Summary
Some abiotic and biotic conditions are known to have a negative impact on post‐transcriptional gene silencing (PTGS), thus representing a potential concern for the production of stable engineered virus resistance traits. However, depending on the strategy followed to achieve PTGS of the transgene, different responses to external conditions can be expected. In the present study, we utilized the Nicotiana benthamiana–Plum pox virus (PPV) pathosystem to evaluate in detail the stability of intron‐hairpin (ihp)‐mediated virus resistance under conditions known to adversely affect PTGS. The ihp plants grown at low or high temperatures were fully resistant to multiple PPV challenges, different PPV inoculum concentrations and even to a PPV isolate differing from the ihp construct by more than 28% at the nucleotide level. In addition, infections of ihp plants with viruses belonging to Cucumovirus, Potyvirus or Tombusvirus, all known to affect PTGS at different steps, were not able to defeat PPV resistance. Low temperatures did not affect the accumulation of transgenic small interfering RNAs (siRNAs), whereas a clear increase in the amount of siRNAs was observed during infections sustained by Cucumber mosaic virus and Potato virus Y. Our results show that the above stress factors do not represent an important concern for the production, through ihp‐PTGS technology, of transgenic plants having robust virus resistance traits.
Keywords: Artichoke mottled crinkle virus (AMCV) p19, Cucumber mosaic virus (CMV) 2b, Plum pox virus (PPV)‐C, Potato virus Y (PVY), helper component proteinase (HC‐Pro), RNA silencing suppressors, temperature
The ectopic expression of plant virus‐derived sequences has been exploited extensively to confer virus resistance traits in plants (Prins et al., 2008). These resistances, collectively known as pathogen‐derived resistances (PDRs), can be divided from a mechanistic point of view into two broad classes: one based on RNA silencing‐mediated resistance (Simon‐Mateo and Garcia, 2011; Ilardi and Di Nicola‐Negri, 2011) and one based on protein‐mediated resistance (e.g. Lucioli et al., 2008, 2014).
RNA silencing, triggered by double‐stranded RNAs (dsRNAs), is an ancient sequence‐specific gene regulation mechanism (Brodersen and Voinnet, 2006), which, in plants, has a major role in defending the host from foreign nucleic acids, such as viruses (Ding and Voinnet, 2007).
During RNA silencing, partially or fully dsRNAs are recognized and diced into short molecules (small interfering RNAs, siRNAs) of 21–24 nucleotides in size by host Dicer‐like enzymes. The siRNAs are then protected from degradation by 2′‐O‐methylation and loaded onto a multi‐subunit RNA‐induced silencing complex, which, in the case of post‐transcriptional gene silencing (PTGS), mostly guides sequence‐specific degradation of homologous RNAs. In addition, RNA‐dependent RNA polymerases, in particular RDR6 in concert with SGS3, amplify the RNA silencing response by recognizing aberrant RNAs and turning them into dsRNA (Mourrain et al., 2000; Qu et al., 2008). Importantly, viruses counterattack this generalized first line of plant defence, encoding proteins, called viral suppressors of RNA silencing (VSRs), able to interfere with different steps of the RNA silencing pathway (Burgyan and Havelda, 2011; Incarbone and Dunoyer, 2013; Wu et al., 2010).
A consequence of the existence of this host‐encoded RNA regulation pathway is that transgenic expression in plants of viral dsRNA sequences (Smith et al., 2000) or aberrant sense viral RNAs activates the RNA silencing pathway, thus conferring resistance to viruses sharing sequence homology with them. In particular, the RNA silencing driven by the expression of intron‐hairpin (ihp) constructs (ihp‐PTGS) has been shown to consistently confer virus resistance in most T0 plants (Di Nicola‐Negri et al., 2005; Smith et al., 2000). Nevertheless, it has been reported that some abiotic and biotic conditions, such as low plant growth temperatures (Szittya et al., 2003; Wu et al., 2008) and mixed viral infections, can break RNA silencing‐mediated virus resistance (Di Serio et al., 2002; Hassani‐Mehraban et al., 2009; Jørgensen and Albrechtsen, 2007; Mitter et al., 2003; Savenkov and Valkonen, 2001; Simon‐Mateo et al., 2003), thus representing a possible limitation for the full applicability of this biotechnological strategy.
Plum pox virus (PPV), genus Potyvirus, is the most devastating disease affecting stone fruits (Garcia et al., 2014). PPV, which has a quarantine status in many countries (Bonants et al., 2013), is subgrouped into seven strains: D, M, Rec, T, EA, W and C. In a previous study, we have shown that Nicotiana benthamiana plants transformed with ihp constructs derived from an Italian PPV‐M isolate (PPV ISPaVe44) are fully resistant to seven PPV isolates of the D, M and Rec strains (Di Nicola‐Negri et al., 2010). In addition, transgenic plants (line 6) for the h‐UTR/P1 construct, which covers the PPV 5′ untranslated region (UTR) and a part of the P1 gene, accumulating large amounts of h‐UTR/P1‐derived siRNAs, are resistant to PPV‐ISPaVe39 and PPV from sweet cherry (PPV‐SwC), which belong to the PPV‐EA and PPV‐C strains, respectively. Notably, the overall nucleotide identity between the h‐UTR/P1 construct and PPV‐SwC is as low as 71.2% (Di Nicola‐Negri et al., 2010).
The ability of the h‐UTR/P1 construct to confer resistance to a wide spectrum of PPV isolates, together with the opportunity to work with the model plant N. benthamiana, in which several PDR studies have been conducted, induced us to verify and compare, in similar controlled laboratory conditions, the impact on h‐UTR/P1 plants of abiotic and biotic conditions previously described to negatively influence RNA silencing‐mediated virus resistance.
As a preparatory step, we first analysed the consequences of different levels of h‐UTR/P1‐derived siRNAs on the ability of the transgenic plants to resist to virus isolates distantly related to PPV‐ISPaVe44. Three h‐UTR/P1 N. benthamiana lines resistant to PPV‐ISPaVe44 and accumulating small (lines 1 and 2) or, similar to line 6 (line 4), large amounts of siRNAs (Di Nicola‐Negri et al., 2005) were challenged with PPV‐SwC and PPV‐ISPaVe39. All transgenic plants of line 4 (72 plants), but none of those of lines 1 and 2 (66 plants), were resistant to the viruses, as evaluated by double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA) (Table S1, see Supporting Information); thus, high levels of h‐UTR/P1‐derived siRNAs were required to confer a broad spectrum of resistance to PPV. Hereafter, if not specifically stated, virus inoculations were performed using 1:10 w/v of fresh virus‐infected N. benthamiana leaves ground in 0.1 m phosphate buffer pH 7.2, and resistance was evaluated by visual inspection of symptoms and by DASI‐ELISA conducted with virus‐specific antibodies (Agritest, Bari, Italy), as described by Di Nicola‐Negri et al. (2010).
We then tested the impact of low temperatures on h‐UTR/P1‐mediated resistance. Nine T2 homozygous single‐locus plants of line 6 (68) and 18 T1 transgenic plants of the multi‐loci line 4 were grown at 25 °C, and then shifted to 15 °C, 3 and 5 days before PPV‐ISPaVe44 inoculation, respectively. All transgenic plants kept at 15 °C were PPV‐ISPaVe44 free until the end of the experiment (51 days post‐inoculation, dpi), whereas all 14 wild‐type control plants were systemically infected between 12 and 15 dpi.
In nature, multiple exposures to viruses can occur during a plant's life span. Therefore, we decided to test the ability of h‐UTR/P1 plants to maintain a high level of resistance when exposed to low temperature and challenged more than once with PPV. 68 plants pre‐adapted at 15 °C for 5 days were challenged with PPV‐ISPaVe44 and then re‐inoculated with the same viral isolate at 14–16 and/or 58 days after the first inoculation. None of the 51 68 plants grown at 15 °C and inoculated more than once with PPV‐ISPaVe44 was susceptible to the virus (Table 1). Immunocapture‐reverse transcription‐polymerase chain reaction (IC‐RT‐PCR) was performed, as described by Di Nicola‐Negri et al. (2010), on five DASI‐ELISA‐negative 68 plants double inoculated with PPV‐ISPaVe44. All tested plants were virus free according to this additional assay (Fig. S1A, see Supporting Information).
Table 1.
Resistance analysis of h‐UTR/P1 68 transgenic plants challenged more than once at 15 °C with Plum pox virus (PPV)‐ISPaVe44
| Virus | Plant line | Inoculation schedule | Number of PPV‐ISPaVe44‐infected/inoculated plantsa | |||||
|---|---|---|---|---|---|---|---|---|
| I | IIb | IIIc | 12–14 dpfid | 26–29 dpfi | 35–47 dpfi | 85 dpfi | ||
| PPV‐ISPaVe44 | h‐UTR/P1 68 | + | + | 0/20 | 0/20 | – | – | |
| Wild‐type | + | 18/19 | 19/19 | – | – | |||
| h‐UTR/P1 68 | + | + | 0/11 | 0/11e | 0/11 | – | ||
| Wild‐type | + | 13/14 | 14/14 | 14/14 | – | |||
| h‐UTR/P1 68 | + | + | 0/10 | 0/10 | 0/10 | 0/10 | ||
| Wild‐type | + | 9/10 | 8/9 (1)f | 9/9 (1) | 9/9 (1) | |||
| h‐UTR/P1 68 | + | + | + | 0/10 | 0/10 | 0/10 | 0/10 | |
| Wild‐type | + | 9/10 | 9/10 | 9/10 | 9/10 | |||
Based on double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA).
Inoculation was performed between 14 and 16 days after the first one.
Inoculation was performed 58 days after the first one.
dpfi, days post‐first inoculation.
Five of the 11 DASI‐ELISA‐negative plants were analysed by immunocapture‐reverse transcription‐polymerase chain reaction (IC‐RT‐PCR). All were virus free.
The number in parentheses represents the number of plants that died during the resistance test.
In addition, as a robust resistance should provide protection against a broad spectrum of viral isolates, we tested whether 68 plants were also resistant to PPV‐SwC at low temperatures. Thus, we challenged 68 plants with different sap dilutions of N. benthamiana leaves infected with PPV‐SwC as inoculum at both 25 °C and 15 °C (Table 2). All transgenic plants were resistant to the virus independent of the inoculum concentration and whether they were grown at 25 °C or 15 °C (Table 2 and Fig. S2, see Supporting Information). Consistently, IC‐RT‐PCR conducted at 50 dpi on all 68 plants grown at 15 °C did not detect the presence of PPV‐SwC (Fig. S1B). In addition, the resistance behaviour conferred by the h‐UTR/P1 construct was further confirmed by the analysis of T1 plants of line 4; all 18 transgenic plants grown at 15 °C were completely resistant to PPV‐SwC, as evaluated by DASI‐ELISA performed at 14, 31 and 41 dpi.
Table 2.
Resistance analyses of h‐UTR/P1 68 transgenic Nicotiana benthamiana plants challenged at 25 and 15 °C with different inoculum concentrations of Plum pox virus from sweet cherry (PPV‐SwC)
| Virus | Inoculum concentrationa | Plant line | Number of PPV‐SwC‐infected/inoculated plantsb | |||
|---|---|---|---|---|---|---|
| 13 dpic | 41 dpi | |||||
| 25 °C | 15 °C | 25 °C | 15 °C | |||
| PPV‐SwC | 1/20 | h‐UTR/P1 68 | 0/5 | 0/5 | 0/5 | 0/4 (1)d |
| Wild‐type | 5/5 | 5/5 | 5/5 | 5/5 | ||
| 1/10 | h‐UTR/P1 68 | 0/5 | 0/5 | 0/5 | 0/5 | |
| Wild‐type | 5/5 | 5/5 | 5/5 | 5/5 | ||
| 1/5 | h‐UTR/P1 68 | 0/5 | 0/5 | 0/5 | 0/5 | |
| Wild‐type | 5/5 | 5/5 | 5/5 | 4/4 (1) | ||
w/v of PPV‐SwC‐infected N. benthamiana leaves ground in 0.1 m phosphate buffer, pH 7.2.
Based on double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA).
dpi, days post‐inoculation.
The numbers in parentheses represent the number of plants that died during the resistance test.
The loss of RNA silencing effectiveness at low temperature is associated with a reduction in transgene‐derived siRNAs (Szittya et al., 2003). As reported above, transgenic plants of lines 68 and 4, accumulating large amounts of h‐UTR/P1 siRNAs, but not those of lines 1 and 2, characterized by small amounts of transgene‐derived siRNAs, were resistant to PPV‐SwC. The fact that PPV‐SwC resistance was maintained at 15 °C suggests that the plant growth temperature should not have a substantial impact on the amount of h‐UTR/P1 siRNAs in these plants. To test this hypothesis, total RNAs from five 68 plants kept at 25 °C and then shifted for 5 days to 15 °C were extracted, separated on polyacrylamide gel and blotted onto nylon membrane, as described by Lucioli et al. (2003), and hybridized with UTR/P1‐specific 32P‐radiolabelled RNA (Di Nicola‐Negri et al., 2005). Consistent with the ability of 68 plants to resist PPV‐SwC, the accumulation of h‐UTR/P1 siRNAs between 25 and 15 °C did not change (Fig. 1).
Figure 1.
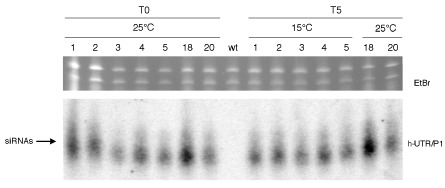
Detection of transgene‐derived h‐UTR/P1 small interfering RNAs (siRNAs) in 68 plants grown at low temperatures. Transgenic plants (1–5) grown at 25 °C were sampled at time 0 (T0) and re‐sampled after they had grown for 5 days (T5) at 15 °C. Transgenic plants 18 and 20 were grown at a uniform temperature of 25 °C; wt, wild‐type Nicotiana benthamiana. Twenty‐five micrograms of total RNA of each sample were loaded onto a 8% polyacrylamide gel electrophoresis (PAGE) gel. Top panel shows a portion of the gel stained with ethidium bromide (EtBr) for loading control. Filter was hybridized with h‐UTR/P1‐specific 32P‐radiolabelled RNA as described by Lucioli et al. (2003).
Overall, the experiments indicate that ihp‐PTGS is able to confer broad‐spectrum resistance/immunity to PPV even at low temperature. Conversely, RNA silencing‐mediated virus resistance driven by aberrant viral sense gene constructs (S‐PTGS) was lost when transgenic plants were challenged at 15 °C (Szittya et al., 2003; Wu et al., 2008). Our results are in accord with and extend the recent findings of Hu et al. (2011), showing that transgenic tobacco plants expressing ihp sequences of Tobacco mosaic virus (TMV) or Cucumber mosaic virus (CMV) are resistant to TMV and CMV, respectively, when grown at low temperatures (Hu et al., 2011). Here, we reported that the virus resistance trait was maintained at 15 °C when plants were repeatedly challenged not only with the homologous virus, but also with an isolate differing from the hairpin construct by more than 28% at the nucleotide level.
It has been shown that the transgenic expression of artificial microRNA (amiRNA) confers resistance to virus infection (Niu et al., 2006; Qu et al., 2007). In addition, it was argued that one advantage of amiRNA technology relative to other RNA silencing approaches is that the resistance trait is maintained at 15 °C (Niu et al., 2006). In this work, we showed that ihp‐PTGS technology can confer broad, stable and robust resistance to viral infection at 15 °C. Although few studies have analysed, at the whole‐plant level, the impact of different plant growth temperatures on RNA silencing‐mediated virus resistance, the available data suggest that RNA silencing obtained through ihp‐PTGS is much more robust than that through S‐PTGS. One possible explanation accounting for this difference is that ihp‐PTGS does not require RDR6 and SGS3 for the production of dsRNA (Vaucheret, 2006). In this context Zhong et al. (2013) have shown recently that an increase in the plant growth temperature from 22 to 30 °C inhibits S‐PTGS, but not ihp‐PTGS, and that S‐PTGS inhibition probably involves SGS3. Consistent with the results of Zhong et al. (2013), when 11 transgenic 68 plants grown at 30 °C were double challenged (0 and 16 dpi) with PPV‐ISPaVe44, they remained virus free until the end of the experiment (51 dpi). Five randomly selected PPV‐ISPaVe44 DASI‐ELISA‐negative plants were further analysed by IC‐RT‐PCR and all were PPV free (Fig. S1C); thus, both low and high temperatures did not break ihp‐PTGS‐mediated PPV resistance.
Among the conditions known to have a potential impact on RNA silencing‐mediated virus resistance are heterologous virus infections (Di Serio et al., 2002; Hassani‐Mehraban et al., 2009; Jørgensen and Albrechtsen, 2007; Mitter et al., 2003; Savenkov and Valkonen, 2001; Simon‐Mateo et al., 2003). In this study, we analysed whether virus infection sustained by Potato virus Y NTN (PVYNTN, GenBank GU550516), CMV (subgroup IA; GenBank JQ074218) and Artichoke mottled crinkle virus (AMCV) (Molinari et al., 1998; Silhavy et al., 2002), which encode helper component proteinase (HC‐Pro), 2b and p19 VSR, respectively, were able to defeat ihp‐PTGS resistance to PPV. Transgenic 68 plants were initially co‐inoculated with PPV‐ISPaVe44 plus PVY or CMV. Wild‐type and 68 N. benthamiana plants were similarly infected with PVY or CMV, whereas only wild‐type plants were also infected by PPV‐ISPaVe44; thus, PVY and CMV, when co‐inoculated with PPV‐ISPaVe44, were unable to break the resistance (Table 3 and Fig. S3, see Supporting Information). Similar to our data, PPV resistance was not broken when recovered transgenic N. benthamiana NIbV3 plants carrying a silenced modified NIb gene of PPV were co‐challenged with CMV (Simon‐Mateo et al., 2003). Conversely, transgenic N. benthamiana expressing a Grapevine virus A (GVA) minireplicon, in which PTGS was active against GVA sequences, became susceptible to GVA infection when co‐challenged with PVY (Brumin et al., 2009). It has been reported recently that N. benthamiana plants transformed with an ihp construct for the N gene of the Tomato yellow ring virus (TYRV‐t) are resistant to TYRV‐t in mixed inoculation with either CMV or PVY. However, the same transgenic plants were susceptible to TYRV‐t when co‐inoculated with the soybean strain of the same virus (TYRV‐s) (Hassani‐Mehraban et al., 2009). Here, to ascertain the stability of hairpin‐mediated PPV resistance in a condition in which virus infection is sustained by a different PPV isolate, we took advantage of the fact that h‐P1/HC‐Pro, h‐HC‐Pro and h‐HC‐Pro/P3 constructs are able to confer resistance to PPV‐ISPaVe44, but not to the distantly related PPV‐SwC (Di Nicola‐Negri et al., 2010). None of the transgenic plants harbouring one of the three hairpin constructs and co‐inoculated with PPV‐ISPaVe44 and PPV‐SwC was infected with PPV‐ISPaVe44, albeit accumulating PPV‐SwC in a similar manner to the double‐infected control plants (Table 3). Thus, contrary to the results described by Hassani‐Mehraban et al. (2009), PPV resistance was not broken by intraspecies RNA silencing suppressors.
Table 3.
Transgenic N icotiana benthamiana plants challenged with Plum pox virus (PPV)‐ISPaVe44 plus other viruses
| Viruses | Plant lines | Number of PPV‐ISPaVe44‐infected/inoculated plantsa | |||
|---|---|---|---|---|---|
| 12–20 dpib | 41–57 dpi | ||||
| I resistance testc | II resistance testd | I resistance test | II resistance test | ||
| CMV+PPV‐ISPaVe44 | h‐UTR/P1 68 | 0/15 | 0/10 | 0/11 (4)e | 0/10 |
| Wild‐type | 6/6 (4) | 7/7 | 3/3 (7) | 6/6 (1) | |
| PVY+PPV‐ISPaVe44 | h‐UTR/P1 68 | 0/13 | 0/12 | 0/10 (3) | 0/7 (5) |
| Wild‐type | 9/10 | 9/9 | 8/8 (2) | (9) | |
| PPV‐SwC+PPV‐ISPaVe44 | h‐P1/HC‐Pro 72 | – | 0/9 | – | 0/9 |
| h‐HC‐Pro 1017 | 0/7 | 0/7 | 0/7 | 0/7 | |
| h‐HC‐Pro/P3 162 | 0/6 | 0/5 | 0/4 (2) | 0/5 | |
| Wild‐type | 4/4 | 12/12 | 4/4 | 12/12 | |
CMV, Cucumber mosaic virus; PVY, Potato virus Y.
Plants analysed at the first time point for the presence of PPV were all infected with the heterologous viruses.
dpi, days post‐inoculation.
Infection test performed in a glasshouse.
Infection test performed in a growth chamber (25 °C).
The numbers in parentheses represent the number of plants that died during the resistance tests.
To further investigate the stability of the PPV resistance trait, we first inoculated 68 and wild‐type plants with PVY, CMV or AMCV and then, when the upper noninoculated leaves had become systemically infected with one of the above viruses, they were super‐inoculated with PPV‐ISPaVe44. Depending on the type of the first viral inoculum and the experiment, 33%–100% of the control plants became infected with PPV‐ISPave44 (Table 4). Conversely, none of the 50 transgenic plants infected with PVY, CMV or AMCV became susceptible to PPV‐ISPaVe44 (Table 4). Similarly, the h‐P1/HC‐Pro, h‐HC‐Pro and h‐HC‐Pro/P3 transgenic plants systemically infected with PPV‐SwC and challenged at 19 dpi with PPV‐ISPaVe44 were resistant to the latter virus until the end of the experiments (Table 4). Total RNAs extracted from 68 plants just prior to PVY, CMV or AMCV inoculation and when 68 plants had become systemically infected with the viruses were analysed for the presence of h‐UTR/P1‐specific small RNAs. Interestingly, PVY and CMV and, to a lesser extent, AMCV induced the accumulation of h‐UTR/P1 siRNAs (Fig. 2). The increase in the amount of transgene‐derived siRNAs was not a generic result of virus infection. In fact, the amount of h‐UTR/P1 siRNAs was similar between healthy uninoculated and African cassava mosaic virus (ACMV, GenBank J02057/J02058; kindly provided by Christina Wege, Universitat Stuttgart, Stuttgart) infected 68 plants (Fig. S4, see Supporting Information). The same RNAs were then re‐hybridized with virus‐specific 32P‐labelled probes. The amounts of PVY and AMCV siRNAs were slightly higher in 68 than in wild‐type plants, whereas CMV siRNAs accumulated to a similar level in both plant types (Fig. S5, see Supporting Information).
Table 4.
Analysis of Plum pox virus (PPV)‐ISPaVe44‐resistant transgenic N icotiana benthamiana plants systemically infected with poty‐, cucumo‐ or tombusviruses and super‐inoculated with PPV‐ISPaVe44
| Number of PPV‐ISPaVe44‐infected/inoculated plantsa | ||||||
|---|---|---|---|---|---|---|
| 12–15 dpib | 21–42 dpi | |||||
| Virus pre‐inoculum | Plant lines | Total numberc | I infection testd | II infection teste | I infection test | II infection test |
| CMV | h‐UTR/P1 68 | 23 | 0/13 | 0/10 | 0/12 (1)f | 0/10 |
| Wild‐type | 14 | 2/10 | 3/4 | 6/9 (1) | 4/4 | |
| PVY | h‐UTR/P1 68 | 22 | 0/13 | – | 0/12 (1) | 0/7 (2) |
| Wild‐type | 18 | 1/8 (1) | – | 4/6 (3) | 3/9 | |
| AMCV | h‐UTR/P1 68 | 5 | 0/5 | – | – | – |
| Wild‐type | 9 | 9/9 | – | – | – | |
| PPV‐SwC | h‐P1/HC‐Pro 72 | 11 | – | 0/11 | – | 0/11 |
| h‐HC‐Pro 1017 | 13 | 0/7 | 0/6 | 0/7 | 0/6 | |
| h‐HC‐Pro/P3 162 | 14 | 0/7 | 0/7 | 0/7 | 0/7 | |
| Wild‐type | 16 | 0/3 | 9/13 | 2/3 | 10/13 | |
AMCV, Artichoke mottled crinkle virus; CMV, Cucumber mosaic virus; PVY, Potato virus Y.
Based on double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA).
dpi, day post‐inoculation with PPV‐ISPaVe44.
Total number of systemically virus‐infected plants that were super‐inoculated with PPV‐ISPaVe44.
Infection test performed in a glasshouse.
Infection test performed in a growth chamber (25 °C).
The numbers in parentheses represent the number of plants that died during the resistance tests.
Figure 2.
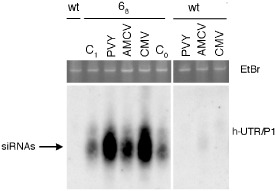
Detection of transgene‐derived h‐UTR/P1 small interfering RNAs (siRNAs) in virus‐infected 68 plants. Twenty‐five micrograms of total RNAs were extracted from leaves of 68 and wild‐type (wt) plants systemically infected with Artichoke mottled crinkle virus (AMCV), Cucumber mosaic virus (CMV) and Potato virus Y (PVY) at 11, 11 and 15 days post‐inoculation (dpi), respectively, and loaded onto an 8% polyacrylamide gel electrophoresis (PAGE) gel. C0, 68 noninoculated control plants at the time of virus inoculation. C1, 68 noninoculated control plants at the time at which AMCV‐ and CMV‐infected plants were sampled (11 dpi). Top panel shows a portion of the gel stained with ethidium bromide (EtBr) for loading control. Filter was hybridized with h‐UTR/P1‐specific 32P‐radiolabelled RNA as described by Lucioli et al. (2003).
The ability of PVY, CMV and AMCV, but not ACMV, to enhance the accumulation of transgene‐derived siRNAs could be related to the ability of HC‐Pro, 2b and p19 VSRs to bind siRNAs (Burgyan and Havelda, 2011; Incarbone and Dunoyer, 2013). Indeed, it could be argued that, if UTR/P1 siRNA homeostasis is influenced by the amount of free UTR/P1 siRNAs, amongst others, VSR‐mediated sequestration of these siRNAs could lead to a compensatory production of additional UTR/P1 siRNAs. However, dedicated experiments will be required to determine whether the enhanced siRNA accumulation observed in virus‐infected UTR/P1 plants is a more general phenomenon and the molecular mechanism behind it. Nevertheless, mixed viral infection experiments showed that ihp constructs conferred stable resistance to PPV, even though the plants were fully infected with viruses encoding VSRs known to influence different steps of the RNA silencing machinery (Burgyan and Havelda, 2011; Incarbone and Dunoyer, 2013; Wu et al., 2010). The PPV‐resistant phenotype of 68 plants could not be explained by a different susceptibility of these plants to PVY, CMV or AMCV. Indeed, wild‐type and 68 plants challenged with PVY, CMV or AMCV accumulated similar amounts of each virus as evaluated by RNA gel analysis and DASI‐ELISA (Fig. S6, see Supporting Information, and data not shown).
In contrast with our data, transgenic N. benthamiana transformed with a sense coat protein gene of Potato virus A (PVA) and characterized by S‐PTGS became susceptible to PVA infection if previously inoculated with PVY (Savenkov and Valkonen, 2001). In accord with the susceptibility of S‐PTGS‐mediated virus resistance to mixed viral infection is the demonstration that transgenic N. benthamiana plants for the sense replicase gene of Cymbidium ringspot virus (CymRSV) became susceptible to CymRSV when pre‐challenged with CMV (Di Serio et al., 2002). Moreover, recovered N. benthamiana NIbV3 plants became susceptible to PPV when previously challenged with CMV (Simon‐Mateo et al., 2003), and transgenic potato plants transformed with the sense coat protein gene of PVY became susceptible to PVY if pre‐challenged with another Potyvirus, Tobacco etch virus (TEV) (Jørgensen and Albrechtsen, 2007). In addition, in one case, CMV was shown to suppress PVY resistance induced by the expression of an inverted repeat separated by a spacer loop sequence (Mitter et al., 2003). In particular, a lag time of 10 days between CMV and PVY inoculation was required to defeat PVY immunity in transgenic tobacco plants of line 16 (Mitter et al., 2003). However, our data show that a lag time of 8 and 16 days (Table 4, first and second infection tests) between CMV and PPV‐ISPaVe44 inoculation (Table 4, first and second infection tests) did not affect ihp‐PTGS‐mediated PPV resistance. According to our observations, CMV infection did not break the resistance to TMV in transgenic tobacco plants expressing a TMV ihp sequence (Hu et al., 2011).
Collectively, our data and literature reports strongly suggest that a clear difference in robustness between S‐PTGS‐ and ihp‐PTGS‐mediated virus resistance exists when plants are exposed to variable abiotic and biotic conditions. In ihp‐PTGS plants accumulating large amounts of transgene‐specific siRNAs, such as lines 68 and 4, the RNA silencing amplification step does not appear to be essential for setting up a highly efficient antiviral state. In addition to RDR6 and SGS3, XRN4, SDE3 and SDE5 have been shown to influence siRNA production from sense, but not inverted, repeat transgenes (Gazzani et al., 2004; Hernandez‐Pinzon et al., 2007; Himber et al., 2003). Thus, if the expression/stability of one of the above gene products is influenced by variable abiotic or biotic conditions, as recently shown in the case of SGS3 (Zhong et al., 2013), the robustness of ihp‐PTGS‐ relative to S‐PTGS‐mediated virus resistance can be easily reconciled.
In conclusion, our data support the notion that low and high temperatures (15 and 30 °C), as well as mixed viral infections sustained by viruses encoding 2b, HC‐Pro and p19 VSRs, do not represent an important concern for the production, through ihp‐PTGS technology, of transgenic plants with stable and robust virus resistance traits.
Supporting information
Fig. S1 Immunocapture‐reverse transcription‐polymerase chain reaction (IC‐RT‐PCR) on double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA) Plum pox virus (PPV)‐negative 68 transgenic plants grown at different temperatures. (A) Plants double challenged [0 and 16 days post‐inoculation (dpi)] with PPV‐ISPaVe44 at 15 °C and analysed at 42 dpi; lanes 1–5 and 9, 68 transgenic plants; lanes 6–8 and 10, wild‐type plants; I, negative control for immunocapture reaction. (B) Plants inoculated with different sap dilutions of Nicotiana benthamiana leaves infected with PPV‐SwC at 15 °C and analysed at 50 dpi; lanes 1–13 and 15, 68 transgenic plants; lanes 14 and 16–18, wild‐type plants. (C) Plants double challenged (0 and 16 dpi) with PPV‐ISPaVe44 at 30 °C and analysed at 51 dpi; lanes 1–5 and 8, 68 transgenic plants; lanes 6, 7, 9 and 10, wild‐type plants.
Fig. S2 h‐UTR/P1 transgenic 68 and wild‐type Nicotiana benthamiana plants challenged at 15 °C with Plum pox virus from sweet cherry (PPV‐SwC).
Fig. S3 h‐UTR/P1 transgenic 68 and wild‐type Nicotiana benthamiana plants co‐inoculated with Cucumber mosaic virus (CMV) and Plum pox virus (PPV)‐ISPaVe44 or Potato virus Y NTN (PVYNTN) and PPV‐ISPaVe44.
Fig. S4 Detection of h‐UTR/P1‐derived small interfering RNAs (siRNAs) in transgenic 68 Nicotiana benthamiana plants infected with African cassava mosaic virus (ACMV).
Fig. S5 Detection of virus‐derived small interfering RNAs (siRNAs) in 68 and wild‐type (wt) plants infected with Potato virus Y NTN (PVYNTN), Cucumber mosaic virus (CMV) or Artichoke mottled crinkle virus (AMCV).
Fig. S6 Gel analysis of total RNA extracted from transgenic 68 and wild‐type Nicotiana benthamiana plants challenged with Artichoke mottled crinkle virus (AMCV) or Potato virus Y NTN (PVYNTN).
Table S1 Plum pox virus (PPV) resistance analysis of h‐UTR/P1 plant lines characterized by different levels of transgene‐derived small interfering RNAs (siRNAs).
Acknowledgements
We are grateful to Andrea Iazzoni for technical support. This study was supported by Research Program MiPAAF—CIPE Project FRUMED, ProViSUD.
References
- Bonants, P. , Edema, M. and Robert, V. (2013) Q‐bank, a database with information for identification of plant quarantine plant pest and diseases. EPPO Bull. 43, 211–215. [Google Scholar]
- Brodersen, P. and Voinnet, O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280. [DOI] [PubMed] [Google Scholar]
- Brumin, M. , Stukalov, S. , Haviv, S. , Muruganantham, M. , Moskovitz, Y. , Batuman, O. , Fenigstein, A. and Mawassi, M. (2009) Post‐transcriptional gene silencing and virus resistance in Nicotiana benthamiana expressing a Grapevine virus A minireplicon. Transgenic Res. 18, 331–345. [DOI] [PubMed] [Google Scholar]
- Burgyan, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Di Nicola‐Negri, E. , Brunetti, A. , Tavazza, M. and Ilardi, V. (2005) Hairpin RNA‐mediated silencing of Plum pox virus P1 and HC‐Pro genes for efficient and predictable resistance to the virus. Transgenic Res. 14, 989–994. [DOI] [PubMed] [Google Scholar]
- Di Nicola‐Negri, E. , Tavazza, M. , Salandri, L. and Ilardi, V. (2010) Silencing of Plum pox virus 5′ UTR/P1 sequence confers resistance to a wide range of PPV strains. Plant Cell Rep. 29, 1435–1444. [DOI] [PubMed] [Google Scholar]
- Ding, S.‐W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Serio, F. , Rubino, L. , Russo, M. and Martelli, G.P. (2002) Homology‐dependent virus resistance against Cymbidium ringspot virus is inhibited by post‐transcriptional gene silencing suppressor viruses. J. Plant Pathol. 84, 121–124. [Google Scholar]
- Garcia, J.A. , Glasa, M. , Cambra, M. and Candresse, T. (2014) Plum pox virus and sharka: a model potyvirus and a major disease. Mol. Plant Pathol. 15, 226–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani, S. , Lawrenson, T. , Woodward, C. , Headon, D. and Sablowski, R. (2004) A link between mRNA turnover and RNA interference in Arabidopsis. Science, 306, 1046–1048. [DOI] [PubMed] [Google Scholar]
- Hassani‐Mehraban, A. , Brenkman, A.B. , van den Broek, N.J.F. , Goldbach, R. and Kormelink, R. (2009) RNAi‐mediated transgenic tospovirus resistance broken by intraspecies silencing suppressor protein complementation. Mol. Plant–Microbe Interact. 22, 1250–1257. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Pinzon, I. , Yelina, N.E. , Schwach, F. , Studholme, D.J. , Baulcombe, D. and Dalmay, T. (2007) SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans‐acting endogenous siRNA. Plant J. 50, 140–148. [DOI] [PubMed] [Google Scholar]
- Himber, C. , Dunoyer, P. , Moissiard, G. , Ritzenthaler, C. and Voinnet, O. (2003) Transitivity‐dependent and ‐independent cell‐to‐cell movement of RNA silencing. EMBO J. 22, 4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Q. , Niu, Y. , Zhang, K. , Liu, Y. and Zhou, X. (2011) Virus‐derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virol. J. 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardi, V. and Di Nicola‐Negri E. (2011) Genetically engineered resistance to Plum pox virus infection in herbaceous and stone fruit hosts. GM Crops, 2, 24–33. [DOI] [PubMed] [Google Scholar]
- Incarbone, M. and Dunoyer, P. (2013) RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B. and Albrechtsen, M. (2007) Stability of RNA silencing‐based traits in potato after virus infection. Mol. Breed. 19, 371–376. [Google Scholar]
- Lucioli, A. , Noris, E. , Brunetti, A. , Tavazza, R. , Ruzza, V. , Castillo, A.G. , Bejarano, E.R. , Accotto, G.P. and Tavazza, M. (2003) Tomato yellow leaf curl Sardinia virus Rep‐derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus‐mediated transgene silencing is activated. J. Virol. 77, 6785–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucioli, A. , Sallustio, D.E. , Barboni, D. , Berardi, A. , Papacchioli, V. , Tavazza, R. and Tavazza, M. (2008) A cautionary note on pathogen‐derived sequences. Nat. Biotechnol. 26, 617–619. [DOI] [PubMed] [Google Scholar]
- Lucioli, A. , Berardi, A. , Gatti, F. , Tavazza, R. , Pizzichini, D. and Tavazza, M. (2014) Tomato yellow leaf curl Sardinia virus‐resistant tomato plants expressing the multifunctional N‐terminal domain of the replication‐associated protein show transcriptional changes resembling stress‐related responses. Mol. Plant Pathol. 15, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter, N. , Sulistyowati, E. and Dietzgen, R.G. (2003) Cucumber mosaic virus infection transiently breaks dsRNA‐induced transgenic immunity to Potato virus Y in tobacco. Mol. Plant–Microbe Interact. 16, 936–944. [DOI] [PubMed] [Google Scholar]
- Molinari, P. , Marusic, C. , Lucioli, A. , Tavazza, R. and Tavazza, M. (1998) Identification of artichoke mottled crinkle virus (AMCV) proteins required for virus replication: complementation of AMCV p33 and p92 replication‐defective mutants. J. Gen. Virol. 79, 639–647. [DOI] [PubMed] [Google Scholar]
- Mourrain, P. , Béclin, C. , Elmayan, T. , Feuerbach, F. , Godon, C. , Morel, J.B. , Jouette, D. , Lacombe, A.M. , Nikic, S. , Picault, N. , Remoue, K. , Sanial, M. , Vo, T.A. and Vaucheret, H. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Niu, Q.W. , Lin, S.S. , Reyes, J.L. , Chen, K.C. , Wu, H.W. , Yeh, S.D. and Chua, N.H. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428. [DOI] [PubMed] [Google Scholar]
- Prins, M. , Laimer, M. , Noris, E. , Schubert, J. , Wassenegger, M. and Tepfer, M. (2008) Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ye, X. and Morris, T.J. (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4‐initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA, 105, 14 732–14 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, J. , Ye, J. and Fang, R. (2007) Artificial microRNA‐mediated virus resistance in plants. J. Virol. 81, 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savenkov, E.I. and Valkonen, J.P.T. (2001) Coat protein gene‐mediated resistance to Potato virus A in transgenic plants is suppressed following infection with another potyvirus. J. Gen. Virol. 82, 2275–2278. [DOI] [PubMed] [Google Scholar]
- Silhavy, D. , Molnar, A. , Lucioli, A. , Szittya, G. , Hornyik, C. , Tavazza, M. and Burgyan, J. (2002) A viral protein suppresses RNA silencing and binds silencing‐generated, 21‐ to 25‐nucleotide double‐stranded RNAs. EMBO J. 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon‐Mateo, C. and Garcia, J.A. (2011) Antiviral strategies in plants based on RNA silencing. Biochim. Biophys. Acta (BBA) Gene Reg. Mech. 1809, 722–731. [DOI] [PubMed] [Google Scholar]
- Simon‐Mateo, C. , Lopez‐Moya, J.J. , Guo, H.S. , Gonzalez, E. and Garcia, J.A. (2003) Suppressor activity of potyviral and cucumoviral infections in potyvirus‐induced transgene silencing. J. Gen. Virol. 84, 2877–2883. [DOI] [PubMed] [Google Scholar]
- Smith, N.A. , Singh, S.P. , Wang, M.‐B. , Stoutjesdijk, P.A. , Green, A.G. and Waterhouse, P.M. (2000) Gene expression: total silencing by intron‐spliced hairpin RNAs. Nature, 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Szittya, G. , Silhavy, D. , Molnar, A. , Havelda, Z. , Lovas, A. , Lakatos, L. , Banfalvi, Z. and Burgyan, J. (2003) Low temperature inhibits RNA silencing‐mediated defence by the control of siRNA generation. EMBO J. 22, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2006) Post‐transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Wang, X. and Ding, S.W. (2010) Viral suppressors of RNA‐based viral immunity: host targets. Cell Host Microbe, 8, 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.L. , Hou, W.C. , Wang, M.M. , Zhu, X.P. , Li, F. , Zhang, J.D. , Li, X.Z. and Guo, X.Q. (2008) RNA silencing‐mediated resistance is related to biotic/abiotic stresses and cellular RdRp expression in transgenic tobacco plants. BMB Rep. 41, 376–381. [DOI] [PubMed] [Google Scholar]
- Zhong, S.H. , Liu, J.Z. , Jin, H. , Lin, L. , Li, Q. , Chen, Y. , Yuan, Y.X. , Wang, Z.Y. , Huang, H. , Qi, Y.J. , Chen, X.Y. , Vaucheret, H. , Chory, J. , Li, J. and He, Z.H. (2013) Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA, 110, 9171–9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Immunocapture‐reverse transcription‐polymerase chain reaction (IC‐RT‐PCR) on double antibody sandwich indirect‐enzyme‐linked immunosorbent assay (DASI‐ELISA) Plum pox virus (PPV)‐negative 68 transgenic plants grown at different temperatures. (A) Plants double challenged [0 and 16 days post‐inoculation (dpi)] with PPV‐ISPaVe44 at 15 °C and analysed at 42 dpi; lanes 1–5 and 9, 68 transgenic plants; lanes 6–8 and 10, wild‐type plants; I, negative control for immunocapture reaction. (B) Plants inoculated with different sap dilutions of Nicotiana benthamiana leaves infected with PPV‐SwC at 15 °C and analysed at 50 dpi; lanes 1–13 and 15, 68 transgenic plants; lanes 14 and 16–18, wild‐type plants. (C) Plants double challenged (0 and 16 dpi) with PPV‐ISPaVe44 at 30 °C and analysed at 51 dpi; lanes 1–5 and 8, 68 transgenic plants; lanes 6, 7, 9 and 10, wild‐type plants.
Fig. S2 h‐UTR/P1 transgenic 68 and wild‐type Nicotiana benthamiana plants challenged at 15 °C with Plum pox virus from sweet cherry (PPV‐SwC).
Fig. S3 h‐UTR/P1 transgenic 68 and wild‐type Nicotiana benthamiana plants co‐inoculated with Cucumber mosaic virus (CMV) and Plum pox virus (PPV)‐ISPaVe44 or Potato virus Y NTN (PVYNTN) and PPV‐ISPaVe44.
Fig. S4 Detection of h‐UTR/P1‐derived small interfering RNAs (siRNAs) in transgenic 68 Nicotiana benthamiana plants infected with African cassava mosaic virus (ACMV).
Fig. S5 Detection of virus‐derived small interfering RNAs (siRNAs) in 68 and wild‐type (wt) plants infected with Potato virus Y NTN (PVYNTN), Cucumber mosaic virus (CMV) or Artichoke mottled crinkle virus (AMCV).
Fig. S6 Gel analysis of total RNA extracted from transgenic 68 and wild‐type Nicotiana benthamiana plants challenged with Artichoke mottled crinkle virus (AMCV) or Potato virus Y NTN (PVYNTN).
Table S1 Plum pox virus (PPV) resistance analysis of h‐UTR/P1 plant lines characterized by different levels of transgene‐derived small interfering RNAs (siRNAs).


