Summary
The responses of two closely related members of Arabidopsis 13‐lipoxygenases (13‐LOXs), LOX3 and LOX4, to infection by the sedentary nematodes root‐knot nematode (Meloidogyne javanica) and cyst nematode (Heterodera schachtii) were analysed in transgenic Arabidopsis seedlings. The tissue localization of LOX3 and LOX4 gene expression using β‐glucuronidase (GUS) reporter gene constructs showed local induction of LOX3 expression when second‐stage juveniles reached the vascular bundle and during the early stages of plant–nematode interaction through gall and syncytia formation. Thin sections of nematode‐infested knots indicated LOX3 expression in mature giant cells, and high expression in neighbouring cells and those surrounding the female body. LOX4 promoter was also activated by nematode infection, although the GUS signal weakened as infection and disease progressed. Homozygous insertion mutants lacking LOX3 were less susceptible than wild‐type plants to root‐knot nematode infection, as reflected by a decrease in female counts. Conversely, deficiency in LOX4 function led to a marked increase in females and egg mass number and in the female to male ratio of M. javanica and H. schachtii, respectively. The susceptibility of lox4 mutants was accompanied by increased expression of allene oxide synthase, allene oxide cyclase and ethylene‐responsive transcription factor 4, and the accumulation of jasmonic acid, measured in the roots of lox4 mutants. This response was not found in lox3 mutants. Taken together, our results reveal that LOX4 and LOX3 interfere differentially with distinct metabolic and signalling pathways, and that LOX4 plays a major role in controlling plant defence against nematode infection.
Keywords: Arabidopsis thaliana, giant cell system, jasmonate pathway, lipoxygenase, Meloidogyne, parasitism, plant response
Introduction
Meloidogyne spp. root‐knot nematodes (RKNs) and cyst nematodes, such as Heterodera schachtii, are destructive pathogens that exhibit a sophisticated interaction with plants governed by continuous signal exchange between the two (Abad et al., 2003; Bird et al., 2004; Sobczak and Golinowski, 2011). Being sedentary parasites, their development is strictly dependent on nematode feeding site (NFS) formation and maintenance, ensuring a continuous nutrient supply (Vanholme et al., 2004). Although the mechanisms involved in the formation of giant cells (GCs) and syncytia by RKNs and cyst nematodes, respectively, during their establishment are not yet well understood, more and more studies have indicated that parasitism effectors secreted into the plant tissues by the nematodes interact with essential plant components, leading to the establishment and maintenance of NFSs (Atkinson, 1994; Niebel et al., 1993; Rosso and Grenier, 2011; Sijmons, 1993; Vanholme et al., 2004). As obligate endoparasites that complete most of their life cycle inside the roots, the ability of RKNs to overcome plant defences and maintain these feeding cells makes them extremely successful at damaging a broad range of hosts. In the last few decades, knowledge derived from intraspecific transcriptomics of Arabidopsis thaliana, Glycine max (soybean) and Solanum lycopersicum (tomato) plants following recognition of plant‐parasitic nematodes has shed light on the complex, multistep signalling cascades that lead to the accommodation of, or defence against, these nematodes (Alkharouf et al., 2006; Dubreuil et al., 2007; Ibrahim et al., 2011; Portillo et al., 2013; Szakasits et al., 2009).
Among the signalling cascades, fatty acid (FA) metabolic pathways mediated by lipid signalling molecules, known as oxylipins, regulate many defence and developmental pathways and present unique shared signalling elements (Feussner and Wasternack, 2002). Catalysed by the activities of the lipoxygenase (LOX: 9‐LOX or 13‐LOX) and α‐dioxygenase enzyme families (Chen et al., 2004; Feussner and Wasternack, 2002; Hamberg et al., 2005; Vicente et al., 2012), oxylipins play a fundamental role in the physiological and pathological responses of plants and vertebrates (Vellosillo et al., 2007). Numerous studies aimed at defining the action of oxylipins have shown that, in addition to their being regulators of developmental stages (Caldelari et al., 2011; Vellosillo et al., 2007), expression of oxylipin biosynthetic genes is specifically induced on inoculation with plant pathogens (Jalloul et al., 2002; Melan et al., 1993; Sanz et al., 1998; Turner et al., 2002). Recent discoveries have demonstrated more direct roles for FAs and their breakdown products, oxylipins, in inducing various modes of plant defence by modulating basal, effector‐triggered and systemic immunity in plants (Kachroo and Kachroo, 2009; Vicente et al., 2012). In Arabidopsis, 13‐LOX‐derived oxylipin biosynthesis is induced on recognition of the avirulent protein Avr‐Rpm1, and is associated with hypersensitive response (HR) induction during race‐specific resistance to Pseudomonas syringae (Andersson et al., 2006). However, despite its widely recognized biological significance, the nature of the oxylipin‐mediated crosstalk between plants and nematodes remains unknown. Several lines of evidence implicate lipid metabolism in plant defence reactions against parasitic nematodes; for example, a temporal correlation between LOX expression and nematode infection was demonstrated in Pisum sativum infected by Heterodera goettingiana (Veronico et al., 2006). Similarly, microarray‐based expression profiling has indicated that several FA metabolism genes, including those encoding LOXs, patatin‐like protein 1 and 12‐oxo‐phytodienoate reductase (OPR), are induced in susceptible soybean roots in response to soybean cyst nematode (SCN) infection (Alkharouf et al., 2006). Microarray analysis of GC‐enriched A. thaliana root tissue revealed that LOX and pathogenesis‐related (PR) proteins are down‐regulated by the nematode during this compatible interaction (Jammes et al., 2005). Up‐regulation of LOX genes was also reported in syncytia induced on soybean resistance genotypes by an avirulent population of SCN (Klink et al., 2009, 2010). Genetic evidence for LOX participation in a Zea mays (maize)–RKN interaction was demonstrated by modification of the plant's response to infection in mutants and transgenic lines altered in the synthesis of oxylipins (Gao et al., 2008).
Among the best‐characterized oxylipins are jasmonic acid (JA) and its immediate precursor 12‐oxo‐phytodienoic acid (OPDA), which are formed enzymatically and accumulate in response to various stresses, in particular wounding and pathogen infection (Block et al., 2005). Although considerable genetic evidence suggests that JA is an important signalling molecule in plant defence (Takahashi et al., 2004), our current understanding of JA signalling in plant–nematode associations is less clear. The exogenous application of JA (as methyl jasmonate) in rice resulted in nematodes being less effective in counteracting root defence pathways, demonstrating a pivotal role for JA in rice defence against the RKN Meloidogyne graminicola (Nahar et al., 2011). Conversely, in maize, gene expression studies, together with genetic results, provided compelling evidence for JA as a negative regulator of plant resistance: a mutant lacking ZmLOX3 function led to increased levels of JA and increased susceptibility to the RKN Meloidogyne incognita (Gao et al., 2008). Similar to maize, the use of JA mutant lines of tomato demonstrated that intact JA signalling is required for tomato susceptibility to the RKN Meloidogyne javanica, although it was not involved in Mi‐gene‐mediated resistance (Bhattarai et al., 2008). In addition, in their review, Gutjahr and Paszkowski (2009) stated that negative pathway crosstalk may occur between the JA and salicylic acid (SA) signalling pathways; thus Mi‐gene‐dependent, SA‐mediated resistance to RKN might be suppressed by JA signalling (Bhattarai et al., 2008). Similarly, cyst nematode parasitism of Arabidopsis is inhibited by SA (Wubben et al., 2008), whereas the role of JA signalling in the promotion or suppression of root colonization by cyst nematodes is still under investigation (Gutjahr and Paszkowski, 2009).
Although the first step in JA biosynthesis is catalysed by a 13‐LOX (Bannenberg et al., 2009), we still do not know which 13‐LOX isoforms contribute to JA biosynthesis during nematode infection. Investigation of the A. thaliana genome has led to the identification of six predicted LOX genes encoding four 13‐LOXs (LOX2, LOX3, LOX4 and LOX6) and two 9‐LOXs (LOX1 and LOX5) (Bannenberg et al., 2009). Given the central regulatory roles of specific 9‐ and 13‐LOX‐derived oxylipins in regulating plant defence responses, an understanding of the function of individual LOX isoforms in plant–nematode interactions is fundamental. Among the 13‐LOXs, A. thaliana LOX2 (AtLOX2) appears to be distantly related to AtLOX‐3, AtLOX‐4 and AtLOX‐6, which are clustered together and have been suggested to be involved in the plant's defence pathway (Bannenberg et al., 2009). Strikingly, LOX3 and LOX4 are considered to be highly similar, sharing 85% overall amino acid identity and 97% in the substrate‐binding pocket sequence (Caldelari et al., 2011). Given that LOX3 and LOX4 are the only 13‐LOXs being expressed in roots (Vellosillo et al., 2007), we pursued and analysed the function of these LOXs in Arabidopsis in response to plant‐parasitic nematodes. Previous research has shown that AtLOX3 is expressed in the lateral root (LR) primordium and during LR development (Vellosillo et al., 2007). Moreover, gene transcription of both AtLOX3 and AtLOX4 has been shown to be activated during leaf senescence (He et al., 2002), as well as by pathogen infection and on exposure to methyl jasmonate, with no detection of AtLOX6 expression in the examined seedlings (Melan et al., 1993, 1994; Vellosillo et al., 2007). Finally, a recent study has shown that Arabidopsis lox3 lox4 double mutants are male sterile with no viable pollen, although fertility in the double mutant is restored genetically by complementation with either LOX3 or LOX4 cDNAs or biochemically with exogenous application of JA (Caldelari et al., 2011).
We studied the role of the two closely related 13‐LOX members, LOX3 and LOX4, in Arabidopsis parasitism by nematodes. Our data indicate that both LOX3 and LOX4 isoforms are triggered in response to nematode infection. However, our findings place LOX4 as the dominant member in regulating the root susceptibility response to plant‐parasitic nematodes, and LOX3 mainly as a player in regulating the developmental alterations induced by the developing nematode.
Results
LOX3 and LOX4 promoter activities are differentially affected by nematode infection
The expression patterns of LOX3 and LOX4 were examined in transgenic Arabidopsis lines containing the LOX3 and LOX4 promoters fused to a β‐glucuronidase (GUS) reporter gene at various time points post‐inoculation. Uninfected roots of each line were examined to localize LOX3 and LOX4 expression. GUS activity of LOX3‐GUS plants was consistent with a previous study (Vellosillo et al., 2007): its expression was primarily restricted to the LR primordium, but it declined during subsequent development of the LR, with no GUS signal detected in the mature root tip (Fig. 1A–C). Stronger staining was observed on RKN infection (Fig. 1). Thus, associated with second‐stage juvenile (J2) migration, GUS activity was observed mainly within the differentiating vascular bundle in which the nematode is located during the early parasitic stages [i.e. 24 h post‐inoculation (hpi)] (Fig. 1D). Within 48 and 72 hpi, root swelling induced by developing J2s was accompanied by a pronounced GUS signal, mainly in the vascular tissue (Fig. 1E–H). A similar, intense GUS signal was observed at later time points throughout the entire galls at 28 days post‐inoculation (dpi) (Fig. 1I).
Figure 1.
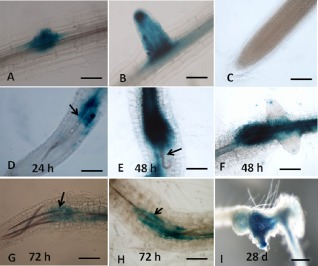
Microscopic analysis of β‐glucuronidase (GUS) expression patterns on root‐knot nematode (RKN) infection in Arabidopsis plants harbouring the lipoxygenase‐3 (LOX3) promoter‐GUS fusion construct. Non‐infested control root harbouring the LOX3‐GUS fusion construct demonstrated strong GUS staining of lateral root primordia (A) and emerging lateral root (B). GUS staining decreased with time for the mature lateral root and was no longer detected in the mature root tip (C). In Arabidopsis plants infested with Meloidogyne javanica, a strong GUS signal was observed during second‐stage juvenile (J2) migration along the vascular bundle at 24 h post‐inoculation (hpi) (D) and during entrance into the sedentary phase at 48 and 72 hpi (E–H). Strong GUS staining was observed in galls at 28 days post‐inoculation (dpi) (I) by M. javanica. (A–H) Light micrographs as viewed under light microscope. (I) Bright‐field image of galls photographed using a stereomicroscope. Bars: (A) 50 μm; (B–H) 100 μm; (I) 500 μm.
We then examined the expression of the closely related member, LOX4, during RKN infection. In non‐infected Arabidopsis plants harbouring the LOX4‐GUS construct, GUS activity was associated with the stem–root junction, suggesting a functional role for LOX4 in the differentiation processes (Fig. 2A). Similar to Vellosillo et al. (2007), no LOX4 promoter activity was detected in the LR primordium, LR or root tip of uninfested plants (Fig. 2B,C). Compared with LOX3 promoter activity, a weak GUS signal was induced by the LOX4 promoter following RKN penetration and migration (Fig. 2D–G). Similarly, galls showed only a weak GUS signal that remained after egg production by the developed RKN female (Fig. 2H,I).
Figure 2.

Microscopic analysis of β‐glucuronidase (GUS) expression patterns on root‐knot nematode (RKN) infection in Arabidopsis plants harbouring the lipoxygenase‐4 (LOX4) promoter‐GUS fusion. Non‐infested control root harbouring the LOX4‐GUS fusion construct demonstrated consistent GUS staining at the stem–root junction (A), whereas no GUS staining was detected in the root primordia, developing lateral roots or mature root tip (B, C). On nematode inoculation, GUS expression was correlated with nematode parasitic stages: a weak GUS signal was observed during the migratory phase of Meloidogyne javanica second‐stage juveniles (J2s) at 24 h post‐inoculation (hpi) (D), 48 hpi (E, F) and 72 hpi (G). The GUS signal remained faint in galls at 21 and 28 days post‐inoculation (dpi) (H, I) by M. javanica. Bars, 100 μm.
LOX3 and LOX4 promoter activity in GCs induced by RKNs
To localize LOX3 and LOX4 gene expression in the different gall tissues, such as GCs and the surrounding vascular elements, thin sections were cut from 28‐day‐old knots developed in Arabidopsis lines harbouring the LOX3‐GUS or LOX4‐GUS construct following infection with M. javanica. Microscopic examination of stained root sections of both plants revealed LOX3 expression in mature GCs, as well as in dividing cells bordering the nematode‐induced GCs and the cells surrounding the female body (Fig. 3A–C). The analysis of root sections for LOX4 promoter activity indicated lower expression than that of LOX3; LOX4 expression was detected as a weak GUS signal within the dividing cells surrounding the GCs and the female body, with no signal in the GCs formed in the LOX4‐GUS line following RKN infection (Fig. 3D–F).
Figure 3.
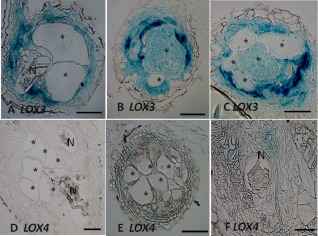
Microscopic analysis of β‐glucuronidase (GUS) activity in cross‐section of Arabidopsis root gall expressing lipoxygenase‐3 (LOX3) and LOX4 promoter‐GUS constructs. At 28 days post‐inoculation (dpi), all giant cells were mature and nematodes developed into the fourth juvenile stage. Histological analysis of roots expressing the LOX3‐GUS construct clearly showed strong GUS expression inside the cells surrounding the female body and the hyperplastic cells surrounding the giant cells inside the vascular cylinder (A), and within the developed giant cell systems (B, C). Arabidopsis roots expressing the LOX4‐GUS construct still showed a very weak signal in a few cells associated with the developed giant cell system (D–F). The female body of the nematode (N) can be seen at the edge of the giant cells (*). Bars, 200 μm.
LOX3 and LOX4 promoter activity after cyst nematode infection
We also analysed LOX3 and LOX4 promoter activity after infection with another group of endoparasitic nematodes, the cyst nematodes. The LOX3‐GUS line infected with H. schachtii J2s was activated as early as 24 hpi and the signal was centred inside the vascular cylinder close to the nematode head (Fig. 4A). At 48 hpi, the signal was very intense and localized around the nematode throughout the induced syncytia and surrounding vascular cylinder (Fig. 4B). At late stages of infection, during mature female development into a cyst, a clear GUS signal remained indicating continuous induction of LOX3 by the cyst nematode (Fig. 4C). The activation pattern of the LOX4 promoter was similar to that of LOX3, but with lower intensity (Fig. 4D–F). It was also activated early, at 24 hpi, with a signal around the nematode body (Fig. 4D) and clear staining at 48 hpi (Fig. 4E). The activity was maintained into the late infection stages, but the staining intensity decreased gradually from the earlier time points to 21 dpi (Fig. 4F).
Figure 4.
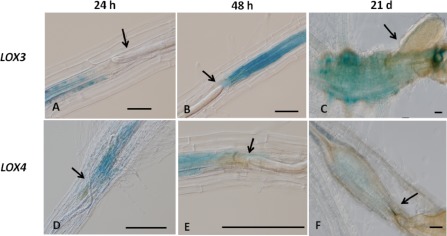
Microscopic analysis of β‐glucuronidase (GUS) expression patterns on cyst nematode infection in Arabidopsis plants harbouring the lipoxygenase‐3 (LOX3) and LOX4 promoter‐GUS fusions. Consistent GUS staining was observed as early as 24 h post‐inoculation (hpi) and the signal was centred inside the vascular cylinder close to the nematode head (A). The signal increased as the infection progressed to 48 hpi (B), and the GUS signal remained at late infection stages at 21 days post‐inoculation (dpi) (C). In Arabidopsis plants harbouring the LOX4 promoter‐GUS fusion, LOX4 was activated at 24 hpi around the nematode body inside the vascular cylinder (D), and a clear signal was also observed at 48 hpi (E). The activity was maintained until the late infection stages, but the intensity decreased markedly at 21 dpi (F). Bars, 100 μm.
LOX3 and LOX4 are induced by mechanical wounding
To determine whether the 13‐LOX members LOX3 and LOX4 play a role in the wound‐induced local or systemic response, root samples of LOX3‐GUS and LOX4‐GUS seedlings were collected at 9 and 24 h after wounding and were assayed for GUS staining. Intense LOX3 promoter activity was revealed on wounding, as reflected by rapid GUS staining at the wounded site which spread further along the axis of the vascular cylinder of the injured LOX3‐GUS line (Fig. 5A,B compared with unwounded control in Fig. 5C). In comparison with LOX3‐GUS roots, LOX4‐GUS wounded roots showed a weak GUS signal that remained confined to the wounded tissue (Fig. 5D,E compared with the unwounded control in Fig. 5F). As JA is well known as an essential signal in wound‐induced gene expression, we assessed whether the LOX3 and LOX4 promoters contain JA‐responsive cis‐elements by scanning 2000 bp of the LOX3 and LOX4 promoter sequences using the PLACE (plant cis‐acting elements) database (Higo et al., 1999). Two JA‐responsive cis‐elements were identified in the AtLOX3 promoter, T/GBOXATPIN2, together with a GCC‐box motif (Brown et al., 2003), but not in LOX4, potentially explaining the intense wound responsiveness of the LOX3 promoter. To evaluate sequence motifs that may be common among promoters of NFS‐expressed genes, we compared the motifs found in the LOX3 and LOX4 promoters with those of other genes known to be regulated in NFS. Analysis of the promoter regions revealed that, of the four motifs that have been shown to be present in NFS‐up‐regulated promoter sequences (Escobar et al., 1999; Fenoll et al., 1997; Mazarei et al., 2002; Puzio et al., 2000; Sukno et al., 2006; Thurau et al., 2003), only the elicitor‐responsive element (EIRE) motif was found in LOX3 promoters, but not in the LOX4 promoter region. In addition, the NEMATODE‐box motif (a nematode‐responsive element) CAATTG, described by Escobar et al. (1999), was found three times in the LOX3 promoter, but only once in the LOX4 promoter. Further promoter scanning for cis‐regulatory elements using PlantCARE, ATHENA and Genomatix Matinspector databases revealed the presence of several motifs associated with root‐specific expression (Fig. S1, see Supporting Information), e.g. a consensus sequence motif of organ‐specific elements (OSEs) activated in infected cells of root nodules was present three times in the LOX3 promoter, but not in the LOX4 promoter (Fig. S1). In addition, the L1‐BOX motif, a cis‐acting regulatory element involved in trichome‐specific expression, and the SORLIP5AT motif, which is over‐represented in root‐specific genes, were all detected in the LOX3 promoter, but not in the LOX4 promoter (Fig. S1). However, an auxin‐responsive element and an ethylene (ET)‐responsive element (ERLEE4) were found in the LOX4 promoter, but not in the LOX3 promoter. These differences in regulatory element composition might account for the seemingly differential gene regulation observed for LOX3 and LOX4 in root tissues on nematode infection.
Figure 5.
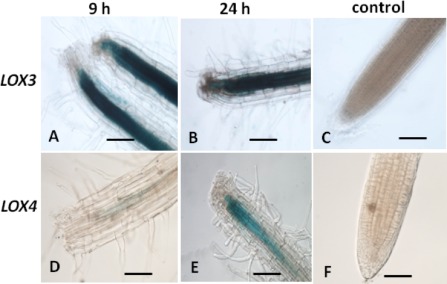
Effect of wounding on lipoxygenase‐3 (LOX3) and LOX4 promoter activity. The spatial expression patterns of LOX3 and LOX4 within 9 and 24 h after wounding were investigated through promoter activity assay. Rapid induction of β‐glucuronidase (GUS) staining was observed in response to tissue damage caused by puncturing the root tip of Arabidopsis plants harbouring LOX3‐GUS within 9 h (A) and 24 h (B), with no signal detected in uninjured root tips (C). In Arabidopsis plants harbouring the LOX4‐GUS construct, at 9 h post‐wounding, the GUS signal was weak and localized to the injured site (D). At 24 h, the GUS signal increased but still remained confined to the injured root site (E). No signal was detected within the uninjured root tip (F). Bars, 100 μm.
Analysis of LOX3 and LOX4 expression in mutants lacking functional LOX3 and LOX4
To explore the inter‐regulation between LOX3 and LOX4, we studied the expression of LOX3 and LOX4 in wild‐type (WT) and mutant lines lox3‐1 and lox4‐1 with and without RKN infection at 5 dpi on the whole root. Similar to the GUS assay, a marked induction in LOX3 expression was observed on infection in WT roots, whereas no significant increase was observed for LOX4 (Fig. 6). As compared with WT plants, increased basal levels of LOX3 transcript were detected in the lox4‐1 mutant (in non‐infested roots compared with non‐infested WT roots), implying an enhanced constitutive level of LOX3 in the lox4‐1 mutant (Fig. 6).
Figure 6.

Expression pattern of lipoxygenase‐3 (LOX3) and LOX4 genes in lox3‐1 and lox4‐1 mutant lines compared with the wild‐type (WT). Total RNA was isolated from each sample at 5 days post‐inoculation (dpi) by Meloidogyne javanica or without infection and used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Transcript levels were normalized using the normalization factor calculated as the geometric mean of the expression levels of three Arabidopsis endogenous reference genes: 18S, UBP22 and EF‐1α. Two biological replicates were taken and three technical repeats were performed per sample. The graph shows the mean and standard deviation (SD) of the amounts of LOX3 (A) and LOX4 (B) transcript relative to those of WT untreated control samples. Error bars correspond to SD (n = 2), and different letters above the bars denote a significant difference (P ≤ 0.05, analysis of variance) between samples amplified with the same primers, as analysed by Tukey–Kramer multiple comparison test.
Mutants lacking functional LOX4 are altered in their response to root‐knot and cyst nematodes
To evaluate the functional roles of the LOX3 and LOX4 isoforms in the root response to RKNs, Arabidopsis lines lacking LOX3 or LOX4 function were analysed for nematode development and reproduction at 6 weeks post‐inoculation (wpi) (Fig. 7). Homozygous T‐DNA insertion mutants that lacked LOX3 (lox3‐1, lox3‐2) or LOX4 (lox4‐1, lox4‐2) function were confirmed by polymerase chain reaction (PCR) prior to the infection studies, as described in Experimental procedures and Fig. S2 (see Supporting Information). Analysis of lines lox3‐1 and lox3‐2 showed that a lack of LOX3 function leads to a lower proportion of females than in control galls, whereas most of the nematodes remain in the J3/4 stage (Fig. 7A,B). Strikingly, analysis of lines lox4‐1 and lox4‐2 showed a much higher proportion of females in mutants lacking LOX4 than in control galls, accompanied by a marked decrease in the percentage of nematodes at the J3/4 stage (Fig. 7A,B). Egg mass production by adult females was used as a measure of nematode reproduction efficiency on the respective mutant line during the infection period. Our results clearly show that a deficiency in LOX4 induces nematode development and reproduction, as indicated by the increased production of females and egg masses (by 1.9‐ and 3.5‐fold, respectively, compared with the WT control), leading to an increased susceptibility response (Fig. 7B,C).
Figure 7.
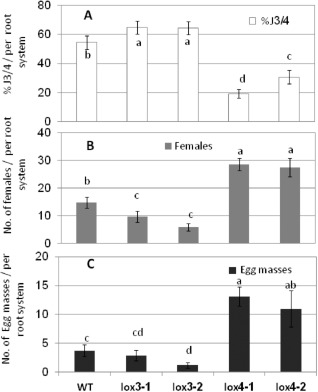
Arabidopsis mutants lacking 13‐lipoxygenase (13‐LOX) isoforms are altered in their response to Meloidogyne javanica root‐knot nematode. Susceptibility/resistance of wild‐type (WT) and transgenic Arabidopsis plants to M. javanica was measured as the percentage of third‐/fourth‐stage juveniles (J3/4s), number of females and number of egg masses produced in each root system. Two‐week‐old seedlings were inoculated with 150 sterile second‐stage juveniles (J2s) and were assessed for J3/4 and adult stages as females and egg mass production at 28 days post‐inoculation (dpi) by observation under a dissecting scope following staining with acid fuchsin dye. (A) Total number of M. javanica J3/4s observed in the root system of each plant used to calculate the percentage of J3/4s of all counted nematode stages. (B) Number of adult females as detected by dissecting each root system. (C) Number of egg masses per root system. Note the significant reduction in the percentage of J3/4s with the increase in female and egg mass counts in mutants lacking LOX4 function. Data are expressed as the means of 25 plants from each line; the experiment was repeated three times, giving consistent results. Error bars correspond to standard deviation (SD) (n = 25). Different letters above the bars denote a significant difference (P ≤ 0.05, analysis of variance) between Arabidopsis lines, analysed by Tukey–Kramer multiple comparison tests.
Cyst nematode infection by H. schachtii was assayed by measuring the development of parasitic J4 females at 14 dpi. We found no significant reduction in nematode development in the lox3‐1 mutant, as measured by the female to male and female to plant ratios (Fig. 8A,B). However, the analysis of the lox4‐1 mutant demonstrated a significant increase in the female to male ratio compared with the WT and lox3‐1 mutant line (Fig. 8A). These results indicate that, although LOX3 has only a minor effect in the defence against nematodes, LOX4 might control a broad defence mechanism with substantial impact on nematode development and reproduction.
Figure 8.

Arabidopsis mutants lacking a functional lipoxygenase‐4 (LOX‐4) isoform are altered in their response to Heterodera schachtii infection. Nematode susceptibility assays of lox3‐1 and lox4‐1 lines relative to wild‐type (WT) lines are presented through the measurement of the female to male ratio (A) and female to plant ratio (B). The lox4‐1 mutant showed a consistent trend of increased susceptibility to H. schachtii over all four experiments, showing statistically significant effects in the female to male ratio. Mean values significantly different from the corresponding WT were determined by Tukey and Duncan post‐hoc tests with P < 0.05. Significant differences are indicated with asterisks. Four independent replicates with at least three independent plates per line per replicate and at least five plants per plate were performed. Eventually, a total of 90 plants per line were tested.
Increased susceptibility of mutants lacking intact LOX4 is associated with the induction of JA and ET‐responsive genes
To evaluate the contribution of the major hormone‐regulated plant defence pathways—JA, SA and ET pathways—to the increased susceptibility of the lox4‐1 mutant, the expression of a set of gene markers and biosynthesis genes involved in these different pathways was investigated using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Figure 9 shows the expression levels of a set of defence‐related genes in uninfected and infected lox3‐1 and lox4‐1 whole‐root systems in comparison with the WT control tissue (whole‐root system of uninfected or infected plants, respectively) at 48 hpi and 5 dpi; transcript levels were normalized for each sample with the geometric mean of the expression of three selected housekeeping genes: 18S rRNA, elongation factor‐1α (EF‐1α) and UBP22 (Fig. 9). RNA extracted from non‐infected roots was used as a negative control. At 48 hpi, the relative expression levels of both marker genes associated with the JA pathway—allene oxide synthase (AOS) and allene oxide cyclase (AOC2)—were significantly up‐regulated in the lox4‐1 mutant line compared with the WT on nematode infection. Similarly, the expression level of ET‐responsive transcription factor 4 (ERF4), involved in the ET pathway, was sharply increased in the lox4‐1 mutant at 48 hpi.
Figure 9.
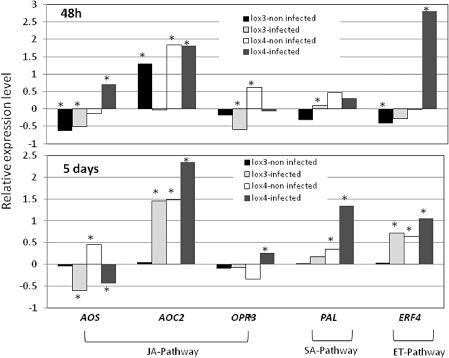
Analysis of the expression levels of five defence‐related target genes in wild‐type (WT) and lox3‐1 and lox4‐1 mutant lines. Arabidopsis WT and mutant lines were inoculated with Meloidogyne javanica at two different time points, 48 h post‐inoculation (hpi) and 5 days post‐inoculation (dpi), and non‐infected WT and mutant lines were grown under the same conditions. RNA was subjected to quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and transcript levels were normalized using the normalization factor calculated as the geometric mean of the expression levels of three Arabidopsis endogenous reference genes: 18S, UBP22 and EF‐1α. Two biological replicates were taken and three technical repeats were performed per sample. The graph shows the mean and standard deviation (SD) of the relative amounts of transcript of these genes in each mutant line in comparison with control tissue grown under the same conditions (control expression level with and without infection was set at zero). The statistical significance of the differences between mutant lines and control tissue from non‐infected and infected plants was determined by Tukey–Kramer multiple comparison tests, and significant differential expression (P ≤ 0.05) is indicated with asterisks. ET, ethylene; JA, jasmonic acid; SA, salicylic acid.
At 5 dpi, the expression of AOC2 was still induced and could also be related to the increased expression of the jasmonate pathway in the lox4‐1 mutant following infection. Interestingly, at this time point, increased expression of the SA marker gene, PAL1, was observed in both the non‐infected and infected lox4‐1 mutant compared with WT roots. Overall, our results present evidence indicating that, in mutants lacking functional LOX4, there is an increase in JA/ET‐regulated transcript accumulation, as well as SA‐regulated transcripts, suggesting a general role of LOX4 in the regulation of the root defence system.
Level of endogenous JA is increased in a mutant lacking functional LOX4
Because LOXs are involved in FA metabolism for the production of oxylipins, we next examined any possible effects of the LOX insertion mutant on JA and OPDA biosynthesis following infection with RKN. For JA and OPDA extraction and quantification, roots of 3‐week‐old seedlings of the corresponding LOX insertion mutants were compared with the WT following infection by liquid chromatography/tandem mass spectrometry (LC/MS/MS). Levels of JA in roots of all the mutant and WT lines were generally low. Nonetheless, lox4‐1 mutants had significantly (P ≤ 0.05) higher levels of JA, nine‐fold higher compared with WT (Fig. 10). These results indicate that LOX4 may play a role in regulating JA levels in WT plants following nematode infection, through either a negative regulatory role of LOX3 or substrate competition. OPDA levels in the roots of all three mutants were low, with no significant differences between them (Fig. 10). However, given that whole‐root tissues were taken for the JA/OPDA analysis, it might be that the JA/OPDA content at the nematode infection sites is diluted, explaining the relatively low level of these metabolites. Although a positive correlation is illustrated between the increased levels of JA displayed in lox4‐1 mutant roots and the high susceptible response, their simultaneous appearance should be studied further.
Figure 10.
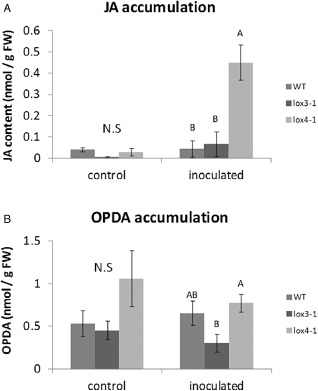
Endogenous jasmonic acid (JA) and 12‐oxo‐phytodienoic acid (OPDA) in Arabidopsis mutants lacking 13‐lipoxygenase (13‐LOX) function and in wild‐type (WT) plants. The production of JA (A) and OPDA (B) was monitored by liquid chromatography/tandem mass spectrometry (LC/MS/MS) in negative‐phase mode in root tissues at 7 days post‐inoculation (dpi) with Meloidogyne javanica second‐stage juveniles (J2s), and the data were expressed as the means of four replicates. Mock treatments consisted of inoculation with H2O only. Error bars correspond to standard deviation (SD) (n = 4), and different letters above the bars denote a significant difference (P ≤ 0.05, analysis of variance) between samples, as analysed by Tukey–Kramer multiple comparison test. N.S, no significant difference.
Discussion
In this study, two closely related 13‐LOX isoforms, LOX3 and LOX4, were found to be stimulated by nematode infection and to play a regulatory role in modulating the response of Arabidopsis to nematode invasion. Although a lack of LOX3 function led to increased resistance, as indicated by the low female and egg mass counts, the absence of LOX4 led to a marked increase in plant susceptibility, as reflected by the larger number of adult females and egg masses, and female to male ratio, of M. javanica and H. schachtii, respectively (Figs 7 and 8).
The data presented demonstrate that LOX3 expression is highly induced during parasitic interactions with both RKNs and cyst nematodes, which might be correlated with the developmental alterations occurring during nematode establishment in planta. These results fit well with the role of the LOX products, oxylipins, in regulating developmental and differentiation processes (Caldelari et al., 2011; Vellosillo et al., 2007). However, functional analysis of LOX4 demonstrated a stronger effect on nematode development, implying that LOX4 exerts distinct defence pathways, thus increasing susceptibility when its function is abolished.
LOX3 is involved in developmental alterations induced during nematode development and reproduction
Despite the well‐characterized enzymatic function of the six LOX isoforms in Arabidopsis (Bannenberg et al., 2009), the physiological roles of each individual Arabidopsis LOX in plant–nematode interactions have not been assigned. Previous analysis of the four Arabidopsis 13‐LOXs indicated that LOX3, LOX4 and LOX6 are components of the same phylogenetic clade, whereas LOX2 is the most divergent isoform (Bannenberg et al., 2009), and LOX3 and LOX4 share high sequence similarity (Caldelari et al., 2011). In the present study, strong promoter activation of LOX3 was observed in the first hours after infection, and continued throughout feeding site establishment during syncytium and gall formation (Figs 1 and 4). Thin sections of the galls in LOX3:GUS plants showed GUS activity in GCs, in neighbouring cells and in cells surrounding the female body (Fig. 3). However, the GUS signal in gall sections from roots harbouring the LOX4:GUS construct showed only a weak signal in the surrounding dividing cells (Fig. 3), suggesting a primary role for LOX3 in root developmental alterations induced by the invading nematode. Interestingly, the expression patterns of both LOX3 and LOX4 on wounding were similar to those following RKN infection, with rapid and intense induction of LOX3 compared with a moderate local response of LOX4 (Fig. 5). Although previous studies have indicated that the RKN migrates between cells without damaging them (Von Mende, 1997), with H. schachtii, tissue damage has been reported as the nematode migrates intracellularly through the root before establishment (reviewed in Sobczak and Golinowski, 2011). Regardless of whether part of the initial triggering signal of LOX3 and LOX4 is related to a wounding response elicited by the migrating nematodes, its continuous expression, especially in the case of LOX3, supports its putative involvement in the developmental alteration induced during nematode infection. Moreover, although different FA‐derived oxylipins might be produced through the 13‐LOX pathway in roots following wounding relative to nematode infection, our results indicate that LOX3 and LOX4 are likely to be not only part of a classical plant wound response, but also associated directly with the modulation of nematode disease progress.
Our expression results on wounding are consistent with the existence of two JA‐responsive cis‐elements in the AtLOX3 promoter, but not in that of AtLOX4. In addition, the AtLOX3 promoter contains cis‐elements that are conserved among promoters of root‐specific genes required for root development processes, such as the SORLIP5AT cis‐element (Jiao et al., 2005), the OSE characteristic of promoters activated in infected cells of root nodules (Ramlov et al., 1993) and the L1‐BOX motif involved in trichome‐specific expression (Fig. S1); this further supports the role of LOX3 in regulating root developmental processes required for nematode development and reproduction. However, the presence of auxin, ET and defence‐responsive cis‐elements in the AtLOX4 promoter might support its role in the suppression of nematode development. This mosaic of regulatory elements among both promoters implies that the expression of both LOX3 and LOX4 is controlled through conserved combinations of different promoter elements.
LOX3 and LOX4 affect parasitism by plant‐parasitic nematodes
The analysis of the lox3‐1 mutant line showed a relatively moderate reduction in disease susceptibility to RKNs and not much effect in the cyst nematode interaction (Figs 7 and 8). In contrast with LOX3, the analysis of mutants lacking LOX4 function showed a considerably increased susceptibility response for RKNs and a similar tendency in cyst nematodes (Figs 7 and 8). These results suggest that LOX3 and LOX4 interfere with different pathways that might affect the plant response to nematode infection. Increased expression of LOX3 in the non‐infected lox4‐1 mutant (Fig. 6) might facilitate the early parasitic stages. In this scenario, the increased susceptibility of lox4 mutants might be partly a result of the enhanced expression of the LOX3 isoform, which, in turn, might contribute to support the developmental alteration required for nematode development. Previous analysis of Arabidopsis LOX3 and LOX4 by Caldelari et al. (2011) showed that these LOX3 and LOX4 isoforms act redundantly in JA synthesis to ensure male fertility. A deficiency of both LOX3 and LOX4 thus results in a sterile male phenotype (Farmer and Ryan, 1992). As opposed to LOX3 and LOX4 function in floral tissues, our results indicate that, in roots, LOX3 and LOX4 isoforms play distinct roles in the plant response to nematodes (Figs 7 and 8).
Moreover, although mutation in LOX4 results in significant changes in expression of defence‐related target genes, such as AOS, AOC2, ERF4 and PAL1, the changes occurring in the lox3‐1 mutant were less remarkable than those in the lox4‐1 mutant. This indicates that these two members in roots interfere with different pathways in a different manner. Thus, the enhanced susceptibility of the lox4‐1 mutant might result from additional direct or indirect effects on either other oxylipins or other defence signalling pathways. In accordance with the clear induction of JA biosynthesis genes observed with the lox4‐1 mutant, our experimental procedure measuring JA in the whole‐root system resulted in a significant increase in JA quantity in lox4‐1 roots following infection (Fig. 10). Our results indicate that OPDA, the JA biosynthetic intermediate, is detected, together with JA, but no significant differences were observed in OPDA levels measured for WT lines compared with lox3‐1 and lox4‐1 mutant lines. In contrast with our observation, a previous study has shown that, OPDA, a potent gene regulator in the wound response and in the protection of plants against pathogens, accumulates in the absence of JA, suggesting strong biological activity for OPDA (Landgraf et al., 2002).
Although the JA signalling pathway is induced in the lox4‐1 mutant, there is also clear evidence for a higher SA level in the roots of this mutant (Fig. 9). These observations contradict any suggested antagonistic interaction between SA and JA (Bostock, 2005; Koornneef et al., 2008, Pieterse et al. 2012). However, given that the antagonistic interaction between SA and JA is concentration and time dependent (Koornneef et al., 2008; Mur et al., 2006) and that ET signalling has been shown to prevent SA‐mediated suppression of the JA pathway (Leon‐Reyes et al., 2010), it is suggested that the up‐regulation of both JA and SA pathways is associated with the increased observed susceptible response.
In summary, our findings indicate that the lox4‐1 mutant of Arabidopsis is more susceptible to RKN and cyst nematodes, as shown by an increase in the number of nematodes that develop into females. Thus, we hypothesize that an increased level of JA acts as a susceptibility factor in the plant–nematode interaction. Our results are in good agreement with those obtained by Bhattarai et al. (2008), who analysed the response of tomato mutants altered in JA signalling and demonstrated that an intact JA signalling pathway is required for tomato susceptibility to RKNs. In maize, Mu‐insertional lox3‐4 mutants displayed increased attractiveness to RKNs, and an increased number of juveniles and eggs were accompanied by elevated levels of JA (Gao et al., 2008). Likewise, the analysis of JA‐deficient double mutants of the two oxo‐phytodienoate reductase genes, OPR7 and OPR8, of maize resulted in increased resistance to RKNs (M.V. Kolomiets and J. Starr, TAMU, College Station, TX, personal communication). However, the analysis of JA function in the rice–nematode interaction through exogenous shoot application and the use of JA biosynthesis mutants demonstrated that JA plays a pivotal role in rice defence against RKNs (Nahar et al., 2011). Similarly, foliar application of JA to tomato resulted in a repressive effect on RKNs (Fujimoto et al., 2011). Indeed, these results reveal the opposite function for JA in the regulation of plant responses to nematodes. However, it is worth noting that the exogenous application of JA to shoots might induce the expression of various pathways in roots that are responsible for the increased resistance observed by Nahar et al. (2011) on rice and by Fujimoto et al. (2011) on tomato. In addition, the differences occurring in these two described systems with regard to the role of JA/ET and SA signalling pathways in the regulation of RKN disease development (Bhattarai et al., 2008 and Nahar et al., 2011) could be part of major differences that exist between monocotyledonous and dicotyledonous plants.
Taken together, our current understanding is that, on nematode infection in roots, LOX3 and LOX4 isoforms produce their own oxylipins that elicit distinct responses to nematode infection, with LOX3 being involved in the regulation of developmental processes that are induced by the parasitic nematodes and LOX4 being a dominant player in the regulation of defence pathways.
Although the functions of JA and OPDA seem to be complex, this study indicates that LOX4 has an important activity in root tissues, thus leading to increased susceptibility of the lox4‐1 mutant. The common transcriptional regulation in response to RKN and H. schachtii infection reinforces an important role for both LOX isoforms in nematode establishment and maintenance of feeding sites (GCs and syncytia) in the roots of host plants. Further studies are required to elucidate the potential role of JA in the repression of relevant defence pathways against plant‐parasitic nematodes in a root‐specific manner, and to determine whether this is conserved for both types of nematode (root‐knot and cyst).
Experimental Procedures
Plant material, growth conditions and mutant characterization
Arabidopsis thaliana WT, transgenic lines and mutants used in this study were derived from ecotype Columbia (Col‐0). In general, Arabidopsis plants were grown in a growth chamber at 24 °C under a photoperiod of 14 h light/10 h dark, with a light intensity of 50–60 μmol/m2/s provided by white fluorescent tubes throughout the growth period. T‐DNA insertion mutants were obtained from the Arabidopsis Biological Resources Center (ABRC), Columbus, OH, USA. Two different T‐DNA insertion mutants were identified for each LOX‐encoding gene: for LOX3 (At1g17420), allele lox3‐1 corresponds to line SALK_119404 and allele lox3‐2 corresponds to line SALK_147830; for LOX4 (At1g72520), allele lox4‐1 corresponds to SALK_071732 and allele lox4‐2 corresponds to SALK_017873. Homozygous insertion mutants were identified by PCR using T‐DNA and gene‐specific primer sets with the SIGnAL T‐DNA Express Arabidopsis gene mapping tool (http://signal.salk.edu/) (Fig. S1). Primers for each gene binding upstream and downstream of the predicted T‐DNA insertion, as well as one primer binding in the left border region of the corresponding T‐DNA, are listed in Table S1 (see Supporting Information). GUS‐transgenic lines of LOX3 and LOX4 were kindly provided by Professor Carmen Castresana (Vellosillo et al., 2007).
Nematode inocula and infection assay
Meloidogyne javanica was propagated on glasshouse‐grown tomato (Lycopersicon esculentum) cv. Avigail 870, and nematode eggs were bulk extracted from the roots with 0.05% (v/v) sodium hypochlorite (NaOCl), followed by sucrose flotation, as described by Hussey and Baker (1973). For nematode sterilization, nematode eggs were placed on a sterile Whatman filter holder (Whatman International Ltd, Dassel, Germany) with a cellulose acetate filter membrane (Sartorius Stedim Biotech GmbH, Goettingen, Germany; pore size, 5 μm). The eggs on the filter were exposed for 10 min to 0.01% (w/v) mercuric chloride (Hg2Cl2) (Sigma‐Aldrich, St Louis, MO, USA), followed by 0.7% streptomycin solution (Sigma‐Aldrich) and three washing steps with 50 mL of sterile distilled water (Jansen van Vuuren and Woodward, 2001). Sterile eggs were then collected from the membrane and placed on 25‐μm pore size sieves in 0.01 m 2‐(N‐morpholino)ethanesulphonic acid (MES) buffer (Sigma‐Aldrich) under aseptic dark conditions for 3 days, and hatching J2s were collected. For nematode infection of plants, seeds of WT and transgenic lines were surface sterilized with 1.4% NaOCl and 0.01% (w/v) Tween‐20 for 10 min, followed by three washing steps with sterile distilled water to remove residual sterilization solution. Seeds were germinated on standard strength Gamborg B5 salts medium (Duchefa, Haarlem, the Netherlands) supplemented with 2% (w/v) sucrose solidified with 0.8% (w/v) Gelrite agar (Duchefa) and containing 50 μg/mL kanamycin monosulphate (Duchefa) for the transgenic lines; seeds were then incubated in the dark at 4 °C for 2 days to synchronize germination (Vieira et al., 2011). One week after germination, transgenic seedlings selected on kanamycin‐containing medium and WT seedlings were gently transferred for nematode infection with a forceps to a Petri dish (Miniplast, Ein Shemer, Israel; one plant per dish) containing B5 medium (Sijmons et al., 1991). For inoculation, 150 freshly hatched sterile M. javanica J2s were applied on each 2‐week‐old seedling, and the dishes were left uncovered in a laminar flow hood until the water had completely soaked into the medium (Sijmons et al., 1991). The inoculated and non‐inoculated plants were left to grow vertically, and root samples were taken for either promoter:GUS bioassay or the assessment of RKN disease progress at the designated time points post‐infection.
Heterodera schachtii was multiplied in vitro on mustard (Sinapsis alba cv. Albatross) roots growing on Gamborg medium (Gamborg et al., 1968) supplemented with 3% sucrose. Hatching of J2s was stimulated by soaking the cysts in sterile 3 mm ZnCl2. The juveniles were washed four times in sterile water for inoculation. Twelve‐day‐old roots of A. thaliana plants were inoculated with about 35 juveniles under axenic conditions.
Evaluation of response to root knot and cyst nematodes
To analyse nematode development in WT and transgenic Arabidopsis lines, root systems grown in mono‐oxenic culture were harvested at 28 dpi and soaked in acid fuchsin solution (Sigma‐Aldrich; 17.5 mg acid fuchsin, 500 mL ethanol and 500 mL acetic acid) overnight. The stained roots were washed three times in distilled water and stored as described previously (Bybd et al., 1983). Stained root systems were then mounted in tap water and dissected under a stereomicroscope (Olympus SZX12, Olympus, Tokyo, Japan) to determine the developmental stage (juveniles J3/4 and adult females) of the nematodes embedded in the roots and galls. The number of egg masses produced during the incubation period was also determined. The mean values of J3/4 juveniles, females and egg mass production were obtained by screening 25 replicates per Arabidopsis line in three independent experiments. Statistical differences were determined by comparison of all means using the Tukey–Kramer test with an α level of 0.05 in the JMP software package (version 3.2.6, SAS Institute, Cary, NC, USA). For cyst nematode (H. schachtii) infection tests, Arabidopsis seeds were planted into Petri dishes containing modified Gamborg medium, and 10‐day‐old plants were inoculated with ∼35 surface‐sterilized H. schachtii J2s per plant. Two weeks after inoculation, the number of fourth‐stage (J4) females per plant, as well as the number of males, was determined. Mean values significantly different from the corresponding WT were determined by Tukey and Duncan post‐hoc tests with P < 0.05, and significant differences are indicated with asterisks. Four independent replicates with at least three independent plates per line per replicate, and with at least five plants per plate, were performed. Eventually, a total of 90 plants per line was tested.
Histochemical localization of GUS activity and microscopic analysis
Two‐week‐old Col‐0 and promoter‐GUS Arabidopsis seedlings were inoculated as described above, and assayed histochemically for GUS activity at the designated times after infection (Favery et al., 1998). A set of non‐infested plates served as the control group. For GUS assays, infected and non‐infected transgenic root tissues were removed from the Petri dishes at specific time points after inoculation and infiltrated with GUS‐staining buffer containing 50 mm sodium phosphate, pH 7.0, 10 mm ethylenediaminetetraacetic acid (EDTA), 5 mm K4[Fe2(CN)6], 5 mm K3[Fe2(CN)6], 0.2% (v/v) Triton X‐100 and 2 mm 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronide (X‐Gluc). GUS staining was performed for 12 h at 37 °C. To stain the nematodes, GUS‐stained roots were placed in 1% NaOCl for 4 min, washed in water and then boiled for 2 min in a 1:10 diluted solution of acid fuchsin, as described above. The roots were then rinsed in water and incubated in acidified glycerol for 2 days to clear root tissue and reveal the stained nematodes (Hansen et al., 1996). For each time point examined, at least 30 infected or wounded and 15 non‐infected or non‐wounded transgenic roots were assayed for GUS expression. To determine the cellular localization of GUS expression in developing GCs, semi‐thin gall sections were investigated microscopically according to Mitchum et al. (2004). For sectioning, GUS‐stained root was fixed in 0.25% (w/v) glutaraldehyde and 4% (v/v) paraformaldehyde in 50 mm phosphate‐buffered saline, pH 7.2, and then dehydrated and embedded in Technovit 7100 (Heraeus Kulzer, Hanau, Germany) according to the manufacturer's instructions. Embedded tissues were sectioned (3 μm) and posteriorly mounted in Depex (Sigma‐Aldrich). For observation and documentation, GUS‐stained roots mounted on microscope slides and semi‐thin sections were photographed with either a Leica DMLB light microscope and a Nikon Eclipse 90i (Leica Microsystems GmbH, Wetzlar, Germany; Nikon Corporation, Tokyo, Japan) or a stereomicroscope (Leica MZFLIII, Leica Microsystems GmbH) equipped with a Nikon DS‐Fi1 camera.
Genomic DNA extraction
Genomic DNA was isolated from 5‐week‐old soil‐grown seedlings of the WT (col‐0) and insertion lines lox3‐1 (SALK_119404), lox3‐2 (SALK_147830), lox4‐1 (SALK_071732) and lox4‐2 (SALK_017873) using the cetyltrimethylammonium bromide (CTAB) plant DNA extraction method, according to Goetz et al. (2001). Individuals homozygous for the respective insertions were selected by PCR (Alonso et al., 2003) using the T‐DNA‐specific primer LBb1.3 and two gene‐specific primers for each SALK line. PCR products generated using LBb1.3 and gene‐specific right genomic primers were used to confirm the T‐DNA insertion location and orientation. The second PCR was performed to identify plants homozygous for the insertions using the SALK‐specific primers. All primers are listed in Table S1.
RNA isolation and qRT‐PCR analysis
Gene transcripts were quantified by real‐time qRT‐PCR on total RNA extracted from infested Arabidopsis roots at different time points as indicated after inoculation (dpi). RNA extracted from non‐infected roots was used as a control. Ten root systems were pooled for each treatment, and RNA was extracted using the InviTrap Spin Plant RNA Mini Kit (Invitek, Berlin, Germany). To remove contaminating genomic DNA, RNA samples were incubated in the presence of 10 units of TURBO DNA‐free DNASE (Applied Biosystems, Foster City, CA, USA). DNA‐free RNA was converted into first‐strand cDNA using the Verso cDNA Synthesis Kit (ABgene, Epsom, UK) and Absolute SYBR green ROX mix (ABgene). qRT‐PCR primers were designed using the Primer Express software (Applied Biosystems; Table S2, see Supporting Information). The subsequent real‐time PCR contained 3.4 μL of the cDNA in a total volume of 10 μL, consisting of 1 × SYBR green mix (ABgene) and 150 nm of each forward and reverse primer in real‐time PCR plasticware (Axygen, Union City, CA, USA). All PCR cycles began with 2 min at 50 °C, then 10 min at 95 °C, followed by 40 two‐step cycles comprising 10 s at 95 °C and 1 min at 60 °C. For each treatment, the significance of the differential expression was evaluated on the basis of two replicates per condition (two biological replicates × three technical replicates) using Rotor‐Gene 6 software. After PCR, a melting curve was generated by gradually increasing the temperature to 95 °C to test for amplicon specificity. For qPCR, a mixture of all cDNAs was used for all treatments, as a template for calibration curves designed for each primer pair. Transcript levels were normalized for each sample with the geometric mean of the three selected housekeeping genes [the Arabidopsis small ribosomal RNA (18S; GenBank accession no. AT5G47230), the Arabidopsis UBP22 (UBP22; GenBank accession no. At5g10790) and EF‐1α (GenBank accession no. AT5G60390)] according to Bustin et al. (2009) and Vandesompele et al. (2002). All of the housekeeping genes were confirmed to display minimal variation across the treatments and were the most stable housekeeping genes from a set of tested genes in a given cDNA sample. The values were expressed as the increase or decrease relative to a calibration sample. Control reactions were a PCR‐negative control without cDNA template to confirm that there were no nonspecific PCR products (NTC), and a second reaction containing mRNA that had not been subjected to RT reaction (NRT control). Statistical differences between treatments and/or root lines were calculated by least significant difference (LSD), according to the Tukey–Kramer multiple comparison test at P ≤ 0.05.
Measurement of endogenous JA and OPDA
For tissue‐specific and RKN‐induced analysis of JA and OPDA levels, 6‐week‐old seedlings grown under 14 h of light (50–60 μmol/m2/s provided by white fluorescent tubes) at 25–28 °C were harvested and immediately frozen in liquid N2. Four biological replicates (three seedlings per replicate) were analysed. JA and OPDA were analysed using an Ascentis Express 18 Column (3 cm × 2.1 mm, 2.7 μm), together with a next‐generation machine API 3200 LC/MS/MS in negative‐phase mode, according to the methods outlined by Pan et al. (2008). For JA internal standard, 15 pmol of deuterated JA was used (https://www.cdnisotopes.com cat# D‐6936). The organic phase was dried under continuous nitrogen and resuspended with 150 μL of methanol. 12‐OPDA levels in the lox3‐1 and lox4‐1 mutant and WT roots infected with RKN and non‐infected were measured as described by Stumpe et al. (2005). Each treatment was replicated four times, and the data were expressed as the means of four replicates.
Bioinformatics analysis
The promoter sequences of Arabidopsis LOX3 and LOX4 genes were subjected to in silico analysis with the databases PLACE (http://www.dna.affrc.go.jp/PLACE/), PlantCARE (http://bioinformatics.SlPSb.ugent.be/wetools/plantcare/html/), the Genomatix software suite (http://www.genomatix.de) and the ATHENA resource http://www.bioinformatics2.wsu.edu/Athena to identify the presence of putative cis‐regulatory elements.
Statistical analysis
Data were analysed using the JMP software package. Mean comparisons of J3/4, adult females and females producing egg masses, as affected by different Arabidopsis insertion lines, as well as the transcript abundance of the LOX‐encoding genes and defence‐related genes, were calculated using LSD, according to the Tukey–Kramer multiple comparison test, with significance set at P ≤ 0.05.
Supporting information
Fig. S1 In silico analysis of cis‐acting regulatory elements in 5′ regulatory regions of LOX3 and LOX4 genes. Regulatory elements in the promoter regions of LOX3 and LOX4 were identified using the plant promoter and regulatory element resources PLACE, PlantCARE, ATHENA and Genomatix Matinspector databases. The 2000‐bp sequence of the 5′ regulatory regions was analysed and promoter elements that differed among sequences are presented.
Fig. S2 Polymerase chain reaction (PCR) confirming the T‐DNA insertion in lipoxygenase (LOX) insertion mutants. Two different T‐DNA homozygous insertion mutants were identified for each LOX‐encoding gene: lox3‐1 corresponds to line SALK_119404, lox3‐2 corresponds to line SALK_147830, lox4‐1 corresponds to line SALK_071732 and lox4‐2 corresponds to line SALK_017873. T‐DNA verification was performed by PCR using three primers per line: two genomic binding primers—left genomic primer (LP) and right genomic primer (RP)—specific for each line and complementary to the flanking sequence of the T‐DNA insertion, and one left border primer (LB), LBb1.3, matching all SALK lines. For each SALK line, the PCR products were run in the following order: SALK line and WT with primer set LP + RP, followed by SALK line and WT with primer set LBb1.3 + RP. The following bands were expected: for WT (no insertion), a single product of 900–1100 bp with the genomic LP and RP primers and no product for the homozygous line; a product of 430–902 bp for the homozygous line with primer set LBb1.3 + RP. See Table S1 for the list of primers.
Table S1 Oligonucleotides used in this study for Arabidopsis SALK line verification. T‐DNA and gene‐specific primers were designed according to the SIGnAL T‐DNA Express Arabidopsis gene mapping tool (http://signal.salk.edu/).
Table S2 Overview of the target genes used in this study, showing the accession number for each target gene primer pair used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), the defence pathway in which each gene is mainly involved and the appropriate reference gene used.
Acknowledgements
Our thanks go to Professor Carmen Castresana for providing LOX3 and LOX4 transgenic lines harbouring the promoter‐GUS constructs. We thank Dr Julia Hoffman for providing the initial population of Heterodera schachtii.
References
- Abad, P. , Favery, B. , Rosso, M.N. and Castagnone‐Sereno, P. (2003) Root‐knot nematode parasitism and host response: molecular basis of a sophisticated interaction. Mol. Plant Pathol. 4, 217–224. [DOI] [PubMed] [Google Scholar]
- Alkharouf, N.W. , Klink, V.P. , Chouikha, I.B. , Beard, H.S. , MacDonald, M.H. , Meyer, S. , Knap, H.T. , Khan, R. and Matthews, B.F. (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta, 224, 838–852. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Andersson, M.X. , Hamberg, M. , Kourtchenko, O. , Brunnstrom, A. , McPhail, K.L. , Gerwick, W.H. , Göbel, C. , Feussner, I. and Ellerström, M. (2006) Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana. Formation of a novel oxo‐phytodienoic acid‐containing galactolipid, arabidopside E. J. Biol. Chem. 281, 31 528–31 537. [DOI] [PubMed] [Google Scholar]
- Atkinson, H.J. (1994) Plant–nematode interactions: molecular and genetic basis In: Pathogenesis and Host–Parasite Specificity in Plant Disease: Histopathological, Biochemical, Genetic and Molecular Basis (Singh S.U., ed.), pp. 355–369. Oxford: Pergamon Press. [Google Scholar]
- Bannenberg, G. , Martinez, M. , Hamberg, M. and Castresana, C. (2009) Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana . Lipids, 44, 85–95. [DOI] [PubMed] [Google Scholar]
- Bhattarai, K.K. , Xie, Q.G. , Mantelin, S. , Bishnoi, U. , Girke, T. , Navarre, D.A. and Kaloshian, I. (2008) Tomato susceptibility to root‐knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant–Microbe Interact. 21, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Bird, D.M. , Weerasinghe, R.R. , Allen, N.S. and Lohar, D.P. (2004) (Abstract) The first 24 hours: primary signaling events between root‐knot nematode and its host. J. Nematol. 36, 306–307. [Google Scholar]
- Block, A. , Schmelz, E. , Jones, J.B. and Klee, H.J. (2005) Coronatine and salicylic acid: the battle between Arabidopsis and Pseudomonas for phytohormone control. Mol. Plant Pathol. 6, 79–83. [DOI] [PubMed] [Google Scholar]
- Bostock, R.M. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu. Rev. Phytopathol. 43, 545–580. [DOI] [PubMed] [Google Scholar]
- Brown, R.L. , Kazan, K. , McGrath, K.C. , Maclean, D.J. and Manners, J.M. (2003) A role for the GCC‐box in jasmonate‐mediated activation of the PDF1.2 gene of Arabidopsis . Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M.W. , Shipley, G.L. , Vandesompele, J. and Wittwer, C.T. (2009) The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin. Chem. 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Bybd, D.W. , Kirkpatrick, T. and Barker, K.R. (1983) An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143. [PMC free article] [PubMed] [Google Scholar]
- Caldelari, D. , Wang, G. , Farmer, E.E. and Dong, X. (2011) Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75, 25–33. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Hackett, R. , Walker, D. , Taylor, A. , Lin, Z. and Grierson, D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid‐derived flavor compounds. Plant Physiol. 136, 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, G. , Magliano, M. , Deleury, E. , Abad, P. and Rosso, M.N. (2007) Transcriptome analysis of root‐knot nematode functions induced in the early stages of parasitism. New Phytol. 176, 426–436. [DOI] [PubMed] [Google Scholar]
- Escobar, C. , De Meutter, J. , Aristizabal, F.A. , Sanz‐Alferez, S. , del Campo, F.F. , Barthels, N. , Van der Eycken, W. , Seurinck, J. , van Montagu, M. , Gheysen, G. and Fenoll, C. (1999) Isolation of the LEMMI9 gene and promoter analysis during a compatible plant–nematode interaction. Mol. Plant–Microbe Interact. 12, 440–449. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. and Ryan, C.A. (1992) Octadecanoid precursors of jasmonic acid activate the synthesis of wound‐inducible proteinase inhibitors. Plant Cell, 4, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B. , Lecomte, P. , Gil, N. , Bechtold, N. , Bouchez, D. , Dalmasso, A. and Abad, P. (1998) RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 17, 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoll, C. , Aristizabal, F.A. , Sanz‐Alferez, S. and del Campo, F.F. (1997) Regulation of gene expression in feeding sites In: Cellular and Molecular Aspects of Plant–Nematode Interactions (Fenoll C., Grundler F.M.W. and Ohl S.A., eds), pp. 133–149. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Feussner, I. and Wasternack, C. (2002) The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Fujimoto, T. , Tomitaka, Y. , Abe, H. , Tsuda, S. , Futai, K. and Mizukubo, T. (2011) Expression profile of jasmonic acid‐induced genes and the induced resistance against the root‐knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J. Plant Physiol. 168, 1084–1097. [DOI] [PubMed] [Google Scholar]
- Gamborg, O.L. , Miller, R.A. and Ojima, K. (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Starr, J. , Gobel, C. , Engelberth, J. , Feussner, I. , Tumlinson, J. and Kolomiets, M. (2008) Maize 9‐lipoxygenase ZmLOX3 controls development, root‐specific expression of defense genes, and resistance to root‐knot nematodes. Mol. Plant–Microbe Interact. 21, 98–109. [DOI] [PubMed] [Google Scholar]
- Goetz, M. , Godt, D.E. , Guivarc'h, A. , Kahmann, U. , Chriqui, D. and Roitsch, T. (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. USA, 98, 6522–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr, C. and Paszkowski, U. (2009) Weights in the balance: jasmonic acid and salicylic acid signaling in root–biotroph interactions. Mol. Plant–Microbe Interact. 22, 763–772. [DOI] [PubMed] [Google Scholar]
- Hamberg, M. , Ponce de Leon, I. , Rodriguez, M.J. and Castresana, C. (2005) Alpha‐dioxygenases. Biochem. Biophys. Res. Commun. 338, 169–174. [DOI] [PubMed] [Google Scholar]
- Hansen, E. , Harper, G. and McPherson, M.J. (1996) Differential expression patterns of the wound‐inducible transgene wun1‐uidA in potato roots following infection with either cyst or root knot nematodes. Physiol. Mol. Plant Pathol. 48, 161–170. [Google Scholar]
- He, Y. , Fukushige, H. , Hildebrand, D.F. and Gan, S. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, R.S. and Baker, K.R. (1973) Comparison of methods of collecting inocula for Meloidogyne spp., including a new technique. Plant Dis. Rep. 57, 1025–1028. [Google Scholar]
- Ibrahim, H.M. , Hosseini, P. , Alkharouf, N.W. , Hussein, E.H. , Gamal El‐Din Ael, K. , Aly, M.A. and Matthews, B.F. (2011) Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics, 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalloul, A. , Montillet, J.L. , Assigbetse, K. , Agnel, J.P. , Delannoy, E. , Triantaphylides, C. , Daniel, J.F. , Marmey, P. , Geiger, J.P. and Nicole, M. (2002) Lipid peroxidation in cotton: Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J. 32, 1–12. [DOI] [PubMed] [Google Scholar]
- Jammes, F. , Lecomte, P. , de Almeida‐Engler, J. , Bitton, F. , Martin‐Magniette, M.L. , Renou, J.P. , Abad, P. and Favery, B. (2005) Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis . Plant J. 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jansen van Vuuren, R. and Woodward, B. (2001) The response of cassava cultivars to root‐knot nematode infestation: an in vitro method. Euphytica, 120, 109–113. [Google Scholar]
- Jiao, Y. , Ma, L. , Strickland, E. and Deng, X.W. (2005) Conservation and divergence of light‐regulated genome expression patterns during seedling development in rice and Arabidopsis . Plant Cell, 17, 3239–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A. and Kachroo, P. (2009) Fatty acid‐derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. [DOI] [PubMed] [Google Scholar]
- Klink, V.P. , Hosseini, P. , Matsye, P. , Alkharouf, N.W. and Matthews, B.F. (2009) A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 71, 525–567. [DOI] [PubMed] [Google Scholar]
- Klink, V.P. , Hosseini, P. , Matsye, P.D. , Alkharouf, N.W. and Matthews, B.F. (2010) Syncytium gene expression in Glycine max ([PI 88788]) roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines . Plant Physiol. Biochem. 48, 176–193. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. , Leon‐Reyes, A. , Ritsema, T. , Verhage, A. , Den Otter, F.C. , Van Loon, L.C. and Pieterse, C.M. (2008) Kinetics of salicylate‐mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf, P. , Feussner, I. , Hunger, A. , Scheel, D. and Rosahl, S. (2002) Systemic accumulation of 12‐oxophytodienoic acid in SAR‐induced potato plants. Eur. J. Plant Pathol. 108, 279–283. [Google Scholar]
- Leon‐Reyes, A. , Du, Y. , Koornneef, A. , Proietti, S. , Korbes, A.P. , Memelink, J. , Pieterse, C.M. and Ritsema, T. (2010) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant–Microbe Interact. 23, 187–197. [DOI] [PubMed] [Google Scholar]
- Mazarei, M. , Puthoff, D.P. , Hart, J.K. , Rodermel, S.R. and Baum, T.J. (2002) Identification and characterization of a soybean ethylene‐responsive element‐binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol. Plant–Microbe Interact. 15, 577–586. [DOI] [PubMed] [Google Scholar]
- Melan, M.A. , Dong, X. , Endara, M.E. , Davis, K.R. , Ausubel, F.M. and Peterman, T.K. (1993) An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 101, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melan, M.A. , Enriquez, A. and Peterman, T.K. (1994) The LOX1 gene of Arabidopsis is temporally and spatially regulated in germinating seedlings. Plant Physiol. 105, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G. , Sukno, S. , Wang, X. , Shani, Z. , Tsabary, G. , Shoseyov, O. and Davis, E.L. (2004) The promoter of the Arabidopsis thaliana Cel1 endo‐1,4‐beta glucanase gene is differentially expressed in plant feeding cells induced by root‐knot and cyst nematodes. Mol. Plant Pathol. 5, 175–181. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , De Vleesschauwer, D. , Hofte, M. and Gheysen, G. (2011) The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 157, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel, A. , De Almeida Engler, J. , Tire, C. , Engler, G. , Van Montagu, M. and Gheysen, G. (1993) Induction patterns of an extensin gene in tobacco upon nematode infection. Plant Cell, 5, 1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Welti, R. and Wang, X. (2008) Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography–electrospray tandem mass spectrometry. Phytochem. 69, 1773–1781. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Portillo, M. , Cabrera, J. , Lindsey, K. , Topping, J. , Andres, M.F. , Emiliozzi, M. , Oliveros, J.C. , Garcia‐Casado, G. , Solano, R. , Koltai, H. , Resnick, N. , Fenoll, C. and Escobar, C. (2013) Distinct and conserved transcriptomic changes during nematode‐induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. [DOI] [PubMed] [Google Scholar]
- Puzio, P.S. , Lausen, J. , Heinen, P. and Grundler, F.M. (2000) Promoter analysis of pyk20, a gene from Arabidopsis thaliana . Plant Sci. 157, 245–255. [DOI] [PubMed] [Google Scholar]
- Ramlov, K.B. , Laursen, N.B. , Stougaard, J. and Marcker, K.A. (1993) Site‐directed mutagenesis of the organ‐specific element in the soybean leghemoglobin lbc3 gene promoter. Plant J. 4, 577–580. [DOI] [PubMed] [Google Scholar]
- Rosso, M.N. and Grenier, E. (2011) Other nematode effectors and evolutionary constraints In: Genomics and Molecular Genetics of Plant–Nematode Interactions (Jones J., Gheysen G. and Fenoll C., eds), pp. 287–307. Heidelberg: Springer‐Verlag GmbH & Co. KG. [Google Scholar]
- Sanz, A. , Moreno, J.I. and Castresana, C. (1998) PIOX, a new pathogen‐induced oxygenase with homology to animal cyclooxygenase. Plant Cell, 10, 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons, P.C. (1993) Plant–nematode interactions. Plant Mol. Biol. 23, 917–931. [DOI] [PubMed] [Google Scholar]
- Sijmons, P.C. , Grundler, F.M.N. , von Mende, N. , Burrows, P.R. and Wyss, U. (1991) Arabidopsis thaliana as new model host for plant parasitic nematodes. Plant J. 1, 245–254. [Google Scholar]
- Sobczak, M. and Golinowski, W. (2011) Cyst nematodes and syncytia In: Genomics and Molecular Genetics of Plant–Nematode Interactions (Jones J., Gheysen G. and Fenoll C., eds), pp. 61–82. Heidelberg: Springer‐Verlag GmbH & Co. KG. [Google Scholar]
- Stumpe, M. , Carsjens, J.G. , Stenzel, I. , Gobel, C. , Lang, I. , Pawlowski, K. , Hause, B. and Feussner, I. (2005) Lipid metabolism in arbuscular mycorrhizal roots of Medicago truncatula . Phytochemistry, 66, 781–791. [DOI] [PubMed] [Google Scholar]
- Sukno, S. , Shimerling, O. , McCuiston, J. , Tsabary, G. , Shani, Z. , Shoseyov, O. and Davis, E.L. (2006) Expression and regulation of the Arabidopsis thaliana cel1 Endo 1,4 beta glucanase gene during compatible plant–nematode interactions. J. Nematol. 38, 354–361. [PMC free article] [PubMed] [Google Scholar]
- Szakasits, D. , Heinen, P. , Wieczorek, K. , Hofmann, J. , Wagner, F. , Kreil, D.P. , Sykacek, P ., Grundler, F.M. and Bohlmann, H. (2009) The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 57, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H. , Kanayama, Y. , Zheng, M.S. , Kusano, T. , Hase, S. , Ikegami, M. and Shah, J. (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene‐for‐gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. [DOI] [PubMed] [Google Scholar]
- Thurau, T. , Kifle, S. , Jung, C. and Cai, D. (2003) The promoter of the nematode resistance gene Hs1pro‐1 activates a nematode‐responsive and feeding site‐specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana . Plant Mol. Biol. 52, 643–660. [DOI] [PubMed] [Google Scholar]
- Turner, J.G. , Ellis, C. and Devoto, A. (2002) The jasmonate signal pathway. Plant Cell, 14 (Suppl), S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. and Speleman, F. (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–research0034.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, B. , De Meutter, J. , Tytgat, T. , Van Montagu, M. , Coomans, A. and Gheysen, G. (2004) Secretions of plant‐parasitic nematodes: a molecular update. Gene, 332, 13–27. [DOI] [PubMed] [Google Scholar]
- Vellosillo, T. , Martinez, M. , Lopez, M.A. , Vicente, J. , Cascon, T. , Dolan, L. , Hamberg, M. and Castresana, C. (2007) Oxylipins produced by the 9‐lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell, 19, 831–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronico, P. , Giannino, D. , Melillo, M.T. , Leone, A. , Reyes, A. , Kennedy, M.W. and Bleve‐Zacheo, T. (2006) A novel lipoxygenase in pea roots. Its function in wounding and biotic stress. Plant Physiol. 141, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente, J. , Cascon, T. , Vicedo, B. , Garcia‐Agustin, P. , Hamberg, M. and Castresana, C. (2012) Role of 9‐lipoxygenase and alpha‐dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol. Plant 5, 914–928. [DOI] [PubMed] [Google Scholar]
- Vieira, P. , Danchin, E.G. , Neveu, C. , Crozat, C. , Jaubert, S. , Hussey, R.S. , Engler, G. , Abad, P. , de Almeida‐Engler, J. , Castagnone‐Sereno, P. and Rosso, M.N. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mende, N. (1997) Invasion and migration behaviour of sedentary nematodes In: Cellular and Molecular Aspects of Plant–Nematode Interactions (Fenoll C., Grundler F.M.W. and Ohl S.A., eds), pp. 51–64. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Wubben, M.J. , Jin, J. and Baum, T.J. (2008) Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA‐independent pathogenesis‐related gene expression in roots. Mol. Plant–Microbe Interact. 21, 424–432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 In silico analysis of cis‐acting regulatory elements in 5′ regulatory regions of LOX3 and LOX4 genes. Regulatory elements in the promoter regions of LOX3 and LOX4 were identified using the plant promoter and regulatory element resources PLACE, PlantCARE, ATHENA and Genomatix Matinspector databases. The 2000‐bp sequence of the 5′ regulatory regions was analysed and promoter elements that differed among sequences are presented.
Fig. S2 Polymerase chain reaction (PCR) confirming the T‐DNA insertion in lipoxygenase (LOX) insertion mutants. Two different T‐DNA homozygous insertion mutants were identified for each LOX‐encoding gene: lox3‐1 corresponds to line SALK_119404, lox3‐2 corresponds to line SALK_147830, lox4‐1 corresponds to line SALK_071732 and lox4‐2 corresponds to line SALK_017873. T‐DNA verification was performed by PCR using three primers per line: two genomic binding primers—left genomic primer (LP) and right genomic primer (RP)—specific for each line and complementary to the flanking sequence of the T‐DNA insertion, and one left border primer (LB), LBb1.3, matching all SALK lines. For each SALK line, the PCR products were run in the following order: SALK line and WT with primer set LP + RP, followed by SALK line and WT with primer set LBb1.3 + RP. The following bands were expected: for WT (no insertion), a single product of 900–1100 bp with the genomic LP and RP primers and no product for the homozygous line; a product of 430–902 bp for the homozygous line with primer set LBb1.3 + RP. See Table S1 for the list of primers.
Table S1 Oligonucleotides used in this study for Arabidopsis SALK line verification. T‐DNA and gene‐specific primers were designed according to the SIGnAL T‐DNA Express Arabidopsis gene mapping tool (http://signal.salk.edu/).
Table S2 Overview of the target genes used in this study, showing the accession number for each target gene primer pair used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), the defence pathway in which each gene is mainly involved and the appropriate reference gene used.


