Summary
Cysteine‐rich proteins (CRPs) encoded by some plant viruses in diverse genera function as RNA silencing suppressors. Within the N‐terminal portion of CRPs encoded by furoviruses, there are six conserved cysteine residues and a Cys–Gly–X–X–His motif (Cys, cysteine; Gly, glycine; His, histidine; X, any amino acid residue) with unknown function. The central domains contain coiled‐coil heptad amino acid repeats that usually mediate protein dimerization. Here, we present evidence that the conserved cysteine residues and Cys–Gly–X–X–His motif in the CRP of Chinese wheat mosaic virus (CWMV) are critical for protein stability and silencing suppression activity. Mutation of a leucine residue in the third coiled‐coil heptad impaired CWMV CRP activity for suppression of local silencing, but not for the promotion of cell‐to‐cell movement of Potato virus X (PVX). In planta and in vitro analysis of wild‐type and mutant proteins indicated that the ability of the CRP to self‐interact was correlated with its suppression activity. Deletion of up to 40 amino acids at the C‐terminus did not abolish suppression activity, but disrupted the association of CRP with endoplasmic reticulum (ER), and reduced its activity in the enhancement of PVX symptom severity. Interestingly, a short region in the C‐terminal domain, predicted to form an amphipathic α‐helical structure, was responsible for the association of CWMV CRP with ER. Overall, our results demonstrate that the N‐terminal and central regions are the functional domains for suppression activity, whereas the C‐terminal region primarily functions to target CWMV CRP to the ER.
Introduction
Virus replication in plant cells triggers an RNA‐based antiviral defence pathway, widely known as RNA silencing (Baulcombe, 2005; Ding, 2010). It is proposed that viral double‐stranded RNA (dsRNA) replication intermediates or highly structured single‐stranded (ss) viral RNAs are processed by a Dicer or Dicer‐like protein into small interfering RNAs (siRNAs) of 21–24 nucleotides, which are subsequently loaded into an effector complex, known as the RNA‐induced silencing complex (RISC), to serve as a guide for the slicing of homologous viral RNA (Ding and Voinnet, 2007). In plants, this defence mechanism operates not only to limit viral accumulation in the initial site of infection, but also to inhibit viral cell‐to‐cell and long‐distance movement (Deleris et al., 2006). To counteract this defence, most plant viruses encode a suppressor protein that interferes with RNA silencing pathways, and these are highly diverse in sequence and structure (Burgyán and Havelda, 2011). Initially, many suppressor proteins were shown to be able to bind long dsRNA or siRNA, and it was therefore thought that they acted by interfering at the step of dsRNA processing or loading of siRNA into RISC (Lakatos et al., 2006, Mérai et al., 2006). However, recent studies have shown that several suppressor proteins can bind a key protein in the silencing pathway or target siRNA for degradation (Giner et al., 2010; Wu et al., 2010).
Chinese wheat mosaic virus (CWMV) is one agent of the yellow mosaic disease in wheat crops in China. CWMV belongs to the genus Furovirus (family Virgaviridae), members of which cause diseases of cereals in the USA, Europe and East Asia (Diao et al., 1999). Other members include Soil‐borne wheat mosaic virus (SBWMV), Soil‐borne cereal mosaic virus (SBCMV), Sorghum chlorotic spot virus (SrCSV) and Oat golden stripe virus (OGSV) (Adams et al., 2011). Furoviruses are naturally transmitted by an obligate root‐infecting organism, Polymyxa graminis (order Plasmodiophorales). Like all furoviruses, CWMV has a bipartite, positive‐sense, ssRNA genome. RNA1 encodes three proteins required for viral replication and cell‐to‐cell movement. RNA2 encodes a 19‐kDa major coat protein (CP), a minor CP of 84 kDa produced by occasional read‐through of the UGA termination codon and a 19‐kDa small cysteine‐rich protein (CWMV CRP) (Diao et al., 1999).
Viruses belonging to the genera Hordeivirus, Tobravirus, Pecluvirus, Benyvirus, Pomovirus, Carlavirus and Vitivirus also encode small CRPs located in a 3′ proximal open reading frame (ORF) of one of their genomic RNAs. The CRPs do not show significant amino acid sequence similarity, but they are similarly characterized by the presence of multiple cysteine residues, usually in their N‐terminal portions (Koonin et al., 1991). CRPs encoded by hordei‐, tobra‐, furo‐ and pecluviruses (all members of the family Virgaviridae) contain a highly conserved Cys–Gly–Xaa–Xaa–His (CGxxH) motif (Cys, cysteine; Gly, glycine; His, histidine; Xaa, any amino acid residue), but its role is still unknown (Te et al., 2005). A number of studies have demonstrated similar roles for CRPs in viral genome accumulation (Dunoyer et al., 2001; Hehn et al., 1995; Liu et al., 2002), efficient virus movement (Gilmer et al., 1992; Yelina et al., 2002), seed transmission (Edwards, 1995; Wang et al., 1997) and symptom modulation (Donald and Jackson, 1994; Liu et al., 2002). Moreover, CRPs encoded by SBWMV and Barley stripe mosaic virus (BSMV; Hordeivirus) can functionally replace the Tobacco rattle virus (TRV; Tobravirus) 16K CRP, indicating that these proteins have a similar function (Liu et al., 2002). Most CRPs have been shown to possess RNA silencing suppression activities (Andika et al., 2012; Dunoyer et al., 2002; Martín‐Hernández and Baulcombe, 2008; Senshu et al., 2011; Te et al., 2005; Yelina et al., 2002).
The functional activities and properties of CRPs encoded by furoviruses have not yet been characterized in detail. In this study, we employed site‐directed mutagenesis and deletion analysis to identify the roles of conserved amino acid residues and domains in the CRP of CWMV in RNA silencing suppression, self‐interaction, subcellular localization and viral pathogenesis.
Results
CWMV CRP activity is weak in the suppression of local silencing, but effective in the promotion of viral cell‐to‐cell spread
An Agrobacterium co‐infiltration assay using green fluorescent protein (GFP)‐transgenic Nicotiana benthamiana line 16c plants (Voinnet et al., 2000) was carried out to investigate whether the CWMV CRP is an RNA silencing suppressor. The CWMV CRP gene was inserted into the expression cassette of the binary vector pBin61, and the resulting plasmid (pBin‐CWMV CRP) was transformed into Agrobacterium and then mixed with Agrobacterium harbouring a GFP‐expressing binary plasmid (pBin‐GFP), before infiltrating the mixture of bacterial cells into the leaves of 16c plants. Binary plasmids carrying two well‐characterized suppressors, HC‐Pro (pBin‐HC‐Pro) and p19 (pBin‐p19), encoded by Potato virus Y (PVY) and Tomato bushy stunt virus (TBSV), respectively, were included in the assay for comparison. As shown in Fig. 1A, stronger green fluorescence was observed in the leaf patches expressing CWMV CRP than in the patches receiving empty vector (pBin61), 3, 4 and 5 days after inoculation (dai), although to a much lesser extent than in those expressing HC‐Pro or p19. In RNA blot analyses, at 3 dai, a moderate level of GFP mRNA accumulation was detected in leaves expressing CWMV CRP, but this was greatly reduced at 4 and 5 dai. By contrast, in the leaves expressing HC‐Pro or p19, GFP mRNA was abundant from 3 to 5 dai (Fig. 1B). At 3 dai, GFP siRNAs were not reduced in leaves expressing CWMV CRP when compared with those receiving empty vector (Fig. 1C). These results show that CWMV CRP possesses a weak activity in the suppression of local silencing and no effect on the production and accumulation of siRNAs.
Figure 1.
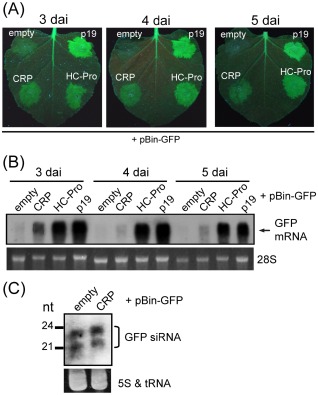
Silencing suppression activity of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) in Agrobacterium co‐infiltration assay. (A) Green fluorescent protein (GFP)‐transgenic Nicotiana benthamiana line 16c leaves were infiltrated with mixtures of Agrobacterium culture harbouring pBin‐GFP plus Agrobacterium harbouring the construct indicated [CWMV CRP, Potato virus Y (PVY) HC‐Pro or Tomato bushy stunt virus (TBSV) p19]. GFP fluorescence was visualized under long‐wavelength UV light and photographed 3, 4 and 5 days after inoculation (dai). (B) Northern blot analysis of GFP mRNA accumulation in agroinfiltrated patches of leaves shown in (A). The RNA gel was stained with ethidium bromide and 28S rRNA is shown as a loading control (bottom panel). (C) Northern blot analysis of GFP siRNA accumulation in agroinfiltrated patches of leaves shown in (A). The RNA gel was stained with ethidium bromide and 5S rRNA and tRNA are shown as loading control (bottom panel).
To further assess the silencing suppression activity of CWMV CRP, we performed a viral movement complementation assay (VMCA) using a Potato virus X (PVX) mutant (Senshu et al., 2011). PVX is the type species of the genus Potexvirus and its movement is facilitated by triple gene block (TGB) proteins (Verchot‐Lubicz et al., 2010). PVX TGBp1 (P25) has dual functions as a movement protein and RNA silencing suppressor (Bayne et al., 2005; Verchot‐Lubicz et al., 2010). In the VMCA assay, the cell‐to‐cell movement of P25‐defective (ΔP25) PVX‐GFP is trans‐complemented with a P25 mutant (contains the substitution of tyrosine‐117 to alanine) defective in silencing suppression, but retaining its virus movement function (Fig. 2A). Because cell‐to‐cell movement of PVX is dependent on the suppression of RNA silencing, the complementation of PVX cell‐to‐cell movement is only observed when a protein with silencing suppression activity is additionally expressed (Bayne et al., 2005). An Agrobacterium culture harbouring pGR106:PVX(ΔP25)‐GFP was highly diluted (10 000 fold) and mixed (1:1:1) with two Agrobacterium cultures, one culture harbouring pBin‐P25/T117A and another one harbouring pBin‐p19, pBin‐HC‐Pro or pBin61. These mixtures were then used to infiltrate N. benthamiana leaves. At 5 dai, numerous bright GFP foci were seen on leaves expressing CWMV CRP, p19 or HC‐Pro, but not on leaves receiving empty vector (Fig. 2B). When observed using confocal laser scanning microscopy (CLSM), most GFP fluorescence was seen as clusters of bright GFP‐expressing cells in leaves expressing CWMV CRP, p19 or HC‐Pro, whereas GFP expression was confined to a single cell in leaves expressing empty vector (Fig. 2C). No difference in the size of GFP foci or intensity of GFP fluorescence was observed between leaves expressing CWMV CRP and those expressing p19 or HC‐Pro, indicating that CWMV CRP is as effective as HC‐Pro or p19 in the promotion of cell‐to‐cell movement of the PVX mutant.
Figure 2.
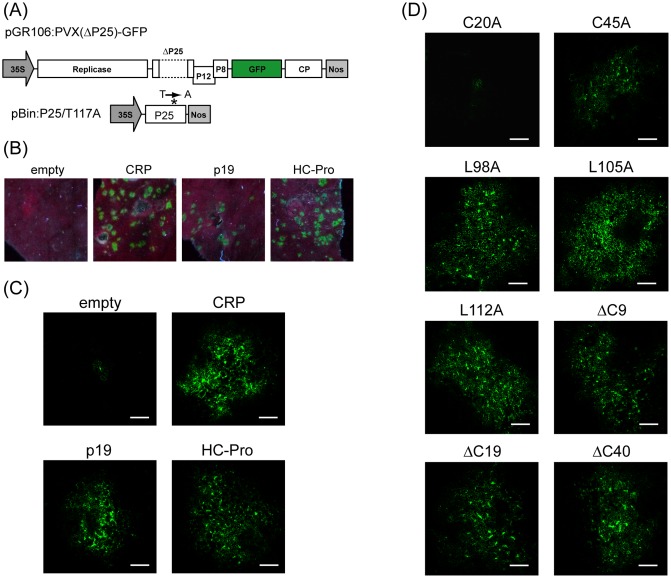
Silencing suppression activity of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) in a viral movement complementation assay. (A) Schematic representation of recombinant Potato virus X‐green fluorescent protein (PVX‐GFP) and P25 mutants used in trans‐complementation assay (not to scale). Broken lines represent the deleted region of the P25 gene (ΔP25). The P25 mutant (P25/T117A) which contains the substitution of tyrosine‐117 by alanine (marked with an asterisk) is defective in silencing suppression, but retains its virus movement function. 35S and Nos represent cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase terminator sequences, respectively. (B–D) Leaves of Nicotiana benthamiana plants infiltrated with mixtures of Agrobacterium cultures (1 : 1 : 1) harbouring pGR106:PVX(ΔP25)‐GFP (diluted 10 000‐fold), pBin‐P25/T117A and pBin‐p19, pBin‐HC‐Pro, pBin‐CRP or CRP mutants (see Fig. 3A). GFP fluorescence was observed using long‐wavelength UV light, and photographed (B) or observed using confocal laser scanning microscopy (C, D) at 5 days after inoculation (dai). Bars, 200 μm.
RNA silencing activity and protein stability of CWMV CRP mutants
The CWMV CRP has 67%, 63%, 60% and 36% amino acid identity to the CRPs encoded by the other furoviruses, OGSV, SBCMV, SBWMV and SrCSV, respectively. Amino acid alignment of these five CRPs shows that the N‐terminal portion (amino acids 1−112) is highly conserved, whereas the C‐terminal portion (amino acids 113−173) is more variable (Fig. 3A). A CGxxH motif is located at amino acid positions 70–74 in CWMV CRP. Seven cysteine residues in the N‐terminal half of CWMV CRP (at positions 8, 11, 20, 39, 45, 49 and 70) are conserved among these five CRPs. A motif comprising four or five heptad (abcdefg) repeats, which is the characteristic of a peptide that can form a coiled‐coil, was similarly detected in the central region of these proteins (coiled‐coil domain) (Fig. 3A). The C‐terminal ends of these five CRPs do not align; the SrCSV 18‐kDa CRP is the smallest and there are an additional 10 (SBCMV) or 19 amino acid residues in the other furoviruses (Fig. 3A).
Figure 3.
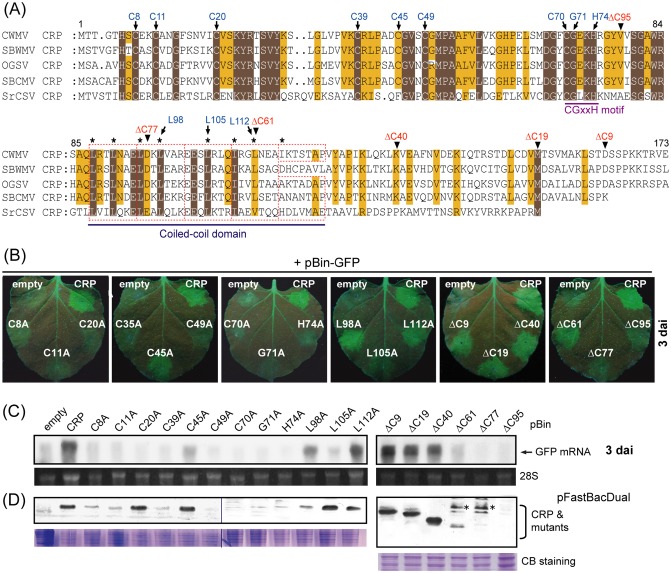
Effects of mutations in conserved amino acid residues and deletion of C‐terminal amino acids on silencing suppression activity and protein stability of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP). (A) Amino acid sequence alignment of the CRPs encoded by furoviruses. The amino acid positions of CWMV CRP are indicated above the alignment. Dots represent gaps. Dark brown or light brown boxes indicate that amino acids are identical or chemically similar, respectively. Broken red rectangles indicate the predicted heptad repeats. Highly hydrophobic residues at positions a and d of the heptad repeat (abcdefg)n are marked with asterisks. Amino acids that were substituted by alanine are marked with arrows and the C‐terminal ends of the deletion mutants are marked with arrowheads. (B) RNA silencing suppression activity of CWMV mutants in an Agrobacterium co‐infiltration assay. Leaves of Nicotiana benthamiana line 16c plants were infiltrated with mixtures of Agrobacterium harbouring pBin‐GFP plus Agrobacterium harbouring the binary vector containing the CWMV CRP mutants indicated in the images. Green fluorescent protein (GFP) fluorescence was visualized under UV light and photographed at 3 days after inoculation (dai). (C) Northern blot analysis of GFP mRNA accumulation in agroinfiltrated patches of leaves shown in (B). (D) Western blot analysis to detect the accumulation of wild‐type and mutants of CWMV CRP in insect cells. Coomassie blue (CB)‐stained total cell proteins are shown as loading controls (bottom panel). Antibody‐reactive bands of unknown origin are marked with asterisks.
To investigate the importance of the conserved cysteine residues and motifs in CWMV CRP for RNA silencing suppression activity, we generated CWMV CRP mutants in which cysteine at positions 8, 11, 20, 39, 45, 49 or 70, glycine at position 71, histidine at position 74 and leucine at positions 98, 105 or 112 (hydrophobic residues at position d of the second, third and fourth heptad repeats) were substituted by alanine. These mutants are referred to as C8A, C11A, C20A, C39A, C45A, C49A, C70A, G71A, H74A, L98A, L105A and L112A. In addition, to examine the role of the CWMV CRP C‐terminal region, we constructed a series of six deletion mutants in which 9, 19, 40, 61, 77 or 95 amino acids at the C‐terminus were removed (mutants ΔC9, ΔC19, ΔC40, ΔC61, ΔC77 and ΔC95, respectively). The CWMV CRP mutant sequences were inserted into the binary vector pBin61 and used in Agrobacterium co‐infiltration assays.
Mutants C8A, C11A, C20A, C39A, C49A, C70A, G71A, H74A and L105A were unable to suppress local silencing. Mutant L98A and L112A retained suppression activity, whereas C45A showed partially reduced activity (Fig. 3B,C). Although most of the conserved cysteine residues and the CGxxH motif were critical for suppressor activity, only one of the three leucine residues tested (at position 105) appeared to be essential. Mutants ΔC9, ΔC19 and ΔC40 displayed suppression activities similar to CWMV CRP, whereas ΔC61 showed very weak activity and none was observed for mutants ΔC77 and ΔC95 (Fig. 3B,C). This result indicates that the C‐terminal 40 amino acids are dispensable for the suppression of RNA silencing.
The accumulation of CWMV CRP or its mutants in agroinfiltrated leaves was below the detection limit of Western blot assays, which is possibly a result of the low levels of protein expression or low sensitivity of the CWMV CRP antiserum. We therefore made use of a baculovirus expression vector to express wild‐type and mutant derivatives of CWMV CRP in insect cells. Western blot analysis showed that C20A, C45A, L105A, L112A, ΔC9, ΔC19 and ΔC40 proteins accumulated at similar levels to the wild‐type protein, whereas the accumulation of C8A, C11A, C39A, C49A, C70A, G71A, H74A, L98A, ΔC61, ΔC77 and ΔC95 was low or undetectable (Fig. 3D). Next, to determine protein stability in planta, wild‐type and mutant derivatives of CWMV CRP were fused to the C‐terminus of enhanced GFP (EGFP) and expressed in leaves by Agrobacterium infiltration. The relative levels of fluorescence of the EGFP fusion proteins in leaves was in close agreement with the protein expression results obtained from the baculovirus system in insect cells. Proteins EGFP‐CRP, EGFP‐C20A, EGFP‐C45A, EGFP‐L105A, EGFP‐L112A, EGFP‐ΔC9, EGFP‐ΔC19, EGFP‐ΔC19 and EGFP‐ΔC40 were expressed at similar levels, whereas EGFP‐L98A and EGFP‐ΔC61 expression was weak and there was no detectable expression of the other fusion proteins (Fig. S1, see Supporting Information). In summary, these observations reveal that the inability of the majority of CWMV CRP mutants to suppress RNA silencing is probably a result of the failure of these proteins to stably accumulate in planta, rather than to a loss of silencing suppression activity per se. The exceptions are the C20A and L105A mutants, which still stably accumulate in planta but are defective in the suppression of local silencing.
Next, we also assessed the silencing suppression activity of CWMV CRP mutants by VMCA as described above. Only mutants that were stably expressed in planta were assayed. In leaves expressing mutants C45A, L98A, L105A, L112A, ΔC9, ΔC19 and ΔC40, GFP was observed in clusters of multiple cells, whereas, in leaves expressing mutant C20A, GFP remained in single cells (Fig. 2D). Except for mutant L105A, the result of analysis by VMCA was in agreement with the result obtained from Agrobacterium co‐infiltration assay (Fig. 3B,C). Thus, leucine‐105 is critical for the suppression of local silencing, but not for the inhibition of the RNA silencing that promotes viral cell‐to‐cell movement.
The ability of CWMV to self‐interact is correlated with its silencing suppression activity
Because other CRPs encoded by Peanut clump virus (PCV, Pecluvirus) and BSMV (Hordeivirus) can self‐interact (Bragg and Jackson, 2004; Dunoyer et al., 2002), we tested the ability of wild‐type and mutants of CWMV CRP to self‐interact using a bimolecular fluorescence complementation (BiFC) assay (Hu and Kerppola, 2003). These proteins were fused to N‐terminal or C‐terminal portions of yellow fluorescent protein (NYFP or CYFP) which had been inserted into the expression cassette of a binary vector (pBin61). The resulting plasmids were transformed into Agrobacterium and used to co‐infiltrate N. benthamiana leaves. The reconstruction of YFP fluorescence, indicating a protein–protein interaction, was visualized using CLSM at 3 dai. The BiFC assay showed that CWMV CRP and mutants C45A, L98A and L112A were able to self‐interact, but mutants C20A and L105A were not (Fig. 4A). To further assess the self‐interaction of CWMV CRP and mutants in vitro, a pull‐down assay was performed. Total protein extracts of Escherichia coli expressing maltose‐binding protein (MBP)‐CRP, MBP‐C20A, MBP‐L105A or unfused MBP (MBP) were mixed and incubated with total protein extracts of E. coli expressing glutathione S‐transferase (GST)‐CRP or unfused GST (GST). Amylose resin was used to pull down the MBP‐tagged recombinant proteins and any complex formed. Western blot analysis showed that GST‐CRP bound strongly to MBP‐CRP and only partially to MBP‐L105A, but not to MBP‐C20A or unfused MBP (Fig. 4B). These results indicate that mutants C20A and L105A are unable, or have reduced capacity, to form dimers. We also examined the self‐binding ability of mutant ΔC40 that retained suppression activity (Fig. 3B,C). Mutant ΔC40 showed self‐interaction in BiFC assay (Fig. 4C), as well as in the pull‐down assay, in which GST‐ΔC40 was co‐purified with MBP‐ΔC40 (Fig. 4D), indicating that the 40‐amino‐acid C‐terminal region is not essential for CWMV CRP self‐interaction. Taken together, these observations suggest that the ability of CWMV CRP to self‐interact is important for its suppression activity.
Figure 4.
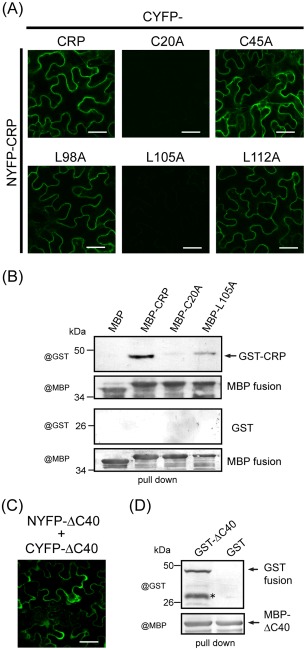
Self‐interaction of wild‐type and mutants of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP). (A, C) Bimolecular fluorescence complementation (BiFC) assay: leaves of Nicotiana benthamiana plants were infiltrated with mixtures of Agrobacterium harbouring constructs indicated above and on the left side of the images. Yellow fluorescent protein (YFP) fluorescence was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 30 μm. (B, D) Maltose‐binding protein (MBP) pull‐down assay: MBP‐CRP, MBP‐C20A, MBP‐L105A or free MBP (MBP) was mixed with glutathione S‐transferase (GST)‐CRP or unfused GST (GST) and an MBP pull‐down was performed (B). MBP‐ΔC40 was mixed with GST‐ΔC40 or unfused GST (GST) and an MBP pull‐down was performed (D). The purified proteins were analysed by Western blot using GST‐ and MBP‐specific antisera. An asterisk marks the signal from the degraded protein.
Deletion of the C‐terminal region disrupts the association of CWMV CRP with endoplasmic reticulum (ER) membranes
CLSM of N. benthamiana epidermal cells expressing EGFP‐CRP showed that the EGFP fusion accumulated in a cytoplasmic network (Fig. 5A) and the perinuclear area [confirmed with 4′,6‐diamidino‐2‐phenylindole (DAPI) staining] (Fig. 5B), whereas EGFP alone was diffusely distributed throughout the cytoplasm and nucleus (data not shown). This result suggests that CWMV CRP associates with ER membranes. To further examine this, CWMV CRP was fused to the C‐terminus of red fluorescent protein (RFP) and transiently expressed in leaves of transgenic N. benthamiana 16c that expressed GFP containing an ER targeting signal (Haseloff et al., 1997; Voinnet et al., 1998). GFP‐ER and RFP‐CRP were co‐localized in the cortical network and nuclear envelope (Fig. 5C), confirming that CWMV CRP associates with the ER. EGFP fused with the C20A, C45A, L98A, L105A and L112A mutants showed a similar localization in the perinuclear and cortical ER [Fig. S2 (see Supporting Information) and data not shown]. Because of the association of CWMV CRP with ER, we analysed the CWMV CRP sequence using several transmembrane (TM) segment prediction programs. Two programs, TMpred and Topred, similarly predicted that a region spanning amino acids 41 to 58 or 61 is a potential TM segment. In contrast, no TM region was predicted in the CRPs of other furoviruses. Examination of this region showed that two hydrophobic residues, serine‐48 and valine‐58, determine the difference in TM prediction between CWMV CRP and other CRPs of furoviruses. Serine‐48 and valine‐58 were substituted by asparagine and glutamic acid (S48N/V58E), respectively, and fused with EGFP. This fusion protein exhibited a weak fluorescence in epidermal cells, possibly as a result of protein instability, but retained its localization in the perinuclear ER (Fig. S2), suggesting that the predicted TM region is not responsible for the association of CWMV with ER membranes.
Figure 5.
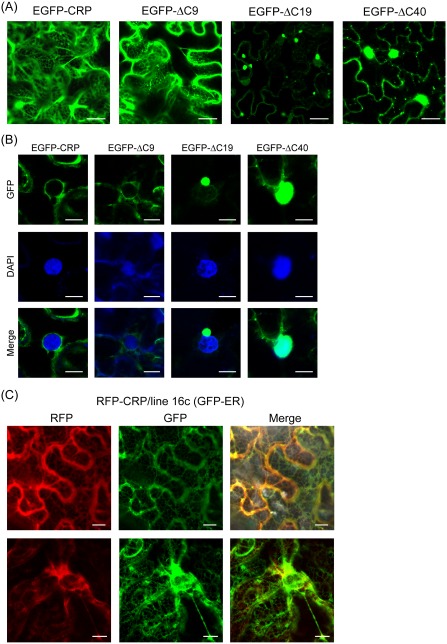
Subcellular distribution of wild‐type and deletion mutants of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP). (A, B) Epidermal cells of Nicotiana benthamiana plants transiently expressing enhanced green fluorescent protein (EGFP) fused with CWMV CRP wild‐type or mutants. Panels in (B) show the perinuclear areas. (C) Subcellular distribution of red fluorescent protein (RFP) fused with CWMV CRP in epidermal cells of transgenic N. benthamiana line 16c expressing GFP. Top and bottom panels show the cytoplasmic and perinuclear areas, respectively. Fluorescent proteins and 4′,6‐diamidino‐2‐phenylindole (DAPI) staining were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Images are derived from single confocal sections. Bars: 20 μm (A); 10 μm (B, C).
To examine the effect of C‐terminal deletion on CWMV CRP localization, the subcellular distribution of EGFP fused with ΔC9, ΔC19 or ΔC40 in leaves was observed by CLSM. EGFP‐ΔC9 associated with ER membranes in a similar manner to EGFP‐ CRP (Fig. 5A,B). EGFP‐ΔC19 accumulated in the cytoplasm and primarily aggregated to form granule‐like structures that sometimes located near the nucleus. No fluorescence signal was observed in the cortical and perinuclear ER, but, instead, a faint signal was observed inside the nucleus, indicating that part of the pool of EGFP‐ΔC19 enters the nucleus (Fig. 5A,B). EGFP‐ΔC40 showed accumulation in both the cytoplasm and nucleus, but not in the cortical ER network. In addition, many punctate structures scattered throughout the cytoplasm were observed (Fig. 5A,B). These observations strongly suggest that the 40‐amino‐acid C‐terminal region (134–173 amino acids) is required for the association of CWMV CRP with ER.
An amphipathic α‐helical domain in the C‐terminal region is responsible for association of CWMV CRP with ER
To further investigate the role of the C‐terminal region in CWMV CRP localization, the C‐terminal 40 amino acids were fused to EGFP and expressed in leaf epidermal cells. EGFP‐134–173aa accumulated in the cortical ER network, similar to EGFP‐CRP (Fig. 6D), indicating that the C‐terminal region contains the ER targeting signal. Because the deletion of nine amino acids at the C‐terminus did not affect the association of CWMV CRP with ER (Fig. 5A), the ER targeting signal possibly lies in a region between amino acids 134 and 164. Protein secondary structure programs predicted an α‐helical region roughly from amino acids 142 to 160 (Fig. 6A). Analysis by a helical wheel and helical net projection revealed that, within this predicted helical region, amino acids 141–158 have the potential to form an amphipathic α‐helix (Fig. 6B,C). The alignment of hydrophobic nonpolar residues (isoleucine‐143, leucine‐150, valine‐153, methionine‐154, valine‐157 and methionine‐158) on one face of the helix and hydrophilic residues on the other suggests an amphipathicity for this helix. In addition, there are two basic residues (lysine‐142 and arginine‐146) that occur in close proximity to the hydrophobic patch (Fig. 6B,C). The hydrophobic residues have the potential to traverse the phospholipid bilayer, whereas the basic residues will interact with acidic phospholipids. Thus, this helical structure possesses similar characteristics to the amphipathic α‐helical membrane‐binding domains of several other proteins (Bernstein et al., 2000; Elazar et al., 2004; Gouttenoire et al., 2009; Liu et al., 2009; Thiyagarajan et al., 2004). As expected, EGFP fused to amino acids 141–158 (EGFP‐141–158aa) associated with the cortical ER network, whereas substitution of methionine‐154 and valine‐157 with alanine (EGFP‐141–158aa M154A/V157A) abolished the association of the fusion protein with ER (Fig. 6D). Together, these observations suggest that the C‐terminal amphipathic α‐helical region is responsible for the association of CWMV CRP with ER.
Figure 6.
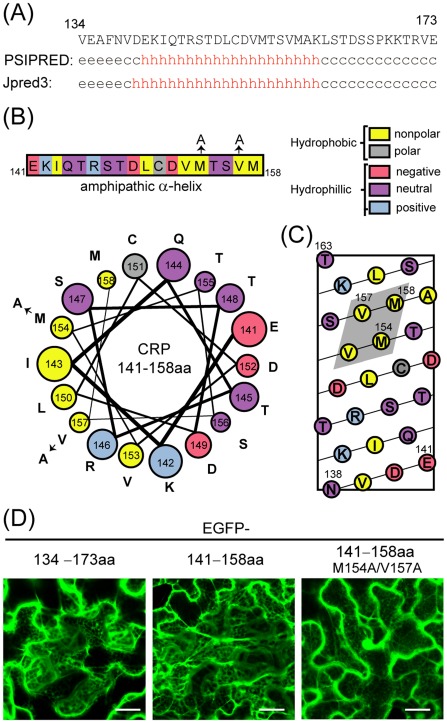
Role of the C‐terminal amphipathic α‐helical region in the association of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) with endoplasmic reticulum (ER). (A) Secondary structure predictions of the C‐terminal 40 amino acids of CWMV CRP using psipred and Jpred3. The predicted structures are indicated as helical (h), strand (e) or undetermined (c, coil). (B) Linear and helical wheel projections of CWMV CRP amino acids 141–158. Colour coding indicates amino acid characteristics. Amino acids that were substituted by alanine are indicated by arrows. (C) A helical net projection of CWMV CRP amino acids 138–163. The potential hydrophobic patch is shaded. (D) Subcellular localization of enhanced green fluorescent protein (EGFP) fused to the C‐terminal 40 amino acids (134–173aa), the predicted α‐helical region (141–158aa) or its substitution mutant (141–158aa M154A/V157A) of CWMV CRP. GFP fluorescence was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 25 μm.
Deletion of the C‐terminal region of CWMV CRP reduces its ability to enhance PVX symptom severity
CRPs have been shown to enhance the pathogenicity of heterologous viruses (Andika et al., 2012; Liu et al., 2002; Lukhovitskaya et al., 2005; Senshu et al., 2011; Te et al., 2005; Yelina et al., 2002). To test whether CWMV CRP has a similar activity, the wild‐type or mutant gene of CWMV CRP was inserted into a PVX vector (Fig. 7A) and the recombinant viruses were inoculated into N. benthamiana plants. From 8 dai, the upper leaves of plants infected with PVX‐CRP developed obvious mosaic symptoms, followed by severe wrinkling of the leaves and the production of small necrotic lesions, and eventually the plant became stunted (Fig. 7B). In contrast, plants infected with PVX carrying the CWMV CRP sequence in the antisense orientation (PVX‐as) or mutant C20A (PVX‐C20A) showed only mild mosaic symptoms and slight downwardly curled leaves (Fig. 7B). Plants infected with PVX‐ΔC9 also had severely wrinkled leaves, although the symptoms were less severe than in those infected with PVX‐CRP, and there were no necrotic lesions, whereas PVX‐ΔC19 and PVX‐ΔC40 induced less wrinkled leaves than PVX‐ΔC9 and the plants were not stunted (Fig. 7B). A Western blot assay detected similar levels of PVX CP accumulation in plants infected with PVX‐CRP, PVX‐ΔC9, PVX‐ΔC19 and PVX‐ΔC40, but lower levels in plants infected with PVX‐as or PVX‐C20A (Fig. 7C). These results suggest that the C‐terminal region contributes to the activity of CRP in enhancing PVX symptoms, but not in PVX accumulation.
Figure 7.
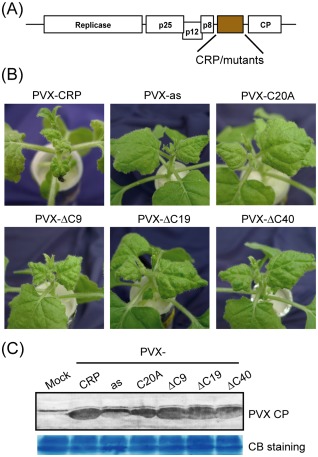
Effects of the expression of wild‐type and mutants of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) on Potato virus X (PVX) symptoms and accumulation. (A) Schematic representation of a PVX vector (pGR106 base) chimera carrying CWMV CRP or mutant genes (not to scale). (B) Nicotiana benthamiana plants infected with PVX vector chimeras carrying the wild‐type, antisense (as) or mutants (C20A, ΔC9, ΔC19 and ΔC40) of CWMV CRP. Plants were photographed at 14 days after inoculation (dai). (C) Western blot analysis of PVX coat protein (CP) accumulation in the upper systemically infected leaves of N. benthamiana. Total protein was extracted from leaves at 10 dai. Coomassie blue (CB)‐stained total cell proteins are shown as loading controls (bottom panel).
Discussion
In this study, we confirmed that the CWMV CRP acts as an RNA silencing suppressor, a characteristic that is shared by many small CRPs encoded by plant RNA viruses. In co‐infiltration assays, CWMV CRP silencing suppression activity is weak and transient compared with those of PVY HC‐Pro and TBSV p19 (Fig. 1). In this regard, CWMV CRP resembles the Beet necrotic yellow vein virus (Benyvirus) p14, TRV 16K and Potato virus M (PVM; Carlavirus) 12‐kDa CRPs, which also exhibit weak silencing suppression activity in the co‐infiltration assay (Andika et al., 2012; Martín‐Hernández and Baulcombe, 2008; Senshu et al., 2011). However, BSMV γb and PCV P15 CRPs possess a strong silencing suppression activity, similar to those of HC‐Pro and p19 (Bragg and Jackson, 2004; Dunoyer et al., 2002), showing that a weak silencing suppression activity is not a general characteristic of CRPs. In VMCA, CWMV CRP showed a similar level of activity to p19 or HC‐Pro in the promotion of PVX cell‐to‐cell spread (Fig. 2), demonstrating that CWMV CRP effectively inhibits the spread of the silencing signals that prevent virus cell‐to‐cell movement. Interestingly, mutant L105A retained the ability to promote the cell‐to‐cell spread of PVX, although impaired in its ability to suppress local silencing (Figs 2 and 3), suggesting that these two functions are not necessarily linked. In line with this observation, the movement protein of PVM (TGBp1) and Apple chlorotic leaf spot virus (Trichovirus), as well as the CP of Citrus tristeza virus (Closterovirus), can suppress the spread of the silencing signal despite being unable to interfere with local silencing (Lu et al., 2004; Senshu et al., 2011; Yaegashi et al., 2007).
The region comprising the N‐terminal half and central region of the five CRPs of furoviruses is highly conserved (Fig. 3A). The N‐terminal half contains seven cysteine residues and the CGxxH motif. Within the central region, a coiled‐coil structure (Lupas and Gruber, 2005), which is generally known to mediate homologous protein interactions, was predicted to exist. In contrast, the C‐terminal regions do not show any significant sequence homology (Fig. 3A). Deletion of up to 40 amino acids at the C‐terminus did not affect CWMV CRP suppression activity, and suppression activity was still partially retained even after deletion of the C‐terminal 61 amino acids (Fig. 3B,C), supporting the notion that the N‐terminal half and central region are the functional regions for CWMV CRP silencing suppression activity. Mutational analysis revealed that cysteine‐8, cysteine‐11 and cysteine‐39, and also cysteine‐70, glycine‐71 and histidine‐74 in the CGxxH motif, are critical for protein stability. By contrast, mutation of cysteine‐20 and cysteine‐45 did not affect protein stability, but abolished or partially reduced CWMV CRP suppression activity. Cysteine‐45 is conserved among four furovirus CRPs, but is not present in SrCSV 18‐kDa CRP (Fig. 3A), and therefore is not structurally and functionally critical for CWMV CRP activity. On the basis of the close phylogenetic relatedness among CRPs encoded by furoviruses, pecluviruses, tobraviruses and hordeiviruses (Te et al., 2005), our data suggest a possible common role for the CGxxH motif in maintaining the structure of these CRPs.
Cysteine residues in the CRPs of hordeiviruses and carlaviruses are arranged in zinc‐finger‐like motifs, and mutation of the cysteine residues in these proteins does not appear to affect protein stability, although it compromises their ability to bind zinc in vitro and attenuates symptoms of the viral disease (Bragg et al., 2004), interferes with RNA silencing suppression (Senshu et al., 2011) and DNA binding, and with the induction of a hypersensitive reaction (Lukhovitskaya et al., 2009). No zinc‐finger motif has been identified in the CRPs of furoviruses, pecluviruses and tobraviruses, and their zinc‐binding activity has not been reported. Given the crucial role of cysteine residues in the stability of CWMV CRP, it seems reasonable to assume that some of these cysteine residues may be required for the formation of disulphide bonds, which are commonly required for the appropriate folding and stability of proteins.
The CRPs of hordeiviruses and pecluviruses also contain coiled‐coil domains, and the roles of this domain in self‐interaction and silencing suppression have been established for BSMV γb and PCV P15 CRPs (Bragg and Jackson, 2004; Dunoyer et al., 2002). In this study, mutational analysis provided evidence that the coiled‐coil structure contributes to CWMV CRP self‐interaction. Moreover, the coiled‐coil structure in CWMV CRP also seems to be important for protein stability, because the C‐terminal deletions that partially or completely removed the coiled‐coil domain drastically reduced protein stability (Fig. 3D). In the coiled‐coil structure, the a and d positions of heptad amino acids (abcdefg) are usually occupied by hydrophobic residues to form the hydrophobic face of the amphipathic α‐helical structure (Lupas and Gruber, 2005). Among three leucine residues at position d of the heptad repeat that were substituted by alanine (less hydrophobic), only the substitution of leucine at the third heptad (leucine‐105) affected CWMV CRP self‐interaction. A polar residue (glutamic acid‐102) is present at position a of the third heptad, instead of the usual highly hydrophobic residue (Fig. 3A). Possibly because of this condition, leucine‐105 plays a critical role in the homologous interaction between the coiled‐coil structures. Mutation of cysteine‐20 also had a deleterious effect on self‐interaction (Fig. 4), which may suggest that cysteine‐20 has a direct role in CWMV CRP self‐interaction. Another possibility is that mutation of cysteine‐20 drastically alters the protein structure, which, in turn, destabilizes the self‐interaction. Studies on the p19 suppressor of tombusviruses and the P1b suppressor of Cucumber vein yellowing virus (Ipomovirus) have indicated that dimerization is important for the activity of these proteins to bind siRNA (Valli et al., 2008; Vargason et al., 2003; Ye et al., 2003). As BSMV γb and PCV P15 CRP can bind siRNA (Mérai et al., 2006), it would be interesting to investigate whether CWMV CRP also has a similar ability, and whether dimer formation is also required for its siRNA binding activity. Dimer forms of BSMV γb and PCV P15 CRPs are stable and persist to some extent under standard denaturing conditions (Bragg and Jackson, 2004; Dunoyer et al., 2002), indicating a strong protein–protein interaction. In contrast, under similar denaturing conditions, no CWMV CRP dimer was detected by Western blot in the total proteins extracted from CWMV‐infected leaves or from E. coli or insect cells expressing CWMV CRP (data not shown). The CWMV CRP dimer may be less stable than those of BSMV γb and PCV P15 CRPs in vivo, which could explain the weak and transient activity of CWMV CRP in Agrobacterium co‐infiltration assays.
The organelle targeting of certain suppressor proteins has not been particularly linked with the suppression function, which is consistent with the notion that RNA silencing is primarily a cytoplasmic event. For example, the localization of Cucumber mosaic virus (Cucumovirus) 2b suppressor and PCV P15 CRP to nuclei and peroxisomes, respectively, is not required for suppression activity (Dunoyer et al., 2002; González et al., 2012). Likewise, our results showed that the C‐terminal ER targeting signal of CWMV CRP is dispensable for suppression activity (Figs 3 and 5). Thus, organelle targeting may be required for an ancillary function of suppressor proteins, as in the case of PCV P15 CRP localization to peroxisomes, which is required for virus systemic movement (Dunoyer et al., 2002). The association of CWMV CRP with ER may contribute to its pathogenic function because deletion of the C‐terminal region reduced its activity in enhancing PVX symptoms (Fig. 7). The replication of many positive‐strand RNA viruses is known to occur on ER membranes (den Boon and Ahlquist, 2010). It is interesting to note that, like CWMV CRP (Fig. 6), the association of some viral replication‐associated proteins with ER is mediated by an amphipathic α‐helical region (Elazar et al., 2004; Gouttenoire et al., 2009; Liu et al., 2009). Future studies will focus on the elucidation of the significance of the association of CWMV CRP with ER in the context of CWMV replication.
Experimental Procedures
Plant materials
GFP‐transgenic N. benthamiana line 16c (Voinnet et al., 1998) was kindly provided by David Baulcombe (University of Cambridge, UK). Nicotiana benthamiana plants were grown on soil or quartz sand supplied daily with Hoagland and Arnon solution, as described previously (Andika et al., 2005). The plants were kept in a growth cabinet at 24 °C with 16 h of daylight.
Plasmid construction
The CWMV CRP gene was amplified by reverse transcription‐polymerase chain reaction (RT‐PCR) using total RNA extracted from leaves of wheat plants infected with CWMV isolate Rongcheng (Shandong Province, China) (Yang et al., 2001). Site‐directed mutagenesis of CWMV CRP and PVX P25 (TGBp1) was carried out by two PCR steps, as described previously (Herlitze and Koenen, 1990). For transient expression experiments, DNA fragments were ligated into the binary vector pBin61 (Voinnet et al., 1998) between XbaI or BamHI and SmaI sites. pBin‐GFP, pBin‐HC‐Pro and pBin‐p19 (Voinnet et al., 1999, 2000, 2003) were provided by David Baulcombe (University of Cambridge, UK). To prepare fluorescent protein fusion constructs, the EGFP or RFP gene was first inserted between the XbaI and BamHI sites, and DNA fragments were then inserted between the BamHI and SmaI sites of pBin‐EGFP or pBin‐RFP. For the BiFC assay, DNA fragments corresponding to amino acids 1−174 and 175–239 of the YFP gene were inserted between the XbaI and BamHI sites of pBin61. Subsequently, DNA fragments were inserted between the BamHI and SmaI restriction sites located downstream of the N‐terminal or C‐terminal portions of the YFP gene in pBin‐NYFP and pBin‐CYFP plasmids. To construct PVX vector chimeras, the DNA fragments were inserted into the NotI site of pGR106 (Lu et al., 2003). PVX(ΔP25)‐GFP was generated by digestion of pGR106‐GFP plasmid with ScaI and ApaI, which deleted the 3′ and 5′ portions of the replicase and P25 ORFs, respectively. Subsequently, a PCR fragment corresponding to the ScaI–ApaI region, but containing 354 base pair deletions in the P25 ORF, was ligated. This deletion introduces an in‐frame 118‐amino‐acid internal deletion in the P25 protein, similar to that described previously (Voinnet et al., 2000). For the expression of recombinant proteins in E. coli, DNA fragments were introduced between the BamHI and NotI sites of pGEX‐6P‐1 (Invitrogen, Grand Island, NY, USA) and between the BamHI and SalI sites of pMBP‐c2X (New England BioLabs, Ipswich, MA, USA) for the expression of GST‐ and MBP‐tagged recombinant proteins, respectively. For baculovirus expression in Spodoptera frugiperda 9 (Sf9) cells, DNA fragments were inserted into the NotI site of the pFastBacDual vector (Invitrogen).
CWMV CRP antiserum production
GST‐tagged CWMV CRP was expressed in E. coli strain BL21 (Novagen, Madison, WI, USA) and purified using the methods described previously (Sun and Suzuki, 2008). Protein was diluted in a buffer containing 50 mm Tris‐HCl (pH 8.0), 75 mm NaCl and 1 × protease inhibitor cocktail (Roche, Mannheim, Germany), and emulsified with Freund's incomplete adjuvant (Difco, Franklin Lakes, NJ, USA). Antigen (1 mg/mL) was subcutaneously injected into New Zealand white rabbits. Antiserum was produced by Shengong Biotech Co. Ltd. (Shanghai, China).
Agrobacterium infiltration and PVX vector inoculation
For Agrobacterium infiltration, plasmid constructs were transformed into Agrobacterium tumefaciens strain C58C1 and used to infiltrate N. benthamiana, as described previously (Voinnet et al., 1998). For inoculation of PVX vector chimeras, plasmid constructs were transformed into A. tumefaciens strain GV3101 harbouring transformation helper plasmid pSup, and used to inoculate N. benthamiana, as described previously (Lu et al., 2003).
Baculovirus expression
Recombinant baculoviruses were prepared using the Bac‐to‐Bac Baculovirus Expression System (Invitrogen). Culture of Sf9 cells and cell lysis were performed as described previously (Sun and Suzuki, 2008).
MBP pull‐down assay
Total crude protein of E. coli (strain BL21) expressing the recombinant protein was extracted by sonication in a binding buffer containing 20 mm Tris‐HCl (pH 7.0), 0.2 m NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 10 mm β‐mercaptoethanol and 1 × protease inhibitor cocktail (Roche). Two protein extracts containing different recombinant proteins (c. 0.05 mg each) were mixed and incubated overnight at 4 °C. MBP pull‐down was performed by the addition of 0.4 mL of amylase resin (New England BioLabs) to each sample, followed by incubation for 4 h at 4 °C with rotation. The incubated resin of each sample was washed five times with binding buffer, resuspended in sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) sample buffer, boiled for 8 min, run on an SDS‐PAGE gel and analysed by Western blotting.
RNA blot analysis
Total RNA was extracted by Trizol (Invitrogen) according to the manufacturer's protocol. Digoxigenin (DIG)‐labelled DNA probes were used for RNA blot hybridization. For the detection of GFP mRNA, GFP siRNA and PVX RNA, DNA probes covering the entire GFP gene, GFP 3′‐terminal 246 nucleotides or PVX CP gene, respectively, were prepared using the PCR DIG Probe Synthesis Kit (Roche). The hybridization conditions and detection of mRNAs were as described in the DIG Application Manual supplied by Roche. Gel electrophoresis, blotting and detection of siRNAs using the DIG system were carried out as described previously (Goto et al., 2003).
Western blot analysis
Preparation of protein samples, SDS‐PAGE, electroblotting and immunodetection were carried out as described previously (Sun and Suzuki, 2008). CWMV CRP, MBP and PVX CP were detected using primary, anti‐CWMV CRP (1:5000), anti‐MBP (1:1:000) (New England BioLabs) and anti‐PVX CP (1:2000) polyclonal sera, respectively, and secondary polyclonal alkaline phosphatase‐conjugated goat anti‐rabbit immunoglobulin G (IgG) (1:10 000) (Sigma, St. Louis, MO, USA). GST was detected using primary (1:10 000) anti‐GST monoclonal serum (Zhongshan Jinqiao, Beijing, China) and secondary (1:10 000) alkaline phosphatase‐conjugated goat anti‐mouse IgG (Sigma).
Fluorescent protein and DAPI imaging
GFP fluorescence in Agrobacterium co‐infiltration assays was visualized using a UV handy lamp (Peking Liuyi model WD‐9403E, Beijing, China). YFP, GFP or RFP expression and DAPI staining in epidermal cells were observed using a Leica TCS SP5 CLSM (Leica Microsystems, Wetzlar, Germany).
Sequence analysis
Alignments of CRPs of furoviruses were prepared in ClustalW2 using sequences derived from CWMV (GenBank Accession No. AJ271839), SBWMV (L07938), SBCMV (AJ132577), OGSV (AJ132579) and SrCSV (AB033692). The coiled‐coil domain was predicted using the coils program (Lupas et al., 1991; http://www.ch.embnet.org/software/COILS_form.html). For the prediction of TM protein segments, the programs TopPred II (Claros and von Heijne, 1994; http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) and TMpred (Hofmann and Stoffel, 1993; http://www.ch.embnet.org/software/TMPRED_form.html) were used. The protein secondary structure was predicted using psipred (Jones, 1999; http://bioinf.cs.ucl.ac.uk/psipred/) and Jpred3 (Cole et al., 2008; http://www.compbio.dundee.ac.uk/www‐jpred/). Helical wheel and helical net projection were generated using genetyx‐mac version 15.01 (Software Development Co. Ltd, Tokyo, Japan).
Supporting information
Fig. S1 Expression of enhanced green fluorescent protein (EGFP) fused to the wild‐type and Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) mutants in epidermal cells. Leaves of Nicotiana benthamiana plants were infiltrated with Agrobacterium harbouring a binary vector containing the constructs indicated above the images. EGFP fluorescence was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 30 μm.
Fig. S2 Accumulation of enhanced green fluorescent protein (EGFP) fused with Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) mutants in the perinuclear region. Leaves of Nicotiana benthamiana plants were infiltrated with Agrobacterium harbouring a binary vector containing the constructs indicated above the images. EGFP fluorescence and 4′,6‐diamidino‐2‐phenylindole (DAPI) staining in epidermal cells were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 10 μm.
Acknowledgements
We thank David Baulcombe for providing the plasmids and plant materials, Yanli Wang for providing the RFP plasmid, Rong Xiang and Zilong Tan for experimental assistance and Mike Adams for critical reading of the manuscript. This study was funded by grants from the Project of New Varieties of Genetically Modified Wheat of China (2008ZX08002‐001) and China Agriculture Research System (CARS‐3‐1) from the Ministry of Agriculture of China, the Project of Molecular Mechanism of Plant Defense to Pest and Disease (2012CB722504) from the Ministry of Science and Technology of China, and Modern Agricultural Biotechnology and Crop Disease Control from the Key Subject Construction Program of Zhejiang Province.
References
- Adams, M.J. , Heinze, C. , Jackson, A.O. , Kreuze, J. , Macfarlane, S.A. and Torrance, L. (2011) Family Virgaviridae In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J., eds), pp. 1139–1162. London: Elsevier Academic Press. [Google Scholar]
- Andika, I.B. , Kondo, H. and Tamada, T. (2005) Evidence that RNA silencing‐mediated resistance to Beet necrotic yellow vein virus is less effective in roots than in leaves. Mol. Plant–Microbe Interact. 18, 194–204. [DOI] [PubMed] [Google Scholar]
- Andika, I.B. , Kondo, H. , Nishiguchi, M. and Tamada, T. (2012) The cysteine‐rich proteins of beet necrotic yellow vein virus and tobacco rattle virus contribute to efficient suppression of silencing in roots. J. Gen. Virol. 93, 1841–1850. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2005) RNA silencing. Trends Biochem. Sci. 30, 290–293. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. and Baulcombe, D.C. (2005) Cell‐to‐cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Bernstein, L.S. , Grillo, A.A. , Loranger, S.S. and Linder, M.E. (2000) RGS4 binds to membranes through an amphipathic α‐helix. J. Biol. Chem. 275, 18 520–18 526. [DOI] [PubMed] [Google Scholar]
- den Boon, J.A. and Ahlquist, P. (2010) Organelle‐like membrane compartmentalization of positive‐strand RNA virus replication factories. Annu. Rev. Microbiol. 64, 241–256. [DOI] [PubMed] [Google Scholar]
- Bragg, J. and Jackson, A. (2004) The C‐terminal region of the Barley stripe mosaic virusγb protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5, 465–481. [DOI] [PubMed] [Google Scholar]
- Bragg, J.N. , Lawrence, D.M. and Jackson, A.O. (2004) The N‐terminal 85 amino acids of the barley stripe mosaic virus γb pathogenesis protein contain three zinc‐binding motifs. J. Virol. 78, 7379–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyán, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Claros, M.G. and von Heijne, G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Cole, C. , Barber, J.D. and Barton, G.J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A. , Gallego‐Bartolome, J. , Bao, J.S. , Kasschau, K.D. , Carrington, J.C. and Voinnet, O. (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science, 313, 68–71. [DOI] [PubMed] [Google Scholar]
- Diao, A. , Chen, J. , Ye, R. , Zheng, T. , Yu, S. , Antoniw, J.F. and Adams, M.J. (1999) Complete sequence and genome properties of Chinese wheat mosaic virus, a new furovirus from China. J. Gen. Virol. 80, 1141–1145. [DOI] [PubMed] [Google Scholar]
- Ding, S. (2010) RNA‐based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, R.G. and Jackson, A.O. (1994) The barley stripe mosaic virus gamma b gene encodes a multifunctional cysteine‐rich protein that affects pathogenesis. Plant Cell, 6, 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P. , Herzog, E. , Hemmer, O. , Ritzenthaler, C. and Fritsch, C. (2001) Peanut clump virus RNA‐1‐encoded P15 regulates viral RNA accumulation but is not abundant at viral RNA replication sites. J. Virol. 75, 1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P. , Pfeffer, S. , Fritsch, C. , Hemmer, O. , Voinnet, O. and Richards, K. (2002) Identification, subcellular localization and some properties of a cysteine‐rich suppressor of gene silencing encoded by peanut clump virus. Plant J. 29, 555–567. [DOI] [PubMed] [Google Scholar]
- Edwards, M.C. (1995) Mapping of the seed transmission determinants of barley stripe mosaic virus. Mol. Plant–Microbe Interact. 8, 906–915. [DOI] [PubMed] [Google Scholar]
- Elazar, M. , Liu, P. , Rice, C.M. and Glenn, J.S. (2004) An N‐terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78, 11 393–11 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmer, D. , Bouzoubaa, S. , Hehn, A. , Guilley, H. , Richards, K. and Jonard, G. (1992) Efficient cell‐to‐cell movement of beet necrotic yellow vein virus requires 3′ proximal genes located on RNA 2. Virology, 189, 40–47. [DOI] [PubMed] [Google Scholar]
- Giner, A. , Lakatos, L. , García‐Chapa, M. , López‐Moya, J. and Burgyán, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, I. , Rakitina, D. , Semashko, M. , Taliansky, M. , Praveen, S. , Palukaitis, P. , Carr, J.P. , Kalinina, N. and Canto, T. (2012) RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA, 18, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, K. , Kanazawa, A. , Kusaba, M. and Masuta, C. (2003) A simple and rapid method to detect plant siRNAs using nonradioactive probes. Plant Mol. Biol. Rep. 21, 51–58. [Google Scholar]
- Gouttenoire, J. , Montserret, R. , Kennel, A. , Penin, F. and Moradpour, D. (2009) An amphipathic α‐helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83, 11 378–11 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J. , Siemering, K. , Prasher, D. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehn, A. , Bouzoubaa, S. , Bate, N. , Twell, D. , Marbach, J. , Richards, K. , Guilley, H. and Jonard, G. (1995) The small cysteine‐rich protein P14 of beet necrotic yellow vein virus regulates accumulation of RNA 2 in cis and coat protein in trans. Virology, 210, 73–81. [DOI] [PubMed] [Google Scholar]
- Herlitze, S. and Koenen, M. (1990) A general and rapid mutagenesis method using polymerase chain reaction. Gene, 91, 143–147. [DOI] [PubMed] [Google Scholar]
- Hofmann, K. and Stoffel, W. (1993) TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe‐Seyler, 374, 166. [Google Scholar]
- Hu, C. and Kerppola, T. (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. (1999) Protein secondary structure prediction based on position‐specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Koonin, E. , Boyko, V. and Dolja, V. (1991) Small cysteine‐rich proteins of different groups of plant RNA viruses are related to different families of nucleic acid‐binding proteins. Virology, 181, 395–398. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y.P. , Dolja, V.V. , Calvino, L.F. , López‐Moya, J.J. and Burgyán, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Reavy, B. , Swanson, M. and MacFarlane, S. (2002) Functional replacement of the tobacco rattle virus cysteine‐rich protein by pathogenicity proteins from unrelated plant viruses. Virology, 298, 232–239. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Westler, W.M. , den Boon, J.A. , Wang, X. , Diaz, A. , Steinberg, H.A. and Ahlquist, P. (2009) An amphipathic α‐helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5, e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D.C. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Folimonov, A. , Shintaku, M. , Li, W.X. , Falk, B.W. , Dawson, W.O. and Ding, S‐W. (2004) Three distinct suppressors of RNA silencing encoded by a 20‐kb viral RNA genome. Proc. Natl. Acad. Sci. USA, 101, 15 742–15 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhovitskaya, N. , Yelina, N. , Zamyatnin, A.J. , Schepetilnikov, M. , Solovyev, A. , Sandgren, M. , Morozov, S.Y. , Valkonen, J.P. and Savenkov, E.L. (2005) Expression, localization and effects on virulence of the cysteine‐rich 8 kDa protein of Potato mop‐top virus . J. Gen. Virol. 86, 2879–2889. [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Ignatovich, I.V. , Savenkov, E.I. , Schiemann, J. , Morozov, S.Y. and Solovyev, A.G. (2009) Role of the zinc‐finger and basic motifs of chrysanthemum virus B p12 protein in nucleic acid binding, protein localization and induction of a hypersensitive response upon expression from a viral vector. J. Gen. Virol. 90, 723–733. [DOI] [PubMed] [Google Scholar]
- Lupas, A. , Van Dyke, M. and Stock, J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Lupas, A.N. and Gruber, M. (2005) The structure of α‐helical coiled coils. Adv. Protein Chem. 70, 37–78. [DOI] [PubMed] [Google Scholar]
- Martín‐Hernández, A. and Baulcombe, D. (2008) Tobacco rattle virus 16‐kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J. Virol. 82, 4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai, Z. , Kerenyi, Z. , Kertesz, S. , Magna, M. , Lakatos, L. and Silhavy, D. (2006) Double‐stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80, 5747–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu, H. , Yamaji, Y. , Minato, N. , Shiraishi, T. , Maejima, K. , Hashimoto, M. , Miura, C. , Neriya, Y. and Namba, S. (2011) A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by potato virus M. J. Virol. 85, 10 269–10 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. and Suzuki, N. (2008) Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1‐EP713. RNA, 14, 2557–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te, J. , Melcher, U. , Howard, A. and Verchot‐Lubicz, J. (2005) Soilborne wheat mosaic virus (SBWMV) 19K protein belongs to a class of cysteine rich proteins that suppress RNA silencing. Virol. J. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiyagarajan, M.M. , Stracquatanio, R.P. , Pronin, A.N. , Evanko, D.S. , Benovic, J.L. and Wedegaertner, P.B. (2004) A predicted amphipathic helix mediates plasma membrane localization of GRK5. J. Biol. Chem. 279, 17 989–17 995. [DOI] [PubMed] [Google Scholar]
- Valli, A. , Dujovny, G. and García, J. (2008) Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from cucumber vein yellowing ipomovirus. J. Virol. 82, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J. , Szittya, G. , Burgyán, J. and Hall, T. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Torrance, L. , Solovyev, A.G. , Morozov, S.Y. , Jackson, A.O. and Gilmer, D. (2010) Varied movement strategies employed by triple gene block‐encoding viruses. Mol. Plant–Microbe Interact. 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Vain, P. , Angell, S. and Baulcombe, D.C. (1998) Systemic spread of sequence‐specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Pinto, Y.M. and Baulcombe, D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA, 96, 14 147–14 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. , Lederer, C. and Baulcombe, D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana . Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Rivas, S. , Mestre, P. and Baulcombe, D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wang, D. , MacFarlane, S.A. and Maule, A.J. (1997) Viral determinants of pea early browning virus seed transmission in pea. Virology, 234, 112–117. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Wang, X. and Ding, S. (2010) Viral suppressors of RNA‐based viral immunity: host targets. Cell Host Microbe, 8, 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegashi, H. , Takahashi, T. , Isogai, M. , Kobori, T. , Ohki, S. and Yoshikawa, N. (2007) Apple chlorotic leaf spot virus 50 kDa movement protein acts as a suppressor of systemic silencing without interfering with local silencing in Nicotiana benthamiana . J. Gen. Virol. 88, 316–324. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Chen, J. , Chen, J. , Jiang, H. , Zhao, Q. and Adams, M.J. (2001) Sequence of a second isolate of Chinese wheat mosaic furovirus. J. Phytopathol. 149, 135–140. [Google Scholar]
- Ye, K. , Malinina, L. and Patel, D. (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature, 426, 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina, N. , Savenkov, E. , Solovyev, A. , Morozov, S. and Valkonen, J. (2002) Long‐distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei‐ and potyviruses: complementary functions between virus families. J. Virol. 76, 12 981–12 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression of enhanced green fluorescent protein (EGFP) fused to the wild‐type and Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) mutants in epidermal cells. Leaves of Nicotiana benthamiana plants were infiltrated with Agrobacterium harbouring a binary vector containing the constructs indicated above the images. EGFP fluorescence was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 30 μm.
Fig. S2 Accumulation of enhanced green fluorescent protein (EGFP) fused with Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP) mutants in the perinuclear region. Leaves of Nicotiana benthamiana plants were infiltrated with Agrobacterium harbouring a binary vector containing the constructs indicated above the images. EGFP fluorescence and 4′,6‐diamidino‐2‐phenylindole (DAPI) staining in epidermal cells were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 10 μm.


