSV, 85677470), Cucumber Bulgarian virus (CBLV, 30018256), Artichoke mottled crinkle virus (AMCV, 9625555), Cucumber leaf spot virus (CLSV, 89888605), Maize white line mosaic virus (MWLMV, 148717972), Johnson grass chlorotic stripe mosaic virus (JCSMV, 39163652), Pothos latent virus (PoLV, 9633617). Secondary structure and solvent accessibility prediction on OuMV MP were performed with PredictProtein (https://www.predictprotein.org /), Sable (http://sable.cchmc.org/) and Minnou (http://minnou.cchmc.org /) tools, and consensus site predictions were chosen for alanine‐scanning site‐directed mutagenesis. Topology prediction was performed using Membrane Protein Explorer v. 3.21 (http://blanco.biomol.uci.edu/mpex/) and TopPred v. 1.10 (Claros and von Heijne, 1994).
Alanine‐scanning site‐directed mutagenesis
Plasmid pGC‐RNA2 (referred to as pGC‐MPwt), containing the OuMV RNA‐2 in a pJL89 vector, between a double Cauliflower mosaic virus 35S promoter and the Hepatitis delta virus (HDV) ribozyme (Crivelli et al., 2011), was used as template for alanine‐scanning site‐directed mutagenesis (Cunningham and Wells, 1989) by PCR with the Q5® High‐Fidelity site‐directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA). Primers to insert the desired mutation(s) were designed using NEBaseChanger™ software v. 1.2.2 (http://nebasechanger.neb.com/), and are available in Table S1 (see Supporting Information). Eleven RNA‐2 mutant plasmids were obtained, coding for single, double or triple alanine mutant MPs (Table 1). The presence of the desired mutation(s) in the MP sequence and the absence of additional nucleotide changes were verified in all the plasmids by Sanger sequencing across the entire sequence of the RNA‐2 cDNA.
GFP:MP fusion cDNA construct generation
A GFP‐encoding sequence with a 5′ and 3′ NcoI restriction site was obtained by digestion from a pGC‐GFP:CP plasmid (Rossi et al., 2014), and inserted at the 5′ end of pGC‐MPwt using an NcoI site at the first ATG, to obtain plasmid pGC‐GFP:MPwt. GFP:MP fusion constructs from plasmids pGC‐MP98, pGC‐MP150 and pGC‐MP169 (selected from the five movement‐defective cDNA constructs because they carry single amino acid substitutions) were also generated, and the derived plasmids were named pGC‐GFP:MP98, pGC‐GFP:MP150 and pGC‐GFP:MP169. Plasmids were checked by restriction enzyme digestion (XbaI and MfeI, New England Biolabs) and by Sanger sequencing for single‐copy GFP insertion and correct orientation.
Leaf agroinfiltration
Plasmid constructs were introduced into Agrobacterium tumefaciens strain C58C1 by electroporation using a gene micropulser (Bio‐Rad, Hercules, CA, USA), and transformed bacterial cultures were grown in Luria–Bertani (LB) medium containing kanamycin and tetracycline at 50 μg/mL and 5 μg/mL concentration, respectively, and stored at −80°C in 25% v/v glycerol. To launch viral infection, bacterial cultures of RNA‐2‐derived clones, and of plasmids pGC‐RNA1 and pGC‐RNA3, coding for RdRp and CP, respectively (Crivelli et al., 2011), were grown in a shaking incubator at 28°C in 2XYT medium (5g/L NaCl, 10g/L yest extract, 16g/L tryptone) supplemented with the appropriate antibiotics (Crivelli et al., 2011). Cultures were collected by centrifugation at 3000 g for 10 min, adjusted to a final optical density at 600 nm (OD600) of 0.5 with agroinfiltration buffer [10 mm MES (2‐N‐morpholino‐ethanesulfonic acid), 10 mm MgCl2 and 100 µm acetosyringone], and left for 3–4 h on ice (Bendahmane et al., 2002, 1999). Leaves were agroinfiltrated using a bacterial mixture consisting of pGC‐RNA1, pGC‐RNA3 and the desired RNA‐2 mutant in equal amounts. RNA‐2 mutations were verified by RNA extraction from locally and systemically infected plant tissue, RT‐PCR and sequencing. The development of symptoms was monitored for a 21‐day period after infiltration.
To investigate the cell‐to‐cell movement of GFP:MP fusions in epidermal cells, Agrobacterium suspensions carrying pGC‐RNA1, pGC‐RNA3 and the desired pGC‐GFP:MP construct were diluted in agroinfiltration buffer at an OD of 0.4 at 600 nm, and agroinfiltrated in 1 : 10 serial dilutions until single‐cell transfection was obtained. Fully expanded N. benthamiana leaves were agroinfiltrated. The localization of GFP:MP was monitored at 3 dpi. A pBin:GFP construct described previously (Yeh et al., 2001) was used as the GFP‐only control.
ELISA
For double antibody sandwich (DAS)‐ELISA, an anti‐CP polyclonal serum described previously (Rastgou et al., 2009) was used to coat NUNC high‐binding plates (Life Technologies, Grand Island, NY, USA). Crude extracts were obtained by mechanical homogenization of leaf samples in phosphate‐buffered saline (PBS)–Tween buffer (10 mg/mL, w/v) containing 2% (w/v) polyvinylpyrrolidone‐40 (PVP‐40) (Sigma‐Aldrich, St. Louis, MO, USA), and incubated overnight at 4 °C with gentle shaking. Plates were washed with PBS–Tween buffer and incubated for 2 h with an alkaline phosphatase‐conjugated anti‐CP antibody (1 : 1000, v/v). Colorimetric absorption at 405 nm was measured following incubation with the 4‐nitrophenyl phosphate disodium salt hexahydrate enzyme substrate (Sigma‐Aldrich).
At least five plants per mutant MP construct were tested. Samples were considered to be positive if the absorbance value was three times higher than that measured for healthy control plants (Table S2).
Immunoblotting analysis
For Western blotting, N. benthamiana or A. thaliana leaves were harvested 3 days after infiltration. Total protein was extracted as described previously (Margaria et al., 2007), and approximately 10 µg of protein per sample were separated by sodium dodecyl sulfate‐12% polyacrylamide gel electrophoresis (SDS‐PAGE). Gels were either stained in a Coomassie‐based staining solution or electroblotted onto Immobilon‐P membrane (Millipore, Watford, UK). The membrane was blocked by incubation in 1 × TBS containing 0.05% (w/v) Tween and 7% (w/v) milk powder (TBST) for 30 min at room temperature. Anti‐CP (A255) or anti‐MP (A314) antisera (Rastgou et al., 2009) were added to the blocking solution at a dilution of 1 : 2000 (v/v), and incubated overnight at 4 °C. After washing in TBST, an anti‐rabbit horseradish peroxidase‐conjugated secondary antibody (NXA931; Sigma‐Aldrich) was used at a dilution of 1 : 20 000 (v/v) for detection, employing the Supersignal West Pico Chemiluminescence Substrate (ThermoScientific, Waltham, MA, USA). Detection of GFP:MP fusions was performed using a mouse‐monoclonal anti‐GFP antibody (Life Technologies), diluted 1 : 1000 (v/v), followed by room temperature incubation with an anti‐mouse horseradish peroxidase‐conjugated secondary antibody (A4416, Sigma), diluted 1 : 10 000.
Protoplast preparation
Protoplasts were isolated from leaves agroinfiltrated with a mixture of pGC‐RNA1, pGC‐RNA3 plus each one of the desired pGC‐GFP:MP constructs at 3 days post‐agroinfiltration, as described previously (Jones et al., 1990). Nicotiana benthamiana and A. thaliana leaves were gently rubbed on the underside using a cotton‐tipped applicator dipped in sterile 0.05 m KH2PO4 (pH 7.0) containing 1% (w/v) celite, and macerated in 20 mL of fresh enzyme solution [10% w/v mannitol, 0.25 g Cellulysin® (EDM Millipore, Billerica, MA, USA), 25 mg Macerase™ Pectinase (EDM Millipore), 25 mg bovine serum albumin (BSA)] at room temperature on a rotary shaker, for 4–5 h and 7–8 h, respectively. Crude suspensions were used directly for laser scanning confocal microscopy.
Membrane fractionation
Approximately 2 g of N. benthamiana leaves agroinfiltrated with a mix of pGC‐RNA1, pGC‐RNA3 and the desired pGC‐RNA2 clone (pGC‐MPwt; pGC‐MP253, as movement‐competent mutant; pGC‐MP98; pGC‐MP169; pGC‐MP150, as single amino acid movement‐defective mutants) were ground in liquid nitrogen, immediately resuspended in lysis buffer (50 mm Tris, pH 7.5, 15 mm MgCl2, 10 mm KCl, 0.3 m sorbitol, 0.1% v/v β‐mercaptoethanol; 1% v/v protease inhibitor) and incubated for 30 min at 30°C. Cellular debris was removed by centrifugation at 500 g for 5 min, and the supernatant was gently transferred to a new tube. Centrifugation at 30 000 g for 1 h at 4°C was performed to obtain the soluble S30 fraction and the membrane‐enriched P30 fraction.
To analyse the solubility properties of membrane‐bound OuMV MP, the P30 fraction was treated with lysis buffer, alkaline buffer (Na2CO3, pH 11.5) or urea 7 m, and incubated for 30 min on ice (Leastro et al., 2015; Navarro et al., 2004). Membrane and soluble fractions were collected by centrifugation at 30 000 g for 30 min at 4°C.
All the fractions were analysed by SDS‐PAGE and immunoblotting as described above.
Leaf staining, plasmolysis and confocal laser scanning microscopy
Imaging was conducted 48 and 72 h following infiltration, with a Zeiss Cell Observer SD microscope equipped with a Yokogawa CSU‐X1 spinning disc head and a 100× 1.4 numerical aperture (NA) oil immersion objective. GFP fluorescence was visualized using a 488‐nm excitation laser. Cell walls were visualized with 561‐nm laser excitation, after immersion of leaf sections in a staining solution [10 ng/mL w/v propidium iodide (Life Technologies P3566) in 1% w/v sucrose Murashige and Skoog (MS) medium] for 30 min. Callose deposits were visualized using 405‐nm laser excitation, following staining with aniline blue fluorochrome (Biosupplies Australia Pty. Ltd., Bundoora, Australia) at 5 µg/mL in 1% w/v sucrose MS medium for 15 min. Both propidium iodide (Prokhnevsky et al., 2005; Vogel et al., 2007) and aniline blue (Chisholm et al., 2001; Iglesias and Meins, 2000; Leastro et al., 2015) staining have been used previously to image the subcellular localization of virus proteins in plant cells.
Plots showing the pixel fluorescence intensity for a subset of foci were generated using Image J (http://imagej.nih.gov/ij/) to extract the intensity values recorded in the images taken by confocal microscopy. Microsoft® Excel® 2010 was used to create the plots.
Plasmolysis was performed by the incubation of N. benthamiana leaf sections at 3 dpi in 1 m mannitol containing 10 ng/mL w/v propidium iodide for 10 min. The Z series was obtained using a fully automated XYZ stage. Images were analysed using a Mac Pro 2 × 2.4‐GHz Intel Xeon E5620 8‐core computer (Apple, Cupertino, CA, USA), an Apple Cinema 27‐in display (Apple, Cupertino, CA, USA) and ImageJ http://imagej.nih.gov/ij and Bitplane Imaris version 7.2 software (Bitplane AG, Zurich, Switzerland) for three‐dimensional image reconstruction and display.
RNA isolation and RT‐PCR analysis
Total RNA was extracted from N. benthamiana leaves using the Spectrum® Plant RNA isolation kit (Life Technologies): a 0.25‐g leaf sample was macerated in 1 mL of lysis buffer containing 1% (v/v) β‐mercaptoethanol, and processed according to protocol A as described in the manufacturer's instructions. RNA pellets were resuspended in 40 µL of diethylpyrocarbonate‐treated (DEPC)–H2O, and the quality was checked using a Nanodrop2000 spectrophotometer (Thermo Scientific). DNAase treatment was performed on 10 µg of RNA using the DNA‐free™ kit, according to the manufacturer's instructions (Life Technologies). First‐strand cDNA synthesis was performed using Superscript IV reverse transcriptase (Life Technologies), according to the manufacturer's protocol, employing the OuMV primer RNA2_3′R (5′‐CCTGACCGAAGCCAGGAGAA‐3′). PCRs were performed with the GoTaq Flexi DNA Polymerase kit (Promega, Madison, WI, USA), using the forward primer MP_1for (5′‐ATGGGGGACAATGCTTTAGA‐3′) or MP_350for (5′‐TCGATTGAAGATCTGATGC‐3′) and the RNA2_3′R reverse primer. The PCR conditions consisted of 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, plus an extension at 72°C for 7 min on a Bio‐Rad thermal cycler. Amplification products were purified using the DNA Clean and Concentrator kit (ZymoResearch, Irvine, CA, USA) and sent for Sanger sequencing (Genomic Core Facility, Penn State University, University Park, PA, USA) using the appropriate forward primer.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1 Topology prediction using (A) TopPred v. 1.10 and (B) Membrane Protein Explorer v. 3.21, showing amino acids localized in putative membrane‐associated regions.
Table S1 Primers used for alanine‐scanning site‐directed mutagenesis of Ourmia melon virus RNA‐2.
Table S2 Enzyme‐linked immunosorbent assay (ELISA) absorbance values of leaf samples from Arabidopsis thaliana plants agroinfiltrated with Agrobacterium suspensions carrying pGC‐RNA1, pGC‐RNA3 and the desired pGC‐MP construct, or mechanically inoculated using, as source for inoculum, sap prepared from Nicotiana benthamiana agroinfiltrated tissue. At least five plants (P) were agroinfiltrated/inoculated for each combination. Absorbance values were recorded using a spectrophotometer following incubation for 10 min with the enzyme substrate. Samples showing absorbance values three times higher than the healthy control were considered to be positive.
Movie S1 Three‐dimensional reconstruction of Nicotiana benthamiana epidermal cells, showing the accumulation of Ourmia melon virus MP‐green fluorescent protein fusions (GFP:MP) in punctate foci at the cell periphery. Stacks of images were taken using a Zeiss Cell Observer SD microscope equipped with a Yokogawa CSU‐X1 spinning disc with excitation at 488 nm, and analysed with Bitplane Imaris version 7.2 software.
Movie S2 Three‐dimensional reconstruction of Arabidopsis thaliana epidermal cells, showing the accumulation of Ourmia melon virus MP‐green fluorescent protein fusions (GFP:MP) in punctate foci at the cell periphery. Stacks of images were taken using a Zeiss Cell Observer SD microscope equipped with a Yokogawa CSU‐X1 spinning disc with excitation at 488 nm, and analysed with Bitplane Imaris version 7.2 software.
References
- Amari, K. , Vazquez, F. and Heinlein, M. (2012) Manipulation of plant host susceptibility: an emerging role for viral movement proteins? Front. Plant Sci. 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. , Farnham, G. , Moffett, P. and Baulcombe, D.C. (2002) Constitutive gain‐of‐function mutants in a nucleotide binding site‐leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez‐Alfonso, Y. , Faulkner, C. , Ritzenthaler, C. and Maule, A.J. (2010) Plasmodesmata: gateways to local and systemic virus infection. Mol. Plant–Microbe Interact. 23, 1403–1412. [DOI] [PubMed] [Google Scholar]
- Bertens, P. , Heijne, W. , Van der Wel, N. , Wellink, J. and Van Kammen, A. (2003) Studies on the C‐terminus of the Cowpea mosaic virus movement protein. Arch. Virol. 148, 265–279. [DOI] [PubMed] [Google Scholar]
- Bertens, P. , Wellink, J. , Goldbach, R. and van Kammen, A. (2000) Mutational analysis of the Cowpea mosaic virus movement protein. Virology, 267, 199–208. [DOI] [PubMed] [Google Scholar]
- Boevink, P. and Oparka, K.J. (2005) Virus–host interactions during movement processes. Plant Physiol. 138, 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto, T. and Palukaitis, P. (1999) Are tubules generated by the 3a protein necessary for Cucumber mosaic virus movement? Mol. Plant–Microbe Interact. 12, 985–993. [Google Scholar]
- Carrington, J.C. , Kasschau, K.D. , Mahajan, S.K. and Schaad, M.C. (1996) Cell‐to‐cell and long‐distance transport of viruses in plants. Plant Cell, 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Parra, M.A. , Anderberg, R.J. and Carrington, J.C. (2001) Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long‐distance movement of Tobacco etch virus . Plant Physiol. 127, 1667–1675. [PMC free article] [PubMed] [Google Scholar]
- Choi, S.K. , Palukaitis, P. , Min, B.E. , Lee, M.Y. , Choi, J.K. and Ryu, K.H. (2005) Cucumber mosaic virus 2a polymerase and 3a movement proteins independently affect both virus movement and the timing of symptom development in zucchini squash. J. Gen. Virol. 86, 1213–1222. [DOI] [PubMed] [Google Scholar]
- Chu, M. , Park, J.W. and Scholthof, H.B. (1999) Separate regions on the Tomato bushy stunt virus p22 protein mediate cell‐to‐cell movement versus elicitation of effective resistance responses. Mol. Plant–Microbe Interact. 12, 285–292. [Google Scholar]
- Claros, M.G. and von Heijne, G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Crivelli, G. , Ciuffo, M. , Genre, A. , Masenga, V. and Turina, M. (2011) Reverse genetic analysis of ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. J. Virol. 85, 5091–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, B.C. and Wells, J.A. (1989) High‐resolution epitope mapping of hGH–receptor interactions by alanine‐scanning mutagenesis. Science, 244, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Haldeman, R. , Robertson, N.L. , Dougherty, W.G. and Carrington, J.C. (1994) Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13, 1482–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja, V.V. , Haldeman‐Cahill, R. , Montgomery, A.E. , Vandenbosch, K.A. and Carrington, J.C. (1995) Capsid protein determinants involved in cell‐to‐cell and long distance movement of tobacco etch potyvirus. Virology, 206, 1007–1016. [DOI] [PubMed] [Google Scholar]
- Fernández‐Calvino, L. , Faulkner, C. and Maule, A. (2011) Plasmodesmata as active conduits for virus cell‐to‐cell movement In: Recent Advances in Plant Virology (Caranta C., Aranda M.A., Tepfer M. and Lopez‐Moya J.J., eds), pp. 47–74. Wymondham, Norfolk: Caister Academic Press. [Google Scholar]
- Fujiki, Y. , Hubbard, A.L. , Fowler, S. and Lazarow, P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell. Biol. 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, J.A. and Pallás, V. (2015) Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 11, 21–30. [DOI] [PubMed] [Google Scholar]
- Gholamalizadeh, R. , Vahdat, A. , Hossein‐Nia, S.V. , Elahinia, A. and Bananej, K. (2008) Occurrence of Ourmia melon virus in the Guilan Province of northern Iran. Plant Dis. 92, 1135. [DOI] [PubMed] [Google Scholar]
- Giesman‐Cookmeyer, D. and Lommel, S.A. (1993) Alanine scanning mutagenesis of a plant virus movement protein identifies three functional domains. Plant Cell, 5, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries, P.A. and Nelson, R.S. (2008) Movement of viruses in plants In: Encyclopedia of Virology (Mahy B.W.J. and Van Regenmortel M.H.V., eds), pp. 348–355. Oxford: Elsevier Ltd. [Google Scholar]
- Hillman, B.I. and Cai, G. (2013) The family Narnaviridae: simplest of RNA viruses. Adv. Virus Res. 86, 149–176. [DOI] [PubMed] [Google Scholar]
- Huang, M. and Zhang, L. (1999) Association of the movement protein of Alfalfa mosaic virus with the endoplasmic reticulum and its trafficking in epidermal cells of onion bulb scales. Mol. Plant–Microbe Interact. 12, 680–690. [Google Scholar]
- Huang, M. , Jongejan, L. , Zheng, H. , Zhang, L. and Bol, J.F. (2001) Intracellular localization and movement phenotypes of Alfalfa mosaic virus movement protein mutants. Mol. Plant–Microbe Interact. 14, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Hull, R. (2014) Movement of viruses within plants In: Plant Virology (Minihane, C. and Mullane, C., eds), pp. 531–603. San Diego, CA: Academic Press. [Google Scholar]
- Iglesias, V.A. and Meins, F. (2000) Movement of plant viruses is delayed in a β‐1,3‐glucanase‐deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 21, 157–166. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. (1999) Protein secondary structure prediction based on position‐specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Jones, R.W. , Jackson, A.O. and Morris, T.J. (1990) Defective‐interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology, 176, 539–545. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. , Dolja, V.V. and Krupovic, M. (2015) Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology, 479, 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte, C. , Vetter, G. , Loudes, A.M. , Robinson, D.G. , Hillmer, S. , Stussi‐Garaud, C. and Ritzenthaler, C. (2003) Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY‐2 cells. Plant Cell, 15, 2058–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leastro, M.O. , Pallás, V. , Resende, R.O. and Sánchez‐Navarro, J.A. (2015) The movement proteins (NSm) of distinct tospoviruses peripherally associate with cellular membranes and interact with homologous and heterologous NSm and nucleocapsid proteins. Virology, 478, 39–49. [DOI] [PubMed] [Google Scholar]
- Li, W. , Lewandowski, D.J. , Hilf, M.E. and Adkins, S. (2009) Identification of domains of the Tomato spotted wilt virus NSm protein involved in tubule formation, movement and symptomatology. Virology, 390, 110–121. [DOI] [PubMed] [Google Scholar]
- Lisa, V. , Milne, R.G. , Accotto, G.P. , Boccardo, G. , Caciagli, P. and Parvizy, R. (1988) Ourmia melon virus, a virus from Iran with novel properties. Ann. Appl. Biol. 112, 291–302. [Google Scholar]
- Lucas, W.J. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. and Gilbertson, R.L. (1994) Plasmodesmata in relation to viral movement within leaf tissues. Annu. Rev. Phytopathol. 32, 387–415. [Google Scholar]
- Margaria, P. , Ciuffo, M. , Pacifico, D. and Turina, M. (2007) Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol. Plant–Microbe Interact. 20, 547–558. [DOI] [PubMed] [Google Scholar]
- Martínez‐Gil, L. , Sánchez‐Navarro, J.A. , Cruz, A. , Pallás, V. , Pérez‐Gil, J. and Mingarro, I. (2009) Plant virus cell‐to‐cell movement is not dependent on the transmembrane disposition of its movement protein. J. Virol. 83, 5535–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, U. (1990) Similarities between putative transport proteins of plant viruses. J. Gen. Virol. 71, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Melcher, U. (2000) The ‘30K’ superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R. and Elena, S.F. (2015) Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology, 476, 304–315. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R. and Koonin, E.V. (1993) Cell‐to‐cell movement of plant viruses. Insights from amino acid sequence comparisons of movement proteins and from analogies with cellular transport systems. Arch. Virol. 133, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, B. , Rubino L. and Russo, M. (2004) Expression of the Cymbidium ringspot virus 33‐kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 78, 4744–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R.S. (2005) Movement of viruses to and through plasmodesmata In: Annual Plant Reviews Plasmodesmata (Oparka K.J., ed), pp. 188–211. Oxford, UK: Blackwell Publishing. [Google Scholar]
- Oparka, K. , Prior, D. and Crawford, J. (1994) Behaviour of plasma membrane, cortical ER and plasmodesmata during plasmolysis of onion epidermal cells. Plant Cell Environ. 17, 163–171. [Google Scholar]
- Pallas, V. and García, J.A. (2011) How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92, 2691–2705. [DOI] [PubMed] [Google Scholar]
- Pei, J. and Grishin, N.V. (2014) PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three‐dimensional structural information. Methods Mol. Biol. 1079, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró, A. , Martínez‐Gil, L. , Tamborero, S. , Pallás, V. , Sánchez‐Navarro, J.A. and Mingarro, I. (2014) The Tobacco mosaic virus movement protein associates with but does not integrate into biological membranes. J. Virol. 88, 3016–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky, A.I. , Peremyslov, V.V. and Dolja, V.V. (2005) Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 79, 14 421–14 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L. and Grantham, G.L. (1995) A spontaneous mutation in the movement protein gene of brome mosaic virus modulates symptom phenotype in Nicotiana benthamiana . J. Virol. 69, 2689–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastgou, M. , Habibi, M.K. , Izadpanah, K. , Masenga, V. , Milne, R.G. , Wolf, Y.I. , Koonin, E.V. and Turina, M. (2009) Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter‐kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90, 2525–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastgou, M. , Turina, M. and Milne, R.G. (2011) Genus Ourmiavirus In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q, Adams M.J., Carstens E.B. and Lefkowitz E.J., eds), pp. 1177–1179. San Diego, CA: Academic Press. [Google Scholar]
- Ritzenthaler, C. and Hofmann, C. (2007) Tubule‐guided movement of plant viruses In Viral Transport in Plants (Waigmann E. and Heinlein M., eds), pp. 63–83. Heidelberg, Berlin: Springer‐Verlag. [Google Scholar]
- Rossi, M. , Genre, A. and Turina, M. (2014) Genetic dissection of a putative nucleolar localization signal in the coat protein of Ourmia melon virus . Arch. Virol. 159, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Rossi, M. , Vallino, M. , Abbà, S. , Ciuffo, M. , Balestrini, R. , Genre, A. and Turina, M. (2015) The importance of the kr‐rich region of the coat protein of Ourmia melon virus for host specificity, tissue tropism, and interference with antiviral defense. Mol. Plant–Microbe Interact. 28, 30–41. [DOI] [PubMed] [Google Scholar]
- Rounds, C.M. , Lubeck, E. , Hepler, P.K. and Winship, L.J. (2011) Propidium iodide competes with Ca2+ to label pectin in pollen tubes and Arabidopsis root hairs. Plant Physiol. 157, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. and Bol, J.F. (2001) Role of the Alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant–Microbe Interact. 14, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Schoelz, J.E. , Harries, P.A. and Nelson, R.S. (2011) Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant, 4, 813–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, H.B. (2005) Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10, 376–382. [DOI] [PubMed] [Google Scholar]
- Singh, P. , Indi, S.S. and Savithri, H.S. (2014) Groundnut bud necrosis virus encoded NSm associates with membranes via its C‐terminal domain. PLoS One, 9, e99370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B.A. , Evans, N.A. , Bonig, I. and Clarke, A.E. (1984) The application of Sirofluor, a chemically defined fluorochrome from aniline blue for the histochemical detection of callose. Protoplasma, 122, 191–195. [Google Scholar]
- Taliansky, M. , Torrance, L. and Kalinina, N.O. (2008) Role of plant virus movement proteins In: Methods in Molecular Biology, Plant Virology Protocols: From Viral Sequence to Function (Foster G.D., Johansen E.I., Hong Y. and Nagy P.D., eds), pp. 33–54. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Tilsner, J. , Taliansky, M.E. and Torrance, L. (2014) Plant virus movement. eLS, Wiley Online Library, Chichester, UK. doi: 10.1002/9780470015902.a0020711.pub2. [DOI]
- Tsai, C.H. and Dreher, T.W. (1993) Increased viral yield and symptom severity result from a single amino acid substitution in the Turnip yellow mosaic virus movement protein. Mol. Plant–Microbe Interact. 6, 268–273. [DOI] [PubMed] [Google Scholar]
- Ueki, S. and Citovsky, V. (2007) Spread throughout the plant: systemic transport of viruses In: Viral Transport in Plants (Waigmann E. and Heinlein M., eds), pp. 85–118. Heidelberg, Berlin: Springer‐Verlag. [Google Scholar]
- Vogel, F. , Hofius, D. and Sonnewald, U. (2007) Intracellular trafficking of potato leafroll virus movement protein in transgenic Arabidopsis. Traffic, 8, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Wang, H.L. , Wang, Y. , Giesman‐Cookmeyer, D. , Lommel, S.A. and Lucas, W.J. (1998) Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long‐distance transport system. Virology, 245, 75–89. [DOI] [PubMed] [Google Scholar]
- Yeh, H.H. , Tian, T.Y. , Medina, V. and Falk B.W. (2001) Green fluorescent protein expression from recombinant lettuce infectious yellows virus‐defective RNAs originating from RNA 2. Virology, 289, 54–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1 Topology prediction using (A) TopPred v. 1.10 and (B) Membrane Protein Explorer v. 3.21, showing amino acids localized in putative membrane‐associated regions.
Table S1 Primers used for alanine‐scanning site‐directed mutagenesis of Ourmia melon virus RNA‐2.
Table S2 Enzyme‐linked immunosorbent assay (ELISA) absorbance values of leaf samples from Arabidopsis thaliana plants agroinfiltrated with Agrobacterium suspensions carrying pGC‐RNA1, pGC‐RNA3 and the desired pGC‐MP construct, or mechanically inoculated using, as source for inoculum, sap prepared from Nicotiana benthamiana agroinfiltrated tissue. At least five plants (P) were agroinfiltrated/inoculated for each combination. Absorbance values were recorded using a spectrophotometer following incubation for 10 min with the enzyme substrate. Samples showing absorbance values three times higher than the healthy control were considered to be positive.
Movie S1 Three‐dimensional reconstruction of Nicotiana benthamiana epidermal cells, showing the accumulation of Ourmia melon virus MP‐green fluorescent protein fusions (GFP:MP) in punctate foci at the cell periphery. Stacks of images were taken using a Zeiss Cell Observer SD microscope equipped with a Yokogawa CSU‐X1 spinning disc with excitation at 488 nm, and analysed with Bitplane Imaris version 7.2 software.
Movie S2 Three‐dimensional reconstruction of Arabidopsis thaliana epidermal cells, showing the accumulation of Ourmia melon virus MP‐green fluorescent protein fusions (GFP:MP) in punctate foci at the cell periphery. Stacks of images were taken using a Zeiss Cell Observer SD microscope equipped with a Yokogawa CSU‐X1 spinning disc with excitation at 488 nm, and analysed with Bitplane Imaris version 7.2 software.


