Summary
The type III secretion system (T3SS) and exopolysaccharide (EPS) amylovoran are two essential pathogenicity factors in Erwinia amylovora, the causal agent of the serious bacterial disease fire blight. In this study, small molecules that inhibit T3SS gene expression in E. amylovora under hrp (hypersensitive response and pathogenicity)‐inducing conditions were identified and characterized using green fluorescent protein (GFP) as a reporter. These compounds belong to salicylidene acylhydrazides and also inhibit amylovoran production. Microarray analysis of E. amylovora treated with compounds 3 and 9 identified a total of 588 significantly differentially expressed genes. Among them, 95 and 78 genes were activated and suppressed by both compounds, respectively, when compared with the dimethylsulphoxide (DMSO) control. The expression of the majority of T3SS genes in E. amylovora, including hrpL and the avrRpt2 effector gene, was suppressed by both compounds. Compound 3 also suppressed the expression of amylovoran precursor and biosynthesis genes. However, both compounds induced significantly the expression of glycogen biosynthesis genes and siderophore biosynthesis, regulatory and transport genes. Furthermore, many membrane, lipoprotein and exported protein‐encoding genes were also activated by both compounds. Similar expression patterns were observed for compounds 1, 2 and 4. Using crab apple flower as a model, compound 3 was capable of reducing disease development in pistils. These results suggest a common inhibition mechanism shared by salicylidene acylhydrazides and indicate that small‐molecule inhibitors that disable T3SS function could be explored to control fire blight disease.
Introduction
Erwinia amylovora is the causative agent of fire blight, a bacterial disease of apples (Malus sylvestris) and pears (Pyrus communis L.). Fire blight is a devastating necrotic disease, resulting in serious economic losses to the apple and pear industry. In the USA alone, average regional losses and cost of fire blight control reach over $100 million annually (Norelli et al., 2003). Since the 1970s, streptomycin applications during bloom time in spring have been the most effective in controlling blossom blight. However, the occurrence of streptomycin‐resistant E. amylovora strains in Washington, New York, Oregon, Missouri, Michigan, Canada and elsewhere has rendered this antibiotic ineffective (Coyier and Covey, 1975; Loper et al., 1991; Manulis et al., 1998; McGhee and Sundin, 2011; Norelli et al., 2003; Russo et al., 2008; Shaffer and Goodman, 1985). Thus, alternative new strategies for the control of fire blight are needed to prevent severe losses in susceptible orchards (Kim et al., 2012).
The discovery of the hypersensitive response (HR) and pathogenicity (hrp) gene cluster, which encodes a type III secretion system (T3SS) common to Gram‐negative bacterial pathogens, is a breakthrough discovery in modern molecular plant bacteriology (He et al., 2004). Like many other Gram‐negative plant‐pathogenic bacteria, E. amylovora also contains an hrp‐T3SS that delivers effector proteins into host plants (Oh and Beer, 2005; Oh et al., 2005). In E. amylovora, the T3SS is required to elicit an HR on nonhost plants and to cause disease on susceptible host plants. The hrp gene cluster of E. amylovora is located within ∼60 kb of pathogenicity island 1 (PAI‐1) that includes T3SS regulatory genes (hrpL, hrpS and hrpXY), genes encoding structural components of T3SS and effectors (Oh and Beer, 2005; Oh et al., 2005; Zhao and Qi, 2011). Based on current knowledge, E. amylovora T3SS secretes at least 15 virulence‐associated proteins, including HrpA, HrpN, HrpW, HrpJ, HrpK, HopAK1 (Eop2), DspE, HopC1, HopX1 (Eop3), AvrRpt2 (Eop4) and Eop1 (EopB) (Bogdanove et al., 1998; Nissinen et al., 2007; Zhao et al., 2005, 2006).
Thus far, two pathogenicity factors, including T3SS and its effectors and the acidic exopolysaccharide (EPS) amylovoran, are required for E. amylovora to cause disease (Zhao and Qi, 2011; Zhao et al., 2009a). Amylovoran biosynthetic genes are located within a 12‐gene amylovoran biosynthetic (ams) operon, designated as amsA to amsL, with amsG as the first gene in the operon (Bernhard et al., 1993; Bugert and Geider, 1995). Two genes adjacent to the ams cluster, galE and galF, and located on the right are involved in amylovoran precursor formation. Amylovoran biosynthesis is strictly regulated by the Rcs phosphorelay system, which is also required for virulence (Wang et al., 2009, 2012; Zhao et al., 2009b). Additional determinants contribute to virulence and plant colonization of E. amylovora, including the extracellular EPS levan, protease, iron‐scavenging siderophore desferrioxamine and genes involved in sorbitol and sucrose metabolism (Aldridge et al., 1997; Bogs and Geider, 2000; Dellagi et al., 1998, 1999; Du and Geider, 2002; Khan et al., 2012; Zhao and Qi, 2011; Zhao et al., 2009b).
As a member of the family Enterobacteriaceae, E. amylovora is closely related to many mammalian pathogens, including Escherichia coli, Salmonella enterica, Shigella flexneri and Yersinia pestis, which also utilize T3SS to deliver effector proteins into their eukaryotic hosts (Büttner and He, 2009; He et al., 2004). The T3SS‐homologous macromolecular apparatus has been identified only in Gram‐negative bacteria and is a key virulence determinant, and many studies have conducted chemical screenings by targeting known T3SS processes, including gene expression, effector secretion and translocation, and symptom development (Baron, 2010; Keyser et al., 2008; Kline et al., 2011). This is based on the speculation that T3SS is a logical target for chemotherapeutic intervention, and disabling or blocking the function of T3SS could serve as a strategy for the control of bacterial diseases. These large‐scale chemical screening studies have led to the discovery of several classes of T3SS‐inhibiting compounds, including salicylidene acylhydrazides (Baron, 2010; Keyser et al., 2008; Kline et al., 2011). Subsequent studies have shown that salicylidene acylhydrazide family compounds broadly interfere with the functionality of T3SS in different mammalian pathogenic bacteria, including Yersinia pseudotuberculosis, S. enterica, Escherichia coli O157: H7, enteropathogenic Escherichia coli (EPEC), Shigella flexneri, Pseudomonas aeruginosa and Chlamydia spp. (Bailey et al., 2007; Hudson et al., 2007; Kauppi et al., 2003; Layton et al., 2010; Muschiol et al., 2006, 2009; Negrea et al., 2007; Nordfelth et al., 2005; Tree et al., 2009; Veenendaal et al., 2009; Wolf et al., 2006). Furthermore, salicylidene acylhydrazides protect mice from vaginal infection by Chlamydia trachomatis and Neisseria gonorrhoeae (Chu et al., 2010; Slepenkin et al., 2011).
Other classes of small‐molecule inhibitors of T3SS have also been reported (Felise et al., 2008; Kimura et al., 2011; Kline et al., 2009; Pan et al., 2009). Tethered thiazolidinone blocked T3SS protein secretion of Salmonella typhimurium and Yersinia enterocolitica, and also prevented HR when Pseudomonas syringae was co‐infiltrated into tobacco (Felise et al., 2008; Kline et al., 2009). Aurodox, a linear polyketide compound isolated from Streptomyces sp. K01‐0509, has been reported recently to inhibit the protein secretion of Citrobacter rodentium (Kimura et al., 2011). The oral administration of aurodox contributed to the survival of mice that had received a lethal dose of C. rodentium. However, it was unclear whether or not these small‐molecule inhibitors were also effective against plant‐pathogenic enterobacteria, such as E. amylovora.
One of the goals of this study was to determine whether or not small‐molecule inhibitors, such as salicylidene acylhydrazides, were effective in inhibiting T3SS gene expression in E. amylovora. Moreover, the global effects of salicylidene acylhydrazide compounds on the transcriptomic profiles of E. amylovora were evaluated. Finally, the efficacy of these compounds in controlling fire blight by either disabling or blocking of T3SS function during blossom infection was also investigated.
Results
Screening of small‐molecule inhibitors that suppress T3SS gene expression in vitro
To define in vitro growth conditions that induce T3SS gene expression in E. amylovora, two media [hrp‐inducing minimal medium (HMM) and M9] amended with different carbon sources, including galactose, glucose, mannitol and fructose, each at 10 and 20 mm, were used. The promoter activities of T3SS genes were determined by flow cytometry (Wang et al., 2009). HMM containing 20 mm galactose elicited the strongest inducing effect on T3SS gene promoter activities (data not shown; Fig. S1A, see Supporting Information) (Gaudriault et al., 1997). Moreover, following a time‐course study of hrp gene promoter activities in HMM containing 20 mm galactose, the expression of hrpL, hrpN, dspE and hrpA genes increased with time and reached their highest levels at 18 h (Fig. S1B). Thus, HMM containing 20 mm galactose was selected as the medium of choice, and bacteria were grown for 18 h at 18 °C in all subsequent chemical screening experiments.
Fifteen small‐molecule inhibitors were selected and tested for the suppression of the promoter activities of hrpN, dspE, hrpA and hrpL genes in E. amylovora. These compounds were selected as either they have been reported to inhibit T3SS gene expression in related enterobacterial pathogens (Felise et al., 2008; Pan et al., 2009; Tree et al., 2009) or they shared similar structures with T3SS inhibitor compounds (chemical compounds 9–12). Based on their abilities to inhibit the promoter activities of the four target T3SS genes, these compounds were divided into three groups. Chemical compounds 1, 2, 3 and 9 strongly inhibited the promoter activities of hrpN, dspE, hrpA and hrpL genes (Fig. 1). However, chemical compounds 4, 12, 14 and 16 had low to modest inhibitory effects on the promoter activities of one to four genes, whereas chemical compounds 5–8, 10, 11 and 15 had either minimal or no inhibitory effects on the promoter activities of these T3SS genes (Fig. 1). Thus, for the remainder of this study, experiments were conducted using chemical compounds 3 and 9, as compound 3 is the most potent in suppressing T3SS gene expression and compound 9 has not been reported previously.
Figure 1.
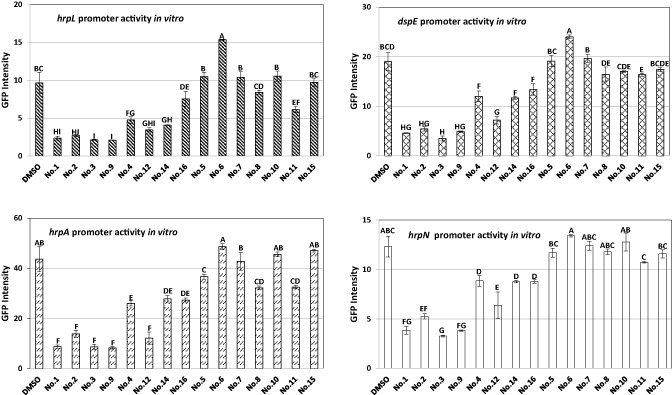
Effect of small‐molecule inhibitors on the promoter activities of hrpN, dspE, hrpL and hrpA genes in Erwinia amylovora strain Ea273. Bacterial strains were grown in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) in the presence of 100 μm of the corresponding compounds at 18 °C for 18 h. The equivalent volume of dimethylsulphoxide (DMSO) was added as a control. Error bars represent the standard deviation. One‐way analysis of variance (ANOVA) and Turkey's W‐test (P = 0.05) were carried out to determine the difference in the means using the SAS program.
Both chemical compounds 3 and 9 exhibited similar inhibitory effects on the promoter activities of the four target T3SS genes at different concentrations (Fig. S2, see Supporting Information). To eliminate the likelihood that these compounds resulted in growth defects of E. amylovora, bacterial growth was monitored in vitro in the presence of these two compounds under hrp‐inducing conditions, and no significant effects on E. amylovora growth were observed (Fig. S3A, see Supporting Information).
Effects of chemical compounds 3 and 9 on the transcriptome of E. amylovora
Microarray analysis was used to evaluate the transcriptomic profiles of E. amylovora following the treatment of bacterial cultures with compounds 3 and 9 relative to dimethylsulphoxide (DMSO), all grown in HMM containing 20 mm galactose (Fig. 2). A total of 588 genes were significantly differentially expressed, of which 320 and 268 genes were suppressed or induced by the compounds, respectively (Table S1, see Supporting Information). For treatment with compound 3, 251 genes were up‐regulated and 279 genes were down‐regulated, whereas, for compound 9 treatment, 112 genes were up‐regulated and 119 genes were down‐regulated when compared with DMSO treatment (Fig. 2, Table S1). Based on the combined results from both compounds, 78 genes were commonly suppressed and 95 genes were commonly induced by compounds 3 and 9 (Fig. 2).
Figure 2.
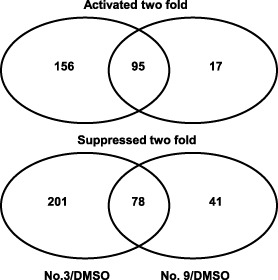
Venn diagrams showing the number of differentially expressed genes in compound 3‐ and 9‐treated samples relative to dimethylsulphoxide (DMSO) control. Differentially expressed genes were those that were activated (>twofold) or suppressed (<twofold) by compounds 3 (50 μm) and 9 (50 μm), respectively. Erwinia amylovora strain Ea273 was grown in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) at 18 °C for 18 h.
Overall, the expression of the majority of the 38 known T3SS genes was down‐regulated or suppressed by compound 3, and some were suppressed by compound 9, when compared with DMSO treatment (Table 1). These genes included regulatory genes (hrpL and hrpS), all known effectors (avrRpt2, hopC1, hopAk1, hopX1, dspE, eop2 and hrpW), chaperones (dspF, esc1 and esc3) and T3SS structural genes (Table 1). However, both hrpN and hrpA hybridization spots on the microarray were saturated, thus resulting in a ratio of 1.0 (Table 1). In addition, compound 3 suppressed the expression of 14 amylovoran precursor and biosynthesis genes (galEF and amsABCDEFGHIJKL) (Table 1). All ams genes also exhibited lower levels of expression following treatment with compound 9 when compared with DMSO treatment. These results indicated that both compounds inhibited T3SS and amylovoran biosynthesis genes. Furthermore, the membrane‐encoding protein operon (EAM_2938‐2935), reported to be positively regulated by HrpL and RcsBC (McNally et al., 2012; Wang et al., 2012), was also inhibited by compounds 3 and 9 (Table 1).
Table 1.
Suppression of type III secretion system (T3SS) and amylovoran biosynthesis genes after treatment with compounds 3 and 9
| Gene ID | Gene | Compound 3/DMSOa | Compound 9/DMSOa | Protein description |
|---|---|---|---|---|
| EAM_0423 | avrRpt2(eop4) | 0.27 | 0.38 | Cysteine protease protein AvrRpt2 |
| EAM_2190 | hopX1(eop3) | 0.24 | 0.55b | Type III effector protein HopC1 |
| EAM_2697 | hopC1 | 0.31 | 0.41 | Putative type III effector protein |
| EAM_2780 | hopAK1(eop2) | 0.23 | 0.52 | Type III effector (pectin lyase) |
| EAM_2871 | dspF | 0.27 | 0.89b | Putative avirulence protein |
| EAM_2872 | dspE | 0.28 | 0.73 | Type III effector protein |
| EAM_2873 | hrpW | 0.37 | 0.70b | Putative pectate lyase |
| EAM_2874 | esc3(orfC) | 0.37 | 0.63b | Chaperone |
| EAM‐2875 | eop1(orfB) | 0.38 | 0.62b | Effector protein Eop1 |
| EAM_2876 | esc1(orfA) | 0.26 | 0.43 | Chaperone for Eop1 |
| EAM_2877 | hrpN | 0.92 b | 1.04 b | Harpin |
| EAM_2878 | hrpV | 0.21 | 0.44 | Type III secretion system protein |
| EAM_2879 | hrpT | 0.25 | 0.54b | Type III secretion system protein |
| EAM_2880 | hrcC | 0.22 | 0.42 | Type III secretion system protein |
| EAM_2881 | hrpG | 0.28 | 0.56 | Type III secretion system protein |
| EAM_2882 | hrpF | 0.18 | 0.43 | Type III secretion system protein |
| EAM_2883 | hrpE | 0.19 | 0.40 | Type III secretion system protein |
| EAM_2884 | hrpD | 0.17 | 0.32 | Type III secretion system protein |
| EAM_2885 | hrcJ | 0.24 | 0.44 | Type III secretion system protein |
| EAM_2886 | hrpB | 0.39 | 0.64 | Type III secretion system protein |
| EAM_2887 | hrpA | 0.77 | 1.11 b | Type III secretion system protein |
| EAM_2891 | hrpS | 0.41 | 0.97b | Sigma 54 enhancer‐binding protein |
| EAM_2894 | hrpL | 0.16 | 0.28 | Sigma factor HrpL |
| EAM_2895 | hrpJ | 0.19 | 0.37 | Type III secretion system protein |
| EAM_2896 | hrpI | 0.18 | 0.37 | Type III secretion system protein |
| EAM_2897 | hrpQ | 0.26 | 0.53 | Type III secretion system protein |
| EAM_2898 | hrcN | 0.21 | 0.41 | Type III secretion system protein |
| EAM_2899 | hrpO | 0.30 | 0.47 | Type III secretion system protein |
| EAM_2900 | hrpP | 0.25 | 0.53b | type III secretion system protein |
| EAM_2901 | hrcQ | 0.18 | 0.48 | type III secretion system protein |
| EAM_2902 | hrcR | 0.23 | 0.53b | type III secretion system protein |
| EAM_2903 | hrcS | 0.17 | 0.44 | type III secretion system protein |
| EAM_2904 | hrcT | 0.32 | 0.57 | type III secretion system protein |
| EAM_2905 | hrcU | 0.19 | 0.45 | type III secretion system protein |
| EAM_2908 | hsvC | 0.06 | 0.35 | Conserved hypothetical protein |
| EAM_2909 | hsvB | 0.04 | 0.22 | Conserved hypothetical protein |
| EAM_2910 | hsvA | 0.06 | 0.30 | Putative amidinotransferase |
| EAM_2911 | hrpK | 0.39 | 0.47 | Type III secretion system protein |
| EAM_2161 | galE | 0.42 | 0.94b | UDP‐glucose 4‐epimerase |
| EAM_2162 | galF | 0.33 | 0.77b | UTP‐glucose‐1‐phosphate uridylyltransferase |
| EAM_2163 | amsL | 0.38 | 0.85b | Amylovoran biosynthesis protein |
| EAM_2164 | amsK | 0.26 | 0.56 | Glycosyltransferase |
| EAM_2165 | amsJ | 0.33 | 0.69b | Amylovoran biosynthesis protein |
| EAM_2166 | amsF | 0.45 | 0.98b | Amylovoran biosynthesis protein |
| EAM_2167 | amsE | 0.26 | 0.65 | Amylovoran glycosyltransferase |
| EAM_2168 | amsD | 0.30 | 0.65 | Glycosyltransferase |
| EAM_2169 | amsC | 0.34 | 0.84b | Oligosaccharide repeat unit polymerase |
| EAM_2170 | amsB | 0.28 | 0.71 | Glycosyltransferase |
| EAM_2171 | amsA | 0.24 | 0.69b | Tyrosine‐protein kinase |
| EAM_2172 | amsI | 0.24 | 0.68b | Protein‐tyrosine‐phosphatase |
| EAM_2173 | amsH | 0.28 | 0.72b | Amylovoran export protein |
| EAM_2174 | amsG | 0.34 | 0.93b | UDP‐galactose‐lipid carrier transferase |
| EAM_2935 | 0.29 | 0.53 | γ‐Glutamyltranspeptidase | |
| EAM_2936 | 0.15 | 0.36 | Putative exported protein | |
| EAM_2937 | 0.26 | 0.5 | Putative disulphide bond formation membrane protein | |
| EAM_2938 | 0.38 | 0.62 | Putative membrane protein |
Expression ratio ≤0.5 indicates genes were suppressed in compound treatments.
P > 0.05; all others with P < 0.05.
Bold type indicates that corresponding spots were saturated in the array.
Among the up‐regulated genes, five glycogen metabolism genes (glgABCPX) and 13 exogenous iron uptake system genes, including siderophore desferrioxamine E biosynthesis (dfoJAC), siderophore‐interacting protein (EAM_3350), TonB‐dependent ferrioxamine receptors (foxR, EAM_1726), TonB‐dependent receptor complex (tonB, exbBD) and TonB‐dependent transporter genes [haemin ABC transporter (hmuVUTS)], were highly induced by both compounds (Table 2). In addition, yet another 35 genes were commonly up‐regulated by both compounds, and these encoded membrane, lipoprotein, exported proteins and oxidative stress‐related proteins and transporters (Table 2). Furthermore, some iron–sulphur cluster genes (sufAC) were also induced by compound 3 (Table S1), suggesting that this compound might also interfere with endogenous iron function.
Table 2.
Genes activated by compounds 3 and 9
| Gene ID | Gene | Compound 3/DMSO* | Compound 9/DMSO* | Protein description |
|---|---|---|---|---|
| EAM_3268 | glgP | 5.58 | 3.32 | Glycogen phosphorylase |
| EAM_3269 | glgA | 6.91 | 3.24 | Glycogen synthase |
| EAM_3270 | glgC | 7.18 | 4.03 | Glucose‐1‐phosphate adenylyltransferase |
| EAM_3271 | glgX | 5.06 | 2.43 | Glycogen debranching enzyme |
| EAM_3272 | glgB | 7.34 | 3.75 | 1,4‐α‐Glucan branching enzyme |
| EAM_0358 | foxR | 13.17 | 7.35 | Ferrioxamine TonB‐dependent receptor |
| EAM_0359 | dfoC | 60.30 | 43.63 | Decarboxylase |
| EAM_0360 | dfoA | 236.45 | 133.50 | Siderophore biosynthesis protein |
| EAM_0361 | dfoJ | 214.85 | 120.14 | Siderophore biosynthesis |
| EAM_1639 | hmuV | 59.27 | 37.76 | Haemin ABC transporter |
| EAM_1640 | hmuU | 64.50 | 46.38 | Haemin ABC transporter |
| EAM_1641 | hmuT | 118.68 | 73.26 | Haemin ABC transporter |
| EAM_1642 | hmuS | 99.19 | 71.24 | Haemin ABC transporter |
| EAM_1726 | 32.4 | 4.87 | TonB‐dependent receptor | |
| EAM_1888 | tonB | 2.92 | 4.02 | TonB protein |
| EAM_2955 | exbD | 4.53 | 4.44 | Biopolymer transport protein |
| EAM_2956 | exbB | 6.20 | 6.13 | Biopolymer transport protein |
| EAM_3350 | 15.4 | 22.4 | Putative siderophore‐interacting protein | |
| EAM_2594 | nrdH | 18.3 | 10.5 | Glutaredoxin‐like protein |
| EAM_2595 | nrdI | 15.3 | 9.1 | Putative flavodoxin |
| EAM_2596 | nrdE | 9.1 | 5.72 | Ribonucleoside‐diphosphate reductase 2 α chain |
| EAM_2597 | nrdF | 5.7 | 4.72 | Ribonucleoside‐diphosphate reductase 2 β chain |
| EAM_0425 | blc | 3.46 | 2.03 | Outer membrane lipoprotein |
| EAM_0561 | 2.15 | 2.03 | Putative exported protein | |
| EAM_0565 | 2.11 | 2.0 | Putative membrane protein | |
| EAM_0611 | osmY | 3.0 | 2.41 | Osmotically inducible protein Y |
| EAM_0612 | 2.11 | 2.0 | Putative membrane protein | |
| EAM_0624 | 6.16 | 2.65 | Putative lipoprotein | |
| EAM_9625 | 5.15 | 2.61 | Putative polypeptide transport protein | |
| EAM_0893 | 6.86 | 2.60 | Putative exported protein | |
| EAM_0927 | 7.55 | 3.65 | Putative membrane protein | |
| EAM_0965 | 10.0 | 3.75 | Putative lipoprotein | |
| EAM_0966 | 8.43 | 3.79 | Putative phospholipid‐binding protein | |
| EAM_1076 | osmC | 7.79 | 2.69 | Peroxiredoxin (osmotically‐inducible protein C) |
| EAM_1077 | 3.26 | 9.37 | Putative membrane protein | |
| EAM_1078 | 3.45 | 12.4 | Putative membrane protein | |
| EAM_1086 | 2.40 | 2.34 | Putative lipoprotein | |
| EAM_1190 | 17.7 | 3.77 | Putative exported protein | |
| EAM_1231 | 9.74 | 5.19 | Putative membrane protein | |
| EAM_1543 | 8.05 | 3.62 | Putative lipoprotein | |
| EAM_1544 | 9.17 | 3.82 | Putative exported protein | |
| EAM_1616 | osmE | 3.40 | 2.14 | Osmotically inducible lipoprotein E |
| EAM_1617 | 6.47 | 10.6 | Putative membrane protein | |
| EAM_1958 | osmB | 4.38 | 1.49 | Osmotically inducible lipoprotein B |
| EAM_1984 | 6.84 | 2.77 | Putative membrane protein | |
| EAM_2038 | 3.46 | 2.04 | Putative membrane protein | |
| EAM_2057 | 5.74 | 7.49 | Putative peroxidase | |
| EAM_2058 | 5.96 | 7.4 | Putative exported protein | |
| EAM_2059 | 6.7 | 7.98 | Putative permease | |
| EAM_2467 | prt1 | 4.91 | 2.82 | Extracellular metalloprotease |
| EAM_2941 | 20.2 | 19.2 | Putative ABC transporter | |
| EAM_3038 | 4.12 | 2.03 | Putative exported protein | |
| EAM_3377 | 5.93 | 3.20 | Putative exported protein |
*Expression ratio ≥2.0 indicates genes were activated in compound treatments; P < 0.05.
Validation of microarray data by quantitative real‐time polymerase chain reaction (qRT‐PCR)
To validate the results of the microarray data, qRT‐PCR was carried out for 10 selected genes, including hrpA, dspE, hrpL, hrpN, avrRpt2, amsG, amsD, glgB, foxR and EAM_3350. The results of qRT‐PCR were mostly consistent with those of microarray analysis, except for hrpN and hrpA, which showed strong inhibition in qRT‐PCR analysis (Fig. 3). The expression of hrpA in qRT‐PCR for compounds 3 and 9 was suppressed by 0.13‐ and 0.66‐fold, respectively, whereas the expression of hrpN was suppressed by fold changes of 0.07 and 0.29, respectively.
Figure 3.
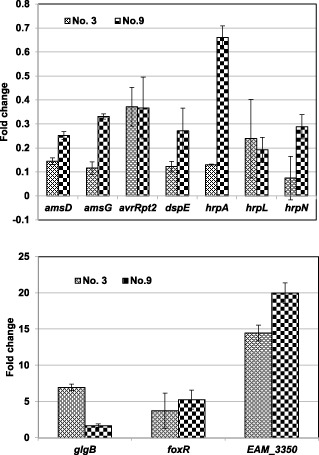
Verification of microarray data by quantitative real‐time polymerase chain reaction (qRT‐PCR). The relative fold change of each gene was derived from the comparison of the compound‐treated versus dimethylsulphoxide (DMSO)‐treated Erwinia amylovora strain Ea273 in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) for compounds 3 and 9 using qRT‐PCR. The 16S rDNA (rrsA) gene was used as a control. The values of the relative fold change were means of three replicates. The experiments were repeated three times with similar results. Error bars indicate the standard deviation.
Other small‐molecule inhibitors exhibit similar gene expression patterns
qRT‐PCR analysis was also performed to evaluate the influence of compounds 1, 2 and 4 on the expression levels of selected target genes, including four hrp genes (hrpA, hrpN, hrpL and dspE), an amylovoran biosynthesis gene (amsG) and two iron uptake genes (EAM_359 and hmuS) in E. amylovora. Similar to the results obtained with compounds 3 and 9, all three compounds, 1, 2 and 4, suppressed both T3SS and ams gene expression, but induced the expression of iron acquisition genes, when compared with DMSO treatment (Fig. 4). For example, hrpL expression levels exhibited fold changes of 0.51, 0.20, 0.24, 0.7 and 0.19 for compounds 1, 2, 3, 4 and 9, respectively. In contrast, expression of the haemin transport protein‐encoding gene hmuS exhibited fold changes of 52.27, 5.09, 50.12, 9.99 and 22.91 for compounds 1, 2, 3, 4 and 9, respectively. These results suggest that the suppression of either T3SS or ams expression, together with the induction of iron uptake gene expression, may represent a common transcriptional effect of these compounds.
Figure 4.
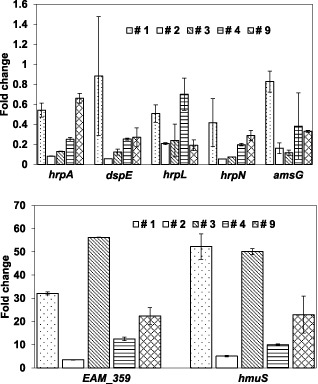
Effect of salicylidene acylhydrazide inhibitors on virulence gene expression. Relative fold changes were determined by quantitative real‐time polymerase chain reaction (qRT‐PCR) for hrpA, dspE, hrpL, hrpN, amsG, EAM_359 and hmuS genes. Erwinia amylovora Ea273 was grown in the presence of compounds 1–4 and 9 (50 μm) in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) at 18 °C for 18 h. The 16S rDNA (rrsA) gene was used as a control. The values of the relative fold change were means of three replicates. The experiments were repeated three times with similar results. Error bars indicate the standard deviation.
Small‐molecule inhibitors suppress amylovoran production
To determine whether small‐molecule inhibitors influence the production of amylovoran, a cetylpyrimidinium chloride (CPC) assay was performed as described previously (Zhao et al., 2009b). At a concentration of 25 μm, the optical densities at 600 nm (OD600), which reflect amylovoran production, were 1.09, 1.09, 0.99, 1.19 and 1.32 for compounds 1, 2, 3, 4 and 9, respectively, when compared with DMSO (1.40) (Fig. 5). This indicates that all of these chemical compounds suppress amylovoran production slightly, although this is not statistically significant for compound 9 relative to DMSO treatment.
Figure 5.
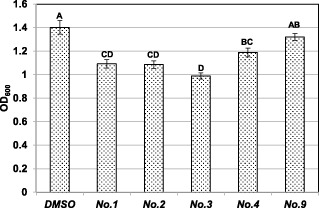
Effect of small‐molecule inhibitors on amylovoran production. Erwinia amylovora strain Ea273 was grown in MBMA medium with 1% sorbitol at 28 °C for 24 h with shaking. Chemical compounds and an equal volume of dimethylsulphoxide (DMSO) were added to the cell culture at a final concentration of 25 μm. Amylovoran concentrations were measured by the cetylpyrimidinium chloride (CPC) method and normalized to a cell density of unity. Data points represent the means of three replicates ± standard deviations. Each experiment was performed at least three times with similar results.
Compound 3 delays disease progress on crab apple flowers
To determine whether small‐molecule inhibitors could prevent fire blight disease development on stigmas, a crab apple flower assay was carried out as described previously (Pusey, 1997). As chemical compound 3 is the most potent in suppressing T3SS gene expression, it was selected for this experiment. Six days post‐inoculation, a significant difference in symptom development in pistils was observed for compound 3‐treated flowers relative to treatment with DMSO alone (Fig. 6). Pistils of flowers inoculated with a dspE mutant of E. amylovora, which is nonpathogenic, remained healthy and showed the least amount of necrosis at the inoculation site. Similarly, compound 3‐treated flowers exhibited slight necrotic symptoms. In contrast, flowers inoculated with either the wild‐type (WT) strain alone or the WT strain treated with DMSO exhibited heavy necrosis along entire pistils (Fig. 6).
Figure 6.

Virulence assay on crab apple (Malus mandshurica) flowers. Symptoms of crab apple flowers at 6 days post‐inoculation with Erwinia amylovora strain Ea273, dspE mutant, Ea273 treated with dimethylsulphoxide (DMSO) or compound 3 at 50 μm. Photographs were taken at 6 days post‐inoculation. The experiments were repeated three times with similar results. Arrow indicates representative inoculation site (stigma) and symptoms below on style and ovary.
To investigate whether compound 3 also affected E. amylovora growth in vivo, bacterial growth was determined using dilution plating. Two and four days post‐inoculation, bacterial populations were found to be comparable between DMSO‐ and compound 3‐treated flowers. By six days post‐inoculation, compound 3‐treated flowers showed about a two‐fold reduction in bacterial population when compared with those inoculated with WT treated with DMSO alone, although this difference was not statistically significant (Fig. S3B).
We also tested whether these compounds could inhibit the elicitation of HR by E. amylovora in tobacco. Surprisingly, none of the 15 compounds inhibited HR when infiltrated with E. amylovora strain Ea273 at levels of 10–100 μm. When E. amylovora strain Ea1189 was used, at 50 μm or higher, three compounds (6, 8 and 11) inhibited HR development at 24 h (Fig. S4, see Supporting Information; data not shown for compound 6); however, normal HR was observed at 48 h for all compounds at all levels.
Discussion
As a landmark discovery, advances in the study of bacterial virulence factors have provided mounting evidence that T3SS is a potent virulence mechanism shared by a broad spectrum of pathogenic Gram‐negative bacteria that infect both plant and mammalian hosts (He et al., 2004). Thus, the T3SS apparatus is essential for bacterial cells to evade host immune defenses, and T3SS could serve as a universal target for the development of novel antibacterial agents (Baron, 2010; Kline et al., 2011). It is likely that agents that inhibit T3SS can result in an antibacterial response without killing bacteria (Keyser et al., 2008). In this study, 15 chemical compounds known to either act or share similar structures to those reported as T3SS inhibitors were evaluated for their suppression of T3SS gene expression in E. amylovora. Among these, four compounds were found to either strongly or moderately suppress T3SS gene expression. Two of these compounds (9 and 12) have not been reported previously to act as T3SS inhibitors. Moreover, five compounds (5–8 and 15), reported previously to inhibit T3SS in other bacteria, were not found to be effective against E. amylovora.
High‐throughput screening is a powerful tool in the identification of small‐molecule inhibitors that disable T3SS function. Recent studies have identified several classes of synthetic compounds, as well as natural products, as active T3SS inhibitors in a wide range of Gram‐negative bacterial pathogens, including Escherichia coli, Salmonella, Yersinia, Shigella and Chlamydia (Baron, 2010; Gauthier et al., 2005; Keyser et al., 2008; Kline et al., 2011; Muschiol et al., 2006). These inhibitors include salicylanilides, salicylidene anilides, salicylidene acylhydrazides, thiazolidinone, sulphonylaminobenzanilides, caminosides and guadinomines, and are likely to target T3SS gene expression, conserved outer membrane proteins, effector secretion and needle assembly. Pan et al. (2009) have also described a group of T3SS‐inhibiting compounds of diverse chemical structures that are distinct from all previously reported T3SS inhibitors.
In this study, all compounds that suppressed the expression of T3SS genes in E. amylovora under hrp‐inducing conditions belonged to the salicylidene acylhydrazides, except for one compound: 16. This is the largest family of small‐molecule inhibitors identified thus far (Keyser et al., 2008; Wang et al., 2011). Recent evidence suggests that this group of compounds interferes with various T3SS functions, including gene expression, effector secretion or translocation, and needle assembly (Kauppi et al., 2003; Muschiol et al., 2006, 2009; Veenendaal et al., 2009; Wolf et al., 2006). In this study, the suppression of T3SS gene expression by compounds 3 and 9 was found to be dose dependent in E. amylovora, and similar results were observed for all five compounds (1–4 and 9) tested for their effect on gene expression.
Global gene expression profiles for compounds 3 and 9 have indicated that the transcriptional profiling for E. amylovora is highly similar to that reported for Escherichia coli O157: H7 and S. typhimurium after salicylidene acylhydrazide treatment (Layton et al., 2010; Tree et al., 2009). Previously, it has been demonstrated that salicylidene acylhydrazides suppress the transcription of all T3SS genes of Escherichia coli O157: H7 and S. typhimurium (Layton et al., 2010; Tree et al., 2009). In this study, compounds 3 and 9 also exhibited strong inhibition of all known T3SS genes in E. amylovora. It is important to note that this is the first report of the T3SS inhibitory effect of compound 9. These findings indicate that salicylidene acylhydrazides broadly inhibit T3SS gene expression not only in mammalian pathogens, but also in plant‐pathogenic enterobacteria. Furthermore, this suggests that the suppression of T3SS by salicylidene acylhydrazide is a conserved transcriptional response among different bacterial species. However, other compounds, such as 6, 7 and 8, also inhibit T3SS in some organisms (Felise et al., 2008, Pan et al., 2009), but are not effective in E. amylovora, thus suggesting that these compounds are likely to have specific targets.
Amylovoran is another essential pathogenicity factor of E. amylovora (Zhao et al., 2009a, b). Transcriptomic analysis has revealed that salicylidene acylhydrazides also inhibit amylovoran production and amylovoran biosynthesis gene expression, but promote glycogen gene expression. These findings demonstrate, for the first time, that salicylidene acylhydrazides may also target other bacterial virulence factors in addition to T3SS. It has been reported that salicylanilide class compounds inhibit two‐component signal transduction systems in bacteria (Macielag et al., 1998). In E. amylovora, the RcsBCD phosphorelay two‐component system plays an essential role in controlling amylovoran and glycogen production, as well as virulence (Wang et al., 2009, 2012). Therefore, it is likely that salicylidene acylhydrazides may act on the RcsBCD system as well.
Another unique feature of the salicylidene acylhydrazide‐induced transcriptome is the activation of the iron uptake system. Layton et al. (2010) have observed that about one‐quarter of genes in S. typhimurium regulated significantly following treatment with 100 μm INP0403 (compound 2 in this study) are genes involved in iron acquisition, including the TonB‐dependent receptor complex (tonB, exbBD), ABC ferric transporter (fepABCDEG), iron transport (feoAB, fhuABCDEF) and iron uptake regulator (fur). Similarly, in this study, most of the activated genes of the E. amylovora transcriptome by compounds 3 and 9 were exogenous iron uptake genes, including genes involved in siderophore desferrioxamine E biosynthesis (dfoJAC), siderophore‐interacting protein (EAM_3350), TonB‐dependent ferrioxamine receptors (foxR, EAM_1726), TonB‐dependent receptor complex (tonB, exbBD), TonB‐dependent transporter genes [haemin ABC transporter (hmuVUTS)] and endogenous iron–sulphur assembly genes (sufAC). These findings further suggest that a common mechanism may be shared by salicylidene acylhydrazides in regulating bacterial transcription. However, both compounds inhibit the expression of the ftnA gene (EAM_2940) by more than five‐fold (Table S1). Ferritin, encoded by ftnA, is a primary intracellular iron storage protein that stores iron and contributes to its controlled release. Reduced levels of ferritin proteins result in reduced iron storage. This indicates that iron uptake/release may be fundamental in protecting bacteria from salicylidene acylhydrazides. As siderophore desferrioxamine is a virulence factor in E. amylovora (Dellagi et al., 1998, 1999), this new finding may suggest a unique role for siderophores in protecting bacteria from stress conditions, in addition to their role in virulence.
Both compounds 9 and 12 suppressed the expression of a membrane‐encoding protein operon (EAM_2938‐2935) (Table 1), previously reported to be positively regulated by HrpL and RcsBC, and to be a novel virulence factor (McNally et al., 2012; Wang et al., 2012). Interestingly, both compounds also activated 35 genes that encode for membrane, lipoprotein, exported proteins, oxidative stress‐related proteins (such as OsmC) and ABC transporters (Table 2). However, the precise role of these membrane and lipoprotein‐encoding genes remains unknown. Recently, thiol peroxidase Tpx and oxidoreductase WrbA have been shown to be putative targets of salicylidene acylhydrazide compounds, and conformational changes of Tpx between reduced and oxidized states affect the binding ability to these compounds (Gabrielsen et al., 2012; Wang et al., 2011). However, in this study, there were no observed differences in expression of tpx and other peroxiredoxin genes, such as ahpC and sodA. However, several genes encoding redox proteins and central metabolism, such as fpr (EAM_0114), encoding ferredoxin‐NADP reductase, the nuo operon (nuoABCEFGHIJKLMN), encoding the energy‐conserving NADH dehydrogenase, the succinate dehydrogenase operon (sdhABCD) and the succinyl‐CoA synthetase operon (sucABCD), are highly suppressed either by both compounds or by compound 3 (Table S1). Recruitment of ‘free’ ferrous iron has been found to generate destructive hydroxyl radicals through the Fenton reaction. The ferrous iron can also be released from Fe–S clusters when superoxide production is induced. As Tpx and WrbA are scavengers of reactive oxygen species (ROS), and alterations in central metabolism related to NADP consumption are critical for superoxide‐mediated Fe–S cluster destabilization and for the stimulation of the Fenton reaction (Davies et al., 2009; Dwyer et al., 2007; Kohanski et al., 2010), it is likely that salicylidene acylhydrazides may increase iron uptake, which, in turn, promotes the formation of ROS. Recent studies have also suggested that ROS can regulate both the structure and function of a variety of biomolecules by altering their oxidation–reduction (redox) states (Fang, 2011). Thus, it is tempting to speculate that exposure to these small molecules could also result in ROS that act as redox signals, thus disrupting T3SS function and amylovoran biosynthesis. Future analysis is necessary to fully address the mode of action of these salicylidene acylhydrazides in inhibiting T3SS functions.
In the spring, blossom blight infection is the main source of dissemination of E. amylovora, and the pathogen must establish large epiphytic populations on stigmas of flowers for further successful infection of host plants. The control of fire blight is achieved mainly following the application of antibiotics to flowers to suppress pathogen infection. Using crab apple flowers as a model, we have shown that compound 3 is capable of reducing disease development in pistils. These results suggest that small‐molecule inhibitors that disable T3SS function could be useful in the control of fire blight disease.
Experimental Procedures
Bacterial strains and growth media
The bacterial strains and plasmids used in this study are listed in Table 3. Luria–Bertani (LB) medium was routinely used for the culture of E. amylovora. Amylovoran production was determined by growing bacterial cells in MBMA medium (3 g KH2PO4, 7 g K2HPO4, 1 g [NH4]2SO4, 2 mL glycerol, 0.5 g citric acid and 0.03 g MgSO4) amended with 1% sorbitol (Zhao et al., 2009b). Bacterial strains were also grown in HMM (1 g [NH4]2SO4, 0.346 g MgCl2.6H2O, 0.099 g NaCl, 8.708 g K2HPO4 and 6.804 g KH2PO4) or M9 minimal medium (12.8 g Na2HPO4.7H2O, 3.0 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 0.24 g MgSO4 and 0.011 g CaCl2) to activate T3SS gene expression by the addition of carbon sources as indicated. When necessary, the following antibiotics were added to the medium: 50 μg/mL kanamycin (Km), 100 μg/mL ampicillin (Ap) and 50 μg/mL rifampicin (Rif).
Table 3.
Bacterial strains, plasmids and small‐molecule inhibitors used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Erwinia amylovora | ||
| Ea273 | Wild‐type, isolated from apple | Wang et al. (2010) |
| Ea1189 | Wild‐type, isolated from apple | Burse et al. (2004) |
| Ea273a | Rifampicin‐resistant derivative of Ea273, RifR | This study |
| ΔdspE | dspE::Km; KmR ‐insertional mutant of dspE of Ea1189, KmR | Zhao et al., 2009b |
| Plasmids | ||
| pFPV25 | ApR , GFP‐based promoter trap vector containing a promoterless gfpmut3a gene | Valdivia and Falkow (1997) |
| pHrpN‐GFP | 735‐bp EcoRI‐BamHI DNA fragment containing promoter sequence of hrpN gene in pFPV25 | This study |
| pHrpA‐GFP | 708‐bp EcoRI‐BamHI DNA fragment containing promoter sequence of hrpA gene in pFPV25 | This study |
| pZW2 | 608‐bp KpnI‐XbaI DNA fragment containing promoter sequence of hrpL gene of Ea273 in pFPV25 | Wang et al. (2010) |
| pZW3 | 570‐bp SmaI DNA fragment containing promoter sequence of dspE gene of Ea273 in pFPV25 in forward orientation | Wang et al. (2010) |
| Small‐molecule inhibitors | ||
| Compound 1 | ME0052, INP0010 | Nordfelth et al. (2005) |
| Compound 2 | ME0053, INP0403 | Nordfelth et al. (2005) |
| Compound 3 | ME0054, INP0401 | Nordfelth et al. (2005) |
| Compound 4 | ME0055, INP0031 | Nordfelth et al. (2005) |
| Compound 5 | INP0002 | Negrea et al. (2007) |
| Compound 6 | Compound 1, TTS29 | Felise et al. (2008) |
| Compound 7 | Compound 2, CAS #3530‐35‐6 | Pan et al. (2009) |
| Compound 8 | Compound 3, CAS #27885‐92‐3 | Pan et al. (2009) |
| Compound 9 | 5277768 | This study |
| Compound 10 | 5472847 | This study |
| Compound 11 | 5551447 | This study |
| Compound 12 | 5569869 | This study |
| Compound 14 | INP0007 | Negrea et al. (2007) |
| Compound 15 | Compound 1, CAS #329057‐04‐7 | Pan et al. (2009) |
| Compound 16 | Compound 3‐didiproprionate, CAS #55750‐06‐6 | Pan et al. (2009) |
ApR, ampicillin resistance; CAS, Chemical Abstracts Service; GFP, green fluorescent protein; KmR, kanamycin resistance; RifR, rifampicin resistance. Compounds 9–12 are from ChemBridge and have unique numbers.
Small‐molecule inhibitors and sources
Chemical compounds 1–4 (Nordfelth et al., 2005), previously reported to inhibit T3SS of enterobacterial pathogens, were prepared according to published procedures (Dahlgren et al., 2010), and kindly supplied by Dr Mikael Elofsson (Fig. S5, see Supporting Information, Table 3). Previously, other names used for these compounds included ME0052–ME0054 and INP0010, INP0403, INP0401 and INP0031, respectively (Negrea et al., 2007; Tree et al., 2009; Veenendaal et al., 2009). Compounds 5–16 were purchased from ChemBridge (San Diego, CA, USA), Timtec (Newark, DE, USA) and Sigma‐Aldrich (St. Louis, MO, USA) (Fig. S5). Compounds 5 and 14 were also known as INP0002 and INP0007, respectively (Negrea et al., 2007). Compound 6 was reported by Felise et al. (2008), and compounds 7, 8, 15 and 16 were reported by Pan et al. (2009) to inhibit T3SS, but referred to as compounds #2, 3, 1 and #3‐diproprionate, respectively (Table 3).
Construction of promoter–green fluorescent protein (GFP) fusions for promoter activity assays
The promoter region of hrpN and hrpA genes was amplified by PCR. Primer pairs hrpN1–hrpN2 and hrpA1–hrpA2, with restriction sites, were used to amplify 735 and 708 bp for hrpN and hrpA promoter sequences from E. amylovora strain, respectively (Table S2, see Supporting Information). PCR products and the promoter trapping vector pFPV25 were both digested with EcoRI and BamHI. The resulting fragments were gel purified, ligated together and cloned upstream of the promoterless GFP gene. The final plasmids were designated as pHrpN‐GFP for hrpN and pHrpA‐GFP for hrpA, which were confirmed by restriction enzyme digestion and sequencing.
GFP reporter assays by flow cytometry
The BD FACSCanto flow cytometer (BD Bioscience, San Jose, CA, USA) was used to monitor the GFP intensities of bacterial strains containing the corresponding promoter–GFP constructs as described previously (Wang et al., 2009). For hrp‐inducing condition assays, E. amylovora strain Ea273, containing GFP–promoter fusion plasmids, was grown overnight in LB medium, and washed twice with phosphate‐buffered saline (PBS). The bacterial suspension was re‐inoculated into either HMM or M9 minimal medium amended with different carbon sources at different levels. Bacterial cultures were harvested by centrifugation at different time points, and resuspended in PBS for flow cytometry.
For chemical screening, bacterial strains containing GFP–promoter fusion plasmids were washed with PBS and resuspended in HMM containing 20 mm galactose. One milliliter of cell suspension was seeded into 24‐well plates at a density of OD600 = 0.2. Compounds were added to each well to yield final concentrations of 10, 25, 50 and 100 μm, and DMSO was added as a control. GFP intensities were measured by flow cytometry after incubation at 18 °C for 18 h.
Flow cytometry was performed on a BD LSRII 10 parameter multilaser analyzer (BD Bioscience). Data were collected for a total of 100 000 events and analysed statistically by gating using flow cytometry software FCS Express V3 (De Novo Software, Los Angeles, CA, USA). A geometric mean was calculated for each sample. Each treatment was performed in triplicate and each experiment was repeated three times.
CPC assays for the determination of amylovoran concentrations
The amylovoran concentration in supernatants of bacterial cultures was determined quantitatively using a turbidity assay with CPC, as described previously (Bellemann et al., 1994; Wang et al., 2009; Zhao et al., 2009b). Briefly, wild‐type E. amylovora Ea273 was grown overnight in LB broth and washed with PBS three times. After the final wash, the bacterial pellet was resuspended in 200 μL of PBS. A total of 100 μL of bacterial cells was inoculated into 10 mL of MBMA medium with 1% sorbitol. A 3‐mL cell suspension was seeded in Falcon tubes. Chemicals were added to the bacterial suspension to a final concentration of 25 μm, and DMSO was added as a control. After incubation for 24 h at 28 °C with shaking, 50 μL of CPC at 50 mg/mL was added to 1 mL of supernatant following centrifugation. After 10 min of incubation at room temperature, the amylovoran concentration was determined by measuring OD600 turbidity. The final concentration of amylovoran production was normalized for a cell density of 1.0. Each treatment was performed in triplicate and each experiment was repeated three times.
Crab apple flower assays
Bacterial suspensions were grown overnight in LB broth, harvested by centrifugation and resuspended in PBS at a density of OD600 = 0.2. Bacterial suspensions were incubated with compounds at 50 μm or an equivalent volume of DMSO for 4 h at room temperature in the dark. One‐day‐old fully opened crab apple (Malus mandshurica) flowers were detached and immediately transferred to a microcentrifuge tube containing 2 mL of 10% sucrose as described previously (Pusey, 1997). Two microlitres of bacterial suspension were evenly delivered to the stigmas per flower by pipette under a microscope. The inoculated flowers were held in a sealed container that was incubated in a growth chamber at 24 °C and 90% relative humidity. Symptoms were recorded at 6 days post‐inoculation. For the bacterial population assays, flowers were homogenized at 0, 2, 4 and 6 days post‐inoculation. Populations were estimated by plating serial dilutions and calculating colony‐forming units (CFU). Each treatment was performed in triplicate and each experiment was repeated three times.
RNA isolation
WT E. amylovora strain Ea273 was grown overnight in LB broth at 28 °C. Bacterial cells were washed and re‐inoculated into HMM after washing twice with PBS. Two millilitres of cell suspension were seeded into 12‐well tissue culture plates at a density of OD600 = 0.3 cells/well. Compounds were added to each well at a final concentration of 50 μm, and an equal volume of DMSO was used as a control. The plates were incubated at 18 °C for 18 h with shaking. The 1.5‐mL cultures were stabilized using 3 mL of RNA protect reagent (Qiagen, Hilden, Germany). Cells were harvested by centrifugation for 10 min at 4000 g, and RNA was extracted using a Qiagen Bacterial RNA Protect Mini Kit as recommended by the manufacturer (Qiagen). On‐column DNA digestion was performed using Qiagen DNase. RNA was quantified using a NanoDrop ND‐100 spectrophotometer (NanoDrop Technologies; Wilmington, DE, USA) and RNA quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Microarray hybridization and data analysis
Microarray hybridization and data analysis were carried out as described previously (McNally et al., 2012; Wang et al., 2012). Briefly, a 60‐mer E. amylovora microarray (8 × 15 K) was designed at the James Hutton Institute, Aberdeen, UK and synthesized by Agilent Technologies. Each slide contains eight arrays, and each array contains ∼15 000 spots, with each probe spotted in triplicate. A detailed description and design of the oligonucleotide microarray is available at the ArrayExpress website (accessions: microarray A‐MEXP‐2000). All microarray data are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo, accession number GSE45602). Four biological replicates for each chemical treatment (two samples were combined) were hybridized to three arrays. Four biological replicates (two each combined) for DMSO treatment were hybridized to two arrays. All arrays were cross‐compared as technical replicates.
Synthesis and labelling of cDNA were performed using a total of 10 μg of total RNA and a FairPlay III microarray labelling kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions. After labelling, the concentration of cDNA was determined by a NanoDrop ND‐100 spectrophotometer. The labelled cDNA (600 ng) was then hybridized to the slide for 17 h at 65 °C in an Agilent rotating oven (10 rpm) in the presence of a 2 × hybridization buffer (Agilent Technologies). After successive washing by gene expression wash buffers (Agilent Technologies), the hybridized slide was scanned using an Axon 4000B array scanner at a resolution of 5 μm (Molecular Devices, Sunnyvale, CA, USA). Microarray images were processed by GenePix Pro 6.0 image analysis software (vs. 6.0.1.26, Molecular Devices). Raw data were processed through logarithmic transformation and normalization using R software (R.2.2.1). Statistical comparisons were performed using multiple testing procedures to evaluate the statistical significance of differentially expressed genes. A modified t‐test was computed to measure the significance associated with each differential expression value. A gene expression value was assumed to be significantly different when the P value was less than 0.05 (except otherwise mentioned) and the expression ratio was ≥2.0 or ≤0.5.
qRT‐PCR
To validate the microarray results, gene expression levels of hrpA, dspE, hrpL, hrpN, avrRpt2, amsG, amsD, glgB, foxR (EAM_0358), hmuS, dfoJ (EAM_359) and EAM_3350 were determined by qRT‐PCR. For each sample, synthesis of cDNA was performed using 1 μg of total RNA and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Primers were designed using Primer3 software with high specificity and are listed in Table S2. qRT‐PCR was conducted in an ABI 7300 System (Applied Biosystems, Foster City, CA, USA) using Fast SYBR Green PCR master mix (Applied Biosystems). All reactions were run on 96‐well optical reaction plates. Thermal cycling conditions included a step of 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, and a final dissociation curve analysis step from 65 °C to 95 °C. Technical replicate experiments were performed for each biological triplicate sample. The amplification specificity for each reaction was confirmed by dissociation curve analysis. The Ct values determined were then exploited for further analysis. The relative quantification method (ΔΔCt method) was used to determine the expression level of selected genes. The expression of the 16S rRNA gene was used to normalize gene expression across samples. The gene expression values obtained from DMSO‐treated samples were used to calculate a relative quantification (RQ) value for each gene (Wang et al., 2012).
Supporting information
Fig. S1 Promoter activities of type III secretion system (T3SS) genes of Erwinia amylovora in M9 and hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) with different concentrations of carbon sources. (A) Promoter activities of hrpN, dspE, hrpL and hrpA in E. amylovora strain Ea273 in M9 and HMM containing 10 and 20 mm of galactose at 18 °C for 18 h. (B) Time course of promoter activities of hrpN, dspE, hrpL and hrpA in E. amylovora strain Ea273 in HMM containing 20 mm galactose at 18 °C.
Fig. S2 Effect of different concentrations of small‐molecule inhibitors 3 and 9 on promoter activities of hrpN, dspE, hrpL and hrpA genes. Erwinia amylovora strain Ea273 was grown in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) at 18 °C for 18 h. The highest volume of dimethylsulphoxide (DMSO) added (equivalent to 100 μm of chemicals) is shown as the control.
Fig. S3 (A) Effect of small molecules 3 and 9 on bacterial growth in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) containing 20 mm galactose at 18 °C. (B) Effect of small molecule 3 on bacterial growth on flowers. Flowers were homogenized at 0, 2, 4 and 6 days post‐inoculation. The population was measured by plating serial dilutions and calculating colony‐forming units (CFU).
Fig. S4 Small‐molecule inhibitors delayed the hypersensitive response (HR) in tobacco. Tobacco leaf was infiltrated with Erwinia amylovora Ea1189 (OD600 = 0.15) in the presence of compounds 8 (left) and 11 (right) at the different concentrations indicated. The same volume of dimethylsulphoxide (DMSO) was added as a control as 100 μm of compounds. Photographs were taken 24 h post infiltration.
Fig. S5 Structures of the small‐molecule inhibitors used in this study. Not shown are compounds 6 (see Felise et al., 2008; compound #1, TTS29), 7, 8, 15 and 16 (see Pan et al., 2009; compounds #2, 1, 3 and 3‐diproprionate, respectively).
Table S1 Differentially expressed genes in all comparisons.
Table S2 Promoter cloning and quantitative real‐time polymerase chain reaction (qRT‐PCR) primers used in this study.
Acknowledgements
We would like to thank Dr Ian Toth, Dr Peter Cock and Dr Pete E. Hedley of the James Hutton Institute, Aberdeen, UK for sharing the microarray. This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant No. 2010‐5110‐20497 from the USDA National Institute of Food and Agriculture (YFZ) and USDA‐SCRI grant AG 2009‐1181‐06023(SSK). The authors have no conflicts of interest to declare.
References
- Aldridge, P. , Metzger, M. and Geider, K. (1997) Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol. Gen. Genet. 256, 611–619. [DOI] [PubMed] [Google Scholar]
- Bailey, L. , Gylfe, A. , Sundin, C. , Muschiol, S. , Elofsson, M. , Nordstrom, P. , Henriques‐Normark, B. , Lugert, R. , Waldenstrom, A. , Wolf‐Watz, H. and Bergstrom, S. (2007) Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumonia infection cycle. FEBS Lett. 581, 587–595. [DOI] [PubMed] [Google Scholar]
- Baron, C. (2010) Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13, 100–105. [DOI] [PubMed] [Google Scholar]
- Bellemann, P. , Bereswill, S. , Berger, S. and Geider, K. (1994) Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 16, 290–296. [DOI] [PubMed] [Google Scholar]
- Bernhard, F. , Coplin, D.L. and Geider, K. (1993) A gene cluster for amylovoran synthesis in Erwinia amylovora: characterization and relationship to cps genes in Erwinia stewartii . Mol. Gen. Genet. 239, 158–168. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Kim, J.F. , We, Z.M. , Kolchinsky, I. , Charkowski, A.O. , Conlin, A.K. , Collmer, A. and Beer, S.V. (1998) Homology and functional similarity of an hrp‐linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc. Natl. Acad. Sci. USA, 95, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs, J. and Geider, K. (2000) Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 182, 5351–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugert, P. and Geider, K. (1995) Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora . Mol. Microbiol. 15, 917–933. [DOI] [PubMed] [Google Scholar]
- Burse, A. , Weingart, H. and Ullrich, M.S. (2004) NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 70, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Splepenkin, A. , Elofsson, M. , Keyser, P. , de la Maza, L. and Peterson, E.M. (2010) Candidate vaginal microbicides with activity against Chlamydia trachomatis and Neisseria gonorrhoeae . Int. J. Antimicrob. Agents, 36, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyier, D.L. and Covey, R.P. (1975) Tolerance of Erwinia amylovora to streptomycin sulfate in Oregon and Washington. Plant Dis. Rep. 59, 849–852. [Google Scholar]
- Dahlgren, M. , Zetterstrom, C.E. , Gylfe, A. , Linusson, A. and Elofsson, M. (2010) Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram‐negative bacterium Yersinia . Bioorg. Med. Chem. 18, 2686–2703. [DOI] [PubMed] [Google Scholar]
- Davies, B.W. , Kohanski, M.A. , Simmons, L.A. , Winkler, J.A. , Collins, J.J. and Walker, G.C. (2009) Hydroxyurea induces hydroxyl radical‐mediated cell death in Escherichia coli . Mol. Cell, 36, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi, A. , Brisset, M.N. , Paulin, J.P. and Expert, D. (1998) Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant–Microbe Interact. 11, 734–742. [DOI] [PubMed] [Google Scholar]
- Dellagi, A. , Reis, D. , Vian, B. and Expert, D. (1999) Expression of the ferrioxamine receptor gene of Erwinia amylovora CFBP1430 during pathogenesis. Mol. Plant–Microbe Interact. 12, 463–466. [DOI] [PubMed] [Google Scholar]
- Du, Z. and Geider, K. (2002) Characterization of an activator gene upstream of lsc, involved in levan synthesis of Erwinia amylovora . Physiol. Mol. Plant Pathol. 60, 9–17. [Google Scholar]
- Dwyer, D.J. , Kohanski, M.A. , Hayete, B. and Collins, J.J. (2007) Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli . Mol. Syst. Biol. 3, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, F.C. (2011) Antimicrobial actions of reactive oxygen species. Mbio 2, e00141–e00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felise, H.B. , Nguyen, H.V. , Pfuetzner, R.A. , Barry, K.C. , Jackson, S.R. , Blanc, M. , Bronstein, P.A. , Kline, T. and Miller, S.I. (2008) An inhibitor of Gram‐negative bacterial virulence protein secretion. Cell Host Microb. 4, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen, M. , Beckham, K.S.H. , Feher, V.A. , Zetterström, C.E. , Wang, D. , Müller, S. , Elofsson, M. , Amaro, R.E. , Byron, O. and Roe, A.J. (2012) Structural characterisation of Tpx from Yersinia pseudotuberculosis reveals insights into the binding of salicylidene acylhydrazide compounds. Plos ONE, 7, e32217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudriault, S. , Malandrin, L. , Paulin, J.P. and Barny, M.A. (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via Hrp secretion pathway in a DspB‐dependent way. Mol. Microbiol. 26, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Gauthier, A. , Robertson, M.L. , Lowden, M. , Ibarra, J.A. , Puente, J.L. and Finlay, B.B. (2005) Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli . Antimicrob. Agents Chemother. 49, 4101–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochem. Biophys. Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hudson, D.L. , Layton, A.N. , Field, T.R. , Bowen, A.J. , Wolf‐Watz, H. , Elofsson, M. , Stevens, M.P. and Galyov, E.E. (2007) Inhibition of type III secretion in Salmonella enteric serovar Typhimurium by small‐molecule inhibitors. Antimicrob. Agents Chemother. 51, 2631–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi, A.M. , Nordfelth, R. , Uvell, H. , Wolf‐Watz, H. and Elofsson, M. (2003) Targeting bacterial virulence: inhibitors of type III secretion in Yersinia . Chem. Biol. 10, 241–249. [DOI] [PubMed] [Google Scholar]
- Keyser, P. , Elofsson, M. , Rosell, S. and Wolf‐Watz, H. (2008) Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram‐negative bacteria. J. Intern. Med. 264, 17–29. [DOI] [PubMed] [Google Scholar]
- Khan, M.A. , Zhao, Y.F. and Korban, S.S. (2012) Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol. Biol. Rep. 30, 247–260. [Google Scholar]
- Kim, I. , Pusey, L. , Zhao, Y.F. , Korban, S.S. , Choi, H. and Kim, K. (2012) Microencapsulation and controlled release of Pantoea agglomerans E325 for biocontrol of fire blight disease of apple. J. Control. Release, 161, 109–115. [DOI] [PubMed] [Google Scholar]
- Kimura, K. , Iwatsuki, M. , Nagai, T. , Matsumoto, A. , Takahashi, Y. , Shiomi, K. , Omura, S. and Abe, A. (2011) A small‐molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium . J. Antibiot. 64, 197–203. [DOI] [PubMed] [Google Scholar]
- Kline, T. , Barry, K.C. , Jackson, S.R. , Felise, H.B. , Nguyen, H.V. and Miller, S.I. (2009) Tethered thiazolidinone dimers as inhibitors of the bacterial type III secretion system. Bioorg. Med. Chem. Lett. 19, 1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, T. , Felise, H.B. , Sanowar, S. and Miller, S.I. (2011) The type III secretion system as a source of novel antibacterial drug targets. Curr. Drug Targets, 13, 338–351. [DOI] [PubMed] [Google Scholar]
- Kohanski, M.A. , Dwyer, D.J. and Collins, J.J. (2010) How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton, A.N. , Hudson, D.L. , Thompson, A. , Hinton, J.C.D. , Stevens, J.M. , Galyov, E.E. and Stevens, M.P. (2010) Salicylidene acylhydrazide‐mediated inhibition of type III secretion system‐1 in Salmonella enteric serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302, 114–122. [DOI] [PubMed] [Google Scholar]
- Loper, J. , Henkels, M.D. , Roberts, R.G. , Grove, G.G. , Willet, M.J. and Smith, T.J. (1991) Evaluation of streptomycin, oxytetracycline, and copper resistance of Erwinia amylovora isolated from pear orchards in Washington State. Plant Dis. 75, 287–290. [Google Scholar]
- Macielag, M.J. , Demers, J.P. , Fraga‐Spano, S.A. , Hlasta, D.J. , Johnson, S.G. , Kanojia, R.M. , Russell, R.K. , Sui, Z. , Weidner‐Wells, M.A. , Werblood, H. , Foleno, B.D. , Goldschmidt, R.M. , Loeloff, M.J. , Webb, G.C. and Barrett, J.F. (1998) Substituted salicylanilides as inhibitors of two‐component regulatory systems in bacteria. J. Med. Chem. 41, 2939–2945. [DOI] [PubMed] [Google Scholar]
- Manulis, S. , Zutra, D. , Kleitman, F. , Drox, O. , David, I. , Zilberstaine, M. and Shabi, E. (1998) Distribution of streptomycin‐resistant strains of Erwinia amylovora in Israel and occurrence of blossom blight in the autumn. Phytoparasitica, 26, 223–230. [Google Scholar]
- McGhee, G.C. and Sundin, G.W. (2011) Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora . Phytopathology, 101, 192–204. [DOI] [PubMed] [Google Scholar]
- McNally, R.R. , Toth, I.K. , Cock, P.J. , Pritchard, L. , Hedley, P.E. , Morris, J.A. , Zhao, Y.F. and Sundin, G.W. (2012) Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Mol. Plant Pathol. 13, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschiol, S. , Bailey, L. , Gylfe, Å. , Sundin, C. , Hultenby, K. , Bergstrom, S. , Elofsson, M. , Wolf‐Watz, H. , Normark, S. and Henriques‐Normark, B. (2006) A small‐molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis . Proc. Natl. Acad. Sci. USA, 103, 14 566–14 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschiol, S. , Normark, S. , Henriques‐Normark, B. and Subtil, A. (2009) Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol. 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrea, A. , Bjur, E. , Ygberg, S.E. , Elofsson, M. , Wolf‐Watz, H. and Rhen, M. (2007) Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enteric serovar typhimurium. Antimicrob. Agents Chemother. 51, 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissinen, R.M. , Ytterberg, A.J. , Bogdanove, A.J. , van Wijk, K. and Beer, S.V. (2007) Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8, 55–67. [DOI] [PubMed] [Google Scholar]
- Nordfelth, R. , Kauppi, A.M. , Norberg, H.A. , Wolf‐Watz, H. and Elofsson, M. (2005) Small‐molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73, 3104–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norelli, J.L. , Jones, A.L. and Aldwinckle, H.S. (2003) Fire blight management in the twenty‐first century: using new technologies that enhance host resistance in apple. Plant Dis. 87, 756–765. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. and Beer, S.V. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Kim, J.F. and Beer, S.V. (2005) The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6, 125–138. [DOI] [PubMed] [Google Scholar]
- Pan, N.J. , Brady, M.J. , Leong, J.M. and Goguen, J.D. (2009) Targeting type III secretion in Yersinia pestis . Antimicrob. Agents Chemother. 53, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey, P.L. (1997) Crab apple blossoms as a model for research on biological control of fire blight. Phytopathology, 87, 1096–1102. [DOI] [PubMed] [Google Scholar]
- Russo, N.L. , Burr, T.J. , Breth, D.I. and Aldwinckle, H.S. (2008) Isolation of streptomycin‐resistant isolates of Erwinia amylovora in New York. Plant Dis. 92, 714–718. [DOI] [PubMed] [Google Scholar]
- Shaffer, W.H. and Goodman, R.N. (1985) Appearance of streptomycin‐resistant Erwinia amylovora in Missouri apple orchards. Phytopathology, 75, 1281. [Google Scholar]
- Slepenkin, A. , Chu, H. , Elofsson, M. , Keyser, P. and Peterson, E.M. (2011) Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J. Infect. Dis. 204, 1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree, J.J. , Wang, D. , McInally, C. , Mahajan, A. , Layton, A. , Houghton, I. , Elofsson, M. , Stevens, M. , Gally, D.L. and Roe, A.J. (2009) Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77, 4209–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia, R.H. and Falkow, S. (1997) Fluorescence‐based isolation of bacterial genes expressed within host cells. Science, 277, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Veenendaal, A.K. , Sundin, C. and Blocker, A.J. (2009) Small‐molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191, 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. and Zhao, Y.F. (2009) The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora . Mol. Plant Pathol. 10, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. and Zhao, Y.F. (2010) Molecular signature of differential virulence in natural isolates of Erwinia amylovora . Phytopathology, 100, 192–198. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Zetterström, C.E. , Gabrielsen, M. , Beckham, K.S. , Tree, J.J. , Macdonald, S.E. , Byron, O. , Mitchell, T.J. , Gally, D.L. , Herzyk, P. , Mahajan, A. , Uvell, H. , Burchmore, R. , Smith, B.O. , Elofsson, M. and Roe, A. (2011) Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence‐blocking compounds. J. Biol. Chem. 286, 29 922–29 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Qi, M. , Calla, B. , Korban, S.S. , Clough, S.J. , Cock, P.J. , Sundin, G.W. , Toth, I. and Zhao, Y.F. (2012) Genome‐wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora . Mol. Plant–Microbe Interact. 25, 6–17. [DOI] [PubMed] [Google Scholar]
- Wolf, K. , Betts, H.J. , Chellas‐Géry, B. , Hower, S. , Linton, C.N. and Fields, K.A. (2006) Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61, 1543–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. and Qi, M.S. (2011) Comparative genomics of Erwinia amylovora and related Erwinia species – What do we learn? Genes, 2, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. , Blumer, S.E. and Sundin, G.W. (2005) Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187, 8088–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. , He, S.Y. and Sundin, G.W. (2006) The Erwinia amylovora avrRpt2EAgene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 644–654. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.F. , Sundin, G.W. and Wang, D. (2009a) Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora . Can. J. Microbiol. 55, 457–464. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.F. , Wang, D. , Nakka, S. , Sundin, G.W. and Korban, S.S. (2009b) Systems level analysis of two‐component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics, 10, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Promoter activities of type III secretion system (T3SS) genes of Erwinia amylovora in M9 and hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) with different concentrations of carbon sources. (A) Promoter activities of hrpN, dspE, hrpL and hrpA in E. amylovora strain Ea273 in M9 and HMM containing 10 and 20 mm of galactose at 18 °C for 18 h. (B) Time course of promoter activities of hrpN, dspE, hrpL and hrpA in E. amylovora strain Ea273 in HMM containing 20 mm galactose at 18 °C.
Fig. S2 Effect of different concentrations of small‐molecule inhibitors 3 and 9 on promoter activities of hrpN, dspE, hrpL and hrpA genes. Erwinia amylovora strain Ea273 was grown in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) at 18 °C for 18 h. The highest volume of dimethylsulphoxide (DMSO) added (equivalent to 100 μm of chemicals) is shown as the control.
Fig. S3 (A) Effect of small molecules 3 and 9 on bacterial growth in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium (HMM) containing 20 mm galactose at 18 °C. (B) Effect of small molecule 3 on bacterial growth on flowers. Flowers were homogenized at 0, 2, 4 and 6 days post‐inoculation. The population was measured by plating serial dilutions and calculating colony‐forming units (CFU).
Fig. S4 Small‐molecule inhibitors delayed the hypersensitive response (HR) in tobacco. Tobacco leaf was infiltrated with Erwinia amylovora Ea1189 (OD600 = 0.15) in the presence of compounds 8 (left) and 11 (right) at the different concentrations indicated. The same volume of dimethylsulphoxide (DMSO) was added as a control as 100 μm of compounds. Photographs were taken 24 h post infiltration.
Fig. S5 Structures of the small‐molecule inhibitors used in this study. Not shown are compounds 6 (see Felise et al., 2008; compound #1, TTS29), 7, 8, 15 and 16 (see Pan et al., 2009; compounds #2, 1, 3 and 3‐diproprionate, respectively).
Table S1 Differentially expressed genes in all comparisons.
Table S2 Promoter cloning and quantitative real‐time polymerase chain reaction (qRT‐PCR) primers used in this study.


