Summary
Sudden death syndrome (SDS), caused by Fusarium virguliforme, is an important yield‐limiting disease of soybean. This soil‐borne fungus colonizes soybean roots causing root rot, and also releases a phytotoxin that is translocated to leaves causing interveinal chlorosis and necrosis leading to defoliation and early maturation. The objective of our study was to compare gene expression profiles during the early response of soybean leaves exposed to sterile culture filtrates of F. virguliforme in soybean genotypes with different levels of resistance to SDS. The analysis identified SDS‐related defence genes that were induced in the most resistant genotype, but not in the other genotypes. Further functional annotations based on sequence homology suggested that some of the induced genes probably encode proteins involved in cell wall modification, detoxification, defence responses, primary metabolism and membrane transport. Quantitative real‐time reverse‐transcribed polymerase chain reaction confirmed the differential transcript accumulation of a subset of these genes. In addition, in silico mapping of differentially expressed genes to SDS‐resistant quantitative trait loci allowed for the identification of new potential defence genes that could be genetically mapped to the soybean genome, and could be used further in a marker‐assisted selection programme. A comparison of the response of soybean to F. virguliforme phytotoxin (Fv toxin) relative to other biotic and abiotic stresses revealed that the resistance response to Fv toxin is quite similar to the response to inoculation with an incompatible Pseudomonas syringae pv. glycinea strain, suggesting that Fv toxin might induce hypersensitive response pathways in soybean leaf tissues in the absence of pathogen in these tissues.
Introduction
Sudden death syndrome (SDS) disease is caused by the soil‐borne fungus Fusarium virguliforme, formally known as Fusarium solani (Mart.) Sacc. f. sp. glycines (Roy et al., 1997; Rupe et al., 1989). Although not receiving much attention until the 1980s, SDS has since reached epidemic proportions in some soybean‐growing areas in the USA, Argentina and Brazil (Rupe and Hartman, 1999). The fungus can be isolated from roots and occasionally from the crown region, but has never been isolated from the leaves. Although colonization of the fungus is restricted to the roots, SDS damage occurs throughout the plant, including the pods, with the most striking symptoms occurring on the leaves. The tissue damage and impressive foliar symptoms are believed to be caused by a phytotoxin(s) that is (are) released by the fungus during its colonization of the roots and translocated through the vascular system (Jin et al., 1996). Several partially characterized compounds with some toxic activity have been isolated from culture filtrates and have been proposed as putative candidates for the F. virguliforme phytotoxin (Fv toxin) responsible for the production of the SDS‐like symptoms (Brar and Bhattacharyya, 2012; Brar et al., 2011; Ji et al., 2006; Jin et al., 1996). Because cell‐free culture filtrates of F. virguliforme can induce SDS foliar symptoms typical of pathogen‐inoculated plants (Li et al., 1999), the culture filtrates provide a practical means to screen large numbers of plants for resistance to SDS that correlates with field resistance (Hartman et al., 2004; Li et al., 1999), indicating that the filtrates contain the Fv toxin(s) required for SDS symptom development.
The use of true resistant cultivars would be the most effective method to control SDS. However, the genetics of SDS resistance is complicated, as it is a polygenic trait and variation for resistance in genetic populations is continuous (Hnetkovsky et al., 1996; Iqbal et al., 2001; Njiti et al., 1996). Although partially resistant lines show good levels of resistance, it is clearly only partial, as SDS symptoms develop with greater inoculum levels (Hartman et al., 1997). The discovery of partially resistant plant introductions (PIs) has allowed the identification of different quantitative trait loci (QTLs) associated with disease resistance (Iqbal et al., 2001). However, many QTLs have been genetically mapped in the soybean genome, with only a few of these QTLs confirmed by mapping the same QTL in different populations. The confirmed QTLs mapped to linkage groups (LGs) C2, D2, G and N (Farias Neto et al., 2007; Hnetkovsky et al., 1996; Iqbal et al., 2001; Lightfoot et al., 2005). In recent work (Kazi et al., 2008) exploring the genetic relationship between root and leaf resistance to F. virguliforme, it was reported that separate loci might control soybean resistance to root infection and leaf symptoms. Because the loci playing a role in resistance in leaves and roots may differ, it has been difficult to identify the exact genes underlying SDS resistance (Kazi et al., 2008; Triwitayakorn et al., 2005).
Although several QTLs conferring SDS resistance have been identified in some soybean lines, the specific genes involved in SDS resistance remain to be determined. To manipulate soybean SDS resistance, an understanding of the precise resistance mechanisms at the genetic level would be beneficial. The characterization of the expression of the genes associated with partial resistance in response to SDS can help to pinpoint target genes for resistance manipulation, in addition to providing information to infer the physiological responses that occur during infection. Soybean microarray tools have been successful in obtaining gene expression profiles for many biological events, such as somatic embryogenesis (Thibaud‐Nissen et al., 2003), pathogen infection (Calla et al., 2009; Choi et al., 2008; van de Mortel et al., 2007; Moy et al., 2004; Panthee et al., 2007, 2009; Radwan et al., 2011; Zou et al., 2005), symbiont inoculation (Brechenmacher et al., 2008; Libault et al., 2010), aphid attack (Li et al., 2008) and response to herbicides (Zhu et al., 2008, 2009). A comparison of the expression results across treatments will assist in the identification of common or unique sets of defence‐responsive pathways that might be triggered during biotic and abiotic stimuli.
To better characterize the response of soybean to F. virguliforme, several biochemical and molecular studies have been completed. In the biochemical study conducted by Lozovaya et al. (2004), the isoflavone phytoalexin glyceollin, lignins and phenolic components were shown to increase in a partially resistant genotype compared with the highly susceptible cultivar Spencer. At the molecular level, Iqbal et al. (2005) investigated the potential mechanisms triggered by F. virguliforme infection within 10 days of inoculation, and proposed a defensive role for genes associated with phenolics, phenopropanoids, general defence, cell wall modification and signal transduction. Using Arabidopsis thaliana as a potential host of F. virguliforme, Yuan et al. (2008) suggested a possible role of SnRK1 and components downstream of phenylalanine ammonia lyase in the defence regulation to the fungal infection. In a recent work focused on soybean–F. virguliforme interactions, Radwan et al. (2011) characterized the molecular mechanisms of soybean roots in response to F. virguliforme infection, and reported that many of the genes with increased expression were common between resistant and susceptible genotypes, but some genotype‐specific expression was documented. Currently, there are no reports that have investigated the gene expression response of soybean leaves to Fv toxin. In this study, resistant and susceptible soybean genotypes were treated with F. virguliforme culture filtrates (referred to as ‘Fv toxin’ throughout the article) and gene expression responses were determined at 8 h post‐treatment (hpt) to identify the genes associated with resistance or susceptibility to Fv toxin. These differentially expressed transcripts were in silico mapped to the soybean genome and compared with soybean responses to other biotic and abiotic stresses.
Results
Symptom development
Three soybean genotypes of varying levels of resistance or susceptibility responded differently to Fv toxin (Fig. 1). The highly susceptible variety, Essex, started to show severe leaf curling and mild mottling within 24 h after stems were placed within Fv toxin. The most resistant genotype used in the study, PI567.374, did not show any visible foliar symptoms at 24 hpt, whereas cultivar Williams 82 (intermediate levels of resistance between PI567.374 and Essex) initiated slight leaf curling at 24 hpt. By 48 hpt, some symptoms were observed on all tested genotypes. By 120 hpt, Essex displayed the typical interveinal chlorosis and necrosis symptoms with some dead tissue along the leaf edges. In contrast, William 82 and PI567.374 did not display these more severe symptoms. At all of these time points, mock‐treated seedlings of the three soybean genotypes appeared very healthy and normal without any of the symptoms mentioned above (data not shown). To validate the results obtained by the fungal culture filtrates, disease symptoms induced by the natural infection of F. virguliforme at 21 days post‐inoculation (dpi) were compared side by side with those induced by the fungal culture filtrates at 5 days post‐treatment (dpt) (Fig. 2). In both experiments, Essex leaves showed typical symptoms of SDS, including interveinal chlorosis and necrosis symptoms with some dead tissue along the leaf edges.
Figure 1.
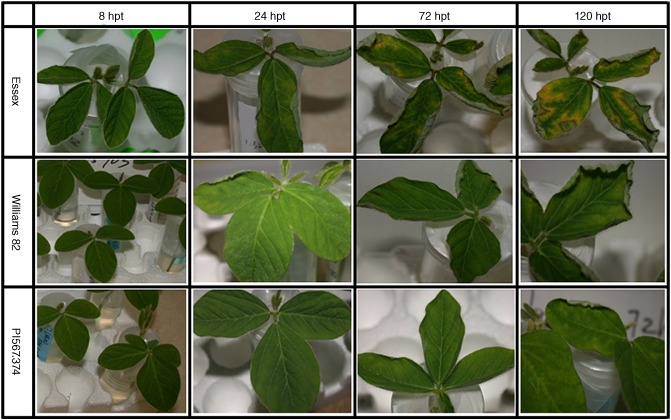
Foliar symptom development of sudden death syndrome (SDS) on three soybean genotypes with different levels of resistance (Essex, Williams 82 and PI567.374) treated using Fusarium virguliforme phytotoxin (Fv toxin) at 8, 24, 72 and 120 h post‐treatment (hpt).
Figure 2.
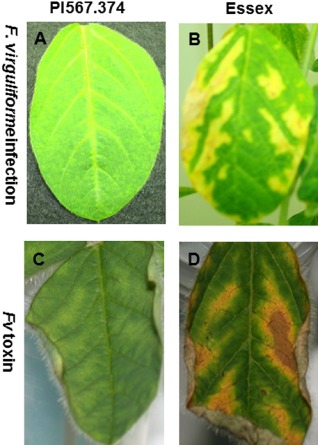
Cross comparison between sudden death syndrome (SDS) foliar symptoms induced by Fusarium virguliforme (Fv) infection at 21 days post‐inoculation (dpi) (A, B) and by the fungal culture filtrates (Fv toxin) at 5 days post‐treatment (dpt) (C, D). The symptoms on the leaf of the resistant genotype (PI567.374) are shown in (A) and (C), and those on the leaf of the susceptible genotype (Essex) are shown in (B) and (D).
Overall gene expression profile and quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) validation
To determine the ideal time point for the investigation of the early response of soybean gene expression profiles following Fv toxin treatment, a pilot microarray experiment was carried out to compare the gene expression profiles at 0, 4, 8 and 12 hpt. The results indicated that there was little detectable gene expression differences at 4 hpt, whereas samples at 8 hpt displayed distinguishable differential expression (data not shown). At 12 hpt, the gene expression profiles followed the same direction as at 8 hpt with a slight increase/decrease in gene expression (data not shown). Therefore, 8 hpt was chosen to monitor gene expression in a replicated study. A loop design was used for the microarray experiment involving six treatments (three genotypes with and without cell‐free culture filtrate; Fig. 3). Comparisons among the three genotypes with and without Fv toxin were carried out to identify differentially expressed genes. In addition, the differences between the two treatments (mock and Fv toxin) were compared for each genotype. The statistical analysis identified 1906 significant differentially expressed genes from the experiment (Table S1, see Supporting Information). The criteria for a gene to be deemed as ‘significantly changed’ were that it had both significance [false discovery rate (FDR)‐corrected P value of ≤0.05] and a fold‐change ratio of ≥1.5 or ≤0.667. These genes were functionally classified into 17 categories (Fig. S1, see Supporting Information) according to their annotations and guidelines of gene categories established by Calla et al. (2009).
Figure 3.
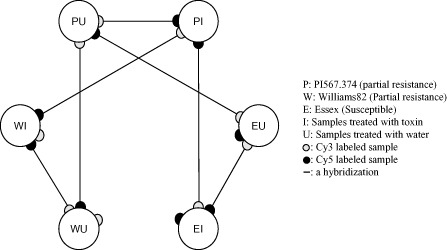
Microarray experimental design. Six mRNA samples, as defined on the right, were used. Each line connecting two samples represents hybridizations of microarray slides. The filled circles at the end of each line represent samples labelled with cyanine 5 (Cy5) dye (red) and Cy3 dye (green).
To further confirm and validate the expression patterns of the microarray data, 14 genes (Table S2, see Supporting Information) were selected for verification using qRT‐PCR. Ten defence‐related genes that increased in expression level and four random genes that decreased were selected. The induced transcripts were selected from different suggested categories of soybean leaf resistance (see details in Discussion section), and the suppressed transcripts were selected from those showing a decrease in abundance. Leaves from three new independent biological replicates were used for RNA extraction, which was employed as a template for RT‐PCR. The expression profiles of the 14 genes were determined by qRT‐PCR at 6, 8 and 24 hpt. The directions of the changes in differential expression determined by qRT‐PCR were the same as those determined in the microarray study, but the magnitudes differed slightly between the two studies (Fig. 4 and Table S3, see Supporting Information). These slight differences in the magnitude of the expression changes can be explained by the fact that different batches of RNA were used to perform the microarray and qRT‐PCR experiments, and by the different sensitivities of the assays. qRT‐PCR confirmed that the results from the microarray experiment were accurate and that the gene list identified by the microarray experiment was a reliable source for further study. In addition, our qRT‐PCR results supported our early selection of 8 hpt as an ideal time point to conduct this experiment.
Figure 4.
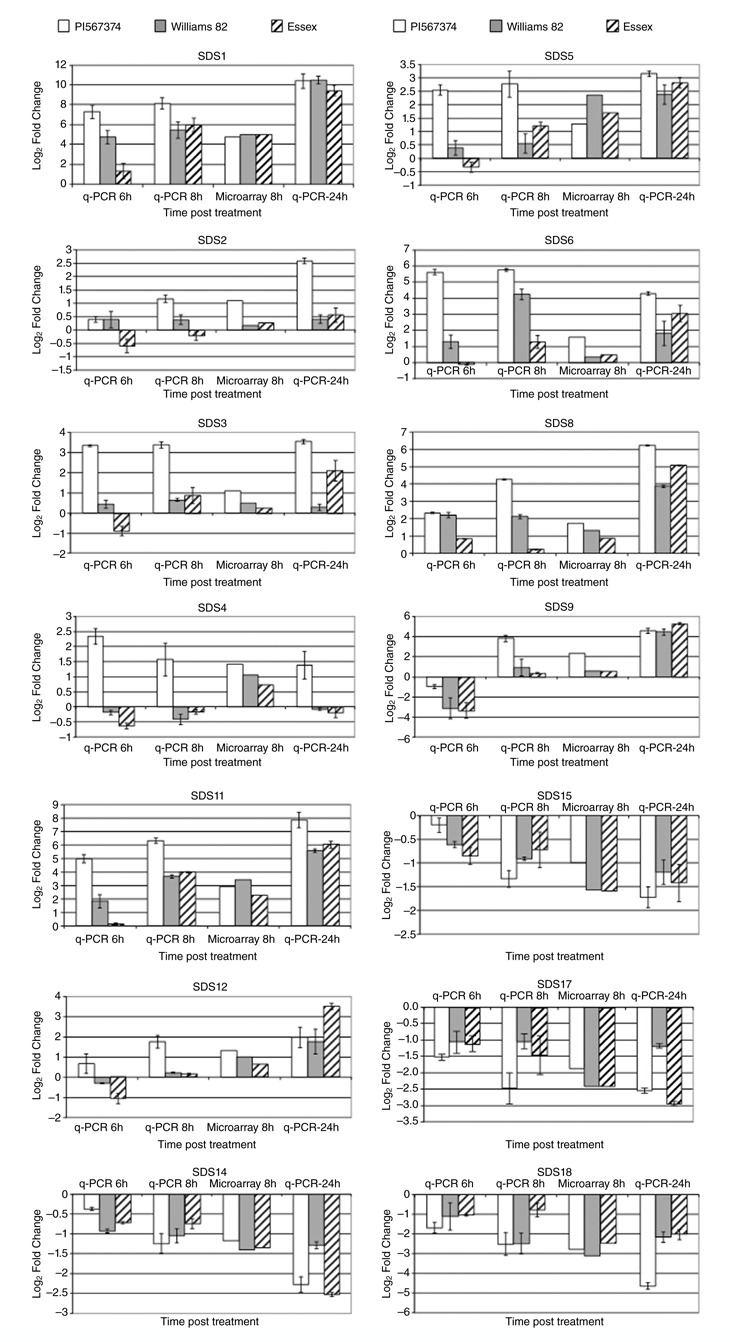
Quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) confirmed the regulation of 14 genes from the microarray results. The expression profiles of these genes were determined by qRT‐PCR at 6, 8 and 24 h post‐treatment (hpt). Expression levels are given as the log2‐transformed fold change of Fusarium virguliforme phytotoxin (Fv toxin)‐treated soybeans compared with mock treatment. In each histogram of qRT‐PCR results, the means of three biological replicates are represented ± standard deviation (SD). Bars from left to right: white bars, PI567.374; grey bars, Williams 82; striped bars, Essex. The soybean actin gene was used as a reference control gene.
Gene expression profiles from comparisons between Fv toxin‐ and mock‐treated soybean
Fv toxin induced a strong response in all genotypes relative to mock treatments, as 1906 genes showed significant changes in expression. Of the significantly differentially expressed genes, 543 genes were up‐regulated and 113 genes were down‐regulated in the PI567.374 Fv toxin‐treated (PI) versus PI567.374 mock (PU) comparison (Table S4A, see Supporting Information), 781 genes were up‐regulated and 909 genes were down‐regulated in the Williams 82 Fv toxin‐treated (WI) versus Williams 82 mock (WU) comparison (Table S4B), and 655 genes were up‐regulated and 445 genes were down‐regulated in the Essex Fv toxin‐treated (EI) versus Essex mock (EU) comparison (Table S4C). Combinations of these up‐ and down‐regulated genes from the three genotypes, in response to Fv toxin, showed that 405 genes increased (Table S5A, see Supporting Information) and 93 genes decreased (Table S5B) in expression level, indicating that they are common Fv toxin‐induced genes in all three genotypes.
From this up‐regulated gene list, 44 genes (Table S6A, Fig. S5, see Supporting Information) were significantly induced only between the resistant genotypes (Fv toxin treated versus mock: PI–PU and WI–WU), but not for the highly susceptible genotype (Fv toxin treated versus mock: EI–EU), suggesting that these are Fv toxin‐induced genes in the two resistant genotypes (PI567.374 and Williams 82). Sixty‐five genes (Table S7A, see Supporting Information) were induced uniquely in PI567.374, but not in the other two varieties, suggesting that transcript induction of these genes on Fv toxin treatment is specific to the most resistant genotype PI567.374. Of the 93 genes found in common across the three comparisons (Table S5B), 14 genes (Table S6B) were shared only between PI–PU and WI–WU, indicating that they are common Fv toxin‐repressed genes in resistant genotypes, and six genes were unique in PI–PU (Table S7B), suggesting their specificity for the resistant PI567.374. The common toxin‐induced genes (405) and common toxin‐repressed genes (93) were classified into different functional categories (Figs S2 and S3, see Supporting Information).
Comparison between the induced or repressed genes from this study (Table S1) with those significantly differentially expressed from soybean root infected by F. virguliforme (Radwan et al., 2011) revealed that both studies shared some common transcripts, but other significant transcripts were different (Table S8, see Supporting Information). Some of the common induced transcripts between the two studies belonged to cell wall and defence‐related genes, whereas some showing differences belonged to hormone signalling pathways.
Specific differences among the three genotypes without Fv toxin treatment
Because the microarrays used for this study covered approximately one‐fifth of the soybean genome, it was not surprising that gene expression differences relative to mock‐treated plants were not significant when the stringent FDR‐adjusted P value was used. Therefore, to identify the genes that had the most evidence for differential expression between genotypes, the less stringent uncorrected raw P value of ≤0.05 was used together with a 1.5 times up or down fold‐change cut‐off. This approach identified 134 genes as significantly different in gene expression as a function of genotype (Table S1). For up‐regulated genes (Table S9, see Supporting Information), seven genes were shared between PU–WU and PU–EU, two genes were shared between PU–EU and WU–EU, and only one gene (Glyma08g45300.1, BG790156), which belonged to the unknown category, was common among the three comparisons. These genes represent candidate defence genes for the most resistant genotype PI567.374, and contained the defence‐associated gene (Glyma06g19890.1, BG790192) EDS1 (for enhanced disease susceptibility when mutated), one heat shock protein (Glyma09g24410.1, BI893938), one sulphate transporter (Glyma02g16370, AW598447) and three unknown genes. For down‐regulated genes (Table S1), four genes were shared between PU–EU and WU–EU and none were common among the three comparisons. These genes, one of which was EDS1::ERD1 (early response to dehydration 1) chimeric (Glyma02g42860.1, BF010313), were expressed more strongly in the susceptible variety Essex. In conclusion, from this group of comparisons, there was minimal variability in gene expression among the three genotypes in the absence of Fv toxin treatment.
Identification of Fv toxin‐induced resistant‐associated genes specific for PI567.374
Eighty‐eight genes were expressed more strongly in Fv toxin‐treated PI567.374 than in the other two varieties, suggesting that some of these 88 genes were potential defence‐related genes specific to PI567.374 (Table S10, see Supporting Information). To further analyse these candidate defence genes, a hierarchical clustering analysis was performed. Our aim in performing this clustering analysis was to identify common or unique sets of defence‐responsive pathways that might be triggered during biotic and abiotic stimuli. Nineteen different treatments were used in this analysis, including nine from this study, three from a Pseudomonas syringae study (Zou et al., 2005), three from soybean roots inoculated by Bradyrhizobium japonicum (Brechenmacher et al., 2008) and four from an atrazine study (Zhu et al., 2009).
As illustrated in Fig. S4 (see Supporting Information), there were clearly two clusters among these 88 genes. Cluster A was composed of 78 genes. They were all toxin‐repressed genes, but showed a lower level of repression in PI567.374 than in Williams 82 and Essex. Only a few of these genes showed significant changes in nodulation, atrazine and compatible P. syringae treatments, but nearly all were significantly down‐regulated in the incompatible P. syringae studies. The expression of these genes seemed to be associated with reduced severity of tissue damage, perhaps by the inhibition of necrosis or programmed cell death (PCD). Cluster B was composed of nine genes, which were all toxin‐induced genes, but showed more significant induction in PI567.374 than in the other two varieties. These nine genes could be further subclustered into three minigroups. The genes in the first minigroup (Glyma06g40790.1, AW756020; Glyma11g19920.1, BF070757; Glyma01g45520.1, AW832575) showed no significant change in either P. syringae study, but were induced significantly in PI567.374, suggesting that they were specific for Fv toxin resistance. Genes in the second minigroup (Glyma16g34070.1, BG510438; Glyma01g02580.1, BG043573; Glyma03g19810.2, BF070234) were induced by Fv toxin in PI567.374, but showed no or less induction in the other varieties. They were also induced by the incompatible P. syringae inoculation, but not by the compatible strain. These data suggest that these three genes are common defence‐related genes. Genes in the third mini‐group (Glyma01g45520.1, AW759226; Glyma08g45300.1, BG790156; Glyma12g08060.1, BF009754) were induced similarly among the three varieties after Fv toxin treatment and in both compatible and incompatible P. syringae studies, indicating that they are involved in common pathogen‐induced responses and should be omitted from the list of potentially specific Fv toxin‐induced resistance genes.
Identification of SDS Fv toxin‐induced defence genes shared between PI567.374 and Williams 82
PI567.374 and Williams 82 showed full and partial resistance responses, respectively, to Fv toxin (Fig. 1 and 2), suggesting that these two genotypes may share some common defence to SDS. Consistent with this hypothesis, through the comparison between PI–EI and WI–EI, we identified 16 potentially shared defence‐related genes (Table S10), and through the comparison between PI–PU and WI–WU, we identified 44 potentially shared defence‐induced genes (Table S6A). Three genes were found to overlap between these two comparisons. These genes were BG509368, BG510920 and BI316566. In addition, BG510438, which was found among the 44 genes, but not among the 16 genes, was statistically grouped into the genes in cluster B of Fig. S4.
To further analyse these candidate genes, we performed a hierarchical clustering analysis of the 16 genes (Fig. S5A) and 44 genes (Fig. S5B) to cross compare the same 19 treatments as described previously. The overall pattern of clustering indicated that these genes were almost all plant pathogen inducible, and the induction level was normally higher in resistant plants than in susceptible plants. In addition, some of the genes were also affected by atrazine treatment. However, few were induced on B. japonicum inoculation.
The identification of four genes (Glyma12g01420.1, BG509368; Glyma12g01420.1, BG510920; Glyma15g42500.1, BI316566; Glyma16g34070.1, BG510438) as significantly differentially expressed in both resistant genotypes strengthens the likelihood that these genes are defence‐related genes. Therefore, it was worth checking these genes one by one to understand their responses in other studies. A list of genes associated with soybean resistance to Fv toxin is summarized in Table 1. BG510438 and BI316566 were induced to higher expression levels in resistant varieties than in the susceptible Essex in response to Fv toxin treatment, and were also induced significantly with incompatible P. syringae inoculation, but not on inoculation with the compatible strain; they were also induced by atrazine treatment at 4 and 8 hpt. The expression levels of these genes were not altered by B. japonicum inoculation. These data suggest that BG510438 and BI316566 are defence‐induced genes shared between PI567.374 and Williams 82. BG509368 and BG510920 showed a similar pattern to the former two, but were not induced on atrazine treatment, suggesting that they are plant pathogen defence specific.
Table 1.
Significant genes associated with soybean resistance to Fusarium virguliforme phytotoxin (Fv toxin)
| Gene ID | Category | Annotation | Fold change | P value | Specificity* | Specificity† | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PI | W82 | PI | W82 | PI | W82 | HR | Vir | At | |||
| Glyma06g40790.1 | Oxidation | Aldo/keto reductase (AKR) | 2.120 | 1.410 | 0.008 | 0.098 | Yes | No | No | No | Yes |
| Glyma11g19920.1 | Cell wall | Xyloglucan endotransglycosylase | 3.000 | 1.290 | 0.016 | 0.486 | Yes | No | No | No | No |
| Glyma01g45520.1 | Amino acid metabolism | S‐Methyl‐5‐thioribose kinase | 2.980 | 1.710 | 0.001 | 0.027 | Yes | No | No | No | No |
| Glyma16g34070.1 | Defence | Soybean R1 protein | 2.330 | 2.230 | 0.028 | 0.018 | Yes | Yes | Yes | No | Yes |
| Glyma12g01420.1 | Defence | R gene | 2.160 | 2.500 | 0.013 | 0.003 | Yes | Yes | Yes | No | No |
| Glyma15g42500.1 | Membrane | ABC‐transporter | 2.030 | 2.370 | 0.034 | 0.008 | Yes | Yes | Yes | No | Yes |
| Glyma01g02580.1 | Secondary metabolism | Cinnamyl alcohol dehydrogenase | 5.130 | 1.520 | 0.042 | 0.542 | Yes | No | Yes | No | No |
*Variety in which this gene was induced significantly and differentially on Fv toxin treatment when compared with Essex (susceptible).
†Treatment for which this gene was found to be induced significantly and differentially when compared with mock treatment.
PI, PI567.374; W82, Williams 82; HR, soybean leaves inoculated by avirulent strain of Pseudomonas syringae pv. glycinea; Vir, soybean leaves inoculated by virulent strain of P. syringae pv. glycinea; At, soybean leaves treated by atrazine herbicide.
Clustering the soybean response to Fv toxin to other biotic and abiotic treatments
Comparisons among the Fv toxin‐treated soybean plants indicated that the three genotypes shared 405 induced genes and 93 repressed genes (Table S5A,B). Hierarchical clustering of these genes among the 19 treatments is shown in Fig. 5. The overall cluster pattern suggested that the three varieties clustered very tightly together. Of the other treatments, inoculation with the compatible or incompatible P. syringae strain showed the most similar expression pattern, with the incompatible strain being closest to Fv toxin treatment. Atrazine treatments were the next closest clusters, followed by the most distant, soybean response to B. japonicum. This clustering analysis showed that the soybean defence response to Fv toxin has gene expression commonality to defence against a hypersensitive response (HR)‐inducing pathogen, P. syringae, but little similarity to the response to atrazine treatment or to symbiotic bacteria (nodulation).
Figure 5.
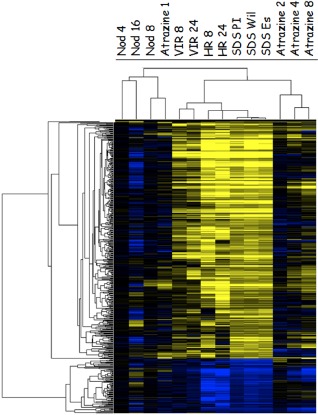
Hierarchical clusters of genes that were significantly induced or repressed in all three genotypes on Fusarium virguliforme phytotoxin (Fv toxin) treatment. Genes were clustered among 14 treatments. Nod, soybean inoculated with Bradyrhizobium japonicum at 4 days (Nod 4), 8 days (Nod 8) and 16 days (Nod 16) after inoculation; VIR and HR, soybean inoculated by compatible (VIR) and incompatible (HR) Pseudomonas syringae pv. glycinae at 8 and 24 h after inoculation; SDS PI, SDS Wil and SDS Es, PI567374, Williams 82 and Essex treated by Fv toxin at 8 h after treatment. Atrazine 1, Atrazine 2, Atrazine 4 and Atrazine 8, soybean treated by atrazine herbicide at 1, 2, 4 and 8 h after treatment. Yellow represents up‐regulated genes and blue represents down‐regulated genes.
In silico mapping of differentially expressed transcripts to the soybean genome
The sequences of all the differentially expressed transcripts identified in this study were aligned to the public soybean 7x genome sequence using blastn to define the exact physical locations of each of the transcript sequences on the soybean genome. Likewise, the positions of 10 simple sequence repeat (SSR) markers on the 7x genome sequence linked to F. virguliforme resistance were also obtained. The locations of the transcript sequences were then compared with those of the SSR markers.
The results (Table S11, see Supporting Information) showed that seven of the 10 SSR markers linked with at least four genes at a distance of <550 kb. The seven SSR markers were located in three different LGs: C2 (chromosome 6), D2 (chromosome 17) and G (chromosome 18). Twenty‐two genes selected as differentially expressed in this study (Table S1) were located close to one of the seven SSR markers (Table S10). In LG D2 (chromosome 17), two SSR markers (Sat_222 and Satt389) were only 874 kb away from each other, and thus one (Glyma17g16850.1, BE608541) of five genes is linked to both SSR markers. It is interesting to note that one of these genes (Glyma17g16440.1, BF010339, unknown) was mapped 2 kb away from Sat_222. Three SSR markers were mapped to LG C2 (chromosome 06) where potential defence signalling components are located. These potential defence signalling components include ATP synthase protein I (Glyma06g43060.1), aspartate transaminase (Glyma06g42830.1), fasciclin‐like (Glyma06g46530.2), aconitate hydratase (Glyma06g46190.1), MYB factor (Glyma06g45940.1) and pectinesterase (Glyma06g47200.1). Two SSR markers (Satt038 and Satt130) were mapped in LG G (chromosome 18) of the soybean genome. The cDNA sequences linked to the SSR markers in chromosome 18 include ethylene‐responsive transcription factor (Glyma18g02170.1), lipoxygenase (Glyma18g02330.1), biotin carboxyl (Glyma18g02670.1), coilin protein (Glyma18g01670.1), peroxidase (Glyma06g02300.1), phototropism protein (Glyma11g31500.1) and NADH‐quinone oxidoreductase (Glyma11g31620.1).
It is interesting to note that the transcripts of lipoxygenase (Glyma18g02330.1) and peroxidase (Glyma06g02300.1) were induced only in PI567.374 in response to Fv toxin. None of the seven genes associated with soybean resistance to Fv toxin (Table 1) mapped to any of the mapped SDS resistance SSR markers. These seven genes were located on chromosomes 01, 11, 12, 15 and 16. Glyma06g40790.1 and Glyma11g19920.1 mapped together on chromosome 11. The mapping of these interesting genes in different chromosomes from those harbouring the resistance SSR used here may reflect, in part, the need to map more QTLs linked to SDS resistance.
Discussion
This is the first report of a gene expression study investigating the early response of soybean leaves affected by Fv toxin. A total of 1906 genes were identified as differentially expressed from the three soybean genotypes used in this study. Some of these transcripts were uniquely induced in the resistant genotype PI567.374. In addition, the expression patterns of several genes of interest were compared with the patterns induced by other biotic and abiotic stresses, as well as B. japonicum inoculation. Hierarchical clustering indicated that the defence response to Fv toxin is similar to that elicited by an incompatible P. syringae inoculation, less similar to that induced by a compatible strain, and quite distant to that produced by atrazine stress and B. japonicum inoculation, suggesting that soybean leaves may be activating HR pathways in response to Fv toxin.
In a recent study focusing on the soybean–F. virguliforme interaction, Radwan et al. (2011) reported that soybean roots employed multiple mechanisms to slow down the fungal infection, including genes involved in defence, signalling, secondary metabolism, cell wall modification and oxidative stress. In addition, it was noted that transcripts associated with the onset of PCD were induced in soybean roots challenged by the fungus (Radwan et al., 2011). The initiation of PCD pathways in the roots might be of benefit to the necrotrophic pathogen and, although the most dramatic symptoms induced by the F. virguliforme toxin are in the leaves, we cannot ignore the possible effects of this toxin on soybean roots. Common transcripts are induced both in roots infected by F. virguliforme and in leaves treated by Fv toxin. Some of these shared transcripts belong to nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) R genes, suggesting a potential functional role(s) of resistance proteins during the soybean response to the fungal infection or its toxin(s). One proposed role of R proteins is that they might function as the host receptor of fungal toxins (Lorang et al., 2007; Nagy and Bennetzen, 2008), where pathogen‐produced toxins that are specific to one host, such as victorin produced by Cochliobolus victorae, lead to the deployment of host signalling pathways (Wolpert et al., 2002) triggering PCD to the benefit of the necrotrophic pathogen.
Because soybean roots and leaves may respond differently when the plants are infected with F. virguliforme (both leaves and roots were exposed to Fv toxin, but the fungus colonized only the roots, not the leaves), it would be expected that different signalling pathways might be distinguishable between these two tissues. Comparing the expression responses in the roots (Radwan et al., 2011) to responses in the leaves (current study), we observed that many genes related to hormones were up‐regulated in the roots, e.g. jasmonic acid (12‐oxophytodienoate reductase), ethylene (1‐aminocyclopropane‐1‐carboxylate oxidase and ethylene‐responsive element binding factor 15), jasmonic acid (allene oxide cyclase), auxin (auxin conjugate hydrolase), cytokinin (cytokinin‐induced message) and gibberellic acid (gibberellin 2‐β‐dioxygenase). These results reflect, in part, how these different tissues may employ different resistance mechanisms. The important role of ethylene and jasmonic acid hormones during plant resistance to necrotrophic pathogens has been reported from different genomics and reverse genetics studies (Abuqamar et al., 2008; Berrocal‐Lobo et al., 2002; Calla et al., 2009; Radwan et al., 2011; Thomma et al., 1999; Zhao et al., 2007).
In this study, we benefited from the sequenced genome of soybean by using in silico mapping of differentially expressed transcripts to known QTLs linked to SDS resistance. These transcripts mapped to the LG C2, D2 and G regions that harbour several QTLs to SDS resistance. Some of the putative SDS defence‐related genes identified in this study (Table 1) were not located near a known SDS resistance QTL, and will need to be converted into markers to determine whether they segregate with resistance. Some of the mapped genes encode proteins involved in defence signalling pathways, such as ATP synthase protein I, aspartate transaminase, fasciclin‐like, aconitate hydratase, MYB transcription factor and pectinesterase. It was noted that none of the mapped transcripts from this leaf study were differentially expressed in the root response to F. virguliforme (Radwan et al., 2011), supporting the possible existence of different resistance mechanisms between soybean roots and leaves.
On the basis of comparisons among different genotypes, we have identified four putative SDS defence genes specific to PI567.374 and three shared between this genotype and Williams 82 (Table 1). Further functional annotations based on sequence homology suggest that these genes may encode proteins involved in cell wall modification, detoxification, the defence response and primary metabolism (Table 1), suggesting that multiple mechanisms may be employed by soybean leaves to reduce the damage caused by Fv toxin. These mechanisms can be summarized as follows.
Resistance mechanism through detoxification
Glyma06g40790.1 (AW756020) has strong homology to an aldo/keto reductase (AKR). An AKR protein has been reported to be involved in cellular detoxification (Gavidia et al., 2002), protecting plants against toxic carbonyls and oxidative stress induced by stress stimuli or phytotoxins. For example, the overexpression of an alfalfa AKR gene in tobacco has been shown to increase the tolerance against drought and UV‐B radiation (Hideg et al., 2003). In addition, an AKR protein, NADPH‐dependent carbonyl reductase, has been found to metabolize the HC toxin, a cyclic tetrapeptide from Cochliobolus carbonum race 1 that parasitizes sensitive maize cultivars (Meeley et al., 1992). The putative AKR identified in this SDS study is a high‐priority candidate SDS defence gene, as it could play a role in F. virguliforme detoxification. Further studies are needed to verify this possibility.
Resistance mechanism through strengthening of the host cell wall
Proteins involved in cell wall strengthening play an important role in plant defence against pathogen attack. For example, xyloglucan endotransglycosylase (XET) is a cell wall‐modifying protein (CWMP) that is uniquely involved in both cell wall elongation and rapid reinforcement (Eckardt, 2004; Fry et al., 1992). Increased levels of this protein have been reported to play a key role in defence reactions associated with the incompatible tomato–Cuscuta interaction and root‐knot nematode‐induced galls in A. thaliana (Albert et al., 2004; Jammes et al., 2005).
Lignin, the most abundant biopolymer on earth following cellulose, serves as a main structural component of the primary cell wall (Walter et al., 1988). Lignin is resistant to degradation by most microbes, and therefore lignin deposition is often employed by plants in response to infection. Lignin deposition can also be stimulated by extracellular elicitors produced by fungi (Robertsen, 1986). Extensins are another class of cell wall‐modifying enzyme that may be induced by plant pathogens. For example, lignin is deposited in wheat leaves challenged by Puccinia graminis (Li et al., 2001; Menden et al., 2006), and extensin is induced in melons on attempted infection by Colletotrichum lindemuthianum (Ahn et al., 1996). In our study, we identified the differential regulation of Glyma11g19920.1 (BF070757), a XET homologue, and one lignin biosynthesis‐related gene, Glyma01g02580.1 (BG043573), encoding cinnamyl alcohol dehydrogenase, in the SDS‐resistant genotype PI567.374 on F. virguliforme filtrate treatment. In addition, two cell wall‐related genes, Glyma02g12140.1 (BG043510) and Glyma01g06030.2 (BE610339), encoding expansin 3 and expansin 4, respectively, exhibited significantly higher levels of expression in resistant PI567.374 than in the other two genotypes on Fv toxin treatment. All of these proteins involved in cell wall strengthening might help resistant line PI567.374 to develop a better defence.
Resistance mechanism through common plant defence responses
Two of the seven genes identified as related to resistance in this study possess sequence homology to NBS‐LRR R genes. Glyma16g34070.1 (BG510438) has high homology to soybean R1, whereas Glyma12g01420.1 (BG510920 and BG509368) has homology to various plant R genes, such as RPP8 and RPP13. These transcripts were induced significantly in resistant lines treated with Fv toxin and also in an incompatible P. syringae inoculation (Zou et al., 2005). All of these R genes are members of the NBS‐LRR class of plant resistance genes (Bittner‐Eddy et al., 2000; Graham et al., 2002; Hammond‐Kosack and Jones, 1997). Proteins of this class recognize either directly or indirectly the effector proteins of a wide range of plant pathogens. Several different R gene‐mediated signalling networks have been proposed through the analysis of A. thaliana mutants (Feys and Parker, 2000). Some R proteins require EDS1 or NDR1 (non‐race specific disease resistance‐1) to mediate a strong resistance response, whereas others can function independently (Bittner‐Eddy and Beynon, 2001). In this study, we identified one highly expressed EDS1 gene, Glyma06g19890.1 (BG790192), in all genotypes treated by fungal filtrates, and this transcript was highly expressed constitutively in PI567.374 relative to the other two varieties on mock treatment. These data support the hypothesis that EDS1 might play a role in the full function of one or more R proteins in the resistance response to SDS toxin.
Although this work focuses on the identification of promising candidate genes that may be involved in multiple mechanisms deployed by soybean to detoxify and/or reduce the damaging effect of Fv toxin, other efforts have been invested to study the chemical nature of the toxin (Ji et al., 2006; Jin et al., 1996). One of these studies (Ji et al., 2006) suggested that Fv toxin and victorin cause damage through a common mechanism. Several similarities were noted between the physiological changes that occurred in their respective hosts, including: (i) degradation of the Rubisco large subunit; (ii) accumulation of free radicals under light conditions; (iii) initiation of PCD; and (iv) triggering of necrosis in leaves. A protein, FvTox1, and its corresponding gene, have been identified from F. virguliforme culture filtrates that induce chlorosis and necrosis (Brar et al., 2011). In an attempt to functionally characterize this protein, Brar et al. (2011) expressed the FvTox1 protein in insect cells. When infiltrated into leaves, the recombinant FvTox1 protein resulted in chlorosis and necrosis in susceptible soybean plants in the presence of light, suggesting the production of free radicals by the interruption of photosynthesis. In addition, the expression of antibodies that bind FvTox1 in transgenic soybean plants led to reduced SDS symptoms, suggesting that FvTox1 is an important virulence factor contributing to foliar SDS development (Brar and Bhattacharyya, 2012). The analysis of an F. virguliforme strain specifically mutated in FvTox1 has yet to be reported. It would be interesting to identify the soybean receptor protein that recognizes FvTox1.
In conclusion, in this work, we identified many candidate SDS defence‐related genes. Seven genes were specific to the more resistant genotypes and are primary candidates for future functional characterization and more refined QTL association mapping. It would be interesting to expand the comparative gene expression profiling of soybean roots and leaves on infection by the pathogen and treatment by pathogen phytotoxins to assess the potential role of the fungal toxins in the initiation and triggering of PCD in soybean roots.
Experimental Procedures
Soybean plant growth and Fv culture filtrate production
Soybean seeds of three genotypes, PI567.374, Williams 82 and Essex, were sown in plastic pots (8.5 × 8.5 × 7.5 cm; Invitrogen/Life Technologies, Grand Island, NY, USA) filled with soil‐less growing medium (Sunshine Mix, LC1, Sun Gro Horticulture Inc., Bellevue, WA, USA). The soybean plants were grown in a growth chamber at 26 °C (day) and 22 °C (night) under a 14‐h day length with an approximate light intensity of 150 μmol photons/m2/s. The culture filtrates of F. virguliforme (isolate Mont‐1) were obtained from cultures grown for 12 days at 25 °C in potato dextrose broth (PDB) liquid medium following the protocol described by Hartman et al. (2004). Cultures were filtered through a 0.22‐μm membrane (Millipore, Billerica, MA, USA) to remove any possible viable fungal structures.
Fungal culture filtrate treatment
Plant treatments involved the immersion of stems of soybean seedlings into cell‐free culture filtrate, as described elsewhere (Hartman et al., 2004) with some minor modifications. In brief, plants were grown in growth chambers until the first trifoliolates were fully expanded (approximately 14 days). Stems of soybean seedlings were cut just above the cotyledonary nodes and unifoliates were removed. Stems of cut soybean seedlings were immersed (3–5 cm) in 50‐mL conical polypropylene centrifuge tubes containing 30 mL of fresh, sterile‐filtered Fv medium diluted 1:30 in sterile water. For mock treatment, stems of cut soybean seedlings were immersed in tubes containing 30 mL of noninoculated PDB medium diluted 1:30 in water. Just after immersion of the cut seedlings into the filtrate, tube openings were wrapped with parafilm to reduce evaporation of the solution, and the plants were placed in the dark for 4 h to prevent severe wilting that would occur if freshly cut plants were placed immediately under bright light.
Plant sample collection and RNA extraction
Samples were collected at 8 hpt, well before symptom development. Therefore, an approach was devised to ensure the efficacy of toxin treatment. The two lateral leaflets of a trifoliate were collected for the microarray study and qRT‐PCR, leaving the middle leaflet to remain on the plant for three additional days to allow the verification of toxin efficacy by observing symptom development. Samples from plants in which the remaining leaflet eventually developed appropriate symptoms were pooled (leaflets from four to five plants per pool) for total RNA isolation. The entire experiment was repeated three times to obtain three independent biological replicates. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and verified for quality as described previously (Zou et al., 2005)
cDNA microarray procedure
The soybean microarray cDNA library Gm‐r1088 (Vodkin et al., 2004), which provided probe sets for a total of 9216 clones and 64 control clones, was used for this study. Library Gm‐r1088 includes 274 cDNAs that originated from leaves of Williams 82 inoculated with P. syringae pv. glycinae carrying avrB for an incompatible HR, and 365 and 324 cDNAs that originated from SDS‐treated leaves of PI567.374 and Williams 82, respectively.
Fluorescently labelled probes were prepared following the protocol employed by Thibaud‐Nissen et al. (2003) using 50 μg of purified total RNA as template for reverse transcription in the presence of cyanine 3‐deoxyuridine triphosphate (Cy3‐dUTP) or Cy5‐dUTP (Invitrogen). Labelled probes were combined based on the microarray experimental design (Fig. 3), and then mixed with hybridization buffer and hybridized to microarray slides, incubated and washed. Finally, the microarray slides were scanned for fluorescence emission with a Perkin‐Elmer (Foster City, CA, USA) ScanArray Express microarray scanner. Images were analysed after scanning with GenePix Pro (version 4.0, Axon Instruments, Union City, CA, USA).
Microarray data analysis
Raw data were normalized, flagged and transformed using in‐house software (Brechenmacher et al., 2008). The merged file was normalized with R‐MAANOVA (Wu et al., 2003) using global lowess or ‘glowess’ (Wu et al., 2003) as the normalization method. After normalization, the weak spots (spots with an average intensity of Cy3 and Cy5 lower than the intensity of the negative control X13988) were replaced with the median of the negative control X13988, thereby reducing the possible effect of the high variability associated with very weak spots on the analysis. Statistical Analysis Software (SAS Institute, Cary, NC, USA) was used to run the analysis of variance (ANOVA) on a per gene basis. The following linear mixed model with array as a random effect and sample as a fixed effect was used to analyse the data (the dye effect was needed in the model as it was ‘unbalanced’ in the design):
where Yijkl is the median of the signal intensity for the individual spot and channel, μ is the average log2 signal intensity, Ai is the array effect (random), Dj is the dye effect (Cy3 or Cy5), Vk is the variety effect (PI567.374, W82 or Essex), Tl is the treatment effect (Fv toxin or water), (DV)jk is the interaction between dye and variety effects, (DT)jl is the interaction between dye and treatment effects, (VT)kl is the interaction between variety and treatment effects, (DVT)jkl is the interaction between dye, variety and treatment effects, and ε is the residual error. The differentially expressed genes were selected on the basis of both the fold‐change ratio (≥1.5‐fold or ≤0.667‐fold) and the FDR‐corrected P value ≤ 0.05.
RT‐PCR and qRT‐PCR procedures
RT‐PCR and qRT‐PCR were performed according to Radwan et al. (2011). In brief, total RNA was treated with DNase I (Invitrogen) to remove genomic DNA contamination. Two micrograms of DNase‐treated RNA were reverse transcribed using the SuperScript III First‐Strand Synthesis System for RT‐PCR (Invitrogen). A ‘minus’ reverse transcriptase PCR, where no reverse transcriptase enzyme was added, was used to ensure that each mRNA sample was free from genomic DNA. The resulting cDNA was adjusted to a final concentration of 30 ng/μL, and 2 μL was used in 20 μL of the PCR mix, which contained 0.2 μm of each primer, 8.9 μL of Brilliant SYBR Green PCR master mix (Stratagene/Agilent, Santa Clara, CA, USA) and 7.5 μL of water. The primers for the target genes (Table S2) were designed using Primer 3.0 software (http://fokker.wi.mit.edu/primer3/input.htm). The expression of a soybean β‐actin gene (Glyma08g19420.1) was used as the internal standard to normalize the small difference in template amounts (Table S2). PCR was carried out in an Mx3005p Thermal Cycler (Stratagene/Agilent) under the following conditions: an initial denaturation at 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C, 20 s at TM (Melting temperature estimation for the primer pairs used in each PCR reaction) (°C) (Table S2) and 30 s at 72 °C. The specificity of the primers was validated by the presence of a single peak in the dissociation curve analyses and by sequencing the PCR products. The relative quantitative abundance of each investigated gene transcript was calculated by comparison of the expression of each targeted gene with the expression of a soybean reference β‐actin gene using the delta–delta method (McMaugh and Lyon, 2003). Three biological and technical replicates were conducted on each treatment and mock control.
In silico mapping of differentially expressed genes in soybean
Differentially expressed genes identified from this study were in silico mapped to the soybean genome (http://www.phytozome.net/soybean) using 10 SSR markers (Satt371, Satt307, Satt277, Satt038, Satt130, Satt528, Satt574, Satt311, Sat_222 and Satt389) These SSR markers were genetically mapped to the confirmed QTLs linked to F. virguliforme resistance (Farias‐Neto et al., 2007; Iqbal et al., 2001; Kazi et al., 2008; Njiti et al., 2002). Bioinformatics analysis followed that described previously (Calla et al., 2009; Radwan et al., 2011). The locations of SSR markers and cDNA sequences were then compared with each other to identify cDNA located on the same chromosome positioned within 550 kb of an SSR marker.
Supporting information
Fig. S1 Bar chart of categories of significantly differentially expressed genes from the experiment; 1906 genes were sorted into 17 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S2 Functional categorization of common toxin‐induced genes in all three genotypes (total of 405 genes, Table S5A). Genes were sorted into 16 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S3 Functional categorization of common toxin‐repressed genes in all three varieties (total of 93 genes, Table S5B). Genes were sorted into 13 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S4 Hierarchical cluster analysis of the differential expression profiles of the genes expressed at a higher level in PI567.734 than in the other two varieties. Sixty‐six genes were expressed at a higher level in PI567.734 than in Williams 82 and Essex, and were clustered among a total of 19 experiments. Nod, nodulation when inoculated with Bradyrhizobium japonicum; VIR and HR, compatible and incompatible Pseudomonas syringae inoculations; PIVEI, PIVWI, WIVEI, PIVPIU, WIVWU, EIVEU, PIUVWU, WUVEU and PIUVEU, PI–EI, PI–WI, WI–EI, PI–PU, WI–WU, EI–EU, PU–WU, WU–EU and PU–EU, respectively; AW, atrazine versus water treatment, 1,2,4 and 8 h post application. A, Cluster A; B, Cluster B. P, PI567.374; W, Williams 82; E, Essex; V, versus; I, plant treated by Fusarium virguliforme phytotoxin (Fv toxin); U, mock.
Fig. S5 Hierarchical cluster analysis of shared genes between PI–EI and WI–EI. (A) Fifteen of the 16 genes that were induced in Williams 82 and PI567.374 on Fusarium virguliforme phytotoxin (Fv toxin) treatment were clustered with the 19 treatments as defined in the legend of Fig. S4. (B) Hierarchical clustering of shared genes between PI–PU and WI–WU. Four of the 44 genes were identified as commonly induced in PI567.374 and Williams 82, and were clustered with the 19 treatments as defined in the legend of Fig. S4. P, PI567374; W, Williams 82; E, Essex, I, plant treated by Fv toxin; U, mock.
Table S1 Significantly differentially expressed genes from the whole experiment. A gene is considered to be significantly differentially expressed if it has both significance [false discovery rate (FDR) P value of ≤0.05] and a fold‐change ratio of ≥1.5 or ≤0.667. P, PI567374; W, Williams 82; E, Essex; I, plant treated with Fusarium virguliforme phytotoxin (Fv toxin); U, mock; v, versus. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S2 Primer name, gene annotation, GenBank ID and matching soybean gene, primer sequence, estimated melting temperature for each primer pair, TM (°C) and amplicon size (bp) of primers used in quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). For, forward primer; Rev, reverse primer.
Table S3 Cross comparison of gene expression profiles between microarrays and quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). The expression profiles of these genes were determined by qRT‐PCR at 6, 8 and 24 h post‐treatment (hpt). Expression levels are given as the log2‐transformed fold change of Fusarium virguliforme (Fv) filtrate‐treated soybeans compared with mock treatment. SE, standard error.
Table S4 Significantly differentially expressed genes from the three genotypes treated with Fusarium virguliforme phytotoxin (Fv toxin). Expression levels are calculated as the ratio of Fv toxin‐treated genotype to the same mock‐treated genotype. PI, PI567.374 plants treated by Fv toxin; PU, PI567.374 mock‐treated plants; WI, Williams 82 plants treated by Fv toxin; WU, Williams 82 mock‐treated plants; EI, Essex plants treated by Fv toxin; EU, Essex mock‐treated plants. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S5 Up‐ and down‐regulated genes shared among the three genotypes treated by Fusarium virguliforme phytotoxin (Fv toxin). Expression levels are calculated as the ratio of filtrate‐treated genotype to the same mock‐treated variety. PI, PI567.374 plants treated by Fv toxin; PU, PI567.374 mock‐treated plants; WI, Williams 82 plants treated by Fv toxin; WU, Williams 82 mock‐treated plants; EI, Essex plants treated by Fv toxin; EU, Essex mock‐treated plants.
Table S6 Toxin‐induced and repressed genes in resistant genotypes (shared only between PI–PU and WI–WU). Expression levels are calculated as the ratio of Fusarium virguliforme phytotoxin (Fv toxin)‐treated genotype to the same mock‐treated genotype. P, PI567374; W, Williams 82; I, plant treated by Fv toxin; U, mock.
Table S7 Toxin‐induced and repressed genes unique to resistant PI567.374 (the most resistant genotype) on treatment with Fusarium virguliforme phytotoxin (Fv toxin).
Table S8 Gene expression comparison between soybean roots infected by Fusarium virguliforme (Fv) (Radwan et al., 2011) at 7 days post‐inoculation (dpi) and soybean leaves treated by Fv toxin at 8 h post‐treatment (hpt) (current study). PI, PI567.374; E, Essex. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S9 Up‐regulated genes in the three genotypes without Fusarium virguliforme phytotoxin (Fv toxin) treatment. P, PI567374; W, Williams 82; E, Essex; U, mock.
Table S10 Resistance‐related genes (up‐regulated genes shared between PI–WI and PI–EI and between PI–EI and WI–EI). Expression levels were calculated as the ratio of Fusarium virguliforme phytotoxin (Fv toxin)‐treated genotype to another different Fv toxin‐treated genotype. P, PI567374; W, Williams 82; E, Essex; I, plant treated by Fv toxin.
Table S11 Physical mapping of simple sequence repeat (SSR) markers linked to Fusarium virguliforme (Fv) resistance and cDNA arrays with differential abundance [false discovery rate (FDR) P value of <0.05]. cDNA sequences mapping <550 kbp away from SSR markers are presented. P, PI567374; W, Williams 82; E, Essex; I, plant treated by Fv toxin; U, mock; v, versus. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Acknowledgements
The authors thank Yinping Qin for assistance with the toxin assay, and David Walker for use of the qRT‐PCR machine. This work was financed by the United States Department of Agriculture‐Agricultural Research Service (USDA‐ARS) Current Research Information System (#3611–21000‐018‐00D) as well as by grants from the University of Illinois Soybean Disease Biotechnology Center and the National Science Foundation Plant Genome Research Program (# DBI‐0421620). The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
References
- Abuqamar, S. , Chai, M.F. , Luo, H. , Song, F. and Mengiste, T. (2008) Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell, 20, 1964–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, J.H. , Choi, Y. , Kwon, Y.M. , Kim, S.G. , Choi, Y.D. and Lee, J.S. (1996) A novel extensin gene encoding a hydroxyproline‐rich glycoprotein requires sucrose for its wound‐inducible expression in transgenic plants. Plant Cell, 8, 1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, M. , Werner, M. , Proksch, A. , Fry, S.C. and Kaldenhoff, R. (2004) The cell wall‐modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa . Plant Biol. (Stuttg.) 6, 402–407. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. , Molina, A. and Solano, R. (2002) Constitutive expression of ETHYLENE‐RESPONSE‐FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Bittner‐Eddy, P.D. and Beynon, J.L. (2001) RPP13‐Nd an Arabidopsis LZ‐NBS‐LRR type resistance gene that is salicylic acid‐independent and that does not require disease resistance signalling pathways defined by EDS1 or NDR1. MPMI, 14, 416–421. [DOI] [PubMed] [Google Scholar]
- Bittner‐Eddy, P.D. , Crute, I.R. , Holub, E.B. and Beynon, J.L. (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica . Plant J. 21, 189–214. [DOI] [PubMed] [Google Scholar]
- Brar, H.K. and Bhattacharyya, M.K. (2012) Expression of a single‐chain variable‐fragment antibody against a Fusarium virguliforme toxin peptide enhances tolerance to sudden death syndrome in transgenic soybean plants. MPMI 25, 817–824. [DOI] [PubMed] [Google Scholar]
- Brar, H.K. , Swaminathan, S. and Bhattacharyya, M.K. (2011) The Fusarium virguliforme toxin FvTox1 causes foliar sudden death syndrome‐like symptoms in soybean. MPMI 24, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Brechenmacher, L. , Kim, M.Y. , Benitez, M. , Li, M. , Joshi, T. , Calla, B. , Lee, M.P. , Libault, M. , Vodkin, L.O. , Xu, D. , Lee, S.H. , Clough, S.J. and Stacey, G. (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum . MPMI 21, 631–645. [DOI] [PubMed] [Google Scholar]
- Calla, B. , Vuong, T. , Radwan, O. , Hartman, G.L. and Clough, S.J. (2009) Gene expression profiling soybean stem tissue early response to Sclerotinia sclerotiorum and in silico mapping in relation to resistance markers. Plant Genome, 2, 149–166. [Google Scholar]
- Choi, J.J. , Alkharouf, N.W. , Schneider, K.T. , Matthews, B.F. and Frederick, R.D. (2008) Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Funct. Integr. Genomics, 8, 341–359. [DOI] [PubMed] [Google Scholar]
- Eckardt, N.A. (2004) Inside the matrix: crystal structure of a xyloglucan endotransglycosylase. The Plant Cell, 16, 792–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias Neto, A.F. , Hashmi, R. , Schmidt, M. , Carlson, S.R. , Hartman, G.L. , Li, S. , Nelson, R.L. and Diers, B.W. (2007) Mapping and confirmation of a new sudden death syndrome resistance QTL on linkage group D2 from the soybean genotypes PI 567374 and ‘Ripley’. Mol. Breeding, 20, 53–62. [Google Scholar]
- Feys, B.J. and Parker, J.E. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Fry, S.C. , Smith, R.C. , Renwick, K.F. , Martin, D.J. , Hodge, S.K. and Matthews, K.J. (1992) Xyloglucan endotransglycosylase, a new wall‐loosening enzyme activity from plants. Biochem. J. 282, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavidia, I. , Pérez‐Bermúdez, P. and Seitz, H.U. (2002) Cloning and expression of two novel aldo‐keto reductases from Digitalis purpurea leaves. Eur. J. Biochem. 269, 2842–2850. [DOI] [PubMed] [Google Scholar]
- Graham, M.A. , Marek, L.F. and Shoemaker, R.C. (2002) Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics, 162, 1961–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K. and Jones, J.D. (1997) Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Hartman, G.L. , Huang, Y.H. , Nelson, R.L. and Noel, G.R. (1997) Germplasm evaluation of Glycine max for resistance to Fusarium solani the causal organism of sudden death syndrome. Plant Dis. 81, 515–518. [DOI] [PubMed] [Google Scholar]
- Hartman, G.L. , Huang, Y.H. and Li, S. (2004) Phytotoxicity of Fusarium solani culture filtrates from soybean and other hosts assayed by stem cuttings. Australas. Plant Pathol. 33, 9–15. [Google Scholar]
- Hideg, E. , Nagy, T. , Oberschall, A. , Dudits, D. and Vass, I. (2003) Detoxification function of aldose/aldehyde reductase during drought and ultraviolet‐B (280–320 nm) stresses. Plant Cell Environ. 26, 513–522. [Google Scholar]
- Hnetkovsky, N. , Chang, S.J.C. , Doubler, T.W. , Gibson, P.T. and Lightfoot, D.A. (1996) Genetic mapping of loci underlying field resistance to soybean sudden death syndrome (SDS). Crop Sci. 36, 393–400. [Google Scholar]
- Iqbal, M.J. , Meksem, K. , Njiti, V.N. , Kassem, M.A. and Lightfoot, D.A. (2001) Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden death syndrome (SDS) in Essex × Forrest RILs. Theor. Appl. Genet. 102, 187–192. [Google Scholar]
- Iqbal, M.J. , Yaegashi, S. , Ahsan, R. , Shopinski, K. and Lightfoot, D.A. (2005) Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 110, 1429–1438. [DOI] [PubMed] [Google Scholar]
- Jammes, F. , Lecomte, P. , Almeida‐Engler, J. , Bitton, F. , Martin‐Magniette, M.L. , Renou, J.P. , Abad, P. and Favery, B. (2005) Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis. Plant J. 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Ji, J. , Scott, M.P. and Bhattacharyya, M.K. (2006) Light is essential for degradation of ribulose‐1,5‐bisphosphate carboxylase‐oxygenase large subunit during sudden death syndrome development in soybean. Plant Biol. 8, 597–605. [DOI] [PubMed] [Google Scholar]
- Jin, H. , Hartman, G.L. , Nickell, C.D. and Widholm, J.M. (1996) Characterization and purification of a phytotoxin produced by Fusarium solani, the causal agent of soybean sudden death syndrome. Phytopathology, 86, 227–282. [Google Scholar]
- Kazi, S. , Shultz, J. , Afzal, J. , Johnson, J. , Njiti, V.N. and Lightfoot, D.A. (2008) Separate loci underlie resistance to root infection and leaf scorch during soybean sudden death syndrome. Theor. Appl. Genet. 116, 967–977. [DOI] [PubMed] [Google Scholar]
- Li, H.P. , Fischer, R. and Liao, Y.C. (2001) Molecular evidence for induction of phenylalanine ammonia‐lyase during Puccinia graminis infection and elicitation in wheat. Can. J. Plant Pathol. 23, 286–291. [Google Scholar]
- Li, S. , Hartman, G.L. and Widholm, J.M. (1999) Viability staining of soybean suspension cultured cells and a stem‐cutting assay to evaluate phytotoxicity of Fusarium solani culture filtrates. Plant Cell Rep. 18, 375–380. [Google Scholar]
- Li, Y. , Zou, J. , Li, M. , Bilgin, D. , Vodkin, L.O. , Hartman, G.L. and Clough, S.J. (2008) Soybean defense responses to the soybean aphid. New Phytol. 179, 185–195. [DOI] [PubMed] [Google Scholar]
- Libault, M. , Farmer, A. , Brechenmacher, L. , Drnevich, J. , Langley, R.J. , Bilgin, D.D. , Radwan, O. , Neece, D.J. , Clough, S.J. , May, G.D. and Stacey, G. (2010) Complete transcriptome of the soybean root hair cell, a single cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot, D.A. , Njiti, V.N. , Gibson, P.T. , Kassem, M.A. , Iqbal, J.M. and Meksem, K. (2005) Registration of the Essex–Forrest recombinant inbred line mapping population. Crop Sci. 45, 1678–1681. [Google Scholar]
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl. Acad. Sci. USA, 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovaya, V.V. , Lygin, A.V. , Zernova, O.V. , Li, S. , Hartman, G.L. and Widholm, J.M. (2004) Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusarium solani . Plant Physiol. Biochem. 42, 671–679. [DOI] [PubMed] [Google Scholar]
- McMaugh, S.J. and Lyon, B.R. (2003) Real‐time quantitative RT‐PCR assay of gene expression in plant roots during fungal pathogenesis. Biotechniques, 34, 982–986. [DOI] [PubMed] [Google Scholar]
- Meeley, R.B. , Johal, G.S. , Briggs, S.P. and Walton, J.D. (1992) A biochemical phenotype for a disease resistance gene of maize. Plant Cell, 4, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B.M. (2006) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. J. Phytochem. 68, 513–520. [DOI] [PubMed] [Google Scholar]
- van de Mortel, M. , Recknor, J.C. , Graham, M.A. , Nettleton, D. , Dittman, J.D. , Nelson, R.T. , Godoy, C.V. , Abdelnoor, R.V. , Almeida, A.M. , Thomas, J. , Baum, T.J. and Whitham, S.A. (2007) Distinct biphasic mRNA changes in response to Asian soybean rust infection. MPMI, 20, 887–899. [DOI] [PubMed] [Google Scholar]
- Moy, P. , Qutob, D. , Chapman, B.P. , Atkinson, I. and Gijzen, M. (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae . MPMI, 17, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Nagy, E.D. and Bennetzen, J.L. (2008) Pathogen corruption and site‐directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiti, V.N. , Shenaut, M.A. , Suttner, R.J. , Schmidt, M.E. and Gibson, P.T. (1996) Soybean response to sudden death syndrome: inheritance influenced by cyst nematode resistance in Pyramid × Douglas progenies. Crop Sci. 36, 1165–1170. [Google Scholar]
- Njiti, V.N. , Meksem, K. , Iqbal, M.J. , Johnson, J.E. , Kassem, M.A. , Zobrist, K.F. , Kilo, V.Y. and Lightfoot, D.A. (2002) Common loci underlie Weld resistance to soybean sudden death syndrome in Forrest, Pyramid, Essex, and Douglas. Theor. Appl. Genet. 104, 294–300. [DOI] [PubMed] [Google Scholar]
- Panthee, D. , Marois, J. , Wright, D. , Narvaez, D. , Yuan, J. and Stewart, C. (2009) Differential expression of genes in soybean in response to the causal agent of Asian soybean rust (Phakopsora pachyrhizi Sydow) is soybean growth stage‐specific. Theor. Appl. Genet. 118, 359–370. [DOI] [PubMed] [Google Scholar]
- Panthee, D.R. , Yuan, J.S. , Wright, D.L. , Marois, J.J. , Mailhot, D. and Stewart, C.N. (2007) Gene expression analysis in soybean in response to the causal agent of Asian soybean rust (Phakopsora pachyrhizi Sydow) in an early growth stage. Funct. Integr. Genomics, 7, 291–301. [DOI] [PubMed] [Google Scholar]
- Radwan, O. , Liu, Y. and Clough, S.J. (2011) Transcriptional analysis of soybean roots response to Fusarium virguliforme, the causal agent of sudden death syndrome. MPMI, 24, 958–972. [DOI] [PubMed] [Google Scholar]
- Robertsen, B. (1986) Elicitors of the production of lignin‐like compounds in cucumber hypocotyls. Physiol. Mol. Plant Pathol. 28, 137–148. [Google Scholar]
- Roy, K.W. , Rupe, J.C. , Herdhman, D.E. and Abney, T.S. (1997) Sudden death syndrome of soybean. Plant Dis. 81, 1100–1111. [DOI] [PubMed] [Google Scholar]
- Rupe, J.C. and Hartman, G.L. (1999) Sudden death syndrome In: Compendium of Soybean Diseases, 4th edn (Hartman G.L., Sinclair J.B. and Rupe J.C., eds), pp. 37–39. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Rupe, J.C. , Hirrel, M.C. and Hershman, D.E. (1989) Sudden death syndrome In: Compendium of Soybean Diseases, 3rd edn (Sinclair J.B. and Backman P.A., eds), pp. 84–85. St. Paul MN: American Phytopathological Society. [Google Scholar]
- Thibaud‐Nissen, F. , Shealy, R.T. , Khanna, A. and Vodkin, L.O. (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol. 132, 118–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Tierens, K.F. and Broekaert, W.F. (1999) Requirement of functional ethylene‐insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea . Plant Physiol. 121, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triwitayakorn, K. , Njiti, V.N. , Iqbal, M.J. , Yaegashi, S. , Town, C.D. and Lightfoot, D.A. (2005) Genomic analysis of a region encompassing QRfs1 and QRfs2, genes that underlie soybean resistance to sudden death syndrome. Genome, 48, 125–138. [DOI] [PubMed] [Google Scholar]
- Vodkin, L.O. , Kahnna, A. , Shealy, R.T. , Clough, S.J. , Gonzales, D.O. , Philip, R. , Zabala, G.C. , Thibaud‐Nissen, F. , Sidarous, M. , Stromvik, M.V. , Shoop, E.G. , Schmidt, C. , Retzel, E. , Erpelding, J. , Shoemaker, R.C. , Rodrizues‐Huete, A.M. , Polacco, J.C. , Coryell, V. , Keim, P. , Gong, G. , Lui, L. , Pardinas, J.L. and Schweitzer, P. (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. Genomics, 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M.H. , Grima‐Pettenati, J. , Grand, C. , Boudet, A.M. and Lamb, C.J. (1988) Cinnamyl‐alcohol dehydrogenase, a molecular marker specific for lignin synthesis: cDNA cloning and mRNA induction by fungal elicitor. Proc. Natl. Acad. Sci. USA, 85, 5546–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert, T.J. , Dunkle, I.D. and Ciuffetti, L.M. (2002) Host‐selective toxin and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Wu, H. , Kerr, M.K. , Cui, X. and Churchill, G.A. (2003) MAANOVA: a software package for the analysis of spotted cDNA microarray experiments In: The Analysis of Gene Expression Data: Methods and Softwares (Zeger S.L., ed.), pp. 313–341. New York: Springer. [Google Scholar]
- Yuan, J. , Mengxia, Z.M. , Lightfoot, D.A. , Iqbal, M.J. , Yang, J.Y. and Meksem, K. (2008) In silico comparison of transcript abundances during Arabidopsis thaliana and Glycine max resistance to Fusarium virguliforme . BMC Genomics, 9 (Suppl. 2), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, J. , An, L. , Doerge, R.W. , Chen, Z.J. , Grau, C.R. , Meng, J. and Osborn, T.C. (2007) Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus . Planta, 227, 13–24. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Patzoldt, W.L. , Shealy, R.T. , Vodkin, L.O. , Clough, S.J. and Tranel, P.J. (2008) Transcriptome response to glyphosate in sensitive and resistant soybean. J. Agric. Food Chem. 56, 6355–6363. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Patzoldt, W.L. , Radwan, O. , Tranel, P.J. and Clough, S.J. (2009) Effects of D1 interfering herbicides atrazine and bentazon on the soybean transcriptome. The Plant Genome, 2, 191–205. [Google Scholar]
- Zou, J. , Rodriguez‐Zas, S. , Aldea, M. , Li, M. , Zhu, J. , Gonzales, D.O. , Vodkin, L.O. , DeLucia, E. and Clough, S.J. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense‐related genes and rapid HR‐specific down regulation of photosynthesis. MPMI, 18, 1161–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Bar chart of categories of significantly differentially expressed genes from the experiment; 1906 genes were sorted into 17 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S2 Functional categorization of common toxin‐induced genes in all three genotypes (total of 405 genes, Table S5A). Genes were sorted into 16 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S3 Functional categorization of common toxin‐repressed genes in all three varieties (total of 93 genes, Table S5B). Genes were sorted into 13 different categories of biological function. Sequences showing no homology to previously described genes were classified into the ‘No hits’ category, sequences with homology to predicted proteins of unknown function were classified into the ‘Unknown’ category, and sequences showing homology with several functions were classified into the ‘Miscellaneous’ category.
Fig. S4 Hierarchical cluster analysis of the differential expression profiles of the genes expressed at a higher level in PI567.734 than in the other two varieties. Sixty‐six genes were expressed at a higher level in PI567.734 than in Williams 82 and Essex, and were clustered among a total of 19 experiments. Nod, nodulation when inoculated with Bradyrhizobium japonicum; VIR and HR, compatible and incompatible Pseudomonas syringae inoculations; PIVEI, PIVWI, WIVEI, PIVPIU, WIVWU, EIVEU, PIUVWU, WUVEU and PIUVEU, PI–EI, PI–WI, WI–EI, PI–PU, WI–WU, EI–EU, PU–WU, WU–EU and PU–EU, respectively; AW, atrazine versus water treatment, 1,2,4 and 8 h post application. A, Cluster A; B, Cluster B. P, PI567.374; W, Williams 82; E, Essex; V, versus; I, plant treated by Fusarium virguliforme phytotoxin (Fv toxin); U, mock.
Fig. S5 Hierarchical cluster analysis of shared genes between PI–EI and WI–EI. (A) Fifteen of the 16 genes that were induced in Williams 82 and PI567.374 on Fusarium virguliforme phytotoxin (Fv toxin) treatment were clustered with the 19 treatments as defined in the legend of Fig. S4. (B) Hierarchical clustering of shared genes between PI–PU and WI–WU. Four of the 44 genes were identified as commonly induced in PI567.374 and Williams 82, and were clustered with the 19 treatments as defined in the legend of Fig. S4. P, PI567374; W, Williams 82; E, Essex, I, plant treated by Fv toxin; U, mock.
Table S1 Significantly differentially expressed genes from the whole experiment. A gene is considered to be significantly differentially expressed if it has both significance [false discovery rate (FDR) P value of ≤0.05] and a fold‐change ratio of ≥1.5 or ≤0.667. P, PI567374; W, Williams 82; E, Essex; I, plant treated with Fusarium virguliforme phytotoxin (Fv toxin); U, mock; v, versus. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S2 Primer name, gene annotation, GenBank ID and matching soybean gene, primer sequence, estimated melting temperature for each primer pair, TM (°C) and amplicon size (bp) of primers used in quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). For, forward primer; Rev, reverse primer.
Table S3 Cross comparison of gene expression profiles between microarrays and quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). The expression profiles of these genes were determined by qRT‐PCR at 6, 8 and 24 h post‐treatment (hpt). Expression levels are given as the log2‐transformed fold change of Fusarium virguliforme (Fv) filtrate‐treated soybeans compared with mock treatment. SE, standard error.
Table S4 Significantly differentially expressed genes from the three genotypes treated with Fusarium virguliforme phytotoxin (Fv toxin). Expression levels are calculated as the ratio of Fv toxin‐treated genotype to the same mock‐treated genotype. PI, PI567.374 plants treated by Fv toxin; PU, PI567.374 mock‐treated plants; WI, Williams 82 plants treated by Fv toxin; WU, Williams 82 mock‐treated plants; EI, Essex plants treated by Fv toxin; EU, Essex mock‐treated plants. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S5 Up‐ and down‐regulated genes shared among the three genotypes treated by Fusarium virguliforme phytotoxin (Fv toxin). Expression levels are calculated as the ratio of filtrate‐treated genotype to the same mock‐treated variety. PI, PI567.374 plants treated by Fv toxin; PU, PI567.374 mock‐treated plants; WI, Williams 82 plants treated by Fv toxin; WU, Williams 82 mock‐treated plants; EI, Essex plants treated by Fv toxin; EU, Essex mock‐treated plants.
Table S6 Toxin‐induced and repressed genes in resistant genotypes (shared only between PI–PU and WI–WU). Expression levels are calculated as the ratio of Fusarium virguliforme phytotoxin (Fv toxin)‐treated genotype to the same mock‐treated genotype. P, PI567374; W, Williams 82; I, plant treated by Fv toxin; U, mock.
Table S7 Toxin‐induced and repressed genes unique to resistant PI567.374 (the most resistant genotype) on treatment with Fusarium virguliforme phytotoxin (Fv toxin).
Table S8 Gene expression comparison between soybean roots infected by Fusarium virguliforme (Fv) (Radwan et al., 2011) at 7 days post‐inoculation (dpi) and soybean leaves treated by Fv toxin at 8 h post‐treatment (hpt) (current study). PI, PI567.374; E, Essex. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.
Table S9 Up‐regulated genes in the three genotypes without Fusarium virguliforme phytotoxin (Fv toxin) treatment. P, PI567374; W, Williams 82; E, Essex; U, mock.
Table S10 Resistance‐related genes (up‐regulated genes shared between PI–WI and PI–EI and between PI–EI and WI–EI). Expression levels were calculated as the ratio of Fusarium virguliforme phytotoxin (Fv toxin)‐treated genotype to another different Fv toxin‐treated genotype. P, PI567374; W, Williams 82; E, Essex; I, plant treated by Fv toxin.
Table S11 Physical mapping of simple sequence repeat (SSR) markers linked to Fusarium virguliforme (Fv) resistance and cDNA arrays with differential abundance [false discovery rate (FDR) P value of <0.05]. cDNA sequences mapping <550 kbp away from SSR markers are presented. P, PI567374; W, Williams 82; E, Essex; I, plant treated by Fv toxin; U, mock; v, versus. Genes with increased and decreased transcripts are highlighted in red and blue, respectively.


