Summary
Taxonomic relationships
Plum pox virus (PPV) is a member of the genus Potyvirus in the family Potyviridae. PPV diversity is structured into at least eight monophyletic strains.
Geographical distribution
First discovered in Bulgaria, PPV is nowadays present in most of continental Europe (with an endemic status in many central and southern European countries) and has progressively spread to many countries on other continents.
Genomic structure
Typical of potyviruses, the PPV genome is a positive‐sense single‐stranded RNA (ssRNA), with a protein linked to its 5′ end and a 3′‐terminal poly A tail. It is encapsidated by a single type of capsid protein (CP) in flexuous rod particles and is translated into a large polyprotein which is proteolytically processed in at least 10 final products: P1, HCPro, P3, 6K1, CI, 6K2, VPg, NIapro, NIb and CP. In addition, P3N‐PIPO is predicted to be produced by a translational frameshift.
Pathogenicity features
PPV causes sharka, the most damaging viral disease of stone fruit trees. It also infects wild and ornamental Prunus trees and has a large experimental host range in herbaceous species. PPV spreads over long distances by uncontrolled movement of plant material, and many species of aphid transmit the virus locally in a nonpersistent manner.
Sources of resistance
A few natural sources of resistance to PPV have been found so far in Prunus species, which are being used in classical breeding programmes. Different genetic engineering approaches are being used to generate resistance to PPV, and a transgenic plum, ‘HoneySweet’, transformed with the viral CP gene, has demonstrated high resistance to PPV in field tests in several countries and has obtained regulatory approval in the USA.
Introduction
Sharka (plum pox), caused by Plum pox virus (PPV), is the most serious viral disease for the stone fruit industry, particularly because it causes severe losses in susceptible cultivars and is spread efficiently by aphids. As a result of domestic and international regulations, the presence of the pathogen in an area greatly complicates stone fruit production and the multiplication and trade of nursery plants. Sharka was first reported in plum trees in Bulgaria in 1917–1918 and was recognized as a viral disease by Atanasoff (1932). Since then, the virus has spread progressively to most of Europe, around the Mediterranean basin and the Near and Middle East. It has also spread to South and North America and Asia (Barba et al., 2011). Despite considerable efforts and quarantine regulations in many countries, sharka has been reported in most of the important Prunus industries worldwide, and is occasionally intercepted in internationally traded Prunus planting material. The disease has not been reported to date in California (USA), Australia, New Zealand and South Africa [European and Mediterranean Plant Protection Organization (EPPO), 2013].
Under natural conditions, the disease affects plants of the genus Prunus, used as commercial cultivars as well as rootstocks: P. armeniaca, P. cerasifera, P. davidiana, P. domestica, P. mahaleb, P. marianna, P. mume, P. persica, P. salicina and interspecific hybrids between these species. Prunus avium, P. cerasus and P. dulcis may be infected occasionally or only by specific PPV strains. In addition, several ornamental and wild Prunus species have been identified as natural or experimental hosts of PPV (Damsteegt et al., 2007; James and Thompson, 2006). Sharka is particularly detrimental in apricots, European plums, peaches and Japanese plums because it can seriously reduce yield and fruit quality. Losses in susceptible cultivars may reach 100% in some cases (Kegler and Hartmann, 1998; Németh, 1994). The alcohol and spirits produced from diseased fruits also see their yield and quality reduced. PPV symptoms may appear on leaves, shoots, bark, petals, fruits and even stones (Fig. 1). They are usually distinct on leaves early in the growing season and include mild light‐green discoloration, chlorotic spots, bands or rings, vein clearing or yellowing and leaf deformation. Flower symptoms can occur on petals (discoloration) of some cultivars. Infected fruits show chlorotic spots or lightly pigmented yellow rings or line patterns. Fruits may become deformed or irregular in shape, and may develop brown or necrotic areas under the discoloured rings. European plums and apricots may also show premature fruit drop, whereas Japanese plums and peaches show ring spotting on fruits. The stones from diseased apricots show typical pale rings or spots. Sweet and sour cherry fruits undergo fruit deformations and premature drop. Infected almond trees generally show no or inconspicuous leaf symptoms. Generally, the fruits of early maturing cultivars of all susceptible species show more marked symptoms than those of late maturing cultivars. PPV also experimentally infects a number of herbaceous hosts (Llácer, 2006; Polák, 2006). Further information about PPV and sharka disease, including illustrations of disease symptoms, can be found in Barba et al. (2011), CABI (2013), EPPO (2004, 2006), García and Cambra (2007), PaDIL (2013) and Sochor et al. (2012).
Figure 1.
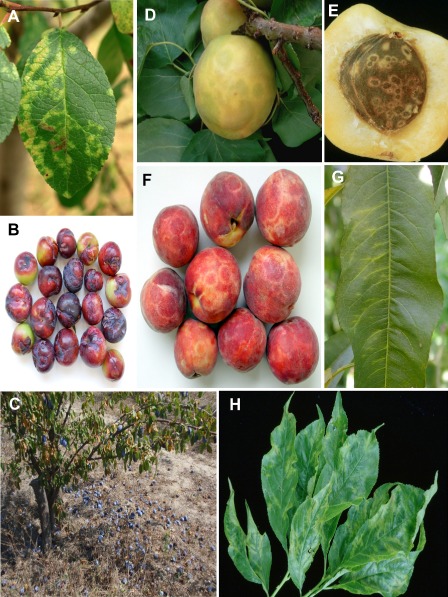
Typical symptoms induced by Plum pox virus on a domestic plum leaf (A), domestic plum fruits (B), premature domestic plum fruit drop (C), an apricot fruit (D), an apricot stone (E), peach fruits (F), a peach leaf (G) and Japanese plum leaves (H). (A, B, D, E and F) were kindly supplied by Dr M. A. Cambra, Centro de Protección Vegetal y Certificación, Diputación General de Aragón, Montañana‐Zaragoza, Spain.
The costs associated with the disease in many countries involve not only direct losses related to yield and quality losses, quarantine, eradication and compensatory measures, but also indirect costs related to preventative measures, inspections, diagnostics and their impact on foreign and domestic trade (Barba et al., 2011). It is estimated that the costs of managing sharka worldwide since the 1970s have exceeded 10 000 million euros (Cambra et al., 2006c).
Epidemiology and Transmission
The illegal traffic and insufficiently controlled exchanges of plant material in a global market are the main pathways for PPV spread over long distances. The introduction of infected propagative plant material is followed by natural and local spread by aphids. PPV is graft transmitted and the vegetative multiplication of infected plants greatly contributes to the spread of the virus from infected areas if certified virus‐free material is not used. Once PPV has become established in an orchard, a number of aphid species with a worldwide distribution may transmit the virus locally in a noncirculative, nonpersistent manner (Ng and Falk, 2006), with Myzus persicae, Aphis spiricola and Hyalopterus pruni being the main vector species (Cambra et al., 2006b; Gildow et al., 2004; Labonne and Dallot, 2006). A single probe of a viruliferous aphid is sufficient to inoculate about 26 000 PPV RNA molecules in a receptor GF305 peach seedling, with a 20% chance of resulting in a systemic infection (Moreno et al., 2009).
The efficiency of natural transmission by aphids and the spatial pattern of spread of sharka may differ for different PPV isolates and host cultivars (Dallot et al., 2003; Sutic et al., 1976). In southern Europe and North America, preferential movement of viruliferous aphids to trees several tree spaces away was observed (Gottwald, 2006; Gottwald et al., 1995). Other virus–host combinations showed a compound contagion process with long‐range (up to 150 m) and short‐range to adjacent tree movements in Spain (Capote et al., 2010). In France, 90% of diseased trees were found within 200 m of previously infected ones, but natural dissemination at distances over 600 m has also been recorded (Labonne and Dallot, 2006). Infections starting with a completely random spatial pattern which finally reaches a uniform distribution in the orchard have also been reported (Varveri, 2006). The application of horticultural mineral oil has been shown to be an efficient control strategy to reduce PPV incidence in nursery plots (Vidal et al., 2013).
Several weed species can be infected with PPV, but the significance of weeds in the epidemiology of the disease is considered to be negligible (Llácer, 2006). There is no confirmed evidence for seed or pollen transmission of PPV in any of its Prunus hosts (Pasquini and Barba, 2006).
Detection and Identification
To avoid PPV spread over long distances by the movement of plant material, reliable detection methods are needed for the accurate detection of the virus in symptomless nursery plants and propagative material. Two official and validated international protocols for the detection and characterization of PPV strains have been developed [EPPO, 2004; International Plant Protection Convention‐Food and Agriculture Organization (IPPC‐FAO), 2012]. An update of these protocols is currently being prepared by EPPO. The recommended methods include biological indexing, serological and molecular assays, as well as sampling, reagents and detailed protocols for each technique. The choice of the most appropriate PPV detection method is crucial and must be adapted to the purpose of the analysis and to the expected prevalence of the disease (Vidal et al., 2012a, b).
Biological indexing based on graft inoculation of GF305 (P. persica seedlings), Nemaguard (P. persica × P. davidiana, hybrid seedling) and/or P. tomentosa is best performed according to Damsteegt et al. (1997) and Gentit (2006). Serological enzyme‐linked immunosorbent analyses (ELISAs) based on the PPV‐specific monoclonal antibody 5B‐IVIA/AMR or on polyclonal antibodies have been used extensively for the universal detection of PPV isolates (Cambra et al., 2006a, 2011). Molecular techniques based on reverse transcription‐polymerase chain reaction (RT‐PCR) assays were first reported for the detection of PPV by Wetzel et al. (1991b). In subsequent years, other RT‐PCR systems, as well as variants based on hemi‐nested, nested RT‐PCR in a single closed tube and co‐operational‐PCR techniques, have been developed to increase sensitivity (García and Cambra, 2007). Nowadays, the technique of choice for nucleic acid‐based PPV detection is real‐time RT‐PCR (Olmos et al., 2005; Schneider et al., 2004), but loop‐mediated isothermal amplification (LAMP) has also been developed into an interesting option (Varga and James, 2006b). Protocols are available for the direct use of plant crude extracts or immobilized tissue prints of plant samples feasible as PCR targets, instead of purified RNA (Capote et al., 2009). Reviews of these user‐friendly methods are available (De Boer and Lopez, 2012; Moreno et al., 2009). In order to estimate diagnostic parameters, such as sensitivity, specificity and likelihood ratios, of different PPV detection methods, latent class models using maximum likelihood functions and a Bayesian approach have been employed by Vidal et al. (2012a). The basic conclusions were as follows: (i) ELISA (5B‐IVIA/AMR based) is highly specific and is recommended when low prevalence of PPV is expected; moreover, it is sufficiently sensitive to consistently detect PPV in composite samples of four plants in spring and summer; and (ii) the highly sensitive spot real‐time RT‐PCR can be successfully used to detect PPV in composite samples (up to 10) in any season of the year, and to assess the PPV‐free status of key material because of its high negative predictive values. The use of sensitive real‐time RT‐PCR is recommended when more than 10% PPV prevalence is expected. The combination of both techniques reaches 100% accuracy in any season of the year (Olmos et al., 2008).
Strain‐specific monoclonal antibodies (Cambra et al., 2006a, 2011; Candresse et al., 1998, 2011) or molecular methods based on RT‐PCR amplification and sequencing (Capote et al., 2006; Glasa et al., 2011, 2013; Olmos et al., 1997, 2002; Šubr et al., 2004; Varga and James, 2005, 2006a) can be used for the identification or characterization of PPV strains. These methods are summarized in the IPPC‐FAO (2012) protocol for PPV diagnosis.
Causative Agent: Genome and Expression
Genome and capsid structure
PPV is a member of the genus Potyvirus of the family Potyviridae (Adams et al., 2012; López‐Moya and García, 2008). Its genome consists of a positive‐sense single‐stranded RNA (ssRNA) of 9741–9795 nucleotides (Fanigliulo et al., 2003; Glasa and Šubr, 2005; Glasa et al., 2011, 2013; James and Varga, 2005; Laín et al., 1989; Maejima et al., 2011; Maiss et al., 1989; Myrta et al., 2006; Palkovics et al., 1993; Schneider et al., 2011; Teycheney et al., 1989; Ulubaş Serçe et al., 2009; SharCo database, http://w3.pierroton.inra.fr:8060/).
The PPV genomic RNA has a protein (viral protein genome‐linked, VPg) linked to its 5′ end and a 3′‐terminal poly A tail (Riechmann et al., 1989), and is encapsidated by a single type of capsid protein (CP) subunit. However, detectable levels of another viral protein, helper component proteinase (HCPro), have been found to be associated with PPV virions (Manoussopoulos et al., 2000). This association could be related to the ability of HCPro to act as a bridge between virus particles and the stylet of aphids which specifically transmit the virus (Blanc et al., 1997; López‐Moya et al., 1995; Roudet‐Tavert et al., 2002). However, roles unrelated to aphid transmission have also been suggested for interactions between HCPro and CP (Roudet‐Tavert et al., 2002).
RNA translation and proteolytic processing
Most of the genomic RNA encodes a long open reading frame (ORF) which is translated into a polyprotein of about 355 kDa, starting from its second AUG codon (nucleotides 147–149) (Riechmann et al., 1991) probably by a leaky scanning mechanism (Simón‐Buela et al., 1997a). This polyprotein is processed by three virus‐encoded proteinases to produce at least 10 mature protein products: P1, HCPro, P3, 6K1, CI, 6K2, VPg, NIapro, NIb and CP (Fig. 2). As reported for other potyviruses (Chung et al., 2008), another PPV protein, P3N‐PIPO, is predicted to be produced by a frameshift into a short ORF embedded within the P3 coding sequence.
Figure 2.
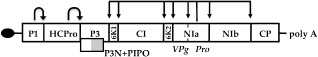
Genomic map of Plum pox virus. The long open reading frame (ORF) is represented by a rectangular box divided into viral products by solid black lines. PIPO ORF, translatable with a frameshift, is indicated by a grey box below the P3 region. Cleavage sites recognized by the indicated proteinases are signalled by arrows. The terminal protein (VPg) is represented as a black ellipse.
The N‐terminal region of the PPV polyprotein is processed by the serine proteinase P1 and the cysteine proteinase HCPro, which cleave at their respective C‐termini (García et al., 1993; Ravelonandro et al., 1993). The proteolytic activity of the C‐terminal catalytic domain of the P1 protease requires the contribution of a host factor present in wheat germ, but not in rabbit reticulocyte lysate (Rodamilans et al., 2013).
NIapro is the protease involved in the cleavage of the central and C‐terminal regions of the PPV polyprotein (García et al., 1989b). It is linked to the protein VPg in the NIa product, which, together with the protein NIb, forms crystalline inclusions, mainly located in the nucleus, but also detected in the cytoplasm of PPV‐infected cells (Martín et al., 1992; van Oosten and van Bakel, 1970). Processing by NIapro takes place at sites characterized by a consensus sequence e/q‐x‐V‐x‐H‐Q/e↓s, and appears to be highly regulated, allowing partially processed products to play functional roles (García et al., 1989a, 1990, 1992). For instance, although mature 6K1 has been detected in PPV‐infected cells (Waltermann and Maiss, 2006), a main functional role has been suggested for the unprocessed P3 + 6K1 protein (Riechmann et al., 1995).
RNA replication, movement and counteraction of host defences
As is a general rule for plus‐strand RNA viruses (Grangeon et al., 2012), PPV RNA replication takes place in association with intracellular membranes (Martín and García, 1991). Leaf extracts in which PPV RNA is synthesized in vitro are enriched in endoplasmic reticulum and tonoplast vesicles, but no in vivo information is available about the PPV replication complexes (Martin et al., 1995). However, they should not differ very much from the membrane vesicles and large perinuclear ring‐like structures in which RNA replication of other potyviruses has been shown to occur (Cotton et al., 2009; Grangeon et al., 2010, 2012; Wei and Wang, 2008; Wei et al., 2010b). In these structures, the potyviral RNA is replicated by the RNA‐dependent RNA polymerase NIb (Hong and Hunt, 1996), using as primer VPg uridylyted by the same polymerase (Anindya et al., 2005; Puustinen and Mäkinen, 2004). Another viral factor required for PPV replication is the CI protein (Fernández et al., 1997), which forms the pinwheel‐shaped inclusions typical of potyviral infections (Martín et al., 1992), and has NTPase and RNA helicase activities (Fernández et al., 1995; Laín et al., 1990, 1991).
Several studies with different potyviruses, including PPV, have shown that the CI protein is also involved in virus movement (Carrington et al., 1998; Gómez de Cedrón et al., 2006). As expected from its ability to form inclusion bodies, PPV CI is able to self‐interact (López et al., 2001); however, CI–CI interactions required for RNA replication and virus movement appear to be different to some extent (Gómez de Cedrón et al., 2006). Results obtained with Turnip mosaic virus suggest that, together with P3N‐PIPO, CI coordinates the formation of conical structures at plasmodesmata for cell‐to‐cell spread (Wei et al., 2010a). Specific interactions of CI with virus particles might be important for virus movement, but also for RNA uncoating and translation initiation (Gabrenaite‐Verkhovskaya et al., 2008).
To be amplified in a host plant, the virus not only has to complete the processes of replication and movement, but also needs to escape the plant antiviral defences. Thus, the proteinase HCPro of PPV is not only required for aphid transmission, but is also essential to counteract antiviral RNA silencing (Tenllado et al., 2003; Varrelmann et al., 2007).
Post‐translational modifications
Given the limited size of the genome of plus‐strand RNA viruses, it is not surprising that many proteins of these viruses are multifunctional and their activities require a meticulous regulation. Post‐translational modifications could contribute to this regulation. The CP protein, which is expected to be involved in the control of the genomic RNA allocated for translation, replication and propagation during potyviral infection (Ivanov et al., 2001), is phosphorylated (Fernández‐Fernández et al., 2002a; Šubr et al., 2007) and O‐N‐acetylglucosylated (O‐GlcNAcylated) by the O‐GlcNAc transferase SECRET AGENT (Chen et al., 2005; Fernández‐Fernández et al., 2002a; Scott et al., 2006). Specific sites of O‐GlcNAc modification (Kim et al., 2011; Pérez et al., 2006, 2013) and a single amino acid mutation that appears to alter the phosphorylation status of the protein (Šubr et al., 2010) have been mapped to the N‐terminal region of PPV CP. Although O‐GlcNAcylation of CP is not essential for PPV infectivity, it plays a relevant role in the infection process (Chen et al., 2005; Pérez et al., 2013).
PPV Diversity
Given its economic importance, much effort has been devoted to the study of the biological, serological and molecular variability of PPV. This effort has revealed that the diversity of PPV is structured into individual monophyletic ensembles of closely related isolates, which have been designated as strains. Currently, eight strains are recognized for PPV, which may be more than for any other potyvirus.
Initially, the existence of two different PPV serotypes, named M (Marcus) and D (Dideron), was reported by Kerlan and Dunez (1979). With the advent of molecular biology, these two serotypes have been confirmed to represent two molecularly distinct strains based on their genome sequences (Laín et al., 1989; Maiss et al., 1989; Palkovics et al., 1993; Teycheney et al., 1989).
PPV‐D is widespread in Europe, whereas PPV‐M is found mainly in southern and central European countries. PPV‐D is also responsible for most outbreaks outside of Europe (Damsteegt et al., 2001; Maejima et al., 2011; Reyes et al., 2003). Although widely present on apricots and plums, this strain is less frequently associated with peach under natural conditions. The PPV‐M strain can be split into two subgroups that show partial geographical separation, but so far have not been reported from outside Europe (Dallot et al., 2011; Myrta et al., 2001). PPV‐M isolates are efficiently aphid transmitted, causing fast epidemics, mainly in peach orchards (Capote et al., 2010; Dallot et al., 2003).
In addition to the two major PPV‐D and PPV‐M strains, two minor strains were identified in the 1990s. The substantial divergence in the genomic sequence of the Egyptian El Amar isolate has led to its classification into the distinct PPV‐EA strain (Glasa et al., 2006; Myrta et al., 2006; Wetzel et al., 1991a), which remains geographically limited to Egypt, where additional isolates have been found on apricot, peach and Japanese plum (Matic et al., 2011; Youssef and Shalaby, 2006).
PPV isolates naturally infecting sour cherries in Moldova were classified into a new, PPV‐C (cherry), strain (Kalashyan et al., 1994; Nemchinov et al., 1996). Later, occasional findings of molecularly similar PPV isolates in sour and sweet cherries were reported from Italy (Crescenzi et al., 1997; Fanigliulo et al., 2003), Hungary (Nemchinov et al., 1998), Belarus (Malinowski et al., 2012) and Croatia (Kajic et al., 2012). Given its restricted natural host range, the actual epidemiological impact of PPV‐C seems to be lower than that of the major PPV strains.
The picture of PPV genetic diversity has changed further in the past 10 years. The development of detection tools targeting different parts of the genome (Glasa et al., 2002) has led to the discovery of a homogeneous group of isolates deriving from a recombination between PPV‐M and PPV‐D. These isolates were classified as the PPV‐Rec (Recombinant) strain and have been found in several European countries, as well as outside Europe, mainly infecting plum and apricot trees (Candresse et al., 2007; Glasa et al., 2002, 2004; Matic et al., 2006; Thompson et al., 2009). The efficient aphid transmission of PPV‐Rec isolates has been demonstrated (Glasa et al., 2004). Given its wide distribution and prevalence, PPV‐Rec is now considered as the third major PPV strain. As the first reported PPV recombinant isolate originated from Serbia (Cervera et al., 1993), the Balkans have been suggested to be the centre of origin of PPV‐Rec, which then spread to other areas through the exchange of infected propagation material of tolerant plum genotypes (Glasa et al., 2005).
A divergent PPV‐W3174 isolate was originally detected in 2003 in a plum tree in Canada (James and Varga, 2005) and, based on its molecular distinctiveness, was assigned to a new strain, PPV‐W (Winona). Later, PPV‐W isolates were recorded in Latvia, Ukraine and Russia (Glasa et al., 2011; Mavrodieva et al., 2013; Sheveleva et al., 2012), confirming the suggestion that the origin of this strain may be found in eastern Europe. Moreover, these new PPV‐W isolates differed from the W3174 Canadian isolate in not being affected by the two recombination events detected in the W3174 genome (Glasa et al., 2011). The W strain has been found in the field on plum, blackthorn, Canadian plum, cherry plum and downy cherry (Mavrodieva et al., 2013). The analysis of partial and complete genome sequences indicates that PPV‐W diversity is greater than that of the other PPV strains (Glasa et al., 2012; Mavrodieva et al., 2013; Sheveleva et al., 2012).
Genome characterization of the atypical Turkish Ab‐Tk isolate has revealed a recombination event affecting its 5′ genomic region (Glasa and Candresse, 2005; Ulubaş Serçe et al., 2009). Further surveys have confirmed the occurrence of closely related isolates in the Ankara region in Turkey, which have been classified into a new strain, PPV‐T (Turkey) (Ulubaş Serçe et al., 2009). PPV‐T isolates have been found to be widely distributed in apricots, peaches and plums in Turkey, and an occasional finding of PPV‐T has been recorded from Albania (unpublished results of the European SharCo FP7 project).
Very recently, unusual PPV isolates recovered from naturally infected sour cherries in the Volga river basin (Russia) have been characterized and proposed to form a second cherry‐adapted strain, PPV‐CR (Cherry Russian) (Glasa et al., 2013). The spread of similar isolates was confirmed in old sour cherry trees in the Moscow region (Chirkov et al., 2013). The epidemiology of this strain remains to be determined.
An additional putative PPV strain (PPV‐An) could be represented by a recently identified isolate from eastern Albania (Palmisano et al., 2012). The full‐length genomic sequence of this isolate fulfils the features of an ancestral PPV‐M isolate previously hypothesized in the PPV evolutionary scenario (Glasa and Candresse, 2005; Fig. 3).
Figure 3.
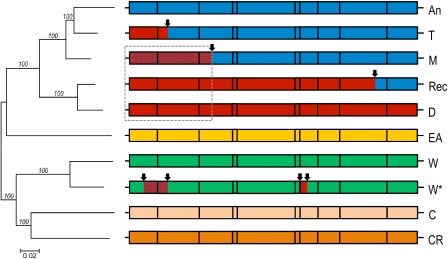
Phylogenetic and recombination analysis of Plum pox virus (PPV) strains. Phylogenetic tree of representative PPV isolates belonging to the known PPV strains (left). The genomic organization and recombination history of the corresponding PPV strains are shown on the right. The phylogenetic tree was reconstructed with the neighbour‐joining technique from full‐length nucleotide sequences and bootstrap (1000 replicates) was used to evaluate branch validity. The following sequences were used: PPV‐An (unpublished sequence; Palmisano et al., 2012); PPV‐T (EU734794); PPV‐M (AJ243957); PPV‐Rec (AY028309); PPV‐D (AY912057); PPV‐EA (DQ431465); PPV‐C (HQ840518); PPV‐CR (KC020126). For PPV‐W, two isolates were used to reflect differences in recombination history between members of this strain: LV‐145bt (HQ670748) and W3174 (AY912055), which is marked by an asterisk. For the right panel, strains are colour coded and arrows mark the recombination breakpoints identified. The 5′ genome portion in PPV‐M, PPV‐D and PPV‐Rec, affected by an ancestral recombination, is boxed and the colour of the affected region in PPV‐M is modified from that of the parental PPV‐D to reflect its divergence posterior to the recombination event.
Full‐length genomic sequences have been determined for PPV isolates representing each of the recognized strains, providing a clear picture of the phylogenetic relationship between strains and of the PPV evolutionary history. PPV strains are characterized by relatively low intrastrain diversity (reaching 1.1%–3.9% at the nucleotide level for full‐length genomes, except for PPV‐W, where the divergence reaches 7.9%) and by comparatively high between‐strain diversity (4.4%–22.8%; Glasa et al., 2012). Despite the extensive exchanges of Prunus propagation material, PPV strains still show, for at least some of them, a partial or complete geographical structure. The analysis of PPV diversity has also provided the first indications that recombination plays a role in the evolution of potyviruses (Cervera et al., 1993). Although forming monophyletic groups, PPV‐M, PPV‐D, PPV‐Rec, PPV‐T and PPV‐W are evolutionarily linked by recombination events, including an ancestral recombination affecting the 5′ part of PPV‐M, PPV‐D and PPV‐Rec strains (Fig. 3).
The possibility that future surveys of PPV variability, in particular in poorly explored areas such as Asia, or employing new unbiased and high‐throughput next‐generation sequencing technologies, may reveal further new, unusual or emerging forms of PPV in the future, cannot be excluded.
Pathogenicity and Host Range Determinants
Although PPV strains are entities clearly differentiated from molecular, serological and evolutionary perspectives, it is much less clear whether they show specific biological features, such as pathogenicity, host range and epidemiological behaviour (Candresse and Cambra, 2006). Under field conditions, PPV‐Rec isolates are rarely found in peach, and experimental transmission to the peach seedling indicator GF305 results in very mild symptoms, suggesting that PPV‐Rec could be poorly adapted to peach (Candresse and Cambra, 2006; Glasa et al., 2004). Moreover, as mentioned above, PPV isolates of strain M seem to spread more readily to peach than isolates of strain D, which is generally considered to be poorly epidemic in peach (Candresse and Cambra, 2006; Llácer and Cambra, 2006). However, this conclusion is challenged by the existence of atypical PPV‐D isolates that efficiently spread in peach, suggesting that some pathogenicity properties could be more dependent on isolate‐specific traits, rather than on strain‐specific ones (Dallot et al., 1998; Glasa et al., 2010; Levy et al., 2000).
The most conspicuous strain‐specific pathogenicity feature of PPV is the ability to infect cherry trees of isolates of the PPV‐C and PPV‐CR strains (Chirkov et al., 2013; Crescenzi et al., 1997; Glasa et al., 2013; Nemchinov and Hadidi, 1996; Nemchinov et al., 1996). However, although PPV‐C isolates appear to be specifically adapted to cherry, they are also able to infect other Prunus species under experimental conditions (Bodin et al., 2003; Crescenzi et al., 1997; Nemchinov and Hadidi, 1996).
The characterization of molecular determinants of specific pathogenicity traits of PPV isolates in the field has been hampered by several factors, such as high within‐strain variability and the differential epidemiological behaviour of an isolate depending on the Prunus host or on local agroecological conditions, etc. In addition, a substantial amount of intra‐isolate variability is observed within single Prunus trees, demonstrating the dynamic structure and heterogeneous nature of PPV populations (Jridi et al., 2006; Predajňa et al., 2012b).
However, some information is available about the determinants of pathogenicity and host range of PPV in experimental conditions, mainly in herbaceous plants. Using a collection of Arabidopsis thaliana accessions, it has been shown that multiple specific interactions between virus and host factors control PPV infection (Decroocq et al., 2006). PPV isolates of the C strain are unable to infect systemically any of the Arabidopsis ecotypes, whereas some Arabidopsis ecotypes are specifically infected by particular PPV isolates. Thus, isolates of the PPV‐EA and PPV‐M strains were able to systemically spread only in Arabidopsis ecotypes or mutants with a dysfunctional resistance to Tobacco etch virus movement (RTM) system, and the viral determinant to overcome the RTM resistance was mapped to the N‐terminal region of the CP (Decroocq et al., 2009).
The analysis of chimeras between PPV isolates of different strains (Dallot et al., 2001; Sáenz et al., 2000) or of the same strain (Salvador et al., 2008a) with diverse biological characteristics has shown that determinants for these properties are largely spread throughout the viral genome, and that, in some cases, optimal adaptation to P. persica or Nicotiana clevelandii is mutually exclusive. In particular, a pathogenicity determinant for infection in herbaceous (Sáenz et al., 2000) and woody (Dallot et al., 2001) hosts was localized in the P3 + 6K1 region. In agreement with this, nucleotide changes in the P3 and 6K1 coding sequences have been associated with adaptation to N. clevelandii (Salvador et al., 2008a). Nucleotide changes in the P1 (Salvador et al., 2008a) and CP (Carbonell et al., 2013) coding sequences have also been detected during adaptation to this host, and a specific mutation occurred consistently when a peach PPV isolate was adapted to pea (Wallis et al., 2007).
The P1 protein appears to be especially relevant for host adaptation (Valli et al., 2007). Replacement of the PPV P1 coding sequence by the corresponding sequence of another potyvirus, Tobacco vein mottling virus, abolished infectivity in P. persica, but enhanced virus competence in N. clevelandii (Salvador et al., 2008b). Moreover, point mutations in the P1 gene causing effects on infectivity, virus accumulation and symptom severity were detected in virus variants that coexisted in a PPV population (Maliogka et al., 2012). Also supporting the importance of P1 for PPV pathogenicity, the 3′ proximal part of the P1 gene was shown to determine the symptomatology of interstrain PPV chimeras (Nagyová et al., 2012). Interestingly, long sequences of the 5′ noncoding region of PPV that are not essential for viral infectivity also contribute to viral competitiveness and pathogenesis (Simón‐Buela et al., 1997b)
HCPro is a known potyviral pathogenicity factor, as a consequence of its ability to suppress RNA silencing (Kasschau et al., 2003) and, probably, because of interactions with other host processes (Eggenberger et al., 2008; Mlotshwa et al., 2005). A contribution of HCPro to PPV pathogenicity in N. clevelandii has also been reported (Sáenz et al., 2001); HCPro defects have been shown to contribute to the restriction of PPV systemic spread in N. tabacum (Sáenz et al., 2002). Moreover, synergistic interactions of PPV HCPro with another virus, Potato virus X, have also been described (González‐Jara et al., 2005; Pacheco et al., 2012).
Information about the biochemical basis of viral symptoms is very scarce. However, results suggest that imbalance in antioxidant systems and increased generation of reactive oxygen species might contribute to the deleterious effects of PPV infection (Díaz‐Vivancos et al., 2006, 2008; Hernández et al., 2006).
Interactome Studies and the Identification of Host Factors Contributing to PPV Infection
Our understanding of the many ways in which potyviruses interact with their host plants has dramatically progressed in recent years, thanks to the convergence of a range of strategies, including biochemical, molecular, genomic and genetic approaches. Although probably still far from complete, our current view of the potyviral interactome is thus far more complex today than it was only a decade ago (Elena and Rodrigo, 2012; Revers et al., 1999). Every single protein encoded by the potyviral genome has several identified viral or host interactors if potyviruses are considered collectively (Elena and Rodrigo, 2012). Although some of these interactions are likely to be virus specific, in many other cases a level of generality is probably associated with these findings. A good example is the finding that all potyviruses analysed to date require (and interact with) one or more isoforms of translation initiation factor 4E (eIF4E), but that different potyviruses may interact with different isoforms (Nicaise et al., 2007). Although PPV is not the most prominent Potyvirus in interactomic studies, PPV research has allowed us to fill several blanks in our growing knowledge of the potyviral interactome.
Systematic efforts have demonstrated the existence of 52 of 100 possible interactions between the various PPV proteins (including self‐interactions) (Zilian and Maiss, 2011), making PPV the best or second best known potyvirus from this perspective and a clear model for other genus members. When it comes to the identification of host plant interactors, work on PPV has allowed the identification of two plant proteins physically interacting with viral proteins. The Arabidopsis RH8 helicase interacts with VPg (Huang et al., 2010) and the Nicotiana benthamiana photosystem I PSI‐K protein interacts with the CI helicase (Jiménez et al., 2006). Reduction of the accumulation of RH8 has a negative effect on PPV infection, demonstrating that it behaves as a susceptibility factor. In contrast, the down‐regulation of PSI‐K leads to higher PPV accumulation, suggesting that it has an antiviral role. The fact that the co‐expression of PPV CI causes a decrease in the accumulation of PSI‐K transiently expressed in N. benthamiana suggests that CI could be involved in counteracting the defensive role of PSI‐K.
Although the physical interactions involved have not been studied in detail, both eIF(iso)4E and eIF(iso)4G1 have been shown to be absolutely required for successful PPV infection in Arabidopsis (Decroocq et al., 2006; Nicaise et al., 2007), a situation that parallels that observed for many other potyviruses. In the specific cases of PPV and Turnip mosaic virus, two further proteins have been shown to partially affect viral accumulation, probably through their effect on eIF(iso)4E accumulation: the DNA‐binding protein phosphatase AtDBP1 (Castelló et al., 2010) and a small interactor of AtDBP1, DIP2 (Castelló et al., 2011).
Although these studies have so far not resulted directly in the identification of further host plant interactors, it is worth noting that PPV is one of the best studied potyviruses when it comes to both transcriptomic studies (Babu et al., 2008; Dardick, 2007; Schurdi‐Levraud Escalettes et al., 2006; Wang et al., 2005) and the genetic dissection of host determinants of the interaction in Arabidopsis (Decroocq et al., 2006; Pagny et al., 2012; Sicard et al., 2008). The latter has allowed the demonstration that PPV is among the potyviruses controlled by the RTM resistance system (Decroocq et al., 2009), and the identification and mapping of various host resistance determinants, including recessive ones, is likely to correspond to susceptibility factors (Pagny et al., 2012). These studies, and the physical mapping of a major resistance locus of P. armeniaca cultivars, suggest that MATH domain proteins could be involved in the control of PPV long‐distance movement (Pagny et al., 2012; Zuriaga et al., 2013).
Approaches to Generate Resistance Against PPV
Conventional breeding
The identification of natural resistance in Prunus germplasm and its introduction into commercial cultivars by conventional breeding is one of the main strategies to control PPV, especially in areas of endemicity (Decroocq et al., 2011). First reports on resistant Prunus genotypes, based on field observations under natural infection pressure, date from the 1940s (Christoff, 1947; Jordovic, 1968; Syrgiannidis, 1980). Later experimental evaluations of Prunus for resistance involved artificial inoculations by grafting, chip‐budding or aphids in the field (Bivol et al., 1987; Minoiu, 1973; Trifonov, 1975; Zawadzka, 1981) or under controlled conditions (Dosba et al., 1991; Martínez‐Gomez and Dicenta, 1999). However, limitations in the reliability of detection methods and differences in the evaluation protocols, PPV isolates used and agroclimatic context resulted in conflicting results in some cases (Kegler et al., 1998).
In spite of many years of extensive efforts, very few natural sources of resistance have been identified so far in Prunus species (Kegler et al., 1998; Martínez‐Gómez et al., 2000). Resistant apricot genotypes (mainly of North American origin) have been used in several breeding programmes (Badenes and Llácer, 2006; Krška et al., 2006). PPV resistance in apricots is believed to be a complex trait controlled by at least two genes (Guillet‐Bellanger and Audergon, 2001; Moustafa et al., 2001; Vilanova et al., 2003). No known source of resistance has been identified in peach, but resistance has been identified in the wild relative P. davidiana, in almond (P. amygdalus) and in almond × peach hybrids (Kegler et al., 1998; Pascal et al., 2002; Rubio et al., 2003).
In the absence of resistant cultivars in domestic plum, tolerant cultivars that do not display fruit symptoms, but do not restrict PPV multiplication and movement, have been used in southern and central Europe (Kegler et al., 1998; Ogašanovic et al., 1994). The hypersensitive response (Kegler et al., 1991, 2001), an active defence response resulting in localized cell death, has been found to be an effective resistance mechanism against PPV under natural or artificial inoculation, and has been used in plum breeding programmes (Hartmann, 1998), although, in rare cases, the response was found to be partial, depending on the PPV isolate (Polák et al., 2005).
Marker‐assisted selection, based on molecular markers associated with resistance, has been used to streamline the lengthy breeding and selection of resistant genotypes (Lalli et al., 2005; Vilanova et al., 2003). In apricot, linkage groups 1 and 3 have been highlighted as bearing PPV resistance quantitative trait loci (QTLs) (Marandel et al., 2009).
Genetic engineering
Given the economic importance of PPV, it is no surprise that, following the initial construction of virus‐resistant transgenic plants, several laboratories embarked on this quest. It was a particularly ambitious goal as this implied both the development of the technology for PPV and the generation of transgenic woody plants. Following initial efforts at genome characterization, early constructs allowed the expression of the PPV CP gene in transgenic herbaceous (Ravelonandro et al., 1992; Regner et al., 1992) and Prunus (Laimer da Camara Machado et al., 1992; Scorza et al., 1994) hosts. Remarkably, among the plum trees produced during these early efforts, one transgenic line, C5, was shown to be highly resistant to PPV (Ravelonandro et al., 1997) as a result of post‐transcriptional gene silencing (PTGS) (Hily et al., 2005; Scorza et al., 2001). The resistance of this C5 clone, later renamed ‘HoneySweet’, has been validated extensively in long‐term field trials in a range of countries and agronomical conditions (Hily et al., 2004; Malinowski et al., 2006; Polák et al., 2008). The biosafety of this transgenic plum line has also been evaluated extensively, in both field and laboratory experiments, in particular within the framework of a collaborative European Union‐funded project (Fuchs et al., 2007). Particular attention was paid to the possibility of the emergence of recombinants between an infecting virus and the transgene (Capote et al., 2008; Zagrai et al., 2011) and to resistance stability after infection with heterologous viruses (Zagrai et al., 2008), but many other aspects were also analysed, culminating in the regulatory approval of the HoneySweet plum in the USA (Scorza et al., 2013). As a consequence of these detailed studies, the HoneySweet plum is probably one of the best studied virus‐resistant transgenic plants (Collinge et al., 2010; Gottula and Fuchs, 2009; Simón‐Mateo and García, 2011).
Efforts to develop PPV‐resistant transgenic plants have by no means been limited to the CP expression strategy. Over the years, a wide range of other approaches have been evaluated, with variable success. Given that the HoneySweet plum resistance is PTGS based, it is no surprise that the expression of a range of other PPV genome regions, in wild‐type or mutated form, has been shown to confer resistance, probably through the same mechanism (Barajas et al., 2004; Guo and García, 1997; Guo et al., 1998a, 1999; Jacquet et al., 1998; Tavert‐Roudet et al., 1998; Wittner et al., 1998). Similarly, the effectiveness of PTGS‐inducing, hairpin‐containing viral transgenes has been confirmed in a wide range of studies (Di Nicola‐Negri et al., 2005; Hily et al., 2007; Pandolfini et al., 2003; Tenllado et al., 2003; Zhang et al., 2006). A potential limitation of resistance conferred by the expression of viral genomic sequences is the possibility that it could be suppressed by infection with a heterologous virus (Simón‐Mateo et al., 2003). The susceptibility of engineered PPV chimeras to endogenous microRNAs suggests that the expression of artificial microRNAs might also be an effective option (Simón‐Mateo and García, 2006). However, the fact that PPV rapidly escaped the silencing mechanism through the accumulation of point mutations poses caution on this antiviral approach.
A wide range of other strategies have been envisioned in an effort to develop virus‐resistant transgenic plants (Prins et al., 2008), but so far these nonconventional approaches have met with only limited success in the case of PPV (Liu et al., 2000; Wen et al., 2004), with the possible exception of the transgenic expression of single‐chain antibodies targeting the viral NIb replicase (Esteban et al., 2003; Gil et al., 2011).
The most recent strategy evaluated with success against PPV brings together interactomics or genetic studies aimed at the identification of host susceptibility factors (see above). In theory, the inactivation of such genes could result in resistance to viral infection, as was demonstrated in Arabidopsis in the case of eIF(iso)4E for several potyviruses (Duprat et al., 2002), including PPV (Decroocq et al., 2006). Several transgenic plum lines in which eIF(iso)4E expression had been knocked down through RNA silencing showed 100% PPV infection evasion, even after two successive vegetative cycles (Wang et al., 2013), demonstrating that this strategy can be used in stone fruits against PPV. In the long run, to avoid public reluctance (at least in Europe) against transgenic plants, the use of this strategy without the need for transgenesis could even be envisioned, either through the targeted screening of the Prunus diversity for suitable null or mutant eIF(iso)4E alleles or through the selection of mutant alleles using TILLING (Targeting Induced Local Lesions IN Genomes) technology (Piron et al., 2010).
PPV as a Tool in Biotechnology
Plant viruses are the object of interest not only because of the harm they cause to crops. Viral infections can enhance the aesthetic value of ornamental plants (Garber, 1989; Saunders et al., 2003) and viruses may even establish interactions mutually beneficial to the virus and the host (Roossinck, 2005). Although there are no reports indicative of the beneficial effects of PPV, genetic engineering has allowed us to modify and use PPV, or parts of it, as a valuable biotechnological tool.
The availability of functional full‐length cDNAs of the PPV genome (López‐Moya and García, 2000; Maiss et al., 1992; Predajňa et al., 2012a; Riechmann et al., 1990; Szathmary et al., 2009) has facilitated the development of PPV‐based vectors to express either small peptides fused to the viral CP or independent proteins (García et al., 2006). Several vectors developed to express epitopes of foreign agents at the surface of PPV virions differed in their tolerance to inserted sequences and in the antigenicity and immunogenicity of the expressed epitopes (Fernández‐Fernández et al., 1998, 2002b).
PPV‐based vectors allowing the expression of whole independent proteins have also been constructed, using as insertion site the P1/HCPro or NIb/CP junction (García et al., 2006). These vectors have been used to express reporters that facilitate monitoring of the viral infection (Dietrich and Maiss, 2003; Guo et al., 1998b; Ion‐Nagy et al., 2006; Lansac et al., 2005), but also antigenic proteins to produce recombinant vaccines (Fernández‐Fernández et al., 2001).
Viral vectors can be expressed in transgenic plants transformed with full‐length cDNA copies of the viral genome. These amplicons combine the genetic stability of transgenic plants with the elevated replication rate of viruses. PPV amplicons have been developed in N. benthamiana, but show important constraints that limit their utility (Calvo et al., 2010). A PPV amplicon has been used to design a method to control virus expression by regulating the temperature during plant transformation and its subsequent culture, which could help to reduce such limitations (Dujovny et al., 2009).
The protease domain of the NIa protein of PPV has demonstrated a notable biotechnological interest as its high efficiency and specificity make it very attractive for the processing of fusion proteins both in vitro (Pérez‐Martín et al., 1997; Zheng et al., 2008) and in vivo (Zheng et al., 2012).
Conclusion
For several decades now PPV has been among a handful of intensively studied potyviruses and, as a consequence, is among the most studied and best understood viruses in this vast, widespread and highly damaging genus. The high visibility of PPV is no doubt a consequence of both its high socioeconomic impact in the affected Prunus crops and its quarantine regulatory status in many countries. These factors have contributed to its inclusion in a recent list of the 10 most significant viruses in molecular plant pathology (Scholthof et al., 2011). Research efforts on PPV have been particularly active and trend‐setting in several areas, including the development of advanced diagnostic and detection techniques (to support quarantine, eradication and certification control strategies), efforts in epidemiology and modelling of disease spread, plant–virus interaction studies and the development of classical or transgenic resistance. In the past few years, many of these research lines have converged under the auspices of the SharCo project supported by the European Union, leading to an exemplary collaborative translational research effort to better control the devastating sharka disease. Further collaborative inputs of the same magnitude are needed today to capitalize on the progress made in our understanding of this virus and to provide the fruit industry with a range of control options, including panels of varieties with high‐level and durable resistance to PPV for all the major affected Prunus crops.
Acknowledgements
We are grateful to F. Palmisano, A. Minafra, D. Boscia, V. Savino and A. Myrta for providing unpublished information about PPV‐An. We acknowledge the support of the European Union for the authors within the framework of the FP7 KBBE‐204429 SharCo project. The research of the authors was also supported by grants BIO2010‐18541 from Ministerio de Economía y Competitividad (MINECO) (JAG), APVV‐0042‐10 and APVV‐0174‐12 from the Slovak Research and Development Agency (MG), AGL2009‐07531 from MINECO (MC), and EU‐FP7 Marie Curie STONE and French FranceAgriMer 2009 0076 020 104 and 2011 0038 012 104 (TC). We would like to apologize to those individuals whose relevant publications could not be cited because of space constraints.
References
- Adams, M.J. , Zerbini, F.M. , French, R. , Rabenstein, F. , Stenger, D.C. and Valkonen, J.P.T. (2012) Family Potyviridae In: Virus Taxonomy (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J., eds), pp. 1069–1090. San Diego: Elsevier. [Google Scholar]
- Anindya, R. , Chittori, S. and Savithri, H.S. (2005) Tyrosine 66 of Pepper vein banding virus genome‐linked protein is uridylylated by RNA‐dependent RNA polymerase. Virology, 336, 154–162. [DOI] [PubMed] [Google Scholar]
- Atanasoff, D. (1932) Plum pox. A new virus disease. Annals of the University of Sofia, Faculty of Agriculture and Silviculture 11, 49–69. [Google Scholar]
- Babu, M. , Griffiths, J.S. , Huang, T.S. and Wang, A. (2008) Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection. BMC Genomics, 9, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenes, M.L. and Llácer, G. (2006) Breeding for resistance: breeding for Plum pox virus resistant apricots (Prunus armeniaca L.) in Spain. EPPO Bull. 36, 323–326. [Google Scholar]
- Barajas, D. , Tenllado, F. , Gonzalez‐Jara, P. , Martinez‐Garcia, B. , Atencio, F.A. and Diaz‐Ruiz, J.R. (2004) Resistance to Plum pox virus (PPV) in Nicotiana benthamiana plants transformed with the PPV HC‐Pro silencing suppressor gene. J. Plant Pathol. 86, 239–248. [Google Scholar]
- Barba, M. , Hadidi, A. , Candresse, T. and Cambra, M. (2011) Plum pox virus In: Virus and Virus‐like Disease of Pome and Stone Fruits (Hadidi A., Barba M., Candresse T. and Jelkmann W., eds), pp. 185–197. St. Paul, Minnesota: APS Press. [Google Scholar]
- Bivol, T. , Ignat, V.F. , Kukurusak, E.A. and Kegler, H. (1987) Experiments on resistance of plum varieties and hybrids to plum pox virus in Moldavia. Arch. Phytopathol. Pflanzenschutz, 23, 443–449. [Google Scholar]
- Blanc, S. , López‐Moya, J.J. , Wang, R.Y. , García‐Lampasona, S. , Thornbury, D.W. and Pirone, T.P. (1997) A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology, 231, 141–147. [DOI] [PubMed] [Google Scholar]
- Bodin, M. , Glasa, M. , Verger, D. , Costes, E. and Dosba, F. (2003) Distribution of the sour cherry isolate of plum pox virus in infected Prunus rootstocks. J. Phytopathol. 151, 625–630. [Google Scholar]
- CABI (2013) Crop protection compendium. Available at http://www.cabi.org/cpc/ [accessed on Oct 12, 2013].
- Calvo, M. , Dujovny, G. , Lucini, C. , Ortuño, J. , Alamillo, J.M. , Simón‐Mateo, C. , López‐Moya, J.J. and García, J.A. (2010) Constraints to virus infection in Nicotiana benthamiana plants transformed with a potyvirus amplicon. BMC Plant Biol. 10, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambra, M. , Boscia, D. , Myrta, A. , Palkovics, L. , Navrátil, M. , Barba, M. , Gorris, M.T. and Capote, N. (2006a) Serological detection and characterisation of Plum pox virus . EPPO Bull. 36, 254–261. [Google Scholar]
- Cambra, M. , Capote, N. , Cambra, M.A. , Llácer, G. , Botella, P. and López‐Quílez, A. (2006b) Epidemiology of sharka disease in Spain. EPPO Bull. 36, 271–275. [Google Scholar]
- Cambra, M. , Capote, N. , Myrta, A. and Llácer, G. (2006c) Plum pox virus and the estimated costs associated with sharka disease. EPPO Bull. 36, 202–204. [Google Scholar]
- Cambra, M. , Boscia, D. , Gil, M. , Bertolini, E. and Olmos, A. (2011) Immunology and immunological assays applied to the detection, diagnosis and control of fruit tree viruses In: Virus and Virus‐like Disease of Pome and Stone Fruits (Hadidi A., Barba M., Candresse T. and Jelkmann W., eds), pp. 303–313. St. Paul, Minnesota: APS Press. [Google Scholar]
- Candresse, T. and Cambra, M. (2006) Causal agent of sharka disease: historical perspective and current status of Plum pox virus strains. EPPO Bull. 36, 239–246. [Google Scholar]
- Candresse, T. , Cambra, M. , Dallot, S. , Lanneau, M. , Asensio, M. , Gorris, M.T. , Revers, F. , Macquaire, G. , Olmos, A. , Boscia, D. , Quiot, J.B. and Dunez, J. (1998) Comparison of monoclonal antibodies and polymerase chain reaction assays for the typing of isolates belonging to the D and M serotypes of plum pox potyvirus. Phytopathology, 88, 198–204. [DOI] [PubMed] [Google Scholar]
- Candresse, T. , Svanella‐Dumas, L. , Gentit, P. , Caglayan, K. and Cevik, B. (2007) First report of the presence of Plum pox virus Rec strain in Turkey. Plant Dis. 91, 331. [DOI] [PubMed] [Google Scholar]
- Candresse, T. , Saenz, P. , García, J.A. , Boscia, D. , Navratil, M. , Gorris, M.T. and Cambra, M. (2011) Analysis of the epitope structure of Plum pox virus coat protein. Phytopathology, 101, 611–619. [DOI] [PubMed] [Google Scholar]
- Capote, N. , Gorris, M.T. , Martinez, M.C. , Asensio, M. , Olmos, A. and Cambra, M. (2006) Interference between D and M types of Plum pox virus in Japanese plum assessed by specific monoclonal antibodies and quantitative real‐time reverse transcription‐polymerase chain reaction. Phytopathology, 96, 320–325. [DOI] [PubMed] [Google Scholar]
- Capote, N. , Perez‐Panades, J. , Monzo, C. , Carbonell, E. , Urbaneja, A. , Scorza, R. , Ravelonandro, M. and Cambra, M. (2008) Assessment of the diversity and dynamics of Plum pox virus and aphid populations in transgenic European plums under Mediterranean conditions. Transgenic Res. 17, 367–377. [DOI] [PubMed] [Google Scholar]
- Capote, N. , Bertolini, E. , Olmos, A. , Vidal, E. , Martinez, M.C. and Cambra, M. (2009) Direct sample preparation methods for the detection of Plum pox virus by real‐time RT‐PCR. Int. Microbiol. 12, 1–6. [PubMed] [Google Scholar]
- Capote, N. , Cambra, M. , Botella, P. , Gorris, M. , Martinez, M. , Lopez‐Quilez, A. and Cambra, M. (2010) Detection, characterization, epidemiology and eradication of Plum pox virus Marcus type in Spain. J. Plant Pathol. 92, 619–628. [Google Scholar]
- Carbonell, A. , Maliogka, V.I. , Pérez, J.J. , Salvador, B. , San León, D. , García, J.A. and Simón‐Mateo, C. (2013) Diverse amino acid changes at specific positions in the N‐terminal region of the coat protein allow Plum pox virus to adapt to new hosts. Mol. Plant–Microbe Interact. 26: 1211–1224. Available at 10.1094/MPMI-04-13-0093-R. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Jensen, P.E. and Schaad, M.C. (1998) Genetic evidence for an essential role for potyvirus CI protein in cell‐to‐cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Castelló, M.J. , Carrasco, J.L. and Vera, P. (2010) DNA‐binding protein phosphatase AtDBP1 mediates susceptibility to two potyviruses in Arabidopsis. Plant Physiol. 153, 1521–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló, M.J. , Carrasco, J.L. , Navarrete, M. , Daniels, J. , Granot, D. and Vera, P. (2011) A plant small polypeptide is a novel component of DNA‐binding protein phosphatase 1 (DBP1)‐mediated resistance to Plum pox virus in Arabidopsis. Plant Physiol. 157, 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera, M.T. , Riechmann, J.L. , Martín, M.T. and García, J.A. (1993) 3′‐Terminal sequence of the plum pox virus PS and ŏ6 isolates: evidence for RNA recombination within the potyvirus group. J. Gen. Virol. 74, 329–334. [DOI] [PubMed] [Google Scholar]
- Chen, D. , Juárez, S. , Hartweck, L. , Alamillo, J.M. , Simón‐Mateo, C. , Pérez, J.J. , Fernández‐Fernández, M.R. , Olszewski, N.E. and García, J.A. (2005) Identification of secret agent as the O‐GlcNAc transferase that participates in Plum pox virus infection. J. Virol. 79, 9381–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirkov, S. , Ivanov, P. and Sheveleva, A. (2013) Detection and partial molecular characterization of atypical plum pox virus isolates from naturally infected sour cherry. Arch. Virol. 158, 1383–1387. [DOI] [PubMed] [Google Scholar]
- Christoff, A. (1947) Sharka disease of plum. Izv. Kamar. Nar. Kyltura. Seria: Biologia, Zemedelie i Lesovadstvo 1, 261–296. [Google Scholar]
- Chung, B.Y.W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge, D.B. , Jorgensen, H.J. , Lund, O.S. and Lyngkjaer, M.F. (2010) Engineering pathogen resistance in crop plants: current trends and future prospects. Annu. Rev. Phytopathol. 48, 269–291. [DOI] [PubMed] [Google Scholar]
- Cotton, S. , Grangeon, R. , Thivierge, K. , Mathieu, I. , Ide, C. , Wei, T.Y. , Wang, A.M. and Laliberte, J.F. (2009) Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83, 10 460–10 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi, A. , d'Aquino, L. , Comes, S. , Nuzzaci, M. , Piazzolla, P. , Boscia, D. and Hadidi, A. (1997) Characterization of the sweet cherry isolate of plum pox potyvirus. Plant Dis. 81, 711–714. [DOI] [PubMed] [Google Scholar]
- Dallot, S. , Labonne, G. , Boeglin, M. , Quiot‐Douine, L. , Quiot, J.B. and Candresse, T. (1998) Peculiar plum pox potyvirus D‐populations are epidemic in peach trees. Acta Hortic. 472, 355–365. [Google Scholar]
- Dallot, S. , Quiot‐Douine, L. , Sáenz, P. , Cervera, M.T. , García, J.A. and Quiot, J.B. (2001) Identification of Plum pox virus determinants implicated in specific interactions with different Prunus spp. Phytopathology, 91, 159–164. [DOI] [PubMed] [Google Scholar]
- Dallot, S. , Gottwald, T. , Labonne, G. and Quiot, J.B. (2003) Spatial pattern analysis of sharka disease (Plum pox virus strain M) in peach orchards of southern France. Phytopathology, 93, 1543–1552. [DOI] [PubMed] [Google Scholar]
- Dallot, S. , Glasa, M. , Jevremovic, D. , Kamenova, I. , Paunovic, S. and Labonne, G. (2011) Mediterranean and central‐eastern European countries host viruses of two different clades of plum pox virus strain M. Arch. Virol. 156, 539–542. [DOI] [PubMed] [Google Scholar]
- Damsteegt, V.D. , Waterworth, H.E. , Mink, G.I. , Howell, W.E. and Levy, L. (1997) Prunus tomentosa as a diagnostic host for detection of Plum pox virus and other Prunus viruses. Plant Dis. 81, 329–332. [DOI] [PubMed] [Google Scholar]
- Damsteegt, V.D. , Stone, A.L. , Luster, D.G. , Levy, L. , Gildow, F.E. and Welliver, R. (2001) Preliminary characterization of a North American isolate of Plum pox virus from naturally infected peach and plum orchards in Pennsylvania, USA. Acta Hortic. 550, 145–152. [Google Scholar]
- Damsteegt, V.D. , Scorza, R. , Stone, A.L. , Schneider, W.L. , Webb, K. , Demuth, M. and Gildow, F.E. (2007) Prunus host range of Plum pox virus (PPV) in the United States by aphid and graft inoculation. Plant Dis. 91, 18–23. [DOI] [PubMed] [Google Scholar]
- Dardick, C. (2007) Comparative expression profiling of Nicotiana benthamiana leaves systemically infected with three fruit tree viruses. Mol. Plant–Microbe Interact. 20, 1004–1017. [DOI] [PubMed] [Google Scholar]
- De Boer, S.H. and Lopez, M.M. (2012) New grower‐friendly methods for plant pathogen monitoring. Annu. Rev. Phytopathol. 50, 197–218. [DOI] [PubMed] [Google Scholar]
- Decroocq, V. , Sicard, O. , Alamillo, J.M. , Lansac, M. , Eyquard, J.P. , García, J.A. , Candresse, T. , Le Gall, O. and Revers, F. (2006) Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 19, 541–549. [DOI] [PubMed] [Google Scholar]
- Decroocq, V. , Salvador, B. , Sicard, O. , Glasa, M. , Cosson, P. , Svanella‐Dumas, L. , Revers, F. , García, J.A. and Candresse, T. (2009) The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N‐terminal region of the coat protein. Mol. Plant–Microbe Interact. 22, 1302–1311. [DOI] [PubMed] [Google Scholar]
- Decroocq, V. , Badenes, M. and Neumuller, M. (2011) Breeding for resistance to Plum pox virus In: Virus and Virus‐like Diseases of Pome and Stone Fruits (Hadidi A., Barba M., Candresse T. and Jelkmann W., eds), pp. 401–406. St. Paul, Minnesota: APS Press. [Google Scholar]
- Di Nicola‐Negri, E. , Brunetti, A. , Tavazza, M. and Ilardi, V. (2005) Hairpin RNA‐mediated silencing of Plum pox virus P1 and HC‐Pro genes for efficient and predictable resistance to the virus. Transgenic Res. 14, 989–994. [DOI] [PubMed] [Google Scholar]
- Díaz‐Vivancos, P. , Rubio, M. , Mesonero, V. , Periago, P.M. , Barceló, A.R. , Martínez‐Gómez, P. and Hernández, J.A. (2006) The apoplastic antioxidant system in Prunus: response to long‐term plum pox virus infection. J. Exp. Bot. 57, 3813–3824. [DOI] [PubMed] [Google Scholar]
- Díaz‐Vivancos, P. , Clemente‐Moreno, M.J. , Rubio, M. , Olmos, E. , García, J.A. , Martínez‐Gómez, P. and Hernández, J. (2008) Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J. Exp. Bot. 59, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. and Maiss, E. (2003) Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84, 2871–2876. [DOI] [PubMed] [Google Scholar]
- Dosba, F. , Denise, F. , Maison, P. , Massonie, G. and Audergon, J.M. (1991) Plum pox virus resistance of apricot. Acta Hortic. 293, 569–579. [Google Scholar]
- Dujovny, G. , Valli, A. , Calvo, M. and García, J.A. (2009) A temperature‐controlled amplicon system derived from Plum pox potyvirus . Plant Biotechnol. J. 7, 49–58. [DOI] [PubMed] [Google Scholar]
- Duprat, A. , Caranta, C. , Revers, F. , Menand, B. , Browning, K.S. and Robaglia, C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. [DOI] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. and Rodrigo, G. (2012) Towards an integrated molecular model of plant–virus interactions. Curr. Opin. Virol. 2, 713–718. [DOI] [PubMed] [Google Scholar]
- EPPO (2004) Diagnostic protocol for regulated pests. Plum pox potyvirus . EPPO Bull. 34, 247–256. [Google Scholar]
- EPPO (2006) Current status of Plum pox virus and sharka disease worldwide. EPPO Bull. 34, 205–218. [Google Scholar]
- EPPO (2013) PQR_EPPO database on quarantine pests. Available at http://www.eppo.int [accessed on Oct 12, 2013].
- Esteban, O. , García, J.A. , Gorris, M.T. , Domínguez, E. and Cambra, M. (2003) Generation and characterisation of functional recombinant antibody fragments against RNA replicase NIb from plum pox virus. Biochem. Biophys. Res. Commun. 301, 167–175. [DOI] [PubMed] [Google Scholar]
- Fanigliulo, A. , Comes, S. , Maiss, E. , Piazzolla, P. and Crescenzi, A. (2003) The complete nucleotide sequence of Plum pox virus isolates from sweet (PPV‐SwC) and sour (PPV‐SoC) cherry and their taxonomic relationships within the species. Arch. Virol. 148, 2137–2153. [DOI] [PubMed] [Google Scholar]
- Fernández, A. , Laín, S. and García, J.A. (1995) RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 23, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, A. , Guo, H.S. , Sáenz, P. , Simón‐Buela, L. , Gómez de Cedrón, M. and García, J.A. (1997) The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25, 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Fernández, M.R. , Martínez‐Torrecuadrada, J.L. , Casal, J.I. and García, J.A. (1998) Development of an antigen presentation system based on plum pox potyvirus. FEBS Lett. 427, 229–235. [DOI] [PubMed] [Google Scholar]
- Fernández‐Fernández, M.R. , Mouriño, M. , Rivera, J. , Rodríguez, F. , Plana‐Durán, J. and García, J.A. (2001) Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus‐based vector. Virology, 280, 283–291. [DOI] [PubMed] [Google Scholar]
- Fernández‐Fernández, M.R. , Camafeita, E. , Bonay, P. , Méndez, E. , Albar, J.P. and García, J.A. (2002a) The capsid protein of a plant single‐stranded RNA virus is modified by O‐linked N‐acetylglucosamine. J. Biol. Chem. 277, 135–140. [DOI] [PubMed] [Google Scholar]
- Fernández‐Fernández, M.R. , Martínez‐Torrecuadrada, J.L. , Roncal, F. , Domínguez, E. and García, J.A. (2002b) Identification of immunogenic hot spots within plum pox potyvirus capsid protein for efficient antigen presentation. J. Virol. 76, 12 646–12 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, M. , Cambra, M. , Capote, N. , Jelkmann, W. , Kundu, J. , Laval, V. , Martelli, G.P. , Minafra, A. , Petrovic, N. , Pfeiffer, P. , Pompe‐Novak, M. , Ravelonandro, M. , Saldarelli, P. , Stussi‐Garaud, C. , Vigne, E. and Zagrai, I. (2007) Safety assessment of transgenic plums and grapevines expressing viral coat protein genes: new insights into real environmental impact of perennial plants engineered for virus resistance. J. Plant Pathol. 89, 5–12. [Google Scholar]
- Gabrenaite‐Verkhovskaya, R. , Andreev, I.A. , Kalinina, N.O. , Torrance, L. , Taliansky, M.E. and Makinen, K. (2008) Cylindrical inclusion protein of potato virus A is associated with a subpopulation of particles isolated from infected plants. J. Gen. Virol. 89, 829–838. [DOI] [PubMed] [Google Scholar]
- Garber, P.M. (1989) Tulipmania. J. Polit. Econ. 97, 535–560. [Google Scholar]
- García, J.A. and Cambra, M. (2007) Plum pox virus and sharka disease. Plant Viruses, 1, 69–79. [Google Scholar]
- García, J.A. , Riechmann, J.L. and Laín, S. (1989a) Artificial cleavage site recognized by plum pox potyvirus protease in Escherichia coli . J. Virol. 63, 2457–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, J.A. , Riechmann, J.L. and Laín, S. (1989b) Proteolytic activity of the plum pox potyvirus NIa‐like protein in Escherichia coli . Virology, 170, 362–369. [DOI] [PubMed] [Google Scholar]
- García, J.A. , Laín, S. , Cervera, M.T. , Riechmann, J.L. and Martín, M.T. (1990) Mutational analysis of plum pox potyvirus polyprotein processing by the NIa protease in Escherichia coli . J. Gen. Virol. 71, 2773–2779. [DOI] [PubMed] [Google Scholar]
- García, J.A. , Martín, M.T. , Cervera, M.T. and Riechmann, J.L. (1992) Proteolytic processing of the plum pox potyvirus polyprotein by the NIa protease at a novel cleavage site. Virology, 188, 697–703. [DOI] [PubMed] [Google Scholar]
- García, J.A. , Cervera, M.T. , Riechmann, J.L. and López‐Otín, C. (1993) Inhibitory effects of human cystatin C on plum pox potyvirus proteases. Plant Mol. Biol. 22, 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, J.A. , Lucini, C. , García, B. , Alamillo, J.M. and López‐Moya, J.J. (2006) Use of Plum pox virus as a plant expression vector. EPPO Bull. 36, 341–345. [Google Scholar]
- Gentit, P. (2006) Detection of Plum pox virus: biological methods. EPPO Bull. 36, 251–253. [Google Scholar]
- Gil, M. , Esteban, O. , García, J.A. , Peña, L. and Cambra, M. (2011) Resistance to Plum pox virus in plants expressing cytosolic and nuclear single‐chain antibodies against the viral RNA NIb replicase. Plant Pathol. 60, 967–976. [Google Scholar]
- Gildow, F. , Damsteegt, V. , Stone, A. , Schneider, W. , Luster, D. and Levy, L. (2004) Plum pox in North America: identification of aphid vectors and a potential role for fruit in virus spread. Phytopathology, 94, 868–874. [DOI] [PubMed] [Google Scholar]
- Glasa, M. and Candresse, T. (2005) Partial sequence analysis of an atypical Turkish isolate provides further information on the evolutionary history of Plum pox virus (PPV). Virus Res. 108, 199–206. [DOI] [PubMed] [Google Scholar]
- Glasa, M. and Šubr, Z.W. (2005) The complete nucleotide sequence of a natural recombinant Plum pox virus (PPV) isolate. Phytopatol. Pol. 36, 41–46. [Google Scholar]
- Glasa, M. , MarieJeanne, V. , Labonne, G. , Šubr, Z. , Kudela, O. and Quiot, J.B. (2002) A natural population of recombinant Plum pox virus is viable and competitive under field conditions. Eur. J. Plant Pathol. 108, 843–853. [Google Scholar]
- Glasa, M. , Palkovics, L. , Komínek, P. , Labonne, G. , Pittnerova, S. , Kudela, O. , Candresse, T. and Šubr, Z. (2004) Geographically and temporally distant natural recombinant isolates of Plum pox virus (PPV) are genetically very similar and form a unique PPV subgroup. J. Gen. Virol. 85, 2671–2681. [DOI] [PubMed] [Google Scholar]
- Glasa, M. , Paunovic, S. , Jevremovic, D. , Myrta, A. , Pittnerová, S. and Candresse, T. (2005) Analysis of recombinant Plum pox virus (PPV) isolates from Serbia confirms genetic homogeneity and supports a regional origin for the PPV‐Rec subgroup. Arch. Virol. 150, 2051–2060. [DOI] [PubMed] [Google Scholar]
- Glasa, M. , Svanella, L. and Candresse, T. (2006) The complete nucleotide sequence of the Plum pox virus El Amar isolate. Arch. Virol. 151, 1679–1682. [DOI] [PubMed] [Google Scholar]
- Glasa, M. , Predajna, L. and Šubr, Z. (2010) Competitiveness of different Plum pox virus isolates in experimental mixed infections reveals rather isolate‐ than strain‐specific behaviour. J. Plant Pathol. 92, 267–271. [Google Scholar]
- Glasa, M. , Malinowski, T. , Predajna, L. , Pupola, N. , Dekena, D. , Michalczuk, L. and Candresse, T. (2011) Sequence variability, recombination analysis, and specific detection of the W strain of Plum pox virus . Phytopathology, 101, 980–985. [DOI] [PubMed] [Google Scholar]
- Glasa, M. , Candresse, T. and The SharCo Consortium (2012) A large scale study of Plum pox virus genetic diversity and of its geographical distribution. In: 22nd International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Rome, Book of Abstracts, p. 38.
- Glasa, M. , Prikhodko, Y. , Predajna, L. , Nagyova, A. , Shneyder, Y. , Zhivaeva, T. , Subr, Z. , Cambra, M. and Candresse, T. (2013) Characterization of sour cherry isolates of Plum pox virus from the Volga basin in Russia reveals a new cherry strain of the virus. Phytopathology, 103, 972–979. [DOI] [PubMed] [Google Scholar]
- Gómez de Cedrón, M. , Osaba, L. , López, L. and García, J.A. (2006) Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res. 116, 136–145. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Atencio, F.A. , Martínez‐García, B. , Barajas, D. , Tenllado, F. and Díaz‐Ruíz, J.R. (2005) A single amino acid mutation in the plum pox virus helper component‐proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology, 95, 894–901. [DOI] [PubMed] [Google Scholar]
- Gottula, J. and Fuchs, M. (2009) Toward a quarter century of pathogen‐derived resistance and practical approaches to plant virus disease control. Adv. Virus Res. 75, 161–183. [DOI] [PubMed] [Google Scholar]
- Gottwald, T.R. (2006) Epidemiology of sharka disease in North America. EPPO Bull. 36, 279–286. [Google Scholar]
- Gottwald, T.R. , Avinent, L. , Llácer, G. , Hermoso de Mendoza, A. and Cambra, M. (1995) Analysis of the spatial spread of sharka (plum pox virus) in apricot and peach orchads in eastern Spain. Plant Dis. 79, 266–278. [Google Scholar]
- Grangeon, R. , Cotton, S. and Laliberté, J.‐F. (2010) A model for the biogenesis of Turnip mosaic virus replication factories. Commun. Integr. Biol. 3, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon, R. , Jiang, J. and Laliberte, J.F. (2012) Host endomembrane recruitment for plant RNA virus replication. Curr. Opin. Virol. 2, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet‐Bellanger, I. and Audergon, J.M. (2001) Inheritance of the stark early orange apricot cultivar resistance to Plum pox virus. Acta Hortic. 550, 111–115. [Google Scholar]
- Guo, H.S. and García, J.A. (1997) Delayed resistance to plum pox potyvirus mediated by a mutated RNA replicase gene: involvement of a gene silencing mechanism. Mol. Plant–Microbe Interact. 10, 160–170. [Google Scholar]
- Guo, H.S. , Cervera, M.T. and García, J.A. (1998a) Plum pox potyvirus resistance associated to transgene silencing that can be stabilized after different number of plant generations. Gene, 206, 263–272. [DOI] [PubMed] [Google Scholar]
- Guo, H.S. , López‐Moya, J.J. and García, J.A. (1998b) Susceptibility to recombination rearrangements of a chimeric plum pox potyvirus genome after insertion of a foreign gene. Virus Res. 57, 183–195. [DOI] [PubMed] [Google Scholar]
- Guo, H.S. , López‐Moya, J.J. and García, J.A. (1999) Mitotic stability of infection‐induced resistance to plum pox potyvirus associated with transgene silencing and DNA methylation. Mol. Plant–Microbe Interact. 12, 103–111. [DOI] [PubMed] [Google Scholar]
- Hartmann, W. (1998) Hypersensitivity—a possibility for breeding sharka resistant plum hybrids. Acta Hortic. 472, 429–432. [Google Scholar]
- Hernández, J.A. , Díaz‐Vivancos, P. , Rubio, M. , Olmos, E. , Ros‐Barceló, A. and Martínez‐Gómez, P. (2006) Long‐term plum pox virus infection produces an oxidative stress in a susceptible apricot, Prunus armeniaca, cultivar but not in a resistant cultivar. Physiol. Plant. 126, 140–152. [Google Scholar]
- Hily, J.M. , Scorza, R. , Malinowski, T. , Zawadzka, B. and Ravelonandro, M. (2004) Stability of gene silencing‐based resistance to Plum pox virus in transgenic plum (Prunus domestica L.) under field conditions. Transgenic Res. 13, 427–436. [DOI] [PubMed] [Google Scholar]
- Hily, J.M. , Scorza, R. , Webb, K. and Ravelonandro, M. (2005) Accumulation of the long class of siRNA is associated with resistance to Plum pox virus in a transgenic woody perennial plum tree. Mol. Plant–Microbe Interact. 18, 794–799. [DOI] [PubMed] [Google Scholar]
- Hily, J.M. , Ravelonandro, M. , Damsteegt, V. , Bassett, C. , Petri, C. , Liu, Z. and Scorza, R. (2007) Plum pox virus coat protein gene Intron‐hairpin‐RNA (ihpRNA) constructs provide resistance to plum pox virus in Nicotiana benthamiana and Prunus domestica . J. Am. Soc. Hortic. Sci. 132, 850–858. [Google Scholar]
- Hong, Y. and Hunt, A.G. (1996) RNA polymerase activity catalyzed by a potyvirus‐encoded RNA‐dependent RNA polymerase. Virology, 226, 146–151. [DOI] [PubMed] [Google Scholar]
- Huang, T.S. , Wei, T. , Laliberte, J.F. and Wang, A. (2010) A host RNA helicase‐like protein, AtRH8, interacts with the potyviral genome‐linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 152, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ion‐Nagy, L. , Lansac, M. , Eyquard, J.P. , Salvador, B. , García, J.A. , Le Gall, O. , Hernould, M. , Schurdi‐Levraud, V. and Decroocq, V. (2006) PPV long‐distance movement is occasionally permitted in resistant apricot hosts. Virus Res. 120, 70–78. [DOI] [PubMed] [Google Scholar]
- IPPC‐FAO (2012) International standards for phytosanitary measures: diagnostic protocols: Plum pox virus . ISPM 27, Annex 2 (DP2).
- Ivanov, K.I. , Puustinen, P. , Merits, A. , Saarma, M. and Mäkinen, K. (2001) Phosphorylation down‐regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 276, 13 530–13 540. [DOI] [PubMed] [Google Scholar]
- Jacquet, C. , Ravelonandro, M. , Bachelier, J.C. and Dunez, J. (1998) High resistance to plum pox virus (PPV) in transgenic plants containing modified and truncated forms of PPV coat protein gene. Transgenic Res. 7, 29–39. [Google Scholar]
- James, D. and Thompson, D. (2006) Hosts and symptoms of Plum pox virus: ornamental and wild Prunus species. EPPO Bull. 36, 222–224. [Google Scholar]
- James, D. and Varga, A. (2005) Nucleotide sequence analysis of Plum pox virus isolate W3174: evidence of a new strain. Virus Res. 110, 143–150. [DOI] [PubMed] [Google Scholar]
- Jiménez, I. , López, L. , Alamillo, J.M. , Valli, A. and García, J.A. (2006) Identification of a Plum pox virus CI‐interacting protein from chloroplast that has a negative effect in virus infection. Mol. Plant–Microbe Interact. 19, 350–358. [DOI] [PubMed] [Google Scholar]
- Jordovic, M. (1968) Recent advances on studies of Sarka virus disease. Acta Hortic. 10, 487–501. [Google Scholar]
- Jridi, C. , Martin, J.F. , Marie‐Jeanne, V. , Labonne, G. and Blanc, S. (2006) Distinct viral populations differentiate and evolve independently in a single perennial host plant. J. Virol. 80, 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajic, V. , Černi, S. and Škoric, D. (2012) Plum pox virus on sour cherry in Croatia. In: 22nd International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Rome, Book of Abstracts, p. 157.
- Kalashyan, Y.A. , Bilkey, N.D. , Verderevskaya, T.D. and Rubina, E.V. (1994) Plum pox virus on sour cherry in Moldava. EPPO Bull. 24, 645–649. [Google Scholar]
- Kasschau, K.D. , Xie, Z.X. , Allen, E. , Llave, C. , Chapman, E.J. , Krizan, K.A. and Carrington, J.C. (2003) P1/HC‐Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell, 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kegler, H. and Hartmann, W. (1998) Present status of controlling conventional strains of plum pox virus In: Plant Virus Disease Control (Hadidi A., Khetarpal R.K. and Koganezawa H., eds), pp. 616–628. St. Paul Minnesota: Phytopathological Society. [Google Scholar]
- Kegler, H. , Grüntzig, M. and Schimansky, H.H. (1991) Zur Resistenz der Pflaumenhybride K4 und ihrer F1‐Nachkommen gegen das Scharka‐Virus der Pflaume (plum pox virus). Nachr.bl. Dtsch. Pflanzenschutzd. 43, 102–106. [Google Scholar]
- Kegler, H. , Fuchs, E. , Gruntzig, M. and Schwarz, S. (1998) Some results of 50 years of research on the resistance to plum pox virus. Acta Virol. 42, 200–215. [PubMed] [Google Scholar]
- Kegler, H. , Gruntzig, M. , Fuchs, E. , Rankovic, M. and Ehrig, F. (2001) Hypersensitivity of plum genotypes to plum pox virus. J. Phytopathol. 149, 213–218. [Google Scholar]
- Kerlan, C. and Dunez, J. (1979) Différenciation biologique et sérologique des souches du virus de la sharka. Ann. Phytopathol. 11, 241–250. [Google Scholar]
- Kim, Y.C. , Udeshi, N.D. , Balsbaugh, J.L. , Shabanowitz, J. , Hunt, D.F. and Olszewski, N.E. (2011) O‐GlcNAcylation of the Plum pox virus capsid protein catalyzed by SECRET AGENT: characterization of O‐GlcNAc sites by electron transfer dissociation mass spectrometry. Amino Acids, 40, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krška, B. , Salava, J. and Polák, J. (2006) Breeding for resistance: breeding for Plum pox virus resistant apricots (Prunus armeniaca L.) in the Czech Republic. EPPO Bull. 36, 330–331. [Google Scholar]
- Labonne, G. and Dallot, S. (2006) Epidemiology of sharka disease in France. EPPO Bull. 36, 267–270. [Google Scholar]
- Laimer da Camara Machado, M. , da Camara Machado, A. , Hanzer, V. , Weiss, H. , Regner, F. , Steinkellner, H. , Mattanovich, D. , Plail, R. , Knapp, E. , Kalthoff, B. and Kattinger, H. (1992) Regeneration of transgenic plants of Prunus armeniaca containing the coat protein of Plum Pox Virus. Plant Cell Rep. 11, 25–29. [DOI] [PubMed] [Google Scholar]
- Laín, S. , Riechmann, J.L. and García, J.A. (1989) The complete nucleotide sequence of plum pox potyvirus RNA. Virus Res. 13, 157–172. [DOI] [PubMed] [Google Scholar]
- Laín, S. , Riechmann, J.L. and García, J.A. (1990) RNA helicase: a novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acids Res. 18, 7003–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laín, S. , Martín, M.T. , Riechmann, J.L. and García, J.A. (1991) Novel catalytic activity associated with positive‐strand RNA virus infection: nucleic acid‐stimulated ATPase activity of the plum pox potyvirus helicase like protein. J. Virol. 63, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli, D.A. , Decroocq, V. , Blenda, A.V. , Schurdi‐Levraud, V. , Garay, L. , Le Gall, O. , Damsteegt, V. , Reighard, G.L. and Abbott, A.G. (2005) Identification and mapping of resistance gene analogs (RGAs) in Prunus: a resistance map for Prunus. Theor. Appl. Genet. 111, 1504–1513. [DOI] [PubMed] [Google Scholar]
- Lansac, M. , Eyquard, J.P. , Salvador, B. , García, J.A. , Le Galla, O. , Decroocq, V. and Escalettes, V.S.L. (2005) Application of GFP‐tagged Plum pox virus to study Prunus–PPV interactions at the whole plant and cellular levels. J. Virol. Methods, 129, 125–133. [DOI] [PubMed] [Google Scholar]
- Levy, L. , Damsteegt, V. and Welliver, R. (2000) First Report of Plum pox virus (Sharka Disease) in Prunus persica in the United States. Plant Dis. 84, 202. [DOI] [PubMed] [Google Scholar]
- Liu, B.L. , Tabler, M. and Tsagris, M. (2000) Episomal expression of a hammerhead ribozyme directed against plum pox virus. Virus Res. 68, 15–23. [DOI] [PubMed] [Google Scholar]
- Llácer, G. (2006) Hosts and symptoms of Plum pox virus: herbaceous hosts. EPPO Bull. 36, 227–228. [Google Scholar]
- Llácer, G. and Cambra, M. (2006) Hosts and symptoms of Plum pox virus: fruiting Prunus species. EPPO Bull. 36, 219–221. [Google Scholar]
- López, L. , Urzainqui, A. , Domínguez, E. and García, J.A. (2001) Identification of an N‐terminal domain of the plum pox potyvirus CI RNA helicase involved in self‐interaction in a yeast two‐hybrid system. J. Gen. Virol. 82, 677–686. [DOI] [PubMed] [Google Scholar]
- López‐Moya, J.J. and García, J.A. (2000) Construction of a stable and highly infectious intron‐containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 68, 99–107. [DOI] [PubMed] [Google Scholar]
- López‐Moya, J.J. and García, J.A. (2008) Potyviruses In: Encyclopedia of Virology, 3rd edn (Mahy B.W.J. and Van Regenmortel M.H.V., eds), pp. 313–322. Oxford: Elsevier. [Google Scholar]
- López‐Moya, J.J. , Canto, T. , Díaz‐Ruíz, J.R. and López‐Abella, D. (1995) Transmission by aphids of a naturally non‐transmissible plum pox virus isolate with the aid of potato virus Y helper component. J. Gen. Virol. 76, 2293–2297. [DOI] [PubMed] [Google Scholar]
- Maejima, K. , Himeno, M. , Komatsu, K. , Takinami, Y. , Hashimoto, M. , Takahashi, S. , Yamaji, Y. , Oshima, K. and Namba, S. (2011) Molecular epidemiology of Plum pox virus in Japan. Phytopathology, 101, 567–574. [DOI] [PubMed] [Google Scholar]
- Maiss, E. , Timpe, U. , Brisske, A. , Jelkmann, W. , Casper, R. , Himmler, G. , Mattanovich, D. and Katinger, H.W.D. (1989) The complete nucleotide sequence of plum pox virus RNA. J. Gen. Virol. 70, 513–524. [DOI] [PubMed] [Google Scholar]
- Maiss, E. , Timpe, U. , Briske‐Rode, A. , Leseman, D.‐E. and Casper, R. (1992) Infectious in vivo transcripts of a plum pox potyvirus full‐length cDNA clone containing the cauliflower mosaic virus 35S RNA promoter. J. Gen. Virol. 73, 709–713. [DOI] [PubMed] [Google Scholar]
- Malinowski, T. , Cambra, M. , Capote, N. , Zawadzka, B. , Gorris, M.T. , Scorza, R. and Ravelonandro, M. (2006) Field trials of plum clones transformed with the Plum pox virus coat protein (PPV‐CP) gene. Plant Dis. 90, 1012–1018. [DOI] [PubMed] [Google Scholar]
- Malinowski, T. , Sowik, I. , Salavei, A.V. and Kukharchyk, N.V. (2012) Partial characterisation of biological properties of PPV‐C isolates found in Belarus and establishment of in vitro cultures of infected L2 and OWP‐6 rootstocks. In: 22nd International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Rome, Book of Abstracts, p. 152.
- Maliogka, V.I. , Salvador, B. , Carbonell, A. , Saénz, P. , San León, D. , Oliveros, J.C. , Delgadillo, M.O. , García, J.A. and Simón‐Mateo, C. (2012) Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host‐dependent pathogenicity features. Mol. Plant Pathol. 13, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoussopoulos, I.N. , Maiss, E. and Tsagris, M. (2000) Native electrophoresis and Western blot analysis (NEWeB): a method for characterization of different forms of potyvirus particles and similar nucleoprotein complexes in extracts of infected plant tissues. J. Gen. Virol. 81, 2295–2298. [DOI] [PubMed] [Google Scholar]
- Marandel, G. , Salava, J. , Abbott, A. , Candresse, T. and Decroocq, V. (2009) Quantitative trait loci meta‐analysis of Plum pox virus resistance in apricot (Prunus armeniaca L.): new insights on the organization and the identification of genomic resistance factors. Mol. Plant Pathol. 10, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.T. , Cervera, M.T. , Bonay, P. and García, J.A. (1995) Properties of the active plum pox potyvirus RNA polymerase complex in defined glycerol gradient fractions. Virus Res. 37, 127–137. [DOI] [PubMed] [Google Scholar]
- Martín, M.T. and García, J.A. (1991) Plum pox potyvirus RNA replication in a crude membrane fraction from infected Nicotiana clevelandii leaves. J. Gen. Virol. 72, 785–790. [DOI] [PubMed] [Google Scholar]
- Martín, M.T. , García, J.A. , Cervera, M.T. , Goldbach, R.W. and van Lent, J.W.M. (1992) Intracellular localization of three non‐structural plum pox potyvirus proteins by immunogold labelling. Virus Res. 25, 201–211. [Google Scholar]
- Martínez‐Gomez, P. and Dicenta, F. (1999) Evaluation of resistance to sharka in the breeding apricot program in CEBAS‐CSIC in Murcia (Spain). Acta Hortic. 488, 731–737. [Google Scholar]
- Martínez‐Gómez, P. , Dicenta, F. and Audergon, J.M. (2000) Behaviour of apricot (Prunus armeniaca L.) cultivars in the presence of sharka (plum pox potyvirus): a review. Agronomiei 20, 407–422. [Google Scholar]
- Matic, S. , Al‐Rwahnih, M. and Myrta, A. (2006) Diversity of Plum pox virus isolates in Bosnia and Herzegovina. Plant Pathol. 55, 11–17. [Google Scholar]
- Matic, S. , Elmaghraby, I. , Law, V. , Varga, A. , Reed, C. , Myrta, A. and James, D. (2011) Serological and molecular characterization of isolates of Plum pox virus strain El Amar to better understand its diversity, evolution, and unique geographical distribution. J. Plant Pathol. 93, 303–310. [Google Scholar]
- Mavrodieva, V. , James, D. , Williams, K. , Negi, S. , Varga, A. , Mock, R. and Levy, L. (2013) Molecular analysis of a Plum pox virus W isolate in plum germplasm hand carried into the USA from the Ukraine shows a close relationship to a Latvian isolate. Plant Dis. 97, 44–52. [DOI] [PubMed] [Google Scholar]
- Minoiu, N. (1973) Vectors transmitting plum pox virus to plum. Anal. Inst. Ceretai Pentru. Prot. Plantelor 9, 45–56. [Google Scholar]
- Mlotshwa, S. , Schauer, S.E. , Smith, T.H. , Mallory, A.C. , Herr, J.M. , Roth, B. , Merchant, D.S. , Ray, A. , Bowman, L.H. and Vance, V.B. (2005) Ectopic DICER‐LIKE1 expression in P1/HC‐Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell, 17, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A. , Fereres, A. and Cambra, M. (2009) Quantitative estimation of plum pox virus targets acquired and transmitted by a single Myzus persicae . Arch. Virol. 154, 1391–1399. [DOI] [PubMed] [Google Scholar]
- Moustafa, T.A. , Badenes, M.L. , Martinez‐Calvo, J. and Llacer, G. (2001) Determination of resistance to sharka (Plum pox) virus in apricot. Sci. Hort. 91, 59–70. [Google Scholar]
- Myrta, A. , Boscia, D. , Potere, O. , Kolber, M. , Nemeth, M. , DiTerlizzi, B. , Cambra, M. and Savino, V. (2001) Existence of two serological subclusters of Plum pox virus, strain M. Eur. J. Plant Pathol. 107, 845–848. [Google Scholar]
- Myrta, A. , Varga, A. and James, D. (2006) The complete genome sequence of an El Amar isolate of plum pox virus (PPV) and its phylogenetic relationship to other PPV strains. Arch. Virol. 151, 1189–1198. [DOI] [PubMed] [Google Scholar]
- Nagyová, A. , Kamencayová, M. , Glasa, M. and Šubr, Z.W. (2012) The 3′‐proximal part of the Plum pox virus P1 gene determinates the symptom expression in two herbaceous host plants. Virus Genes, 44, 505–512. [DOI] [PubMed] [Google Scholar]
- Nemchinov, L. and Hadidi, A. (1996) Characterization of the sour cherry strain of plum pox virus. Phytopathology, 86, 575–580. [Google Scholar]
- Nemchinov, L. , Hadidi, A. , Maiss, E. , Cambra, M. , Candresse, T. and Damsteegt, V. (1996) Sour cherry strain of plum pox potyvirus (PPV): molecular and serological evidence for a new subgroup of PPV strains. Phytopathology, 86, 1215–1221. [Google Scholar]
- Nemchinov, L. , Hadidi, A. , Kölber, M. and Németh, M. (1998) Molecular evidence for the occurrence of plum pox virus—cherry subgroup in Hungary. Acta Hortic. 472, 503–510. [Google Scholar]
- Németh, M. (1994) History and importance of plum pox in stone‐fruit production. EPPO Bull. 24, 525–536. [Google Scholar]
- Ng, J.C. and Falk, B.W. (2006) Virus–vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44, 183–212. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Gallois, J.L. , Chafiai, F. , Allen, L.M. , Schurdi‐Levraud, V. , Browning, K.S. , Candresse, T. , Caranta, C. , Le Gall, O. and German‐Retana, S. (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana . FEBS Lett. 581, 1041–1046. [DOI] [PubMed] [Google Scholar]
- Ogašanovic, D. , Rankovic, M. , Plazinic, R. and Papic, V. (1994) Performance of newly‐bred Cacak plum cultivars and current breeding tendencies. Acta Hortic. 359, 75–81. [Google Scholar]
- Olmos, A. , Cambra, M. , Dasi, M.A. , Candresse, T. , Esteban, O. , Gorris, M.T. and Asensio, M. (1997) Simultaneous detection and typing of plum pox potyvirus (PPV) isolates by heminested‐PCR and PCR‐ELISA. J. Virol. Methods, 68, 127–137. [DOI] [PubMed] [Google Scholar]
- Olmos, A. , Bertolini, E. and Cambra, M. (2002) Simultaneous and co‐operational amplification (Co‐PCR): a new concept for detection of plant viruses. J. Virol. Methods, 106, 51–59. [DOI] [PubMed] [Google Scholar]
- Olmos, A. , Bertolini, E. , Gil, M. and Cambra, M. (2005) Real‐time assay for quantitative detection of non‐persistently transmitted Plum pox virus RNA targets in single aphids. J. Virol. Methods, 128, 151–155. [DOI] [PubMed] [Google Scholar]
- Olmos, A. , Bertolini, E. , Capote, N. and Cambra, M. (2008) An evidence‐based approach to Plum pox virus detection by DASI‐ELISA and RT‐PCR in dormant period. Virology, 1, 1–8. [Google Scholar]
- van Oosten, H.J. and van Bakel, C.H.J. (1970) Inclusion bodies in plants infected with sharka (plum pox) virus. Neth. J. Plant Pathol. 76, 313–319. [Google Scholar]
- Pacheco, R. , Garcia‐Marcos, A. , Manzano, A. , de Lacoba, M.G. , Camanes, G. , Garcia‐Agustin, P. , Diaz‐Ruiz, J.R. and Tenllado, F. (2012) Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant–virus interactions that lead to cell death. Mol. Plant–Microbe Interact. 25, 709–723. [DOI] [PubMed] [Google Scholar]
- PaDIL (2013) Available at http://old.padil.gov.au/pbt/ [accessed on Oct 12, 2013].
- Pagny, G. , Paulstephenraj, P.S. , Poque, S. , Sicard, O. , Cosson, P. , Eyquard, J.P. , Caballero, M. , Chague, A. , Gourdon, G. , Negrel, L. , Candresse, T. , Mariette, S. and Decroocq, V. (2012) Family‐based linkage and association mapping reveals novel genes affecting Plum pox virus infection in Arabidopsis thaliana . New Phytol. 196, 873–886. [DOI] [PubMed] [Google Scholar]
- Palkovics, L. , Burgyán, J. and Balázs, E. (1993) Comparative sequence analysis of four complete primary structures of plum pox virus strains. Virus Genes, 7, 339–347. [DOI] [PubMed] [Google Scholar]
- Palmisano, F. , Boscia, D. , Minafra, A. , Myrta, A. and Candresse, T. (2012) An atypical Albanian isolate of Plum pox virus could be the progenitor of the Marcus strain. In: 22nd International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, June 3–8, Rome, Book of Abstracts, p. 33.
- Pandolfini, T. , Molesini, B. , Avesani, L. , Spena, A. and Polverari, A. (2003) Expression of self‐complementary hairpin RNA under the control of the rolC promoter confers systemic disease resistance to plum pox virus without preventing local infection. BMC Biotechnol. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal, T. , Pfeiffer, F. and Kervella, J. (2002) Preliminary observations on the resistance to sharka in peach and related species. Acta Hortic. 592, 699–704. [Google Scholar]
- Pasquini, G. and Barba, M. (2006) The question of seed transmissibility of Plum pox virus . EPPO Bull. 36, 287–292. [Google Scholar]
- Pérez, J.D.J. , Udeshi, N.D. , Shabanowitz, J. , Ciordia, S. , Juárez, S. , Scott, C.L. , Olszewski, N.E. , Hunt, D.F. and García, J.A. (2013) O‐GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology, 442, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez, J.J. , Juárez, S. , Chen, D. , Scott, C.L. , Hartweck, L.M. , Olszewski, N.E. and García, J.A. (2006) Mapping of two O‐GlcNAc modification sites in the capsid protein of the potyvirus Plum pox virus . FEBS Lett. 580, 5822–5828. [DOI] [PubMed] [Google Scholar]
- Pérez‐Martín, J. , Cases, I. and de Lorenzo, V. (1997) Design of a solubilization pathway for recombinant polypeptides in vivo through processing of a bi‐protein with a viral protease. Protein Eng. 10, 725–730. [DOI] [PubMed] [Google Scholar]
- Piron, F. , Nicolai, M. , Minoia, S. , Piednoir, E. , Moretti, A. , Salgues, A. , Zamir, D. , Caranta, C. and Bendahmane, A. (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE, 5, e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polák, J. (2006) Hosts and symptoms of Plum pox virus: woody species other than fruit and ornamental species of Prunus . EPPO Bull. 36, 225–226. [Google Scholar]
- Polák, J. , Pívalová, J. and Svoboda, J. (2005) Preliminary observations on the resistance to sharka in peach and related species. Plant Prot. Sci. 41, 47–51. [Google Scholar]
- Polák, J. , Pívalová, J. , Kundu, J.K. , Jokeš, M. , Scorza, R. and Ravelonandro, M. (2008) Behaviour of transgenic Plum pox virus‐resistant Prunus domestica L. clone C5 grown in the open field under a high and permanent infection pressure of the PPV‐Rec strain. J. Plant Pathol. 90, 33–36. [Google Scholar]
- Predajňa, L. , Nagyova, A. , Glasa, M. and Šubr, Z. (2012a) Cloning of the complete infectious cDNA of the plum pox virus strain PPV‐Rec. Acta Virol. 56, 129–132. [DOI] [PubMed] [Google Scholar]
- Predajňa, L. , Šubr, Z. , Candresse, T. and Glasa, M. (2012b) Evaluation of the genetic diversity of Plum pox virus in a single plum tree. Virus Res. 167, 112–117. [DOI] [PubMed] [Google Scholar]
- Prins, M. , Laimer, M. , Noris, E. , Schubert, J. , Wassenegger, M. and Tepfer, M. (2008) Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen, P. and Mäkinen, K. (2004) Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 279, 38 103–38 110. [DOI] [PubMed] [Google Scholar]
- Ravelonandro, M. , Monsion, M. , Teycheney, P.Y. , Delbos, R. and Dunez, J. (1992) Construction of a chimeric viral gene expressing plum pox virus coat protein. Gene, 120, 167–173. [DOI] [PubMed] [Google Scholar]
- Ravelonandro, M. , Peyruchaud, O. , Garrigue, L. , de Marcillac, G. and Dunez, J. (1993) Immunodetection of the plum pox virus helper component in infected plants and expression of its gene in transgenic plants. Arch. Virol. 130, 251–268. [DOI] [PubMed] [Google Scholar]
- Ravelonandro, M. , Scorza, R. , Bachelier, J.C. , Labonne, G. , Levy, L. , Damsteegt, V. , Callahan, A.M. and Dunez, J. (1997) Resistance of transgenic Prunus domestica to plum pox virus infection. Plant Dis. 81, 1231–1235. [DOI] [PubMed] [Google Scholar]
- Regner, F. , da Camara Machado, A. , Laimer da Camara Machado, M. , Steinkellner, H. , Mattanovich, D. , Hanzer, V. , Weiss, H. and Kattinger, H. (1992) Coat protein mediated resistance to Plum Pox Virus in Nicotiana clevelandii and Nicotiana benthamiana . Plant Cell Rep. 11, 30–33. [DOI] [PubMed] [Google Scholar]
- Revers, F. , Le Gall, O. , Candresse, T. and Maule, A.J. (1999) New advances in understanding the molecular biology of plant/potyvirus interactions. Mol. Plant–Microbe Interact. 12, 367–376. [Google Scholar]
- Reyes, F. , Fiore, N. , Reyes, M.A. , Sepúlveda, P. , Paredes, V. and Prieto, H. (2003) Biological behavior and partial molecular characterization of six Chilean isolates of Plum pox virus. Plant Dis. 87, 15–20. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1989) The genome‐linked protein and 5′ end RNA sequence of plum pox potyvirus. J. Gen. Virol. 70, 2785–2789. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1990) Infectious in vitro transcripts from a plum pox potyvirus cDNA clone. Virology, 177, 710–716. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1991) Identification of the initiation codon of plum pox potyvirus genomic RNA. Virology, 185, 544–552. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Cervera, M.T. and García, J.A. (1995) Processing of the plum pox virus polyprotein at the P3–6K1 junction is not required for virus viability. J. Gen. Virol. 76, 951–956. [DOI] [PubMed] [Google Scholar]
- Rodamilans, B. , Valli, A. and García, J.A. (2013) Mechanistic divergence between P1 proteases of the family Potyviridae. J. Gen. Virol. 94, 1407–1414. [DOI] [PubMed] [Google Scholar]
- Roossinck, M.J. (2005) Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 3, 917–924. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , German‐Retana, S. , Delaunay, T. , Delécolle, B. , Candresse, T. and Le Gall, O. (2002) Interaction between potyvirus helper component‐proteinase and capsid protein in infected plants. J. Gen. Virol. 83, 1765–1770. [DOI] [PubMed] [Google Scholar]
- Rubio, M. , Martinez Gomez, P. and Dicenta, F. (2003) Resistance of almond cultivars to Plum pox virus (Sharka). Plant Breed. 122, 462–464. [Google Scholar]
- Sáenz, P. , Cervera, M.T. , Dallot, S. , Quiot, L. , Quiot, J.B. , Riechmann, J.L. and García, J.A. (2000) Identification of a pathogenicity determinant of Plum pox virus in the sequence encoding the C‐terminal region of protein P3+6K1 . J. Gen. Virol. 81, 557–566. [DOI] [PubMed] [Google Scholar]
- Sáenz, P. , Quiot, L. , Quiot, J.‐B. , Candresse, T. and García, J.A. (2001) Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Mol. Plant–Microbe Interact. 14, 278–287. [DOI] [PubMed] [Google Scholar]
- Sáenz, P. , Salvador, B. , Simón‐Mateo, C. , Kasschau, K.D. , Carrington, J.C. and García, J.A. (2002) Host‐specific involvement of the HC protein in the long‐distance movement of potyviruses. J. Virol. 76, 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, B. , Delgadillo, M.O. , Saénz, P. , García, J.A. and Simón‐Mateo, C. (2008a) Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant–Microbe Interact. 21, 20–29. [DOI] [PubMed] [Google Scholar]
- Salvador, B. , Sáenz, P. , Yanguez, E. , Quiot, J.B. , Quiot, L. , Delgadillo, M.O. , García, J.A. and Simón‐Mateo, C. (2008b) Host‐specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 9, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, K. , Bedford, I.D. , Yahara, T. and Stanley, J. (2003) The earliest recorded plant virus disease. Nature, 422, 831. [DOI] [PubMed] [Google Scholar]
- Schneider, W.L. , Sherman, D.J. , Stone, A.L. , Damsteegt, V.D. and Frederick, R.D. (2004) Specific detection and quantification of Plum pox virus by real‐time fluorescent reverse transcription‐PCR. J. Virol. Methods, 120, 97–105. [DOI] [PubMed] [Google Scholar]
- Schneider, W.L. , Damsteegt, V.D. , Gildow, F.E. , Stone, A.L. , Sherman, D.J. , Levy, L.E. , Mavrodieva, V. , Richwine, N. , Welliver, R. and Luster, D.G. (2011) Molecular, ultrastructural, and biological characterization of Pennsylvania isolates of Plum pox virus . Phytopathology, 101, 627–636. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.‐B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurdi‐Levraud Escalettes, V. , Hullot, C. , Wawrzy'nczak, D. , Mathieu, E. , Eyquard, J.P. , Le Gall, O. and Decroocq, V. (2006) Plum pox virus induces differential gene expression in the partially resistant stone fruit tree Prunus armeniaca cv. Goldrich. Gene, 374, 96–103. [DOI] [PubMed] [Google Scholar]
- Scorza, R. , Ravelonandro, M. , Callahan, A.M. , Cordts, J.M. , Fuchs, M. , Dunez, J. and Gonsalves, D. (1994) Transgenic plums (Prunus domestica L.) express the plum pox virus coat protein gene. Plant Cell Rep. 14, 18–22. [DOI] [PubMed] [Google Scholar]
- Scorza, R. , Callahan, A. , Levy, L. , Damsteegt, V. , Webb, K. and Ravelonandro, M. (2001) Post‐transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum pox potyvirus coat protein gene. Transgenic Res. 10, 201–209. [DOI] [PubMed] [Google Scholar]
- Scorza, R. , Callahan, A. , Ravelonandro, M. and Braverman, M. (2013) Development and regulation of the Plum pox virus resistant transgenic plum ‘HoneySweet’ In: Regulation of Agricultural Biotechnology: The United States and Canada (Wozniak C.A. and McHughen A., eds), pp. 269–280. Dordrecht, The Netherlands: Springer. doi: 10.1007/1978-1094-1007-2156-1002_1012. [DOI] [Google Scholar]
- Scott, C.L. , Hartweck, L.M. , Pérez, J.D.J. , Chen, D. , García, J.A. and Olszewski, N.E. (2006) SECRET AGENT, an Arabidopsis thaliana O‐GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 580, 5829–5835. [DOI] [PubMed] [Google Scholar]
- Sheveleva, A. , Ivanov, P. , Prihodko, Y. , James, D. and Chirkov, S. (2012) Occurrence and genetic diversity of Winona‐like Plum pox virus isolates in Russia. Plant Dis. 96, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Sicard, O. , Loudet, O. , Keurentjes, J.J. , Candresse, T. , Le Gall, O. , Revers, F. and Decroocq, V. (2008) Identification of quantitative trait loci controlling symptom development during viral infection in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 21, 198–207. [DOI] [PubMed] [Google Scholar]
- Simón‐Buela, L. , Guo, H.S. and García, J.A. (1997a) Cap‐independent leaky scanning as the mechanism of translation initiation of a plant viral genomic RNA. J. Gen. Virol. 78, 2691–2699. [DOI] [PubMed] [Google Scholar]
- Simón‐Buela, L. , Guo, H.S. and García, J.A. (1997b) Long sequences in the 5′ noncoding region of plum pox virus are not necessary for viral infectivity but contribute to viral competitiveness and pathogenesis. Virology, 233, 157–162. [DOI] [PubMed] [Google Scholar]
- Simón‐Mateo, C. and García, J.A. (2006) MicroRNA‐guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 80, 2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón‐Mateo, C. and García, J.A. (2011) Antiviral strategies in plants based on RNA silencing. Biochim. Biophys. Acta, 1809, 722–731. [DOI] [PubMed] [Google Scholar]
- Simón‐Mateo, C. , López‐Moya, J.J. , Guo, H.S. , González, E. and García, J.A. (2003) Suppressor activity of potyviral and cucumoviral infections in potyvirus‐induced transgene silencing. J. Gen. Virol. 84, 2877–2883. [DOI] [PubMed] [Google Scholar]
- Sochor, J. , Babula, P. , Adam, V. , Krska, B. and Kizek, R. (2012) Sharka: the past, the present and the future. Viruses, 4, 2853–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šubr, Z. , Pittnerova, S. and Glasa, M. (2004) A simplified RT‐PCR‐based detection of recombinant Plum pox virus isolates. Acta Virol. 48, 173–176. [PubMed] [Google Scholar]
- Šubr, Z. , Ryšlavá, H. and Kollerová, E. (2007) Electrophoretic mobility of the capsid protein of the Plum pox virus strain PPV‐Rec indicates its partial phosphorylation. Acta Virol. 51, 135–138. [PubMed] [Google Scholar]
- Šubr, Z.W. , Kamencayová, M. , Nováková, S. , Nagyová, A. , Nosek, J. and Glasa, M. (2010) A single amino acid mutation alters the capsid protein electrophoretic double‐band phenotype of the Plum pox virus strain PPV‐Rec. Arch. Virol. 155, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Sutic, D. , Babovic, M. and Markovic, S. (1976) Transmissibility of some sharka virus strains by Mizus persicae, depending on various infection sources. Acta Hortic. 67, 171–175. [Google Scholar]
- Syrgiannidis, G.O. (1980) Selection of two apricot varieties resistant to sharka virus. Acta Phytopathol. Hung. 15, 85–88. [Google Scholar]
- Szathmary, E. , Nadudvari, J.N. , Szabo, L. , Tobias, I. , Balazs, E. and Palkovics, L. (2009) Characterization of a natural Plum pox virus isolate bearing a truncated coat protein. Arch. Virol. 154, 141–145. [DOI] [PubMed] [Google Scholar]
- Tavert‐Roudet, G. , Ravelonandro, M. , Bachelier, J.C. and Dunez, J. (1998) Transgenic Nicotiana benthamiana plants containing the P1 gene of plum pox virus are resistant to virus challenge. Eur. J. Plant Pathol. 104, 103–107. [Google Scholar]
- Tenllado, F. , Barajas, D. , Vargas, M. , Atencio, F.A. , González‐Jara, P. and Díaz‐Ruíz, J.R. (2003) Transient expression of homologous hairpin RNA causes interference with plant virus infection and is overcome by a virus encoded suppressor of gene silencing. Mol. Plant–Microbe Interact. 16, 149–158. [DOI] [PubMed] [Google Scholar]
- Teycheney, P.Y. , Tavert, G. , Delbos, R. , Ravelonandro, M. and Dunez, J. (1989) The complete nucleotide sequence of plum pox virus RNA (strain D). Nucleic Acids Res. 17, 10 115–10 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, D. , Varga, A. , De Costa, H. , Birch, C. , Glasa, M. and James, D. (2009) First report of Plum pox virus recombinant strain on Prunus spp. in Canada. Plant Dis. 93, 674. [DOI] [PubMed] [Google Scholar]
- Trifonov, D. (1975) Susceptibility of plum varieties to Plum pox virus. Acta Hortic. 44, 163–164. [Google Scholar]
- Ulubaş Serçe, C. , Candresse, T. , Svanella‐Dumas, L. , Krizbai, L. , Gazel, M. and Çağlayan, K. (2009) Further characterization of a new recombinant group of Plum pox virus isolates, PPV‐T, found in orchards in the Ankara province of Turkey. Virus Res. 142, 121–126. [DOI] [PubMed] [Google Scholar]
- Valli, A. , López‐Moya, J.J. and García, J.A. (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae . J. Gen. Virol. 88, 1016–1028. [DOI] [PubMed] [Google Scholar]
- Varga, A. and James, D. (2005) Detection and differentiation of Plum pox virus using real‐time multiplex PCR with SYBR Green and melting curve analysis: a rapid method for strain typing. J. Virol. Methods, 123, 213–220. [DOI] [PubMed] [Google Scholar]
- Varga, A. and James, D. (2006a) Real‐time RT‐PCR and SYBR Green I melting curve analysis for the identification of Plum pox virus strains C, EA, and W: effect of amplicon size, melt rate, and dye translocation. J. Virol. Methods, 132, 146–153. [DOI] [PubMed] [Google Scholar]
- Varga, A. and James, D. (2006b) Use of reverse transcription loop‐mediated isothermal amplification for the detection of Plum pox virus. J. Virol. Methods, 138, 184–190. [DOI] [PubMed] [Google Scholar]
- Varrelmann, M. , Maiss, E. , Pilot, R. and Palkovics, L. (2007) Use of pentapeptide‐insertion scanning mutagenesis for functional mapping of the plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Varveri, C. (2006) Epidemiology of Plum pox virus strain M in Greece. EPPO Bull. 36, 276–278. [Google Scholar]
- Vidal, E. , Moreno, A. , Bertolini, E. and Cambra, M. (2012a) Estimation of the accuracy of two diagnostic methods for the detection of Plum pox virus in nursery blocks by latent class models. Plant Pathol. 61, 413–422. [Google Scholar]
- Vidal, E. , Yokomi, R.K. , Moreno, A. , Bertolini, E. and Cambra, M. (2012b) Calculation of diagnostic parameters of advanced serological and molecular tissue‐print methods for detection of Citrus tristeza virus: a model for other plant pathogens. Phytopathology, 102, 114–121. [DOI] [PubMed] [Google Scholar]
- Vidal, E. , Zagrai, L. , Milusheva, S. , Bozhkova, V. , Tasheva‐Terzieva, E. , Kamenova, I. , Zagrai, I. and Cambra, M. (2013) Horticultural mineral oil treatments in nurseries during aphid flights reduce Plum pox virus incidence under different ecological conditions. Ann. Appl. Biol. 162, 299–308. [Google Scholar]
- Vilanova, S. , Romero, C. , Abbott, A.G. , Llacer, G. and Badenes, M.L. (2003) An apricot (Prunus armeniaca L.) F2 progeny linkage map based on SSR and AFLP markers, mapping plum pox virus resistance and self‐incompatibility traits. Theor. Appl. Genet. 107, 239–247. [DOI] [PubMed] [Google Scholar]
- Wallis, C.M. , Stone, A.L. , Sherman, D.J. , Damsteegt, V.D. , Gildow, F.E. and Schneider, W.L. (2007) Adaptation of plum pox virus to a herbaceous host (Pisum sativum) following serial passages. J. Gen. Virol. 88, 2839–2845. [DOI] [PubMed] [Google Scholar]
- Waltermann, A. and Maiss, E. (2006) Detection of 6K1 as a mature protein of 6 kDa in plum pox virus‐infected Nicotiana benthamiana . J. Gen. Virol. 87, 2381–2386. [DOI] [PubMed] [Google Scholar]
- Wang, A. , Chapman, P. , Chen, L. , Stobbs, L.W. , Brown, D.C.W. and Brandle, J.E. (2005) A comparative survey, by expressed sequence tag analysis, of genes expressed in peach leaves infected with Plum pox virus (PPV) and free from PPV. Can. J. Plant Pathol. 27, 410–419. [Google Scholar]
- Wang, X. , Kohalmi, S.E. , Svircev, A. , Wang, A. , Sanfacon, H. and Tian, L. (2013) Silencing of the host factor eIF(iso)4E gene confers plum pox virus resistance in plum. PLoS ONE, 8, e50627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. and Wang, A. (2008) Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI‐ and COPII‐dependent manner. J. Virol. 82, 12 252–12 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hong, J. , Xiong, R. , Kasschau, K.D. , Zhou, X. , Carrington, J.C. and Wang, A. (2010a) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N‐PIPO. Plos Pathog. 6, e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T.Y. , Huang, T.S. , McNeil, J. , Laliberte, J.F. , Hong, J. , Nelson, R.S. and Wang, A.M. (2010b) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R. , Zhang, S.C. , Michaud, D. and Sanfacon, H.N. (2004) Inhibitory effects of cystatins on proteolytic activities of the Plum pox potyvirus cysteine proteinases. Virus Res. 105, 175–182. [DOI] [PubMed] [Google Scholar]
- Wetzel, T. , Candresse, T. , Ravelonandro, M. , Delbos, R.P. , Mazyad, H. , Aboul‐Ata, A.E. and Dunez, J. (1991a) Nucleotide sequence of the 3′ terminal region of the RNA of the El Amar strain of plum pox potyvirus. J. Gen. Virol. 72, 1741–1746. [DOI] [PubMed] [Google Scholar]
- Wetzel, T. , Candresse, T. , Ravelonandro, M. and Dunez, J. (1991b) A polymerase chain reaction assay adapted to plum pox virus detection. J. Virol. Methods, 33, 355–365. [DOI] [PubMed] [Google Scholar]
- Wittner, A. , Palkovics, L. and Balazs, E. (1998) Nicotiana benthamiana plants transformed with the plum pox virus helicase gene are resistant to virus infection. Virus Res. 53, 97–103. [DOI] [PubMed] [Google Scholar]
- Youssef, S.A. and Shalaby, A. (2006) Plum pox virus (PPV) in Egypt. EPPO Bull. 36, 208. [Google Scholar]
- Zagrai, I. , Capote, N. , Ravelonandro, M. , Cambra, M. , Zagrai, L. and Scorza, R. (2008) Plum pox virus silencing of C5 transgenic plums is stable under challenge inoculation with heterologous viruses. J. Plant Pathol. 90, 63–71. [Google Scholar]
- Zagrai, I. , Ravelonandro, M. , Gaboreanu, I. , Ferencz, B. , Scorza, R. , Zagrai, L. , Kelemen, B. , Pamfil, D. and Popescu, O. (2011) Transgenic plums expressing Plum pox virus coat protein gene do not assist the development of virus recombinants under field conditions. J. Plant Pathol. 93, 159–165. [Google Scholar]
- Zawadzka, B. (1981) The response of several plum cultivars to infection with Plum pox virus. Acta Hortic. 94, 215–222. [Google Scholar]
- Zhang, S.C. , Tian, L.M. , Svircev, A. , Brown, D.C.W. , Sibbald, S. , Schneider, K.E. , Barszcz, E.S. , Malutan, T. , Wen, R. and Sanfacon, H. (2006) Engineering resistance to Plum pox virus (PPV) through the expression of PPV‐specific hairpin RNAs in transgenic plants. Can. J. Plant Pathol. 28, 263–270. [Google Scholar]
- Zheng, N. , Pérez, J.D. , Zhang, Z. , Domínguez, E. , García, J.A. and Xie, Q. (2008) Specific and efficient cleavage of fusion proteins by recombinant plum pox virus NIa protease. Protein Expr. Purif. 57, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N. , Huang, X. , Yin, B. , Wang, D. and Xie, Q. (2012) An effective system for detecting protein–protein interaction based on in vivo cleavage by PPV NIa protease. Protein Cell, 3, 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilian, E. and Maiss, E. (2011) Detection of plum pox potyviral protein–protein interactions in planta using an optimized mRFP‐based bimolecular fluorescence complementation system. J. Gen. Virol. 92, 2711–2723. [DOI] [PubMed] [Google Scholar]
- Zuriaga, E. , Soriano, J.M. , Zhebentyayeva, T. , Romero, C. , Dardick, C. , Cañizares, J. and Badenes, M.L. (2013) Genomic analysis reveals MATH gene(s) as candidate(s) for Plum pox virus (PPV) resistance in apricot (Prunus armeniaca L.). Mol. Plant Pathol. 14, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]


