Summary
Salicylic acid (SA) acts as a signalling molecule in plant defence against biotrophic and hemibiotrophic phytopathogens. The biosynthesis of SA on pathogen detection is essential for local and systemic acquired resistance, as well as the accumulation of pathogenesis‐related (PR) proteins. SA biosynthesis can occur via several different substrates, but is predominantly accomplished by isochorismate synthase (ICS1) following pathogen recognition. The roles of BTB domain‐containing proteins, NPR1, NPR3 and NPR4, in SA binding and signal transduction have been re‐examined recently and are elaborated upon in this review. The pathogen‐mediated manipulation of SA‐dependent defences, as well as the crosstalk between the SA signalling pathway, other plant hormones and defence signals, is also discussed in consideration of recent research. Furthermore, the recent links established between SA, pathogen‐triggered endoplasmic reticulum stress and the unfolded protein response are highlighted.
Introduction
The medicinal effects of salicylic acid (SA) have been studied in humans for well over two centuries. It is well known that chewing the leaves or bark of the willow tree (Salix), rich in SA, can relieve fevers, pain and inflammation (Maclagan, 1876; Vlot et al., 2009). However, the roles of SA in the plant system were only described about two decades ago. SA is one of many phenolic compounds produced by plants, and is involved in a multitude of regulatory pathways. Although SA has been shown to regulate cell growth, stomatal aperture, respiration, seed germination, seedling development, thermotolerance, fruit yield, nodulation in legumes and the expression of senescence‐related genes, it is mostly known for its central role in defence responses (Spoel and Dong, 2012; Vlot et al., 2009).
In the continued battle for dominance in the plant–pathogen struggle, pathogens have devised multiple ways to overcome plant innate immunity (Bozkurt et al., 2012). This commonly involves the delivery of effector proteins from pathogens into the host plant. Effectors detrimentally affect plants by suppressing immunity or modifying growth, metabolism or physiology. This often leads to effector‐triggered susceptibility (ETS), a condition in which a plant is left vulnerable to pathogen parasitism (Bozkurt et al., 2012; Jones and Dangl, 2006; Win et al., 2012). The transfer of effectors into plant cells may occur via several different methods. The bacterium Pseudomonas syringae utilizes a type III secretion system (T3SS), which provides a molecular route for the translocation of effectors. These type III effectors (T3Es) may target either the plant apoplast or cytoplasm, and generally act as immunosuppressors (Alfano, 2009; Win et al., 2012). These effectors have recently been shown to target the SA signalling pathway and have been discussed in brief (Jelenska et al., 2007, 2010). To inhibit immunosuppression by effectors, plants have evolved resistance (R) proteins to detect effectors or their modified targets (Jones and Dangl, 2006; Lewis et al., 2009). Nearly all R proteins are composed of nucleotide‐binding leucine‐rich repeat (NB‐LRR) domains. These NB‐LRR proteins can be further subdivided into the toll interleukin‐1 receptor (TIR) and coiled‐coil (CC)‐NB‐LRRs. NB‐LRR proteins act as plant immune receptors and are responsible for the initiation of effector‐triggered immunity (ETI), often in terms of cell death, known as the hypersensitive response (HR) (Win et al., 2012).
The HR is characterized by localized necrosis and tissue lesions as part of an attempt to ensure pathogen containment (Mur et al., 2008). However, cell death does not guarantee that a pathogen will not spread beyond localized lesions. This is demonstrated by strains of Tobacco mosaic virus (TMV) resistant to N gene‐mediated HR (Padgett and Beachy, 1993). In addition to local necrosis, pathogen detection leads to the establishment of both local acquired resistance (LAR) and systemic acquired resistance (SAR) (Fu and Dong, 2013). Both defence responses are characterized by an increase in pathogenesis‐related (PR) protein accumulation and SA biosynthesis (Durrant and Dong, 2004). SAR provides a long‐lasting, system‐wide immunity to a broad spectrum of pathogens (Conrath, 2006). In addition, studies have indicated that SA‐dependent defences may also be transgenerationally primed by hypomethylated genes, resulting in improved resistance to pathogen infection in subsequent generations (Fu and Dong, 2013; Luna et al., 2012). Moreover, a number of studies have indicated considerable crosstalk between the SA defence pathway with other plant hormone pathways, such as the jasmonic acid (JA), ethylene (ET) and abscisic acid (ABA) pathways (Seo and Park, 2010).
SA Biosynthesis: Chorismate Versus Phenylalanine
The requirement for SA in plant defence has been verified for both Eudicotyledonae and Monocotyledonae, with higher background levels of SA obscuring SA induction in monocots (Umemura et al., 2009). In these higher plants, SA biosynthesis is derived from the shikimate–phenylpropanoid pathway, and may occur via two distinct branches. One of these routes, known as the cinnamic acid pathway, requires the compound phenylalanine, whereas the other occurs via isochorismate production (Chen et al., 2009; Vlot et al., 2009).
The conversion of phenylalanine to cinnamic acid is catalysed by phenylalanine ammonia lyase (PAL). Cinnamic acid can undergo hydroxylation to ortho‐coumaric acid with subsequent oxidation of the side chain to produce SA. The production of SA from phenylalanine may also occur by an initial oxidation of the cinnamic acid side chain to produce benzoic acid, which subsequently undergoes hydroxylation at the ortho position (Metraux, 2002). In both pathways, PAL is responsible for the initial catalytic reaction (Vlot et al., 2009).
However, the majority of pathogen‐induced SA production occurs via a distinct pathway. This is evident in plants with the SA induction‐deficient 2 (sid2) mutation. These mutants only exhibit 5%–10% of the pathogen‐induced SA quantities of the wild‐type (Wildermuth et al., 2001). The sid2 mutation has been traced to the isochorismate synthase 1 (ICS1) gene encoding a chloroplastic ICS (Fig. 1), which, together with isochorismate pyruvate lyase (IPL), is responsible for the conversion of chorismate to isochorismate and, ultimately, SA (Shah, 2003; Vlot et al., 2009). Indeed, SA produced via ICS1 has been shown to be necessary for the establishment of both LAR and SAR (Wildermuth et al., 2001). Additional experiments are required to tease apart the differential requirement for various SA biosynthesis pathways under different types of biotic stress.
Figure 1.
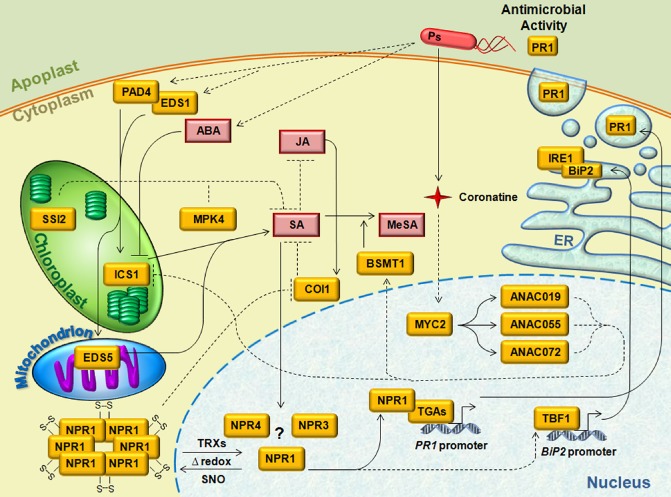
An overview of the salicylic acid (SA) signalling pathways. Pathogen secretion of the phytotoxin coronatine indirectly promotes MYC2 activation of NAC genes, which inhibit SA accumulation through the down‐regulation of isochorismate synthase 1 (ICS1) expression. Indirect activation of BENZOIC ACID/SALICYLIC ACID CARBOXYL METHYLTRANSFERASE 1 (BSMT1) expression by NAC genes may also result in BSMT1‐mediated conversion of SA into methyl salicylate (MeSA). Pathogens may also indirectly promote abscisic acid (ABA) accumulation to inhibit SA production through ICS1. Pathogen detection elicits SA biosynthesis via PHYTOALEXIN DEFICIENT 4 (PAD4) and ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) interactions with ICS1 and EDS5, whereas pathogens may activate other antagonistic phytohormone pathways via effectors. The accumulation of SA results in changes in the cellular redox potential and facilitates thioredoxin (TRX)‐mediated NON‐EXPRESSOR OF PATHOGENESIS‐RELATED GENES 1 (NPR1) deoligomerization; conversely, NPR1 re‐oligomerization requires S‐nitrosothiol (SNO), a nitric oxide donor. A recent study has indicated that NPR3 and NPR4 act as SA receptors and regulate NPR1 functions. NPR1 and TGAs directly regulate PATHOGENESIS‐RELATED 1 (PR1) expression, which results in PR1 protein production and secretion into the apoplast, where it exerts its antimicrobial activity on the proliferating pathogens. NPR1 also positively regulates TBF1 expression and, in turn, TBF1 promotes SA‐dependent BiP2 expression. The resulting BiP2 protein binds to Unfolded Protein Response (UPR) regulatory proteins, such as IRE1, to prevent activation of the UPR in the absence of biotic stress. IRE1, an endoplasmic reticulum (ER)‐bound, transmembrane protein with kinase/endonuclease activity, orchestrates the coordinated expression of UPR genes following SA or pathogen treatment. Examples of inhibitory effects between jasmonic acid (JA) and SA pathways include the indirect negative regulation of the JA pathway by SA, such as indirect inhibition of CORONATINE‐INSENSITIVE 1 (COI1) by cytosolic NPR1, and JA interaction with COI1, which indirectly inhibits the SA pathway. In addition, JA signalling proteins MITOGEN‐ACTIVATED PROTEIN KINASE 4 (MPK4) and SUPPRESSOR OF SA INSENSITIVITY 2 (SSI2) indirectly regulate SA‐mediated defence. Yellow boxes indicate proteins. Red boxes indicate phytohormones. Solid lines indicate direct causation/interaction. Dotted lines indicate indirect causation/interaction. S–S, disulfide bridges; Ps, Pseudomonas syringae.
Zhang et al. (2010) determined that NON‐EXPRESSOR OF PATHOGENESIS‐RELATED GENES 1 (NPR1), which facilitates a large part of SA downstream signalling, is involved in the down‐regulation of ICS1 upstream of SA. In this negative feedback loop, the activation of ICS1 leads to SA production. SA accumulation results in NPR1 deoligomerization and translocation to the nucleus, where NPR1 suppresses ICS1 gene expression. When NPR1 is unable to localize to the nucleus, continued ICS1 expression results in ICS1 transcript over‐accumulation and toxic levels of SA, indicating that NPR1 acts as a negative regulator of SA biosynthesis and ICS1 expression (Zhang et al., 2010).
SA Derivatives
Although unmodified SA can be found within plant tissues, SA also exists in several different conjugated forms. Many of these conjugates have been identified as forms necessary for increased temporal and spatial manipulation of regulatory processes, as well as possible pathogen‐specific defence responses. Conjugate formation occurs via methylation, glucosylation and amino acid conjugation (Loake and Grant, 2007). Indeed, the majority of SA derived from pathogen recognition is glucosylated by UDP‐glucosyltransferase (UGT), also known as SA glucosyltransferase (SAGT) (Vlot et al., 2009), which forms inactive SA 2‐O‐β‐d‐glucoside (SAG) (Loake and Grant, 2007). SAG, a theoretically functional form of SA upon hydrolysis, is collected in vacuoles, where it is stored until needed (Loake and Grant, 2007).
Pathogen detection may also lead to increased production of the volatile ester methyl salicylate (MeSA). The synthesis of MeSA is dependent on a SABATH methyltransferase, known as SA carboxyl methyltransferase (SAMT), which utilizes S‐adenosyl‐1‐methionine as a methyl donor and substrates containing a carboxyl group (Loake and Grant, 2007). The SABATH family of enzymes is named after three of the earliest identified genes in this family, and is not merely an acronym. Thus, letters were used from the genes SAMT, BAMT (BENZOIC ACID CARBOXYL METHYLTRANSFERASE) and THEOBROMINE SYNTHASE to construct the SABATH family name (Eckardt, 2007). Transgenic Arabidopsis overexpressing OsBSMT1, a rice SA methyltransferase (Attaran et al., 2009), also accumulates higher levels of MeSA and MeBA (methyl benzoic acid) (Loake and Grant, 2007).
In Arabidopsis, MeSA synthesis induced by P. syringae carrying an avirulent effector avrRpm1 requires a functional JA pathway. This may be a result of JA acting as a regulator and promoting the conversion of SA into the volatile MeSA. Interestingly, SAR is not dependent on either JA biosynthesis or JA downstream signalling. In addition, in the absence of MeSA, SAR may still be mounted, as shown in a study with an Arabidopsis bsmt1 (benzoic acid/salicylic acid carboxyl methyltransferase 1) mutant. Thus, SAR in Arabidopsis does not require the production of MeSA (Attaran et al., 2009).
During infection by P. syringae, the presence of the pathogen toxin coronatine (COR) is indirectly responsible for MeSA volatilization outside the leaf tissue. It has been speculated that COR may volatilize MeSA from the leaves in an effort to decelerate the induction of the SA‐mediated defence pathway by slowing the accumulation of SA (Attaran et al., 2009). This volatilization is further exemplified by the discovery that overexpressors of OsBSMT1 actually induce PR1 transcript production in adjacent wild‐type plants in an ICS1‐independent and NPR1‐dependent manner on P. syringae infection (Koo et al., 2007). Thus, MeSA may participate in the induction of defences in systemic tissue or even nearby plants (Spoel and Dong, 2012).
It has been determined recently that the Arabidopsis mutants bsmt1 and sagt1 (SA glucosyltransferase 1) fail to establish enhanced local resistance, whereas corresponding overexpressor lines exhibit increased susceptibility to P. syringae and decreased accumulation of SA in local tissue (Zheng et al., 2012). Zheng et al. (2012) also demonstrated that BSMT1, operating as a regulator of plant defence, can be exploited by pathogens to promote virulence because of its ability to convert SA into the volatile MeSA. Indeterminately, preliminary studies have suggested that the converting ability of SAGT1 may be similarly utilized by pathogens to promote susceptibility. As BSMT1 can effectively convert even low levels of SA into MeSA, the induction of BSMT1 by COR can suppress SA accumulation, thereby limiting plant defences. Nonetheless, the direct role of COR in the suppression of MeSA esterase activity or its expression has not been established (Zheng et al., 2012).
SA is also known to conjugate with certain amino acids to activate defence responses (Loake and Grant, 2007). More recently, acyl acid amido synthetases of the GH3 family have been identified as crucial prereceptor modulators of plant hormone action. In addition, a structural basis for the functions of these modulators has been elucidated (Westfall et al., 2012). For example, the acyl‐adenylate/thioester‐forming enzyme (GH3.5) has been shown to function in conjugate‐dependent defence. GH3.5 conjugates amino acids to SA and acetic acid, and mutations in GH3.5 are known to detrimentally affect disease resistance (Vlot et al., 2009). In addition, jasmonate resistant 1 (JAR1), another member of the GH3 acyl‐adenylate/thioesterase family, is responsible for the catalysis of the conjugation of JA to isoleucine (Guranowski et al., 2007).
In Arabidopsis, PBS3 (GH3.12) has been shown to function in phytohormone–amino acid conjugation and is active in the absence of a thioester intermediate (Okrent et al., 2009). PBS3 is required for resistance to P. syringae, SAG accumulation following pathogen induction and defence activation (Dempsey et al., 2011). However, SA can decrease this enzyme's activity in vitro and may act as a competitive inhibitor, as low levels of SA inhibit PBS3 activity. This may allow for quick, reversible adjustments in phytohormone activity or the promotion of rapid crosstalk between various phytohormone signalling pathways (Okrent et al., 2009). Interestingly, although the pbs3 mutant accumulates double the SA levels relative to the wild‐type, it still displays enhanced disease susceptibility. Thus, it appears that SA cannot adequately activate PR1 expression alone or that PR1 expression is dependent on particular levels of both free SA and SAG (Nobuta et al., 2007).
SA Signalling
A considerable body of work has identified SA as an important signalling molecule for the activation of plant defences. The inability to synthesize SA and the tendency to exhibit enhanced susceptibility to pathogen infection are well correlated, and have been demonstrated in several Arabidopsis mutants [such as phytoalexin deficient 4 (pad4), enhanced disease susceptibility (eds)‐1, ‐4 and ‐5, and sid2] (Fig. 1), in addition to transgenic lines, such as NahG, expressing the bacterial enzyme salicylate hydroxylase which degrades SA into catechol (van Wees and Glazebrook, 2003). Similarly, the npr1 mutants exhibit enhanced disease susceptibility (Cao et al., 1994). NPR1 is a vital part of one of the SA‐mediated defence signalling pathways. As a central transcriptional regulator, NPR1 is responsible for controlling approximately 95% of SA‐dependent genes (Wang et al., 2006). Moreover, a recent report has suggested that NPR1 may also play a central role in SA perception as a bona fide receptor protein (Wu et al., 2012). However, another recent study has provided evidence for a different SA sensing mechanism that takes place via the NPR1‐like proteins NPR3 and NPR4 (Fu et al., 2012). The evidence supporting the roles of NPR family proteins as possible SA receptors is discussed in subsequent sections of this review article. Moreover, NPR1 has demonstrated the ability to interact differentially with multiple members of the TGA family of basic leucine zipper transcription factors via direct binding to an as‐1 cis‐regulatory element present in promoters of PR genes (Jakoby et al., 2002).
Signalling Components Upstream of SA
To elucidate the SA signalling pathways in Arabidopsis, many genetic screens have been conducted to identify genes that are involved in SA synthesis and signal transduction. These screens have yielded the identification of numerous mutants, both upstream and downstream of the SA signal. Examples of upstream SA signalling components include the aforementioned PAD4, SID2, EDS1, EDS4 and EDS5 proteins (Fig. 1). EDS1 is a lipase‐like protein that interacts with PAD4, a TIR‐NBS‐LRR upstream of SA, and functions in activated ETI and basal immunity against biotrophic pathogens (Falk et al., 1999; Vlot et al., 2009). Notably, the presence of EDS1 and PAD4 is required for TIR‐NB‐LRR‐triggered HR. Conversely, EDS1 and PAD4 must interact directly to facilitate basal resistance to virulent pathogens. Direct interaction between EDS1 and PAD4 also coincides with increased expression of PAD4 and activation of the SA defence pathway (Rietz et al., 2011) (Fig. 1). These data suggest various associations between EDS1 and PAD4 in the regulation of either basal immunity or pathogen containment and HR.
In a yeast three‐hybrid assay, interactions between EDS1 and PAD4 were weak, as indicated by the inability of PAD4 to compete with EDS1–EDS1 or EDS1–SAG101 (SENESCENCE‐ASSOCIATED GENE 101) interactions. Interestingly, in another yeast three‐hybrid assay, free PAD4 promoted EDS1–SAG101 interactions. As EDS1 did not experimentally facilitate the formation of ternary complexes between PAD4 and SAG101, it is believed to transition between the partners (Rietz et al., 2011). Both EDS1 and PAD4 are further postulated to work in a positive feedback loop that is regulated by SA. This is demonstrated by SA's ability to rescue defence induction in eds1 and pad4 mutants and to activate EDS1 and PAD4 expression in wild‐type plants (Vlot et al., 2009).
EDS1 has been identified recently as a pivotal effector target that molecularly connects RPS4 (RECOGNITION OF BACTERIAL EFFECTOR AVRRPS4), a TIR‐NB‐LRR disease resistance protein, to plant defence pathways (Wirthmueller et al., 2007). Complexes of EDS1 with an effector protein AvrRps4 and the plant resistance protein RPS4 were detectable within Arabidopsis leaf extracts after activation of resistance, as well as within the nuclei of tobacco cells subjected to transient expression assays. Translocation of AvrRps4 to the host nucleus or cytoplasm by EDS1, as an RPS4–EDS1 receptor complex, induces defence signalling pathways that are cell compartment specific. Although bacterial growth is suppressed via nuclear processes, nucleocytoplasmic coordination is necessary for both the transcriptional regulation of enhanced resistance as well as HR. In this manner, EDS1 functions as a TIR‐NB‐LRR signal transducer and effector target that is responsible for the activation of defence responses across cellular compartments (Bhattacharjee et al., 2011; Heidrich et al., 2011).
The regulation of signalling downstream of the CC‐NB‐LRR subset of R proteins is mainly regulated by Non‐specific Disease Resistance 1 (NDR1) instead of EDS1. NDR1, a glycophosphatidyl‐inositol‐anchored plasma membrane protein, is believed to act upstream of SA (Century et al., 1997), as benzothiadiazole (BTH), a biologically active SA analogue, can rescue the SAR‐deficient ndr1 mutants (Vlot et al., 2009).
NPR1‐Dependent SA Signalling and SA Receptors
NPR1 is a key regulator of SA‐dependent defence signalling pathways and a suppressor of HR. SA is known to regulate the shift of oligomerized NPR1 into its monomeric (Mou et al., 2003) and dimeric (Boyle et al., 2009) forms. A previous study elegantly showed that the phosphorylation of NPR1 leads to its polyubiquitinylation via Cullin3 (CUL3) E3 ligase, followed by subsequent degradation by the 26S proteasome (Spoel et al., 2009). NPR1 performs its signalling functions as a cofactor in association with transcription factors regulating the expression of plant defence genes. The ability of TGA transcription factors to interact with NPR proteins has been documented, and TGAs, excluding TGA2, appear to facilitate redundant roles in NPR1‐ and SA‐mediated PR gene expression and subsequent activation of defence genes (Boyle et al., 2009; Despres et al., 2000; Fu and Dong, 2013).
Although it has been repeatedly shown that SA has the ability to modify NPR1 activity and localization, inconsistent results have arisen with regard to NPR1's direct response to SA. Maier et al. (2011) first reported NPR1 and some NPR1‐like proteins to be sensitive to SA treatment. Subsequently, a recent study has described NPR3 and NPR4 as novel SA receptors (Fu et al. 2012) (Fig. 2a–d). Both NPR3 and NPR4 were also identified as possible candidate CUL3 adaptors responsible for NPR1 degradation because of their BTB (bric à brac, tramtrack, broad‐complex) domains, which are found in some CUL3 mediators, and distinctive ankyrin repeats, which are responsible for protein–protein interactions, and are typically found in various CUL3 substrate adaptors (Fu et al., 2012).
Figure 2.
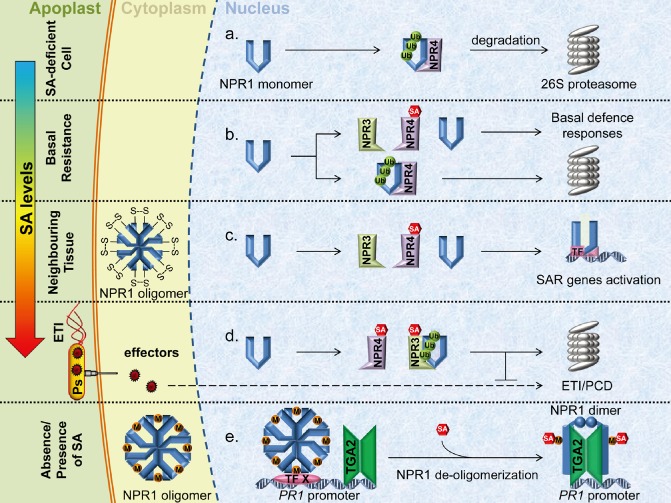
Models for salicylic acid (SA) perception in planta. (a–d) The data of Fu et al. (2012) indicate that NON‐EXPRESSOR OF PATHOGENESIS‐RELATED 3 (NPR3) and NPR4 function as SA receptors. (a) Binding of NPR1 by NPR4 in the absence of SA leads to NPR1 degradation via the 26S proteasome. The Cullin3 (CUL3) adaptor protein is omitted for simplicity. (b) Basal SA levels allow for binding of SA to NPR4, thereby limiting the ability of NPR4 to act as a CUL3 substrate adaptor and binding NPR1 for degradation. Low levels of NPR1 accumulate and subsequently activate basal resistance responses, whilst some NPR4‐dependent NPR1 degradation continues. (c) Moderate SA levels experienced in effector‐triggered immunity (ETI) in neighbouring cells (systemic tissue) allow for SA binding to NPR4, limit NPR4–NPR1 interaction and, in turn, permit NPR1‐dependent expression of systemic acquired resistance (SAR) genes. A pool of NPR1 undergoes degradation via NPR3 interaction. (d) Cells subjected to direct avirulent pathogen attack experience high SA accumulation, leading to subsequent NPR3‐dependent NPR1 degradation and ETI/programmed cell death (PCD) inhibition. (e) Wu et al. (2012) postulate that NPR1 functions as the SA receptor. The NPR1 oligomer contains transitional metal ions (M), such as copper, to facilitate the binding of SA. Reducing conditions in the cell begin the de‐oligomerization of NPR1, but SA is required for complete oligomer disassembly. The nuclear NPR1 oligomer interacts with the PATHOGENESIS‐RELATED 1 (PR1) promoter via an unknown transcription factor (TF X) and binds with a TGA2 dimer on SA induction.
Previously, an npr3 knockout line was shown to exhibit increased basal PR1 expression, together with enhanced resistance to the oomycete Hyaloperonospora arabidopsidis isolate Noco; however, no defect in resistance to P. syringae was demonstrated in these plants (Zhang Y. et al., 2006). Conversely, npr4 mutants show decreased PR gene expression and compromised resistance to P. syringae pv. tomato DC3000 (Pst DC3000) (Liu et al., 2005). npr3 npr4 double mutants demonstrated constitutive PR1 gene expression and enhanced disease resistance, which was partially NPR1 dependent and not caused by increased SA accumulation (Zhang Y. et al., 2006).
In the scenario proposed by Fu et al. (2012), detection of pathogen ingress activates SA accumulation, with higher concentrations occurring locally at the infection site. At higher SA concentrations, NPR3, a protein with low SA affinity, binds SA and facilitates NPR1 degradation (Fig. 2d). As NPR1 is suggested to be an anti‐apoptotic protein (Fu et al., 2012; Rate and Greenberg, 2001), its degradation promotes local ETI and programmed cell death (PCD). In systemic tissues with lower SA concentrations, SA does not bind to the low‐affinity NPR3. Instead, SA binds to NPR4, a high‐affinity SA receptor, and blocks the degradation of NPR1, thereby promoting continued suppression of HR. This allows SA‐mediated defence gene expression and permits the establishment of SAR (Fu et al., 2012; Gust and Nurnberger, 2012) (Fig. 2b,c). Incidentally, Fu et al. (2012) did not observe direct binding between NPR1 and SA under the conditions tested.
In contrast with the report by Fu et al. (2012), Wu et al. (2012) suggested that NPR1 is responsible for direct binding of SA through Cys521/529 via the transition metal copper (Fig. 2e). This binding instigates a conformational change in NPR1 in addition to releasing the C‐terminal transactivation domain from the N‐terminal autoinhibitory BTB/POZ domain. Wu et al. (2012) proposed that SA quickly re‐equilibrates with the mobile phase of NPR1, producing a highly labile NPR1–SA intermediate. Biologically, high lability would promote the rapid detection of SA and allow NPR1 to respond quickly to changes in SA concentration. Stoichiometric results from untreated plants expressing NPR1's C‐terminal transactivation domain (Δ513), as well as the SA‐dependent redistributed form of Δ513, support the presence of an active NPR1 dimeric form. However, elution volumes of the dimer differed between the Δ513 samples, which may support the presence of differing NPR1 conformations (Wu et al., 2012).
In previous reports from the same laboratory, it had been proposed that NPR1 could be detected in both the cytoplasm and the nucleus prior to SA induction, and that nuclear NPR1 dimers associated with TGA2 dimers on the PR1 gene promoter in an SA‐dependent manner to promote gene expression (Boyle et al., 2009; Despres et al., 2000; Rochon et al., 2006). Wu et al. (2012) additionally revealed that NPR1 is present as an oligomer (probably composed of more than four NPR1 molecules) before SA induction, and that this oligomeric structure is stabilized by noncovalent interactions (Fig. 2e).
This model, however, is somewhat in disagreement with previous studies from another laboratory, which suggest that SA manipulates the translocation of NPR1 to the nucleus via cellular redox reactions (Mou et al., 2003). In the inactive state of this scenario, NPR1 resides within the cytoplasm as an oligomer bound by redox‐sensitive disulphide bonds (Kinkema et al., 2000; Mou et al., 2003). Following induction, S‐nitrosothiol (SNO) and cytosolic thioredoxins (TRX) catalyse redox changes in NPR1 from oligomeric to monomeric forms, with SA inducing TRX‐5h to catalyse NPR1 monomer release and possibly to prevent re‐oligomerization (Tada et al., 2008). S‐Nitrosoglutathione (GSNO) is responsible for the donation of nitric oxides and their covalent attachment to reactive cysteine thiols, forming SNOs and promoting monomerization (Tada et al., 2008). These active monomers are then translocated into the nucleus, where NPR1 assists in the binding of transcription factors, such as TGAs, to regulate the expression of defence genes.
Although Fu et al. (2012) detected no SA‐binding activity to NPR1 based on a conventional ligand‐binding assay, Wu et al. (2012) employed equilibrium dialysis, which they claimed is a better suited experimental approach, as it prevents re‐equilibration of SA between mobile and solid phases. High lability of the SA–NPR1 intermediate would make detection via nonequilibrium approaches difficult, and could potentially explain previously reported difficulties in isolating an NPR1–SA complex and discrepancies between the results reported by Fu et al. (2012) and Wu et al. (2012). It is possible that NPR1 activation, deoligomerization, nuclear translocation, phosphorylation and targeted degradation may function according to the model of Fu et al. (2012), and yet some aspects of activation and deoligomerization may be catalysed by direct NPR1–SA binding according to the model of Wu et al. (2012). It also appears plausible that all three NPR proteins function as SA sensors when assembled into various homo‐ and heteromeric protein complexes. As each of these probable SA receptors possesses a different binding affinity, each NPR protein may allow for the differential regulation of defence responses under various SA concentrations. The proposed roles of NPR3 and NPR4 as novel SA receptors regulating the levels of NPR1 via targeted proteolysis could be biologically complemented by a direct SA–NPR1 interaction, providing an additional level of fine‐tuning control to SA‐dependent responses. In the future, it would also be interesting to investigate the potential biological role of the NPR1–NPR2 interaction, which is SA independent, as well as to assay the SA sensitivity of the npr3 npr4 double mutant (Fu et al., 2012).
NPR3, in addition to functioning as an SA‐binding protein, has been shown recently to have repressor activity within both NPR1‐dependent and NPR1‐independent pathways in floral tissue. The npr3 mutants demonstrated the accumulation of PR1 transcript on bacterial infection and enhanced resistance to pathogen infection in immature flowers (Shi et al., 2012). However, NPR1 gene expression was unchanged on either infection by Pst DC3000 or npr3 mutation. Thus, NPR1 appears to be differentially regulated in leaves and flowers. Furthermore, the npr1 npr3 double mutant exhibits intermediate levels of susceptibility to pathogen challenge, supporting NPR3 repressor activity (Shi et al., 2012). Using bimolecular fluorescence complementation (BiFC) assays, Shi et al. (2012) additionally demonstrated that NPR3 and TGA2 interact within both the nucleus and cytoplasm. Similarly, NPR1 and NPR3 have been shown to associate within the cytoplasm in a BiFC assay. Thus, they argued that NPR3 may act as a repressor of NPR1 activity during TGA2 and NPR1 interaction. These additional cytosolic associations present important future questions that will further address the structure and dynamics of the NPR1 oligomer.
Crosstalk Between SA and other Phytohormones
To date, numerous interactions have been detailed between the plant hormone defence pathways (Robert‐Seilaniantz et al., 2011). Some commonly studied pathways include JA, ET and ABA pathways, which are known modulators of defence responses and pathogen resistance (Spoel and Dong, 2008). In addition to these well‐established signalling molecules, several other phytohormones have demonstrated effects on plant defence signalling, including gibberellins, cytokinins, brassinosteroids and auxins (Leon‐Reyes et al., 2010; Wang et al., 2007). Although a definitive relationship between these plant signalling pathways and the SA pathway still remains obscure, the importance of balancing phytohormones is becoming increasingly apparent (Robert‐Seilaniantz et al., 2011; de Torres Zabala et al., 2009). Both positive and negative regulators of various hormone signalling pathways are crucial regulatory targets of hormonal crosstalk in disease and defence. It is vital, therefore, to consider interactions between these, and other, defence signalling pathways.
SA–JA and SA–ET
Although the antagonistic effects of the JA pathway on SA signalling are well recognized, emerging data suggest a more convoluted network of interactions between the two pathways than previously thought (Loake and Grant, 2007). Pharmacological experiments in Arabidopsis have revealed strong antagonistic effects exerted by SA on JA‐responsive genes, such as Plant Defensin 1.2 (PDF1.2), as demonstrated by a simultaneous infection with a biotrophic and necrotrophic pathogen. Similarly, under these conditions, the Arabidopsis wild‐type plants demonstrated enhanced susceptibility to the necrotrophic pathogen, which indicates the ability of the plant to prioritize the SA pathway over the JA pathway (Koornneef et al., 2008; Spoel et al., 2007). In another instance, P. syringae infiltration locally facilitated increased susceptibility to necrotrophic Alternaria brassicicola, but this effect was not observed in systemic tissues (Spoel et al., 2007). However, trade‐offs in defence are not always the result of antagonistic crosstalk. Indeed, low levels of SA and JA are known to act synergistically, suggesting a requirement for threshold levels of hormones for antagonistic effects (Spoel and Dong, 2008).
NPR1 has also demonstrated a critical role in mediating crosstalk between the SA and JA pathways, where SA‐mediated suppression of JA‐inducible genes is prevented in npr1 plants. Interestingly, the nuclear localization of NPR1 is not required for SA‐mediated suppression of the JA‐responsive genes. Thus, suppression of the JA response by SA has been proposed to occur via a novel function of NPR1 in the cytosol (Koornneef and Pieterse, 2008). The JA derivative methyl jasmonate has conversely demonstrated the ability to work in concert with SA in the activation of PR gene expression (Klessig et al., 2000). Accordingly, moderate levels of both JA and SA, applied concurrently, result in antagonistic interactions with progression to tissue necrosis. Lower concentrations of JA and SA alternatively produce the concerted expression of established JA defence markers and PR1 (Mur et al., 2006). It has also been proposed that JA may play important roles in SAR signalling; however, a recent study has provided conclusive evidence against a direct requirement for JA in SAR, whilst indicating that a functional JA biosynthetic pathway is required for MeSA production (Attaran et al., 2009).
Induction of the SA pathway via infection by P. syringae or the application of exogenous SA suppresses the JA signalling pathway, leaving plants more vulnerable to necrotrophic fungi, such as A. brassicicola (Leon‐Reyes et al., 2010). A recent paper by Wathugala et al. (2012) identified SENSITIVE TO FREEZING 6, which is a subunit of the multiprotein transcriptional co‐activator complex, known as Mediator (SFR6/MED16), and is required for both SA‐ and JA‐mediated defences and resistance to P. syringae and UV‐C irradiation (Wathugala et al., 2012).
In addition to the extensively studied SA–JA crosstalk, current data also suggest the existence of an elaborate network of interactions between the SA and ET pathways (Loake and Grant, 2007). Although ET has been shown to work in synergy with SA in the activation of PR gene expression, ET also engages in its own, distinct, defence signalling pathway (van Loon et al., 2006).
There are multiple points of convergence between the often synergistic JA and ET signalling pathways, several of the most interesting being APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF), ETHYLENE RESPONSE FACTOR 1 (ERF1), OCTADECANOID RESPONSIVE ARABIDOPSIS 59 (ORA59) and CONSTITUTIVE EXPRESSOR OF VSP1 (CEV1). The cev1 mutants demonstrate insensitivity to SA‐mediated suppression or a constitutive expression of multiple JA‐ and ET‐dependent marker genes. In this manner, strong induction of both ET and JA pathways prior to SA treatment repressed SA‐dependent suppression of the JA pathway. ORA59 has been proposed to function as a mediator of this process (Leon‐Reyes et al., 2010).
SA–ABA
Bacterial instigation of pathogenesis via molecular determinants is traditionally separated into three groups: T3Es, pathogen‐associated molecular patterns (PAMPs) and toxins, such as COR (de Torres Zabala et al., 2009), with the production of phytotoxins being one of the main methods used to increase pathogen virulence (Feys et al., 1994). COR, a bacterial toxin that induces chlorosis, is produced by several P. syringae pathovars (Feys et al., 1994) and is a structural mimic of jasmonyl‐l‐isoleucine (JA‐Ile). COR advances bacterial expansion by opening stomata, promoting growth in the apoplast and suppressing SA accumulation, which leads to enhanced disease susceptibility (Kazan and Manners, 2012). SA synthesis is known to occur in response to both T3Es and PAMPs, and is predominantly dependent on ICS1. Recent studies have demonstrated that phytopathogens may utilize the ABA signalling pathways to promote virulence (Seo and Park, 2010). Indeed, COR demonstrates the ability to increase ABA levels, which antagonize SA synthesis (de Torres Zabala et al., 2009). As such, ABA synthesis coincides with increased COR levels, and mutants deficient in ABA production demonstrate higher steady‐state levels of ICS1 mRNA and elevated levels of SA. The ability of pathogens to disrupt plant defences by manipulating hormonal signalling is not limited to the JA, SA and ET pathways (Seo and Park, 2010). Antagonism between ABA and SA has also been demonstrated in response to water stress (Mosher et al., 2010).
Although adverse effects of ABA on the SA‐mediated defence pathway have been described, a more complex set of interactions is now coming to light. Studies now suggest that positive interactions between the biotically induced SA, JA and ET signalling pathways and the ABA signalling pathway improve responses to both biotic and abiotic stresses. Seo and Park (2010) demonstrated that the transcription factor MYB96 is responsible for interactions between the SA and ABA signals by functioning as a signalling link, as well as regulating synergistic interactions. Indeed, in the sid2 mutants infected with virulent Pst DC3000, ABA synthesis was decreased compared with that in wild‐type plants. This suggests that SA may be responsible for the positive regulation of ABA levels (de Torres Zabala et al., 2009).
Negative Regulation of SA Signalling by Cor and Pathogenicity Determinants
MITOGEN‐ACTIVATED PROTEIN KINASE 4 (MPK4), together with SUPPRESSOR OF SA INSENSITIVITY 2 (SSI2) and CORONATINE‐INSENSITIVE 1 (COI1), encode important JA signalling proteins, all of which are negative regulators of SA‐mediated defence (Fig. 1). COI1, an F‐box protein, is theorized to negatively regulate suppressors of JA‐mediated defences (Chini et al., 2007; Kunkel and Brooks, 2002; Thines et al., 2007). The coi1 mutants are insensitive to the phytotoxin COR (Feys et al., 1994), and demonstrate heightened resistance to P. syringae and inducible expression of SA‐dependent defences. Contrary to coi1, the mpk4 and ssi2 mutants demonstrate constitutive expression of SA‐dependent defences (Kunkel and Brooks, 2002).
COR‐mediated virulence occurs by the activation of three NAC transcription factor genes, ANAC019, ANAC055 and ANAC072, via the transcription factor MYC2 (Kazan and Manners, 2012; Zheng et al., 2012). A recent study unravelled that these three NAC transcription factors contribute to increased susceptibility by repressing ICS1 and activating the basal expression of BSMT1, a gene involved in the conversion of SA into MeSA (Zheng et al., 2012) (Fig. 1).
Additional pathogen counter‐measures include the use of effectors in the targeting of specific sections of SA signalling. The chloroplast‐localized T3E HopI1, which has been found in all P. syringae strains analysed to date, has recently been suggested to regulate chloroplast‐mediated defences. HopI1 targets the pathways responsible for SA biosynthesis, transport or antagonism through the activation of inhibitory pathways. HopI1 is thought to do this via a J domain, which interacts with Hsp70 in the chloroplast. This is further supported by data indicating that Hsp70 has been shown to catalytically activate many cellular processes involved in client protein folding, assembly and degradation after interacting with the J proteins (Jelenska et al., 2007, 2010).
Effector proteins HopF2 and HopAI1 have also been shown to negatively regulate SA biosynthesis. Rather than regulating SA production in the chloroplast, these effectors target the MPK cascades responsible for the activation of PAMP‐triggered immunity (PTI), supporting research which indicates that MPK activation is a key regulatory event in PTI. Further, HopF2 has been shown to interact with MAP kinase kinase 5 (MKK5) in the inhibition of both PTI and MPK cascades (Wang et al., 2010; Zhang et al., 2007), whereas HopAI1 similarly suppresses MPK3 and MPK6 through direct interaction, thereby inhibiting PTI and cell wall reinforcement (Zhang et al., 2007).
In addition to pathogen effector suppression of plant defence, some virus‐encoded proteins have demonstrated the ability to inhibit or enhance SA‐dependent signalling. In this scenario, the Cucumber mosaic virus (CMV) encoding the 2b counter‐defence protein (CMV 2b) exhibits a complex regulatory effect by interfering with JA‐dependent signalling, RNA silencing, SA biosynthesis and SA‐mediated gene expression. Some of this activity may be explained through effects of CMV 2b on two ARGONAUTE (AGO) family proteins, which compose the Arabidopsis RNA‐induced silencing complex (RISC) (Zhang X. et al., 2006). Inhibition of AGO1 activity would disrupt microRNA (miRNA)‐directed cleavage of mRNAs, such as AGO2 mRNA by miR403, consequently leading to AGO2 transcript accumulation. CMV 2b also demonstrates small RNA (sRNA) binding, which may disrupt RNA silencing and phytohormone signalling. It is also plausible that CMV 2b facilitation of SA biosynthesis may inhibit JA signalling in a more direct manner (Lewsey et al., 2010).
Lastly, the Cauliflower mosaic virus (CaMV) encodes a protein, P6, which functions as a pathogenicity determinant and inhibitor of SA‐dependent defences. P6 functions in RNA silencing and can affect the regulation of both SA‐ and JA‐dependent responses, often by the suppression of SA signalling and enhancement of JA signalling. Interestingly, inactive NPR1 becomes more concentrated in the nucleus in the presence of P6. However, a direct interaction between P6 and NPR1 has not yet been established, and it has therefore been suggested that P6 may regulate NPR1 nuclear localization indirectly via small interfering RNAs (siRNAs) or miRNAs. As NPR1 is a key regulator of SA accumulation, and the JA signalling pathway acts antagonistically to the SA pathway, it is possible that P6 increases biotrophic susceptibility through the promotion of inactive NPR1 nuclear localization and enhancement of the JA signalling pathway (Love et al., 2012).
SA, The Endoplasmic Reticulum (Er) and the Unfolded Protein Response (Upr)
NPR1 regulation of PR gene expression is a well‐documented part of SAR establishment. PR genes encode antimicrobial proteins that are either secreted or destined for vacuoles. Folding of nascent PR peptides is facilitated by ER‐resident chaperones, including BiP (luminal binding protein). Strikingly, NPR1 is also responsible for direct control of the expression of a number of protein secretory pathway genes, including BiP2 (Wang et al., 2005). Arabidopsis bip2 mutants exhibit a moderate reduction in secreted PR1 accumulation following BTH induction and, consequently, are impaired in chemically induced SAR (Wang et al., 2005).
If the translation of pro‐defence peptides in the cell exceeds the folding capacity, a protective cellular signalling pathway, termed the Unfolded Protein Response (UPR), is activated (Ye et al., 2011). During UPR, correct protein folding, modification and secretion are necessary to ameliorate the accumulation of unfolded proteins within the ER.
In an uninduced state, BiPs bind to UPR regulatory proteins, such as the N‐termini of IRE1 (a transmembrane kinase/endonuclease), and effectively prevent activation of the UPR (Iwata and Koizumi, 2012) (Fig. 1). However, following ER stress induction, BiP proteins dissociate from the membrane‐bound proteins to assist in protein folding (Wang et al., 2005). The IRE1 branch of the UPR exhibits roles in plant immunity, as ire1 and bip2 mutants are, to various extents, hypersusceptible to P. syringae and defective in SAR (Moreno et al., 2012; Wang et al., 2005).
An additional gap in the SA–UPR puzzle was recently closed when a novel heat shock‐like transcription factor, TBF1, was identified as a direct regulator of secretory pathway genes in an SA‐dependent manner (Fig. 1). TBF1 and NPR1, although not shown to physically interact, are involved in an intricate transcriptional relationship, as both molecules reciprocally influence each other's expression (Pajerowska‐Mukhtar et al., 2012).
Deficient ER quality control (QC) gene expression in developing rice seeds leads to defects in secretory pathways associated with protein storage (Hayashi et al., 2012). Similarly, defective ER QC leads to impaired expression of secretory proteins that are required for plant defence. A recent study by Hayashi et al. (2012) proposed a model connecting ER stress responses to the SA pathway. In this model, under nonstress conditions, PR proteins are continuously synthesized and secreted at low concentrations. Following early stress induction, OsIRE1–OsbZIP50, OsbZIP39 and OsbZIP60 pathways induce ER QC factors to alleviate ER stress (Hayashi et al., 2012). PR gene expression was reduced in an OsIRE1‐dependent manner, whereas OsWRKY45 expression was up‐regulated in an OsbZIP50‐dependent manner. The SA response, occurring in tandem with ER stress induction, promotes the accumulation and activation of OsWRKY45 which, in turn, induces SA‐responsive gene expression (Hayashi et al., 2012). Notably, Arabidopsis does not possess an OsWRKY45 equivalent, as OsWRKY45 acts upstream of the rice NPR1 orthologue NH1, and all of the potential functional orthologues of OsWRKY45 in Arabidopsis are placed downstream of NPR1 (Shimono et al., 2007). Thus, in this case, parallels cannot be drawn between monocots and dicots, and the mechanistic underpinnings of the connections between the SA pathway and UPR in Arabidopsis will need to be addressed separately (Hayashi et al., 2012).
Conclusions
Although great strides have been made in understanding the SA signalling pathway over the last two decades, much still remains to be elucidated. Significant progress has been made concerning the biosynthesis of SA; however, the extended importance of SA conjugation is continuously expanding. The convoluted nature of the SA signalling cascades with other defence pathways and the oxidative burst in response to pathogen infection indicates a central importance of the SA pathway to plant survival. As progress has been made in clarifying the molecular underpinnings of the SA pathway, our understanding of the central signalling components, such as NPR1, has been brought into question. Three members of the NPR family, NPR1, NPR3 and NPR4, have emerged as potential SA receptors, making SA the final phytohormone for which cognate binding protein(s) have been firmly identified in planta. The link between the SA pathway and UPR, recently established in Arabidopsis and rice, still requires more in‐depth studies. Collectively, the SA signalling pathway constitutes a massive body of molecular regulators and myriad other biological connections. Future studies on the extent of the SA pathway would be greatly facilitated by applications of modern techniques, integrating wet laboratory work with bioinformatics‐aided analyses, using network biology and systems‐level approaches.
Acknowledgements
We gratefully acknowledge support from the University of Alabama (UAB) Biology Department to JLB, the UAB Gulf Oil Response Pilot Grant and the UAB Faculty Development Grant to K.P.‐M. The authors thank Dr Shahid Mukhtar and Mrs Kristin Rockett for critical reading of the manuscript. The authors declare no conflicts of interest.
References
- Alfano, J.R. (2009) Roadmap for future research on plant pathogen effectors. Mol. Plant Pathol. 10, 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran, E. , Zeier, T.E. , Griebel, T. and Zeier, J. (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell, 21, 954–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Halane, M.K. , Kim, S.H. and Gassmann, W. (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science, 334, 1405–1408. [DOI] [PubMed] [Google Scholar]
- Boyle, P. , Le Su, E. , Rochon, A. , Shearer, H.L. , Murmu, J. , Chu, J.Y. , Fobert, P.R. and Despres, C. (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell, 21, 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Banfield, M.J. and Kamoun, S. (2012) Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. [DOI] [PubMed] [Google Scholar]
- Cao, H. , Bowling, S.A. , Gordon, A.S. and Dong, X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S. , Shapiro, A.D. , Repetti, P.P. , Dahlbeck, D. , Holub, E. and Staskawicz, B.J. (1997) NDR1, a pathogen‐induced component required for Arabidopsis disease resistance. Science, 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zheng, Z. , Huang, J. , Lai, Z. and Fan, B. (2009) Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernandez, G. , Adie, B. , Chico, J.M. , Lorenzo, O. , Garcia‐Casado, G. , Lopez‐Vidriero, I. , Lozano, F.M. , Ponce, M.R. , Micol, J.L. and Solano, R. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Conrath, U. (2006) Systemic acquired resistance. Plant Signal. Behav. 1, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D.A. , Vlot, A.C. , Wildermuth, M.C. and Klessig, D.F. (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book, 9, e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres, C. , DeLong, C. , Glaze, S. , Liu, E. and Fobert, P.R. (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell, 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Eckardt, N.A. (2007) Gibberellins are modified by methylation in planta. Plant Cell, 19, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A. , Feys, B.J. , Frost, L.N. , Jones, J.D. , Daniels, M.J. and Parker, J.E. (1999) EDS1, an essential component of R gene‐mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA, 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B. , Benedetti, C.E. , Penfold, C.N. and Turner, J.G. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell, 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. , Yan, S. , Saleh, A. , Wang, W. , Ruble, J. , Oka, N. , Mohan, R. , Spoel, S.H. , Tada, Y. , Zheng, N. and Dong, X. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature, 486, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guranowski, A. , Miersch, O. , Staswick, P.E. , Suza, W. and Wasternack, C. (2007) Substrate specificity and products of side‐reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 581, 815–820. [DOI] [PubMed] [Google Scholar]
- Gust, A.A. and Nurnberger, T. (2012) Plant immunology: a life or death switch. Nature, 486, 198–199. [DOI] [PubMed] [Google Scholar]
- Hayashi, S. , Wakasa, Y. and Takaiwa, F. (2012) Functional integration between defence and IRE1‐mediated ER stress response in rice. Sci. Rep. 2, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich, K. , Wirthmueller, L. , Tasset, C. , Pouzet, C. , Deslandes, L. and Parker, J.E. (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment‐specific immune responses. Science, 334, 1401–1404. [DOI] [PubMed] [Google Scholar]
- Iwata, Y. and Koizumi, N. (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 17, 720–727. [DOI] [PubMed] [Google Scholar]
- Jakoby, M. , Weisshaar, B. , Droge‐Laser, W. , Vicente‐Carbajosa, J. , Tiedemann, J. , Kroj, T. , Parcy, F. and b Z.I.P.R.G. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Jelenska, J. , Yao, N. , Vinatzer, B.A. , Wright, C.M. , Brodsky, J.L. and Greenberg, J.T. (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska, J. , van Hal, J.A. and Greenberg, J.T. (2010) Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA, 107, 13 177–13 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31. [DOI] [PubMed] [Google Scholar]
- Kinkema, M. , Fan, W. and Dong, X. (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell, 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig, D.F. , Durner, J. , Noad, R. , Navarre, D.A. , Wendehenne, D. , Kumar, D. , Zhou, J.M. , Shah, J. , Zhang, S. , Kachroo, P. , Trifa, Y. , Pontier, D. , Lam, E. and Silva, H. (2000) Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA, 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, Y.J. , Kim, M.A. , Kim, E.H. , Song, J.T. , Jung, C. , Moon, J.K. , Kim, J.H. , Seo, H.S. , Song, S.I. , Kim, J.K. , Lee, J.S. , Cheong, J.J. and Choi, Y.D. (2007) Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid‐mediated pathogen resistance in Arabidopsis thaliana . Plant Mol. Biol. 64, 1–15. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, A. , Verhage, A. , Leon‐Reyes, A. , Snetselaar, R. , Van Loon, L. and Pieterse, C.M. (2008) Towards a reporter system to identify regulators of cross‐talk between salicylate and jasmonate signaling pathways in Arabidopsis. Plant Signal. Behav. 3, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Leon‐Reyes, A. , Du, Y. , Koornneef, A. , Proietti, S. , Korbes, A.P. , Memelink, J. , Pieterse, C.M. and Ritsema, T. (2010) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant–Microbe Interact. 23, 187–197. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D. , Guttman, D.S. and Desveaux, D. (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin. Cell Dev. Biol. 20, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Lewsey, M.G. , Murphy, A.M. , Maclean, D. , Dalchau, N. , Westwood, J.H. , Macaulay, K. , Bennett, M.H. , Moulin, M. , Hanke, D.E. , Powell, G. , Smith, A.G. and Carr, J.P. (2010) Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol. Plant–Microbe Interact. 23, 835–845. [DOI] [PubMed] [Google Scholar]
- Liu, G. , Holub, E.B. , Alonso, J.M. , Ecker, J.R. and Fobert, P.R. (2005) An Arabidopsis NPR1‐like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. [DOI] [PubMed] [Google Scholar]
- Loake, G. and Grant, M. (2007) Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Geraats, B.P.J. and Linthorst, H.J.M. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. [DOI] [PubMed] [Google Scholar]
- Love, A.J. , Geri, C. , Laird, J. , Carr, C. , Yun, B.W. , Loake, G.J. , Tada, Y. , Sadanandom, A. and Milner, J.J. (2012) Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS ONE, 7, e47535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, E. , Bruce, T.J. , Roberts, M.R. , Flors, V. and Ton, J. (2012) Next‐generation systemic acquired resistance. Plant Physiol. 158, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclagan, T. (1876) The treatment of rheumatism by salicin and salicylic acid. Br. Med. J. 1, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, F. , Zwicker, S. , Huckelhoven, A. , Meissner, M. , Funk, J. , Pfitzner, A.J.P. and Pfitzner, U.M. (2011) NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) and some NPR1‐related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12, 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metraux, J.P. (2002) Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7, 332–334. [DOI] [PubMed] [Google Scholar]
- Moreno, A.A. , Mukhtar, M.S. , Blanco, F. , Boatwright, J.L. , Moreno, I. , Jordan, M.R. , Chen, Y. , Brandizzi, F. , Dong, X. , Orellana, A. and Pajerowska‐Mukhtar, K.M. (2012) IRE1/bZIP60‐mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE, 7, e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher, S. , Moeder, W. , Nishimura, N. , Jikumaru, Y. , Joo, S.H. , Urquhart, W. , Klessig, D.F. , Kim, S.K. , Nambara, E. and Yoshioka, K. (2010) The lesion‐mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid‐dependent manner. Plant Physiol. 152, 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z. , Fan, W. and Dong, X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell, 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Lloyd, A.J. , Ougham, H. and Prats, E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. [DOI] [PubMed] [Google Scholar]
- Nobuta, K. , Okrent, R.A. , Stoutemyer, M. , Rodibaugh, N. , Kempema, L. , Wildermuth, M.C. and Innes, R.W. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid‐dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okrent, R.A. , Brooks, M.D. and Wildermuth, M.C. (2009) Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4‐substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, H.S. and Beachy, R.N. (1993) Analysis of a tobacco mosaic virus strain capable of overcoming N gene‐mediated resistance. Plant Cell, 5, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska‐Mukhtar, K.M. , Wang, W. , Tada, Y. , Oka, N. , Tucker, C.L. , Fonseca, J.P. and Dong, X. (2012) The HSF‐like transcription factor TBF1 is a major molecular switch for plant growth‐to‐defense transition. Curr. Biol. 22, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N. and Greenberg, J.T. (2001) The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. [DOI] [PubMed] [Google Scholar]
- Rietz, S. , Stamm, A. , Malonek, S. , Wagner, S. , Becker, D. , Medina‐Escobar, N. , Vlot, A.C. , Feys, B.J. , Niefind, K. and Parker, J.E. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rochon, A. , Boyle, P. , Wignes, T. , Fobert, P.R. and Despres, C. (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C‐terminal cysteines. Plant Cell, 18, 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. and Park, C.M. (2010) MYB96‐mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2003) The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. [DOI] [PubMed] [Google Scholar]
- Shi, Z. , Maximova, S. , Liu, Y. , Verica, J. and Guiltinan, M.J. (2012) The salicylic acid receptor NPR3 is a negative regulator of the transcriptional defense response during early flower development in Arabidopsis. Mol. Plant. doi: 10.1093/mp/sss091. [DOI] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. and Dong, X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe, 3, 348–351. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. and Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Johnson, J.S. and Dong, X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA, 104, 18 842–18 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. , Mou, Z. , Tada, Y. , Spivey, N.W. , Genschik, P. and Dong, X. (2009) Proteasome‐mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell, 137, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G.H. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala, M. , Bennett, M.H. , Truman, W.H. and Grant, M.R. (2009) Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. [DOI] [PubMed] [Google Scholar]
- Umemura, K. , Satou, J. , Iwata, M. , Uozumi, N. , Koga, J. , Kawano, T. , Koshiba, T. , Anzai, H. and Mitomi, M. (2009) Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J. 57, 463–472. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Weaver, N.D. , Kesarwani, M. and Dong, X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science, 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Amornsiripanitch, N. and Dong, X. (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. Plos Pathog. 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Pajerowska‐Mukhtar, K. , Culler, A.H. and Dong, X.N. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, J. , Hou, S. , Wang, X. , Li, Y. , Ren, D. , Chen, S. , Tang, X. and Zhou, J.M. (2010) A Pseudomonas syringae ADP‐ribosyltransferase inhibits Arabidopsis mitogen‐activated protein kinase kinases. Plant Cell, 22, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala, D.L. , Hemsley, P.A. , Moffat, C.S. , Cremelie, P. , Knight, M.R. and Knight, H. (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195, 217–230. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C.M. and Glazebrook, J. (2003) Loss of non‐host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742. [DOI] [PubMed] [Google Scholar]
- Westfall, C.S. , Zubieta, C. , Herrmann, J. , Kapp, U. , Nanao, M.H. and Jez, J.M. (2012) Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science, 336, 1708–1711. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Win, J. , Chaparro‐Garcia, A. , Belhaj, K. , Saunders, D.G. , Yoshida, K. , Dong, S. , Schornack, S. , Zipfel, C. , Robatzek, S. , Hogenhout, S.A. and Kamoun, S. (2012) Effector biology of plant‐associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 1–13. [DOI] [PubMed] [Google Scholar]
- Wirthmueller, L. , Zhang, Y. , Jones, J.D. and Parker, J.E. (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1‐dependent defense. Curr. Biol. 17, 2023–2029. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Zhang, D. , Chu, J.Y. , Boyle, P. , Wang, Y. , Brindle, I.D. , De Luca, V. and Despres, C. (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. [DOI] [PubMed] [Google Scholar]
- Ye, C. , Dickman, M.B. , Whitham, S.A. , Payton, M. and Verchot, J. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Cui, H. , Chen, L.J. , Li, H.T. , Zou, Y. , Long, C.Z. , Lan, L.F. , Chai, J.J. , Chen, S. , Tang, X.Y. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Chen, S. and Mou, Z. (2010) Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J. Plant Physiol. 167, 144–148. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Cheng, Y.T. , Qu, N. , Zhao, Q. , Bi, D. and Li, X. (2006) Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. [DOI] [PubMed] [Google Scholar]
- Zheng, X.Y. , Spivey, N.W. , Zeng, W. , Liu, P.P. , Fu, Z.Q. , Klessig, D.F. , He, S.Y. and Dong, X. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe, 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]


