SUMMARY
In an environment that is rich in potentially pathogenic microorganisms, the survival of higher eukaryotic organisms depends on efficient pathogen sensing and rapidly mounted defence responses. Such protective mechanisms are found in all multicellular organisms, and are collectively referred to as ‘innate immunity’. Innate immunity is the first line of defence against invading microorganisms in vertebrates and the only line of defence in invertebrates and plants. Bacterial glycoconjugates, such as lipopolysaccharides (LPSs) from the outer membrane of Gram‐negative bacteria and peptidoglycan (PGN) from the cell walls of both Gram‐positive and Gram‐negative bacteria, have been found to act as elicitors of plant innate immunity. These conserved, indispensable, microbe‐specific molecules are also referred to as ‘microbe‐associated molecular patterns’ (MAMPs). MAMPs are recognized by the plant innate immune system through the action of pattern recognition receptors (PRRs). A greater insight into the mechanisms of MAMP recognition and the description of PRRs for different microbial glycoconjugates will have considerable impact on the improvement of plant health and disease resistance. Here, the current knowledge about LPS and PGN as MAMPs is reviewed.
INTRODUCTION
Plants detect several general elicitors from both host and nonhost pathogens. Microbe‐associated molecular patterns (MAMPs) are conserved and generally indispensable microbial structures. In plants, MAMPs are perceived by the innate immune system pathogen recognition receptors (PRRs). This MAMP recognition by their cognate PRRs is referred to as ‘microbe‐ or pathogen‐triggered immunity’ (MTI or PTI) (Jones and Dangl, 2006). Pathogens have evolved a number of methods to suppress the action of MAMPs in the elicitation of basal defences. One key strategy is the delivery into the plant cell of effector proteins which, in bacterial pathogens, is mediated by type III secretion systems. These effectors function to suppress basal defences and to alter host metabolism in order to promote disease (He et al., 2006; Jamir et al., 2004; Nomura et al., 2006), resulting in effector‐triggered susceptibility (ETS). However, recognition of a given effector through a set of resistance (R) gene products results in effector‐triggered immunity (ETI), i.e. disease resistance or the hypersensitive response (HR) (Jones and Dangl, 2006).
The induction of MTI in plants has been most extensively studied using the small peptides flg22 and elf18 derived from the bacterial MAMPs flagellin and the translation elongation factor EF‐Tu, respectively (Felix and Boller, 2003; Zipfel et al., 2006). The minimal structure(s) required for induction of immune responses in plants using bacterial glycopolymers, such as lipopolysaccharide (LPS) and peptidoglycan (PGN), is still not known. LPSs from the outer membrane of Gram‐negative bacteria are amphiphilic molecules that can form aggregates in aqueous suspensions and are heterogeneous in both size and composition. Both LPS and PGN are macromolecules that can interact with the host PRRs. Mammals are extremely sensitive to these MAMPs, even at low doses (Alexander and Rietschel, 2001), compared with plant cells. The recognition of MAMPs in mammals and insects is often mediated by leucine‐rich repeat (LRR) proteins, such as Toll in Drosophila and the Toll‐like receptors (TLRs) in mammals (Hashimoto et al., 1988; Lemaitre et al., 1996; Medzhitov et al., 1997). On LPS recognition in mammals, soluble LPS‐binding protein (LBP) binds LPS and catalyses the movement of LPS to CD14, a soluble glycosylphosphatidyl inositol (GPI)‐anchored glycoprotein (Wright et al., 1990), which is known to facilitate the transfer of LPS to the TLR4–MD‐2 receptor complex in the plasma membrane and triggers an intracellular signalling cascade. LPS can cause septic shock in mammals; nonmammalian vertebrates, such as fish, are immune to the toxic effect of LPS as they lack several of the co‐stimulatory molecules involved in TLR4‐mediated endotoxin recognition and signalling in mammals (Iliev et al., 2005). Recently, it was revealed that, in a preparation of crude Escherichia coli LPS, it was PGN contamination, and not LPS, that was the active component responsible for gene activation in cell cultures of rainbow trout macrophages (MacKenzie et al., 2010). In animals, several PGN recognition molecules are known, including CD14 (also known to bind the LPS–LBP complex) (Dziarski et al., 1998), NOD (nucleotide‐binding oligomerization domain)‐containing proteins (Franchi et al., 2006), TLR2, even though this is controversial (Dziarski and Gupta, 2005; Travassos et al., 2004), and the PGN recognition proteins (PGRPs) (reviewed by Guan and Mariuzza, 2007). Like the complex recognition of LPS and PGN in mammals, in contrast with the more straightforward recognition of, for example, flagellin by its PRR TLR5, a more complex perception system could also be expected for these macromolecules in plants. So far, no PRRs have been identified in plants for bacterial glycoconjugates. Examples of LRR‐receptor kinase PRRs that have been identified in plants are FLS2, EFR and XA21, which recognize bacterial flagellin, EF‐Tu and the sulphated protein AX21, respectively (Felix and Boller, 2003; Lee et al., 2009; Zipfel et al., 2006).
In this review, we focus on the glycosylated bacterial MAMPs LPS and PGN. We provide an overview of a range of responses induced by LPS, the substructures within LPS that are recognized by plants and variations within the LPS structure that can alter its activity as a MAMP. Furthermore, we include the current, still rather limited, knowledge about PGN's role as a MAMP in plants.
LIPOPOLYSACCHARIDES
LPS and plant innate immunity
LPSs are outer membrane glycolipids of Gram‐negative bacteria known to induce the innate immune response in mammals, insects and plants (reviewed by Alexander and Rietschel, 2001; Newman et al., 2007; Silipo et al., 2005). LPS is composed of a hydrophobic lipid part, referred to as lipid A, which is embedded in the outer part of the phospholipid bilayer. Lipid A is linked to the core oligosaccharide, usually by the sugar 3‐deoxy‐d‐manno‐2‐octulosonate (KDO). The core oligosaccharide consists of a short series of sugars and ends in the O‐antigen, which is composed of repeating oligosaccharide units (Raetz and Whitfield, 2002) (Fig. 1). The main surface component of the bacterial cell envelope LPS is thought to contribute to the restrictive Gram‐negative outer membrane permeability, allowing bacterial growth in unfavourable environments, such as those that may be encountered within or on plants. The exclusion of antimicrobial substances of plant origin probably contributes to the ability of pathogenic bacteria to parasitize plants. LPS‐defective mutants show increased in vitro sensitivity to antibiotics and antimicrobial peptides, and the numbers of viable bacteria often decline very rapidly on introduction into plants (Deng et al., 2010; Dow et al., 2000; Newman et al., 2007).
Figure 1.
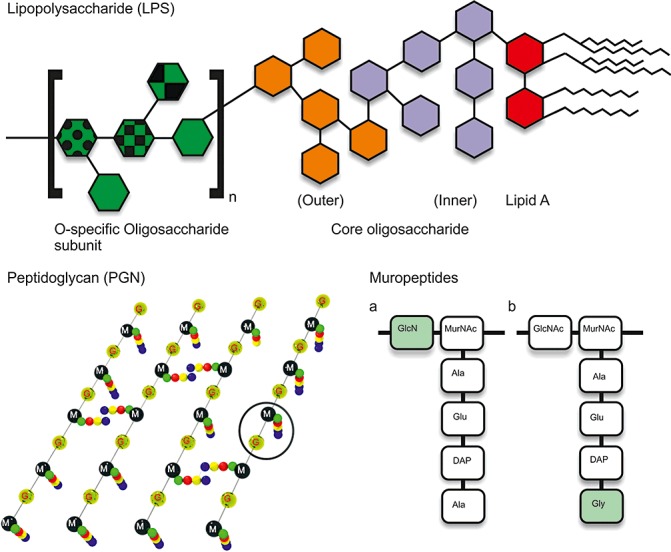
General structural architecture of lipopolysaccharide (LPS) and peptidoglycan (PGN). LPS is a tripartite molecule comprising a membrane‐anchored lipid A moiety, a core oligosaccharide and an O‐antigen polysaccharide made up of repeating units. 3‐deoxy‐d‐manno‐2‐octulosonate (KDO) residues link lipid A to the core oligosaccharide, which can also be decorated with other (often nonstoichiometric) substituents, such as phosphate and phosphoethanolamine. LPS is only found in Gram‐negative bacteria. PGN, found in both Gram‐positive and Gram‐negative bacteria, has a glycan backbone made up of a repeat polymer of two amino sugars, N‐acetylglucosamine (G) and N‐acetylmuramic acid (M). Attached to the N‐acetylmuramic acid is a peptide side chain consisting of a peptide moiety displaying considerable diversity. In general, the third position amino acid in Gram‐positive bacteria is l‐lysine (Lys), whereas, in Gram‐negative bacteria, it is meso‐2,6‐diaminopimelic acid (DAP). Furthermore, Gram‐positive bacteria have peptide stems usually cross‐linked through an interpeptide bridge (generally glycine), whereas Gram‐negative bacteria peptide stems are usually directly cross‐linked. Muropeptides derived from PGN of Gram‐negative pathogens: (a) Xanthomonas campestris pv. campestris (Xcc); (b) Agrobacterium tumefaciens (At). Differences between the structures are indicated in the green boxes. In At, a glycine‐containing muropeptide was observed, whereas, in Xcc, alanine is present. In Xcc, an N‐deacetylated GlcN is present rather than the GlcNAc found in At.
In contrast with this role in promoting plant disease, i.e. protection and barrier function against host compounds, there have been various reports detailing the effects of LPS on the induction of plant innate immunity, consistent with its designation as a MAMP and the definition of a MAMP as a conserved structure that is essential for microbial survival, not only among pathogens, but also in nonpathogenic and saprophytic microorganisms.
LPS as an inducer of plant defences
LPS has been reported to have a number of effects on the induction of immune responses in plants, including the oxidative burst, NO production, calcium influx, the induction of pathogenesis‐related (PR) gene expression and cell wall alterations that include the deposition of callose and phenolics. These observations have been made with LPS preparations from a range of bacteria used in cell suspension cultures or leaves from various plants, which may explain some variations in outcome. For example, LPS can induce the production of active oxygen species (Albus et al., 2001; Braun et al., 2005; Desaki et al., 2006; Gerber et al., 2004; Meyer et al., 2001), but this is not always observed (Desender et al., 2006; Dow et al., 2000). LPSs from a range of bacteria induced NO synthesis in suspension cultures and leaves of the crucifer Arabidopsis thaliana (Zeidler et al., 2004). This common effect of LPSs from diverse bacteria suggests the involvement of a shared molecular determinant, the lipid A moiety, and indeed isolated lipid A is also active (Zeidler et al., 2004). In these experiments, LPS also activated the accumulation of transcripts of defence‐related genes, including PR1, an effect mediated by NO (Zeidler et al., 2004). Lipo‐oligosaccharides (LOSs) from the crucifer pathogen Xanthomonas campestris pv. campestris (Xcc) were active in inducing the expression of the defence‐related genes PR1 and PR2 in A. thaliana (Silipo et al., 2005). In some cases, effects on plant gene induction that are specific to a particular LPS have been observed. This may reflect the ability of particular plants to recognize structural features within LPS that are not widely conserved. For example, in turnip (Brassica campestris), LOS of Xcc induced the expression of a gene encoding a defence‐related β‐(1–3)‐glucanase when applied to leaves at 1 µg/mL (Newman et al., 1995). In contrast, LOSs from E. coli and Salmonella enterica sv. minnesota were ineffective at concentrations up to 50 µg/mL (Newman et al., 1995). Nevertheless, LPSs from these enteric bacteria can elicit defence‐related gene induction in different plants at 50 µg/mL.
The concentrations of LPS required to elicit most of the effects described above are in the 5–100 µg/mL range, which suggests that plants do not have the exquisite sensitivity to LPSs shown by mammalian cells, which can respond at concentrations in the pg/mL to ng/mL range. These considerations have led to suggestions that plants possess only low‐affinity systems to detect LPS (Zeidler et al., 2004), although plants can detect other bacterial MAMPs, such as peptides derived from flagellin and Ef‐Tu elongation factor, at subnanomolar levels. One complicating factor is the aggregation of LPS molecules within the purified preparations, which may affect the ability of LPS to cross the matrix of the plant cell wall to reach presumed membrane‐associated receptors (Aslam et al., 2009).
Several attempts have been made to identify plant components involved in LPS recognition and perception. Interestingly, Livaja et al. (2008) found that, in Arabidopsis cells, Burkholderia cepacia LPS induced an LRR receptor‐like kinase At5g45840 by nearly 17‐fold after 30 min. Furthermore, in a proteomic analysis of the changes following the perception of LPS from an endophytic strain of B. cepacia in Nicotiana tabacum BY‐2 cells, 88 LPS‐induced/regulated proteins and phosphoproteins were identified, many of which were found to be involved in metabolism and energy‐related processes. Moreover, proteins were found that are known to be involved in protein synthesis, protein folding, vesicle trafficking and secretion (2006, 2008). Livaja et al. (2008) performed transcription profiling of A. thaliana cells treated with 100 µg/mL LPS from B. cepacia or 50 µg/mL harpin from Pseudomonas syringae. The transcriptional changes in the treated and nontreated cells were monitored at 0.5, 1, 2, 4, 8 and 24 h after elicitor treatment. Focusing on changes induced by B. cepacia LPS, the authors surprisingly did not find any genes involved in callose synthesis. Furthermore, genes involved in reactive oxygen species (ROS) production were found to be upregulated at a very low level by B. cepacia LPS, except after 8 h, when a superoxide dismutase (SOD) and a ferritin 1 precursor gene were strongly induced. In addition, Livaja et al. (2008) found that B. cepacia LPS only induced the PR genes PR3 and PR4, whereas studies in B. cepacia LPS‐treated Arabidopsis leaves revealed the induction of several PR genes (Zeidler et al., 2004). Other LPS preparations, from Pseudomonas aeruginosa and E. coli, respectively, induce PR1 and PR5 in Arabidopsis leaves (Mishina and Zeier, 2007). The conflict in results reflects both the different plant systems (A. thaliana cell cultures vs. the whole plant) and the origin of the LPS. All the above very specific effects show the ability of particular plants to recognize structural features within LPS that are not necessarily widely conserved.
Prevention of HR and priming of plant defence by LPS
Perhaps the first effect of LPS on plants to be described was the ability to prevent the programmed cell death response (HR) in leaves (reviewed by Erbs and Newman, 2003). These findings present a conundrum. If basal resistance responses and HR both contribute to plant defence, why does LPS activate the former but block the latter? One possible answer is that the effects of LPS in the prevention of HR and the triggering of basal defences may allow the plant to express resistance without catastrophic tissue collapse. This contention is supported by observations that the prevention of HR does not apparently lead to an increased susceptibility of the tissue. The onset of HR is generally associated with a decline in the number of bacteria that can be recovered from the leaf. In contrast, in leaf tissue pretreated with LPS, Newman et al. (2000) found that bacterial counts declined 10‐fold over the first 24 h, but then were maintained at the same level throughout a 72‐h experiment.
In a number of cases, LPS does not act in the direct induction of plant defence responses, but increases the speed and/or degree of induction on subsequent pathogen inoculation (Newman et al., 2002), an effect named priming, also seen with a number of other biological agents and synthetic compounds (Conrath et al., 2006). The molecular basis of these effects is unknown. Intriguingly, the application of LPSs and other MAMPs to plants can lead to an increased expression of surveillance systems for bacterial type III‐secreted effectors and viral proteins (2004, 2006). This activation of additional surveillance mechanisms may be related to the priming phenomenon.
LPS recognition by plants
The recognition of LPS/LOS in mammals is rather complex; how complex this recognition is in plants is still unknown, and the mechanism of this recognition and the consequent transduction steps in plants remains obscure.
Gross et al. (2005) showed that, in tobacco cells, Xcc LPS was internalized 2 h after its introduction to a cell suspension, where it co‐localized with Ara6, a plant homologue of Rab5, which is known to regulate early endosomal functions in mammals. It was speculated that this endocytosis in tobacco cells was, in correlation with the mammalian system, part of a downregulation of defence responses. In a recent study by Zeidler et al. (2010), the localization and mobilization of fluorescein‐labelled S. minnesota LPS was studied in Arabidopsis. Leaves of A. thaliana were pressure infiltrated with 100 µg/mL of fluorescein‐labelled S. minnesota LPS and the mobility of LPS was studied over time by fluorescence microscopy. After 1 h, a fluorescent signal was observed in the intercellular space of the infiltrated leaf. The labelled LPSs were visible in the midrib of the leaves after 4 h, whereas this fluorescence had spread to the smaller leaf veins near the midrib after 6 h. After 24 h, it was detectable in the lateral veins. Moreover, cross‐sections of the midrib 3 h after supplementation with fluorescein‐labelled LPS revealed a fluorescent signal in the xylem. Using capillary zone electrophoresis, a distribution of fluorescein‐labelled S. minnesota LPS was found in treated as well as in systemic leaves (Zeidler et al., 2010). In contrast with the results reported by Gross et al. (2005), no intracellular accumulation of the labelled LPS was observed in Arabidopsis. This conflict could reflect the different LPS and plant systems used.
Several laboratories have investigated the contribution of the different moieties within LPS to the MAMP elicitor activity. Silipo et al. (2005) determined the complete structure of purified LOS from Xcc, the lipid A and core oligosaccharides derived from it by mild acid hydrolysis and, in parallel, examined the activity of these (structurally defined) components in defence gene induction in Arabidopsis. Xcc LOS was found to be a unique molecule with a high negative charge density and a phosphoramide group, which has not been found previously as a component of LPS (Silipo et al., 2005). Xcc LOS induced the defence‐related PR1 and PR2 genes in Arabidopsis leaves in two temporal phases: the core oligosaccharide induced only the early phase and the lipid A moiety only the later phase. These findings suggest that, although both Xcc lipid A and the Xcc core oligosaccharide are active in defence gene induction, they may be recognized by different plant receptors (Silipo et al., 2005). This elicitor activity of Xcc lipid A correlates with earlier studies by Zeidler et al. (2004), who showed that lipid A preparations from various bacteria induced a rapid burst of NO production that was associated with the induction of defence‐related genes in Arabidopsis. In a recent study by Madala et al. (2011), in which the structure of B. cepacia strain ASP B 2D lipid A was determined, the role of lipid A as a MAMP in Arabidopsis was confirmed, and it was found to induce transcriptional changes associated with plant defence responses.
Interestingly, the core oligosaccharide from E. coli and Ralstonia solanacearum does not prevent HR or induce defence‐related genes (Newman et al., 1997), indicating that the effect of the Xcc core oligosaccharide could be a result of the unique phosphoramide group in this particular LPS molecule (Silipo et al., 2005). In contrast, in tobacco cells, Xcc lipid A could not induce the oxidative burst, but rather it was the inner core part of the LPS molecule that was responsible (Braun et al., 2005). This disparity in outcomes might again be a reflection of the use of different plants, the difference in the age of the plants used (plant cell cultures vs. seedlings vs. fully developed plants) and the different defence responses measured after treatment with LPS and its derivatives. Furthermore, the purity of the MAMPs used is crucial; there are many documented cases in which biological activity resulted from co‐purification, in particular PGN associating with LPS, or LPS associating with PGN (Girardin et al., 2003; Leulier et al., 2003; MacKenzie et al., 2010; McDonald et al., 2005). The contamination is probably greatest in commercial products of LPS (and PGN), but will, at times, probably also be found in purified LPS/PGN from laboratories, when the product is not thoroughly checked for contaminants.
Evidence for a role of the O‐antigen in eliciting defence responses stems from the different ability of LPS derived from wild‐type Pseudomonas fluorescens and a mutant lacking the O‐antigen to produce induced systemic resistance (ISR) (Leeman et al., 1995; Van Loon et al., 1998). More recently, the role of the O‐antigen was examined directly by studies of the biological activity of synthetic O‐antigen polysaccharides. Structural studies of LPS from many phytopathogenic bacteria have revealed that the O‐antigen often comprises a rhamnan backbone with the trisaccharide repeating unit [α‐l‐Rha‐(1–3)‐α‐l‐Rha‐(1–2)‐α‐l‐Rha‐(1–3)] and single monosaccharide branches from the backbone that differentiate the structure of the O‐chain from each serotype (Bedini et al., 2002). The trisaccharide was synthesized and the trimer oligomerized to generate a set of oligosaccharides of increasing chain length. The tri‐, hexa‐ and nonasaccharide synthetic O‐antigens were found to suppress the HR and induce PR1 and PR2 transcript accumulation in Arabidopsis. Interestingly, the efficiency of HR suppression and PR gene induction improved with increasing chain length (Bedini et al., 2005). Moreover, this increasing chain length was associated with the formation of a coiled structure, suggesting a role for this structure as a MAMP. By extension, these findings suggest a role for the O‐antigen from many phytopathogenic bacteria in triggering plant innate immunity (Bedini et al., 2005).
In mammals, it is well established that electrostatic interactions involving phosphate groups are required for the biological activity of LPS (1998, 2000). Ionic interactions also seem to participate in the plant recognition of Xcc LOS, as LOS and derivatives were unable to induce localized induced responses in Arabidopsis when all of the phosphate groups, together with the phosphodiester‐linked galacturonyl residues and the phosphoramide group, were removed from these molecules (Silipo et al., 2005).
Variations in LPS structure
Alterations in lipid A or other structures within LPS are known to occur during symbiotic interactions with plants (Kannenberg and Carlson, 2001) and in response to compounds in plant root exudates (Fischer et al., 2003), and may occur during plant pathogenesis. These alterations may serve both to increase the resistance of the bacteria against host defences and to attenuate the activity of lipid A or LPS in triggering these defences. Regulated palmitoylation of lipid A has been shown to have such a dual role in the pathogenesis of bacterial pathogens of mammals (Bishop et al., 2005; Bishop, 2005). Palmitoylation occurs by transfer of a palmitate chain from a phospholipid to lipid A, catalysed by an outer membrane enzyme PagP. Homologues of pagP occur in the plant pathogens Erwinia chrysanthemi and E. carotovora, although it is not yet known whether they have a role in virulence (Bishop et al., 2005).
The characterization of the structure and function of LOS from a nonpathogenic Xcc mutant strain 8530, which carries a Tn5 insertion in a gene of unknown function (Dow et al., 1995), revealed that this mutant had a truncated core region. The fact that Xcc strain 8530 was defective in core completion led to significant modifications in the acylation and phosphorylation patterns of its lipid A, and these changes had an influence on its ability to trigger innate immune responses in Arabidopsis (Silipo et al., 2008). The core sugars provide protection against antimicrobial compounds and attenuate the endotoxic properties of lipid A, similar to the lipid A modifications seen in mammalian pathogens (Raetz et al., 2007). These findings indicate that Xcc has the capacity to modify the structure of lipid A and thus reduce its activity as a MAMP in plants (Silipo et al., 2008). The acyl chains of lipid A can vary, as can their number and length, depending on the growth conditions and bacterial species. For instance, changes in the structure and activity of Yersinia pestis (an obligate parasite of mammals and insects) lipid A have been studied, and have revealed that Y. pestis switches from the predominantly hexa‐acylated lipid A at low temperatures, when its hosts are insects, to the tetra‐acylated lipid A at higher temperatures, when its hosts are mammals (Kawahara et al., 2002). Furthermore, recent studies in mammalian cells have shown that LPS from Shigella flexneri elicits a weaker TLR4‐mediated response than E. coli LPS as a result of differences in the acylation status of their lipid A moieties (Rallabhandi et al., 2008).
Lipid A from Halomonas magadiensis, an extremophilic and alkaliphilic Gram‐negative bacterium, isolated from a soda lake in an East African Rift Valley, has been found to act as an LPS antagonist in human cells (Silipo et al., 2004). Halomonas magadiensis lipid A, characterized by an unusual and very low degree of acylation, was verified to inhibit E. coli lipid A‐induced immune responses in human cells (Ialenti et al., 2006). Escherichia coli lipid A, which is an effective agonistic structure of immune responses in mammalian cells, is composed of a bis‐phosphorylated hexa‐acylated disaccharide backbone with an asymmetric distribution of the acyl residues. Studies have revealed that structural differences in the lipid A skeleton, e.g. acylation, can affect its agonist/antagonist activity (Munford and Varley, 2006). In agreement with the ability to block enteric LPS‐induced human monocyte activation, our laboratory found that H. magadiensis lipid A was able to antagonize the action of E. coli lipid A when inducing PR1 gene expression in A. thaliana. Although the mode of perception of LPS in plants is far less understood than in mammals and insects, these results indicate that A. thaliana is sensitive to the same structures of lipid A that determine biological activity in humans (Erbs et al., 2008a).
Thus far, LPS preparations used for the analysis of plant responses and for structural studies have been derived from bacteria grown in culture. We know almost nothing about the alterations in LPS that occur when bacteria are within plants, although this may be highly relevant to signalling. Changes could occur in both the size distribution of LPS (alteration in the ratio of LOS to LPS) and/or in the decoration of LPS with saccharide, fatty acid, phosphate or other constituents. Increases in the sensitivity of mass spectrometric methodologies may allow the development of micro‐methods to analyse such changes in bacteria isolated from plants. Transcriptome or proteome profiling of bacteria isolated from plants may also provide clues to possible LPS modifications.
PEPTIDOGLYCAN
PGN and plant innate immunity
PGN, which consists of glycan strands that are cross‐linked by peptide bridges, provides rigidity and structure to the cells of both Gram‐positive and Gram‐negative bacteria (Fig. 1). PGN is found as a thick layer in Gram‐positive bacteria, whereas only a thin layer is present in Gram‐negative bacteria. The carbohydrate backbone of PGN is conserved in all bacteria and made up of alternating N‐acetylmuramic acid (MurNAc) and N‐acetylglucosamine (GlcNAc) linked by β‐1,4‐glycosidic bonds. The carboxyl group of MurNAc is the point of linkage to the peptide. In contrast with the glycan backbone, considerable variations are found in the peptide moieties. Several types of PGN, classified by the nature of the third residue of the stem peptide, are commonly found. Typically, this is meso‐2,6‐diaminopimelic acid (DAP) PGN in Gram‐negative bacteria and in some Gram‐positive bacilli (genera Bacillus and Clostridium), whereas, in most other Gram‐positive bacteria, it is l‐lysine (Lys) PGN. PGN, a molecule never found in eukaryotes, is an essential and unique cell wall component of all bacteria, making it an excellent target for the eukaryotic innate immune system (reviewed by Dziarski and Gupta, 2005; McDonald et al., 2005).
The first evidence that PGN interacts with plant cells was published in 2003 by Felix and Boller. They showed that PGN from the Gram‐positive human pathogen Staphylococcus aureus was active as an elicitor in inducing extracellular alkalization of cultured tobacco cells, whereas no response was observed in cultured tomato cells, indicating a different perception system for PGN within the Solanaceae. In contrast in a recent study in tomato, Nguyen et al. (2010) showed that pre‐inoculation with S. aureus PGN reduced the growth of subsequent bacterial infection in PGN‐treated tissue. This priming of defence with a MAMP is similar to that described for LPS (Newman et al., 2002). Another plant study examining Arabidopsis challenged with S. aureus PGN showed that the PGN sugar backbone was responsible for triggering immune responses (Gust et al., 2007), and not the breakdown product of PGN, the muramyl dipeptide or the muropeptide dimer, which is known to be the minimal chemical structure required for triggering of the innate immune system in vertebrates and insects (reviewed by Traub et al., 2006).
In our laboratory, we isolated PGN from two Gram‐negative bacterial plant pathogens, Xcc and Agrobacterium tumefaciens (At) strain DSM 30204, hydrolysed the PGN to their related muropeptides and elucidated the structure of the main component (Erbs et al., 2008b) (Fig. 1). We chose these two bacteria because of their different infection strategies: Xcc causes necrosis and tissue breakdown in its host, whereas At is a pathogen that depends on the maintenance of the viability of host cells for the transfer of its T‐DNA. Using highly purified PGN and muropeptides from Xcc and At, we found that PGN and its constituents function as MAMPs in Arabidopsis and induce immune responses, such as the generation of ROS, extracellular pH increase, PR1 gene expression and callose deposition (Erbs et al., 2008b). Muropeptides were significantly more effective than the intact PGN molecule. PGN and fragments from Xcc were more potent defence elicitors than those from At, possibly reflecting the biotrophic mode of parasitism of the latter. Differences in the structures of Xcc and At muropeptides were observed; an acetyl group was missing in the PGN of Xcc and, very unusually (Schleifer and Kandler, 1972), a glycine residue was replaced by an alanine in At PGN (Erbs et al., 2008b) (Fig. 1). Listeria monocytogenes, a human intracellular pathogen, has been found to N‐deacetylate its PGN and, in this way, escape recognition and killing by host cells (Boneca et al., 2007). The N‐deacetylation of Xcc PGN had the opposite effect and elicited immune responses in Arabidopsis (Erbs et al., 2008b). The variations in At and Xcc PGN structures might explain their different eliciting activities; the chemical synthesis of these and related compounds could be a way of addressing this question. The alterations of PGN in At to reduce its effectiveness as a MAMP are reminiscent of the alterations in lipid A known to occur during symbiotic interactions with plants, alterations which are thought to have a similar effect on the ability of lipid A to induce defence responses (Kannenberg and Carlson, 2001). Furthermore, although flagellin from many bacteria is a strong MAMP, flagellin from Agrobacterium is not recognized in Arabidopsis (Zipfel et al., 2006).
The greater activity in Arabidopsis of the muropeptides than of native PGN from Xcc contrasts with the perception by Arabidopsis of S. aureus PGN, where the opposite effect was seen. These observations could be indicative of different perception systems for PGN from Gram‐positive and Gram‐negative bacteria.
PGN recognition by plants
At least six PGN perception systems in humans (TLR2, NOD1, NOD2) and Drosophila immune cells (PGRP‐SA, PGRP‐LC, PGRP‐SC1B) are known that recognize different fragments of bacterial PGNs (Akira et al., 2006; Ferrandon et al., 2007). The PGRPs that are ubiquitous in most animals, from insects to mammals, do not exist in plants. Insect and mammalian PGRPs probably bind PGN in a similar fashion (the mammalian PGRPs are far less understood than the insect PGRPs), but their roles in innate immunity differ. The mammalian PGRPs have been found to be directly bactericidal, whereas the insect PGRPs activate the signal transduction pathways (Dziarski and Gupta, 2006). Moreover, the ability of Drosophila to distinguish Gram‐positive from Gram‐negative bacteria is based on the recognition of specific forms of PGN, and not, as earlier thought, on the recognition of LPS (Leulier et al., 2003). In addition, based on findings in rainbow trout, where PGN was found to be the active component, in crude LPS preparations, responsible for gene activation, it was hypothesized that the recognition of Gram‐negative bacteria in rainbow trout is similar to that described for Drosophila (MacKenzie et al., 2010).
No evidence is available for how PGN is perceived in plants. There have been suggestions that CERK1, a plasma membrane protein with three LysM motifs in the extracellular domain, might be involved in the binding of PGN in Arabidopsis. CERK1 is required for all defence responses induced by chitin, an N‐acetyl‐d‐glucosamine polymer constituting the main component of fungal cell walls. LysM motifs have been studied extensively and are regarded as carbohydrate‐binding moieties, and some LysM domains are known to bind PGN (Buist et al., 2008). Recently, affinity enrichment studies with PGN from both Gram‐positive and Gram‐negative bacteria have revealed that Arabidopsis CERK1, the chitin elicitor receptor kinase 1 that is essential for perception of the fungal cell wall component chitin (Miya et al., 2007) as well as for the restriction of bacterial growth on plants (Gimenez‐Ibanez et al., 2009a), is unlikely to play a role in PGN recognition. CERK1 was unable to bind PGN from S. aureus or E. coli. Moreover, when Arabidopsis leaves were vacuum infiltrated with chitin, Western blotting experiments with CERK1 revealed that chitin treatment induced a transient bandshift of the CERK1 protein (Petutschnig et al., 2010). In order to test the specificity of the bandshift response, Arabidopsis cell cultures were treated with a range of commercially available MAMPs, among them PGN from different source organisms (Invitrogen) and LPS (Sigma), and none of these MAMPs was able to induce a CERK1 bandshift (Petutschnig et al., 2010).
In another study, PGN from the virulent bacterial pathogen P. syringae pv. tomato DC3000 (Pto DC3000) was tested to determine whether it constituted a ligand for any of the three LysM motifs of CERK1. Pto DC3000 PGN was found to induce the generation of ROS in Arabidopsis cerk1 mutant plants, indicating that PGN perception is independent of CERK1 (Gimenez‐Ibanez et al., 2009b). In contrast, Segonzac and Zipfel (2011) reported the as yet unpublished finding from Professor Nürnberger's laboratory that a LysM‐RLP (receptor‐like protein) is required for PGN perception and subsequent resistance to Pto DC3000 in Arabidopsis.
Millet et al. (2010) generated promoter:β‐glucuronidase (GUS) transgenic lines of the four genes CYP71A12, MYB51, WRKY11 and At5g25260, known to be upregulated in Arabidopsis seedlings treated with flg22 (Denoux et al., 2008). PGN (Sigma‐Aldrich) from the Gram‐positive root‐colonizing bacterium Bacillus subtilis was tested for GUS reporter gene activation and callose deposition in these Arabidopsis seedling roots. Bacillus subtilis PGN was able to induce callose deposition in the elongation zone of Arabidopsis seedling roots. In addition, CYP71A12 and MYB51 were strongly activated in the elongation zone of seedling roots by PGN treatment, a response similar to that observed for flg22 treatment, whereas WRKY11 and At5g25260 were upregulated to a lesser extent by PGN. In a bak1‐3 mutant, the GUS response to PGN was abolished, indicating that BAK1 (BRI1‐associated receptor kinase 1) is involved in PGN‐mediated signalling (Millet et al., 2010). BAK1, an LRR‐receptor kinase that has been shown previously to control plant growth by hormone‐dependent heterodimerization with the plant brassinosteroid (BR) hormone receptor BRI1 (an LRR‐receptor kinase itself) (Wang et al., 2001), is implicated in flagellin and Ef‐Tu recognition (Chinchilla et al., 2007). In addition to its role as a positive regulator of MTI and plant growth, BAK1 appears to fulfil additional functions. Bak1 mutants have also been shown recently to have altered disease‐resistance phenotypes to biotrophic and necrotrophic pathogens, that are probably the consequence of infection‐induced deregulated cell death control (Kemmerling et al., 2007). Thus, in addition to its role as a positive regulator of MTI, BAK1 may further act as a negative regulator of plant cell death.
Furthermore, the hypothesis that the P. syringae phytotoxin coronatine (COR) is the effector that suppresses MAMP‐activated responses in roots was tested. COR was found to suppress MAMP‐elicited GUS reporter gene activation when Arabidopsis seedlings were treated with both purified COR and one of the following MAMPs: flg22, PGN (Sigma‐Aldrich) or chitin (Millet et al., 2010).
CONCLUDING REMARKS
The effect of MAMPs, such as LPS and PGN, on the induction of basal plant defences raises the issue of how bacteria can ever cause disease in plants. Successful pathogens have evolved mechanisms to subvert or suppress MTI. Many type III‐secreted effectors act to block the induction of basal defences, thus promoting disease (He et al., 2006; Nomura et al., 2006). Other bacterial products, such as extracellular cyclic glucans and extracellular polysaccharides, have also been shown to suppress defences (Aslam et al., 2008; Yun et al., 2006). Extracellular polysaccharides may exert their suppressive effect through the sequestration of Ca2+ ions, thus preventing influx from the extracellular apoplastic pool (Aslam et al., 2008; Yun et al., 2006). Ca2+ influx occurs as an early local response to pathogen attack and is thought to act as a signal and to activate callose synthetase. The mechanistic basis for the suppression of defences by cyclic glucan is unknown.
Although plant receptors for the bacterial proteinaceous MAMPs flagellin and EF‐Tu have been identified, those involved in the perception of LPS and PGN are still obscure. The cloning and characterization of these genes remain major goals. The development of a range of molecular genetic tools for model plants, such as A. thaliana, affords more opportunities for success. The generally indispensable nature of many MAMPs precludes a similar genetic analysis using bacterial null mutants. In this case, one approach is through the identification of genes encoding plant receptors for MAMPs, followed by the assessment of effects of mutagenesis or silencing of these genes on pathogen virulence. This has been performed, for example, to assess the role of flagellin recognition by FLS2 in disease resistance in A. thaliana. The knowledge obtained from such experiments may also have a bearing on other aspects of plant–microbe interactions, such as the induction of systemic resistance by beneficial (nonpathogenic) bacteria, such as P. fluorescens (in which LPS is implicated), and signalling between bacterial symbionts, such as rhizobial species and their plant hosts. The further understanding of the molecular basis of plant perception of glycoconjugates may have substantial practical impact for the improvement of plant health through the creation of new plant varieties, either through transgenic technology or guidance of breeding programmes.
ACKNOWLEDGEMENTS
Gitte Erbs and Mari‐Anne Newman acknowledge funding by The Danish Council for Independent Research, Technology and Production Sciences (FTP) and Villum Fonden, Denmark.
REFERENCES
- Akira, S. , Uematsu, S. and Takeuchi, O. (2006) Pathogen recognition and innate immunity. Cell, 24, 783–801. [DOI] [PubMed] [Google Scholar]
- Albus, U. , Baier, R. , Holst, O. , Pühler, A. and Niehaus, K. (2001) Suppression of an elicitor‐induced oxidative burst reaction in Medicago sativa cell cultures by Sinorhizobium meliloti lipopolysaccharides. New Phytol. 151, 597–606. [DOI] [PubMed] [Google Scholar]
- Alexander, C. and Rietschel, E.T. (2001) Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7, 167–202. [PubMed] [Google Scholar]
- Aslam, S.N. , Newman, M.A. , Erbs, G. , Morrissey, K.L. , Chinchilla, D. , Boller, T. , Jensen, T.T. , De Castro, C. , Ierano, T. , Molinaro, A. , Jackson, R.W. , Knight, M.R. and Cooper, R.M. (2008) Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 18, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Aslam, S.N. , Erbs, G. , Morrissey, K.L. , Newman, M.‐A. , Chinchilla, D. , Boller, T. , Molinaro, A. , Jackson, R.W. and Cooper, R.M. (2009) MAMPs signatures, synergy, size and charge: influences on perception or mobility and host defence responses. Mol. Plant Pathol. 10, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedini, E. , Parrilli, M. and Unverzagt, C. (2002) Oligomerization of a rhamnanic trisaccharide repeating unit of O‐chain polysaccharides from phytopathogenic bacteria. Tetrahedron Lett. 43, 8879–8882. [Google Scholar]
- Bedini, E. , De Castro, C. , Erbs, G. , Mangoni, L. , Dow, J.M. , Newman, M.‐A. , Parrilli, M. and Unverzagt, C. (2005) Structure‐dependent modulation of a pathogen response in plants by synthetic O‐antigen polysaccharides. J. Am. Chem. Soc. 127, 2414–2416. [DOI] [PubMed] [Google Scholar]
- Bishop, R.E. (2005) The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912. [DOI] [PubMed] [Google Scholar]
- Bishop, R.E. , Kim, S.H. and El Zoeiby, A. (2005) Role of lipid A palmitoylation in bacterial pathogenesis. J. Endotoxin Res. 11, 174–180. [DOI] [PubMed] [Google Scholar]
- Boneca, I.G. , Dussurget, O. , Cabanes, D. , Nahori, M.A. , Sousa, S. , Lecuit, M. , Psylinakis, E. , Bouriotis, V. , Hugot, J.P. , Giovannini, M. , Coyle, A. , Bertin, J. , Namane, A. , Rousselle, J.‐C. , Cayet, N. , Prévost, M.‐C. , Balloy, V. , Chignard, M. , Philpott, D.J. , Cossart, P. and Girardin, S.E. (2007) A critical role for peptidoglycan N‐deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA, 104, 997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S.G. , Meyer, A. , Holst, O. , Pühler, A. and Niehaus, K. (2005) Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol. Plant–Microbe Interact. 18, 674–681. [DOI] [PubMed] [Google Scholar]
- Buist, G. , Steen, A. , Kok, J. and Kuipers, O.P. (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nürnberger, T. , Jones, J.D. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J.M. , Flors, V. , García‐Agustín, P. , Jakab, G. , Mauch, F. , Newman, M.‐A. , Pieterse, C.M.J. , Benoit Poinssot, M.J.P. , Pozo, M.J. , Pugin, A. , Schaffrath, U. , Ton, J. , Wendehenne, D. , Zimmerli, L. and Mauch‐Mani, B. (2006) Priming: getting ready for battle. Mol. Plant–Microbe Interact. 19, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Deng, W.‐L. , Lin, Y.‐C. , Lin, R.‐H. , Wei, C.‐F. , Huang, Y.‐C. , Peng, W.‐L. and Huang, H.‐C. (2010) Effects of galU mutation on Pseudomonas syringae–plant interactions. Mol. Plant–Microbe Interact. 23, 1184–1196. [DOI] [PubMed] [Google Scholar]
- Denoux, C. , Galletti, R. , Mammarella, N. , Gopalan, S. , Werck, D. , De Lorenzo, G. , Ferrari, S. , Ausubel, F.M. and Dewdney, J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki, Y. , Miya, A. , Venkatesh, B. , Tsuyumu, S. , Yamane, H. , Kaku, H. , Minami, E. and Shibuya, N. (2006) Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 47, 1530–1540. [DOI] [PubMed] [Google Scholar]
- Desender, S. , Klarzynski, O. , Potin, P. , Barzic, M.R. , Andrivon, D. and Val, F. (2006) Lipopolysaccharides of Pectobacterium atrosepticum and Pseudomonas corrugata induce different defence response patterns in tobacco, tomato, and potato. Plant Biol. 8, 636–645. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Osbourn, A.E. , Wilson, T.J.G. and Daniels, M.J. (1995) A locus determining pathogenicity of Xanthomonas‐campestris is involved in lipopolysaccharide biosynthesis. Mol. Plant–Microbe Interact. 8, 768–777. [DOI] [PubMed] [Google Scholar]
- Dow, M. , Newman, M.‐A. and von Roepenack, E. (2000) The induction and modulation of plant defence responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. [DOI] [PubMed] [Google Scholar]
- Dziarski, R. and Gupta, D. (2005) Peptidoglycan recognition in innate immunity. J. Endotoxin Res. 11, 304–310. [DOI] [PubMed] [Google Scholar]
- Dziarski, R. and Gupta, D. (2006) The peptidoglycan recognition proteins (PGRPs). Genome Biol. 7, 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski, R. , Tapping, R.I. and Tobias, P.S. (1998) Binding of bacterial peptidoglycan to CD14. J. Biol. Chem. 273, 8680–8690. [DOI] [PubMed] [Google Scholar]
- Erbs, G. and Newman, M.‐A. (2003) The role of lipopolysaccharides in induction of plant defence responses. Mol. Plant Pathol. 4, 421–425. [DOI] [PubMed] [Google Scholar]
- Erbs, G. , Jensen, T.T. , Silipo A. Grant, W. , Dow, J.M. , Molinaro, A. , Parrilli, M. and Newman, M.‐A. (2008a) An antagonist of lipid A action in mammals has complex effects on lipid A induction of defence responses in the model plant Arabidopsis thaliana . Microbes Infect. 10, 571–574. [DOI] [PubMed] [Google Scholar]
- Erbs, G. , Silipo, A. , Aslam, S. , De Castro, C. , Liparoti, V. , Flagiello, A. , Pucci, P. , Lanzetta, R. , Parrilli, M. , Molinaro, A. , Newman, M.‐A. and Cooper, R.M. (2008b) Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomanas elicit innate immunity: structure and activity. Chem. Biol. 15, 438–448. [DOI] [PubMed] [Google Scholar]
- Felix, G. and Boller, T. (2003) Molecular sensing of bacteria in plants. J. Biol. Chem. 278, 6201–6208. [DOI] [PubMed] [Google Scholar]
- Ferrandon, D. , Imler, J.L. , Hetru, C. and Hoffmann, J.A. (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874. [DOI] [PubMed] [Google Scholar]
- Fischer, S.E. , Miguel, M.J. and Mori, G.B. (2003) Effect of root exudates on the exopolysaccharide composition and the lipopolysaccharide profile of Azospirillum brasilense Cd under saline stress. FEMS Microbiol. Lett. 219, 53–62. [DOI] [PubMed] [Google Scholar]
- Franchi, L. , McDonald, C. , Kanneganti, T.D. , Amer, A. and Nunez, G. (2006) Nucleotide‐binding oligomerization domain‐like receptors: intracellular pattern recognition molecules for pathogen detection and host defense. J. Immunol. 177, 3507–3513. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Zeidler, D. , Durner, J. and Dubery, I.A. (2004) Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia . Planta, 218, 647–657. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Laukens, K. , Witters, E. and Dubery, I.A. (2006) Lipopolysaccharide‐responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol. Biochem. 44, 369–379. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Laukens, K. , De Vijlder, T. , Witters, E. and Dubery, I.A. (2008) Proteomic profiling of cellular targets of lipopolysaccharide‐induced signaling in Nicotiana tabacum BY‐2 cells. Biochim. Biophys. Acta, 1784, 1750–1762. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009a) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Ntoukakis, V. and Rathjen, J.P. (2009b) The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis . Plant Signal. Behav. 4, 539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin, S.E. , Boneca, I.G. , Carneiro, L.A.M. , Antignac, A. , Jehanno, M. , Viala, J. , Tedin, K. , Taha, M.‐K. , Labigne, A. , Sansonetti, P.J. and Philpott, D.J. (2003) Nod1 detects a unique muropeptide from Gram‐negative bacterial peptidoglycan. Science, 300, 1584–1587. [DOI] [PubMed] [Google Scholar]
- Gross, A. , Kapp, D. , Nielsen, T. and Niehaus, K. (2005) Endocytosis of Xathomonas campestris pathovar campestris lipopolysaccharides in non‐host plant cells of N. benthamiana . New Phytol. 165, 215–226. [DOI] [PubMed] [Google Scholar]
- Guan, R. and Mariuzza, R.A. (2007) Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 15, 127–134. [DOI] [PubMed] [Google Scholar]
- Gust, A. , Biswas, R. , Lenz, H.D. , Rauhut, T. , Ranf, S. , Kemmerling, B. , Götz, F. , Glawischnig, E. , Lee, J. , Felix, G. and Nürnberger, T. (2007) Bacteria‐derived peptidoglycans constitute pathogen‐associated molecular patterns triggering innate immunity in Arabidopsis . J. Biol. Chem. 282, 32 338–32 348. [DOI] [PubMed] [Google Scholar]
- Hashimoto, C. , Hudson, K.L. and Anderson, K.V. (1988) The Toll gene of Drosophila, required for dorsal–ventral embryonic polarity, appears to encode a transmembrane protein. Cell, 52, 269–279. [DOI] [PubMed] [Google Scholar]
- He, P. , Shan, L. , Lin, N.‐C. , Martin, G.B. , Kemmerling, B. , Nürnberger, T. and Sheen, J. (2006) Specific bacterial suppressors of MAMP signalling upstream of MAPKKK in Arabidopsis innate immunity. Cell, 125, 563–575. [DOI] [PubMed] [Google Scholar]
- Ialenti, A. , Di Meglio, P. , Grassia, G. , Maffia, P. , Di Rosa, M. , Lanzetta, R. , Molinaro, A. , Silipo, A. , Grant, W. and Ianaro, A. (2006) A novel lipid A from Halomonas magadiensis inhibits enteric LPS‐induced human monocyte activation. Eur. J. Immunol. 36, 354–360. [DOI] [PubMed] [Google Scholar]
- Iliev, D.B. , Roach, J.C. , Mackenzie, S. , Planas, J.V. and Goetz, F.W. (2005) Endotoxin recognition: in fish or not in fish? FEBS Lett. 579, 6519–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir, Y. , Guo, M. , Oh, H.‐S. , Petnicki‐Ocwieja, T. , Chen, S. , Tang, X. , Dickman, M.B. , Collmer, A. and Alfano, J.R. (2004) Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kannenberg, E.L. and Carlson, R.W. (2001) Lipid A and O‐chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39, 379–391. [DOI] [PubMed] [Google Scholar]
- Kawahara, K. , Tsukano, H. , Watanabe, H. , Lindner, B. and Matsuura, M. (2002) Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70, 4092–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling, B. , Schwedt, A. , Rodriguez, P. , Mazzotta, S. , Frank, M. , Qamar, S.A. , Mengiste, T. , Betsuyaku, S. , Parker, J.E. , Müssig, C. , Thomma, B.P.H. , Albrecht, C. , de Vries, S.C. , Hirt, H. and Nürnberger, T. (2007) The BRI1‐associated kinase 1, BAK1, has a brassinolide‐independent role in plant cell‐death control. Curr. Biol. 17, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Lee, S.W. , Han, S.W. , Sririyanum, M. , Park, C.J. , Seo, Y.S. and Ronald, P.C. (2009) A type I‐secreted, sulfated peptide triggers XA21‐mediated innate immunity. Science, 326, 850–853. [DOI] [PubMed] [Google Scholar]
- Leeman, M. , Vanpelt, J.A. , Denouden, F.M. , Heinsbroek, M. , Pahm, B. and Schippers, B. (1995) Induction of systemic resistance against fusarium‐wilt of radish by lipopolysaccharides of Pseudomonas fluorescens . Phytopathology, 85, 1021–1027. [Google Scholar]
- Lemaitre, B. , Nicolas, E. , Michaut, L. , Reichhart, J.‐M. and Hoffmann, J.A. (1996) The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell, 86, 973–983. [DOI] [PubMed] [Google Scholar]
- Leulier, F. , Parquet, C. , Pili‐Floury, S. , Ryu, J.‐H. , Caroff, M. , Lee, W.‐J. , Mengin‐Lecreulx, D. and Lemaitre, B. (2003) The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4, 478–484. [DOI] [PubMed] [Google Scholar]
- Livaja, M. , Zeidler, D. , von Rad, U. and Durner, J. (2008) Transcriptional responses of Arabidopsis thaliana to the bacteria‐derived PAMPs harpin and lipopolysaccharide. Immunobiology, 213, 161–171. [DOI] [PubMed] [Google Scholar]
- MacKenzie, S.A. , Roher, N. , Boltaña, S. and Goetz, F.W. (2010) Peptidoglycan, not endotoxin, is the key mediator of cytokine gene expression induced in rainbow trout macrophages by crude LPS. Mol. Immunol. 47, 1450–1457. [DOI] [PubMed] [Google Scholar]
- Madala, N.E. , Leone, M.R. , Molinaro, A. and Dubery, I.A. (2011) Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana . Glycobiology, 21, 184–194. [DOI] [PubMed] [Google Scholar]
- McDonald, C. , Inohara, N. and Nuñez, G. (2005) Peptidoglycan signalling in innate immunity and inflammatory disease. J. Biol. Chem. 280, 20 177–20 180. [DOI] [PubMed] [Google Scholar]
- Medzhitov, R. , Preston‐Hurlburt, P. and Janeway Jr, C.A. (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature, 388, 394–397. [DOI] [PubMed] [Google Scholar]
- Meyer, A. , Pühler, A. and Niehaus, K. (2001) The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum . Planta, 213, 214–222. [DOI] [PubMed] [Google Scholar]
- Millet, Y.A. , Danna, C.H. , Clay, N.K. , Songnuan, W. , Simon, M.D. , Werck‐Reichhart, D. and Ausubel, F.M. (2010) Innate immune responses activated in Arabidopsis roots by microbe‐associated molecular patterns. Plant Cell, 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis . Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis . Proc. Natl. Acad. Sci. USA, 104, 19 613–19 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford, R.S. and Varley, A.W. (2006) Shield as signal: lipopolysaccharide and the evolution of immunity to Gram‐negative bacteria. Plos. Pathog. 2, 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.‐A. , Daniels, M.J. and Dow, J.M. (1995) Lipopolysaccharide from Xanthomonas campestris induces defence‐related gene expression in Brassica campestris . Mol. Plant–Microbe Interact. 8, 778–780. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Daniels, M.J. and Dow, J.M. (1997) The activity of lipid A and core components of bacterial lipopolysaccharides in the prevention of the hypersensitive response in pepper. Mol. Plant–Microbe Interact. 10, 926–928. [DOI] [PubMed] [Google Scholar]
- Newman, M.‐A. , von Roepenack, E. , Daniels, M.J. and Dow, J.M. (2000) Lipopolysaccharides and plant responses to phytopathogenic bacteria. Mol. Plant Pathol. 1, 25–31. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , von Roepenack‐Lahaye, E. , Parr, A. , Daniels, M.J. and Dow, J.M. (2002) Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J. 29, 487–495. [DOI] [PubMed] [Google Scholar]
- Newman, M.‐A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 68–79. [DOI] [PubMed] [Google Scholar]
- Nguyen, H.P. , Chakravarthy, S. , Velásquez, A.C. , McLane, H.L. , Zeng, L. , Nakayashiki, H. , Park, D.‐H. , Collmer, A. and Martin, G. (2010) Methods to study PAMP‐triggered immunity using tomato and Nicotiana benthamiana . Mol. Plant–Microbe Interact. 23, 991–999. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , DebRoy, S. , Lee, Y.H. , Pumplin, N. , Jones, J. and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science, 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Petutschnig, E.K. , Jones, A.M.E. , Serazetdinova, L. , Lipka, U. and Lipka, V. (2010) The LysM‐RLK is a major chitin binding protein in Arabidopsis thaliana and subject to chitin‐induced phosphorylation. J. Biol. Chem. 285, 28 902–28 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz, C.R.H. and Whitfield, C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz, C.R.H. , Reynolds, C.M. , Trent, M.S. and Bishop, R.E. (2007) Lipid A modification systems in Gram‐negative bacteria. Annu. Rev. Biochem. 76, 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallabhandi, P. , Awomoyi, A. , Thomas, K.E. , Phalipon, A. , Fujimoto, Y. , Fukase, K. , Kusumoto, S. , Qureshi, N. , Sztein, M.B. and Vogel, S.N. (2008) Differential activation of human TLR4 by Escherichia coli and Shigella flexneri 2a lipopolysaccharide: combined effects of lipid A acylation state and TLR4 polymorphisms on signaling. J. Immunol. 180, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer, K.H. and Kandler, O. (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schromm, A. , Brandenburg, K. , Loppnow, H. , Zahringer, U. , Rietschel, E. , Carroll, S. , Koch, M. , Kusumoto, S. and Seydel, U. (1998) The charge of endotoxin molecules influences their conformation and IL‐6‐inducing capacity. J. Immunol. 161, 5464–5471. [PubMed] [Google Scholar]
- Schromm, A. , Brandenburg, K. , Loppnow, H. , Moran, A. , Koch, M. , Rietschel, E. and Seydel, U. (2000) Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 267, 2008–2013. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. and Zipfel, C. (2011) Activation of plant pattern‐recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. [DOI] [PubMed] [Google Scholar]
- Silipo, A. , Sturiale, L. , Garozzo, D. , de Castro, C. , Lanzetta, R. , Parrilli, M. , Grant, W.D. and Molinaro, A. (2004) Structure elucidation of the highly heterogeneous lipid A from the lipopolysaccharide of the Gram‐negative extremophile bacterium Halomonas magadiensis strain 21 M1. Eur. J. Org. Chem. 2004, 2263–2271. [Google Scholar]
- Silipo, A. , Molinaro, A. , Sturiale, L. , Dow, J.M. , Erbs, G. , Lanzetta, R. , Newman, M.A. and Parrilli, M. (2005) The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris . J. Biol. Chem. 280, 33 660–33 668. [DOI] [PubMed] [Google Scholar]
- Silipo, A. , Sturiale, L. , Garozzo, D. , Erbs, G. , Tandrup Jensen, T. , Lanzetta, R. , Dow, J.M. , Parrilli, M. , Newman, M.‐A. and Molinaro, A. (2008) The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in Arabidopsis . ChemBioChem 9, 896–904. [DOI] [PubMed] [Google Scholar]
- Traub, S. , von Aulock, S. , Hartung, T. and Herman, C. (2006) MDP and other muropeptides—direct and synergistic effects on the immune system. J. Endotoxin Res. 12, 69–85. [DOI] [PubMed] [Google Scholar]
- Travassos, L.H. , Girardin, S.E. , Philpott, D.J. , Blanot, D. , Nahori, M.‐A. , Werts, C. and Boneca, I.G. (2004) Toll‐like receptor 2‐dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO J. 5, 1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C. , Bakker, P.A. and Pieterse, C.M. (1998) Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y. , Seto, H. , Fujioka, S. , Yoshida, S. and Chory, J. (2001) BRI1 is a critical component of a plasma‐membrane receptor for plant steroids. Nature, 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Wright, S.D. , Ramos, R.A. , Tobias, P.S. , Ulevitch, R.J. and Mathison, J.C. (1990) CD14 serves as the cellular receptor for complexes of lipopolysaccharides with lipopolysaccharide binding protein. Science, 249, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Yun, M.H. , Torres, P.S. , El Oirdi, M. , Rigano, L.A. , Gonzalez‐Lamothe, R. , Marano, M.R. , Castagnaro, A.P. , Dankert, M.A. , Bouarab, K. and Vojnov, A. (2006) Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 141, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler, D. , Zähringer, U. , Gerber, I. , Dubery, I. , Hertung, T. , Bors, W. , Hutzler, P. and Durner, J. (2004) Innate immunity in Arabidopsis thaliana lipopolysaccharides activates nitric oxide synthase NOS and induces defense genes. Proc. Natl. Acad. Sci. USA, 101, 15 811–15 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler, D. , Dubery, I.A. , Schmitt‐Kopplin, P. , Von Rad, U. and Durner, J. (2010) Lipopolysaccharide mobility in leaf tissue of Arabidopsis thaliana . Mol. Plant Pathol. 11, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]


