Summary
Movement proteins (MPs) encoded by plant viruses interact with host proteins to facilitate or interfere with intra‐ and/or intercellular viral movement. Using yeast two‐hybrid and bimolecular fluorescence complementation assays, we herein present in vivo evidence for the interaction between Alfalfa mosaic virus (AMV) MP and Arabidopsis Patellin 3 (atPATL3) and Patellin 6 (atPATL6), two proteins containing a Sec14 domain. Proteins with Sec14 domains are implicated in membrane trafficking, cytoskeleton dynamics, lipid metabolism and lipid‐mediated regulatory functions. Interestingly, the overexpression of atPATL3 and/or atPATL6 interfered with the plasmodesmata targeting of AMV MP and correlated with reduced infection foci size. Consistently, the viral RNA levels increased in the single and double Arabidopsis knockout mutants for atPATL3 and atPATL6. Our results indicate that, in general, MP–PATL interactions interfere with the correct subcellular targeting of MP, thus rendering the intracellular transport of viral MP‐containing complexes less efficient and diminishing cell‐to‐cell movement.
Keywords: movement protein, patellin, intercellular movement, AMV, ilarvirus
Introduction
To establish systemic infection, plant viruses must traffic from initially infected cells to neighbouring cells through plasmodesmata (PD) channels until they reach the vascular system (Fernandez‐Calviño et al., 2011; Pallas et al., 2011). Such intercellular movement is an active process that requires one or more viral‐encoded movement proteins (MPs) to interact with other viral factors (genome and other proteins) and with host proteins to alter, in some instances, plant physiology (Pallas and Garcia, 2011; Whitham and Wang, 2004). In the last few years, different approaches have permitted the identification of host proteins that interact with several MPs of the 30K superfamily (Melcher, 2000) which, in some cases, affect viral movement (reviewed in Boevink and Oparka, 2005; Lucas, 2006; Whitham and Wang, 2004). Thus, Tomato spotted wilt virus MP interacts with a DnaJ‐like protein (Soellick et al., 2000) and with At‐4/1, a protein showing homology to the myosin and kinesin motor proteins, which has been proposed to be a component of the PD transport machinery (von Bargen et al., 2001; Paape et al., 2006). The MP of Tobacco mosaic virus (TMV) interacts with not only several cytoskeleton components, such as microtubule‐associated protein MPB2C (Kragler et al., 2003), microtubule end‐binding protein 1 (EB1) (Brandner et al., 2008) and actin filaments (McLean et al., 1995), but also with cell wall‐associated proteins, such as pectin methylesterase (PME) (Chen et al., 2000) and calreticulin (Chen et al., 2005). Moreover, TMV MP also interacts with a protein kinase associated with PD (Lee et al., 2005), a DnaJ‐like protein (Shimizu et al., 2009), a plant ankyrin repeat‐containing protein (ANK) (Ueki and Citovsky, 2011) and synaptotagmin, a calcium sensor that regulates vesicle endo‐ and exocytosis (Lewis and Lazarowitz, 2010). Interaction with ANK and PME positively contributes to TMV intercellular movement and systemic movement, respectively. It has been found that the interaction with ANK decreases callose deposition, whereas PME regulates viral unloading from the phloem (Chen and Citovsky, 2003; Ueki and Citovsky, 2011). Synaptotagmin is also required for TMV systemic spread (Lewis and Lazarowitz, 2010). In contrast, calreticulin, MPB2C and EB1 negatively regulate the targeting of TMV MP to PD (Brandner et al., 2008; Chen et al., 2005; Curin et al., 2007; Kragler et al., 2003). An interaction has also been described between the MP of Brome mosaic virus and NbNaCa1, a protein that is similar to the α‐chain of the nascent polypeptide‐associated complex, which is involved in regulating the localization of MP at PD (Kaido et al., 2007). The MP of Cauliflower mosaic virus (CaMV) has been reported to interact with an Arabidopsis protein, related to mammalian proteins, described as rab acceptors (Huang et al., 2001). The Tomato mosaic virus MP has been shown to interact with putative transcriptional co‐activators (KELP and MBF1) and with protein kinases (Matsushita et al., 2001, 2002, 2003; Yoshioka et al., 2004). The overexpression of KELP interferes with viral cell‐to‐cell movement (Sasaki et al., 2009). A yeast‐based approach allowed the expression of the Prunus necrotic ringspot virus (PNRSV) MP, which triggers the general control of amino acid biosynthesis pathway through Gcn2p kinase activation (Aparicio et al., 2011).
Alfalfa mosaic virus (AMV) is the only member of the Alfamovirus genus in the Bromoviridae family. The AMV genome consists of three single‐stranded RNAs of plus‐sense polarity. Replicase subunits P1 and P2 are encoded by monocistronic RNAs 1 and 2, respectively, whereas RNA 3 encodes MP and serves as a template for the synthesis of the nonreplicating subgenomic RNA 4 (sgRNA4) from which the coat protein (CP) is translated. AMV MP belongs to the 30K family and is implicated in intercellular viral movement (reviewed in Bol, 2005). A mutational analysis has shown that AMV MP is able to form tubular structures in protoplasts, which correlate with cell‐to‐cell movement capacity (Sánchez‐Navarro and Bol, 2001). However, the host factors interacting with AMV MP have not yet been identified.
In this work, we report the interaction between AMV MP and two members of the Arabidopsis Patellin (PATL) family: Patellins 3 and 6 (atPATL3 and atPATL6). PATLs are related to Sec14 (Peterman et al., 2004), which is the defining member of a family of phosphatidylinositol transfer proteins (Allen‐Baume et al., 2002). The proteins related to Sec14 play a role in lipid signalling and metabolism, and in membrane trafficking (Routt and Bankaitis, 2004). Biochemical fractioning and intracellular localization experiments have demonstrated that PATLs 1 from Arabidopsis (atPATL1) and zucchini (Cucurbita pepo) are peripheral membrane‐associated proteins, suggesting that PATLs can be implicated in vesicle/membrane trafficking events (Peterman et al., 2004, 2006). Indeed, atPATL1 is critical in cell plate formation and maturation in the late telophase in Arabidopsis root cells (Peterman et al., 2004). Our analysis of the subcellular localization of AMV MP in the presence of either atPATL3 or atPATL6 indicated that these host proteins would diminish viral cell‐to‐cell movement by interfering with MP targeting to PD. Accordingly, we found that the transient overexpression of both atPATLs reduced the infection foci size, whereas viral RNA accumulation increased in the single and double Arabidopsis atPATL knockouts.
Results
AMV MP interacts with atPATL3 and atPATL6 in yeast and in vivo
In order to shed light on the molecular mechanism driving the intercellular movement of the virus, we decided to search for host proteins that interact with AMV MP. Previous analyses have been conducted to identify the host proteins involved in AMV transport by yeast two‐hybrid (Y2H) screens with full‐length MP as the bait, but they yielded inconclusive results (Zuidmeer‐Jongejan, 2002). We reasoned that the characteristic hydrophobic domain (HD) of the 30K family of MPs (Pallas et al., 2013; Sánchez‐Navarro and Pallás, 1997), which, in some viruses, has been shown to be implicated in MP membrane association (Fujiki et al., 2006; Martínez‐Gil et al., 2009), would probably interfere with the protein–protein interactions screened by a conventional GAL 4‐based Y2H system (MATCHMAKER Two‐Hybrid System 3, Clontech, Palo Alto, CA, USA). Therefore, we decided to use a deleted version of AMV MP, lacking the HD, as a bait to screen a cDNA library of mRNA from Arabidopsis leaves. From the 3 × 106 yeast transformants, we identified diverse potential interacting partners (A. Peiro et al., unpublished results). These included deleted versions of atPATL3 (at1g72160) (three clones) and atPATL6 (at3g51670) (two clones), which lacked the N‐terminal 285 and 210 residues, respectively (Fig. S1A, atPATL3‐ΔNter and atPATL6‐ΔNter, see Supporting Information). Attempts to corroborate these interactions with full‐length MP revealed that the viral protein interacted with atPATL6‐ΔNter and, more weakly, with atPATL3‐ΔNter, but not with full‐length atPATL3 (Fig. S1B). Interestingly, the full‐length MP of the related PNRSV (MPp) also interacted with atPATL3‐ΔNter (Fig. S1C).
In order to confirm the interaction of AMV MP with the entire atPATLs, we decided to use an alternative split‐protein sensor system, which was specially designed to detect the interactions between putative membrane‐associated proteins. In this system, the two interacting partners are expressed as fusion proteins with the N‐ and C‐terminal fragments of (β/α)8‐barrel enzyme N‐(5‐phosphoribosyl)‐anthranilate isomerase (Trp1p) from Saccharomyces cerevisiae. The interaction between both fusion proteins reconstitutes Trp1 activity and allows yeast cells to grow on medium lacking tryptophan (Tafelmeyer et al., 2004). For this purpose, the N‐terminal Trp1 fragment (NTrp) was fused to the N‐terminus of full‐length atPATL3 or atPATL6 to create NTrp:atPATL3 and NTrp:atPATL6, respectively, whereas the C‐terminal Trp1 fragment (CTrp) was fused to the C‐terminus of full‐length AMV MP, which resulted in MP:Ctrp (see Fig. 1). Yeast cells were co‐transformed with the corresponding plasmids, and positive transformants were selected after incubation at 28 °C for 3 days on synthetic medium with tryptophan (SD‐UL). Positive protein interactions were detected under the same growth conditions, but using synthetic medium lacking tryptophan (SD‐ULW). As shown in Fig. 1, yeast cells co‐transformed with MP:CTrp1 and NTrp:atPATL3 or with NTrp:atPATL6 grow in the interaction selective medium (SD‐ULW), whereas no growth is observed in the negative interaction controls: NTrp:atPATL3 and NTrp:atPATL6 co‐transformed with p53 protein (p53:CTrp) and MP:CTrp plus NTrp:eCFP (this plasmid expressed the NTrp1 fragment fused to the N‐terminus of the enhanced cyan fluorescent protein, eCFP).
Figure 1.
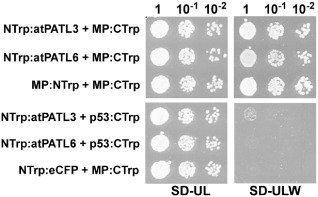
Trp1 yeast two‐hybrid assays. Different dilutions (top) of yeast cells co‐transformed with the indicated pair of plasmids (left) were spotted onto synthetic medium containing (SD‐UL) or lacking (SD‐ULW) tryptophan to confirm the correct transformation or positive interactions, respectively. Self‐interaction of the movement protein (MP) (MP:NTrp + MP:CTrp) was used as a positive interaction. Cells co‐transformed with NTrp:atPATL3 or NTrp:atPATL6 plus p53:CTrp or MP:CTrp plus NTrp:eCFP were used as negative controls.
Previously, it has been shown that AMV MP accumulates at PD (van der Wel et al., 1998). Indeed, the transient expression of the MP fused to the green fluorescent protein (GFP) (MP:GFP) with Agrobacterium tumefaciens C58 cells in Nicotiana benthamiana leaves displayed a punctate structure pattern on the cell wall (Herranz et al., 2005; Sánchez‐Navarro and Bol, 2001). To confirm that this pattern corresponds to PD, we labelled the callose‐rich neck regions of PD with aniline blue in the leaves that transiently expressed MP:GFP. The confocal laser scanning microscopy (CLSM) images depicted a clear co‐localization of MP:GFP with callose deposits, which indicates that these green fluorescent punctate structures are indeed PD with associated MP:GFP (Fig. 2, overlay panel, arrows show examples of the co‐localization at PD).
Figure 2.
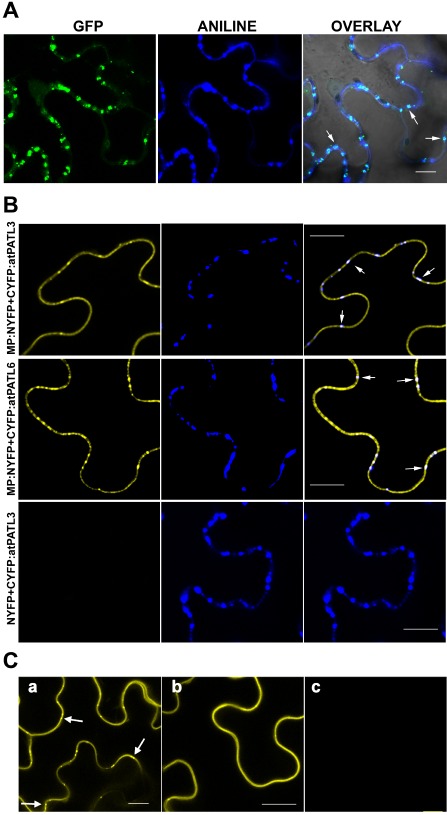
Localization of Alfalfa mosaic virus (AMV) movement protein (MP) at plasmodesmata (PD) and bimolecular fluorescence complementation (BiFC) analysis of the MP–Patellin (MP–PATL) interactions. (A) Confocal laser scanning microscopy (CLSM) images of epidermal cells expressing MP:GFP [green fluorescent protein (GFP) panel] and stained with aniline blue (ANILINE panel) showing MP:GFP and callose localization, respectively. OVERLAY panel is the superposition of GFP, ANILINE and the corresponding bright field image. Arrows indicate PD labelled with both MP:GFP and aniline blue. (B) BiFC analysis to corroborate AMV MP–atPATL interaction in planta. CLSM images of epidermal cells co‐infiltrated with MP:NYFP and CYFP:atPATL3 or CYFP:atPATL6 (indicated on the left) and stained with aniline blue solution. Panels on the right are the superposition of the yellow fluorescent protein (YFP) fluorescence and Aniline staining images (panels on the left and centre, respectively). Arrows indicate reconstituted fluorescence co‐localizing with callose‐rich PD. Leaves infiltrated with NYFP and CYFP:atPATL3 are the negative interaction controls. (C) BiFC analysis to analyse the implication of the GOLD domain in the interaction between AMV MP and atPATL3. CLSM images of epidermal leaves co‐infiltrated with MP:NYFP and CYFP:atPATL3 (a), MP:NYFP and CYFP:atPATLP3‐ΔGOLD (b) or MP:NYFP and CYFP:GOLD‐P3 (c) are shown. Arrows indicate fluorescence spots representing PD. Bar, 10 μm.
In order to corroborate the atPATL–MP interactions in planta, we used bimolecular fluorescence complementation (BiFC) analysis (Aparicio et al., 2006; Hu et al., 2002). Thus, the N‐terminal fragment of the yellow fluorescent protein (YFP) (NYFP) was fused to the C‐terminus of AMV MP, whereas the C‐terminal YFP fragment (CYFP) was fused to the N‐terminus of atPATL3 and atPATL6 (Fig. S2A, see Supporting Information). Agrobacterium C58 mixtures of cultures, MP:NYFP plus CYFP:atPATL3 or CYFP:atPATL6, were infiltrated in N. benthamiana leaves. Two days later, leaves were stained with aniline blue, 10 min before monitoring reconstituted YFP fluorescence by CLSM. We found that, when AMV MP was co‐expressed with both atPATL3 and atPATL6, fluorescence was detected all around the cell periphery, but also in the MP‐characteristic cell‐wall punctate pattern (Fig. 2, panels on the left). Moreover, this pattern co‐localized with aniline‐labelled PD (Fig. 2, panels on the right). No fluorescence was detected when atPATL3 and atPATL6 were co‐infiltrated with NYFP (Fig. 2, bottom panels and not shown, respectively). These results indicate that atPATL3 and atPATL6 interact in planta with AMV MP, and this confirms not only the interactions revealed by the Y2H system, but also that a pool of MP–atPATL complexes accumulates at PD.
In Arabidopsis, the PATL family comprises six members characterized by a variable N‐terminal domain, followed by a Sec14 lipid‐binding domain and a C‐terminal GOLD domain (Fig. S2A) (Peterman et al., 2004). As the GOLD domain is believed to be implicated in protein–protein interactions (Anantharaman and Aravind, 2002), we studied by BiFC whether the GOLD domain of atPATL3 is involved in the interaction with AMV MP. The atPATL3 GOLD domain comprises amino acid positions 353–487 (see http://www.uniprot.org/uniprot/Q56Z59). Therefore, the CYFP fragment was fused to the N‐terminal region of atPATL3 lacking the GOLD domain (CYFP:atPATL3‐ΔGOLD) and also to the GOLD domain alone (CYFP:GOLD‐P3) (see Fig. S2A). Each construct was co‐infiltrated together with MP:NYFP. Unexpectedly, fluorescence was reconstituted only in the cells expressing CYFP:atPATL3‐ΔGOLD plus MP:NYFP (Fig. 2, panel b). This finding indicates that the GOLD domain is not required to establish an interaction between MP and atPATL3. The co‐infiltration of MP:NYFP with CYFP:atPATL3 was used as a positive interaction (Fig. 2, panel a). Western blot analysis confirmed that all the fusion proteins were correctly expressed ( Fig. S2B).
The observation that a pool of MP–atPATL complexes accumulated at PD led us to wonder whether atPATL3 and atPATL6 would locate in these structures. We analysed the subcellular localization of atPATL3 and atPATL6 by the transient agro‐expression of these proteins fused with GFP at its C‐terminus (atPATL3:GFP and atPATL6:GFP). The CLSM images showed that both proteins accumulated in the cellular periphery (Fig. 3, panels a and c). Magnification of the wall periphery showed that neither of the two atPATLs specifically labelled PD (Fig. 3, panels b and d). Hence, the relocation of atPATL3 and atPATL6 at PD observed in the BiFC assay suggests that this rearrangement is driven by MP throughout the MP–atPATL complexes formed in vivo.
Figure 3.
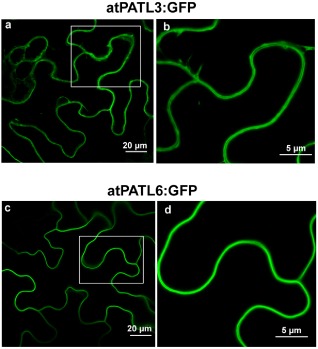
Subcellular localization of Arabidopsis Patellin3 (atPATL3) and atPATL6. Confocal laser scanning microscopy (CLSM) images of epidermal cells infiltrated with Agrobacterium C58 expressing atPATL3 and atPATL6 with the green fluorescent protein (GFP) fused at their C‐terminus (atPATL3:GFP and atPATL6:GFP, respectively). Both fusion proteins present a strong signal at the cell periphery. (b) and (d) show enlarged images of the boxed areas.
AMV MP–atPATL interaction interferes with viral infection
The next step was to examine whether the MP–atPATL interactions can affect viral infection. We first tested the effect of atPATL3 and atPATL6 overexpression. For this purpose, two leaves from three plants per construct of the transgenic Nicotiana tabacum plants expressing P1 and P2 proteins of AMV (P12 plants, Taschner et al., 1991) were inoculated with the RNA transcripts from a modified AMV RNA 3 clone, which expresses GFP together with MP and CP (Sánchez‐Navarro et al., 2001) (Fig. 4A, R3‐GFP). This construct permits the infection foci area to be visualized and measured (Fig. 4B). At 24 h post‐inoculation (hpi), leaves were infiltrated with the Agrobacterium C58 cultures expressing atPATL3, atPATL6, a mixture of both (atPATL3–6) or luciferase (LUC) as a negative control.
Figure 4.
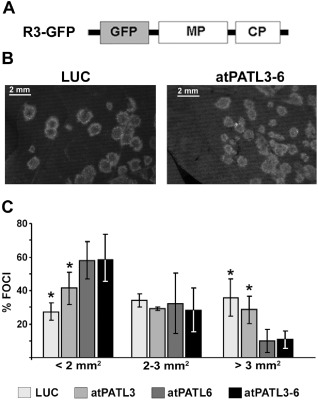
Effect of Arabidopsis Patellin (atPATL) over‐expression on the viral infection. (A) Schematic representation showing the modified Alfalfa mosaic virus (AMV) RNA 3 expressing the green fluorescent protein (GFP) used in this study (R3‐GFP). The open reading frames corresponding to GFP, movement protein (MP) and coat protein (CP) are shown as boxes. (B) Representative images of the foci induced by R3‐GFP in leaves infiltrated with Agrobacterium expressing luciferase (LUC) or a mixture of both atPATLs (atPATL3–6). (C) Graphic showing the average percentage of foci grouped into three different categories according to size area in leaves overexpressing LUC, atPATL3, atPATL6 or a mixture of both (atPATL3–6). Standard deviation values are shown. Significant differences are indicated by *P < 0.05.
By fluorescence microscopy, the images of 50 individual infection foci/leaves were acquired at 4 days post‐inoculation (dpi) and their areas were measured using Image J software (Wayne, Rasband, National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij). Foci were grouped into three different categories according to area size. The graph in Fig. 4C shows the average of three repetitions, where the percentage of fluorescent foci with an area smaller than 2 mm2 was 27.3% ± 5.3% in the leaves infiltrated with LUC, increasing to 41.3% ± 9.6%, 57.9% ± 11.0% and 58.4% ± 15.0% in the corresponding leaves infiltrated with atPATL3, atPATL6 and atPATL3–6, respectively. In contrast, the percentage of foci with an area larger than 3 mm2 was 35.7% ± 11.2% in the LUC leaves, which decreased to 28.5% ± 8.2%, 9.9% ± 6.9% and 10.8% ± 5.2% in the different atPATL‐infiltrated leaves. Overall, these results indicate that the overexpression of these atPATLs hinders cell‐to‐cell movement and has a more pronounced effect when both are expressed simultaneously.
After taking into account that atPATL3‐ΔNter also interacts with the MP of PNRSV (MPp) in Y2H ( Fig. S1C), we wondered whether the overexpression of atPATL3 could also have an effect on MPp function. We first used BiFC assays to confirm that MPp interacted with the entire atPATL3 in planta. The reconstitution of YFP fluorescence was detected in the cells co‐expressing MPp:NYFP with CYFP:atPATL3, but not in cells expressing the NYFP pairs plus CYFP:atPATL3, or MPp:NYFP plus CYFP:GOLD‐P3 (Fig. S3A, panel YFP, see Supporting Information). In accordance with the previous results showing that MPp accumulates at PD (Aparicio et al., 2010), we found that a pool of the MPp–atPATL3 complexes also co‐localized with the aniline blue‐stained PD (Fig. S3A, overlay panel). To investigate the effect on MPp function, we used chimeric AMV RNA 3 which, in addition to the extra GFP gene, harbours the PNRSV MP fused in frame to the C‐terminal 44 amino acids of AMV MP and AMV CP (Fig. S3B, construct R3‐GFP‐MPp). The 44 AMV MP residues are required for specific interactions with the AMV CP to render functional RNA 3, which replicates and moves in P12 plants (Sánchez‐Navarro et al., 2006). Tobacco P12 leaves inoculated with the R3‐GFP‐MPp transcript were infiltrated at 24 hpi with Agrobacterium C58 expressing atPATL3 or LUC. As before, the images of 50 individual infection foci/leaves were acquired at 4 dpi and were grouped into three different categories according to size area. The percentage of fluorescent foci with an area smaller than 2 mm2 was 20% in the control LUC and 55% in the atPATL3 infiltrated leaves. However, the percentage of the foci with an area larger than 3 mm2 was 40% for the LUC leaves, which decreased to 20% in atPATL3 (Fig. S3C). This experiment was repeated with similar results (Fig. S3D). These results demonstrate that the overexpression of atPATL3 also reduces the capability of PNRSV MP to facilitate the viral movement of the chimeric RNA 3.
Finally, we analysed how the absence of atPATL3 and atPATL6 affected the viral accumulation of AMV. Thus, the Arabidopsis T‐DNA insertion mutants for atPATL3 (atpatl3, SALK093994) and atPATL6 (atpatl6, SAIL_284_B11) were isolated, and the double mutant was constructed (atpatl3–6). A polymerase chain reaction (PCR) analysis using atPATLs and the T‐DNA left border‐specific primers was carried out to verify the homozygosity of the mutants. The reverse transcription‐ polymerase chain reaction (RT‐PCR) analysis confirmed the absence of detectable atPATL3 and atPATL6 mRNAs, which corroborates that the mutations result in loss of expression (data not shown). The germination ratio of the double mutant in Murashige & Skoog (MS) medium was decreased slightly (Fig. 5A), although growth in soil had a similar phenotype to the wild‐type (wt). Both mutants and wt plants were inoculated with compatible AMV PV0196 isolate (DSMZ GmbH, Plant Virus Collection, Braunschweig, Germany) virions. At 4 dpi, the total RNA extracted from the inoculated leaves was analysed by Northern blot to detect the accumulation of viral RNAs 3 and 4 using a digoxigenin‐labelled probe corresponding to the AMV CP open reading frame (ORF) (Herranz et al., 2012). The Northern blot in Fig. 5B shows the RNA 3 and 4 accumulation levels of five independent plants from one experiment. The Northern blot signal was quantified using Image J software. The graph in Fig. 5C illustrates that AMV RNAs accumulated at higher levels in atpatl3, atpatl6 and atpatl3–6 than in wt plants. This experiment was repeated with similar results. Overall, our results indicate that the interaction between atPATL3 or atPATL6 and MP negatively affects AMV accumulation in Arabidopsis plants.
Figure 5.
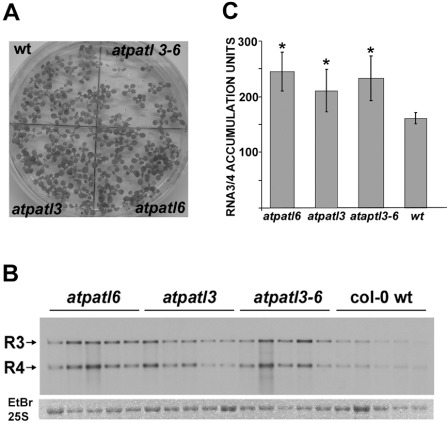
Viral accumulation in Arabidopsis Patellin (atPATL) knockouts. (A) Image of Arabidopsis wild‐type (wt) and knockout seedlings germinated on Murashige and Skoog medium. (B) Detection of Alfalfa mosaic virus (AMV) RNA 3 and 4 accumulation (indicated on the left) in wt and knockout lines by Northern blot analysis of five infected plants. Bottom panel shows the ethidium bromide (EtBr)‐stained gel as a loading control (it only shows the band corresponding to the 25S ribosomal RNA). (C) Graphic showing the average of viral RNA accumulation measured from the Northern blot in (B). Standard deviation values are shown. Significant differences are indicated by *P < 0.05.
AtPATL3 and AtPATL6 interfere with the targeting of AMV MP to PD
The reduced foci size of AMV RNA 3 observed when atPATL3 and atPATL6 were overexpressed could be caused by diminished cell‐to‐cell movement capacity as a result of impaired MP targeting to PD. In order to determine whether AMV MP subcellular localization was affected by the overexpression of these atPATLs, we compared the distribution pattern of MP:GFP alone or in the presence of atPATL3 and atPATL6. N. benthamiana leaves were agro‐infiltrated with MP:GFP plus LUC, MP:GFP plus atPATL3 or MP:GFP plus atPATL6, and were stained with aniline blue to label PD. Five CLSM images/leaves from three leaves were taken for each co‐infiltration (Fig. 6), from which the number of callose‐labelled PDs with associated MP:GFP was counted. The average PD/mm2 was 3.487 in the leaves co‐infiltrated with MP:GFP and LUC, whereas this density dropped to 2.832 and 2.692 PD/mm2 in the leaves co‐expressing atPATL3 and atPATL6, respectively. These results suggest that atPATL3 and atPATL6 negatively influence the PD targeting of the AMV MP.
Figure 6.
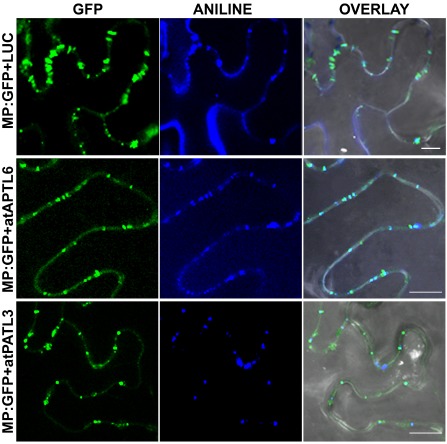
Alfalfa mosaic virus (AMV) movement protein (MP) subcellular localization in the presence of Arabidopsis Patellins (atPATLs). Images of epidermal cells co‐infiltrated with Agrobacterium expressing the proteins indicated on the left and stained with aniline blue. Overlay panels correspond to the superposition of green fluorescent protein (GFP), ANILNE and the corresponding bright field images. Bar, 10 μm.
Discussion
The Arabidopsis PATL family comprises six members (designated as PATL1–6), characterized by a variable N‐terminal region, followed by a Sec14‐like domain and a C‐terminal GOLD domain (Peterman et al., 2004). The yeast protein Sec14 is a prototype module known to be a lipid‐binding domain. Proteins with a Sec14 domain are involved in membrane trafficking, cytoskeleton dynamics, lipid metabolism and lipid‐mediated regulatory functions (reviewed in Bankaitis et al., 2007; Mousley et al., 2007; Phillips et al., 2006). GOLD domains are present in several of the proteins involved in Golgi functioning and vesicle trafficking, and are presumed to act as protein–protein interaction domains (Anantharaman and Aravind, 2002). Despite PATLs being distributed across the plant kingdom, very little is known about their in vivo functions. Database mining indicates that, in Arabidopsis, atPATL3 and atPATL6 are expressed in the whole plant to some degree, including the roots, at the vegetative stage, in the entire rosette and internodes after plant transition to flowering, and in flowers, siliques and seeds (Winter et al., 2007). The biochemical and intracellular localization experiments carried out with PATL1 from Arabidopsis and zucchini have reported that this protein binds phosphatidylinositol and exists in a cytoplasmic pool, which can be associated with cellular membranes to play a critical role in cell plate formation and maturation during the late telophase (Peterman et al., 2004, 2006). PATLs interact with membranes through the Sec14 domain by acting as adaptors to recruit GOLD domain‐binding proteins to specific membrane sites. The cell‐to‐cell trafficking of viral genomes requires the interaction of MPs with host cytoskeleton components and the endomembrane system to reach PD (reviewed in Boevink and Oparka, 2005; Hofmann et al., 2007; Lucas, 2006). Recent data on closely related PNRSV MP have revealed that this protein is peripherally associated with the cytosolic face of the endoplasmic reticulum membrane (Martínez‐Gil et al., 2009).
We have shown that atPATL3 and atPATL6 interact with AMV MP in yeast and in planta. Moreover, the BiFC results reproduce the typical punctate accumulation pattern in the cell periphery of the viral protein, which suggests that some MP–atPATL complexes accumulate at PD. The observation that both atPATL3 and atPATL6 do not accumulate at PD when expressed alone suggests that this PD localization pattern probably results from the interaction with AMV MP which, during its transport towards PD, drags atPATL molecules, modifying their subcellular localization.
Surprisingly, the BiFC analysis also demonstrated that a deleted atPATL3 version lacking the GOLD domain (atPATLP3‐ΔGOLD) is still able to interact with MP, whereas the GOLD domain alone is not. The GAL 4‐based Y2H system showed that a truncated version of atPATL3 and atPATL6, containing the C‐terminal part of Sec14 and the entire GOLD domain, is also able to interact with AMV MP (Fig. S1). In general, these observations indicate that the GOLD domain is not necessary to establish the MP–atPATL3 interaction, whereas part of the Sec14 domain is required to determine this interaction. However, we cannot rule out the possibility that the construct containing the GOLD domain alone can encode a protein with inefficient folding that proves unsuitable to interact with MP. The presence of part of the Sec14 C‐terminal domain may enable correct GOLD folding, thus rendering a functional domain. Another possibility is that the MP–atPATL3 interaction may require the previous interaction of both proteins with the endomembrane system, a process in which the Sec14 domain would play a critical role.
The overexpression and absence of atPATL3 and atPATL6 induce an opposite effect on AMV infection. In general terms, our results reveal that their overexpression impairs cell‐to‐cell movement that reduces AMV accumulation. Moreover, we found that both atPATLs interfere with AMV MP targeting to PD. Taking all our results together, it is tempting to speculate that the interaction of atPATL3 and atPATL6 with AMV MP interferes with intercellular viral movement by negatively affecting the transport of viral complexes towards and through PD. Thus, atPATLs would operate as a defensive barrier against viral infection. This hypothesis is reinforced by the observation that atPATL3 also interacts with the MP of PNRSV, and that it negatively affects its intercellular transport capability. Impairment of viral spread has been reported as a result of the overexpression of different plant proteins which interact directly with viral MPs. In most cases, this interference has been found to be associated with changes in MP intracellular localization patterns, including impaired PD targeting (Brandner et al., 2008; Chen et al., 2005; Curin et al., 2007; Kaido et al., 2007, Kragler et al., 2003; Sasaki et al., 2009; see Pallas and Garcia, 2011 for a review; Fajardo et al., 2013). Future experiments will unravel the mechanism by which PATLs interfere with AMV transport and subcellular localization, and also how specific is the interaction among the MPs from the 30 K family and PATLs.
Experimental Procedures
Plasmid construction
Full‐length ORFs of atPATL3 and atPATL6 were amplified by RT‐PCR from Arabidopsis total RNA with specific sense and antisense primers containing the restriction site SfiI. Amplified fragments were exchanged by the eCFP gene in the pNtrp‐eCFP plasmid digested with SfiI. The resultant clones, pNtrp‐atPATL3 and pNtrp‐atPATL6, contain the 42 N‐terminal amino acids of the (β/α)8‐barrel enzyme N‐(5‐phosphoribosyl)‐anthranilate isomerase (Trp1p) from S. cerevisiae fused at the N‐terminus of both atPATLs. The AMV MP gene was amplified with specific primers containing the SfiI sequence from a previously described clone (Sánchez‐Navarro et al., 2001; 2006). The PCR product was inserted into the pI‐Ctrp3 plasmid, previously digested with SfiI. The resultant clone contains the 179 C‐terminal amino acids of the Trp1p protein fused at the C‐terminus of the MP.
A modified pSK plasmid (psk35S) containing the 35S promoter of CaMV, followed by a multiple cloning site which included NcoI and NheI restriction sites and the potato proteinase inhibitor terminator (Popit), was used to generate GFP and BiFC fusion proteins. Full‐length GFP, NYFP and CYFP fragments were PCR amplified with specific pairs of sense/antisense primers containing PagI (compatible with NcoI)‐NcoI or NheI‐XbaI (compatible with NheI) restriction sites. PCR products were cloned into NcoI‐linearized psk35S plasmid to obtain psk35S:NYFP‐(NcoI‐NheI)‐Popit and psk35S:CYFP‐(NcoI‐NheI)‐Popit, or into NheI‐linearized pks35S plasmid to obtain psk35S:(NcoI‐NheI)‐GFP‐Popit, psk35S:(NcoI‐NheI)‐NYFP‐Popit and psk35S:(NcoI‐NheI)‐CYFP‐Popit. atPATL3, atPATL6, atPATLP3‐ΔGOLD and GOLD‐P3 ORFs were PCR amplified with specific primers containing NcoI and NheI restriction sites, and cloned in either psk35S:CYFP‐(NcoI‐NheI)‐Popit or psk35S:(NcoI‐NheI)‐GFP‐Popit. AMV MP was PCR amplified and cloned into psk35S:(NcoI‐NheI)‐NYFP‐Popit. Finally, the resultant expression cassettes derived from the psk35S constructs were introduced into the XhoI‐digested pMOG800 binary vector. Plasmids px032/GFP‐MP‐CP, px032/GFP‐MPp‐CP and binary plasmids expressing the AMV MP fused to the GFP (MP:GFP) have been described previously (Herranz et al., 2005; Sánchez‐Navarro et al., 2001; 2006).
Split‐protein sensor Y2H‐based system
Mixtures of pNTrp and pCTrp fusion proteins were co‐transformed into yeast CRY1 cells as described previously (Gietz and Woods, 2002). Yeast CRY1 strain shows tryptophan (W), histidine (H), adenine (A), uracil (U) and leucine (L) auxotrophies. Positive transformants were selected after incubation at 28 °C for 3 days in synthetic medium lacking uracil and leucine (SD‐UL). Positive protein interactions were detected under the same growth conditions and synthetic medium, but lacking tryptophan (SD‐ULW).
Plant inoculation
Arabidopsis or N. tabacum P12 plants were maintained in pots in a growth chamber at 24 °C with a photoperiod of 16 h light/8 h dark. For Arabidopsis inoculation, three leaves of 4‐week‐old plants were mechanically inoculated with purified virions of AMV PV0196 isolate (DSMZ GmbH, Plant Virus Collection) in 30 mm sodium phosphate buffer, pH 7. In the case of tobacco P12, two leaves were mechanically inoculated with 5 μg/leaf of R3‐GFP or R3‐GFP‐MPp transcripts. For transcription purposes, plasmids px032/GFP‐MP‐CP and px032/GFP‐MPp:A44‐CP were linearized with PstI and transcribed with T7 RNA polymerase (Roche, Basel, Switzerland) following the manufacturer's recommendations.
Agroinfiltration procedures
Binary plasmids were transformed into Agrobacterium tumefaciens C58 cells by electroporation and spread onto Luria Broth (LB) plates containing 50 μg/mL of kanamycin and 25 μg/mL of rifampicin (LBkr). Positive colonies were grown in liquid LBkr at 28 °C for 24 h, and the bacterial cultures were resuspended in infiltration buffer [10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6] at an optical density at 600 nm (OD600) of 0.4 and 0.2 for BiFC and subcellular localization analysis, respectively. In all cases, cultures were infiltrated into 3‐week‐old N. benthamiana plants.
For transient overexpression experiments in tobacco P12 plants, Agrobacterium C58 cultures expressing LUC or atPATLs were prepared in infiltration buffer at OD600 = 0.4 and infiltrated at 24 hpi. GFP fluorescence of the infection sites was visualized and photographed at 4 dpi with a Leica MZ16F fluorescence stereomicroscope (Leica, Solsm, Germany). The area of foci was measured using Image J software (Wayne Rasband, National Institutes of Health; http://rsbweb.nih.gov/ij).
Callose staining
Leaves were infiltrated with aniline blue solution [0.005% aniline blue (Merck, Darmstadt, Germany) in sodium phosphate buffer, 70 mm, pH 9.0] 10 min before visualization.
CLSM images
Images were taken with a Zeiss LSM 780 AxiObserver (Zeiss, Oberkochen, Germany) or Leica TCS SL microscope. In all cases, images correspond to single slices of 1.8 μm thickness from epidermal cells. Excitation and emission wavelengths were 488 and 508 nm for GFP, 514 and 527 nm for YFP and 405 and 460–535 nm for aniline blue, respectively.
Northern and Western blots
Inoculated leaves were harvested at the indicated times. Total RNA was extracted from 0.1 g of leaves using Trizol Reagent (Sigma, St Louis, MO, USA). RNAs were denatured by formaldehyde treatment and analysed by Northern blot hybridization, as described previously (Sambrook et al., 1989). Viral RNAs were visualized on blots using a digoxigenin‐labelled riboprobe corresponding to the AMV CP gene. Synthesis of the digoxigenin‐labelled riboprobe, hybridization and digoxigenin detection procedures were carried out as described previously (Pallas et al., 1998).
For Western blot analysis, 50 mg of leaves were homogenized with 100 μL of Laemmli buffer (Laemmli, 1970), boiled for 3 min and centrifuged for 3 min at 15.800 × g to pellet cellular debris; 25 μL of extracts were resolved by 14% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE). Gels were electrotransferred to poly(vinylidene difluoride) (PVDF) membranes (Amersham, Pittsburgh, PA, USA) following the manufacturer's recommendations. Detection of NYFP‐fused proteins was carried out with anti‐GFP N‐terminal antibody (Sigma; product number G1544), whereas CYFP‐fusion proteins were detected using anti‐GFP antibody (Roche; Cat No. 11814460001). Detection procedures were carried out following the manufacturer's recommendations.
Supporting information
Fig. S1 Identification of movement protein–Patellin (MP–PATL) interactions by the conventional GAL 4‐based yeast two‐hybrid (Y2H) system (MATCHMAKER Two‐Hybrid System 3, Clontech). (A) Scheme showing the domain architecture of the full‐length atPATL3 and C‐terminal fragments of atPATL3 and atPATL6 (atPATL3‐ΔNter and atPATL6‐ΔNter lacking the N‐terminal 285 or 210 amino acids, respectively), which correspond to the protein fragments found as interacting partners of the Alfalfa mosaic virus (AMV) MP (see http://www.uniprot.org/uniprot/Q56Z59 and http://www.uniprot.org/uniprot/Q9SCU1). (B, C) AH109 yeast strain cells were co‐transformed with empty pGBKT7 plasmid (pBD) or plasmid containing the full‐length AMV MP (pBD:MP) and the full‐length Prunus necrotic ringspot virus (PNRSV) MP (pBD:MPp) plus plasmid pGADT7 containing the full‐length atPATL3 (pAD:atPATL3) or the C‐terminal atPATL3 and atPATL6 fragments shown in (A) (pAD:atPATL3‐ΔNter and pAD: atPATL6‐ΔNter). Transformants were spotted onto minimal synthetic dropout (SD) medium containing (SD‐LW) or lacking (SD‐LWHA) histidine and adenine to confirm appropriate co‐transformation or positive interactions, respectively. Cells were grown at 28 °C for 4 days. Interaction with the empty pBD vector was used as a negative control.
Fig. S2 Western analysis of the bimolecular fluorescence complementation (BiFC) interactions between Alfalfa mosaic virus (AMV) movement protein (MP) and the deleted Arabidopsis Patellins (atPATLs). (A) Schematic depiction of atPATL3 and atPATL6 fused to the C‐terminal fragment of the yellow fluorescent protein (YFP) (CYFP:atPATL3 and CYFP:atPATL6). The first two constructs represent the full‐length atPATL3 and atPATL6 with the characteristic C‐terminal GOLD domain. The other two constructs show a deleted version of atPATL3 lacking the GOLD domain (CYFP: atPATLP3‐ΔGOLD) and a construct with the GOLD domain alone (CFP:GOLD‐P3). Numbers correspond to amino acid residue positions in the original sequences. (B) Western analysis to confirm the expression of the fusion proteins analysed in Fig. 2. Detection was carried out with specific antibodies recognizing NYFP and CYFP fusion proteins (Nter and Cter panels, respectively). Lanes 1–4 correspond to leaves infiltrated with MP:NYFP plus CYFP:atPATL3, CYFP:atPATL6, CYFP:atPATLP3‐ΔGOLD or CYFP:GOLD‐P3, respectively. Lane 5 is the noninfiltrated leaves. Asterisk denotes a nonspecific host protein.
Fig. S3 Effect of Arabidopsis Patellin3 (atPATL3) overexpression in R3‐GFP‐MPp accumulation. (A) Bimolecular fluorescence complementation (BiFC) analysis to confirm Prunus necrotic ringspot virus (PNRSV) movement protein (MPp)–atPATL3 interaction in planta. Nicotiana benthamiana leaves were co‐infiltrated with the pairs indicated on the left and stained with aniline blue. Yellow fluorescent protein (YFP) fluorescence was reconstituted only in cells co‐expressing MPp:NYFP and CYFP:atPATL3. OVERLAY panels show the superposition of YFP and ANILNE image. Arrows denote aniline blue‐labelled plasmodesmata (PD) showing fluorescence reconstitution. (B) Schematic representation showing the chimeric RNA 3 with the Alfalfa mosaic virus (AMV) MP replaced by PNRSV MP (MPp). The open reading frames corresponding to green fluorescent protein (GFP), MPp, A44 and coat protein (CP) are shown as large boxes. To render a functional RNA 3, the PNRSV MP is fused in frame to the C‐terminal 44 amino acids of the AMV MP (A44). (C, D) Graphics showing the average percentage of foci grouped into three different categories according to size area in leaves from two experiments. Nicotiana tabacum P12 leaves were inoculated with R3‐GFP‐MPp and infiltrated at 24 h post‐inoculation (hpi) with Agrobacterium expressing luciferase (LUC) or atPATL3. Foci areas were measured at 4 days post‐inoculation.
Acknowledgements
AP was a recipient of a Pre‐Doctoral Fellowship from the program JAE Pre‐Doc of Consejo superior de Investigaciones Científicas. ACI‐G was a recipient of a Pre‐Doctoral Fellowship associated with the project BFU2008‐00604. FA was a recipient of a contract Ramón y Cajal (RYC‐2010–06169) Program of the Ministerio de Educación y Ciencia of Spain. We thank L. Corachan for excellent technical assistance. This work was supported by grants BIO2011‐25018 from the Direccion General de Investigacion Cientifica y Tecnica, the Prometeo Program GV2011/003 from the Generalitat Valenciana and PAID‐06‐10‐1496 from the Universitat Politecnica de Valencia (Spain).
References
- Allen‐Baume, V. , Segui, B. and Cockcroft, S. (2002) Current thoughts on the phosphatidylinositol transfer protein family. FEBS Lett. 531, 74–80. [DOI] [PubMed] [Google Scholar]
- Anantharaman, V. and Aravind, L. (2002) The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 3, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, F. , Sánchez‐Navarro, J.A. and Pallas, V. (2006) In vitro and in vivo mapping of the Prunus necrotic ringspot virus coat protein C‐terminal dimerization domain by bimolecular fluorescence complementation. J. Gen. Virol. 87, 1745–1750. [DOI] [PubMed] [Google Scholar]
- Aparicio, F. , Sánchez‐Navarro, J.A. and Pallas, V. (2010) Implication of the C terminus of the Prunus necrotic ringspot virus movement protein in cell‐to‐cell transport and in its interaction with the coat protein. J. Gen. Virol. 91, 1865–1870. [DOI] [PubMed] [Google Scholar]
- Aparicio, F. , Aparicio‐Sanchis, R. , Gadea, J. , Sánchez‐Navarro, J.A. , Pallás, V. and Murguía, J.R. (2011) A plant virus movement protein regulates the Gcn2p kinase in budding yeast. PLoS ONE, 6, e27409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis, A.V. , Vincent, P. , Merkulova, M. , Tyeryar, K. and Liu, Y. (2007) Phosphatidylinositol transfer proteins and functional specification of lipid signaling pools. Adv. Enzyme Regul. 47, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bargen, S. , Salchert, K. , Paape, M. , Piechulla, B. and Kellmann, J.W. (2001) Interactions between the tomato spotted wilt virus movement protein and plant proteins showing homologies to myosin, kinesin and DnaJ‐like chaperones. Plant Physiol. Biochem. 39, 1083–1093. [Google Scholar]
- Boevink, P. and Oparka, K.J. (2005) Virus–host interactions during movement processes. Plant Physiol. 138, 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol, J.F. (2005) Replication of alfamo‐ and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43, 39–62. [DOI] [PubMed] [Google Scholar]
- Brandner, K. , Sambade, A. , Boutant, E. , Didier, P. , Mély, Y. , Ritzenthaler, C. and Heinlein, M. (2008) Tobacco mosaic virus movement protein interacts with green fluorescent protein‐tagged microtubule end‐binding protein 1. Plant Physiol. 147, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H. and Citovsky, V. (2003) Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 35, 386–392. [DOI] [PubMed] [Google Scholar]
- Chen, M.H. , Sheng, J. , Hind, G. , Handa, A.K. and Citovsky, V. (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell‐to‐cell movement. EMBO J. 19, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H. , Tian, G.W. , Gafni, Y. and Citovsky, V. (2005) Effects of calreticulin on viral cell‐to‐cell movement. Plant Physiol. 138, 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curin, M. , Ojangu, E.L. , Trutnyeva, K. , Llau, B. , Truve, E. and Waigmann, E. (2007) MPB2C, a microtubule‐associated plant factor, is required for microtubular accumulation of tobacco mosaic virus movement protein in plants. Plant Physiol. 143, 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo, T.V. , Peiro, A. , Pallas, V. and Sanchez‐Navarro, J.A. (2013) Systemic transport of Alfalfa mosaic virus can be mediated by the movement proteins of several viruses assigned to five genera of the 30K family. J. Gen. Virol. 94, 677–681. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Calviño, L. , Faulkner, C. and Maule, A. (2011) Plasmodesmata as active conduits for virus cell‐to‐cell movement In: Recent Advances in Plant Virology (Caranta C., Aranda M.A., Tepfer M. and Lopez‐Moya J.J., eds), pp. 47–74. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Fujiki, M. , Kawakami, S. , Kim, R.W. and Beachy, R.N. (2006) Domains of tobacco mosaic virus movement protein essential for its membrane association. J. Gen. Virol. 87, 2699–2770. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. and Woods, R.A. (2002) Transformation of yeast by Liac/SS carrier DNA/PEG method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Herranz, M.C. , Sanchez‐Navarro, J.A. , Sauri, A. , Mingarro, I. and Pallas, V. (2005) Mutational analysis of the RNA‐binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell‐to‐cell movement. Virology, 339, 31–41. [DOI] [PubMed] [Google Scholar]
- Herranz, M.C. , Pallas, V. and Aparicio, F. (2012) Multifunctional roles for the N‐terminal basic motif of Alfalfa mosaic virus coat protein: nucleolar/cytoplasmic shuttling, modulation of RNA‐binding activity, and virion formation. Mol. Plant–Microbe Interact. 25, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Hofmann, C. , Sambade, A. and Heinlein, M. (2007) PD and intercellular transport of viral RNA. Biochem. Soc. Trans. 35, part 1. [DOI] [PubMed] [Google Scholar]
- Hu, C.D. , Chinenov, Y. and Kerppola, T.K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell, 9, 789–798. [DOI] [PubMed] [Google Scholar]
- Huang, M. , Jongejan, L. , Zheng, H. , Zhang, L. and Bol, J.F. (2001) Intracellular localization and movement phenotypes of Alfalfa mosaic virus movement protein mutants. Mol. Plant–Microbe Interact. 14, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Kaido, M. , Inoue, Y. , Takeda, Y. , Takeda, A. , Mori, M. , Tamai, A. , Meshi, T. , Okuno, T. and Mise, K. (2007) Downregulation of the NbNACa1 gene encoding a movement‐protein‐interacting protein reduces cell‐to‐cell movement of Brome mosaic virus in Nicotiana benthamiana . Mol. Plant–Microbe Interact. 20, 671–681. [DOI] [PubMed] [Google Scholar]
- Kragler, F. , Curin, M. , Trutnyeva, K. , Gansch, A. and Waigmann, E. (2003) MPB2C, a microtubule‐associated plant protein binds to and interferes with cell‐to‐cell transport of tobacco mosaic virus movement protein. Plant Physiol. 132, 1870–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y. , Taoka, K. , Yoo, B.C. , Ben‐Nissan, G. , Kim, D.J. and Lucas, W.J. (2005) Plasmodesmal‐associated protein kinase in tobacco and Arabidopsis recognizes a subset of non‐cell‐autonomous proteins. Plant Cell, 17, 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D. and Lazarowitz, S.G. (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell‐to‐cell transport. Proc. Natl. Acad. Sci. USA, 107, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Martínez‐Gil, L. , Sánchez‐Navarro, J.A. , Cruz, A. , Pallás, V. , Pérez‐Gil, J. and Mingarro, I. (2009) Plant virus cell‐to‐cell movement is not dependent on the transmembrane disposition of its movement protein. J. Virol. 83, 5535–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita, Y. , Deguchi, M. , Youda, M. , Nishiguchi, M. and Nyunoya, H. (2001) The tomato mosaic tobamovirus movement protein interacts with a putative transcriptional coactivator KELP. Mol. Cells, 12, 57–66. [PubMed] [Google Scholar]
- Matsushita, Y. , Miyakawa, O. , Deguchi, M. , Nishiguchi, M. and Nyunoya, H. (2002) Cloning of a tobacco cDNA coding for a putative transcriptional coactivator MBF1 that interacts with the tomato mosaic virus movement protein. J. Exp. Bot. 53, 1531–1532. [PubMed] [Google Scholar]
- Matsushita, Y. , Ohshima, M. , Yoshioka, K. , Nishiguchi, M. and Nyunoya, H. (2003) The catalytic subunit of protein kinase CK2 phosphorylates in vitro the movement protein of Tomato mosaic virus. J. Gen. Virol. 84, 497–505. [DOI] [PubMed] [Google Scholar]
- McLean, B.G. , Zupan, J. and Zambryski, P.C. (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell, 7, 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, U. (2000) The 30K superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Mousley, C.J. , Tyeryar, K.R. , Vincent‐Pope, P. and Bankaitis, V.A. (2007) The Sec14‐superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim. Biophys. Acta, 1771, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape, M. , Solovyev, A.G. , Erokhina, T.N. , Minina, E.A. , Schepetilnikov, M.V. , Lesemann, D.E. , Schiemann, J. , Morozov, S.Y. and Kellmann, J.W. (2006) At‐4/1, an interactor of the Tomato spotted wilt virus movement protein, belongs to a new family of plant proteins capable of directed intra‐ and intercellular trafficking. Mol. Plant–Microbe Interact. 19, 874–883. [DOI] [PubMed] [Google Scholar]
- Pallas, V. and Garcia, J.A. (2011) How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92, 2691–2705. [DOI] [PubMed] [Google Scholar]
- Pallas, V. , Mas, P. and Sanchez‐Navarro, J.A. (1998) Detection of plant RNA viruses by nonisotopic dot‐blot hybridization. Methods Mol. Biol. 81, 461–468. [DOI] [PubMed] [Google Scholar]
- Pallas, V. , Genoves, A. , Sanchez‐Pina, M.A. and Navarro, J.A. (2011) Systemic movement of viruses via the plant phloem In: Recent Advances in Plant Virology (Caranta C., Aranda M.A., Tepfer M. and López‐Moya J.J., eds), pp. 75–102. Norfolk, UK: Caister Academic Press. [Google Scholar]
- Pallas, V. , Aparicio, F. , Herranz, M.C. , Sanchez‐Navarro, J.A. and Scott, S.W. (2013) The molecular biology of ilarviruses. Adv. Virus Res. 87, 130–181. [DOI] [PubMed] [Google Scholar]
- Peterman, T.K. , Ohol, Y.M. , McReynolds, L.J. and Luna, E.J. (2004) Patellin1, a novel Sec14‐like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136, 3080–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman, T.K. , Sequeira, A.S. , Samia, J.A. and Lunde, E.E. (2006) Molecular cloning and characterization of patellin1, a novel sec14‐related protein, from zucchini (Cucurbita pepo). J. Plant Physiol. 163, 1150–1158. [DOI] [PubMed] [Google Scholar]
- Phillips, S.E. , Vincent, P. , Rizzieri, K.E. , Bankaitis, V.A. and Gaucher, E.A. (2006) The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 41, 21–49. [DOI] [PubMed] [Google Scholar]
- Routt, S.M. and Bankaitis, V.A. (2004) Biological functions of phosphatidylinositol transfer proteins. Biochem. Cell Biol. 82, 254–262. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E. F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, U.S.A.
- Sánchez‐Navarro, J.A. and Bol, J.F. (2001) Role of the Alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant–Microbe Interact. 14, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. and Pallás, V. (1997) Evolutionary relationships in the ilarviruses: nucleotide sequence of prunus necrotic ringspot virus RNA 3. Arch. Virol. 142, 749–763. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Herranz, M.C. and Pallás, V. (2006) Cell‐to‐cell movement of Alfalfa mosaic virus can be mediated by the movement proteins of Ilar‐, bromo‐, cucumo‐, tobamo‐ and comoviruses, and does not require virion formation. Virology, 346, 66–73. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Miglino, R. , Ragozzino, A. and Bol, J.F. (2001) Engineering of Alfalfa mosaic virus RNA 3 into an expression vector. Arch. Virol. 146, 923–939. [DOI] [PubMed] [Google Scholar]
- Sasaki, N. , Ogata, T. , Deguchi, M. , Nagai, S. , Tamai, A. , Meshi, T. , Kawakami, S. , Watanabe, Y. , Matsushita, Y. and Nyunoya, H. (2009) Over‐expression of putative transcriptional coactivator KELP interferes with Tomato mosaic virus cell‐to‐cell movement. Mol. Plant Pathol. 10, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T. , Yoshii, A. , Sakurai, K. , Hamada, K. , Yamaji, Y. , Suzuki, M. , Namba, S. and Hibi, T. (2009) Identification of a novel tobacco DnaJ‐like protein that interacts with the movement protein of tobacco mosaic virus. Arch. Virol. 154, 959–967. [DOI] [PubMed] [Google Scholar]
- Soellick, T.R. , Uhrig, J.F. , Bucher, G.L. , Kellmann, J.W. and Schreier, P.H. (2000) The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad. Sci. USA, 97, 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafelmeyer, P. , Johnsson, N. and Johnsson, K. (2004) Transforming a (β/?)8‐barrel enzyme into a split‐protein sensor through directed evolution. Chem. Biol. 11, 681–689. [DOI] [PubMed] [Google Scholar]
- Taschner, P.E. , Van der Kuyl, A.C. , Neeleman, L. and Bol, J.F. (1991) Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology, 181, 445–450. [DOI] [PubMed] [Google Scholar]
- Ueki, S. and Citovsky, V. (2011) To gate, or not to gate: regulatory mechanisms for intercellular protein transport and virus movement in plants. Mol. Plant, 4, 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel, N.N. , Goldbach, R.W. and van Lent, J.W.M. (1998) The movement protein and coat protein of Alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology, 244, 322–329. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. and Wang, Y.Z. (2004) Roles for host factors in plant viral pathogenicity. Curr. Opin. Plant. Biol. 7, 365–371. [DOI] [PubMed] [Google Scholar]
- Winter, D. , Vinega, B. , Naha, H. , Ammar, R. , Wilson, G.V. and Provart, N.J. (2007) An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large‐scale biological data sets. PLoS ONE, 8, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, K. , Matsushita, Y. , Kasahara, M. , Konagaya, K. and Nyunoya, H. (2004) Interaction of Tomato mosaic virus movement protein with tobacco RIO kinase. Mol. Cells, 17, 223–229. [PubMed] [Google Scholar]
- Zuidmeer‐Jongejan, L. (2002) Interactions of plant proteins with alfalfa mosaic virus movement protein. PhD Thesis, University of Leiden, Leiden.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of movement protein–Patellin (MP–PATL) interactions by the conventional GAL 4‐based yeast two‐hybrid (Y2H) system (MATCHMAKER Two‐Hybrid System 3, Clontech). (A) Scheme showing the domain architecture of the full‐length atPATL3 and C‐terminal fragments of atPATL3 and atPATL6 (atPATL3‐ΔNter and atPATL6‐ΔNter lacking the N‐terminal 285 or 210 amino acids, respectively), which correspond to the protein fragments found as interacting partners of the Alfalfa mosaic virus (AMV) MP (see http://www.uniprot.org/uniprot/Q56Z59 and http://www.uniprot.org/uniprot/Q9SCU1). (B, C) AH109 yeast strain cells were co‐transformed with empty pGBKT7 plasmid (pBD) or plasmid containing the full‐length AMV MP (pBD:MP) and the full‐length Prunus necrotic ringspot virus (PNRSV) MP (pBD:MPp) plus plasmid pGADT7 containing the full‐length atPATL3 (pAD:atPATL3) or the C‐terminal atPATL3 and atPATL6 fragments shown in (A) (pAD:atPATL3‐ΔNter and pAD: atPATL6‐ΔNter). Transformants were spotted onto minimal synthetic dropout (SD) medium containing (SD‐LW) or lacking (SD‐LWHA) histidine and adenine to confirm appropriate co‐transformation or positive interactions, respectively. Cells were grown at 28 °C for 4 days. Interaction with the empty pBD vector was used as a negative control.
Fig. S2 Western analysis of the bimolecular fluorescence complementation (BiFC) interactions between Alfalfa mosaic virus (AMV) movement protein (MP) and the deleted Arabidopsis Patellins (atPATLs). (A) Schematic depiction of atPATL3 and atPATL6 fused to the C‐terminal fragment of the yellow fluorescent protein (YFP) (CYFP:atPATL3 and CYFP:atPATL6). The first two constructs represent the full‐length atPATL3 and atPATL6 with the characteristic C‐terminal GOLD domain. The other two constructs show a deleted version of atPATL3 lacking the GOLD domain (CYFP: atPATLP3‐ΔGOLD) and a construct with the GOLD domain alone (CFP:GOLD‐P3). Numbers correspond to amino acid residue positions in the original sequences. (B) Western analysis to confirm the expression of the fusion proteins analysed in Fig. 2. Detection was carried out with specific antibodies recognizing NYFP and CYFP fusion proteins (Nter and Cter panels, respectively). Lanes 1–4 correspond to leaves infiltrated with MP:NYFP plus CYFP:atPATL3, CYFP:atPATL6, CYFP:atPATLP3‐ΔGOLD or CYFP:GOLD‐P3, respectively. Lane 5 is the noninfiltrated leaves. Asterisk denotes a nonspecific host protein.
Fig. S3 Effect of Arabidopsis Patellin3 (atPATL3) overexpression in R3‐GFP‐MPp accumulation. (A) Bimolecular fluorescence complementation (BiFC) analysis to confirm Prunus necrotic ringspot virus (PNRSV) movement protein (MPp)–atPATL3 interaction in planta. Nicotiana benthamiana leaves were co‐infiltrated with the pairs indicated on the left and stained with aniline blue. Yellow fluorescent protein (YFP) fluorescence was reconstituted only in cells co‐expressing MPp:NYFP and CYFP:atPATL3. OVERLAY panels show the superposition of YFP and ANILNE image. Arrows denote aniline blue‐labelled plasmodesmata (PD) showing fluorescence reconstitution. (B) Schematic representation showing the chimeric RNA 3 with the Alfalfa mosaic virus (AMV) MP replaced by PNRSV MP (MPp). The open reading frames corresponding to green fluorescent protein (GFP), MPp, A44 and coat protein (CP) are shown as large boxes. To render a functional RNA 3, the PNRSV MP is fused in frame to the C‐terminal 44 amino acids of the AMV MP (A44). (C, D) Graphics showing the average percentage of foci grouped into three different categories according to size area in leaves from two experiments. Nicotiana tabacum P12 leaves were inoculated with R3‐GFP‐MPp and infiltrated at 24 h post‐inoculation (hpi) with Agrobacterium expressing luciferase (LUC) or atPATL3. Foci areas were measured at 4 days post‐inoculation.


