Summary
Aspergillus flavus is an opportunistic fungal pathogen that infects maize kernels pre‐harvest, creating major human health concerns and causing substantial agricultural losses. Improved control strategies are needed, yet progress is hampered by the limited understanding of the mechanisms of infection. A series of studies were designed to investigate the localization, morphology and transcriptional profile of A. flavus during internal seed colonization. Results from these studies indicate that A. flavus is capable of infecting all tissues of the immature kernel by 96 h after infection. Mycelia were observed in and around the point of inoculation in the endosperm and were found growing down to the germ. At the endosperm–germ interface, hyphae appeared to differentiate and form a biofilm‐like structure that surrounded the germ. The exact nature of this structure remains unclear, but is discussed. A custom‐designed A. flavus Affymetrix GeneChip® microarray was used to monitor genome‐wide transcription during pathogenicity. A total of 5061 genes were designated as being differentially expressed. Genes encoding secreted enzymes, transcription factors and secondary metabolite gene clusters were up‐regulated and considered to be potential effector molecules responsible for disease in the kernel. Information gained from this study will aid in the development of strategies aimed at preventing or slowing down A. flavus colonization of the maize kernel.
Introduction
The infection of maize kernels by Aspergillus flavus, a fungal pathogen of maize, results in kernel deterioration and contamination with the carcinogenic mycotoxin aflatoxin (AF). Public exposure to feed and food containing AF in the USA is prevented through a rigorous, but expensive ($30–50 million per year), testing process mandated by the Grain Inspection, Packers and Stockyard Administration (GIPSA) (Robens and Cardwell, 2003). Diseased grain has a lower market value and may be destroyed if the AF concentration exceeds the action levels set by the US Food and Drug Administration (2000). In developing countries, AF poisoning is more common because of limited regulations and improper storage techniques (CDC, 2004).
Current management recommendations for reducing A. flavus disease are to maintain plant health and reduce plant stress throughout the growing season, especially during pollination and grainfill, and to plant regionally appropriate hybrids (Koenning and Payne, 1999; White, 1999). Although these practices are effective, often growers are faced with adverse environmental conditions, resulting in less than ideal growing conditions. New methods are needed that are more cost‐effective and provide more proactive forms of resistance against seed pathogens. The goal of this study was to examine the response by A. flavus to interactions with the maize kernel to identify the mechanisms important in A. flavus pathogenesis.
The morphological and molecular changes that occur in A. flavus, an opportunistic pathogen, during maize kernel pathogenesis are poorly understood. Evidence suggests that the pathogen enters a nondamaged kernel through the pedicel end as the ear approaches maturity (Marsh and Payne, 1984; Payne et al., 1988b; Smart et al., 1990; Windham and Williams, 1998). However, A. flavus has been detected in younger, insect‐damaged kernels (Anderson et al., 1975; Taubenhaus, 1920). After gaining entrance, A. flavus preferentially colonizes tissues high in oil, such as the germ and aleurone, with the highest concentration of AF occurring in the germ of infected kernels (Fennell et al., 1973; Jones et al., 1980; Keller et al., 1994; Smart et al., 1990). There has been some debate about the extent of endosperm colonization (Brown et al., 1995; Keller et al., 1994; Lillehoj et al., 1976; Smart et al., 1990).
The aim of this study was to investigate the localization, morphology and transcriptional profile of A. flavus during seed colonization. We addressed the first of these objectives by identifying the kernel tissue types infected by A. flavus over time, and the second by looking at the morphology of infectious hyphae within the different tissues of the kernel. Lastly, global gene expression was monitored using a custom‐designed A. flavus Affymetrix GeneChip® microarray during in vivo growth. We conclude that A. flavus is capable of infecting all tissue types within the immature maize kernel, and find that a change in fungal morphology occurs at the endosperm–germ interface. An alteration in fungal gene expression was induced in A. flavus during the colonization of the living kernel. Those genes found to be up‐regulated are considered as potential effectors. Collectively, these studies show that A. flavus infection of the maize kernel is more complex than previously suspected.
Results
Aspergillus flavus colonizes all kernel tissues within 96 h of inoculation
Despite the efforts of several investigators, there is some uncertainty as to which tissue types of the maize kernel can be infected by A. flavus and how rapidly infection proceeds. To determine both the tissue specificity and rate of A. flavus infection after pin‐mediated endosperm inoculation, the presence of the pathogen was monitored using polymerase chain reaction (PCR). Aspergillus flavus growth was successfully tracked inside the kernel when fungal DNA was positively detected by PCR in the endosperm, germ and pedicel tissues of inoculated kernels. DNA isolated from uninfected maize tissue and A. flavus grown in potato dextrose broth (PDB) were used to test the species specificity of the primers and served as positive controls (Fig. 1, Ctrl).
Figure 1.
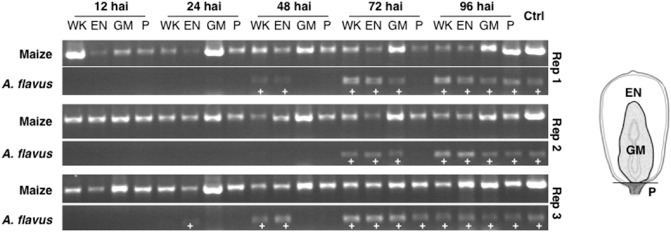
An ethidium bromide‐stained agarose gel of polymerase chain reaction (PCR) products amplified from DNA isolated from Aspergillus flavus‐infected maize tissue. The DNA was tested for the presence of A. flavus from inoculated whole kernels (WK), endosperm (EN), germ (GM) and pedicel (P) tissues at 12, 24, 48, 72 and 96 h after inoculation (hai). The location of the maize tissues analysed is shown in the schematic drawing on the right. Maize‐specific catalase primers effectively amplified the targeted PCR fragment from uninfected maize tissue DNA (Ctrl column, maize row), but failed to produce a band from DNA isolated from A. flavus grown in shake cultures (data not shown). The A. flavus‐specific gpdA primers were capable of amplifying the product from the A. flavus shake culture DNA (Ctrl column, A. flavus row), but not from noninfected maize kernel DNA (data not shown). Positive detection of A. flavus within the diseased maize tissue is marked with a white ‘+’. The lack of amplicons in the A. flavus gpdA primer reactions (A. flavus rows, missing a ‘+’ sign) is caused by the absence of fungal DNA in the samples assayed.
Aspergillus flavus was first detected in the endosperm tissue of diseased kernels as early as 24 h after inoculation (hai) and in the germ by 72 hai (Fig. 1, A. flavus rows). The removal of the germ from surrounding endosperm tissue became more difficult over time and, in some cases, A. flavus was macroscopically observed growing on the surface of the germ. Aspergillus flavus was detected in all three germ samples by 72 hai; however, it is unclear on the basis of these data alone whether the fungus was confined to the germ surface or had successfully penetrated and begun colonization. Although the pedicel tissue, which includes the kernel base, was the region farthest away from the site of inoculation, this region of the kernel was readily infected with A. flavus at 72–96 hai.
Aspergillus flavus undergoes a morphological change at the endosperm–germ interface
We performed a histological investigation to examine growth and morphological changes in A. flavus associated with kernel infection. Aspergillus flavus conidia were introduced into starchy endosperm tissue by inserting the inoculation pin into the crown of the kernel (Fig. 2a). Four days after inoculation, growth of A. flavus in vivo was observed by fungal‐specific histological stains on 10‐μm sections of diseased kernels, and hyphae could be differentiated from host cells. Endosperm colonization was predominantly at or around the site of inoculation. Often the process of inoculation created a cavity where the pin was inserted and a mass of mycelium was found surrounding or growing within this cavity (Fig. 2b). Conidiation was observed in a few of the cavities. The integrity of endosperm tissue was diminished in and around the infected area, but it is unclear whether this was a product of fungal degradation or part of the host's reaction to infection.
Figure 2.
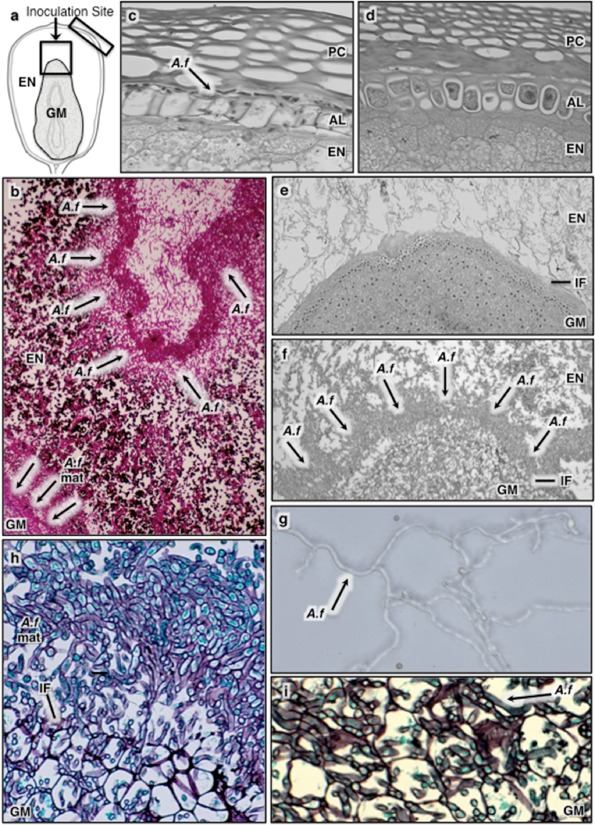
Histology of the developing maize kernel 4 days after Aspergillus flavus pin‐bar inoculation. The schematic drawing of the kernel (a) depicts the location of pin‐bar inoculation (black arrow) and the kernel region (black box, □) from which the histology panels originate. White was added to the letters and arrow margins for easier viewing in the panels. Aspergillus flavus was observed (b) in the endosperm (EN) surrounding a cavity created by the inoculation pin and outside the germ (GM) in a unique A. flavus mat‐like (A.f mat) structure. Hyphae were also found growing between the pericarp (PC) and aleurone (AL) (c) in which the AL cells in contact with A. flavus appear to be altered compared with cells in the noninoculated control (d). (e) Endosperm–germ interface (IF) in a noninoculated kernel. In the infected kernel, the A.f mat structure developed at the IF (f) and covered the germ tip. Hyphae within the A.f mat were morphologically distinct from A. flavus vegetative hyphae (g) and were highly branched, tightly intertwined and extended into the GM (h). Colonization of the GM by A. flavus was observed in highly infected kernels (i).
On the basis of the growth pattern observed at 96 hai, it can be inferred that A. flavus spread from the site of inoculation down to the germ through the starchy endosperm, although this was not always apparent as few kernel sections contained the narrow infection pathway extending from endosperm to the germ. Earlier studies have led to speculation that the fungus uses aleurone tissue to reach the germ. In this study, A. flavus was found between the aleurone and the pericarp cell layers (Fig. 2c) with infection heaviest near the inoculation site, but pockets of mycelium also occurred in more distal regions. Our observations were unable to confirm that A. flavus uses the aleurone as a means to access the germ; however, there was no evidence to indicate otherwise. Aleurone cells proximal to A. flavus hyphae lacked cytoplasm (Fig. 2c vs. 2d).
Hyphae at the endosperm–germ interface (Fig. 2e, noninoculated interface) were morphologically different from vegetative hyphae (Fig. 2f, h vs. 2g). Aspergillus flavus was not present at the endosperm–germ interface in every kernel surveyed, but, in kernels in which the fungus had made contact with the germ, it formed a mat‐like structure composed of highly branched, tightly intertwined hyphae pressed up against the scutellum (Fig. 2f, h). This mat‐like structure was seen predominantly at the germ apex, but, in heavily infected kernels, it extended from the tip to the base of the germ. This distinctive structure was observed in all kernels in which hyphae were able to penetrate and colonize germ tissue, although its formation did not guarantee germ infection. Hyphae within the germ appeared to originate from the mat‐like structure (Fig. 2h). Invasion of the germ was mostly confined to the scutellum tissue (Fig. 2i), but, on occasion, A. flavus was also found in the embryo (not shown).
Aspergillus flavus undergoes significant transcriptional changes during the pathogenesis of maize kernels
As a part of this study, we performed global gene expression profiling of A. flavus during the pathogenesis of maize kernels. Reese et al. (2011), using detached kernels, reported significant transcriptional differences in A. flavus grown on kernels at different stages of development. To limit any effects of kernel age on fungal gene expression, we grouped expression values from A. flavus‐inoculated blister (R2), milk (R3), dough (R4) and dent (R5) kernels into a single treatment before comparing them with the expression values from A. flavus inoculated into nonliving autoclaved kernels. This allowed for the identification of potential pathogenic effectors expressed in maize kernels regardless of the kernel's age.
An analysis of variance (ANOVA) identified 5061 of the 12 197 predicted A. flavus genes represented on the Affymetrix GeneChip® microarray to be differentially expressed during kernel pathogenesis (Table S1, see Supporting Information). Approximately 30% had a fold change of two or greater and, of these genes, 682 were up‐regulated and 944 were down‐regulated. Ninety of the differentially expressed genes from our study were the same A. flavus genes that had been designated previously by Reese et al. (2011) as being differentially expressed during the colonization of kernels at different stages of development.
The function of many of the differentially expressed gene products (>40%) is unknown and their annotation is listed as hypothetical. In all, 459 of the genes encoded a secretory signal peptide (bold; Table S1). Included amongst these were a cerato‐platanin‐like gene, three necrosis and ethylene‐inducing protein (NEP)‐like genes, an SMK (salt‐mediated killer)‐like toxin gene and a polygalacturonase pecA (P2c). Of these, 93 were annotated as extracellular hydrolytic enzymes. Predicted secretory proteins were both up‐ and down‐regulated.
Other fungal genes differentially expressed in the A. flavus–maize interaction included 20 transcription factors (TFs), eight of which were up‐regulated and 12 down‐regulated (Table 1). Many were similar in sequence to other known fungal TFs, and they were found scattered throughout the eight A. flavus chromosomes (Fig. 3).
Table 1.
Aspergillus flavus transcription factors differentially expressed in kernel pathogenesis
| Up‐regulated | Down‐regulated | ||||
|---|---|---|---|---|---|
| NCBI gene identifier | Annotation | Map ID* | NCBI gene identifier | Annotation | Map ID* |
| AFLA_050250 | bZIP transcription factor (CpcA) | a | AFLA_052030 | Developmental regulatory protein (WetA) | i |
| AFLA_073870 | C6 transcription factor (RegA) | b | AFLA_076320 | Fungal‐specific TF domain‐containing protein | j |
| AFLA_031340 | bZIP transcription factor (AtfA) | c | AFLA_082850 | C2H2 transcription factor (BrlA) | k |
| AFLA_136410 | Transcriptional regulator Medusa (MedA) | d | AFLA_087080 | Fungal‐specific TF domain‐containing protein | l |
| AFLA_139360 | AF transcription activator (AflR) | e | AFLA_124290 | Fungal‐specific TF domain‐containing protein | m |
| AFLA_069140 | SIR2 family histone deacetylase (Hst4) | f | AFLA_131640 | HLH transcription factor (Hpa3) | n |
| AFLA_122570 | C6 transcription factor | g | AFLA_112830 | Fungal‐specific TF domain‐containing protein | o |
| AFLA_116680 | PAS domain S‐box family protein (white collar) | h | AFLA_005460 | Cytochrome P450 monooxygenase (SirB‐like) | p |
| AFLA_001570 | Fungal‐specific TF domain‐containing protein | q | |||
| AFLA_093040 | C6 transcription factor (NirA) | r | |||
| AFLA_067910 | HLH transcription factor | s | |||
| AFLA_101920 | Extracellular developmental signal biosynthesis protein (FluG) | t | |||
NCBI, National Center for Biotechnology Information.
*Map ID denotes the chromosomal location of the TF in Fig. 3.
Figure 3.
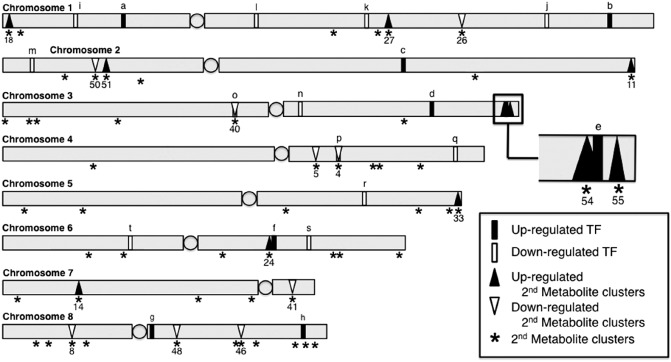
The diagram illustrates the chromosomal location of the differentially expressed Aspergillus flavus transcription factors (TFs) (Table 1) and secondary metabolite gene clusters up‐ and down‐regulated (Table 2, up and down arrows) during maize kernel colonization. Up‐regulated TFs (map ID: a–h) are shown as filled vertical bars, and down‐regulated TFs (map ID: i–t) as open vertical bars. All 55 predicted secondary metabolite clusters are depicted on the map as asterisks (*) positioned under the grey chromosome bars. Centromeres are represented as shaded circles on each of the chromosomes. Up‐regulated secondary metabolite clusters and down‐regulated clusters are denoted by filled and open triangles, respectively. The aflatoxin regulator aflR (map ID: e) is found within cluster 54, TF AFLA_112830 (map ID: o) in cluster 40 and TF AFLA_005460 (map ID: p) is part of cluster 4.
Next, we examined the expression profile for the 55 A. flavus secondary metabolite gene clusters to determine which clusters were expressed in A. flavus under pathogenic conditions. Secondary metabolite genes are located together on the chromosome as a cluster, typically near the telomeric ends. Each cluster contains a ‘backbone’ enzyme responsible for the first step in the biosynthesis of the metabolite. (Consult the A. flavus genome browser (http://www.Aspergillusflavus.org) for more information on the secondary metabolite clusters.)
Fifty of the 55 predicted A. flavus secondary metabolite gene clusters had one or more genes differentially expressed during pathogenesis (Table 2). Of these clusters, 18 had ≥50% of the genes within the cluster, including the backbone enzyme, differentially expressed: clusters 11, 14, 18, 24, 27, 33, 51, 54 (AF) and 55 (cyclopiazonic acid) were up‐regulated, and clusters 4, 5, 8, 26, 40, 41, 46, 48 and 50 were down‐regulated (chromosome locations, Fig. 3). Clusters with ≥50% but near equal numbers of up‐ to down‐regulated genes were not considered to be differentially expressed. The AF gene cluster (54) had 50% of the 34 predicted AF genes, including the backbone enzyme, differentially expressed as part of A. flavus disease development in the kernel. All 17 of the differentially regulated AF genes were up‐regulated. AF was detected in inoculated kernels at all four developmental stages (data not shown).
Table 2.
Percentage of genes within the 55 Aspergillus flavus secondary metabolite clusters differentially expressed in kernel pathogenesis
| Cluster ID | Number of genes in cluster | Total % | Differentially expressed | ||||
|---|---|---|---|---|---|---|---|
| Up‐regulated (%) | + | Down regulated (%) | Backbone enzyme | Change in expression* | |||
| 1 | 9 | 22 | 22 | 0 | ✗ | ||
| 2 | 9 | 33 | 11 | 22 | ✗ | ||
| 3 | 2 | 50 | 0 | 50 | ✗ | ||
| 4 | 4 | 100 | 0 | 100 | ✓ | ↓ | |
| 5 | 3 | 67 | 0 | 67 | ✓ | ↓ | |
| 6 | 12 | 50 | 33 | 17 | ✓ | ||
| 7 | 10 | 50 | 30 | 20 | ✓ | ||
| 8 | 9 | 78 | 11 | 67 | ✓ | ↓ | |
| 9 | 10 | 30 | 30 | 0 | ✗ | ||
| 10 | 3 | 0 | NA | NA | ✗ | ||
| 11 | 12 | 75 | 50 | 25 | ✓ | ↑ | |
| 12 | 3 | 33 | 33 | 0 | ✗ | ||
| 13 | 4 | 50 | 0 | 50 | ✗ | ||
| 14 | 2 | 50 | 50 | 0 | ✓ | ↑ | |
| 15 | 12 | 58 | 33 | 25 | ✓ | ||
| 16 | 2 | 0 | NA | NA | NA | ||
| 17 | 11 | 27 | 27 | 0 | ✓ | ||
| 18 | 6 | 50 | 50 | 0 | ✓ | ↑ | |
| 19 | 6 | 0 | NA | NA | NA | ||
| 20 | 10 | 30 | 10 | 20 | ✓ | ||
| 21 | 38 | 24 | 0 | 24 | ✗ | ||
| 22 | 6 | 17 | 17 | 0 | ✗ | ||
| 23 | 17 | 47 | 12 | 35 | ✓ | ||
| 24 | 3 | 100 | 100 | 0 | ✓ | ↑ | |
| 25 | 9 | 11 | 11 | 0 | ✗ | ||
| 26 | 12 | 58 | 17 | 42 | ✓ | ↓ | |
| 27 | 5 | 60 | 60 | 0 | ✓ | ↑ | |
| 28 | 6 | 50 | 33 | 17 | ✗ | ||
| 29 | 2 | 50 | 0 | 50 | ✗ | ||
| 30 | 6 | 50 | 17 | 33 | ✓ | ||
| 31 | 15 | 13 | 0 | 13 | ✗ | ||
| 32 | 21 | 62 | 52 | 10 | ✗ | ||
| 33 | 8 | 50 | 38 | 13 | ✓ | ↑ | |
| 34 | 8 | 63 | 13 | 50 | ✗ | ||
| 35 | 5 | 40 | 20 | 20 | ✗ | ||
| 36 | 7 | 29 | 29 | 0 | ✓ | ||
| 37 | 3 | 0 | NA | NA | NA | ||
| 38 | 3 | 0 | NA | NA | NA | ||
| 39 | 5 | 40 | 20 | 20 | ✗ | ||
| 40 | 9 | 56 | 11 | 44 | ✓ | ↓ | |
| 41 | 3 | 67 | 0 | 67 | ✓ | ↓ | |
| 42 | 11 | 36 | 9 | 27 | ✓ | ||
| 43 | 19 | 53 | 32 | 21 | ✓ | ||
| 44 | 6 | 67 | 33 | 33 | ✓ | ||
| 45 | 16 | 31 | 13 | 19 | ✗ | ||
| 46 | 4 | 50 | 0 | 50 | ✓ | ↓ | |
| 47 | 6 | 50 | 33 | 17 | ✓ | ||
| 48 | 20 | 65 | 5 | 60 | ✓ | ↓ | |
| 49 | 6 | 67 | 33 | 33 | ✓ | ||
| 50 | 8 | 75 | 13 | 63 | ✓ | ↓ | |
| 51 | 5 | 60 | 60 | 0 | ✓ | ↑ | |
| 52 | 7 | 14 | 0 | 14 | ✗ | ||
| 53 | 8 | 63 | 38 | 25 | ✓ | ||
| 54 | 34 | 50 | 50 | 0 | ✓ | ↑ | |
| 55 | 4 | 75 | 75 | 0 | ✓ | ↑ | |
*Clusters with ≥50% of the genes differentially expressed during infection (including the backbone enzyme) are denoted with an arrow. An up arrow indicates the cluster was up‐regulated and a down arrow that the cluster was down‐regulated. Clusters with equal or near‐equal percentages of up‐ and down‐regulated genes were not marked even though greater than 50% of the cluster was differentially expressed. Cluster 15, aflatrem. Cluster 54, aflatoxin. Cluster 55, cyclopiazonic acid.
Some genes from the aflatrem clusters were differentially expressed, but collectively failed to meet the above‐mentioned criteria to be considered as differentially expressed. Aflatrem is a tremorgenic toxin that is produced by A. flavus (Wilson and Wilson, 1964) Two clusters (15 and 32) are necessary for the synthesis of aflatrem (Nicholson et al., 2009). In this study, cluster 15 (aflatrem) and cluster 32 had 58% and 62% of the cluster differentially expressed, respectively. The backbone enzyme for cluster 15, dimethylallyl tryptophan synthase, was down‐regulated during infection. The other differentially expressed genes in cluster 15 (aflatrem) were an even mixture of both up‐regulated (four genes) and down‐regulated (two genes + backbone). As there were almost equal numbers of up‐ and down‐ regulated genes, cluster 15 was not considered to be differentially expressed. In contrast, all but two of the 13 differentially expressed genes in cluster 32 were up‐regulated, including a NEP‐like gene that had a fold change of 12.1 (Georgianna et al., 2010; Nicholson et al., 2009). Cluster 32 does not have a backbone enzyme. The aflatrem genes atmG and atmC, which are needed for aflatrem synthesis, were not differentially expressed.
Discussion
Discrepancy exists in the literature as to whether or not A. flavus effectively colonizes all tissue types within the maize kernel (Brown et al., 1995; Keller et al., 1994; Lillehoj et al., 1976; Smart et al., 1990). We found that A. flavus is readily detected by PCR in the endosperm, germ and pedicel tissues of susceptible B73 kernels. The endosperm samples used in this study also contained aleurone tissue, but closer examination, as discussed later, revealed that the heaviest concentration of A. flavus was in the endosperm and not the aleurone.
These data largely support the findings of Brown et al. (1995) and Lillehoj et al. (1976). However, neither Smart et al. (1990) nor Keller et al. (1994) saw substantial endosperm colonization or mycotoxin production in the endosperm, respectively. We speculate that the incongruities between these studies are caused by variations in experimental design.
Differences in inoculation techniques would influence the route of infection and, potentially, disease severity by altering how the fungus enters the kernel and which tissue it comes into contact with initially. Although pin‐bar inoculation ensures a higher success rate of infection in the field, it should be recognized that this method is invasive. Inoculation by pin‐bar compromises the kernel's natural defences by penetrating the pericarp and aleurone layers, and forcing conidia directly into the endosperm. Although this method of inoculation may not reflect the method of hyphal entry in intact kernels, it closely simulates the infection process after insect damage, which is highly relevant in field‐grown plants (Parsons and Munkvold, 2010).
Keller et al. (1994) inoculated in a manner more similar to that used in our study, but infected mature instead of developing kernels. As maize kernels mature, the endosperm undergoes desiccation, which makes infection of the endosperm difficult by lowering water activity to a level unfavourable for fungal growth (Anderson et al., 1975; Gibson et al., 1994; Payne et al., 1988a). Higher amounts of water in developing kernels, such as those used in our study, are more conducive for endosperm colonization than the smaller amounts found in mature kernels. Also, Keller et al. (1994) followed growth by expression of the AF pathway intermediate, norsolorinic acid (NOR), and thus may not have observed NOR production in the endosperm, which is less conducive for AF formation.
These observational differences in tissue colonization emphasize how the method of entry and the developmental age of the kernel may significantly affect the route of colonization. Additional studies testing a range of inoculation techniques on different aged kernels would further confirm this assumption. Other variables, such as the overall health of the kernel and maize kernel genotype, should also be considered.
Results from the histological studies showed that A. flavus colonizes and uses endosperm tissue as a medium for growth down to the germ. These observations are in accordance with the findings of our PCR localization study.
Aspergillus flavus was also observed growing between the aleurone and pericarp layers, but growth was restricted to isolated patches of mycelium scattered throughout the kernel. The heaviest concentration of these mycelial patches was near the site of inoculation, and growth did not appear to extend down to the germ. In contrast, fungal hyphae traversing the endosperm from the point of inoculation to the germ were consistently observed in infected kernels, although colonization of the endosperm outside the site of inoculation was not extensive. This suggests that endosperm tissue supports the spread of A. flavus in the kernel, but might not be a preferred host tissue (compared with germ). Another possibility is that, after initial infection, the surrounding endosperm tissue becomes resistant to infection. Endosperm‐specific resistance‐associated proteins have been identified (Chen et al., 2007). However, it should be reiterated that the route of infection is probably influenced by the local environmental conditions under which infection takes place.
On reaching the germ, A. flavus undergoes dramatic morphological changes and forms a distinct mat‐like structure of highly branched, intertwined hyphae at the germ–endosperm interface. We hypothesize that this structure contributes to germ infection, but its presence does not guarantee germ infection. Although this structure has not been described previously in A. flavus infection of the kernel, there is some previous evidence that the endosperm–germ interface is an important region of the kernel in the interaction with seed‐colonizing fungi (Anderson et al., 1975; Fennell et al., 1973; Freeman, 1904; Lillehoj et al., 1976; Manns and Adams, 1923). We suggest that this structure is analogous to the biofilm produced by Aspergillus fumigatus during in vivo infection of lung tissue (Loussert et al., 2010). Aspergillus fumigatus is a human pathogen capable of causing aspergillosis in immunosuppressed individuals. Biofilms are defined as groupings of microbes anchored to a nonmotile surface held together by an extracellular matrix (Ramage et al., 2009). Although biofilm formation is not exclusive to pathogens, it contributes to pathogenicity by enabling the pathogen to adhere tightly to the host and protects against host immune responses (Hall‐Stoodley and Stoodley, 2009; Jabra‐Rizk et al., 2004; Ramage et al., 2009).
Both physical and molecular evidence supports the conclusion that changes in fungal morphology during pathogenesis include biofilm‐like formation. Physically, it was observed that separation of the germ from the kernel became more difficult as the severity of infection increased. The formation of this ‘biofilm’ could be responsible for the increased adhesion between these two tissues. Changes in fungal gene expression also support this theory. The TF medusa (medA) was found to be up‐regulated during infection of maize kernels, and is discussed in further detail later. This TF is known to be a major regulator of biofilm production in A. fumigatus (Gravelat et al., 2010).
It is unclear at which stage of kernel development A. flavus infects maize. Reese et al. (2011) reported that kernel age significantly influences A. flavus gene expression. To minimize this effect, living maize kernels at different stages of development were inoculated. Aspergillus flavus expression values from these kernels were grouped into a single treatment and compared with gene expression from fungus grown in nonliving autoclaved kernels. Over 40% of the monitored A. flavus genes were differentially expressed in response to interactions with the living kernel. All but 90 of the fungal genes were expressed during infection, regardless of the kernel's age. Differentially expressed genes were broadly scattered across the genome and contained representatives from multiple functional categories. Unfortunately, a large portion of the transcripts found to be differentially expressed have not yet been annotated and their functions remain unknown.
Secretory proteins
Pathogens secrete a barrage of toxins and proteins, termed effectors, that help to establish and maintain disease in the host plant (de Jonge et al., 2011; Kale and Tyler, 2011). The examination of pathogen secretomes has been useful in identifying potential effectors (Guttman et al., 2002; Kamoun, 2006). Aspergillus flavus possesses several genes encoding extracellular secretory proteins expressed during kernel colonization. These include the up‐regulation of two toxin genes: a cerato‐platanin‐like gene and an SMK‐like toxin gene. Cerato‐platanin is a phytotoxin produced by Ceratocystis fimbriata that elicits the synthesis of antimicrobial substances in the host (Pazzagli et al., 1999).
Also up‐regulated were three NEP‐like genes. These small proteins are expressed in pathogens during infection and can cause cellular death in plants (Dong et al., 2012; Küfner et al., 2009; Santhanam et al., 2013). It was unexpected to see increased transcription of NEP‐like genes in A. flavus considering that these proteins are associated with necrosis in dicotyledonous, but not monocotyledonous, plants, such as maize (Bailey, 1995). The exact role of NEP proteins in the pathogenesis of monocotyledonous plants remains unclear (de Jonge et al., 2011). Curiously, one of the NEP‐like genes up‐regulated lies within the A. flavus secondary metabolite gene cluster 32, and another lies next to cluster 18 on chromosome 1.
A number (20%) of the secretory genes were annotated as encoding hydrolytic enzymes. Aspergillus flavus secretes hydrolases during saprobic growth to break down complex food sources (Luo et al., 2009). The up‐regulation of secreted A. flavus hydrolase genes indicates that their production continues during pathogenesis. This was expected as hydrolases facilitate nutrient acquisition. It has long been suspected that extracellular hydrolases assist in the spread of disease by deconstructing host tissue (Kikot et al., 2009; Mellon et al., 2007; Schaller et al., 2005). Fakhoury and Woloshuk (1999) identified an α‐amylase in A. flavus that was needed for endosperm colonization and allowed for growth down to the germ. In addition, Shieh et al. (1997) found an A. flavus polygalacturonase (pecA), a plant cell wall‐degrading enzyme, important in the colonization of cotton bolls. The pecA gene, as well as several α‐amylase and other cell wall‐degrading enzyme‐encoding genes, were up‐regulated in this study. Yet, the down‐regulation of some hydrolases during pathogenesis probably indicates that only specific enzymes or isozymes from this functional category are necessary for infection, or that the host's defence is repressing expression.
Transcription factors
Transcription factors (TFs) have a sizable impact on fungal development and metabolism as they regulate the transcription of multiple genes. They can also contribute to pathogenicity by inducing the expression of effector genes (Michielse et al., 2009; Zahiri et al., 2010). Our study identified A. flavus TFs differentially expressed during maize kernel pathogenesis. Several of these TFs have been well characterized in other Aspergillus species, which allows us to extrapolate their potential roles in A. flavus disease development.
Plant pathogens encounter a variety of environmental stressors and nutritional conditions during infection. For example, in response to pathogen attack, the plant produces antimicrobial reactive oxygen species (ROS) to prevent the spread of disease (Lamb and Dixon, 1997). In order to successfully infect the host, the pathogen must overcome this and other confronted obstacles. Two TFs previously shown to be involved in Aspergillus stress tolerance were found to be up‐regulated in this study, and may indicate that A. flavus encounters and deals with environmental stressors during the colonization of the living maize kernel.
In A. nidulans and A. oryzae, the TF AtfA is important in stress tolerance, especially against ROS, by regulating the expression of stress‐responsive genes (Balázs et al., 2010; Sakamoto et al., 2009). As mentioned, ROS are an important part of the plant defence response. Moreover, in A. nidulans, amino acid starvation activates the transcription of cpcA, the central regulator for the cross‐pathway control system, and this, in turn, causes an increase in amino acid biosynthetic gene expression (Busch et al., 2003; Eckert et al., 2000; Hoffmann et al., 2001). This is relevant to A. flavus kernel colonization as amino acid availability is limited after the blister stage of development.
Increased transcription of atfA and cpcA would conceivably give A. flavus the ability to thrive in an environment that might otherwise be inhospitable. This ability to adapt to a rapidly changing local environment is probably very important in pathogenesis. In support of this, the A. fumigatus cpcA deletion mutant was less virulent in a murine model of pulmonary aspergillosis (Krappmann et al., 2004).
Changes in the environment can also trigger reproduction in fungi. Asexual sporulation in A. nidulans is influenced by nutrient starvation and other stressors (Adams et al., 1998). The TFs fluG, brlA and wetA, whose expression is required for asexual reproduction in A. nidulans, were down‐regulated during A. flavus pathogenesis (Adams et al., 1998; Clutterbuck, 1969; Etxebeste et al., 2010). These data suggest that asexual reproduction is suppressed in A. flavus. Yet, the developmental modulator medA, which is also required for sporulation in A. nidulans, was, in contrast, more highly expressed in the colonization of the living kernel (Adams et al., 1998; Busby et al., 1996).
Gravelat et al. (2010) discovered that MedA is required for biofilm formation and full virulence in A. fumigatus. It is tempting to speculate that MedA serves a similar function in A. flavus during kernel pathogenesis and is involved in the formation of the mat‐like structure at the endosperm–germ interface. At this time, it is unclear what is the role of this biofilm‐like structure in kernel pathogenesis, but it may be important in germ infection. A medA homologue in the maize pathogen Ustilago maydis was also found to be required for full virulence (Chacko and Gold, 2012). Additional studies are needed to further examine the relationship between medA and the other asexual regulatory genes during the in vivo growth of A. flavus, especially during ‘biofilm’ formation.
Secondary metabolites
Fungal secondary metabolites are energetically expensive to synthesize and are not required for primary growth (Calvo et al., 2002; Keller et al., 2005). It is unclear what is the functional role of most fungal secondary metabolites, although it is thought that they protect the fungus against stressors encountered during growth.
This study identified 18 of the 55 predicted A. flavus secondary metabolite clusters as having the majority of the genes within the cluster (≥50% including the backbone enzyme) differentially expressed. Because of the condition under which the up‐regulated clusters were found to be more highly expressed, it is tempting to assume that the up‐regulated clusters (11, 14, 18, 24, 27, 33, 51, 54, and 55) aid in the colonization of the maize kernel.
Several fungal pathogens produce secondary metabolites when infecting a host (Osbourn, 2010). However, which secondary metabolites are produced in vivo and whether or not their presence contributes to pathogenicity are difficult to ascertain. The aforementioned increase in A. flavus secondary metabolite gene expression observed during pathogenesis may not be a pathogen‐associated reaction, but rather a physiological response to changes in the environment. The environment within the plant is dynamic and changes in response to infection. Exposure to the new surroundings of the kernel may trigger the production of secondary metabolite expression in A. flavus. For example, AF synthesis is known to be influenced by many environmental variables, including temperature and pH (Calvo et al., 2002; Price et al., 2005).
Yet, secondary metabolites can function as effectors (Kwon‐Chung and Sugui, 2009; Thines et al., 2006). Of the A. flavus clusters up‐regulated during kernel pathogenesis, cluster 24 contains the pes1 homologue, a known virulence factor in A. fumigatus (Reeves et al., 2006). Pes1 is a nonribosomal peptide synthetase and absence of expression leads to a reduction in A. fumigatus tolerance of oxidative stress. Clusters 18, 33 and 51 were also up‐regulated. A previous report noted that these clusters are not expressed under standard laboratory testing conditions (Georgianna et al., 2010). This suggests that these metabolites serve a more specialized biological role in the fungus, which has yet to be determined.
It is unclear whether secondary metabolite gene clusters in A. flavus are involved in pathogenicity. However, this study has made known which clusters are expressed under pathogenic conditions. This information is important for future studies investigating A. flavus secondary metabolism (Brakhage and Schroeckh, 2011).
Conclusion
The experiments presented in this article expand our understanding of the localization, morphology and transcriptional state of A. flavus during seed pathogenicity. This study has shown that A. flavus infection of the kernel is more complex and specialized than previously thought. It was observed that A. flavus undergoes a series of physical and transcriptional changes during pathogenicity. The formation of a biofilm‐like structure shows that A. flavus is capable of responding to interactions with the living maize kernel. The benefits of this transformation are as yet unknown. The monitoring of the transcriptome during pathogenesis has provided a better understanding of the signals to which A. flavus is responding and how the fungus responds back. Changes in transcription would suggest that A. flavus encounters and deals with stressors during colonization. In addition, several types of effectors (i.e. toxins, secretory proteins, secondary metabolites) appear to be secreted as part of pathogenesis, which probably help to establish and maintain disease in the kernel. Together, these data indicate that A. flavus behaves similarly to other plant pathogens. Information acquired from these studies gives us a better understanding of A. flavus infection and can aid in the development of strategies aimed at reducing or eliminating A. flavus infection of maize.
Experimental Procedures
Fungal growth and maize kernels
For inoculum, A. flavus strain NRRL 3357 was grown on potato dextrose agar (PDA) medium at 28 °C for 7–10 days. Conidial suspensions in 0.05% (v/v) Triton X‐100 were adjusted to 1 × 106 spores/mL.
The maize inbred line B73 was grown in fields at the Central Crops Research Station in Clayton, NC, USA. Plants were hand pollinated and the dates of pollination were recorded to ensure that ears used in the study were at the same stage of development. At selected developmental stages, husks were pulled away from the ears and the kernels were inoculated with a pin‐bar consisting of 18 straight pins embedded in an epoxy block. Pins were dipped into the A. flavus conidial suspension and inserted into the crown of the exposed kernels, introducing approximately 13 spores into the endosperm tissue. For each ear, four rows of kernels were inoculated and the husk was repositioned around the ear and held in place with a rubber band. A paper pollination bag (Lawson 402, Northfield, IL, USA) was placed over the inoculated ear and left until the ear was harvested.
Tracking A. flavus growth in the maize kernel
For this study, we used kernels inoculated at the late milk (R3) to early dough (R4) stage of development (approximately 22 days after pollination). Kernels were harvested 12, 24, 48, 72 and 96 hai. The kernels were carefully removed from the ear and stored on ice until the tissues were separated. Under sterile conditions, the pedicel regions of approximately 20 kernels, including the kernel base, were removed. The remainder of the kernel was processed further by separating the endosperm from the germ. The tissues were weighed, placed in a 20‐mL vial (2.5 cm × 5 cm) and 4 mL of chilled methanol–water (60:40, v/v) per gram were added together with ceramic beads. A retrofitted reciprocating saw was used to macerate the tissues by two 30‐s pulses. DNA was isolated using a cetyltrimethylammonium bromide (CTAB)‐based method as follows. Five hundred microlitres of CTAB buffer (He et al., 2007) were added to 200 μL of kernel slurry and incubated at 65 °C for 30 min with occasional vortexing. Five hundred microlitres of chloroform–isoamyl alcohol (24:1, v/v) were added to the kernel mixture and the tube was subjected to centrifugation at 17 900 g for 5 min. The supernatant was transferred to a new tube containing 32 μL of 7.5 m ammonium acetate and 233 μL of 100% (v/v) isopropanol, and subjected to centrifugation as described previously. The DNA pellet was washed with 70% (v/v) ethanol, dried and resuspended in 30 μL of water. PCR was used to monitor the progression of A. flavus colonization within the different tissues of the kernel. Primers specific for A. flavus glyceraldehyde‐3‐phosphate dehydrogenase A (gpdA, AFLA_025100) were designed: gpdA_F, 5′‐TCTGTTGTCGACCTCACCTG‐3′; gpdA_R, 5′‐GTCAATTTCAAGGGGTGGTG‐3′; primers specific to a maize catalase gene (CAT1_F, 5′‐GTCCAGACACCTGTTATTGTCCGT; CAT1_R, 5′‐GAGGAAGGTGAACATGTGTAGGCT) were used to test maize DNA quality. The PCR programme was as follows: 95 °C for 4 min, [96 °C for 10 s, 55 °C for 10 s, 72 °C for 2.5 min] 40 cycles, 72 °C for 5 min. PCR products were analysed by agarose gel electrophoresis and stained with ethidium bromide.
Histological studies
Late milk (R3) to early dough (R4) kernels harvested at 96 hai were cut longitudinally and placed in a fixation solution of 10% (w/v) neutral buffered formalin (Fisher Scientific, Pittsburgh, PA, USA, Cat. # SF100‐4). A microtome was used to obtain 10‐μm sections, which were stained according to Grocott's methenamine silver (GMS) or the periodic acid‐Schiff (PAS) method (Grocott, 1955; Kligman and Mescon, 1950).
Whole‐genome expression analysis
For this study, we used kernels inoculated at the blister (R2), milk (R3), dough (R4) and dent (R5) stages of development. To ensure that infection had occurred, only kernels with a mound of sporulating mycelium at the inoculation site were harvested. The kernels were harvested at 4 dai, except for dent kernels which were left in the field until sporulation occurred (6 dai). Approximately 100 inoculated kernels were carefully removed from five ears, flash frozen and stored at −80 °C until RNA could be extracted. Three replicates, each with 100 kernels, were collected at the four stages of development. AF was extracted and quantified using the protocol from Georgianna et al. (2010). Mature autoclaved B73 kernels severed as a nonliving control to the field kernels. These kernels were inoculated with a straight pin dipped into a suspension of 1 × 105 conidia/mL and placed in 20‐mL vials (2.5 cm × 5 cm), incubated at 29 °C for 5 days and then stored at −80 °C until RNA could be extracted.
RNA was isolated as outlined in Smith et al. (2008) and the quality was checked before processing for array hybridization. A custom‐designed A. flavus Affymetrix GeneChip® DNA microarray (AFAVUSa520391F), containing probe sets representing the 12 197 predicted A. flavus genes, was used to monitor A. flavus gene expression during kernel pathogenesis. Standard Affymetrix protocols were followed in the generation of biotinylated cRNA, the hybridization of the cRNA with the single‐stranded DNA oligonucleotide probes and the scanning of the array. This procedure was carried out at the Purdue Genomic Core Facility (http://www.genomics.purdue.edu) in West Lafayette, IN, USA.
Microarray analysis
The .CEL files generated from the Affymetrix GeneChip® DNA microarray scans were imported into dChip to calculate and extract expression values for each of the A. flavus probe sets (Li and Wong, 2001). The probe expression values were analysed without the inclusion of the mismatched probes to reduce variance and inaccurate intensity values associated with mismatched probes (Irizarry et al., 2003; Naef et al., 2002). Raw intensity values were imported into JMP Genomics (SAS, Cary, NC, USA) and log2 transformed. The expression profile for each of the arrays was reviewed using parallel plots. Based on the observed variation in distribution patterns amongst the arrays, the data were normalized using Loess normalization. The arrays were grouped into autoclaved kernel and field kernel treatment groups, and an ANOVA was performed to determine which genes were differentially expressed between treatment groups. A positive false discovery rate (pFDR) of 0.05 was used to control for the family‐wise error rate (Storey, 2002).
Supporting information
Table S1 Aspergillus flavus genes differentially expressed during maize seed colonization.
Acknowledgements
This project was supported by the United States Department of Agriculture, National Institute of Food and Agriculture (NIFA) award number 2010‐65108. Thanks are due to Dr Max Feldman for helpful discussions and suggestions on the paper, and to C. A. Smith, D. R. Georgianna and J. L. Burroughs for their assistance in the field.
References
- Adams, T.H. , Wieser, J.K. and Yu, J.‐H. (1998) Asexual sporulation in Aspergillus nidulans . Microbiol. Mol. Biol. Rev. 62, 35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, H.W. , Nehring, E.W. and Wichser, W.R. (1975) Aflatoxin contamination of corn in the field. J. Agric. Food Chem. 23, 775–782. [DOI] [PubMed] [Google Scholar]
- Bailey, B. (1995) Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca . Phytopathology, 85, 1250–1255. [Google Scholar]
- Balázs, A. , Pócsi, I. , Hamari, Z. , Leiter, E. , Emri, T. , Miskei, M. , Oláh, J. , Tóth, V. , Hegedus, N. , Prade, R.A. , Molnár, M. and Pócsi, I. (2010) AtfA bZIP‐type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans . Mol. Genet. Genomics, 283, 289–303. [DOI] [PubMed] [Google Scholar]
- Brakhage, A.A. and Schroeckh, V. (2011) Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. [DOI] [PubMed] [Google Scholar]
- Brown, R.L. , Cleveland, T.E. , Payne, G. , Woloshuk, C. , Campbell, K. and White, D. (1995) Determination of resistance to aflatoxin production in maize kernels and detection of fungal colonization using an Aspergillus flavus transformant expressing Escherichia coli β‐glucuronidase. Phytopathology, 85, 983–989. [Google Scholar]
- Busby, T.M. , Miller, K.Y. and Miller, B.L. (1996) Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics, 143, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, S. , Bode, H.B. , Brakhage, A.A. and Braus, G.H. (2003) Impact of the cross‐pathway control on the regulation of lysine and penicillin biosynthesis in Aspergillus nidulans . Curr. Genet. 42, 209–219. [DOI] [PubMed] [Google Scholar]
- Calvo, A. , Wilson, R. , Bok, J. and Keller, N. (2002) Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2004) Outbreak of aflatoxin poisoning—Eastern and Central provinces, Kenya, January–July 2004. MMWR Recomm. Rep. 53, 790–793. [PubMed] [Google Scholar]
- Chacko, N. and Gold, S. (2012) Deletion of the Ustilago maydis ortholog of the Aspergillus sporulation regulator medA affects mating and virulence through pheromone response. Fungal Genet. Biol. 49, 426–432. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Damann, K.E. and Cleveland, T.E. (2007) Identification of maize kernel endosperm proteins associated with resistance to aflatoxin contamination by Aspergillus flavus . Phytopathology, 97, 1094–1103. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A.J. (1969) A mutational analysis of conidial development in Aspergillus nidulans . Genetics, 63, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Kong, G. , Qutob, D. , Yu, X. , Tang, J. , Kang, J. , Dai, T. , Wang, H. , Gijzen, M. and Wang, Y. (2012) The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis‐inducing activity. Mol. Plant–Microbe Interact. 25, 896–909. [DOI] [PubMed] [Google Scholar]
- Eckert, S.E. , Kübler, E. , Hoffmann, B. and Braus, G.H. (2000) The tryptophan synthase‐encoding trpB gene of Aspergillus nidulans is regulated by the cross‐pathway control system. Mol. Gen. Genet. 263, 867–876. [DOI] [PubMed] [Google Scholar]
- Etxebeste, O. , Garzia, A. , Espeso, E.A. and Ugalde, U. (2010) Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18, 569–576. [DOI] [PubMed] [Google Scholar]
- Fakhoury, A.M. and Woloshuk, C.P. (1999) Amy1, the α‐amylase gene of Aspergillus flavus: involvement in aflatoxin biosynthesis in maize kernels. Phytopathology, 89, 908–914. [DOI] [PubMed] [Google Scholar]
- Fennell, D. , Bothast, R. , Lillehoj, E. and Peterson, R. (1973) Bright greenish‐yellow fluorescence and associated fungi in white corn naturally contaminated with aflatoxin. Cereal Chem. 50, 404–414. [Google Scholar]
- Freeman, E. (1904) The seed‐fungus of Lolium temulentum, L., the Darnel. Philos. Trans. R. Soc. London B, 196, 1–27. [Google Scholar]
- Georgianna, D. , Fedorova, N. , Burroughs, J. , Dolezal, A. , Bok, J. , Horowitz‐Brown, S. , Woloshuk, C.P. , Yu, J. , Keller, N.P. and Payne, G.A. (2010) Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11, 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, A.M. , Baranyi, J. , Pitt, J.I. , Eyles, M.J. and Roberts, T.A. (1994) Predicting fungal growth: the effect of water activity on Aspergillus flavus and related species. Int. J. Food Microbiol. 23, 419–431. [DOI] [PubMed] [Google Scholar]
- Gravelat, F. , Ejzykowicz, D. , Chiang, L. , Chabot, J. , Urb, M. , Macdonald, K. , al‐Bader, N. , Filler, S.G. and Sheppard, D.C. (2010) Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell. Microbiol. 12, 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocott, R. (1955) A stain for fungi in tissue sections and smears using Gomori's methenamine‐silver nitrate technique. Am. J. Clin. Pathol. 8, 975–979. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S. , Vinatzer, B.A. , Sarkar, S.F. , Ranall, M.V. , Kettler, G. and Greenberg, J.T. (2002) A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae . Science, 295, 1722–1726. [DOI] [PubMed] [Google Scholar]
- Hall‐Stoodley, L. and Stoodley, P. (2009) Evolving concepts in biofilm interactions. Cell. Microbiol. 11, 1034–1043. [DOI] [PubMed] [Google Scholar]
- He, Z.‐M. , Price, M.S. , OBrian, G.R. , Georgianna, D.R. and Payne, G.A. (2007) Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus . BMC Microbiol. 7, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, B. , Valerius, O. , Andermann, M. and Braus, G.H. (2001) Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans . Mol. Biol. Cell, 12, 2846–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A. , Hobbs, B. , Collin, F. , Beazer‐Barclay, Y.D. , Antonellis, K.J. , Scherf, U. and Speed, T. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jabra‐Rizk, M. , Falkler, W. and Meiller, T. (2004) Fungal biofilms and drug resistance. Emerg. Infect. Dis. 10, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. , Duncan, H. , Payne, G. and Leonard, K. (1980) Factors influencing infection by Aspergillus flavus in silk‐inoculated Corn. Plant Dis. 64, 859–863. [Google Scholar]
- de Jonge, R. , Bolton, M.D. and Thomma, B. (2011) How filamentous pathogens co‐opt plants: the ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14, 400–406. [DOI] [PubMed] [Google Scholar]
- Kale, S.D. and Tyler, B.M. (2011) Entry of oomycete and fungal effectors into plant and animal host cells. Cell. Microbiol. 13, 1839–1848. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Keller, N.P. , Butchko, R. , Sarr, B. and Phillips, T.D. (1994) A visual pattern of mycotoxin production in maize kernels by Aspergillus spp. Phytopathology, 84, 483–488. [Google Scholar]
- Keller, N.P. , Turner, G. and Bennett, J.W. (2005) Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. [DOI] [PubMed] [Google Scholar]
- Kikot, G.E. , Hours, R.A. and Alconada, T.M. (2009) Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: a review. J. Basic Microbiol. 49, 231–241. [DOI] [PubMed] [Google Scholar]
- Kligman, A.M. and Mescon, H. (1950) The periodic acid‐Schiff stain for the demonstration of fungi in animal tissue. J. Bacteriol. 60, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning, S. and Payne, G. (1999) Mycotoxins in corn. NCSU Plant Pathology Extension . Raleigh, NC: North Carolina Cooperative Extension; Available at: http://www.ces.ncsu.edu/depts/pp/notes/Corn/corn001.htm [accessed on Jun 26, 2013]. [Google Scholar]
- Krappmann, S. , Bignell, E. , Reichard, U. , Rogers, T. , Haynes, K. and Braus, G. (2004) The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 52, 785–799. [DOI] [PubMed] [Google Scholar]
- Küfner, I. , Ottmann, C. , Oecking, C. and Nürnberger, T. (2009) Cytolytic toxins as triggers of plant immune response. Plant Signal Behav. 4, 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon‐Chung, K.J. and Sugui, J.A. (2009) What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Med. Mycol. 47 (Suppl 1), S97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, C. and Dixon, R.A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Li, C. and Wong, W.H. (2001) Model‐based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2, RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj, E. , Kwolek, W. , Peterson, R. , Shotwell, O. and Hesseltine, C. (1976) Aflatoxin contamination, fluorescence, and insect damage in corn infected with Aspergillus flavus before harvest. Cereal Chem. 53, 505–512. [Google Scholar]
- Loussert, C. , Schmitt, C. , Prevost, M. , Balloy, V. , Fadel, E. , Philippe, B. , Kauffmann‐Lacroix, C. , Latge, J. and Beauvais, A. (2010) In vivo biofilm composition of Aspergillus fumigatus . Cell. Microbiol. 12, 405–410. [DOI] [PubMed] [Google Scholar]
- Luo, M. , Brown, R.L. , Chen, Z.‐Y. and Cleveland, T.E. (2009) Host genes involved in the interaction between Aspergillus flavus and maize. Toxin Rev. 28, 118–128. [Google Scholar]
- Manns, T.F. and Adams, J.F. (1923) Parasitic fungi internal of seed corn. J. Agric. Res. 23, 495–524. [Google Scholar]
- Marsh, S. and Payne, G. (1984) Preharvest infection of corn silks and kernels by Aspergillus flavus . Phytopathology, 74, 1284–1289. [Google Scholar]
- Mellon, J.E. , Cotty, P.J. and Dowd, M.K. (2007) Aspergillus flavus hydrolases: their roles in pathogenesis and substrate utilization. Appl. Microbiol. Biotechnol. 77, 497–504. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Manders, E.M.M. , Boas, S. , Olivain, C. , Alabouvette, C. and Rep, M. (2009) The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. Plos Pathog. 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef, F. , Hacker, C.R. , Patil, N. and Magnasco, M. (2002) Empirical characterization of the expression ratio noise structure in high‐density oligonucleotide arrays. Genome Biol. 3, RESEARCH0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, M.J. , Koulman, A. , Monahan, B.J. , Pritchard, B.L. , Payne, G.A. and Scott, B. (2009) Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75, 7469–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn, A. (2010) Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 26, 449–457. [DOI] [PubMed] [Google Scholar]
- Parsons, M. and Munkvold, G. (2010) Associations of planting date, drought stress, and insects with Fusarium ear rot and fumonisin B1 contamination in California maize. Food Addit. Contam. Part A, 27, 591–607. [DOI] [PubMed] [Google Scholar]
- Payne, G. , Hagler, W., Jr and Adkins, C. (1988a) Aflatoxin accumulation in inoculated ears of field‐grown maize. Plant Dis. 72, 422–424. [Google Scholar]
- Payne, G. , Thompson, D. , Lillehoj, E. , Zuber, M. and Adkins, C. (1988b) Effect of temperature on the preharvest infection of maize kernels by Aspergillus flavus . Phytopathology, 78, 1376–1380. [Google Scholar]
- Pazzagli, L. , Cappugi, G. , Manao, G. , Camici, G. , Santini, A. and Scala, A. (1999) Purification, characterization, and amino acid sequence of cerato‐platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani . J. Biol. Chem. 274, 24 959–24 964. [DOI] [PubMed] [Google Scholar]
- Price, M. , Conners, S. , Tachdjian, S. , Kelly, R. and Payne, G. (2005) Aflatoxin conducive and non‐conducive growth conditions reveal new gene associations with aflatoxin production. Fungal Genet. Biol. 42, 506–518. [DOI] [PubMed] [Google Scholar]
- Ramage, G. , Mowat, E. , Jones, B. , Williams, C. and Lopez‐Ribot, J. (2009) Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 35, 340–355. [DOI] [PubMed] [Google Scholar]
- Reese, B.N. , Payne, G.A. , Nielsen, D.M. and Woloshuk, C.P. (2011) Gene expression profile and response to maize kernels by Aspergillus flavus . Phytopathology, 101, 797–804. [DOI] [PubMed] [Google Scholar]
- Reeves, E. , Reiber, K. , Neville, C. , Scheibner, O. , Kavanagh, K. and Doyle, S. (2006) A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus . FEBS J. 273, 3038–3053. [DOI] [PubMed] [Google Scholar]
- Robens, J. and Cardwell, K. (2003) The costs of mycotoxin management to the USA: management of aflatoxins in the United States. Toxin Rev. 22, 139–152. [Google Scholar]
- Sakamoto, K. , Iwashita, K. , Yamada, O. , Kobayashi, K. , Mizuno, A. , Akita, O. , Mikami, S. , Shimoi, H. and Gomi, K. (2009) Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 46, 887–897. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , van Esse, H. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 3, 278–286. [DOI] [PubMed] [Google Scholar]
- Schaller, M. , Borelli, C. , Korting, H.C. and Hube, B. (2005) Hydrolytic enzymes as virulence factors of Candida albicans . Mycoses, 48, 365–377. [DOI] [PubMed] [Google Scholar]
- Shieh, M. , Brown, R. , Whitehead, M. , Cary, J. , Cotty, P. , Cleveland, T. and Dean, R. (1997) Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl. Environ. Microbiol. 63, 3548–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, M. , Wicklow, D. and Caldwell, R. (1990) Pathogenesis in Aspergillus ear rot of maize: light microscopy of fungal spread from wounds. Phytopathology, 80, 1287–1294. [Google Scholar]
- Smith, C. , Robertson, D. , Yates, B. , Nielsen, D. , Brown, D. , Dean, R. and Payne, G.A. (2008) The effect of temperature on Natural Antisense Transcript (NAT) expression in Aspergillus flavus . Curr. Genet. 54, 241–269. [DOI] [PubMed] [Google Scholar]
- Storey, J.D. (2002) A direct approach to false discovery rates. J. R. Stat. Soc. B, 64, 479–498. [Google Scholar]
- Taubenhaus, J. (1920) A study of the black and the yellow molds of ear corn. Tex. Agric. Exp. Sta. Bull. 270, 1–51. [Google Scholar]
- Thines, E. , Aguirre, J. , Foster, A. and Deising, H. (2006) Genetics of phytopathology: secondary metabolites as virulence determinants of fungal plant pathogens In: Progress in Botany, Vol. 67 (Esser K., Luttge U., Beyschlag W. and Murata J., eds), pp. 134–161. New York: Springer Publishing. [Google Scholar]
- US Food and Drug Administration (2000) Guidance for industry: action levels for poisonous or deleterious substances in human food and animal feed. Available at: http://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/ChemicalContaminantsMetalsNaturalToxinsPesticides/ucm077969.htm [accessed on Jun 26, 2013].
- White, D. (1999) Compendium of Corn Diseases. St. Paul, MN: APS Press. [Google Scholar]
- Wilson, B.J. and Wilson, C.H. (1964) Toxin from Aspergillus flavus: production on food materials of a substance causing tremors in mice. Science, 144, 177–178. [DOI] [PubMed] [Google Scholar]
- Windham, G. and Williams, W. (1998) Aspergillus flavus infection and aflatoxin accumulation in resistant and susceptible maize hybrids. Plant Dis. 82, 281–284. [DOI] [PubMed] [Google Scholar]
- Zahiri, A. , Heimel, K. , Wahl, R. , Rath, M. and Kämper, J. (2010) The Ustilago maydis forkhead transcription factor Fox1 is involved in the regulation of genes required for the attenuation of plant defenses during pathogenic development. Mol. Plant–Microbe Interact. 23, 1118–1129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Aspergillus flavus genes differentially expressed during maize seed colonization.


