SUMMARY
Lima bean is an important vegetable processing crop to the mid‐Atlantic USA and is highly susceptible to the oomycete pathogen Phytophthora phaseoli, which causes downy mildew. Genetic resistance and fungicides are used to manage P. phaseoli and often fail. Currently, the molecular basis of the interaction between this host and pathogen is unknown. To begin to rectify this situation, we used Illumina RNA‐Seq to perform a global transcriptome analysis comparing P. phaseoli growing in culture with P. phaseoli infecting its host. Sequence reads from a total of six libraries mapped to gene models from the closely related late blight pathogen, Phytophthora infestans, resulting in 10 427 P. phaseoli genes with homology to P. infestans and expression in at least one library. Of these, 318 P. phaseoli homologues matched known or putative virulence genes in P. infestans. Two well‐studied classes, RxLRs and elicitins, were up‐regulated in planta, whereas the reverse was true for another class, called crinklers. These results are discussed with respect to the differences and similarities in the pathogenicity mechanisms of P. phaseoli and P. infestans.
INTRODUCTION
Oomycete pathogens are fungal‐like organisms that can cause tens of billions of dollars of damage each year (French and Mackay, 1996; Fry, 2008) to a wide range of agriculturally and ornamentally important crops (Fry, 2008). They also cause severe damage in forests, threatening entire ecosystems (Rizzo et al., 2005).
The oomycete Phytophthora phaseoli has a narrow host range and is restricted primarily to lima bean. It is homothallic and the causal agent of downy mildew on lima bean, which has been reported to cause three million dollars worth of losses, or half the profits, in the state of Delaware (Evans et al., 2007). Optimal harvesting of lima bean pods occurs late in the autumn, when cooler, wetter conditions prevail, which also favour P. phaseoli infection. The first commercially released lima bean cultivar with resistance to downy mildew, Thaxter, was developed by App (1959) and was later overcome by the evolution of new races. Over the years, different physiological pathogen races, A, B, C, D, E and F, have been reported, in 1904, 1958, 1969, 1976, 1995 and 2000, respectively (Evans et al., 2002). These races are distinguished on the basis of their reaction to different cultivars of lima bean (Evans et al., 2007), and resistance to races E and F is controlled by single dominant nonallelic genes (Ernest et al., 2006). Currently, there are no cultivars resistant to both of the prevalent field races E and F, and the only effective control measure is fungicide sprays. In order to develop improved disease strategies, it is essential to understand the molecular mechanisms by which these pathogens break down or avoid plant defences.
Research on pathogenic Phytophthora species has demonstrated that these pathogens secrete hundreds of proteins that act on their hosts, driving the outcome of either disease or resistance depending on the host genotype (Haas et al., 2009; Kamoun, 2006; Tyler et al., 2006). These secreted proteins include a type of pathogen‐associated molecular pattern (PAMP), called elicitins (reviewed in Nurnberger and Brunner, 2002), RxLR effectors (genes that encode an RxLR motif), crinklers (CRN) and NPP1 (necrosis‐inducing Phytophthora protein). Elicitins encode putative extracellular proteins that can trigger plant cell death; they share a 98‐amino‐acid domain with a core of six conserved cysteines in the C‐terminal domain (Kamoun, 2006). RxLR‐type effectors share an N‐terminal amino acid motif (RxLR) that is conserved throughout the Phytophthora species currently being studied (Birch et al., 2006), where ‘x’ denotes a nonconserved amino acid position. This motif aids in the translocation of the protein into the host's cytoplasm, where many function as avirulence factors, interacting with the plant's resistance (R) gene and triggering defences (Dou et al., 2008; Whisson et al., 2007). A structural dissection of these proteins has revealed a signal peptide and an RxLR motif at the N‐terminus, which are important for secretion and targeting of the protein (Bos et al., 2006), whereas the C‐terminus carries the actual effector activity (Dou et al., 2008; Van Poppel et al., 2009). The CRN family represents a third, well‐studied class of effector, named for its ability to trigger host necrosis and crinkling of leaves, as well as to induce host defences (Torto et al., 2003). NPP1 proteins cause plant cell death and are identified by the presence of a common NPP1 domain, collectively named NLPs (Nep1‐like proteins) (Gijzen and Nurnberger, 2006; Rudd et al., 2009).
The completion of the Phytophthora infestans genome has provided a rich resource to mine for genes in related oomycetes (Haas et al., 2009), such as P. phaseoli. Phylogenetic analysis of the Phytophthora genus has shown that P. phaseoli lies with P. infestans in the same clade, and is therefore more closely related to the late blight pathogen than to other sequenced species (Blair et al., 2008). We employed the P. infestans genome as a reference for a transcriptional study using Illumina RNA‐Seq (Asmann et al., 2008). We compared transcriptional abundance between three samples [P. phaseoli race F growing on culture medium (hereafter referred to as P18), P. phaseoli growing on lima bean hypocotyls at 3 days post‐inoculation (dpi) (hereafter referred to as T3) and P. phaseoli growing on lima bean hypocotyls at 6 dpi (hereafter referred to as T6)] to determine which genes were more highly expressed during the infection process. We specifically focused on the identification of RxLRs, elicitins and CRNs, predicting that they would be more highly expressed in plant infected tissue versus tissue grown on plates. Our results partially supported our prediction, revealing more transcriptional changes in P. phaseoli during the later infection time point (T6), including the induction of RxLRs and elicitins, and the repression of CRNs. Our study presents a critical and necessary first step to characterize the virulence mechanisms employed by P. phaseoli.
RESULTS
Gene expression profiling using Illumina sequencing
We used Illumina sequencing to perform transcriptional profiling (RNA‐Seq). We generated and sequenced two biological replicates for each sample (see Table 1 for descriptions of each library). These data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (Edgar et al., 2002), accessible through accession number GSE23453. In order to analyse the relatedness of the six libraries, we generated multidimensional scaling (MDS) plots using edgeR (empirical analysis of DGE in R program; Robinson et al., 2010; Fig. 1). The distance between the biological replicates of P18 and T6 is less than the distance between the replicates of T3.
Table 1.
Description of each RNA‐Seq library.
| Library name | Description | RNA‐Seq location | Cycle number* | Sequence reads† |
|---|---|---|---|---|
| P18‐rep1 | Plate‐grown mycelium | NCGR‡ | 36 | 9.75 |
| P18‐rep2 | Plate‐grown mycelium | University of Delaware | 42 | 28.24 |
| T3‐rep1 | Infected plant 3 dpi§ | University of Delaware | 42 | 30.53 |
| T3‐rep2 | Infected plant 3 dpi | University of Delaware | 42 | 34.22 |
| T6‐rep1 | Infected plant 6 dpi | NCGR | 36 | 15.14 |
| T6‐rep2 | Infected plant 6 dpi | University of Delaware | 42 | 34.65 |
Refers to the length of the short read generated from Illumina sequencing.
Total number of sequence reads for each library in millions.
NCGR, National Center for Genome Research.
dpi, days post‐inoculation.
Figure 1.
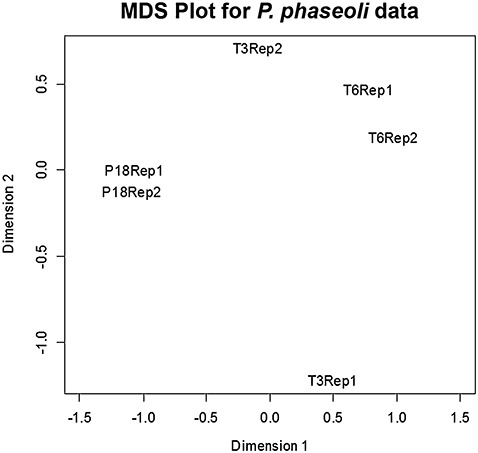
Multidimensional scaling (MDS) showing the relationship between replicates in two dimensions (dimensions 1 and 2), generated by edgeR. Two replicates (Rep1 and Rep2) for the samples P18 and T6 are closely related in both dimensions, whereas the T3 replicates are closely related in dimension 1 and separated in dimension 2. Axes x and y are representations of all the gene expression levels between groups based on tissue type and replicates.
Phytophthora phaseoli contigs mapped mostly to P. infestans genes
The total Illumina reads of over 150 million sequence tags from the six samples were pooled for contig assembly using Velvet (Schmidt et al., 2009), resulting in 301 868 contigs with an average length of 150 bp (Table S1, see Supporting Information), and showing a normal distribution (Fig. S1, see Supporting Information). For each contig, we generated a count of the number of sequence reads (or tags) in each sample used to generate that contig. Contigs were mapped to the P. infestans reference gene sequences by performing blast searches, and 117 702 mapped to 12 456 different PITG (PITG: Phytophthora infestans, strain T30‐4) genes of P. infestans (Table S1). After normalization (see Experimental procedures), the number of genes was reduced to 10 427 (Table S2, see Supporting Information), which were used in all further analyses.
We mapped approximately 39% of the contigs to P. infestans genes, leaving 184 166 that did not match to P. infestans (hereafter referred to as non‐Pi). To determine their origin, we generated blast‐able databases of six plant genomes, Phaseolus species (Phaseolus coccineus, Phaseolus vulgaris, Phaseolus acutifolius and Phaseolus angustissimus), Glycine max (soybean) and Medicago truncatula (barrel medic), and two additional oomycete genomes, Phytophthora sojae and Phytophthora ramorum. The majority of the non‐Pi contigs mapped to these six plant genomes, whereas approximately1% mapped to each of the two oomycete genomes (Table 2). We performed the same analysis for contigs from the P18, T3 and T6 samples and again found that, for the last two, the majority of non‐Pi contigs mapped to the plant genomes. The P18 sample showed 31 124 non‐Pi contigs that did not map to P. infestans; of these, 8089 mapped to one of the eight other genomes, indicating that 23 035 contigs are either exclusive to P. phaseoli or map to some other organism not tested in this study.
Table 2.
Total number of contigs matching to various species.
| Number of contigs* | Phytophthora infestans † | Non‐Pi‡ | Non‐Pi maps to§ | ||||
|---|---|---|---|---|---|---|---|
| Phas | Gm | Mt | Ps | Pr | |||
| Full dataset | |||||||
| 301 868 | 117 702 | 184 116 | 127 566 | 117 420 | 63 478 | 2 209 | 1 920 |
| % of 184 116 | 69.3 | 63.8 | 34.5 | 1.2 | 1.0 | ||
| P18 dataset | |||||||
| 142 089 | 110 965 | 31 124 | 3 201 | 2294 | 1 580 | 1 961 | 1 714 |
| % of 31 124 | 10.3 | 7.4 | 5.1 | 6.3 | 5.5 | ||
| T3 dataset | |||||||
| 147 564 | 17 721 | 129 843 | 108 522 | 99 897 | 55 239 | 534 | 442 |
| % of 129 843 | 83.6 | 76.9 | 42.5 | 0.4 | 0.3 | ||
| T6 dataset | |||||||
| 236 139 | 99 941 | 136 198 | 96 955 | 86 628 | 48 376 | 1 927 | 1 681 |
| % of 136 198 | 71.2 | 63.6 | 35.5 | 1.4 | 1.2 | ||
Number of contigs assembled for the total experiment, and for each sample.
Number of contigs that mapped to the P. infestans genome (with an e‐value of ≤e−10).
Number of contigs that did not map to the P. infestans genome (‘non‐Pi’).
Non‐Pi contigs that mapped to one of the following genomes: Phas, Phaseolus species (Phaseolus coccineus, Phaseolus vulgaris, Phaseolus acutifolius and Phaseolus angustissimus); Gm, Glycine max; Mt, Medicago truncatula; Ps, Phytophthora sojae; Pr, Phytophthora ramorum.
Gene expression during P. phaseoli infection
We used edgeR to identify which of the 10 427 genes were differentially expressed in the P18 condition versus T3 and T6. We examined sequence abundance levels and found that the majority were differentially expressed in T3 versus P18 (Fig. 2A, Tables 3 and S3, see Supporting Information). Similarly, a larger number of genes were expressed in T6 versus P18 (Fig. 2B, Tables 3 and S4, see Supporting Information). In Fig. 2, red dots that fall outside the blue lines [log fold change (FC) =−2 and log FC = 2] represent tags that are differentially expressed [false discovery rate (FDR) < 0.05 and P < 0.01] and that have a greater than two‐fold change over P18 (Fig. 2A,B and Table 3). This resulted in eight and 1284 unique genes that were differentially expressed in comparisons between the pairs P18 versus T3 and P18 versus T6, respectively (Fig. 2, Tables 3, S3 and S4).
Figure 2.
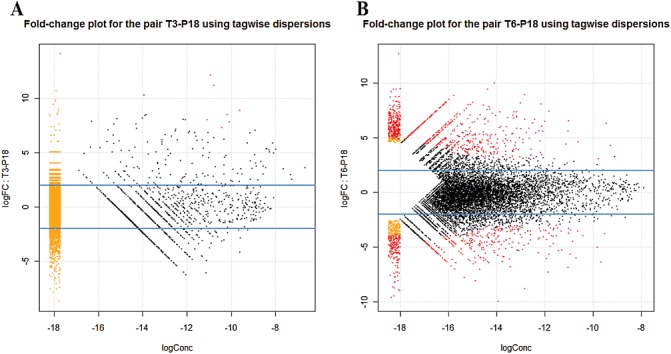
Smear plot generated from edgeR showing the log fold change (FC) against the log concentration (Conc) (a summary measure of the average concentration for each tag over all treatment conditions) for each tag, using tagwise dispersion. The most differentially expressed tags are highlighted in red, showing eight for P18–T3 (A) and 1284 for P18–T6 (B). The smear of dots (orange and red) on the left side signifies that genes were observed in only groups of replicate samples.
Table 3.
Gene expression between the pairs P18–T3 and P18–T6.
| Gene class | Total* | Differentially expressed genes | |||||
|---|---|---|---|---|---|---|---|
| Pair P18–T3 | Pair P18–T6 | ||||||
| P18† | T3† | No change‡ | P18† | T6† | No change‡ | ||
| RxLR effectors | 66 | 0 | 0 | 66 | 10 | 14 | 42 |
| Elicitin‐like | 19 | 0 | 0 | 19 | 2 | 4 | 13 |
| Crinkler (CRN) | 107 | 0 | 0 | 107 | 18 | 2 | 87 |
| NPP1 (necrosis‐inducing Phytophthora protein) | 6 | 0 | 0 | 6 | 0 | 1 | 5 |
| Enzyme inhibitors | 13 | 0 | 0 | 13 | 2 | 1 | 10 |
| PcF/SCR‐like | 3 | 0 | 0 | 3 | 0 | 1 | 2 |
| ABC superfamily | 104 | 0 | 0 | 104 | 8 | 18 | 78 |
| Sporangial development | 46 | 0 | 0 | 46 | 0 | 26 | 20 |
| Hypothetical proteins | 5 911 | 0 | 0 | 5 911 | 278 | 492 | 5 141 |
| Others | 4 152 | 0 | 8 | 4 144 | 142 | 265 | 3 745 |
| Total | 10 427 | 0 | 8 | 10 419 | 460 | 824 | 9 143 |
Bold type indicates genes in families probably involved in pathogenicity.
Total number of Phytophthora phaseoli genes, homologous to Phytophthora infestans genes.
Genes had a false discovery rate (FDR) < 0.05 and P < 0.01.
Genes had FDR > 0.05.
We identified 318 P. infestans genes with known or putative roles in virulence (reviewed in Hogenhout et al., 2009; Kamoun, 2006; Tyler et al., 2006) for both pairs of samples. These genes grouped into classes defined as RxLR effectors, elicitin, CRN, NPP1, enzyme inhibitors, PcF/SCR‐like (secreted 52‐amino‐acid peptide of P. cactorum) and ABC transporter superfamily (Table 3, bold type). None of the genes in these families were differentially expressed with a significant P value (P < 0.01) in T3 versus P18, whereas 81 genes were significantly expressed in T6, 41 of which increased in T6, whereas 40 decreased (Tables 3 and S4).
Closer analysis of these 81 genes revealed significant expression of 24, six, 20, 26, three, one and one RxLRs, elicitins, crinklers, ABC transporters, enzyme inhibitors, NPP1 and PcF/SCR‐like, respectively. Of the RxLRs and elicitins, 14 and four showed increased expression at T6, whereas ten and two decreased, compared with P18. Of the 20 crinklers, two increased at T6, whereas 18 decreased, and, for the 26 ABC transporter genes, eight increased at T6 and 18 decreased (Table 3).
Confirmation of gene expression profiles
Fifteen genes representing the families described above were selected for confirmation with reverse transcription‐polymerase chain reaction (RT‐PCR), and a subset with real‐time quantitative RT‐PCR (Fig. 3). Hereafter, we refer to them by their Broad Institute P. infestans designations, as that is what they most closely resemble, with ‘Pp’ (for P. phaseoli) at the beginning of each name. Although the majority of the genes chosen showed increased expression in the T6 library, two genes, PpPITG_12562 and PpPITG_00058, showed unchanged expression and one gene, PpPITG_04378, showed higher expression in P18. To determine whether any of the 15 genes were induced at earlier time points, we carried out an expression time course. Most genes were expressed most highly at 5 dpi, except PpPITG_04378, which showed no expression (Fig. S2, see Supporting Information). Two genes with low expression, PpPITG_15039 and PpPITG_20413, were included in further analyses as their sequences revealed them to be a candidate RxLR effector and an elicitin, respectively.
Figure 3.
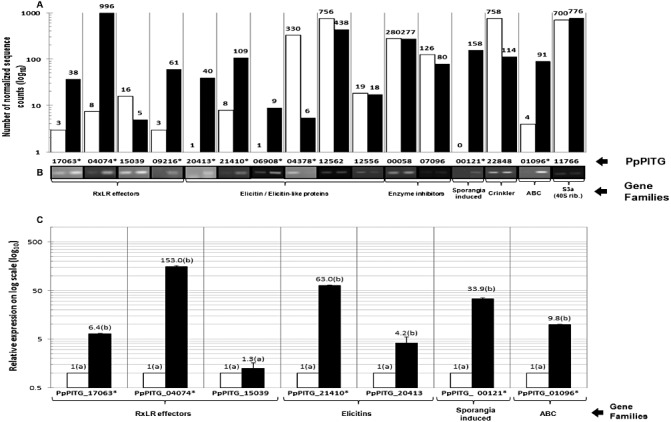
Confirmation of selected genes in Phytophthora phaseoli for the pair P18–T6. (A) The number of normalized sequence counts (numbers on the respective bars) that mapped to P. infestans genes is shown on a logarithmic y axis. (B) Reverse transcription‐polymerase chain reaction (RT‐PCR) confirmation of genes in (A). PpPITG_11766 encodes a 40S ribosomal protein, and was used as a housekeeping gene (S3a). (C) Expression levels were calculated by the 2(–ΔΔCT) method using plate‐grown P. phaseoli as the calibrator and S3a as the housekeeping gene. Error bars indicate the standard error and the letters ‘a’ and ‘b’ indicate the significance in Student's t‐test. White and black bars represent plate‐grown (P18) and plant‐grown (6 days post‐inoculation) (T6) P. phaseoli, respectively. The genes that were differentially expressed (P < 0.01) are marked with an asterisk. This experiment was performed with three biological replicates.
Sequence alignments
Full‐length copies of selected genes were obtained by designing primers with an adapter sequence (restriction site) flanking the PITG gene, and then cloned into a pCambia vector and sequenced. The resulting translated amino acid sequences were used for alignments. A single species was chosen to represent each of six Phytophthora clades (Blair et al., 2008). Although none of the six libraries had reads that mapped to the well‐characterized P. infestans elicitin INF1 (PITG_12551) (Jiang and Govers, 2006; Jiang et al., 2006; 1997a, 1997b), we nonetheless amplified the full‐length copy of the P. phaseoli homologue for comparison and to serve as a control in additional experiments. blast results with the predicted proteins showed that P. phaseoli homologues PpPITG_12551 (hereafter PpINF1), PpPITG_21410 (hereafter PpINF4), PpPITG_06908, PpPITG_20413, PpPITG_04074, PpPITG_17063, PpPITG_15039 and PpPITG_22848 were more than 90% identical to P. infestans (Table 4). PpINF1 and PpINF4 showed 66 and 59 of 118 amino acids, respectively, conserved across seven different Phytophthora species, whereas 114 of PpINF1 and PpINF4 amino acids were shared with P. infestans INF1 and INF4 (Fig. 4A,B), corresponding to 96% identity (Table 4).
Table 4.
Comparison of putative RxLR and elicitin protein sequences from Phytophthora phaseoli with Phytophthora infestans.
| Gene | % ID* | Number of amino acids (total) | Number of amino acids (conserved)† | |
|---|---|---|---|---|
| P. phaseoli | P. infestans | |||
| PpPITG_12551 ‡ | 96 | 118 | 118 | 114 |
| PpPITG_21410 ‡ | 96 | 118 | 118 | 114 |
| PpPITG_06908 | 96 | 174 | 173 | 167 |
| PpPITG_20413 | 96 | 335 | 335 | 324 |
| PpPITG_04074 ‡ | 91 | 185 | 176 | 171 |
| PpPITG_17063 ‡ | 97 | 160 | 160 | 156 |
| PpPITG_15039 ‡ | 94 | 505 | 505 | 477 |
| PpPITG_22848 | 95 | 381 | 381 | 362 |
blasted against the P. infestans genome sequence.
Conserved between both P. phaseoli and P. infestans.
Indicates that these amino acid sequences were used for alignments in Fig. 4.
Figure 4.
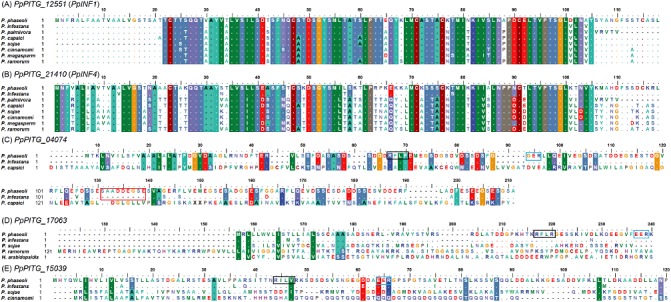
Alignment of five protein sequences from Phytophthora phaseoli against different Phytophthora species in which a homologue was present, and one additional oomycete, Hyaloperonospora arabidopsidis. Full‐length P. phaseoli homologous sequences were translated using BioEdit. (A) PpINF1; (B) PpINF4; (C) PpPITG_04074, insertion of nine amino acids (red box); (D) PpPITG_17063 with RxLR; (E) PpPITG_15039. Black and blue boxes denote putative RxLR and dEER motifs, respectively.
The predicted RxLR PpPITG_04074 shared 171 amino acids with P. infestans. We noted an insertion of nine amino acids (SAADDEGSE; red outlined box in Fig. 4C) in PpPITG_04074, corresponding to the different sequence lengths of 176 and 185 amino acids in P. infestans and P. phaseoli, respectively (Table 4). Another predicted RxLR, PpPITG_17063, revealed that, of 160 amino acids in P. phaseoli, only one was conserved across three different Phytophthora species and an additional oomycete, Hyaloperonospora arabidopsidis. Conversely, 156 amino acids were conserved when aligned with only P. infestans (Fig. 4D). One final RxLR effector of interest, PpPITG_15039, showed seven of 505 amino acids conserved across three different Phytophthora species, whereas 477 were shared with P. infestans (Fig. 4E).
Gene expression during hypocotyl infection is similar to field infection
We tested the expression of ten of the 16 genes described above to determine how closely expression during hypocotyl infection mimicked pod infection in the field. We also examined expression in other Phytophthora species on their respective hosts to obtain more information regarding the importance of these particular genes. Conditions included P. phaseoli‐infected lima bean hypocotyls (growth chamber), and pods (field), P. capsici‐infected lima bean pods (field) and P. infestans‐infected tomato plants (growth chamber). We designed primers for specific homologous genes in the P. phaseoli sequence. Except for PpPITG_17063, all genes showed stronger expression during the infection of lima bean hypocotyls and pods versus plate‐grown P. phaseoli. Five genes, including one RxLR (PpPITG_09216) and one elicitin (PpPITG_21410), also showed greater expression in P. infestans during tomato infection. None of the genes showed expression in any of the P. capsici samples. For the housekeeping gene S3a (Judelson et al., 2008; Yan and Liou, 2006), we expected and obtained two different gene lengths depending on whether the template was cDNA or genomic, indicating no genomic contamination in our cDNA template (Fig. 5).
Figure 5.

Reverse transcription‐polymerase chain reaction (RT‐PCR) expression of ten genes in Phytophthora phaseoli, P. infestans and P. capsici. Lanes: 1, plate‐grown P. phaseoli mycelium (cDNA); 2, P. phaseoli mycelium on lima bean hypocotyls (6 days post‐inoculation); 3, P. phaseoli mycelium on lima bean pods (field‐collected); 4, plate‐grown P. infestans mycelium; 5, P. infestans mycelium on tomato; 6, plate‐grown P. capsici; 7, P. capsici mycelium on lima bean pods (field‐collected); 8, lima bean pods challenged with water; 9, lima bean hypocotyls challenged with water; 10, P. phaseoli genomic DNA (gDNA); 11, negative control (water). Gene names are listed on the right and sizes are listed on the left. All sizes were as expected, and the experiment was performed with at least three biological replicates.
Phytophthora phaseoli putative elicitins cause cell death in Nicotiana benthamiana
Expression studies and sequence alignments indicated that we had identified potentially important infection‐related genes in P. phaseoli, including putative elicitins. To determine functionality, we performed transient assays on 5‐week‐old N. benthamiana plants with four predicted elicitins: PpINF1, PpINF4, PpPITG_20413 and PpPITG_06908 (Fig. 6A). We used P. infestans INF1 (PGR106INF1; provided by Dennis Halterman, University of Wisconsin, Madison, WI, USA) as a control. Agro‐infiltration of PGR106INF1 and the P. phaseoli INF1 homologue induced hypersensitive response (HR)‐like cell death within 3 days (data not shown), whereas PpINF4 and PpPITG_06908 showed an HR at 7 days post‐infiltration (Fig. 6A), as indicated by the congruence of macroscopic necrosis with microscopic trypan blue‐stained cell death. PpPITG_20413 induced an HR at 18 days post‐infiltration. An empty pCAMBIA vector (1301s) and a green fluorescent protein (GFP)‐containing PGR vector (PGR106GFP) were used as infiltration controls, and neither revealed macroscopic HR or blue staining (Fig. 6).
Figure 6.
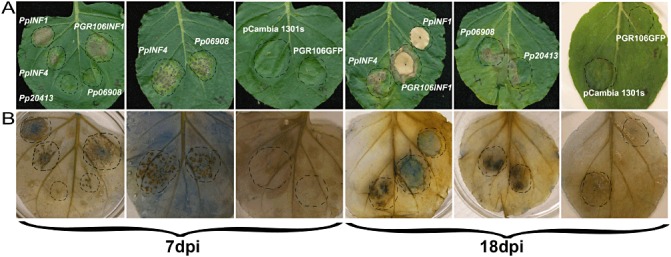
Transient assays of four elicitins from Phytophthora phaseoli showing the induction of hypersensitive response (HR)‐like host cell death. (A) Leaves of Nicotiana benthamiana were infiltrated with Agrobacterium tumefaciens (strain GV3101) to express candidate elicitins. Agrobacterium containing INF1 from P. infestans (PGR106INF1) was used as a positive control, and pCambia 1301s and PGR106GFP were used as negative controls. (B) Agrobacterium‐infiltrated leaves from (A) were stained with trypan blue. This experiment was performed with at least three biological replicates and these are representative images taken at 7 and 18 days post‐inoculation (dpi).
DISCUSSION
This is the first transcriptional study of the interaction between lima bean and the oomycete P. phaseoli, a pathogen of interest because of its damage to lima bean crops and its genetic relatedness to the potato late blight pathogen (Blair et al., 2008). We anchored the P. phaseoli transcriptome using the P. infestans genome as a reference (Haas et al., 2009), and identified a total of 10 427 genes in P. phaseoli (uniquely expressed in either plant or plate‐grown samples, combined with those expressed in both libraries) that mapped to P. infestans genes, out of a total of 17 797 predicted genes (Haas et al., 2009). Contigs that did not match the P. infestans genome were subjected to blastn searches against databases designed from six plant (four Phaseolus spp., soybean and Medicago) and two Phytophthora (P. sojae and P. ramorum) genomes. The majority of contigs from T3 and T6 mapped to the plant genomes, indicating that we had sequenced a mixed population, as expected. Interestingly, 8089 unique contigs from the P18 sample mapped to either the six plant or the two Phytophthora genomes; this may indicate highly conserved genes between plants and oomycetes, or random chance that these short sequences exist in both genomes. There were 23 035 contigs that did not map to any of the eight genomes in this study; these contigs may represent genes involved in the adaptation of the pathogen to lima bean. They may include effector genes, which are now known to populate gene‐poor, repeat‐rich regions in genomes of related Phytophthora, such as P. infestans, and may undergo hundreds of mutations that help the pathogen adapt to new hosts (Haas et al., 2009; Raffaele et al., 2010). Additional time and experimentation will allow the exploration of these ∼23 000 contigs to reveal whether they reside in gene‐rich or gene‐sparse regions of P. phaseoli (Raffaele et al., 2010). Smaller numbers of contigs mapped to the P. sojae and P. ramorum genomes, and it will be interesting to explore whether these represent conserved infection pathways among oomycetes.
Preliminary studies of P. phaseoli indicate that it is roughly 220 Mb in size, compared with a size of 240 Mb for P. infestans (Raffaele et al., 2010). However, the short read P. phaseoli sequences require substantial assembly before we can be certain of its exact size (S. Kamoun, personal communication). A more complete assembly of the genome will also allow us to determine whether the 23 035 contigs belong exclusively to P. phaseoli, and whether they include a suite of effectors unique to this pathogen. Unlike its sister species P. infestans, which infects many tissues of its hosts, P. phaseoli signs and symptoms are only seen primarily on floral parts in the field (Fig. S3, see Supporting Information) and, unlike broad host range Phytophthora species, such as ramorum, capsici and infestans, P. phaseoli only infects lima bean. A closer examination of the ∼23 000 contigs may reveal insights into these unique characteristics of P. phaseoli.
Phytophthora phaseoli gene expression during late stages of infection
We studied the expression of P. phaseoli genes at 3 and 6 dpi compared with expression of plate‐grown P. phaseoli (P18). Of the 10 427 genes, eight and 1284 were differentially expressed at T3 and T6, respectively, when compared with P18. The small number of significantly expressed genes and the difference between the replicates at the earlier time point can probably be attributed to sampling challenges at T3. Neither signs nor symptoms of the pathogen are seen on lima bean plants in hypocotyl assays at T3 (Fig. S3). Therefore, we sampled from areas in which, based on experience, we estimated infection to occur. This does not rule out the possibilities that there might be: (i) very few infection structures in the hypocotyl at this time point; or (ii) having no visual marker, we might have missed the exact location in which the majority of infection was taking place. Future studies will include a microscopic examination of infection during a time course that will aid in our understanding of exactly where infection occurs before signs or symptoms are seen.
Phytophthora phaseoli expresses RxLRs, but not many elicitins or crinklers, during infection
We predicted that RxLRs and CRNs (effectors), and elicitins (PAMPs), would be highly expressed during infection. The results partially supported our hypothesis, in that RxLRs and elicitins were mostly induced, whereas CRNs were mostly repressed, in planta. A similar study by Haas et al. (2009), in which the authors examined genome‐wide expression in P. infestans over four time points from 2 to 5 dpi of potato, revealed different results. In general, RxLRs showed earlier expression in the study by Haas et al. (2009), and later expression in our study (Table 5). Phytophthora phaseoli showed the expression of zero and 14 candidate RxLRs at T3 and T6, respectively, whereas P. infestans showed the expression of 83, 13 and zero RxLRs at 2, 3 and 5 dpi, respectively. Interestingly, the elicitin class showed the induction of zero and four genes at T3 and T6 for P. phaseoli, whereas there were no elicitins induced at all during infection by P. infestans. In a transcriptional study on P. sojae showed induction of RxLR's, NPP1, CRN's and elicitin was high at the germinated‐cyst stage, and another peak later in infection stage on soybean (Ye et al., 2011). Although we need to examine gene expression over a longer time course of infection by P. phaseoli for conclusive statements, at present these data suggest that P. phaseoli expresses some genes in common with P. infestans, but the timing may differ.
Table 5.
Number of genes differentially expressed (P < 0.01) during Phytophthora phaseoli infection of lima bean (this study) compared with Phytophthora infestans infection of potato (Haas et al., 2009).
| Gene family | Number of genes differentially expressed | ||||
|---|---|---|---|---|---|
| Pp3* | Pp6 | Pi2† | Pi3 | Pi5 | |
| RxLR effectors | 0 | 14 | 83 | 13 | 0 |
| Elicitin‐like | 0 | 4 | 0 | 0 | 0 |
| ABC transporter | 0 | 18 | 6 | 4 | 1 |
| Crinkler (CRN) | 0 | 2 | 10 | 2 | 0 |
| NPP1 (necrosis‐inducing Phytophthora protein)‐like | 0 | 1 | 8 | 3 | 0 |
| PcF/SCR‐like | 0 | 1 | 1 | 0 | 0 |
| Enzyme inhibitors | 0 | 1 | 11 | 1 | 0 |
| Sporangia development | 0 | 26 | 1 | 0 | 0 |
| Hypothetical proteins | 0 | 492 | 216 | 25 | 0 |
| Others | 8 | 265 | 133 | 24 | 3 |
| Total | 8 | 824 | 469 | 72 | 4 |
Pp3, P. phaseoli at 3 days post‐inoculation of lima bean.
Pi2, P. infestans at 2 days post‐inoculation of potato (Haas et al., 2009).
Our RxLR dataset did not include several known avirulence genes: Avr3a, Avr4 and Avrblb1 (Armstrong et al., 2005; Van Poppel et al., 2008; Vleeshouwers et al., 2008). To explore this further, we tested for Avr3a with RT‐PCR and saw no evidence of the expression of this gene, nor were we able to detect it in genomic DNA using degenerate primers (data not shown). This is not surprising as data from P. sojae indicate that, of the many strains examined, only those that were avirulent on a particular cultivar of soybean expressed Avr3a (Qutob et al., 2009). The authors suggest that this could be a result of the absence of the gene in this particular strain or gene silencing.
The CRNs are a complex and unique family, expanded in P. infestans (196) relative to P. phaseoli (107; this study), P. sojae (100) and P. ramorum (19) (Haas et al., 2009). CRNs cause crinkling and necrosis of leaf tissue, and two, CRN1 and CRN2, are constitutively active during the colonization of tomato (Torto et al., 2003). Data from several transcriptional studies to date indicate that CRNs are induced during infection; Haas et al. (2009) reported that the few CRN‐encoding genes up‐regulated during the infection of potato at 2–3 dpi were among those most highly expressed. Similarly, a P. sojae transcriptional analysis found that CRNs consistently showed higher expression than either RxLR or NPP‐like effector families (Ye et al., 2011) and, in a separate study, a P. sojae CRN2 homologue was induced early during soybean infection (Moy et al., 2004). Of the 107 CRNs in our dataset, only two showed induction during infection, whereas 18 were repressed and the remainder were unchanged. The reason for the small numbers of CRN genes showing altered expression and the repression of CRNs during infection could be either the infection time points chosen (CRNs might act early during infection) or, simply, that CRNs might not be necessary for P. phaseoli to infect the hypocotyl. Future experiments to determine the role of CRNs will include the analysis of their expression during pod infection, which, unlike the hypocotyl, does show signs of necrosis.
We observed a small group of elicitins with differential expression: four increased, 13 did not change and two decreased. The same explanation as stated above for the CRNs could also apply here to explain the small numbers; briefly, P. phaseoli might not require elicitin induction during the infection of hypocotyls, but might require it to infect pods. Future studies will involve an exploration of whether these elicitins are required for pod infection, or perhaps utilized mainly as PAMPs during an incompatible response.
CONCLUSIONS
We have successfully used the P. infestans genome as a reference for the identification of genes expressed in its close relative, P. phaseoli. Our dataset helps to rectify the paucity of molecular data on this pathosystem, and shows robust pathogen gene expression at 6 dpi, including a handful of RxLR effectors and elicitins, well‐studied gene families known to be involved in virulence in other Phytophthora species.
EXPERIMENTAL PROCEDURES
Phytophthora phaseoli races and growth conditions
Isolate F of P. phaseoli (UD culture collection, strain number PhyP18) is stored on sterilized corn or cucumber seeds and reactivated by transfer onto the semi‐synthetic medium frozen pea agar (Calvert et al., 1960). This medium was also used to grow P. phaseoli for harvesting for RNA. Monthly transfers onto lima bean agar were made to maintain pathogen viability. Pure cultures of P. phaseoli were incubated at their optimum temperature for growth and sporulation of 18 ± 1 °C, following Evans et al. (2007).
Plant inoculations and harvesting of infected tissues
The susceptible lima bean cultivar Concentrated Fordhook (CF) was used for all experiments (Evans et al., 2007). Briefly, actively growing 2‐week‐old plate cultures were used for the inoculation of hypocotyls. Once downy mildew was established, inocula were prepared by cutting the infected hypocotyls into small pieces of 2–4 mm in length, and stirred in reverse osmotic (RO) water in a 250‐mL conical flask to release hyphae and sporangia into the supernatant. The supernatant was then poured directly over 6‐day‐old CF hypocotyls. Inoculated plants were moved to a digitally controlled dew chamber (Percival Scientific, Inc., Perry, IA, USA). Chambers were set to a temperature of 20 ± 1 °C, a relative humidity (RH) of 98% ± 1%, with one 30‐min dew cycle per day. A sterile razor blade was used to scrape only the mycelia from infected plant tissue, thereby reducing the plant tissue. For the second biological replicate that included an earlier time point, a sterile razor blade was used to cut the infected portion of the inoculated hypocotyls. We did not see any signs of P. phaseoli at 3 dpi; therefore, we cut the portion of the hypocotyl that had been in direct contact with the inoculum.
Phytophthora infestans (strain PhyI‐8) was isolated from a commercial potato field in Delaware. Briefly, tomato plants developing their first true leaves were inoculated with an 8‐day‐old pure culture of P. infestans on V‐8 agar. Inoculated plants were incubated at 21 °C and covered with an alcohol‐sterilized plastic bag. Signs were observed at 6 dpi, and infected leaf and petiole were harvested and frozen in liquid nitrogen for nucleic acid extraction.
Fresh lima bean pods infected by P. capsici were provided by T. A. Evans from infected Delaware fields. Pods were photographed and then used for RNA isolation. For comparison, P. capsici mycelium growing on potato dextrose agar plates was also harvested.
Isolation of total RNA
RNA was extracted from all samples described above using Tri reagent (Ambion Inc., Austin, TX, USA) according to the manufacturer's instructions, and resuspended in RNAse‐free water. All samples were measured with a Nano‐Drop spectrophotometer (Wilmington, DE, USA) for concentration and purity. All samples were stored at −80 °C until further use.
RT‐PCR and real‐time quantitative RT‐PCR
RNA extracted from at least two biological replicates was used for cDNA synthesis employing the Quantitect kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. Primers complementary to the DNA of the infection‐related genes were designed for P. phaseoli based on genomic data from the P. infestans database (Table S5, see Supporting Information). RT‐PCR was run on an Eppendorf Mastercycler (Hamburg, Germany) under the following conditions: 95 °C for 2 min, 95 °C for 15 s, 58 °C for 15 s and 68 °C for 20 s for 40 cycles, and, finally, melting curve analysis. For real‐time qRT‐PCR, SYBR Green dye was used (PerfeCTa; VWR, Gaithersburg, MD, USA) according to the instructions. In each case, the internal control was ribosomal protein‐encoding gene S3a and, for quantitative RT‐PCR, plate‐grown mycelium was used as the calibrator. To minimize the misinterpretation of results, water controls were run for all PCR samples, and no products were amplified.
Library preparation and sequencing
Total RNA was extracted from six libraries and was processed for Illumina RNA‐Seq according to the manufacturer's instructions (Illumina, San Diego, CA, USA). The first set was sequenced at the National Center for Genomic Research (NCGR, Santa Fe, NM, USA: http://www.ncgr.org/) and comprised mycelia from infected hypocotyls a 6 dpi (T6‐rep1) and plate‐grown mycelia (P18‐rep1). The second biological replicate was sequenced at the University of Delaware and included tissue from the same conditions described above, together with an additional infection time point taken at 3 dpi (T3‐reps 1 and 2; Table 1). Total RNA from each sample was used to prepare SBS libraries and run on an Illumina Genome AnalyzerII (GAII) for 36 or 42 cycles (Table 1). The first dataset (four lanes of plate‐grown library and three lanes of plant‐grown library) resulted in a total of 9.75 and 28.24 million reads for plate‐grown (P18‐rep1) RNA and plant‐grown (T6‐rep1) RNA, respectively. The second dataset was processed on one flow cell on an Illumina GAII (one lane for each sample), which resulted in a total of 28.24, 30.53, 34.22 and 34.65 million reads PF (passed filter) for one plate‐grown (P18) and three plant‐grown (T3‐rep1, T3rep‐2 and T6‐rep1) RNA samples, respectively. The data were deposited in NCBI's Gene Expression Omnibus, accessible through accession number GSE23453 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23453).
Sequence bioinformatics
Illumina sequence reads were filtered for quality at a 98% confidence level per nucleotide position with a minimum acceptance threshold of 36 bp in length. The seq tags were combined from all treatments into an assembly using Velvet, kmer = 31, to produce a collection of contigs representing the pool of cDNA fragments found in this experiment. A simple blast alignment (blastn) of these cDNA contigs against the known P. infestans gene database was performed using the blastall package (version 2.2.22). The blast results yielded highly positive identification of 117 702 P. phaseoli cDNAs (out of 361 868 contigs; identity > 98%). Of the contigs that did not match to the P. infestans genome, we employed a similar methodology to blast these sequences against eight other genomes: P. sojae (http://genome.jgi‐psf.org/Physo1_1/Physo1_1.download.ftp.htm), P. ramorum (http://genome.jgi‐psf.org/Phyra1_1/Phyra1_1.download.ftp.html), Medicago truncatula (http://www.medicago.org/genome/downloads/Mt3/), Glycine max (http://www.phytozome.net/soybean.php) and four Phaseolus species (Phaseolus coccineus, Phaseolus vulgaris, Phaseolus acutifolius and Phaseolus angustissimus) (http://www.ncbi.nlm.nih.gov/nucest). Transcriptome representation of each cDNA contig was scored by matching each filtered sequence read by treatment sample to the contigs and counting 100% matches. Essentially, this approach produced a metric that was proportional to the sequence coverage across each contig and within each sample. The coverage score for each contig that mapped to P. infestans genes (12 456) was then normalized by the sequence length of each contig to account for the size‐dependent nature of coverage representation in seq tag sequencing strategies. Normalization also involved averaging the reads of each gene, and those with less than 10 reads among all six libraries were filtered out, yielding a final set of 10 427. These data were compiled into the gene expression table that served as input into the edgeR program.
Differential expression analysis
The differential expression analysis was performed using edgeR, a Bioconductor software package for the examination of the differential expression of replicated count data (Robinson and Oshlack, 2010; Robinson et al., 2010). All the steps were followed as described for RNA‐seq data in the edgeR user guide. Briefly, the expression table containing the normalized counts for all six libraries (from above) was loaded into an edgeR library in R. These data were used as the source from which a DGEList (Digital Gene Expression List) object was created, and the library sizes were calculated.
This package calculates the differential expression between two groups only. The three groups were P18, T3 and T6. To identify the genes that were differentially expressed between the groups P18 and T3, the table with normalized counts for these two groups was loaded into edgeR, and then analysed using common dispersion and/or tagwise dispersion. Next, exactTest was used to determine the differentially expressed genes and the table containing the gene ID with the log Conc, log FC, P and FDR values was exported into Microsoft Excel using comma separated files. This table was used to identify the list of effector genes for further analysis. Further, plotSmear was generated using DGE results, wherein the top most differentially expressed genes (P < 0.01 and FDR < 0.05) were identified using tagwise dispersions and highlighted in red on the plot (Fig. 3A). Similar steps were followed to identify the genes that were significantly and differentially expressed for the P18 and T6 pair of groups, and these genes were also highlighted in red on the smear plot (Fig. 3B).
Full‐length isolation of the genes
To amplify the full‐length gene, the P. infestans genome sequence was used as a reference to design primers employing two of the three restriction enzymes in combination (BamHI, PstI, KpnI; Table S5). Primers were designed with the respective adapter sequences and restriction sites (Table S5), both upstream and downstream of the start and stop codons, and high‐fidelity polymerase (KOD polymerase) was used to amplify the full‐length gene in P. phaseoli. The amplified product was cloned into pCambia 1301s (provided by Yinong Yang, Pennsylvania State University, University Park, PA, USA) after digesting with the appropriate restriction enzyme. The resultant clones were sequenced to identify PCR‐derived errors, and a clone with the expected sequence was used for alignments and future experiments. Sequences were assembled and analysed using the Molecular Evolutionary Genetic Analysis program (mega 4.0; (Tamura et al., 2007).
Multiple sequence alignment of P. phaseoli
Multiple sequence alignment was performed to investigate the relationship of five selected predicted effector genes with other Phytophthora spp. using the publicly available program BioEdit (Hall, 1999). Initially, eight clones for each gene were sequenced by Sanger‐based methods using primers complementary to the vector sequence and flanking the insert. For genes with more than 1 kb of readable sequence, gene‐specific primers were designed at an interval of 700 bp. The full‐length sequence, translated using BioEdit, was used to perform a protein–protein blast (blastp) and position‐specific iterated blast (PSI‐blast) search in NCBI, and the protein sequences of other Phytophthora species were downloaded into text files. These files were then employed for multiple sequence alignments using BioEdit.
Construct development for agro‐infiltration
To construct a binary expression vector, we cloned several of the effector genes (four elicitins: PpINF1, PpINF4, PpPITG_06908 and PpPITG_20413) into the binary vector pCambia 1301s as described above. The ligated vector (constructs) was transformed independently into Agrobacterium tumefaciens strain GV3101 using electroporation. These constructs were sequenced to confirm the full‐length sequences and later used for transient assay (infiltration) on N. benthamiana leaves.
Agrobacterium tumefaciens‐mediated transient expression
The recombinant A. tumefaciens containing the different binary plasmids was grown as described previously (Kanneganti et al., 2006; Van der Hoorn et al., 2000). Briefly, recombinant A. tumefaciens was grown overnight (28–30 °C, 150 rpm; ThermoScientific Max Q 6000 incubator, Dubuque, IA, USA) in tubes containing 5 mL of medium with 50 µg/mL of kanamycin (Fisher, Fair Lawn, NJ, USA). Cells were harvested at an optical density at 600 nm (OD600) of 0.8–1.2 by centrifugation (8 min, 4000 g) and then resuspended in MMA induction buffer (Induction buffer=1 L MMA: 5 g Murashige and Skoog (MS) salts, 1.95 g 2‐(N‐morpholino)ethanesulphonic acid, 20 g sucrose, 200 µm acetosyringone, pH 5.6) to a final OD600 of 0.3–0.5. All suspensions were incubated for 3–4 h prior to infiltration onto 5–6‐day‐old N. benthamiana leaves. Symptom development was monitored 3–19 days after infiltration. Two constructs, PGR106INF1 and PGR106GFP, received from Dennis Halterman (University of Wisconsin, Madison, WI, USA), were used as controls. Infiltrations of these recombinants were performed independently onto the abaxial side of the leaf with a 1‐mL disposable syringe without a needle. Leaves were superficially wounded with a needle to improve infiltration.
Hypersensitive cell death assay
To better visualize plant cells that were undergoing an HR caused by the infiltrated elicitins, leaves that showed macroscopic cell death were stained and cleared with lactophenol–trypan blue (Tang et al., 1999). Briefly, we boiled the leaves in lactophenol (lactic acid : phenol : glycerol, 1:1:1) containing 0.01% trypan blue for 10 min and rinsed them in 50% ethanol and then in water.
Supporting information
Fig. S1 Distribution of the contig length to the contig coverage. Contig length and coverage are shown on the x axis and y axis, respectively.
Fig. S2 Expression of 16 candidate virulence genes during a time course of Phytophthora phaseoli on lima bean. Lane 1, 12 h post‐inoculation (hpi). Lane 2, 48 hpi. Lane 3, 72 hpi. Lane 4, 120 hpi. Lane 5, PCR water control. This experiment was performed with at least two biological replicates.
Fig. S3 (A–F) Signs and symptoms of downy mildew on lima bean caused by Phytophthora phaseoli. (A–C) Signs of the pathogen in the field: (A) shoot tips; (B) floral raceme; (C) old pod. (D–F) Signs of the pathogen in the glasshouse: (D) lima bean hypocotyl inoculated with water at 6 days post‐inoculation (dpi); (E) hypocotyl inoculated with P. phaseoli at 3 dpi; (F) mycelia and sporangia at 6 dpi on hypocotyl and leaf. Signs and symptoms of P. infestans potato blight in the glasshouse (G, H) and field (I): (G) healthy potato plant inoculated with water at 7 dpi; (H) advanced symptoms on potato seedling at 7 dpi; (I) early infection symptoms on potato. (J, K) Signs and symptoms of P. infestans tomato blight: (J) tomato plant inoculated with water at 7 dpi; (K) advanced symptoms on tomato seedling at 7 dpi. Images G–K were taken by Nancy Gregory.
Table S1 Table showing the list of contigs formed by pooling six libraries. The table contains the length of the contigs and the coverage and the raw counts of the sequences from six libraries that matched to each contigs. The last column contains the PITG gene number from the P. infestans database that matched to the contigs.
Table S2 Table showing the normalized counts (see materials and methods for details) for all the six libraries that macthed to the PITG genes of P. infestans database.
Table S3 Table showing the results of differential gene analysis using edgeR for the pair T3–P18. It has the list of PITG genes with logFC (fold‐change), P‐value, false discovery rate (FDR), and normalised counts of replicates for the samples (T3 and P18). The average of the counts for replicates were considered to identify if the genes were up‐regulated in one or the other sample. The Benjamini and Hochberg's (BH) method was used to adjust P‐values for multiple testing and for controlling the false discovery rate (FDR).
Table S4 Table showing the results of differential gene analysis using edgeR for the pair T6–P18. It has the list of PITG genes with logFC (fold‐change), P‐value, false discovery rate (FDR), and normalized counts of replicates for the samples (T6 and P18). The average of the counts for replicates were considered to identify if the genes were up‐regulated in one or the other sample. The Benjamini and Hochberg's (BH) method was used to adjust P‐values for multiple testing and for controlling the false discovery rate (FDR).
Table S5 List of primers used for RT‐PCR, qRT‐PCR and full‐length isolation of the genes.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The authors wish to thank the University of Delaware's College of Agriculture and Natural Resources for two seed grants to NMD (SHD CANR SEED 07) and TAE (SHD CANR SEED 08), which largely funded this project. The authors would also like to thank Dr Greg May at the National Center for Genomic Research (Santa Fe, NM, USA) for sequencing the first biological replicate set of samples. We also gratefully acknowledge Sophien Kamoun and Liliana Cano at the Sainsbury Laboratory (Norwich, UK) for helpful discussions, and Brett Tyler and Sucheta Tripathy at the Virginia Bioinformatics Institute, Blacksburg, VA, USA for assistance with P. sojae and P. ramorum bioinformatics. Thanks are due to Dennis Halterman (University of Wisconsin, Madison, WI, USA) and Y. Yang (Pennsylvania State University, University Park, PA, USA) for kindly providing PGR106INF and control vectors and for the pCAMBIA expression vectors, respectively.
REFERENCES
- App, F. (1959) The history and economic importance of lima bean downy mildew disease. Proc. Am. Soc. Hortic. Sci. 33, 473–476. [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I. , Venter, E. , Avrova, A.O. , Rehmany, A.P. , Böhme, U. , Brooks, K. , Cherevach, I. , Hamlin, N. , White, B. , Fraser, A. , Lord, A. , Quail, M.A. , Churcher, C. , Hall, N. , Berriman, M. , Huang, S. , Kamoun, S. , Beynon, J.L. and Birch, P.R. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmann, Y.W. , Wallace, M.B. and Thompson, E.A. (2008) Transcriptome profiling using next‐generation sequencing. Gastroenterology, 135, 1466–1468. [DOI] [PubMed] [Google Scholar]
- Birch, P.R. , Rehmany, A.P. , Pritchard, L. , Kamoun, S. and Beynon, J.L. (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 14, 8–11. [DOI] [PubMed] [Google Scholar]
- Blair, J.E. , Coffey, M.D. , Park, S.‐Y. , Geiser, D.M. and Kang, S. (2008) A multi‐locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 45, 266–277. [DOI] [PubMed] [Google Scholar]
- Bos, J.I.B. , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. , Armstrong, M.R. , Birch, P.R.J. and Kamoun, S. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Calvert, O.H. , Williams, L.F. and Whitehead, M.D. (1960) Frozen‐Lima‐Bean agar for culture and storage of Phytophthora sojae . Phytopathology, 50, 136–137. [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest, E.G. , Kee, E. , Santamaria, L. and Evans, T.A. (2006) Inheritance of resistance to lima bean downy mildew (Phytophthora phaseoli) and preliminary lima improvement efforts. Annu. Rep. Bean Improv. Coop. 49, 37–38. [Google Scholar]
- Evans, T.A. , Davidson, C.R. , Dominiak, J.D. , Mulrooney, R.P. , Carroll, R.B. and Antonius, S.H. (2002) Two new races of Phytophthora phaseoli from lima bean in Delaware. Plant Dis. 86, 813. [DOI] [PubMed] [Google Scholar]
- Evans, T.A. , Mulrooney, R.P. , Gregory, N.F. and Kee, E. (2007) Lima bean downy mildew: impact, etiology, and management strategies for Delaware and the Mid‐Atlantic Region, U.S. Plant Dis. 91, 128–135. [DOI] [PubMed] [Google Scholar]
- French, E.R. and Mackay, G.R. (1996) Enhancing the global late blight network. Report of the project design meeting on the global initiative on late blight. CIP, Lima, Peru, Centro Internacional de la papa. 17–20.
- Fry, W. (2008) Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9, 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen, M. and Nurnberger, T. (2006) Nep1‐like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M.V. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I.B. , Bulone, V. , Cai, G.H. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grunwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M.E. , Jones, J.D.G. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z.Y. , Ma, L.J. , MacLean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J.G. , Morgan, W. , Morris, P.F. , Munro, C.A. , O’Neill, K. , Ospina‐Giraldo, M. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q.H. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J.I. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S.G. , Fry, W. , Meyers, B.C. , van West, P. , Ristaino, J. , Govers, F. , Birch, P.R.J. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009)Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature,461,393–398. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A.L. , Terauchi, R. and Kamoun, S. (2009) Emerging Concepts in Effector Biology of Plant‐Associated Organisms. Mol. Plant-Microbe Interact . 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H. and Govers, F. (2006) Nonneutral GC3 and retroelement codon mimicry in Phytophthora . J. Mol. Evol. 63, 458–472. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H. , Tyler, B.M. , Whisson, S.C. , Hardham, A.R. and Govers, F. (2006) Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 23, 338–351. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. , Ah‐Fong, A.M.V. , Aux, G. , Avrova, A.O. , Bruce, C. , Calkir, C. , da Cunha, L. , Grenville‐Briggs, L. , Latijnhouwers, M. , Ligterink, W. , Meijer, H.J. , Roberts, S. , Thurber, C.S. , Whisson, S.C. , Birch, P.R. , Govers, F. , Kamoun, S. , van West, P. and Windass, J. (2008) Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol. Plant–Microbe Interact. 21, 433–447. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Lindqvist, H. and Govers, F. (1997a) A novel class of elicitin‐like genes from Phytophthora infestans . Mol. Plant–Microbe Interact. 10, 1028–1030. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van West, P. , de Jong, A.J. , de Groot, K.E. , Vleeshouwers, V.G. and Govers, F. (1997b) A gene encoding a protein elicitor of Phytophthora infestans is down‐regulated during infection of potato. Mol. Plant–Microbe Interact. 10, 13–20. [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.D. , Huitema, E. , Cakir, C. and Kamoun, S. (2006) Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl‐like protein PiNPP1.1 and INF1 elicitin. Mol. Plant–Microbe Interact. 19, 854–863. [DOI] [PubMed] [Google Scholar]
- Moy, P. , Qutob, D. , Chapman, B.P. , Atkinson, I. and Gijzen, M. (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae . Mol. Plant–Microbe Interact. 17, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T. and Brunner, F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen‐associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Tedman‐Jones, J. , Dong, S. , Kuflu, K. , Pham, H. , Wang, Y. , Dou, D. , Kale, S.D. , Arredondo, F.D. , Tyler, B.M. and Gijzen, M. (2009)Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS ONE,4,e5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S. , Farrer, R.A. , Cano, L.M. , Studholme, D.J. , MacLean, D. , Thines, M. , Jiang, R.H. , Zody, M.C. , Kunjeti, S.G. , Donofrio, N.M. , Meyers, B.C. , Nusbaum, C. and Kamoun, S. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science, 330, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Rizzo, D.M. , Garbelotto, M. and Hansen, E.M. (2005) Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 43, 309–335. [DOI] [PubMed] [Google Scholar]
- Robinson, M.D. and Oshlack, A. (2010) A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. and Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Motteram, J. , Kufner, I. , Deller, S. , Brunner, F. , Hammond‐Kosack, K.E. and Nürnberger, T. (2009) Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain‐containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 22, 790–799. [DOI] [PubMed] [Google Scholar]
- Schmidt, B. , Sinha, R. , Beresford‐Smith, B. and Puglisi, S.J. (2009) A fast hybrid short read fragment assembly algorithm. Bioinformatics, 25, 2279–2280. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Xie, M. , Kim, Y.J. , Zhou, J. , Klessig, D.F. and Martin, G.B. (1999) Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell, 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Benynon, J.L. , Chapman, J. , Damasceno, C.M. , Dorrance, A.E. , Dou, D. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grunwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. , Kamoun, S. , Krampis, K. , Lamour, K.H. , Lee, M.K. , McDonald, W.H. , Medina, M. , Meijer, H.J. , Nordberg, E.K. , Maclean, D.J. , Ospina‐Giraldo, M.D. , Morris, P.F. , Phuntumart, V. , Putnam, N.H. , Rash, S. , Rose, J.K. , Sakihama, Y. , Salamov, A.A. , Savidor, A. , Scheuring, C.F. , Smith, B.M. , Sobral, B.W. , Terry, A. , Torto‐Alalibo, T.A. , Win, J. , Xu, Z. , Zhang, H. , Grigoriev, I.V. , Rokhsar, D.S. and Boore, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A. , Laurent, F. , Roth, R. and De Wit, P.J. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf‐9‐induced and Avr4/Cf‐4‐induced necrosis. Mol. Plant–Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Van Poppel, P.M. , Guo, J. , van de Vondervoort, P.J. , Jung, M.W. , Birch, P.R. , Whisson, S.C. and Govers, F. (2008) The Phytophthora infestans avirulence gene Avr4 encodes an RXLR‐dEER effector. Mol. Plant–Microbe Interact. 21, 1460–1470. [DOI] [PubMed] [Google Scholar]
- Van Poppel, P.M.J.A. , Jiang, R.H.Y. , ŚLiwka, J. and Govers, F. (2009) Recognition of Phytophthora infestans Avr4 by potato R4 is triggered by C‐terminal domains comprising W motifs. Mol. Plant Pathol. 10, 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G.F. , Jacobsen, E. , Govers, F. , Kamoun, S. and Van der Vossen, E.A.G. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Ye, W. , Wang, X. , Tao, K. , Lu, Y. , Dai, T. , Dong, S. , Dou, D. , Gijzen, M. and Wang, Y. (2011) Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant–Microbe Interact. DOI: 10.1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Distribution of the contig length to the contig coverage. Contig length and coverage are shown on the x axis and y axis, respectively.
Fig. S2 Expression of 16 candidate virulence genes during a time course of Phytophthora phaseoli on lima bean. Lane 1, 12 h post‐inoculation (hpi). Lane 2, 48 hpi. Lane 3, 72 hpi. Lane 4, 120 hpi. Lane 5, PCR water control. This experiment was performed with at least two biological replicates.
Fig. S3 (A–F) Signs and symptoms of downy mildew on lima bean caused by Phytophthora phaseoli. (A–C) Signs of the pathogen in the field: (A) shoot tips; (B) floral raceme; (C) old pod. (D–F) Signs of the pathogen in the glasshouse: (D) lima bean hypocotyl inoculated with water at 6 days post‐inoculation (dpi); (E) hypocotyl inoculated with P. phaseoli at 3 dpi; (F) mycelia and sporangia at 6 dpi on hypocotyl and leaf. Signs and symptoms of P. infestans potato blight in the glasshouse (G, H) and field (I): (G) healthy potato plant inoculated with water at 7 dpi; (H) advanced symptoms on potato seedling at 7 dpi; (I) early infection symptoms on potato. (J, K) Signs and symptoms of P. infestans tomato blight: (J) tomato plant inoculated with water at 7 dpi; (K) advanced symptoms on tomato seedling at 7 dpi. Images G–K were taken by Nancy Gregory.
Table S1 Table showing the list of contigs formed by pooling six libraries. The table contains the length of the contigs and the coverage and the raw counts of the sequences from six libraries that matched to each contigs. The last column contains the PITG gene number from the P. infestans database that matched to the contigs.
Table S2 Table showing the normalized counts (see materials and methods for details) for all the six libraries that macthed to the PITG genes of P. infestans database.
Table S3 Table showing the results of differential gene analysis using edgeR for the pair T3–P18. It has the list of PITG genes with logFC (fold‐change), P‐value, false discovery rate (FDR), and normalised counts of replicates for the samples (T3 and P18). The average of the counts for replicates were considered to identify if the genes were up‐regulated in one or the other sample. The Benjamini and Hochberg's (BH) method was used to adjust P‐values for multiple testing and for controlling the false discovery rate (FDR).
Table S4 Table showing the results of differential gene analysis using edgeR for the pair T6–P18. It has the list of PITG genes with logFC (fold‐change), P‐value, false discovery rate (FDR), and normalized counts of replicates for the samples (T6 and P18). The average of the counts for replicates were considered to identify if the genes were up‐regulated in one or the other sample. The Benjamini and Hochberg's (BH) method was used to adjust P‐values for multiple testing and for controlling the false discovery rate (FDR).
Table S5 List of primers used for RT‐PCR, qRT‐PCR and full‐length isolation of the genes.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


