SUMMARY
Coffee (Coffea arabica L.), one of the key export and cash crops in tropical and subtropical countries, suffers severe losses from the rust fungus Hemileia vastatrix. The transcriptome of H. vastatrix was analysed during a compatible interaction with coffee to obtain an exhaustive repertoire of the genes expressed during infection and to identify potential effector genes. Large‐scale sequencing (454‐GS‐FLEX Titanium) of mixed coffee and rust cDNAs obtained from 21‐day rust‐infected leaves generated 352 146 sequences which assembled into 22 774 contigs. In the absence of any reference genomic sequences for Coffea or Hemileia, specific trinucleotide frequencies within expressed sequence tags (ESTs) and blast homology against a set of dicots and basidiomycete genomes were used to distinguish pathogen from plant sequences. About 30% (6763) of the contigs were assigned to H. vastatrix and 61% (13 951) to C. arabica. The majority (60%) of the rust sequences did not show homology to any genomic database, indicating that they were potential novel fungal genes. In silico analyses of the 6763 H. vastatrix contigs predicted 382 secreted proteins and identified homologues of the flax rust haustorially expressed secreted proteins (HESPs) and bean rust transferred protein 1 (RTP1). These rust candidate effectors showed conserved amino‐acid domains and conserved patterns of cysteine positions suggestive of conserved functions during infection of host plants. Quantitative reverse transcription‐polymerase chain reaction profiling of selected rust genes revealed dynamic expression patterns during the time course of infection of coffee leaves. This study provides the first valuable genomic resource for the agriculturally important plant pathogen H. vastatrix and the first comprehensive C. arabica EST dataset.
INTRODUCTION
The basidiomycete Hemileia vastatrix Berk. & Broome 1869 (Berkeley, 1869) (order Pucciniales) is an obligatory parasite specific to coffee plants (Coffea spp.) (Bettencourt and Rodrigues, 1988) and causes coffee leaf rust, one of the most destructive diseases of the cash crop Coffea arabica L. The fungus is widely distributed in all regions of the world in which coffee is grown and can represent a serious limitation to economically sustainable coffee production (Bettencourt and Rodrigues, 1988). The natural resistance of coffee plants to leaf rust is conditioned by gene‐for‐gene interactions, with at least nine plant resistance factors that are implicated in the recognition of the corresponding virulence genes in H. vastatrix isolates (Bettencourt and Rodrigues, 1988; Rodrigues et al., 1975). Little information is available on the fungal genetic factors that condition coffee–rust interactions. To date, 45 H. vastatrix races have been characterized according to their virulence spectrum on a set of 18 coffee differentials, with the number and pattern of occurrence varying by region (Várzea and Marques, 2005). In the last few years, new rust races have overcome the resistance of some improved cultivars grown in several countries (Silva et al., 2006; Várzea and Marques, 2005). Because the life cycle of H. vastatrix is essentially asexual, via cyclic uredospore production on host leaves (Rodrigues et al., 1975), no genetic assumption could be derived for the identification of loci carrying rust avirulence genes.
The initiation of the dikaryotic phase of H. vastatrix on coffee leaves involves specific events, including appressorium formation over stomata, prior to progression through a defined set of developmental stages required to enter stomatal openings and to form the first haustoria within stomatal subsidiary cells. Further on, the infection process involves the colonization of mesophyll cells by intercellular hyphal growth and intense haustoria formation, and sporulation can start as early as 20 days after infection (Coutinho et al., 1993; Martins and Moraes, 1996; Ramiro et al., 2009; Rodrigues et al., 1975; 1999, 2002). Histological observations have shown that C. arabica resistance is expressed by a hypersensitive response (HR) with cell death of stomatal and mesophyll cells (Martins and Moraes, 1996; 2002, 2008). Molecular analyses have indicated that perception of the fungus, whether virulent or avirulent, occurs when the pathogen enters the stomata (Ganesh et al., 2006; Ramiro et al., 2009). However, specific host resistance responses, including hypersensitive cell death, H2O2 production and defence‐related gene expression patterns, are associated with the production of haustoria in mesophyll cells, and not with the production of haustoria in stomatal cells, suggesting that specific recognition of H. vastatrix occurs at a later stage (Ramiro et al., 2009).
Recently, major insights have emerged from studies on biotrophic fungi, indicating that they secrete effector proteins, including virulence and avirulence proteins, that alter host physiology and defence responses (for review, see Stergiopoulos and de Wit, 2009). Effector proteins may be subdivided into two broad categories depending on whether they are secreted in the apoplasm or delivered into the cytoplasm of the host cell. In the flax rust fungus Melampsora lini, the avirulent genes identified so far encode small secreted proteins, are highly expressed in the haustoria and their products are recognized by flax (Linus usitatissimum) resistance proteins inside plant cells (Catanzariti et al., 2006; Dodds et al., 2004; Rafiqi et al., 2010). Flax rust avirulence proteins have no homology to any known protein (Catanzariti et al., 2006), but homologues have been identified recently in the related species Melampsora larici‐populina (Hacquard et al., 2010). In the same way, a family of rust transferred proteins (RTPs) has been identified that is conserved in rust fungi (Joly et al., 2010; Puthoff et al., 2008). The RTP1 protein was primarily identified in Uromyces fabae, where it translocates from the haustoria into the host and accumulates in the nucleus (Kemen et al., 2005). Searches in public protein databases for other rust secreted proteins identified the Cronartium ribicola secreted protein CrorI (Ekramoddoullah et al., 1999) and several Melampsora spp. proteins (Feau et al., 2007; Hacquard et al., 2010; Joly et al., 2010).
Until recently, the identification of in planta‐expressed transcripts was a major limitation to studies of biotrophic pathogens. Sequencing through the Sanger approach has led to the identification of several hundreds of genes and a few secreted proteins (Catanzariti et al., 2006; Hahn and Mendgen, 1997; Hu et al., 2007; Joly et al., 2010; Puthoff et al., 2008; Yin et al., 2009). The fundamental knowledge of the functional genome is now being enhanced by the ability to deeply probe an organism's transcriptome using high‐throughput sequencing data production. In this study, we used 454 massively parallel sequencing to gain new insights into the in planta transcriptome of the coffee rust parasite. These data reveal the first features of the H. vastatrix transcriptional landscape to establish a catalogue of putative fungal effector genes and to significantly advance our understanding of the coffee–rust fungus interaction.
RESULTS
454‐pyrosequencing of transcripts from Coffea leaves infected by H. vastatrix
A total of 352 146 nucleotide reads (118 113 878 nucleotides) was generated by single read 454‐pyrosequencing and assembled into 22 774 contigs (Table 1; Data S1, see Supporting Information) by Newbler (v2.0.00.22). Remaining unassembled reads (65 428) correspond to low‐quality sequences rejected during the assembly process and putative singleton sequences. These singletons were not considered further in this study. Assembled contigs ranged between 85 and 4589 base pairs (bp) and consisted of 2–421 assembled reads. Contigs were commonly smaller than 500 bp (64%), but 11% were larger than 1 kb, indicating that the 454‐approach was useful for the generation of large transcript sequences. In addition, 30% of the contigs (6804) contained 10 reads or more. An estimation of the relative abundance of contigs (number of reads relative to the contig length) showed that 10% (2365 contigs) represented transcripts with a medium to high rate of expression (Ra > 0.05) from rust‐infected Coffea leaf tissues (Table 1).
Table 1.
Number of 454‐reads and contigs generated by 454‐GS‐FLEX Titanium pyrosequencing from rust‐infected Coffea arabica leaves.
| Total | Fungal | Plant | |
|---|---|---|---|
| Contig no. | 22 774 | 6 763 | 13 951 |
| Read no. | 286 718 | 57 332 | 205 089 |
| Average size (bp) | 542 | 426 | 631 |
| Contig no. with: | |||
| Size ≥ 1 kb | 2 593 | 373 | 2 198 |
| Content ≥ 10 reads | 6 804 | 1 168 | 5 120 |
| Content ≥ 50 reads | 1 038 | 163 | 790 |
| Content ≥ 200 reads | 53 | 8 | 38 |
| Ra ≥ 1 | 73 | 9 | 49 |
| Ra ≥ 0.1 | 1 134 | 259 | 643 |
| Ra ≥ 0.05 | 2 381 | 533 | 1 418 |
Ra, relative abundance (Ra = number of reads/contig length).
Origin of expressed sequence tags (ESTs) from infected leaf tissues
A strategy based on parallel prediction approaches (Fig. 1) was set to determine the origin of plant and fungal transcripts from rust‐infected C. arabica leaf tissues. In total, six different prediction methods were used to determine the plant or fungal origin of the Coffea/Hemileia contig sequences. In a first step, specific trinucleotide frequencies were estimated for Coffea and Hemileia transcribed sequences with the MIPS EST3 program (Emmersen et al., 2007). The EST3 program was specifically trained with an intrinsic training set of Coffea and Pucciniales sequences from public databases [see Experimental procedures and Fig. S1 (Supporting Information)]; however, the program indicated that the dinucleotide bias between Coffea and Pucciniales was not sufficiently high to provide the most accurate prediction. Such a low bias between plant and fungal sequences has been reported previously between the model species Arabidopsis thaliana and Saccharomyces cerevisiae (Gentles and Karlin, 2001). A second step involved homology searches against public databases or selected sequence databases generated for this study [see Experimental procedures and Fig. S2 (Supporting Information)]. blast homology searches always predicted a larger number of plant contigs, whereas trinucleotide frequency‐based predictions always identified a larger number of fungal sequences (Figs S1 and S2), which could reflect the smaller number of genetic resources for rusts relative to plants in databases. The final set of tentative C. arabica and H. vastatrix contigs was determined on the basis of the number of plant or fungus predictions (see Experimental procedures). At the end, 13 951 contigs were attributed to the plant host C. arabica, and 6763 contigs were attributed to the rust fungus H. vastatrix (Tables 1 and S1, see Supporting Information). An additional set of 2060 contigs (9% of the total number of contigs) could not be attributed to C. arabica or H. vastatrix because they either showed a similar number of plant and fungus predictions (1358 contigs not resolved) or none of the prediction methods was able to determine their origin (702 contigs not determined).
Figure 1.
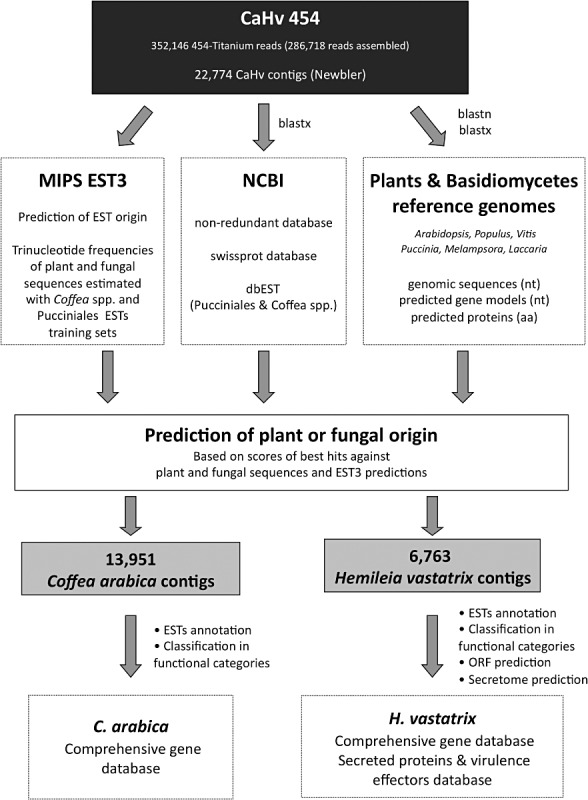
Bioinformatic procedure used to determine the origin of sequences produced by the 454‐GS‐FLEX Titanium pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix.
Cellular category distribution of plant and fungal contigs
Contigs attributed to C. arabica and H. vastatrix were compared with the euKaryotic Orthologous Group (KOG) database in order to classify sequences into functional cellular categories (Fig. 2). Almost half of the sequences (43% and 46% of plant and fungus, respectively) had no significant matches at the cut‐off E‐value of <10−5. In the plant contigs, the most important cellular categories corresponded to the following: signal transduction mechanisms (7.2%); post‐translational modification, protein turnover and chaperones (7.1%); carbohydrate transport and metabolism (3.8%); and intracellular trafficking, secretion and vesicular transport (3.7%). The proportion of plant transcripts falling into these categories probably reflects the strong impact of rust growth in plant living tissues, including remobilization of plant photoassimilates and manipulation of host defence. In the fungal contigs, the most important cellular categories corresponded to the following: post‐translational modification, protein turnover and chaperones (6.6%); translation, ribosomal structure and biogenesis (5.8%); energy production and conversion (4.3%); intracellular trafficking, secretion and vesicular transport (3.9%); and signal transduction mechanisms (3.7%). The proportions of fungal sequences falling into amino acid transport and metabolism, lipid transport and metabolism, and carbohydrate transport and metabolism categories were also notable (Fig. 2). The high representation of transcripts falling into translation and post‐translation categories highlights the fact that fungal cells are highly active at this stage of infection, which probably reflects the switch from an exclusively biotrophic‐oriented phase to a sporulation phase devoted to both the functioning of the haustorial biotrophic structures and the production of uredospores in uredinia, as observed in other rust fungi (Hacquard et al., 2010).
Figure 2.
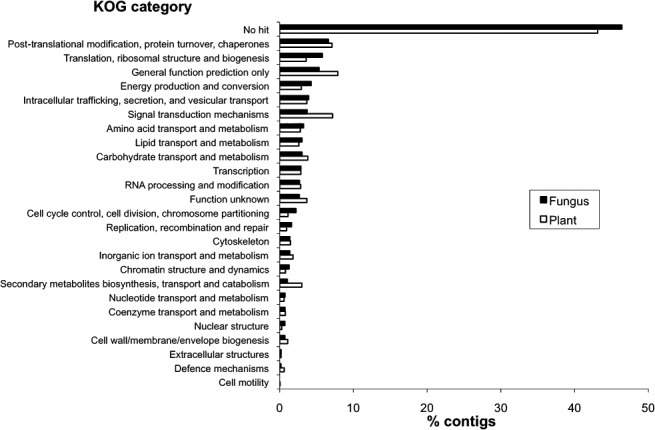
Assignment of fungal and plant sequences to cellular categories based on their KOG (euKaryotic Orthologous Group) classification.
Coffea arabica transcripts expressed during the late compatible interaction with the coffee rust fungus
The 13 951 C. arabica contigs represented 71% of the reads, with an average length of 631 bp (Table 1). Indeed, most of the larger contigs (85%) identified in the assembly were attributed to the plant host. Thirty eight contigs contained more than 200 reads, representing highly expressed plant transcripts (Table 2). Interestingly, 95% of the plant contigs showed homology in the GenBank nonredundant nucleotide (nr) database, but only 55% had a significant blast hit against Coffea ESTs deposited in dbEST. As the contigs were generally shorter than the complete coding sequence, distinct contigs matching the same gene were commonly found. In total, 6202 plant contigs may correspond to novel coffee transcripts not yet described. For instance, 13 new WRKY transcription factor genes were found (data not shown) that have not been reported previously in coffee (Ramiro et al., 2010). To further investigate the extent of transcriptomic coverage in our experiment, we investigated the presence of known genes in various metabolic pathways, such as the caffeine or chlorogenic acid biosynthesis pathways, which are known to be specific for coffee and a few plant species (Ashihara et al., 2008; Mahesh et al., 2007), and the photosynthesis and respiratory pathways, known to be altered in plant–rust interactions (Major et al., 2010). All the genes involved in the metabolic pathways were retrieved in the C. arabica contigs, whereas only 70% of the caffeine or chlorogenic acid biosynthesis pathways were found. Genes involved in glycolysis and the tricarboxylic acid (TCA) cycle were represented by several abundant contigs, with up to 250 reads (Ra = 0.75) for contigs encoding the glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) (Tables 2 and S1), indicating that glycolysis is probably activated in rust‐infected leaves. A total of 532 contigs fell into the carbohydrate transport and metabolism category and 390 contigs fell into the amino acid transport and metabolism category, which highlights that plant metabolism is highly active at this stage of rust infection. In addition, contigs encoding functions related to the plant defence response were among the most abundant contigs (Tables 2 and S1), such as the pathogenesis‐related (PR) proteins (1,3‐β‐glucanases, PR1b, PR‐5 of the thaumatin‐like protein family and chitinases) known to accumulate at a late stage in compatible plant–rust interactions (Duplessis et al., 2009; Fernandez et al., 2004; Ramiro et al., 2009).
Table 2.
Most abundant 454‐contigs of Coffea arabica and Hemileia vastatrix expressed in rust‐infected Coffea arabica leaves at 21 days post‐inoculation.
| Contig ID | Length* | Reads* | Best blast hit (species)† | Accession no. | E‐value | Best rpsblast hit (KOG)‡ | KOG category‡ |
|---|---|---|---|---|---|---|---|
| Fungal contigs | |||||||
| Contig00342 | 124 | 407 | No hit | No hit | No hit | ||
| Contig19064 | 150 | 298 | No hit | No hit | No hit | ||
| Contig00300 | 1769 | 247 | Translation elongation factor EF‐1α (Candida albicans) | XP_711899.1 | 0.0 | Translation elongation factor EF‐1α/Tu | Translation, ribosomal structure and biogenesis |
| Contig18071 | 513 | 245 | No hit | No hit | No hit | ||
| Contig19429 | 256 | 231 | No hit | RhoA GTPase effector DIA/diaphanous | Signal transduction mechanisms, cytoskeleton | ||
| Contig20014 | 256 | 212 | No hit | No hit | No hit | ||
| Contig04616 | 1604 | 202 | Aryl‐alcohol dehydrogenase (Aspergillus terreus) | XP_001217691.1 | E‐108 | Voltage‐gated shaker‐like K+ channel, subunit β/KCNAB | Energy production and conversion |
| Contig19637 | 1576 | 201 | Hypothetical protein AAur_0776 (Arthrobacter aurescens) | YP_946576.1 | 4.00E‐26 | No hit | No hit |
| Contig00263 | 339 | 197 | No hit | No hit | No hit | ||
| Contig19425 | 352 | 197 | No hit | No hit | No hit | ||
| Contig00144 | 479 | 189 | No hit | No hit | No hit | ||
| Contig00025 | 278 | 186 | No hit | No hit | No hit | ||
| Contig17859 | 114 | 182 | No hit | No hit | No hit | ||
| Contig00063 | 263 | 182 | No hit | No hit | No hit | ||
| Contig20059 | 1031 | 179 | No hit | No hit | No hit | ||
| Contig19461 | 422 | 178 | No hit | No hit | No hit | ||
| Contig20013 | 611 | 178 | No hit | No hit | No hit | ||
| Contig00486 | 574 | 174 | No hit | No hit | No hit | ||
| Contig21472 | 918 | 172 | Unknown protein (Populus trichocarpa×Populus deltoides) | ABK96247.1 | 1.00E‐36 | No hit | No hit |
| Contig16131 | 398 | 171 | No hit | RNA polymerase II, large subunit | Transcription | ||
| Contig18436 | 117 | 169 | No hit | No hit | No hit | ||
| Contig17838 | 366 | 166 | No hit | No hit | No hit | ||
| Contig00104 | 264 | 164 | No hit | No hit | No hit | ||
| Contig18196 | 1345 | 164 | Thiamine biosynthesis protein NMT1, plant‐induced rust protein 1 (Uromyces fabae) | O00057 | E‐162 | No hit | No hit |
| Contig00082 | 126 | 159 | No hit | No hit | No hit | ||
| Plant contigs | |||||||
| Contig21649 | 2095 | 421 | Os03g0708600 (Oryza sativa) | Nuclear pore complex, Nup98 component | Nuclear structure, intracellular trafficking, secretion and vesicular transport | ||
| Contig00026 | 123 | 393 | β‐1,3‐Glucanase, basic (Coffea arabica×Coffea canephora) | AAQ90286.1 | 2.00E‐16 | No hit | No hit |
| Contig21461 | 440 | 392 | Metallothionein‐like protein MT‐1 (Mimulus guttatus) | P20238 | 2.00E‐18 | No hit | No hit |
| Contig21497 | 1471 | 349 | Os03g0708600 (Oryza sativa) | CAO23566.1 | 0.0 | No hit | No hit |
| Contig17133 | 389 | 310 | Pathogenesis‐related thaumatin‐like protein (Coffea arabica) | ABW76502.1 | 2.00E‐75 | No hit | No hit |
| Contig19448 | 100 | 299 | Thaumatin‐like protein (Sambucus nigra) | AAK59275.1 | 3.00E‐07 | No hit | No hit |
| Contig18008 | 157 | 290 | Asparagine synthetase (Helianthus annuus) | AAF02775.1 | 1.00E‐12 | Asparagine synthase (glutamine hydrolysing) | Amino acid transport and metabolism |
| Contig18133 | 802 | 285 | Os03g0708600 (Oryza sativa) | Q42679| | E‐112 | S‐Adenosylmethionine decarboxylase | Signal transduction mechanisms |
| Contig19562 | 632 | 280 | Polyubiquitin [Pinus sylvestris] | CAA66667.1 | E‐107 | Ubiquitin/40S ribosomal protein S27a fusion | Translation, ribosomal structure and biogenesis |
| Contig18010 | 115 | 276 | Unknown [Populus trichocarpa] | ABK95431.1 | 4.00E‐14 | Asparagine synthase (glutamine hydrolysing) | Amino acid transport and metabolism |
| Contig17878 | 1169 | 276 | Photosystem II CP47 chlorophyll apoprotein (Dioscorea elephantipes) | YP_001294378.1 | 7.00E‐43 | No hit | No hit |
| Contig01991 | 479 | 269 | Os03g0708600 (Oryza sativa) | CAO62471.1 | 8.00E‐28 | No hit | No hit |
| Contig18005 | 135 | 268 | Asparagine synthetase (Elaeis guineensis) | ACF06573.1 | 1.00E‐05 | No hit | No hit |
| Contig17908 | 130 | 266 | B‐Xylosidase (Camellia sinensis) | ACD93208.1 | 2.00E‐13 | No hit | No hit |
| Contig00069 | 107 | 260 | Glucan endo‐1,3‐β‐d‐glucosidase (Solanum lycopersicum) | CAA52871.1 | 1.00E‐10 | No hit | No hit |
| Contig19395 | 344 | 256 | Hypothetical protein OsI_015840 (Oryza sativa) | EAY94607.1 | 7.00E‐52 | Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) | Carbohydrate transport and metabolism |
| Contig17879 | 361 | 256 | Photosystem II CP47 chlorophyll apoprotein (Coffea arabica) | YP_817508.1 | 8.00E‐66 | No hit | No hit |
| Contig18062 | 575 | 252 | Os03g0708600 (Oryza sativa) | BAD91081.1 | 4.00E‐61 | β‐Galactosidase | Carbohydrate transport and metabolism |
| Contig21900 | 1869 | 250 | Hypothetical protein (Vitis vinifera) | CAN81585.1 | 0.0 | 3‐oxoacyl CoA thiolase | Lipid transport and metabolism |
| Contig18097 | 103 | 248 | Unknown (Populus trichocarpa) | ABK94055.1 | 5.00E‐08 | No hit | No hit |
| Contig20631 | 728 | 244 | Os03g0708600 (Oryza sativa) | CAO43774.1 | E‐112 | Asparaginyl peptidases | Post‐translational modification, protein turnover, chaperones |
| Contig21491 | 218 | 237 | Unknown (Populus trichocarpa) | ABK95810.1 | 4.00E‐24 | No hit | No hit |
| Contig21248 | 747 | 237 | Os03g0708600 (Oryza sativa) | CAO39242.1 | 2.00E‐88 | Asparagine synthase (glutamine hydrolysing) | Amino acid transport and metabolism |
| Contig17869 | 274 | 235 | Unnamed protein product (Vitis vinifera) | CAO45976.1 | 1.00E‐17 | Iron/ascorbate family oxidoreductases | Secondary metabolite biosynthesis, transport and catabolism |
| Contig00669 | 140 | 234 | Unknown protein (Arabidopsis thaliana) | NP_194974.1 | 6.00E‐05 | No hit | No hit |
Length of contigs in nucleotides and number of reads in the assembled contigs.
blastx searches were carried out against the National Center for Biotechnology Information (NCBI) nonredundant nucleotide (nr) database.
rpsblast searches were carried out against the euKaryotic Orthologous Genes (KOG) database.
Hemileia vastatrix transcripts expressed during late infection of C. arabica leaf tissues
The 6763 H. vastatrix contigs showed an average length of 426 bp, but only a few contigs (5.5%) were larger than 1 kb, indicating that fungal contigs were enriched in smaller sequences relative to the total set of contigs (Table 1). About 17% of the contigs were composed of a minimum of 10 reads, indicating that the infected C. arabica leaf tissues contained a number of rust transcripts that were substantially expressed (Table 2). A small number of H. vastatrix contigs had hits in the nr or Swiss‐Prot databases (24%), but 33% showed homology to gene models predicted in M. larici‐populina or Puccinia graminis f.sp. tritici, the two rust genomes sequenced so far. Only 24% of the H. vastatrix contigs presented a hit against Laccaria bicolor, the basidiomycete with the largest number of genes predicted so far (Martin et al., 2008). Among the contigs that showed homology with a rust gene, but not with L. bicolor genes, 521 showed no homology against any other database, probably representing Pucciniales lineage‐specific genes not shared across the Basidiomycota phyllum. Interestingly, 26 contigs did not show homology with predicted rust gene models, but presented a significant match with M. larici‐populina and/or P. graminis f.sp. tritici genome sequences. These H. vastatrix contigs probably correspond to mis‐predicted rust genes in the corresponding genomes, and may be helpful in annotating new genes in the reference rust genomes. Finally, among the 6763 H. vastatrix contigs, 60% showed no homology to the databases used in this study and may correspond to genes unique to coffee rust. In contrast, several homologues of previously described rust transcripts associated with the haustorial infection structure were found among the most abundant H. vastatrix transcripts (Table 2), such as the NMT1 protein, a NADP‐dependent mannitol dehydrogenase, the thiamine synthesis‐related enzyme THI4 and a plasma membrane (H+)‐ATPase described in U. fabae (Hahn and Mendgen, 1997). Other homologues of in planta bean rust transcripts previously reported to be expressed in haustoria were detected among H. vastatrix transcripts of lower abundance, notably several amino acid transporters and the hexose transporter HXT1 (Voegele et al., 2001). Several secreted proteins of low molecular weight previously described in rust fungi were also among the most abundant H. vastatrix transcripts (see below). Signalling genes were also present among abundant contigs, encoding 14‐3‐3 proteins, calcium and calmodulin‐related protein kinases or G‐proteins. All these results tend to indicate that H. vastatrix is transcriptionally very active at this stage of coffee leaf infection, including both biotrophic structures (i.e. haustoria) and the onset of sporulation.
A total of 22 contigs predicted to be of H. vastatrix origin, including 12 genes with known homology [heterotrimeric G‐protein α subunit (Gp‐α); mitogen‐activated protein kinase (MAPK); chitin deacetylases (CD1 and CD2); amino acid transporters (AAT1_2 and AAT3); aspartate amino‐transferase (Asp_AT); invertase (Inv); mannitol dehydrogenase (MAD); translation elongation factor (TEF); β‐tubulin (β‐Tub); cytochrome B (CytB)], three Pucciniales‐conserved genes (RTP; haustorially expressed secreted proteins HESP‐379 and HESP‐C49) and seven new rust genes with no known homology in international databases, was selected in order to test the accuracy of the bioinformatic selection and confirm the fungal origin of the contigs. Polymerase chain reaction (PCR) analyses conducted on C. arabica and H. vastatrix DNA confirmed the fungal origin of the contigs (Fig. S3, see Supporting Information).
Prediction of secreted proteins encoded by H. vastatrix transcripts
Using stringent criteria, bioinformatic analyses predicted a total of 382 rust contigs encoding putative secreted proteins in the H. vastatrix sequence dataset. In detail, 142 showed homology against basidiomycete predicted gene models or nr and Swiss‐Prot databases, 86 against Pucciniales ESTs and 154 had no homology in selected databases. Among the 382 H. vastatrix contigs encoding secreted proteins, 50 contained 30 or more reads supportive of a high expression in infected leaf tissues (Tables 3 and S2, see Supporting Information). Among them, a homologue of the previously described U. fabae secreted protein RTP1 (Kemen et al., 2005) was found, and some genetic determinants often related to the virulence of fungal pathogens, such as cyclophilin or different types of carbohydrate active enzyme (chitinases, endoglucanases) (Table 3). It is tempting to speculate that specific coffee rust effectors reside in the set of predicted secreted proteins that showed no homology with any other rusts or fungi. Among the 382 predicted protein sequences, 20 encoded small proteins of less than 200 amino acids highly enriched in cysteine (Cys) residues (between 5% and 10% of all amino acids) (Table S2).
Table 3.
Fifty most abundant rust transcripts of Hemileia vastatrix encoding putative secreted proteins.
| Contig ID | Length* | Reads* | Best blast hit (species)† | Accession no. | E‐value | Best blast hit against Mlp SP (protein ID)‡ | E‐value |
|---|---|---|---|---|---|---|---|
| Contig20014 | 256 | 212 | No hit | No hit | |||
| Contig19637 | 1576 | 201 | Hypothetical protein AAur_0776 (Arthrobacter aurescens) | YP_946576.1 | 4.00E‐26 | 112808 | 8.00E‐22 |
| Contig00486 | 574 | 174 | No hit | No hit | |||
| Contig21472 | 918 | 172 | Unknown (Populus trichocarpa×Populus deltoides) | ABK96247.1 | 1.00E‐36 | 91075 | 1.00E‐44 |
| Contig00104 | 264 | 164 | No hit | No hit | |||
| Contig00329 | 1293 | 149 | No hit | 85997 | 1.00E‐17 | ||
| Contig22018 | 1021 | 148 | Rust transferred protein (Melampsora occidentalis) | ABS86407.1 | 3.00E‐13 | No hit | |
| Contig00379 | 943 | 140 | Galactan 1,3‐β‐galactosidase (Phanerochaete chrysosporium) | BAD98241.1 | 2.00E‐66 | 62953 | 4.00E‐25 |
| Contig23035 | 551 | 123 | Hypothetical protein (Cryptococcus neoformans) | XP_569628.1 | 2.00E‐18 | 78206 | 4.00E‐43 |
| Contig00017 | 706 | 121 | No hit | No hit | |||
| Contig01344 | 1133 | 120 | No hit | 77195 | 2.00E‐16 | ||
| Contig19484 | 2442 | 115 | AF353616_1 Cro r II (Cronartium ribicola) | AAK28629.1 | 0.0 | 43507 | 0.0 |
| Contig20033 | 189 | 115 | No hit | No hit | |||
| Contig21820 | 1318 | 101 | No hit | No hit | |||
| Contig21571 | 850 | 100 | No hit | No hit | |||
| Contig17858 | 1053 | 97 | No hit | No hit | |||
| Contig20611 | 1593 | 94 | Hypothetical protein UM02377.1 (Ustilago maydis) | XP_758524.1 | 9.00E‐87 | No hit | |
| Contig18013 | 1370 | 82 | Phospholipase/lecithinase/haemolysin (Burkholderia dolosa) | YP_002099763.1 | 4.00E‐14 | 68520 | 2.00E‐39 |
| Contig21062 | 105 | 78 | No hit | No hit | |||
| Contig22188 | 1339 | 75 | No hit | No hit | |||
| Contig03126 | 1280 | 70 | Predicted protein (Coprinopsis cinerea) | XP_001830463.1 | 8.00E‐37 | No hit | |
| Contig22364 | 748 | 64 | No hit | No hit | |||
| Contig00659 | 1891 | 63 | Hypothetical protein An14g01620 (Aspergillus niger) | XP_001400789.1 | 2.00E‐69 | 95653 | 8.00E‐43 |
| Contig18607 | 1578 | 58 | Endopeptidase (Cryptococcus neoformans) | XP_566887.1 | E‐137 | 42703 | 0.0 |
| Contig04624 | 1283 | 56 | No hit | 104797 | 6.00E‐09 | ||
| Contig04096 | 803 | 56 | No hit | 104797 | 1.00E‐17 | ||
| Contig05042 | 1348 | 53 | Hypothetical protein CIMG_08209 (Coccidioides immitis) | XP_001241046.1 | 4.00E‐75 | No hit | |
| Contig02757 | 1368 | 51 | Predicted protein (Laccaria bicolor) | XP_001874996.1 | 2.00E‐30 | 72374 | 7.00E‐41 |
| Contig18226 | 1260 | 50 | Pectinesterase family protein (Neosartorya fischeri) | XP_001262221.1 | 3.00E‐22 | 75334 | 2.00E‐18 |
| Contig00575 | 739 | 49 | No hit | No hit | |||
| Contig03936 | 1515 | 48 | Hypothetical protein (Melampsora medusae f.sp. deltoidis) | ABS86355.1 | 5.00E‐29 | 71512 | 3.00E‐35 |
| Contig03932 | 2212 | 48 | Predicted protein (Laccaria bicolor) | XP_001878325.1 | 4.00E‐07 | 70885 | 7.00E‐54 |
| Contig02027 | 2008 | 46 | Hypothetical protein CC1G_00344 (Coprinopsis cinerea) | XP_001837208.1 | E‐127 | 116816 | E‐146 |
| Contig06274 | 1187 | 45 | Unknown (Populus trichocarpa×Populus deltoides) | ABK96247.1 | 2.00E‐31 | 86274 | 3.00E‐35 |
| Contig04146 | 665 | 45 | No hit | No hit | |||
| Contig20800 | 1878 | 44 | Hypothetical protein UM04400.1 (Ustilago maydis) | XP_760547.1 | E‐108 | 44205 | E‐163 |
| Contig18836 | 1255 | 42 | Endoglucanase (Phanerochaete chrysosporium) | AAU12275.2 | 1.00E‐49 | 47207 | 6.00E‐71 |
| Contig00581 | 1427 | 42 | Hypothetical protein PTRG_11055 (Pyrenophora tritici‐repentis) | XP_001941386.1 | 7.00E‐13 | 73154 | 5.00E‐43 |
| Contig22633 | 366 | 41 | No hit | No hit | |||
| Contig22182 | 1413 | 38 | Predicted protein (Laccaria bicolor) | XP_001881269.1 | 2.00E‐63 | 34172 | 7.00E‐78 |
| Contig00191 | 850 | 37 | No hit | No hit | |||
| Contig18344 | 1278 | 36 | No hit | 72270 | 1.00E‐24 | ||
| Contig06220 | 1362 | 34 | Protein disulphide isomerase (Laccaria bicolor) | XP_001888384.1 | 2.00E‐36 | 117675 | 2.00E‐59 |
| Contig19568 | 1331 | 34 | Unknown (Populus trichocarpa×Populus deltoides) | ABK96247.1 | 2.00E‐10 | 77357 | 5.00E‐24 |
| Contig01984 | 1291 | 32 | RE56164p (Drosophila melanogaster) | AAN71436.1 | 4.00E‐05 | No hit | |
| Contig18144 | 108 | 32 | No hit | No hit | |||
| Contig19643 | 880 | 31 | No hit | No hit | |||
| Contig04210 | 1667 | 30 | Glycoside hydrolase family 61 protein (Laccaria bicolor) | XP_001883194.1 | 9.00E‐13 | 78608 | 3.00E‐68 |
| Contig03336 | 1183 | 30 | Expansin family protein (Laccaria bicolor) | XP_001877082.1 | 9.00E‐22 | 71932 | 3.00E‐47 |
| Contig18608 | 1065 | 30 | No hit | 104797 | 3.00E‐16 | ||
Length of contigs in nucleotides and number of reads in the assembled contigs.
blastx searches were carried out against the National Center for Biotechnology Information (NCBI) nonredundant nucleotide (nr) database.
blastx searches were carried out against predicted secreted proteins (SP) in the M. larici‐populina (Mlp) Joint Genome Institute (JGI) genome sequence.
Identification of conserved rust candidate effectors
Hemileia vastatrix transcripts encoding secreted proteins were compared across the order Pucciniales (rust ESTs and genome sequences) and against the Plant–Host Interaction database (PHI‐base) to determine conserved virulence‐related genes in plant pathogens and candidate effectors among rust fungi (Table S3, see Supporting Information). The absence of homology in a given EST set may be related to a lack of expression in a particular physiological state and not to the absence of the corresponding gene in the rust species. Interestingly, several in planta‐expressed secreted proteins reported in U. fabae and M. lini were identified in several rust species, including H. vastatrix and/or M. larici‐populina and P. graminis f.sp. tritici (Table S3). The sequences of the homologues of U. fabae RTP1 (Kemen et al., 2005) and M. lini HESP‐C49, HESP‐178 and HESP‐379 (Catanzariti et al., 2006), representing haustorially expressed rust effectors found in several rust fungi, were selected and aligned. The alignment of RTP1 homologues (Fig. 3), including the RTP homologues described previously in Uromyces spp. and P. graminis (Puthoff et al., 2008), three H. vastatrix RTP homologues and 10 Melampsora spp. RTP homologues, highlighted species‐related divergence with a rather conserved region of 130 amino acids at the C‐terminus. In contrast, the N‐terminal region of RTP‐like proteins was relatively divergent between rust species (Fig. 3). HESP homologues identified in several rust species, including H. vastatrix, did not present strong similarity along the protein sequence; however, the Cys positions were highly conserved in homologues of M. lini HESP‐C49 and HESP‐178, with 11 and seven conserved Cys residues, respectively (Fig. 4). HESP‐178 contains the 8‐Cys CFEM (common in fungal and extracellular membrane) domain previously described in the G‐protein‐coupled receptor Pth11 and required for virulence in Magnaporthe oryzae (Kulkarni et al., 2003); however, the 8‐Cys composing the CFEM domain was only found in M. lini, M. larici‐populina and H. vastatrix homologues (Fig. 4). Rust homologues of HESP‐379 (Fig. 4) showed a highly conserved region of 100 amino acids following the peptide signal and a less conserved C‐terminal region enriched in serines that contains a predicted site for the glycosylphosphatidylinositol (GPI) anchor, which indicates that this protein probably remains attached to the exterior leaflet of the fungal plasma membrane (Pierleoni et al., 2008). Finally, comparison with the PHI‐base identified nine contigs for which homologous genes in other plant or animal pathogenic fungi had a referenced phenotype related to virulence (Table S3). For instance, two contigs matched the Botrytis cinerea bcpme1 gene encoding a pectin methylesterase gene, the disruption of which results in reduced virulence on apple fruits, grapevine and A. thaliana leaves (Valette‐Collet et al., 2003). Another contig showed homology to the Magnaporthe grisea GAS1 gene encoding a small protein expressed specifically during appressorium formation, with a role in rice leaf penetration (Xue et al., 2002).
Figure 3.
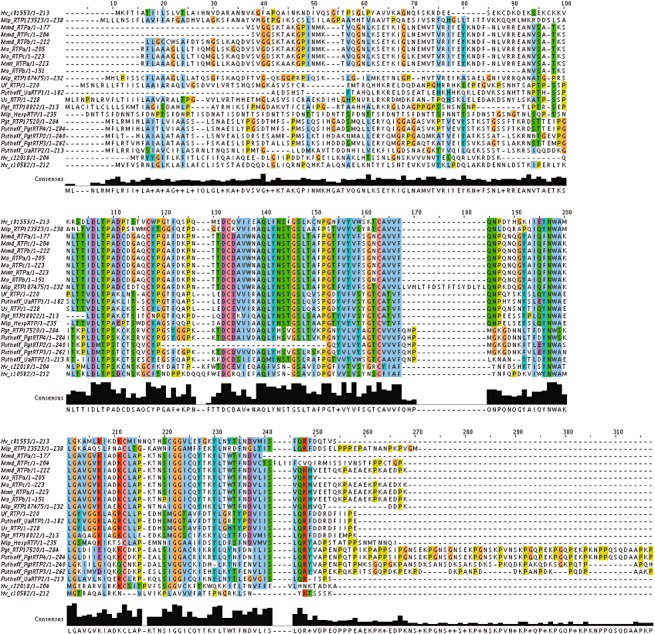
ClustalW alignment of rust proteins homologous to the Uromyces fabae rust transferred protein 1 (RTP1). Proteins were retrieved from the literature and international databases. Hv, Hemileia vastatrix; Mlp, Melampsora larici‐populina; Mmd, Melampsora medusae f.sp. deltoidae; Mmt, Melampsora medusae f.sp. tremuloidae; Mo, Melampsora occidentalis; Pgt, Puccinia graminis f.sp. tritici; Ua, Uromyces appendiculatus; Uf, Uromyces fabae; Us, Uromyces striatus. Sequences retrieved from Puthoff et al. (2008) are indicated. Mmd, Mmt and Mo RTP1‐like proteins were predicted from expressed sequence tags (ESTs) and lacked the N‐terminal region. Mlp and Pgt sequences were retrieved in the genome sequences available at the Joint Genome Institute and the Broad Institute, respectively. Mlp_HespRTP1 is a fusion protein with homology to a haustorially expressed secreted protein (HESP) in the N‐terminal region, and only the C‐terminal region presenting homology to RTP1 was used here to perform the alignment.
Figure 4.

ClustalW alignments of conserved rust secreted protein sequences presenting homology with Melampsora lini haustorially expressed secreted proteins (HESPs). (A) Rust homologues of HESP‐C49. (B) Rust homologues of HESP‐178. (C) Rust homologues of HESP‐379. Hv, Hemileia vastatrix; Ml, Melampsora lini; Mlp, Melampsora larici‐populina; Mmd, Melampsora medusae f.sp. deltoidae; Pgt, Puccinia graminis f.sp. tritici; Pp, Phakopsora pachyrhizi, Ps, Puccinia striiformis; Ptr, Puccinia triticina; Ua, Uromyces appendiculatus; Uf, Uromyces fabae. MlpG and PgtG indicate that sequences were retrieved from the genome sequences, whereas MlpE indicates sequences deduced from expressed sequence tags (ESTs). Dotted rectangles indicate the N‐terminal regions predicted to contain the signal peptides for secretion. In (A) and (B), conserved cysteines are indicated by red stars below the consensus sequence. In (C), the red arrow indicates the glycosylphosphatidylinositol (GPI) anchor location determined by the PredGPI prediction server for all HESP‐379 homologues except Pp_gi120516459.
Expression profiling of H. vastatrix genes during infection of coffee leaves
Taking advantage of the genomic resource generated for H. vastatrix, specific primers were designed from the 22 rust gene sequences selected previously (Fig. S3) to quantify their transcript accumulation by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) during the time course of infection of C. arabica leaves. Fungal gene expression profiles in coffee leaves were assessed at 18 and 48 h post‐inoculation (hpi) and at 7 and 21 days post‐inoculation (dpi). The later time point corresponds to the infection stage used for 454‐pyrosequencing. Microscopic evaluation of fungal differentiation indicated that, at 18 hpi, the fungus had germinated and differentiated appressoria. At 48 hpi, most infection sites were at the penetration hyphae stage, with the first haustoria formed in stomatal cells. At 7 dpi, all structures, including haustoria in the host mesophyll, had differentiated and no major changes in relative proportions were recorded from this point onwards; at 21 dpi, the leaves were heavily colonized. RT‐qPCR expression profiling was conducted in two independent time‐course replicates.
Fold‐change expression levels measured for the 22 genes at each time point, compared with the average expression level measured along the time course, are presented in Fig. 5. Half of the H. vastatrix genes showed almost no or moderate regulation of transcript accumulation during plant infection. By contrast, other genes presented a strong induction of expression at 18 hpi (Gp‐α, MAPK, CytB, AAT3, HESP‐379 and Hv00571), 7 dpi (HESP‐379, Hv00147, Hv00162, Hv00628) or 21 dpi (CD1, MAD and RTP). Interestingly, distinct expression profiles were obtained for the CD1 and CD2 genes, as well as for the AAT1_2 and AAT3 genes, indicating a specificity of expression during the interaction with the plant host for these rust multigene families. Amplification cycle values (Cq) measured at 21 dpi for H. vastatrix genes were consistent with the number of reads in the corresponding 454‐contigs, most abundant contigs presenting lower Cq values than genes with lower expression levels (Fig. S4, see Supporting Information).
Figure 5.
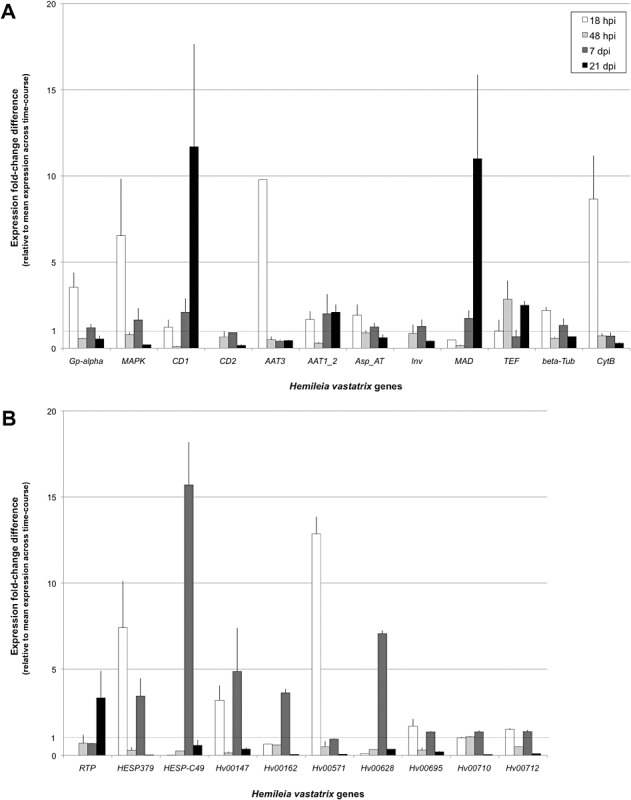
Expression profiles for 22 Hemileia vastatrix transcripts during the time course of infection of Coffea arabica var. H147/1 leaves by H. vastatrix strain 178a at 18 and 48 h post‐inoculation (hpi) and 7 and 21 days post‐inoculation (dpi). (A) Hemileia vastatrix genes with known homology in databases and encoding an heterotrimeric G‐protein α subunit (Gp‐alpha), a mitogen‐activated protein kinase (MAPK), two chitin deacetylases (CD1 and CD2), two amino acid transporters (AAT1_2 and AAT3), an aspartate amino‐transferase (Asp_AT), an invertase (Inv), a mannitol dehydrogenase (MAD), a translation elongation factor (TEF), a β‐tubulin (beta‐Tub) and a cytochrome B (CytB). (B) Ten H. vastatrix genes of no known function and showing homology to known rust effectors (rust transferred protein RTP; haustorially expressed secreted proteins HESP379 and HESP‐C49), or showing no homology in international databases and representing new rust genes (Hv00147, Hv00162, Hv00571, Hv00628, Hv00695, Hv00710, Hv00712). Transcript levels were assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) using three H. vastatrix reference genes (cytochrome oxidase subunit III, contig19515; ribosomal protein 40S, contig01333; hypothetical protein Hv00099, contig21737) and, for each gene, fold‐change differences correspond to the expression level measured at a given time point relative to the mean level calculated across the time course (note the one‐fold line indicated). Data were obtained from two independent time‐course replicates.
DISCUSSION
For years, avirulence genes mediating resistance in host plants have been searched for in fungal pathogens through direct genetic approaches (Stergiopoulos and de Wit, 2009). The concept of effector triggered immunity (ETI) (Jones and Dangl, 2006) has led plant pathologists to enlarge the search to so‐called effectors and not only avirulence genes (Dodds and Rathjen, 2010). With the huge collection of genomic data now available for the research community, which is increasing each year, the description of plant pathogen secretomes and of putative virulence effectors detected among secreted proteins has dramatically increased (Lebrun and Kamoun, 2010), and shows evidence that fungal genomes encode a large reservoir of effector‐like proteins (Choi et al., 2010; Soanes et al., 2008). In this article, we report the use of the Roche 454‐pyrosequencing technology to establish a repertoire of H. vastatrix transcripts expressed in coffee leaves to search for candidate virulence effectors of the coffee orange rust fungus. More than 350 000 reads were produced and assembled into 22 774 contigs from plant or fungal transcripts. A common strategy for the identification of transcript origin in EST sequencing projects based on mixed plant tissues and fungal infection structures usually involves the performance of homology searches using the blastx algorithm against the nr databases at GenBank, although such an approach has clear limitations because of the over‐representation of plant sequences relative to fungal ones (e.g. 12 578 963 Eudicotyledon ESTs and 629 036 Basidiomycota ESTs in dbEST in August 2010). Predictive methods, based on GC percentage, codon usage or di‐, tri‐ or hexa‐nucleotide frequencies in ESTs, have also been used, and programs have been designed to help define the plant or fungal origin of ESTs in plant–pathogen interaction studies (Bowen et al., 2009; Emmersen et al., 2007). In the absence of the availability of extensive Coffea and Hemileia genomic data, an original predictive approach was selected that combined the two methods described above. The prediction of plant or fungal origin of rust‐infected coffee leaf 454‐contigs was completed by systematic comparison of the sequences with genomic data available for three dicots and three basidiomycete species, as well as plant and fungal sequences deposited in GenBank, and Coffea and Pucciniales ESTs deposited in dbEST. Finally, the combined prediction approach resulted in about 30% fungal contigs and 61% plant contigs, and the remaining sequences were considered as not determined. Coffea arabica is an allotetraploid species that contains two subgenomes related to the Coffea canephora and Coffea eugenioides species (Lashermes et al., 1999; Petitot et al., 2008). As of August 2010, 43 619 C. arabica ESTs were available in public EST databases. As reported, the C. arabica genome is complex and no sequencing project is yet planned for this economically important crop species. The discovery of around 14 000 novel C. arabica transcripts provides new opportunities to decipher the allotetraploid coffee genome and may enhance our knowledge of the genetics and physiology of this crop species. When the C. canephora genome is made publicly available (http://www.coffeegenome.org/), it will be possible to compare the contigs produced in the present study to update the accuracy of contig selection and to aid in the annotation of gene prediction. Although fungal contigs represented 30% of the contigs, the proportion of 454‐sequences was much less in the rust contigs than in the coffee contigs (i.e. 20% of the total number of reads). A high proportion of H. vastatrix proteins without any known function was found, which highlights the need for the annotation of fungal genome sequences.
Among the most important cellular categories represented in the fungal contigs were translational‐ and post‐translational‐related functions, as well as energy production and conversion. By 21 dpi, a large number of H. vastatrix haustoria and infection hyphae had formed in Coffea‐infected coffee leaves (Silva et al., 2006). Rust haustorial structures are involved in the derivation of nutrients from the host tissues to the fungus (Voegele et al., 2009). Several transcripts corresponded to transporters previously reported to be specifically or predominantly expressed in haustoria of the bean rust fungus (Struck et al., 2002; 2001, 2009), as well as at the uredinial stage of the poplar rust (Hacquard et al., 2010). The gene expression patterns observed here indicate that the fungal structures are highly active at this stage of the interaction with the plant host. Carbohydrate, lipid and amino acid transport and metabolism categories were also supported by a large number of rust contigs, supportive of metabolite fluxes between H. vastatrix and its host. Expression profiling of selected H. vastatrix genes mostly confirmed the expression assessed by read abundance in 454‐contigs (Fig. S4). Distinct patterns of rust gene expression were observed during the time course of infection of coffee leaves (Fig. 5), indicative of a highly dynamic transcription activity during plant colonization and biotrophy. Signalling genes encoding a Gp‐α and a MAPK showed peak expression at early stages when the fungus had differentiated appressoria, as reported previously for other pathogenic fungi (Li et al., 2007), indicating that these genes might also be required for appressorium differentiation in the coffee rust fungus. Interestingly, two genes encoding amino acid transporters homologous to U. fabae transporter genes (Voegele et al., 2009) showed distinct patterns of expression, AAT3 being highly expressed at 18 hpi, and AAT1_2 being slightly induced at 18 hpi and also at 7 and 21 dpi. The latter gene was also expressed in U. fabae haustoria (Hahn et al., 1997) and at late stages of infection of poplar leaves by M. larici‐populina (Hacquard et al., 2010). The profiles observed here suggest the specific expression of transporter genes at different stages of host colonization. The CD1 gene showed a strong peak of expression at 21 dpi, consistent with the late expression of a chitin deacetylase encoding gene of the poplar leaf rust fungus in uredinia (Hacquard et al., 2010). Strong MAD transcript accumulation in haustoria and protein localization in haustoria and uredospores in planta have been reported for the bean rust fungus (Voegele et al., 2005), and mannitol was described as a possible major source of storage. The strong expression of the H. vastatrix MAD gene homologue at 21 dpi suggests that mannitol might play a general role in rust biology, and it remains to be determined whether this polyol serves as a source of storage or is used by the fungus during sporulation.
Using programs predictive for signal peptide secretion signatures with stringent criteria, 382 transcripts encoding putative secreted proteins were identified within the 6763 fungal contigs (Table 3), consisting of 5.6% of the rust contigs in the dataset. Comparison between the genomes of the poplar rust and the wheat stem rust fungi revealed that they encode important reservoirs of secreted proteins, several hundreds being specific to each rust species (Choi et al., 2010; S. Duplessis et al., unpublished data). The present analysis identified several H. vastatrix transcripts homologous to other rust transcripts previously described in rust fungi and related to the biotrophic phase. In contrast, 154 contigs did not show any homology in databases, and may be H. vastatrix‐specific secreted proteins. Interestingly, 3.9% of the total number of fungal contigs were classified into the intracellular trafficking, secretion and vesicular transport category, among the most important cellular class expressed in the coffee rust fungus, which is supportive of an important secretion activity at this stage of the compatible coffee–rust interaction. Previously, consistent amounts of secreted proteins have been reported from in planta‐expressed rust transcripts, particularly from purified haustoria (Catanzariti et al., 2006; Jakupovic et al., 2005; Link and Voegele, 2008; Puthoff et al., 2008). More recently, a combined laser capture microdissection (LCM) and transcriptome analysis of poplar rust uredinia has shown that the fungal cells present in uredinia subjacent zones (i.e. haustoria and infection hyphae in the plant palisade mesophyll) are also marked by the accumulation of secreted proteins coding transcripts (Hacquard et al., 2010). This suggests that effector‐like rust secreted proteins are not only produced at early stages of infection in haustoria, but all along the infection process up to uredospore formation. The important proportion of H. vastatrix transcripts homologous to rust secreted proteins detected at this late stage of coffee leaf infection is in accordance with these observations.
In U. fabae, the translocation of the secreted protein RTP1 to the plant nucleus has been shown by immunolocalization (Kemen et al., 2005), indicating that rust proteins may not be secreted only to the apoplast, but could target plant subcellular locations. Interestingly, this RTP1 protein, although of unknown function, has homologues in H. vastatrix and in other rusts, and these are organized in multigene families. The N‐terminal part of the protein is quite diverse, whereas the C‐terminal part of the protein is relatively well conserved among Pucciniales (Fig. 3), suggesting that it may be related to the function of the protein in the host plant. This result is in accordance with the data of Puthoff et al. (2008), who reported that rust putative effectors may be conserved throughout the order Pucciniales, although differences in protein composition could reflect diversification driven by host–rust co‐evolution. An H. vastatrix RTP gene was strongly induced at a late stage of infection, and not expressed at 18 hpi (Fig. 5), which is consistent with the expression profiles reported previously in U. fabae (Kemen et al., 2005) and U. appendiculatus (Puthoff et al., 2008). The identification of the putative function of RTP will help to clarify this domain organization in the different rust species, and will be of interest to determine whether rust effectors interact with host plant proteins through their C‐terminal part, as reported for oomycete effectors (Schornack et al., 2009). With regard to H. vastatrix transcript homologues of M. lini HESPs, although poorly conserved at the global sequence level, a high conservation of Cys positions was found between homologues of HESP‐C49 and the CFEM‐containing HESP‐178 (Fig. 4). This result suggests that these fungal proteins may share a role in pathogenicity or in enabling infection of the host plant. It remains to be determined whether the role of these conserved Cys residues is in coping with the apoplastic environment enriched in plant proteases or in a more particular function during the interaction with the host targets. By contrast, HESP‐379 homologues are highly conserved in their N‐terminal region and contain a predicted GPI anchor in the C‐terminal region, indicating that this protein might reside at the exterior of the rust cell attached to the plasma membrane. A HESP‐379 homologue from U. fabae (Uf_HSP5_Uf047_gi164277360 in Fig. 4C) has been described previously in the haustorial secretome of the bean rust fungus (Link and Voegele, 2008), suggesting that this conserved rust protein could be secreted by H. vastatrix haustoria during the interaction with C. arabica. Distinct patterns of expression were observed for H. vastatrix HESP‐379 and HESP‐C49 genes, with expression peaks at 18 hpi and 7 dpi, respectively (Fig. 5). Interestingly, several new rust genes of unknown function, including the Hv00628 gene encoding a secreted protein, were highly induced at 18 hpi or 7 dpi, highlighting the dynamic expression of rust genes during host infection (Fig. 5). Strikingly, although most selected genes assessed by RT‐qPCR were primarily identified at 21 dpi, only four showed their highest expression level at this stage compared with earlier time points. This result indicates the need to dissect the plant infection process for a better understanding of the rust genetic programs expressed in planta.
With the use of a new‐generation sequencing technology in a nonmodel rust species, we have been able to report the expression of novel genetic determinants, particularly rust candidate effectors. However, it is not possible here to detail in which rust subcompartment the detected transcripts were expressed. The LCM technique, which allows the isolation of subareas in tissues, represents an interesting methodology to dissect rust biology at the uredinial stage. Recently, LCM has been applied to separate Phakopsora pachyrhizi uredospores from emerging uredinia (Tremblay et al., 2009), and has helped to identify 925 ESTs expressed in planta in this structure. Similarly, LCM has been used to dissect different fungal structures at the uredinial stage of poplar leaf infection by the rust fungus M. larici‐populina and has revealed, by custom oligoarray‐transcriptome profiling, that distinct genetic programs related to biotrophy and sporulation are expressed by the rust structures (Hacquard et al., 2010). Combining such an LCM approach with new‐generation sequencing will probably provide major improvements in our understanding of rust biology.
The data reported here are new and represent the first steps towards a molecular understanding of the pathogen by providing a global view of the H. vastatrix and C. arabica interacting transcriptomes. Using data collected at an advanced stage of the fungal developmental cycle in the leaf, we have detected novel coffee and fungal gene transcripts and predicted putative rust pathogenicity genes. Approximately 60% of the H. vastatrix contigs did not map to any previously annotated protein coding genes and represent novel and interesting sequences. Currently, more than 45 H. vastatrix races have been described (Rodrigues et al., 1975; Várzea and Marques, 2005). By identifying H. vastatrix genes linked to pathogenicity, a genetic survey of the different races could be undertaken to determine the level of variability and the genetic relationships among the virulent populations. Insights into allelic diversity in H. vastatrix effector gene loci will reveal the mechanisms underlying the dynamics of adaptation to plant defences in populations of the rust fungus. In addition, gene data may help in the design of molecular diagnostic tools to identify H. vastatrix races.
EXPERIMENTAL PROCEDURES
Biological material and infection procedures
Coffea arabica CIFC H147/1 plants (carrying the resistance factors SH2, SH3, SH4 and SH5) were grown in the glasshouse in 70% relative humidity at 25 °C and under 12 h light. Plants were inoculated with H. vastatrix as described in Silva et al. (1999) with fresh uredospores of H. vastatrix isolate 178a (bearing the virulence factors v2, v3, v4 and v5), eliciting a compatible interaction. In this H147/1 × 178a pathosystem, sporulation occurred at 25–26 dpi (data not shown). For 454‐pyrosequencing, leaves were collected at 21 dpi, ahead of sporulation, frozen by immersion in liquid nitrogen and stored at −80 °C until RNA extraction. For the time‐course infection experiment, leaves of C. arabica H147/1 inoculated with H. vastatrix isolate 178a were collected at different time points after inoculation (18 hpi, 48 hpi, 7 dpi and 21 dpi) in order to represent the progression of infection and host tissue colonization. Successful uredospore germination, appressoria formation and fungal growth in host tissues were confirmed by light microscopy after staining with cotton blue lactophenol. This was performed either by the observation of leaf surface moulds (18 hpi) or of leaf sections, as described previously (Silva et al., 1999). Material was immediately frozen in liquid nitrogen and stored at −80 °C.
RNA extraction and cDNA synthesis
Total RNAs were extracted from coffee leaves using the RNeasy Plant Minikit (Qiagen, Courtaboeuf, France) completed by a DNase treatment. One microgram of total RNA was used to produce cDNA using the SMART™ PCR cDNA Synthesis Kit (Clontech, Palo Alto, CA, USA). RT and PCRs were carried out with PrimeScript™ Reverse Transcriptase (Takara Bio, Shiga, Japan) and Advantage® 2 Polymerase Mix (Clontech), respectively. The cDNA purification steps were performed using a Qiaquick PCR Purification Kit (Qiagen).
Pyrosequencing and assembly of 454‐reads
Sequences were generated from 5 µg of full‐length cDNA using the 454‐GS‐FLEX Titanium pyrosequencing technology on a Roche 454 Titanium sequencer (single read strategy) at the Genoscope (Centre National de Séquençage, Evry, France; http://www.genoscope.cns.fr/spip/) employing a half‐plate and following standard procedures recommended by Roche. The assembly of the 352 146 pyrosequencing reads was performed at the Genoscope using the Roche Newbler assembler (software release 2.0.00.22) following recommended parameters provided by Roche. Raw 454‐sequences are available upon request. The 22 774 assembled CaHv (C. arabica leaves infected by the rust fungus H. vastatrix) contig sequences are provided in Data S1. The relative abundance of transcripts was estimated according to Vega‐Arreguín et al. (2009) with the following index: Ra =N/L, where Ra is the relative abundance, N is the number of 454‐sequences per contig and L is the length of the assembled contig.
PCR and RT‐qPCR
In order to determine the origin of the 22 selected 454‐contigs, PCR amplifications were carried out on an MyCycler thermocycler (Bio‐Rad, Hercules, CA, USA), using specific primers designed after the 454‐contig sequences, with Dream Taq DNA polymerase (MBI Fermentas, Vilnius, Lithuania), following the manufacturer's protocol for a total of 40 cycles. Hemileia vastatrix appressoria cDNA, rust‐infected C. arabica leaf cDNA and healthy C. arabica leaf cDNA were used as template DNA, and control amplifications with no template DNA were also performed. Amplification products were separated by electrophoresis on 3% agarose gels using a Gene Ruler Low Range marker (MBI Fermentas).
In order to assess transcript levels by RT‐qPCR, the specific primers of the 22 H. vastatrix genes were used and expression analysis was conducted along a H. vastatrix differentiation/infection time course at 18 and 48 hpi and at 7 and 21 dpi in two independent time‐course replicates. First‐strand cDNAs were synthesized from 1 µg of total RNA in a final volume of 20 µL using an Omniscript Reverse Transcriptase Kit (Qiagen) and oligo(dT)18 primer (MBI Fermentas) following the manufacturer's instructions. RT‐qPCR was performed on a MJ Mini thermocycler (Bio‐Rad) equipped with a MiniOpticon Real Time PCR System (Bio‐Rad) using EvaGreen Supermix (Bio‐Rad), following the manufacturer's recommendations. Three H. vastatrix genes validated as reference genes (A. Vieira et al., unpublished results) were used to normalize in planta expression (cytochrome oxidase subunit III, contig19515; ribosomal protein 40S, contig01333; hypothetical protein Hv00099, contig 21737). For each gene, fold‐change expression levels were obtained using the relative expression formula of Pfaffl (2001), where the average expression level across all sampled time points was used as control.
Sequence databases used and EST annotation
In the absence of extensive genomic data for both C. arabica and H. vastatrix (43 619 ESTs and no EST, respectively, in dbEST as of August 2010), sequence homology searches were performed against public databases [nr and Swiss‐Prot databases at NCBI (http://www.ncbi.nlm.nih.gov/); PHI‐base v3.2], a reference database (Winnenburg et al., 2008; http://www.phi‐base.org/) and three distinct database sets generated for this study. The first set (database I) was composed of the three dicot genomic databases (A. thaliana; Populus trichocarpa cv. Nisqually; Vitis vinifera, http://www.phytozome.net/) and three basidiomycete genomic databases [L. bicolor and M. larici‐populina at the Joint Genome Institute (JGI), http://www.jgi.doe.gov/; P. graminis f.sp. tritici at the Broad Institute, http://www.broadinstitute.org/]. The second set (database II) included only Coffea spp. and Pucciniales spp. ESTs built from sequences retrieved from dbEST at GenBank. Reference ESTs were cleaned on the basis of systematic searches against nr DB using the blastn algorithm (E‐value ≤ 10−10) and by removing hits corresponding to plasmid vectors, bacteria and animal sequences, and fungal sequences in the Coffea EST set or plant sequences in the Pucciniales EST set. ESTs with no homology in nr were not considered. Totals of 41 241 and 23 168 reliable Coffea reference ESTs and Pucciniales reference ESTs, respectively, were retained in database II. Finally, the third set (database III) was narrowed to Pucciniales sequences, with a total of 155 280 transcribed sequences retrieved from the NCBI database (May 2010), and classified into 10 groups based on the corresponding rust species of origin (i.e. 203 P. graminis f.sp. tritici ESTs; six Puccinia coronata var. lolii ESTs; 2850 Puccinia striiformis f.sp. tritici ESTs; 47 466 P. triticina ESTs; 27 M. lini cDNA and DNA sequences; 49 017 M. larici‐populina ESTs; 34 394 Phakopsora pachyrhizi ESTs; 601 U. fabae ESTs; 19 480 Uromyces appendiculatus ESTs; and 1236 Cronartium quercuum f.sp. fusiforme ESTs). Homology searches were performed using blast algorithms (Altschul et al., 1997) against genome sequences and predicted transcripts (blastn), deduced protein sequences (blastx) and rust ESTs (tblastx) with a cut‐off criterion (E‐value < 10−5). For each search against a given database, only the best blast hit was considered and the corresponding E‐value, scores and sequence match length were extracted using local scripts. The assignment of 454‐contig sequences into KOG functional categories was obtained using Reverse psi‐blast (rpsblast; Altschul et al., 1997) against the KOG database (Tatusov, 2003).
Identification of plant and fungal transcripts from 454‐derived contigs
Contigs of assembled 454‐reads were further considered for the analysis, whereas singletons were not included. Two main approaches were used to help distinguish fungal sequences from plant host sequences, and are described here. In total, six predictions were made (Table 2; Figs S1 and S2) and the number of plant (nP) or fungus (nF) predictions for each contig was used to distinguish between contigs. Contigs that were only predicted as plant (nP > 2 and nF = 0) or fungus (nF > 2 and nP = 0) were considered as plant or fungus, respectively. When contigs had been alternatively attributed to plant or fungal origins, the following codes were used: likely plant (nP ≥ 4 and nF = 2; or nP > 2 and nF = 1; or nP = 1 and nF = 0); dubious plant (nP > nF and nF > 1); likely fungus (nF ≥ 4 and nP = 2; or nF > 2 and nP = 1; or nF = 1 and nP = 0); dubious fungus (nF > nP and nP > 1); not resolved (nP = nF). Finally, when none of the six predictions identified a sequence as P or F, the sequence was registered as ‘not determined’.
Species‐specific trinucleotide frequencies were considered using the EST3 program from MIPS trained with reference sequences (Emmersen et al., 2007). A first training set (intrinsic training set) was constructed on the basis of homology searches against database II for the contigs obtained after the assembly of the 454‐reads (Fig. S1). A second EST3 training set (extrinsic training set of 300 plant and 300 fungal sequences) was built from database II sequences, based on the Coffea and Pucciniales ESTs homologies to the nr database (i.e. highest blast scores only to plant or fungal sequences in nr). The EST3 program trained with the extrinsic training set predicted a number of plant and fungal sequences from the contigs obtained after the assembly of the 454‐reads. From this prediction, three sets of 300 predicted Coffea and 300 predicted Hemileia contigs were selected at random to build three intrinsic training sets to improve EST3 prediction. The EST3 program trained with these random sets of intrinsic Coffea/Hemileia sequences predicted different numbers of plant and fungal sequences. The contigs that were always predicted as plant or fungus by EST3 using either intrinsic or extrinsic training sequences were considered as tentative plant and tentative fungal contigs, respectively.
Homology searches (blastn and blastx) were performed against the public databases or the database sets generated for this study (Fig. S2). For the homology searches performed against the public databases and database II, contigs were predicted to be of plant or fungal origin, respectively, based on the best blast hit. For database I searches, average blast scores were calculated for positive hits against dicots and basidiomycetes, and were then subtracted (i.e. mean plant scores subtracted by mean fungal scores). If the value obtained was higher than 100 (arbitrary cut‐off), the contig was considered to be of plant origin, whereas a value below −100 (arbitrary cut‐off) was considered to be of fungal origin. Genes of known plant or fungal origin were manually inspected and the {−100;100} cut‐off values were validated for origin detection (data not shown). The contigs obtaining a value between 100 and −100 were systematically manually inspected considering the length and percentage of identity between the query and subject sequences to determine the eventual origin. When all blast result parameters were too close to clearly define the origin, the contigs were considered as ‘not determined’ (nd). All prediction and homology search results for the 22 774 CaHv contigs are detailed in Table S1.
Bioinformatic analysis of fungal transcripts
Contigs assigned to the rust fungus H. vastatrix were systematically compared with database III and PHI‐base using the blastx algorithm. Comparisons were also extended to the proteomes of ascomycete fungi, including hemibiotroph and necrotroph plant pathogens, on the MycorWeb portal (data not shown; http://mycor.nancy.inra.fr/IMGC/TuberGenome/blast.php). In order to identify fungal sequences encoding secreted proteins, open reading frames (ORFs) were predicted with the translation tool FrameFinder of the ESTate package (http://www.ebi.ac.uk/~guy/estate/) using a codon usage table defined with the tblastx alignment results of 2461 reliable H. vastatrix contigs (F_contigs, Table S1, column L). The accuracy of the ORF prediction was assessed by comparing the results of homology searches against the nr and Swiss‐Prot databases with the protein sequence translated from the predicted ORF (blastp) and the initial best blast hit obtained with the contig nucleotide sequence (blastx). ORFs below 25 amino acids were not considered. A secretome bioinformatic pipeline previously used to detect secreted proteins in fungal genome sequences (2008, 2010) was employed to define a tentative set of secreted proteins encoded by H. vastatrix transcripts. Briefly, the pipeline is composed of the SignalP v3.0 and the TargetP 1.1 programs (Bendtsen et al., 2004; Emanuelsson et al., 2000) for signal peptide and/or subcellular localization predictions, and the TMHMM v2.0 program (Krogh et al., 2001) to predict transmembrane (TM) helices. Stringent criteria were used to consider a protein as secreted: (i) secretion prediction by TargetP; (ii) all SignalP scores (i.e. Smean, Smax, D_score, hmm_pred) positive; (iii) no TM helix or, in the case of prediction of a single TM, overlap with the signal peptide. Deduced protein sequences and secretion prediction results are detailed in Table S2. Alignments of RTP and HESP rust homologues were performed with the ClustalW2 program available at the European Bioinformatics Institute website (Larkin et al., 2007) (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The identification of GPI anchor sites in sequences of HESP‐379 homologues was performed using the PredGPI prediction server (Pierleoni et al., 2008) (http://gpcr2.biocomp.unibo.it/predgpi/).
Supporting information
Data S1 Complete list of 22 774 CaHv contigs assembled from 454‐GS‐FLEX Titanium reads sequenced from rust‐infected Coffea leaf tissues in FASTA format. The contig sequences are tagged according to their origin (Coffea or Hemileia). Contigs not resolved or not attributed are also included in the list and tagged accordingly. Before each sequence, the FASTA definition line also includes the length of contig (length=) and the number of reads in the contig (numreads=).
Fig. S1 Bioinformatic procedure used to determine the origin of the sequences produced by the 454‐GS‐FLEX Titanium pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv 454) using the difference in the trinucleotide frequencies of plant and fungal sequences with the EST3 program at MIPS (Emmersen et al., 2007; http://mips.helmholtz‐muenchen.de/proj/est3/).
Fig. S2 Bioinformatic procedure used to determine the origin of sequences produced by the 454‐GS‐FLEX Titanium pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv 454) using BLAST searches in selected Coffea spp. and Pucciniales expressed sequence tags (ESTs), nonredundant nucleotide (nr) and Swiss‐Prot databases at the National Center for Biotechnology Information (NCBI) and genomic resources (genome sequences, predicted gene models, deduced proteins). Final CaHv 454 contig prediction is based on both EST3 prediction (detailed in Fig. S1) and BLAST searches.
Fig. S3 Confirmation of fungal origin by polymerase chain reaction (PCR) amplification of fungal and plant cDNA samples for 22 selected contigs predicted to be of fungal origin by the bioinformatic approach. Electrophoretic separation on 3% agarose gels of PCR amplification products after 40 cycles is presented. Gel lanes are as follows: HvA, Hemileia vastatrix appressoria cDNA; iL, infected leaf cDNA; hL, healthy leaf cDNA; N, no‐template control; M, gene ruler low‐range marker (MBI Fermentas; detailed next to Hv00695 gel). The following is the list of genes assessed with their corresponding contigID and expected amplicon size: Hv00162 (contigID_02863, 178 bp); Hv00571 (contigID_15588, 192 bp); Hv00710 (contigID_14082, 209 bp); Hv00712 (contigID_06198, 162 bp); HESP379 (contigID_00177, 197 bp); HESP‐C49 (contigID_07838, 176 bp); AAT1_2 (contigID_15333, 220 bp); AAT3 (contigID_01489, 246 bp); MAD (contigID_01480, 94 bp); RTP (contigID_05622, 75 bp); Inv (contigID_17450, 76 bp); Hv00628 (contigID_02411, 169 bp); Gp‐alpha (contigID_00973, 146 bp); TEF (contigID_00300, 128 bp); CytB (contigID_19719, 120 bp); beta‐Tub (contigID_06023, 196 bp); Hv00695 (contigID_12408, 215 bp); Asp_AT (contigID_05141, 72 bp); CD1 (contigID_14071, 350 bp); CD2 (contigID_07966, 160 bp); MAPK (contigID_11618, 204 bp); Hv00147 (contigID_04496, 222 bp).
Fig. S4 Relationships between the number of reads in 454‐contigs and the average Cq value measured at 21 days post‐inoculation for 22 genes assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) in this study and three reference fungal genes. Lower Cq values indicate higher transcript levels.
Table S1 Complete list of homologies found for the 22 774 454‐contigs of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv contigs) against the various databases tested in this study. Columns A–C detail the contig_ID, length of contig and number of reads in the contig; columns D–I detail the results of the distinct predictions carried out (F, fungus; P, plant; nd, not determined); columns J–L detail the number of P and F predictions and the final prediction made for contig origin (F, likely F and dubious F for fungal sequences and P, likely P and dubious P for plant sequences); columns N–T detail the results of the different EST3 predictions; columns V–AE detail the BLAST hits obtained against selected Coffea spp. and Pucciniales expressed sequence tags (ESTs); columns AG–CZ detail the BLAST hits obtained against three selected dicots and three selected basidiomycete genomes (BLASTN), predicted gene models (BLASTN) and deduced proteins (BLASTX); columns DB–DN detail the BLAST hits obtained against the nonredundant (nr) and Swiss‐Prot databases (National Center for Biotechnology Information, NCBI). Lines 2–6764 correspond to contigs attributed to H. vastatrix; lines 6767–20 717 correspond to contigs attributed to C. arabica; lines 20 720–22 077 correspond to contigs not resolved; lines 22 078–22 779 correspond to contigs not attributed. The different categories of contigs are sorted by read number.
Table S2 Prediction of in planta‐expressed secreted proteins in Hemileia vastatrix 454‐contigs (Hv contigs). Protein sequences were predicted from Hv contigs using framefinder (columns D–I), and secretion prediction (column J) was based on predictions of secretion and transmembrane domains with SignalP, TargetP and TMHMM (columns K–AB). Columns AC–AS detail the best hits against euKaryotic Orthologous Group (KOG) categories (RPSBLAST against KOG database), Melampsora larici‐populina secreted proteins [BLASTX against predicted proteins, Joint Genome Institute genome website], nonredundant and Swiss‐Prot protein databases (BLASTP; National Center for Biotechnology Information) with corresponding accession numbers, E‐values and scores. Hv contigs in the table are sorted by secretion prediction (column J) and by contig read number (column C).
Table S3 Hemileia vastatrix 454‐contig homologies with Pucciniales expressed sequence tags (ESTs) and the Pathogen–Host Interaction database (PHI‐base). TBLASTX searches were performed against nucleotide sequences of PHI‐base (v3.2) and ESTs of the following Pucciniales species retrieved from dbEST at the National Center for Biotechnology Information: Cronartium quercuum f.sp. fusiforme, Melampsora larici‐populina, Melampsora lini, Phakopsora pachyrhizi, Puccinia triticina, Puccinia striiformis, Puccinia graminis, Puccinia coronata var. lolii, Uromyces appendiculatus, Uromyces fabae.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We would like to thank our colleague, Francis Martin, for fruitful discussions, support and access to the bioinformatic facilities of the EcoGenomic team at INRA Nancy supported by the IFR110 Génomique, Ecophysiologie et Ecologie Fonctionnelles. Ana Cabral, Mariana Mota (Instituto Superior de Agronomia, Portugal), Paula Leandro and Johannes Thuerich (CIFC/IICT) are acknowledged for their technical support in RT‐qPCR experiments. Sequencing was supported by IRD (Action Incitative 2009 DRV) and performed at Genoscope within the framework of a CEA/Genoscope‐INRA‐IRD collaborative project (http://www.genoscope.cns.fr/spip/Identification‐of‐virulence.html). This work was partly supported by an INRA innovative research grant (EFPA, 2009) attributed to Sébastien Duplessis, and by Fundação para a Ciência e a Tecnologia (FCT; Portugal) project PTDC/AGR‐AAM/71866/2006. Emilie Tisserant was supported by a Doctoral Fellowship from the European project EnergyPoplar and the Région Lorraine. Andreia Loureiro was supported by FCT grant SFRH/BPD/47008/2008. The bioinformatic resources used in the study were partly supported by the LORIA MBI project (Région Lorraine). This work was undertaken through a French–Portuguese collaborative project (Partenariat Hubert Curien PHC‐Pessoa 22583XM) funded by the Ministère des Affaires Etrangères et Européennes of France and FCT of Portugal.
REFERENCES
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashihara, H. , Sano, H. and Crozier, A. (2008) Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry, 69, 841–856. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Berkeley, M.J. (1869) Note. Gard. Chron. 45, 1157. [Google Scholar]
- Bettencourt, A.J. and Rodrigues, C.J. Jr (1988) Principles and practice of coffee breeding for resistance to rust and other disease In: Coffee, Volume 4, Agronomy (Clarke R.J. and Macrae R., eds), pp. 199–234. London: Elsevier Applied Science. [Google Scholar]
- Bowen, J.K. , Mesarich, C.H. , Rees‐George, J. , Cui, W. , Fitzgerald, A. , Win, J. , Plummer, K.M. and Templeton, M.D. (2009) Candidate effector gene identification in the ascomycete fungal phytopathogen Venturia inaequalis by expressed sequence tag analysis. Mol. Plant Pathol. 10, 431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.‐M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially‐expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Park, J. , Kim, D. , Jung, K. , Kang, S. and Lee, Y.H. (2010) Fungal Secretome Database: integrated platform for annotation of fungal secretomes. BMC Genomics, 11, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho, T.A. , Rijkenberg, F.H.J. and van Asch, M.A.J. (1993) Development of infection structures by Hemileia vastatrix in resistant and susceptible selections of Coffea and in Phaseolus vulgaris . Can. J. Bot. 71, 1001–1008. [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Major, I. , Martin, F. and Seguin, A. (2009) Poplar and pathogen interactions: insights from genome‐wide analyses of resistance and defense gene families and expression profiling of poplar leaves upon infection with rust. Crit. Rev. Plant Sci. 28, 309–334. [Google Scholar]
- Ekramoddoullah, A.K.M. , Tan, Y. , Taylor, D.W. , Yu, X. and Misra, S. (1999) Identification of a protein secreted by the blister rust fungus Cronartium ribicola in infected white pines, cloning and characterization of its cDNA. Can. J. Bot. 77, 800–808. [Google Scholar]
- Emanuelsson, O. , Nielsen, H. , Brunak, S. and von Heijne, G. (2000) Predicting subcellular localization of proteins based on their N‐terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emmersen, J. , Rudd, S. , Mewes, H.‐W. and Tetko, I.V. (2007) Separation of sequences from host–pathogen interface using triplet nucleotide frequencies. Fungal Genet. Biol. 44, 231–241. [DOI] [PubMed] [Google Scholar]
- Feau, N. , Bergeron, M.‐J. , Joly, D.L. , Roussel, F. and Hamelin, R.C. (2007) Detection and validation of EST‐derived SNPs for poplar leaf rust Melampsora medusae f. sp. deltoidae . Mol. Ecol. Notes, 7, 1222–1228. [Google Scholar]
- Fernandez, D. , Santos, P. , Agostini, C. , Bon, M.C. , Petitot, A.S. , Silva, M.C. , Guerra‐Guimarães, L. , Ribeiro, A. , Argout, X. and Nicole, M. (2004) Coffee (Coffea arabica L.) genes early expressed during infection by the rust fungus (Hemileia vastatrix). Mol. Plant Pathol. 5, 527–536. [DOI] [PubMed] [Google Scholar]
- Ganesh, D. , Petitot, A.S. , Silva, M.C. , Alary, R. , Lecouls, A.C. and Fernandez, D. (2006) Monitoring of the early molecular resistance responses of coffee (Coffea arabica L) to the rust fungus (Hemileia vastatrix) using real‐time quantitative RT‐PCR. Plant Sci. 170, 1045–1051. [Google Scholar]
- Gentles, A.J. and Karlin, S. (2001) Genome‐scale compositional comparisons in Eukaryotes. Genome Res. 11, 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard, S. , Delaruelle, C. , Legue, V. , Tisserant, E. , Kohler, A. , Frey, P. , Martin, F. and Duplessis, S. (2010) Laser capture microdissection of uredinia formed by Melampsora larici‐populina revealed a transcriptional switch between biotrophy and sporulation. Mol. Plant–Microbe Interact. 23, 1275–1286. [DOI] [PubMed] [Google Scholar]
- Hahn, M. and Mendgen, K. (1997) Characterization of in planta‐induced rust genes isolated from a haustorium‐specific cDNA library. Mol. Plant–Microbe Interact. 10, 427–437. [DOI] [PubMed] [Google Scholar]
- Hahn, M. , Neef, U. , Struck, C. , Göttfert, M. and Mendgen, K. (1997) A putative amino acid transporter is specifically expressed in haustoria of the rust fungus Uromyces fabae . Mol. Plant–Microbe Interact. 10, 438–445. [DOI] [PubMed] [Google Scholar]
- Hu, G.G. , Linning, R. , Mccallum, B. , Banks, T. , Cloutier, S. , Butterfield, Y. , Liu, J. , Kirkpatrick, R. , Stott, J. , Yang, G. , Smailus, D. , Jones, S. , Marra, M. , Schein, J. and Bakkeren, G. (2007) Generation of a wheat leaf rust, Puccinia triticina, EST database from stage‐specific cDNA libraries. Mol. Plant Pathol. 8, 451–467. [DOI] [PubMed] [Google Scholar]
- Jakupovic, M. , Heintz, M. , Reichmann, P. , Mendgen, K. and Hahn, M. (2005) Microarray analysis of expressed sequence tags from haustoria of the rust fungus Uromyces fabae . Fungal Genet. Biol. 43, 8–19. [DOI] [PubMed] [Google Scholar]
- Joly, D.L. , Feau, N. , Tanguay, P. and Hamelin, R.C. (2010) Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.). BMC Genomics, 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant–Microbe Interact. 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , von Heijne, G. and Sonnhammer, E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kulkarni, R.D. , Kelkar, H.S. and Dean, R.A. (2003) An eight‐cysteine‐containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lashermes, P. , Combes, M.C. , Robert, J. , Trouslot, P. , D'hont, A. , Anthony, F. and Charrier, A. (1999) Molecular characterisation and origin of the Coffea arabica L. genome. Mol. Gen. Genet. 261, 259–266. [DOI] [PubMed] [Google Scholar]
- Lebrun, M.‐H. and Kamoun, S. (2010) Effectors, effectors et toujours des effectors. New Phytol. 186, 290–292. [DOI] [PubMed] [Google Scholar]
- Li, L. , Wright, S.J. , Krystofova, S. , Park, G. and Borkovich, K.A. (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61, 423–452. [DOI] [PubMed] [Google Scholar]
- Link, T.I. and Voegele, R.T. (2008) Secreted proteins of Uromyces fabae: similarities and stage specificity. Mol. Plant Pathol. 9, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesh, V. , Million‐Rousseau, R. , Ullmann, P. , Chabrillange, N. , Bustamante, J. , Mondolot, L. , Morant, M. , Noirot, M. , Hamon, S. , de Kochko, A. , Werck‐Reichhart, D. and Campa, C. (2007) Functional characterization of two p‐coumaroyl ester 3′‐hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol. Biol. 64, 145–159. [DOI] [PubMed] [Google Scholar]
- Major, I. , Nicole, M.‐C. , Duplessis, S. and Séguin, A. (2010) Photosynthetic and respiratory changes in leaves of poplar elicited by rust infection. Photosynth. Res. 104, 41–48. [DOI] [PubMed] [Google Scholar]
- Martin, F. , Aerts, A. , Ahrén, D. , Brun, A. , Danchin, E.G. , Duchaussoy, F. , Gibon, J. , Kohler, A. , Lindquis, t.E. , Pereda, V. , Salamov, A. , Shapiro, H.J. , Wuyts, J. , Blaudez, D. , Buée, M. , Brokstein, P. , Canbäck, B. , Cohen, D. , Courty, P.E. , Coutinho, P.M. , Delaruelle, C. , Detter, J.C. , Deveau, A. , DiFazio, S. , Duplessis, S. , Fraissinet‐Tachet, L. , Lucic, E. , Frey‐Klett, P. , Fourrey, C. , Feussner, I. , Gay, G. , Grimwood, J. , Hoegger, P.J. , Jain, P. , Kilaru, S. , Labbé, J. , Lin, Y.C. , Legué, V. , Le Tacon, F. , Marmeisse, R. , Melayah, D. , Montanini, B. , Muratet, M. , Nehls, U. , Niculita‐Hirzel, H. , Oudot‐Le Secq, M.P. , Peter, M. , Quesneville, H. , Rajashekar, B. , Reich, M. , Rouhier, N. , Schmutz, J. , Yin, T. , Chalot, M. , Henrissat, B. , Kües, U. , Lucas, S. , Van de Peer, Y. , Podila, G.K. , Polle, A. , Pukkila, P.J. , Richardson, P.M. , Rouzé, P. , Sanders, I.R. , Stajich, J.E. , Tunlid, A. , Tuskan, G. and Grigoriev, I.V. (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature, 452, 88–92. [DOI] [PubMed] [Google Scholar]
- Martin, F. , Kohler, A. , Murat, C. , Balestrini, R. , Coutinho, P.M. , Jaillon, O. , Montanini, B. , Morin, E. , Noel, B. , Perducani, R. , Porcel, B. , Rubini, A. , Amicucci, A. , Amselem, J. , Anthouard, V. , Arcioni, S. , Artiguenave, F. , Aury, J.M. , Ballario, P. , Bolchi, A. , Brenna, A. , Brun, A. , Buée, M. , Cantarel, B. , Chevalier, G. , Couloux, A. , Da Silva, C. , Denoeud, F. , Duplessis, S. , Ghignone, S. , Hilselberger, B. , Iotti, M. , Marçais, B. , Mello, A. , Miranda, M. , Pacioni, G. , Quesneville, H. , Riccioni, C. , Ruotolo, R. , Splivallo, R. , Stocchi, V. , Tisserant, E. , Viscomi, A.R. , Zambonelli, A. , Zampieri, E. , Henrissat, B. , Lebrun, M.‐H. , Paolocci, F. , Bonfante, P. , Ottonello, S. and Wincker, P. (2010) Périgord Black Truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature, 464, 1033–1038. [DOI] [PubMed] [Google Scholar]
- Martins, E.M.F. and Moraes, W.B.C. (1996) Development of Hemileia vastatrix in coffee plants with genetic and induced resistance. J. Phytopathol. 144, 519–526. [Google Scholar]
- Petitot, A.‐S. , Lecouls, A.‐C. and Fernandez, D. (2008) Sub‐genomic origin and regulation patterns of a duplicated WRKY gene in the allotetraploid species Coffea arabica . Tree Genet. Genomes, 4, 379–390. [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni, A. , Martelli, P.L. and Casadio, R. (2008) PredGPI: a GPI‐anchor predictor. BMC Bioinformatics, 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthoff, D.P. , Neelam, A. , Ehrenfried, M.L. , Scheffler, B.E. , Ballard, L.L. , Campbell, K.B. , Cooper, B. and Tucker, M.L. (2008) Analysis of expressed sequence tags from Uromyces appendiculatus hyphae and haustoria and their comparison to sequences from other rust fungi. Phytopathology, 98, 1126–1135. [DOI] [PubMed] [Google Scholar]
- Rafiqi, M. , Gan, P.H.P. , Ravensdale, M. , Lawrence, G.J. , Ellis, J.G. , Jones, D.A. , Hardham, A.R. and Dodds, P.N. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro, D.A. , Escoute, J. , Petitot, A.S. , Nicole, M. , Maluf, M.P. and Fernandez, D. (2009) Biphasic haustorial differentiation of coffee rust (Hemileia vastatrix race II) associated with differential defence responses in resistant and susceptible coffee cultivars. Plant Pathol. 58, 944–955. [Google Scholar]
- Ramiro, D. , Jalloul, A. , Petitot, A.‐S. , Grossi‐de‐Sá, M.F. , Maluf, M. and Fernandez, D. (2010) Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genet. Genomes, 6, 767–781. [Google Scholar]
- Rodrigues C.J., Jr , Bettencourt, A.J. and Rijo, L. (1975) Races of the pathogen and resistance to coffee rust. Annu. Rev. Phytopathol. 13, 49–70. [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , Oliva, R. , Van Damme, M. , Schwizer, S. , Raffaele, S. , Chaparro‐Garcia, A. , Farrer, R. , Segretin, M.E. , Bos, J. , Haas, B.J. , Zody, M.C. , Nusbaum, C. , Win, J. , Thines, M. and Kamoun, S. (2009) Ten things to know about oomycete effectors. Mol. Plant Pathol. 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, M.C. , Nicole, M. , Rijo, L. , Geiger, J.‐P. and Rodrigues, C.J. Jr (1999) Cytochemical aspects of the plant–rust fungus interface during the compatible interaction Coffea arabica (cv. Caturra)–Hemileia vastatrix (race III). Int. J. Plant Sci. 160, 79–91. [Google Scholar]
- Silva, M.C. , Nicole, M. , Guerra‐Guimarães, L. and Rodrigues, C.J. Jr (2002) Hypersensitive cell death and post‐haustorial defence responses arrest the orange rust (Hemileia vastatrix) growth in resistant coffee leaves. Physiol. Mol. Plant Pathol. 60, 169–183. [Google Scholar]
- Silva, M.C. , Várzea, V. , Guerra‐Guimarães, L. , Azinheira, H.G. , Fernandez, D. , Petitot, A.‐S. , Bertrand, B. , Lashermes, P. and Nicole, M. (2006) Coffee resistance to the main diseases: leaf rust and coffee berry disease (CBD). Braz. J. Plant Physiol. 18, 119–147. [Google Scholar]
- Silva, M.C. , Guerra‐Guimarães, L. , Loureiro, A. and Nicole, M. (2008) Involvement of peroxidases in the hypersensitive reaction of coffee plants to orange rust (Hemileia vastatrix). Physiol. Mol. Plant Pathol. 72, 29–38. [Google Scholar]
- Soanes, D.M. , Alam, I. , Cornell, M. , Wong, H.M. , Hedeler, C. , Paton, N.W. , Rattray, M. , Hubbard, S.J. , Oliver, S.G. and Talbot, N.J. (2008) Comparative genomic analysis of plant pathogenic and saprophytic fungi reveals gene families that are potentially important for fungal pathogenesis. PLoS ONE, 3, e2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Struck, S. , Ernst, M. and Hahn, M. (2002) Characterization of a developmentally regulated amino acid transporter (AAT1p) of the rust fungus Uromyces fabae . Mol. Plant Pathol. 3, 23–30. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, A. , Li, S. , Scheffler, B.E. and Matthews, B.F. (2009) Laser capture microdissection (LCM) and expressed sequence tag analysis of uredinia formed by Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Physiol. Mol. Plant Pathol. 73, 163–174. [Google Scholar]
- Valette‐Collet, O. , Cimerman, A. , Reignault, P. , Levis, C. and Boccara, M. (2003) Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant–Microbe Interact. 16, 360–367. [DOI] [PubMed] [Google Scholar]
- Várzea, V.M. and Marques, D.V. (2005) Population variability of Hemileia vastatrix versus coffee durable resistance. In: Durable Resistance to Coffee Leaf Rust (Zambolim L., Zambolim E.M. and Várzea V.M.P., eds), pp. 285–304. Viçosa: Universidade Federal de Viçosa. [Google Scholar]
- Vega‐Arreguín, J.C. , Ibarra‐Laclette, E. , Jiménez‐Moraila, B. , Martínez, O. , Vielle‐Calzada, J.P. , Herrera‐Estrella, L. and Herrera‐Estrella, A. (2009) Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics, 10, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Struck, C. , Hahn, M. and Mendgen, K. (2001) The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae . Proc. Natl. Acad. Sci. USA, 98, 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Hahn, M. , Lohaus, G. , Link, T. , Heiser, I. and Mendgen, K. (2005) Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae . Plant Physiol. 137, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Hahn, M. and Mendgen, K. (2009) The uredinales: cytology, biochemistry, and molecular biology In: The Mycota, 5. Plant Relationships Volume (Deising H.B., ed.), pp. 69–98. Berlin: Springer. [Google Scholar]
- Winnenburg, R. , Urban, M. , Beacham, A. , Baldwin, T.K. , Holland, S. , Lindeberg, M. , Hansen, H. , Rawlings, C. , Hammond‐Kosack, K.E. and Kohler, J. (2008) PHI‐base update: additions to the pathogen host interaction database. Nucleic Acids Res. 36, D572–D576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, C. , Park, G. , Choi, W. , Zheng, L. , Dean, R.A. and Xu, J.R. (2002) Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell, 14, 2107–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, C.T. , Chen, X.M. , Wang, X.J. , Han, Q.M. , Kang, Z.S. and Hulbert, S. (2009) Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . BMC Genomics, 10, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Complete list of 22 774 CaHv contigs assembled from 454‐GS‐FLEX Titanium reads sequenced from rust‐infected Coffea leaf tissues in FASTA format. The contig sequences are tagged according to their origin (Coffea or Hemileia). Contigs not resolved or not attributed are also included in the list and tagged accordingly. Before each sequence, the FASTA definition line also includes the length of contig (length=) and the number of reads in the contig (numreads=).
Fig. S1 Bioinformatic procedure used to determine the origin of the sequences produced by the 454‐GS‐FLEX Titanium pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv 454) using the difference in the trinucleotide frequencies of plant and fungal sequences with the EST3 program at MIPS (Emmersen et al., 2007; http://mips.helmholtz‐muenchen.de/proj/est3/).
Fig. S2 Bioinformatic procedure used to determine the origin of sequences produced by the 454‐GS‐FLEX Titanium pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv 454) using BLAST searches in selected Coffea spp. and Pucciniales expressed sequence tags (ESTs), nonredundant nucleotide (nr) and Swiss‐Prot databases at the National Center for Biotechnology Information (NCBI) and genomic resources (genome sequences, predicted gene models, deduced proteins). Final CaHv 454 contig prediction is based on both EST3 prediction (detailed in Fig. S1) and BLAST searches.
Fig. S3 Confirmation of fungal origin by polymerase chain reaction (PCR) amplification of fungal and plant cDNA samples for 22 selected contigs predicted to be of fungal origin by the bioinformatic approach. Electrophoretic separation on 3% agarose gels of PCR amplification products after 40 cycles is presented. Gel lanes are as follows: HvA, Hemileia vastatrix appressoria cDNA; iL, infected leaf cDNA; hL, healthy leaf cDNA; N, no‐template control; M, gene ruler low‐range marker (MBI Fermentas; detailed next to Hv00695 gel). The following is the list of genes assessed with their corresponding contigID and expected amplicon size: Hv00162 (contigID_02863, 178 bp); Hv00571 (contigID_15588, 192 bp); Hv00710 (contigID_14082, 209 bp); Hv00712 (contigID_06198, 162 bp); HESP379 (contigID_00177, 197 bp); HESP‐C49 (contigID_07838, 176 bp); AAT1_2 (contigID_15333, 220 bp); AAT3 (contigID_01489, 246 bp); MAD (contigID_01480, 94 bp); RTP (contigID_05622, 75 bp); Inv (contigID_17450, 76 bp); Hv00628 (contigID_02411, 169 bp); Gp‐alpha (contigID_00973, 146 bp); TEF (contigID_00300, 128 bp); CytB (contigID_19719, 120 bp); beta‐Tub (contigID_06023, 196 bp); Hv00695 (contigID_12408, 215 bp); Asp_AT (contigID_05141, 72 bp); CD1 (contigID_14071, 350 bp); CD2 (contigID_07966, 160 bp); MAPK (contigID_11618, 204 bp); Hv00147 (contigID_04496, 222 bp).
Fig. S4 Relationships between the number of reads in 454‐contigs and the average Cq value measured at 21 days post‐inoculation for 22 genes assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) in this study and three reference fungal genes. Lower Cq values indicate higher transcript levels.
Table S1 Complete list of homologies found for the 22 774 454‐contigs of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix (CaHv contigs) against the various databases tested in this study. Columns A–C detail the contig_ID, length of contig and number of reads in the contig; columns D–I detail the results of the distinct predictions carried out (F, fungus; P, plant; nd, not determined); columns J–L detail the number of P and F predictions and the final prediction made for contig origin (F, likely F and dubious F for fungal sequences and P, likely P and dubious P for plant sequences); columns N–T detail the results of the different EST3 predictions; columns V–AE detail the BLAST hits obtained against selected Coffea spp. and Pucciniales expressed sequence tags (ESTs); columns AG–CZ detail the BLAST hits obtained against three selected dicots and three selected basidiomycete genomes (BLASTN), predicted gene models (BLASTN) and deduced proteins (BLASTX); columns DB–DN detail the BLAST hits obtained against the nonredundant (nr) and Swiss‐Prot databases (National Center for Biotechnology Information, NCBI). Lines 2–6764 correspond to contigs attributed to H. vastatrix; lines 6767–20 717 correspond to contigs attributed to C. arabica; lines 20 720–22 077 correspond to contigs not resolved; lines 22 078–22 779 correspond to contigs not attributed. The different categories of contigs are sorted by read number.
Table S2 Prediction of in planta‐expressed secreted proteins in Hemileia vastatrix 454‐contigs (Hv contigs). Protein sequences were predicted from Hv contigs using framefinder (columns D–I), and secretion prediction (column J) was based on predictions of secretion and transmembrane domains with SignalP, TargetP and TMHMM (columns K–AB). Columns AC–AS detail the best hits against euKaryotic Orthologous Group (KOG) categories (RPSBLAST against KOG database), Melampsora larici‐populina secreted proteins [BLASTX against predicted proteins, Joint Genome Institute genome website], nonredundant and Swiss‐Prot protein databases (BLASTP; National Center for Biotechnology Information) with corresponding accession numbers, E‐values and scores. Hv contigs in the table are sorted by secretion prediction (column J) and by contig read number (column C).
Table S3 Hemileia vastatrix 454‐contig homologies with Pucciniales expressed sequence tags (ESTs) and the Pathogen–Host Interaction database (PHI‐base). TBLASTX searches were performed against nucleotide sequences of PHI‐base (v3.2) and ESTs of the following Pucciniales species retrieved from dbEST at the National Center for Biotechnology Information: Cronartium quercuum f.sp. fusiforme, Melampsora larici‐populina, Melampsora lini, Phakopsora pachyrhizi, Puccinia triticina, Puccinia striiformis, Puccinia graminis, Puccinia coronata var. lolii, Uromyces appendiculatus, Uromyces fabae.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


