SUMMARY
Xanthomonas axonopodis pv. citri, the bacterium responsible for citrus canker, uses effector proteins secreted by a type III protein secretion system to colonize its hosts. Among the putative effector proteins identified for this bacterium, we focused on the analysis of the roles of AvrXacE1, AvrXacE2 and Xac3090 in pathogenicity and their interactions with host plant proteins. Bacterial deletion mutants in avrXacE1, avrXacE2 and xac3090 were constructed and evaluated in pathogenicity assays. The avrXacE1 and avrXacE2 mutants presented lesions with larger necrotic areas relative to the wild‐type strain when infiltrated in citrus leaves. Yeast two‐hybrid studies were used to identify several plant proteins likely to interact with AvrXacE1, AvrXacE2 and Xac3090. We also assessed the localization of these effector proteins fused to green fluorescent protein in the plant cell, and observed that they co‐localized to the subcellular spaces in which the plant proteins with which they interacted were predicted to be confined. Our results suggest that, although AvrXacE1 localizes to the plant cell nucleus, where it interacts with transcription factors and DNA‐binding proteins, AvrXacE2 appears to be involved in lesion‐stimulating disease 1‐mediated cell death, and Xac3090 is directed to the chloroplast where its function remains to be clarified.
INTRODUCTION
Citrus canker is a worldwide distributed citrus disease caused by the bacterium Xanthomonas axonopodis pv. citri (Xac). The pathogen enters host plant tissues through stomata and wounds, and multiplies into the intercellular spaces, leading to raised necrotic corky lesions on leaves, stems and fruits, which reduce fruit quality and quantity (Brunings and Gabriel, 2003; Graham et al., 2004). The pathogen uses several mechanisms to interact with the plant and establish the disease in the host tissue. These mechanisms include a type III protein secretion system (Dunger et al., 2005), the secretion of xanthan exopolysaccharide (Dunger et al., 2007) and the production of a filamentous haemagglutinin‐like adhesin that allows bacterial attachment to the plant surface (Gottig et al., 2009). Recently, we have also characterized a plant natriuretic peptide‐like protein, uniquely present in Xac, which improves host homeostasis and, in this way, increases bacterial survival in the plant tissue (Gottig et al., 2008).
Plants are constantly exposed to bacterial pathogens and thus have evolved different mechanisms to counteract them. At an early stage, plants recognize pathogen‐associated molecular patterns (PAMPs) and trigger a basal immune response. Nevertheless, pathogens may attenuate this response by translocating effector proteins into the plant cell which may produce modifications in the resistance signalling pathway. As a consequence, plants have evolved mechanisms to prevent effector action by detecting these effector proteins through resistance (R) proteins (Chang et al., 2004; Chisholm et al., 2006; Jones and Dangl, 2006). However, direct binding between effector proteins and their corresponding R proteins has rarely been demonstrated. As many effector [formerly known as avirulence (Avr)] proteins are known to increase pathogen virulence, an alternative model that has been proposed suggests that effector proteins are able to interact and modify a specific target in the plant to promote disease in the absence of the corresponding R proteins, and that R proteins may function in guarding these targets and activating the resistance signalling pathway (Chang et al., 2004; Chisholm et al., 2006). However, little is known about these target proteins and how they are modified. Several genes coding for bacterial effector proteins have been isolated, but only a few have been characterized biochemically. Some are reported to be small ubiquitin‐like modifier (SUMO)‐proteases, cysteine proteases, tyrosine phosphatases and ubiquitin‐ligases, which may be able to modify different targets and, consequently, plant cell defence signalling (Chang et al., 2004; Chisholm et al., 2006; Dangl and McDowell, 2006). Recently, two excellent reviews on Xanthomonas type III secreted effectors have been published (Buttner and Bonas, 2010; White et al., 2009). In this context, one major challenge is to identify plant proteins interacting with bacterial effectors that could lead to the identification of pathogenicity targets and/or of R proteins, and therefore open up a way to identify R genes. In the case of citrus canker, this information may be even more important, as no genetically resistant cultivars have been found in nature (Brunings and Gabriel, 2003; Graham et al., 2004). Thus, any information that can lead to an understanding of the molecular mechanisms underlying disease development brings the possibility of disease control closer.
The genes of many type III effector proteins possess a conserved cis‐regulatory element, the plant‐inducible promoter (PIP) box, with the consensus sequence TTCGC–N15–TTCGC (although imperfect PIP boxes exist in which the size of the N15 linker may also vary) (Koebnik et al., 2006). These sites are recognized by the product of the gene hrpX, an AraC‐type regulator which is activated in planta by HrpG, a member of the OmpR family of two‐component response regulators (Wengelnik et al., 1996). The Xac genome has been completely sequenced and several genes potentially coding for effector proteins that contain a PIP box in their promoter region have been identified (da Silva et al., 2002). In Xac, several candidate effectors have been identified and, among them, the few characterized belong to the AvrBs3 family of effector proteins that contain repeats of 34 amino acids. PthA was the first member of this family to be identified and it was able to induce citrus canker when expressed in citrus leaves (Duan et al., 1999); other members of this family have been characterized since then, including Apl1 (Fujikawa et al., 2006), HssB3.0 (Shiotani et al., 2007) and AvrTaw (Rybak et al., 2009). Other effector candidates are AvrXacE1 and AvrXacE2 (da Silva et al., 2002) and, despite the fact that little homology has been found with proteins of known structure, they cluster with putative Avr proteins of other phytopathogens and group into the XopE effector family (White et al., 2009). AvrXacE1 and AvrXacE2 possess the catalytic triad of cysteine, histidine and aspartic acid, and have been grouped with peptide N‐glycanases (PNGases) (Nimchuk et al., 2007). Moreover, they possess, in their N‐terminal regions, N‐myristoylation motifs (Thieme et al., 2007). Another Xac gene that encodes a leucine‐rich repeat (LRR) effector protein is xac3090, which groups into the XopL effector family (White et al., 2009). The LRR motif, observed in eukaryotic proteins, is involved in protein–protein interactions (Kobe and Kajava, 2001) and is found in a number of R proteins that mediate the defence response (Jones and Dangl, 2006).
The aim of this study was to investigate the role of AvrXacE1, AvrXacE2 and Xac3090 in pathogenicity and their possible interactions with citrus leaf proteins. Initially, the selection of these effector proteins was based on their presence in the published Xac genome (da Silva et al., 2002) and the lack of information about them. Recently, the fact that avrXacE1, avrXacE2 and xac3090 display a variable distribution among different Xanthomonas pathovar strains suggests that they play crucial roles in host specificity (Hajri et al., 2009) and supports the importance of understanding their participation in the pathogenic process. In this study, we constructed and characterized Xac deletion mutants in avrXacE1, avrXacE2 and xac3090 and evaluated their roles in the development of citrus canker symptoms. We also performed yeast two‐hybrid studies using AvrXacE1, AvrXacE2 and Xac3090 as baits against a Citrus sinensis cDNA library in order to investigate their possible interactions with proteins of the host plant, and evaluated their localization in the plant cell.
RESULTS
Analysis of AvrXacE1, AvrXacE2 and Xac3090 sequences
The Xac genome sequence was searched for putative bacterial effector proteins and, among those identified (da Silva et al., 2002), we decided to study further the avrXacE1 (XAC0286), avrXacE2 (XAC3224) and xac3090 (XAC3090) gene products. These genes code for proteins of high homology with Avr proteins identified in other phytopathogens. AvrXacE1 and AvrXacE2 possess 401 and 356 amino acids, respectively, and present a single N‐myristoylation motif in their N‐terminal regions (Fig. 1). Xanthomonas campestris pv. vesicatoria, the causal agent of bacterial spot of pepper, has proteins orthologous to AvrXacE1 and AvrXacE2: XopE1 and XopE2, which are 92% and 97% identical, respectively. XopE1 and XopE2 are targeted to the plant cell plasma membrane, probably as a result of N‐myristoylation which myristoyl anchors these proteins to the membranes (Thieme et al., 2007). AvrXacE1 and AvrXacE2 also show similarity to AvrPphE, an effector protein of Pseudomonas syringae, AvrXccE1 of Xanthomonas campestris pv. campestris and other putative effector proteins of phytopathogenic bacteria, which bear the catalytic triad of cysteine, histidine and aspartic acid present in PNGase (Nimchuk et al., 2007). Xac3090 is a protein of 497 amino acids and possesses LRR domains that have previously been shown to be involved in protein–protein interactions (Kobe and Deisenhofer, 1995).
Figure 1.
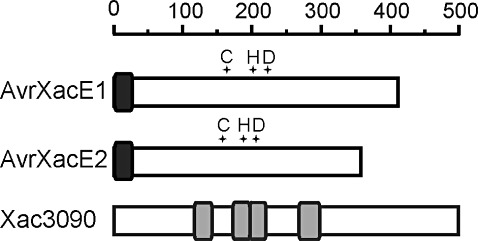
Schematic representation of Xanthomonas axonopodis pv. citri (Xac) effector proteins. Rule indicates amino acid numbers. Putative cysteine–histidine–aspartic catalytic triads (crosses) are shown. Dark grey boxes indicate the conserved N‐termini of AvrXac proteins harbouring a putative N‐myristoylation signal. Light grey boxes in Xac3090 indicate the locations of leucine‐rich repeats.
Analysis of the roles of AvrXacE1, AvrXacE2 and Xac3090 in citrus canker and in nonhost response
To characterize the role of these Xac proteins in pathogenicity, deletion mutant strains, constructed by marker exchange with double crossover in these genes, were tested for their ability to cause disease in citrus leaves. The wild‐type strain belongs to group A (formerly Asiatic group) and typical citrus canker disease symptoms are observed when infiltrated or sprayed onto citrus leaves. When wild‐type bacteria are infiltrated in citrus leaves at 107 colony‐forming units (cfu)/mL, the infection is first visualized as a water‐soaking phenotype and, later, as a result of cell hypertrophy and hyperplasia, erupted tissue, which may be accompanied by some necrotic spots, is observed. At lower bacterial concentrations (104–106 cfu/mL), characteristic raised necrotic corky lesions are observed (Gottig et al., 2009). Wild‐type and mutant strains were infiltrated into the abaxial side of citrus leaves. Lesions caused by ΔavrXacE1 and ΔavrXacE2 mutants showed more extensive necrotic areas relative to those caused by wild‐type bacteria (Fig. 2A). In order to verify the observed phenotypes, ΔavrXacE1 and ΔavrXacE2 were complemented with plasmids bearing a copy of avrXacE1 and avrXacE2, respectively. The inoculation of these strains in citrus leaves decreased the degree of necrosis observed with the mutant strains, confirming the complementation. We also constructed a double mutant strain in avrXacE1 and avrXacE2 (named ΔavrXacE1/E2). This double mutant caused a phenotype similar to that observed for the single mutants, also showing more extensive necrotic lesions (Fig. 2A). These results suggest that these two effectors act separately, affecting different signalling pathways that modify plant tissue necrosis, as the absence of either gives the same degree of necrosis. Conversely, the ΔXac3090 mutant strain produced lesions similar to those obtained with the wild‐type strain, as well as the ΔXac3090 complemented strain (Fig. 2A). The lack of difference in the plant response to the ΔXac3090 strain may suggest that this effector subtly modifies the host cell, but not sufficiently strongly to cause visible changes in the infected tissue. Nevertheless, the redundancy hypothesis, which proposes that effector proteins are often functionally redundant (Kvitko et al., 2009), may explain the lack of differences observed.
Figure 2.
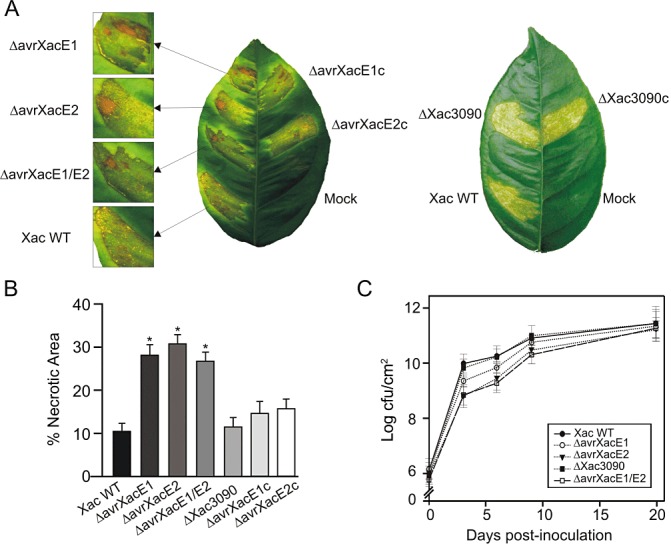
Characterization of Xanthomonas axonopodis pv. citri (Xac) mutants on pathogenicity. (A) Xac wild‐type (Xac WT), mutants ΔAvrXacE1 and ΔAvrXacE2, their complemented strains ΔAvrXacE1c and ΔAvrXacE2c, and ΔAvrXacE1/E2 (left leaf) and Xac wild‐type, ΔXac3090 and ΔXac3090c (right leaf) were inoculated at 107 colony‐forming units (cfu)/mL in 10 mm MgCl2 into the intercellular spaces of fully expanded citrus leaves. Mock was infiltrated with 10 mm MgCl2 . A representative leaf is shown 8 days after inoculation. (B) The percentage necrotic areas in lesions at day 8 after inoculation were calculated as the necrotic area per infected area. Areas were measured from digitalized images of infected leaves using Adobe Photoshop software. Mean ratings ± standard deviations were calculated from 20–30 infiltrated areas per strain. Asterisks indicate P < 0.01. (C) Bacterial growth of Xac wild‐type and mutants in citrus leaves. Bacterial multiplication was monitored over a period of 20 days. Values represent the mean of three samples from three different plants, and the experiment was repeated three times.
In order to quantify the observed variations in the extent of the necrotic areas among tissues inoculated with the different bacteria, 30 infected leaves were photographed and the necrotic areas were measured from the digitalized images using Adobe Photoshop software. Lesions caused by the infiltration of the deletion mutants ΔavrXacE1, ΔavrXacE2 and ΔavrXacE1/E2 presented a more than two‐fold increase in necrotic area relative to wild‐type infection. No significant difference (P < 0.01) was observed in ΔXac3090 inoculation when compared with the wild‐type (Fig. 2B).
To evaluate the role of these predicted Xac effectors on bacterial growth during citrus canker infection, we quantified the populations of the wild‐type and the different mutants in infected citrus leaves. In the first week of infection, Xac wild‐type and ΔXac3090 grew more rapidly, showing values of 2 × 1010 cfu/cm2 at 6 days post‐inoculation. The mutants ΔavrXacE1, ΔavrXacE2 and ΔavrXacE1/E2 grew more slowly, reaching significantly different values (P < 0.05) of 7 × 109, 3 × 109 and 2 × 109 cfu/cm2, respectively (Fig. 2C). After 20 days, all strains attained a cell density of 2 × 1011 cfu/cm2 (Fig. 2C), after which all strains decayed slightly (data not shown). Considering that Xac is a biotrophic pathogen that requires living plant tissue to grow, the results of the growth curves are consistent with the phenotypes observed, given that an increased necrotic area supports less bacterial growth.
To assess whether AvrXacE1, AvrXacE2 and Xac3090 contribute towards the development of the hypersensitive response (HR) in nonhost plants, we infiltrated wild‐type, ΔavrXacE1, ΔavrXacE2 and ΔXac3090 strains into the leaves of tomato, tobacco and cotton plants. In all the different infiltrated nonhost plants, a similar and typical HR phenotype was observed for each one of the tested strains (data not shown).
Identification of citrus proteins interacting with AvrXacE1, AvrXacE2 and Xac3090
In order to identify possible protein targets of these bacterial proteins in the plant cell, we performed a yeast two‐hybrid assay using AvrXacE1, AvrXacE2 and Xac3090 as bait against a prey library derived from C. sinensis cDNA. The prey obtained in the screens were sequenced and compared with the complete GenBank nonredundant database using blastx. The bait interacted with a large number of prey and the results are presented in Table 1. Interactions detailed in Table 1 were confirmed, first by independent yeast two‐hybrid assays between bait and the specific prey, and by the use of pOBD and pOAD empty vectors as controls. We considered that interactions were significant when they were observed in more than one prey derived from the same protein. However, we also included light‐harvesting complex I protein as an interacting protein with Xac3090, which only appeared once, because of its similar localization to other prey observed, thus suggesting a true interaction. We also found some prey that ubiquitously interacted with all the bait tested and also with unrelated bait used previously (C. S. Farah, personal observation); these were disregarded. Specifically for AvrXacE1, we obtained interactions with a citrus protein homologous to retinoblastoma‐binding protein (RBBP), an RNA‐binding protein which possesses an RNA recognition motif (RRM), a homologue of a heterogeneous nuclear ribonucleoprotein A1, an RNA‐directed DNA polymerase, a pentatricopeptide repeat‐containing protein and the nuclear protein SKIP. In the case of AvrXacE2, the principal interaction was with the DNAj domain of the Heat shock protein 40 (Hsp40) chaperone protein, the lesion stimulating disease 1 (LSD1) protein and other chaperones with homology to small Hsps and Hsp70‐interacting protein. Finally, Xac3090 was observed to interact with a protein homologous to the inhibitor of apoptosis‐like protein (IAP), a RING‐type zinc‐finger protein, a chloroplast ribosomal protein, phospho‐2‐dehydro‐3‐deoxyheptonate aldolase 1, the auxin responsive protein IAA13 and the light‐harvesting complex I protein.
Table 1.
Summary of protein–protein interactions involving Xanthomonas axonopodis pv. citri effector proteins.
| Bait | Specific prey (name, gene number; scientific name) | Initial codons of prey (number of times observed) | Number of times observed | % Identity with the sequenced prey |
|---|---|---|---|---|
| AvrXacE1 (XAC0286) | Retinoblastoma‐binding protein, putative, XP_002530663; [Ricinus communis] | 210(5),239 | 6 | 91 |
| Heterogeneous nuclear ribonucleoprotein A1, putative, XP_002522793.1; [Ricinus communis] | 66(4),115 | 5 | 87 | |
| RNA‐directed DNA polymerase (reverse transcriptase), ABN06064; [Medicago truncatula] | 783(3),869 | 4 | 58 | |
| Pentatricopeptide repeat‐containing protein, putative, XP_002514151; [Ricinus communis] | 78(3) | 3 | 61 | |
| Nuclear protein skip, putative, XP_002530607; [Ricinus communis] | 151(2) | 2 | 90 | |
| AvrXacE2 (XAC3224) | Chaperone protein DNAj, putative, XP_002513190.1; [Ricinus communis] | 49(4),131(16),144 | 21 | 65 |
| Zinc‐finger protein LSD1, putative, XP_002276731.2 [Vitis vinifera] | 14(2),23(2) | 4 | 80 | |
| Small heat shock protein, BAK61831.1; [Citrus unshiu] | 1,27(3) | 4 | 94 | |
| SEU1 protein, CAF18247.1; [Antirrhinum majus] | 46(2) | 2 | 66 | |
| Heat shock protein 70 (HSP70)‐interacting protein, putative, XP_002533498.1; [Ricinus communis] | 8(2) | 2 | 76 | |
| XAC3090 | Inhibitor of apoptosis‐like protein, AAM65075; [Arabidopsis thaliana] | 195(5) | 5 | 55 |
| Chloroplast 30S ribosomal protein, putative, AED93316.1; [Arabidopsis thaliana] | 39(4) | 4 | 80 | |
| Phospho‐2‐dehydro‐3‐deoxyheptonate aldolase 1, chloroplast precursor, putative, XP_002531676; [Ricinus communis] | 27(3),82 | 4 | 64 | |
| Auxin‐responsive protein IAA13, putative, XP_002526541; [Ricinus communis] | 70(3) | 3 | 67 | |
| Light‐harvesting complex I protein Lhca2, XP_002303791; [Populus trichocarpa] | 21 | 1 | 76 |
Localization of AvrXacE1, AvrXacE2 and Xac3090 bacterial proteins in the plant cell
To analyse the subcellular localization of these bacterial proteins, plasmid constructions encoding their sequences without stop codons were fused in frame to the N‐terminus of green fluorescent protein (GFP) and transferred into Nicotiana benthamiana using Agrobacterium‐mediated transient transformation. Confocal microscopy imaging of N. benthamiana cells expressing the fusions AvrXacE1‐GFP and AvrXacE2‐GFP showed a fluorescence pattern mainly in the plant cell membrane and in the nucleus, whereas Xac3090‐GFP‐expressing plants displayed nuclear and chloroplast fluorescence (Fig. 3). To verify nucleus localization, a co‐localization assay using Hoechst solution, a nuclear stain, was performed. Furthermore, to confirm that Xac3090‐GFP localizes to the chloroplast, we identified chloroplasts by the red autofluorescence of chlorophyll, and corroborated a co‐localization of Xac3090‐GFP with chloroplast structures (Fig. 4A).
Figure 3.
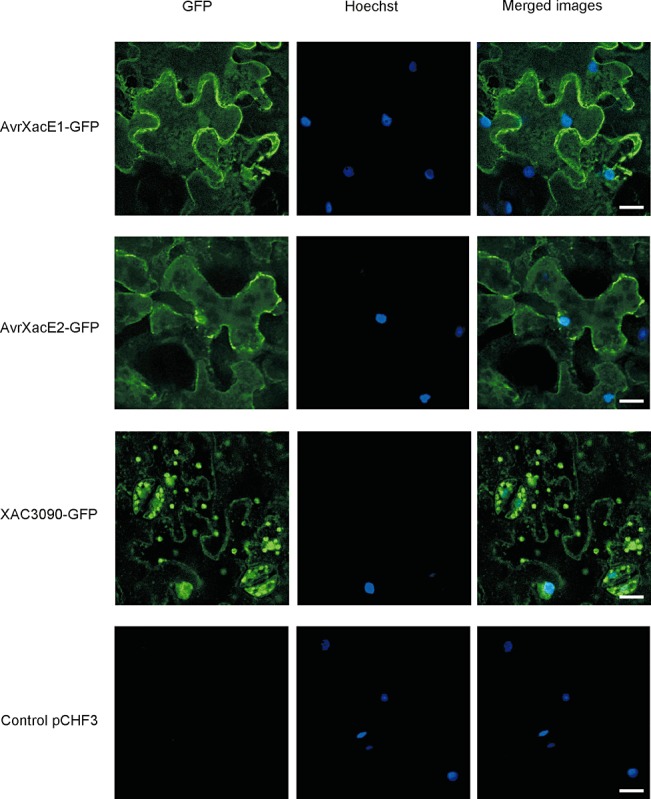
Detection and localization of avirulence–green fluorescent proteins (Avr‐GFPs). Confocal microscopic localization of AvrXacE1, AvrXacE2 and Xac3090 proteins in Nicotiana benthamiana leaves transiently agroinfiltrated. The cells were transfected with one of the following pCHF3 plasmids: pCHF3286, pCHF33224, pCHF33090 and the empty plasmid (pCHF3). Cell nuclei were counterstained with Hoechst 33258. The localization of GFP fusion proteins was visualized by confocal laser scanning microscopy 24 h after transformation. Bar, 15 µm.
Figure 4.
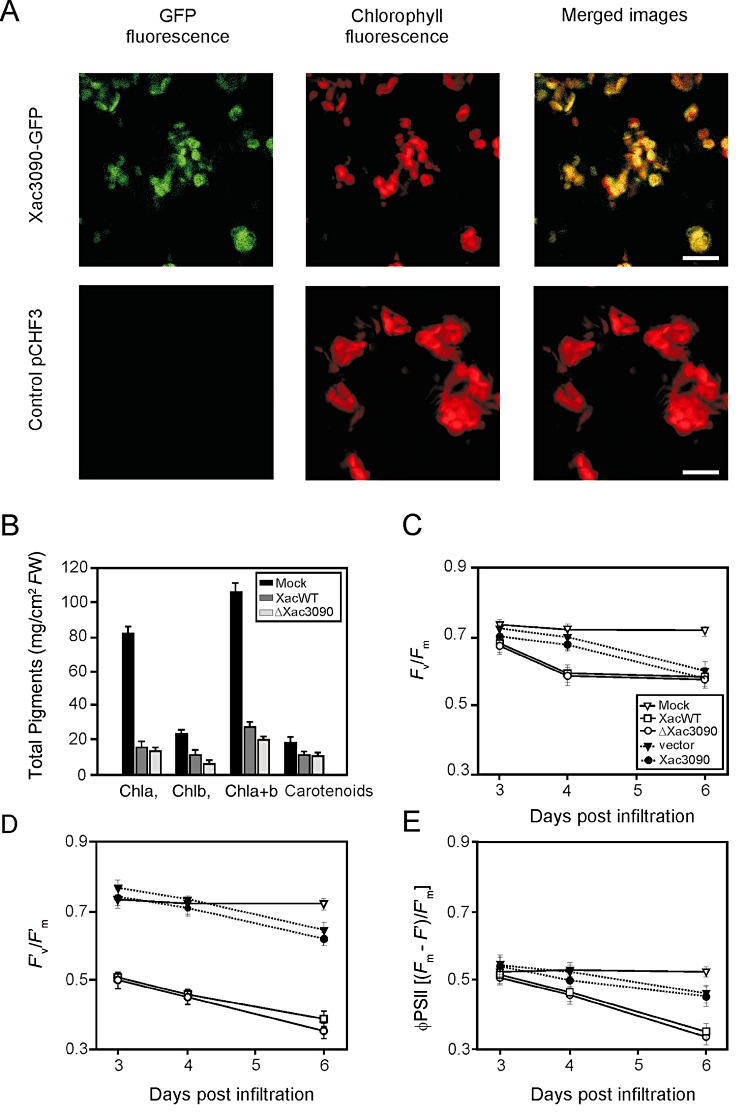
Localization of Xac3090‐green fluorescent protein (Xac3090‐GFP) and photosynthetic‐related parameters. (A) Confocal microscopic localization of Xac3090‐GFP in Nicotiana benthamiana leaves transiently agroinfiltrated. The localization of GFP fusion proteins was visualized by confocal laser scanning microscopy 24 h after agroinfiltration with pCHF33090 and pCHF3 (empty plasmid). Bar, 15 µm. (B) Pigment determination in citrus leaves. Xanthomonas axonopodis pv. citri (Xac) wild‐type and ΔXac3090 were infiltrated at 107 colony‐forming units (cfu)/mL in 10 mm MgCl2 into citrus leaves. Pigment chlorophyll a (Chl a), chlorophyll b (Chl b) and carotenoids were determined. FW, fresh weight. (C) Potential quantum efficiency of photosystem II (PSII) (F v/F m). (D) Effective quantum efficiency of PSII (F′v/F′m□). (E) PSII operating efficiency (φ PSII). (C–E) Chlorophyll fluorescence parameters in citrus leaves infiltrated with 10 mm MgCl2 (open triangles), Xac wild‐type (open squares), ΔXac3090 (open circles) and A. tumefaciens bearing the empty vector (filled triangles) and Xac3090‐GFP (filled circles). The results are the mean of five replicates and error bars represent the standard deviations.
Analysis of the role of Xac3090 in chloroplast photosynthetic efficiency
The previous localization results suggested that physiological chloroplast parameters might be altered by this protein. We therefore characterized photosynthetic parameters, specifically chlorophyll a, chlorophyll b and carotenoid content, in citrus leaves infiltrated with the wild‐type and ΔXac3090 strains. We observed a large decrease in pigments in tissue infected with both strains relative to mock‐infiltrated tissue. No significant differences (P < 0.05) were observed between the strains (Fig. 4B).
Next, we tested whether the photosynthetic efficiency was altered in tissue infected with ΔXac3090. Photosynthetic performance in the wild‐type and mutant strain was evaluated by determining chlorophyll fluorescence parameters after 72, 96 and 144 h. The maximum quantum efficiency of photosystem II (PSII) (F v/F m), maximum efficiency of PSII at a given light intensity (F′v/F′m), measured at 100 µmol quanta/m2/s, and PSII operating efficiency (φ PSII), were calculated as described previously (Baker and Rosenqvist, 2004). A noticeable decrease in these parameters was observed for the leaves infiltrated with both strains relative to the mock infiltration, but no significant differences (P < 0.05) were observed between the strains (Fig. 4C–E).
To study whether Xac3090 may affect the photosynthetic efficiency in citrus leaves, Xac3090‐GFP was transferred into citrus leaves via Agrobacterium tumefaciens. This technique has been used recently to evaluate the effect of an Avr protein in Citrus paradise (Figueiredo et al., 2011). Figure 4C–E shows that the chlorophyll fluorescence parameters were not significantly different (P < 0.05) from those observed for control leaves infiltrated with A. tumefaciens carrying the empty vector pCHF3. However, in both A. tumefaciens‐mediated inoculations, impaired fluorescence parameters were observed, probably as a result of A. tumefaciens infiltration. Therefore, although the results of the localization and protein–protein interaction studies indicate that Xac3090 localizes to the chloroplast, no direct effect on the photosynthetic efficiency could be attributed to this protein. A possible explanation may be that the function of Xac3090 on this organelle may depend on other effector proteins acting in a concerted manner.
DISCUSSION
Like many other Gram‐negative bacterial pathogens, Xac uses a type III secretion system to deliver effector proteins into host cells. Although the roles of the majority of these effector proteins remain unknown, some can suppress innate immune responses (White et al., 2009). Only a limited number of studies have been carried out on the biochemical and functional characterization of phytopathogen effector proteins (Grant et al., 2006; White et al., 2009). The fact that mutations in one single effector protein in Xanthomonas campestris pv. campestris (Castaneda et al., 2005; Jiang et al., 2009) and Pseudomonas syringae pv. tomato (Kvitko et al., 2009) do not have an effect on the virulence phenotype suggests that type III effector proteins may have redundant functions in the host cell. In this work, we found differences in virulence phenotypes for two of the three effector proteins analysed. Mutants in avrXacE1 and avrXacE2 displayed lesions with larger necrotic areas than those of the wild‐type strain in citrus leaves, whereas no phenotypical differences were observed for the xac3090 mutant. Nevertheless, the three mutants induced an HR similar to that obtained on infiltration with the wild‐type strain in all the nonhost plants assayed, suggesting that the HR response is rapid and sufficiently strong to hide possible subtle effects of these effectors. Interestingly, mutants in XopE1 and XopE2, the orthologues of AvrXacE1 and AvrXacE2 from X. campestris pv. vesicatoria, showed no significant differences in disease symptoms or HR induction when compared with the wild‐type strain (Thieme et al., 2007), suggesting that a variable set of complex host–pathogen interactions takes place in each particular plant–pathogen interaction.
Xac is a biotrophic pathogen and therefore requires living plant tissue to provide favourable conditions to multiply and colonize other niches. Therefore, the temporary delay of host necrosis may be advantageous to Xac. The necrosis progression observed in infections with mutants in avrXacE1 and avrXacE2 suggests that they may be involved in the slowing or suppression of necrosis in the initial stages of the infection process to allow a more rapid bacterial growth, and thus to ensure colonization for a biotrophic pathogen.
AvrXacE1. In the yeast two‐hybrid assay, AvrXacE1 interacted with DNA‐ and RNA‐binding proteins involved in RNA metabolism, such as RBBP. This protein bears the N‐terminal domains present in the animal orthologue RBBP6, which have been suggested to be involved in pre‐mRNA processing and ubiquitin‐like protein modification, possibly playing a role in the regulation of the splicing machinery (Pugh et al., 2006), although, in plants, its role remains to be elucidated. In addition, AvrXacE1 interacted with a heterogeneous nuclear ribonucleoprotein (hnRNP) which bears an RRM found in proteins implicated in the regulation of alternative splicing and protein components of small nuclear ribonucleoproteins (snRNPs) (Fu et al., 2007). An Arabidopsis thaliana RNA‐binding protein with RRM has been identified as a target of the effector protein HopU1 from Pseudomonas syringae. HopU1 is able to ADP‐ribosylate RNA‐binding proteins and therefore to interfere in host immunity by altering RNA metabolism. Another protein identified that interacts with AvrXacE1 is the nuclear protein SKIP. SKIP has been well characterized as a transcriptional regulator, as well as a spliceosome component, in humans. In rice, SKIPa positively modulates stress resistance through transcriptional regulation of diverse stress‐related genes (Hou et al., 2009). AvrXacE1 was observed in both the nucleus and plant cell membrane. As this protein possesses an N‐myristoylation motif, myristoylation could anchor this effector protein in the membrane in a similar mechanism to its XopE1 homologue (Thieme et al., 2007). The protein Snf1 from Saccharomyces cerevisiae is localized to the plasma membrane when it is N‐myristoylated, but is shifted to the nucleus when N‐myristoylation is blocked in response to a specific signal (Lin et al., 2003). Further analysis will be needed to clarify whether this kind of spatial regulation also occurs in plant infected cells.
AvrXacE2. Bacterial mutants in the avrXacE2 gene produced larger numbers of necrotic lesions than did the wild‐type strain, suggesting that this protein may function to attenuate cell death. Interestingly, one of the plant proteins with which this effector interacts is LSD1. In Arabidopsis, LSD1 encodes a plant‐specific zinc finger‐containing transcription factor that plays a role in the negative regulation of cell death, regulating the translocation of the transcription factor AtbZIP10 (a positive regulator of cell death) between the nucleus and the cytoplasm (Kaminaka et al., 2006). It can therefore be suggested that AvrXacE2 may modulate the LSD1 pathway and thus the outcome of cell death. The dual localization of AvrXacE2 in the plant cell membranes and in the nucleus may be explained by the rate at which the translocation of this effector occurs in plant cells, and needs to be studied further.
Xac3090. This bacterial effector protein interacted with chloroplast proteins, such as a specific chloroplast ribosomal protein and a light‐harvesting complex I protein. This result prompted us to speculate that Xac3090 may alter the photosynthetic machinery; however, no significant differences were observed in pigment content or chlorophyll fluorescence parameters when a bacterial strain lacking xac3090 was infiltrated into citrus leaves, or even after transient expression of Xac3090‐GFP in citrus leaves, suggesting that the effect is too subtle to be detected with the techniques used, or that the spatiotemporal coordinated actions of other effectors are needed to cause modifications in this specific plant organelle. Xac3090 interacted with a phospho‐2‐dehydro‐3‐deoxyheptonate aldolase involved in the synthesis of secondary phenylpropanoid compounds (Cho et al., 2007), IAP and a homologue to the auxin responsive protein IAA13, suggesting that Xac3090 may participate in these pathways.
Interestingly, Xac3090 localizes in both the nucleus and chloroplasts of N. benthamiana cells. There are some nuclear transcription factors, named Whirly proteins, which, in addition to the nucleus, are targeted to chloroplasts and mitochondria, and may be involved in the communication between the nucleus and organelles in plant cells (Schwacke et al., 2007). Moreover, it has been proposed recently that proteins may be relocated from one compartment to another in response to environmental changes, and that dual targeting may function for the storage or sequestration of transcription factors inside the organelles until specific conditions require their activity in the nucleus (Krause and Krupinska, 2009).
The genus Xanthomonas comprises a group of plant pathogenic bacteria causing wilts, cankers, leaf spots and blights. Although the host range of this genus is wide, particular pathovars can be confined to a specific host plant, and the variety of effector proteins present in a given pathogen may determine the host and tissue specificity (White et al., 2009). In particular, for Xanthomonas axonopodis, a study of 132 strains representative of 18 different pathovars displaying different host ranges with wide geographic distribution found a correspondence between the composition of effector repertoires and pathovars, thus supporting the idea that host specificity results from the interaction between repertoires of bacterial virulence genes and repertoires of genes involved in host defences (Hajri et al., 2009). Interestingly, all of the characterized Xac strains possess the three genes analysed in this study, whereas two of the characterized X. axonopodis pv. aurantifolii strains, which also cause citrus canker in only lemons and Mexican lime (Brunings and Gabriel 2003), lack a functional copy of AvrXacE1 (Hajri et al., 2009). Moreover, in the strains of the phylogenetically related bacterium X. axonopodis pv. citrumelo, which causes citrus bacterial spots (Brunings and Gabriel, 2003; Graham et al., 2004), AvrXacE2 was present in three of six strains analysed, whereas Xac3090 and AvrXacE1 were absent (Hajri et al., 2009). Thus, the host range may be explained in part by these complex effector repertoires and, specifically in the case of citrus canker, an understanding of Xac bacterial effector protein localization in the plant cell, and the plant proteins with which they interact, may be relevant in designing future control strategies.
EXPERIMENTAL PROCEDURES
Bacterial strains, culture conditions and media
Escherichia coli strain DH5α was used for DNA subcloning. Cells were cultivated at 37 °C in Luria–Bertani (LB) medium. Xac wild‐type and mutant strains were grown at 28 °C in Silva Buddenhagen (SB) medium (Dunger et al., 2007). Antibiotics were used at the following concentrations: ampicillin (Ap), 100 µg/mL for E. coli and 25 µg/mL for Xac; streptomycin (Sm), 100 µg/mL for E. coli and 50 µg/mL for Xac; spectinomycin (Sp), 50 µg/mL for E. coli and 25 µg/mL for Xac; gentamycin (Gm), 20 µg/mL; kanamycin (Km), 40 µg/mL. Xac wild‐type (strain Xcc99‐1330) was kindly provided by Blanca I. Canteros (INTA Bella Vista, Argentina). Agrobacterium tumefaciens strain GV3101 was cultured at 28 °C in LB medium using: rifampicin (Rf), 10 µg/mL; Gm, 25 µg/mL; Sm, 100 µg/mL; Sp, 50 µg/mL.
Mutant strain constructions
Mutant strains in avrXacE1 (XAC0286), avrXacE2 (Xac3224) and xac3090 genes were constructed by marker exchange with double crossover. avrXacE1 was amplified from Xac genomic DNA with XAC0286U and XAC0286D oligonucleotides (primer sequences are provided in Table 2) and cloned into pKmobGII previously digested with the restriction enzymes EcoRI and SalI, generating pKmobXAC0286. A 2‐kb DNA fragment coding for Sm/Sp resistance from pKRP13 (Reece and Phillips, 1995) was subcloned into the BamHI site of pKmobXAC0286, generating pKmobXAC0286Ω. avrXacE2 was amplified from Xac genomic DNA with XAC3224U and XAC3224D oligonucleotides (Table 2) and cloned into pET32a previously digested with the restriction enzymes NcoI and SalI, generating pET323224. The aacC1 gene coding for Gm resistance was subcloned into the EcoRI site of pET323224, generating pET323224Gm. pET323224Gm was digested with restriction enzymes BamHI and SalI, and the fragment XAC3224Gm was subcloned into pKmobGII previously digested with BamHI and SalI, generating pKmobXAC3224Gm. xac3090 was amplified with XAC3090H and XAC3090E oligonucleotides (Table 2) and cloned into pKmobGII previously digested with the restriction enzymes EcoRI and HindIII, generating pKmobXAC3090, and the aacC1 gene coding for Gm resistance was subcloned into the PstI site of pKmobXAC3090, rendering pKmobXAC3090Gm. Escherichia coli strain S17‐1 cells transformed with pKmobXAC0286Ω, pKmobXAC3224Gm and pKmobXAC3090Gm were conjugated to Xac. Following selection for antibiotic resistance and sensitivity (Sp and Sm resistance for ΔavrXacE1; Gm resistance for ΔavrXacE2 and ΔXac3090; Km sensitivity for all), mutant strains were verified by polymerase chain reaction (PCR). The double mutant strain ΔavrXacE1/E2 was obtained by conjugation of pKmobXAC3224Gm into ΔavrXacE1.
Table 2.
Primers used in this study.
| Primer | Primer sequences 5′–3′ |
|---|---|
| XAC0286U | GTCCGAATTCTCGGAGAGCGACATGGGACTAT |
| XAC0286D | CGGTGTCGACTCGCGCAGCAAAGGTGTTCGG |
| XAC0286UK | GTCCGGTACCATCGGAGAGCGACATGGGA |
| XAC0286D2 | CGGTGTCGACATACCAGAAAGCCTGCCTTG |
| XAC3224U | ACAGTCCATGGAAATGGGTTGCACTATCTCAAC |
| XAC3224D | CGGTGTCGACCATCACCGGTTACTGGCTCTG |
| XAC3224UK | ACATGGTACCAATGGGTTGCACTATCTCAAC |
| XAC3224D2 | CGGTGTCGACGCTCTGCTCGCACAGCTG |
| XAC3090H | CTGCAAGCTTCGAGCGACAGCCAAGACGAGGAAT |
| XAC3090E | CAGTGAATTCGCGCGGGTACAACGGATGGTGGAC |
| XAC3090THU | CTGCACCATGGTGGGCGCTTCACCATTAC |
| XAC3090THD | CGGTGTCGACTCGTGTTTGCCCTTTACTGA |
| XAC3090UK | GTCCGGTACCGGGCGCTTCACCATTAC |
| XAC3090D2 | CGGTGTCGACAGGTGCCGGGCTTGTTT |
| XAC3090SL | ACATCCCGGGTGTTGTGTGGAATTGTGAGC |
| HIS | AGTTGAGCTCCTTTGTTAGCAGCCGGATC |
| HIS2 | AGTTGAATTCCAGCAGCCAACTCAGCT |
| GFPSU | ACATGTCGACATGGTGAGCAAGGGCGAG |
| GFPSD | CGGTCTCGAGTTACTTGTACAGCTCGTCCA |
Complementation of Xac mutant strains
Xac mutant strains were complemented with the coding sequence of each effector protein in the plasmid pBBR‐MCS2 (Kovach et al., 1995). avrXacE1 was amplified from Xac genomic DNA with XAC0286UK and XAC0286D2 oligonucleotides (Table 2) and cloned into pET32a previously digested with the restriction enzymes KpnI and SalI, generating pET0286. AvrXacE1‐His was amplified from pET0286 with XAC0286UK and HIS oligonucleotides (Table 2) and cloned into pBBR‐MCS5 previously digested with the restriction enzymes KpnI and SacI, generating pBBR0286H. Restriction enzyme digestion of the avrXacE1 fragment from pBBR0286H with KpnI and SacI, and subsequent cloning into pBBR‐MCS2, generated pBBR2‐0286. avrXacE2 was amplified from Xac wild‐type genomic DNA with XAC3224U and XAC3224D oligonucleotides (Table 2) and cloned into pET32a previously digested with the restriction enzymes NcoI and SalI, generating pET3224. AvrXacE2‐His was amplified from pET3224 with XAC3224UK and HIS oligonucleotides (Table 2) and cloned into pBBR‐MCS5 previously digested with KpnI and SacI, generating pBBR3224H. Restriction enzyme digestion of the avrXacE2 fragment from pBBR3224H with KpnI and SacI, and subsequent cloning into pBBR‐MCS2, generated pBBR2‐3224. xac3090 was amplified with XAC3090UK and XAC3090D2 oligonucleotides (Table 2) and cloned into pET32a previously digested with the restriction enzymes KpnI and SalI, generating pET3090. xac3090‐His was amplified with XAC3090UK and HIS2 oligonucleotides (Table 2) and cloned into pBBR‐MCS5 previously digested with KpnI and EcoRI, generating pBBR3090H. Restriction enzyme digestion of the Xac3090 fragment from pBBR3090H with KpnI and EcoRI, and subsequent cloning into pBBR‐MCS2, generated pBBR2‐3090. Escherichia coli strain S17‐1 cells transformed with pBBR2‐0286, pBBR2‐3224 and pBBR2‐3090 were conjugated to Xac mutant strains ΔavrXacE1, ΔavrXacE2 and ΔXac3090, respectively. Following selection for kanamycin antibiotic resistance, complemented mutant strains were verified by PCR.
Plant material and inoculations
Citrus (Citrus sinensis cv. valencia) plants were grown in a glasshouse at 23–25 °C with a photoperiod of 16 h. Bacteria were cultured in SB broth to an optical density at 600 nm (OD600 nm) of 1.0, harvested by centrifugation and resuspended in 10 mm MgCl2 at 104–107 cfu/mL. For disease symptom assays, bacterial suspensions were infiltrated into leaves with needleless syringes. In planta growth assays were performed by grinding 0.8‐cm‐diameter leaf discs from infiltrated leaves in 1 mL of 10 mm MgCl2, followed by serial dilutions and plating onto SB agar plates with Ap. Colonies were counted after 48 h of incubation at 28 °C, and the results are presented as cfu per square centimetre of leaf tissue. The in planta growth assays were repeated at least in triplicate and the results were analysed by one‐way analysis of variance (anova). The percentages of necrotic areas in the lesions were calculated as the necrotic area per infected area. Areas were measured from digitalized images of 30 infected leaves using Adobe Photoshop software and analysed using one‐way anova.
Citrus sinensis cDNA prey library
The C. sinensis cDNA library containing approximately 0.8 × 106 independent clones in the pOAD vector (Uetz et al., 2000) was produced and kindly donated by Raúl Andrés Cernadas, Cássia Docena and Celso Eduardo Benedetti at the Laboratório Nacional de Luz Síncrotron (Campinas, Brazil), and has been described elsewhere (Domingues et al., 2010).
Construction of bait vectors
Xac DNA sequences coding for proteins AvrXacE1, AvrXacE2 and Xac3090 were amplified by PCR using the primers XAC0286U and XAC0286D, XAC3224U and XAC3224D, and XAC3090THU and XAC3090THD, respectively (Table 2). The primers contained unique restriction sites (EcoRI and SalI in XAC0286; NcoI and SalI in XAC3224 and XAC3090) to facilitate cloning into the EcoRI and SalI sites of the pOBD vector (Uetz et al., 2000), generating pOBDXAC0286, and into the NcoI and SalI sites of pOBD, generating pOBDXAC3090 and pOBDXAC3224, all downstream of and in frame with the Gal4 DNA‐binding domain.
Growth of yeast strains and transformation
Saccharomyces cerevisiae strain PJ694‐a (MATa trp1‐901 leu2‐3112 ura3‐52 his3‐200 gal4Δgal80ΔLYS2::GAL1‐HIS3 GAL2‐ADE2 met2::GAL7‐lacZ) was grown in the yeast medium YAPD or synthetic complete (SC) medium as described previously (Alegria et al., 2004). When indicated, SC medium was prepared lacking one or more specific components: adenine (–Ade), histidine (–His), tryptophan (–Trp) and leucine (–Leu). In the case of growth on solid medium, 1.6% Bacto Agar and 3‐aminotriazole (3AT) (see below) were added. Rapid transformations with pOBD‐bait plasmids were carried out using the PEG3350–lithium acetate protocol described by Gietz et al. (1998) and selected on SC–Trp plates at 30 °C for 2–4 days. These cells were then employed in high‐efficiency transformations with the pOAD library using 30 µg of plasmid DNA and the 30× scale‐up procedure described by Gietz and Woods (2002), which resulted, on average, in 0.5 × 107 to 1 × 107 transformants on SC–Trp–Leu plates. To determine the amount of 3AT to be used for each bait, c. 103 yeast cells transformed with the pOBD‐bait plasmid were plated onto SC–Trp–His medium containing 0, 1, 5, 10, 25 or 50 mm 3AT and incubated for 5 days at 30 °C. The pOAD library showed no clones able to auto‐activate the His or Ade gene reporters on their own.
Yeast two‐hybrid assays and DNA sequencing
After transformation of yeast cells with the pOAD library, cells were resuspended in 5 mL of sterile water and spread onto 10 150‐mm plates with SC–Trp–Leu–His–Ade plus 3AT. The amount of 3AT used was 3 mm for the three baits assayed. Plates were incubated at 30 °C for up to 14 days. Colonies that grew in the absence of His and Ade were transferred to fresh plates with SC–Trp–Leu–His–Ade plus 3AT. Plasmid DNAs were isolated from yeast colonies and used to transform DH10B E. coli cells. The prey DNA sequences were sequenced using a pOAD‐specific primer. Sequences were analysed by comparison with the available C. sinensis expressed sequence tag (EST) database. Some prey observed to interact with a wide variety of physiologically unrelated bait were considered to be false positives and were not included in Table 1.
Construction of GFP expression vectors
The gene coding for GFP was amplified from pMP2444 by PCR with GFPSU and GFPSD oligonucleotides (Table 2) and cloned into pBBR0286H, pBBR3224H and pBBR3090H, described above, previously digested with XhoI and SalI, generating pBBR0286GFP, pBRR3224GFP and pBBR3090GFP, respectively. Then, avrXacE1 was amplified from pBBR0286GFP by PCR with XAC0286UK and GFPSD oligonucleotides (Table 2), digested with the restriction enzymes KpnI and XhoI and cloned into the pCHF3 plasmid (Jarvis et al., 1998) previously digested with KpnI and SalI, generating pCHF3286. avrXacE2 was amplified from pBBR3224GFP by PCR with XAC3224UK and GFPSD oligonucleotides (Table 2), digested with the restriction enzymes KpnI and XhoI and cloned into pCHF3 previously digested with the same enzymes, generating pCHF33224. xac3090 was amplified from pBBR3090GFP by PCR with XAC3090SL and GFPSD oligonucleotides (Table 2), digested with the restriction enzymes SmaI and XhoI and cloned into pCHF3 previously digested with SmaI and SalI, generating pCHF33090.
In planta expression of effector proteins
Plasmid vectors pCHF3286, pCHF33224 and pCHF33090, carrying the genes for AvrXacE1, AvrXacE2 and Xac3090, respectively, were electroporated into A. tumefaciens strain GV3101 with a Gene Pulser II (Bio‐Rad, Hercules, CA, USA) according to the manufacturer's instructions. For infiltration assays, A. tumefaciens strains were grown in LB broth at 28 °C for 18 h, diluted 1:2000 in LB plus 20 µm acetosyringone and incubated at 28 °C to OD600 nm of 1.0. The resulting cultures were harvested by centrifugation, resuspended in sterile buffer [10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES) and 100 µm acetosyringone] to a final OD600 nm of 0.5 and incubated for 3 h at 25 °C. These cells were used to infiltrate leaves of 4–5‐week‐old N. benthamiana and citrus using needleless syringes. Infiltrated leaves were observed under a hand‐held UV lamp. Fluorescence by GFP and Hoechst 33258 in plant cells was assessed at 24, 48 and 72 h post‐infiltration. Briefly, leaf sections were excised and fixed in ice‐cold 100% methanol for 1 min, rinsed in phosphate‐buffered saline (PBS) for 1 min and then stained for 4 min with Hoechst 33258 reagent (1 µg/mL). The leaf pieces were washed for 2 min in PBS and mounted in glycerol 50% (v/v) for observation under a Nikon Eclipse TE‐2000‐E2 (Nikon Instruments Inc., Melville, NY, USA) confocal laser scanning microscope. Images are displayed as maximum projections of picture stacks. GFP was excited with a 40‐mW argon laser at 488 nm and the emission filter wavelengths were 497–526 nm. Hoechst 33258 was excited with a 17‐mW Blue diode at 408 nm, and the emission filter wavelengths were 405–565 nm.
Plant pigment determination
The chlorophyll extraction method was performed as described by Lichtenthaler (1987). Briefly, citrus leaves were infiltrated with wild‐type and mutant Xac strains and, after 4 days, 1‐cm2 discs were excised and placed into plastic tubes with 1 mL of ethanol 100% (v/v), sealed and incubated in the dark at 60 °C for 48 h. Absorbances of the clear extract at 649, 665 and 750 nm were recorded, and concentrations of chlorophylls a, b, a + b and carotenoids were computed for 1 cm2 of infected area.
Chlorophyll fluorescence parameter determination
Citrus leaves were infiltrated with Xac wild‐type and mutant strains and kept in a glasshouse for 3, 4 and 6 days post‐inoculation. The chlorophyll fluorescence measurements were performed using a portable pulse amplitude modulation fluorometer (Qubit Systems Inc., Kingston, ON, Canada), as described by Baker and Rosenqvist (2004). Minimal fluorescence F 0 was measured in 120‐min dark‐adapted leaves using weak modulated light of <0.15 µmol/m2/s. Maximal fluorescence F m was measured after a 0.8‐s saturating white light pulse (5000 µmol/m2/s) on the same leaves. The maximal variable fluorescence (F v = F m – F 0) and photochemical efficiency of PSII (F v/F m) for dark‐adapted leaves were calculated. In light‐adapted leaves, the steady‐state fluorescence yield (F s), maximal fluorescence (F′m) after a 0.8‐s saturating white light pulse and minimal fluorescence (F′0) were measured when actinic light was turned off.
ACKNOWLEDGEMENTS
We thank Raúl Andrés Cernadas and Celso Eduardo Benedetti (both at the Laboratório Nacional de Luz Síncrotron, Campinas, Brazil) and Cássia Docena (Instituto de Química, Universidade de Sâo Paulo, Sâo Paulo, Brazil) for donating the Citrus sinensis cDNA library cloned in the pOAD vector, Microquin for culture media and Sonia Scarpeci for assistance with microscopy. This work was supported by grants from the Argentine Federal Government (ANPCyT, PICT2010‐1507 to NG and PICT2010‐0300 to JO), the Fundación Josefina Prats to GD and CGG, and Fundação de Amparo à Pesquisa do Estado de São Paulo (#2005/59243‐3) to CSF. NG, EGO and JO are staff members and CGG and BSG are Fellows of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
REFERENCES
- Alegria, M.C. , Docena, C. , Khater, L. , Ramos, C.H. , da Silva, A.C. and Farah, C.S. (2004) New protein–protein interactions identified for the regulatory and structural components and substrates of the type III secretion system of the phytopathogen Xanthomonas axonopodis Pathovar citri . J. Bacteriol. 186, 6186–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N.R. and Rosenqvist, E. (2004) Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J. Exp. Bot. 55, 1607–1621. [DOI] [PubMed] [Google Scholar]
- Brunings, A.M. and Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Buttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Castaneda, A. , Reddy, J.D. , El‐Yacoubi, B. and Gabriel, D.W. (2005) Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant–Microbe Interact. 18, 1306–1317. [DOI] [PubMed] [Google Scholar]
- Chang, J.H. , Goel, A.K. , Grant, S.R. and Dangl, J.L. (2004) Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr. Opin. Microbiol. 7, 11–18. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cho, M.H. , Corea, O.R. , Yang, H. , Bedgar, D.L. , Laskar, D.D. , Anterola, A.M. , Moog‐Anterola, F.A. , Hood, R.L. , Kohalmi, S.E. , Bernards, M.A. , Kang, C. , Davin, L.B. and Lewis, N.G. (2007) Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 282, 30 827–30 835. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and McDowell, J.M. (2006) Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA, 103, 8575–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues, M.N. , De Souza, T.A. , Cernadas, R.A. , de Oliveira, M.L. , Docena, C. , Farah, C.S. and Benedetti, C.E. (2010) The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11, 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y.P. , Castañeda, A. , Zhao, G. , Erdos, G. and Gabriel, D.W. (1999) Expression of a single, host‐specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol. Plant–Microbe Interact. 12, 556–560. [Google Scholar]
- Dunger, G. , Arabolaza, L.N. , Gottig, N. , Orellano, E.G. and Ottado, J. (2005) Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and in non‐host plant responses. Plant Pathol. 54, 781–788. [Google Scholar]
- Dunger, G. , Relling, V.M. , Tondo, M.L. , Barreras, M. , Ielpi, L. , Orellano, E.G. and Ottado, J. (2007) Xanthan is not essential for pathogenicity in citrus canker but contributes to Xanthomonas epiphytic survival. Arch. Microbiol. 188, 127–135. [DOI] [PubMed] [Google Scholar]
- Figueiredo, J.F. , Romer, P. , Lahaye, T. , Graham, J.H. , White, F.F. and Jones, J.B. (2011) Agrobacterium‐mediated transient expression in citrus leaves: a rapid tool for gene expression and functional gene assay. Plant Cell Rep. 30, 1339–1345. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. , Guo, M. , Jeong, B.R. , Tian, F. , Elthon, T.E. , Cerny, R.L. , Staiger, D. and Alfano, J.R. (2007) A type III effector ADP‐ribosylates RNA‐binding proteins and quells plant immunity. Nature, 447, 284–288. [DOI] [PubMed] [Google Scholar]
- Fujikawa, T. , Ishihara, H. , Leach, J.E. and Tsuyumu, S. (2006) Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol. Plant–Microbe Interact. 19, 342–349. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. and Woods, R.A. (2002) Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. , Woods, R.A. , Manivasakam, P. and Schiestl, R.H. (1998) Growth and transformation of Saccharomyces cerevisiae In: Cells: A Laboratory Manual, Vol. I. Culture and Biochemical Analysis of Cells (Spector D., Goldman R. and Leinwand L., eds), pp. 87–96. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Daurelio, L.D. , Valentine, A. , Gehring, C. , Orellano, E.G. and Ottado, J. (2008) Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide‐like protein to modify host homeostasis. Proc. Natl. Acad. Sci. USA, 105, 18 631–18 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Garofalo, C.G. , Orellano, E.G. and Ottado, J. (2009) A filamentous hemagglutinin‐like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE, 4, e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. , Fisher, E.J. , Chang, J.H. , Mole, B.M. and Dangl, J.L. (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. [DOI] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. , Boureau, T. and Poussier, S. (2009) A ‘repertoire for repertoire’ hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS ONE, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Xie, K. , Yao, J. , Qi, Z. and Xiong, L. (2009) A homolog of human ski‐interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. USA, 106, 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P. , Chen, L.J. , Li, H. , Peto, C.A. , Fankhauser, C. and Chory, J. (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science, 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.L. , Xu, R.Q. , Huang, J.D. , Wei, H.Y. , Jiang, G.F. , Cen, W.J. , Liu, J. , Ge, Y.Y. , Li, G.H. , Su, L.L. , Hang, X.H. , Tang, D.J. , Lu, G.T. , Feng, J.X. , He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the pip box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaminaka, H. , Nake, C. , Epple, P. , Dittgen, J. , Schutze, K. , Chaban, C. , Holt, B.F. III , Merkle, T. , Schafer, E. , Harter, K. and Dangl, J. (2006) bZIP10‐LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25, 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B. and Deisenhofer, J. (1995) Proteins with leucine‐rich repeats. Curr. Opin. Struct. Biol. 5, 409–416. [DOI] [PubMed] [Google Scholar]
- Kobe, B. and Kajava, A.V. (2001) The leucine‐rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- Koebnik, R. , Kruger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Krause, K. and Krupinska, K. (2009) Nuclear regulators with a second home in organelles. Trends Plant Sci. 14, 194–199. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velasquez, A.C. , Wei, C.F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. [Google Scholar]
- Lin, S.S. , Manchester, J.K. and Gordon, J.I. (2003) Sip2, an N‐myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278, 13 390–13 397. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z.L. , Fisher, E.J. , Desveaux, D. , Chang, J.H. and Dangl, J.L. (2007) The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N‐terminal domain for function. Mol. Plant–Microbe Interact. 20, 346–357. [DOI] [PubMed] [Google Scholar]
- Pugh, D.J. , Eiso, A.B. , Faro, A. , Lutya, P.T. , Hoffmann, E. and Rees, D.J. (2006) DWNN, a novel ubiquitin‐like domain, implicates RBBP6 in mRNA processing and ubiquitin‐like pathways. BMC Struct. Biol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece, K.S. and Phillips, G.J. (1995) New plasmids carrying antibiotic‐resistance cassettes. Gene, 165, 141–142. [DOI] [PubMed] [Google Scholar]
- Rybak, M. , Minsavage, G.V. , Stall, R.E. and Jones, J.B. (2009) Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 10, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R. , Fischer, K. , Ketelsen, B. , Krupinska, K. and Krause, K. (2007) Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics, 277, 631–646. [DOI] [PubMed] [Google Scholar]
- Shiotani, H. , Fujikawa, T. , Ishihara, H. , Tsuyumu, S. and Ozaki, K. (2007) A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host‐specific suppression of virulence. J. Bacteriol. 189, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos, S.M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Thieme, F. , Szczesny, R. , Urban, A. , Kirchner, O. , Hause, G. and Bonas, U. (2007) New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N‐myristoylation motif. Mol. Plant–Microbe Interact. 20, 1250–1261. [DOI] [PubMed] [Google Scholar]
- Uetz, P. , Giot, L. , Cagney, G. , Mansfield, T.A. , Judson, R.S. , Knight, J.R. , Lockshon, D. , Narayan, V. , Srinivasan, M. , Pochart, P. , Qureshi‐Emili, A. , Li, Y. , Godwin, B. , Conover, D. , Kalbfleisch, T. , Vijayadamodar, G. , Yang, M. , Johnston, M. , Fields, S. and Rothberg, J.M. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , Van, A.G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]


