Abstract
The fungal pathogen Candida albicans frequently forms drug-resistant biofilms in hospital settings and in chronic disease patients. Cell adhesion and biofilm formation involve a family of cell surface Als (agglutinin-like sequence) proteins. It is now well documented that amyloid-like clusters of laterally arranged Als proteins activate cell—cell adhesion under mechanical stress, but whether amyloid-like bonds form between aggregating cells is not known. To address this issue, we measure the forces driving Als5-mediated intercellular adhesion using an innovative fluidic force microscopy platform. Strong cell—cell adhesion is dependent on expression of amyloid-forming Als5 at high cell surface density and is inhibited by a short antiamyloid peptide. Furthermore, there is greatly attenuated binding between cells expressing amyloid-forming Als5 and cells with a nonamyloid form of Als5. Thus, homophilic bonding between Als5 proteins on adhering cells is the major mode of fungal aggregation, rather than protein—ligand interactions. These results point to a model whereby amyloid-like β-sheet interactions play a dual role in cell—cell adhesion, that is, in formation of adhesin nanoclusters (cis-interactions) and in homophilic bonding between amyloid sequences on opposing cells (trans-interactions). Because potential amyloid-forming sequences are found in many microbial adhesins, we speculate that this novel mechanism of amyloid-based homophilic adhesion might be widespread and could represent an interesting target for treating biofilm-associated infections.
Keywords: Biofilms, nanoscale forces, homophilic adhesion, functional amyloids, Candida albicans, fluidic force microscopy
Graphical Abstract
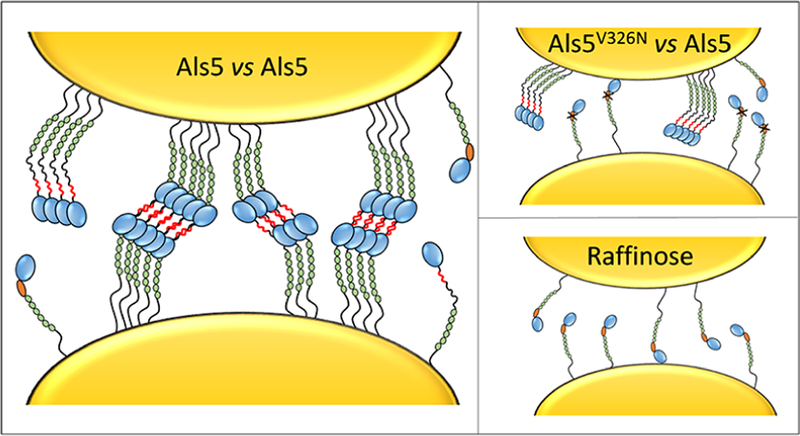
The fungal pathogen Candida albicans forms biofilm-like abscesses in invasive fungal disease and also attaches strongly to implanted medical devices, onto which they form biofilms.1–3 Such multicellular communities are difficult to treat because they provide protection from host defenses and are resistant to drugs.4–6 In C. albicans, cell adhesion, cell aggregation, and biofilm formation depend on multifunctional cell adhesion glycoproteins known as Als (agglutinin-like sequence) proteins.3,7–9 The tandem N-terminal immunoglobulin (Ig)-like regions of Als adhesins show broad substrate specificity for ligand—receptor interactions in cell adhesion. A threonine-rich region (T) contains a short amyloid-forming core sequence and strengthens cell—cell adhesion through noncovalent bonding that is yet not fully understood.8,9 The central region of the protein contains a variable number of tandem repeat (TR) domains that bind to each other and to substrates through hydrophobic interactions.
Amyloids are protein aggregates made up of ordered β-sheet structures that result from periodic interactions between identical short sequences of amino acids in multiple molecules. These structures form around and include amyloid core sequences of 5–7 residues. Identical core sequences in different molecules of the same protein form β-strands in stacked sheets that pack into a characteristic fibrillar geometry called cross-β.10,11 While pathogenic amyloid fibrils involved in neurodegenerative diseases in humans are well-known, it has become clear that some amyloids also have functional consequences.8,12,13 A widely investigated family of such functional microbial amyloids are curli, i.e., amyloid fibers that promote biofilm assembly and host colonization in Escherichia coli.13,14 In the dimorphic fungus C. albicans, a large body of evidence based on genomics, biochemistry, cell biology, and single-molecule experiments supports the involvement of Als amyloid core sequences in cell adhesion and biofilm formation.9 Although Als proteins form amyloids in vitro, in vivo the proteins are anchored to the fungal cell wall and so are geometrically constrained in a way that prevents fiber formation.9 Instead, amyloid-like β-sheet interactions of the core sequence mediate formation of cell surface nanodomains, that is, patches of aggregated adhesins. These amyloid-like structures are found in fungal abscesses, which means that fungal amyloids occur during colonization of human tissues and can reduce macrophage-dependent inflammatory reactions.15–19
A unique feature of Als amyloid-like nanodomains is that they are induced by physical force, such as hydrodynamic flow or cell-surface contacts at extension forces similar to those encountered in vivo.9,20,21 Mechanical force induces conformational changes in Als molecules resulting in the exposure of hidden amyloid sequences. This structural change triggers the formation of nanoscale amyloid-like clusters made of hundreds of proteins on the cell surface, which mediate high-avidity interactions between cells. While it is well-established that fungal aggregation involves amyloid-like interactions between laterally arranged Als proteins (cis), there is no direct evidence that such bonds form between aggregating cells (trans).
Pioneered 20 years ago, single-cell atomic force microscopy (AFM)22,23 has become a valuable tool to probe forces in living cells. A crucial step for AFM-based single-cell force spectroscopy (SCFS) experiments is to immobilize the probing cell on the AFM cantilever, which is straightforward with animal cells, but delicate and time-consuming with microbial cells.24 Bacteria and yeasts are typically attached using chemical fixation, glue, drying, electrostatic interactions or bioadhesives.24 Most of these protocols are invasive and labor-intensive, meaning performing biologically relevant and statistically significant measurements is very challenging. A promising method to achieve this goal is fluidic force microscopy (FluidFM),25–28 a recent technology in which microchanneled cantilevers with micro- or nanosized apertures allow for fast manipulation of single cells under physiological conditions. By means of a pressure controller, FluidFM enables to quickly and reversibly immobilize cells on the cantilever tip for SCFS measurements.
Here we combine FluidFM with sequence manipulation to measure the molecular forces involved in Als-mediated aggregation, with the aim to identify whether amyloid-like interactions form between cells. We study the most extensively investigated adhesin, Als5, in the absence of other C. albicans adhesive molecules, by expressing the protein in a S. cerevisiae surface display model.9 Optimizing the FluidFM methodology for the reliable and serial measurement of forces between yeast cells enables us to address a crucial question: is Als5 engaged in homophilic adhesion, and if so, is homophilic binding mediated by the amyloid-forming region of the adhesin? The results show that intercellular adhesion involves not only cis amyloid-based interactions between lateral proteins but also trans amyloid-like bonding between proteins on opposing cells. We suggest that a variety of microbes may use similar β-sheet interactions to guide adhesion and that this new mode of cell aggregation might represent an interesting target for treating biofilm-associated infections.
Results.
Probing Cell—Cell Adhesion Forces by Means of FluidFM.
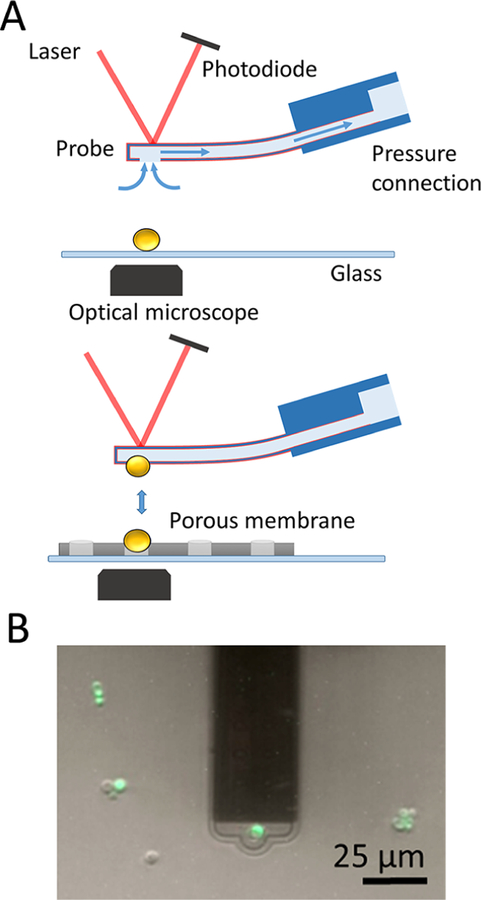
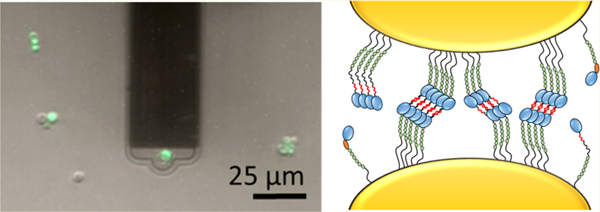
Unique to FluidFM is the possibility to rapidly attach (detach) single cells to (from) the cantilever by applying a negative (positive) pressure, thus eliminating the labor intensive cell probe preparation step (Figure 1A). During preliminary experiments, two problems were encountered; i.e., multiple cells were sticking to the microchanneled cantilever, and application of large positive pressures was not sufficient to remove the probing cell from the cantilever. To avoid these issues, cantilevers were coated with a solution of poly-L-lysine-grafted-polyethylene glycol (PLL-g-PEG),25–27 a polymer that electrostatically interacts with the silicon nitride cantilever through the positive charges of PLL while exposing antifouling PEG chains toward the solution. The cantilever is filled with and immersed in a PLL-g-PEG solution while applying an overpressure to flow the liquid into the microchannel and then rinsed with buffer. A yeast cell is immobilized on the cantilever aperture by applying a negative pressure (Figure 1A, upper panel). Staining with a live/dead indicator dye confirmed that the protocol did not alter the cell integrity (Figure 1B, green color). To immobilize target cells on solid substrates, PLL adsorption was not effective, as PLL fingerprints were observed in force curves, and cells were often detaching from the substrate during the analysis. Firm immobilization was achieved by trapping the cells mechanically into porous membranes (Figure 1A, bottom panel).29 This FluidFM setup enabled us to measure the forces between yeast cells at higher throughput (at least ∼20 cells per day) than with conventional AFM in native conditions, i.e., without the need of any chemical to attach the probing and target cells.
Figure 1.
Probing yeast—yeast interaction forces using FluidFM. (A) FluidFM setup. A single cell is immobilized on the cantilever aperture by applying a negative pressure. PLL-g-PEG coating (in red) is used to prevent contamination of the cantilever (upper panel). The cell probe is moved toward a target cell immobilized in a porous membrane and force—distance curves are recorded (lower panel). (B) Labeling of the attached cell (in green) demonstrates that cell integrity is preserved, thus that the method is nondestructive.
Forces in Cell—Cell Adhesion.
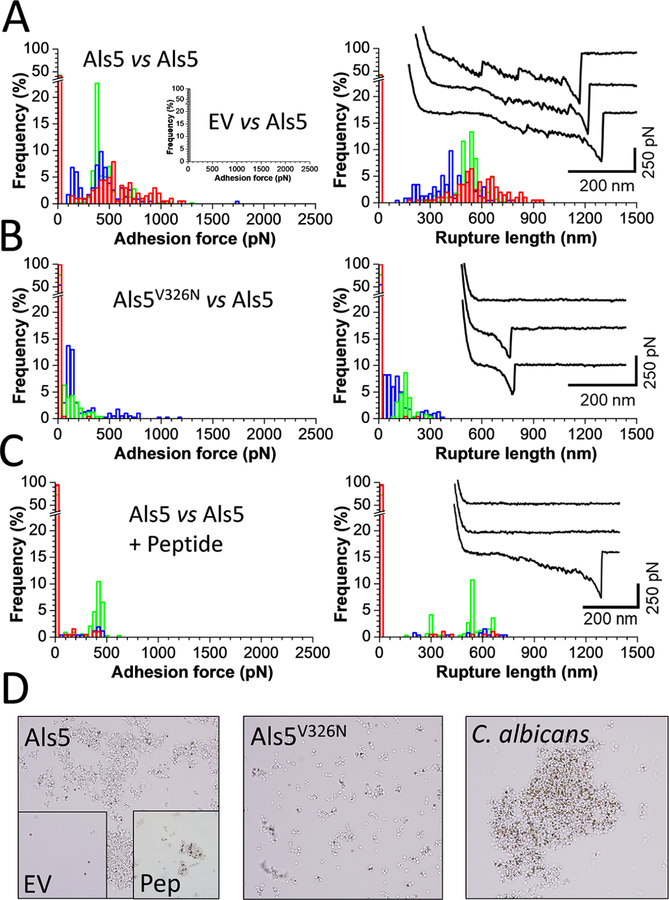
We initially measured the interaction forces between Als5 yeast cells grown in galactose, which induces maximal expression of Als5 from the GAL1 promoter in plasmid pJL1.30–32 Cell pairs were first mechanically activated by bringing them into contact for a short period of time, a procedure that exposes the amyloid core regions and leads to strong adhesion.20,30,33 Adhesion forces, rupture lengths, and typical force signatures obtained for three representative cell pairs are shown in Figure 2A (see Figure 4 for data on more cell pairs). Most curves (adhesion frequency of 73 ± 23%; mean ± sd) featured large and complex adhesion signatures of 558 ± 178 pN magnitude and 593 ± 91 nm rupture length (mean ± sd from n = 1617 adhesive curves from 10 cell pairs). Cell to cell variations were observed (Figure 4) that are likely to reflect differences in adhesion expression in the cell population.30 Rupture forces for cell—cell interactions were 1–3-fold the values typically seen in in single-molecule experiments.9,33 A large fraction of the curves featured sawtooth patterns with multiple consecutive peaks followed by a last peak of ∼500 pN magnitude. Such adhesive events agree well with force values expected for β-sheet-rich structures,34 largely present in the Ig, T, and TR regions.35,36 Force signatures between Als5 cells were similar to those measured using conventional SCFS (Supporting Figure S1), except that sawtooth patterns were better resolved in FluidFM. This finding further validates FluidFM for the reliable quantitation of cell—cell adhesion forces.
Figure 2.
Als5-dependent cell—cell adhesion forces. (A) Maximum adhesion force (left) and rupture length (right) histograms with representative retraction force profiles (insets) obtained by recording force—distance curves in PBS for three Als5 cell pairs (see three colors). The left inset shows results obtained for the interaction between empty vector (EV) and Als5 cells. (B) Results obtained for the interaction between Als5V326N cells expressing Als5 proteins with a single site mutation in the amyloid-forming region and Als5 cells. (C) Results obtained for the interaction between Als5 cells after addition of the SNGINIVATTRTV peptide (200 μg/mL final concentration). For data on more cells, see Figure 4. All curves were obtained using an applied force of 500 pN, a 1 s contact time, and an approach and retraction speed of 1.5 μm/s. (D) Yeast aggregation assays with BSA-coated beads. Magnetic beads (brown-gold, 1 μm diameter) were agitated with the designated yeast strains (yeast cells are gray), the beads and associated cells were separated with a magnet, washed, and observed using phase contrast microscopy. EV: aggregation of cells not expressing Als5; Pep: aggregation of Als5 cells in the presence of antiamyloid peptide, 200 μg/ml.
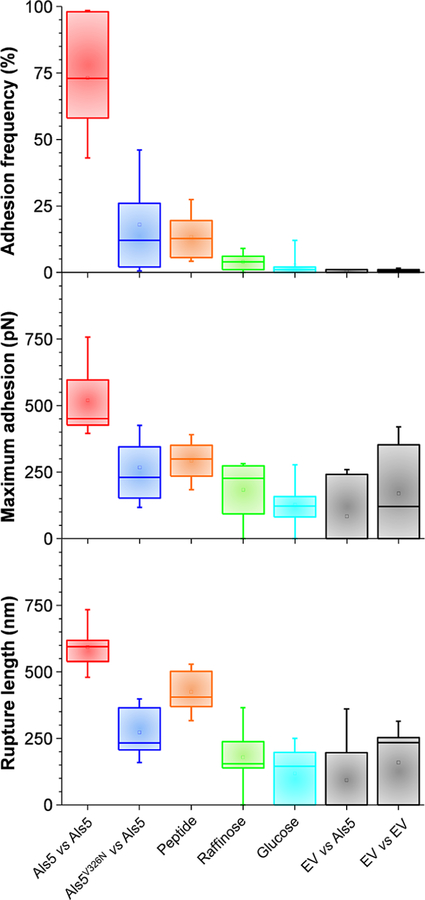
Figure 4.
Role of amyloid-like region and protein density in cell—cell adhesion. (A)—(C) Boxplots of adhesion frequency (A), maximum adhesion force (B), and rupture length (C) obtained by measuring forces for multiple cell pairs: Als5 vs Als5 (9 cells; 2022 curves), Als5V326N vs Als5 (15 cells; 3900 curves), Als5 vs Als5-raffinose (5 cells; 1219 curves), Als5 vs Als5-glucose (6 cells; 1859 curves), EV vs Als5 (4 cells; 1551 curves), EV vs EV (10 cells; 2529 curves). Boxplots represent the mean values (squares), medians (horizontal lines), 25 and 75% quartiles (box limits), and outliers (whiskers).
The force signatures we measured here are consistent with formation of homophilic bonds between Als5 molecules. Our rupture distances (∼500 nm) are indeed twice as long that those observed for single Als5 molecules,33 indicating that cell—cell detachment involves the stretching and unfolding of two (or more) interacting Als5 proteins on opposing cells. Under force, we expect that the stalk region will extend up to 100 nm, that the T region (108 residues yielding a 30 nm length increment) and 6 TR repeats (6 × 36 residues, 55 nm increment) will completely unfold, and that the 2 Ig domains will deform only modestly due to disulfide bonding (50 nm).33,36 This means that a fully stretched adhesin should be ∼235 nm long, thus that Als5—Als5 bonds should rupture at around 470 nm, which is in the range of our data.
In addition, force curves showed characteristics of Als5 unfolding.33,37 Some sequential peaks were separated by ∼9 nm, the length increment representing the characteristic fingerprint of TR repeats, as we previously showed (a single TR repeat is made up of 36 amino acids, each amino acid contributes about 0.36 nm to the extended polypeptide chain, and the folded length of a repeat is 4.3 nm).37 The last peak at 500 pN is associated with the binding and rupture of Ig—Ig complexes between cells. Sawtooth profiles were preceded by less defined multiple peaks varying in strength and spacing, which we believe are associated with the T regions because molecular modeling shows that T regions fold into structures of variable sizes, and, under mechanical stress, conformational changes in the adhesins lead to the exposure of amyloid core sequences of the T region which favor amyloid formation.9 Therefore, our force signatures are as expected for a system in which the primary mode of cell—cell adhesion is Als5—Als homophilic binding.
Als5 Homophilic Binding Involves Amyloid-Like Interactions.
Cell—cell adhesion forces could result from heterophilic receptor—ligand interactions between the Ig region on one molecule and a ligand sequence on another molecule or from homophilic interactions between identical sequence segments, as in amyloid-like bonds. To distinguish these mechanisms, we measured the forces between cells lacking Als5 adhesins (empty vector, EV) and Als5 cells (Figure 2A, inset, and Figure 4). Adhesion forces were abrogated (adhesion frequency <1%; same result for EV-EV adhesion forces), demonstrating that Als5 homophilic binding is the major mode of cell—cell adhesion, rather than Ig-dependent interactions with heterologous ligands.38,39
In view of the importance of functional amyloids in fungal adhesion,9 we hypothesized that homophilic adhesion involves trans interactions between Als5 amyloid forming sequences on neighboring cells. To test this model, we first analyzed a strain expressing Als5 proteins with the V326N single site mutation in the amyloid-forming region (Als5V326N), which inhibits cell aggregation21,30 and prevents the formation of nanoclusters on the cell surface.30,33,40 Als5V326N has ligand binding activity and secondary structure similar to wild type Als5.30 We found that the adhesion probability, maximum adhesion force and rupture lengths (28 ± 32%; 218 ± 101 pN; 257 ± 103 nm; n = 1080 adhesive curves from 15 cell pairs) between Als5V326N and Als5 cells were markedly reduced (Figure 2B and Figure 4). Force signatures featured mostly single rupture events, indicating that Als5 domains were not unfolded before rupture. Together with the short extensions (∼200 nm) these data show that the interaction between Als5V326N and Als5 cells involves receptor—ligand binding too weak to allow unfolding of protein domains. We also tested the ability of a short Als-specific antiamyloid peptide to block cell—cell adhesion. We found that Als5—Als5 adhesion was strongly inhibited in the presence of the peptide (Figure 2C and Figure 4; adhesion probability, maximum adhesion force and rupture lengths of 13 ± 8%, 293 ± 75 pN and 425 ± 77 nm; n = 321 adhesive curves from 8 cell pairs). Unlike with Als5V326N cells, the remaining protein extensions were long (∼400 nm), in the range of those observed for Als5—Als5 pairs, meaning residual adhesion involved binding of two unfolded adhesins.
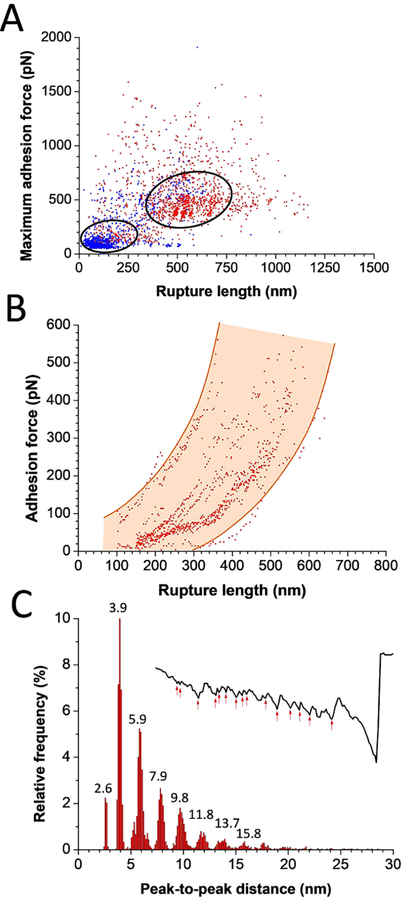
Detailed analysis of force profiles further supported the notion that Als5 homophilic adhesion relies on amyloid-like interactions (Figure 3). Scatter plots of the strongest adhesion events versus rupture distances (n = 1329 curves from 9 cell pairs) show the frequent detection of strong, long-range force peaks for Als5—Als5 cell pairs, associated with the stretching and unfolding of the T, TR, and to some extent Ig domains (Figure 3A). By contrast, mostly weak, short-range peaks were observed on Als5V326N—Als5 cell pairs (n = 1091 curves from 14 cell pairs), meaning domain unfoldings were infrequent. In addition, analysis of all peak-to-peak distances for the Als5—Als5 curves revealed two remarkable features. First, scatter plots of the adhesion force versus rupture distances of all peaks (Figure 3B; 868 data points) documented a strong, nonlinear correlation between force and extension, supporting the notion that most pulling experiments lead to the stretching and unfolding of two (or more) adhesins in trans interaction. Scattering of the data points results from the complex geometry of the cell—cell interface; i.e., all Als5 molecules are not loaded with the same pulling geometry and do not have the same molecular environment. Second, Figure 3C shows that a large proportion of Als5—Als5 curves (>50%) displayed recurring serial length increments of 2.0 ± 0.1 nm (n = 4791 peaks from 7 cell pairs). This is approximately the expected length increase that would result from separation of a β-strand in the amyloid core from a β-sheet and its subsequent extension under force. Such periodic features were hardly observed for the interaction of Als5 cells with Als5V326N cells (<3% of all curves, perhaps due to rare other binding modes). Thus, Als5—Als5 sawtooth profiles are consistent with homophilic bonding through extended β-sheet interactions. We speculate that interstrand H bonds form between parallel β-strand arrays on opposing cells. Upon pulling they break one at a time, after which TRs on 2 (or more) Als unfold, then the Ig still holds the cells together until final cell—cell separation.
Figure 3.
Detailed analysis of Als5—Als5 sawtooth force profiles. (A) Scatter plots of the maximum adhesion forces versus rupture length for Als5—Als5 cell pairs (in red, n = 1329 curves from 9 cell pairs) and Als5V326N—Als5 cell pairs (in blue, n = 1091 curves from 14 cell pairs). (B) Scatter plot of the adhesion force versus rupture length of all peaks estimated from Als5—Als5 force profiles (868 data points). (C) Distribution of peak-to-peak distances estimated from Als5—Als5 force profiles (n = 4791 peaks from 7 cell pairs). Inset: a representative force—distance curve with multiple peaks marked.
We asked whether our unfolding patterns followed by strong adhesion events were correlated with yeast aggregation at the microscale. BSA-coated beads and yeast cells were mixed and agitated, leading to adhesion of single yeast cells to the beads, followed by cell-to-cell aggregation, eventually forming micro-colonies. S. cerevisiae expressing Als5 formed large aggregates, as did C. albicans, whereas control empty vector (EV) S. cerevisiae cells formed no aggregates (Figure 2D). Cells expressing Als5 bearing the V326N single site mutation in the amyloid-forming region (Als5V326N cells) or incubated with antiamyloid peptide adhered well to the beads, but failed to form large aggregates. These results show that our cell—cell interaction forces correlate with the level of yeast cell aggregation, and suggest that the mechanical response of interacting Als5 plays an important role in the process.
Cis and Trans: Dual Activity of Amyloid-Like Interactions in Fungal Adhesion.
The formation of amyloid-like nanodomain clusters between laterally arranged Als5 proteins represents an important driving force for cellular aggregation.9 We therefore hypothesized that cell adhesion should strongly depend on the density of adhesins exposed at the cell surface. To test this, the protein density was tuned by changing the sugar source in the growth medium (Supporting Figure S2; Figure 4). While galactose activates the expression of Als5, glucose actively represses the galactose promoter. When glucose-grown Als5 cells were probed with galactose-grown Als5 cells, there was almost no adhesion (adhesion probability <1%), thus confirming that adhesion of galactose cells is solely mediated by Als5. Cells grown in the presence of raffinose, which allows low levels of proteins to be produced, showed reduced adhesion (adhesion frequency of 4 ± 4%), yet some adhesive events, 273 ± 244 pN in magnitude and 267 ±198 nm in length (from n = 1219 curves from 14 cells), were observed. So Als5 mediates weak cell—cell adhesion at low protein density. The mean rupture distance was much shorter than for galactose-grown cells and unfolding patterns were missing, which indicates that single adhesins were involved in receptor—ligand interactions that are too weak to trigger the unfolding the β-sheet-rich domains. Our finding that Als5 density has a pronounced effect on cell—cell adhesion forces suggests that protein clustering and strong adhesion only occur at high protein concentration. Accordingly, the Als amyloid forming region mediates fungal aggregation through protein clustering and homophilic binding. During biofilm formation, such amyloid-like β-sheet interactions will be additive and favor long-lived multicellular associations.
Discussion.
Functional amyloids play essential roles in cell adhesion, aggregation and biofilm formation but the molecular forces involved are poorly understood. By means of FluidFM single-cell experiments we have identified a novel form of fungal cell—cell adhesion based on a functional amyloid. Our data imply that Als5-mediated homophilic adhesion in C. albicans involves β-sheet interactions between amyloid forming sequences on neighboring cells. There are multiple lines of evidence that these interactions share the characteristics of the bonding in amyloid interactions.
We have shown that cell—cell adhesion is dependent on the Als5 small amyloid core sequence, 325IVIVATT331, which must be expressed on both interacting cells at high surface density. The cell-to-cell binding is inhibited by a short antiamyloid peptide. We argue that these sequences mediate the formation of amyloid-like steric zipper β sheet structures.41,42 As in amyloids, these multiple interactions are cooperative, and therefore high local concentrations lead to formation of highly stable structures.43,44 In Als5, both activities are abrogated in the nonamyloid-forming adhesin Als5V326N (Figure 2B) or by treatment with antiamyloid compounds.8,9 Consequently, amyloidogenic T regions are under strong positive evolutionary selection.40 Amyloid-like clustering and strong cellular aggregation are also observed in Als5Aβ, a form of adhesin in which LVFFA, an amyloid core sequence from human Aβ is substituted for 325IVIVA329.45 Other cell—cell adhesion-dependent amyloid-like properties of Als5 include the development of surface birefringence and enhanced binding of amyloidophilic dyes.8,9,30,46
Our results highlight the complex role of functional amyloids in fungal adhesion and lead us to propose a more detailed molecular model, whereby force-dependent Als5 amyloid core sequences mediate cell aggregation via an interplay of cis and trans interactions (Figure 5, left). In amyloid structures, H bonding between β-strands forms β-sheets, and interdigitation of side chains of homologous amino acids is required to associate β-sheets and stabilize the structure.41 The orthogonal spatial orientation of these interactions is mirrored in the cis and trans interactions on the cell surface. In Als5-mediated cell—cell adhesion, the presence of the amyloid core sequence leads to a characteristic recurring ∼2 nm extension motif in the force profiles. This length increment is consistent with rupture of H-bonds between multiple β-strands in a β-sheet, as expected from an amyloid-like structure.41 Altering the amyloid core sequence (Figure 5, upper right) or lowering the protein density (Figure 5, lower right) leads to poor adhesion, indicating that strong homophilic adhesion relies on intra- and inter-β-sheet bonds and requires protein clustering. As a consequence, in Als5 cells with low surface density (Figure S2) or Als5V326N cells (Figure 2B) there is only receptor—ligand binding, which is significantly weaker than the homophilic amyloid-like mode. Therefore, FluidFM experiments support the notion that strong homophilic adhesion is mediated through amyloid-like β-sheet interactions.
Figure 5.
Mechanism of amyloid-like homophilic adhesion in Candida albicans. (Left) Force-induced unfolding of Als5 proteins leads to the exposure of hidden amyloid sequences, which trigger the lateral assembly (cis interactions) of the proteins on the cell surface. Strong homophilic adhesion (trans interactions) between adhesins on opposing cells is mediated by amyloid-like bonds. Green, orange, and blue colors represent the folded TR, T, and Ig regions. Shown in red is the amyloid sequence of the force-induced unfolded T region. Altering the amyloid sequence (upper right) or lowering the protein density (lower right) abrogates strong homophilic adhesion between adhesins, emphasizing the key role of cis and trans amyloid bonding in fungal aggregation.
In conclusion, we have shown that FluidFM is a powerful tool to quantify interaction forces in single fungal cells, at increased throughput and without the need of chemical fixation. Our cell—cell experiments biophysically probe detailed interactions of functional adhesins in their native cellular environment. This technology enabled us to discover a previously undescribed function for C. albicans Als proteins, i.e., mediating amyloid-like homophilic adhesion. Our findings favor a novel aggregation mechanism, in which force-dependent amyloid core sequences in Als proteins play a dual role, by triggering the formation of cell surface adhesion nanoclusters, which in turn facilitate high-avidity, strong homophilic interactions between adhesins on opposing cells. As potential amyloid-forming sequences are common in fungi and bacteria,8,9,13 it is possible that cell—cell association through such amyloid-like homophilic bonding might be a widespread adhesion mechanism among microbial species.
Methods.
Microorganisms, Cultures, and Aggregation Assay.
S. cerevisiae W303–1B strains (Rodney Rothstein, Columbia University) harboring empty vector (pJL1-EV) or expressing Als5p or Als5pV326N,30 were cultivated on Complete Supplement Mixture minus tryptophan with adenine (CSM-Trp+Ade) galactose agar plates at 30 °C. One colony was inoculated in liquid CSM-Trp+Ade medium. Als5p and Als5pV326N are under galactose promoter; therefore, the growth medium included 2% galactose as C source to promote the expression of adhesins, or 2% glucose to inhibit their expression, or 2% raffinose (neutral for expression). The cultures were incubated at 30 °C under gentle agitation (170 rpm) for about 24 h, until an optical density at λ = 680 nm of 0.5–1.0 was reached (mid-to-late log phase). Cells were harvested by centrifugation at 1500 rpm, washed three times with phosphate buffered saline (PBS) and resuspended in PBS. Cell suspensions were diluted 10 times before use to limit cell aggregation and enable manipulation of single cells.
Cell Labeling.
Cell viability during FluidFM experiments was estimated using BacLight (Invitrogen, kitL7012), a kit traditionally used for bacteria but which has also been shown to work efficiently with yeasts.47 A staining solution was prepared by mixing 1.5 μL of Syto9 stain and 1.5 μL of propidium iodide (PI) and 3 μL of this solution were added to 3 mL of fungal suspension diluted in PBS. Stained cells were imaged with an inverted fluorescence microscope (Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600). Images were recorded with a 10× objective lens and recorded with a CCD camera. Fluorochromes were, respectively, excited and detected at 455–495 and 505–555 nm (for Syto9) and at 533–558 and 570–640 nm (for PI). To demonstrate our ability to record signal from both fluorochromes, a disinfectant solution (neoform MED rapid, VWR) was added to the suspension at the end of the experiment to kill the cells which fluorescent signal turned red as expected.
Cell Aggregation Assay.
We assessed yeast aggregation at the microscale using a bead adherence assays similar to that described elsewhere.48 Briefly, yeast cells were mixed with BSA-coated magnetic beads at a cell-to-bead ratio of 100:1 in Tris—EDTA buffer, vortexed, and incubated at room temperature with gentle shaking for 30–45 min. Each tube was vortexed briefly and immediately placed into a magnetic separator (Dynal). Adherent and aggregated cells were gently washed three times with buffer, while the tube remained within the magnet. The cells were resuspended in buffer, and the sample was placed onto a microscope slide for examination.
FluidFM Measurements.
Target cells were immobilized mechanically into porous membranes. Ten milliliters of cell suspension were filtered through 5 μm pore size polycarbonate membrane (it4ip, Belgium). The filter was then rinsed with buffer, carefully cut (1 cm × 1 cm), and stuck with double face adhesive tape on the microscope Petri dish while avoiding dewetting. Cell—cell force spectroscopy experiments were performed at room temperature (20 °C) in buffered solution (PBS), using a Bioscope Resolve AFM (Bruker Corporation) combined with an inverted optical microscope (Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600, Zeiss AG) and connected to a pressure pump unit and a pressure controller through a microfluidic tubing system (Cytosurge AG). Cell probes were prepared using rectangular, hollow silicon nitride cantilever with an aperture of 4 μm and a spring constant of 0.3 N m−1 (Cytorsurge AG). The micropipets were coated using a filtered solution of 0.5 mg mL−1 PLL(20)-g[3.5]-PEG(2) (SuSos AG, Dübendorf, Switzerland) to limit protein adsorption and biofouling.26 The solution was filled in the probe reservoir and was pressed through the cantilever by applying an overpressure (100 kPa); meanwhile, the probe was immersed in the same PLL-g-PEG solution for 1 h. The probe was then rinsed in ultrapure water for 5 min. The FluidFM probes were calibrated using the thermal noise method prior to measurement. A single yeast cell was then picked up from the glass surface of the Petri dish by approaching the FluidFM probe and applying a negative pressure (−80 kPa). The transfer of the cell on the probe was verified by optical microscopy.
The obtained yeast probe was then transferred over the porous membrane and precisely positioned over a single target yeast cell using optical microscopy. The negative pressure applied was decreased throughout the experiment to reduce the pump work and avoid noise on the force curves. The contact between the two cells was done by engaging the probe with a set point of 500 pN, and then force measurements were performed in the ramping mode. Cell pairs were first activated by bringing them into contact five times during 1 min. This process maximizes cell—cell adhesion by allowing force-dependent secondary structure changes in the adhesins.9 Then multiple force curves (>200) were recorded, using a 1 s contact time,37 a maximum applied force of 500 pN, a ramp size of 1.5 μm and tip approach and pulling velocities of 1500 nm s−1. Data were analyzed using the Nanoscope Analysis 1.9 software from Bruker. Adhesion force and distance rupture histograms were obtained by calculating the maximum adhesion force and rupture distance of the last peak for each curve. At the end of each experiment, contaminants and cell debris were washed from the cantilevers, by using solutions of Terg-a-zyme and NaOH and applying sequential negative and positive pressures. The cleaned probes were stored in a 2% antibiotic antimycotic solution until the next analysis.
In some experiments, we used a short antiamyloid peptide, sequence SNGINIVATTRTV.30 Galactose-grown Als5 cells were used on the cantilever and as the membrane-immobilized cells. A cell was selected and activated, and then 30 cycles of attachment and detachment were monitored to ensure strong bonding. The cell probe was then retracted and peptide added to 200 μg/mL. Adhesion of the same cell pair was then repeatedly measured in the presence of peptide.
Conventional AFM Measurements.
Cells were immobilized on hydrophobic alkanethiol-modified gold substrates. Gold-coated glass coverslips were immersed overnight in an ethanol solution containing 1 mM 1-dodecanethiol (Sigma-Aldrich, 98%), rinsed with ethanol, and dried with nitrogen (N2). This substrate was then stuck with double-face adhesive tape in the AFM Petri dish and a 100 μL drop of yeast suspension was deposited on it, incubated for 10 min, and rinsed. Measurements were performed at room temperature (20 °C) in PBS, using a Bioscope Catalyst (Bruker Corporation) combined with an inverted optical microscope (Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600, Zeiss AG). Cell probes were prepared using triangular shaped tipless canti-levers (NP-O10, Microlevers, Bruker Corporation) coated with bioinspired polydopamine wet adhesives.49 The cantilever was approached to a single cell for 3 min and then retracted. The proper attachment of the cell to the probe was verified by optical microscopy. The force measurements were then performed and analyzed as described above for FluidFM measurements.
Supplementary Material
ACKNOWLEDGMENTS
Work at the Université catholique de Louvain was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. [693630]), the National Fund for Scientific Research (FNRS), the FNRS-WELBIO (grant no. WELBIO-CR-2015A-05), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at Brooklyn College was supported by awards SC1 GM083756 and R01 GM098616 from the US National Institutes of Health. Y.F.D. is a Research Director at the FNRS. We thank David Alsteens for help with data interpretation and Cytosurge for thechnical support with the FluidFM technology.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.9b01010.
Figures showing FluidFM and conventional SCFS reveal similar adhesion forces and yeast-yeast adhesion forces strongly depend on Als5 density(PDF)
REFERENCES
- (1).Finkel JS; Mitchell AP Nat. Rev. Microbiol 2011, 9, 109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Verstrepen KJ; Reynolds TB; Fink GR Nat. Rev. Microbiol 2004, 2, 533–40. [DOI] [PubMed] [Google Scholar]
- (3).Dranginis AM; Rauceo JM; Coronado JE; Lipke PN Microbiol Mol. Biol. Rev 2007, 71, 282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lohse MB; Gulati M; Johnson AD; Nobile CJ Nat. Rev. Microbiol 2017, 16, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mitchell KF; Taff HT; Cuevas MA; Reinicke EL; Sanchez H; Andes DR Antimicrob. Agents Chemother 2013, 57, 1918–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Smukalla S; Caldara M; Pochet N; Beauvais A; Guadagnini S; Yan C; Vinces MD; Jansen A; Prevost MC; Latge JP; Fink GR; Foster KR; Verstrepen KJ Cell 2008, 135, 726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hoyer LL Trends Microbiol 2001, 9, 176–80. [DOI] [PubMed] [Google Scholar]
- (8).Lipke PN; Garcia MC; Alsteens D; Ramsook CB; Klotz SA; Dufrene YF Trends Microbiol 2012, 20, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lipke PN; Klotz SA; Dufrene YF; Jackson DN; Garcia-Sherman MC Microbiol. Mol. Biol. Rev 2017, 82, e00035–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Greenwald J; Riek R Structure 2010, 18, 1244–60. [DOI] [PubMed] [Google Scholar]
- (11).Riek R; Eisenberg DS Nature 2016, 539, 227–235. [DOI] [PubMed] [Google Scholar]
- (12).Fowler DM; Koulov AV; Balch WE; Kelly JW Trends Biochem. Sci 2007, 32, 217–24. [DOI] [PubMed] [Google Scholar]
- (13).Blanco LP; Evans ML; Smith DR; Badtke MP; Chapman MR Trends Microbiol 2012, 20, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chapman MR; Robinson LS; Pinkner JS; Roth R; Heuser J; Hammar M; Normark S; Hultgren SJ Science 2002, 295, 851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Klotz SA; Sobonya RE; Lipke PN; Garcia-Sherman MC Open Forum Infect Dis 2016, 3, ofw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Garcia-Sherman MC; Lundberg T; Sobonya RE; Lipke PN; Klotz SA NPJ. Biofilms Microbiomes 2015, 1, 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Garcia-Sherman MC; Lysak N; Filonenko A; Richards H; Sobonya RE; Klotz SA; Lipke PN PLoS One 2014, 9, No. e86067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gilchrist KB; Garcia MC; Sobonya R; Lipke PN; Klotz SA J. Infect. Dis 2012, 206, 1473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Behrens NE; Lipke PN; Pilling D; Gomer RH; Klotz SA mBio 2019, 00218–19. [DOI] [PMC free article] [PubMed]
- (20).Chan CX; Lipke PN Eukaryotic Cell 2014, 13, 1136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chan CX; Joseph IG; Huang A; Jackson DN; Lipke PN PLoS One 2015, 10, No. e0129152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Benoit M; Gabriel D; Gerisch G; Gaub HE Nat. Cell Biol 2000, 2, 313–7. [DOI] [PubMed] [Google Scholar]
- (23).Helenius J; Heisenberg CP; Gaub HE; Muller DJ J. Cell Sci 2008, 121, 1785–91. [DOI] [PubMed] [Google Scholar]
- (24).Dufrene YF Trends Microbiol 2015, 23, 376–82. [DOI] [PubMed] [Google Scholar]
- (25).Dörig P; Stiefel P; Behr P; Sarajlic E; Bijl D; Gabi M; Vörös J; Vorholt JA; Zambelli T Appl. Phys. Lett 2010, 97, 023701. [Google Scholar]
- (26).Potthoff E; Guillaume-Gentil O; Ossola D; Polesel-Maris J; LeibundGut-Landmann S; Zambelli T; Vorholt JA PLoS One 2012, 7, No. e52712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Guillaume-Gentil O; Potthoff E; Ossola D; Franz CM; Zambelli T; Vorholt JA Trends Biotechnol 2014, 32, 381–8. [DOI] [PubMed] [Google Scholar]
- (28).Potthoff E; Ossola D; Zambelli T; Vorholt JA Nanoscale 2015, 7, 4070–9. [DOI] [PubMed] [Google Scholar]
- (29).Kasas S; Ikai A Biophys. J 1995, 68, 1678–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Garcia MC; Lee JT; Ramsook CB; Alsteens D; Dufrene YF; Lipke PN PLoS One 2011, 6, No. e17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Johnston M Microbiol. Rev 1987, 51, 458–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Weinhandl K; Winkler M; Glieder A; Camattari A Microb. Cell Fact 2014, 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Alsteens D; Garcia MC; Lipke PN; Dufrene YF Proc. Natl. Acad. Sci. U. S. A 2010, 107, 20744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rief M; Gautel M; Oesterhelt F; Fernandez JM; Gaub HE Science 1997, 276, 1109–12. [DOI] [PubMed] [Google Scholar]
- (35).Rauceo JM; De Armond R; Otoo H; Kahn PC; Klotz SA; Gaur NK; Lipke PN Eukaryotic Cell 2006, 5, 1664–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Salgado PS; Yan R; Taylor JD; Burchell L; Jones R; Hoyer LL; Matthews SJ; Simpson PJ; Cota E Proc. Natl. Acad. Sci. U. S. A 2011, 108, 15775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Alsteens D; Dupres V; Klotz SA; Gaur NK; Lipke PN; Dufrene YF ACS Nano 2009, 3, 1677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Klotz SA; Gaur NK; De Armond R; Sheppard D; Khardori N; Edwards JE Jr.; Lipke PN; El-Azizi M Med. Mycol 2007, 45, 363–70. [DOI] [PubMed] [Google Scholar]
- (39).Klotz SA; Gaur NK; Lake DF; Chan V; Rauceo J; Lipke PN Infect. Immun 2004, 72, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Otoo HN; Lee KG; Qiu W; Lipke PN Eukaryotic Cell 2008, 7, 776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Eisenberg DS; Sawaya MR Annu. Rev. Biochem 2017, 86, 69–95. [DOI] [PubMed] [Google Scholar]
- (42).Shewmaker F; McGlinchey RP; Wickner RB J. Biol. Chem 2011, 286, 16533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Urbic T; Najem S; Dias CL Biophys. Chem 2017, 231, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wei G; Su Z; Reynolds NP; Arosio P; Hamley IW; Gazit E; Mezzenga R Chem. Soc. Rev 2017, 46, 4661–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Rameau RD; Jackson DN; Beaussart A; Dufrene YF; Lipke PN mBio 2016, 7, e01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ramsook CB; Tan C; Garcia MC; Fung R; Soybelman G; Henry R; Litewka A; O’Meally S; Otoo HN; Khalaf RA; Dranginis AM; Gaur NK; Klotz SA; Rauceo JM; Jue CK; Lipke PN Eukaryotic Cell 2010, 9, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhang T; Fang HHP Biotechnol. Lett 2004, 26, 989–992. [DOI] [PubMed] [Google Scholar]
- (48).Gaur NK; Klotz SA Infect. Immun 1997, 65, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Beaussart A; Herman P; El-Kirat-Chatel S; Lipke PN; Kucharikova S; Van Dijck P; Dufrene YF Nanoscale 2013, 5, 10894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.