SUMMARY
A recombinase‐based in vivo expression technology (RIVET) approach with Xanthomonas campestris pv. vesicatoria (Xcv) revealed that lipA, annotated as putative secreted lipase, is expressed during the interaction between this pathogen and tomato. Here, the tnpR and uidA reporter genes were used to show that lipA is strongly induced in XVM2 minimal medium and during the early stages of tomato infection by Xcv. A mutant strain impaired in lipA was generated by insertional mutagenesis. This mutant grew in a similar manner to the wild‐type in rich medium, but its growth was significantly compromised in a medium containing olive oil as a single carbon source. The lipolytic activity of the extracellular fraction of the lipA mutant was reduced significantly relative to that of the wild‐type strain, thus confirming that lipA indeed encodes a functional secreted enzyme with lipolytic activity. A plasmid carrying a wild‐type copy of lipA complemented the lipA mutant for extracellular lipolytic activity. Dip inoculation experiments with tomato lines Hawaii 7998 (H7998) and Micro Tom showed that the lipA mutant grew to a lesser extent than the wild‐type in tomato leaves. Following leaf syringe infiltrations, the mutant strain induced disease symptoms that were less severe than those induced by the wild‐type strain, supporting a significant role of lipA in the pathogenicity of Xcv.
INTRODUCTION
The Gram‐negative Gammaproteobacterium Xanthomonas campestris pv. vesicatoria (Xcv) is the causal agent of bacterial spot disease of tomato and pepper. Xcv is able to infect all above‐ground parts of the host plants (leaves, stems, fruits and flowers), with typical symptoms including necrotic lesions that are often surrounded by chlorotic haloes. High temperature and relative humidity make for optimal conditions for the breakout of this world‐wide disease (Jones and Stall, 1998). It has been proposed that Xcv should be reclassified into four different species: X. vesicatoria, X. euvesicatoria, X. gardneri and X. perforans (Jones et al., 2004). Nevertheless, in reports focusing on the basic aspects of the pathogenicity and pathogen–host interactions, the new nomenclature has not been widely used by the scientific community; therefore, in this article, we use the classical nomenclature (namely Xcv).
Xcv utilizes a conserved type III secretion system (T3SS), encoded by the so‐called hrp genes (for hypersensitivity response and pathogenicity), to secrete protein effectors into the host cells. These effectors interact with the host cellular metabolism to promote disease or to elicit a defence response on susceptible or resistant plants, respectively (Alfano and Collmer, 2004; Mudgett, 2005). During the last two decades, significant efforts have been made to understand the role played by the Hrp‐T3SS and its secreted effectors in the pathogenicity of Xcv and other Gram‐negative plant pathogenic bacteria. However, the ability to infect a plant expands beyond the ability of these pathogens to form a T3SS and to transfer effectors through it. The successful establishment of pathogenic bacteria in the host tissue requires the coordinated activity of a plethora of genes, many of which are still unknown.
In a recent study, we reported a recombinase‐based in vivo expression technology (RIVET) approach that identified several genes from Xcv strain 97‐2 that are specifically induced or overexpressed during its interaction with tomato (Tamir‐Ariel et al., 2007). The rationale behind this approach was that these in vivo‐induced (ivi) genes have the potential to contribute to the ability of the pathogen to establish in the host and to promote disease. RIVET allowed us to identify 61 unique Xcv ivi genes, several of which were mutated to further assess their contribution to Xcv virulence (2007, 2011).
In the present study, we focus on an additional ivi gene: lipA. In the Xcv 85‐10 genome (Thieme et al., 2005), lipA (Xcv0536) is annotated as a putative secreted lipase, and encodes a protein similar to lipases (EC 3.1.1.3) and esterases (EC 3.1.1.1). These hydrolases, collectively known as ‘lipolytic enzymes’, are characterized by their ability to catalyse the hydrolysis of ester bonds from diverse substrates. While esterases preferentially break ester bonds of shorter chain fatty acids, lipases display a much broader substrate range than esterases. However, distinguishing between lipases and esterases is not a simple task as they are similar in both sequence and functional properties (Fojan et al., 2000).
Lipases/esterases are ubiquitous enzymes found in animals, plants, fungi and bacteria (Borgstrom and Brockman, 1984; Jaeger et al., 1994; Mukherjee and Hills, 1994). The role of secreted lipases/esterases in the virulence of animal and plant pathogens has been demonstrated (1995, 1998; Gacser et al., 2007; Jaeger et al., 1994; Schaller et al., 2005; Stehr et al., 2004; Voigt et al., 2005), although few studies have been performed with plant pathogenic fungi and bacteria. Importantly, the orthologue lipA gene from Xanthomonas oryzae pv. oryzae (Xoo) has been suggested to act as a cell wall‐degrading enzyme, and has been shown to contribute to Xoo virulence and induce innate defence responses in rice (Jha et al., 2007; Rajeshwari et al., 2005). Further characterization of the Xoo LipA suggested that this protein possesses esterase activity (Aparna et al., 2009). In the present study, we report that, in Xcv, lipA also encodes a secreted enzyme with lipolytic activity, and that this enzyme is required for wild‐type levels of virulence of Xcv on tomato. Importantly, using reporter fusions with the lipA promoter, we demonstrate that lipA is expressed in planta from the early stages of infection of tomato leaves by Xcv. Moreover, we show that the expression of lipA in planta is comparable to or, at some time points, even higher than that of hrpA, a gene encoding a T3SS component that is required for Xcv pathogenicity.
RESULTS
Sequence analysis of the Xcv lipA gene
One of the ivi genes identified in the RIVET approach with Xcv strain 97‐2 (Tamir‐Ariel et al., 2007) matched significantly (99% identity for both the nucleotide and amino acid sequences) with gene Xcv0536 (lipA, putative secreted lipase) from Xcv 85‐10 (Thieme et al., 2005; GenBank NC_007508). As in strain 85‐10, the 97‐2 lipA gene has an open reading frame of 1263 bp. Sequence analysis of the Xcv 97‐2 lipA gene and its proximate region [following polymerase chain reaction (PCR) amplification with primer sets beflipA and aftlipA; Table S1, see Supporting Information] showed that lipA is surrounded by the same genes as in strain 85‐10. In addition, lipA appears to be a single gene locus, as the distance between the stop codon of lipA and the putative start codon of the downstream gene Xcv0537 is 272 bp (Fig. 1A). Reverse transcriptase (RT)‐PCR confirmed this hypothesis: cDNA prepared from XVM2‐grown Xcv 97‐2 was PCR amplified with different primer sets corresponding to lipA and Xcv0537 regions. Although PCR products could be obtained using internal primers of both genes, no fragment corresponding to a hypothetical transcript containing both lipA and Xcv0537 could be amplified (Fig. 1B), thus supporting the notion that lipA is indeed part of a single gene locus.
Figure 1.

The genomic region around lipA and lipA expression as a single gene transcript. (A) Genomic location of lipA (Xcv0536) according to the Xanthomonas campestris pv. vesicatoria (Xcv) 85‐10 annotation (GenBank accession AM039952; Thieme et al., 2005). Gene numbers are according to the 85‐10 genome annotation. Sequence analysis revealed a similar genomic structure for this region in strain 97‐2. Xcv0535 and Xcv0537 encode a hypothetical protein and a NUDIX hydrolase family protein, respectively. (B) Polymerase chain reaction (PCR) using cDNA prepared from an overnight XVM2‐grown culture of strain 97‐2 or genomic DNA (gDNA) of this strain as template. Products 1, 2 and 3 were amplified using primer sets 468lipF/905lipR, Xcv0536‐7F/Xcv0536‐7R and Xcv0537F/Xcv0537R (Table S1), respectively. The locations of these primers and products are shown at the top of (A). Although PCR products of the expected sizes were obtained with all primer sets with gDNA as template, cDNA was detected only for products 1 and 3, confirming that lipA is part of a single gene locus.
BlastN revealed that 97‐2 lipA matches significantly (77%–99% identity) several orthologous genes from various Xanthomonas species, mainly annotated as conserved hypothetical proteins. Among these orthologues is the lipA gene from Xoo (86% identity at the amino acid level with LipA from Xoo BXO43; GenBank accession 3H2G_A), which has been shown to contribute to the virulence of this pathogen (Rajeshwari et al., 2005). A high identity was also found between Xcv LipA and the protein products of genes from sequenced Xylella fastidiosa strains (strains Ann‐1, 9a5c, M23, Dixon, Temecula1, GB514 and EB92.1; 66%–70% identity at the amino acid level). In X. fastidiosa M23 and GB514, the matching proteins (GenBank accessions YP_001830473 and ADN62559, respectively) are also annotated as putative secreted lipases.
BlastP revealed that the Xcv lipA product matches several proteins from various bacterial species annotated as secreted lipases/esterases. The predicted 420‐amino‐acid LipA protein possesses a LIP domain (Pfam03583) between positions 132 and 202. The LIP domain is typical of lipases that are expressed and secreted during the infection cycle of several pathogens, with special reference to the fungus Candida albicans, for which the involvement of secreted lipases in pathogenesis has been demonstrated (Schaller et al., 2005; Stehr et al., 2004).
In Gram‐negative bacteria, extracellular lipolytic enzymes, as well as other secreted enzymes, are generally secreted via the type II secretion system (T2SS), with the secreted proteins carrying an N‐terminal signal peptide, which is cleaved on secretion (Rosenau and Jaeger, 2000). The Xoo LipA has also been shown to be secreted via type II secretion (Jha et al., 2007). The analysis of Xcv LipA by SignalP 3.0 (Emanuelsson et al., 2007) predicted the presence of a signal peptide in this protein with a probability of 100%, with a probability of 99.8% of cleavage between positions 29 and 30.
The lipA promoter is activated during growth of Xcv in minimal medium
We have already shown that the lipA gene is expressed in XVM2 (Fig. 1B), a minimal medium that mimics, to some extent, the apoplast environment (Wengelnik et al., 1996). To further characterize the expression of lipA in Xcv 97‐2, we exploited the RIVET technique to monitor the activity of the lipA promoter. We generated a reporter strain carrying a chromosomal tnpR‐uidA transcriptional fusion under the control of the lipA promoter (strain ResPlipA). The transcriptional fusion was inserted into the chromosome of strain XcvRes (strain 97‐2 carrying a cassette composed of markers flanked by res sites, which are the substrates for the tnpR‐encoded recombinase) using plasmid pPlipA (Table 1). The recognition and action of TnpR on the res sites lead to excision of the flanked markers in a reaction called resolution, producing a phenotypic switch (Fig. 2A). In our case, the res cassette (res‐KmR‐sacB‐res) confers resistance to kanamycin (Km) and sensitivity to sucrose (Tamir‐Ariel et al., 2007). Thus, the activity of the lipA promoter can be detected by Km sensitivity. As a control, strain ResPlipArev, carrying the res cassette and the same transcriptional fusion under the control of the lipA promoter, but in the reverse orientation (plasmid pPlipArev; Table 1), was utilized.
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant characteristics* | Reference or source |
|---|---|---|
| Xanthomonas campestris pv. vesicatoria | ||
| 97‐2 | Wild‐type, Xanthomonas perforans according to the new nomenclature; belongs to race T3 | Astua‐Monge et al. (2000) |
| XcvRes | KmR; reporter strain containing the res cassette inserted within hpaG in 97‐2 | Tamir‐Ariel et al. (2007) |
| ResPlipA | KmR, GmR; PlipA (lipA promoter)::tnpR::uidA in XcvRes background | This work |
| ResPlipArev | KmR, GmR; PlipArev (lipA promoter in reverse orientation)::tnpR::uidA in XcvRes | This work |
| ResPhrpA | KmR, CmR; PhrpA (hrpA promoter)::tnpR::uidA in XcvRes | Tamir‐Ariel et al. (2007) |
| lipA mutant | KmR; strain 97‐2 disrupted in the lipA open reading frame (ORF) by insertional mutagenesis with plasmid pJPlipA | This work |
| lipA –/comp | KmR, GmR; lipA mutant carrying plasmid pBBR1‐lipA (complemented strain) | This work |
| lipA –/pBBR1 | KmR, GmR; lipA mutant carrying plasmid pBBR1MCS‐5 | This work |
| wt/pBBR1 | GmR; wild‐type 97‐2 carrying plasmid pBBR1MCS‐5 | This work |
| Escherichia coli | ||
| S17‐1 λ pir | λ lysogenic S17‐1 derivative producing π protein for replication of plasmids carrying oriR6K; recA pro hsdR RP4‐2‐Tc::Mu‐Km::Tn7 λ‐ pir | Simon et al. (1983) |
| Plasmids | ||
| pJP5603 | KmR; R6K‐based suicide vector; requires the pir‐encoded π protein for replication | Penfold and Pemberton (1992) |
| pJPGm | GmR; a pJP5603‐derived vector, in which the KmR gene was replaced by a GmR gene, excised from pML122 (Labes et al., 1990) | This work |
| pJPGmtnpR | GmR; pJPGm carrying a promoterless tnpR::uidA | This work |
| pPlipA | GmR; pJPGmtnpR with the lipA promoter cloned upstream and in the same orientation as tnpR::uidA to create a transcriptional fusion | This work |
| pPlipArev | GmR; pJPGmtnpR with the lipA promoter cloned upstream and in the opposite orientation to tnpR::uidA | This work |
| pJPlipA | KmR; pJP5603 carrying a 614‐bp internal fragment of the lipA ORF; used to generate the lipA mutant by insertional mutagenesis | This work |
| pBBR1MCS‐5 | GmR; RK2 broad‐host‐range cloning vector | Kovach et al. (1995) |
| pBBR1‐lipA | GmR; pBBR1MCS‐5 carrying wild‐type (97‐2) lipA with its native promoter region | This work |
In strains and plasmids, KmR, CmR and GmR represent kanamycin, chloramphenicol and gentamicin resistance, respectively.
Figure 2.
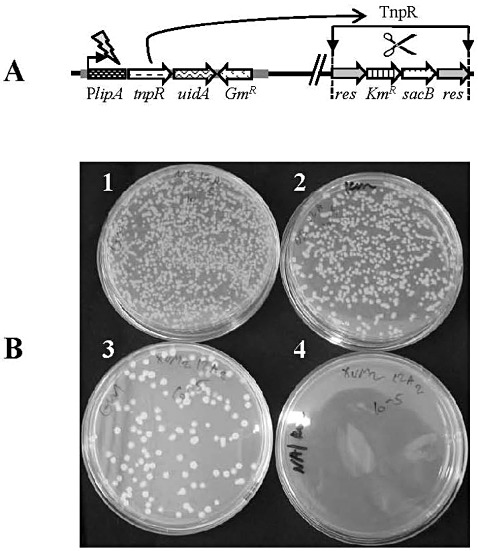
Assessment of TnpR‐mediated resolution in strain ResPlipA during growth in rich (nutrient broth, NB) and minimal (XVM2) media. (A) Diagram illustrating the recombinase‐based in vivo expression technology (RIVET)‐based transcriptional fusion and the resolution event in strain ResPlipA. This strain contains a tnpR‐uidA transcriptional function under the control of the lipA promoter (PlipA) and a res cassette with genes conferring kanamycin resistance (KmR) and sucrose sensitivity (sacB). PlipA activation leads to the expression of tnpR and the consequent excision of the res cassette (resolution event), making the cells susceptible to Km and resistant to sucrose. PlipA activation also leads to the expression of uidA, which can be measured by β‐glucuronidase (GUS) assays. (B) After growth in NB or XVM2 medium, ResPlipA cells were serially diluted and plated onto nutrient agar/gentamicin (NA/Gm) and nutrient agar/gentamicin + kanamycin (Na/Gm + Km) plates. All cells are able to grow in the presence of Gm, but only nonresolved cells are able to grow in the presence of Km. Plates corresponding to 10−5 serial dilutions are shown: 1, NA/Gm plate after growth in NB; 2, NA/Gm + Km plate after growth in NB; 3, NA/Gm plate after growth in XVM2; 4, NA/Gm + Km plate after growth in XVM2. This experiment was performed three times with similar results.
Strains ResPlipA and ResPlipArev were grown in rich (nutrient broth, NB) and XVM2 media. To assess TnpR‐mediated resolution of the res cassette, dilutions of the cultures were plated onto nutrient agar (NA) plates supplemented with Km and/or gentamicin (Gm). Although Km resistance (KmR) is conferred by the res cassette, Gm resistance (GmR) is mediated by the GmR gene from the pPlipA or pPlipArev plasmid. Colonies of strain ResPlipArev proceeding from both XVM2 and NB media grew to similar numbers on NA/Gm and NA/Gm + Km plates, indicating that no resolution occurred in this strain in either rich or minimal medium (not shown). In contrast with this strain, after growth in XVM2, colonies of ResPlipA could be detected after plating dilutions on NA/Gm, but not on NA/Gm + Km (Fig. 2B). After growth in NB medium, colonies of strain ResPlipA were formed on both NA/Gm and NA/Gm + Km (Fig. 2B). Consistently, slightly greater numbers of colonies (10%–15% more in different experiments) developed on NA/Gm than on NA/Gm + Km plates, suggesting a low activation of the lipA promoter, or residual resolution of the res cassette in rich medium.
The above results were confirmed by qualitative β‐glucuronidase (GUS) (uidA) assays on colonies of both strains grown on solid XVM2 and NA. In these assays, only XVM2‐grown ResPlipA colonies were GUS positive (stained blue) (not shown). Together, these results showed that significant promoter activation occurred only in the ResPlipA strain grown in XVM2 medium, thus demonstrating that the lipA promoter is activated in minimal but not, or to a very low extent, in rich medium.
The lipA gene is expressed during Xcv growth in planta
We further characterized the expression of lipA in planta. In these experiments, strain ResPhrpA was used as an additional control. This strain contains the tnpR‐uidA transcriptional fusion under the control of the hrpA promoter, which is known to be expressed in planta (Wengelnik et al., 1996). Tomato plants were inoculated by dip inoculation with bacterial concentrations of 105 colony‐forming units (cfu)/mL. Bacteria were extracted from leaves at several times after inoculation and extract dilutions were plated onto NA/Gm + Km and NA/Gm to assess the percentage of tnpR‐mediated resolution. Figure S1 (Supporting Information) shows that the level of in planta resolution of the ResPlipA strain was similar to that of strain ResPhrpA. No significant differences (P= 0.05) were observed between these strains at any time after inoculation, despite a consistent trend of higher resolution of the ResPlipA strain at the earliest assessed time: 2 days after inoculation (dai).
In the different experiments (two with similar results), 60%–70% of the ResPlipA cells were resolved at 2 dai and, after 4–5 days, complete resolution was achieved (similar to the ResPhrpA strain). In contrast, the control strain ResPlipArev showed very low levels of resolution, which were significantly (P= 0.05) lower than those of the other two strains at all tested times (Fig. S1). The residual resolution of this strain, similar to the residual resolution observed for the ResPlipA strain during growth in rich medium, could be explained by the high sensitivity of the resolution assay (Angelichio and Camilli, 2002; Rediers et al., 2005).
The resolution assay is very sensitive and provides valuable information about the relative amounts of cells in which the promoter being studied is activated. However, this assay cannot provide clues about the intensity of promoter activity, as weakly and strongly activated promoters lead to similar resolution events. To quantify the expression of the lipA gene in planta, we performed quantitative GUS assays of leaf extracts at several times after inoculation. These experiments confirmed that lipA is expressed in planta (Fig. 3). Moreover, lipA expression was shown to occur at least 24 h after inoculation (hai). During a period of 24–96 hai, a consistent trend was observed, indicating that lipA expression is higher than the expression of hrpA, although these differences were not always significant (Fig. 3). GUS expression driven by the lipA promoter in the reverse orientation (strain ResPlipArev) was clearly lower than expression under the lipA and hrpA promoters, and differences between ResPlipA and ResPlipArev were significant (P= 0.05) in most cases. These results are in agreement with the findings from resolution assays and support the notion that the low levels of resolution observed for the ResPlipArev strain were probably a result of a very low residual expression of tnpR.
Figure 3.

β‐Glucuronidase (GUS) assays for the determination of the quantitative evaluation of in planta promoter activity of strains ResPlipA, ResPlipArev and ResPhrpA. Tomato plants were inoculated by the dip method with inoculum concentrations of 108 colony‐forming units (cfu)/mL (A) or 106 cfu/mL (B). GUS activity in leaf extracts was determined 24, 48 and 72 h after inoculation (hai) in (A), and 48, 72 and 96 hai in (B), using the fluorometric substrate 4‐methylumbelliferyl‐β‐d‐glucuronide (MUG). Data represent averages and standard deviations (SDs) of GUS activity per cfu (three replicates per treatment per time point, each replicate being composed of five leaf discs) of one experiment of two with similar results. Different letters in each graph indicate significant differences (P= 0.05) among treatments according to Tukey–Kramer test and analysis of variance (anova). MU, 4‐methylumbelliferine.
lipA contributes to Xcv virulence
To assess whether lipA has a role in Xcv virulence, we generated a 97‐2 lipA mutant by insertional mutagenesis, and compared its performance relative to the wild‐type in inoculation experiments. First, we verified that the mutant strain was not impaired in its growth ability. Indeed, no differences in growth ability were found between the lipA mutant and the wild‐type strain in NB (Fig. S2, see Supporting Information).
No clear differences could be observed between the wild‐type and lipA mutant in their ability to induce symptoms on tomato Hawaii 7998 (H7998) following dip inoculation (at 5 × 105 cfu/mL). In these experiments, a trend for a slight reduction in growth in planta of the lipA mutant relative to the wild‐type was observed, although significant differences (P= 0.05) between the strains were observed at only a few sampling times, generally at advanced stages after inoculation. Results from a representative experiment (of three experiments with similar results) are shown in Fig. 4A.
Figure 4.
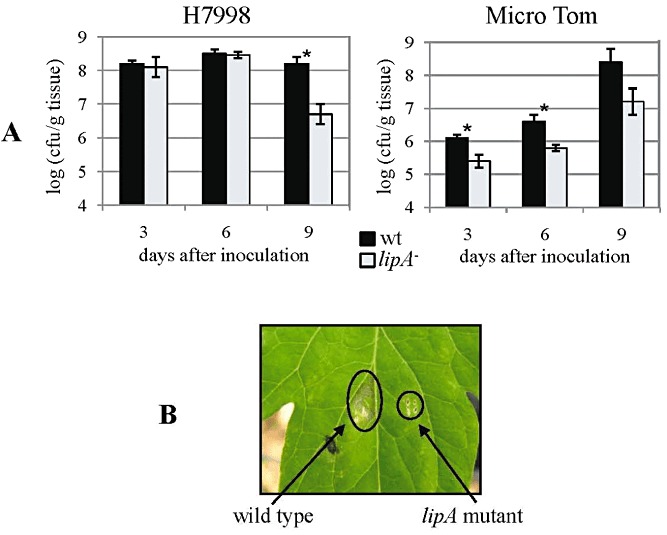
Growth in planta and symptom induction ability of the lipA mutant. (A) Growth in planta of the lipA mutant (lipA –) relative to the wild‐type strain (wt) in tomato Hawaii 7998 (H7998) (left) and Micro Tom (right) following dip inoculation with 5 × 105 colony‐forming units (cfu)/mL bacterial suspensions. Results represent averages and standard deviations (SDs) of one experiment (of three or two with similar results for cultivars H7998 and Micro Tom, respectively). In each experiment, five replicates (each composed of five leaf discs) were used per treatment/time point. Asterisks represent significant differences between strains (P= 0.05) at the given time point according to Student's t‐test. (B) Symptom induction of the lipA mutant compared with the wild‐type strain in tomato H7998 following syringe infiltration with bacterial suspensions of 106 cfu/mL. In each experiment (three with similar results), each strain was used to inoculate at least 30 leaflets. The photograph was taken 7 days after infiltration.
In preliminary experiments, we observed that Micro Tom plants were less susceptible than H7998 to Xcv. We hypothesized that the use of a less susceptible cultivar could facilitate the detection of differences in in planta growth ability between these strains. Indeed, following dip inoculation of Micro Tom plants, disease developed more slowly and both strains multiplied at lower rates than in H7998 plants (Fig. 4A). On Micro Tom plants, a clear trend of reduced growth in planta was observed for the lipA mutant relative to the wild‐type and, in these experiments, the differences were significant (P= 0.05) at most sampling times (Fig. 4A).
In support of the above results, when H7998 leaves were inoculated by syringe infiltration (at 106 cfu/mL), the symptoms induced by the lipA mutant were clearly less severe than those induced by the wild‐type (Fig. 4B), although no differences between these strains were observed for growth in planta under these conditions (not shown). However, when syringe infiltration was performed at 107 cfu/mL, a slightly reduced growth in planta was observed for the lipA mutant relative to the wild‐type at 4 dai (Fig. 5). Although not significant in most experiments, these differences were consistent in three different experiments with similar results. In these experiments, leaves were also co‐inoculated with the two strains to assess whether the lipA mutant could be complemented for growth in planta by the wild‐type strain. Indeed, co‐inoculation allowed the mutant to grow at wild‐type levels in planta (Fig. 5), suggesting that LipA secreted by the wild‐type restored full growth ability to the lipA mutant.
Figure 5.
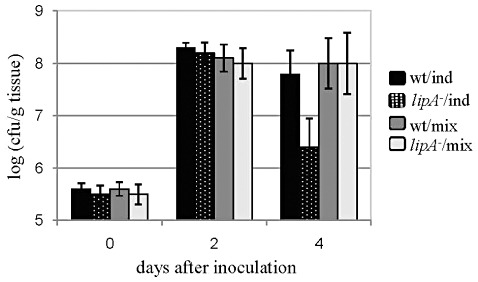
Growth in planta of the lipA mutant (lipA –) and the wild‐type strain (wt) in tomato H7998 following syringe infiltration with bacterial suspensions of 107 colony‐forming units (cfu)/mL. Treatments included individual inoculation with each strain (ind) or co‐inoculation with both strains (mix). In co‐inoculation, to determine cfus of the lipA mutant and the wild‐type, dilutions of leaf extracts were plated on both nutrient agar/cephalexin (NA/Cp) (total bacteria) and nutrient agar/cephalexin + kanamycin (NA/Cp + Km) (lipA mutant only). Results represent averages and standard deviations (SDs) of one experiment of three with similar results. In each experiment, five replicates (each consisting of five leaf discs) were used per treatment/time point. Despite consistently lower counts of the lipA mutant inoculated individually relative to the other treatments at 4 days after inoculation (dai), only in one experiment were these differences statistically significant (P= 0.05) according to Tukey–Kramer test and analysis of variance (anova).
We also questioned whether the mutant strain could be compromised in its epiphytic ability. To assess this, nonhost common bean plants were dip inoculated with both strains (at 105 cfu/mL) and the bacterial populations on the leaves were assessed at different time points. No differences were found between mutant and wild‐type in three different experiments (not shown), thus suggesting that, at least under these experimental conditions, lipA does not contribute to the epiphytic ability of Xcv.
The lipA product is secreted and possesses lipolytic activity
To verify whether lipA encodes a secreted protein with lipolytic activity, as inferred from its annotation and sequence analysis, we assessed the growth ability of the lipA mutant in a liquid medium containing olive oil as carbon source. Olive oil is mainly composed of triglycerides of oleic and palmitic acid. As triglycerides are substrates of lipolytic enzymes, differences in growth between strains in such a medium are positively correlated with differences in their lipolytic activity (Beven et al., 2001). In addition, the presence of neutral red in this medium allows an estimation of its acidification (which correlates with the development of a strong red coloration) as a consequence of lipolytic activity which releases free fatty acids.
These experiments included the utilization of strain lipA –/comp, the lipA mutant carrying plasmid pBBR1‐lipA (pBBR1MCS‐5 with wild‐type lipA; Table 1). For these experiments, the lipA mutant and the wild‐type were transformed with pBBR1MCS‐5, rendering strains lipA –/pBBR1 and wt/pBBR1, respectively, and Gm (resistance conferred by both pBBR1MCS‐5 and pBBR1‐lipA) was added to the growth media of the three strains. The lipA mutant was compromised in its ability to grow on olive oil as sole carbon source when compared with the wild‐type, and clear differences in red coloration were observed between cultures, with wild‐type cultures being more acidic (stronger red coloration) than those of the mutant (Fig. 6A). Quantitative estimation of these experiments by optical density measurements showed that the lipA mutant possesses significantly (P= 0.05) reduced lipolytic activity relative to the wild‐type (Fig. 6B). Despite clear differences in turbidity between the lipA mutant and wild‐type cultures, an accurate estimation of differences in growth ability between the strains in these experiments was affected by the presence of neutral red in the medium. Therefore, we performed growth curves of the strains in the same medium without the inclusion of neutral red, which demonstrated that the lipA mutant is affected in its ability to grow on olive oil as carbon source relative to the wild‐type (Fig. 6C). Transformation of the lipA mutant with plasmid pBBR1‐lipA (complemented strain lipA –/comp) restored both the growth and lipolytic activity of the mutant under these conditions. Moreover, the complemented strain showed higher lipolytic and growth abilities than the wild‐type in this medium, which could be a result of the presence of multiple copies of pBBR1‐lipA in the former (Fig. 6).
Figure 6.
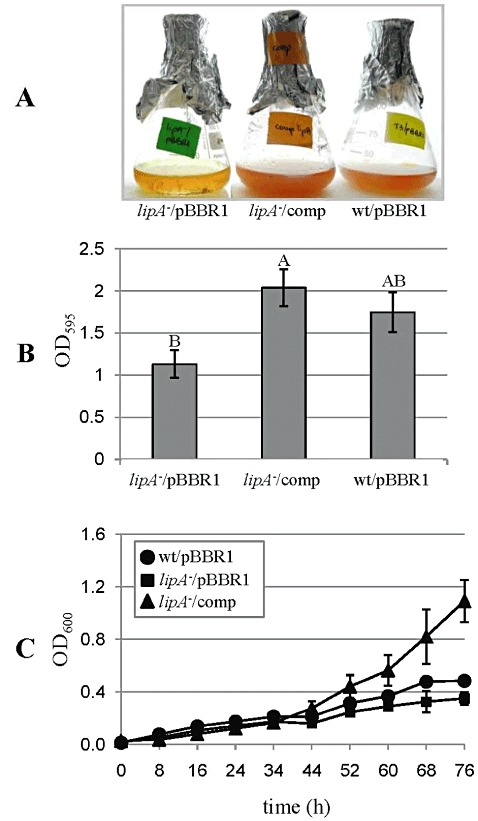
Growth of Xanthomonas campestris pv. vesicatoria (Xcv) strains in a medium containing olive oil as sole carbon source with gentamicin (Gm), including (A and B) or not including (C) neutral red. Strains were the lipA mutant and the wild‐type, both carrying pBBR1MCS‐5 (lipA –/pBBR1 and wt/pBBR1, respectively), and the lipA mutant carrying pBBR1‐lipA (complemented strain, lipA –/comp). (A) Photograph taken after 3 days of growth at 28 °C with shaking. In addition to enhanced growth, a stronger red coloration in the wild‐type and the complemented strain relative to the mutant indicates enhanced medium acidification as a consequence of higher lipolytic activity. (B) Quantification of lipolytic activity by measurement of the optical density of the cultures at 595 nm (OD595). Results represent averages and standard deviations (SDs) (three replicates per treatment) of one experiment of three with similar results. Different letters indicate statistical significance at P= 0.05 according to Tukey–Kramer test and analysis of variance (anova). (C) Growth curves of the strains in olive oil medium without neutral red. Each point represents the average and SD of three replicates of one experiment (of two with similar results).
We further evaluated the extracellular lipolytic activity of the lipA mutant relative to the wild‐type and the complemented strain by lipolytic assays of culture supernatants, using the lipolytic substrate 4‐methylumbelliferyl oleate (MUO). For these experiments, the same strains were grown in XVM2/Gm, as no differences in growth were observed between mutant and wild‐type strains in this medium (not shown), and the lipA gene was shown to be expressed in XVM2 (1, 2). Two experiments yielded similar results, showing a significantly reduced lipolytic activity in the supernatant of the lipA mutant relative to that of the wild‐type and the complemented strain (Fig. 7). In agreement with the results of growth assays in olive oil medium, the supernatants of the complemented strain showed significantly higher (P= 0.05) lipolytic activity than that of the wild‐type (Fig. 7). Importantly, lipolytic activity was still detected in the mutant supernatant, which could be a result of the activity of additional secreted lipases/esterases and/or of intracellular enzymes that may be released to the supernatant from lysed cells.
Figure 7.
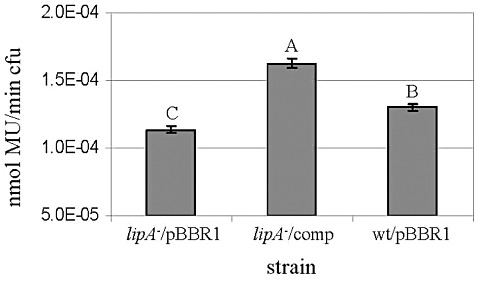
Lipolytic activity of supernatants of Xanthomonas campestris pv. vesicatoria (Xcv) strains grown in XVM2/gentamicin (XVM2/Gm) medium. Strains were the lipA mutant and the wild‐type, both carrying pBBR1MCS‐5 (lipA –/pBBR1 and wt/pBBR1, respectively), and the lipA mutant carrying pBBR1‐lipA (complemented strain, lipA –/comp). Results represent averages and standard deviations (SDs) (five replicates per treatment) of one experiment (of two with similar results). Different letters indicate statistical significance at P= 0.05 according to Tukey–Kramer test and analysis of variance (anova). cfu, colony‐forming unit; MU, 4‐methylumbelliferine.
DISCUSSION
Despite significant advances in the understanding of the Xcv–host interactions during the last two decades, and particularly of the involvement of bacterial type III secreted effectors in disease promotion, many bacterial determinants that contribute to Xcv virulence and fitness are yet to be identified. To deepen our understanding of the gene machinery required by Xcv to successfully infect its host plants, we generated a RIVET approach, which lead to the discovery of several in planta‐induced genes (Tamir‐Ariel et al., 2007).
In the present study, we aimed to characterize one of these Xcv in planta‐induced genes which, in the genome of Xcv 85‐10, is annotated as lipA, a putative secreted lipase. Interestingly, two RIVET approaches, using a pathogen–mouse system, identified ivi genes encoding secreted lipases from the pathogens Vibrio cholerae (Camilli and Mekalanos, 1995) and Staphylococcus aureus (Lowe et al., 1998). In this article, we demonstrated that lipA is expressed at the early stages of tomato infection by Xcv, and that impairment of lipA significantly reduces the virulence and ability of this pathogen to grow in planta. Co‐inoculation experiments revealed that the presence of the wild‐type restores the growth ability of the lipA mutant in planta. In addition, we showed that lipA is part of a single gene locus rather than a multigene operon. Together, these findings demonstrate that the reduction of virulence in the mutant is a result of impairment of the lipA gene and not of a polar effect of the mutation. The lipA mutant was also affected in its ability to grow in a medium with olive oil as sole carbon source, and the extracellular fraction of this mutant possessed significantly lower lipolytic activity than that of the wild‐type. Moreover, transformation of the lipA mutant with a plasmid carrying wild‐type lipA restored its lipolytic activity, thus confirming that the Xcv lipA gene indeed encodes a functional extracellular lipolytic enzyme.
Secreted lipolytic enzymes have been reported to contribute to the virulence of bacterial and fungal pathogens of animals and, more recently, of plants. Although it is not fully understood how these enzymes contribute to virulence, it is hypothesized that they may promote host tissue colonization, with putative roles including nutrient acquisition through the digestion of lipids, synergistic interactions with other enzymes, unspecific hydrolysis, promotion of inflammatory processes by affecting immune cells (in the case of animal hosts) and defence against antimicrobial compounds and microfloral competitors (Rediers et al., 2005; Stehr et al., 2004).
Propionibacterium acnes is the causal agent of acne vulgaris in humans. It has been suggested that lipolytic activity by this bacterium increases the amount of free fatty acids in the skin, predisposing human carriers to acne, probably through the promotion of colonization and persistence of P. acnes (Gribbon et al., 1993; Ingham et al., 1981). Several extracellular enzymes, including lipases/esterases, have also been implicated in the pathogenicity of S. aureus (Novick, 1993; Rollof et al., 1988), Pseudomonas aeruginosa (1992, 1994; Konig et al., 1994) and Burkholderia cepacia (Mullen et al., 2007), among other bacterial pathogens of humans. Secreted lipases have also been associated with the pathogenicity of fungal pathogens of humans, such as Candida albicans (Hube et al., 2000; Schaller et al., 2005; Stehr et al., 2004), Candida parapsilosis (Gacser et al., 2007) and Malassezia furfur (Brunke and Hube, 2006).
The first evidence for the possible involvement of pathogen‐secreted lipolytic enzymes in the infection of plants came from the studies of 1995, 1998), in which the utilization of polyclonal antibodies against an extracellular lipase of the grey mould fungus Botrytis cinerea suppressed lesion formation on detached tomato leaves by this pathogen. Further, Nasser‐Eddine et al. (2001) reported that a secreted lipase was expressed in planta by the pea pathogen Nectria haematococca. Voigt et al. (2005) demonstrated a virulence role for the secreted lipase encoded by the FGL1 gene of Fusarium graminearum: in this fungus, disruption of FGL1 led to reduced extracellular lipolytic activity and to reduced virulence on both wheat and maize. In contrast, Feng et al. (2005) showed that LIP1, a secreted lipase of F. graminearum, is expressed in planta, is needed for efficient utilization of saturated triglyceride lipids, but does not contribute significantly to the virulence of this pathogen. Similarly, in Botrytis cinerea, knockout of LIP1, encoding a secreted lipase, led to the abolishment of lipolytic activity; however, the mutant strain retained full pathogenicity on various hosts (Reis et al., 2005).
Little is known about the relevance of secreted lipolytic enzymes during the infection process of plants by bacterial pathogens. However, in line with our findings with Xcv, Rajeshwari et al. (2005) reported that the orthologous lipA gene from Xoo contributes slightly to the virulence of this pathogen. In the study by Rajeshwari et al. (2005), Xoo mutants of lipA and xynB, the latter encoding a secreted xylanase, were analysed. Although single mutants of these genes were only partially affected, a double lipA‐xynB mutant was severely affected in its virulence on rice. It was proposed that LipA and XynB act as cell wall‐degrading enzymes, and that the slight effects on virulence observed for each mutation could be explained by the redundancy of multiple cell wall‐degrading enzymes in Xoo. Although the actions of the lipA and xynA products could be unrelated, the authors suggested the possibility of a synergistic action, by which LipA could be involved in the hydrolysis of ester bonds of xylan, thus promoting xylan degradation by Xoo xylanases (Rajeshwari et al., 2005). A synergistic action of a xylanase and an esterase from Schizophyllum commune has been demonstrated in the degradation of birchwood xylan (Biely et al., 1986).
In Xoo, LipA, as well as other cell wall‐degrading enzymes, is secreted via type II secretion (Rajeshwari et al., 2005). Using an Hrp‐type III secretion mutant, Jha et al. (2007) showed that type II secreted LipA, ClsA (cellulase) and CbsA (putative cellobiosidase) induce innate defence responses in rice, including callose deposition and programmed cell death. These defence responses are suppressed when type III secretion is functional, allowing successful infection by this pathogen (Jha et al., 2007). Recently, Aparna et al. (2009) have solved the high‐resolution crystal structure of Xoo LipA. LipA was shown to bind a glycoside ligand, β‐octyl glucoside, through a distinct ligand‐binding module comprising carbohydrate‐specific and acyl chain recognition sites. The combination of point mutations in this module with in vitro and in planta assays demonstrated the role of this module in ligand binding, virulence and elicitation of host defence responses. Moreover, the assessment of LipA activity with various substrates suggested that this protein exhibits esterase activity, but no or little lipase activity, and that the natural ligand of LipA probably contains aliphatic chains (Aparna et al., 2009).
LipA is highly conserved in Xanthomonas species and a high level of identity exists between Xoo and Xcv lipA orthologues. Therefore, based on the aforementioned study of Aparna et al. (2009), it is likely that the Xcv LipA is primarily an esterase, with limited lipase activity. This may explain, at least in part, why the extracellular fraction of the Xcv lipA mutant showed significantly, but not dramatically, reduced lipolytic activity relative to that of the wild‐type when MUO (mainly a substrate of lipase activity) was used as a substrate. In addition, the genome of Xcv 85‐10, as well as of other xanthomonads, contain tens of genes annotated as lipases or esterases. Thus, the presence of additional lipolytic enzymes in the assessed extracellular fractions could have masked the negative effects on lipolytic activity caused by the absence of LipA in the extracellular fraction of the lipA mutant. In support of this possibility, substrate clearance assays on tributyrin, performed as described by Aparna et al. (2009), did not reveal detectable differences in lipolytic activity between the crude fractions of the lipA mutant and the wild‐type (not shown).
When referring to the contribution of lipA to Xcv virulence, it should be stressed that ‘slight’ does not mean unimportant. The infection process of plant pathogenic bacteria has a multifactorial nature, in which many virulence‐associated genes contribute to a given functionality only slightly and quantitatively. This, together with the insufficient sensitivity of some pathogenicity assays, may explain why several mutations of putative virulence genes often render non‐defective or only partially defective phenotypes (Alfano and Collmer, 1996). Here, we showed that, in most samplings, no significant differences were observed in bacterial growth in planta between the lipA mutant and the wild‐type, following dip inoculation with cultivar H7998. In contrast, dip inoculation using Micro Tom plants discriminated between these strains for their ability to grow in planta. Moreover, relative to the wild‐type, the lipA mutant was clearly compromised in its ability to induce symptoms following syringe infiltrations at 106 cfu/mL, although no significant differences in growth could be observed between these strains. Other studies with several plant–pathogen systems, including with Xcv, have shown that reduced symptom severity does not always correlate with a reduction in the pathogen population in the plant (Balaji et al., 2008; Lund et al., 1998). Together, these results emphasize the importance of utilizing assays that are sufficiently sensitive to reveal slight phenotypes of many bacterial virulence determinants. In support of this, syringe infiltrations at 107 cfu/mL allowed the detection of a slight, but consistent, reduced growth in planta of the lipA mutant in comparison with the wild‐type, and showed an apparent complementation of the lipA mutant growth ability by co‐inoculation with the wild‐type strain.
Functional redundancy may also explain the observed slight effect of the lipA mutation on Xcv virulence in some of the assays. Here, we showed that, although reduced, lipolytic activity is still retained in the lipA mutant. As mentioned above, the Xcv genome contain tens of genes putatively encoding lipases or esterases, confirming that, collectively, lipolytic enzymes play an important role in Xcv pathogenicity.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and media
The bacterial strains and plasmids used in this study are listed in Table 1. Wild‐type Xcv strain 97‐2 (race T3; X. perforans according to the new nomenclature) (Astua‐Monge et al., 2000) and mutant strains were routinely grown on NB (Becton, Dickinson and Co., Sparks, MS, USA) or XVM2 minimal medium (Wengelnik et al., 1996) at 28 °C. For growth in solid medium, agar was added at 16 g/L. Escherichia coli strains were grown on Luria–Bertani medium at 37 °C. For the determination of bacterial extracellular lipolytic activity, a minimal medium containing olive oil as the sole carbon source (lipid medium) was used according to Kagami et al. (1998). This medium contained 50 mm 3(N‐morpholino)propanesulphonic acid (MOPS) (pH 7.5), 40 mm K2HPO4, 25 mm NaH2PO4, 7.5 mm NH4SO4, 0.4 mm MgSO4, 0.5% (v/v) olive oil and 40 µg/mL neutral red. Neutral red was excluded from the medium for the performance of growth curves. Antibiotics were added, when required, at the indicated concentrations: cephalexin (Cp), 50 µg/mL; Km, 50 µg/mL; chloramphenicol (Cm), 25 µg/mL; Gm, 15 µg/mL.
General molecular techniques
Routine molecular manipulations were carried out as described by Sambrook et al. (1989). All enzymes were purchased from Fermentas (Burlington, ON, Canada). Kits for plasmid and PCR product extraction and purification were purchased from Real Biotech Corporation (Taipei, Taiwan). Genomic DNA from Xcv was prepared with the GenElute™ Bacterial Genomic DNA Kit (Sigma‐Aldrich, St. Louis, MO, USA). Southern blot hybridization was performed using the ECL Direct Nucleic Acid Labelling and Detection System (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). DNA sequencing was performed at Hy Laboratories (Rehovot, Israel). Oligonucleotide primers used in this study were purchased from Hy Laboratories and are listed in Table S1.
Assessment of lipA expression by RT‐PCR
Xcv 97‐2 was grown overnight in 5 mL XVM2 at 28 °C with shaking (150 rpm). Total RNA was extracted using the MasterPure™ RNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA), according to the manufacturer's instructions, with the exception that TURBO DNA‐free DNase (Ambion, Austin, TX, USA) was used for DNA degradation. cDNA was then generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions.
Generation of ResPlipA, ResPlipArev and lipA mutants
A 711‐bp DNA fragment from strain 97‐2, upstream of the lipA open reading frame (thus containing the lipA promoter), was amplified using primers FBamSacP4d22 and RBam4d22, and cloned upstream of promoterless tnpR‐uidA, to create a transcriptional fusion, as follows. The amplified PCR product was digested with BamHI and cloned into pJPGmtnpR, which was cut with the same enzyme, and dephosphorylated. The orientation of the cloned fragment was determined by restriction analysis, as well as by PCR and sequencing of the cloned fragment. In pLipA, the lipA promoter was cloned in the same orientation as the transcription of tnpR‐uidA, whereas, in pLipArev, the lipA promoter was cloned in the opposite orientation to tnpR‐uidA. These two vectors were conjugated (using E. coli S17‐1 λ pir) into the XcvRes reporter strain (carrying the res cassette) to obtain ResPlipA and ResPlipArev strains, respectively. A lipA mutant strain was generated in the background of wild‐type 97‐2. Primers FSac4d22 and RSac4d22 were designed to amplify an internal fragment of 614 bp within the lipA open reading frame. The resulting PCR product was cut with SacI and cloned into pJP5603 previously cut with the same enzyme, and dephosphorylated. The resulting vector, pJPlipA, was transformed into strain 97‐2 to generate the lipA mutant by insertional mutagenesis. All mutants were selected on the basis of antibiotic resistance and were verified by PCR and Southern blot analyses (not shown).
Inoculation techniques
Tomato (Solanum lycopersicum L.) cultivars H7998 (Yu et al., 1995) and Micro Tom (Scott and Harbaugh, 1989) were grown from seeds in a glasshouse (25–28 °C). The inoculation of plants for the assessment of growth in planta of the lipA mutant relative to the wild‐type was carried out by dipping 4–5‐week‐old plants into bacterial suspensions of 5 × 105 cfu/mL containing 0.02% of the surfactant Silwet L‐77 and 10 mm MgCl2 (Kunkel et al., 1993). After dipping, plants were covered with plastic bags for 48 h to keep them in a moist environment. At least four plants per treatment were inoculated in each experiment. Leaf discs (0.8 cm in diameter) were extracted from the first three fully expanded leaves, macerated, serial diluted and plated onto NA plates with Cp (for wild‐type) or Cp + Km (for the lipA mutant) to determine the bacterial concentration in inoculated leaves. For each treatment/time point, five replicates composed of five leaf discs were used. Inoculations were also performed by syringe infiltration of leaves of 4–5‐week‐old plants with bacterial suspensions of 106 or 107 cfu/mL in 1 mm MgCl2. Growth in planta was determined as described above. Epiphytic assays were performed following inoculation of 5‐week‐old plants of common bean (Phaseolus vulgaris L.) cultivar Savana by the dip inoculation method with a few modifications: in these experiments, plants were not covered with plastic bags after inoculation. Plants were maintained in a glasshouse (25–28 °C), and 10 leaves of each treatment were sampled at different time points. The leaves were weighed and placed in 15‐mL Falcon tubes containing 10 mL of 10 mm MgCl2. A 7‐min sonication treatment was applied with Transsonic 310/H (Elma, Singen, Germany) to detach bacterial cells from the leaf surface. The tubes were then vortexed and 1‐mL samples were taken for serial dilutions and plating to determine bacterial counts.
Assessment of promoter activity by resolution assays
For the assessment of resolution in culture, strains ResPlipA and ResPlipArev were first grown in 5 mL of NB supplemented with Km for 48 h at 28 °C, with shaking. Then, a 1 : 500 dilution was made into 5 mL of fresh medium to be tested (NB and XVM2), and cells were grown at similar conditions for 24 h. Three replicates were made for each growth medium. Serial dilutions were plated from each replicate onto NA/Gm plates (total cells) and on NA supplemented with Gm + Km (unresolved cells). In planta resolution was assessed following the dip inoculation method with cultivar H7998 as described above (but with inocula at 105 cfu/mL). The percentage of resolution at different time points after inoculation was determined following extractions of cells from 0.8‐cm‐diameter leaf discs (as described above for the assessment of growth in planta) and plating of serial dilutions onto NA supplemented with Gm (total cells) and Gm + Km (unresolved cells). For each treatment/time point, four replicates composed of five leaf discs were used. In these experiments, an additional control was strain ResPhrpA, for which Cm was used instead of Gm.
Analysis of promoter activity by GUS assays
For quantitative assays in planta, 5‐week‐old H7998 plants were inoculated with strains ResPlipA, ResPlipArev and ResPhrpA (at 106 or 108 cfu/mL in 1 mm MgCl2) by dip inoculation. Each mutant was inoculated into at least three plants, and samples (leaf discs) from each plant were collected at 48, 72 and 96 hai with a 0.8‐cm‐diameter corkscrew, and macerated (three replicates per treatment, each being composed of five leaf discs). Macerated tissues were centrifuged at low speed (1000 g) for 0.5 min to remove plant material, and supernatants were centrifuged again at 12 000 g for 2 min to pellet bacterial cells. The pellets were then resuspended in double‐distilled water, and samples from each treatment were diluted and plated for cfu counts. The remaining cells were stored at −80 °C until use for GUS activity measurements, employing the fluorometric substrate 4‐methylumbelliferyl‐β‐d‐glucuronide (MUG), as described by Jefferson (1987). Qualitative GUS assays of bacteria grown in culture (NA and XVM2‐agar) were performed using the X‐GlucA cyclohexylammonium substrate (Duchefa Biochemie, Haarlem, the Netherlands), as described by Tamir‐Ariel et al. (2007).
Lipolytic activity assays
For the assessment of the lipolytic activity of growing bacteria, Xcv strains were grown for 24 h in 5 mL of NB/Gm at 28 °C with shaking (150 rpm). Then, 1 mL was transferred to 100‐mL Erlenmeyer flasks containing 20 mL of lipid medium (as described above) with Gm. Qualitative and quantitative [optical density at 595 nm (OD595)] estimations of growth and lipolytic activity were performed after 3 days of incubation (three replicates per treatment). For the determination of lipolytic activity of culture extracellular fractions, strains were grown in NB/Gm as described above, but 1 mL was transferred to 250‐mL Erlenmeyer flasks containing 50 mL of XVM2/Gm medium. Cultures were grown for 48 h at 28 °C with shaking (150 rpm), and cells were removed by centrifugation (4500 g, 5 min, 4 °C; twice). Pelleted cells were resuspended in 10 mm MgCl2, and serial diluted and plated on NA/Gm for cfu determination. The supernatants were assessed fluorometrically for quantitative lipolytic activity using the MUO substrate (Sigma‐Aldrich), as described by Beven et al. (2001). Briefly, 1.5‐mL supernatant samples were used, with seven measurements being taken every 10 min. Time ‘zero’ was measured before the addition of MUO, and each treatment included five replicates. Standard curves were drawn using various concentrations of the fluorescent standard 4‐methylumbelliferine (MU; Sigma‐Aldrich).
Statistics
All experiments were performed at least twice. Experiments were statistically analysed by Student's t‐test (for comparison between two treatments) or by one‐way analysis of variance (anova) and Tukey–Kramer test (for comparison between more than two treatments) using JMP software (SAS Institute, Cary, NC, USA).
Nucleotide sequence
The nucleotide sequence of the lipA gene from Xcv strain 97‐2 was deposited in the GenBank database under the accession number FJ599764.
Supporting information
Fig. S1 Assessment of TnpR‐mediated resolution of strains ResPlipA (●), ResPlipArev (■) and ResPhrpA (▲) in planta. Tomato plants were inoculated by the dip inoculation method with bacterial concentrations of 105 colony‐forming units (cfu)/mL. The percentage resolution was determined at several time points following the plating of serial dilutions of leaf extracts on nutrient agar/gentamicin (NA/Gm) (total cells) and nutrient agar/gentamicin + kanamycin (NA/Gm + Km) (unresolved cells). Results from one experiment (with four replicates per treatment per time point, each replicate being composed of five leaf discs) of two with similar results are shown. Significant differences (P = 0.05) were obtained at all time points between ResPlipArev and the other strains based on Tukey–Kramer test and analysis of variance (ANOVA). No significant differences were observed between strains ResPlipA and ResPhrpA.
Fig. S2 Growth curves of the lipA mutant and wild‐type strain in nutrient broth (NB) medium. Bars represent the standard deviation of three independent measurements at each time. One representative growth curve experiment of two with similar results is shown.
Table S1 Primers used in this study.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by grant #975/07 from the Israel Science Foundation (ISF).
REFERENCES
- Alfano, J.R. and Collmer, A. (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell, 8, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Angelichio, M.J. and Camilli, A. (2002) In vivo expression technology. Infect. Immun. 70, 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparna, G. , Chatterjee, A. , Sonti, R.V. and Sankaranarayanan, R. (2009) A cell wall‐degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. Plant Cell, 21, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Minsavage, G.V. , Stall, R.E. , Davis, M.J. , Bonas, U. and Jones, J.B. (2000) Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant‐inducible avirulence gene. Mol. Plant–Microbe Interact. 13, 911–921. [DOI] [PubMed] [Google Scholar]
- Balaji, V. , Mayrose, M. , Sherf, O. , Jacob‐Hirsch, J. , Eichenlaub, R. , Iraki, N. , Manulis‐Sasson, S. , Rechavi, G. , Barash, I. and Sessa, G. (2008) Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven, C.A. , Dieckelmann, M. and Beacham, I.R. (2001) A strain of Pseudomonas fluorescens with two lipase‐encoding genes, one of which possibly encodes cytoplasmic lipolytic activity. J. Appl. Microbiol. 90, 979–987. [DOI] [PubMed] [Google Scholar]
- Biely, P. , MacKenzie, C.R. , Pulls, J. and Schneider, H. (1986) Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Biotechnology, 4, 731–733. [Google Scholar]
- Borgstrom, B. and Brockman, H.L. (1984) Lipases. Amsterdam: Elsevier. [Google Scholar]
- Brunke, S. and Hube, B. (2006) MfLIP1, a gene encoding an extracellular lipase of the lipid‐dependent fungus Malassezia furfur . Microbiology, 152, 547–554. [DOI] [PubMed] [Google Scholar]
- Camilli, A. and Mekalanos, J.J. (1995) Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commenil, P. , Belingheri, L. , Sancholle, M. and Dehorter, B. (1995) Purification and properties of an extracellular lipase from the fungus Botrytis cinerea . Lipids, 30, 351–356. [DOI] [PubMed] [Google Scholar]
- Commenil, P. , Belingheri, L. and Dehorter, B. (1998) Antilipase antibodies prevent infection of tomato leaves by Botrytis cinerea . Physiol. Mol. Plant Pathol. 52, 1–14. [Google Scholar]
- Emanuelsson, O. , Brunak, S. , von Heijne, G. and Nielsen, H. (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Feng, J. , Liu, G. , Selvaraj, G. , Hughes, G.R. and Wei, Y. (2005) A secreted lipase encoded by LIP1 is necessary for efficient use of saturated triglyceride lipids in Fusarium graminearum . Microbiology, 151, 3911–3921. [DOI] [PubMed] [Google Scholar]
- Fojan, P. , Jonson, P.H. , Petersen, M.T.N. and Petersen, S.B. (2000) What distinguishes an esterase from a lipase: a novel structural approach. Biochimie, 82, 1033–1041. [DOI] [PubMed] [Google Scholar]
- Gacser, A. , Trofa, D. , Schafer, W. and Nosanchuk, J.D. (2007) Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J. Clin. Invest. 117, 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribbon, E.M. , Cunliffe, W.J. and Holland, K.T. (1993) Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 139, 1745–1751. [DOI] [PubMed] [Google Scholar]
- Hube, B. , Stehr, F. , Bossenz, M. , Mazur, A. , Kretschmar, M. and Schäfer, W. (2000) Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 174, 362–374. [DOI] [PubMed] [Google Scholar]
- Ingham, E. , Holland, K.T. , Gowland, G. and Cunliffe, W.J. (1981) Partial purification and characterization of lipase (EC 3.1.1.3) from Propionibacterium acnes . J. Gen. Microbiol. 124, 393–401. [DOI] [PubMed] [Google Scholar]
- Jaeger, K.E. , Kinscher, D.A. , Konig, B. and Konig, W. (1992) Extracellular lipase of Pseudomonas aeruginosa: biochemistry and potential role as a virulence factor In: Cystic Fibrosis, Basic and Clinical Research (Hoiby N. and Winkler U.K., eds), pp. 113–119. Amsterdam: Elsevier. [Google Scholar]
- Jaeger, K.E. , Ransac, S. , Dijkstra, B.W. , Colson, C. , van Heuvel, M. and Misset, O. (1994) Bacterial lipases. FEMS Microbiol. Rev. 15, 29–63. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jha, G. , Rajeshwari, R. and Sonti, R.V. (2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant–Microbe Interact. 20, 31–40. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. and Stall, R. (1998) Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36, 41–58. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Lacy, G.H. , Bouzar, H. , Stall, R.E. and Schaad, N.W. (2004) Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27, 755–762. [DOI] [PubMed] [Google Scholar]
- Kagami, Y. , Ratliff, M. , Surber, M. , Martinez, A. and Nunn, D.N. (1998) Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin‐like component XcpT by the cytoplasmic component XcpR. Mol. Microbiol. 27, 221–233. [DOI] [PubMed] [Google Scholar]
- Konig, B. , Jaeger, K.E. and Konig, W. (1994) Induction of inflammatory mediator release (12‐hydroxyeicosatetraenoic acid) from human platelets by Pseudomonas aeruginosa . Int. Arch. Allergy Immunol. 104, 33–41. [DOI] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M., II and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. , Bent, A.F. , Dahlbeck, D. , Innes, R.W. and Staskawicz, B.J. (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2 . Plant Cell, 5, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labes, M. , Puhler, A. and Simon, R. (1990) A new family of RSF1010‐derived expression and lac‐fusion broad‐host‐range vectors for gram‐negative bacteria. Gene, 89, 37–46. [DOI] [PubMed] [Google Scholar]
- Lowe, A.M. , Beattie, D.T. and Deresiewicz, R.L. (1998) Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27, 967–976. [DOI] [PubMed] [Google Scholar]
- Lund, S.T. , Stall, R.E. and Klee, H.J. (1998) Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell, 10, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Mukherjee, K.D. and Hills, M.J. (1994) Lipases from plants In: Lipases: Their Structure, Biochemistry and Application (Woolwey P. and Petersen S.B., eds), pp. 49–75. Cambridge: Cambridge University Press. [Google Scholar]
- Mullen, T. , Market, K. , Murphy, P. , McClean, S. and Callaghan, M. (2007) Role of lipase in Burkholderia cepacia complex (Bcc) invasion of lung epithelial cells. Eur. J. Clin. Microbiol. Infect. Dis. 26, 869–877. [DOI] [PubMed] [Google Scholar]
- Nasser‐Eddine, A. , Hannemann, F. and Schafer, W. (2001) Cloning and expression analysis of NhL1, a gene encoding an extracellular lipase from the fungal pea pathogen Nectria haematococca MP VI (Fusarium solani f. sp. pisi) that is expressed in planta. Mol. Genet. Genomics, 265, 215–224. [DOI] [PubMed] [Google Scholar]
- Novick, R. (1993) Staphylococcus In: Bacillus Subtilis and Other Gram‐Positive Bacteria. Biochemistry, Physiology and Molecular Genetics (Sonsheim A.L., ed.), pp. 17–33. Washington, DC: American Society for Microbiology. [Google Scholar]
- Penfold, R.J. and Pemberton, J.M. (1992) An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene, 118, 145–146. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. and Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant–Microbe Interact. 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Rediers, H. , Rainey, P.B. , Vanderleyden, J. and De Mot, R. (2005) Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche‐specific gene expression. Microbiol. Mol. Biol. Rev. 69, 217–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, H. , Pfiffi, S. and Hahn, M. (2005) Molecular and functional characterization of a secreted lipase from Botrytis cinerea . Mol. Plant Pathol. 6, 257–267. [DOI] [PubMed] [Google Scholar]
- Rollof, J. , Braconier, J.H. , Soderstrom, C. and Nilsson‐Ehle, P. (1988) Interference of Staphylococcus aureus lipase with human granulocyte function. Eur. J. Clin. Microbiol. Infect. Dis. 7, 505–510. [DOI] [PubMed] [Google Scholar]
- Rosenau, F. and Jaeger, K.E. (2000) Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie, 82, 1023–1032. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schaller, M. , Borelli, C. , Korting, H.C. and Hube, B. (2005) Hydrolytic enzymes as virulence factors of Candida albicans . Mycoses, 48, 365–377. [DOI] [PubMed] [Google Scholar]
- Scott, J.W. and Harbaugh, B.K. (1989) Micro‐Tom: a miniature dwarf tomato. Florida Agric. Exp. Sta. Circ. 370, 1–6. [Google Scholar]
- Simon, R. , Priefer, U. and Puhler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram‐negative bacteria. Biotechnology, 1, 784–791. [Google Scholar]
- Stehr, F. , Felk, A. , Gácser, A. , Kretschmar, M. , Mähnß, B. , Neuber, K. , Hube, B. and Schäfer, W. (2004) Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res. 4, 401–408. [DOI] [PubMed] [Google Scholar]
- Tamir‐Ariel, D. , Navon, N. and Burdman, S. (2007) Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 189, 6359–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir‐Ariel, D. , Rosenberg, T. and Burdman, S. (2011) The Xanthomonas campestris pv. vesicatoria citH gene is expressed early in the infection process of tomato and is positively regulated by the TctDE two‐component regulatory system. Mol. Plant Pathol. 12, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Buttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorholter, F.J. , Weber, E. , Puhler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, C.A. , Schäfer, W. and Salomon, S. (2005) A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 42, 364–375. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , Marie, C. , Russel, M. and Bonas, U. (1996) Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178, 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z.H. , Wang, J.F. , Stall, R.E. and Vallejos, C.E. (1995) Genomic localization of tomato genes that control a hypersensitive reaction to Xanthomonas campestris pv. vesicatoria (Doidge) Dye. Genetics, 141, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Assessment of TnpR‐mediated resolution of strains ResPlipA (●), ResPlipArev (■) and ResPhrpA (▲) in planta. Tomato plants were inoculated by the dip inoculation method with bacterial concentrations of 105 colony‐forming units (cfu)/mL. The percentage resolution was determined at several time points following the plating of serial dilutions of leaf extracts on nutrient agar/gentamicin (NA/Gm) (total cells) and nutrient agar/gentamicin + kanamycin (NA/Gm + Km) (unresolved cells). Results from one experiment (with four replicates per treatment per time point, each replicate being composed of five leaf discs) of two with similar results are shown. Significant differences (P = 0.05) were obtained at all time points between ResPlipArev and the other strains based on Tukey–Kramer test and analysis of variance (ANOVA). No significant differences were observed between strains ResPlipA and ResPhrpA.
Fig. S2 Growth curves of the lipA mutant and wild‐type strain in nutrient broth (NB) medium. Bars represent the standard deviation of three independent measurements at each time. One representative growth curve experiment of two with similar results is shown.
Table S1 Primers used in this study.
Supporting info item
Supporting info item
Supporting info item


