Summary
Zoospore chemotaxis to soybean isoflavones is essential in the early stages of infection by the oomycete pathogen Phytophthora sojae. Previously, we have identified a G‐protein α subunit encoded by PsGPA1 which regulates the chemotaxis and pathogenicity of P. sojae. In the present study, we used affinity purification to identify PsGPA1‐interacting proteins, including PsHint1, a histidine triad (HIT) domain‐containing protein orthologous to human HIT nucleotide‐binding protein 1 (HINT1). PsHint1 interacted with both the guanosine triphosphate (GTP)‐ and guanosine diphosphate (GDP)‐bound forms of PsGPA1. An analysis of the gene‐silenced transformants revealed that PsHint1 was involved in the chemotropic response of zoospores to the isoflavone daidzein. During interaction with a susceptible soybean cultivar, PsHint1‐silenced transformants displayed significantly reduced infectious hyphal extension and caused a strong cell death in plants. In addition, the transformants displayed defective cyst germination, forming abnormal germ tubes that were highly branched and exhibited apical swelling. These results suggest that PsHint1 not only regulates chemotaxis by interacting with PsGPA1, but also participates in a Gα‐independent pathway involved in the pathogenicity of P. sojae.
Keywords: chemotaxis, G‐protein α subunit, HIT domain‐containing protein, interacting proteins, oomycete pathogen Phytophthora sojae, pathogenicity
Introduction
In an ever‐changing environment, it is essential for organisms to sense such changes and to respond appropriately (Wadhams and Armitage, 2004). The detection of plant‐specific molecules may be critical for microbes to recognize and subsequently colonize potential hosts (Tyler et al., 1996). For example, Rhizobium bacteria swim towards potential colonization sites after sensing chemoattractants exuded by plants (Bauer and Caetano‐Anollés, 1990). Pseudomonas fluorescens WCS365 exhibits chemotaxis towards root exudates, including organic and amino acids (de Weert et al., 2002). Parasitic fungal species of the genus Escovopsis are attracted to their hosts via host‐specific chemotaxis (Gerardo et al., 2006). In the genus Phytophthora of the oomycetes, chemotaxis has been observed not only on filamentous hyphae, but also on motile zoospores produced under flooding conditions (Hosseini et al., 2014; Morris et al., 1998; Morris and Ward, 1992). One interesting example is that the zoospores of the soybean pathogen Phytophthora sojae are able to respond to the soybean isoflavones daidzein (4′,7‐dihydroxy‐isoflavone) and genistein (4′,5,7‐trihydroxy‐isoflavone) at a concentration as low as 0.25 nm, whereas zoospores of other oomycetes do not respond to such low concentrations (Tyler et al., 1996).
Phytophthora sojae is a soil‐borne pathogen which specifically infects soybean (Tyler, 2007). The pathogen exhibits a hyphal growth pattern, and produces sporangia that can differentiate into zoospores under conditions of flooding and nutrient deprivation (Tyler et al., 1996). The infection cycle of P. sojae normally begins with motile unicellular zoospores that lack a cell wall and have two flagella, allowing them to swim towards healthy plants by sensing plant root exudates, including isoflavones (Morris and Ward, 1992). When attached to the root surfaces, zoospores lose their flagella, form walled cysts and germinate (Hardham, 2001). Germinating hyphae penetrate roots directly through the anticlinal walls between epidermal cells (Hardham, 2001). After penetration, infectious hyphae overcome plant defences to achieve colonization (Rose et al., 2002; Stintzi et al., 1993). Subsequently, intracellular aseptate hyphae ramify through all plant tissues, forming an absorptive mycelium to collect host nutrients (van West et al., 2003). Thus, zoospore chemotaxis and early infection are key stages in the interaction between P. sojae and soybean (Tyler, 2007).
Previously, in work exploring the mechanism of P. sojae chemotaxis, we performed a series of studies using gene silencing. First, we found that the heterotrimeric G‐protein α subunit PsGPA1 controlled zoospore behaviour, including chemotaxis to the isoflavone daidzein (Hua et al., 2008). Zoospores of PsGPA1‐silenced transformants failed to attach to soybean roots and exhibited changes in the expression levels of genes encoding putative downstream Gα effectors, including calcium‐binding proteins and ‘regulator of G‐protein signalling’ (RGS) proteins (Hua et al., 2008). In addition, a functional analysis of a novel group of receptor‐like proteins, termed G‐protein‐coupled receptors with a phosphatidylinositol phosphate kinase domain (GPCR‐PIPKs), revealed that one member, PsGK4, was also involved in chemotaxis and plant infection (Yang et al., 2013). Thus, it is believed that Gα‐mediated signalling is essential in the early stages of interaction between P. sojae and soybean.
A heterotrimeric G‐protein complex exists in all eukaryotes and functions as a molecular switch. A G‐protein is composed of a Gα subunit and a Gβγ dimer which is attached to the plasma membrane (Ross and Gilman, 1980). Signal molecules are perceived by the extracellular domains of seven‐transmembrane receptors (GPCRs) to activate such receptors (Neer, 1995). Ligand binding to the GPCR leads to the exchange of guanosine triphosphate (GTP) for guanosine diphosphate (GDP) on the Gα protein and the dissociation of the Gα and Gβγ dimer. Both the Gα‐GTP and Gβγ moieties regulate downstream effector proteins in various systems, including adenylyl cyclases, phospholipases and ion channels (McCudden et al., 2005). Gα activity is regulated by RGS proteins (McCudden et al., 2005). Many G‐protein α subunits have been identified in filamentous fungi, and regulatory pathways involving these proteins have now been described, including pathways involved in nutrient sensing, pheromone responses, chemotaxis, mating and pathogenesis (Li et al., 2007; Liu and Dean, 1997; Willard and Devreotes, 2006). In contrast with yeast and many filamentous fungi that contain two or three Gα proteins (Bolker, 1998; Li et al., 2007), the genome of each Phytophthora species contains only one copy of the Gα gene (Hua et al., 2008). To date, no Gα‐interacting proteins have been found in any Phytophthora species, although some genes seem to participate in the G‐protein pathway (Hua et al., 2008). It remains unknown how the Gα subunit acts during P. sojae development and infection. To identify the molecular components of the G‐protein pathway and to unravel the signalling networks underlying chemotaxis and pathogenicity, additional downstream targets of Gα should be identified.
In the present study, we used affinity purification and mass spectrometry (MS) to identify proteins physically associated with PsGPA1 of P. sojae. One such protein, PsHint1, was functionally analysed, as the homologous protein histidine triad (HIT) nucleotide‐binding protein 1 (HINT1) of humans has been reported to interact with RGSZ1, a regulator of G‐protein signalling (Ajit et al., 2007). The functions of PsHint1 in zoospore chemotaxis and during infection were analysed and compared with those of PsGPA1.
Results
PsHint1 is associated with PsGPA1 both in vivo and in vitro
To identify PsGPA1‐interacting proteins, we generated P. sojae transformants expressing 3 × FLAG‐tagged PsGPA1. Transformants were chosen for total protein isolation and further analysis. Phytophthora sojae transformed with a 3 × FLAG‐expressing tag (only) in the vector pTOR was used as a control. Western blot with anti‐FLAG antibodies revealed a band of 42 kDa, indicating that the PsGPA1‐3 × FLAG fusion protein was present in the total protein lysates of the transformants, but not in the control (Fig. S1A, see Supporting Information). This protein, isolated from sporulating hyphae and zoospores of the transformants, was next purified by admixing total protein lysates with anti‐FLAG M2 beads (Fig. S1B). The bead‐bound proteins were subsequently eluted and analysed by MS analysis (see Experimental procedures). Four proteins, a calcium‐dependent protein kinase (Ps555885; JGI V3.0, Protein ID), an acetyl‐CoA synthetase (Ps507774), a WD repeat‐containing protein (Ps359158) and a HIT motif‐containing protein (Ps492857; hereafter PsHint1) co‐precipitated with 3 × FLAG‐tagged PsGPA1 (Table S1, see Supporting Information). The gene encoding PsHint1 was cloned because the HIT motif‐containing protein HINT1 of humans interacts with RGSZ1, but no homologue thereof has yet been functionally studied in fungal or oomycete pathogens. The corrected PsHint1 gene model is 483 bp in length [scaffold_3: 4205502–4205984 (+)] (Fig. S2, see Supporting Information), intronless and encodes a 160‐amino‐acid protein with a HIT motif at the C‐terminus (Fig. 1A).
Figure 1.
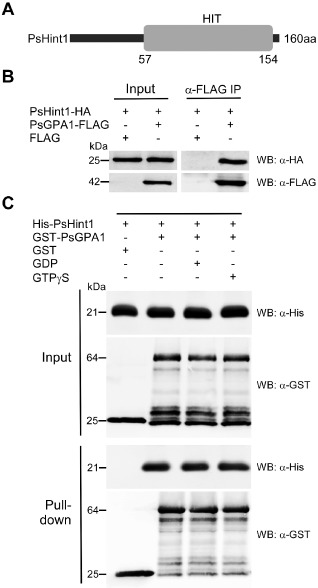
PsGPA1 can interact with PsHint1 in vivo and in vitro. (A) The domain architecture of PsHint1. The numbers under the histidine triad (HIT) domain are the amino acid positions in the full‐length protein. (B) Confirmation of PsGPA1 association with PsHint1 in vivo. PsHint1‐HA (haemagglutinin) was expressed with FLAG or with FLAG‐tagged PsGPA1 in Phytophthora sojae. Immunoprecipitation was performed with anti‐FLAG M2 beads and immunoblotted with anti‐HA and anti‐FLAG antibodies. (C) Both guanosine diphosphate (GDP)‐ and guanosine triphosphate (GTP)γS‐bound forms of PsGPA1 interacted with PsHint1 in vitro. Glutathione S‐transferase (GST)‐fused PsGPA1 and histidine (His)‐tagged PsHint1 were expressed in Escherichia coli and used for further analysis. The addition of GST or GST‐PsGPA1 is indicated in each lane. GST‐PsGPA1 and His‐PsHint1 were incubated in buffer with or without GDP or GTPγS, followed by the addition of glutathione–agarose beads. The antibodies used for Western blot are abbreviated as WB:α‐His and WB:α‐GST. The top panels show the proteins from whole‐cell extracts, and the bottom panels show the pull‐down proteins.
To confirm and further study the associations, PsHint1 was cloned and fused to a 6 × HA (haemagglutinin) tag, and expressed with 3 × FLAG or with 3 × FLAG‐tagged PsGPA1 in P. sojae. Using co‐immunoprecipitation (CO‐IP), we validated the association between PsHint1 and PsGPA1 (Fig. 1B). We also used in vitro glutathione S‐transferase (GST) pull‐down assay to confirm that PsGPA1 interacts with PsHint1 (Fig. 1C). GST‐PsGPA1 and 6 × His‐PsHint1 were expressed separately in Escherichia coli, purified and used for a pull‐down experiment. As shown in Fig. 1C, 6 × His‐PsHint1 bound specifically to GST‐PsGPA1, but not to GST, suggesting that these two proteins interacted directly. Furthermore, PsHint1 interacted with PsGPA1 to the same extent in the presence of either GDP or GTPγS (Fig. 1C), indicating that Gα activity did not influence the interaction.
Phylogenetic and expression analysis of PsHint1
Based on blast searches with representative Hint proteins from well‐characterized model organisms, Hint homologues were identified from P. sojae and 19 other eukaryotic or prokaryotic genomes. At least one Hint was present in each genome, and the P. sojae genome contained four separate Hint‐encoding genes. Conserved domain searches of Hint proteins revealed that each had a HIT motif (Fig. S3A, see Supporting Information). Alignment of the HIT motif amino acid sequences revealed that PsHint1 exhibited a highly conserved sequence (Fig. S3B). To test whether the other P. sojae Hint homologues interact with PsGPA1, we checked and corrected the corresponding gene models for PsHint2 (Ps311252), PsHint3 (Ps383555) and PsHint4 (Ps506062) proteins, which were 24%, 15% and 9% identical to PsHint1, respectively (Fig. S4A–C, see Supporting Information). The encoded proteins were assayed for interactions with PsGPA1. We found no evidence of any interaction of PsGPA1 with PsHint2, PsHint3 or PsHint4 (Fig. S4D).
A phylogenetic relationship was established using protein sequences of the HIT motifs (Fig. 2). The Hint proteins were separated into two distinct clades. The three proteins encoded by Homo sapiens are HINT1 (NP005331), HINT2 (NP115982) and HINT3 (NP612638). PsHint1 (Ps492857) fell into the same clade as HINT1 (NP005331) and HINT2 (NP115982), being most closely related to HINT1 (44% identity) and HINT2 (47% identity). In addition, PsHint1 was very similar to Pr52195 of P. ramorum (85% identity), Pc574037 of P. capsici (81% identity) and PITG_21011 of P. infestans (79% identity).
Figure 2.
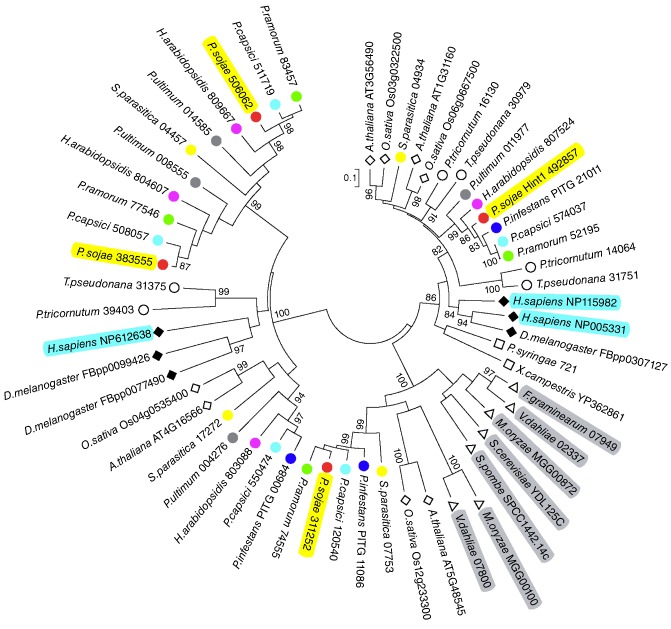
Phylogenetic analysis of putative Hint proteins. Neighbour‐joining tree was constructed based on the amino acid sequences of histidine triad (HIT) motifs. The coloured circles represent Hint proteins from oomycetes. Filled rhombi, mammals; open rhombi, plants; open circles, diatoms; open squares, bacteria; open triangles, fungi.
To further explore the functions of PsHint1, we employed semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) (Fig. 3A) and quantitative real‐time PCR (Fig. 3B) to assay the expression levels of PsHint1 at distinct developmental stages, including mycelia, sporulating hyphae, mixtures of zoospores and cysts, cysts, germinated cysts and infected plant tissue. PsHint1 was up‐regulated during five stages of infection; the expression level was threefold higher at 6 h post‐infection (hpi; IF6) compared with that in mycelia, suggesting that PsHint1 plays a role during infection.
Figure 3.
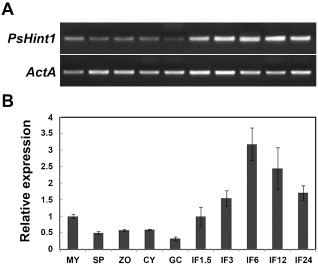
Expression of PsHint1 during the asexual life cycle and stages of infection. Expression levels were determined by reverse transcription‐polymerase chain reaction (RT‐PCR) (A) and quantitative real‐time PCR (B) using RNAs extracted from mycelia (MY), zoosporangia (SP), zoospores (ZO), cysts (CY), germinating cysts (GC) and ‘IF1.5 to IF24’ materials (samples taken 1.5, 3, 6, 12 and 24 h after the infection of soybean leaves). Relative expression levels were calculated using the MY values as references.
Silencing of PsHint1 in P. sojae
For a functional analysis of PsHint1, gene‐silenced transformants were generated using polyethylene glycol (PEG)‐mediated protoplast transformation of P. sojae (Hua et al., 2008; McLeod et al., 2008). Phytophthora sojae strain P6497 was transformed with a pTOR‐based plasmid expressing antisense PsHint1 under the control of the HAM34 promoter and terminator, and with the geneticin‐resistance gene neomycin phosphotransferase (NPT) under the control of the heat shock protein 70 (HSP70) promoter and terminator (Ah‐Fong and Judelson, 2011; Hua et al., 2008; Judelson et al., 1991). Transformants that grew stably on 10% (v/v) V8 plates containing 50 μg/mL geneticin (G8160; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) were selected to evaluate the PsHint1 mRNA levels. Strains in which PsHint1 was expressed at a level similar to that in the wild‐type served as controls (CK). There were three transformants showing significant reductions in PsHint1 expression compared with that in wild‐type and control transformants (Fig. 4). The extent of reduction was 88% for T14, 76% for T45 and 40% for T22. Genomic PCR analysis confirmed that the pTOR‐based plasmid was stably transformed into the P6497 strain (Fig. S5, see Supporting Information). To confirm that PsHint1 silencing was gene specific, we analysed the expression levels of the other three Hint‐encoding genes: PsHint2 (Ps311252), PsHint3 (Ps383555) and PsHint4 (Ps506062). In PsHint1‐silenced transformants, no significant change in the expression levels of any of these genes was observed when compared with the control (Fig. S6, see Supporting Information). Thus, the PsHint1‐silenced transformants T14, T22 and T45 were used in phenotypic analyses.
Figure 4.
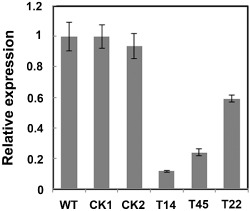
Expression analysis of PsHint1‐silenced transformants. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) evaluation of the expression levels of the gene encoding PsHint1 in hyphae of the initial control strain P6497, the control transformants (CK) and three PsHint1‐silenced transformants (T14, T45 and T22). Relative expression levels were calculated using that of the wild‐type strain P6497 as the reference. All qRT‐PCR experiments were repeated three times using independent RNA isolates.
The chemotropic response of zoospores is affected in PsHint1‐silenced transformants
Although the three PsHint1‐silenced transformants grew 15–35% more slowly than the wild‐type and CK on 10% (v/v) V8 plates (the growth reductions correlated with the silencing efficiencies), colony morphology, sporangia formation, zoospore release and oospore formation were unaffected (data not shown). As PsHint1 interacted with PsGPA1, which controls P. sojae zoospore chemotaxis (Hua et al., 2008), we tested the chemotaxis by zoospores of the silenced transformants using both soybean roots and the isoflavone daidzein, as described previously (Morris et al., 1998; Yang et al., 2013). As shown in Fig. 5A, within 10 min of the addition of soybean root material to a suspension of swimming wild‐type zoospores, encystment occurred both on and adjacent to the root elongation region, whereas zoospores of the PsHint1‐silenced transformants failed to aggregate to this region.
Figure 5.
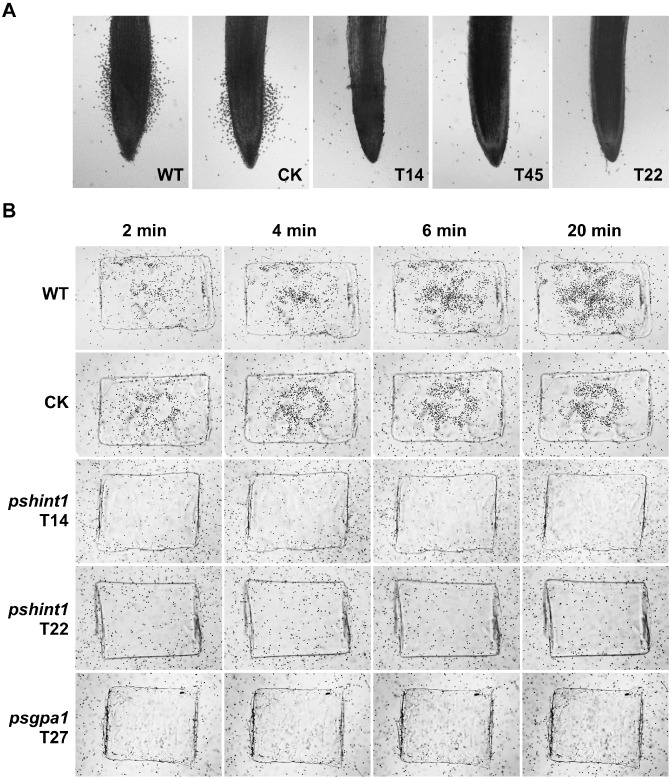
Zoospores of PsHint1‐silenced transformants do not exhibit chemotaxis towards soybean root tips or the isoflavone daidzein. (A) Root tips were placed in microscopic chambers and photographs were taken after 10 min of incubation at room temperature. (B) Agarose plugs containing 15 μm daidzein were placed in microscopic chambers and photographs were taken after 2, 4, 6 and 20 min of incubation at room temperature. The chambers were filled with equal amounts of zoospore suspensions of wild‐type, CK, PsHint1‐ or PsGPA1‐silenced transformants. WT, wild‐type strain P6497; CK, control transformant; T14, T45 and T22, PsHint1‐silenced transformants; T27, PsGPA1‐silenced transformants.
Next, soybean root material was replaced with an agarose plug containing the isoflavone daidzein, which is naturally exuded by soybean roots (Morris and Ward, 1992). Zoospores of the wild‐type swam rapidly towards the plug and began to encyst within 2 min. In contrast, zoospores of the PsHint1‐silenced transformants (T14, T22 and T45) and PsGPA1‐silenced transformants (T27) swam around the agarose plug, but did not encyst, even when the experimental period was extended to 20 min (Fig. 5B).
Interestingly, zoospores of the PsHint1‐silenced transformants encysted very quickly, like the PsGPA1‐silenced transformants. In the absence of either physical or chemical stimuli, 31%–37% of zoospores from the three PsHint1‐silenced transformants encysted within 1.5 h after their release from sporangia, whereas 80% of the wild‐type and CK zoospores swam for hours before encysting. These results suggest that PsHint1 is a Gα co‐regulator involved in zoospore chemotaxis and encystment in P. sojae.
PsHint1 is required for pathogenicity and invasive hyphal growth
To determine whether PsHint1 was involved in the infective process, the infection assay described by Yang et al. (2013) was used. Etiolated seedlings of the soybean cultivar Hefeng 47 were inoculated with zoospore suspensions from wild‐type, CK and PsHint1‐silenced transformants, and incubated under dark and wet conditions at room temperature. Disease symptoms were monitored for 3 days. Seedlings inoculated with the wild‐type strain P6497 developed lesions and died within 3 days (Fig. 6A). However, seedlings inoculated with PsHint1‐silenced transformants developed only small necrosis‐like lesions at the sites of inoculation within 3 days. Such lesions did not grow further when the incubation time was extended to 7 days (Fig. 6A), excluding the possibility that the loss of lesion development was attributable to the reduced growth rates of the PsHint1‐silenced transformants. The infection assay was also performed using wounded seedlings; similar results were obtained. Seedlings inoculated at wound sites with PsHint1‐silenced transformants did not develop disease symptoms and were still healthy at 7 days post‐infection (dpi) (Fig. S7, see Supporting Information). We thus concluded that the loss of pathogenicity was not directly associated with penetration.
Figure 6.
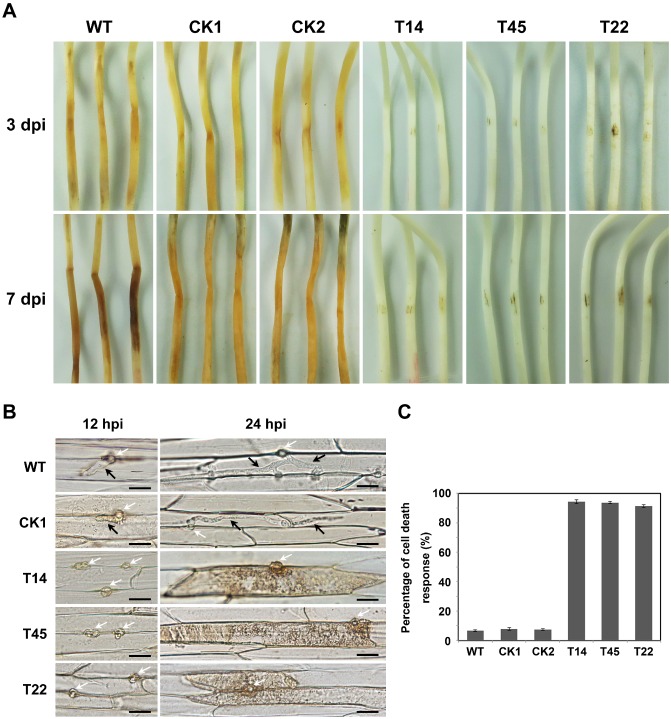
PsHint1‐silenced transformants exhibit impaired pathogenicity. (A) Pathogenicity tests (using the susceptible soybean cultivar Hefeng47) using zoospores from wild‐type (WT), control transformants (CK1 and CK2) and PsHint1‐silenced transformants (T14, T45 and T22). Etiolated hypocotyls were drop inoculated with equal numbers of zoospores (100 zoospores/5–10 μL) and incubated at 25 °C in the dark. (B) Microscopic observations of invasive hyphae in epidermal cells of soybean hypocotyls. White arrows, infectious ostioles; black arrows, invasive hyphae. Bar, 20 μm. (C) Percentage of cell death response of inoculated soybean hypocotyls of WT, CK and PsHint1‐silenced transformants. Epidermal cells were harvested and observed at 24 h after inoculation.
To explore why pathogenicity was lost, epidermal cells from the seedling inoculation sites were excised at different time points and examined microscopically. At 12 hpi, the wild‐type strain P6497, CK and PsHint1‐silenced transformants had penetrated the epidermal cells. Although the invasive hyphae of the wild‐type expanded freely into adjacent epidermal cells, the hyphae of transformants were restricted to the region around the penetration site (Fig. 6B). At 24 hpi, infectious P6497 hyphae extended into neighbouring cells. In contrast, 90% of the plant cells penetrated by PsHint1‐silenced transformants developed a cell death response (Fig. 6B,C), which may contribute to the suppression of P. sojae infections.
Germinated cysts of PsHint1‐silenced transformants exhibit impaired polarized growth
Apart from changes in chemotaxis and pathogenicity, the PsHint1‐silenced transformants exhibited phenotypic features that may not reflect Gα silencing. As PsGPA1 silencing impairs cyst germination (Hua et al., 2008), we explored this phenotype in PsHint1‐silenced transformants. In 5% (v/v) V8 liquid medium, 90% of the cysts from the wild‐type strain and CK transformant germinated within 1.5 h, whereas 70% or fewer cysts from the PsHint1‐silenced transformants formed germination tubes within 1.5 h (64.2% for T14, 66.6% for T45 and 69.5% for T22). In the wild‐type and CK strain, 90% of germ tubes maintained the characteristic shape (a tapering tip and a constant tube diameter) up to 9 h after germination (Fig. 7A). However, in PsHint1‐silenced transformants, 52%–90% of the germ tubes exhibited branches and apical swelling at 9 h after germination (Fig. 7A), indicating that polarized growth was affected relatively early. The proportions of abnormally germinated cysts of P6497, CK1, CK2, T14, T45 and T22 were 10%, 7%, 8%, 90%, 75% and 52%, respectively (Fig. 7B), and the differences were statistically significant.
Figure 7.
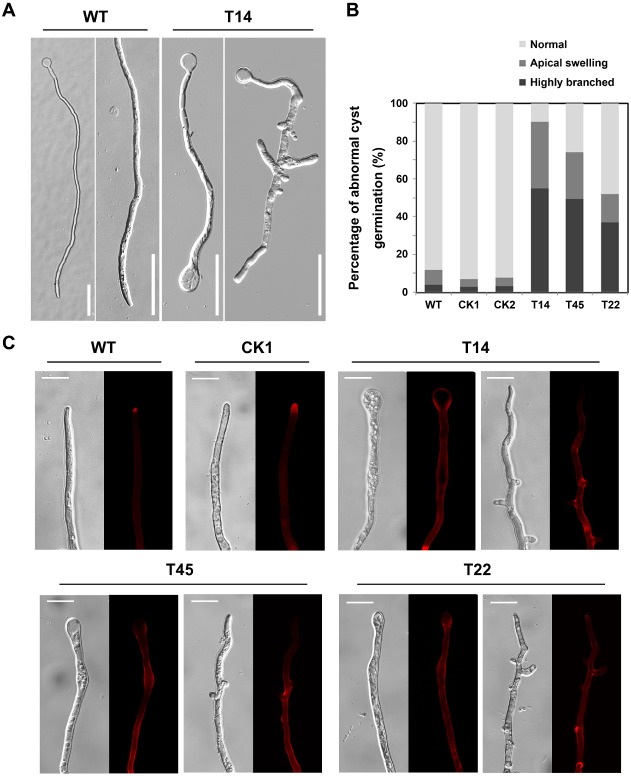
PsHint1 is required for normal cyst germination. (A) Germ tubes of the transformants were abnormal in terms of morphology. Cysts from the wild‐type strain and the PsHint1‐silenced transformant T14 were incubated at 25 °C for 9 h, after which photographs were taken. The results obtained for T45 and T22, the other two PsHint1‐silenced transformants, were similar to those obtained for T14. Bar, 50 μm. (B) Numbers of abnormally germinated cysts (with apical swelling or highly branched) were counted under a microscope, and the ratios of abnormally germinated cysts to the total number of germinated cysts were calculated. (C) Germinated cysts of the wild‐type strain, control transformant and PsHint1‐silenced transformants were stained with Congo Red and examined via epifluorescence microscopy. In silenced transformants, stained patches were detected along the hyphae, indicating abnormal polysaccharide accumulation. Bar, 20 μm.
To explore the cause of abnormal germ tube formation, we stained the cell walls of silenced transformants with Congo Red. As the cell walls of oomycetes consist essentially of β‐1,3‐polysaccharides, β‐1,6‐polysaccharides and cellulose (Guerriero et al., 2010; Lin and Aronson, 1970), changes in the levels and positions of polysaccharides during cyst germination can be observed on staining. After 5 min of staining, red fluorescence accumulated at the hyphal tips of the wild‐type and CK strain, whereas, in the PsHint1‐silenced transformants, fluorescence was evident along the hyphae and at branch sites (Fig. 7C), indicating that not only did PsHint1 interact with the Gα subunit, but was also involved in polysaccharide transport influencing the polarized growth of hyphae.
Discussion
PsHint1 is an interacting partner of PsGPA1
Phytophthora sojae is a soil‐borne oomycete pathogen which specifically infects soybean (Tyler, 2007). Zoospore chemotaxis in response to low levels of soybean isoflavones seems to contribute to host specificity (Morris and Ward, 1992). Previous work has found that the zoospore motility and chemotaxis of Phytophthora species are controlled by the G‐protein α subunit GPA1 (Hua et al., 2008; Latijnhouwers et al., 2004), and that the response of P. sojae to the isoflavone daidzein also involves the action of a novel oomycete membrane receptor, PsGK4 (Yang et al., 2013). However, no protein has previously been shown to interact directly with GPA1, although certain genes were up‐ or down‐regulated in Gα‐silenced transformants (Hua et al., 2008). In the present study, a HIT nucleotide‐binding motif‐containing protein, PsHint1, was identified as a PsGPA1‐interacting protein via affinity purification coupled with MS, and the interaction was confirmed using CO‐IP and GST pull‐down assays. HIT proteins, so‐termed because of the presence of a motif related to the sequence HφHφHφφ (where φ is a hydrophobic amino acid), form a superfamily of nucleotide hydrolases and transferases. In humans, five such families are recognized. HINT proteins form the first branch of the HIT superfamily, and at least one Hint protein is present in all fully sequenced genomes (Martin et al., 2011). Previous work described an interaction between human HINT1 and RGSZ1, and showed that the major function of HINT1 was to modulate the mu opioid receptor signalling pathway, together with RGSZ1 (Ajit et al., 2007). In E. coli, the Hint orthologue echinT is essential for growth under high‐salt conditions (Chou et al., 2005). In oomycetes and fungi, the functions of Hint homologues have not been studied. Thus, this is the first report to show that the Gα subunit in an oomycete interacts directly with a Hint protein, and describes a novel G‐protein signalling pathway.
The similar effects on zoospore chemotaxis of PsGPA1‐ and PsHint1‐silenced transformants indicate that PsHint1 participates in the control of P. sojae chemotaxis, like PsGPA1. However, the impaired pathogenicity of the PsGPA1‐silenced transformants is attributable to a disturbance in pre‐infective events, including cyst germination, and not to an effect on virulence per se or vegetative growth (Hua et al., 2008). Yet, the PsHint1‐silenced transformants were defective in terms of invasive hyphal growth, but not cyst germination. The expression pattern of PsHint1 also differed from that of PsGPA1 at various developmental stages. PsHint1 was up‐regulated during five stages of infection (Fig. 3). PsGPA1 mRNAs were also detectable in stages of infection, but the highest levels of expression were found in sporulating hyphae and zoospores (Hua et al., 2008).
Interestingly, our GST pull‐down assay showed that PsHint1 interacted with both the GDP‐ and GTP‐bound forms of PsGPA1 (Fig. 1C), implying that PsHint1 does not preferentially interact with the activated form of the Gα subunit. This indicates that PsHint1 might be a component of the heterotrimeric G‐protein complex, perhaps functioning as a scaffold protein. We suspect that PsHint1 acts as a co‐regulator of PsGPA1 in the context of zoospore chemotaxis; thus, PsHint1 may not be a Gα downstream effector. As the GST pull‐down assay does not explore kinetic features, the PsHint1 affinities to PsGPA1 bound to either GDP or GTP could differ, but differences have not yet been detected. During interactions between human Gα subunits and downstream effectors, small differences in affinity may be allosterically amplified to exert large effects on the ratios of GTP‐ and GDP‐bound protein states (Hatley et al., 2003). Therefore, kinetic analysis‐based modelling, which may reveal small affinity differences (Huang et al., 2006), should be conducted to measure the affinities of PsHint1 for the two nucleotide‐bound states of PsGPA1.
It is possible that PsHint1 is also involved in pathways other than those associated with heterotrimeric G‐proteins. Ongoing work on other putative PsGPA1‐interacting proteins, including calcium‐dependent protein kinases, acetyl‐CoA synthetase and WD repeat‐containing proteins, will complement our work on the Gα signalling network.
PsHint1 function affects zoospore chemotaxis more specifically than motility
Zoospores play key roles in the dispersal of, and initiation of infection by, P. sojae in the field. Zoospore chemotaxis is thought to be important to allow the pathogen to locate to soybean roots, but how external stimuli are perceived and actioned remains poorly understood. Earlier studies on G‐protein α subunits of Phytophthora species clearly showed that zoospore motility and chemotaxis were controlled by Gα‐mediated signalling (Hua et al., 2008; Latijnhouwers et al., 2004). In comparison with wild‐type zoospores, PsGPA1‐ or PiGPA1‐silenced transformant zoospores swam more slowly and changed direction more frequently. Zoospores of PsGPA1‐silenced transformants were defective in chemotaxis to the isoflavone daidzein (Hua et al., 2008), whereas PiGPA1‐silenced zoospores were defective in chemotaxis to glutamic acid (Latijnhouwers et al., 2004). In the present study, we found that PsHint1 is required for a normal chemotactic response to isoflavones (Fig. 5B). In contrast with GPA1‐silenced zoospores, PsHint1‐silenced transformants did not exhibit changes in motility under normal conditions.
Zoospores sense gradients of specific chemical signals released from hosts, locate the hosts and aggregate at sites of infection (Horio et al., 1992; Morris and Ward, 1992). Such aggregated zoospores become sluggish, spin in tight circles and rapidly shed their flagella before becoming round cytospores (this is the encystment process) (Islam et al., 2002, 2003; Latijnhouwers et al., 2002). Two basic mechanisms are employed to target zoospores to their host: zoospore taxis and the induction of zoospore immobilization (van West et al., 2003). It is possible that a signal inducing zoospore immobilization is constitutively activated in PsGPA1‐ or PiGPA1‐silenced transformants, under which circumstances the Gα subunit would serve as a negative regulator. In the present study, PsHint1‐silenced transformants were affected in terms of chemotaxis, as were Gα‐silenced transformants, revealing that cross‐talk between the G‐protein signalling pathway and a PsHint1‐mediated pathway positively regulates zoospore chemotaxis. Our data also indicate that zoospore chemotaxis and motility are controlled by two different pathways, both of which are regulated by the Gα subunit, whereas PsHint1 is involved in only one of the pathways.
In previous work with peronosporomycetes (oomycetes), selective inhibitors of protein kinase C (PKC) affected zoospore swimming behaviour, suggesting that PKC activity is indispensable for normal swimming (Islam et al., 2011). The phenotype was similar to that of the PsGPA1‐silenced transformant, indicating that a relationship may exist between PKC and Gα. Human studies have suggested that purified PKCα can phosphorylate purified Gαz in vitro (Ajit et al., 2007), and that PKCI‐1 (HINT1) indeed inhibits PKC, but such activity may require the presence of transcription factor 14‐3‐3 proteins (Robinson and Aitken, 1994). No direct interaction, including phosphorylation, has been shown among human HINT1, PKC and Gα proteins, and any such interaction in P. sojae remains to be explored. Our current results suggest that PsHint1 is associated with PsGPA1 in a scaffolding context, and another protein, perhaps analogous to PKC in humans, functions to control zoospore chemotaxis. Therefore, proteins interacting with PsGPA1 and PsHint1 should continue to be investigated.
PsHint1 is essential for full virulence
As expected, the silencing of PsHint1 reduced P. sojae pathogenicity. However, the extent of the reduction was much greater in PsHint1‐silenced transformants than in PsGPA1‐silenced strains. The former transformants seemed to have lost all virulence, as necrosis‐like lesions on susceptible soybeans that developed after zoospore inoculation did not expand by 7 dpi (Fig. 6A), reminiscent of the phenotype when incompatibility is present. To explore this phenomenon, inoculated soybean seedlings were examined microscopically at various times after inoculation. Germ tubes of PsHint1‐silenced transformants could penetrate plant cells, but infectious hyphal growth was absent (Fig. 6B). This was distinct from that observed with PsGPA1‐silenced transformants; pre‐infection events, such as encystment and cyst germination, were disturbed (Hua et al., 2008). Although cyst germination was abnormal and the germ tubes lacked growth polarity in PsHint1‐silenced transformants (Fig. 7A), these phenotypic features do not explain why infectious hyphal growth was restricted.
Apart from the hyphal growth defect, the subsequent death of penetrated plant cells may have contributed to the suppression of invasive growth (Fig. 6B). It is possible that a powerful plant defence system was induced by PsHint1 gene silencing, or that the PsHint1‐silenced transformant failed to suppress the basal defences of soybean (those induced by the recognition of microbe‐associated molecules). Such defences often include cell wall strengthening, rapid reactive oxygen species (ROS) production (Chen et al., 2008) and the transcriptional activation of pathogenesis‐related (PR) genes (Nurnberger et al., 2004). On Phytophthora–host plant interaction, plant cell death could be suppressed or accelerated by RXLR effectors (Doehlemann and Hemetsberger, 2013; Giraldo and Valent, 2013; Kamoun, 2006; Wang et al., 2011). In addition to RXLR effectors, there are a variety of secreted proteins from Phytophthora that are capable of causing plant cell death, including elicitins, crinkling‐ and necrosis‐inducing proteins (CRNs) and necrosis‐ and ethylene‐inducing‐like proteins (NLPs) (Dong et al., 2012; Kamoun et al., 1997; Qutob et al., 2002; Torto et al., 2003). PsHint1 might be involved in the transcription or secretion of these proteins. Therefore, further work should focus on the analysis of the cell death response induced by PsHint1 silencing, and the expression, secretion and mode of action of RXLR effectors and other Phytophthora secreted proteins in PsHint1‐silenced transformants.
Experimental Procedures
Phytophthora sojae strains and culture conditions
The genome‐sequenced P. sojae strain P6497 (Race 2), generously provided by Dr Brett Tyler (Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA), served as the wild‐type strain. The wild‐type strain and silenced transformants thereof were routinely grown on 10% (v/v) V8 medium at 25 °C in the dark. We obtained asexual samples, including hyphae, zoospores and germinated cysts, as described previously (Hua et al., 2008). The growth rates of the transformants were calculated using cultures growing on 10% (v/v) V8 agar medium. Sporulating hyphae or zoospores of P. sojae were prepared by repeatedly washing 7‐day‐old hyphae with water, followed by incubation for 4–8 h at 25 °C until most hyphae developed sporangia, which then released zoospores.
Plasmid construction
For the affinity purification assay, the full‐length coding region of PsGPA1 was PCR amplified using P. sojae cDNA with the primers PsGPA‐FLAG‐F/R (Table S2, see Supporting Information) , and Primer‐Star polymerase (Takara Bio Inc., Otsu, Japan). Similarly, the 3 × FLAG fragment was amplified using the primers FLAG‐overlap‐F and FLAG‐R (Table S2, see Supporting Information). Overlapping fragments of PsGPA1 and 3 × FLAG were inserted between the EcoRI and SacII sites of pTOR to form the fusion construct, and the 3 × FLAG fragment (FLAG‐F/R) was inserted into pTOR as an empty vector control. For the co‐expression of HA‐tagged PsHint1 and FLAG‐tagged PsGPA1 in P. sojae, PsHint1 and PsGPA1‐flag were PCR amplified using the primers PsHint1‐GG‐F/R and PsGPA‐flag‐GG‐F/R (Table S2, see Supporting Information), which had BsaI recognition sites in their respective fusion sites and in their 5′ extensions. The PCR fragments were cloned with a BsaI‐based Golden Gate cloning reaction (Engler et al., 2008) in the pTOR‐GG vector. The expression vector is based on pTOR vector and has two BsaI sites compatible with the entry clone.
For the in vitro GST pull‐down assay, full‐length PsGPA1 cDNA was obtained using the primers PsGPA‐GST‐F/R (Table S2, see Supporting Information) and inserted between the EcoRI and XhoI sites of pGEX‐4T‐1 (GE Healthcare, Little Chalfont, Buckinghamshire, UK) in‐frame with the sequence encoding GST. The sequence encoding PsHint1 was amplified using P. sojae cDNA with the primers PsHint1‐his‐F/R (Table S2, see Supporting Information) and inserted between the BamHI and HindIII sites of pET28a(+) (Novagen, Madison, WI, USA).
To silence PsHint1 of P. sojae, PsHint1 cDNA (PsHint1‐STR‐anti‐F/R) in the antisense orientation was inserted into pTOR prior to P. sojae transformation.
Phylogenetic analysis
GenBank accession numbers are shown in Fig. 2. Multiple alignments of the amino acid sequences of HIT motifs were initially performed using Clustal X (Thompson et al., 1997) (Table S3, see Supporting Information). Phylogenetic trees were constructed using mega 4.0 (Tamura et al., 2007), employing the neighbour‐joining method. All sequence alignments were tested by bootstrapping (10 000 repetitions). Conserved domain searches were performed using the smart (smart.embl‐heidelberg.de/) and Pfam (pfam.sanger.ac.uk/search) search programs.
Transformation of P. sojae
We transformed P. sojae with various plasmids using a PEG‐mediated protoplast transformation system (Hua et al., 2008). Putative transformants were identified by growth on 10% (v/v) V8 medium containing 50 μg/mL geneticin. Stable transformation of gentamycin‐resistant colonies by pTOR‐based plasmids was evaluated by PCR using the primers pTOR00F and pTOR00R. To select transformants suitable for affinity purification, proteins isolated from sporulating hyphae of genomically PCR‐positive transformants were analysed by Western blot using anti‐FLAG antibodies (Abmart Inc., Shanghai, China). To screen PsHint1‐silenced transformants, genomically PCR‐positive transformants were subjected to qRT‐PCR using the primers PsHint1‐QRT‐F/R. Transformants in which the PsHint1 level was similar to that in the wild‐type served as controls.
RNA extraction and gene expression analysis
Total RNAs from distinct stages of the life cycle of P. sojae (vegetative hyphae, sporulating hyphae, zoospores, cysts, germinated cysts and stages of infection) were extracted as described previously (Hua et al., 2008; Yang et al., 2013). RNA integrity was tested via agarose gel electrophoresis. Prior to cDNA synthesis, all RNA samples were DNase I‐treated using a DNase kit, following the manufacturer's protocol (Takara Bio Inc.). First‐strand cDNA was synthesized from 1–5 μg of total RNA using oligo (dT) priming with an M‐MLV reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA), following the manufacturer's protocol. Semi‐RT‐PCR was performed to amplify the actin A gene (ACT‐RTF/R) and PsHint1 (PsHint1‐QRT‐F/R). SYBR green qRT‐PCR assays were used to measure PsHint1 expression with the aid of the primer pair PsHint1‐QRT‐F/R. All reactions were performed on an ABI 7500 Fast Real‐Time PCR system (Applied Biosystems Inc., Foster City, CA, USA); the results were analysed using ABI 7500 sequence detection software. Relative gene expression levels were determined using actin A (ACT‐RTF/R) levels as internal controls. Means and standard deviations were calculated using data from three replicates.
CO‐IP and GST pull‐down assay
Plasmids pTOR::GPA1‐FLAG and pTOR::FLAG were (separately) transformed into P. sojae strain P6497. Sporulating hyphae were prepared by repeatedly washing 2‐day‐old mycelia grown in 10% (v/v) V8 broth with sterile distilled water (SDW), followed by incubation in the dark at 25 °C for 4–8 h, until sporangia developed. Zoospores were produced as described above. Harvested sporulating hyphae and zoospores were ground to a powder using a pestle and mortar, and resuspended in lysis buffer [1% (v/v) protease inhibitor cocktail (Sigma‐Aldrich, St. Louis, MO, USA) with 1% (v/v) 0.1 m phenylmethylsulfonyl fluoride (PMSF) (Beyotime, Shanghai, China) in 1 × phosphate‐buffered saline (PBS)]. Cell debris was removed by centrifugation for 20 min at 20 000 g at 4 °C. Anti‐FLAG M2 beads (1 U; Sigma‐Aldrich) were added to 40 U of lysate, followed by incubation for 5 h at 4 °C with gentle shaking. Proteins binding to the beads were eluted into elution buffer (7 m urea, 2 m thiourea, 65 mm dithiothreitol and 200 mm PMSF) and analysed via nanoflow liquid chromatography‐tandem MS.
The eluted proteins were digested with trypsin at an enzyme‐to‐protein ratio of 1:50 (w/w) in a 37 °C water bath for 16 h. Tryptic peptides were analysed by nanoflow liquid chromatography‐tandem MS on a high‐resolution hybrid linear ion trap orbitrap mass spectrometer (LTQ‐Orbitrap; ThermoFisher, San Jose, CA, USA) equipped with a nanoelectrospray ion source. An Agilent (Santa Clara, CA, USA) 1100 series liquid chromatography system equipped with a reverse‐phase microcapillary column (0.075 mm × 150 mm, Acclaim® PepMap100 C18 column, 3 μm, 100 Å; DIONEX, Sunnyvale, CA, USA) was used, with the mobile phase consisting of buffer A (0.1% formic acid in H2O) and buffer B (0.1% formic acid in acetonitrile). The analytical condition was set at a linear gradient from 0 to 60% buffer B in 60 min, and the flow rate was adjusted to 200 nL/min. The column was re‐equilibrated at initial conditions for 10 min (Wang et al., 2013). The tandem MS spectra acquired from precursor ions were submitted to Maxquant (version 1.2.2.5) using the following search parameters: the data were queried against the National Center for Biotechnology Information (NCBI) non‐redundant P. sojae protein database; the enzyme was trypsin (KR/P); tandem MS tolerance was set at 20 ppm; the minimum peptide length was 6; the false detection rates for peptides and proteins were all set to less than 0.01. The peptides which were unique when analysing the genome were used for further analysis. The data were analysed against a background list of proteins that bound non‐specifically to the anti‐FLAG antibody (as revealed by the empty vector control).
Plasmids pGEX‐4T‐1::GPA1 and pET28a::PsHint1 were introduced (separately) into the E. coli strain BL21 (DE3). Large‐scale bacterial sonicates and a glutathione Sepharose 4B slurry were prepared as described in the Handbook of the GST Gene Fusion System (GE Healthcare). Equal amounts of GST‐PsGPA1‐ and His‐PsHint1‐containing lysates were mixed in the presence of 5 mm MgSO4 and 100 mm GDP or GTPγS, incubated at 25 °C for 1 h in the presence of glutathione–agarose, washed six times in 1 × PBS, and the bound proteins were eluted into 5 × sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) sample buffer. The proteins were separated by SDS‐PAGE and immunoblotted with anti‐His and anti‐GST (Abmart Inc.) antibodies.
Chemotaxis assay
Chemotropism assays were performed in an assay chamber as described previously (Morris et al., 1998). Chemotaxis of zoospores towards soybean roots was examined by incubating 500‐μL aliquots of zoospore suspensions (104 mL−1) in microscopic chambers that contained soybean root tips. After 10 min at 25 °C, photographs were taken of zoospores and cysts surrounding the root tips. To evaluate chemotaxis towards daidzein, 2% (w/v) agarose plugs containing 15 μm daidzein (Sigma‐Aldrich) were placed in chambers, and photographs were taken every 2 min thereafter. Each strain was tested using at least two different preparations of zoospores. All assays were repeated at least three times.
Virulence assays
The soybean cultivar Hefeng 47, which is susceptible to P. sojae strain P6497, was grown in plastic pots containing vermiculite at 25 °C for 4 days in the dark. Zoospores were retrieved as described above and the numbers thereof in 10‐μL amounts of water were counted under a microscope. Wounded or unwounded hypocotyls of etiolated seedlings were inoculated with 100 zoospores, and the seedlings were maintained in a climate‐controlled room at 25 °C and 80% relative humidity in the dark. Manifestations of pathogenicity were evaluated at 3 and 7 dpi, and photographs were taken. To allow for microscopic observation of the penetration of, and infectious hyphal expansion within, soybean tissue, infected epidermal cells were collected at 12 and 24 hpi and examined under a light microscope. Each strain was tested using at least two different preparations of zoospores, and five plants. All assays were independently repeated at least four times.
Cyst germination and Congo Red staining
Tubes containing 500 μL of zoospore suspensions and equal volumes of clarified 10% (v/v) V8 liquid medium were vortexed to induce encystment and then incubated for 2 h at 25 °C in the dark. Drops of cyst suspensions were transferred to glass slides prior to microscopy. Cysts were scored as germinated if the germ tube length equalled or exceeded the cyst diameter (10 μm). To allow the numbers of abnormally germinated cysts to be counted, the tubes were incubated for 9 h. At least 100 cysts were examined in each sample, and all treatments were replicated three times.
To explore cyst morphology, cyst wall polysaccharides were stained with Congo Red [Sigma‐Aldrich; 0.5% (w/v) in water] for 5–10 min, washed in SDW (Siriputthaiwan et al., 2005), mounted on microscope slides and observed using a Zeiss microscope (Jena, Germany).
Supporting information
Fig. S1 Immunoblot analysis of the expression and immunoprecipitation of PsGPA1‐3 × FLAG fusion proteins with the anti‐FLAG antibody. A 42‐kDa band was detected in total proteins (A) and proteins eluted from anti‐FLAG M2 beads (B) in transformants expressing the PsGPA1‐3 × FLAG fusion protein.
Fig. S2 Correction of PsHint1 gene model. (A) The automatic model was suggested to be 408 bp (grey blocks). According to the distribution of transcript tags (Ye et al., 2011) and the alignments against homologues from other oomycetes, the automatic model is incorrect. The new gene model was suggested to be 483 bp in length. Four primer pairs were designed to verify this model, i.e. SF1‐SR, SF2‐SR, SF3‐SR and SF4‐SR. (B) Polymerase chain reaction (PCR) products could be obtained from Phytophthora sojae cDNA using SF2‐SR, SF3‐SR and SF4‐SR, but not SF1‐SR. The SF2‐SR product was sequenced and matched perfectly to the corrected gene model. (C, D) Gene model for Hint1 protein of P. infestans was also checked and reanalysed, and the results were similar to PsHint1.
Fig. S3 Histidine triad (HIT) motifs of Hint proteins from different organisms. (A) Physical maps of Hint proteins showing conserved HIT motifs. (B) Amino acid sequence alignments of HIT motifs from different species, performed using the Clustal W program. Identical amino acid residues are shown with a dark grey background, and similar amino acid residues with a light grey background. Conserved histidine residues are indicated by asterisks at the bottom.
Fig. S4 Other Hint proteins of Phytophthora sojae, PsHint2, PsHint3 and PsHint4, do not interact with PsGPA1.
Fig. S5 Genomic polymerase chain reaction (PCR) analysis of plasmid stably transformed into the PsHint1‐silenced transformants. Genomic PCR screens were performed using genomic DNA as template and the primers pTOR00F and pTOR00R on the indicated strains. WT, wild‐type strain P6497; T14, T45 and T22, PsHint1‐silenced transformants. Product size is shown on the right.
Fig. S6 Expression of PsHint2, PsHint3 and PsHint4 in PsHint1‐silenced transformants. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) evaluation of gene expression levels using hyphal RNA from the recipient strain P6497, a control transformant (CK) and PsHint1‐silenced transformants (T14, T45 and T22). Relative expression levels were calculated using data from the wild‐type strain P6497 as the reference.
Fig. S7 Pathogenicity tests on the wounded susceptible soybean cultivar Hefeng47 using zoospores from wild‐type, control transformants (CK1 and CK2) and PsHint1‐silenced transformants (T14, T45 and T22). Wounded seedlings were drop inoculated with equal numbers of zoospores (100 zoospores/5–10 μL) and incubated at 25 °C in the dark. All experiments were repeated at least three times.
Table S1 Putative PsGPA1‐interacting genes identified by affinity purification.
Table S2 Primers used in the present study.
Table S3 Sequences of the histidine triad (HIT) motifs from putative Hint proteins.
Acknowledgements
This work was supported by the China National Funds for Distinguished Young Scientists (31225022) , the China Netherlands Joint Scientific Thematic Research Programme (JSTP, 2013DFG32030) and Special Fund for Agro‐scientific Research in the Public Interest (201303018) to Y.W.
References
- Ah‐Fong, A.M. and Judelson, H.S. (2011) Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biol. 115, 882–890. [DOI] [PubMed] [Google Scholar]
- Ajit, S.K. , Ramineni, S. , Edris, W. , Hunt, R.A. , Hum, W.T. , Hepler, J.R. and Young, K.H. (2007) RGSZ1 interacts with protein kinase C interacting protein PKCI‐1 and modulates mu opioid receptor signaling. Cell. Signal. 19, 723–730. [DOI] [PubMed] [Google Scholar]
- Bauer, W. and Caetano‐Anollés, G. (1990) Chemotaxis, induced gene expression and competitiveness in the rhizosphere. Plant Soil, 129, 45–52. [Google Scholar]
- Bolker, M. (1998) Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25, 143–156. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Wang, X. , Zhang, Z. , Zheng, X. and Wang, Y. (2008) Differences in the induction of the oxidative burst in compatible and incompatible interactions of soybean and Phytophthora sojae . Physiol. Mol. Plant Pathol. 73, 16–24. [Google Scholar]
- Chou, T.F. , Bieganowski, P. , Shilinski, K. , Cheng, J. , Brenner, C. and Wagner, C.R. (2005) 31P NMR and genetic analysis establish hinT as the only Escherchia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J. Biol. Chem. 280, 15 356–15 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann, G. and Hemetsberger, C. (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198, 1001–1016. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Kong, G. , Qutob, D. , Yu, X. , Tang, J. , Kang, J. , Dai, T. , Wang, H. , Gijzen, M. and Wang, Y . (2012) The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis‐inducing activity. Mol. Plant–Microbe Interact. 25, 896–909. [DOI] [PubMed] [Google Scholar]
- Engler, C. , Kandzia, R. and Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE, 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo, N.M. , Jacobs, S.R. , Currie, C.R. and Mueller, U.G. (2006) Ancient host–pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoS Biol. 4, e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo, M.C. and Valent, B. (2013) Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Guerriero, G. , Avino, M. , Zhou, Q. , Fugelstad, J. , Clergeot, P.H. and Bulone, V. (2010) Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti‐oomycete drugs. PLoS Pathog. 6, e1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham, A.R. (2001) The cell biology behind Phytophthora pathogenicity. Australas. Plant Pathol. 30, 91–98. [Google Scholar]
- Hatley, M.E. , Lockless, S.W. , Gibson, S.K. , Gilman, A.G. and Ranganathan, R. (2003) Allosteric determinants in guanine nucleotide‐binding proteins. Proc. Natl. Acad. Sci. USA, 100, 14 445–14 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T. , Kawabata, Y. , Takayama, T. , Tahara, S. , Kawabata, J. , Fukushi, Y. , Nishimura, H. and Mizutani, J. (1992) A potent attractant of zoospores of Aphanomyces cochlioides isolated from its host, Spinacia oleracea . Experientia, 48, 410–414. [Google Scholar]
- Hosseini, S. , Heyman, F. , Olsson, U. , Broberg, A. , Jensen, D.F. and Karlsson, M. (2014) Zoospore chemotaxis of closely related legume‐root infecting Phytophthora species towards host isoflavones. Plant Pathol. 63, 708–714. [Google Scholar]
- Hua, C. , Wang, Y. , Zheng, X. , Dou, D. , Zhang, Z. and Govers, F. (2008) A Phytophthora sojae G‐protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryotic Cell, 7, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Taylor, J.P. , Chen, J.G. , Uhrig, J.F. , Schnell, D.J. , Nakagawa, T. , Korth, K.L. and Jones, A.M . (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G‐protein GPA1 interact in a novel sugar‐signaling mechanism in Arabidopsis. Plant Cell, 18, 1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.T. , Ito, T. and Tahara, S. (2002) Microscopic studies on attachment and differentiation of zoospores of the Phytopathogenic fungus Aphanomyces cochlioides . J. Gen. Plant Pathol. 68, 111–117. [Google Scholar]
- Islam, M.T. , Ito, T. and Tahara, S. (2003) Host‐specific plant signal and G‐protein activator, mastoparan, trigger differentiation of zoospores of the phytopathogenic oomycete Aphanomyces cochlioides . Plant Soil, 255, 131–142. [Google Scholar]
- Islam, M.T. , von Tiedemann, A. and Laatsch, H. (2011) Protein kinase C is likely to be involved in zoosporogenesis and maintenance of flagellar motility in the peronosporomycete zoospores. Mol. Plant–Microbe Interact. 24, 938–947. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. , Tyler, B.M. and Michelmore, R.W. (1991) Transformation of the oomycete pathogen, Phytophthora infestans . Mol. Plant–Microbe Interact. 4, 602–607. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van West, P. , de Jong, A.J. , de Groot, K.E. , Vleeshouwers, V.G. and Govers, F. (1997) A gene encoding a protein elicitor of Phytophthora infestans is down‐regulated during infection of potato. Mol. Plant–Microbe Interact. 10, 13–20. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M. , Munnik, T. and Govers, F. (2002) Phospholipase D in Phytophthora infestans and its role in zoospore encystment. Mol. Plant–Microbe Interact. 15, 939–946. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M. , Ligterink, W. , Vleeshouwers, V.G. , van West, P. and Govers, F. (2004) A G alpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans . Mol. Microbiol. 51, 925–936. [DOI] [PubMed] [Google Scholar]
- Li, L. , Wright, S.J. , Krystofova, S. , Park, G. and Borkovich, K.A. (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61, 423–452. [DOI] [PubMed] [Google Scholar]
- Lin, C.C. and Aronson, J.M. (1970) Chitin and cellulose in the cell walls of the oomycete, Apodachlya sp. Arch. Mikrobiol. 72, 111–114. [DOI] [PubMed] [Google Scholar]
- Liu, S. and Dean, R.A. (1997) G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea . Mol. Plant–Microbe Interact. 10, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Martin, J.S. , Pierre, M.V. and Dufour, J.F. (2011) Hit proteins, mitochondria and cancer. Biochim. Biophys. Acta, 1807, 626–632. [DOI] [PubMed] [Google Scholar]
- McCudden, C.R. , Hains, M.D. , Kimple, R.J. , Siderovski, D.P. and Willard, F.S. (2005) G‐protein signaling: back to the future. Cell. Mol. Life Sci. 62, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, A. , Fry, B.A. , Zuluaga, A.P. , Myers, K.L. and Fry, W.E. (2008) Toward improvements of oomycete transformation protocols. J. Eukaryot. Microbiol. 55, 103–109. [DOI] [PubMed] [Google Scholar]
- Morris, P.F. and Ward, E.W.B. (1992) Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol. Mol. Plant Pathol. 40, 17–22. [Google Scholar]
- Morris, P.F. , Bone, E. and Tyler, B.M. (1998) Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol. 117, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer, E.J. (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell, 80, 249–257. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Robinson, K. and Aitken, A. (1994) Identification of a new protein family which includes bovine protein kinase C inhibitor‐1. Biochem. J. 304 (Pt 2), 662–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J.K. , Ham, K.S. , Darvill, A.G. and Albersheim, P. (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell, 14, 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, E.M. and Gilman, A.G. (1980) Biochemical properties of hormone‐sensitive adenylate cyclase. Annu. Rev. Biochem. 49, 533–564. [DOI] [PubMed] [Google Scholar]
- Siriputthaiwan, P. , Jauneau, A. , Herbert, C. , Garcin, D. and Dumas, B. (2005) Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum . J. Cell Sci. 118, 323–329. [DOI] [PubMed] [Google Scholar]
- Stintzi, A. , Heitz, T. , Prasad, V. , Wiedemannmerdinoglu, S. , Kauffmann, S. , Geoffroy, P. , Legrand, M. and Fritig, B . (1993) Plant pathogenesis‐related proteins and their role in defense against pathogens. Biochimie, 75, 687–706. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Wu, M. , Wang, J. , Cheung, W. and Morris, P.F. (1996) Chemotactic preferences and strain variation in the response of Phytophthora sojae zoospores to host isoflavones. Appl. Environ. Microbiol. 62, 2811–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams, G.H. and Armitage, J.P. (2004) Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. [DOI] [PubMed] [Google Scholar]
- Wang, F. , Shi, Z. , Hu, F. , Xia, Z. and Wang, L. (2013) Tuning of Ti‐doped mesoporous silica for highly efficient enrichment of phosphopeptides in human placenta mitochondria. Anal. Bioanal. Chem. 405, 1683–1693. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. , Kale, S.D. , Gu, B. , Sheng, Y. , Sui, Y. , Wang, X. , Zhang, Z. , Cheng, B. , Dong, S. , Shan, W. , Zheng, X. , Dou, D. , Tyler, B.M. and Wang, Y . (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weert, S. , Vermeiren, H. , Mulders, I.H. , Kuiper, I. , Hendrickx, N. , Bloemberg, G.V. , Vanderleyden, J. , De Mot, R. and Lugtenberg, B.J . (2002) Flagella‐driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens . Mol. Plant–Microbe Interact. 15, 1173–1180. [DOI] [PubMed] [Google Scholar]
- van West, P. , Appiah, A.A. and Gow, N.A.R. (2003) Advances in research on oomycete root pathogens. Physiol. Mol. Plant Pathol. 62, 99–113. [Google Scholar]
- Willard, S.S. and Devreotes, P.N. (2006) Signaling pathways mediating chemotaxis in the social amoeba, Dictyostelium discoideum . Eur. J. Cell Biol. 85, 897–904. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Zhao, W. , Hua, C. , Zheng, X. , Jing, M. , Li, D. , Govers, F. , Meijer, H.J. and Wang, Y. (2013) Chemotaxis and oospore formation in Phytophthora sojae are controlled by G‐protein‐coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol. Microbiol. 88, 382–394. [DOI] [PubMed] [Google Scholar]
- Ye, W. , Wang, X. , Tao, K. , Lu, Y. , Dai, T. , Dong, S. , Dou, D. , Gijzen, M. and Wang, Y. (2011) Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol Plant Microbe Interact. 24, 1530–1539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Immunoblot analysis of the expression and immunoprecipitation of PsGPA1‐3 × FLAG fusion proteins with the anti‐FLAG antibody. A 42‐kDa band was detected in total proteins (A) and proteins eluted from anti‐FLAG M2 beads (B) in transformants expressing the PsGPA1‐3 × FLAG fusion protein.
Fig. S2 Correction of PsHint1 gene model. (A) The automatic model was suggested to be 408 bp (grey blocks). According to the distribution of transcript tags (Ye et al., 2011) and the alignments against homologues from other oomycetes, the automatic model is incorrect. The new gene model was suggested to be 483 bp in length. Four primer pairs were designed to verify this model, i.e. SF1‐SR, SF2‐SR, SF3‐SR and SF4‐SR. (B) Polymerase chain reaction (PCR) products could be obtained from Phytophthora sojae cDNA using SF2‐SR, SF3‐SR and SF4‐SR, but not SF1‐SR. The SF2‐SR product was sequenced and matched perfectly to the corrected gene model. (C, D) Gene model for Hint1 protein of P. infestans was also checked and reanalysed, and the results were similar to PsHint1.
Fig. S3 Histidine triad (HIT) motifs of Hint proteins from different organisms. (A) Physical maps of Hint proteins showing conserved HIT motifs. (B) Amino acid sequence alignments of HIT motifs from different species, performed using the Clustal W program. Identical amino acid residues are shown with a dark grey background, and similar amino acid residues with a light grey background. Conserved histidine residues are indicated by asterisks at the bottom.
Fig. S4 Other Hint proteins of Phytophthora sojae, PsHint2, PsHint3 and PsHint4, do not interact with PsGPA1.
Fig. S5 Genomic polymerase chain reaction (PCR) analysis of plasmid stably transformed into the PsHint1‐silenced transformants. Genomic PCR screens were performed using genomic DNA as template and the primers pTOR00F and pTOR00R on the indicated strains. WT, wild‐type strain P6497; T14, T45 and T22, PsHint1‐silenced transformants. Product size is shown on the right.
Fig. S6 Expression of PsHint2, PsHint3 and PsHint4 in PsHint1‐silenced transformants. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) evaluation of gene expression levels using hyphal RNA from the recipient strain P6497, a control transformant (CK) and PsHint1‐silenced transformants (T14, T45 and T22). Relative expression levels were calculated using data from the wild‐type strain P6497 as the reference.
Fig. S7 Pathogenicity tests on the wounded susceptible soybean cultivar Hefeng47 using zoospores from wild‐type, control transformants (CK1 and CK2) and PsHint1‐silenced transformants (T14, T45 and T22). Wounded seedlings were drop inoculated with equal numbers of zoospores (100 zoospores/5–10 μL) and incubated at 25 °C in the dark. All experiments were repeated at least three times.
Table S1 Putative PsGPA1‐interacting genes identified by affinity purification.
Table S2 Primers used in the present study.
Table S3 Sequences of the histidine triad (HIT) motifs from putative Hint proteins.


