Summary
The genus Striga comprises about 30 obligate root‐parasitic plants, commonly known as witchweeds. In particular, S. hermonthica, S. asiatica and S. gesnerioides cause immense losses to major stable crops in sub‐Saharan Africa. Most Striga species parasitize grass species (Poaceae), but Striga gesnerioides has evolved to parasitize dicotyledonous plants. Aspects of phylogeny, economic impact, parasitic life style and molecular discoveries are briefly reviewed to profile one of the main biotic constraints to African agriculture.
Taxonomy
Striga Lour.; Kingdom Plant; Division Angiospermae; Clade Eudicots; Order Laminales; Family Orobanchaceae.
Important hosts
Sorghum Moench., maize (Zea mays L.), rice (Oryza L.), sugarcane (Saccharum L.), pearl millet [Pennisetum glaucum (L.) R. Br.], cowpea [Vigna unguiculata (L.) Walp.].
Disease symptoms
Stunted growth, drought‐stressed‐like appearance, in severe cases chlorosis and necrosis.
Economic importance
1 billion $US per annum.
Disease control
Hand weeding, breeding, chemical control, intercropping with catch or trap crops.
Useful webpages
Introduction
Parasitic plants are a major threat to today's agriculture and provide an intriguing case of pathogenesis between species of relatively close evolutionary ancestry. Almost all crop species are potential hosts for parasitic plants, but severe disease outbreaks are usually restricted to certain host–pathogen combinations. The evolutionary strategy of exchanging autotrophy for dependence on host plants (parasitism) may seem odd, but it has proven to be evolutionarily successful for several plant species. Plant parasitism has arisen at least 12 times independently, generating more than 4000 parasitic dicotyledonous plant species (Westwood et al., 2010). Although some parasitic plants are still photosynthetically active (hemiparasitic), others are not, and depend entirely on a host (holoparasitic). The establishment of parasitism is essential for holoparasites and several hemiparasites, and therefore these species are called obligate parasites. Depending on which host organ is infected, parasitic plants are grouped into stem or root parasites. In both cases, the parasite connects to the host vascular system via a specialized feeding organ, the haustorium. Unlike the haustoria of plant‐pathogenic fungi or oomycetes, plant haustoria are always multicellular organs with complex anatomies and multiple cell types (Mayer, 2006). The genus Striga consists of obligate hemiparasitic root parasites, some of which are serious agricultural pests (Parker, 2009). This pathogen profile aims to give readers a brief overview of the biology of Striga and how this relates to current control strategies.
The Genus Striga—Plant Parasites among Plant Parasites
‘Striga’ is the Latin word for ‘witch’. Witchweed, Yan maemod (Thai), Buta (Kiswahili) and other common names for Striga often refer to the word ‘witch’, presumably because plants diseased by Striga display stunted growth and an overall drought‐like phenotype long before Striga plants appear. Striga species are annual plants and most of their life cycle occurs underground. The genus Striga was previously grouped within the family Scrophulariaceae, but more recent analyses have placed Striga as a monophyletic group in the family Orobanchaceae Vent. The family Orobanchaceae contains the highest number of parasitic species (Bennett and Mathews, 2006). Although most Orobanchaceae species are root parasites, ranging from facultative hemiparasitic plants, such as Triphysaria Fisch. & C.A. Mey., to holoparasitic Orobanche L. (broomrapes), 12 known species in the genus Lindenbergia Lehm. are not parasitic (Bennett and Mathews, 2006; See Table S1). This offers an opportunity to study successive stages in plant parasitism within the relatively confined evolutionary boundaries of one plant family (Westwood et al., 2010). Parasitism is believed to have evolved once within this family and the divergence of the Lindenbergia linage predates this event. Specialization towards holoparasitism then followed in several genera independently, often leading to closely related species with different degrees of parasitism (Bennett and Mathews, 2006).
Approximately 30 Striga species have been described and most parasitize grass species (Poaceae). Striga gesnerioides (Willd.) Vatke is the only Striga species that is virulent to dicots (Mohamed and Musselman, 2008). Striga possibly originates from a region between the Semien Mountains of Ethiopia and the Nubian Hills of Sudan (Atera and Itoh, 2011). This region is also the birthplace of domesticated sorghum (Sorghum bicolor L.), which is a major host species for several Striga species, including S. hermonthica (Delile) Benth. and S. asiatica (L.) Kuntze, and is believed to be the host on which monocot‐parasitizing Striga species have evolved and spread throughout Africa and Asia (Vasudeva‐Rao and Musselman, 1987).
Striga asiatica is morphologically similar to S. hirsuta Benth., S. lutea Lour. and S. elegans Benth., and therefore they are grouped into one Striga cluster. A few S. asiatica races or ecotypes occur outside Africa, mainly in Asia (Mohamed and Musselman, 2008). Because the evolutionary relationship between African and Asian S. asiatica populations is not well understood, the populations are often treated separately. Striga hirsuta, S. lutea and S. elegans are not considered to be serious agricultural pests (Mohamed and Musselman, 2008). Striga asiatica is autogamous like most Striga species, but S. hermonthica and S. aspera (Willd.) Benth. are obligate outcrossers and occasionally hybridize (Mohamed and Musselman, 2008). Allogamy probably contributes to the genetic variation between subpopulations of S. hermonthica, and also restricts spread outside the geographical distribution of available pollinators (Berner et al., 1997).
A recent phylogenetic analysis using six chloroplastic loci has suggested a closer relationship of S. gesnerioides to S. aspera and S. hermonthica than S. asiatica to S. hermonthica and S. aspera, despite the similar host specificities of S. asiatica, S. hermonthica and S. aspera. Striga gesnerioides is morphologically distinct relative to other Striga species (Estep et al., 2012). The haustoria differ especially in size and morphology from those of monocot‐parasitizing Striga species. The haustoria of S. gesnerioides, in contrast with those of other Striga species, such as S. hermonthica, exhibit a branched vascular system and lack the so‐called hyaline body (Ba, 1979), which is a specialized tissue surrounding the xylem bridge connecting the vascular systems of host and parasite.
The Impact—Distribution and Host Range
Striga is an ‘Old World’ parasite, and several species were already recognized as cereal pests in Africa and India at the beginning of the last century. Roughly 80% of the described Striga species are endemic to Africa, nine species are found outside Africa and three species, S. curvilflora Benth., S. multiflora Benth. and S. parviflora Benth., are present on the Australian continent (Berner et al., 1995). Striga species are predominantly found on open grasslands and savannahs in semi‐arid tropical regions. Infestations are more pronounced in infertile soils, but S. asiatica can grow in a wide range of different soils (Cochrane and Press, 1997). An increase in monoculture in some parts of Africa has led to reduced soil fertility, thus further worsening the situation with regard to Striga infestations (Berner et al., 1997). In addition to the presence of host‐derived germination stimulants, temperature is an important factor affecting the distribution of Striga, as prolonged exposure to high temperatures and humid conditions is required to break seed dormancy in Striga (Ejeta and Gressel, 2007).
An estimated cereal production area of 50 million hectares, approximately the size of Spain, shows different levels of Striga infestation in Africa (Westwood et al., 2010). In total, 25 African countries reported Striga infestations in 2005 (De Groote et al., 2008). The socioeconomic consequences are difficult to measure, but a few estimations have suggested that Striga affects the life of more than 100 million people in Africa and causes economic damage equivalent to approximately 1 billion $US per year (Labrada, 2008; Waruru, 2013). Host plants include sorghum, millet, maize, upland rice, sugarcane, cowpeas—representing the most important stable crops grown by subsistence farmers in affected areas. Farmers have reported losses between 20% and 80%, and are eventually forced to abandon highly infested fields (Atera and Itoh, 2011). The extent of yield losses cannot be explained solely by competition for nutrients and water (Berner et al., 1995). When disease progresses, very severe symptoms, such as water‐soaked leaf lesions, chlorosis, necrosis and leaf desiccation, occur (Berner et al., 1997). An unknown phytotoxin has been proposed to at least partially contribute to the disease phenotype, but still awaits biochemical identification. Interestingly, Striga extracts are rich in secondary metabolites and find broad use in traditional medicine, especially as a result of their antimicrobial activity (Koua and Babiker, 2011).
Only five Striga species are currently of economic importance, with S. hermonthica causing by far the most serious damage to sub‐Saharan cereal production, followed by S. asiatica, S. gesnerioides and, to a far lesser extent, S. aspera and S. forbesi Benth. (Parker, 2009). Facultative parasitic plants of the sister genus Buchnera L. are sometimes mistaken for Striga, but cause far less damage on host plants such as sorghum, maize or millet (Berner et al., 1997). The obligate parasitic species Alectra vogeli (Benth., Orobanchaceae) is commonly also referred to as yellow witchweed and, similar to S. gesnerioides, is a major biological constraint to cowpea production in eastern and southern Africa (Musselman, 1980).
Striga hermonthica is widespread in sub‐Saharan Africa, and is found throughout West Africa to Ethiopia, Uganda and Kenya in East Africa (Fig. 1A; Mohamed et al., 2001). Occurrences of S. hermonthica have also been reported from south‐east Africa. Striga hermonthica is particularly harmful to sorghum, maize and millet, but is also increasingly being found in sugarcane and rice fields (Atera and Itoh, 2011). Upland rice is becoming more and more important for African agriculture, not least because it can sustain more people per crop area than can maize or sorghum (Atera and Itoh, 2011). In an ongoing effort to breed New Rice for Africa (NERICA), several NERICA lines have been tested for Striga resistance. Some cultivars show good resistance to S. hermonthica and S. asiatica. Susceptible NERICA varieties display biomass reduction of up to 81% when infected with either S. hermonthica or S. asiatica (Cissoko et al., 2011). Notably, even resistant varieties showed a biomass reduction of 46% with S. hermonthica and 24% with S. asiatica in this study. In addition, some susceptible rice varieties perform better than others, a phenomenon called ‘tolerance’. Tolerance is so far the only genetic resource for resistance against Striga in maize (Cardoso et al., 2011). Ideally, breeding for Striga resistance or tolerance should be linked with other favourable traits, such as high yield and drought resistance. Drought is the major constraint to agriculture in semi‐arid zones. Promising results were obtained in sorghum when both traits, Striga and drought resistance, were combined by classical breeding. This effort was internationally recognized when Professor Gebisa Ejeta received the World Food Prize in 2009 (http://www.worldfoodprize.org).
Figure 1.
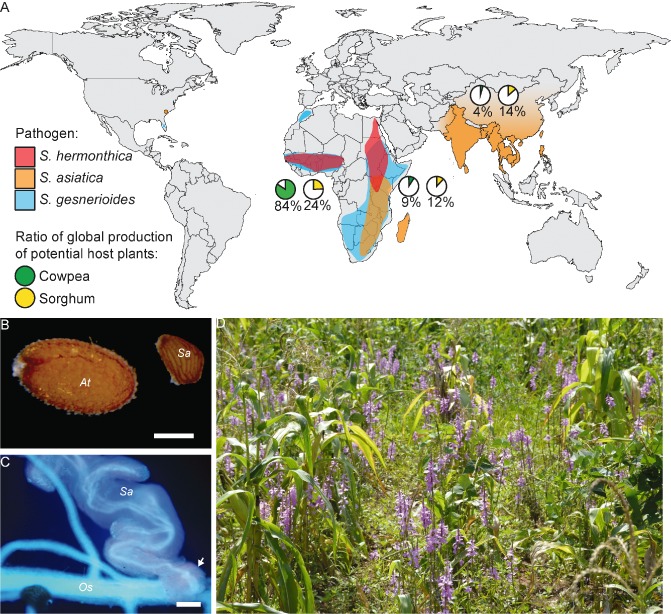
(A) Global distribution of the economically most destructive Striga species and two major host plants, cowpea and sorghum, according to Mohamed and Musselman (2008) and Musselman (1980). The relative production of sorghum and cowpea in West Africa, East Africa and South‐East Asia is based on values published by the Food and Agriculture Organization of the United Nations (FAO, http://www.fao.org/corp/statistics/en) for 2011. (B) Striga asiatica (Sa) seed next to an Arabidopsis thaliana (At) seed. (C) Two‐week‐old S. asiatica (Sa) haustorium (arrow) on rice (Oryza sativa, Os). (D) Striga hermonthica‐infested sorghum field in Kismu (Kenya, December 2012). (B, C) Scale bars represent 200 μm.
Striga asiatica is the most widespread Striga species (Fig. 1A), with a geographical distribution ranging from South Africa to East Africa and from the Arabian Peninsula to Far East Asia, including India and Pakistan (mainly on sorghum and millet), Cambodia, China, Thailand (maize in the 1970s), Vietnam, Malaysia, Indonesia and the Philippines (mainly on rice) (Musselman, 1980). Asian S. asiatica occurs mainly in the form of two morphotypes: white‐flowered S. asiatica, which is found in India and Pakistan, and a yellow‐flowered race which is predominant in Thailand and Indonesia (Vasudcva Rao, 1984). African S. asiatica plants have mainly red flowers. Striga asiatica infestation is less severe in Asia relative to Africa. Until the start of the 1990s, African S. asiatica was mainly restricted to South and Central Africa (Mohamed and Musselman, 2008). Although S. asiatica is now increasingly being found in other parts of Africa, it is most problematic south of the Equator in East and South Africa (Fig. 1A). Tanzania marks a transition zone between S. hermonthica and S. asiatica, with S. asiatica becoming more problematic in countries such as Tanzania, Malawi, Mozambique and Madagascar (Parker, 2009). In addition to numerous previous studies of host specialization (for a review, see Musselman, 1980), a study in Benin has discovered a high degree of host specialization within 14 analysed S. asiatica populations (Botanga et al., 2002). For example, some S. asiatica isolates collected from wild grass species are unable to successfully parasitize sorghum and maize plants, which are susceptible to other S. asiatica. In addition, a significant linear relationship between genetic and geographical distance was observed. Mostly unstudied populations of S. asiatica can be found outside the usual distribution, for example in the Nile delta. Striga asiatica was also accidentally introduced to North and South Carolina (USA) in the 1950s (Hood et al., 1998). The US population of S. asiatica is highly monomorphic, which strongly supports the theory of a single introduction event from Africa. An immense effort costing about 250 million $US was undertaken to eradicate S. asiatica from the USA. Although Striga currently does not pose a high risk for modern high‐input agricultural systems, such as those in the south‐eastern USA, it remains a significant problem for African farmers with no or only limited access to fertilizers, herbicides and modern mechanical tillage equipment (Berner et al., 1997). The introduction of new farming systems into rural societies takes time, such that hand weeding often remains the only technique to control Striga.
Striga gesnerioides is extremely widespread on the African continent (Mohamed et al., 2001). It is mainly a problem to cowpea farming in West Africa (Mali, Burkina Faso, Niger and Benin) and also to tobacco plantations in Zimbabwe. Cowpea is one of the most important food legumes in semi‐arid regions, and is considered to be a subsistence crop in sub‐Saharan Africa. Striga gesnerioides was also introduced to Florida, USA, in 1979. Based on the genetic variation observed in US strains, a single introduction effect is also most likely for S. gesnerioides (Musselman, 1980). Other Striga species are of lesser economic importance, but locally can cause significant yield losses. For example, S. angustifolia (Don) Saldhana has been reported to infect sorghum, rice and sugarcane in India, and S. aspera causes significant losses on maize plantations in Nigeria, Cameroon, Ivory Coast and Ethiopia and on rice plantations in Ivory Coast and Senegal (Kroschel, 1993).
The Parasitic Life Cycle of Striga
Parasitic plants have evolved specific traits which allow parasitism or reflect the consequences of adaptation to a parasitic life style. Critical stages in the life of an obligate root parasite are as follows: (i) the identification of a suitable host, thus coupling germination and seedling growth with the presence and direction of a potential host; (ii) gain of access to the host's nutrients and water supply; this process involves the production of a functional haustorium; (iii) completion of the life cycle on the host; this includes the establishment of parasitism and its maintenance until seeds are set.
Germination—location of a host root
Striga and other root‐parasitic plants have evolved highly efficient strategies to ensure successful reproduction. Key strategies include the dispersal of an enormous amount of tiny seeds (Fig. 1B) of high longevity to establish an extremely persistent seed bank. These dust‐like seeds are easily dispersed by wind, crop seeds, water and people. In addition, Striga seeds can survive for more than 10 years before germination (Atera and Itoh, 2011). Germination is linked to the presence of a nearby host, because the endosperm of Striga seeds can sustain growth/life only for the first 3–7 days (Berner et al., 1995). Within that time, Striga must successfully establish a parasitic relationship with the host plant or otherwise die. This aspect was successfully exploited during an S. asiatica eradication programme in the USA, when so‐called ‘suicide germination’ was induced by fuming farmland with ethylene to trigger Striga germination in the absence of host plants (Parker, 2009).
The germination of Striga depends on the perception of germination stimulants released by host roots. In order to be responsive to germination stimulants, Striga seeds must go through a phase of moisture and high temperatures for 7–14 days, called ‘conditioning’. If, during that time, no germination stimulant is perceived, Striga seeds fall into a secondary dormancy (Cardoso et al., 2011). Several germination stimulants have been isolated and include strigolactones, dihydrosorogoleone, sesquiterpene, kinetin, coumarin, jasmonate, ethylene and fungal metabolites (reviewed in Cardoso et al., 2011). Strigolactones are certainly the best studied and extremely potent inducers of Striga germination. Strigolactones are associated with the negative regulation of root and shoot branching (tillering). They also induce hyphal branching of arbuscular mycorrhizal (AM) fungi, presumably to attract them in low‐nutrient environments (Xie and Yoneyama, 2010). Major discoveries in biosynthesis and perception have been made in recent years, and key players have also been predicted to be present in Striga (Cardoso et al., 2011; Yoshida and Shirasu, 2012). Strigolactones have been shown to induce the germination of Striga at concentrations as low as 10−16 m (Musselman, 1980). The first strigolactone was interestingly isolated from the root exudates of a nonhost plant, cotton (Cook et al., 1966); indeed, the use of nonhost plants producing high levels of Striga germination stimulants is a promising strategy in Striga control. In particular, intercropping with the legume Desmodium has been proven to be successful in some parts of Africa (Khan et al., 2006). Alternatively, low strigolactone‐producing host plants reduce Striga germination and thus infection (Umehara et al., 2008). Low Striga germination stimulant activity is controlled in sorghum by one single recessively inherited gene, lgs (low germination stimulant) (Satish et al., 2012). Lines showing low germination‐inducing activity have been shown to have good tolerance towards S. asiatica and S. hermonthica, but tolerance mediated by low strigolactone production is less reliable when the Striga seed pool in the soil is high (Atera and Itoh, 2011).
Haustorium development
The radical tip grows chemotropically towards potential host roots after germination. On contact, Striga radicals stop growing, attach to host roots, form a haustorium and penetrate into the root cortex of potential hosts. Most plants, including many nonhost plants, do not resist attachment and penetration. An exception to this is Phtheirospermum japonicum (Thunberg) Kanitz, a hemiparasitic plant commonly found in East Asia and relatively closely related to Striga. The root exudate from P. japonicum is able to induce the germination of S. hermonthica, but S. hermonthica radicals rarely penetrate to P. japonicum roots (Yoshida and Shirasu, 2009). It is currently unknown whether P. japonicum actively inhibits the attachment of Striga or whether it is lacking a factor required for Striga penetration.
Within 12 h of attachment, reorganization of the S. asiatica meristem is initiated. Essential for this step is the perception of haustoria‐inducing factors. Several naturally occurring haustoria‐inducing factors have been isolated and their mode of action is best studied by following 2,6‐dimethoxy‐p‐benzoquinone (DMBQ; Chang and Lynn, 1986). DMBQ is a product of lignin oxidation and decarboxylation of phenolic acids found in plant cell walls. The current model of DMBQ perception is mainly based on work performed on S. asiatica and Triphysaria versicolor Fisch. & C.A. Mey. (Bandaranayake et al., 2010; Keyes et al., 2000). In summary, this model proposes that DMBQ is released from host cell walls and eventually enters parasite cells. The NAD(P)H‐dependent quinone reductase QR1 reduces DMBQ to produce an unstable semiquinone intermediate, which is required for haustorium development. Triphysaria QR1 is transcriptionally up‐regulated in response to host root extracts and QR1 knock‐down roots are compromised in haustoria formation. A second quinone reductase (QR2) does not respond to host root extracts, but to DMBQ, and could act as a parallel detoxification pathway. A balance between the detoxification and accumulation of the haustorium‐inducing semi‐quinone might create an equilibrium‐dependent threshold mechanism, whereby a continuous exposure to DMBQ is required for haustoria formation.
In addition to chemical signals, a thigmotropic response is required for Striga to produce morphologically normal haustoria (Wolf and Timko, 1991). Within 24 h after contact, rapid cell division of the radical tip stops and a hypertrophic growth phase begins (Hood et al., 1998). Penetration of the host epidermis is mediated by the elongation of distal cells in the protoderm or epidermis and underlying ground tissue, followed by rounds of periclinal and anticlinal divisions of these cells, leading to growth into the cortex of host plants. When the host endodermis is reached, the most distal cells of the haustorium elongate and divide, thus forming a palisade arrangement of cells. Break through the endodermis is often delayed, but, once accomplished, vascular connections are established. In general, penetration is completed 48–72 h after contact with a host root (Hood et al., 1998). A detailed scanning electron microscopy study by Dorr (1997) showed that invading Striga cells perforate the host vascular system using a specialized structure, the osculum. Interestingly, no phloem‐to‐phloem connections have been observed between Striga and host plants. Once xylem‐to‐xylem connections are established, the cotyledons of Striga enlarge and break free from the seed coat within 24 h (Hood et al., 1998).
Many nonhost plants allow the penetration of S. hermonthica and the early events of haustorium formation. Although infection is mainly terminated in the cortex of Lotus japonicus (Regel) K. Larsen, S. hermonthica reaches the stele in Arabidopsis and cowpea, but fails to develop beyond the six‐leaf‐pair stage (Yoshida and Shirasu, 2009). Nonhost resistance in lettuce, marigold and cowpea against S. asiatica is typically established in the cortex within 72 h post‐infection (Hood et al., 1998). Resistance to S. hermonthica and S. asiatica in rice (Oryza sativa L.) cultivar Nipponbare also occurs post‐attachment (Gurney et al., 2006). Using an inbred line between Nipponbare and the susceptible rice line Kasalath, Gurney et al. (2006) located seven putative quantitative trait loci (QTLs), which explained 31% of the overall resistance phenotype. Although resistance to monocot‐infecting Striga seems to be polygenetic, two types of resistance are known in cowpea towards S. gesnerioides: a hypersensitive reaction (HR)‐type response, causing the death of the parasite within 3–4 days, and a second type of resistance, allowing S. gesnerioides to establish xylem‐to‐xylem connections, but not supporting further growth of the parasite (Li and Timko, 2009). The former type of resistance is race specific and mediated by the RSG3‐301 gene product in cowpea cultivar B301 against S. gesnerioides race 3. Li and Timko (2009) cloned the RSG3‐301 gene and showed that it encodes a coiled‐coil nuclear‐binding site leucine‐rich repeat (CC‐NBS‐LRR) type of resistance (R) protein. CC‐NBS‐LRR proteins are known to confer resistance to a wide variety of plant pathogens in many other plant species.
It is currently unknown which Striga genes are required to successfully infect susceptible host plants. Haustorium development uses cellular processes similar to organogenesis processes known in other autotrophic plants. For example, cycline promoter B1 is activated within 24 h after DMBQ treatment in P. japonicum, and localized auxin and ethylene accumulation are important for haustoria formation in T. versicolor (Ishida et al., 2011; Tomilov et al., 2005).
Haustoria constitute the interface between host and parasite. Although all parasitic plants develop haustoria, haustoria differ anatomically between different species. Although Striga lacks phloem‐to‐phloem connections, direct connections between sieve elements of Orobanche crenata (Forsk.) and Vicia narbonensis (L.) were observed by electron microscopy (Dorr and Kollmann, 1995). The transmission of phloem‐localized viruses or RNA molecules has been reported for several parasitic plants, but not for Striga species (Leblanc et al., 2012). However, interspecies plasmodesmata between S. gesnerioides and pea (Dorr, 1996) raise the possibility of symplastic transport of nutrients and signalling molecules between Striga and host plants. The movement of DNA molecules across graft junctions also occurs via cell‐to‐cell movement and does not involve phloem connections (Stegemann and Bock, 2009). So far, there is no direct evidence of mRNA transit between Striga species and host plants. However, host‐induced silencing of β‐glucuronidase (GUS) gene expression in T. versicolor and the identification of several horizontal gene transfer (HGT) events between S. hermonthica and monocot hosts suggest that mRNA and other RNA molecules could travel between host and root parasite (Tomilov et al., 2008; Yoshida et al., 2010b). A degenerated poly‐A sequence in a horizontally transferred S. hermonthica gene, ShContig9483, supports an mRNA‐related origin (Yoshida et al., 2010b). A putative homologue of ShContig9483 was found in S. gesnerioides but not Orobanche minor Sm., suggesting that this HGT event might have occurred after the genera split, but presumably before S. gesnerioides evolved host specificity towards dicot plants. Mechanisms underlying the integration of host genes into the germline of parasitic plants remain elusive. Nevertheless, HGT has also been reported in several other parasitic plants and seems to be more frequent in parasitic plants than in nonparasitic plants (Leblanc et al., 2012). It remains to be shown whether and to what extent RNA molecules travel between Striga and host plants and, if so, whether these molecules can function in trans.
Establishment of parasitism and life cycle completion
After xylem‐to‐xylem connections have been established, Striga grows upwards and adventitious roots are produced. These adventitious roots are able to form lateral (secondary) haustoria on the same or other host plants. Facultative hemiparasitic plants, such as Triphysaria or Phtheirospermum, produce exclusively lateral haustoria. Secondary haustoria are believed to be evolutionarily older than primary or terminal haustoria (Westwood et al., 2010). Under natural conditions, host plants are usually parasitized by several Striga plants, and the parasites quickly become a metabolic sink for photoassimilates and nutrients. Nitrogen levels are at least twice as high in Striga as in host plants (Agabawi and Younis, 1965). Depletion of nitrogen almost certainly affects host physiology and provokes lower host photosynthesis rates, which are frequently associated with Striga infections. Several photosynthetic parameters are reduced in sorghum plants infected with S. hermonthica, including the electron transport rate through photosystem II and photochemical quenching (Rodenburg et al., 2008). Frost et al. (1997) have shown that the negative effect on photosynthesis correlates with reduced stomatal conductance, which is possibly the consequence of elevated abscisic acid (ABA) levels of sorghum plants infected by S. hermonthica. Not only ABA, but also other plant hormones, such as cytokines and gibberellic acid levels, are altered in sorghum plants infected with Striga relative to control plants (Musselman, 1980; Taylor et al., 1996). It is not known whether Striga manipulates host hormone homeostasis directly and how these changes contribute to parasitism.
After emergence from the soil, Striga plants begin to photosynthesize. However, the low CO2 fixation and high dark respiration rates of S. asiatica result in a negative carbon gain over the 24‐h period, thus making Striga still host dependent when growing above ground (Press et al., 1987). In addition, Striga leaves are characterized by a degenerated palisade cell layer and a relatively small number of chloroplasts per cell. Low photosynthesis in Striga is supported by transcriptome data from RNA isolated from the above‐grown S. hermonthica tissue. A relatively low expression of chlorophyll biosynthesis‐ and photosynthesis‐related genes was observed when compared with the expression of these genes in the facultative hemiparasitic plant T. versicolor (Wickett et al., 2011). The high transpiration rates of Striga suggest that most host photoassimilates are obtained by transpirational pull, explaining why high humidity is inhibitory to Striga growth. Indeed, Striga stomata show high conductance and respiration rates and little response to dark‐induced closure (Press et al., 1987). Relative to the host plant, Striga has a disadvantageous leaf surface ratio and might compensate for this with higher stomatal conductance (Press et al., 1987). Surprisingly, when water depletion was investigated under controlled experimental conditions of Striga‐infected maize plants, no significant increase in water use was observed until the very late stage of infection (63 days post‐infection) (Taylor and Seel, 1998). Before that time, maize plants had already established disease symptoms and showed stunted growth. However, in the final stage of infection, maize plants used nearly 50% more water than control plants.
The fact that disease symptoms appear before Striga emerges illustrates how ineffective the biocontrol of above ground‐grown Striga by hand weeding or herbicides is likely to be. Nevertheless, these techniques are important to avoid the reproduction of Striga. Striga asiatica and S. hermonthica flower about 4 weeks after emergence. Striga gesnerioides has been reported to flower earlier (Berner et al., 1995). Inflorescences are arranged in spikes or racemes, each carrying several flowers. Flower colour varies between species and sometimes within species from blue and pink (e.g. S. hermonthica and S. gesnerioides) to white, yellow or red (e.g. S. asiatica). After pollination, seeds mature within 4 weeks in seed pods, which contain 250–500 dust‐like seeds of 200–300 μm in size. Under optimal conditions, each Striga plant can produce up to 50 000–500 000 seeds (Berner et al., 1995). When the seed pods crack, seeds are spread into the soil and quickly build up in numbers. Striga seeds require a certain time of after‐ripening, about 6 months at elevated temperatures. According to Berner et al. (1997), this could be an adaptation to prevent germination during the last rains of the seasons, when no hosts are in the field.
Next‐Generation Striga Research
In recent years, efforts have been undertaken to elucidate the molecular events underlying Striga infections using next‐generation and conventional sequencing technology (Yoshida et al., 2010a). For example, comparative studies on repetitive regions in five Striga species generated a total of about 2200 Sanger sequence reads and about 10 000 454 reads (Estep et al., 2012). Partially assembled and identified repeats were most similar to the most closely related plant species. Overall, the authors came to the conclusion that the analysed Striga genomes have a rather typically complex angiosperm genome. Estimated haploid genome sizes range from 615 Mb for S. asiatica to 1425 Mb for S. hermonthica and 2460 Mb for S. forbesii, suggesting several polyploidization events. Polyploidization is also an important factor for speciation in the sister genus Orobanche (Schneeweiss et al., 2004). No evidence of large transfers of repetitive DNA regions from the host genomes was observed, which is in contrast with the observed HGT events between monocot genes and S. hermonthica (Yoshida et al., 2010b), and favours the hypothesis that HGT events originate from mRNA species rather than from large pieces of genomic DNA.
Next‐generation sequencing technology has led to an increase in available transcriptional data for S. hermonthica and related species. For example, Wickett et al. (2011) analysed sequence data obtained from Illumina short reads of mRNA isolated from above‐ground tissue of three Orobanchaceae species: the facultative hemiparasite T. versicolor, S. hermonthica and Phelipanche aegyptiaca (pers.) Pomel. The expression of photosynthesis‐related genes was much lower in S. hermonthica than in Triphysaria, and no expression of these genes was detected in Phelipanche. This study also revealed that chlorophyll a synthesis gene expression was conserved and detectable in all three species, even in the nonphotosynthetically active Ph. aegyptiaca.
Next‐generation sequencing technology will almost certainly provide detailed transcriptional information for Striga at different stages of infection and on different hosts, and will allow the simultaneous detection of host and pathogen transcriptomes. So far, host transcriptome data are mainly based on microarray studies or similar methods. Hiraoka et al. (2009) used a suppression subtractive hybridization strategy of mRNA isolated from Lotus japonicus to investigate differences when infected with S. hermonthica (resistant) or Ph. aegyptiaca. Several jasmonic acid‐regulated genes showed higher expression, with some being systemically induced. The incompatible interaction with S. hermonthica was characterized by higher expression of genes in the biosynthetic pathway for vestitol. Vestitol is a phytoalexin with insect repellent activity. It is not clear whether vestitol concentrations indeed rise after Striga infection and whether vestitol is phytotoxic to S. hermonthica, but the up‐regulation of defence‐associated genes suggests that S. hermonthica is actively recognized as a pathogen and elicits a defence response in L. japonicus. The accumulation of cytotoxic material is also probably the cause of nonhost resistance to S. hermonthica in Tripsacum dactyloides, a wild relative of maize. In contrast with Z. mays, haustoria formation is impaired on T. dactyloides plants by an unknown factor. This factor is also able to suppress haustoria formation on Z. mays, when Striga plants are attached to T. dactyloides at the same time (Gurney et al., 2003).
Microarray analysis on cowpea cultivar B301 challenged with either a virulent (SG4z) or avirulent (SG3) race of S. gesnerioides has provided further insight into the transcriptional response during the early stages of plant parasitism. Infected tissue, sampled 6 days post‐infection with SG3, was enriched with genes associated with apoptosis, programmed cell death and responses to biotic and abiotic stresses (Huang et al., 2012). At a later stage (13 days post‐infection), enhanced expression of defence‐related genes and genes involved in lignification processes was observed. At the same time, the expression of multiple defence‐related genes, including genes associated with lignification and secondary cell wall modifications, was repressed in compatible interactions with SG4z. SG4z‐infected plants also showed higher expression of genes coding for proteins involved in the cellular transport of nitrogen and sulphur.
A similar tendency was observed in rice, when one susceptible rice variety (IAC 45) and one resistant variety (Nipponbare) were infected with S. hermonthica and analysed 2, 4 and 11 days after infection using whole‐genome microarrays (Swarbrick et al., 2008). The incompatible interaction between S. hermonthica and Nipponbare showed enhanced expression of defence‐related genes, such as genes encoding pathogenesis‐related (PR) proteins, WRKY transcription factors and pleiotropic ABC transporters, whereas the compatible (susceptible) interaction was characterized by large‐scale down‐regulation of genes associated with growth regulation, metabolism, biogenesis of cellular components and cell division. Several genes coding for nutrient transporters, enzymes involved in amino acid metabolism, were up‐regulated at the same time in the susceptible rice cultivar.
Overall, these data, although sometimes very difficult to compare, draw a common picture, in which Striga is actively recognized by resistant plants and triggers a defence‐like response. This response appears to be very similar to that observed for other nonhost or race‐specific resistance responses to other plant pathogens. It also shows that Striga actively manipulates host transcription to foster parasitism by either up‐regulating host genes associated with nutrient supply or by down‐regulating defence‐related genes. It is not known how Striga manipulates transcription in host plants. Avirulence gene products are interesting candidates, but difficult to isolate as a result of limited genetic resources in parasitic plants. Ultimately, candidate genes will need to be tested in planta. Several hairy root transformation systems for members of the Orobanchaceae family, including T. versicolor, P. japonicum and Ph. Aegyptiaca, are available (Fernandez‐Aparicio et al., 2011; Ishida et al., 2011; Tomilov et al., 2007).
Summary and Perspectives
Many genes essential for plant parasitism are yet to be identified and characterized. The identification of these genes will eventually help to answer fundamental questions in plant‐parasitic interactions, such as: Which genetic modifications are required to enable a parasitic life cycle? What is the role of HGT in parasitic plant–plant interactions? Which parasite gene products are recognized by resistant plants and which gene products help to resist being detected by the host immune system? Which molecules are exchanged at the haustorium interface? What is the molecular basis of tolerance and can all this information be used to breed Striga‐resistant crops? Ongoing whole‐genome sequence projects of parasitic plants and related nonparasitic species, such as Lindenbergia philippensis (Cham. & Schltdl.) Benth. (http://ppgp.huck.psu.edu), will certainly provide new insights into the evolution of Striga species and facilitate the identification of genes important for plant parasitism.
Supporting information
Table S1 Overview of Striga species and related Orobanchaceae species discussed in the main text.
Acknowledgements
TS and MM are supported by grants from the Japanese Society for Promotion of Science (JSPS) and KS by MEXT KAKENHI 24228008. The authors thank Satoko Yoshida and Luke Braidwood for helpful comments on the manuscript.
References
- Agabawi, K.A. and Younis, A.E. (1965) Effect of nitrogen application on growth and nitrogen content of Striga hermonthica, Benth. and Sorghum vulgare, Lur. grown for forage. Plant Soil, 23, 295–304. [Google Scholar]
- Atera, E. and Itoh, K. (2011) Evaluation of ecologies and severity of Striga weed on rice in sub‐Saharan Africa. Agric. Biol. J. N. Am. 2, 752–760. [Google Scholar]
- Ba, A.T. (1979) Notes on the histochemistry of the haustorium of Striga hermonthica. Proceedings, Second International Symposium on Parasitic Weeds, North Carolina, pp. 128–131.
- Bandaranayake, P.C.G. , Filappova, T. , Tomilov, A. , Tomilova, N.B. , Jamison‐McClung, D. , Ngo, Q. , Inoue, K. and Yoder J.I. (2010) A single‐electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria . Plant Cell, 22, 1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J.R. and Mathews, S. (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93, 1039–1051. [DOI] [PubMed] [Google Scholar]
- Berner, D.K. , Kling, J.G. and Singh, B.B. (1995) Striga research and control – a perspective from Africa. Plant Dis. 79, 652–660. [Google Scholar]
- Berner, D.K. , Winslow, M.D. , Cardwell, K.F. , Raj, D.R.M. and Kim, S.K. (1997) Striga Research Methods—A Manual. Ibadan: International Institute of Tropical Agriculture. [Google Scholar]
- Botanga, C. , Kling, J. , Berner, D. and Timko, M. (2002) Genetic variability of Striga asiatica (L.) Kuntz based on AFLP analysis and host–parasite interaction. Euphytica, 128, 375–388. [Google Scholar]
- Cardoso, C. , Ruyter‐Spira, C. and Bouwmeester, H.J. (2011) Strigolactones and root infestation by plant‐parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 180, 414–420. [DOI] [PubMed] [Google Scholar]
- Chang, M. and Lynn, D.G. (1986) The haustorium and the chemistry of host recognition in parasitic angiosperms. J. Chem. Ecol. 12, 561–579. [DOI] [PubMed] [Google Scholar]
- Cissoko, M. , Boisnard, A. , Rodenburg, J. , Press, M.C. and Scholes, J.D. (2011) New Rice for Africa (NERICA) cultivars exhibit different levels of post‐attachment resistance against the parasitic weeds Striga hermonthica and Striga asiatica . New Phytol. 192, 952–963. [DOI] [PubMed] [Google Scholar]
- Cochrane, V. and Press, M.C. (1997) Geographical distribution and aspects of the ecology of the hemiparasitic angiosperm Striga asiatica (L) Kuntze: a herbarium study. J. Trop. Ecol. 13, 371–380. [Google Scholar]
- Cook, C.E. , Whichard, L.P. , Turner, B. and Wall, M.E. (1966) Germination of witchweed (Striga lutea Lour) – isolation and properties of a potent stimulant. Science, 154, 1189–1190. [DOI] [PubMed] [Google Scholar]
- De Groote, H. , Wangare, L. , Kanampiu, F. , Odendo, M. , Diallo, A. , Karaya, H. et al (2008) The potential of a herbicide resistant maize technology for Striga control in Africa. Agric. Syst. 97, 83–94. [Google Scholar]
- Dorr, I. (1996) New results on interspecific bridges between parasites and their hosts In: Advances in Parasitic Plants Reseach (Moreno M.T., Cubero J.I., Berber D., Joel D., Musselmann L.J. and Parker C., eds), pp. 196–201. Seville: Junta de ; Andalucia. [Google Scholar]
- Dorr, I. (1997) How Striga parasitizes its host: a TEM and SEM study. Ann. Bot. (Lond.) 79, 463–472. [Google Scholar]
- Dorr, I. and Kollmann, R. (1995) Symplastic sieve element continuity between Orobanche and host. Bot. Acta, 108, 47–55. [Google Scholar]
- Ejeta, G. and Gressel, J. (2007) Integrating New Technologies for Striga Control: Towards Ending the Witch‐Hunt. Singapore; Hackensack, NJ: World Scientific. [Google Scholar]
- Estep, M.C. , Gowda, B.S. , Huang, K. , Timko, M.P. and Bennetzen, J.L. (2012) Genomic characterization for parasitic weeds of the genus Striga by sample sequence analysis. Plant Genome‐Us, 5, 30–41. [Google Scholar]
- Fernandez‐Aparicio, M. , Rubiales, D. , Bandaranayake, P.C.G. , Yoder, J.I. and Westwood, J.H. (2011) Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca . Plant Methods, 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, D.L. , Gurney, A.L. , Press, M.C. and Scholes, J.D. (1997) Striga hermonthica reduces photosynthesis in sorghum: the importance of stomatal limitations and a potential role for ABA? Plant Cell Environ. 20, 483–492. [Google Scholar]
- Gurney, A.L. , Grimanelli, D. , Kanampiu, F. , Hoisington, D. , Scholes, J.D. and Press, M.C. (2003) Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytol. 160, 557–568. [DOI] [PubMed] [Google Scholar]
- Gurney, A.L. , Slate, J. , Press, M.C. and Scholes, J.D. (2006) A novel form of resistance in rice to the angiosperm parasite Striga hermonthica . New Phytol. 169, 199–208. [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y. , Ueda, H. and Sugimoto, Y. (2009) Molecular responses of Lotus japonicus to parasitism by the compatible species Orobanche aegyptiaca and the incompatible species Striga hermonthica . J. Exp. Bot. 60, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M.E. , Condon, J.M. , Timko, M.P. and Riopel, J.L. (1998) Primary haustorial development of Striga asiatica on host and nonhost species. Phytopathology, 88, 70–75. [DOI] [PubMed] [Google Scholar]
- Huang, K. , Mellor, K.E. , Paul, S.N. , Lawson, M.J. , Mackey, A.J. and Timko, M.P. (2012) Global changes in gene expression during compatible and incompatible interactions of cowpea (Vigna unguiculata L.) with the root parasitic angiosperm Striga gesnerioides . BMC Genomics, 13, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, J.K. , Yoshida, S. , Ito, M. , Namba, S. and Shirasu, K. (2011) Agrobacterium rhizogenes‐mediated transformation of the parasitic plant Phtheirospermum japonicum . PLoS ONE, 6, e25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes, W.J. , O'Malley, R.C. , Kim, D. and Lynn, D.G. (2000) Signaling organogenesis in parasitic angiosperms: xenognosin generation, perception, and response. J. Plant Growth Regul. 19, 217–231. [DOI] [PubMed] [Google Scholar]
- Khan, Z.R. , Pickett, J.A. , Wadhams, L.J. , Hassanali, A. and Midega, C.A.O. (2006) Combined control of Striga hermonthica and stemborers by maize–Desmodium spp. intercrops. Crop Prot. 25, 989–995. [Google Scholar]
- Koua, F.H.M. and Babiker, H.A.A. (2011) Phytochemical and biological study of Striga hermonthica (Del.) Benth callus and intact plant. Res. Pharm. Biotechnol. 3, 85–92. [Google Scholar]
- Kroschel, J. (1993) Review of Parker C., and Riches C. R. ‘Parasitic Weeds of the World: Biology and Control’. Exp. Agric. 30, 490. [Google Scholar]
- Labrada, R. (2008) Farmer training on parasitic weed management In: Progress on Farmer Training in Parasitic Weed Management (Labrada R., ed.), pp. 1–5. Rome: FAO. [Google Scholar]
- Leblanc, M. , Kim, G. and Westwood, J.H. (2012) RNA trafficking in parasitic plant systems. Front. Plant Sci. 3, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.X. and Timko, M.P. (2009) Gene‐for‐gene resistance in Striga–cowpea associations. Science, 325, 1094. [DOI] [PubMed] [Google Scholar]
- Mayer, A.M. (2006) Pathogenesis by fungi and by parasitic plants: similarities and differences. Phytoparasitica, 34, 3–16. [Google Scholar]
- Mohamed, K.I. and Musselman, L.J. (2008) Taxonomy of agronomically important Striga and Orobanche species In: Progress on Farmer Training in Parasitic Weed Management (Labrada R., ed.), pp. 7–14. Rome: FAO. [Google Scholar]
- Mohamed, K.I. , Musselman, L.J. and Riches, C.R. (2001) The genus Striga (Scrophulariaceae) in Africa. Ann. Missouri Bot. Gard. 88, 60–103. [Google Scholar]
- Musselman, L.J. (1980) The biology of Striga, Orobanche, and other root‐parasitic weeds. Annu. Rev. Phytopathol. 18, 463–489. [Google Scholar]
- Parker, C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. 65, 453–459. [DOI] [PubMed] [Google Scholar]
- Press, M.C. , Tuohy, J.M. and Stewart, G.R. (1987) Gas‐exchange characteristics of the sorghum–Striga host–parasite association. Plant Physiol. 84, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg, J. , Bastiaans, L. , Schapendonk, A.H.C.M. , van der Putten, P.E.L. , van Ast, A. , Dingemanse, N.J. et al (2008) CO‐assimilation and chlorophyll fluorescence as indirect selection criteria for host tolerance against Striga . Euphytica, 160, 75–87. [Google Scholar]
- Satish, K. , Gutema, Z. , Grenier, C. , Rich, P.J. and Ejeta, G. (2012) Molecular tagging and validation of microsatellite markers linked to the low germination stimulant gene (lgs) for Striga resistance in sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet. 124, 989–1003. [DOI] [PubMed] [Google Scholar]
- Schneeweiss, G.M. , Palomeque, T. , Colwell, A.E. and Weiss‐Schneeweiss, H. (2004) Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am. J. Bot. 91, 439–448. [DOI] [PubMed] [Google Scholar]
- Stegemann, S. and Bock, R. (2009) Exchange of genetic material between cells in plant tissue grafts. Science, 324, 649–651. [DOI] [PubMed] [Google Scholar]
- Swarbrick, P.J. , Huang, K. , Liu, G. , Slate, J. , Press, M.C. and Scholes, J.D. (2008) Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica . New Phytol. 179, 515–529. [DOI] [PubMed] [Google Scholar]
- Taylor, A. and Seel, W.E. (1998) Do Striga hermonthica‐induced changes in soil matric potential cause the reduction in stomatal conductance and growth of infected maize plants? New Phytol. 138, 67–73. [Google Scholar]
- Taylor, A. , Martin, J. and Seel, W.E. (1996) Physiology of the parasitic association between maize and witchweed (Striga hermonthica): is ABA involved? J. Exp. Bot. 47, 1057–1065. [Google Scholar]
- Tomilov, A. , Tomilova, N. and Yoder, J.I. (2007) Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta, 225, 1059–1071. [DOI] [PubMed] [Google Scholar]
- Tomilov, A.A. , Tomilova, N.B. , Abdallah, I. and Yoder, J.I. (2005) Localized hormone fluxes and early haustorium development in the hemiparasitic plant Triphysaria versicolor . Plant Physiol. 138, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilov, A.A. , Tomilova, N.B. , Wroblewski, T. , Michelmore, R. and Yoder, J.I. (2008) Trans‐specific gene silencing between host and parasitic plants. Plant J. 56, 389–397. [DOI] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. , Akiyama, K. , Arite, T. , Takeda‐Kamiya, N. et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Vasudcva Rao, M.J.V. (1984) Patterns of resistance to Striga asiatica in sorghum and millets, with special reference to Asia In: Striga: Biology and Control. Workshop on the Biology and control of Striga, (Ayensu E.S., Docgett H., Keynes R.D., Marton‐Lefevre J., Musselman L.J., Parker C. and Pickering A., eds), pp. 93–112. Dakar, Senegal: ICSU Press. [Google Scholar]
- Vasudeva‐Rao, M.J. and Musselman, L.J. (1987) Host Specificity in Striga Spp. and Physiological ‘Strains’. Boca Raton, FL: CRC Press. [Google Scholar]
- Waruru, M. (2013) Deadly Striga weed spreading across Eastern Africa. Available at: http://www.scidev.net/en/sub‐suharan‐africa/news/deadly‐striga‐weed‐spreading‐across‐eastern‐africa.html: SciDev.Net [accessed on July 2, 2013].
- Westwood, J.H. , Yoder, J.I. , Timko, M.P. and dePamphilis, C.W. (2010) The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235. [DOI] [PubMed] [Google Scholar]
- Wickett, N.J. , Honaas, L.A. , Wafula, E.K. , Das, M. , Huang, K. , Wu, B.A. et al (2011) Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr. Biol. 21, 2098–2104. [DOI] [PubMed] [Google Scholar]
- Wolf, S.J. and Timko, M.P. (1991) In vitro root culture – a novel‐approach to study the obligate parasite Striga asiatica (L) Kuntze. Plant Sci. 73, 233–242. [Google Scholar]
- Xie, X. and Yoneyama, K. (2010) The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. and Shirasu, K. (2009) Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica . New Phytol. 183, 180–189. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. and Shirasu, K. (2012) Plants that attack plants: molecular elucidation of plant parasitism. Curr. Opin. Plant Biol. 15, 708–713. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Ishida, J.K. , Kamal, N.M. , Ali, A.M. , Namba, S. and Shirasu, K. (2010a) A full‐length enriched cDNA library and expressed sequence tag analysis of the parasitic weed, Striga hermonthica . BMC Plant Biol. 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. , Maruyama, S. , Nozaki, H. and Shirasu, K. (2010b) Horizontal gene transfer by the parasitic plant Striga hermonthica . Science, 328, 1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Overview of Striga species and related Orobanchaceae species discussed in the main text.


