Summary
The cucumber mosaic virus (CMV) 2b protein is an RNA silencing suppressor protein that can also play direct and indirect roles in symptom induction. Previous work has shown that a hybrid virus, FRad352b‐CMV (renamed here as CMV‐FRad2b‐Pro), generated by replacement of the 2b gene of strain Fny‐CMV with that from Rad35‐CMV, displays markedly lower pathogenicity than Fny‐CMV on N icotiana species. However, the replacement of proline with leucine at position 55 of the 2b protein of CMV‐FRad2b‐Pro (protein Rad2b‐Pro) created a virus (CMV‐FRad2b‐Leu) that induced severe symptoms. Infection of A rabidopsis thaliana mutants defective in the expression of DICER‐like (DCL) endoribonucleases 2 and 4, which mediate antiviral RNA silencing, as well as of dcl3 and dcl2/3/4 triple‐mutant plants, indicated that Rad2b‐Pro was a weaker RNA silencing suppressor than the protein Rad2b‐Leu. This was confirmed in N icotiana benthamiana using agroinfiltration assays, showing that, compared with either Rad2b‐Leu or the Fny2b protein, Rad2b‐Pro was ineffective at inhibiting local or systemic silencing of expression of a green fluorescent protein reporter gene. Transgenic expression of Rad2b‐Leu, but not of Rad2b‐Pro, in A rabidopsis induced symptom‐like phenotypes and rescued the accumulation of the 2b‐deletion mutant Fny‐CMVΔ2b. Bimolecular fluorescent complementation indicated that, in planta, Rad2b‐Leu, but not Rad2b‐Pro, self‐interacts. Thus, self‐interaction is crucial to the ability of the 2b protein to suppress silencing and induce a symptom‐like phenotype, and is dependent on the properties of the residue at position 55.
Introduction
Cucumber mosaic virus (CMV) infects a remarkably broad range of wild and cultivated plants, and is an increasingly important model for studies of plant–virus interactions (Jacquemond, 2012; Palukaitis and Garcia‐Arenal, 2003; Scholthof et al., 2011). The CMV genome comprises three single‐stranded plus‐sense RNAs. RNA1 acts as the mRNA for the 1a protein, a component of the viral replicase complex with methyltransferase and helicase activities. RNA2 has two open reading frames that encode the 2a protein (a replicase component with RNA‐dependent RNA polymerase activity) and the 2b suppressor of RNA silencing. RNA3 also has two open reading frames encoding the 3a movement protein and the coat protein (CP). RNAs 2 and 3 are the translation templates for proteins 2a and 3a, respectively, but the 2b and CP proteins are translated from the subgenomic mRNAs 4A and 4 (Ding et al., 1994; Palukaitis and Garcia‐Arenal, 2003).
In Arabidopsis thaliana, 21‐ and 22‐nucleotide short‐interfering (si)RNAs, generated by DICER‐like (DCL) endoribonucleases 2 and 4, act redundantly to confer antiviral RNA silencing against CMV (Deleris et al., 2006; Diaz‐Pendon et al., 2007; Ziebell and Carr, 2009). The CMV 2b protein is an effective viral suppressor of RNA silencing (VSR), preventing the initiation of gene silencing and the systemic spread of silencing (Brigneti et al., 1998; Guo and Ding, 2002). Diaz‐Pendon et al. (2007) provided evidence that the subgroup II strain Q‐CMV 2b protein efficiently suppresses 21‐ and 22‐nucleotide viral siRNA‐mediated antiviral silencing by reducing the production of viral siRNA in Arabidopsis ecotype Col‐0. Zhang et al. (2006) initially found that, by direct interaction, the 2b protein of the subgroup IA strain Fny‐CMV weakens in vitro slicer activity of Argonaute 1 (AGO1), a core component of the RNA‐induced silencing complex (RISC). Consistent with a previous analysis of the Fny‐CMV 2b protein by Gonzalez et al. (2010), Duan et al. (2012) reported that a domain of the 2b protein from another subgroup IA strain SD‐CMV, which is thought to be required for interaction with AGO1, is spatially separated from the double‐stranded (ds)RNA binding domain, and interaction of 2b with AGO1 is dispensable for the suppression of siRNA‐mediated RNA silencing. Very recently, they found that the 2b–AGO1 interaction affects the biogenesis of trans‐acting siRNA by inhibiting AGO1 slicer activity in vivo (Feng et al., 2013). Moreover, the inhibition of AGO1 by the 2b protein triggers increased expression and activity of AGO2 (Harvey et al., 2011), but the 2b protein's ability to inhibit antiviral silencing is most probably mediated through the sequestration of siRNAs rather than the binding of AGO1 (Gonzalez et al., 2010, 2012). In Arabidopsis, the third class of siRNA with a size of 24 nucleotides, produced in vivo via cleavage of dsRNA by DCL3, is incorporated into AGO4 in the cytoplasm to guide RNA‐dependent DNA methylation (RdDM) in the nucleus (Ye et al., 2012). CMV 2b proteins can also interfere with RdDM by sequestering the 24‐nucleotide siRNA, as well as by binding the AGO4 protein to reduce this factor's slicer activity (Duan et al., 2012; Gonzalez et al., 2010; Guo and Ding, 2002; Hamera et al., 2011).
Another important biological effect of the CMV 2b protein is the disruption of the micro (mi)RNA‐mediated regulation of the turnover and translation of cellular mRNAs (Lanet et al., 2009; Lewsey et al., 2007). In transgenic Arabidopsis, constitutive expression of the Fny2b gene, but not of the 2b genes from milder subgroup II strains Q or LS, caused symptom‐like developmental defects (Lewsey et al., 2007; Zhang et al., 2006). These effects may be mediated by the effects of the Fny2b protein on AGO1, by direct binding of 2b to miRNA duplexes or through both activities (Gonzalez et al., 2010, 2012; Goto et al., 2007; Zhang et al., 2006).
The molecular basis of how the CMV 2b protein binds siRNA and miRNA duplexes is still unclear. The tombusviral P19 protein, a well‐studied silencing suppressor, forms head‐to‐tail homodimers that sequester siRNA duplexes to prevent siRNA recruitment into RISC (Ye et al., 2003). The orthologous 2b protein encoded by another Cucumovirus, Tomato aspermy virus (TAV), forms hook‐like homodimers in vitro that bind siRNA duplexes (Chen et al., 2008). Subsequently, Gonzalez et al. (2010) demonstrated the self‐interaction of the Fny2b protein in Nicotiana benthamiana using bimolecular fluorescence complementation (BiFC) assays, consistent with the idea that self‐interaction is required for the silencing of suppressor activity in vivo.
Experiments with CMVΔ2b mutants, CMV‐derived constructs lacking either the 2b open reading frame or the ability to express it, have shown that the 2b protein is a pathogenicity determinant (Ding et al., 1996; Shi et al., 2002; Soards et al., 2002). Furthermore, differential virulence exhibited by subgroup IA, IB or II CMV strains can be attributed in part to their respective 2b genes (Du et al., 2007; Shi et al., 2002). Specific domains within the 2b protein affecting viral symptoms have been documented (Ding et al., 1996; Goto et al., 2007; Lewsey et al., 2009). Previously, we found that, when the 2b gene of the subgroup IB CMV strain Rad35 was substituted for the Fny‐CMV 2b gene, the resulting chimeric virus (FRad352b‐CMV, but referred to in the present study as CMV‐FRad2b‐Pro) induced milder symptoms than Fny‐CMV on plants of several Nicotiana species (Du et al., 2007). Subsequently, it was found that replacement of a proline residue by leucine at position 55 of the Rad35 2b protein produced a chimeric CMV of increased pathogenicity (originally called FR/2b‐T2582, but referred to in this study as CMV‐FRad2b‐Leu) (Du et al., 2008). In this work, we further investigate the effects of amino acid position 55 and its effects on silencing suppression, pathogenicity and 2b self‐association.
Results
Either DCL2‐ or DCL4‐dependent antiviral RNA silencing is sufficient to inhibit the production of viral symptoms induced by CMV‐FRad2b‐Pro
The effects of CMV‐FRad2b‐Pro and CMV‐FRad2b‐Leu on plants of wild‐type Arabidopsis ecotype Col‐0 were consistent with those seen previously with N. glutinosa (Du et al., 2008). Specifically, CMV‐FRad2b‐Pro induced no obvious signs of disease, whereas CMV‐FRad2b‐Leu induced severe symptoms of stunting and distortion of rosette leaves (Fig. 1A). We tested the induction of symptoms by CMV‐FRad2b‐Pro and CMV‐FRad2b‐Leu in mutant Arabidopsis plants deficient in siRNA biosynthesis. These were dcl2, dcl3 and dcl4 single‐mutant and dcl2/3/4 triple‐mutant plants. Infection of CMV‐FRad2b‐Leu caused severe symptoms on plants of all mutant lines. However, only dcl2/3/4 triple‐mutant plants displayed severe symptoms after infection with CMV‐FRad2b‐Pro, which were similar to the symptoms induced by CMV‐FRad2b‐Leu in dcl2/3/4 triple‐mutant plants, and slightly more severe than the symptoms induced by CMV‐FRad2b‐Leu in wild‐type or single‐mutant plants. Furthermore, infection of CMV‐FRad2b‐Leu caused slightly more severe disease symptoms in all single mutants than in wild‐type plants (Fig. 1A). RNA gel blot analysis showed that CMV‐FRad2b‐Pro accumulated at a much lower level than CMV‐FRad2b‐Leu in systemic leaves of wild‐type Arabidopsis at 14 days post‐inoculation (dpi) (Fig. 1B), which is consistent with that observed on N. glutinosa (Du et al., 2008). We observed that the accumulation of CMV‐FRad2b‐Pro increased to a limited extent in dcl2, dcl3 and dcl4 single‐mutant compared with wild‐type plants. However, the accumulation of CMV‐FRad2b‐Pro increased dramatically in dcl2/3/4 triple‐mutant plants, which was much higher than the accumulation of CMV‐FRad2b‐Leu in wild‐type plants, and similar to that of CMV‐FRad2b‐Leu in dcl2/3/4 triple‐mutant plants. We noted that the accumulation of CMV‐FRad2b‐Leu increased to a different extent in each of the three dcl single mutants compared with wild‐type plants (Fig. 1B). These results demonstrate that the low virulence of CMV‐FRad2b‐Pro is associated with failure to inhibit siRNA‐mediated antiviral defence, and suggest that the high virulence of CMV‐FRad2b‐Leu results from an ability to strongly, but not completely, suppress silencing, even in wild‐type plants. The results indicate that symptom severity is related to virus titre, mainly depending on 2b's capability to suppress RNA silencing.
Figure 1.

Rad2b‐Pro did not suppress efficiently either Dicer‐like 2‐ (DCL2‐) or DCL4‐dependent short‐interfering (si)RNA‐guided antiviral RNA silencing. A rabidopsis thaliana Col‐0 wild‐type (WT) and dcl mutants were inoculated with the Fny‐CMV/Rad35‐CMV chimera CMV‐FRad2b‐Pro, its derivative CMV‐FRad2b‐Leu or mock inoculated (Mock). (A) Viral disease symptoms on A rabidopsis WT, dcl2, dcl3, dcl4 single‐mutant and dcl2/3/4 triple‐mutant plants with virus infection as indicated. Plants were photographed at 14 days post‐inoculation (dpi). (B) Northern blotting analysis of accumulation of viral progeny RNAs. Total RNA samples were extracted from a pool of five plant individuals at 14 dpi. Five micrograms per RNA sample were separated on 1.5% agarose gel with formaldehyde. rRNAs were used as a loading control. M indicates Mock, P indicates CMV‐FRad2b‐Pro and L indicates CMV‐FRad2b‐Leu. The numbers below lanes represent the relative accumulation (RA) of CMV. The RA value of CMV from the wild‐type plant with infection of CMV‐FRad2b‐Pro was set as unity.
Rad2b‐Leu, but not Rad2b‐Pro, causes developmental defects in A rabidopsis
To test the effects of the 2b proteins Rad2b‐Pro and Rad2b‐Leu on plants, but in isolation from other CMV‐encoded factors, we generated lines of transgenic Arabidopsis plants harbouring either the Rad2b‐Pro or Rad2b‐Leu gene sequence under the transcriptional control of the constitutive 35S promoter from cauliflower mosaic virus. Of the 35 primary transformed lines of Rad2b‐Pro‐transgenic plants, none exhibited any alteration in phenotype (Fig. 2A). We confirmed the expression of Rad2b‐Pro in these lines by Western blot using anti‐2b serum (Fig. S1, see Supporting Information). Moreover, variation in the expression level of the Rad2b‐Pro protein was observed among these lines. However, primary transformed lines of Rad2b‐Leu‐transgenic plants showed pleomorphic phenotypes, which were classified into three tiers (Fig. 2A). Of 36 primary transformed lines, 16 showed no obvious phenotypic changes (Tier I), nine were smaller than nontransgenic plants (Tier II), and showed obvious serration at the edge of rosette leaves, and 11 exhibited extreme developmental defects, such as narrow, serrated and strongly upward‐curled leaves (Tier III) (Fig. 2A, Table 1). Furthermore, we found that the intensity of developmental defects caused by Rad2b‐Leu was related to the expression level of the 2b protein (Fig. 2B). As 2b proteins are known to induce changes in plant phenotype by disrupting miRNA‐regulated gene expression, these results suggest that the ability of the Rad2b‐Leu protein to interact with AGO1 or with miRNA duplexes in Arabidopsis is superior to that of the protein Rad2b‐Pro, from which it was derived.
Figure 2.
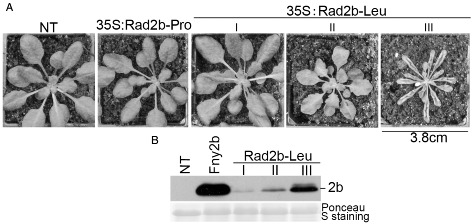
Constitutive expression of Rad2b‐Leu, but not Rad2b‐Pro, caused morphological changes to phenotype in transgenic A rabidopsis. Pleomorphic phenotypes of R ad2b‐Leu‐transgenic plants could be classified into three tiers (I, II, III; Table 1). Four‐week‐old plants were photographed. (B) Levels of Rad2b‐Leu protein in transgenic lines with different phenotypes. Polyclonal antibody against Fny2b was used. A line (3.13F) of F ny2b‐transgenic Arabidopsis with a severe phenotype was used as a control. NT indicates nontransgenic plant.
Table 1.
Alteration of developmental phenotype in transgenic A rabidopsis caused by constitutive expression of Rad2b‐Leu, but not Rad2b‐Pro, in transgenic lines
| Transgene | Number of primary transformed lines | Plants with no obvious defects* | Plants with moderate defects† | Plants with severe defects‡ |
|---|---|---|---|---|
| Rad2b‐Pro | 35 | 35 | 0 | 0 |
| Rad2b‐Leu | 36 | 16 | 9 | 11 |
See Fig. 2 for examples of plants expressing no (*Tier I), moderate (†Tier II) or severe (‡Tier III) Rad2b‐Leu‐induced effects on phenotype.
Constitutive expression of the Rad2b‐Leu, but not Rad2b‐Pro, protein in transgenic plants rescues the accumulation of a CMV mutant lacking the 2b gene
As shown above, CMV‐FRad2b‐Pro could not overcome either DCL2‐ or DCL4‐dependent antiviral silencing sufficiently to allow the virus to induce viral symptoms (Fig. 1). This suggests that Rad2b‐Pro is a weak silencing suppressor. To determine the relative strengths of Rad2b‐Pro and Rad2b‐Leu as RNA silencing suppressors, we tested the effect of both 2b proteins, when expressed in transgenic Arabidopsis, on the accumulation of the 2b‐deletion mutant Fny‐CMVΔ2b. First, we screened five and four putative transformed lines for the expression of Rad2b‐Pro and Rad2b‐Leu, respectively, by western blot analysis using anti‐2b serum. Four Rad2b‐Pro‐transgenic (3.3a, 3.6b, 3.8d, 3.12c) and four Rad2b‐Leu‐transgenic (4.2a, 4,3d, 4.4d, 4.7e) lines were identified by the presence of detectable levels of 2b protein (Fig. S2, see Supporting Information). The Rad2b‐Pro‐transgenic lines 3.3a, 3.6b and 3.12c, and the Rad2b‐Leu‐transgenic lines 4.2a and 4.4d, were subsequently used for virus challenge. The Rad2b‐Leu‐transgenic lines 4.2a and 4.4d were selected as they exhibited no obvious developmental anomalies that could confound the identification of any symptoms induced by virus infection (Fig. S3, see Supporting Information). Fny‐CMVΔ2b was inoculated onto the lower leaves of nontransgenic plants and the transgenic lines 3.3a, 3.6b, 3.12c, 4.2a and 4.4d. None of these lines showed viral symptoms, even by 30 dpi. Virus titres were estimated by western immunoblot using anti‐CMV CP at 14 dpi. The results showed that, in plants of all three Rad2b‐Pro‐transgenic lines, similar levels of CMV CP accumulated, and this level was similar to that which accumulated in nontransgenic plants. In contrast, higher levels of CP accumulated in plants of both Rad2b‐Leu‐transgenic lines (Fig. 3). These results further demonstrated that the presence of proline at residue 55 was responsible for the weak silencing suppressor activity of Rad2b‐Pro.
Figure 3.
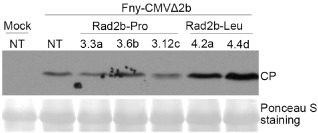
Accumulation of CMVΔ2b in 2b‐transgenic A rabidopsis. Three lines expressing Rad2b‐Pro and two lines expressing Rad2b‐Leu, expressing a similar level of respective 2b proteins, were inoculated with purified virions of Fny‐CMVΔ2b. Virus accumulation was analysed by immunoblotting using polyclonal anti‐coat protein (CP) serum in the upper noninoculated leaves of virus‐inoculated or mock‐inoculated plants at 14 days post‐inoculation. Ponceaus S staining was used to monitor equivalence of protein loading and transfer. NT indicates nontransgenic plant.
The residue at position 55 is critical to the ability of a IB subgroup CMV 2b protein to inhibit induction of local and systemic RNA silencing
The 2b protein of the subgroup II CMV strain Q (Q2b) possesses the ability to block the systemic spread of silencing (Guo and Ding, 2002), and both Q2b and the 2b protein of the subgroup IA strain Fny (Fny2b) have been shown to inhibit the initiation of local RNA silencing (Gonzalez et al., 2010). To assess the effectiveness of 2b proteins derived from CMV subgroup IB strains as inhibitors of localized silencing, we used the co‐infiltration of Agrobacterium tumefaciens cells harbouring a plasmid encoding a green fluorescent protein (GFP) reporter (p35S:GFP), together with A. tumefaciens cells transformed with plasmids carrying Rad2b‐Pro, Rad2b‐Leu or Fny2b coding sequences and a control plasmid carrying the gene for the bacterial β‐glucuronidase (p35S:GUS) (Fig. 4). Agroinfiltration was carried out using plants of the GFP‐transgenic N. benthamiana 16c line. These plants express GFP and are therefore susceptible to the induction of systemic silencing of GFP expression (Brigneti et al., 1998), allowing us to examine the effects of 2b proteins on local and systemic silencing in the same plants.
Figure 4.
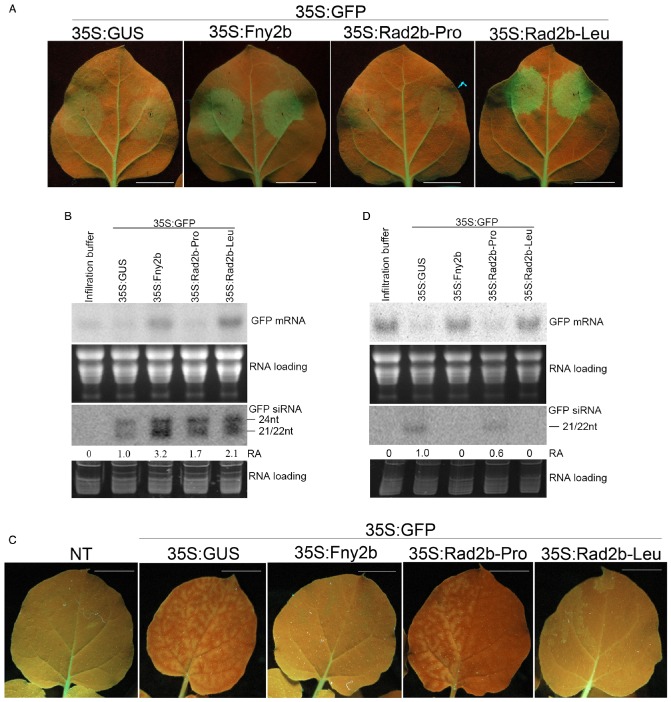
Comparison of Rad2b‐Pro and Rad2b‐Leu in suppression of local and systemic silencing of green fluorescence protein. (A) GFP fluorescence (5 days post‐infiltration) in leaves of N icotiana benthamiana GFP transgenic line 16c with agroinfiltration of reporter plasmid p35S:GFP, together with control plasmid p35S:GUS, or a pBI121‐derived vector encoding 2b proteins, as indicated. (B) Northern blot analyses of GFP mRNA and GFP‐derived short‐interfering (si)RNA from the agroinfiltrated patches, and the patch treated with infiltration solution. Total RNA was extracted at 5 days post‐infiltration. (C) GFP fluorescence (14 days post‐infiltration) in the upper noninfiltrated leaves of N . benthamiana GFP transgenic line 16c with agroinfiltration as in (A). Photographs were taken under UV light. (D) Northern blot analyses of GFP mRNA and siRNAs from the upper noninfiltrated leaves in (C). Total RNA was extracted at 14 days post‐infiltration. Total RNAs for detection of GFP mRNA and GFP‐derived siRNA were visualized by staining with ethidium bromide. The numbers above the detection of GFP siRNA represent the relative accumulation (RA) of GFP siRNA. The RA value of GFP siRNA from the patch expressing β‐glucuronidase (GUS) was set as unity.
As expected, at 5 days post‐infiltration, leaf patches co‐infiltrated with A. tumefaciens carrying the control plasmid p35S:GUS and bacteria carrying p35S:GFP showed weak green fluorescence, whereas patches co‐infiltrated with bacteria harbouring p35S:Fny2b exhibited strong green fluorescence. Patches co‐agroinfiltrated with p35S:Rad2b‐Leu plus p35S:GFP displayed strong GFP fluorescence, comparable with that seen in the presence of p35S:Fny2b. However, when co‐agroinfiltrated with p35S:GFP, p35S:Rad2b‐Pro did not enhance fluorescence, indicating that Rad2b‐Pro was a poor inhibitor of silencing induction (Fig. 4A). This conclusion was supported by northern blot analysis of GFP transcript accumulation in these agroinfiltrated patches (Fig. 4B). Steady‐state accumulation of GFP was enhanced to a similar extent in patches co‐agroinfiltrated with p35S:GFP plus p35S:Rad2b‐Leu or p35S:Fny2b, whereas, in areas co‐agroinfiltrated with p35S:GFP, together with p35S:Rad2b‐Pro, the accumulation of the GFP transcript appeared to be no greater than that observed in patches agroinfiltrated with the control plasmid pBI121 (for clarity, referred to here as p35S:GUS) or patches infiltrated with infiltration solution alone (Fig. 4). Presumably, CMV 2b suppresses RNA silencing by the sequestration and stabilization of small RNAs in planta. To further understand the differential effects of Rad2b‐Pro and Rad2b‐Leu on the suppression of local RNA silencing, we monitored GFP‐derived siRNA accumulation in patch agroinfiltration experiments. The results showed that GFP‐specific siRNA accumulated at a slightly higher level in patches co‐agroinfiltrated with p35S:Rad2b‐Pro than those co‐agroinfiltrated with p35S:GUS, but at a lower level than in patches co‐agroinfiltrated with p35S:Rad2b‐Leu or p35S:Fny2b (Fig. 4B). This suggests that, compared with either Rad2b‐Leu or Fny2b, Rad2b‐Pro has a relatively weak ability to bind siRNA, which is not sufficient to suppress local GFP silencing. However, Rad2b‐Leu, as well as Fny2b, could efficiently bind GFP siRNA, leading to relatively high accumulations of GFP siRNA and mRNA.
As shown above, Rad2b‐Pro appeared to have little or no ability to suppress the initiation of local silencing of GFP in N. benthamiana, but its mutant variant Rad2b‐Leu did (Fig. 4). We found that Rad2b‐Pro and Rad2b‐Leu also have different impacts on systemic silencing, which is mediated by the spread of small RNA duplexes (Dunoyer et al., 2010; Molnar et al., 2010). We examined the induction of systemic silencing of green fluorescence and GFP transcript accumulation in the upper, noninfiltrated leaves of plants of the GFP‐transgenic N. benthamiana line 16c, which is primed for the induction of silencing of GFP gene expression (Brigneti et al., 1998). Fourteen days following co‐agroinfiltration of the lower leaves with p35S:GFP, plus either p35S:Fny2b or p35S:Rad2b‐Leu, there was no diminution of green fluorescence in the upper leaves. However, the upper leaves of plants previously co‐agroinfiltrated in the lower leaves with p35S:GFP, plus either p35S:GUS or p35S:Rad2b‐Pro, showed decreased green fluorescence by this time point (Fig. 4C). Silencing was confirmed by northern blot analysis of GFP transcript and siRNA accumulation in the upper leaves (Fig. 4D). Thus, in plants co‐agroinfiltrated in the lower leaves with p35S:GFP plus either p35S:Rad2b‐Leu or p35S:Fny2b, steady‐state accumulation of GFP transcripts in the upper leaves was undiminished and no GFP‐specific siRNAs were evident (Fig. 4D). However, in plants co‐agroinfiltrated with p35S:GFP and either the control plasmid p35S:GUS or p35S:Rad2b‐Pro, systemic silencing was induced, resulting in increased detectable accumulation of GFP‐specific siRNAs and decreased accumulation of GFP transcripts (Fig. 4D). Taken together, these results demonstrate that the Rad2b‐Pro protein has no ability to inhibit either the initiation of localized RNA silencing or the systemic transmission of the silencing signal. This suggests that, normally, subgroup IB strains would have a proline at position 55 of 2b proteins. However, as stated later here and also shown by Du et al. (2008) and Ye et al. (2009), Rad35 was the only strain (within subgroup IB or among CMV strains in general) that had a proline at position 55 of the 2b protein. Thus, Rad35 is itself a natural mutant variant.
Rad2b‐Leu, but not Rad2b‐Pro, interacts itself in planta
The TAV 2b forms homodimers in vitro which possess a pair of hook‐like structures that provide a perfect domain for the binding of siRNA duplexes (Chen et al., 2008). Furthermore, Chen et al. (2008) established a correlation between dsRNA binding and suppression of RNA silencing by TAV 2b. Subsequently, Gonzalez et al. (2010) demonstrated self‐interaction in vivo of the CMV 2b protein in N. benthamiana using BiFC, and proposed that CMV2b, like TAV2b, functions as a suppressor of RNA silencing in the form of a homodimer or higher order oligomer. Based on the findings that Rad2b‐Pro is unable to bind sRNAs, whereas Rad2b‐Leu works efficiently, we hypothesized that the amino acid at position 55 affects silencing suppression by governing the ability of the Rad2b‐Pro protein to self‐interact in vivo. To test the hypothesis using BiFC, we constructed plasmids encoding fusion proteins of Rad2b‐Pro and Rad2b‐Leu with either the N‐ or C‐terminal domain of a split yellow fluorescent protein (sYFP), termed sYFPN and sYFPC, respectively. We carried out BiFC experiments in transgenic N. benthamiana RDR6i plants (Schwach et al., 2005). These plants are compromised in NbRDR6 expression and silencing, and allow maximal expression of our constructs following introduction by agroinfiltration. Results from the BiFC experiments showed that no fluorescence was observable by microscopic imaging in cells within leaf patches co‐infiltrated with A. tumefaciens cells expressing pRad2b‐Pro‐YFPN and pRad2b‐Pro‐YFPC, but strong YFP fluorescence was detected in both the cytoplasm and nucleus when transiently co‐expressing Rad2b‐Leu‐YFPN and Rad2b‐Pro‐YFPC. When co‐expressing Rad2b‐Leu‐YFPN with YFPC or co‐expressing YFPN with Rad2b‐Leu‐YFPC, no fluorescence was detected. Moreover, we also did not detect fluorescence by co‐expression of Rad2b‐Pro‐YFPN with Rad2b‐Leu‐YFPC (Fig. 5). This observation confirms that the amino acid residue at position 55 controls the self‐association of the IB 2b protein, and suggests that self‐association is a requirement for effective 2b‐mediated silencing suppressor activity.
Figure 5.
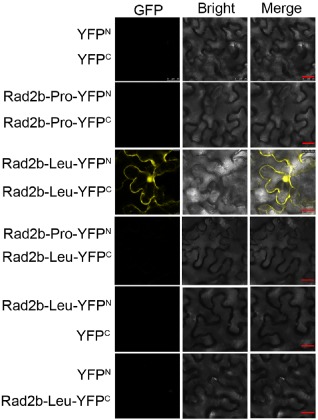
Self‐interaction of 2b proteins in vivo. N icotiana benthamiana RDR 6i was co‐agroinfiltrated to transiently express together 2b proteins (Rad2b‐Pro and Rad2b‐Leu) fused to either the N (Rad2b‐Pro‐YFPN and Rad2b‐Leu‐YFPN) or C (Rad2b‐Pro‐YFPC and Rad2b‐Leu‐YFPC) terminus of yellow fluorescent protein (YFP). Also shown are controls in which the unfused YFP fragments (YFPC and YFPN) were co‐expressed, or expressed with one or other of the 2b‐YFP fusion proteins. Cells in infiltrated tissues were imaged at 5 days after agroinfiltration by confocal scanning laser microscopy for YFP fluorescence and under bright field microscopy. The bar represents 25 μm.
Discussion
Previously, we found that the 2b gene of the subgroup IB strain Rad35‐CMV mediates low virulence on Nicotiana species when exchanged for the homologous gene of the subgroup IA strain Fny‐CMV (Du et al., 2007). In this work, we have shown that Rad2b‐Pro is an extremely weak VSR (Figs 2, 3, 4), resulting in the low virulence of viruses carrying this 2b gene in A. thaliana and Nicotiana species (Fig. 1 and Du et al., 2007). Furthermore, the weak VSR activity of this 2b protein is attributed to the absence of self‐interaction in planta (Fig. 5).
In Arabidopsis, the DCL2, 3 and 4 endoribonucleases are responsible for the production of various siRNAs in vivo (Xie et al., 2004, 2005). siRNA‐mediated antiviral defence against CMV mainly depends on DCL4 and DCL2 in a hierarchical manner (Deleris et al., 2006; Diaz‐Pendon et al., 2007; Ziebell and Carr, 2009). Infection of the CMV 2b deletion mutant CMVΔ2b in the Arabidopsis dcl2/4 double‐mutant plants caused severe symptoms, which were similar to those in wild‐type Arabidopsis plants with infection of wild‐type CMV (Diaz‐Pendon et al., 2007; Ziebell and Carr, 2009). In dcl2 and dcl4 single‐mutant plants, CMV‐FRad2b‐Pro caused symptoms similar to that in wild‐type plants, and its accumulation was increased to a very limited extent (Fig. 1). In transgenic Arabidopsis, constitutive expression of the Fny2b protein, an effective VSR, rescued the accumulation of the 2b‐deletion mutant Fny‐CMVΔ2b by inhibiting antiviral silencing (Lewsey et al., 2009). Similarly, the virus titre of Fny‐CMVΔ2b was increased in Rad2b‐Leu‐transgenic Arabidopsis plants, compared with that in wild‐type plants (Fig. 3). However, transgenic expression of Rad2b‐Pro did not rescue the accumulation of CMV‐FRad2b‐Pro (Fig. 3). All of these results indicate that Rad2b‐Pro could not efficiently suppress antiviral silencing governed by DCL2 or DCL4.
In independent studies, the 2b proteins from a subgroup IA strain (Fny‐CMV) and the subgroup II strains LS‐CMV and Q‐CMV have both been shown to inhibit effectively the initiation of silencing (Guo and Ding, 2002; Lewsey et al., 2007). Likewise, Ye et al. (2009) found that SD2b (subgroup IB) is an effective VSR, which exhibits stronger suppression of post‐transcriptional gene silencing (PTGS) than Q2b. The present work showed that Fny2b exhibits much stronger suppression of PTGS than does Rad2b‐Pro. All of these comparisons demonstrate that differential CMV 2b genes possess variable VSR activities. Although Rad2b‐Pro did not show effective suppression of antiviral RNA silencing in Nicotiana species or A. thaliana when expressed in the heterologous Fny‐CMV background (Fig. 1; Du et al., 2007), we cannot rule out the possibility that it may have VSR activity in its parent Rad35‐CMV context. Indeed, the infection of Rad35‐CMV caused obvious viral symptoms in the Nicotiana species tested, and these were more severe than symptoms caused by the hybrid virus CMV‐FRad2b‐Pro (Du et al., 2007). If we assume that virulence is linked with effective VSR activity (either directly or indirectly by increasing the viral titre), this suggests that, for Rad2b‐Pro to function as an effective VSR, it may need to interact with other viral components encoded by its parent, Rad35‐CMV. Potential interactions of CMV 2b with other CMV components have been proposed previously (Ding et al., 1996; Shi et al., 2002).
Gonzalez et al. (2010) provided evidence that Fny2b self‐interacts in planta, but that the deletion of domains affecting nuclear localization, including NLS1, NLS2 and KSPSE, does not affect Fny2b self‐association. Here, we showed that the substitution of proline with leucine of Rad2b‐Pro at position 55 rescued the self‐interaction of the 2b protein (Fig. 5), indicating that position 55 of CMV 2b proteins is of pivotal importance to 2b self‐interaction. All CMV 2b genes identified to date encode proteins with a leucine residue at position 55, with the exception of the Rad35‐CMV 2b gene (Du et al., 2008). The high conservation at position 55 demonstrates the importance of CMV2b self‐interaction. It seems unlikely that proline at position 55 affects the local secondary structure of Rad2b‐Pro because the residue at position 56 is also proline. We speculate that it alters higher order structures, which are responsible for 2b self‐interaction. The TAV 2b forms homodimers in vitro, producing a pair of hook‐like structures that provide a perfect domain for the binding of siRNA duplexes, which correlates with the suppression of RNA silencing (Chen et al., 2008). Gonzalez et al. (2010) provided evidence that Fny2b self‐interacts in planta, which was supported by subsequent experimental data showing that Fny2b binds small RNA with high affinity at a 2:1 molar ratio (Gonzalez et al., 2012). The substitution of proline with leucine renders Rad2b‐Pro self‐interactive and dramatically enhances its VSR activity, indicating strongly that self‐interaction of CMV 2b is a requirement for VSR activity.
In addition to controlling physical 2b self‐interaction, the amino acid at position 55 could be involved in 2b function. One of the biological functions of CMV 2b is the inhibition of AGO1 activity by direct interaction (Duan et al., 2012; Feng et al., 2013; Gonzalez et al., 2010; Zhang et al., 2006). The 2b–AGO1 interaction is needed to affect trans‐acting siRNA by the inhibition of AGO1 (Feng et al., 2013), but is dispensable for CMV 2b to suppress PTGS (Duan et al., 2012; Gonzalez et al., 2010). Moreover, position 55 is not covered by the AGO binding and nucleolar localization signal domains, which are required for the inhibition of in vivo AGO1 slicer activity (Duan et al., 2012; Feng et al., 2013). These findings allow us to conclude that the change in amino acid at position 55 does not affect the 2b–AGO1 interaction or inhibition of AGO1 slicer function. Recently, several studies have demonstrated the importance of binding to small RNA for 2b to suppress RNA silencing, as 2b VSR activity is compromised when mutations or deletions are created in the small RNA binding domain, the N‐terminal 61‐amino‐acid sequence of CMV 2b (Duan et al., 2012; Gonzalez et al., 2010, 2012; Goto et al., 2007). Our present data do not allow us to completely rule out the possibility that the effects of a single amino acid change at position 55 could be pleiotropic, for example by affecting small RNA sequestration, as well as controlling dimerization.
Experimental Procedures
Plant materials and viruses
Arabidopsis thaliana Heyn. ecotype Col‐0 wild‐type and lines carrying single dcl2‐1, dcl3‐1, dcl4‐2 or triple dcl2/3/4 mutations have been described previously (Deleris et al., 2006; Henderson et al., 2006; Xie et al., 2004, 2005), as has Fny2b‐transgenic Arabidopsis line 3.13F (Lewsey et al., 2007). Nicotiana benthamiana transgenic line 16c, constitutively expressing enhanced GFP, has been described previously (Ruiz et al., 1998), as has N. benthamiana transgenic line RDR6i (Schwach et al., 2005). Arabidopsis plants were grown under an 8‐h photoperiod and a light intensity of 150–200 μE/m2/s at 22 °C. N. benthamiana plants were grown under a 16‐h photoperiod with a light intensity of 150–200 μE/m2/s at 25 °C.
CMV‐FRad2b‐Pro and CMV‐FRad2b‐Leu, which have been described previously (Du et al., 2008), were propagated in N. clevelandii, and purified using the method described by Ng and Perry (2004). Purified virions at a concentration of 100 ng/μL were rub inoculated onto Arabidopsis seedlings at the five to six true‐leaf stage. Successful infection was confirmed by symptom observation for CMV‐FRad2b‐Leu, and by double antibody sandwich enzyme‐linked immunosorbent assay (DAS‐ELISA) detection for CMV‐FRad2b‐Pro.
Plasmid construction and A rabidopsis transformation
A pBI121‐derived plasmid with the GFP sequence under the control of the cauliflower mosaic virus 35S promoter (p35S:GFP) has been described previously (Brigneti et al., 1998). For transient and stable expression of 2b‐derived transgenes, plasmid constructs p35S:Fny2b, p35S:Rad2b‐Pro and p35S:Rad2b‐Leu were generated by replacement of the GUS gene in the binary plasmid pBI121 by Fny2b, Rad2b‐Pro and Rad2b‐Leu, respectively, which were obtained by polymerase chain reaction (PCR) from the corresponding infectious cDNA clones (Du et al., 2007, 2008) using 2b gene‐specific primers (Table S1, see Supporting Information). For BiFC assays, Rad2b‐Pro and Rad2b‐Leu sequences were PCR amplified using 2b gene‐specific primers (Table S1), and then introduced into plasmids pSPYNE‐35S and pSPYCE‐35S (Walter et al., 2004) to generate pRad2b‐Pro‐YFPN, pRad2b‐Pro‐YFPC, pRad2b‐Leu‐YFPN and pRad2b‐Leu‐YFPC. Prior to their use in plant transformation, plasmid construction was authenticated by DNA sequencing. Plasmids were transformed into A. tumefaciens GV3101 by electroporation using a Gene Pulser (Bio‐Rad, Hercules, CA, USA) according to the manufacturer's instructions, and positive transformants were selected on appropriate antibiotic‐containing medium. Transformation was performed by floral dipping (Clough and Bent, 1998), and transformed Arabidopsis plant lines were selected on kanamycin‐containing solid Murashige and Skoog medium and checked by western blotting using anti‐2b serum (Gonzalez et al., 2010).
Assay of suppressor activity
To test the suppressor activities of wild‐type Rad35‐2b and its mutant, A. tumefaciens cells carrying p35S:GFP were equally mixed with an equal volume of cells carrying p35S:Rad2b‐Pro, p35S:Rad2b‐Leu, p35S:Fny2b or p35S:GUS (control plasmid: for clarity, referred to in this work as p35S:GUS). A. tumefaciens (at a final cell density equivalent to A 600 = 1.0) was infiltrated into the fifth and sixth true leaves of GFP‐transgenic line 16c N. benthamiana plants. The infiltration solution employed was composed of 10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES) and 100 μm acetosyringone, and was used alone as a control in some experiments. GFP fluorescence in the infiltrated and noninfiltrated leaves was recorded under UV light using a Nikon Coolpix digital camera (Nikon, Tokyo, Japan) at 5 and 14 days post‐infiltration, respectively.
BiFC assays
All plasmids used for BiFC assays were separately transformed into A. tumefaciens GV3101. A. tumefaciens cultures of cells carrying plasmids with the corresponding sYFPN and YFPC fusions to the same 2b genes were co‐infiltrated together into the fifth and sixth true leaves of N. benthamiana RDR6i plants (Schwach et al., 2005). At 5 days post‐infiltration, YFP fluorescence was observed using a Leica SP5 confocal laser scanning microscope (Leica Microsystems, Mannheim, German).
Western immunoblotting
To solubilize 2b proteins from the nuclear/cytoskeletal fraction in one step, leaf tissue was pulverized in liquid nitrogen and the proteins were extracted in phosphate‐buffered saline (0.14 m NaCl, 0.010 m potassium phosphate, pH 7.4) supplemented with 2% (v/v) 2‐mercaptoethanol. CMV CP was extracted as described previously (Naylor et al., 1998). Protein extracts were separated by electrophoresis on sodium dodecylsulphate (SDS)‐containing 15% polyacrylamide gels and transferred electrophoretically to nitrocellulose membranes (Whatman, Ltd., Maidstone, UK). Protein transfer and lane loadings were checked by Ponceau S staining. Membranes were probed using polyclonal anti‐2b mouse (Gonzalez et al., 2010) or anti‐CP rabbit sera, and primary antibody binding was detected using horseradish peroxidase (HRP)‐conjugated anti‐mouse or anti‐rabbit immunoglobulin G (IgG) and a chemiluminescence reagent kit (Perkin‐Elmer, Norwalk, CT, USA), as described previously (Lewsey and Carr, 2009; Naylor et al., 1998).
Northern blot analysis
Total RNA was extracted from leaf tissues using TRIzol (Invitrogen‐Life Technologies, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. Northern blotting for the analysis of CMV genomic RNAs and GFP mRNA was carried out according to the procedure described previously (Du et al., 2007). The DNA oligonucleotide ProbeI‐40, described previously (Du et al., 2007), was labelled with biotin at its 3′ end to probe CMV genomic RNAs. The biotin‐labelled probe was detected using a Biotin‐Labelling Detection Kit (Beyotime, Beijing, China). 32P‐Labelled probe for GFP transcripts was made using a random‐priming DNA labelling kit (Takara, Dalian, China) and α[32P]‐dCTP. Signal intensities of CMV‐specific bands were quantified using the program Gel Pro Analyzer 4.0 (Media Cybernetics, Rockville, MD, USA), and employed to calculate the relative accumulations of CMV. For the analysis of GFP‐specific siRNA, northern blotting was performed according to the protocol described in the instruction manual of the miRVana miRNA Isolation Kit (Ambion: http://products.invitrogen.com/ivgn/product/AM1560, Austin, TX, USA). GFP‐derived siRNAs were detected using a mixture of four DNA oligonucleotides (Table S2, see Supporting Information), 32P‐labelled at their 5′ ends using T4 polynucleotide kinase (NEB, Inc., Beverly, MA, USA) and γ[32P]‐ATP. The signal intensities of bands corresponding to GFP siRNAs were quantified using the program Gel Pro Analyzer 4.0 (Media Cybernetics).
Supporting information
Fig. S1 Identification of primary Rad35‐Pro transgenic lines using immunoblot detection of 2b proteins employing a polyclonal antibody raised against Fny2b. NT indicates nontransgenic plant.
Fig. S2 Phenotypes of plants of transgenic lines expressing similar levels of the 2b proteins Rad2b‐Pro and Rad2b‐Leu. Plants were photographed when they were 4 weeks old.
Fig. S3 Comparison of expression levels of 2b proteins in transgenic lines harbouring Rad2b‐Pro and Rad2b‐Leu transgenes. Four‐week‐old plants were used for immunoblot detection of 2b proteins employing a polyclonal antibody raised against Fny2b.
Table S1 Primers used for the generation of plasmids.
Table S2 Oligonucleotide probes used for the detection of green fluorescent protein (GFP)‐derived short‐interfering (si)RNAs.
Acknowledgements
We thank Susan V. Howroyd for assistance with Arabidopsis transformation, David Baulcombe for the plasmid p35S:GFP and seeds of N. benthamiana lines RDR6i and 16c, Tao Zhou for BiFC plasmids, Jim Carrington for providing seeds for dcl mutant lines, and Tomas Canto for anti‐2b serum. This work was financially supported by the National Natural Science Foundation of China (30800043, 31170141), Zhejiang Natural Science Foundation of China (Y3080315, Y3090657), the Zhejiang Provincial 151 Talents Development Project (11610632501102) and a Marie Curie International Incoming Fellowship (PIIF‐GA‐2009–236443). Work in JPC's laboratory was supported by grants from the Leverhulme Trust (F/09741/F, RPG‐2012‐667) and the UK Biotechnology and Biological Sciences Research Council (BB/D014376/1, BB/J011762/1).
References
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen, H.Y. , Yang, J. , Lin, C. and Yuan, Y.A. (2008) Structural basis for RNA‐silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deleris, A. , Gallego‐Bartolome, J. , Bao, J. , Kasschau, K.D. , Carrington, J.C. and Voinnet, O. (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science, 313, 68–71. [DOI] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. , Li, F. , Li, W.X. and Ding, S.W. (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell, 19, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. , Anderson, B.J. , Haase, H.R. and Symons, R.H. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology, 198, 593–601. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. , Shi, B.J. , Li, W.X. and Symons, R.H. (1996) An interspecies hybrid RNA virus is significantly more virulent than either parental virus. Proc. Natl. Acad. Sci. USA, 93, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z. , Chen, F. , Zhao, Z. , Liao, Q. , Palukaitis, P. and Chen, J. (2008) The 2b protein and the C‐terminus of the 2a protein of cucumber mosaic virus subgroup I strains both play a role in viral RNA accumulation and induction of symptoms. Virology, 380, 363–370. [DOI] [PubMed] [Google Scholar]
- Du, Z.Y. , Chen, F.F. , Liao, Q.S. , Zhang, H.R. , Chen, Y.F. and Chen, J.S. (2007) 2b ORFs encoded by subgroup IB strains of cucumber mosaic virus induce differential virulence on Nicotiana species. J. Gen. Virol. 88, 2596–2604. [DOI] [PubMed] [Google Scholar]
- Duan, C.G. , Fang, Y.Y. , Zhou, B.J. , Zhao, J.H. , Hou, W.N. , Zhu, H. , Ding, S.W. and Guo, H.S. (2012) Suppression of Arabidopsis ARGONAUTE1‐mediated slicing, transgene‐induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell, 24, 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P. , Schott, G. , Himber, C. , Meyer, D. , Takeda, A. , Carrington, J.C. and Voinnet, O. (2010) Small RNA duplexes function as mobile silencing signals between plant cells. Science, 328, 912–916. [DOI] [PubMed] [Google Scholar]
- Feng, L. , Duan, C.G. and Guo, H.S. (2013) Inhibition of in vivo Slicer activity of Argonaute protein 1 by the viral 2b protein independent of its dsRNA‐binding function. Mol. Plant Pathol. DOI: 10.1111/mpp.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, I. , Martinez, L. , Rakitina, D.V. , Lewsey, M.G. , Atencio, F.A. , Llave, C. , Kalinina, N.O. , Carr, J.P. , Palukaitis, P. and Canto, T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant–Microbe Interact. 23, 294–303. [DOI] [PubMed] [Google Scholar]
- Gonzalez, I. , Rakitina, D. , Semashko, M. , Taliansky, M. , Praveen, S. , Palukaitis, P. , Carr, J.P. , Kalinina, N. and Canto, T. (2012) RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA, 18, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, K. , Kobori, T. , Kosaka, Y. , Natsuaki, T. and Masuta, C. (2007) Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA‐binding abilities. Plant Cell Physiol. 48, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Guo, H.S. and Ding, S.W. (2002) A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamera, S. , Song, X. , Su, L. , Chen, X. and Fang, R. (2011) Cucumber mosaic virus suppressor 2b binds to AGO4‐related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115. [DOI] [PubMed] [Google Scholar]
- Harvey, J.J. , Lewsey, M.G. , Patel, K. , Westwood, J. , Heimstadt, S. , Carr, J.P. and Baulcombe, D.C. (2011) An antiviral defense role of AGO2 in plants. Plos ONE 6, e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R. , Zhang, X. , Lu, C. , Johnson, L. , Meyers, B.C. , Green, P.J. and Jacobsen, S.E. (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38, 721–725. [DOI] [PubMed] [Google Scholar]
- Jacquemond, M. (2012) Cucumber mosaic virus. Adv Virus Res. 84, 439–504. [DOI] [PubMed] [Google Scholar]
- Lanet, E. , Delannoy, E. , Sormani, R. , Floris, M. , Brodersen, P. , Crete, P. , Voinnet, O. and Robaglia, C. (2009) Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell, 21, 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey, M. , Robertson, F.C. , Canto, T. , Palukaitis, P. and Carr, J.P. (2007) Selective targeting of miRNA‐regulated plant development by a viral counter‐silencing protein. Plant J. 50, 240–252. [DOI] [PubMed] [Google Scholar]
- Lewsey, M. , Surette, M. , Robertson, F.C. , Ziebell, H. , Choi, S.H. , Ryu, K.H. , Canto, T. , Palukaitis, P. , Payne, T. , Walsh, J.A. and Carr, J.P. (2009) The role of the Cucumber mosaic virus 2b protein in viral movement and symptom induction. Mol. Plant–Microbe Interact. 22, 642–654. [DOI] [PubMed] [Google Scholar]
- Lewsey, M.G. and Carr, J.P. (2009) Effects of DICER‐like proteins 2, 3 and 4 on cucumber mosaic virus and tobacco mosaic virus infections in salicylic acid‐treated plants. J. Gen. Virol. 90, 3010–3014. [DOI] [PubMed] [Google Scholar]
- Molnar, A. , Melnyk, C.W. , Bassett, A. , Hardcastle, T.J. , Dunn, R. and Baulcombe, D.C. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science, 328, 872–875. [DOI] [PubMed] [Google Scholar]
- Naylor, M. , Murphy, A.M. , Berry, J.O. and Carr, J.P. (1998) Salicylic acid can induce resistance to plant virus movement. Mol. Plant–Microbe Interact. 11, 860–868. [Google Scholar]
- Ng, J.C. and Perry, K.L. (2004) Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5, 505–511. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. and Garcia‐Arenal, F. (2003) Cucumoviruses. Adv. Virus Res. 62, 241–323. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T. , Voinnet, O. and Baulcombe, D.C. (1998) Initiation and maintenance of virus‐induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, K.B. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, B.J. , Palukaitis, P. and Symons, R.H. (2002) Differential virulence by strains of Cucumber mosaic virus is mediated by the 2b gene. Mol. Plant–Microbe Interact. 15, 947–955. [DOI] [PubMed] [Google Scholar]
- Soards, A.J. , Murphy, A.M. , Palukaitis, P. and Carr, J.P. (2002) Virulence and differential local and systemic spread of cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol. Plant–Microbe Interact. 15, 647–653. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schutze, K. , Batistic, O. , Weckermann, K. , Nake, C. , Blazevic, D. , Grefen, C. , Schumacher, K. , Oecking, C. , Harter, K. and Kudla, J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Johansen, L.K. , Gustafson, A.M. , Kasschau, K.D. , Lellis, A.D. , Zilberman, D. , Jacobsen, S.E. and Carrington, J.C. (2004) Genetic and functional diversification of small RNA pathways in plants. Plos Biol. 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z. , Allen, E. , Wilken, A. and Carrington, J.C. (2005) DICER‐LIKE 4 functions in trans‐acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 102, 12 984–12 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Qu, J. , Zhang, J.F. , Geng, Y.F. and Fang, R.X. (2009) A critical domain of the Cucumber mosaic virus 2b protein for RNA silencing suppressor activity. FEBS Lett. 583, 101–106. [DOI] [PubMed] [Google Scholar]
- Ye, K. , Malinina, L. and Patel, D.J. (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature, 426, 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, R. , Wang, W. , Iki, T. , Liu, C. , Wu, Y. , Ishikawa, M. , Zhou, X. and Qi, Y. (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell, 46, 859–870. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell, H. and Carr, J.P. (2009) Effects of dicer‐like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by cucumber mosaic virus and a mutant virus lacking the 2b counter‐defence protein gene. J. Gen. Virol. 90, 2288–2292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of primary Rad35‐Pro transgenic lines using immunoblot detection of 2b proteins employing a polyclonal antibody raised against Fny2b. NT indicates nontransgenic plant.
Fig. S2 Phenotypes of plants of transgenic lines expressing similar levels of the 2b proteins Rad2b‐Pro and Rad2b‐Leu. Plants were photographed when they were 4 weeks old.
Fig. S3 Comparison of expression levels of 2b proteins in transgenic lines harbouring Rad2b‐Pro and Rad2b‐Leu transgenes. Four‐week‐old plants were used for immunoblot detection of 2b proteins employing a polyclonal antibody raised against Fny2b.
Table S1 Primers used for the generation of plasmids.
Table S2 Oligonucleotide probes used for the detection of green fluorescent protein (GFP)‐derived short‐interfering (si)RNAs.


