Summary
Zymoseptoria tritici causes the major fungal wheat disease septoria tritici blotch, and is increasingly being used as a model for transmission and population genetics, as well as host–pathogen interactions. Here, we study the biological function of ZtWor1, the orthologue of Wor1 in the fungal human pathogen Candida albicans, as a representative of a superfamily of regulatory proteins involved in dimorphic switching. In Z. tritici, this gene is pivotal for pathogenesis, as ZtWor1 mutants were nonpathogenic and complementation restored the wild‐type phenotypes. In planta expression analyses showed that ZtWor1 is up‐regulated during the initiation of colonization and fructification, and regulates candidate effector genes, including one that was discovered after comparative proteome analysis of the Z. tritici wild‐type strain and the ZtWor1 mutant, which was particularly expressed in planta. Cell fusion and anastomosis occur frequently in ZtWor1 mutants, reminiscent of mutants of MgGpb1, the β‐subunit of the heterotrimeric G protein. Comparative expression of ZtWor1 in knock‐out strains of MgGpb1 and MgTpk2, the catalytic subunit of protein kinase A, suggests that ZtWor1 is downstream of the cyclic adenosine monophosphate (cAMP) pathway that is crucial for pathogenesis in many fungal plant pathogens.
Introduction
The co‐evolution of plants and their pathogens has resulted in complex interactions in which both pathogens and hosts have evolved elaborate mechanisms resulting in either compatible interactions, in which pathogens successfully invade plants, or incompatible interactions, in which host defences restrict pathogen growth (Dodds and Rathjen, 2010). Plant pathogenic fungi secrete a repertoire of effector proteins that facilitate infection by interfering with host defence mechanisms, whereas most host plants have developed receptors that mediate resistance against these fungi after recognition of these effectors (De Wit et al., 2009). The unravelling of molecular networks involved in pathogenicity provides crucial information which may lead to the development of effective disease control strategies (Lucas, 2011).
Zymoseptoria tritici (Desm.) Quaedvlieg & Crous (Quaedvlieg et al., 2011), formerly known as Mycosphaerella graminicola, the causal agent of septoria tritici blotch (STB) of wheat, is one of the most destructive fungal wheat diseases. Currently, disease management is achieved mainly through fungicide applications, but this is a costly and unsustainable strategy because of the development of fungicide resistance in the pathogen (Cools and Fraaije, 2008; Fraaije et al., 2007; Stergiopoulos et al., 2003). Introgression of resistance genes into commercial wheat cultivars is considered to be a cost‐effective and environmentally safe alternative to the application of fungicides. However, relatively few resistance genes have been characterized (Arraiano et al., 2007; Tabib Ghaffary et al., 2011, 2012) and provide limited efficacy against the complex natural Z. tritici populations. Moreover, Z. tritici has the potential to rapidly evolve new virulence patterns that reduce the durability of resistance, as exemplified by the cultivars Gene, carrying Stb4, and Madsen, with partial resistance, whose resistances declined within 5 years after their release in Oregon (USA) (Cowger et al., 2000; Wittenberg et al., 2009). A better understanding of Z. tritici biology and the molecular mechanisms underlying the infection process is crucial to the design of novel effective approaches for STB management. The availability of the genome sequence of Z. tritici (Goodwin et al., 2011) provides an excellent opportunity for gene discovery and functional analyses elucidating developmental networks and pathogenicity processes in this fungal pathogen.
Zymoseptoria tritici is a model pathogen to study hemibiotrophy and is considered to be among the top 10 most important plant pathogens worldwide (Dean et al., 2012). Unlike other fungal plant pathogens, such as Magnaporthe oryzae (Dean et al., 2005), the fungus does not form appressoria or specialized structures to penetrate the foliage, but enters the leaves through stomata and subsequently colonizes the mesophyll tissue, where it grows in the intercellular space without producing haustoria. The initial biotrophic phase is followed by a rapid switch to necrotrophy, resulting in chlorotic lesions that eventually coalesce into larger necrotic blotches bearing numerous pycnidia, the asexual fructifications that contain the splash‐borne pycnidiospores. The switch from biotrophy to necrotrophy is not well understood, but an active role of toxic compounds has been suggested (Cohen and Eyal, 1993; Duncan and Howard, 2000; Kema et al., 1996).
A suite of genes that is involved in virulence or pathogenicity has been functionally characterized by a variety of targeted gene replacement approaches (Orton et al., 2011). Some of them belong to mitogen‐activated protein (MAP) kinase pathways that affect, among others, penetration and host colonization. For instance, MgSlt2 encodes a MAP kinase that is essential for colonization and fungal cell wall integrity (Mehrabi and Kema, 2006), whereas MgSte12 regulates the ability to form filaments on the plant surface, which is crucial for successful infection. MgGpb1 regulates the cyclic adenosine monophosphate (cAMP) pathway, is required for pathogenicity and negatively controls anastomosis, a phenomenon that is rare in Z. tritici (Mehrabi et al., 2009).
Recently, the transcription factor Wor1, which regulates phase‐specific gene expression and controls the white–opaque switch in the human fungal pathogen Candida albicans, has been functionally analysed as a representative of the WOPR superfamily (Huang et al., 2006; Lohse et al., 2010; Srikantha et al., 2006; Zordan et al., 2006). Members of this transcriptional regulator family also have a role in the transition from yeast‐like to filamentous growth in Histoplasma capsulatum (Nguyen and Sil, 2008). In both pathogens, this morphological transition is correlated with pathogenicity (Cain et al., 2012). Furthermore, targeted gene deletion of the Wor1 orthologues Sge1 and Reg1 in the fungal plant pathogens Fusarium oxysporum f. sp. lycopersici (Fol) and Botrytis cinerea, respectively, revealed their involvement in pathogenicity or virulence, conidiogenesis and the expression of phase‐specific genes, including effectors and genes implemented in the production of secondary metabolites, such as mycotoxins (Michielse et al., 2009, 2011). This indicates that the WOPR gene family may specifically target a cellular function required for different biological and developmental processes in fungal plant pathogens.
In this study, we investigated the role of the Wor1 orthologue ZtWor1 in Z. tritici, and our results show that it is involved in pathogenicity, regulates the expression of small secreted proteins (SSPs) and is most likely part of the cAMP signalling pathway which plays a pivotal role in many cellular processes.
Results
Identification and characterization of ZtWor1
A blastp search of the Z. tritici genomic database using C. albicans Wor1 (CaWor1) as query resulted in the identification of two significant hits, Mycgr3_46572 and Mycgr3_72926, with E values of 1.43E–28 and 1.52E–12, which are located on chromosomes 8 and 6, respectively. Amino acid alignments revealed that Mycgr3_46572 showed the highest homology with the CaWor1 orthologue (Lohse et al., 2010), and phylogenetic tree analysis revealed that Mycgr3_46572 was clustered in the same clade as CaWor1. We therefore designated it as ZtWor1 and studied it in more detail (Fig. 1A). ZtWor1 has an open reading frame of 1614 bp, without introns as verified by reverse‐transcription polymerase chain reaction (RT‐PCR), encoding a protein of 537 amino acids (Fig. 1B). The aforementioned phylogenetic analysis grouped ZtWor1 with FoSge1 from Fol (Fig. 1A), suggesting that it may play a role in the regulation of effector encoding genes in the Z. tritici–wheat pathosystem. Similar to other members of the WOPR superfamily, the ZtWor1 N‐terminus is more conserved than its C‐terminus. Amino acid alignment of ZtWor1 with the four characterized orthologues from Fol (FoSge1), B. cinerea (BcReg1), C. albicans (CaWor1) and H. capsulatum (HcRyp1) revealed the presence of a gluconate transport‐inducing protein domain, called Gti1_Pac2 (Pfam09729), which is present across these fungal lineages. Finally, ZtWor1 contains a potential protein kinase A (PKA) phosphorylation site (KRWTDS/G) and a nuclear localization site (+94 to +101), which are also conserved among members of WOPR (Fig. 1B), suggesting that the ZtWor1 protein is localized in the nucleus, as has also been demonstrated for Sge1 and Ryp1 (Michielse et al., 2009; Nguyen and Sil, 2008).
Figure 1.
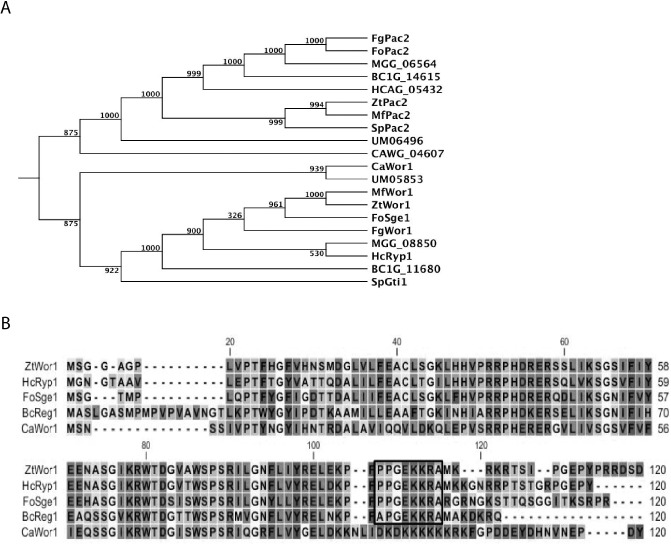
Phylogenetic comparison of Zymoseptoria tritici Wor1 (ZtWor1) with members of the WOPR superfamily based on amino acid sequence alignments. (A) The tree shows the phylogenetic relationship of ZtWor1 with Wor1 and Pac2 orthologues in other fungi, including BcReg1, BC1G_14615, HcRyp1, HCAG_05432, CaWor1, CAWG_04607, SpGti1, SpPac2, MGG_08850, MGG_06564, FgWor1, FgPac2, FoSge1, FoPac2, MfWor1, MfPac2, UM05853 and UM06496 from Botrytis cinerea, Histoplasma capsulatum, Candida albicans, Schizosaccharomyces pombe, Magnaporthe grisea, Fusarium graminearum, Fusarium oxysporum f. sp. lycopersici, Mycosphaerella fijiensis and Ustilago maydis, respectively, using CLC genomics software. The bootstrap values (1000 replicates) are shown above the branches. (B) Alignment of the first 120 deduced amino acid sequences of ZtWor1 and its orthologue members of the WOPR superfamily in other fungi. The nuclear localization motif is boxed.
Deletion and complementation of ZtWor1
In order to evaluate the biological function of ZtWor1 during the infection process, gene knock‐out and complementation mutants were generated based on homologous recombination (Fig. S1A, see Supporting Information). Three independent transformants with similar morphology, IPO323ΔZtWor1 1, 29 and 26, were obtained. The latter, coded ΔZtWor1‐26, was used for all subsequent analyses. As ΔZtWor1‐26 was unable to produce yeast‐like spores, Agrobacterium tumefaciens‐mediated transformation was performed on fragmented mycelial tissue using a construct harbouring the ZtWor1 wild‐type (WT) allele, which resulted in ΔZtWor1‐com7 (Fig. S1B).
ZtWor1 regulates fungal development
In order to assess the role of ZtWor1 in fungal growth and development, Z. tritici IPO323 (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 were compared in liquid yeast glucose broth (YGB) and on solid potato dextrose agar (PDA), Aspergillus nidulans minimal medium (MM) and V8 over a period of 10 days at 20 °C. In YGB, the WT and ΔZtWor1‐com7 produced abundant yeast‐like cells (Fig. 2Aa,Ab), but ΔZtWor1‐26 did not produce any spores and exclusively produced a dense, extensive mycelial network with abundant abnormally swollen cell structures (Fig. 2Ac). Microscopic comparison showed uncontrolled cell fusions or anastomosis in ΔZtWor1‐26 that was rare in the WT (Figs 2Bd, 3 and S2, see Supporting Information). On PDA, we did not observe any effect of the deletion of ZtWor1 on germination and early colony development during the first 48 h after inoculation (data not shown). On MM, the growth pattern started to differ at 5 days after inoculation (dai), as ΔZtWor1‐26 grew significantly more slowly than the WT, as well as ΔZtWor1‐com7, resulting in more compact colonies. Comparative scanning electron microscopy revealed significant differences (P < 0.05) in hyphal diameter between the WT, ΔZtWor1‐com7 and ΔZtWor1‐26 (Fig. S3, see Supporting Information). On V8, the WT and ΔZtWor1‐com7 strains abundantly produced yeast‐like spores, whereas ΔZtWor1‐26 hardly produced any spores, even after prolonged incubation (>14 days). At 5 dai, the WT and ΔZtWor1‐com7 turned black, probably as a result of melanization, but ΔZtWor1‐26 produced an additional mass of aerial hyphae covering the dark colonies that was absent in the WT (Fig. 4).
Figure 2.
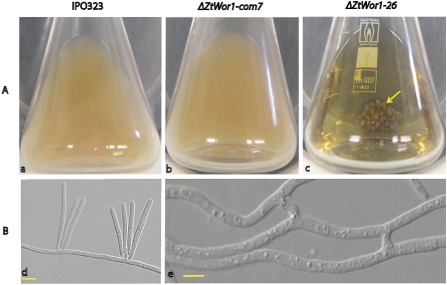
Deletion of Zymoseptoria tritici Wor1 (ZtWor1) affects yeast‐like cell production and early development. (Aa, Ab) The Z. tritici wild‐type (WT) strain and ΔZtWor1‐com7 produce abundant yeast‐like cells resulting from blastic conidiogenesis in yeast glucose broth medium. (Ac) ΔZtWor1‐26 is blocked in sporulation and exclusively produces compact hyphal networks resulting in a distinct bulbous mycelial mass (marked with a yellow arrow). (B) Comparative light microscopy of early Z. tritici development. (Bd) Production of yeast‐like cells on the hyphae of the WT on water agar. (Be) Unique cell fusion or anastomosis events that occur frequently in ΔZtWor1‐26. Scale bars, 5 μm.
Figure 3.

Comparative scanning electron micrographs of hyphae of the Zymoseptoria tritici wild‐type (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 growing on minimal medium (MM) at 10 days after inoculation at 20 °C. Infrequent cell fusion/anastomosis events occur in the Z. tritici WT strain and ΔZtWor1‐com7 (A, B; arrows), but cell fusion/anastomosis frequently happens in ΔZtWor1‐26 (C; arrows). Scale bars, 10 μm.
Figure 4.
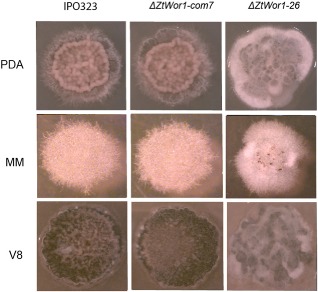
The in vitro effect of ZtWor1 deletion and complementation in Zymoseptoria tritici at 10 days after inoculation on three different media at 20 °C. Comparison of the wild‐type (WT) strain and ΔZtWor1‐26 shows that the latter exclusively produces strongly melanized mycelial cultures without any spores. This is particularly evident on V8, but also on minimal medium (MM). ΔZtWor1‐26 exclusively produces hyphae. The WT phenotype is restored in ΔZtWor1‐com7. PDA, potato dextrose agar.
ZtWor1 expression relies on MgGpb1 and MgTpk2
Overall, signal transduction pathways, including the MAP kinase and cAMP‐dependent PKA pathways, play a crucial role in the sensing of and responding to environmental stimuli, and represent important cascades in the regulation of development in eukaryotes. Previously, we have shown that these pathways are also involved in the pathogenicity and development of Z. tritici (Cousin et al., 2006; Mehrabi et al., 2006a,b). As ΔZtWor1‐26 showed abundant anastomosis, we tested whether the cAMP signalling pathway, which controls a similar phenotype in Z. tritici Gβ mutants (Mehrabi et al., 2009), is also involved in ZtWor1 regulation, and determined the relative expression level of ZtWor1 in MgGpb1 and MgTpk2 mutants (Mehrabi and Kema, 2006; Mehrabi et al., 2009). In both mutants, ZtWor1 expression was severely reduced compared with the WT (Fig. 5A), whereas the expression levels of MgGpb1 and MgTpk2 in ΔZtWor1‐26 were the same as in the WT strain (Fig. 5B). Taken together, it can be concluded that ZtWor1 and the PKA pathway function in parallel to regulate various developmental processes, such as cell fusion, in Z. tritici. Alternatively, ZtWor1 is downstream of the β‐subunit of the heterotrimeric G protein and the cAMP pathway as shown by the expression analysis.
Figure 5.

Expression analysis of Zymoseptoria tritici Wor1 (ZtWor1) in the disrupted mutants MgGpb1 and MgTpk2. (A) In vitro expression level of ZtWor1 in disrupted mutants MgGpb1 and MgTpk2 compared with expression in the Z. tritici wild‐type (WT) strain. (B) Comparative in vitro expression of MgGpb1 and MgTpk2 in the Z. tritici WT and ΔZtWor1‐26 strains.
ZtWor1 is up‐regulated at early and late stages of infection
As the orthologues of ZtWor1 in other fungal plant pathogens are implicated in pathogenicity, we analysed the expression levels of ZtWor1 in vitro and in planta (Fig. 6). ZtWor1 has a bimodal expression profile; it is up‐regulated during the early stage of infection (2 dai), gradually down‐regulated until 16 dai and then significantly up‐regulated again at 20 dai, the stage of infection that coincides with pycnidial formation. The expression level of ZtWor1 in an axenic mycelial culture was comparable with the in planta expression at 2 dai, whereas the expression level in yeast‐like cells was similar to the in planta expression at 20 dai during abundant asexual fructification (Fig. 6).
Figure 6.
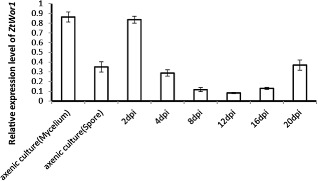
In vitro and in planta expression levels of Zymoseptoria tritici Wor1. In vitro conditions (18 °C and 25 °C to induce yeast‐like cells and mycelia formation, respectively) were chosen to compare the expression levels of Ztwor1 in mycelial and conidial cultures with in planta conditions. The susceptible cv. Obelisk was inoculated with the Z. tritici wild‐type (WT) strain and infected leaves were harvested at 2, 4, 8, 12, 16 and 20 days after inoculation, followed by RNA isolation and cDNA synthesis, which showed that Ztwor1 is particularly expressed at the onset of colonization (switch from yeast‐like spores to hyphae) and conidiogenesis (pycnidia production at the later phase of pathogenesis). The expression of Ztwor1 was normalized with the constitutively expressed Z. tritici β‐tubulin gene.
ZtWor1 is required for pathogenicity
To assess the biological function of ZtWor1 during pathogenesis, WT strain, ΔZtWor1‐com7 and ΔZtWor1‐26 were used to inoculate the susceptible wheat cv. Obelisk and disease development was monitored over time. Small chlorotic flecks appeared at 9 dai, especially at the leaf tips, which expanded over time and eventually coalesced into large necrotic blotches containing numerous pycnidia in the control strains. Occasionally, a few chlorotic and necrotic lesions were observed after inoculation with the deletion mutant, which sometimes contained a few immature pycnidia in a limited number of lesions (Fig 7).
Figure 7.

The effect of Zymoseptoria tritici Wor1 (ZtWor1) deletion on disease development in the susceptible wheat cv. Obelisk. First leaves were inoculated with the Z. tritici wild‐type (WT) strain (2), ΔZtwor1‐com7 (3) and ΔZtwor1‐26 (4), with water as a control (1). Photographs were taken at 21 days after inoculation.
ZtWor1 regulates the expression of specific SSPs
Considering that ΔZtWor1‐26 was significantly reduced in pathogenicity and that orthologues in other fungal plant pathogens regulate the expression of effector genes, we hypothesized a similar role for ZtWor1 in Z. tritici. First, quantitative RT‐PCR was used to determine the role of ZtWor1 in the in vitro expression of several SSPs that are candidate effectors in the Z. tritici–wheat pathosystem based on bioinformatics analyses (Morais do Amaral et al., 2012) (Table S1, see Supporting Information) and in planta expression profiling (G. H. J. Kema et al., Plant Science Group, Plant Research International BV, Wageningen University, Droevendaalsesteeg, unpublished data). We determined that ZtWor1 either positively or negatively regulates SSPs. For instance, the expression level of SSP60 was down‐regulated more than 20‐fold compared with the WT ( Fig. 8 and Table S2, see Supporting Information). Second, we compared the in vitro proteome of the WT strain and ΔZtWor1‐26. Overall, 125 Z. tritici proteins were identified from all conditions [three minimal media including MM, Cladosporium fulvum B5 (B5) and dextrose broth], 18 of which were unique to Z. tritici, as no homologues could be identified in fungal databases. One hundred and fourteen proteins possessed a SignalP motif, indicating that they were secreted, and only one (SSP127) of the 114 proteins was not expressed in ΔZtWor1‐26 (Fig. 9). In vitro expression on MM indicated that SSP127 may have an important role during the early stages of infection. This was confirmed by the relative in planta expression of SSP127, which was highly up‐regulated until 4 dai. (Fig. 10). Subsequently, two independent knock‐out strains of SSP127 were generated and phenotyped on a range of 12 unrelated wheat cultivars that are parents of mapping populations, as well as the suite of wheat cultivars with mapped resistance (Stb) genes (Tabib Ghaffary et al., 2011); however, surprisingly, no significant differences in disease development were observed between the knock‐out strains and the WT (Fig. S4, see Supporting Information). In summary, our data suggest that ZtWor1 is much more involved in developmental processes than as a specific regulator of effector genes.
Figure 8.

Comparative in vitro expression analysis of 29 small secreted proteins (SSPs) in the Zymoseptoria tritici wild‐type (WT) strain and ΔZtwor1‐26 grown in yeast glucose broth (YGB) medium for 7 days at 18 °C. Expression levels were normalized with the constitutively expressed Z. tritici β‐tubulin gene and plotted on a log10 scale.
Figure 9.
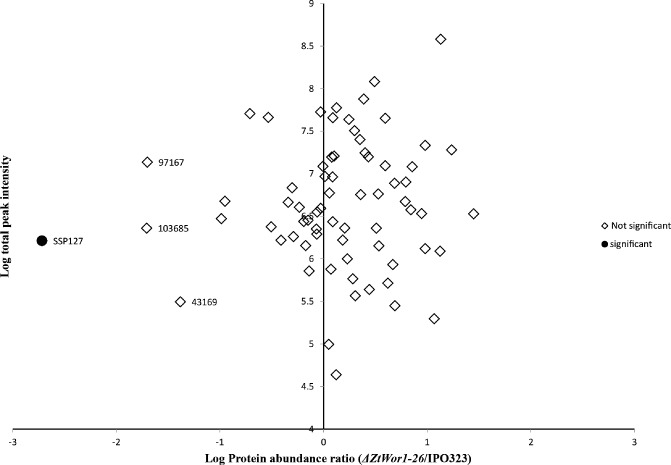
Comparative discriminative in vitro proteome analysis of the Zymoseptoria tritici wild‐type (WT) strain and ΔZtWor1‐26. Plot of the normalized ΔZtWor1‐26/IPO323 intensity ratio against the total measured protein intensity. Proteins not significantly different between mutant and WT are depicted in white, whereas the significantly different protein is shown as a black circle. SSP127 is the only identified protein that showed significant expression differences between the two strains (ratio = 0.002, P = 3.1 × 10–5). Proteins 43169, 97167 and 103685 showed P values larger than 0.05 and are considered to be nonsignificant by the Perseus software, which takes both the ratio and P value into account.
Figure 10.
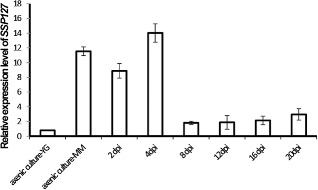
In vitro and in planta expression levels of small secreted protein (SSP) 127 of Zymoseptoria tritici. In vitro conditions represented mycelium production [on minimal medium (MM)] and blastic conidiogenesis [in yeast glucose broth (YGB) medium]. In planta expression profiles were measured during a time course (2–20 days after inoculation) experiment, using the susceptible wheat cv. Obelisk. The relative expression of SSP127 was normalized with the constitutively expressed Z. tritici β‐tubulin gene.
Discussion
For successful infection and completion of its life cycle on wheat, Z. tritici employs a variety of mechanisms to penetrate, colonize and kill host tissue. To date, several pathogenicity factors, such as MgSlt2 and MgGpb1, have been identified and, to some extent, it has been shown how they contribute to the infection process, evade host defence responses and enable disease establishment (Orton et al., 2011).
Here, we analysed the biological function of the regulatory gene ZtWor1 and showed that it is required for the pathogenicity of Z. tritici, possibly through the regulation of effector genes, as it controls the expression levels of a suite of genes encoding SSPs in vitro, which should be corroborated in future experiments. Members of the conserved WOPR family of regulatory proteins, such as the master regulator Wor1, are involved in the dimorphic switch of the human fungal pathogen C. albicans. Similar to other well‐characterized family members in both fungal human and plant pathogens, such as HcRyp1, FoSge1, BcReg1, Schizosaccharomyces pombe (GtiI) and Fusarium graminearum (Fgp1) (Caspari, 1997; Jonkers et al., 2012; Michielse et al., 2009 , 2011; Nguyen and Sil, 2008), this putative transcriptional regulator possesses two globular domains, the WOPR box1 (amino acids 16–107) and the WOPR box2 (amino acids 160–250) located at the N‐terminal region, which is highly conserved across fungal lineages. In contrast, the C‐terminus regions are rich in glutamine amino acids and very divergent among family members. Another common feature of WOPR family members is the presence of a highly conserved amino acid motif (PPGEKKRA) that has been shown to be involved in the nuclear localization of Ryp1 and Sge1. This motif (+94 to +101) is also present in ZtWor1, and probably serves the same role in Z. tritici.
Candida albicans Wor1 is a master regulator of ‘white to opaque switching’, which refers to the development and transition between two distinctive in vitro cell types. Strains deleted for Wor1 cannot form opaque cells, but this phenotype can be rescued by the ectopic expression of Wor1 (Huang et al., 2006; Ohara and Tsuge, 2004; Srikantha et al., 2006; Zordan et al., 2006). Furthermore, it has been shown that Wor1 regulates white–opaque switching through phase‐specific expression of the genes Wor2, Czf1 and Efg1 (Huang et al., 2006; Morschhäuser, 2010). The Efg1 orthologue in Fol is required for conidiogenesis (Ohara and Tsuge, 2004). In H. capsulatum, Ryp1 is a master transcriptional regulator that controls the transition from filamentous growth to the pathogenic budding‐yeast form. Nguyen and Sil (2008) showed that Ryp1 is involved in the expression of yeast‐specific genes, including two genes that are linked to virulence. In both aforementioned human pathogens, the up‐regulation of Wor1 (45×) and Ryp1 (4×) is correlated with dimorphism and with pathogenicity through the regulation of cell type‐specific genes (Nguyen and Sil, 2008; Tsong et al., 2003).
Our analyses showed that ZtWor1 expression oscillates with distinct phases of pathogenesis; up‐regulation during initial disease establishment (2 dai), down‐regulation during colonization (until 12 dai) and up‐regulation again during conidiogenesis (20 dai). In addition, we showed that in vitro ZtWor1 expression in the WT strain correlates with the transition from yeast‐like cells to filamentous growth that occurs during the early stage of infection (∼2 dai). During these respective in planta and in vitro conditions, Z. tritici undergoes extreme morphological changes (Goodwin et al., 2011). Thus, the inability of ΔZtWor1‐26 to develop or differentiate the required appropriate cell types may abolish pathogenicity. However, in addition, we provide evidence that ZtWor1 regulates a suite of genes encoding SSPs in vitro that probably have effector functions by acting as virulence or avirulence determinants in the Z. tritici–wheat pathosystem. In Fol, Sge1 regulates the expression of Six (secreted in xylem) effectors during the colonization of the vascular tissue of tomato plants (Michielse et al., 2009). Recently, Jonkers et al. (2012) showed that the ZtWor1 orthologue Fgp1 in F. graminearum is required for the infection process and the in vitro and in planta expression of genes involved in the trichothecene biosynthetic (TRI) pathway. Thus, in addition to the role of ZtWor1 in morphological changes that possibly affect pathogenicity, it is probable that ZtWor1 globally regulates various virulence factors, which requires further investigation.
The dimorphic switch involved in ZtWor1 expression and the comparison with knock‐out strains in other fungi demonstrate its global involvement in developmental morphogenesis. Functional analysis of BcReg1 in B. cinerea revealed that knock‐out strains produce aberrant nonconidia‐bearing conidiophores during pathogenesis (Michielse et al., 2011), and Sge1 and Fgp1 also affect conidia formation in Fol (Michielse et al., 2009) and F. graminearum (Jonkers et al., 2012), respectively. We observed that ZtWor1 mutants do not sporulate in vitro. Each and every effort to induce sporulation of ZtWor1 mutants using different conditions and (liquid) media was unsuccessful, but complementation of ΔZtWor1‐26 restored the WT, and hence in vitro and in planta conidiogenesis, suggesting that Wor1 is a crucial factor in yeast‐like cell formation.
Complementation also restores pathogenicity, whereas heterologous complementation with Sge1 from Fol did not (data not shown), indicating that Wor1 orthologues in various fungal human and plant pathogens have evolved divergently to regulate pathogenicity through different mechanisms; this has also been shown in F. graminearum, where the interchange of the N‐ and C‐terminal portions of the Wor1 homologues from Fol and F. graminearum did not mutually restore loss of function (Jonkers et al., 2012).
The current study suggests that ZtWor1 may be positioned downstream of two important components of the cAMP pathway, MgGpb1 and MgTpk2, which play important roles in cell differentiation and pathogenicity (Mehrabi and Kema, 2006; Mehrabi et al., 2009). This is a unique hypothesis that requires further investigation, but, interestingly, the phenotypes of the ZtWor1, MgTpk2 and MgGpb1 mutants share several common features. First, they are hampered in pathogenicity; second, MgTpk2 and MgGpb1 mutants penetrate the host and colonize the mesophyll, but cannot differentiate cells towards fructification; third, ZtWor1 and MgGpb1 mutants show an intriguing cell fusion or anastomosis phenotype that is unique in Z. tritici. MgGpb1 negatively regulates anastomosis, and this gene is upstream of MgTpk2 and positively regulates the cAMP pathway, as exogenous cAMP restores the WT phenotype (Mehrabi et al., 2009). Our current data suggest that the previously characterized cAMP genes (MgTpk2 and MgGpb1) and ZtWor1 may be three components of the cAMP pathway controlling different aspects of differentiation and infection. Interestingly, all family members of WOPR contain a PKA phosphorylation site, and the functionality of this well‐conserved motif was determined by mutation, resulting in nonpathogenic phenotypes in Fol, indicating that FoSge1 is pivotal for pathogenicity (Michielse et al., 2009). Furthermore, it was demonstrated that the CaWor1 protein was phosphorylated by Tpk2, thus regulating the ‘white to opaque’ switch (Huang et al., 2010). Our data suggest that ZtWor1 is regulated by the Z. tritici homologue of Tpk2, MgTpk2, as shown by the expression analyses conducted.
In summary, we conclude that ZtWor1 is a putative transcriptional regulator in the dimorphic fungal plant pathogen Z. tritici and plays an essential role in differentiation, asexual fructification, conidiogenesis and the regulation of SSPs, which may act as putative effector genes. In addition, we suggest that ZtWor1 may be regulated by two upstream key genes, MgGpb1 and MgTpk2, indicating that the functionality of ZtWor1 is controlled through the cAMP pathway; hence, ZtWor1 may be considered as a key transcriptional regulator downstream of this pathway. Further research into the cAMP signalling network and the exact role of ZtWor1 in this pathway is required to elucidate how these components regulate the morphopathogenic behaviour of Z. tritici. The data presented show that a comprehensive understanding of the regulatory function of ZtWor1 may lead to the identification of key pathogenicity factors or effector proteins, which will contribute to the further understanding of the complex Z. tritici–wheat interaction.
Experimental Procedures
Strains, media and growth conditions
The sequenced Z. tritici reference strain IPO323, which is highly virulent on the susceptible wheat cv. Obelisk, was used as WT and recipient strain for gene deletion. The WT and all developed strains were stored at −80 °C and recultured on PDA (Sigma‐Aldrich Chemie, Steinheim, Germany) at 18 °C. Yeast‐like spores were produced on V8 juice medium (Campbell Foods, Puurs, Belgium) or in YGB medium (yeast extract, 10 g/L; glucose, 20 g/L) placed in an orbital shaker (Innova 4430; New Brunswick Scientific, Nijmegen, the Netherlands) at 18 °C. To induce mycelial growth, all Z. tritici strains were grown under the same conditions, but at 25 °C. Aspergillus nidulans MM and B5 medium were prepared and used for morphological charecterization experiments and proteomic assays, respectively (Ackerveken et al., 1994; Barratt et al., 1965).
Nucleotide sequence data of ZtWor1 are available at GenBank under accession number BK008803. Zymoseptoria tritici strain IPO323 is available at the Centraal Bureau voor Schimmelcultures, Utrecht, the Netherlands: http://www.cbs.knaw.nl/
Phylogenetic tree construction
Phylogenetic analysis of ZtWor1 with homologues from other fungal pathogens was conducted using the CLC genomics workbench package (Aarhus, Denmark). All Wor1 and Pac2 fungal proteins were retrieved from public databases and aligned using the aforementioned software, considering a gap opening cost and gap extension penalty of 10 and 1, respectively. The phylogenetic tree was constructed based on the unweighted pair group method with arithmetic average (UPGMA) algorithm, and the statistical accuracy of the tree was tested by bootstrap analysis (1000 repetitions).
Generation of gene deletion and complementation constructs
To generate the ZtWor1 deletion construct, pZtWor1KO, the USER friendly cloning method was used with minor modifications (Frandsen et al., 2008). Briefly, ZtWor1‐PRF‐F1, R1 and ZtWor1‐PRF‐F2, R2 primer pairs were used to amplify about 2000 bp upstream and downstream of ZtWor1 using PfuTurbo® Cx Hotstart DNA polymerase (Stratagene, Cedar Creek, TX, USA). In parallel, the pRF‐HU2 vector possessing the hygromycin phosphotransferase (hph) gene as a selection marker was digested with two restriction enzymes, PacI and a nicking enzyme Nt.BbvCI, to generate a compatible overhang with the PCR amplicons. Subsequently, the PCR amplicons and the digested vector were mixed and treated with the USER enzyme mix (New England Biolabs, Ipswich, MA, USA) and incubated at 37 °C for 30 min followed by 25 °C for 30 min. The resulting reaction was directly transformed into Escherichia coli strain DH5α and subsequently cultured on selective kanamycin medium. In order to identify bacterial colonies carrying the construct with the insertions in the expected positions, colony PCR was conducted using User‐F and User‐R primers (located in the middle of the hph gene) in combination with ZtWor1‐R and ZtWor1‐F, respectively (Table 1).
Table 1.
Primers used in this study
| Name | Sequence (5–3) | Location |
|---|---|---|
| ZtWor1‐PRF‐F1 | GGTCTTAAUTGGACGGGCACCTGTACTATTGGCCG | Upstream of ZtWor1 |
| ZtWor1‐PRF‐R1 | GGCATTAAUGAGAGATCGAACACACAGCGGCGCAC | Upstream of ZtWor1 |
| ZtWor1‐PRF‐F2 | GGACTTAAUCCGAGCACTACGCCATTGACGGCC | Downstream of ZtWor1 |
| ZtWor1‐PRF‐R2 | GGGTTTAAUGTTTCGCCTGCCTGCGTTGCCGAG | Downstream of ZtWor1 |
| ZtWor1‐F1 | ATGAGCGGGGGAGCCGGA | ZtWor1 |
| ZtWor1‐R1 | CTCCTCAACCGGCGCGC | ZtWor1 |
| ZtWor1‐F2 | GTGCTCACCGCCTGGACGACTAAAC | Middle of hph gene |
| ZtWor1‐R2 | ACCTTGCTAATAACCCAAACGCC | Downstream of ZtWor1 |
To generate the ZtWor1 complementation construct, pZtWor1com, the multisite Gateway® three‐fragment vector construction kit was used, enabling us to clone three fragments into the destination vector, which was compatible with the A. tumefaciens‐mediated transformation procedure. The full open reading frame of ZtWor1, including 1200 bp upstream as promoter and 500 bp downstream as terminator, were cloned into pDONR™P2R‐P3 (Invitrogen, Carlsbad, CA, USA) resulting in the generation of p2‐ZtWor1com. Furthermore, the green fluorescent protein (GFP) gene and neomycin phosphotransferase gene (known as geneticin selection marker) were cloned into pDONR™221 and pDONR™P4‐P1R, resulting in p221‐GFP and p4‐geneticin, respectively. Finally, three entry vectors were used to clone these three fragments into the destination vector, pPm43GW, through the LR reaction.
Fungal transformation
The gene deletion construct, pZtWor1KO, was cloned into A. tumefaciens strain LBA1100 via electroporation. Agrobacterium tumefaciens‐mediated transformation was carried out to delete ZtWor1 in the WT strain, as described previously (Zwiers and De Waard, 2001). Genomic DNAs of stable transformants were extracted using a KingFisher robot (Thermo Scientific, Hudson, NH, USA) and used in PCR screen.
For complementation, the same procedure was applied with minor modifications. As a result of the lack of spore production in ΔZtWor1‐26, small pieces of hyphal fragments—adjusted to 105 per mL—were used in A. tumefaciens‐mediated transformation, and putatively complemented strains were selected on plates with 250 μg/mL geneticin.
Cell biology assay
Cell biology assays were performed using mycelial fragments as starting material that were generated in YGB at 25 °C for 12 days and subsequently blended and passed through a Miracloth filter (Merck Millipore, Darmstadt, Germany) and finally adjusted to 105 hyphal fragments/mL to monitor anastomosis in the WT strain and ΔZtWor1‐26. Approximately 10 μL of each sample were spotted onto 1% water agar plugs, which were placed on a glass slide and covered with a cover slip. The samples were kept in Petri plates containing a piece of wet cotton to maintain high humidity and were incubated at 20 °C for 7 days. The samples were monitored using an Olympus IX81 microscope (Olympus, Hamburg, Germany), equipped with a 100×/1.45 Oil TIRF or 60×/1.35 Oil objective and a VS‐LMS4 Laser‐Merge‐System with solid state lasers (488 nm/70 mW and 561 nm/70 mW; Visitron System, Munich, Germany). The images were taken using a Photometrics CoolSNAP HQ2 camera (Roper Scientific, Planegg/Martinsried, Germany) and processed by MetaMorph (Molecular Devices, Downingtown, PA, USA) software.
Pathogenicity assay
The susceptible wheat cv. Obelisk was grown in a glasshouse until the first leaves were fully unfolded. As ΔZtWor1‐26 did not sporulate, we used mycelial fragments for all strains. Inoculum was produced by blending mycelia 24 h before inoculation, which were subsequently maintained in YGB at 25 °C for cell recovery, passed through Miracloth to remove large mycelial fragments and adjusted to 105 hyphal fragments/mL for spray inoculation. Knock‐out strains of SSP127 and the WT strain were inoculated on a wide variety of wheat germplasm that was grown and inoculated according to standard procedures (Tabib Ghaffary et al., 2011). Inoculated plants were incubated in black plastic bags for 48 h and then transferred to a glasshouse compartment (22 °C, relative humidity >90% and 16 h light). Disease development was monitored every 3 days and final scoring was performed at 20 dai.
RNA isolation and quantitative RT‐PCR
In vitro and in planta expression analyses of selected genes were conducted using quantitative real‐time RT‐PCR. Plants of cv. Obelisk were inoculated with the WT strain as described previously (Mehrabi et al., 2006a) and leaf samples were collected in three biological replications, flash frozen and ground in liquid nitrogen using a mortar and pestle. Total RNA was extracted either from ground leaves or fungal biomass produced in YGB using the RNeasy plant mini kit (Qiagen, Valencia, CA, USA), and subsequently DNA contamination was removed using the DNAfree kit (Ambion, Huntingdon, Cambridgeshire, UK). First‐strand cDNA was synthesized from 2 μg of total RNA primed with oligo(dT) using SuperScript III according to the manufacturer's instructions. One microlitre of resulting cDNA was employed in a 25‐μL PCR using a QuantiTect SYBR Green PCR Kit (Applied Biosystems, Warrington, UK), and run and analysed using an ABI 7500 Real‐Time PCR System. The relative expression level of each gene was initially normalized with the constitutively expressed Z. tritici β‐tubulin gene (Keon et al., 2007; Motteram et al., 2009) and then calculated on the basis of a comparative C(t) method described previously (Schmittgen and Livak, 2008).
Secretome analysis of WT strain and ΔZtWor1‐26
The WT and ΔZtWor1‐26 strains were grown in YGB (125 rpm, 25 °C, for 5 days) to obtain adequate fungal biomass. Afterwards, fungal mycelia were passed through Miracloth and washed three times with sterile water to remove residual medium. Subsequently, the resulting mycelial fragments were inoculated in three minimal media, including MM, B5 and dextrose broth (30 g dextrose/L), in four biological replications for 48 h. After recovery from these media, mycelia were removed by centrifugation (Beckman, Pleasanton, CA, USA) at 10 000 rpm and the supernatants were applied to filters (0.45 μm). Proteins were precipitated with 10% trichloroacetic acid (TCA) and dissolved in 1 m Tris, pH 8.3. Two‐hundred microlitres of the crude protein extracts were applied to Nanosep 3 K Omegacentrifuge filters (Pall Corporation, Ann Arbor, MI, USA) and centrifuged at 5000 g for 30 min at room temperature (20 °C). Thereafter, the Filter Aided Sample Preparation (FASP) method (Manza et al., 2005; Wisniewski et al., 2009) was used to generate tryptic peptides for liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) analysis. The peptide solutions were acidified by adding 3.5 μL of 0.1% trifluoroacetic acid and analysed by LC‐MS/MS as described previously (Lu et al., 2011). LC‐MS runs with all MS/MS spectra obtained were analysed with MaxQuant 1.1.1.36 (Cox and Mann, 2008) using default settings for the Andromeda search engine (Cox et al., 2011), except that extra variable modifications were set for de‐amidation of N and Q. The Z. tritici database stored at the JGI Genome Portal (http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html) was used together with a database that contains sequences of common contaminants, such as, for instance, BSA (P02769, bovine serum albumin precursor), trypsin (P00760, bovine), trypsin (P00761, porcine), keratin K22E (P35908, human), keratin K1C9 (P35527, human), keratin K2C1 (P04264, human) and keratin K1CI (P35527, human). The ‘label‐free quantification’ (LFQ) as well as the ‘match between runs’ (set to 2 min) options were enabled. De‐amidated peptides were allowed to be used for protein quantification and all other quantification settings were kept at default. Filtering and further bioinformatic analysis of the MaxQuant/Andromeda workflow output and the analysis of the abundances of the identified proteins were performed with the Perseus module (available at the MaxQuant suite) as described previously (Kariithi et al., 2012). Peptides and proteins with a false discovery rate (FDR) of less than 1% and proteins with at least two identified peptides, one of which should be unique and one of which should be unmodified, were accepted. Reversed hits and contaminants were deleted from the MaxQuant result table. The relative protein quantification of WT to mutant was performed with Perseus by applying a two‐sample t‐test using the ‘LFQ intensity’ columns obtained with threshold 0.05 and S 0 = 1. The normal logarithm was taken from normalized LFQ protein MS1 intensities (LFQ) as obtained from MaxQuant. Zero values for one of the two LFQ columns were replaced by a value of 2.4 to make sensible ratio calculations possible.
Supporting information
Fig. S1 Replacement strategy for ZtWor1 in Zymoseptoria tritici. (A) Diagram showing the replacement by the hygromycin phosphotransferase (hph) resistance cassette through homologous recombination. The broken line depicts the flanking regions used for homologous recombination. (B) Identification of replacement mutants by polymerase chain reaction (PCR). Lane M, 1‐kb‐plus ladder marker. Lanes 1, 4 and 7 show three independent replacement mutants (ΔZtWor1‐1, ΔZtWor1‐26 and ΔZtWor1‐29) with no amplicon using primers ZtWor1‐F1 and ZtWor1‐R1, whereas the wild‐type (WT) strain (lane C1) and the complemented strain ΔZtWor1‐com7 (lane10) show the expected amplicon of 650 bp with the same primers. Primers ZtWor1‐F2 and ZtWor1‐R2, which are located in the middle of the hph gene and downstream of the ZtWor1 open reading frame (ORF), produced an amplicon of 2000 bp (lanes 2, 5 and 8), but did not result in amplification in the WT strain (C1) and the complemented strain (lane 11).
Fig. S2 Number of cell fusion events, counted in 0.016 mm2, in colonies of Zymoseptoria tritici wild‐type (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 grown on minimal medium (MM) for 10 days at 20 °C.
Fig. S3 Differences in hyphal diameters of Zymoseptoria tritici wild‐type (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 grown on minimal medium (MM) for 10 days. n = 100 for each strain. The difference is significant at P < 0.05.
Fig. S4 Disease development in 12 wheat cultivars that are parents of mapping populations after inoculation with Zymoseptoria tritici wild‐type (WT) strain compared with two independent knock‐out strains of SSP127.
Table S1 Putative small secreted proteins (SSPs) and their corresponding primers used in this study.
Table S2 Comparative expression profiling of small secreted proteins (SSPs) in the Zymoseptoria tritici wild‐type (WT) strain versus ΔZtWor1‐26.
Acknowledgements
AMG was financially supported by the Ministry of Research and Technology of Iran. We would like to thank Dr. Martijn Rep, University of Amsterdam, for providing the sge1 construct used in complementation experiments and T. Franssen‐Verheijen, Wageningen Electron Microscopy Centre, for analysing the scanning electron micrographs.
References
- Ackerveken, G.F.J.M. , Dunn, R.M. , Cozijnsen, A.J. , Vossen, J.P.M.J. , Broek, H.W.J. and Wit, P.J.G.M. (1994) Nitrogen limitation induces expression of the avirulence gene avr 9 in the tomato pathogen Cladosporium fulvum . Mol. Gen. Genet. 243, 277–285. [DOI] [PubMed] [Google Scholar]
- Arraiano, L.S. , Chartrain, L. , Bossolini, E. , Slatter, H.N. , Keller, B. and Brown, J.K.M. (2007) A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola . Plant Pathol. 56, 73–78. [Google Scholar]
- Barratt, R.W. , Johnson, G.B. and Ogata, W.N. (1965) Wild‐type and mutant stocks of Aspergillus nidulans . Genetics, 52, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain, C.W. , Lohse, M.B. , Homann, O.R. , Sil, A. and Johnson, A.D. (2012) A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics, 190, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari, T. (1997) Onset of gluconate‐H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1+ gene product. J. Cell Sci. 110, 2599–2608. [DOI] [PubMed] [Google Scholar]
- Cohen, L. and Eyal, Z. (1993) The histology of processes associated with the infection of resistant and susceptible wheat cultivars with Septoria tritici . Plant Pathol. 42, 737–743. [Google Scholar]
- Cools, H.J. and Fraaije, B.A. (2008) Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola . Pest Manag. Sci. 64, 681–684. [DOI] [PubMed] [Google Scholar]
- Cousin, A. , Mehrabi, R. , Guilleroux, M. , Dufresne, M. , Van Der Lee, T. , Waalwijk, C. , Langin, T. and Kema, G.H.J. (2006) The MAP kinase‐encoding gene MgFus3 of the non‐appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7, 269–278. [DOI] [PubMed] [Google Scholar]
- Cowger, C. , Hoffer, M.E. and Mundt, C.C. (2000) Specific adaptation by Mycosphaerella graminicola to a resistant wheat cultivar. Plant Pathol. 49, 445–451. [Google Scholar]
- Cox, J. and Mann, M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat. Biotechnol. 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox, J. , Neuhauser, N. , Michalski, A. , Scheltema, R.A. , Olsen, J.V. and Mann, M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. , Mehrabi, R. , Van Den Burg, H.A. and Stergiopoulos, I. (2009) Fungal effector proteins: past, present and future. Mol. Plant Pathol. 10, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.‐R. , Pan, H. , Read, N.D. , Lee, Y.‐H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.‐H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.‐J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Duncan, K.E. and Howard, R.J. (2000) Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola . Mycol. Res. 104, 1074–1082. [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Kim, S.H. , Motteram, J. , Clark, W.S. and Lucas, J.A. (2007) A novel substitution I381V in the sterol 14α‐demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol. Plant Pathol. 8, 245–254. [DOI] [PubMed] [Google Scholar]
- Frandsen, R. , Andersson, J. , Kristensen, M. and Giese, H. (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, S.B. , Ben M'Barek, S. , Dhillon, B. , Wittenberg, A.H.J. , Crane, C.F. , Hane, J.K. , Foster, A.J. , Van der Lee, T.A.J. , Grimwood, J. , Aerts, A. , Antoniw, J. , Bailey, A. , Bluhm, B. , Bowler, J. , Bristow, J. , van der Burgt, A. , Canto‐Canché, B. , Churchill, A.C.L. , Conde‐Ferràez, L. , Cools, H.J. , Coutinho, P.M. , Csukai, M. , Dehal, P. , De Wit, P. , Donzelli, B. , van de Geest, H.C. , van Ham, R.C.H.J. , Hammond‐Kosack, K.E. , Henrissat, B. , Kilian, A. , Kobayashi, A.K. , Koopmann, E. , Kourmpetis, Y. , Kuzniar, A. , Lindquist, E. , Lombard, V. , Maliepaard, C. , Martins, N. , Mehrabi, R. , Nap, J.P.H. , Ponomarenko, A. , Rudd, J.J. , Salamov, A. , Schmutz, J. , Schouten, H.J. , Shapiro, H. , Stergiopoulos, I. , Torriani, S.F.F. , Tu, H. , de Vries, R.P. , Waalwijk, C. , Ware, S.B. , Wiebenga, A. , Zwiers, L.‐H. , Oliver, R.P. , Grigoriev, I.V. and Kema, G.H.J. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. , Wang, H. , Chou, S. , Nie, X. , Chen, J. and Liu, H. (2006) Bistable expression of WOR1, a master regulator of white–opaque switching in Candida albicans . Proc. Natl. Acad. Sci. USA, 103, 12 813–12 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. , Yi, S. , Sahni, N. , Daniels, K.J. , Srikantha, T. and Soll, D.R. (2010) N‐Acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans . PLoS Pathog. 6, e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers, W. , Dong, Y. , Broz, K. and Corby Kistler, H. (2012) The Wor1‐like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum . PLoS Pathog. 8, e1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariithi, H.M. , van Lent, J.W.M. , Boeren, S. , Abd‐Alla, A.M.M. , Ince, I.A. , van Oers, M.M. and Vlak, J.M. (2012) Correlation between structure, protein composition, morphogenesis and cytopathology of Glossina pallidipes salivary gland hypertrophy virus. J. Gen. Virol. 94, 193–208. [DOI] [PubMed] [Google Scholar]
- Kema, G.H.J. , Yu, D. , Rijkenberg, F.H.J. , Shaw, M.W. and Baayen, R.P. (1996) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. , Beale, M.H. , Hammond‐Kosack, K. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Lohse, M.B. , Zordan, R.E. , Cain, C.W. and Johnson, A.D. (2010) Distinct class of DNA‐binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans . Proc. Natl. Acad. Sci. USA, 107, 14 105–14 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Boeren, S. , de Vries, S.C. , van Valenberg, H.J.F. , Vervoort, J. and Hettinga, K. (2011) Filter‐aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Proteomics, 75, 34–43. [DOI] [PubMed] [Google Scholar]
- Lucas, J.A. (2011) Advances in plant disease and pest management. J. Agric. Sci. 149, 91–114. [Google Scholar]
- Manza, L.L. , Stamer, S.L. , Ham, A.‐J.L. , Codreanu, S.G. and Liebler, D.C. (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics, 5, 1742–1745. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. and Kema, G.H.J. (2006) Protein kinase A subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Mol. Plant Pathol. 7, 565–577. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , van der Lee, T. , Waalwijk, C. and Kema, G.H.J. (2006a) MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant–Microbe Interact. 19, 389–398. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , Zwiers, L.‐H. , de Waard, M.A. and Kema, G.H.J. (2006b) MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 19, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , M'Barek, S.B. , van der Lee, T.A.J. , Waalwijk, C. , de Wit, P.J.G.M. and Kema, G.H.J. (2009) Gα and Gβ proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola . Eukaryot. Cell, 8, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Manders, E.M.M. , Boas, S. , Olivain, C. , Alabouvette, C. and Rep, M. (2009) The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. Plos Pathog. 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. , Becker, M. , Heller, J. , Moraga, J. , Collado, I.G. and Tudzynski, P. (2011) The Botrytis cinerea Reg1 protein, a putative transcriptional regulator, is required for pathogenicity, conidiogenesis, and the production of secondary metabolites. Mol. Plant–Microbe Interact. 24, 1074–1085. [DOI] [PubMed] [Google Scholar]
- Morais do Amaral, A. , Antoniw, J. , Rudd, J.J. and Hammond‐Kosack, K.E. (2012) Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola . PLoS ONE, 7, e49904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhäuser, J. (2010) Regulation of white–opaque switching in Candida albicans . Med. Microbiol. Immunol. (Berl.) 199, 165–172. [DOI] [PubMed] [Google Scholar]
- Motteram, J. , Küfner, I. , Deller, S. , Brunner, F. , Hammond‐Kosack, K.E. , Nürnberger, T. and Rudd, J.J. (2009) Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain‐containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 22, 790–799. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.Q. and Sil, A. (2008) Temperature‐induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. USA, 105, 4880–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara, T. and Tsuge, T. (2004) FoSTUA, encoding a basic Helix‐Loop‐Helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum . Eukaryot. Cell, 3, 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton, E.S. , Deller, S. and Brown, J.K.M. (2011) Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 12, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg, W. , Kema, G.H.J. , Groenewald, J.Z. , Verkley, G.J.M. , Seifbarghi, S. , Razavi, M. , Gohari, A.M. , Mehrabi, R. and Crous, P.W. (2011) Zymoseptoria gen. nov.: a new genus to accommodate Septoria‐like species occurring on graminicolous hosts. Persoonia, 26, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protocols, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Srikantha, T. , Borneman, A.R. , Daniels, K.J. , Pujol, C. , Wu, W. , Seringhaus, M.R. , Gerstein, M. , Yi, S. , Snyder, M. and Soll, D.R. (2006) TOS9 regulates white–opaque switching in Candida albicans . Eukaryot. Cell, 5, 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. , van Nistelrooy, J.G.M. , Kema, G.H.J. and De Waard, M.A. (2003) Multiple mechanisms account for variation in base‐line sensitivity to azole fungicides in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 59, 1333–1343. [DOI] [PubMed] [Google Scholar]
- Tabib Ghaffary, S. , Robert, O. , Laurent, V. , Lonnet, P. , Margalé, E. , van der Lee, T. , Visser, R. and Kema, G. (2011) Genetic analysis of resistance to septoria tritici blotch in the French winter wheat cultivars Balance and Apache. Theor. Appl. Genet. 123, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabib Ghaffary, S. , Faris, J. , Friesen, T. , Visser, R. , van der Lee, T. , Robert, O. and Kema, G. (2012) New broad‐spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 124, 125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong, A.E. , Miller, M.G. , Raisner, R.M. and Johnson, A.D. (2003) Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell, 115, 389–399. [DOI] [PubMed] [Google Scholar]
- Wisniewski, J.R. , Zougman, A. , Nagaraj, N. and Mann, M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- Wittenberg, A.H.J. , van der Lee, T.A.J. , Ben M'Barek, S. , Ware, S.B. , Goodwin, S.B. , Kilian, A. , Visser, R.G.F. , Kema, G.H.J. and Schouten, H.J. (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola . PLoS ONE, 4, e5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan, R.E. , Galgoczy, D.J. and Johnson, A.D. (2006) Epigenetic properties of white–opaque switching in Candida albicans are based on a self‐sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA, 103, 12 807–12 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.‐H. and De Waard, M.A. (2001) Efficient Agrobacterium tumefaciens‐mediated gene disruption in the phytopathogen Mycosphaerella graminicola . Curr. Genet. 39, 388–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Replacement strategy for ZtWor1 in Zymoseptoria tritici. (A) Diagram showing the replacement by the hygromycin phosphotransferase (hph) resistance cassette through homologous recombination. The broken line depicts the flanking regions used for homologous recombination. (B) Identification of replacement mutants by polymerase chain reaction (PCR). Lane M, 1‐kb‐plus ladder marker. Lanes 1, 4 and 7 show three independent replacement mutants (ΔZtWor1‐1, ΔZtWor1‐26 and ΔZtWor1‐29) with no amplicon using primers ZtWor1‐F1 and ZtWor1‐R1, whereas the wild‐type (WT) strain (lane C1) and the complemented strain ΔZtWor1‐com7 (lane10) show the expected amplicon of 650 bp with the same primers. Primers ZtWor1‐F2 and ZtWor1‐R2, which are located in the middle of the hph gene and downstream of the ZtWor1 open reading frame (ORF), produced an amplicon of 2000 bp (lanes 2, 5 and 8), but did not result in amplification in the WT strain (C1) and the complemented strain (lane 11).
Fig. S2 Number of cell fusion events, counted in 0.016 mm2, in colonies of Zymoseptoria tritici wild‐type (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 grown on minimal medium (MM) for 10 days at 20 °C.
Fig. S3 Differences in hyphal diameters of Zymoseptoria tritici wild‐type (WT) strain, ΔZtWor1‐com7 and ΔZtWor1‐26 grown on minimal medium (MM) for 10 days. n = 100 for each strain. The difference is significant at P < 0.05.
Fig. S4 Disease development in 12 wheat cultivars that are parents of mapping populations after inoculation with Zymoseptoria tritici wild‐type (WT) strain compared with two independent knock‐out strains of SSP127.
Table S1 Putative small secreted proteins (SSPs) and their corresponding primers used in this study.
Table S2 Comparative expression profiling of small secreted proteins (SSPs) in the Zymoseptoria tritici wild‐type (WT) strain versus ΔZtWor1‐26.


