Summary
The control of rhizomania, one of the most important diseases of sugar beet caused by the Beet necrotic yellow vein virus, remains limited to varietal resistance. In this study, we investigated the putative action of Bacillus amylolequifaciens lipopeptides in achieving rhizomania biocontrol through the control of the virus vector Polymyxa betae. Some lipopeptides that are produced by bacteria, especially by plant growth‐promoting rhizobacteria, have been found to induce systemic resistance in plants. We tested the impact of the elicitation of systemic resistance in sugar beet through lipopeptides on infection by P. betae. Lipopeptides were shown to effectively induce systemic resistance in both the roots and leaves of sugar beet, resulting in a significant reduction in P. betae infection. This article provides the first evidence that induced systemic resistance can reduce infection of sugar beet by P. betae.
Polymyxa betae Keskin (Keskin, 1964) belongs to the Plasmodiophorids, which is now included in the Phytomyxea in the phylum Cercozoa (Adl et al., 2005; Bass et al., 2009). It is an obligate biotrophic parasite of the roots of sugar beet, acting as a vector of the rhizomania disease of sugar beet, caused by Beet necrotic yellow vein virus (BNYVV). Until now, the control of this major disease has been managed through varietal resistance to BNYVV. The control of P. betae in the field remains limited to cultural practices, such as the management of the pH of the soil (Goffart and Maraite, 1991) because effective chemical treatments, such as methyl bromide, have been forbidden (United Nations Environment Programme (UNEP), 1987). Barr et al. (1995) and Asher et al. (2009) have shown the potential of breeding sugar beet for protist resistance from the resistance genes of wild Beta species, but this approach is still not used by breeders because of lower productivity and the unknown long‐term impact of these varieties on the disease. In this context, biological control of the protist vector of rhizomania could be an interesting alternative to breeding to reduce disease pressure.
In many instances, biological control of microbial diseases is obtained after the inoculation of plant beneficial organisms that directly hamper the development of the targeted pathogens. Another interesting biocontrol mechanism relies on the stimulation of the natural defences of the host plant by these beneficial microbes or their products. Plants have developed various strategies to combat aggressors (Van Loon et al., 1998). One of these strategies is a defence reaction in the tissues surrounding the initial infection site, a phenomenon known as ‘localized acquired resistance’. Such an elevated resistance level, however, can spread throughout the plant via the emission of molecular signals that will reach distal tissues, rendering the whole plant less susceptible to subsequent pathogen attack. This phenomenon, widely reviewed in recent years, is known as ‘systemic acquired resistance’ (SAR). It is commonly triggered by the elicitors of avirulent pathogens, such as microbial‐associated molecular patterns (MAMPs) (Abramovitch et al., 2006), but it can also be induced by biological (nonmicrobial) and chemical compounds. In the biocontrol context, another interesting form of systemic resistance in plants is that which is referred to here as “induced systemic resistance (ISR)”, and is induced by nonpathogenic but plant growth‐promoting microorganisms, including fungi (plant growth‐promoting fungi, PGPFs) and rhizobacteria (plant growth‐promoting rhizobacteria, PGPRs). Among the PGPRs, species in the Pseudomonas and Bacillus genera are the most well known (Raaijmakers et al., 2010). Phenotypically, ISR is quite similar to SAR, making the plant resistant to subsequent attacks of pathogenic organisms, such as viruses, bacteria and fungi (Bakker et al., 2007). The signalling of these systemic resistances is controlled by salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), with SA involved in the case of SAR, and JA and ET associated mainly with the signalling of ISR. These two apparently independent signalling routes contain cross‐talks and converge through the same transcriptional regulator, nonexpressor of pathogenesis‐related (PR) genes 1 (NPR‐1) (Katagiri and Tsuda, 2010; Shah, 2009).
The systemic resistances do not confer total resistance to a pathogen, but they provide long‐lasting increased resistance in a large number of plants against a broad range of pathogens. Some chemicals, such as SA or analogues [benzothiadiazole (BTH) and its derivatives, e.g. 2,6‐dichloronicotinic acid], are known to induce SAR and have been successfully used in the field to control diseases (Vallad and Goodman, 2004).
With regard to ISR, field or glasshouse trials with inducing organisms have shown its potential to control several diseases (Bent, 2005; Kloepper et al., 2004). For sugar beet, systemic resistance induced by nonpathogenic species was tested successfully in two pathosystems: Pseudomonas fluorescens enabled the control of Heterodera schachtii (Bargabus et al., 2002), whereas Bacillus mycoides and B. pumilus efficiently controlled Cercospora beticola (Bargabus et al., 2004).
Based on many promising results, research was conducted on the development of microbial formulations that could be used in agriculture. Two classes of bacterial biosurfactant were found to be elicitors of ISR: rhamnolipids and cyclic lipopeptides (cLPs). cLPs produced by Pseudomonas and Bacillus were shown to elicit ISR. Massetolide A from Pseudomonas fluorescens elicited ISR and enabled Phytophthora infestans on tomato to be controlled (Tran et al., 2007). Pure fengycins and surfactins triggered a significant protective effect, similar to that induced by the producing Bacillus strains. In addition, the overexpression of surfactin and fengycin genes in poor cLP‐producing strains was associated with a higher level of resistance (Ongena et al., 2007). The ISR activity of surfactin was associated, in treated plants, with the accumulation of antifungal compounds (phytoalexins) (Adam, 2008) and with the stimulation of the lipoxygenase pathway, leading to the synthesis of fungitoxic oxylipins (Ongena et al., 2007). The mechanism of ISR by cLPs is not yet clear, but a recent study strongly suggests that the plant cell recognition of surfactin is mediated through interaction with lipids at the plasma membrane level, rather than through specific protein receptors (Henry et al., 2011). This lipid bilayer perturbation does not affect cell viability, but is sufficient to trigger a cascade of molecular events leading to an increased defence response (Jourdan et al., 2009). In order to prevent rhizomania in sugar beet more efficiently, it is necessary to combine classical strategies, such as the breeding of varieties that can resist the virus, with control of the vector of the disease. This study sought to evaluate the potential of controlling P. betae by inducing systemic resistance in sugar beet using Bacillus cLPs.
The potential of using Bacillus cLPs to induce systemic resistance in sugar beet against P. betae infection was tested in a bioassay conducted under controlled conditions. Pre‐germinated seeds of sugar beet (var. Cadyx) were transferred in individual glass tubes containing sterilized quartz, and were incubated in a growth chamber with a 14‐h day/10‐h night photoperiod, with temperatures of 25 °C and 20 °C, respectively. The plants were watered every 2 days with Hoagland solution, pH 7.2. Semi‐purified cLPs [80% purity established by reverse‐phase high‐performance liquid chromatography (HPLC) coupled with a single quadrupole mass spectrometer] produced by Bacillus amylolequifaciens strain S499 were used for plant treatment. This extract contained a mixture of compounds belonging to the three cLP families—surfactin, iturin and fengycin—produced by strain S499 in the relative proportions of 55/22/23 (v/v/v), respectively. The extract was produced after growth under laboratory conditions in a medium that had been established for the enhanced production of such compounds (Nihorimbere et al., 2011). Secreted cLPs were submitted to acid precipitation and solid phase extraction on a C18 cartridge in order to recover an 80% pure solution, as determined by reverse‐phase HPLC coupled with electrospray ionization mass spectrometry, following a procedure described by Nihorimbere et al. (2011). The solution of cLPs for plant treatment was prepared by dilution of the methanolic cLP extract with sterile MilliQ water. Two treatments were applied 3 weeks and 1 month after plant germination: each plant was watered with 5 mL of cLP solution (60 mg/L) and 5 mL of Hoagland solution, pH 7.2. The final concentration of cLPs in the solution surrounding the roots was 30 mg/L. The control plants were treated with 5 mL of sterile MilliQ water and 5 mL of Hoagland water, pH 7.2. Twenty‐four plants were treated with each concentration [0 mg/L (controls) and 30 mg/L].
Fourteen days after the first treatment with cLPs or water, 20 plants per treatment were inoculated with P. betae zoospores. The sugar beet age at inoculation was chosen to assess the effect of cLPs during an active growth stage of the plant: at BBCH stage 14–15 during the development of leaves. At this stage, the primary root and numerous secondary roots are present and likely to be infected after P. betae inoculation. The cLP treatments preceding P. betae inoculation were made 7 and 14 days before inoculation, according to time periods commonly and successfully used to test the ISR‐eliciting potential of cLPs and other elicitors on other plant species (Henry et al., 2011; Ongena et al., 2005, 2007). In order to obtain the required mobile stage, the aviruliferous monosporosorus P. betae strain A26‐41, collected from a rhizomania‐free field at Opprebais in Belgium in 1987, was used (Legrève et al., 1998). The multiplication of this strain was achieved by growing sugar beet (Beta vulgaris var. Cadyx) plants on a quartz–sporosori mixture using an automatic immersion system in an environmental cabinet at 20–25 °C, as described by Legrève et al. (1998). Large quantities of zoospores were obtained from the roots of young plants, as described by Desoignies et al. (2011). Two suspensions of 100 and 1000 zoospores/mL were prepared for the inoculation of plants treated with cLPs or water. Ten plants per treatment were inoculated with 500 zoospores each and another 10 with 5000 zoospores each. After inoculation, the quartz substrate was saturated with Hoagland solution, pH 7.2. The elicited, but uninoculated, plants were harvested on the day on which the other plants were inoculated in order to assess whether ISR was effectively induced by cLP treatment at the time of inoculation. The inoculated plants (cLP‐ and water‐treated) were collected 7 days after inoculation with P. betae zoospores. For the harvest, each root system was rinsed in demineralized water, and 200 mg of fresh tissues were collected from each root system. Nucleic acids were extracted in 1 mL of polysomes buffer (Jupin et al., 1990) in 2‐mL microtubes with 0.25‐in ceramic spheres using the FastPrep© instrument (Qbiogene, Irvine, CA, USA). This step was followed by phenol extraction and ethanol precipitation.
We wanted to test whether the cLP treatment could have an impact on the infection of sugar beet by P. betae. Using quantitative polymerase chain reaction (qPCR), the infection was evaluated 7 days after inoculation with two concentrations of zoospores (500 and 5000 per plant). A qPCR was conducted on the root extracts of P. betae‐inoculated plants with or without previous cLP treatment. SYBR‐Green chemistry was used in this study. Two repetitions of the qPCR were made for each sample, standard, control or blank. The reaction components for each sample were 20 μL of iQ SYBR Green Supermix, 0.8 μL of each primer (primers used are shown in Table 1) , 14.4 μL of diethylpyrocarbonate (DEPC)‐treated water and 4 μL of the 20‐fold‐diluted DNA extract. An amplification reaction was performed using the iCycler iQ Real Time detection system (Biorad, Hercules, CA, USA), as follows: first, a denaturation step at 95 °C for 3 min; then 40 cycles of 30 s at 95 °C, 30 s at 62 °C and 30 s at 72 °C; and, finally, melting curve analysis. The DNA standards for Beta vulgaris glutamine synthetase were obtained from serial dilutions of a phenol–chloroform extraction of 500 mg of fresh foliar tissues. For P. betae standards, PCR products were cloned in PGEM‐T vector (Promega, Madison, WI, USA). Serial dilutions of these two standards were used to construct the standard curves for the different experiments. The detection threshold was fixed at a fluorescence of 150 for P. betae quantification and at a fluorescence of 250 for sugar beet glutamine synthetase. The quantification cycles (C q) were used to compare the different samples. A mean was calculated for the two replicates of each reaction. The relative C q was calculated, corresponding to the quantity of P. betae per sugar beet cell unit: for the same extract, the C q value corresponding to the two replicates of qPCR targeting the P. betae sequence was divided by the C q value corresponding to the two replicates of qPCR for sugar beet glutamine synthetase. For each plant tested, the relative value of C q was then obtained. For each concentration of zoospores, an ANOVA‐1 was applied, with the concentration in cLPs as the explicative variable. Various statistical tests of mean comparison (Tukey, Dunett, Scheffe and Student–Newman–Keuls) were applied between the elicited and nonelicited plants. All these tests were performed using SAS enterprise guide 2.0 (SAS, Cary, NC, USA). A replication of the experiment was performed. In this replication, three additional plants were added per treatment and grown until 15 days post‐infection in order to assess P. betae infection in roots. The roots of these plants were harvested and stained with blue lactophenol. Five roots of 3 cm each were randomly collected from each plant and P. betae infection was quantified through microscopic observation.
Table 1.
Primers used. The primers were designed with ePrimer (National Center for Biotechnology Information, NCBI)
| Organism | Targeted region | Use | Primer name | Sequence | Reference |
|---|---|---|---|---|---|
| Polymyxa betae | Internal transcribed spacer 1 | qPCR |
Pb1 Psp2rev |
5′‐GGAATTTGAACAAGTGACTTGG‐3′ 5′‐AGGGCTCTCGAAAGCGCAA‐3′ |
Legrève et al., 2003 |
| Beta vulgaris | Glutamine synthetase gene | qPCR |
GSBvFor GSBvRev |
5′‐AGGGTGATTGGAATGGTGCT‐3′ 5′‐ACTTCTCGATGGCAGCCTTT‐3′ |
This study |
| Beta vulgaris | NPR‐1 gene | RT‐PCR |
NPR1BvF NPR1BvR |
5′‐TCATGAAGCTTGTCGTCCTG‐3′ 5′‐ATACACCTTGCCAGCAATCC‐3′ |
This study |
| Beta vulgaris | PR‐8 gene (class III chitinase) | RT‐PCR |
Chit3BvF Chit3BvR |
5′‐GCTGAACTTAGCTGGGCACT‐3′ 5′‐CTGGACTGACCCCCAAGATA‐3′ |
This study |
qPCR, quantitative polymerase chain reaction; RT‐PCR, reverse transcription‐polymerase chain reaction.
Our data showed that treatment with cLPs reduced significantly the infection of plants by P. betae compared with the negative controls (water‐treated plants). The normalized C q value increased from 1.131 to 1.326 (repetition 1) and from 1.444 to 1.536 (repetition 2) when 500 zoospores were inoculated, and from 1.018 to 1.169 (repetition 1) and from 1.352 to 1.522 (repetition 2) when 5000 zoospores were inoculated. All comparisons of means using Student–Newman–Keuls, Tukey, Scheffe and t‐test showed a significant difference. A boxplot of the dispersion of the relative C q for the first repetition is shown in Fig. 1. These results indicate that infection was reduced by more than 93% (repetition 1) and 74% (repetition 2) when 500 zoospores were inoculated, and by more than 88% (repetition 1) and 93% (repetition 2) when 5000 zoospores were inoculated (Table 2, Fig. 2). The reduction in P. betae infection assessed by qPCR was confirmed by microscopy. No infection was visible in the roots inoculated with 500 zoospores, but a difference was observed in the plants inoculated with 5000 zoospores. Eleven of the 15 nonelicited root fragments collected from plants inoculated with 5000 zoospores were infected by P. betae plasmodia and zoosporangia, whereas only two fragments of the 15 from cLP‐treated plants were infected. Zoosporangia and plasmodia were observed in unelicited fragments, only plasmodia in elicited ones.
Figure 1.
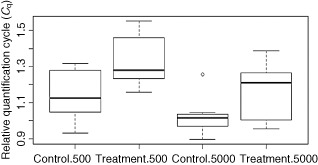
Boxplot of relative C q (C q Polymyxa betae/C q Beta vulgaris) obtained by quantitative polymerase chain reaction (qPCR) for the quantification of the infection rate of sugar beet by P. betae in the first repetition. Control refers to unelicited plants; treatment refers to elicitation with lipopeptides (30 mg/L); 500 and 5000 are the numbers of P. betae zoospores inoculated per plant.
Table 2.
Reduction in Polymyxa betae infection rates in lipopeptides (cLP)‐treated plants
| Experiment | Zoospores inoculated | Difference in P. betae‐normalized C q | Mean sugar beet C q | ΔCycles | Reduction factor | Reduction rate |
|---|---|---|---|---|---|---|
| 1 | 500 | 0.195 | 20.436 | 3.98 | 15.78 | 93.66 |
| 1 | 5000 | 0.151 | 20.32 | 3.07 | 8.39 | 88.08 |
| 2 | 500 | 0.092 | 21.6 | 1.99 | 3.97 | 74.82 |
| 2 | 5000 | 0.169 | 21.61 | 3.65 | 15.67 | 93.61 |
The reduction rate is reported as the difference between the P. betae‐normalized C q of treated plants and the P. betae‐normalized C q of the controls multiplied by the mean of the Beta vulgaris C q. It represents the standardized difference in C q (ΔCycles). At each amplification cycle, the DNA quantity is multiplied by two. The reduction factor can be obtained as 2ΔCycles. Finally, the reduction rate is obtained b: reduction rate = 1 − 1/reduction factor.
Figure 2.
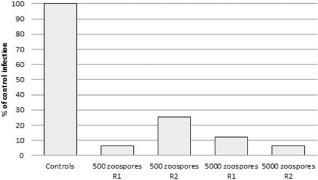
Relative infection of sugar beet by Polymyxa betae after elicitation by cyclic lipopeptides (cLPs) for the two repetitions, R1 and R2. Controls were taken as reference (100%). Values were calculated according to Table 2.
Therefore, cLPs are active against P. betae infection, but their mode of action has not yet been shown. The most probable mode is the cLP elicitation of plant systemic resistance. In order to obtain evidence for this hypothesis, the expression of genes coding for the NPR‐1 transcription factor (roots and leaves) and for a PR‐8 protein (roots) was assessed by reverse transcription‐polymerase chain reaction (RT‐PCR) on control plants harvested on the day of zoospore inoculation. NPR‐1 was selected because it plays a key role in signalling in both the SAR and ISR pathways. PR‐8 is a chitinase class III defence enzyme that can antagonize the growth of P. betae containing chitin. In addition, this gene is controlled by NPR‐1, confirming the action of NPR‐1 as a transcription activator. The expression of these genes was assessed at the time of inoculation with P. betae in order to check whether or not systemic resistance was effective in cLP‐ and water‐treated plants. RT‐PCR was performed on cLP‐ and water‐treated control plants and the other plants were then inoculated. Nucleic acids from root and leaf tissues of control plants (cLP‐ and water‐treated, but uninoculated) were extracted as described previously. DNAse treatment was applied to prevent false positives in RT‐PCR. Then, 7 μL of 20‐fold‐diluted samples were digested with 2.5 μL RQ1 DNAse (Promega), following the manufacturer's protocol. cDNA synthesis was performed in two steps: first, a mixture of 1 μL reverse primer (primers used are shown in Table 1), 8.5 μL of DEPC‐treated water and 1 μL RNA was incubated for 10 min at 65 °C. Second, a reaction mixture of 4 μL of M‐MLV RT buffer, 0.25 μL of M‐MLV reverse transcriptase (200 U/μL) (Promega), 2 μL of deoxynucleoside triphosphate (dNTP) (20 nmol) and 3.5 μL of DEPC‐treated water was added to the first reaction mixture and incubated at 42 °C for 60 min. The cycling times and temperatures for the RT‐PCR detection were 94 °C for 2 min (one cycle), 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s (35 cycles) and 72 °C for 1 min (one cycle). The reaction components for each sample were 13.25 μL DEPC‐treated water, 2.5 μL MgCl2 (25 mm), 5 μL of Green GoTaq Flexi buffer (Promega), 0.75 μL dNTP (20 nmol), 0.5 μL of each primer (20 pmol), 0.125 μL of GoTaq Polymerase (Promega) and 2.5 μL cDNA. After ethidium bromide staining, DNA bands were visualized using Gel Doc 2000 (Biorad, Hercules, CA, USA).
cLP‐treated plants showed stronger expression of the tested genes compared with controls. The results showed that the two tested genes were overexpressed in roots after treatment with cLPs compared with nonelicited (water‐treated) plants (Fig. 3). NPR‐1 was not expressed in untreated roots, whereas PR‐8 was detected, but the semi‐quantitative RT‐PCR showed that the presence of cLPs led to a much greater expression of PR‐8. The strong accumulation of NPR‐1 transcripts in the leaves of plants with cLP‐treated roots clearly demonstrated the systemic nature of cLP‐induced resistance in sugar beet.
Figure 3.
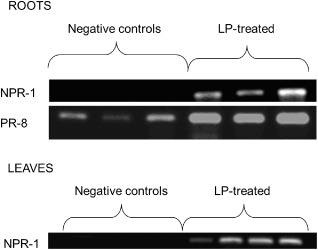
Expression of nonexpressor of pathogenesis‐related (PR) genes 1 (NPR‐1) and PR‐8 in roots and leaves of sugar beet plants, treated or not treated (negative control) with lipopeptides (LP) 7 and 14 days previously. Three plants (roots) and four plants (leaves) were tested per treatment.
All of these data strongly suggest that the penetration or further multiplication of P. betae in the root tissues of elicited plants was severely impaired. As the addition of cLPs and infection by P. betae were performed on the same plant organ, however, these biosurfactants may have inhibited zoospore performance directly. In order to test this hypothesis, the viability of the zoospores was measured after 3 h of contact with varying concentrations of cLPs. The concentration of a freshly prepared zoospore suspension was adjusted to 75 000 zoospores/mL just before the addition of cLPs at final concentrations of 0 (control), 30, 60 and 120 mg/L. Zoospore integrity was measured in five replicates of 1 mL of each suspension by the quantification of ATP via a luminescence assay (Table 3). After 3 h of incubation, the viability of P. betae zoospores was assessed by ATP quantification of each sample with the Cell‐Titer Glo Kit (Promega), following the manufacturer's protocol. The luciferase activity was measured with a Varioskan Flash Multimode Reader (Thermo Scientific, Waltham, MA, USA). No significant difference (Student–Newman–Keuls, t, Tukey and Scheffe tests) was evident among the luciferase activities from the zoospore suspensions prepared at the three cLP concentrations and the control, suggesting that the zoosporicidal activity of cLPs is very low and cannot explain the protective effect observed in biocontrol assays on whole plants. Although occasionally suggested, there are few reports showing the direct lytic activity of rhizobacterial cLPs on the zoospores of soil‐inhabiting protists in general (Jousset et al., 2006; Nielsen et al., 1999).
Table 3.
Quantification of zoospore viability in the presence of cyclic lipopeptides (cLPs)
| Concentration of cLPs | n | Luciferase activity (relative quantification of luminescence) | |
|---|---|---|---|
| Mean | Standard deviation | ||
| 0 mg/L | 5 | 31 272.31 | 8735.19 |
| 30 mg/L | 5 | 40 317.13 | 8243.71 |
| 60 mg/L | 5 | 31 469.70 | 7926.97 |
| 120 mg/L | 5 | 32 416.49 | 6708.03 |
The viability of zoospores was measured throughout the ATP quantification process in a solution containing the same number of zoospores, but increasing concentrations of lipopeptides.
The results obtained in this study show that the elicitation of sugar beet with cLPs from B. amylolequifaciens prior to inoculation with P. betae reduces infection by this parasite. As far as we know, this antagonistic effect on plasmodiophorids has not been demonstrated previously.
cLP treatment confers partial resistance to P. betae infection, probably by inducing systemic resistance in sugar beet. Our results indicate that this involves the transcription activator NPR‐1, which does not allow the discrimination between SAR‐ and ISR‐type responses stimulated by these compounds. Stimulation of the PR chitinase PR‐8 indicates that a SAR‐like reaction could be involved, but more work is needed to better understand the molecular basis of this protective effect. Indeed, there is no clear boundary between ISR and SAR, which are connected by cross‐talk, and the expressed PR proteins could be a sign of induced SAR or, in contrast, of a primed defence that is often the result of ISR (Conrath et al., 2002).
This study also extends the known range of plant species in which Bacillus cLPs are active at stimulating a systemic resistance response, already observed in tomato, bean and tobacco (Ongena and Jacques, 2008). This therefore also reinforces the notion that such biosurfactant compounds constitute a new class of MAMPs, recognized by plant root cells (Ongena et al., 2007). Moreover, Henry et al. (2011) showed that surfactin is involved in the elicitation process, through a lipid‐driven process at the plasma membrane level.
We can conclude that systemic resistance, induced by Bacillus cLPs, drastically reduces the infection of sugar beet by P. betae. Barr et al. (1995) and McGrann et al. (2009) have shown that the partial resistance of sugar beet to P. betae is linked to reduced virus levels. These conclusions indicate that disease pressure could be reduced by decreasing the infection pressure of the BNYVV vector. cLPs appear to offer a new method for the biocontrol of rhizomania. The effect of cLPs on the incidence of rhizomania on sugar beet grown on BNYVV–P. betae‐infested soils or under field conditions will be tested in further assays.
Acknowledgements
Nicolas Desoignies is a Fellow of the Fonds de Formation à la Recherche en Industrie et Agriculture (FRIA). M. Ongena is a Research Associate at the National Funds for Scientific Research (FNRS).
References
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.M. (2006) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, A. (2008) Elicitation of induced systemic resistance in tomato and cucumber and activation of the lipoxygenase pathway by non‐pathogenic rhizobacteria. PhD Thesis, University of Liège, Belgium.
- Adl, S.M. , Simpson, A.G.B. , Spiegel, F.W. and Taylor, M.F.J.R. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451. [DOI] [PubMed] [Google Scholar]
- Asher, M.J.C. , Grimmer, M.K. and Mutasa‐Goettgens, E.S. (2009) Selection and characterization of resistance to Polymyxa betae, vector of Beet necrotic yellow vein virus, derived from wild sea beet. Plant Pathol. 58, 250–260. [Google Scholar]
- Bakker, P.A.H. , Pieterse, C.M.J. and Van Loon, L.C. (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology, 97, 239–243. [DOI] [PubMed] [Google Scholar]
- Bargabus, R. , Zidack, N. , Sherwood, J. and Jacobsen, B. (2002) Characterisation of systemic resistance in sugar beet elicited by a non‐pathogenic, phyllosphere‐colonizing Bacillus mycoides, biological control agent. Physiol. Mol. Plant Pathol. 61, 289–298. [Google Scholar]
- Bargabus, R.L. , Zidack, N.K. , Sherwood, J.E. and Jacobsen, B.J. (2004) Screening for the identification of potential biological control agents that induce systemic acquired resistance in sugar beet. Biol. Control, 30, 342–350. [Google Scholar]
- Barr, K.J. , Asher, M.J. and Lewis, B.G. (1995) Resistance to Polymyxa betae in wild Beta species. Plant Pathol. 44, 301–307. [Google Scholar]
- Bass, D. , Chao, E.E.‐Y. , Nikolaev, S. , Yabuki, A. , Ishida, K. , Berney, C. , Pakzad, U. , Wylezich, C. and Cavalier‐Smith, T. (2009) Phylogeny of novel naked filose and reticulose Cercozoa: Granolosea cl. n. and Proteomyxidea revised. Protist, 160, 75–109. [DOI] [PubMed] [Google Scholar]
- Bent, E. (2005) Induced systemic resistance mediated by plant growth‐promoting rhizobacteria (PGPR) and fungi (PGPF) In: Multigenic and Induced Systemic Resistance in Plants (Tuzun S. and Bent E., eds), pp. 225–258. New York: Springer. [Google Scholar]
- Conrath, U. , Pieterse, C.M.J. and Mauch‐Mani, B. (2002) Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Desoignies, N. , Stocco, C. , Bragard, C. and Legrève, A. (2011) A new phenotype of Polymyxa betae in Arabidopsis thaliana . Eur. J. Plant Pathol. 131, 27–38. [Google Scholar]
- Goffart, J.P. and Maraite, H. (1991) Soil and agronomic factors affecting the inoculum potential of Polymyxa betae Keskin in Belgium. Parasitica, 47, 165–192. [Google Scholar]
- Henry, G. , Deleu, M. , Jourdan, E. , Thonart, P. and Ongena, M. (2011) The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune‐related defence responses. Cell. Microbiol. 13, 1824–1837. [DOI] [PubMed] [Google Scholar]
- Jourdan, E. , Henry, G. , Duby, F. , Dommes, J. , Barthélemy, J. , P., Thonart, P. and Ongena, M. (2009) Insights into the defense‐related events occurring in plant cells following perception of surfactin‐type lipopeptide from Bacillus subtilis . Mol. Plant–Microbe Interact. 22, 456–468. [DOI] [PubMed] [Google Scholar]
- Jousset, A. , Lara, E. , Wall, L.G. and Valverde, C. (2006) Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl. Environ. Microbiol. 72, 7083–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupin, I. , Richards, K. , Jonard, G. , Guilley, H. and Pleij, C.W. (1990) Mapping sequences required for productive replication of Beet necrotic yellow vein virus RNA 3. Virology, 178, 273–280. [DOI] [PubMed] [Google Scholar]
- Katagiri, K. and Tsuda, K. (2010) Understanding the plant immune system. Mol. Plant–Microbe Interact. 23, 1531–1536. [DOI] [PubMed] [Google Scholar]
- Keskin, B. (1964) Polymyxa betae n.sp., ein Parasit in den Wurzeln von Beta vulgaris Tournefort, besonders während der Jugendentwicklung der Zuckerrübe. Arch. Microbiol. 49, 348–374. [DOI] [PubMed] [Google Scholar]
- Kloepper, J.W. , Ryu, C.M. and Zhang, S.A. (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology, 94, 1259–1266. [DOI] [PubMed] [Google Scholar]
- Legrève, A. , Delfosse, P. , Vanpee, B. , Goffin, A. and Maraite, H. (1998) Differences in temperature requirements between Polymyxa sp. of Indian origin and Polymyxa graminis and Polymyxa betae from temperate areas. Eur. J. Plant Pathol. 104, 195–205. [Google Scholar]
- Legrève, A. , Delfosse, P. , Van Hese, V. , Bragard, C. and Maraite, H. (2003) Broad‐spectrum detection of Polymyxa species and form species by polymerase chain reaction In: Proceedings of the Fifth Symposium of the International Working Group on Plant Viruses with Fungal Vectors, Zurich, Switzerland (Rush C.M. and Merz U., eds), pp. 40–43. Denver, CO: American Society of Sugar Beet Technologists. [Google Scholar]
- McGrann, G.R.D. , Townsend, B.J. , Antoniw, J.F. , Asher, M.J.C. and Mutasa‐Göttgens, E.S. (2009) Barley elicits a similar early basal defence response during host and non‐host interactions with Polymyxa root parasites. Eur. J. Plant Pathol. 123, 5–15. [Google Scholar]
- Nielsen, T.H. , Christopheresen, C. , Anthoni, U. and Sørensen, J. (1999) Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 87, 80–90. [DOI] [PubMed] [Google Scholar]
- Nihorimbere, V. , Ongena, M. , Smargiassi, M. and Thonart, P. (2011) Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. 15, 327–337. [Google Scholar]
- Ongena, M. and Jacques, P. (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Schafer, M. , Kech, C. , Budzikiewicz, H. , Luxen, A. and Thonart, P. (2005) Isolation of an N‐alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant–Microbe Interact. 18, 562–569. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Adam, A. , Paquot, M. , Brans, A. , Joris, B. , Aripigny, J.‐L. and Thonart, P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , De Bruijn, I. , Nybroe, O. and Ongena, M. (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2009) Plants under attack: systemic signals in defence. Curr. Opin. Plant Biol. 12, 459–464. [DOI] [PubMed] [Google Scholar]
- Tran, H. , Ficke, A. , Asiimwe, T. , Höfte, M. and Raaijmakers, J.M. (2007) Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens . New Phytol. 175, 731–742. [DOI] [PubMed] [Google Scholar]
- United Nations Environment Programme (UNEP) (1987) Montreal Protocol on substances that deplete the ozone layer. UNEP Service number 87‐6106. Nairobi, Kenya: UNEP. [Google Scholar]
- Vallad, G.E. and Goodman, R.M. (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 44, 1920–1934. [Google Scholar]
- Van Loon, L.C. , Bakker, P.A. and Pieterse, C.M. (1998) Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. [DOI] [PubMed] [Google Scholar]


