Summary
Ralstonia solanacearum is a soil‐borne bacterium causing the widespread disease known as bacterial wilt. Ralstonia solanacearum is also the causal agent of Moko disease of banana and brown rot of potato. Since the last R. solanacearum pathogen profile was published 10 years ago, studies concerning this plant pathogen have taken a genomic and post‐genomic direction. This was pioneered by the first sequenced and annotated genome for a major plant bacterial pathogen and followed by many more genomes in subsequent years. All molecular features studied now have a genomic flavour. In the future, this will help in connecting the classical field of pathology and diversity studies with the gene content of specific strains. In this review, we summarize the recent research on this bacterial pathogen, including strain classification, host range, pathogenicity determinants, regulation of virulence genes, type III effector repertoire, effector‐triggered immunity, plant signalling in response to R. solanacearum, as well as a review of different new pathosystems.
Taxonomy
Bacteria; Proteobacteria; β subdivision; Ralstonia group; genus Ralstonia.
Disease symptoms
Ralstonia solanacearum is the agent of bacterial wilt of plants, characterized by a sudden wilt of the whole plant. Typically, stem cross‐sections will ooze a slimy bacterial exudate. In the case of Moko disease of banana and brown rot of potato, there is also visible bacterial colonization of banana fruit and potato tuber.
Disease control
As a soil‐borne pathogen, infected fields can rarely be reused, even after rotation with nonhost plants. The disease is controlled by the use of resistant and tolerant plant cultivars. The prevention of spread of the disease has been achieved, in some instances, by the application of strict prophylactic sanitation practices.
Useful websites
Stock centre: International Centre for Microbial Resources—French Collection for Plant‐associated Bacteria CIRM‐CFBP, IRHS UMR 1345 INRA‐ACO‐UA, 42 rue Georges Morel, 49070 Beaucouzé Cedex, France, http://www.angers‐nantes.inra.fr/cfbp/. Ralstonia Genome browser: https://iant.toulouse.inra.fr/R.solanacearum. GMI1000 insertion mutant library: https://iant.toulouse.inra.fr/R.solanacearumGMI1000/GenomicResources. MaGe Genome Browser: https://www.genoscope.cns.fr/agc/microscope/mage/viewer.php?
A Classification into Four Phylotypes
In pace with the development of molecular tools, the classification of R. solanacearum has undergone many changes during the past 10 years. In 2005, Fegan and Prior proposed a new hierarchical classification based on the analysis of the sequence of the internal transcribed spacer (ITS) region, the hypersensitive response and pathogenesis B (hrpB) and endoglucanase (egl) genes (Fegan and Prior, 2005). The analysis of 140 R. solanacearum strains isolated from all over the world revealed a subdivision of the species into four phylotypes, which were correlated with the geographical origins of the strains. Phylotype I included strains originating primarily from Asia, phylotype II those from America, phylotype III those from Africa and phylotype IV those from Indonesia, Australia and Japan. Phylotype IV also contained the two close relatives of R. solanacearum: Ralstonia syzygii and the blood disease bacterium (BDB) strains. A multiplex polymerase chain reaction (PCR) based on sequence information from the ITS region has been developed to rapidly identify the phylotype to which a strain belongs (Fegan and Prior, 2005). Each phylotype can be further subdivided into groups of strains, named sequevars, or sequence variants, according to the egl nucleotide sequence. More than 50 sequevars have been defined so far.
This classification was confirmed by comparative genomic hybridization of a set of 18 strains, representing the biodiversity of R. solanacearum on a microarray representative of the GMI1000 reference strain genome (Guidot et al., 2007). Genomic data for nine new R. solanacearum strains also confirmed this classification (Remenant et al., 2010, 2011, 2012). Thanks to the overwhelming phylogenetic data on phylotype II strains, it has been suggested that this phylotype should be divided into two subgroups IIA and IIB (Castillo and Greenberg, 2007; Cellier et al., 2012). It cannot be excluded that deeper sampling in the other phylotypes may also result in a similar refinement of classification.
The geographical isolation and nonhost preference have been the main drivers of the separation of R. solanacearum strains into four phylotypes (Castillo and Greenberg, 2007; Wicker et al., 2012). Using coalescent genealogy reconstruction, Wicker et al. (2012) suggested that R. solanacearum originated from the Australian/Indonesian region in which phylotype IV strains are found. A subgroup of the ancestral strains from this region probably spread throughout present Austral‐Eastern Africa and Madagascar, and differentiated later into phylotypes III and I (predicted with an East African/Asian origin). Another subgroup of ancestral strains migrated to Brazil and differentiated later into the subgroups IIA and IIB, at a time similar to that of the phylotype I/III differentiation, possibly before the fragmentation of Gondwana (Castillo and Greenberg, 2007; Wicker et al., 2012).
A Species Complex, Let's Keep It Simple
A species complex is defined as a cluster of closely related isolates whose individual members may represent more than one species. The term ‘species complex’ was first applied to R. solanacearum by Gillings et al. (1993) to reflect the phenotypic and genotypic variability within the species. Taghavi et al. (1996) then confirmed the concept of the R. solanacearum species complex by including R. syzygii and BDB strains into the R. solanacearum phylogeny. Studies of DNA–DNA similarity revealed that the relatedness between R. solanacearum isolates is often just under the 70% threshold level commonly expected within a species (Roberts et al., 1990). By comparing different strains with the phylotype I strain GMI1000 by microarray hybridization, the most divergent strains still have 68%–69% of their genes hybridizing with the GMI1000 oligonucleotides (Guidot et al., 2007). More recently, Remenant et al. (2010, 2011) used average nucleotide identity (ANI) to evaluate genetic distances between the eight sequenced genomes from this species complex. From the results, based on the ANI > 95% cut‐off (Konstantinidis and Tiedje, 2005; Konstantinidis et al., 2006), the authors suggested that the R. solanacearum species complex should be restructured into three different species: one containing phylotypes I and III, a second containing phylotype II, and a third containing phylotype IV, including R. syzygii and BDB strains (Remenant et al., 2011). ANI provides a more robust and accurate measurement of the genetic distance than does DNA–DNA hybridization (Konstantinidis and Tiedje, 2005). However, as pointed out by Konstantinidis and Tiedje (2005), ANI should not be considered as the sole argument for species definition. Ecological niche occupation, which is a justifiable measurement of the phenotypic potential of a bacterial strain, is another important argument for species definition (Konstantinidis and Tiedje, 2005; Konstantinidis et al., 2006). A simple inspection of the ecological niches occupied by strains from the R. solanacearum species complex indicates that all strains share phenotypic potential, as they are all soil‐borne and plant xylem‐infecting bacteria. In addition, R. solanacearum strains from the four phylotypes are able to infect tomato plants and cause the same symptoms (Remenant et al., 2010). Another important consideration is the (likely allopatric) divergence of related strains accompanied by important genome reduction and ecological specialization, as is the case for R. syzygii, the BDB strains and R. solanacearum phylotype IV strains (i.e. PSI07) (Remenant et al., 2011). Despite a high ANI score (>98%), we could argue that these strains should not be the same species, in the same way as Burkholderia mallei and B. pseudomallei are justified as different species (Konstantinidis et al., 2006).
The renaming proposition of Remenant and colleagues is interesting, but should be re‐evaluated in the light of more whole‐genome sequence data to clearly evaluate whether there is a continuum or clustered genetic relatedness in this species complex. For instance, the sequence of strain ACH0732, which is not clearly associated with a specific phylotype (Fegan and Prior, 2005), would be especially informative.
Host Range
Host range specificity in R. solanacearum is unclear. The diverse strains in the R. solanacearum species complex exhibit an unusually large host range, being able to infect more than 250 plant species in 54 monocot and dicot botanical families (Elphinstone, 2005) (Fig. 1). Host specialization has been reported for some strains, for example the Moko strains infecting banana and Heliconia and the brown‐rot strains infecting potato and tomato. However, host specialization in the R. solanacearum species complex has rarely been described in detail. For example, pathogenicity tests under controlled conditions found that most Moko strains are also virulent to susceptible tomato and potato (Cellier et al., 2012). Many studies have been conducted to tentatively identify the genes associated with host specificity. For this purpose, the methodology of comparative genomic hybridization on pangenomic microarrays representative of R. solanacearum genes was used to compare gene repertories of hundreds of strains for which pathogenicity traits had been defined (Cellier et al., 2012; Guidot et al., 2007). These studies were conducted with potato pathogenic strains and banana pathogenic strains, but did not find any genes associated with these pathogenicity traits (Cellier et al., 2012; Guidot et al., 2007).
Figure 1.

Macro‐ and microscopic views of Ralstonia solanacearum and illustration of associated symptoms on plant bioassays. (a) Ralstonia solanacearum growing on complete BG medium (Boucher et al., 1985). The pink colour of the colonies is caused by the presence of triphenyl tetrazolium chloride in the medium. (b) Electron microscopy image of R. solanacearum rod‐shaped cells under division displaying pili structures (by the late Jacques Vasse). (c) Symptoms of bacterial wilt on Medicago truncatula plants. Inoculation in Jiffy pots with two wild‐type strains (top part). Gnotobiotic inoculation with a wild‐type strain and an hrp mutant (bottom part). (d) Symptoms of bacterial wilt on Arabidopsis thaliana plants. (e) Symptoms of bacterial wilt on tomato plants. (f) One eggplant with symptoms of bacterial wilt, and a healthy control plant.
In order to better characterize the specificity of the interaction between R. solanacearum and solanaceous plants, a recent analysis has been conducted in controlled conditions to study the pathogen interaction between a collection of three Solanaceae (tomato, eggplant and pepper) representative of the bacterial wilt resistance genetic resources and a collection of 12 strains representative of the known phylogenetic diversity of R. solanacearum (Lebeau et al., 2011). Interestingly, although all plants belong to the same family, they interact differently with the 12 R. solanacearum strains. Six interaction phenotypes were defined and were used to name pathoprofiles based on the aggressiveness of the strains on the host plants. Intermediate phenotypes correspond to latent infection of the plants (bacterial colonization of the xylem tissue with few or no wilting symptoms). No pathoprofile is phylotype specific and none of the plants of this collection were resistant to all tested R. solanacearum strains (Lebeau et al., 2011).
Emergence of Strains With a New Host Range
Ralstonia solanacearum is described as a highly flexible organism capable of rapid adaptation to environmental changes and new hosts and of counteracting plant resistance. However, the characterization of emerging strains in R. solanacearum is difficult and has rarely been reported. The most studied recently emerging strains are the phylotype IIB‐4NPB (nonpathogenic on banana) strains in Martinique (Wicker et al., 2007, 2009). These strains belong to the phylotype IIB‐4 group in which Moko disease‐causing strains also cluster, but they are not pathogenic to banana. The epidemiological data demonstrate that phylotype IIB‐4NPB strains constitute an emerging population in Martinique. This genetic group was absent in R. solanacearum collections from the French West Indies until the first strain was isolated in 1999. These strains show a previously unknown host range in R. solanacearum, including cucurbits, ornamental plants and Solanaceae. Importantly, they seem to have expanded their host range from Anthurium–Cucurbitaceae in 1999–2002 to Anthurium–Cucurbitaceae–Solanaceae in 2002–2003 (Wicker et al., 2007). Moreover, they were recovered from solanaceous wild species and several weeds, as well as in the water, throughout Martinique, demonstrating their rapid spread over the island. The factors that have favoured this emergence of strains with novel host specificity are still unclear. The banana/vegetable rotations in Martinique fields probably have a role to play. Indeed, the isolation of IIB‐4NPB strains from wilted tomatoes or wilted cucurbits was only reported on fields that followed a previous banana crop (Wicker et al., 2009). Interestingly, IIB‐4NPB strains were also isolated in Brazil from cucurbits (Cellier et al., 2012). Because Brazil is a Moko disease area, it is also possible that IIB‐4NPB strains emerged in Brazil and established in Martinique through movement of contaminated ornamental material, such as Anthurium.
Generation of Biodiversity
In bacteria, polymorphism created by mutations is redistributed among strains by recombination and horizontal gene transfer. The major contribution of recombination in the evolutionary dynamics of R. solanacearum has recently been demonstrated by Wicker et al. (2012). The authors conducted multilocus sequence analysis (MLSA) with nine loci on a worldwide collection of 89 R. solanacearum strains representative of the four phylotypes and the 51 egl‐based sequevars, and concluded that recombination has played a major role in R. solanacearum genome evolution. They detected phylotype IV as a gene donor for the majority of the recombination events. Interestingly, phylotype I, which is known to affect the largest number of hosts, appeared to be the most recombinogenic lineage. The only clonal group was phylotype IIB. These findings contrast with the conclusions reached by Castillo and Greenberg (2007). Using MLSA and estimations of linkage disequilibrium between eight loci in 58 strains from the four phylotypes, Castillo and Greenberg (2007) concluded that R. solanacearum is an essentially clonal organism. However, most (24 of 58) of the strains analysed by Castillo and Greenberg (2007) belonged to phylotype IIB (phylotype II, group A in Castillo and Greenberg, 2007), which is a clonal group according to Wicker et al. (2012). This could be part of the reason why Castillo and Greenberg (2007) arrived at this conclusion.
Another important mechanism in the evolution of R. solanacearum genomes is horizontal gene transfer (Coupat et al., 2008; Fall et al., 2007; Guidot et al., 2009; Remenant et al., 2010). Analysis of the genomic sequences of nine R. solanacearum genomes revealed numerous genomic islands, many surrounded by mobile elements, such as insertion sequences (ISs) or bacteriophages, suggesting a horizontal acquisition (Remenant et al., 2010, 2011, 2012). Hierarchical clustering based on the variable genes within the genomic islands among 18 R. solanacearum strains indicated that they were acquired by ancestral strains and were then transmitted vertically within phylotypes (Guidot et al., 2007). Methods based on phylogenetic reconstruction of gene families with prokaryote homology detected 151 genes (13.3%) of foreign origin in the R. solanacearum GMI1000 genome (Fall et al., 2007). The small plasmid carrying the type IV secretion system detected in the genome of the CMR15 strain was possibly acquired from Xanthomonas euvesicatoria, another tomato pathogen prevalent in Cameroon (Remenant et al., 2010). Interestingly, recombination ‘hotspots’ were detected in the GMI1000 genome correlating with the presence of Chi‐like signature sequences (Fall et al., 2007).
The frequency of gene transfer between phylogenetically distant bacteria is expected to be low. Nonetheless, horizontal gene transfer between strains from the four phylotypes has been shown to be possible in the laboratory (Coupat et al., 2008; Guidot et al., 2009). Coupat et al. (2008) demonstrated that 80% of R. solanacearum strains are naturally transformable by plasmids and/or genomic DNA, and that large DNA fragments ranging from 30 to 90 kb can be transferred between strains. The potential to exchange virulence genes by horizontal gene transfer could play a major role in rapid pathogenicity evolution of R. solanacearum strains. The role of horizontal gene transfer in enhancing the aggressiveness on tomato of R. solanacearum strains has been demonstrated experimentally (Coupat‐Goutaland et al., 2011).
Pathogenicity Determinants
The main pathogenicity determinant in R. solanacearum is the type III secretion system (T3SS) (Boucher et al., 1985; Coll and Valls, 2013), a syringe‐like membrane appendix that injects the so‐called ‘effector proteins’ (type III effector proteins, or T3E hereafter) into the plant cell cytosol to favour infection (Erhardt et al., 2010; Tampakaki et al., 2010). Mutants defective in any of the >20 hrp or hrp‐conserved (hrc) genes, encoding structural or regulatory proteins of this molecular syringe, are nonpathogenic (Boucher et al., 1985). Exopolysaccharide (EPS), a loose slime of heterogeneous composition (Orgambide et al., 1991), also plays an important role in R. solanacearum pathogenicity. EPS strongly contributes to the occlusion of the xylem vessels that eventually causes the plant wilting symptoms. EPS is also important for plant colonization (Araud‐Razou et al., 1998; Denny and Baek, 1991; Husain and Kelman, 1958; Kao et al., 1992). In addition to these two major virulence determinants mentioned, R. solanacearum produces an array of other factors that also contribute to colonization and/or to symptom appearance. These have been exhaustively reviewed (Genin and Denny, 2012) and include, among others, type II‐secreted plant cell wall‐degrading enzymes, motility or attachment appendages, aerotaxis transducers, cellulases and pectinases. For instance: type 4 pili, involved in twitching motility, biofilm formation and root attachment, and the flagella, responsible for swimming motility, have been shown to contribute to virulence on tomato (Kang et al., 2002; Tans‐Kersten et al., 2001). Interestingly, motility and attachment seem to play a role during plant invasion, as most mutants affected in these capacities are hypopathogenic when inoculated in the soil, but behave like wild‐type strains when inoculated directly in plant stems (Meng et al., 2011; Yao and Allen, 2007). A recent report has suggested that aggregation caused by Flp pili also plays a role in pathogenicity, as a mutant deficient in these pili displays wild‐type swimming or twitching motility, but is impaired in its ability to cause wilting of potato plants (Wairuri et al., 2012).
Regulation of Virulence Genes
Bacterial plant pathogens possess sophisticated regulatory circuits to finely control the energy‐consuming expression of virulence determinants. An exhaustive comparative review on virulence regulatory modules in different bacterial pathogens can be found elsewhere (Mole et al., 2007). The pathways controlling the transcription of the main R. solanacearum virulence genes are well known (Genin, 2010; Genin and Denny, 2012; Schell, 2000). The LysR family transcriptional regulator PhcA plays a central role, as it regulates directly or indirectly many of these genes (Fig. 2). PhcA activates EPS and cellulase‐encoding genes and represses swimming motility, T3SS and siderophore expression (Genin and Denny, 2012). PhcA represses the transcription of a pectinase‐encoding gene via PehR, although it also activates slightly the expression of other pectinase genes (Fig. 2). Interestingly, transcription of the global regulator PhcA is controlled by a Ralstonia‐specific cell density‐dependent mechanism that involves 3‐hydroxy palmitic acid methyl ester (3‐OH PAME) produced by the inner membrane protein PhcB (Flavier et al., 1997; Genin and Denny, 2012). At low cell densities, PhcA expression is repressed by PhcR by a post‐transcriptional mechanism (Fig. 2). When the amounts of 3‐OH PAME build up as a result of confinement or bacterial densities above 107 cells/mL, the molecule activates the two‐component system PhcS/PhcR. PhcR is then phosphorylated and PhcA expression is de‐repressed (Clough et al., 1997; Schell, 2000). One of the main outcomes of PhcA activation in high‐cell‐density conditions is the production of large amounts of EPS. This control is exerted through the induction of XpsR, which activates directly the transcription of the eps operon (Huang et al., 1995). Interestingly, it has been shown that the two‐component regulatory system VsrA/VsrD is also required to fully activate xpsR transcription and, consequently, EPS synthesis (Huang et al., 1998; Schell et al., 1994). In addition, VsrD affects swimming motility directly by repressing the transcription of the flagellum genes (Fig. 2). Another two‐component system, VsrB/VsrC, has also been described to control EPS synthesis and to repress the transcription of the pectinase pglA, adding another layer of control on EPS synthesis (Huang et al., 1995). It is remarkable that members of both systems (vsrB and vsrD) were identified in a genetic screen amongst 153 R. solanacearum K60 genes induced during growth in tomato (Brown and Allen, 2004). XpsR is thus a central switch in EPS regulation, as it integrates inputs from both VsrAD and PhcA to regulate directly the eps promoter, and is required for both its negative control by EpsR and its positive control by VsrC (Garg et al., 2000; Huang et al., 1995).
Figure 2.
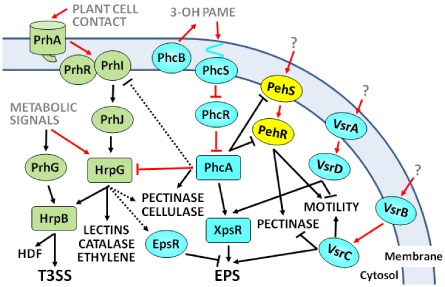
Major pathways controlling the expression of Ralstonia solanacearum virulence genes. Ovals and squares indicate regulatory proteins, the latter representing the main regulatory hubs. Grey, regulatory inputs sensed by the bacterium; black, pathogenic activities controlled by this regulatory network. Arrows and T‐bars indicate activation or repression, respectively. Black lines denote control at the transcriptional level and red lines indicate post‐transcriptional effects. Full lines represent major effects and dotted lines slight transcriptional influences (modulation). For detailed explanations, see text. EPS, exopolysaccharide; HDF, Hrp‐dependent factor; 3‐OH PAME, 3‐hydroxy palmitic acid methyl ester; T3SS, type III secretion system.
Regulation of the T3SS exemplifies that, during evolution, horizontally transferred operons can co‐opt transcriptional regulators present in the recipient bacterium (Cases and de Lorenzo, 2001). For instance, although the genes encoding the T3SS are highly conserved across species, the pathways controlling their transcription in R. solanacearum and Xanthomonas spp. are totally unrelated to those found in Pectobacterium spp. and Pseudomonas spp. (Tang et al., 2006). In R. solanacearum, HrpB, an AraC‐type regulator, and HrpG, its upstream OmpR‐like two‐component response regulator, control hrp/hrc gene expression. HrpB triggers directly the transcription of T3SS genes, probably binding to the so‐called hrpII box found in promoter regions (Cunnac et al., 2004a, b; Genin et al., 1992), and its expression is controlled by HrpG (Brito et al., 1999) (Fig. 2). HrpG and HrpB are both genetically and functionally conserved in Xanthomonas spp. (Li et al., 2011; Wengelnik and Bonas, 1996; Zou et al., 2006), but unique to R. solanacearum are the upstream regulators that trigger the specific induction of HrpG when the bacterium detects a plant cell wall component (Aldon et al., 2000). PrhA is the outer membrane receptor that perceives this signal (Aldon et al., 2000) and transfers it to the membrane‐associated proteins PrhI and PrhR to trigger hrp/hrc expression through the consecutively induced transcriptional regulators PrhJ, HrpG and HrpB (Brito et al., 2002) (Fig. 2). In vitro transcriptomic studies have revealed that the hrp regulators control additional functions other than the T3SS and most of its associated effectors. It has been shown that HrpB is also involved in the regulation of chemotaxis and the biosynthesis of various low‐molecular‐weight chemical compounds, such as the Hrp‐dependent factor (HDF), which may induce a cell density‐dependent LuxR system (Delaspre et al., 2007; Occhialini et al., 2005). HrpG has been found to control an even larger set of T3SS‐unrelated genes independent of HrpB (Fig. 2). These encoded genes are probably involved in plant–pathogen interactions, including adhesion factors (lectins), the only predicted catalase enzyme in the genome and an ethylene‐forming enzyme that produces this plant hormone (Valls et al., 2006). HrpG has also been found to affect slightly known virulence determinants, such as pectinolytic and cellulase activities, some of which are common targets of PhcA (Fig. 2). A recent study has also shown that HrpG is involved in the control of the last step of methionine synthesis (Plener et al., 2012). It has been proposed that HrpG promotes the production of MetE, which synthesizes methionine without the need of vitamin B12, as a way to ensure the biosynthesis of this amino acid in the vitamin‐poor environment encountered in planta. Thus, HrpG occupies a central node in pathogenicity regulation as, in addition to controlling a panoply of virulence genes, it integrates both plant cell‐dependent induction and metabolic cues that affect the transcription of the T3SS (Fig. 2). Examples of the latter metabolic signals are the repression of T3SS genes by casamino acids and their induction during growth in minimal medium (Arlat et al., 1992; Genin et al., 2005). Recently, PrhG has been identified in strain GMI1000 as a HrpG paralogue that also activates HrpB expression. Interestingly, prhG is induced during growth in minimal medium, but not by plant cells, so that this regulator controls the expression of T3SS under minimal medium conditions (Plener et al., 2010). Additional inputs influencing the expression of T3SS have also been described, but the regulators involved and their mechanisms of action remain poorly understood. Examples of these are LrpE, a leucine‐rich repeat protein found to negatively regulate the expression of hrp genes by three‐ to five‐fold (Murata et al., 2006). Similarly, the absence of any of the three genes encoded in the prKLM operon in strain OE1‐1 decreases prhG expression and, consequently, that of hrpB and the PopA effector by 10‐fold (Zhang et al., 2011). However, these genes do not encode transcriptional regulators and must influence the hrp regulon indirectly by an unknown mechanism.
As mentioned previously, the global regulator PhcA modulates T3SS gene expression and does so at two different levels: by slightly inhibiting prhI/R transcription and by strongly inhibiting hrpB gene expression through an unknown mechanism acting on HrpG (Genin et al., 2005; Yoshimochi et al., 2009a, b). Interestingly, EPS production is also slightly downregulated by HrpG through an increase in the levels of EpsR (Valls et al., 2006) (Fig. 2). These examples illustrate that cross‐talk occurs between regulatory cascades at various levels. All of this knowledge has led to the corollary that, in R. solanacearum, the infection process takes place in two steps (Brito et al., 2002; Mole et al., 2007; Schell, 2000): First, early in colonization, expression of the T3SS is induced by plant cell contact and PhcA is not induced because of the low bacterial density, allowing swimming motility to be active. In the second step, when bacterial numbers increase inside the xylem, the PhcA regulator is expressed, triggering EPS and cellulase production and repressing T3SS and the siderophore. However, recent in planta expression data using promoter::reporter fusions integrated in the bacterial chromosome and quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) have challenged this model (Monteiro et al., 2012a, b). These studies showed that the hrpB and T3E transcripts are abundant at late stages of plant colonization, when bacterial numbers are high and plants are already wilted. More recently, transcriptome analyses have confirmed this finding, showing that one‐half of the hrpB‐regulated genes are induced in bacteria recovered from the xylem of wilting tomato plants (Jacobs et al., 2012). All of these observations illustrate our limitations to predict the behaviour of bacterial virulence genes in real field conditions and suggest the existence of still unknown inducing signals.
T3E Repertoire
Over the last 10 years, T3E biology has boomed. Different methods have been applied to define the set of T3Es in R. solanacearum: (i) the search for orthologues of already known T3Es; (ii) the identification of T3Es through gene regulation studies (Cunnac et al., 2004a, b; Occhialini et al., 2005); (iii) the search for atypical protein motifs indicating a potential function in eukaryotic host cells (Angot et al., 2006); and finally (iv) a functional screen for type III injected protein (Mukaihara et al., 2010). Altogether, these efforts generated a comprehensive list of T3Es for two closely related phylotype I strains (Mukaihara et al., 2010; Poueymiro and Genin, 2009). This latter work could now be expanded with the availability of several new genomic sequences spanning the four phylotypes representing the R. solanacearum diversity (Gabriel et al., 2006; Remenant et al., 2010, 2011; Salanoubat et al., 2002; Xu et al., 2011). This would allow the evaluation of common and specific Rip [‘Ralstonia injected proteins’ (Cunnac et al., 2004b; Mukaihara et al., 2010)]. As already pointed out when the first R. solanacearum genome was published (Salanoubat et al., 2002), there are some striking features specific to this bacterium. First, it seems that the T3E repertoire is larger than in other plant pathogenic bacteria; second, R. solanacearum seems to be the recipient of a diverse set of T3Es probably acquired by horizontal gene transfer. Indeed, the GALA T3E (Kajava et al., 2008) and the pentatricopeptide repeat (PPR)‐containing T3E (Salanoubat et al., 2002), harbouring typical eukaryotic features, such as the F‐box domain (Ho et al., 2008) and PPR motifs (Delannoy et al., 2007), could originate from an ancestral eukaryotic donor. However, many T3Es could have originated from horizontal gene transfer from other pathogenic bacteria, as homologues exist in various other animal and plant bacterial pathogens (Poueymiro and Genin, 2009). This is very likely to be the case for the Xanthomonas spp.‐specific transcriptional activator‐like (TAL) T3Es (Fall et al., 2007). Recently, a detailed characterization of these R. solanacearum TAL‐like T3Es showed that they are indeed nuclear targeted and can function as transcriptional activators in plant cells (Li et al., 2013). Another particular feature of the R. solanacearum effector repertoire is the abundance of duplicated T3E genes. Indeed, several T3E genes are present as gene families (Poueymiro and Genin, 2009; Remigi et al., 2011; Sole et al., 2012). Interestingly, most of these gene families are conserved among the different R. solanacearum strains (Remigi et al., 2011; Sole et al., 2012) (see also the MaGe genome browser displaying the sequenced R. solanacearum strains: https://www.genoscope.cns.fr/agc/microscope/mage). These gene families, arising from gene duplications in a common ancestor, are likely to undergo functional diversification to provide adaptation on different host plants, as has been shown for the GALA family (Remigi et al., 2011).
Effector‐Triggered Immunity (ETI)
Historically, the first biological function identified for T3Es was their contribution to ETI (Jones and Dangl, 2006). The T3E PopP1 induces a cultivar‐specific hypersensitive response (HR)‐like response on petunia (Arlat et al., 1994; Lavie et al., 2002). Furthermore, PopP1 (Robertson et al., 2004) and, recently, PopP1 together with AvrA (Poueymiro et al., 2009) have been shown to contribute to HR‐mediated resistance in tobacco. Indeed, on tobacco root inoculation, the double PopP1 AvrA mutant in GMI1000 causes wilting and is indistinguishable from the K60 tobacco pathogenic strain (Poueymiro et al., 2009). The closely related T3E PopP2 has been shown to be responsible for RRS1‐R‐mediated resistance in the Nd‐1 Arabidopsis ecotype (Deslandes et al., 1998). PopP2 interacts directly with RRS1‐R in the nucleus of plant cells, leading to an asymptomatic and much reduced bacterial colonization (Deslandes et al., 2002, 2003). PopP2 has been shown to interact with the Arabidopsis thaliana protein RD19, redirecting its localization from lytic vacuoles to the nucleus, where both physically interact. Although its exact role has yet to be defined, RD19 is required for PopP2/RRS1‐R‐mediated resistance (Bernoux et al., 2008). Furthermore, it has been shown that this T3E, belonging to the widespread YopJ/Avrxv family, displays acetyltransferase activity. This activity results in the autoacetylation of PopP2 required for an effective PopP2/RRS1‐R resistance (Tasset et al., 2010). Ralstonia solanacearum could harbour other T3Es triggering plant immunity. Indeed, Agrobacterium tumefaciens‐mediated expression of AWR5 induces HR‐like symptoms in Nicotiana tabacum (Sole et al., 2012). Interestingly, in the same T3E family, the multiple mutant awr(1–5) displays an increased pathogenicity on Arabidopsis Col‐0, suggesting the possibility that at least one of these AWRs is actually recognized by an R gene, triggering a weak ETI in this host (Sole et al., 2012).
T3E Virulence Functions
Other T3Es have been characterized on the basis of their contribution to disease. Although the exact molecular mechanisms have yet to be described, different T3Es have different contributions to disease on different host plants. On tomato, mutants in the T3Es RSp0304 (HopD1 homologue) and AWR2 show decreased disease progression (Cunnac et al., 2004b). The AWR T3E family is collectively needed on both tomato and eggplant for full pathogenicity. Interestingly, AWR2 can restore the wild‐type phenotype of the multiple awr(1–5) mutant on eggplant (Sole et al., 2012). For the GALAs, another well‐studied T3E family, it has been demonstrated that they are collectively required on tomato and A. thaliana for a full disease phenotype (Angot et al., 2006). More recently, this result has been shown to be more complex, with a redundancy of GALA2, GALA3, GALA6 and GALA7 on Arabidopsis, whereas only GALA7 and GALA3 seem to be able to restore full virulence of the quadruple mutant gala2 gala3 gala6 gala7 on tomato (Remigi et al., 2011). In this same family, it has also been shown that the single gala7 mutant (and none of the other single mutants) is avirulent on the legume host Medicago truncatula (Angot et al., 2006). GALA T3Es could potentially control host protein stability as they probably form E3‐ubiquitin ligases inside the host cell (Angot et al., 2007). Although structurally different, several other T3Es have similarities with ubiquitin ligases. This is the case for RSc1349, the homologue of the Shigella flexneri IpaH ubiquitin ligase (Singer et al., 2008), and the MolK2‐specific RSMK00763, the T3E homologue of the Pseudomonas syringae AvrPtoB (Poueymiro and Genin, 2009). On M. truncatula, GALA7 and AvrA are both required for the early infection steps of intact roots (Turner et al., 2009). GALA7, but not AvrA, is also required for disease development when colonization is facilitated by cutting the root tips (Turner et al., 2009). Another early/late disease development differential role has been shown for the T3SS‐secreted harpin PopA. Indeed, in the Japanese strain OE1‐1, constitutive early expression of this T3E prevents the natural root infection of N. tabacum, but not bacterial multiplication inside stem‐inoculated plants (Kanda et al., 2003).
Classical plant pathoassays have enabled the identification of only a few T3Es with a virulence function (Cunnac et al., 2004b). This low yield of T3Es with virulence functions compared with the large T3E repertoire (Poueymiro and Genin, 2009) could be explained by functional redundancy (Angot et al., 2006; Sole et al., 2012), but also by the fact that some T3Es have only a marginal contribution to virulence, and hence are undetectable with classical wilt scoring. For this purpose, a novel assay based on mixed inoculations was developed (Macho et al., 2010). The principle is to compare the ability of two strains to multiply in the host when they are co‐inoculated, compared with their ability to multiply when inoculated individually. Interestingly, two T3Es, Rsp0304 and PopP2, have been shown to be involved in efficient bacterial multiplication in three host plants: eggplant, tomato and bean (Macho et al., 2010).
Complex Mechanisms Underlie Bacterial Wilt Resistance and Tolerance
The genetic analysis of resistance to bacterial wilt has been developed on both model and cultivated plants. In A. thaliana, the study of the interaction between two ecotypes, Nd‐1 and Kil‐0, and two strains of R. solanacearum, GMI1000 and BCCF402 (both phylotype I strains), respectively, revealed the involvement of RRS1‐R. RRS1‐R, a Toll/interleukin‐1 receptor‐nucleotide‐binding site‐leucine‐rich repeat (TIR‐NBS‐LRR) gene with a WRKY C‐terminal domain, has been described as a single recessive resistance gene against strain GMI1000, through the direct recognition of the PopP2 effector (Deslandes et al., 1998, 2002, 2003). Interestingly, it has been demonstrated recently that the gene‐for‐gene interaction RRS1‐R–PopP2 is also involved in Kil‐0 tolerance (Van der Linden et al., 2013). Indeed, Kil‐0 does not exhibit wilting symptoms after its inoculation with strain BCCF402 of R. solanacearum, despite a high bacterial multiplication in planta. The catalytic triad and the autoacetylated lysine are conserved in the BCCF402 popP2 allele, but some allelic variations in both BCCF402 PopP2 and Kil‐0 RRS1‐R could account for altered protein interactions and/or signal transduction in the plant cell. It cannot be excluded either that this RRS1‐R‐dependent tolerance in Kil‐0 could be dependent on other plant or bacterial factors. A quantitative resistance mechanism has also been described in A. thaliana against R. solanacearum (Godiard et al., 2003). Among the three quantitative trait loci (QTLs) identified, one is associated with ERECTA, an LRR receptor‐like kinase (LRR‐RLK) involved in development (Godiard et al., 2003; Torii et al., 1996). This suggests that cross‐talk can occur between resistance to R. solanacearum and developmental pathways (Godiard et al., 2003). In the model legume M. truncatula, recombinant inbred line (RIL) population A17 × F83005.5 enabled the identification of three QTLs for resistance to R. solanacearum strain GMI1000 (Vailleau et al., 2007). The fine mapping of the major QTL located on chromosome 5 of M. truncatula, MtQRRS1, allowed the identification of a 64‐kb region with a cluster of seven putative R genes among 15 candidate genes (Ben et al., 2013) Tomato cultivar ‘Hawaii 7996’ has been described as resistant to bacterial wilt (Danesh et al., 1994; Mangin et al., 1999; Thoquet et al., 1996a, b; Wang et al., 2000). The polygenic resistances identified are phylotype and strain specific (Carmeille et al., 2006). Recently, Wang et al. (2013a) have identified two major QTLs for resistance to R. solanacearum (Bwr‐6 and Bwr‐12) using the tomato cross ‘Hawaii 7996’ × ‘West Virginia 700’. The Bwr12 QTL is specific for resistance to phylotype I, whereas the Bwr‐6 QTL is associated with resistance to phylotype I and II strains. Interestingly, the presence of both QTLs has an additive effect, displaying enhanced resistance. In tobacco, Qian et al. (2012) identified four QTLs associated with resistance to noncharacterized ‘naturally occurring strains’ from bacterial wilt‐affected areas of China. In eggplant, four QTLs have been identified in a RIL population that exhibited resistance to different strains of R. solanacearum (Lebeau et al., 2013). Among them, the ERs1 QTL has been described as a major dominant source of resistance towards three phylotype I strains of R. solanacearum (Lebeau et al., 2013). The further characterization of the genetic components underlying these different resistances to R. solanacearum will prove to be very useful for the plant breeding community.
Plant Signalling In Response to R. solanacearum
A link has been described between A. thaliana secondary cell walls and the outcome of the interaction with R. solanacearum (Hernandez‐Blanco et al., 2007). Indeed, a mutation in any of the three secondary cell wall‐specific cellulose synthase genes led to a complete resistance to the bacterium. Furthermore, abscisic acid (ABA) signalling has been demonstrated to be involved in this cellulose synthase‐dependent enhanced resistance (Hernandez‐Blanco et al., 2007). The role of the cell walls in A. thaliana as barriers against R. solanacearum colonization has also been studied through the wat1 (walls are thin1) mutant (Denance et al., 2012). The WAT1 gene is required for secondary cell wall deposition, and the corresponding mutant shows enhanced resistance to R. solanacearum (Denance et al., 2012). Comparing two different inoculation methods of R. solanacearum in A. thaliana leaves, by piercing in the central leaf vein or infiltrating a bacterial suspension in the mesophyll, the authors concluded that wat1 resistance is localized to the vascular system. Moreover, salicylic acid (SA) has been identified as a key element of wat1‐mediated resistance to R. solanacearum (Denance et al., 2012). Several other studies have identified WRKY transcription factors as important players in modulating the plant response towards R. solanacearum attack. Indeed, A. thaliana plants lacking a functional WRKY27 gene exhibited an enhanced tolerance to R. solanacearum strains GMI1000 and Rd‐15 (Mukhtar et al., 2008). Similar situations have been observed previously for the ethylene‐insensitive EIN2‐1 gene (Hirsch et al., 2002) and the NWS1 gene (Feng et al., 2004), which appeared to be required for full virulence of the bacteria. Recently, two WRKY transcription factors of pepper have been identified as important positive and negative contributors to R. solanacearum resistance. The overexpression of CaWRKY40 in tobacco enhanced resistance to R. solanacearum, whereas the silencing of CaWRKY40 in pepper attenuated the resistance (Dang et al., 2012). However, CaWRKY58‐overexpressing tobacco plants showed an enhanced susceptibility to the same strain, and CaWRKY58‐silenced pepper plants displayed an enhanced resistance (Wang et al., 2013b). In another study, Feng et al. (2012) identified the ABA signalling pathway as important for the biological control of bacterial wilt in A. thaliana. This ABA‐dependent defence mechanism was shown to be independent of SA, jasmonic acid and ethylene in the biological control exerted by an hrpB mutant of R. solanacearum (Feng et al., 2012).
Gnotobiotic Pathosystems to Study Root Infection
As R. solanacearum infects plants via their roots, gnotobiotic artificial inoculation systems constitute useful tools to enable access to the host roots. Such bioassays, in which axenic plantlets are inoculated with R. solanacearum, have been described on tomato (Monteiro et al., 2012b; Vasse et al., 1995), petunia (Zolobowska and Van Gijsegem, 2006), M. truncatula (Vailleau et al., 2007) and A. thaliana (Digonnet et al., 2012). They enable the study of the early steps of infection, step‐by‐step colonization and the observation of ‘root symptoms’. Ralstonia solanacearum–plant interaction at the root level was first studied in tomato (Monteiro et al., 2012b; Vasse et al., 1995). The authors observed bacterial penetration via the root extremities and at the axils of the secondary root (Vasse et al., 1995). Ten years later, gnotobiotic experiments on petunia allowed the identification of new root lateral structures (RLSs) after the inoculation with R. solanacearum (Zolobowska and Van Gijsegem, 2006). These structures are T3SS dependent and were demonstrated to be highly efficient colonization sites. However, the involvement of specific T3Es in the formation of these RLSs has not been proven (Zolobowska and Van Gijsegem, 2006). In the M. truncatula gnotobiotic pathosystem, it has been described that R. solanacearum penetrates via the root tips (Turner et al., 2009). Bacterial inoculation leads to an arrest of root hair elongation and a reduction in root growth, associated with a browning and a loss of viability of the root tip epidermal cells (Turner et al., 2009). Two T3Es, GALA7 and AvrA, have been demonstrated to be partially involved in the induction of the root epidermal cell death phenotype (Turner et al., 2009). For the interaction involving A. thaliana and R. solanacearum, which has been well described and characterized at the whole plant level (Deslandes et al., 1998), an axenic plant bioassay has been developed recently to study the early steps of root colonization by R. solanacearum (Digonnet et al., 2012). The authors showed that the bacteria penetrate A. thaliana plantlets at the root apex. Bacteria induced a plasmolysis of epidermal and cortical root cells, accompanied by pectin degradation. The bacteria then move preferentially via intercellular spaces to invade the vascular cylinder directly (Digonnet et al., 2012). Contrary to the M. truncatula system (Vailleau et al., 2007), an important bacterial surface colonization can be seen alongside the inoculated plantlets. Thanks to these gnotobiotic systems, it has been shown that R. solanacearum disturbs normal root growth on host infection. Preferential zones of bacterial penetration are the root tips, correlating with the zone in which plant exudates are produced, attracting R. solanacearum (Yao and Allen, 2006). In the future, these systems will prove to be important to better characterize the early events of infection and the chronology of plant colonization.
Perspectives
In this short review, we have tried to encapsulate all the recent research on R. solanacearum, ranging from strain diversity to gene regulation, T3E biology and plant resistance. We would like to finish by emphasizing the fact that this plant pathogenic bacterium is a research model with interesting specificities, i.e. root infection, vascular colonization, large T3E repertoire and broad host range. We believe that further research will shed light on the specificities as well as the general infection strategies common between R. solanacearum and other parasites.
In this post‐genomic era, the following challenges lie ahead: further strain sampling for a better description of the species complex; deciphering of the role of T3Es during infection; analysis of the in planta expression of bacterial genes (RNAseq); metabolic modelling towards a systems biology model; and experimental evolution to study adaptation to different plant hosts.
Acknowledgements
We thank Freddy Monteiro and Irene Van Dijk for their assistance in the elaboration of Fig. 2. We wish to apologize to colleagues whose work could not be cited because of space limitations. We also thank our colleagues and two anonymous reviewers for their constructive comments. This work was supported by the Laboratoire d'Excellence (LABEX) entitled TULIP (ANR‐10‐LABX‐41).
References
- Aldon, D. , Brito, B. , Boucher, C. and Genin, S. (2000) A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19, 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot, A. , Vergunst, A. , Genin, S. and Peeters, N. (2007) Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araud‐Razou, I. , Vasse, J. , Montrozier, H. , Etchebar, C. and Trigalet, A. (1998) Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol. 104, 795–809. [Google Scholar]
- Arlat, M. , Gough, C.L. , Zischek, C. , Barberis, P.A. , Trigalet, A. and Boucher, C.A. (1992) Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 5, 187–193. [DOI] [PubMed] [Google Scholar]
- Arlat, M. , Van Gijsegem, F. , Huet, J.C. , Pernollet, J.C. and Boucher, C.A. (1994) PopA1, a protein which induces a hypersensitivity‐like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum . EMBO J. 13, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben, C. , Debelle, F. , Berges, H. , Bellec, A. , Jardinaud, M.F. , Anson, P. , Huguet, T. , Gentzbittel, L. and Vailleau, F. (2013) MtQRRS1, a R‐locus required for Medicago truncatula quantitative resistance to Ralstonia solanacearum . New Phytol. doi: 10.111/nph.12299. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , de Wit, P.J. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, C. , Barberis, P.A. , Trigalet, A.P. and Demery, D.A. (1985) Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5‐induced avirulent mutants. J. Gen. Microbiol. 131, 2449–2457. [Google Scholar]
- Brito, B. , Marenda, M. , Barberis, P. , Boucher, C. and Genin, S. (1999) prhJ and hrpG, two new components of the plant signal‐dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum . Mol. Microbiol. 31, 237–251. [DOI] [PubMed] [Google Scholar]
- Brito, B. , Aldon, D. , Barberis, P. , Boucher, C. and Genin, S. (2002) A signal transfer system through three compartments transduces the plant cell contact‐dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant–Microbe Interact. 15, 109–119. [DOI] [PubMed] [Google Scholar]
- Brown, D.G. and Allen, C. (2004) Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53, 1641–1660. [DOI] [PubMed] [Google Scholar]
- Carmeille, A. , Caranta, C. , Dintinger, J. , Prior, P. , Luisetti, J. and Besse, P. (2006) Identification of QTLs for Ralstonia solanacearum race 3‐phylotype II resistance in tomato. Theor. Appl. Genet. 113, 110–121. [DOI] [PubMed] [Google Scholar]
- Cases, I. and de Lorenzo, V. (2001) The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, J.A. and Greenberg, J.T. (2007) Evolutionary dynamics of Ralstonia solanacearum . Appl. Environ. Microbiol. 73, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier, G. , Remenant, B. , Chiroleu, F. , Lefeuvre, P. and Prior, P. (2012) Phylogeny and population structure of brown rot‐ and Moko disease‐causing strains of Ralstonia solanacearum phylotype II. Appl. Environ. Microbiol. 78, 2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. , Lee, K.E. , Schell, M.A. and Denny, T.P. (1997) A two‐component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA‐regulated virulence factors in response to 3‐hydroxypalmitic acid methyl ester. J. Bacteriol. 179, 3639–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. and Valls, M. (2013) Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. doi: 10.1111/1751-7915.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupat, B. , Chaumeille‐Dole, F. , Fall, S. , Prior, P. , Simonet, P. , Nesme, X. and Bertolla, F. (2008) Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred. FEMS Microbiol. Ecol. 66, 14–24. [DOI] [PubMed] [Google Scholar]
- Coupat‐Goutaland, B. , Bernillon, D. , Guidot, A. , Prior, P. , Nesme, X. and Bertolla, F. (2011) Ralstonia solanacearum virulence increased following large interstrain gene transfers by natural transformation. Mol. Plant–Microbe Interact. 24, 497–505. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Boucher, C. and Genin, S. (2004a) Characterization of the cis‐acting regulatory element controlling HrpB‐mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum . J. Bacteriol. 186, 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac, S. , Occhialini, A. , Barberis, P. , Boucher, C. and Genin, S. (2004b) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- Danesh, D. , Aarons, S. , McGill, G.E. and Young, N.D. (1994) Genetic dissection of oligogenic resistance to bacterial wilt in tomato. Mol. Plant–Microbe Interact. 7, 464–471. [DOI] [PubMed] [Google Scholar]
- Dang, F.F. , Wang, Y.N. , Yu, L. , Eulgem, T. , Lai, Y. , Liu, Z.Q. , Wang, X. , Qiu, A.L. , Zhang, T.X. and Lin, J. (2012) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. [DOI] [PubMed] [Google Scholar]
- Delannoy, E. , Stanley, W.A. , Bond, C.S. and Small, I.D. (2007) Pentatricopeptide repeat (PPR) proteins as sequence‐specificity factors in post‐transcriptional processes in organelles. Biochem. Soc. Trans. 35, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Delaspre, F. , Nieto Penalver, C.G. , Saurel, O. , Kiefer, P. , Gras, E. , Milon, A. , Boucher, C. , Genin, S. and Vorholt, J.A. (2007) The Ralstonia solanacearum pathogenicity regulator HrpB induces 3‐hydroxy‐oxindole synthesis. Proc. Natl. Acad. Sci. USA, 104, 15 870–15 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denance, N. , Ranocha, P. , Oria, N. , Barlet, X. , Riviere, M.P. , Yadeta, K.A. , Hoffmann, L. , Perreau, F. , Clément, G. , Maia‐Grondard, A. , van den Berg, G.C. , Savelli, B. , Fournier, S. , Aubert, Y. , Pelletier, S. , Thomma, B.P. , Molina, A. , Jouanin, L. , Marco, Y. and Goffner, D. (2012) Arabidopsis wat1 (walls are thin1)‐mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross‐regulation of salicylic acid and tryptophan metabolism. Plant J. 73, 225–239. [DOI] [PubMed] [Google Scholar]
- Denny, T.P. and Baek, S.R. (1991) Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 4, 198–206. [Google Scholar]
- Deslandes, L. , Pileur, F. , Liaubet, L. , Camut, S. , Can, C. , Williams, K. , Holub, E. , Beynon, J. , Arlat, M. and Marco, Y. (1998) Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum . Mol. Plant–Microbe Interact. 11, 659–667. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulieres, F. , Hirsch, J. , Feng, D.X. , Bittner‐Eddy, P. , Holub, E. , Beynon, J. , Arlat, M. and Marco, Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digonnet, C. , Martinez, Y. , Denance, N. , Chasseray, M. , Dabos, P. , Ranocha, P. , Marco, Y. , Jauneau, A. and Goffner, D. (2012) Deciphering the route of Ralstonia solanacearum colonization in Arabidopsis thaliana roots during a compatible interaction: focus at the plant cell wall. Planta, 236, 1419–1431. [DOI] [PubMed] [Google Scholar]
- Elphinstone, J.G. (2005) The current Bacterial Wilt situation: a global overview In: Bacterial Wilt Disease and the Ralstonia Solanaceaum Species Complex (Allen C., Prior P. and Hayward A.C., eds), pp. 9–28. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Erhardt, M. , Namba, K. and Hughes, K.T. (2010) Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2, a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall, S. , Mercier, A. , Bertolla, F. , Calteau, A. , Gueguen, L. , Perriere, G. , Vogel, T.M. and Simonet, P. (2007) Horizontal gene transfer regulation in bacteria as a ‘spandrel’ of DNA repair mechanisms. PLoS ONE, 2, e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fegan, M. and Prior, P. (2005) How complex is the ‘Ralstonia solanacearum Species Complex’ In: Bacterial Wilt Disease and the Ralstonia Solanaceaum Species Complex (Allen C., Prior P. and Hayward A.C., eds), pp. 449–461. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Feng, D.X. , Deslandes, L. , Keller, H. , Revers, F. , Favery, B. , Lecomte, P. , Hirsch, J. , Olivier, J. and Marco, Y. (2004) Isolation and characterization of a novel Arabidopsis thaliana mutant unable to develop wilt symptoms after inoculation with a virulent strain of Ralstonia solanacearum . Phytopathology, 94, 289–295. [DOI] [PubMed] [Google Scholar]
- Feng, D.X. , Tasset, C. , Hanemian, M. , Barlet, X. , Hu, J. , Trémousaygue, D. , Deslandes, L. and Marco, Y. (2012) Biological control of bacterial wilt in Arabidopsis thaliana involves abscisic acid signalling. New Phytol. 194, 1035–1045. [DOI] [PubMed] [Google Scholar]
- Flavier, A.B. , Clough, S.J. , Schell, M.A. and Denny, T.P. (1997) Identification of 3‐hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum . Mol. Microbiol. 26, 251–259. [DOI] [PubMed] [Google Scholar]
- Gabriel, D.W. , Allen, C. , Schell, M. , Denny, T.P. , Greenberg, J.T. , Duan, Y.P. , Flores‐Cruz, Z. , Huang, Q. , Clifford, J.M. , Presting, G. , González, E.T. , Reddy, J. , Elphinstone, J. , Swanson, J. , Yao, J. , Mulholland, V. , Liu, L. , Farmerie, W. , Patnaikuni, M. , Balogh, B. , Norman, D. , Alvarez, A. , Castillo, J.A. , Jones, J. , Saddler, G. , Walunas, T. , Zhukov, A. and Mikhailova, N. (2006) Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant–Microbe Interact. 19, 69–79. [DOI] [PubMed] [Google Scholar]
- Garg, R.P. , Huang, J.Z. , Yindeeyoungyeon, W. , Denny, T.P. and Schell, M.A. (2000) Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum . J. Bacteriol. 182, 6659–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin, S. (2010) Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum . New Phytol. 187, 920–928. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Genin, S. , Gough, C.L. , Zischek, C. and Boucher, C.A. (1992) Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum . Mol. Microbiol. 6, 3065–3076. [DOI] [PubMed] [Google Scholar]
- Genin, S. , Brito, B. , Denny, T.P. and Boucher, C. (2005) Control of the Ralstonia solanacearum Type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. [DOI] [PubMed] [Google Scholar]
- Gillings, M. , Fahy, P. and Davies, C. (1993) Restriction analysis of an amplified polygalacturonase gene fragment differentiates strains of the phytopathogenic bacterium Pseudomonas solanacearum. Lett. Appl. Microbiol. 1, 44–48. [DOI] [PubMed] [Google Scholar]
- Godiard, L. , Sauviac, L. , Torii, K.U. , Grenon, O. , Mangin, B. , Grimsley, N.H. and Marco, Y. (2003) ERECTA, an LRR receptor‐like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36, 353–365. [DOI] [PubMed] [Google Scholar]
- Guidot, A. , Prior, P. , Schoenfeld, J. , Carrere, S. , Genin, S. and Boucher, C. (2007) Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidot, A. , Coupat, B. , Fall, S. , Prior, P. and Bertolla, F. (2009) Horizontal gene transfer between Ralstonia solanacearum strains detected by comparative genomic hybridization on microarrays. ISME J. 3, 549–562. [DOI] [PubMed] [Google Scholar]
- Hernandez‐Blanco, C. , Feng, D.X. , Hu, J. , Sanchez‐Vallet, A. , Deslandes, L. , Llorente, F. , Berrocal‐Lobo, M. , Keller, H. , Barlet, X. , Sánchez‐Rodríguez, C. , Anderson, L.K. , Somerville, S. , Marco, Y. and Molina, A. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell, 19, 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, J. , Deslandes, L. , Feng, D.X. , Balague, C. and Marco, Y. (2002) Delayed symptom development in ein2‐1, an Arabidopsis ethylene‐insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum . Phytopathology, 92, 1142–1148. [DOI] [PubMed] [Google Scholar]
- Ho, M.S. , Ou, C. , Chan, Y.R. , Chien, C.T. and Pi, H. (2008) The utility F‐box for protein destruction. Cell. Mol. Life Sci. 65, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Carney, B.F. , Denny, T.P. , Weissinger, A.K. and Schell, M.A. (1995) A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum . J. Bacteriol. 177, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Yindeeyoungyeon, W. , Garg, R.P. , Denny, T.P. and Schell, M.A. (1998) Joint transcriptional control of xpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR‐type transcriptional activator. J. Bacteriol. 180, 2736–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain, A. and Kelman, A. (1958) Relation of slime production to mechanism of wilting and pathogenicity of Pseudomonas solanacearum . Phytopathology, 48, 155–164. [Google Scholar]
- Jacobs, J.M. , Babujee, L. , Meng, F. , Milling, A. and Allen, C. (2012) The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. Mbio, 3, e00114–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kajava, A.V. , Anisimova, M. and Peeters, N. (2008) Origin and evolution of GALA‐LRR, a new member of the CC‐LRR subfamily: from plants to bacteria? PLoS ONE, 3, e1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda, A. , Yasukohchi, M. , Ohnishi, K. , Kiba, A. , Okuno, T. and Hikichi, Y. (2003) Ectopic expression of Ralstonia solanacearum effector protein PopA early in invasion results in loss of virulence. Mol. Plant–Microbe Interact. 16, 447–455. [DOI] [PubMed] [Google Scholar]
- Kang, Y.W. , Liu, H.L. , Genin, S. , Schell, M.A. and Denny, T.P. (2002) Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46, 427–437. [DOI] [PubMed] [Google Scholar]
- Kao, C.C. , Barlow, E. and Sequeira, L. (1992) Extracellular polysaccharide is required for wild‐type virulence of Pseudomonas solanacearum . J. Bacteriol. 174, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis, K.T. and Tiedje, J.M. (2005) Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA, 102, 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis, K.T. , Ramette, A. and Tiedje, J.M. (2006) The bacterial species definition in the genomic era. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 361, 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M. , Shillington, E. , Eguiluz, C. , Grimsley, N. and Boucher, C. (2002) PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host‐specificity factor and modulates aggressiveness of Ralstonia solanacearum . Mol. Plant–Microbe Interact. 15, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Lebeau, A. , Daunay, M.C. , Frary, A. , Palloix, A. , Wang, J.F. , Dintinger, J. , Chiroleu, F. , Wicker, E. and Prior, P. (2011) Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology, 101, 154–165. [DOI] [PubMed] [Google Scholar]
- Lebeau, A. , Gouy, M. , Daunay, M.C. , Wicker, E. , Chiroleu, F. , Prior, P. , Frary, A. and Dintinger, J. (2013) Genetic mapping of a major dominant gene for resistance to Ralstonia solanacearum in eggplant. Theor. Appl. Genet. 126, 143–158. [DOI] [PubMed] [Google Scholar]
- Li, L. , Atef, A. , Piatek, A. , Ali, Z. , Piatek, M. , Aouida, M. , Sharakuu, A. , Mahjoub, A. , Wang, G. , Khan, S. , Fedoroff, N.V. , Zhu, J.K. and Mahfouz, M.M. (2013) Characterization and DNA‐binding specificities of Ralstonia TAL‐like effectors. Mol. Plant doi: 10.1093/mp/sst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.R. , Zou, H.S. , Che, Y.Z. , Cui, Y.P. , Guo, W. , Zou, L.F. , Chatterjee, S. , Biddle, E.M. , Yang, C.H. and Chen, G.Y. (2011) A novel regulatory role of HrpD6 in regulating hrp‐hrc‐hpa genes in Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 24, 1086–1101. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. , Guidot, A. , Barberis, P. , Beuzon, C.R. and Genin, S. (2010) A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant–Microbe Interact. 23, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Mangin, B. , Thoquet, P. , Olivier, J. and Grimsley, N.H. (1999) Temporal and multiple quantitative trait loci analyses of resistance to bacterial wilt in tomato permit the resolution of linked loci. Genetics, 151, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F. , Yao, J. and Allen, C. (2011) A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J. Bacteriol. 193, 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole, B.M. , Baltrus, D.A. , Dangl, J.L. and Grant, S.R. (2007) Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15, 363–371. [DOI] [PubMed] [Google Scholar]
- Monteiro, F. , Genin, S. , van Dijk, I. and Valls, M. (2012a) A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology, 158, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Monteiro, F. , Sole, M. , van Dijk, I. and Valls, M. (2012b) A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum . Mol. Plant–Microbe Interact. 25, 557–568. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. and Iwabuchi, M. (2010) Genome‐wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant–Microbe Interact. 23, 251–262. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Deslandes, L. , Auriac, M.C. , Marco, Y. and Somssich, I.E. (2008) The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum . Plant J. 56, 935–947. [DOI] [PubMed] [Google Scholar]
- Murata, Y. , Tamura, N. , Nakaho, K. and Mukaihara, T. (2006) Mutations in the IrpE gene of Ralstonia solanacearum affects Hrp Pili production and virulence. Mol. Plant–Microbe Interact. 19, 884–895. [DOI] [PubMed] [Google Scholar]
- Occhialini, A. , Cunnac, S. , Reymond, N. , Genin, S. and Boucher, C. (2005) Genome‐wide analysis of gene expression in Ralstonia solanacearum reveals that the hrpB gene acts as a regulatory switch controlling multiple virulence pathways. Mol. Plant–Microbe Interact. 18, 938–949. [DOI] [PubMed] [Google Scholar]
- Orgambide, G. , Montrozier, H. , Servin, P. , Roussel, J. , Trigalet‐Demery, D. and Trigalet, A. (1991) High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI 1000 and the complete structure of the major polysaccharide. J. Biol. Chem. 266, 8312–8321. [PubMed] [Google Scholar]
- Plener, L. , Manfredi, P. , Valls, M. and Genin, S. (2010) PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum . J. Bacteriol. 192, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener, L. , Boistard, P. , Gonzalez, A. , Boucher, C. and Genin, S. (2012) Metabolic adaptation of Ralstonia solanacearum during plant infection: a methionine biosynthesis case study. PLoS ONE, 7, e36877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. and Genin, S. (2009) Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12, 44–52. [DOI] [PubMed] [Google Scholar]
- Poueymiro, M. , Cunnac, S. , Barberis, P. , Deslandes, L. , Peeters, N. , Cazale‐Noel, A.C. , Boucher, C. and Genin, S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host‐range specificity on tobacco. Mol. Plant–Microbe Interact. 22, 538–550. [DOI] [PubMed] [Google Scholar]
- Qian, Y.L. , Wang, X.S. , Wang, D.Z. , Zhang, L.N. , Zu, C.L. , Gao, Z.L. , Zhang, H.J. , Wang, Z.Y. , Sun, X.Y. and Yao, D.N. (2012) The detection of QTLs controlling bacterial wilt resistance in tobacco (N. tabacum L.). Euphitica, doi: 10.1007/s10681-012-0846-2. [DOI] [Google Scholar]
- Remenant, B. , Coupat‐Goutaland, B. , Guidot, A. , Cellier, G. , Wicker, E. , Allen, C. , Fegan, M. , Pruvost, O. , Elbaz, M. , Calteau, A. , Salvignol, G. , Mornico, D. , Mangenot, S. , Barbe, V. , Médigue, C. and Prior, P. (2010) Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics, 11, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant, B. , de Cambiaire, J.C. , Cellier, G. , Jacobs, J.M. , Mangenot, S. , Barbe, V. et al (2011) Ralstonia syzygii, the blood disease bacterium and some Asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS ONE, 6, e24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant, B. , Babujee, L. , Lajus, A. , Medigue, C. , Prior, P. and Allen, C. (2012) Sequencing of K60, type strain of the major plant pathogen Ralstonia solanacearum . J. Bacteriol. 194, 2742–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi, P. , Anisimova, M. , Guidot, A. , Genin, S. and Peeters, N. (2011) Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.J. , Edengreen, S.J. , Jones, P. and Ambler, D.J. (1990) Pseudomonas syzygii, sp‐nov, the cause of Sumatra disease of cloves. Syst. Appl. Microbiol. 13, 34–43. [Google Scholar]
- Robertson, A.E. , Wechter, W.P. , Denny, T.P. , Fortnum, B.A. and Kluepfel, D.A. (2004) Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial wilt incidence. Mol. Plant–Microbe Interact. 17, 1376–1384. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thébault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Schell, M.A. (2000) Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38, 263–292. [DOI] [PubMed] [Google Scholar]
- Schell, M.A. , Denny, T.P. and Huang, J. (1994) VsrA, a second two‐component sensor regulating virulence genes of Pseudomonas solanacearum . Mol. Microbiol. 11, 489–500. [DOI] [PubMed] [Google Scholar]
- Singer, A.U. , Rohde, J.R. , Lam, R. , Skarina, T. , Kagan, O. , Dileo, R. , Chirgadze, N.Y. , Cuff, M.E. , Joachimiak, A. , Tyers, M. , Sansonetti, P.J. , Parsot, C. and Savchenko, A. (2008) Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat. Struct. Mol. Biol. 15, 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole, M. , Popa, C. , Mith, O. , Sohn, K.H. , Jones, J.D. , Deslandes, L. and Valls, M. (2012) The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant–Microbe Interact. 25, 941–953. [DOI] [PubMed] [Google Scholar]
- Taghavi, M. , Hayward, C. , Sly, I. and Fegan, M. (1996) Analysis of the phylogenetic relationships of straons of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46, 10–15. [DOI] [PubMed] [Google Scholar]
- Tampakaki, A.P. , Skandalis, N. , Gazi, A.D. , Bastaki, M.N. , Sarris, P.F. , Charova, S.N. , Kokkinidis, M. and Panopoulos, N.J. (2010) Playing the ‘Harp’: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Xiao, Y. and Zhou, J.M. (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant–Microbe Interact. 19, 1159–1166. [DOI] [PubMed] [Google Scholar]
- Tans‐Kersten, J. , Huang, H.Y. and Allen, C. (2001) Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183, 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset, C. , Bernoux, M. , Jauneau, A. , Pouzet, C. , Briere, C. , Kieffer‐Jacquinod, S. , Rivas, S. , Marco, Y. and Deslandes, L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1‐R‐mediated immunity in Arabidopsis. PLoS Pathog. 6, e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet, P. , Olivier, J. , Sperisen, C. , Rogowsky, P. , Laterrot, H. and Grimsley, N. (1996a) Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol. Plant–Microbe Interact. 9, 826–836. [Google Scholar]
- Thoquet, P. , Olivier, J. , Sperisen, C. , Rogowsky, P. , Prior, P. , Anaïs, G. , Mangin, B. , Bazin, B. , Nazer, R. and Grimsley, N. (1996b) Polygenic resistance of tomato plants to bacterial wilt in the French West Indies. Mol. Plant–Microbe Interact. 9, 837–842. [Google Scholar]
- Torii, K.U. , Mitsukawa, N. , Oosumi, T. , Matsuura, Y. , Yokoyama, R. , Whittier, R.F. and Komeda, Y. (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine‐rich repeats. Plant Cell, 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, M. , Jauneau, A. , Genin, S. , Tavella, M.J. , Vailleau, F. , Gentzbittel, L. and Jardinaud, M.F. (2009) Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 150, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau, F. , Sartorel, E. , Jardinaud, M.F. , Chardon, F. , Genin, S. , Huguet, T. , Gentzbittel, L. and Petitprez, M. (2007) Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula . Mol. Plant–Microbe Interact. 20, 159–167. [DOI] [PubMed] [Google Scholar]
- Valls, M. , Genin, S. and Boucher, C. (2006) Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum . PLoS Pathog. 2, 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Linden, L. , Bredenkamp, J. , Naidoo, S. , Fouché‐Weich, J. , Denby, K.J. , Genin, S. , Marco, Y. and Berger, D.K. (2013) Gene‐for‐gene tolerance to bacterial wilt in Arabidopsis. Mol. Plant–Microbe Interact. 26, 398–406. [DOI] [PubMed] [Google Scholar]
- Vasse, J. , Frey, P. and Trigalet, A. (1995) Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 8, 241–251. [Google Scholar]
- Wairuri, C.K. , van der Waals, J.E. , van Schalkwyk, A. and Theron, J. (2012) Ralstonia solanacearum needs Flp pili for virulence on potato. Mol. Plant–Microbe Interact. 25, 546–556. [DOI] [PubMed] [Google Scholar]
- Wang, J.F. , Olivier, J. , Thoquet, P. , Mangin, B. , Sauviac, L. and Grimsley, N.H. (2000) Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain‐specific locus. Mol. Plant–Microbe Interact. 13, 6–13. [DOI] [PubMed] [Google Scholar]
- Wang, J.F. , Ho, F.I. , Truong, H.T.H. , Huang, S.M. , Balatero, C.H. , Dittapongpitch, V. and Hidayati, N. (2013a) Identification of major QTLs associated with stable resistance of tomato cultivar ‘Hawaii 7996’ to Ralstonia solanacearum . Euphytica, 190, 241–252. [Google Scholar]
- Wang, K. , Dang, F. , Liu, Z. , Wang, X. , Eulgem, T. , Lai, Y. , Yu, L. , She, J. , Shi, Y. , Lin, J. , Chen, C. , Guan, D. , Qiu, A. and He, S. (2013b) CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, E. , Grassart, L. , Coranson‐Beaudu, R. , Mian, D. , Guilbaud, C. , Fegan, M. and Prior, P. (2007) Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73, 6790–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, E. , Grassart, L. , Coranson‐Beaudu, R. , Mian, D. and Prior, P. (2009) Epidemiological evidence for the emergence of a new pathogenic variant of Ralstonia solanacearum in Martinique (French West Indies). Plant Pathol. 58, 853–861. [Google Scholar]
- Wicker, E. , Lefeuvre, P. , de Cambiaire, J.C. , Lemaire, C. , Poussier, S. and Prior, P. (2012) Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 6, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Zheng, H.J. , Liu, L. , Pan, Z.C. , Prior, P. , Tang, B. , Xu, J.S. , Zhang, H. , Tian, Q. , Zhang, L.Q. and Feng, J. (2011) Complete genome sequence of the plant pathogen Ralstonia solanacearum Strain Po82. J. Bacteriol. 193, 4261–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2006) Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum . J. Bacteriol. 188, 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2007) The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi, T. , Hikichi, Y. , Kiba, A. and Ohnishi, K. (2009a) The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signalling proteins. J. Bacteriol. 1, 3424–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi, T. , Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2009b) Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum . J. Gen. Plant Pathol. 75, 196–204. [Google Scholar]
- Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2011) prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS Microbiol. Lett. 317, 75–82. [DOI] [PubMed] [Google Scholar]
- Zolobowska, L. and Van Gijsegem, F. (2006) Induction of lateral root structure formation on petunia roots: a novel effect of GMI1000 Ralstonia solanacearum infection impaired in Hrp mutants. Mol. Plant–Microbe Interact. 19, 597–606. [DOI] [PubMed] [Google Scholar]
- Zou, L.F. , Wang, X.P. , Xiang, Y. , Zhang, B. , Li, Y.R. , Xiao, Y.L. , Wang, J.S. , Walmsley, A.R. and Chen, G.Y. (2006) Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72, 6212–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]


